ABSTRACT
The recently emerged fungal pathogen Candida auris often displays resistance to one or more antifungal drugs. Its infections have been identified in at least 40 countries on six continents to date. Here we report a case of C. auris candidemia in a patient in Xiamen, a city in south China. We also review currently reported cases of C. auris infection in China and compare the genetic and biological features of C. auris strains isolated from this country. Our phylogenetic analysis indicates that there are at least two C. auris genetic clades present in China (the South African clade and the south Asian clade) that display opposite mating type loci (one is MTLa and the other is MTLα). We also found that there are several distinct features among the clinical isolates studied, including the expression of virulence factors, antifungal susceptibilities, and cellular morphologies, and that these features could be associated with the mating-type of the isolate. For example, C. auris MTLa isolates generally secreted higher levels of secreted aspartyl proteases (Saps) at ambient environmental temperatures. Taken together, this study demonstrates that C. auris clinical isolates from China exhibit diversity in both biological and genetic features.
KEYWORDS: Candida auris, antifungal resistance, mating type locus, virulence, morphology
1. Introduction
The “superbug” fungus Candida auris is becoming a serious global public health threat (Sardi et al. 2018; Du et al. 2020). It was first identified as a novel fungal species and reported in 2009 by a Japanese group (Satoh et al. 2018). Due to its multidrug resistance and rapid prevalence in clinical or healthcare settings, the CDC has issued a couple of alerts to inform the awareness of C. auris infection. In the past decade, great progresses have been made on the study of this species, yet many aspects of its biology, genetics, and epidemiology remain to be investigated. As of February 2021, C. auris infections have been identified in at least 40 countries on six continents. Whole-genome sequencing (WGS) analyses indicate that four major clades of C. auris have emerged independently in four distinct geographical regions (Africa, South America, east Asia, and south Asia) (Lockhart et al. 2017; Du et al. 2020; Ahmad and Alfouzan 2021). In addition, a potential new clade was recently isolated from Iran. Over 200,000 SNPs had been identified between the Iran clade and the other clades (Chow et al. 2019).
The first case of C. auris infection in China was reported in 2018 by us (Wang et al. 2018). Subsequently, seventeen additional cases have been reported in Beijing and Shenyang, China (Chen et al. 2018; Tian et al. 2018). Thus far, of the reported C. auris strains in mainland China, only a single strain (BJCA001) belongs to the South Asian clade, and the other strains belong to the South African clade. Interestingly, strain BJCA001 is susceptible to all antifungal drugs tested, while the other seventeen strains are resistant to fluconazole based on their minimum inhibitory concentration (MIC) breakpoints (Chen et al. 2018; Tian et al. 2018; Wang et al. 2018). Here we report the first case of C. auris candidemia from a patient in Xiamen, a city in south China. We also reviewed all reported cases of C. auris infection to date in mainland China and performed genetic and biological comparative analyses of all known C. auris clinical isolates in China.
2. Materials and methods
2.1. Culture conditions
Candida auris strains were routinely grown in YPD medium supplemented with 5 μg/mL of phloxine B. The mating-type loci (MTL) of all C. auris strains used in this study were determined by PCR chemotyping assays using primers MTLa (5ʹ-TGACTCTTGAACAGCTTACG-3ʹ and 5ʹ-ATCTCGCAATGACCTCGTAC) and MTLα: (5ʹ-ATGCGATGCTAGTATGGATG-3ʹ and 5ʹ-ATGGTGCTTCCTTTTCTGTG-3ʹ).
Minimal inhibitory concentration (MIC) assays were performed according to the CLSI M27 (third edition) and our previous publication (Bing et al. 2020).
2.2. Phylogenetic analyses
The internal transcribed spacer (ITS) ribosomal regions of C. auris strains were aligned using mafft v7.015b26 (Katoh and Standley 2013). The ITS sequences were amplified from C. auris genomic DNA using primers ITS-F 5ʹ-GTCGTAACAAGGTTTCCGTAGGTG-3ʹ and ITS-R 5ʹ-GGTCCGTGTTTCAAGACGG. As described previously (Wang et al. 2018), based on General Time Reversible (GTR) and Gamma distribution with Invariant sites (G + I) models, we generated the maximum-likelihood phylogenetic tree using the tool RAxML v7.3.227 (Stamatakis 2014). The ITS sequences of the previously reported representative strains were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/) based on previous publications (Lockhart et al. 2017; Chen et al. 2018; Tian et al. 2018; Wang et al. 2018; Chow et al. 2019).
