Abstract
Current targeted cancer therapies are largely guided by mutations of a single gene, which overlooks concurrent genomic alterations. Here, we show that RNASEH2B, RB1, and BRCA2, three closely located genes on chromosome 13q, are frequently deleted in prostate cancer individually or jointly. Loss of RNASEH2B confers cancer cells sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition due to impaired ribonucleotide excision repair and PARP trapping. When co-deleted with RB1, however, cells lose their sensitivity, in part, through E2F1-induced BRCA2 expression, thereby enhancing homologous recombination repair capacity. Nevertheless, loss of BRCA2 resensitizes RNASEH2B/RB1 co-deleted cells to PARP inhibition. Our results may explain some of the disparate clinical results from PARP inhibition due to interaction between multiple genomic alterations and support a comprehensive genomic test to determine who may benefit from PARP inhibition. Last, we show that ATR inhibition can disrupt E2F1-induced BRCA2 expression and overcome PARP inhibitor resistance caused by RB1 loss.
PARP inhibitor sensitivity may depend on interaction between multiple genomic alterations.
INTRODUCTION
Alterations of DNA damage response (DDR) are associated with genomic instability, a hallmark of cancer, including prostate cancer (PCa). Genomic studies have revealed that approximately 10% primary and 27% metastatic prostate tumors have genomic loss (mutation or deletion) of at least one gene involved in DDR, with BRCA2 being the most frequently mutated gene (1, 2). These alterations have been correlated with therapeutic vulnerabilities in PCa cells. Specifically, defects in homologous recombination repair (HRR) would predict the response to poly(adenosine diphosphate–ribose) polymerase (PARP) inhibition. PARP is a family of enzyme involved in various cellular processes, notably DNA damage repair and genomic stability. PARP inhibitors (PARPis) are a different type of targeted therapy, which works by preventing PARP1 and PARP2 from repairing DNA single-strand breaks and resulting in stalled replication fork by trapping PARP1 and PARP2 on the DNA breaks (3, 4). These effects contribute to accumulation of DNA double-strand breaks (DSBs) that HRR-deficient cells cannot repair efficiently, causing overwhelming DNA damage and apoptotic cell death. The BRCA1 and BRCA2 genes encode proteins essential for HRR. Cancer cells lacking BRCA1/2 depend instead on PARP-regulated DNA repair and are highly sensitive to PARP inhibition (5, 6). Four PARPis (olaparib, NCT02987543; rucaparib, NCT02975934; niraparib, NCT02854436; and talazoparib, NCT03148795) are under clinical investigation in PCa, leading to regulatory approvals of olaparib and rucaparib for the treatment of metastatic castration-resistant PCa (mCRPC) patients with HRR deficiencies or BRCA1/2 mutations (7–12). While the results from these clinical trials have shown that patients with tumors harboring BRCA1/2 mutations benefit from PARP inhibition with a high response rate, the degree to which patients with non-BRCA genomic alterations respond to PARPis remains unclear after gene-by-gene analysis.
To expand the efficacy of PARPis to tumors with non-BRCA alterations, efforts have been made to find new vulnerabilities for PARP inhibition in different cell models. Clustered regularly interspersed short palindromic repeat (CRISPR)–Cas9 loss-of-function genetic screen is a powerful approach to identify genes that, once deleted, make cells more sensitive to PARP inhibition. Using this approach, recent studies have found that inactivation of enzymes involved in excision of genomic ribonucleotides or aberrant nucleotides may create vulnerability of cancer cells to PARP trapping (13, 14). The alteration of genes encoding these enzymes are potential genomic biomarkers or actionable targets for PARPis. RNASEH2B is one of these genes, which is particularly intriguing for PCa because it is frequently deleted in both primary and metastatic prostate tumors. The protein encoded by RNASEH2B is one of the three subunits comprising ribonuclease (RNase) H2 complex that cleaves the RNA strand of RNA:DNA heteroduplexes, as well as single ribonucleotides embedded in DNA, and plays an important role in DNA replication (15). It has been reported that inactivation of RNase H2 confers sensitivity to olaparib due to its function in ribonucleotide excision repair, loss of which leads to PARP trapping on DNA lesions (13). However, after investigating the publicly available PCa genomic data, we have found that RNASEH2B is commonly co-deleted with two physically close genes RB1 and BRCA2. While deletion of RNASEH2B may confer PCa cells sensitive to PARP inhibition, the response may vary when RB1 and BRCA2 are co-deleted.
Targeted cancer therapies are increasingly being guided by tumor DNA sequencing. However, current genomically driven clinical decision-making is largely based on mutations of a single gene. The potential impact of concurrent genomic alterations on therapeutic response has been overlooked. We speculate that combinatorial effects of compound genomic alterations may sway the synthetic lethality of a single-gene deletion with PARP inhibition. Here, we investigate PARPi response of PCa cells after RNASEH2B deletion and co-deletion with RB1 and BRCA2 in preclinical models. Our study demonstrates that concurrent genomic deletions may have opposing impacts on PARPi response, supporting the utility of a comprehensive genomic test instead of a single gene–based prediction in future clinical practice.
RESULTS
Compound deletions of RNASEH2B, RB1, and BRCA2 genes in PCa
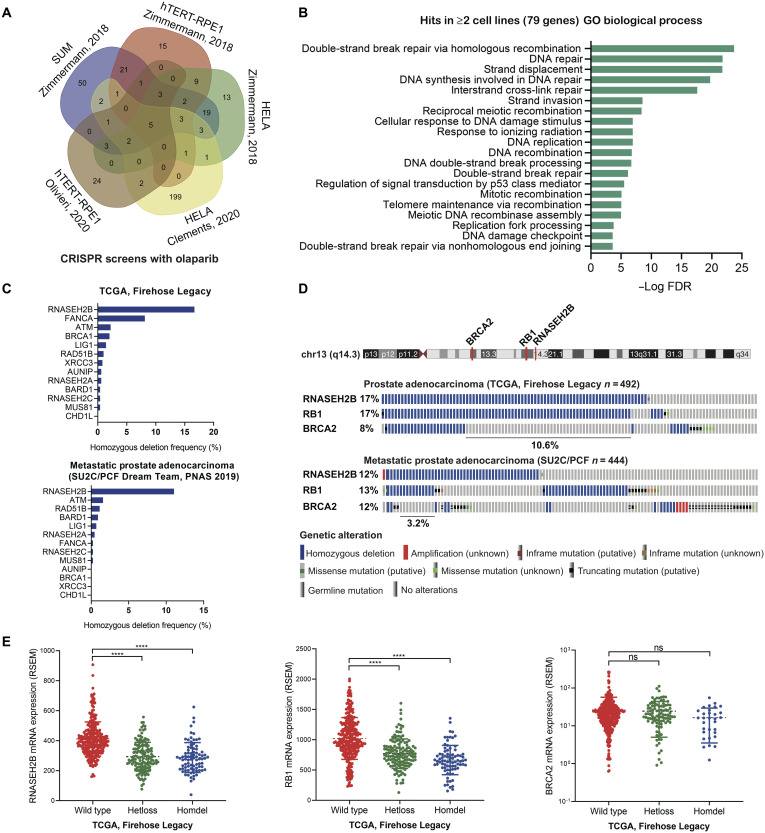
To determine genes associated with PARPi response, we analyzed five publicly available datasets of genome-wide CRISPR-Cas9 screens under the treatment with olaparib in hTERT-RPE1, HELA, and SUM cells (13, 16, 17). We found a total of 79 genes common in at least two screens (Fig. 1A and table S1), loss of which sensitizes cells to olaparib. We analyzed these genes for Gene Ontology (GO) term enrichment using a web-based gene annotation tool, DAVID 6.8 (18). As expected, DNA repair processes were overrepresented, with “double-strand break via homologous recombination” being the most enriched function (Fig. 1B). Out of this list, 13 genes are common to four screens, among which RNASEH2B is the most frequently deleted in both primary (17% homozygous deletion) and metastatic (12% homozygous deletion) prostate tumors (Fig. 1C), followed by FANCA, ATM, and BRCA1 in primary tumors and ATM, RAD51B, and BARD1 in metastatic tumors. The frequency of these genomic alterations was markedly increased when heterozygous deletions were counted as well (fig. S1A). The proteins encoded by RNASEH2A, RNASEH2B, and RNASEH2C are three subunits of the RNase H2 enzyme complex (19). Deletion of any single subunit sensitizes cells to olaparib due to impaired RNase H2 function in ribonucleotide excision repair, creating PARP-trapping lesions (13). While all three subunits are required for the function of the RNase H2 enzyme, the prevalence of RNASEH2B deletion makes it an attractive biomarker to predict PARPi response in PCa.
Fig. 1. Identification of RNASEH2B loss as a potential biomarker to predict PARPi response in PCa.
(A) Venn diagram showing the overlap between identified genes from five CRISPR-Cas9 screens with olaparib treatment. (B) Gene Ontology (GO) terms enriched among identified genes common in at least two CRISPR-Cas9 screens. FDR, false discovery rate. (C) Homozygous deletion frequency of 13 identified genes common in at least four CRISPR-Cas9 screens in primary (TCGA cohort) and metastatic (SU2C/PCF cohort) prostate tumors. (D) Genomic alterations of RNASEH2B, RB1, and BRCA2 genes on chromosome 13q in primary (TCGA cohort) and metastatic (SU2C/PCF cohort) prostate tumors. The RNASEH2B/RB1 co-deletion accounts for 10.6 and 3.2% of cases in each cohort, respectively. (E) mRNA levels of RNASEH2B, RB1, and BRCA2 in primary prostate tumors (TCGA cohort) harboring wild-type RNASEH2B and heterozygous (Hetloss) and homozygous (Homdel) RNASEH2B deletions. P values were determined by two-tailed t test. ****P < 0.0001; ns, not significant; RSEM, RNA-seq by expectation-maximization.
