Abstract
The identification of new pathways supporting the myelodysplastic syndrome (MDS) primitive cells growth is required to develop targeted therapies. Within myeloid malignancies, men have worse outcomes than women, suggesting male sex hormone–driven effects in malignant hematopoiesis. Androgen receptor promotes the expression of five granulocyte colony-stimulating factor receptor–regulated genes. Among them, CCRL2 encodes an atypical chemokine receptor regulating cytokine signaling in granulocytes, but its role in myeloid malignancies is unknown. Our study revealed that CCRL2 is up-regulated in primitive cells from patients with MDS and secondary acute myeloid leukemia (sAML). CCRL2 knockdown suppressed MDS92 and MDS-L cell growth and clonogenicity in vitro and in vivo and decreased JAK2/STAT3/STAT5 phosphorylation. CCRL2 coprecipitated with JAK2 and potentiated JAK2-STAT interaction. Erythroleukemia cells expressing JAK2V617F showed less effect of CCRL2 knockdown, whereas fedratinib potentiated the CCRL2 knockdown effect. Conclusively, our results implicate CCRL2 as an MDS/sAML cell growth mediator, partially through JAK2/STAT signaling.
The chemokine receptor CCRL2 is up-regulated in MDS, promotes cancer cell proliferation, and activates JAK2/STAT signaling.
INTRODUCTION
Myelodysplastic syndromes (MDSs) arise from hematopoietic stem and progenitor cells that acquire somatic mutations (1). Although they can respond to chemotherapy and hypomethylating agents, MDSs are only cured by allogeneic transplantation (1, 2). Oncogenic pathways involving granulocyte colony-stimulating factor receptor (GCSF-R) and Janus kinase 2 (JAK2)/signal transducer and activator of transcription (STAT) signaling have been implicated in the progression of myeloproliferative neoplasms (MPNs), MDS/MPN, and acute myeloid leukemia (AML) (3, 4), but their roles in MDS are less well understood. A deeper understanding of the molecular pathways providing survival and growth advantage to MDS cells over healthy hematopoietic cells is critical for improving the outcomes of patients with these diseases.
Recent studies support that men with myeloid neoplasms generally exhibit higher disease burdens and experience worse outcomes compared with women, independent of other known prognostic markers (5–7). However, the underlying molecular mechanisms responsible for these observations are unclear. Hormonal receptors are expressed in hematopoietic cells (8), and estrogen receptor signaling induces the apoptosis of malignant stem cells (9). The androgen receptor (AR) promotes normal granulopoiesis by activating in myeloid precursors the GCSF-R, with five of the most up-regulated GCSF-R target genes being CCRL2, C3AR1, GYK, SOCS3, and FLOT2 (10). Among these, C3AR1 is overexpressed in AML cells (11), and this has been associated with worse outcomes in patients with AML (12). Similarly, FLOT2 promotes the homing of chronic myeloid leukemia–initiating cells in a mouse model (13), and SOCS3 suppresses the CD33-mediated inhibition of cytokine-induced proliferation of AML cells (14).
CCRL2 encodes for one of the atypical chemokine receptors (15) and is expressed in granulocytes, monocytes, and natural killer cells, playing a role in the migration of these cells to sites of inflammation (16, 17). CCRL2 is required for the activation of CXCR2, the chemokine receptor for interleukin-8 (IL-8) (18). Notably, the IL-8/CXCR2 pathway is critical for the growth of MDS and secondary AML (sAML) cells (19). However, little is known about the regulation and signaling role of CCRL2 in myeloid neoplasms.
Here, we show that CCRL2 is up-regulated in stem and progenitor cells from patients with MDS. It promotes the growth and clonogenicity of MDS and sAML cells and activates JAK2/STAT signaling. CCRL2 knockdown inhibits the growth of MDS and sAML cells and alters the effect of the JAK2 inhibitor fedratinib.
RESULTS
CCRL2 is up-regulated in MDS and MDS-related AML
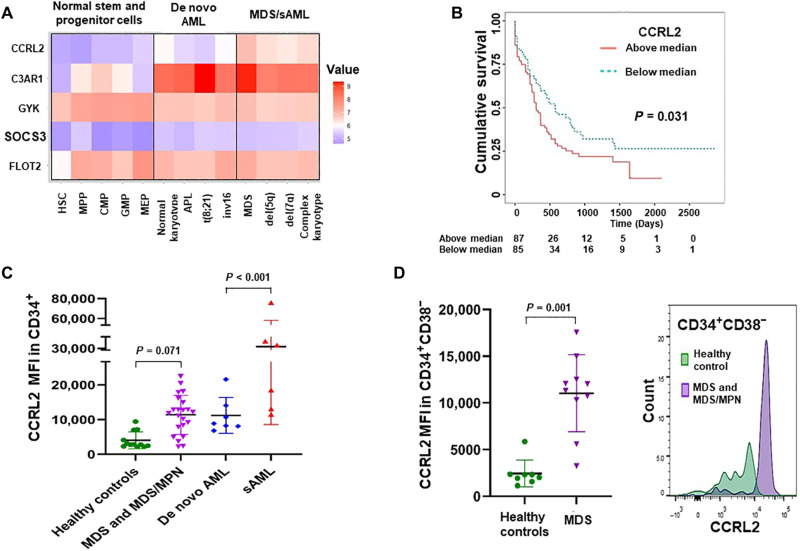
Expression of the most AR–up-regulated GCSF-R target genes (10) in healthy stem and progenitor cells, as well as in subtypes of AML and MDS, was analyzed using the BloodSpot dataset [GSE42519, GSE13159, GSE15434, GSE61804, GSE14468, and The Cancer Genome Atlas (TCGA)] (20). AML/MDS cells express higher levels of C3AR1, CCRL2, and SOCS3 compared with healthy cells (Fig. 1A). CCRL2 was the only one of the five genes found to be up-regulated in MDS and AML with MDS-related chromosomal changes (which will be referred to as sAML), compared with de novo AMLs (Fig. 1A). Analysis of data derived from the Beat AML dataset (21) revealed that CCRL2 expression is higher in CD34+ AMLs compared with healthy CD34+ cells (fig. S1A).