2.3. Secreted aspartyl protease (Sap) activity assay
As described previously (Wang et al. 2018), Sap activity of C. auris strains was determined using YCP-BSA assays at 25°C or 37°C. Approximately 5 × 106 cells of C. auris in 5 μL ddH2O were spotted and cultured on YCB-BSA medium plates for seven days. The width of the white halos (BSA precipitation), which reflect the activity of Saps, was measured.
3. Results and discussion
3.1. Case presentation
A 67‐year‐old man with a 10-year history of gastric ulcers, gastric mucosal haemorrhaging and diabetes was admitted to the First Affiliated Hospital of Xiamen University. He presented with abdominal pain and melaena and was faecal occult blood test positive (OB+). A routine blood culture examination and a computerised tomography (CT) scan were performed. His level of C-reactive protein (CRP) was 16.7 mg/L. He was then diagnosed with gastric mucosal haemorrhaging, peritonitis, and bronchitis. Haemostasis was achieved and piperacillin/tazobactam and fluconazole were given prophylactically to prevent bacterial and fungal infections, respectively. On day six of admission, the patient presented with serious abdominal pain and an elevated body temperature of 39.2°C. Increased redness and swelling around the abdominal drainage tube was observed. Fungal cultures indicated that both the blood and drainage tube tip were positive for yeast species. The isolates were identified as C. auris by MALDI-TOF MS (Girard et al. 2016) and verified by sequencing of the ITS ribosomal region. The ITS sequences of the C. auris isolates from the blood and drainage tube tip were identical, suggesting that they were likely derived from the same clonal strain. The C. auris strain isolated from the blood was called XM1805 and the C. auris strain isolated from the drainage tube tip was called XM1803. Both isolates were not susceptible to fluconazole (MIC ≥128 mg/L) but showed a low MIC to caspofungin (≤0.25 mg/L). MIC assays for additional antifungal drugs were also performed, and these results are presented in supplementary Table 1. Since the C. auris isolates identified were resistant to fluconazole, the administration of fluconazole was stopped and caspofungin (50 mg/day) was administered to the patient for fourteen days. On days ten and twenty-two of admission, blood culture test results were negative for fungi. On day 31, the patient was discharged from the hospital. We note that the patient had a recent travel history to Japan but had not travelled to Shenyang or Beijing, China, two cities where C. auris had been previously isolated.
3.2. Phylogenic and MIC analyses of C. auris clinical isolates from China
A total of twenty known clinical isolates from China, including fifteen from Shenyang, three from Beijing, and two from Xiamen (XM1803 and XM1805), were obtained. We performed comparative biological analyses of all isolates and found that there are several distinct features among the different strains, including the expression of virulence factors, antifungal susceptibilities, and cellular morphologies.
A summary of clinical information of the two C. auris isolates from Xiamen and the previously reported isolates from mainland China is presented in supplementary Table 2. Phylogenic analyses were performed based on ITS sequences of all C. auris isolates from China. Twenty-one C. auris strains from other countries representing the five reported genetic clades were also included in the phylogenic tree. As demonstrated in Figure 1, only strain BJCA001 from Beijing belongs to the South Asian clade, while the other nineteen isolates from China are closely related to the South African clade. This finding suggests that the South African clade is the predominant C. auris genetic clade in China.
Table 2.
