Abstract
We analyzed a 402-bp sequence of the mitochondrial cytochrome b gene of 34 strains of Exophiala jeanselmei and 16 strains representing 12 related species. The strains of E. jeanselmei were classified into 20 DNA types and 17 amino acid types. The differences between these strains were found in 1 to 60 nucleotides and 1 to 17 amino acids. On the basis of the identities and similarities of nucleotide and amino acid sequences, some strains were reidentified: i.e., two strains of E. jeanselmei var. hetermorpha and one strain of E. castellanii as E. dermatitidis (including the type strain), three strains of E. jeanselmei as E. jeanselmei var. lecanii-corni (including the type strain), three strains of E. jeanselmei as E. bergeri (including the type strain), seven strains of E. jeanselmei as E. pisciphila (including the type strain), seven strains of E. jeanselmei as E. jeanselmei var. jeanselmei (including the type strain), one strain of E. jeanselmei as Fonsecaea pedrosoi (including the type strain), and one strain of E. jeanselmei as E. spinifera (including the type strain). Some E. jeanselmei strains showed distinct nucleotide and amino acid sequences. The amino-acid-based UPGMA (unweighted pair group method with the arithmetic mean) tree exhibited nearly the same topology as those of the DNA-based trees obtained by neighbor joining, maximum parsimony, and maximum likelihood methods.
Exophiala species, opportunistic pathogens found in humans and other animals, cause cutaneous, subcutaneous, and systemic infections. These fungi are being recognized with increasing frequency as a cause of human disease, especially in immunocompromised patients. Clinical case reports are suggestive of the importance of Exophiala species as a major emerging group of fungal pathogens. They have been isolated from the environment, for example, the soil, decaying wood, pulp, bathwater, and even a humidifier (8, 10, 14, 19).
Until McGinnis and Padhye (15) described the annellation of the conidiogenous cells in Exophiala jeanselmei, this species was represented by numerous synonyms because of its pleomorphism (4, 15, 27). In the past, taxonomy and identification of Exophiala were mainly based on morphological characteristics. The types of propagation vary widely in their anamorphic life cycles (2). Because this genus is highly pleomorphic, generic and species delimitations are difficult with classical methods. Frequent revisions have been made in the classification and nomenclature of these fungi, in large part because of the polymorphism of the organism and its microstructural similarities to related genera. Uijthof considered the taxonomy and identification of Exophiala to be complicated by its pleoanamorphism and morphological variability (25). Nishimura and Miyaji studied the conidiogenesis of E. dermatitidis, E. jeanselmei, and E. spinifera by scanning electron microscopy and reported that the conidial ontogeneses of these three species are only annellidic and difficult to differentiate (18, 20). Masuda et al. reported that E. dermatitidis and E. jeanselmei share similar morphological features and have been confused (13), and some researchers previously reported E. jeanselmei to be a heterogeneous complex (4, 11, 13).
We are in urgent need of a reliable method of identification of the black yeasts (3). At present, several molecular methods such as DNA-DNA hybridization, restriction fragment length polymorphism (RFLP), randomly amplified polymorphic DNA, restriction enzyme mapping, and DNA sequencing have been used to supplement and aid in taxonomic and phylogenetic studies of the Hyphomycetes, including many studies on Exophiala, its relatives, and teleomorphs (6, 11, 13, 22, 24–26). Results of studies using these methods showed some species, such as E. moniliae and E. jeanselmei, to be complex taxa with high intraspecific variability.
The structure and function of mitochondrial (mt) cytochrome b protein are retained, and its substitution rates are in proportion to evolutionary time. Therefore, this gene has been used often in the identification, classification, and phylogenetic analysis of animals and plants (1, 5, 9). We previously reported the mt cytochrome b gene to be useful for identification, classification, and phylogenetic analysis of pathogenic species of fungi (28, 29, 30, 32, 33). In the present study, we used the mt cytochrome b gene to study E. jeanselmei and related species.
MATERIALS AND METHODS
Fungal strains.
Fifty strains were used in this study (Table 1).
TABLE 1.