Fig. 1. CCRL2 is up-regulated in MDS and sAML cells.
(A) Comparison of the mRNA levels of the five AR-regulated genes CCRL2, C3AR1, GYK, SOCS3, and FLOT2 between healthy stem and progenitor cells, de novo AML (AMLs without MDS-related chromosomal abnormalities), MDS, and sAML (AMLs with MDS-related chromosomal abnormalities) based on data extracted from the BloodSpot database. CCRL2 is the only gene that is up-regulated in MDS and sAML compared with both healthy cells and de novo AMLs (P < 0.001). (B) Patients with AML with high CCRL2 expression (based on median expression) have significantly worse overall survival (P = 0.031) compared with patients with AML with low CCRL2 expression based on the TCGA dataset. (C) Analysis of the CCRL2 protein levels by flow cytometry showed that CD34+ cells from patients with MDS and MDS/MPN express relatively higher levels of CCRL2 (P = 0.071) compared with CD34+ cells from healthy controls. CD34+ blasts from patients with sAML express higher levels of CCRL2 compared with CD34+ blasts from patients with de novo AML (P < 0.001). (D) CCRL2 expression is higher in CD34+CD38− cells from patients with MDS compared with CD34+CD38− cells from healthy individuals (P = 0.001). Graphs show the mean value and SD of the mean value.
Analysis of overall survivals of patients with AML based on median gene expression measured in TCGA dataset (20) showed that CCRL2 expression was the only identified gene among the most AR–up-regulated genes with a statistically significant survival impact. Patients whose AMLs are exhibiting CCRL2 expression above the median had poorer overall survivals (Fig. 1B).
CCRL2 is up-regulated in progenitor cells from patients with MDS and AML
To confirm the results from publicly available databases, the expression of CCRL2 in CD34+ and CD34+CD38− bone marrow cells from 12 healthy controls and 22 patients with either MDS or MDS/MPNs as well as CD34+ blasts from 13 patients with AML was analyzed by flow cytometry (fig. S1, B and C). Patient and healthy control demographic data are summarized in table S1.
CCRL2 was up-regulated in CD34+ cells from patients with MDS and MDS/MPN compared with healthy controls (Fig. 1C and fig. S1D). sAML CD34+ cells also expressed higher CCRL2 compared with de novo AML CD34+ cells (Fig. 1C). No significant difference was identified between patients with MDS with <5% or ≥5% blasts or between patients with MDS and MDS/MPN (fig. S1D). Across patients with MDS and patients with MDS/MPN with excess blasts (≥5%) and sAML younger age, higher blast percentage and FLT3 mutation were associated with higher CCRL2 expression (table S2). Patients with myeloid malignancies were significantly older than healthy controls (P < 0.001 for MDS and MDS/MPN and P = 0.020 for AML), and as there was no significant age overlap between the groups, age adjustment could not be done. However, the CCRL2 expression in CD34+ cells showed a negative correlation with age among healthy controls (Coef, −128.98; P = 0.018), suggesting that older individuals may have lower CCRL2 expression in their CD34+ cells. The analysis of 8 healthy controls and 10 patients with MDS (table S3) revealed that CCRL2 is also up-regulated in CD34+CD38− cells from patients with MDS compared with healthy controls (Fig. 1D).
CCRL2 influences the growth and clonogenicity of MDS cell lines
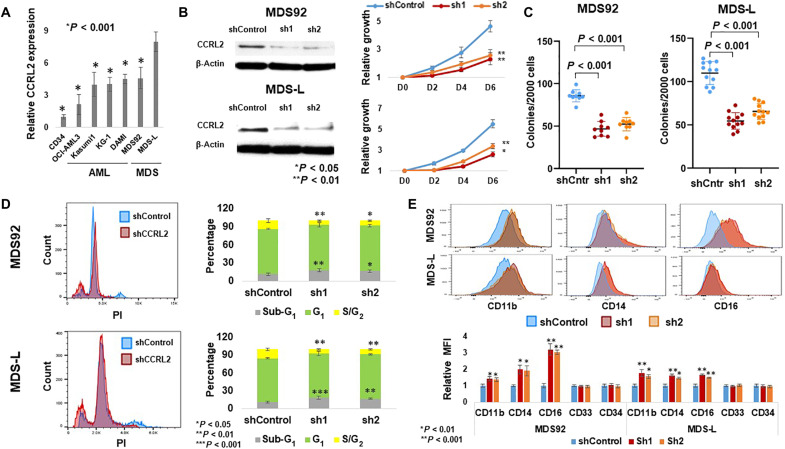
The expression of CCRL2 protein in AML and MDS cell lines and sorted CD34+ cells from three healthy individuals was assessed by flow cytometry. MDS-L cells, a leukemic subline derived from the MDS92 cell line established from a patient with MDS with 5q deletion, monosomy 7, and NRAS mutation (22, 23), expressed the highest levels of CCRL2 from a panel of six MDS and AML cell lines (Fig. 2A and fig. S2A). This finding was confirmed by Western blotting (fig. S2A).
