Abstract
The recently described genus Pandoraea contains five named species (Pandoraea apista, Pandoraea pulmonicola, Pandoraea pnomenusa, Pandoraea sputorum, and Pandoraea norimbergensis) and four unnamed genomospecies. Pandoraea spp. have mainly been recovered from the respiratory tracts of cystic fibrosis (CF) patients. Accurate genus- and species-level identification by routine clinical microbiology methods is difficult, and differentiation from Burkholderia cepacia complex organisms may be especially problematic. This can have important consequences for the management of CF patients. On the basis of 16S ribosomal DNA sequences, PCR assays for the identification of Pandoraea spp. were developed. A first PCR assay was developed for the identification of Pandoraea isolates to the genus level. PCR assays for the identification of P. apista and P. pulmonicola as a group, P. pnomenusa, P. sputorum, and P. norimbergensis were also developed. All five assays were evaluated with a panel of 123 bacterial isolates that included 69 Pandoraea sp. strains, 24 B. cepacia complex strains, 6 Burkholderia gladioli strains, 9 Ralstonia sp. strains, 5 Alcaligenes xylosoxidans strains, 5 Stenotrophomonas maltophilia strains, and 5 Pseudomonas aeruginosa strains. The use of these PCR assays facilitates the identification of Pandoraea spp. and avoids the misidentification of a Pandoraea sp. as a B. cepacia complex isolate.
Chronic microbial colonization of the large airways, leading to exacerbations of pulmonary infection, is the major cause of morbidity and mortality in patients with cystic fibrosis (CF). Typical pathogens of CF patients include Burkholderia cepacia complex organisms, Staphylococcus aureus, Pseudomonas aeruginosa, and Haemophilus influenzae (4, 7, 8). Other glucose nonfermenters such as Stenotrophomonas maltophilia, Alcaligenes xylosoxidans, Ralstonia pickettii, and Burkholderia gladioli can frequently be found as well (1, 4).
A recent polyphasic taxonomic study (2) showed that a number of bacterial isolates cultured from respiratory secretions of CF patients belonged to the new genus Pandoraea. Originally, this genus contained five named species (Pandoraea apista [the type species], Pandoraea norimbergensis, Pandoraea pnomenusa, Pandoraea pulmonicola, and Pandoraea sputorum) and one unnamed genomic species (2). More recently, three new (yet unnamed) Pandoraea genomospecies, previously classified as CDC weak oxidizer group 2, were added (3). Pandoraea spp. have mainly been isolated from the respiratory secretions of CF patients but have also been found in other clinical samples (including blood), soil, water, and food (2, 3, 11).
In the clinical microbiology laboratory, identification to the species level and differentiation of Pandoraea species from organisms belonging to the B. cepacia complex, R. pickettii, or Ralstonia paucula may be problematic (2, 5, 11). To aid in the identification of these organisms, we developed PCR-based identification strategies based on the 16S rRNA gene (rDNA). A first PCR assay was designed to identify all Pandoraea spp. Subsequent PCR assays were designed to identify individual Pandoraea species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All isolates were obtained from the Burkholderia cepacia Research Laboratory and Repository (University of Michigan, Ann Arbor), the BCCM/LMG Bacteria Collection (Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium), or the Centers for Disease Control and Prevention (Atlanta, Ga.). Strains were grown aerobically on Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.) supplemented with 1.8% (wt/vol) agar and were incubated overnight at 32°C. For evaluation of the PCR assays, 69 Pandoraea isolates were used. Isolates were identified by a polyphasic taxonomic approach or DNA-DNA hybridizations, as described previously (2, 3). If serial isolates from a patient were available, only one isolate was included in the present study. We included 29 P. apista isolates (LMG 16407T, LMG 18818, LMG 18089, AU003, AU01287, AU0170, AU0871, AU1330, AU1430, AU1434, AU1465, AU1596, AU1656, AU1728, AU1776, AU2028, AU2160, AU2172, AU2303, AU2347, AU2377, AU2405, AU2513, AU2593, HI2743, HI2744, HI2782, CDC-G3307, and CDC-G9278), 5 P. norimbergensis isolates (LMG 18379T, LMG 13019, LMG 16603, R-4026, and AU1290), 3 P. pulmonicola isolates (LMG 18106T, LMG 18107, and LMG 18108), 15 P. pnomenusa isolates (LMG 18087T, LMG 18817, LMG 18820, AU1039, AU2170, AU2268, HI2279, HI2344, HI2778, HI2779, HI2780, HI2781, CDC-G5056, CDC-G7835, and CDC-G8107), 13 P. sputorum isolates (LMG 18819T, LMG 18100, AU0012, AU0103, AU0125, AU1359, AU1570, AU2075, AU2224, AU2269, AU2302, AU2389, and HI2742), and 1 isolate of each unnamed genomic species (Pandoraea genomospecies 1 R-5199, Pandoraea genomospecies 2 CDC-G5084, Pandoraea genomospecies 3 CDC-G9805, and Pandoraea genomospecies 4 CDC-H652).