RNASEH2B resides on chromosome13q14, which is a genomic region with frequent focal and arm-level deletion or loss of heterozygosity in PCa (20–22). In primary prostate tumors from The Cancer Genome Atlas (TCGA) cohort (23, 24), we found that RNASEH2B is often co-deleted with the well-known tumor suppressor gene RB1 proximally located within a distance of 2.5 Mb (Fig. 1D and fig. S1B). In a small fraction of tumors, RNASEH2B and RB1 are co-deleted together with BRCA2, which is located about 18.5 Mb from RNASEH2B on chromosome 13q. In metastatic prostate tumors from the SU2C/PCF cohort (25), compound genomic alterations comprise single deletions and double/triple co-deletions of these three genes. In addition, we observed a positive correlation of the copy number values between these three genes in the TCGA cohort (fig. S2). An almost perfect correlation between RNASEH2B and RB1 genes indicated a potential focal deletion on chromosome13q14. Furthermore, we found that tumors with RNASEH2B and RB1 heterozygous or homozygous deletions exhibit significantly lower transcript levels in comparison with the wild-type tumors (Fig. 1E). It should be noted that lower levels of mRNA molecules detected in tumors with RNASEH2B and RB1 homozygous deletion are likely from surrounding noncancerous cells due to imperfect tumor purity. The decrease of mRNA levels was not observed in tumors with BRCA2 deletion, indicating more complex transcriptional regulation at the BRCA2 locus.
Deletion of RNASEH2B renders PCa cells sensitive to PARP inhibition
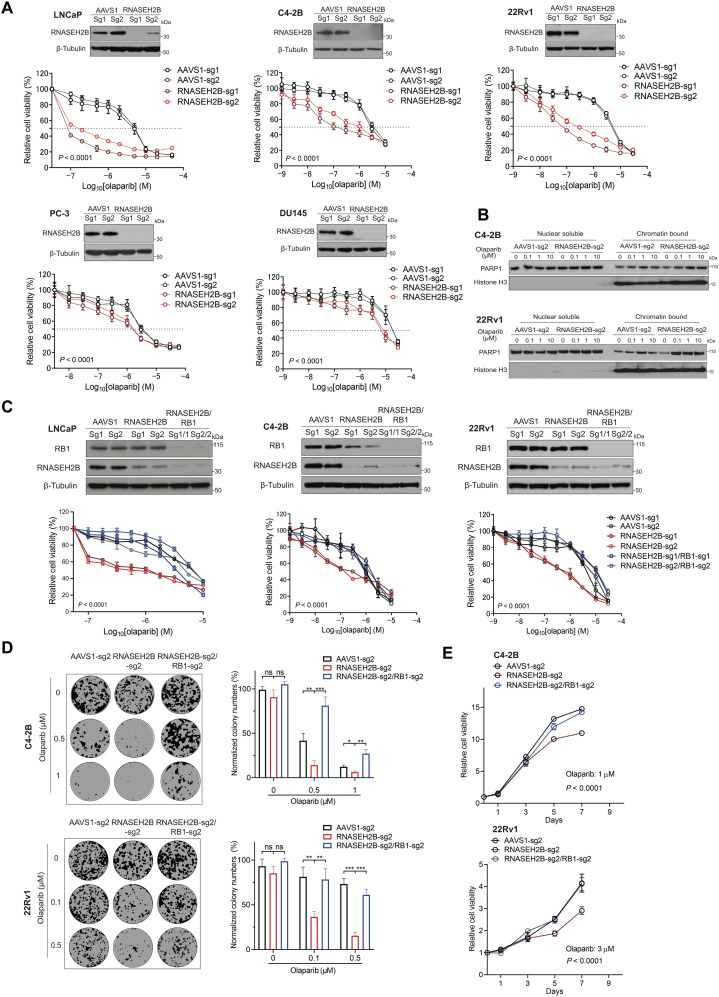
While previous studies have demonstrated that RNASEH2B genetic deletion sensitizes cells to PARP inhibition (13), to what extent loss of RNASEH2B increases PARPi response in PCa cells remains unclear. Using CRISPR-Cas9 gene editing, we deleted RNASEH2B in PCa cell lines LNCaP, C4-2B, 22Rv1, PC-3, and DU145. Two different single-guide RNAs (sgRNAs) were used for RNASEH2B knockout (KO) in each cell line, and two sgRNAs against adeno-associated virus integration site 1 (AAVS1) were used to generate corresponding control cell lines. RNASEH2B deletion was confirmed by Western blot (Fig. 2A). Genetic deletion of RNASEH2B significantly increased cell sensitivity to olaparib across all five cell lines, more so in androgen receptor (AR)–positive LNCaP, C4-2B, and 22Rv1 cells, in contrast to AR-negative PC-3 and DU145 cells. The sensitivity was assessed by the half-maximal inhibitory concentration (IC50) (table S2). We observed 253-, 30-, and 103-fold change in LNCaP, C4-2B, and 22Rv1 cells, in contrast to 3- and 8-fold change in PC-3 and DU145 cells, respectively. Deletion of RNASEH2B had a more modest effect on PARPi response in PC-3 and DU145 cells, likely due to their unique genetic background. Similarly, not all BRCA1/2-mutant tumors respond to PARP inhibition. However, increased sensitivity to olaparib after RNASEH2B deletion is comparable to that after BRCA2 deletion in C4-2B cells (fig. S3), indicating a similar impact of both genes on PARPi response. We showed that PARPi sensitivity was significantly reduced when RNASEH2B was reintroduced into RNASEH2B-deleted C4-2B and 22Rv1 cells (fig. S4), indicating that the response to PARP inhibition is specifically due to RNASEH2B loss. Previous studies have revealed that loss of RNASEH2B creates more DNA lesions for PARP trapping (13). We examined PARP1 protein levels in both nuclear soluble and chromatin fractions after olaparib treatment. We observed increased PARP1 protein trapped onto the chromatin in RNASEH2B-KO C4-2B and 22Rv1 cells compared to AAVS1 control cells (Fig. 2B). Furthermore, we found that RNASEH2B-KO cells were also sensitive to PARPis rucaparib and talazoparib [with strong trapping ability (3, 26)], but to a lesser extent to veliparib (with poor trapping ability) (fig. S5). These results suggest that PARP-trapping ability is critical for PARPi-mediated cell death in PCa cells with RNASEH2B deletion.
Fig. 2. Impacts of RNASEH2B deletion or RNASEH2B/RB1 co-deletion on PCa cell response to PARP inhibition.
(A) The RNASEH2B gene was deleted in LNCaP, C4-2B, 22Rv1, PC-3, and DU145 cells using two different sgRNAs (sg1 and sg2). Corresponding control cell lines were established using two sgRNAs against AAVS1 (sg1 and sg2). Western blots show RNASEH2B protein levels in KO and control cells. β-Tubulin serves as a loading control. Cells were treated with the indicated doses of olaparib for 7 days. Cell viability was determined using alamarBlue assay (mean ± SD; n = 3). (B) The protein level of PARP1 in nuclear soluble and chromatin-bound fractions of RNASEH2B-KO and AAVS1 control cells after olaparib treatment was determined by Western blot. (C) AAVS1 control, RNASEH2B single-gene KO (SKO), and RNASEH2B/RB1 double-gene KO (DKO) LNCaP, C4-2B, and 22Rv1 cells were treated with the indicated doses of olaparib for 7 days. Cell viability was determined using alamarBlue assay (mean ± SD; n = 3). Western blots show RNASEH2B and RB1 protein levels in AAVS1 control, SKO, and DKO cells. β-Tubulin serves as a loading control. (D) The growth of AAVS1 control, SKO, and DKO C4-2B and 22Rv1 cells was determined using colony formation assay after olaparib treatment for 14 days. (E) AAVS1 control, SKO, and DKO C4-2B and 22Rv1 cells were treated with olaparib for the indicated days. Cell proliferation was determined using alamarBlue assay. P values were determined by two-tailed t test or two-way analysis of variance (ANOVA). *P < 0.05; **P < 0.01; ***P < 0.001.
Loss of RB1 diminishes the sensitivity of RNASEH2B-deleted PCa cells to PARP inhibition
To determine whether co-deletion of RNASEH2B and RB1 affects PARPi response, we deleted the RB1 gene in RNASEH2B single-gene KO (SKO) LNCaP, C4-2B, and 22Rv1 cells to generate RNASEH2B/RB1 double-gene KO (DKO) cells (Fig. 2C). We found that the sensitivity of SKO cells to olaparib was completely abolished by concurrent RB1 deletion. In colony formation assays, we also observed that co-deletion of RB1 and RNASEH2B in C4-2B and 22Rv1 cells significantly reduced cell sensitivity to olaparib (Fig. 2D). DKO cells showed significantly increased proliferation in comparison to SKO cells under olaparib treatment (Fig. 2E). Notably, deletion of RB1 alone reduced parental C4-2B cell sensitivity to olaparib (fig. S6). Conversely, overexpression of RB1 increased PCa cell sensitivity to olaparib (fig. S7), suggesting a potential intrinsic PARPi resistance mechanism rising from RB1 loss.