Strains used in this studya
| Species | Cluster | IFM no. | DNA type | Amino acid type | mt RFLP type | Other designation(s) and source | Accession no. |
|---|---|---|---|---|---|---|---|
| E. jeanselmei | I | 4876 | D-19 | A-1 | R-3 | CBS 577.76; var. heteromorpha; human chromomycosis | AB025868 |
| 4877 | D-19 | A-1 | R-3 | CBS 578.76; var. heteromorpha; human chromomycosis | AB025869 | ||
| 4874 | D-20 | A-2 | ND | CBS 232.33; var. heteromorpha; T of Trichosporium heteromorphum; wood pulp | AB025874 | ||
| II | 4973 | D-2 | A-3 | R-9 | Sludge in bathroom drainpipe | AB055588 | |
| 4975 | D-2 | A-3 | R-9 | House bath | AB055589 | ||
| 4978 | D-2 | A-3 | R-9 | Sludge in bathroom drainpipe | AB055590 | ||
| 4996 | D-2 | A-3 | R-9 | Sludge in bathroom drainpipe | AB055591 | ||
| 4971 | D-1 | A-4 | R-11 | Sludge in bathroom drainpipe | AB025895 | ||
| 4970 | D-3 | A-5 | R-8 | Sludge in bathroom drainpipe | AB025894 | ||
| 4981 | D-3 | A-5 | R-8 | Sludge in bathroom drainpipe | AB055592 | ||
| 47649 | D-4 | A-5 | ND | ATCC 12734, CBS 123.33, IMI 062462; var. lecanii-corni; T of Pullularia fermentans var. benedekii | AB025893 | ||
| 4976 | D-5 | A-6 | R-8 | Sludge in bathroom drainpipe | AB055593 | ||
| III | 5393 | D-11 | A-7 | ND | MTU 23016; clinical isolate | AB025834 | |
| 40919 | D-11 | A-7 | ND | NHL; clinical isolate | AB025832 | ||
| 41494 | D-11 | A-7 | R-2 | DCU 419; clinical isolate | AB025833 | ||
| 4857 | D-12 | A-8 | R-12 | CDC B-1003; clinical isolate | AB025842 | ||
| 4855 | D-9 | A-9 | R-6 | DCU Chr-25; clinical isolate | AB025843 | ||
| 40921 | D-9 | A-9 | ND | NHL B-368; clinical isolate | AB025844 | ||
| 4863 | D-10 | A-9 | R-1 | KUM 1881; clinical isolate | AB055594 | ||
| 4865 | D-10 | A-9 | R-1 | KUM 2010; clinical isolate | AB055595 | ||
| 4867 | D-10 | A-9 | R-1 | KUM 2091; clinical isolate | AB055596 | ||
| 5394 | D-10 | A-9 | R-1 | MTU 23019; clinical isolate | AB055597 | ||
| 4853 | D-8 | A-10 | R-15 | KUM 1171; clinical isolate | AB025846 | ||
| 4858 | D-13 | A-11 | R-10 | CDC B-1768; clinical isolate | AB055598 | ||
| 4859 | D-13 | A-11 | R-10 | CDC B-1770; clinical isolate | AB025859 | ||
| 40920 | D-13 | A-11 | ND | NHL; clinical isolate | AB025860 | ||
| 4982 | D-14 | A-12 | R-10 | Public bath | AB055599 | ||
| 4852 | D-16 | A-13 | R-5 | ATCC 34123, CBS 507.90, CBS 664.76, IAM 14677; T of Torula jeanselmei; human | AB025862 | ||
| 4861 | D-16 | A-13 | R-5 | CDC B-2760; clinical isolate | AB025861 | ||
| 48159 | D-17 | A-13 | ND | Nagasaki Medical School, Dermatology Department, clinical isolate | AB025863 | ||
| 4983 | D-15 | A-14 | R-4 | Public bath | AB025858 | ||
| 4860 | D-18 | A-15 | R-14 | CDC B-1773; clinical isolate | AB055600 | ||
| 4856 | D-7 | A-16 | R-7 | DCU 677; clinical isolate | AB025886 | ||
| 41493 | D-6 | A-17 | R-13 | DCU 418; clinical isolate | AB055601 | ||
| E. alcalophila | 4823 | CBS 520.82, IAM 12519; T; soil | AB025887 | ||||
| 41478 | AB025888 | ||||||
| E. angulospora | 47656 | CBS 482.92, NHL 3101; T; water from drinking well | AB025889 | ||||
| E. castellanii | 47653 | CBS 292.49; E. mansonii; T of Mycotorula schawii; human chromomycosis | AB025867 | ||||
| 47654 | CBS 353.52; E. mansonii; T of Exophiala bergeri; human chromomycosis | AB055602 | |||||
| E. dermatitidis | 4827 | ATCC 28869, CBS 207.35, IFO 6421, IMI 093967; T; human chromomycosis | AB025870 | ||||
| E. moniliae | 41500 | CBS 520.76; T; twig of Quercus sp. | AB025877 | ||||
| E. pisciphila | 4882 | ATCC 24901, CBS 537.73, IMI 176060; T; systemic mycosis of fish | AB025847 | ||||
| E. psychrophila | 47651 | CBS 191.87; T; Salmo salar in fish farm | AB025898 | ||||
| E. salmonis | 41053 | ATCC 16986, CBS 157.67, IFM 4884, IMI 124165; T; cerebral mycetoma of Salmo clarkii | AB025897 | ||||
| E. spinifera | 4883 | ATCC 18218, CBS 899.68; T of Phialophora spinifera; human nasal granuloma | AB025855 | ||||
| Capronia mansonii | 4879 | ATCC 18659, CBS 101.67, IMI 134456, NRRL Y-7857; HT; Populus tremula | AB025878 | ||||
| Fonsecaea compacta | 41704 | Human | AB025882 | ||||
| 41931 | MTU | AB025883 | |||||
| Fonsecaea pedrosoi | 4887 | ATCC 18658; IMI 134458, CBS 271.37; NT; human | AB025884 | ||||
| 4889 | ATCC 44356 | AB025885 |
Abbreviations: ND, not determined; ATCC, American Type Culture Collection, Manassas, Va.; CDC, Centers for Disease Control and Prevention, Atlanta, Ga.; DCU, Department of Dermatology, School of Medicine, Chiba University, Chiba, Japan; IAM, Institute of Applied Microbiology, University of Tokyo, Tokyo, Japan; IFM, Institute for Food Microbiology (now the Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University), Chiba, Japan; IFO, Institute for Fermentation, Osaka, Japan; IMI, CAB International Mycological Institute (formerly the Imperial Mycological Institute), Egham, United Kingdom; KUM, Department of Dermatology, School of Medicine, Kanazawa University, Kanazawa, Japan; MTU, Department of Bacteriology, Faculty of Medicine, University of Tokyo; NHL, National Institute of Hygienic Sciences, Tokyo, Japan; NRRL, Northern Regional Research Laboratory (now the National Center for Agricultural Utilization Research), Peoria, Ill.; HT, holotype; NT, neotype; T, ex-type.