Clinical information of the 20 C. auris clinical isolates from mainland China
| Location | Isolates name |
Source | Year of Identification |
Age/Gender | Reference | |
|---|---|---|---|---|---|---|
| Xiamen | XM1803 | Clade III | Drainage tube tip | 2018 | 67‐year‐old, male | This study |
| XM1805 | Clade III | Blood | 2018 | 67‐year‐old, male | This study | |
| Beijing | BJCA001 | Clade I | BALF | 2018 | 76-year-old, female | 1 |
| C1921 | Clade III | Blood | 2018 | preterm male infants | 2 | |
| C1922 | Clade III | Blood | 2018 | Preterm male infants | 2 | |
| Shenyang | RICU1 | Clade III | Urine | 2017 | 70-year-old, male | 3 |
| RICU2 | Clade III | Urine | 2017 | 69-year-old, female | 3 | |
| RICU3 | Clade III | Sputum | 2017 | 69-year-old, female | 3 | |
| RICU4 | Clade III | Blood | 2017 | 56-year-old, male | 3 | |
| RICU5 | Clade III | Catheter | 2017 | 82-year-old, female | 3 | |
| RICU6 | Clade III | Catheter | 2017 | 70-year-old, male | 3 | |
| RICU7 | Clade III | Urine | 2017 | 63-year-old, female | 3 | |
| RICU8 | Clade III | Urine | 2017 | 73-year-old, female | 3 | |
| NICU1 | Clade III | Urine | 2017 | 60-year-old, female | 3 | |
| NICU2 | Clade III | Urine | 2017 | 58-year-old, male | 3 | |
| NICU3 | Clade III | Urine | 2017 | 86-year-old, male | 3 | |
| NICU4 | Clade III | Urine | 2017 | 49-year-old, male | 3 | |
| NICU5 | Clade III | Urine | 2017 | 86-year-old, female | 3 | |
| NICU6 | Clade III | Drainage | 2017 | 82-year-old, female | 3 | |
| NSICU1 | Clade III | Urine | 2017 | 53-year-old, male | 3 | |
| BALF, bronchoalveolar lavage fluid | ||||||
Reference
- Wang XJ, Bing J, Zheng QS, Zhang FF, Liu JB, Yue HZ, Tao L, Du H, Wang YN, Wang H, et al. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infec. 7(1): 93.
- Chen Y, Zhao JY, Han L, Qi LH, Fan WH, Liu J, Wang ZG, Xia X, Chen J, Zhang LL. 2018. Emergency of fungemia cases caused by fluconazole-resistant Candida auris in Beijing, China. Journal of Infection. 77(6): 569–571.
- Tian S, Rong C, Nian H, Li F, Chu Y, Cheng S, Shang H. 2018. First cases and risk factors of super yeast Candida auris infection or colonisation from Shenyang, China. Emerg Microbes Infect. 7(1): 128.
Figure 1.
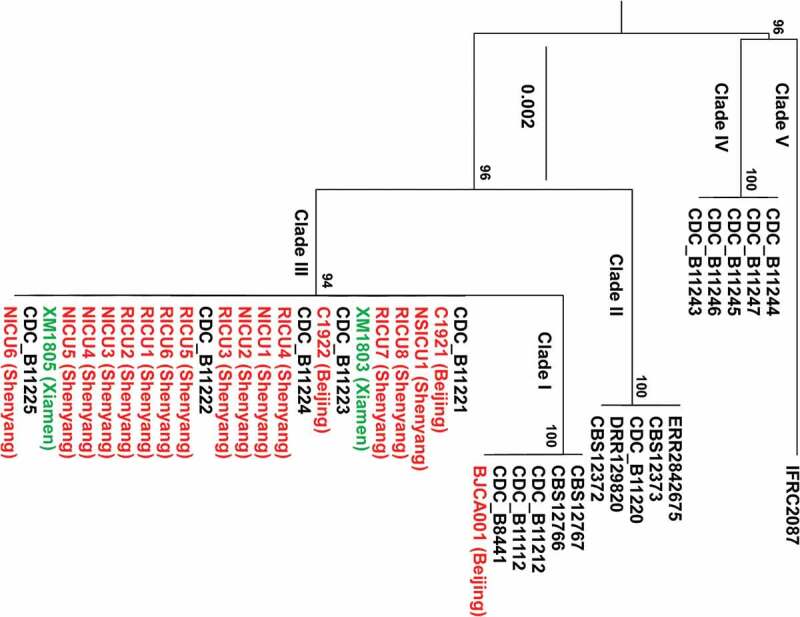
Phylogenic analyses of the C. auris strains isolated from China compared to other C. auris strains. The two strains (XM1803 and XM1805) isolated from Xiamen, China, are highlighted in green and the other isolates from China are highlighted in red. Twenty-one additional C. auris isolates representing the five genetic clades are also included (highlighted in black). The maximum-likelihood phylogenetic tree was generated based on ITS sequences. The scale bar indicates the number of nucleotide substitutions per site.