Fig. 2. CCRL2 promotes the growth and clonogenicity of MDS cell lines.
(A) MDS-L cells express higher levels of CCRL2 compared with CD34+ cells from three separate healthy controls (P < 0.001), de novo AML [OCI-AML3 (P < 0.001) and Kasumi-1 (P < 0.001)], sAML cell lines [KG-1 (P < 0.001) and DAMI (P < 0.001)], and MDS92 cells (P < 0.001), n = 3. (B) CCRL2 knockdown suppressed the growth rate at 2 days (MDS92: P = 0.004-sh1, P = 0.040-sh2; MDS-L: P = 0.007-sh1, P = 0.038-sh2), 4 days (MDS92: P = 0.013-sh1, P = 0.033-sh2; MDS-L: P = 0.002-sh1, P = 0.007-sh2), and 6 days (MDS92: P = 0.003-sh1, P = 0.008-sh2; MDS-L: P = 0.009-sh1, P = 0.016-sh2), n = 3. (C) CCRL2 knockdown suppresses the clonogenic capacity of MDS92 (P < 0.001; n = 9) and MDS-L (P < 0.001; n = 12). Results show the mean value and SD of the mean value. (D) CCRL2 knockdown increases the percentage of apoptotic (sub-G1) MDS92 (P = 0.006-sh1, P = 0.021-sh2) and MDS-L cells (P = 0.002-sh1, P = 0.007-sh2) and decreases the percentage of MDS92 (P = 0.006-sh1, P = 0.017-sh2) and MDS-L cells (P < 0.001-sh1, P = 0.001-sh2) in the S-G2 phase. (E) CCRL2 knockdown increases the CD11b expression in MDS92 (P = 0.006-sh1, P = 0.004-sh2) and MDS-L cells (P < 0.001-sh1, P = 0.004-sh2), the CD14 expression in MDS92 (P = 0.003-sh1, P = 0.004-sh2) and MDS-L cells (P < 0.001-sh1, P < 0.001-sh2), and the CD16 expression in MDS92 (P < 0.001) and MDS-L cells (P < 0.001). n = 3.
To further characterize the role of CCRL2 in the survival and growth of MDS cells, MDS92 and MDS-L cells were transduced with either empty vector or two short hairpin RNA (shRNA) targeting CCRL2 (sh1 and sh2). CCRL2 knockdown was confirmed at both the RNA (fig. S2B) and protein levels (Fig. 2B and fig. S2B). MDS92 and MDS-L cells with suppressed CCRL2 exhibited significantly decreased growth rate at 2, 4, and 6 days (Fig. 2B). Similarly, the clonogenic capacity of MDS92 and MDS-L transduced with shCCRL2 was suppressed (Fig. 2C).
To determine the underlying biologic effect of CCRL2 knockdown, the analysis of cell cycle in cells with intact and suppressed CCRL2 was performed. CCRL2 knockdown in MDS92 and MDS-L cells increased the percentage of apoptotic cells and decreased the percentage of cells in the G2-S phase (Fig. 2D). Similarly, evaluation of apoptosis with annexin V/propidium iodide (PI) staining showed that CCRL2 knockdown in the MDS cell lines increases the percentage of apoptotic cells (fig. S2C). The effect of CCRL2 knockdown in MDS cell differentiation was also assessed using a number of surface markers previously measured in MDS92 and MDS-L cells (24, 25). MDS cells transduced with shCCRL2 showed an increase in CD11b, CD14, and CD16 expression (Fig. 2E). To investigate whether CCRL2 expression affects healthy progenitors, CD34+ cells were sorted from bone marrow aspirates derived from two healthy individuals and transduced with shControl or shCCRL2 (sh1) (fig. S3). No significant alterations were observed in the clonogenicity of cells with suppressed CCRL2 (fig. S3).
CCRL2 promotes the growth of MDS-L cells in vivo
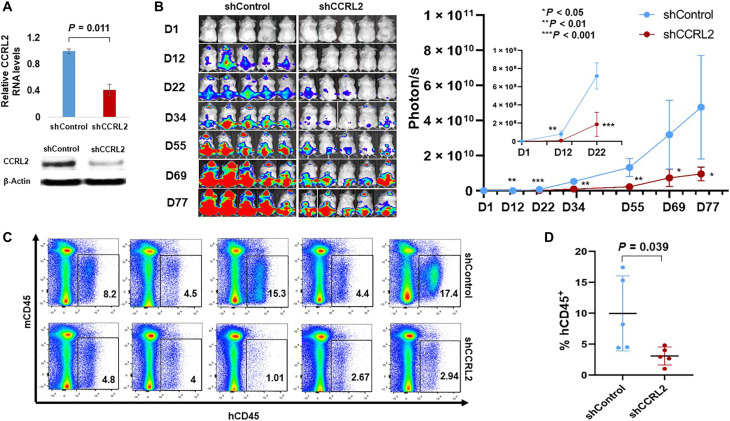
The impact of CCRL2 knockdown in an MDS-L xenograft mouse model was evaluated. MDS-L shCCRL2 and control cells were cultured for 10 days in puromycin, and the suppression of CCRL2 expression was assessed at the RNA and protein levels (Fig. 3A and fig. S4A). The cells were then transduced with a GFP+/luciferase+ dual reporter retrovirus, and GFP+ cells were injected intravenously in NSG-hSCF/hGM-CSF/hIL3 (NSGS) mice 48 hours after intraperitoneal injection of Clophosome-A clodronate liposomes (26). MDS-L shCCRL2 cells demonstrated a slower proliferative rate than control cells (Fig. 3B). No significant differences in peripheral blood counts were observed (fig. S4B). At day 78, all mice were euthanized, and a higher percentage of human CD45+ cells was found in the bone marrow (Fig. 3C) and the spleen (fig. S4C) of control mice compared with those that received MDS-L shCCRL2.