Isolation of fungal DNA.
Fungal mycelia that had been grown for 3 to 5 days at 27°C were harvested from potato dextrose broth. Fungal DNA was extracted according to the method of Wang et al. (28).
PCR amplification.
The primer E1m (5′-TGAGGTGCTACAGTTATTAC-3′) was used as the forward primer, and primer E2 (5′-GGTATAGMTCTTAAWATAGC-3′) or rEME2 (5′-AAAATAGCATAGAAAGGTAA-3′) was used as the reverse primer (28). The PCR amplification protocol consisted of 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 50°C, and extension for 2 min at 72°C (28).
Sequencing.
Both strands of the PCR products were sequenced using the Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Japan Co., Ltd., Tokyo, Japan) on an ABI Prism 377 DNA sequencer with primer E1m and primer rEME2 or E2 (28).
Computer analysis.
DNA and amino acid sequences derived from the yeast mt genetic code were aligned and compared with the GENETYX-MAC program (version 10.1.2), which is part of the Genetic Information Processing software (Software Development Co., Ltd., Tokyo, Japan). Phylogenetic trees were generated by the unweighted pair group method with the arithmetic mean (UPGMA) (16). Neighbor joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) methods were performed with the PAUP program (version 4.0 b6; beta version).
RESULTS
Among the strains of E. jeanselmei that we studied, the 402-bp sequences of the mt cytochrome b gene were aligned, and a high degree of variability was found. The pairwise comparisons of the numbers of differences in the nucleotide and estimated amino acid sequences between E. jeanselmei strains are shown in Table 2. They differed from each other in 1 to 60 nucleotides and 1 to 17 amino acids. The strains identified as E. jeanselmei were classified into 20 DNA types (D-1 to D-20) and 17 amino acid types (A-1 to A-17).
TABLE 2.
Numbers of nucleotide and amino acid sequence differences between different strains of E. jeanselmei
| DNA type | No. of nucleotide or amino acid sequence differencesa
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D-1 | D-2 | D-3 | D-4 | D-5 | D-6 | D-7 | D-8 | D-9 | D-10 | D-11 | D-12 | D-13 | D-14 | D-15 | D-16 | D-17 | D-18 | D-19 | D-20 | |
| D-1 | 39 | 46 | 45 | 45 | 50 | 47 | 41 | 41 | 41 | 43 | 36 | 44 | 45 | 46 | 44 | 44 | 51 | 54 | 51 | |
| D-2 | 8 | 52 | 51 | 53 | 52 | 48 | 47 | 46 | 48 | 46 | 47 | 48 | 49 | 47 | 44 | 44 | 54 | 58 | 56 | |
| D-3 | 12 | 10 | 1 | 1 | 53 | 51 | 49 | 51 | 50 | 46 | 46 | 49 | 50 | 53 | 51 | 51 | 59 | 57 | 57 | |