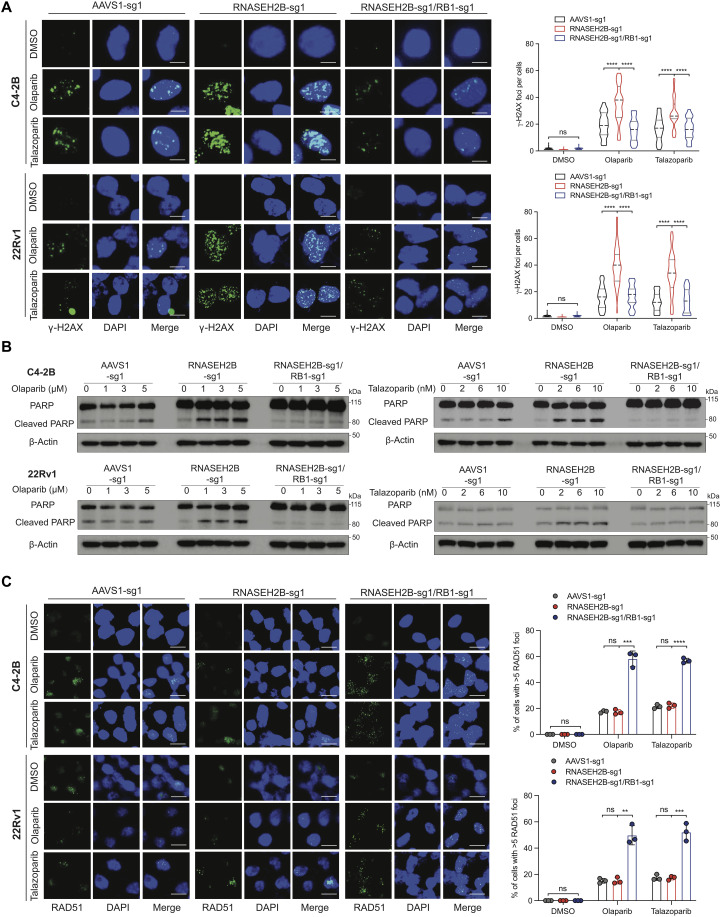
Because PARP inhibition has become a therapeutic option for mCRPC patients, we next carried out functional assays largely in CRPC C4-2B and 22Rv1 cells. Using immunofluorescence analysis of γ-H2AX foci, a marker for DNA DSBs, we detected significantly increased DNA damage in the nucleus of SKO C4-2B and 22Rv1 cells after olaparib or talazoparib treatment for 24 hours compared to their corresponding control cells (Fig. 3A). The increase of DNA DSBs was not observed in DKO cells. Accordingly, the cleaved PARP was also increased in SKO cells compared to control and DKO cells, indicating undergoing apoptosis in RNASEH2B-deleted cells after olaparib or talazoparib treatment (Fig. 3B). Olaparib-induced DNA damage and apoptosis were confirmed independently in SKO cells compared to DKO cells, both of which were generated with a different set of sgRNAs (fig. S8, A and B). Increased apoptosis in SKO cells was further confirmed using caspase-3/7 activity assay (fig. S9).
Fig. 3. Impacts of RNASEH2B deletion and RNASEH2B/RB1 co-deletion on DNA damage, apoptotic cell death, and HRR function in PCa cells.
(A) Representative images of immunofluorescence staining for γ-H2AX foci in AAVS1 control, SKO, and DKO C4-2B and 22Rv1 cells after olaparib (10 μM) or talazoparib (20 nM) treatment for 24 hours. KO cell lines were established using sgRNA #1 (sg1) for both RNASEH2B and RB1 genes. γ-H2AX foci were counted in at least 50 cells under each condition. Three independent experiments were performed. Scale bars, 10 μm. (B) PARP and cleaved PARP protein levels were determined using Western blot in AAVS1 control, SKO, and DKO cells after olaparib or talazoparib treatment as indicated for 24 hours. (C) Representative images of immunofluorescence staining for RAD51 foci in AAVS1 control, SKO, and DKO cells after olaparib (10 μM) or talazoparib (20 nM) treatment for 24 hours. RAD51 foci were counted in at least 50 cells for each replicate under each condition (n = 3 biological replicates). Scale bars, 20 μm. P values were determined by two-tailed t test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next asked whether HRR function was enhanced after RB1 loss in DKO cells. RAD51 is central to HRR, as it mediates DNA homologous pairing and strand invasion (27). We assessed the formation of RAD51 foci, a marker for HRR competence (28), using immunofluorescence staining. We found that RAD51 foci were slightly increased after olaparib or talazoparib treatment for 24 hours in SKO C4-2B and 22Rv1 cells as well as in their corresponding AAVS1 control cells (Fig. 3C and fig. S8C), indicating activation of HRR not affected by RNASEH2B deletion. However, this preserved baseline HRR function in SKO cells was not sufficient to repair DNA DSBs, as we observed accumulation of γ-H2AX foci (Fig. 3A and fig. S8A). On the other hand, we detected significantly increased RAD51 foci and less DNA DSBs in DKO cells, indicating much improved HRR capacity after RB1 loss. These results suggest that PCa cells become insensitive to PARP inhibition likely due to more efficient DNA damage repair after RB1 loss.
Loss of RB1 up-regulates HRR gene expression through E2F1 activation
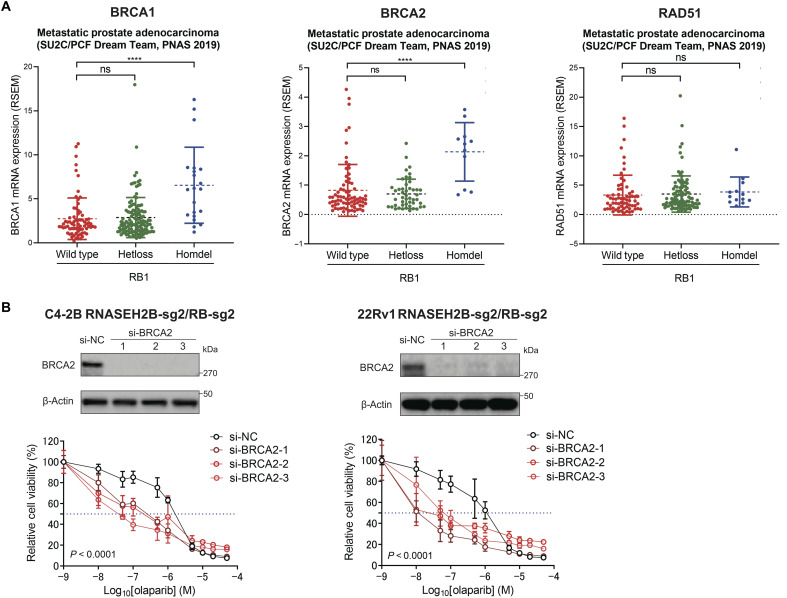
We next investigated the mechanism by which HRR function was enhanced after RB1 loss. It is well known that active form of RB1 interacts with transcription factor E2F1 and restrains its transcription activity (29). Loss of RB1 derepresses E2F1 activity and induces the expression of E2F1 target genes involving cell cycle progression and DNA repair (30). Therefore, we speculated that HRR gene expression might be up-regulated through E2F1 transcriptional activation, which, in turn, enhanced HRR function and rendered cells resistant to PARP inhibition. In line with previous studies (31), we found that the transcript level of E2F1 itself was up-regulated in primary and metastatic prostate tumors with RNASEH2B/RB1 co-deletion (Fig. 4A), likely due to a positive feedback loop. This was supported by the data from publicly available E2F1 chromatin immunoprecipitation sequencing (ChIP-seq) (32, 33), showing strong E2F1 binding at its own promoter region (fig. S10). Using an E2F1 reporter assay, we detected significantly higher E2F1 transcriptional activity in RNASEH2B/RB1 DKO cells compared to RNASEH2B SKO cells (Fig. 4B). The E2F1 transcriptional activity remained at a high level after olaparib treatment. We further analyzed publicly available E2F1 ChIP-seq data and found strong E2F1 binding at the promoter regions of BRCA1/2 and RAD51 genes in PCa LNCaP cells (Fig. 4C). Notably, robust E2F1 ChIP-seq peaks are located immediately upstream of the transcription start sites, indicating a direct transcriptional regulation. Furthermore, the E2F1-mediated BRCA1/2 and RAD51 transcriptional regulation appears to be conserved across different cell types (fig. S11). We then performed E2F1 ChIP combined with quantitative polymerase chain reaction (ChIP-qPCR) and detected enriched E2F1 binding at the promoter regions of BRCA1/2 and RAD51 genes in parental C4-2B and 22Rv1 cells (Fig. 4D). To further demonstrate E2F1-mediated up-regulation of BRCA1/2 and RAD51 in DKO cells, we knocked down E2F1 expression using RNA interference and observed significantly decreased BRCA1/2 and RAD51 protein levels (Fig. 4E). In addition, treatment of DKO cells with the pan-E2F inhibitor HLM006474 reduced BRCA1/2 and RAD51 protein expression. We next compared gene expression changes in RB1-deleted DKO cells relative to RB1-intact SKO. We found that the mRNA levels of BRCA1/2 and RAD51 were significantly up-regulated in DKO cells compared to SKO and corresponding control cells (Fig. 4F). We further compared their protein levels in the absence and presence of olaparib (Fig. 4G). We detected much higher protein levels of BRCA1/2 in DKO cells, while the RAD51 protein level remained unchanged, indicating posttranscriptional regulation involved after RB1 loss in these cells. Notably, olaparib treatment suppressed BRCA1/2 expression in SKO cells, which might contribute to PARPi response in these cells. This is in agreement with the results from previous studies, showing that PARP1 functions as a E2F1 cofactor and regulates DNA repair gene expression (34–36). Nevertheless, BRCA1/2 protein levels were restored and remained at a high level after olaparib treatment in DKO cells. Considering the role of RB1/E2F1 signaling in cell cycle regulation (33), we performed cell cycle analysis and found a negligible change across AAVS1 control, SKO, and DKO cells (Fig. 4H). Therefore, up-regulation of BRCA1/2 expression is largely due to transcriptional regulation rather than cell cycle alteration after RB1 loss, although BRCA1/2 expression is cell cycle dependent. Last, in the SU2C/PCF cohort, we observed that metastatic prostate tumors with homozygous RB1 deletions had significantly higher transcript levels of BRCA1/2 (Fig. 5A), which might have the potential to repair damaged DNA more effectively and survive PARP inhibition. Together, our results suggest that loss of RB1 up-regulates BRCA1/2 gene expression through E2F1 transcriptional activation. The expression of BRCA1/2 remains at a high level after PARP inhibition, leading to proficient DNA DSB repair and PARPi resistance.