C. auris is predominantly a haploid fungus. PCR analyses indicate that only BJCA001 is an MTLa strain, while the isolates from Xiamen (XM1803 and XM1805) and other Chinese hospitals are MTLα strains (Figure 2). Interestingly, only strain BJCA001 was susceptible to all antifungal drugs tested; the other nineteen strains exhibited resistance to one antifungal drug (fluconazole) or additional antifungal drugs based on their MIC breakpoints (Table 1). Previous studies in C. auris indicate that there is an association between fluconazole resistance and the presence of hotspot mutations in the Erg11 lanosterol demethylase (e.g. Y132F, K143R, and VF125AL) (Lockhart et al. 2017; Du et al. 2020). Consistently, no hotspot mutations were found in strain BJCA001, while a hotspot mutation in Erg11 (VF125AL) was found in all of the other nineteen strains from China (Table 1).
Figure 2.
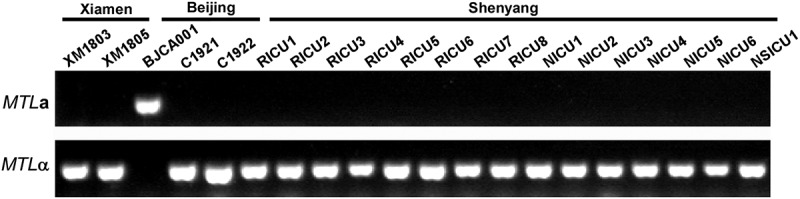
PCR analyses of the MTL loci of the different C. auris strains isolated from China. The MTL loci (MTLa or MTLα) of the twenty C. auris strains isolated from China were determined by PCR chemotyping.
Table 1.
MICs and Erg11 mutations of the 20 C. auris clinical isolates from China
| MIC (μg/mL) |
FLC | ITC | POS | VRC | AMB | 5-FC | AFG | CAS | MFG | Data adapted from |
Erg11 mutation hot spot |
|---|---|---|---|---|---|---|---|---|---|---|---|
| XM1803 | 128 | 0.5 | 0.125 | 0.5 | 2 | 0.125 | 0.5 | 0.25 | 0.125 | This study | VF125AL |
| XM1805 | 128 | 0.5 | 0.25 | 0.5 | 2 | 0.125 | 0.5 | 0.5 | 0.25 | This study | VF125AL |
| BJCA001 | 2 | 0.03 | 0.02 | 0.0 | 0.25 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 1 | No mutation |
| C1921 | 128 | 0.25 | 0.25 | 0.5 | 2 | 0.125 | 0.5 | 0.5 | 0.25 | This study | VF125AL |
| C1922 | 128 | 0.5 | 0.25 | 0.5 | 2 | 0.125 | 1 | 0.5 | 0.25 | This study | VF125AL |
| RICU1 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.03 | 0.06 | 0.06 | Reference 2 | VF125AL |
| RICU2 | 256 | 0.12 | 0.06 | 1.0 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| RICU3 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
| RICU4 | 256 | 0.12 | 0.06 | 1.0 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| RICU5 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| RICU6 | 128 | 0.12 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| RICU7 | 256 | 0.12 | 0.06 | 1.0 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| RICU8 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
| NICU1 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
| NICU2 | 256 | 0.12 | 0.06 | 1.0 | 1 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| NICU3 | 256 | 0.12 | 0.06 | 0.5 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| NICU4 | 256 | 0.12 | 0.06 | 1.0 | 0.5 | <0.06 | 0.12 | 0.12 | 0.12 | Reference 2 | VF125AL |
| NICU5 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
| NICU6 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
| NSICU1 | 256 | 0.06 | 0.03 | 0.5 | 0.5 | <0.06 | 0.12 | 0.06 | 0.06 | Reference 2 | VF125AL |
FLC fluconazole, ITC itraconazole, POS posaconazole, VRC voriconazole, AMB amphotericin B, 5-FC 5-flucytosine, AFG anidulafungin, CAS caspofungin, MFG micafungin, MIC minimal inhibitory concentration (μg/mL).