| D-4 | 12 | 10 | 0 | 2 | 52 | 52 | 48 | 50 | 49 | 45 | 45 | 48 | 49 | 52 | 50 | 50 | 58 | 54 | 54 | |
| D-5 | 11 | 11 | 1 | 1 | 54 | 52 | 50 | 52 | 51 | 47 | 47 | 50 | 51 | 54 | 52 | 52 | 60 | 56 | 56 | |
| D-6 | 15 | 15 | 14 | 14 | 15 | 33 | 45 | 43 | 44 | 44 | 42 | 47 | 48 | 49 | 45 | 46 | 49 | 46 | 46 | |
| D-7 | 12 | 12 | 13 | 13 | 14 | 9 | 42 | 43 | 45 | 44 | 41 | 47 | 48 | 49 | 45 | 45 | 50 | 48 | 48 | |
| D-8 | 7 | 9 | 11 | 11 | 12 | 11 | 10 | 9 | 6 | 22 | 25 | 25 | 26 | 29 | 30 | 31 | 34 | 44 | 46 | |
| D-9 | 8 | 10 | 12 | 12 | 13 | 12 | 11 | 1 | 3 | 22 | 24 | 26 | 27 | 31 | 30 | 31 | 33 | 41 | 45 | |
| D-10 | 8 | 10 | 12 | 12 | 13 | 12 | 11 | 1 | 0 | 22 | 26 | 25 | 26 | 31 | 32 | 33 | 34 | 42 | 45 | |
| D-11 | 7 | 8 | 10 | 10 | 11 | 12 | 11 | 5 | 6 | 6 | 23 | 33 | 34 | 33 | 32 | 33 | 37 | 43 | 39 | |
| D-12 | 7 | 9 | 9 | 9 | 10 | 11 | 10 | 4 | 5 | 5 | 3 | 30 | 31 | 30 | 26 | 27 | 34 | 41 | 42 | |
| D-13 | 9 | 8 | 8 | 8 | 9 | 10 | 9 | 5 | 6 | 6 | 6 | 5 | 1 | 8 | 12 | 13 | 16 | 42 | 46 | |
| D-14 | 10 | 9 | 9 | 9 | 10 | 11 | 10 | 6 | 7 | 7 | 7 | 6 | 1 | 9 | 13 | 14 | 17 | 48 | 47 | |
| D-15 | 12 | 10 | 11 | 11 | 12 | 13 | 12 | 8 | 9 | 9 | 7 | 6 | 3 | 4 | 10 | 11 | 18 | 47 | 47 | |
| D-16 | 10 | 9 | 9 | 9 | 10 | 11 | 10 | 6 | 7 | 7 | 5 | 4 | 1 | 2 | 2 | 1 | 14 | 40 | 45 | |
| D-17 | 10 | 9 | 9 | 9 | 10 | 11 | 10 | 6 | 7 | 7 | 5 | 4 | 1 | 2 | 2 | 0 | 15 | 40 | 46 | |
| D-18 | 16 | 15 | 15 | 15 | 16 | 17 | 15 | 12 | 13 | 13 | 11 | 10 | 7 | 8 | 8 | 6 | 6 | 46 | 52 | |
| D-19 | 16 | 14 | 15 | 15 | 16 | 14 | 12 | 13 | 14 | 14 | 12 | 13 | 8 | 9 | 11 | 9 | 9 | 14 | 22 | |
| D-20 | 14 | 11 | 14 | 14 | 15 | 14 | 12 | 13 | 14 | 14 | 11 | 11 | 11 | 12 | 13 | 12 | 12 | 17 | 6 | |
The numbers of nucleotide differences are above the diagonal, and the numbers of amino acid differences are below the diagonal.
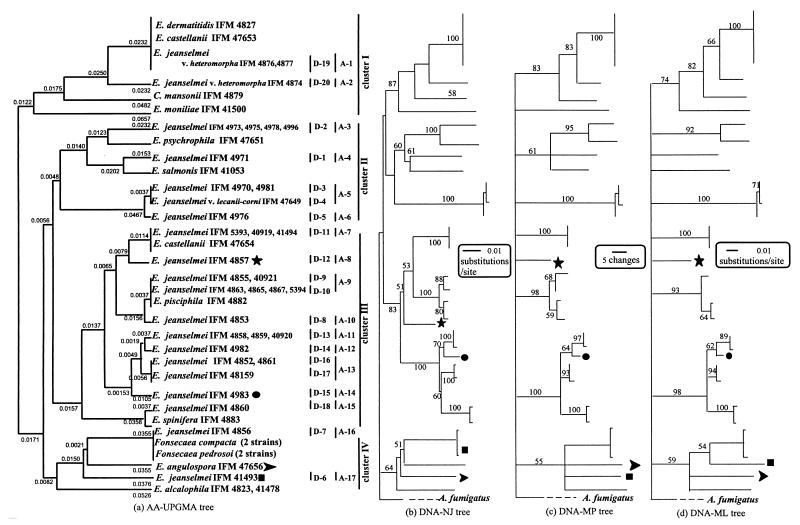
Phylogenetic trees were constructed by UPGMA, NJ, MP, and NL methods (Fig. 1). For each analysis method, the resulting DNA-based and amino-acid-based trees did not show the same topologies (data not shown). The amino-acid-based UPGMA tree and the DNA-based NJ, MP, and ML trees showed a high degree of similarity in the cluster formations and topologies (Fig. 1). Four clusters (I, II, III, and IV) were formed (Fig. 1). A few differences in topology were found in four strains: E. jeanselmei IFM 4857 (D-12, A-8), E. jeanselmei IFM 4983 (D-15, A-14), E. jeanselmei IFM 41493 (D-6, A-17), and E. angulospora IFM 47656 (Fig. 1).