We also included 54 isolates representing phylogenetically related species and other species that may be encountered in the sputum of CF patients: B. cepacia genomovar I (three isolates), Burkholderia multivorans (two isolates), B. cepacia genomovar III (seven isolates), Burkholderia stabilis (two isolates), Burkholderia vietnamiensis (two isolates), B. cepacia genomovar VI (five isolates), Burkholderia ambifaria (three isolates), B. gladioli (six isolates), R. picketii (five isolates), R. paucula (two isolates), Ralstonia gilardii (two isolates), P. aeruginosa (five isolates), A. xylosoxidans (five isolates), and S. maltophilia (five isolates).
DNA preparation.
DNA was prepared by heating one or two colonies (picked from a culture grown overnight) at 95°C for 15 min in 20 μl of lysis buffer containing 0.25% (vol/vol) sodium dodecyl sulfate and 0.05 M NaOH. Following lysis, 180 μl of sterile distilled water was added to the lysis buffer and the DNA solutions were stored at −20°C.
Development of primers for genus- and species-specific PCR assays.
Available 16S rDNA sequences of all Pandoraea, Burkholderia, and Ralstonia species were retrieved from the GenBank database and were aligned by using the MegAlign software package (DNAStar Inc., Madison, Wis.). On the basis of this alignment, the primers shown in Table 1 were developed.
TABLE 1.
16S rDNA-derived oligonucleotide primers used for PCR
| Primer | Sequence (5′-3′) | Target | Annealing temp (°C) | Nucleotide position | Product size (bp) |
|---|---|---|---|---|---|
| panF | GGGCTYAACCTGGGAACTGCATTC | Genus Pandoraea | 68 | 584–607a | 645a |
| panR | CGRYTTGGCRRCCCTCTGTACCG | Genus Pandoraea | 1207–1229a | ||
| appuFb | CAGTGGGGAATTTTGGACAATGGGCGCA | P. apista, P. pulmonicola | 71 | 324–351a | 905a |
| spuF | CTCGCGCTACAAGAGCGGCCGATA | P. sputorum | 63 | 184–207c | 813c |
| spuR | CGAGCACATCCGCCTCTCAGCAGAC | P. sputorum | 973–997c | ||
| pnoF | CAGTGGGGAATTTTGGACAATGGGCGA | P. pnomenusa | 71 | 313–339d | 673d |
| pnoR | CGAGCACTCCCACCTCTCAGCAGGA | P. pnomenusa | 962–986d | ||
| norF | CAAGAGCAGCCGATGTCAGATTAGCTT | P. norimbergensis | 66 | 193–219e | 804e |
| norR | CGAGCACTCTCACCTTTCAGCAA | P. norimbergensis | 975–997e |
Position and size relative to 16S rDNA sequence of P. apista LMG 18089 (AF139172).
Used in combination with oligonucleotide primer panR.
Position and size relative to 16S rDNA sequence of P. sputorum LMG 18819T (AF139176).
Position and size relative to 16S rDNA sequence of P. pnomenusa LMG 18087T (AF139174).
Position and size relative to 16S rDNA sequence of P. norimbergensis LMG 13019 (AF139171).
PCR.