References
- Wang XJ, Bing J, Zheng QS, Zhang FF, Liu JB, Yue HZ, Tao L, Du H, Wang YN, Wang H, et al. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infec. 7(1): 93.
- Tian S, Rong C, Nian H, Li F, Chu Y, Cheng S, Shang H. 2018. First cases and risk factors of super yeast Candida auris infection or colonisation from Shenyang, China. Emerg Microbes Infect. 7(1): 128.
3.3. Morphological and Sap expression analyses
We next performed morphological analyses on seven representative strains isolated from different hospitals in China (including six MTLα strains and one MTLa strain). Two additional MTLa strains (CBS12766 from India and B11245 from the United States (Chowdhary et al. 2013; Lockhart et al. 2017; Du et al. 2020)) were also included. As shown in Figure 3, the nine strains exhibited similar cellular morphologies when cultured at 30°C on YPD liquid medium containing the red dye phloxine B. These strains, however, differed in colony appearance on agar plates of the same medium. The colonies of the MTLα strains appeared white on YPD + phyloxine B plates, whereas those of the MTLa strains appeared pink. These differences could be due to alterations in their cell wall structures or in their abilities to exclude the red dye. It remains to be investigated whether these differences are associated with antifungal resistance or virulence.
Figure 3.
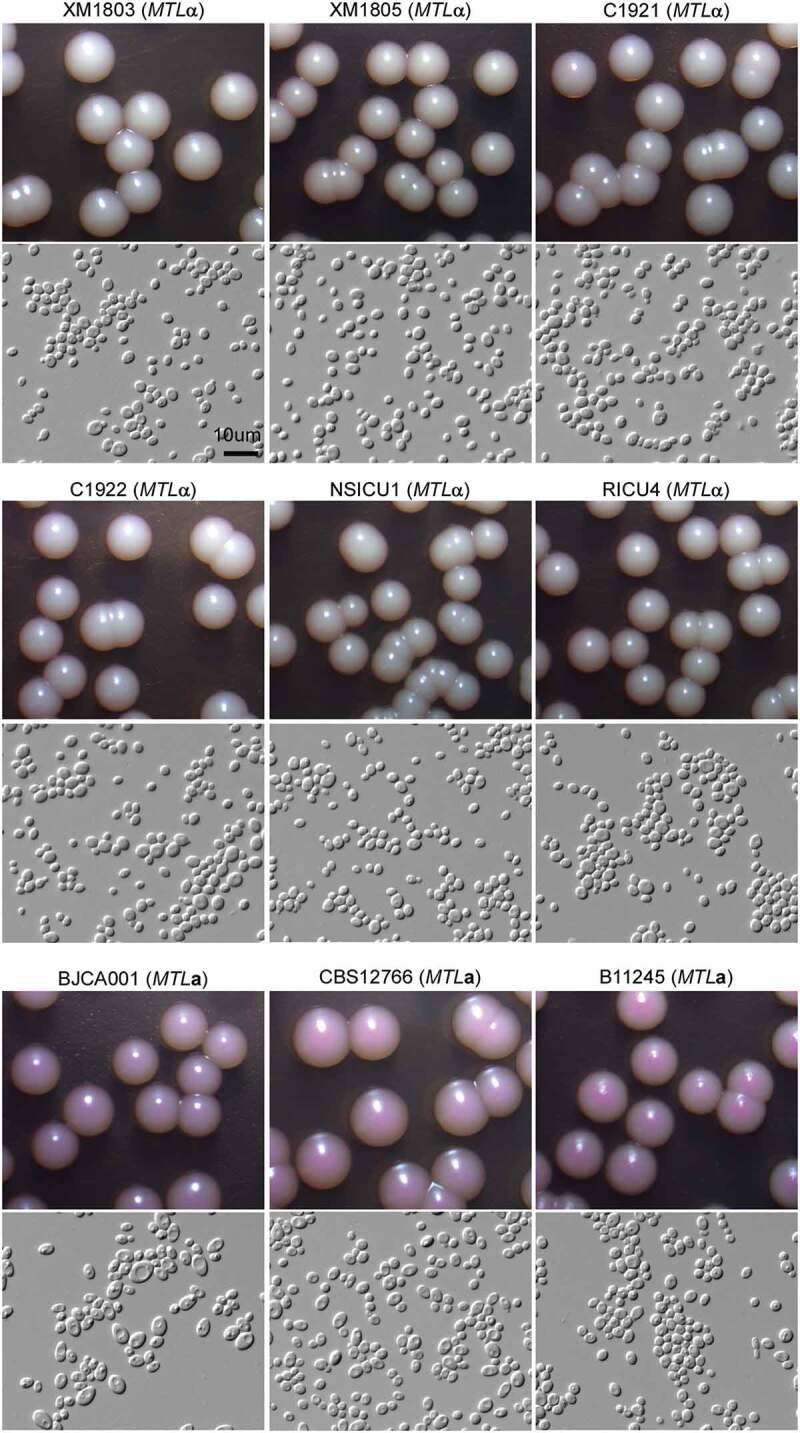