Fig. 3. CCRL2 knockdown suppresses the engraftment and growth of MDS-L cells in NSGS mice.
(A) MDS-L cells transduced with shControl or shCCRL2 shRNAs were treated for 10 days with puromycin, and their CCRL2 expression was assessed by RNA and protein. MDS-L cells transduced with shCCRL2 expressed significantly lower levels of CCRL2 RNA (P = 0.011) and protein (P = 0.006). (B) MDS-L cells with normal or suppressed CCRL2 were transduced with a GFP+/Luciferase dual reporter retrovirus and injected to NSGS mice. The bioluminescence signal in mice injected with MDS-L with suppressed CCRL2 was significantly lower at 12 (P = 0.0016), 22 (P = 0.0002), 34 (P = 0.0010), 55 (P = 0.0017), 69 (P = 0.0259), and 77 days (P = 0.0212) after the injection. (C) MDS-L burden in the bone marrow of NSGS mice was significantly lower in mice injected with MDS-L cells with suppressed CCRL2 (P = 0.039). Results depict the mean value and SD of the mean value.
CCRL2 affects JAK2/STAT signaling in MDS cells
CCRL2 appears to be involved in the activation of cytokine-mediated pathways such as IL-8/CXCR2, which, in turn, activates AKT and extracellular signal–regulated kinase (ERK) signaling in differentiated myeloid cells (18). Moreover, MDS92 and MDS-L cells are dependent on IL-3, which signals through JAK2/STAT (22, 23), suggesting that this cytokine-mediated pathway is also critical for the growth of these cells. Thus, the effect of CCRL2 knockdown in the phosphorylation of AKT, ERK, JAK2, and STAT proteins was assessed.
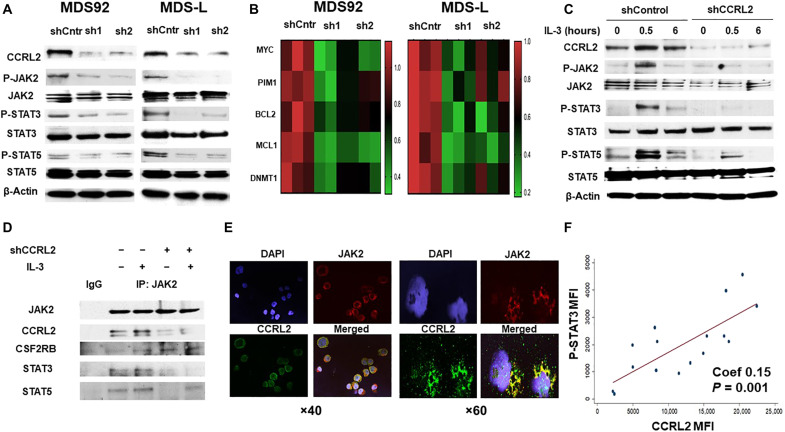
The knockdown of CCRL2 in MDS cells had no impact on the phosphorylation of AKT or ERK (fig. S5, A and B). However, CCRL2 knockdown suppressed the phosphorylation of JAK2, STAT3, and STAT5 in both MDS92 and MDS-L cells (Fig. 4A and fig. S5B). The effect of CCRL2 on the expression of JAK2/STAT target genes in MDS cells was also evaluated. Knockdown of CCRL2 in MDS92 and MDS-L cells decreased the expression of MYC, PIM1, BCL2, MCL1, and DNMT1 at the RNA (Fig. 4B) and protein levels (fig. S5, C and D), suggesting that these genes are regulated by CCRL2 likely at the transcriptional level.
Fig. 4. CCRL2 affects JAK2/STAT signaling in MDS cells.
(A) Representative Western blotting showing CCRL2 knockdown effect by two different shRNAs (sh1 and sh2) on JAK2 (Tyr1007/1008), STAT3 (Tyr105), and STAT5 (Tyr694) phosphorylation in MDS92 and MDS-L. (B) CCRL2 knockdown decreases the RNA levels of the JAK2/STAT target genes: MYC (MDS92: P = 0.001-sh1, P = 0.002-sh2; MDS-L: P = 0.001-sh1, P = 0.010-sh2), PIM1 (MDS92: P = 0.003-sh1, P = 0.010-sh2; MDS-L: P = 0.002-sh1, P = 0.040-sh2), BCL2 (MDS92: P = 0.020-sh1, P = 0.060-sh2; MDS-L: P = 0.008-sh1, P = 0.005-sh2), MCL1 (MDS92: P = 0.025-sh1, P = 0.004-sh2; MDS-L: P = 0.004-sh1, P = 0.019-sh2), and DNMT1 (MDS92: P = 0.009-sh1, P = 0.010-sh2; MDS-L: P = 0.008-sh1, P = 0.040-sh2), n = 3. (C) Western blotting showing that CCRL2 knockdown suppresses the phosphorylation of JAK2, STAT3, and STAT5 at 30 min and 6 hours of IL-3 (20 ng/ml) treatment following 48 hours of IL-3 starvation. (D) Coimmunoprecipitation assay showing that CCRL2 precipitates with JAK2 and that CCRL2 knockdown does not affect the interaction between JAK2 and the common β signal transducing subunit of CD123 (CSF2RB) but decreases the interaction between JAK2 and STAT3/5 proteins. IgG, immunoglobulin G. (E) Representative images from immunofluorescence staining (×40 and ×60 magnification) showing localization of CCRL2 (green) in the cytoplasm and membrane of MDS-L cells. Confocal microscopy reveals areas of colocalization with JAK2 (red). DAPI, 4′,6-diamidino-2-phenylindole. (F) The mean fluorescence intensity (MFI) of phosphorylated STAT3 (P-STAT3) is positively associated with the MFI of CCRL2 in CD34+ cells from patients with MDS and CD34+ blasts from patients with AML (Coef, 0.15; P = 0.001), n = 16.