PCR assays were performed in 25-μl reaction mixtures, containing 2 μl of DNA solution, 1 U of Taq polymerase (Gibco BRL, Gaithersburg, Md.), 250 mM (each) deoxynucleoside triphosphate (Gibco BRL), 5 μl of 5 M betaine (Sigma, St. Louis, Mo.), 1× PCR buffer (Qiagen, Valencia, Calif.), and 20 pmol of each oligonucleotide primer. Amplification was carried out with a PTC-100 programmable thermal cycler (MJ Research, Incline Village, Nev.). After initial denaturation for 2 min at 94°C, 20 amplification cycles were completed, each consisting of 1 min at 94°C, 45 s at the appropriate annealing temperature (Table 1), and 1 min at 72°C. A final extension of 10 min at 72°C was applied. Negative control PCRs with all components of the reaction mixture except template DNA were used for every experiment.
RESULTS AND DISCUSSION
Primer design.
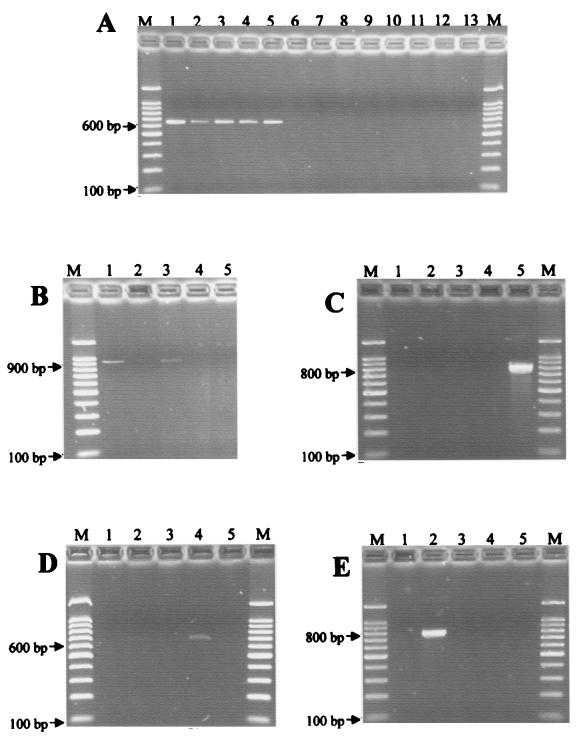
Multiple-sequence alignment of the 16S rRNA genes of the different Pandoraea species revealed high similarity values. Several genus-level sequence signatures were detected, and these were incorporated into genus-specific primers panF (forward primer) and panR (reverse primer). Species-level sequence signatures were also detected, and specific PCR primers were developed for the identification of P. apista and P. pulmonicola (as a group) (forward primer appuF used in combination with reverse primer panR), P. pnomenusa (forward primer pnoF used in combination with reverse primer pnoR), P. sputorum (forward primer spuF used in combination with reverse primer spuR), and P. norimbergensis (forward primer norF used in combination with reverse primer norR) (Table 1). Figure 1 illustrates the results of the PCRs with these various primer pairs.
FIG. 1.
PCR analysis of Pandoraea sp. type strains and other isolates with primer pairs panF-panR (A), appuF-panR (B), spuF-spuR (C), pnoF-pnoR (D), and norF-norR (E). Lanes: M, 100-bp DNA ladder (Gibco BRL); 1, P. apista LMG 16407T; 2, P. norimbergensis LMG 18379T; 3, P. pulmonicola LMG 18106T; 4, P. pnomenusa LMG 18087T; 5, P. sputorum LMG 18819T; 6, B. cepacia genomovar I LMG 1222T; 7, B. cepacia genomovar III HI2711; 8, B. gladioli AU1750; 9, R. gilardii LMG 5886T; 10, R. paucula LMG 3244T; 11, P. aeruginosa AU0225; 12, S. maltophilia AU0026; 13, A. xylosoxidans AU2116.
Sensitivities and specificities of PCR assays.
Each of the 123 strains included in the present study was examined by PCR with the primer pairs mentioned above. The results are detailed in Table 2.
TABLE 2.