Fig. 4. RB1 loss up-regulates HRR gene expression by activating E2F1 transcriptional activity.
(A) Comparison of E2F1 transcript levels between RNASEH2B/RB1 co–wild-type (co-WT) and co-deletion (co-DEL) tumors in three PCa clinical cohorts in cBioPortal. (B) Comparison of E2F1 transcriptional activity between SKO and DKO cells in the presence or absence of olaparib as indicated for 24 hours using E2F1 luciferase reporter assay. (C) Publicly available E2F1 ChIP-seq data showing E2F1 binding capacity at the promoters of BRCA1/2 and RAD51 genes in LNCaP cells. The E2F1 ChIP-seq peaks were observed in the UCSC Genome Browser. Red arrows indicate the qPCR regions. (D) E2F1 binding was determined by ChIP-qPCR at the promoters of BRCA1/2 and RAD51 genes in C4-2B and 22Rv1 cells. An irrelevant genomic region was used as a control. Normal IgG and anti-E2F1 antibody were used for immunoprecipitation. (E) Western blots show protein levels of indicated genes in DKO C4-2B and 22Rv1 cells after E2F1 siRNA knockdown or the treatment with pan-E2F inhibitor HLM006474 for 24 hours. β-Actin serves as a loading control. The intensity of protein bands was quantified using ImageJ software. The first band was defined as 1. (F) BRCA1/2 and RAD51 mRNA levels were determined by RT-qPCR in DKO C4-2B and 22Rv1 cells in comparison to control and SKO cells. (G) Western blots show protein levels of BRCA1/2 and RAD51 in SKO and DKO cells after olaparib treatment for 24 hours. Western blot quantification is described in (E). (H) Cell cycle distribution was analyzed in AAVS1 control, SKO, and DKO C4-2B and 22Rv1 cells under regular cell culture condition. P values were determined by two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Fig. 5. Loss of BRCA2 resensitizes RNASEH2B/RB1 DKO cells to PARP inhibition.
(A) mRNA expression levels of BRCA1/2 and RAD51 in metastatic prostate tumors (SU2C/PCF cohort) harboring wild-type RB1 and heterozygous (Hetloss) and homozygous (Homdel) RB1 deletions. Tumors with BRCA1/2 and RAD51 deletions were excluded in each analysis, respectively. P values were determined by two-tailed t test. ****P < 0.0001. (B) RNASEH2B/RB1 DKO C4-2B and 22Rv1 cells were transfected with three different BRCA2 siRNA (si-BRCA2) or a negative control siRNA (si-NC) at a final concentration of 10 nM for 2 days, followed by the treatment with the indicated doses of olaparib for additional 7 days. Cell viability was determined using alamarBlue assay (mean ± SD; n = 3). Western blots show BRCA2 protein levels 48 hours after siRNA transfection. P values were determined by two-way ANOVA.
PCa cells with co-loss of RNASEH2B/RB1/BRCA2 are sensitive to PARP inhibition
While BRCA1 is critical in HRR, genomic alterations in PCa involve BRCA2 more commonly than BRCA1. Clinical next-generation sequencing analyses of both primary and metastatic prostate tumors have revealed that BRCA2 is co-deleted with RNASEH2B and RB1 in a small portion of patients (Fig. 1D). Our data have suggested that up-regulation of BRCA2 through the RB1/E2F1 pathway likely contributes to PARPi resistance in RB1-deleted cells. We next asked whether deletion of BRCA2 can resensitize DKO cells to PARP inhibition. Here, we knocked down BRCA2 expression in RNASEH2B/RB1 DKO C4-2B and 22Rv1 cells using RNA interference. Three different small interfering RNAs (siRNAs) against BRCA2 completely abolished BRCA2 protein expression determined by Western blot (Fig. 5B). We found that depletion of BRCA2 renders DKO C4-2B and 22Rv1 cells sensitive to olaparib, indicating that elevated BRCA2 expression after RB1 loss is likely one of the mechanisms for PARPi resistance. RNASEH2B/RB1 DKO cells also respond to other PARPis (veliparib, rucaparib, and talazoparib) following BRCA2 depletion (fig. S12). These results suggest that BRCA2-deficient tumors may respond to PARPis regardless of RB1 status.
ATR inhibition overcomes PARPi resistance of PCa tumors with RNASEH2B/RB1 co-deletion
Because PCa cells with RNASEH2B single-gene deletion or RNASEH2B/RB1/BRCA2 three-gene co-deletion are sensitive to PARP inhibition, we next asked how to overcome PARPi resistance for cells with RNASEH2B/RB1 co-deletion. Patients with tumors harboring RNASEH2B/RB1 co-deletion account for 10.6 and 3.2% in all primary and metastatic PCa cases, respectively (Fig. 1D). Emerging evidence has shown that PARP inhibition may activate ATR, which phosphorylates and activates CHK1 and allows cells to survive PARPi-induced replication stress (37). Previous studies have also demonstrated that ATR-CHK1 signaling controls E2F-dependent transcription of HRR genes (38–40). Here, we found that ATR activity was elevated in RNASEH2B/RB1 DKO cells after olaparib treatment, as evidenced by increased CHK1 phosphorylation in a dose-dependent manner (Fig. 6A). We hypothesized that DKO cells relied on ATR activity to survive PARPi-induced DNA damage. We therefore sought to investigate the effect of PARPi and ATR inhibitor (ATRi) either alone or in combination on the growth of DKO cells. To achieve ATR inhibition, we used the clinically used ATRi VE-822. Both SKO and DKO cells failed to show increased response to VE-822 as a single agent in comparison to AAVS1 control cells (Fig. 6B). We treated PARPi-insensitive DKO cells with olaparib combined with VE-822 and found that cotreatment diminished the growth of these cells (Fig. 6C). We observed that DKO C4-2B and 22Rv1 cells were resensitized to olaparib in the context of ATR inhibition (Fig. 6D). Using the Loewe and Bliss synergy analysis (41, 42), we found a synergistic interaction between olaparib and VE-822, with a high synergy score for DKO 22Rv1 (Loewe: 13.263; Bliss: 16.347) and C4-2B (Loewe: 8.314; Bliss: 13.502) cells (Fig. 6E). Synergistic effects were also observed in the same cells using colony formation assay (Fig. 6F).
Fig. 6. ATR inhibition overcomes PARPi resistance in RNASEH2B/RB1 DKO cells.
(A) Western blots show phosphorylated CHK1 and total CHK1 protein levels in RNASEH2B SKO and RNASEH2B/RB1 DKO cells after the treatment with the indicated doses of olaparib for 24 hours. (B) Cell viability of AAVS1 control, SKO, and DKO cells was determined using alamarBlue assay after the treatment with ATR inhibitor VE-822 as indicated for 7 days. (C) RNASEH2B/RB1 C4-2B and 22Rv1 DKO cells were treated with olaparib, VE-822, and olaparib + VE-822 as indicated. Cell proliferation was determined using alamarBlue assay. (D) RNASEH2B/RB1 DKO cells were treated with olaparib alone or in combination with VE-822 as indicated for 7 days. Cell viability was determined using alamarBlue assay. (E) The synergistic score between olaparib and VE-822 was determined using Loewe and Bliss synergy analysis. (F) The growth of RNASEH2B/RB1 DKO cells after the treatment with olaparib and/or VE-822 for 14 days was determined using colony formation assay. Colony number was quantified using ImageJ software. P values were determined by two-tailed t test. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we tested combination treatment in vivo using PARPi-insensitive DKO 22Rv1 cells. After xenograft tumors established in immunodeficient mice, animals were divided into four groups and treated with vehicle, olaparib, VE-822, or olaparib in combination with VE-822 for three cycles as indicated (Fig. 7A). We found that tumor growth was significantly inhibited by combination treatment, while both olaparib and VE-822 had little effect as a single agent. No significant mouse weight loss was observed in all four groups, indicating the combination treatment is tolerable.