To further study JAK2/STAT regulation by CCRL2, MDS-L cells were deprived of IL-3 for 48 hours and then were treated with IL-3 (20 ng/ml). CCRL2 knockdown suppressed IL-3–mediated JAK2 autophosphorylation, as well as the phosphorylation of STAT3 and STAT5 (Fig. 4C), suggesting that CCRL2 has a role in the IL-3 receptor (CD123), JAK2, and STAT interaction. Coimmunoprecipitation showed that CCRL2 is associated with JAK2, and CCRL2 knockdown suppresses the JAK2/STAT interaction following IL-3 treatment (Fig. 4D). In contrast, CCRL2 knockdown did not affect the interaction between JAK2 and the common β signal transducing subunit of the IL-3 receptor (CSF2RB) (Fig. 4D). Immunofluorescence analysis revealed the localization of CCRL2 in the membrane and cytoplasm of MDS-L cells, and confocal microscopy showed areas of colocalization with JAK2 (Fig. 4E).
CCRL2 and STAT3 and STAT5 phosphorylation was assessed by flow cytometry in primary CD34+ cells from nine healthy controls, 11 patients with MDS, and 5 patients with AML (fig. S6A). CCRL2 expression was positively associated with the phosphorylation of STAT3 (Fig. 4F) but not with the phosphorylation of STAT5 (fig. S6B) in CD34+ cells from patients with MDS and CD34+ AML blasts. Notably, the correlation between CCRL2 expression and STAT3 phosphorylation was independent of the blast percentage [Coef, 0.144; 95% confidence interval (CI), 0.075 to 0.220] and the highest mutation allele frequency (Coef, 0.149; 95% CI, 0.071 to 0.230).
CCRL2 has differential effects on JAK2 wild-type and mutated cell lines
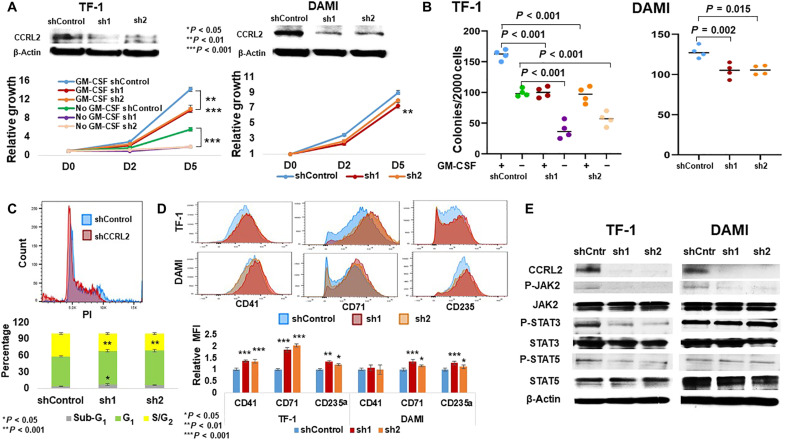
Given the observation that CCRL2 affects JAK2/STAT signaling, the effect of CCRL2 knockdown was assessed in two erythroleukemia cell lines with and without mutational JAK2 activation. Granulocyte-macrophage colony-stimulating factor (GM-CSF)–dependent TF-1 cells are JAK2 wild type, while DAMI cells carry a JAK2V617F mutation and are cytokine independent (27, 28). CCRL2 knockdown significantly decreased the growth of TF-1 cells in the presence and absence of GM-CSF (Fig. 5A), while its effect on the DAMI cell growth was less prominent (Fig. 5A). Similarly, CCRL2 knockdown had a more marked effect on the clonogenicity of TF-1 cells in the presence and particularly in the absence of GM-CSF compared with DAMI cells (Fig. 5B). Consistently, CCRL2 knockdown in TF-1 cells increased the percentage of apoptotic cells (Fig. 5C and fig. S6D) and decreased the percentage of cells in the G2-S phase (Fig. 5C). On the contrary, CCRL2 knockdown did not significantly affect the apoptotic rate and cell cycle progression in DAMI cells (fig. S6, C and D). CCRL2 knockdown increased the expression of CD71 and CD235a, two markers of erythroid differentiation (29), in both TF-1 and DAMI cells with a more prominent effect in TF-1 cells (Fig. 5D). Last, CCRL2 knockdown increased the expression of CD41, a marker of megakaryocytic differentiation (30), only in TF-1 cells (Fig. 5D). CCRL2 knockdown suppressed JAK2 phosphorylation in both cell lines, but STAT3 and STAT5 phosphorylation was decreased only in TF-1 (Fig. 5E).
Fig. 5. CCRL2 knockdown affects the growth and clonogenicity of erythroleukemia cell lines.