Results of PCR assays with different primer pairs
| Primer pair and species tested | Sensitivity (%)a | Specificity (%)a | No. of strains for which result wasb:
|
|
|---|---|---|---|---|
| Positive | Negative | |||
| panF-panR | ||||
| Pandoraea sp. | 99 | 100 | 68/69 | 1/69 |
| All other species | 0/54 | 54/54 | ||
| spuF-spuR | ||||
| P. sputorum | 100 | 98 | 13/13 | 0/13 |
| All other species | 2/110 | 108/110 | ||
| norF-norR | ||||
| P. norimbergensis | 100 | 100 | 5/5 | 0/5 |
| All other species | 0/118 | 118/118 | ||
| appuF-panR | ||||
| P. apista and P. pulmonicola | 97 | 99 | 31/32 | 1/32 |
| All other species | 1/91 | 90/91 | ||
| pnoF-pnoR | ||||
| P. pnomenusa | 100 | 82 | 15/15 | 0/15 |
| Other Pandoraea species | 4/54 | 50/54 | ||
| All other species | 16c/54 | 38/54 | ||
Overall values for each primer pair are given. The specificity of the assay with primer pair pnoF-pnoR for the genus Pandoraea was 93%.
Values are numbers of strains for which result was obtained/numbers of strains tested.
All 16 isolates were B. cepacia complex isolates.
Use of PCR assays for the identification of Pandoraea spp.
B. cepacia complex organisms are capable of chronic colonization of the respiratory tracts of CF patients and are associated with increased rates of morbidity and mortality. To prevent the interpatient spread of B. cepacia complex organisms, strict infection control measures which have a significant impact on the quality of life of CF patients are recommended (7, 8). Accurate identification of B. cepacia complex bacteria is essential for infection control but is far from straightforward, and taxa that are frequently misidentified as belonging to the B. cepacia complex include B. gladioli, R. pickettii, S. maltophilia, and Pandoraea spp. (5, 6, 10, 12). Species belonging to the genus Pandoraea have mainly been isolated from CF patients worldwide, but other sources of isolation include blood, powdered milk, water, and sludge (2, 3, 11). Correct identification of these organisms is difficult and requires extensive biochemical testing or a polyphasic taxonomic approach, or both (2, 3, 5). Recently, several rRNA gene-based PCR assays have been developed for the identification of B. cepacia complex organisms (9, 15) and other pathogens of CF patients, including S. maltophilia (14) and B. gladioli (13).
To design PCR tests that would allow the identification of Pandoraea species, we sought genus-level and species-level signature sequences in the 16S rRNA gene. When available Pandoraea sp. 16S rRNA gene sequences were compared with sequences from representatives of the phylogenetically closely related genera Burkholderia and Ralstonia, several regions within the 16S rRNA gene were identified that showed enough diversity to allow the design of a primer pair (panF-panR) specific for the genus Pandoraea. As reported previously, comparison of the 16S rRNA gene sequences from all Pandoraea species revealed a high degree of identity (2, 3), resulting in few opportunities to design species-specific primers. The norF-norR primer pair allowed the specific amplification of a 16S rDNA fragment for all P. norimbergensis isolates investigated. Primer pair spuF-spuR allowed the amplification of a 16S rDNA fragment of all P. sputorum isolates investigated; this primer pair also reacted with Pandoraea genomospecies 2 and 3. By focusing on sequences shared by P. apista and P. pulmonicola, forward primer appuF was designed. When used in combination with genus-specific reverse primer panR, this primer was specific for P. apista and P. pulmonicola isolates (this primer pair also reacted with one B. cepacia genomovar VI strain investigated). Primer pair pnoF-pnoR was designed for the specific amplification of a 16S rDNA fragment of P. pnomenusa isolates. Although this PCR assay shows excellent sensitivity, it also amplified a fragment of the 16S rDNAs of two P. norimbergensis isolates, one P. apista isolate, and Pandoraea genomospecies 4. Several isolates belonging to the B. cepacia complex reacted with this primer pair as well. However, since the genus-specific PCR assay and the assays for P. apista and P. pulmonicola as a group and for P. norimbergensis all displayed excellent specificities, the combined use of these assays will provide accurate identification of P. pnomenusa (the genus-specific primers will not react with B. cepacia complex isolates, while the primers for P. apista-P. pulmonicola and P. norimbergensis will not react with P. pnomenusa isolates). Strain R-5199 (Pandoraea genomospecies 1) reacted only with the genus-specific primer pair panF-panR. We have not developed PCR assays specific for the unnamed Pandoraea genomic species, pending the availability of similar isolates.