Fig. 7. Combination therapy with ATR and PARP inhibition suppresses RNASEH2B/RB1 DKO cell growth in vivo.
(A) RNASEH2B/RB1 DKO 22Rv1 cells were injected subcutaneously into ICR-SCID mice. Mice were randomly assigned into four groups (n = 5 animals per group) and treated with vehicle, olaparib (50 mg/kg), VE-822 (25 mg/kg), or olaparib in combination with VE-822 for three cycles as indicated. Both drugs were administered by oral gavage once a day. Tumor volume and mouse weight were recorded and analyzed across four groups as indicated. (B) DKO C4-2B and 22Rv1 cells were treated with DMSO, olaparib, VE-822, or olaparib + VE-822 for 24 hours. E2F1 activity was detected using E2F1 luciferase reporter assay. (C) Western blots show protein levels of BRCA1/2 and RAD51 in DKO cells after the treatment with DMSO, olaparib, VE-822, or olaparib + VE-822 for 24 hours. The intensity of protein bands was quantified using ImageJ software. The first band was defined as 1. (D) Representative images of immunofluorescence staining for RAD51 foci in DKO C4-2B and 22Rv1 cells after the treatment with DMSO, olaparib (10 μM), VE-822 (1000 nM), or olaparib + VE-822 for 24 hours. RAD51 foci were counted in at least 50 cells for each replicate under each condition (n = 3 biological replicates). Scale bars, 20 μm. (E) Schematic model depicting the mechanism by which concurrent deletions of RNASEH2B, RB1, and BRCA2 genes affect the response to PARP inhibition. P values were determined by two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We then asked whether the combination of PARP and ATR inhibition affects E2F1-mediated BRCA1/2 and RAD51 expression and HRR function. Using an E2F1 reporter assay, we observed significantly decreased E2F1 activity in DKO C4-2B and 22Rv1 cells after the combination treatment (Fig. 7B). The protein expression levels of BRCA1/2 and RAD51 were also decreased after combination treatment (Fig. 7C). Last, we examine HRR function in DKO cells using RAD51 foci formation assay. We observed increased RAD51 foci after olaparib treatment, which was diminished by combined treatment with VE-822 (Fig. 7D). The loss of RAD51 foci after ATR inhibition is likely due to reduced BRCA1/2 expression (Fig. 7C) and disrupted BRCA-independent RAD51 loading to DSBs as previously reported (43). Together, our results support the notion that the combined therapy with PARPis and ATRis may overcome PARPi resistance in PCa cells with RNASEH2B/RB1 co-deletion through inhibition of E2F1-mediated augmentation of HRR capacity.
DISCUSSION
It has been a great challenge to determine which patients are most likely to benefit from PARP inhibition. Clinical investigation has demonstrated that CRPC patients with tumors harboring deleterious germline or somatic BRCA1/2 alterations have a high likelihood of response to PARPis. However, alterations in other HRR genes [known as BRCAness genes (44)], such as ATM and CHEK2, are not associated with response to the same extent. Furthermore, PARPi response for tumors harboring genomic alterations in non-HRR DDR genes remains largely unknown. RNASEH2B is not a BRCAness gene. Instead, it is one of three genes encoding RNase H2 protein complex, which is critical in ribonucleotide excision repair. In this study, we show that RNASEH2B is frequently deleted in both primary and metastatic prostate tumors, which creates DNA lesions and increases PARP trapping after the treatment with PARPis, leading to accumulation of DNA DSBs and apoptotic cell death. While RNASEH2B deletion is an attractive biomarker to predict PARPi response in PCa, co-deletion with RB1 counteracts the cytotoxic effect of PARP trapping, at least in part, by up-regulation of E2F1-mediated BRCA1/2 expression, thereby enhancing HRR capacity (Fig. 7E). Subsequently, we show that deletion of BRCA2 resensitizes RNASEH2B/RB1 co-deleted cells to PARPis. We further demonstrate that the combination of PARP and ATR inhibition can overcome intrinsic PARPi resistance rising from RB1 loss. Given the interaction between multiple genomic alterations in tumors, these results provide a basis of clinical application of PARPi either alone or in combination with ATRi in PCa. Patients will likely benefit from PARP inhibition if their tumors harbor RNASEH2B single-gene deletion or RNASEH2B/RB1/BRCA2 co-deletion, whereas patients with tumors harboring RNASEH2B/RB1 co-deletion may respond to combined PARP and ATR inhibition.
Loss of RB1 has been shown to be strongly associated with poor clinical outcomes in advanced PCa by facilitating lineage plasticity in the context of concurrent loss of TP53 (45–48). RB1/TP53-deficient tumors are resistant to a wide range of single-agent therapeutics, including PARPis (31). These results are consistent with our finding of relatively lower PARPi sensitivity in PC-3 and DU145 cells despite RNASEH2B deletion because PC-3 cells do not express p53 (p53-null) and DU145 cells have dominant-negative TP53 mutations and RB1 loss (49, 50). Furthermore, studies have shown that the combination of PARP inhibition and RB1-associated cyclin-dependent kinase (CDK) inhibition may be a viable strategy for neuroendocrine PCa treatment (51), supporting an important role of RB1 in PARP inhibition. Our present work does not exclude the possibility that PARPi resistance results from lineage plasticity driven by epigenetic reprogramming or alterations in cell metabolism after RB1 loss (52, 53). Nevertheless, tumors with RB1 loss express significantly higher levels of E2F1, which directly up-regulates HRR genes, most notably BRCA1/2. The RB1/E2F1-mediated HRR gene expression pathway is highly conserved across different cell types based on the E2F1 ChIP-seq results from multiple databases. Accordingly, our data strongly support the notion that RB1 loss renders PARP inhibition inefficient for tumors with non-BRCA genomic alterations, because E2F1-induced BRCA1/2 expression enhances HRR capacity. Considering that RB1 loss is commonly observed as a late subclonal event in mCRPC (54), this may partially explain why mCRPC patients with tumors harboring alterations in non-BRCA HRR genes have a lower response rate. On the other hand, tumors with BRCA1/2 alterations remain sensitive to PARP inhibition regardless of RB1 status (55). While PCa cells with RB1 loss are resistant to PARPis, their response to other DNA damaging agents or radiation therapy may vary. It was reported that loss of RB1 conferred radiosensitivity to PCa cells (56). Further investigations are needed to understand agent-specific sensitivity and resistance mechanisms beyond HRR capacity.
The landscapes of cancer genome are complex including base changes, indels, copy number changes, and structural rearrangements. Genomic deletion is common in cancer and ranges from focal deletions affecting a few genes to arm-level deletions affecting hundreds to thousands of genes (57). Little is known about the functional consequences of large-scale genomic deletions, and it is difficult to determine the specific genes responsible for the biological effects. One of the limitations in our studies is that we did not test whether co-deletions of other protein-coding genes, let alone noncoding RNAs, on chromosome 13q may also influence PARPi sensitivity. While CRISPR screens have not identified any proximal genes at the RNASEH2B/RB1/BRCA2 loci, which, when deleted, alter PARPi response, a further investigation is needed by creating isogenic cell lines with engineered large-scale deletions instead of a gene-by-gene approach. Accordingly, there is a rationale for examining copy number changes and structural rearrangements of PCa tumors, which are not captured by targeted next-generation sequencing tests being implemented in current clinical practice.
The mechanisms of acquired PARPi resistance have been heavily studied in BRCA-deficient cells. A key mechanism appears to be the restoration or bypass of HRR and fork protection functions, which can be overcome by ATR inhibition (43). In this study, we propose an intrinsic resistance mechanism through the RB1-E2F1-BRCA pathway in non–BRCA-deficient cells. We demonstrate that ATR inhibition may impair E2F1-induced BRCA1/2 expression and resensitize cells to PARPis. The combination therapy with PARPi and ATRi is being evaluated in clinical trials for mCRPC patients (NCT03787680). Therefore, it is conceivable to develop predictive biomarkers based on a comprehensive genomic test and explore the combination of PARP and ATR inhibition as a promising strategy for advanced PCa patients when a single agent fails.
MATERIALS AND METHODS
Cell lines and materials
Human PCa cell lines LNCaP, C4-2B, 22Rv1, PC-3, and DU145 (American Type Culture Collection) were cultured in RPMI 1640 medium (Thermo Fisher Scientific), while 293FT cells (Thermo Fisher Scientific) were maintained in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific). Media were supplemented with 10% fetal bovine serum (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), and 1% Hepes (Sigma-Aldrich). All cell lines were authenticated using high-resolution small tandem repeats profiling at Dana-Farber Cancer Institute Molecular Diagnostics Core Laboratory and were tested mycoplasma-free before experiments. The small-molecule inhibitors are listed in table S3.