(A) CCRL2 knockdown with two different lentiviruses significantly suppresses the growth of TF-1 cells in the presence (P < 0.001) and absence of GM-CSF (P < 0.001). CCRL2 knockdown with two different lentiviruses suppresses at a lower extent the growth of DAMI cells (P = 0.009-sh1, P = 0.058-sh2). (B) CCRL2 knockdown with two different lentiviruses decreases the colony formation of TF-1 cells in the presence (P < 0.001) and absence of GM-CSF (P < 0.001). CCRL2 knockdown with two different lentiviruses decreases the colony formation of DAMI cells (P = 0.002-sh1, P = 0.015-sh2) at a lower extent compared with TF-1 cells. (C) CCRL2 knockdown with sh1 increased the percentage of apoptotic TF-1 cells (P = 0.015-sh1, P = 0.097-sh2), and CCRL2 knockdown with the two lentiviruses decreased the percentage of TF-1 cells in the G2-S phase (P < 0.001). (D) CCRL2 knockdown with two different lentiviruses increases the expression of CD41 (P < 0.001), CD71 (P < 0.001), and CD235a (P = 0.002-sh1, P = 0.025-sh2). CCRL2 knockdown does not affect the expression of CD41 (P = 0.230-sh1, P = 0.912-sh2). CCRL2 knockdown increases the expression of CD71 (P < 0.001-sh1, P = 0.026-sh2) and CD235a (P = 0.002-sh1, P = 0.067-sh2). (E) Western blotting showing the effect of CCRL2 knockdown in the JAK2/STAT signaling in TF-1 and DAMI cells. CCRL2 knockdown suppresses the phosphorylation of JAK2, STAT3, and STAT5 in TF-1 cells. CCRL2 knockdown decreases the phosphorylation of JAK2 but does not affect the phosphorylation of STAT3 and STAT5 in DAMI cells.
CCRL2 knockdown alters the effect of JAK2 inhibition
JAK2 inhibition induces the autophosphorylation of JAK2, which has been associated with a withdraw phenomenon caused by the activation of downstream signaling in patients with myelofibrosis (31–33), suggesting that inhibition of JAK2 autophosphorylation could increase the activity of JAK2 inhibition. The effect of CCRL2 knockdown on the efficacy of JAK2 inhibition by the selective JAK2 inhibitor fedratinib (34) was studied. MDS/AML cell lines were treated with fedratinib, and the methylene tetrazolium (MTT) assay was used to assess the half-maximal inhibitory concentration dose (IC50) of the drug at 72 hours of treatment. MDS92 and MDS-L had the lowest IC50 value compared with all the other lines, including JAK2-mutated DAMI cells and erythroleukemia TF-1 cells (fig. S7, A and B).
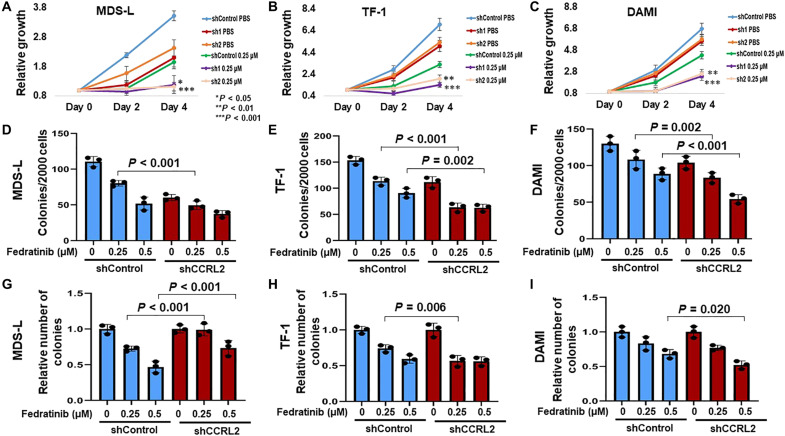
MDS-L, TF-1, and DAMI cells with normal or suppressed CCRL2 expression were treated with phosphate-buffered saline (PBS) or 0.25 μΜ fedratinib, a dose that is lower than the IC50 dose for these three lines. CCRL2 knockdown further suppressed the growth of cells treated with 0.25 μΜ fedratinib at 4 days of treatment (Fig. 6, A to C). Similarly, the combination of CCRL2 knockdown and fedratinib led to more prominent clonogenicity inhibition in MDS-L, TF-1, and DAMI cells (Fig. 6, D to F). Given the prominent negative impact of CCRL2 suppression in the clonogenicity of MDS/AML cells, to study the effect of CCRL2 knockdown in the sensitivity of these cells to fedratinib, the relative number of colonies was assessed. MDS-L cells with suppressed CCRL2 were less sensitive to fedratinib compared with wild-type cells (Fig. 6G), while TF-1 and DAMI cells with suppressed CCRL2 appeared to be overall more sensitive to fedratinib than wild-type cells (Fig. 6, H and I).
Fig. 6. CCRL2 knockdown alters the efficacy of JAK2 inhibition by the selective inhibitor fedratinib.