FIG. 1.
(a) Phylogenetic tree obtained by use of the amino acid sequences estimated from the 402-bp nucleotide sequences of the mt cytochrome b genes. UPGMA was used. (b to d) DNA-based tree of the mt cytochrome b genes. NJ, MP, and ML were used. These trees were subjected to 100 bootstrap replications. Aspergillus fumigatus was selected as the outgroup.
Several strains had completely identical nucleotide sequences (Fig. 1). One identical group included E. castellanii IFM 47653 (CBS 292.49, type strain of Mycotorula schawii, renamed E. dermatitidis by Haase et al. [7]), E. dermatitidis IFM 4827 (CBS 207.35, type strain), and two strains of “E. jeanselmei var. heteromorpha” (IFM 4876 [CBS 577.76] and IFM 4877 [CBS 578.76], renamed E. dermatitidis by the Centraalbureau voor Schimmelcultures [CBS], Baarn, The Netherlands). Another group was E. jeanselmei IFM 4973, IFM 4975, IFM 4978, and IFM 4996 (D-2). E. jeanselmei IFM 4970 and IFM 4981 (D-3) comprised a third group. Several identical E. jeanselmei strains, IFM 5393, IFM 40919, and IFM 41494 (D-11), were also identical with E. castellanii IFM 47654 (CBS 353.52, determined to be the type strain of E. bergeri by CBS). E. jeanselmei IFM 4855 and IFM 40921 (D-9) were identical, as were four E. jeanselmei strains in D-10: IFM 4863, IFM 4865, IFM 4867, and IFM 5394. In D-13, E. jeanselmei IFM 4858, IFM 4859, and IFM 40920 were the same, and in D-16, E. jeanselmei IFM 4852 (CBS 507.90 and CBS 664.76, type strain) and IFM 4861 (D-16) were the same.
Other strains showing different DNA sequences shared identical amino acid sequences. They were E. jeanselmei IFM 4970 and IFM 4981 and E. jeanselmei var. lecanii-corni IFM 47649 (CBS 123.33) (A-5); E. jeanselmei IFM 4855, IFM 40921, IFM 4863, IFM 4865, IFM 4867, and IFM 5394 (A-9) and E. pisciphila IFM 4882 (CBS 537.73, type strain); E. jeanselmei IFM 4852 (type strain), IFM 4861, and IFM 48159 (A-13); and E. jeanselmei IFM 4856 (A-16), Fonsecaea pedrosoi (two strains, including type strain), and two strains of Fonsecaea compacta.
Several strains had high similarities in amino acid sequences. The strains in A-5 (D-3, D-4) differed from the strains in A-6 (D-5) by one amino acid. The same was true between strains in A-9 (D-9, D-10) and A-10 (D-8) and between those in A-15 (D-18) and E. spinifera IFM 4883. The strains in A-11 (D-13), A-12 (D-14), and A-13 (D-16, D-17) had one or two amino acid differences between them.
The two strains of E. castellanii (E. mansonii) were significantly different from Capronia mansonii, the teleomorphic species of E. castellanii; one had 35 nucleotide sequence differences and 14 amino acid sequence differences, and the other had 45 nucleotide sequence differences and 16 amino acid sequence differences.
DISCUSSION
E. jeanselmei is known to be composed of a genetically heterogeneous group (11, 13, 24, 25). Masuda et al. (13) used DNA-DNA hybridization to classify E. jeanselmei into six groups. DNA similarities between one group and another (relative values, 30% or less) seem too low for these groups to comprise a single species (13). Kawasaki et al. classified 45 strains of E. jeanselmei into 18 types according to mt restriction profiles and suggested that it is a complex organism (11). When we used the same strains as Kawasaki and colleagues did, the DNA and amino acid types by the mt cytochrome b were identical with mt RFLP typing (Table 1). The identification and classification of such “species” with classical methods are difficult to accomplish. Therefore, molecular methods are important for identification, reidentification, reclassification, and phylogenetic analysis. The mt cytochrome b as well as other genes will be useful, important tools in future studies.
In A-1 (D-19), E. castellanii IFM 47653 (CBS 292.49, type strain of M. schawii) is now know to have ITS1-ITS4 gene and small-subunit ribosomal DNA identical to those of E. dermatitidis CBS 207.35 (7, 24, 25); thus, the E. castellanii strain has been reclassified as the E. dermatitidis strain by CBS. Two strains of E. jeanselmei var. heteromorpha (IFM 4876 and IFM 4877) also were renamed as the species E. dermatitidis by CBS. Our results confirmed that these three were strains of E. dermatitidis.