Establishment of CRISPR-Cas9 KO cell lines
CRISPR guides targeting RNASEH2B were cloned into lentiGuide-Puro vector (#52963, Addgene), while CRISPR guides targeting RB1 were cloned into lenti-sgRNA hygro vector (#104991, Addgene). The lentiCas9-Blast vector that expresses Cas9 was obtained from Addgene (#52962). Lentiviruses were generated using packaging vectors pMD2.G (#12259, Addgene) and psPAX2 (#12260, Addgene) with Lipofectamine 3000 Transfection Reagent (#L3000015, Invitrogen) in 293FT cells. PCa cells were initially infected with lentiviruses of Cas9 and selected with blasticidin (10 μg/ml) for 2 weeks. Polybrene was added at a final concentration of 8 μg/ml to increase transduction efficiency. To generate RNASEH2B-KO cells, PCa cells were infected with lentiviruses containing specific sgRNAs and selected with puromycin (3 μg/ml) for 2 weeks. The RNASEH2B-KO cells were infected with lentiviruses with RB1 sgRNAs to generate RNASEH2B/RB1 co-deletion cells. Cells were further selected using hygromycin (300 μg/ml) for 2 weeks. sgRNA sequences are listed in table S3.
Cell viability assay
PCa cells were seeded in 96-well plates (1 × 103 to 2 × 103 cells per well) and treated with inhibitors as indicated. Cell viability was measured using alamarBlue Cell Viability Reagent (DAL1100, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Colony formation assay
PCa cells were seeded in 12-well plates (3000 cells per well) at low density to avoid contact between clones. Subsequently, cells were treated with inhibitors as indicated 18 hours after attachment and allowed to grow for additional 14 days. Colonies were fixed with paraformaldehyde (4%) for 10 min and stained with crystal violet (1%) for 15 min. Colony images were quantified using ImageJ software (National Institutes of Health).
Western blot assay
PCa cells were treated as indicated and harvested for protein extraction. Cells were rinsed with phosphate-buffered saline (PBS), scraped, and lysed in cold radioimmunoprecipitation assay (RIPA) Lysis and Extraction Buffer (#89900, Thermo Fisher Scientific) containing Protease and Phosphatase Inhibitor Cocktail (#78447, Thermo Fisher Scientific). Protein concentration was quantified using the Pierce BCA Protein Assay Kit (#23225, Thermo Fisher Scientific) and measured with a spectrophotometer. Western blot was performed as previously described (58) and repeated at least two times. Molecular weight markers were used to determine the size of proteins. Protein bands were quantified using ImageJ software. Antibodies were listed in table S3.
ChIP-qPCR assay
ChIP experiments were performed as previously described (59). Briefly, PCa cells were grown in RPMI 1640 medium with 10% fetal bovine serum for 2 days before ChIP. Cells were cross-linked by formaldehyde (1%) at room temperature (RT) for 10 min. After washing with ice-cold PBS, cells were collected and lysed. The soluble chromatin was purified and fragmented by sonication. Immunoprecipitation (IP) was performed using normal immunoglobulin G (IgG) or E2F1 antibody (2 μg per IP). ChIP DNA was extracted and analyzed by qPCR using iTaq Universal SYBR Green Supermix (#1725120, Bio-Rad). The antibodies and primer sequences are listed in table S3.
RT-qPCR assay
Total RNA was extracted using TRIzol Reagent (#15596026, Thermo Fisher Scientific) according to the manufacturer’s protocol. Reverse transcription qPCR (RT-qPCR) assay was performed as previously described (59). Primer sequences are listed in table S3.
Caspase-3/7 activity assay
PCa cells were seeded in 96-well plates and treated with dimethyl sulfoxide (DMSO) or specific inhibitors as indicated. Caspase-3/7 activity was measured using Caspase-Glo 3/7 Assay Systems (G8091, Promega) according to the manufacturer’s protocol.
E2F1 reporter activity assay
The E2F1 luciferase reporter plasmid was described previously (58). The reporter construct contains three tandem E2F1 consensus elements— TGCAATTTCGCGCCAAACTTG (60)—subcloned into Sac I/Xho I sites of the pGL4.26 vector (Promega) upstream of a minimal promoter. Cells (1 × 104 cells per well) were seeded in 96-well plates and transfected with E2F1 luciferase reporter plasmids (50 ng per well) using X-tremeGENE HP DNA Transfection Reagent (#06366236001, Sigma-Aldrich). After 12 hours, cells were treated with DMSO or specific inhibitors as indicated for additional 24 hours. The luciferase activity was measured using One-Glo Luciferase Assay System (E6110, Promega) according to the manufacturer’s protocol.
RNA interference
PCa cells were transfected with siRNAs at a final concentration of 10 nM using Lipofectamine RNAiMAX Transfection Reagent (#13778150, Thermo Fisher Scientific) according to the manufacturer’s instructions. All siRNAs were purchased from Sigma-Aldrich and listed in table S3. Cell viability and Western blot assays were performed 2 days after siRNA transfection.
Gene overexpression
PCa cells were seeded in 6-well (1 × 106 cells per well) or 96-well (5 × 103 cells per well) plates for 24 hours. Subsequently, cells were transfected with pEGFP-RNASEH2B (#108697, Addgene) or GFP-RB FL (#16004, Addgene) plasmids using Lipofectamine 3000 Transfection Reagent (L3000015, Thermo Fisher Scientific) according to the manufacturer’s instructions. For Western blot, cells were harvested from 6-well plates 24 hours after plasmid (2.5 μg per well) transfection. For cell viability assay, cells in 96-well plates were treated with olaparib for additional 72 hours after plasmid (0.1 μg per well) transfection, followed by alamarBlue cell viability assay.
Immunofluorescence staining
PCa cells were seeded onto the Millicell EZ SLIDE 4-well glass slides (PEZGS0496, Millipore) precoated with poly-l-lysine (P4707, Sigma-Aldrich) and then processed with treatments as indicated. After 24 hours, cells were washed with PBS and fixed in 4% formaldehyde at RT for 10 min. Fixed cells were washed with PBS for three times and extracted with 0.2% Triton X-100 for 10 min. Subsequently, cells were blocked in blocking buffer (5% bovine serum albumin in PBS) for 1 hour and incubated with the primary antibody RAD51 (ab133534, Abcam) or phospho-histone H2AX (#05-636, Sigma-Aldrich) at 1:200 dilution. Following an overnight incubation at 4°C, slides were washed with PBS and incubated with Alexa Fluor 488–conjugated anti-mouse (A28175, Life Technologies) or anti-rabbit (A27034, Life Technologies) secondary antibody at 1:1000 dilution at RT for 1 hour. After washing, the Mounting Medium with DAPI (ab104139, Abcam) was applied onto the slides. The slides were imaged under fluorescence microscope and quantified using ImageJ software. γ-H2AX was quantified by counting the number of foci per cell, while RAD51 was quantified by scoring the percentage of cells with ≥5 foci per cell. Each experiment was performed in triplicate, and at least 50 cells were counted for each replicate under each condition. Immunofluorescence staining was performed in two sets of KO cell lines (sg1 and sg2) independently by two investigators in a blind manner.
Xenograft tumor assay
RNASEH2B/RB1 DKO 22Rv1 cells were used to generate xenograft tumors. Cells (2.5 × 106 cells/50 μl per mouse with additional 50 μl of Matrigel) were subcutaneously injected into the right flank of male ICR-SCID mice (Taconic Laboratories) at the age of 4 to 5 weeks. All procedures were performed in compliance with the guidelines from the Institutional Animal Care and Use Committee at the Brigham and Women’s Hospital. The tumor growth and mouse body weight were monitored twice a week. Tumor volume was measured using a Vernier caliper and calculated according to the following formula: volume = ½(length × width2). Mice bearing about 150-mm3 tumors were randomized into four groups and treated with vehicle, olaparib (50 mg/kg), VE-822 (25 mg/kg), or the combination of olaparib and VE-822. Olaparib was formulated in 5% dimethylacetamide/10% Solutol HS 15/85% PBS; VE-822 was formulated in 10% vitamin E d-alpha tocopheryl polyethylene glycol 1000 succinate. Both drugs were administered by oral gavage once a day, with olaparib 5 days on/2 days off and VE-822 four consecutive days a week starting next day after olaparib treatment. Animals were euthanized after 3-week drug treatment or when tumors exceeded 1000 mm3.
Statistical analysis
Quantitative measurements are presented as means ± SD from at least three biological replicates unless stated otherwise. Statistical analyses were performed using an unpaired two-tailed Student’s t test or a two-way analysis of variance (ANOVA) with a post hoc Tukey’s honest significant difference test when comparing at least three groups. P < 0.05 was considered as statistically significant.
Clinical cohort analysis
Bioinformatic analysis of genomic deletion, mRNA expression, and copy number variations of related genes was performed using publicly available clinical datasets in cBioPortal (23, 24). Statistical analyses were performed using an unpaired two-tailed Student’s t test.
Acknowledgments
We thank B. Gui, C. Feng, X. Bai, T. Tian, K. Jia, and J. Geng for insightful discussion and technical support.
Funding: This work was supported by the NIH (grants 1R21CA252578-01 and 1R01CA262524-01 to L.J.).