(A) CCRL2 knockdown increases the antiproliferative effect of 0.25 μΜ fedratinib (P = 0.033-sh1, P = 0.001-sh2) in MDS-L cells. (B) CCRL2 knockdown increases the antiproliferative effect of 0.25 μΜ fedratinib in TF-1 cells (P < 0.001-sh1, P = 0.002-sh2). (C) CCRL2 knockdown increases the antiproliferative effect of 0.25 μΜ fedratinib (P < 0.001-sh1, P = 0.001-sh2) in DAMI cells. (D) CCRL2 knockdown increases the anticlonogenic effect of 0.25 μΜ fedratinib (P < 0.001) in MDS-L cells. (E) CCRL2 knockdown increases the anticlonogenic effect of 0.25 μΜ (P < 0.001) and 0.5 μΜ fedratinib (P < 0.002) in TF-1 cells. (F) CCRL2 knockdown increases the anticlonogenic effect of 0.25 μΜ (P = 0.002) and 0.5 μΜ fedratinib (P < 0.001) in DAMI cells. (G) The clonogenicity inhibitory effect of fedratinib is decreased in MDS-L cells with suppressed CCRL2 (P < 0.001). (H) The clonogenicity inhibitory effect of 0.25 μΜ fedratinib is increased in TF-1 cells with suppressed CCRL2 (P = 0.006). (I) The clonogenicity inhibitory effect of 0.25 μΜ fedratinib is increased in TF-1 cells with suppressed CCRL2 (P = 0.020). *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
Our findings suggest that CCRL2, one of the most AR–up-regulated genes implicated in the later stages of normal granulopoiesis, is overexpressed in stem and progenitor cells from patients with MDS and CD34+ AML blasts. Knockdown of CCRL2 inhibited the growth and clonogenicity of MDS cell lines both in vitro and in vivo. Down-regulation of CCRL2 decreased the phosphorylation of JAK2/STAT and the expression of STAT target genes. CCRL2 modulated the IL-3–mediated phosphorylation of JAK2/STAT and the physical interaction between JAK2 and STAT3/5 proteins. CCRL2 knockdown inhibited the growth of the erythroleukemia cell lines TF-1 and DAMI, with the effect on DAMI cells likely mitigated by constitutively active JAK2/STAT signaling due to the JAK2V617F mutation. Furthermore, CCRL2 knockdown potentiated the antiproliferative and clonogenicity inhibitory effect of the selective JAK2 inhibitor fedratinib in erythroleukemia cell lines.
Men with chronic myeloid neoplasms have worse outcomes compared with women (5–7). However, the underlying biology, including the role of AR signaling, remains poorly understood (35). Chuang et al. (10) reported that mice lacking AR develop severe neutropenia with deficient granulopoiesis, characterized by down-regulation of five GCSF-R target genes. On the basis of our results, it could be hypothesized that the up-regulation of CCRL2 may be implicated in the gender-related differences in chronic myeloid diseases. The evaluation of a possible role of AR in the regulation of CCRL2 in malignant myeloid cells is required to address this question.
CCRL2 is up-regulated in a subset of malignancies including prostate (36) and colon (37) cancer. In contrast, down-regulation of CCRL2 promotes lung cancer progression in animal models, apparently through suppressed immune surveillance (38). To our knowledge, the role of CCRL2 in myeloid malignancies has not been previously studied. On the basis of our findings, CCRL2 is implicated in the regulation of MDS/AML cellular proliferation with a less prominent effect in the inhibition of differentiation, leading to induction of malignant growth and clonogenicity.
CCRL2 is critical for the activation of IL-8/CXCR2 signaling in neutrophils (18), and this pathway has been implicated in the growth of MDS cells via AKT and ERK signaling (19). Our analysis highlighted a more prominent role of CCRL2 in JAK2/STAT regulation. This pathway induces AML cell proliferation (39), while STAT3 inhibition impairs the growth of MDS/AML cells in vitro and in vivo (40). MDS92 and MDS-L cells are IL-3 dependent (22). This cytokine signals through CD123 promoting JAK2 autophosphorylation (41), leading to receptor phosphorylation attracting STAT proteins that then get phosphorylated and transduce the signal to the nucleus (41, 42). The exact mechanisms affecting JAK2 autophosphorylation and induction of STAT phosphorylation are not fully understood (41), but various molecules including protein tyrosine phosphatases (43) and the lymphocyte adaptor protein LNK (44) are known regulators of these events. Similarly, the JAK2V617F mutation suppresses the inhibitory effect of the JAK2 pseudokinase domain (JH2), leading to JAK2 dimerization and constitutive activation of JAK2/STAT signaling (42). Given that CCRL2 promotes JAK2 autophosphorylation and STAT phosphorylation without affecting the interaction of JAK2 and CD123, it is possible that it intensifies the activity of the tyrosine kinase domain (JH1) of JAK2, which includes Tyr1007/1008 and catalyzes the STAT phosphorylation (41, 42). The decreased effect of CCRL2 in cells carrying the JAK2V617F mutation could be explained by the fact that these cells exhibit increased activity of JH1 at baseline. However, it is possible that other cell-specific alterations could be associated with this observation. Similarly, further study is needed for the understanding of the exact mechanism implicated in the interaction of CCRL2 with JAK2.
JAK2 inhibition has been routinely used in patients with JAK2-mutated myeloid malignancies and erythroleukemias (4). JAK2 inhibition by fedratinib shows promising efficacy in MDS cell lines and JAK2 wild-type erythroleukemia cells. The combination of CCRL2 knockdown and JAK2 inhibition by fedratinib led to a more prominent suppression of cell proliferation and clonogenicity independent of the presence of a JAK2V617F mutation. CCRL2 knockdown increased the sensitivity of erythroleukemia cell lines TF-1 and DAMI to fedratinib, suggesting a possible implication of CCRL2 activity upstream of JAK2. JAK2 inhibition suppressed the dephosphorylation of JAK2, leading to hyperphosphorylation of its cryptic site involving Tyr1007/1008 (31, 33). This phenomenon has been associated with the JAK2 inhibitor withdrawal syndrome observed in a subset of patients with myelofibrosis (31, 32). Thus, it is possible that CCRL2 knockdown increases the efficacy of fedratinib by suppressing JAK2 autophosphorylation caused by JAK2 inhibition. Alternatively, the additive effect of CCRL2 knockdown to fedratinib efficacy could suggest that CCRL2 may be implicated in the regulation of other growth-inducing pathways in MDS and AML.