Colony and cellular morphologies of the different C. auris strains. Strains XM1803, XM1805, C1921, C1922, NSICU1, RICU4, and BJCA001 were isolated from China. The MTLa strains, CBS12766 and B11245, were isolated from India and Venezuela, respectively. Yeast-form cells were plated onto YPD medium supplemented 5 μg/mL of phloxine B and incubated at 25°C for seven days. Scale bar = 10 μm.
To compare expression of the Sap virulence factors among the different C. auris isolates from China, we examined Sap expression levels using YCB-BSA assays (Wang et al. 2018). As shown in Figure 4, MTLα strains secreted a significantly lower level of Saps than MTLa strains at 25°C. The expression of Saps in MTLα strains was significantly increased at 37°C, but it was still lower than that of CBS12766 and B11245 (MTLa strains) at 37°C. Interestingly, strain BJCA001 exhibited a comparatively lower level of Sap expression relative to the other strains at 37°C. These results imply that the expression of Saps in C. auris could be associated with the mating type and regulated by environmental temperatures.
Figure 4.
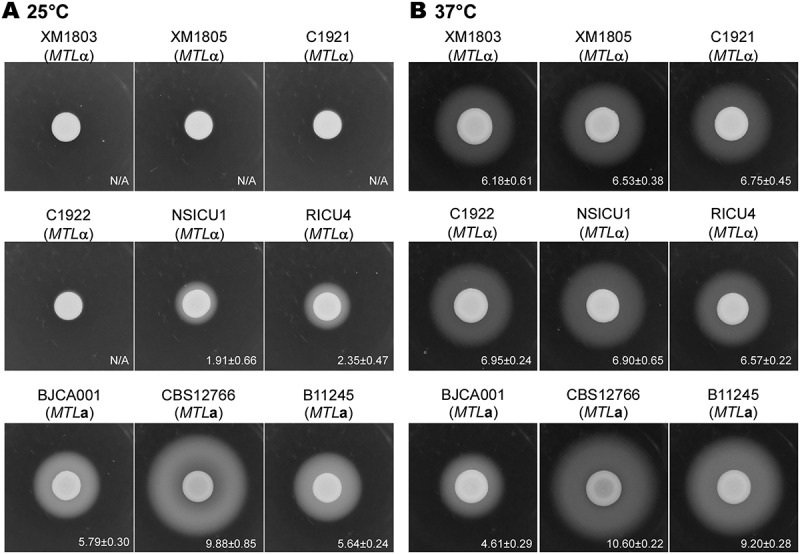
Sap expression for the different C. auris strains. Strains XM1803, XM1805, C1921, C1922, NSICU1, RICU4, and BJCA001 were isolated from China. The MTLa strains, CBS12766 and B11245, were isolated from India and Venezuela, respectively. Approximately 5 × 106 yeast-form cells of each strain in 5 μL ddH2O were spotted onto YCB-BSA medium plates and incubated at 25°C or 37°C for seven days. The white precipitation zones (halos) indicate the activity of the Saps. The widths (mm) of the zones were measured and shown below the corresponding image (n = 3). N/A = no obvious halos were observed.