Some researchers (7, 12, 25) considered three varieties of E. jeanselmei to deserve the status of separate species. Our results showed considerable differences between the three in both nucleotide sequences (49 to 54 differences) and amino acid sequences (9 to 14 differences [Table 2]). Haase et al. (7) proposed E. jeanselmei var. lecanii-cornii to be a distinct species, Exophiala lecanii-cornii. Our results support the proposal that these varieties are in fact separate species.
Through investigation of ribosomal DNA, Uijthof showed that the three species originating from fish (E. salmonis CBS 157.67, E. pisciphila CBS 537.73, and E. psychrophila CBS 191.87) formed a single cluster (25). Our results, by contrast, showed that although E. salmonis and E. psychrophila were in the same cluster, cluster II, E. pisciphila was in cluster III.
E. jeanselmei strains studied were classified according to 20 DNA types and 17 amino acid types. In this study, several E. jeanselmei strains were reidentified because of identical (100%) or very similar (approximately 99%) amino acid sequences. Three strains in D-3 and D-5 (IFM 4970, IFM 4981, and IFM 4976) were considered to belong to E. lecanii-corni (D-4) (type strain). Three strains in D-11 (IFM 5393, IFM 40919, and IFM 41494) belonged to E. bergeri (type strain). Seven strains in D-8, D-9, and D-10 (IFM 4853, IFM 4855, IFM 40921, IFM 4863, IFM 4865, IFM 4867, and IFM 5394) belonged to E. pisciphila (type strain). One strain in D-18 (IFM 4860) belonged to E. spinifera (type strain), and one strain in D-7 (IFM 4856) belonged to F. pedrosoi (neotype). Six strains in D-13, D-14, D-16, and D-17 (IFM 4858, IFM 4859, IFM 40920, IFM 4982, IFM 4861, and IFM 48159) had amino acid sequences identical or highly similar to that of the type strain of E. jeanselmei var. jeanselmei (D-16) and thus were considered to be appropriately classified as E. jeanselmei strains.
Other strains located in D-2 (A-3), D-1 (A-4), D-12 (A-8), D-15 (A-14), and D-6 (A-17) had many differences in amino acid sequences and formed distinct groups (Fig. 1 and Table 2). They may represent new species or species not examined in this study. Further investigations will be necessary to determine their identity.
The strains isolated from clinical materials including 18 clinical isolates identified as “E. jeanselmei,” E. bergeri (chromomycosis of humans), E. spinifera (nasal granuloma of humans), and the fish-pathogenic species E. pisciphila mainly were located in cluster III. The fish-pathogenic species E. pisciphila was in cluster III together with human-pathogenic strains. This may mean that the fish pathogens are implicated in human chromomycosis. In addition, two strains isolated from public bathwater also were in cluster III. The strains isolated from sludge in a bathroom drainpipe mainly were included in cluster II together with two species of fish pathogens. The strains isolated from clinical materials and the environment were apparently monophyletic.
We found that the two strains of E. castellanii (E. mansonii) differed significantly from the teleomorphic species C. mansonii. Small-subunit ribosomal DNA analysis and morphology showed that E. castellanii IFM 47653 (CBS 292.49) should be renamed E. dermatitidis and that IFM 47654 (CBS 353.52) should be E. bergeri (7).
We compared the substitution differences (percent) from the complete region (approximately 1,160 bp) of the cytochrome b gene in three species of fungi (Aspergillus nidulans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe) and a 402-bp selected region. Similar percentages of substitution were found in the pairs A. nidulans-S. cerevisiae, A. nidulans-S. pombe, and S. cerevisiae-S. pombe. The substitution rates for the complete region and selected region in each pair were 38.1 and 41.6%, 43.4 and 45.6%, and 43.4 and 42.7%, respectively. Consequently, the analysis of 402 bp is more convenient, since 402-bp sequences can sufficiently reflect the complete region. The mt cytochrome b gene has proven a valuable tool for use in identification, classification, and phylogeny studies, because it is a functional gene and its substitution rate is in proportion to evolutionary time. DNA sequencing techniques have recently become easier and faster. Researchers can now perform DNA extraction, capillary PCR, and capillary DNA sequencing in only 8 h. This process not only saves time but also provides accurate, rapid identification that can aid diagnosis of patient conditions.