Primer pair RHG-F (5′-GGGATTCATTTCCTTAGTAAC-3′) and RHG-R (5′-GCGATTACTAGCGATTCCAGC-3′), described by LiPuma et al. (9), was previously shown to be specific for Burkholderia and Ralstonia species. Since the genus Pandoraea occupies a phylogenetic position intermediate between the genera Burkholderia and Ralstonia (2), it is not surprising that all Pandoraea isolates investigated reacted positively with this primer pair (J. J. LiPuma, unpublished data). Isolates from the sputa of CF patients that are PCR positive with primer pair RHG-F–RHG-R but negative in PCR assays for B. cepacia complex organisms (9, 15) could belong to the genus Pandoraea and can be further investigated with primers specific for the genus Pandoraea described in this report.
In summary, the combined use of the PCR assays described here allows the separation of members of the genus Pandoraea from closely related genera. It also allows the accurate identification of all five named Pandoraea species. The use of these assays can significantly reduce the misidentification of Pandoraea spp. as B. cepacia complex isolates and by so doing will be an important adjunct in the evaluation of isolates found in cultures of sputa from CF patients.
ACKNOWLEDGMENTS
This work was supported by a grant from the Cystic Fibrosis Foundation (United States) (to J.J.L.). T.C. is supported by the Caroll Haas Research Fund in Cystic Fibrosis.
We thank Severine Laevens and Theodore Spilker for excellent technical assistance. We are also grateful to R. S. Weyant for kindly providing strains.
REFERENCES
- 1.Burns J L, Emerson J, Stapp J R, Yim D L, Krzewinski J, Louden L, Ramsey B W, Clausen C R. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 2.Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan J R W, Gillis M, Vandamme P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol. 2000;50:887–899. doi: 10.1099/00207713-50-2-887. [DOI] [PubMed] [Google Scholar]
- 3.Daneshvar M I, Hollis D G, Steigerwalt A G, Whitney A M, Spangler L, Douglas M P, Jordan J G, MacGregor J P, Hill B C, Tenover F C, Brenner D J, Weyant R S. Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomospecies. J Clin Microbiol. 2001;39:1819–1826. doi: 10.1128/JCM.39.5.1819-1826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry D A, Mahenthiralingam E, Vandamme P, Coenye T, Speert D P. Biochemical and molecular approaches for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiska D L, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative, nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LiPuma J J. Burkholderia cepacia: management issues and new insights. Clin Chest Med. 1998;19:473–486. doi: 10.1016/s0272-5231(05)70094-0. [DOI] [PubMed] [Google Scholar]
- 8.LiPuma J J. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Curr Opin Pulm Med. 1998;4:337–441. doi: 10.1097/00063198-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 9.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMenamin J D, Zaccone T M, Coenye T, Vandamme P, LiPuma J J. Misidentification of Burkholderia cepacia in U.S. cystic fibrosis treatment centers: an analysis of 1051 recent sputum isolates. Chest. 2000;117:1661–1665. doi: 10.1378/chest.117.6.1661. [DOI] [PubMed] [Google Scholar]
- 11.Moore J E, Coenye T, Vandamme P, Elborn J S. First report of Pandoraea norimbergensis isolated from food—potential clinical significance. Food Microbiol. 2001;18:113–114. [Google Scholar]
- 12.Shelly D B, Spilker T, Gracely E J, Coenye T, Vandamme P, LiPuma J J. Utility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum culture. J Clin Microbiol. 2000;38:3112–3115. doi: 10.1128/jcm.38.8.3112-3115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitby P W, Pope L C, Carter K B, LiPuma J J, Stull T L. Species-specific PCR as a tool for the identification of Burkholderia gladioli. J Clin Microbiol. 2000;38:282–285. doi: 10.1128/jcm.38.1.282-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitby P W, Carter K B, Burns J L, Royall J A, LiPuma J J, Stull T L. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol. 2000;38:4305–4309. doi: 10.1128/jcm.38.12.4305-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitby P W, Carter K B, Hatter K L, LiPuma J J, Stull T L. Identification of members of the Burkholderia cepacia complex by species-specific PCR. J Clin Microbiol. 2000;38:2962–2965. doi: 10.1128/jcm.38.8.2962-2965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]