Author contributions: L.J. conceived the project, designed the experiments, analyzed the data, and wrote the paper. C.M. and T.Tsuj. performed the experiments, analyzed the data, and wrote the paper. T.Ta., F.G., T.Tsut., and Z.Szt. performed the experiments. Z.W. and H.A. analyzed the data. Z.Sza., K.W.M., L.Z., and A.S.K. analyzed the data and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 to S3
REFERENCES AND NOTES
- 1.Robinson D., van Allen E. M., Wu Y. M., Schultz N., Lonigro R. J., Mosquera J. M., Montgomery B., Taplin M. E., Pritchard C. C., Attard G., Beltran H., Abida W., Bradley R. K., Vinson J., Cao X., Vats P., Kunju L. P., Hussain M., Feng F. Y., Tomlins S. A., Cooney K. A., Smith D. C., Brennan C., Siddiqui J., Mehra R., Chen Y., Rathkopf D. E., Morris M. J., Solomon S. B., Durack J. C., Reuter V. E., Gopalan A., Gao J., Loda M., Lis R. T., Bowden M., Balk S. P., Gaviola G., Sougnez C., Gupta M., Yu E. Y., Mostaghel E. A., Cheng H. H., Mulcahy H., True L. D., Plymate S. R., Dvinge H., Ferraldeschi R., Flohr P., Miranda S., Zafeiriou Z., Tunariu N., Mateo J., Perez-Lopez R., Demichelis F., Robinson B. D., Sboner A., Schiffman M., Nanus D. M., Tagawa S. T., Sigaras A., Eng K. W., Elemento O., Sboner A., Heath E. I., Scher H. I., Pienta K. J., Kantoff P., de Bono J. S., Rubin M. A., Nelson P. S., Garraway L. A., Sawyers C. L., Chinnaiyan A. M., Integrative clinical genomics of advanced prostate cancer. Cell 162, 454 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Armenia J., Wankowicz S. A. M., Liu D., Gao J., Kundra R., Reznik E., Chatila W. K., Chakravarty D., Han G. C., Coleman I., Montgomery B., Pritchard C., Morrissey C., Barbieri C. E., Beltran H., Sboner A., Zafeiriou Z., Miranda S., Bielski C. M., Penson A. V., Tolonen C., Huang F. W., Robinson D., Wu Y. M., Lonigro R., Garraway L. A., Demichelis F., Kantoff P. W., Taplin M.-E., Abida W., Taylor B. S., Scher H. I., Nelson P. S., de Bono J. S., Rubin M. A., Sawyers C. L., Chinnaiyan A. M.; PCF/SU2C International Prostate Cancer Dream Team, Schultz N., Van Allen E. M., The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murai J., Huang S. Y. N., Das B. B., Renaud A., Zhang Y., Doroshow J. H., Ji J., Takeda S., Pommier Y., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helleday T., The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 5, 387–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Farmer H., McCabe N., Lord C. J., Tutt A. N. J., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M. B., Jackson S. P., Smith G. C. M., Ashworth A., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 7.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K. N., Sartor O., Agarwal N., Olmos D., Thiery-Vuillemin A., Twardowski P., Mehra N., Goessl C., Kang J., Burgents J., Wu W., Kohlmann A., Adelman C. A., Hussain M., Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 382, 2091–2102 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Mateo J., Porta N., Bianchini D., McGovern U., Elliott T., Jones R., Syndikus I., Ralph C., Jain S., Varughese M., Parikh O., Crabb S., Robinson A., McLaren D., Birtle A., Tanguay J., Miranda S., Figueiredo I., Seed G., Bertan C., Flohr P., Ebbs B., Rescigno P., Fowler G., Ferreira A., Riisnaes R., Pereira R., Curcean A., Chandler R., Clarke M., Gurel B., Crespo M., Nava Rodrigues D., Sandhu S., Espinasse A., Chatfield P., Tunariu N., Yuan W., Hall E., Carreira S., de Bono J. S., Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 21, 162–174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abida W., Campbell D., Patnaik A., Sautois B., Shapiro J., Vogelzang N. J., Bryce A. H., Dermott R. M., Ricci F., Rowe J., Zhang J., Simmons A. D., Despain D., Dowson M., Golsorkhi T., Chowdhury S., Preliminary results from the TRITON2 study of rucaparib in patients (pts) with DNA damage repair (DDR)-deficient metastatic castration-resistant prostate cancer (mCRPC): Updated analyses. Ann. Oncol. 30, 846PD (2019). [Google Scholar]
- 10.Clovis Oncology Inc., Clovis Oncology’s Rubraca (rucaparib) granted FDA priority review for advanced prostate cancer [news release] (Clovis Oncology Inc., 2020).
- 11.Marshall C. H., Sokolova A. O., McNatty A. L., Cheng H. H., Eisenberger M. A., Bryce A. H., Schweizer M. T., Antonarakis E. S., Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur. Urol. 76, 452–458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., Boysen G., Porta N., Flohr P., Gillman A., Figueiredo I., Paulding C., Seed G., Jain S., Ralph C., Protheroe A., Hussain S., Jones R., Elliott T., McGovern U., Bianchini D., Goodall J., Zafeiriou Z., Williamson C. T., Ferraldeschi R., Riisnaes R., Ebbs B., Fowler G., Roda D., Yuan W., Wu Y. M., Cao X., Brough R., Pemberton H., A’Hern R., Swain A., Kunju L. P., Eeles R., Attard G., Lord C. J., Ashworth A., Rubin M. A., Knudsen K. E., Feng F. Y., Chinnaiyan A. M., Hall E., de Bono J. S., DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann M., Murina O., Reijns M. A. M., Agathanggelou A., Challis R., Tarnauskaitė Ž., Muir M., Fluteau A., Aregger M., McEwan A., Yuan W., Clarke M., Lambros M. B., Paneesha S., Moss P., Chandrashekhar M., Angers S., Moffat J., Brunton V. G., Hart T., de Bono J., Stankovic T., Jackson A. P., Durocher D., CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugger K., Bajrami I., Silva Dos Santos M., Young S. J., Kunzelmann S., Kelly G., Hewitt G., Patel H., Goldstone R., Carell T., Boulton S. J., MacRae J., Taylor I. A., West S. C., Targeting the nucleotide salvage factor DNPH1 sensitizes BRCA-deficient cells to PARP inhibitors. Science 372, 156–165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reijns M. A., Jackson A. P., Ribonuclease H2 in health and disease. Biochem. Soc. Trans. 42, 717–725 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Olivieri M., Cho T., Álvarez-Quilón A., Li K., Schellenberg M. J., Zimmermann M., Hustedt N., Rossi S. E., Adam S., Melo H., Heijink A. M., Sastre-Moreno G., Moatti N., Szilard R. K., Ewan A. M., Ling A. K., Serrano-Benitez A., Ubhi T., Feng S., Pawling J., Delgado-Sainz I., Ferguson M. W., Dennis J. W., Brown G. W., Cortés-Ledesma F., Williams R. S., Martin A., Xu D., Durocher D., A genetic map of the response to DNA damage in human cells. Cell 182, 481–496.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clements K. E., Schleicher E. M., Thakar T., Hale A., Dhoonmoon A., Tolman N. J., Sharma A., Liang X., Imamura Kawasawa Y., Nicolae C. M., Wang H. G., de S., Moldovan G. L., Identification of regulators of poly-ADP-ribose polymerase inhibitor response through complementary CRISPR knockout and activation screens. Nat. Commun. 11, 6118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D. W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyjek M., Figiel M., Nowotny M., RNases H: Structure and mechanism. DNA Repair 84, 102672 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Kluth M., Scherzai S., Büschek F., Fraune C., Möller K., Höflmayer D., Minner S., Göbel C., Möller-Koop C., Hinsch A., Neubauer E., Tsourlakis M. C., Sauter G., Heinzer H., Graefen M., Wilczak W., Luebke A. M., Burandt E., Steurer S., Schlomm T., Simon R., 13q deletion is linked to an adverse phenotype and poor prognosis in prostate cancer. Genes Chromosomes Cancer 57, 504–512 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Brookman-Amissah N., Nariculam J., Freeman A., Willamson M., Kirby R. S., Masters J. R., Feneley M. R., Allelic imbalance at 13q14.2 approximately q14.3 in localized prostate cancer is associated with early biochemical relapse. Cancer Genet. Cytogenet. 179, 118–126 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Dong J. T., Chen C., Stultz B. G., Isaacs J. T., Frierson H. F. Jr., Deletion at 13q21 is associated with aggressive prostate cancers. Cancer Res. 60, 3880–3883 (2000). [PubMed] [Google Scholar]