Theoretically, when the constancy of evolutionary rate, or a molecular clock, is assumed, UPGMA can give correct topology, and this method has been advocated for use in reconstructing phylogenetic trees (17, 21), while NJ, ML, and MP methods are suitable for lack of constancy in the evolutionary rate. Takezaki and Nei (23), and Zhakikh and Li (34) have shown elsewhere that the MP method can result in an incorrect topology in the case of a constant rate of evolution. The mt gene is favored as a molecular clock for animals and plants for which fossil remains exist (31). Although fungi have no fossil remains, we assume that fungal mt DNA is similar to that of animals and plants. The amino-acid-altering substitutions of nucleotides are more proportional to evolutionary time than is synonymous substitution. We compared the UPGMA tree and other trees (NJ, ML, and MP). DNA-based and amino acid-based trees constructed by the same methods showed different topologies (data not shown). However, the amino-acid-based UPGMA tree exhibited nearly the same topology as those of the DNA-based trees obtained by the other methods (NJ, ML, and MP). This confirmed that the amino acid sequences of mt cytochrome b are more constant than are the nucleotide sequences. Our previous results also indicated that the amino-acid-based UPGMA trees were more correct than the others were (28–30, 32, 33). Therefore, we suggest that, for phylogenetic analysis in the case of the mt cytochrome b gene, UPGMA is suitable for amino acid sequences and NJ, MP, and ML are suitable for nucleotide sequences.
In conclusion, mt cytochrome b gene analysis is useful for identification, classification, and phylogenetic analysis of the black yeast-like fungus E. jeanselmei and related species.
ACKNOWLEDGMENTS
We thank the Honor Scholarship (Association of International Education, Tokyo, Japan), the Sumitomo Scholarship (Social Welfare Business Group of Sumitomo Life Assurance Company, Osaka, Japan), OSF (Okamoto Scholarship Foundation, Chiba, Japan), Yonnmaru Scholarship (Yonnmaru alumni association, Chiba University, School of Medicine, Chiba, Japan), and Goho Life Science International Foundation (Japan) for providing scholarships to L. Wang.
REFERENCES
- 1.Brown W M, Jr, George M, Wilson A C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Hoog G S, Takeo K, Yoshida S, Gottlich E, Nishimura K, Miyaji M. Pleoanamorphic life cycle of Exophiala (Wangiella) dermatitidis. Antonie Leeuwenhoek. 1994;65:143–153. doi: 10.1007/BF00871755. [DOI] [PubMed] [Google Scholar]
- 3.de Hoog G S, Bowman B, Graser Y, Haase G, Elfari M, Gerrits van den Ende A H G, Melzer-Krick B, Untereiner W A. Molecular phylogeny and taxonomy of medically important fungi. Med Mycol. 1998;36(Suppl. 1):52–56. [PubMed] [Google Scholar]
- 4.Dixon D M, Polak-Wyss A. The medically important dematiaceous fungi and their identification. Mycoses. 1991;34:1–18. doi: 10.1111/j.1439-0507.1991.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S V, Wilson A C. Phylogenetically informative length polymorphism and sequence variability in mitochondrial DNA of Australian songbirds (Pomatostomus) Genetics. 1990;126:695–711. doi: 10.1093/genetics/126.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase G, Sonntag L, van de Peer Y, Uijthof J M J, Podbielski A, Melzer-Krick B. Phylogenetic analysis of ten black yeast species using nuclear small subunit rRNA gene sequences. Antonie Leeuwenhoek. 1995;68:19–33. doi: 10.1007/BF00873289. [DOI] [PubMed] [Google Scholar]
- 7.Haase G, Sonntag L, Melzer-Krick B, de Hoog G S. Phylogenetic inference by SSU-gene analysis of members of the Herpotrichiellaceae with special reference to human pathogenic species. Stud Mycol. 1999;43:80–97. [Google Scholar]
- 8.Hironaga M, Mochizuki T, Watanabe S. Cutaneous phaeohyphomycosis of the sole caused by Exophiala jeanselmei and its susceptibility to amphotericin B, 5-FC and ketoconazole. Mycopathologia. 1982;79:101–104. doi: 10.1007/BF00468086. [DOI] [PubMed] [Google Scholar]
- 9.Irwin D M, Kocher T D, Wilson A C. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 10.Katz B, McGinnis M R. A new species of Exophiala recovered from loblolly pine litter. Mycotaxon. 1980;11:182–184. [Google Scholar]
- 11.Kawasaki M, Ishizaki H, Nishimura K, Miyaji M. Mitochondrial DNA analysis of Exophiala jeanselmei and Exophiala dermatitidis. Mycopathologia. 1990;110:107–112. doi: 10.1007/BF00446999. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki M, Ishizaki H, Matsumoto T, Matsuda T, Nishimura K, Miyaji M. Mitochondrial DNA analysis of Exophiala jeanselmei var. lecanii-corni and Exophiala castellanii. Mycopathologia. 1999;146:75–77. doi: 10.1023/a:1007057713782. [DOI] [PubMed] [Google Scholar]