In the current study, we report a case of a C. auris candidemia in a patient in Xiamen, China. This represents the first emergence of this fungal pathogen in south China. The patient had a travel history to Japan; however, it is unlikely that the Xiamen strain originated from Japan given that Japan has only reported the presence of C. auris isolates of the East Asian clade. The Xiamen strain is closely related to the Shenyang and Beijing isolates of the South African clade; however, the patient had no travel history to these two cities. Therefore, the source of this infection remains to be determined.
We also compared the genetic and biological characteristics of existing C. auris strains isolated from China and found that there are several distinct features among the different strains, including the expression of virulence factors, antifungal susceptibilities, and cellular morphologies. Our findings indicate that the majority of clinical isolates (19/20, 95%) belong to the South African clade and are MTLα in mating type, whereas a single strain is associated with the South Asian clade and is MTLa in mating type. Recently, additional C. auris MTLα isolates have been reported in Shenyang, China (Tian et al. 2021), while MTLa isolates have been reported in Hong Kong, China (Tse et al. 2021). The Erg11 hotspot mutation (VF125AL) was found in all nineteen isolates of the South African clade, but not in the isolate of the South Asian clade (BJCA001). Interestingly, these clinical isolates all exhibited distinct cellular morphologies and differences in the levels of secretion of Saps. Taken together, there are two reported genetic clades of C. auris present in China and the different clinical isolates exhibit diversity in both biological and genetic features.
Funding Statement
This work was supported by the National Natural Science Foundation of China awards (32000018 to JB; 82002123 to HD; 31930005 and 31625002 to GH), Natural Science Foundation of Shanghai award 19ZR1405100 (to HD), and funding from the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) award R35GM124594 and the Kamangar family in the form of an endowed chair to CJN.
Disclosure statement
Clarissa J. Nobile is a cofounder of BioSynesis, Inc., a company developing inhibitors and diagnostics of biofilm formation. All other authors declare no conflicts of interest.
Data availability
All data generated or analysed during this study are included in this published article.
References
- Ahmad S, Alfouzan W.. 2021. Candida auris: epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms. 9(4):807. doi: 10.3390/microorganisms9040807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing J, Hu T, Zheng Q, Munoz JF, Cuomo CA, Huang G.. 2020. Experimental evolution identifies adaptive aneuploidy as a mechanism of fluconazole resistance in Candida auris. Antimicrob Agents Chemother. 65(1):e01466–01420. doi: 10.1128/AAC.01466-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao JY, Han L, Qi LH, Fan WH, Liu J, Wang ZG, Xia X, Chen J, Zhang LL. 2018. Emergency of fungemia cases caused by fluconazole-resistant Candida auris in Beijing, China. J Infect. 77(6):569–571. doi: 10.1016/j.jinf.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. 2019. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 25(9):1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, et al. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 19(10):1670–1673. doi: 10.3201/eid1910.130393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Bing J, Hu TR, Ennis CL, Nobile CJ, Huang GH. 2020. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 16(10):e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard V, Mailler S, Chetry M, Vidal C, Durand G, Van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 59(8):535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, et al. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 64(2):134–140. doi: 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi JDO, Silva DR, Mendes-Giannini MJS, Rosalen PL. 2018. Candida auris: epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb Pathog. 125:116–121. doi: 10.1016/j.micpath.2018.09.014. [DOI] [PubMed] [Google Scholar]
- Satoh K, Tamura T, Makimura K. 2018. Errata of Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 53(1):41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Rong C, Nian H, Li F, Chu Y, Cheng S, Shang H. 2018. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerg Microbes Infect. 7(1):128. doi: 10.1038/s41426-018-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian SF, Bing J, Chu YZ, Chen JJ, Cheng ST, Wang QH, Zhang JP, Ma XC, Zhou BS, Liu L, et al. 2021. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerging Microbes Infect. 10(1):1088–1096. doi: 10.1080/22221751.2021.1934557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H, Tsang AKL, Chu YW, Tsang DNC. 2021. Draft genome sequences of 19 clinical isolates of Candida auris from Hong Kong. Microbiol Resour Announc. 10(1):e00308–00320. doi: 10.1128/MRA.00308-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Bing J, Zheng QS, Zhang FF, Liu JB, Yue HZ, Tao L, Du H, Wang YN, Wang H, et al. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 7(1):93. doi: 10.1038/s41426-018-0095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