- 23.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I., Cieslik M., Benelli M., Robinson D., van Allen E. M., Sboner A., Fedrizzi T., Mosquera J. M., Robinson B. D., de Sarkar N., Kunju L. P., Tomlins S., Wu Y. M., Nava Rodrigues D., Loda M., Gopalan A., Reuter V. E., Pritchard C. C., Mateo J., Bianchini D., Miranda S., Carreira S., Rescigno P., Filipenko J., Vinson J., Montgomery R. B., Beltran H., Heath E. I., Scher H. I., Kantoff P. W., Taplin M. E., Schultz N., deBono J. S., Demichelis F., Nelson P. S., Rubin M. A., Chinnaiyan A. M., Sawyers C. L., Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai J., Huang S. Y. N., Renaud A., Zhang Y., Ji J., Takeda S., Morris J., Teicher B., Doroshow J. H., Pommier Y., Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inano S., Sato K., Katsuki Y., Kobayashi W., Tanaka H., Nakajima K., Nakada S., Miyoshi H., Knies K., Takaori-Kondo A., Schindler D., Ishiai M., Kurumizaka H., Takata M., RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66, 622–634.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Graeser M., McCarthy A., Lord C. J., Savage K., Hills M., Salter J., Orr N., Parton M., Smith I. E., Reis-Filho J. S., Dowsett M., Ashworth A., Turner N. C., A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res. 16, 6159–6168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyson N. J., RB1: A prototype tumor suppressor and an enigma. Genes Dev. 30, 1492–1502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas A. K., Johnson D. G., Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 72, 13–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyquist M. D., Corella A., Coleman I., de Sarkar N., Kaipainen A., Ha G., Gulati R., Ang L., Chatterjee P., Lucas J., Pritchard C., Risbridger G., Isaacs J., Montgomery B., Morrissey C., Corey E., Nelson P. S., Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep. 31, 107669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos-Montoya A., Lamb A. D., Russell R., Carroll T., Jurmeister S., Galeano-Dalmau N., Massie C. E., Boren J., Bon H., Theodorou V., Vias M., Shaw G. L., Sharma N. L., Ross-Adams H., Scott H. E., Vowler S. L., Howat W. J., Warren A. Y., Wooster R. F., Mills I. G., Neal D. E., HES6 drives a critical AR transcriptional programme to induce castration-resistant prostate cancer through activation of an E2F1-mediated cell cycle network. EMBO Mol. Med. 6, 651–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNair C., Xu K., Mandigo A. C., Benelli M., Leiby B., Rodrigues D., Lindberg J., Gronberg H., Crespo M., de Laere B., Dirix L., Visakorpi T., Li F., Feng F. Y., de Bono J., Demichelis F., Rubin M. A., Brown M., Knudsen K. E., Differential impact of RB status on E2F1 reprogramming in human cancer. J. Clin. Invest. 128, 341–358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiewer M. J., Mandigo A. C., Gordon N., Huang F., Gaur S., Leeuw R., Zhao S. G., Evans J., Han S., Parsons T., Birbe R., McCue P., McNair C., Chand S. N., Cendon-Florez Y., Gallagher P., McCann J. J., Poudel Neupane N., Shafi A. A., Dylgjeri E., Brand L. J., Visakorpi T., Raj G. V., Lallas C. D., Trabulsi E. J., Gomella L. G., Dicker A. P., Kelly W. K., Leiby B. E., Knudsen B., Feng F. Y., Knudsen K. E., PARP-1 regulates DNA repair factor availability. EMBO Mol. Med. 10, e8816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byers L. A., Wang J., Nilsson M. B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y. H., Diao L., Masrorpour F., Shen L., Liu W., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B. S., Kalhor N., Wistuba I. I., Girard L., Lippman S. M., Mills G. B., Coombes K. R., Weinstein J. N., Minna J. D., Heymach J. V., Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2, 798–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simbulan-Rosenthal C. M., Rosenthal D. S., Luo R. B., Samara R., Espinoza L. A., Hassa P. O., Hottiger M. O., Smulson M. E., PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene 22, 8460–8471 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Kim H., Xu H., George E., Hallberg D., Kumar S., Jagannathan V., Medvedev S., Kinose Y., Devins K., Verma P., Ly K., Wang Y., Greenberg R. A., Schwartz L., Johnson N., Scharpf R. B., Mills G. B., Zhang R., Velculescu V. E., Brown E. J., Simpkins F., Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat. Commun. 11, 3726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buisson R., Boisvert J. L., Benes C. H., Zou L., Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell 59, 1011–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertoli C., Herlihy A. E., Pennycook B. R., Kriston-Vizi J., de Bruin R. A. M., Sustained E2F-dependent transcription is a key mechanism to prevent replication-stress-induced DNA damage. Cell Rep. 15, 1412–1422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Liu Y., Oberly S., Freire R., Smolka M. B., ATR-mediated proteome remodeling is a major determinant of homologous recombination capacity in cancer cells. Nucleic Acids Res. 46, 8311–8325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianevski A., He L., Aittokallio T., Tang J., SynergyFinder: A web application for analyzing drug combination dose-response matrix data. Bioinformatics 33, 2413–2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yazinski S. A., Comaills V., Buisson R., Genois M. M., Nguyen H. D., Ho C. K., Todorova Kwan T., Morris R., Lauffer S., Nussenzweig A., Ramaswamy S., Benes C. H., Haber D. A., Maheswaran S., Birrer M. J., Zou L., ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord C. J., Ashworth A., BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Ku S. Y., Rosario S., Wang Y., Mu P., Seshadri M., Goodrich Z. W., Goodrich M. M., Labbé D. P., Gomez E. C., Wang J., Long H. W., Xu B., Brown M., Loda M., Sawyers C. L., Ellis L., Goodrich D. W., Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu P., Zhang Z., Benelli M., Karthaus W. R., Hoover E., Chen C. C., Wongvipat J., Ku S. Y., Gao D., Cao Z., Shah N., Adams E. J., Abida W., Watson P. A., Prandi D., Huang C. H., de Stanchina E., Lowe S. W., Ellis L., Beltran H., Rubin M. A., Goodrich D. W., Demichelis F., Sawyers C. L., SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W. S., Aggarwal R., Zhang L., Zhao S. G., Thomas G. V., Beer T. M., Quigley D. A., Foye A., Playdle D., Huang J., Lloyd P., Lu E., Sun D., Guan X., Rettig M., Gleave M., Evans C. P., Youngren J., True L., Lara P., Kothari V., Xia Z., Chi K. N., Reiter R. E., Maher C. A., Feng F. Y., Small E. J., Alumkal J. J.; West Coast Prostate Cancer Dream Team , Genomic drivers of poor prognosis and enzalutamide resistance in metastatic castration-resistant prostate cancer. Eur. Urol. 76, 562–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamid A. A., Gray K. P., Shaw G., MacConaill L. E., Evan C., Bernard B., Loda M., Corcoran N. M., van Allen E. M., Choudhury A. D., Sweeney C. J., Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur. Urol. 76, 89–97 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Tan H. L., Sood A., Rahimi H. A., Wang W., Gupta N., Hicks J., Mosier S., Gocke C. D., Epstein J. I., Netto G. J., Liu W., Isaacs W. B., de Marzo A. M., Lotan T. L., Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 20, 890–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chappell W. H., Lehmann B. D., Terrian D. M., Abrams S. L., Steelman L. S., McCubrey J. A., p53 expression controls prostate cancer sensitivity to chemotherapy and the MDM2 inhibitor Nutlin-3. Cell Cycle 11, 4579–4588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B., Li L., Yang G., Geng C., Luo Y., Wu W., Manyam G. C., Korentzelos D., Park S., Tang Z., Wu C., Dong Z., Sigouros M., Sboner A., Beltran H., Chen Y., Corn P. G., Tetzlaff M. T., Troncoso P., Broom B., Thompson T. C., PARP inhibition suppresses GR-MYCN-CDK5-RB1-E2F1 signaling and neuroendocrine differentiation in castration-resistant prostate cancer. Clin. Cancer Res. 25, 6839–6851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boumahdi S., de Sauvage F. J., The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 19, 39–56 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Mandigo A. C., Yuan W., Xu K., Gallagher P., Pang A., Guan Y. F., Shafi A. A., Thangavel C., Sheehan B., Bogdan D., Paschalis A., McCann J. J., Laufer T. S., Gordon N., Vasilevskaya I. A., Dylgjeri E., Chand S. N., Schiewer M. J., Domingo-Domenech J., den R. B., Holst J., McCue P. A., de Bono J. S., McNair C., Knudsen K. E., RB/E2F1 as a master regulator of cancer cell metabolism in advanced disease. Cancer Discov. 11, 2334–2353 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nava Rodrigues D., Casiraghi N., Romanel A., Crespo M., Miranda S., Rescigno P., Figueiredo I., Riisnaes R., Carreira S., Sumanasuriya S., Gasperini P., Sharp A., Mateo J., Makay A., McNair C., Schiewer M., Knudsen K., Boysen G., Demichelis F., de Bono J. S., RB1 heterogeneity in advanced metastatic castration-resistant prostate cancer. Clin. Cancer Res. 25, 687–697 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Chakraborty G., Armenia J., Mazzu Y. Z., Nandakumar S., Stopsack K. H., Atiq M. O., Komura K., Jehane L., Hirani R., Chadalavada K., Yoshikawa Y., Khan N. A., Chen Y., Abida W., Mucci L. A., Lee G. S. M., Nanjangud G. J., Kantoff P. W., Significance of BRCA2 and RB1 co-loss in aggressive prostate cancer progression. Clin. Cancer Res. 26, 2047–2064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thangavel C., Boopathi E., Ciment S., Liu Y., O’Neill R., Sharma A., McMahon S. B., Mellert H., Addya S., Ertel A., Birbe R., Fortina P., Dicker A. P., Knudsen K. E., den R. B., The retinoblastoma tumor suppressor modulates DNA repair and radioresponsiveness. Clin. Cancer Res. 20, 5468–5482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M. S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gui B., Gui F., Takai T., Feng C., Bai X., Fazli L., Dong X., Liu S., Zhang X., Zhang W., Kibel A. S., Jia L., Selective targeting of PARP-2 inhibits androgen receptor signaling and prostate cancer growth through disruption of FOXA1 function. Proc. Natl. Acad. Sci. U.S.A. 116, 14573–14582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decker K. F., Zheng D., He Y., Bowman T., Edwards J. R., Jia L., Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 40, 10765–10779 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiebert S. W., Blake M., Azizkhan J., Nevins J. R., Role of E2F transcription factor in E1A-mediated trans activation of cellular genes. J. Virol. 65, 3547–3552 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3