- 13.Masuda M, Naka W, Tajima S, Harata T, Nishikawa T, Kaufman L, Standard P. Deoxyribonucleic acid hybridization studies of Exophiala dermatitidis and Exophiala jeanselmei. Microbiol Immunol. 1989;33:631–639. doi: 10.1111/j.1348-0421.1989.tb02013.x. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto T, Matsuda T, McGinnis M R, Ajello L. Clinical and mycological spectra of Wangiella dermatitidis. Mycoses. 1993;36:145–155. doi: 10.1111/j.1439-0507.1993.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 15.McGinnis M R, Padhye A A. Exophiala jeanselmei, a new combination for Phialophora jeanselmei. Mycotaxon. 1977;5:341–352. [Google Scholar]
- 16.Nei M. Human evolution at the molecular level. In: Ohta T, Aoki K, editors. Population genetics and molecular evolution. Berlin, Germany: Japan Scientific Society Press/Springer-Verlag; 1985. pp. 41–64. [Google Scholar]
- 17.Nei M. Phylogenetic trees. In: Nei M, editor. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. pp. 287–326. [Google Scholar]
- 18.Nishimura K, Miyaji M. Studies on the phylogenesis of pathogenic ‘black yeasts.’. Mycopathologia. 1983;81:135–144. doi: 10.1007/BF00436818. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura K, Miyaji M. Pathogenicity of Exophiala jeanselmei for ddY mice. Mycopathologia. 1985;91:29–33. doi: 10.1007/BF00437283. [DOI] [PubMed] [Google Scholar]
- 20.Nishimura K, Miyaji M. Further studies on the phylogenesis of the genus Exophiala and Hortaea. Mycopathologia. 1985;92:101–109. doi: 10.1007/BF00444091. [DOI] [PubMed] [Google Scholar]
- 21.Saito N. Reconstruction of gene trees from sequence data. Methods Enzymol. 1996;266:427–449. doi: 10.1016/s0076-6879(96)66027-3. [DOI] [PubMed] [Google Scholar]
- 22.Spatafora J W, Mitchell T G, Vilgalys R. Analysis of genes coding for small-subunit rRNA sequences in studying phylogenetics of dematiaceous fungal pathogens. J Clin Microbiol. 1995;33:1322–1326. doi: 10.1128/jcm.33.5.1322-1326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takezaki N, Nei M. Inconsistency of the maximum parsimony method when the rate of nucleotide substitution is constant. J Mol Evol. 1994;39:210–218. doi: 10.1007/BF00163810. [DOI] [PubMed] [Google Scholar]
- 24.Uijthof J M J, de Hoog G S. PCR-ribotyping of type isolates of currently accepted Exophiala and Phaeococcomyces species. Antonie Leeuwenhoek. 1995;68:35–42. doi: 10.1007/BF00873290. [DOI] [PubMed] [Google Scholar]
- 25.Uijthof J M J. Relationships within the black yeast genus Exophiala based on ITS1 sequences. Mycol Res. 1996;100:1265–1271. [Google Scholar]
- 26.Untereiner W A. Fruiting studies in species of Capronia (Herpotrichiellaceae) Antonie Leeuwenhoek. 1995;68:3–17. doi: 10.1007/BF00873288. [DOI] [PubMed] [Google Scholar]
- 27.Vakili N G. Exophiala jeanselmei, a pathogen of earthworm species. J Med Vet Mycol. 1993;31:343–346. [Google Scholar]
- 28.Wang L, Yokoyama K, Miyaji M, Nishimura K. The identification and phylogenetic relationship of pathogenic species of Aspergillus based on the mitochondrial cytochrome b gene. Med Mycol. 1998;36:153–164. [PubMed] [Google Scholar]
- 29.Wang L, Yokoyama K, Miyaji M, Nishimura K. Mitochondrial cytochrome b gene analysis of Aspergillus fumigatus and related species. J Clin Microbiol. 2000;38:1352–1358. doi: 10.1128/jcm.38.4.1352-1358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Yokoyama K, Takahashi H, Kase N, Hanya Y, Yashiro K, Miyaji M, Nishimura K. Identification of species in Aspergillus section Flavi based on sequencing of the mitochondrial cytochrome b gene. Int J Food Microbiol. 2001;71:75–86. doi: 10.1016/s0168-1605(01)00597-9. [DOI] [PubMed] [Google Scholar]
- 31.Watson J D, Gilman M, Witkowski J, Zoller M. Mitochondrial DNA as a molecular clock. In: Watson J D, Gilman M, Witkowski J, Zoller M, editors. Recombinant DNA. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1991. pp. 446–447. [Google Scholar]
- 32.Yokoyama K, Biswas S K, Miyaji M, Nishimura K. Identification and phylogenetic relationship of the most common pathogenic Candida species inferred from mitochondrial cytochrome b gene sequences. J Clin Microbiol. 2000;38:4503–4510. doi: 10.1128/jcm.38.12.4503-4510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama K, Wang L, Miyaji M, Nishimura K. Identification, classification and phylogeny of the Aspergillus section Nigri inferred from mitochondrial cytochrome b gene. FEMS Microbiol Lett. 2001;200:241–246. doi: 10.1111/j.1574-6968.2001.tb10722.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhakikh A, Li W H. Inconsistency of the maximum-parsimony method: the case of five taxa with a molecular clock. Syst Biol. 1993;42:113–125. [Google Scholar]