In conclusion, our data suggest that CCRL2, a target gene of AR in normal granulopoiesis, is up-regulated in stem and progenitor cells from patients with MDS and influences the growth of MDS and AML cells. CCRL2 appears to promote JAK2/STAT signaling potentially by increasing JAK2 autophosphorylation and its interaction with STAT proteins. Last, CCRL2 suppression increased the efficacy of fedratinib particularly in erythroleukemia cells, independent of the presence of a JAK2V617F mutation.
MATERIALS AND METHODS
Patients and sample processing
Bone marrow samples were procured from bone marrow aspirations of patients with MDS, MDS/MPN, and CD34+ AML, and normal marrow was obtained as excess material from the harvests of normal donors for allogeneic bone marrow transplantation. Specimens were collected by the Johns Hopkins Kimmel Cancer Center Specimen Accessioning Core. Appropriate informed consent was obtained from all donors before specimen collection in accordance with the Declaration of Helsinki and under a research protocol approved by the Johns Hopkins Institutional Review Board. CD34+ cell subsets were isolated using the CD34 MicroBead kit (Miltenyi Biotec) as previously described (45).
Cell lines and reagents
Human MDS92 and MDS-L cells (22, 23) were a gift from D. T. Starczynowski, University of Cincinnati (25). MDS92 and MDS-L cells were cultured in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA) with 10% fetal bovine serum (MilliporeSigma, Burlington, MA) and IL-3 (10 ng/ml; PeproTech). OCI-AML3, DAMI, TF-1 Kasumi-1, and KG-1 cells were purchased from the American Type Culture Collection and cultured under previously described conditions (46–48). All the cell lines were cultured with 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2. Primary CD34+ cells were cultured as previously described (49). Chlophosome-A chlodronate liposomes were purchased from FormuMax Scientific (#F70101C-A). Fedratinib was purchased from Selleckchem (TG101348) and was diluted in dimethyl sulfoxide.
CCRL2 knockdown
Lentiviral vectors expressing CCRL2-targeting shRNA (RHS3979-201740504 and RHS39379-201740506) (Horizon) or empty pLKO.1-puro lentiviral vector was transfected together with pCMV-dR8.9 and vesicular stomatitis virus G–expressing plasmids into 293T cells using Lipofectamine 2000 (Thermo Fisher Scientific) for lentiviral supernatant production as previously described (50). MDS cells were incubated with the viral supernatant and polybrene (8 μg/ml; MilliporeSigma) for transduction. After at least 48 hours, cells were treated with puromycin (2 μg/ml; MilliporeSigma) for 4 days to select for resistant cells. Primary CD34+ cells from healthy controls following the transduction in the presence of low concentration of ristocetin were treated with puromycin (0.5 μg/ml) for 2 days to select for resistant cells.
Flow cytometry analysis
Cells from healthy controls and patients with MDS/AML were labeled with the following fluorescently conjugated antibodies: 7AAD (BD Biosciences, #559925) and fluorescein isothiocyanate (FITC)–conjugated anti-CD34 (BD Biosciences, #555821). Cells from patients with AML were also labeled with BV510-conjugated anti-CD45 (BD Biosciences, #5632204). CD34+CD38− cells from healthy controls and cells from patients with MDS were also labeled with the BV605-conjugated anti-CD38 (BD Biosciences, #562666). Cells were then fixed in 4% formaldehyde for 15 min and permeabilized in 100% methanol for 30 min. Subsequently, cells were labeled with phycoerythrin (PE)–conjugated anti-CCRL2 (BioLegend #358303). For the analysis of STAT3/5 phosphorylation, cells were also labeled with PE–cyanine 7 (Cy7)–conjugated anti–phospho-STAT3 (Tyr705) (Thermo Fisher Scientific, #25-9033-42) and allophycocyanin (APC)–conjugated anti–phospho-STAT5 (Tyr694) (Thermo Fisher Scientific, #17-9010-42). For the assessment of MDS cell differentiation, MDS92 and MDS-L cells were labeled with the PE-Cy7–conjugated CD11b (BioLegend, #557743), PE-conjugated CD14 (BioLegend, #301805), PE-conjugated CD16 (BioLegend, #302007), FITC-conjugated anti-CD34 (BD Biosciences, #555821), and Alexa Fluor 700–conjugated anti-CD33 (BioLegend, #303435). For the assessment of erythroleukemia cell differentiation, TF-1 and DAMI cells were labeled with the FITC-conjugated anti-CD41 (BioLegend, #303703), PE-Cy7–conjugated CD71 (BioLegend, #113811), and APC-conjugated anti-CD235a (#349113). Gating was based on clearly distinguishable populations or, in the absence of such, the negative antibody control as previously described (45). For the assessment of apoptosis, cells were stained for annexin V and PI (BD Biosciences, #556547) as previously described (51). After staining, cells were analyzed using the BD LSR II (BD Biosciences). Mean fluorescence intensity (MFI) was determined for each marker using FlowJo analysis software version 10.0.8 (FlowJo, Ashland, CO, USA).
Cell cycle analysis
Cells were washed with PBS and fixed in 70% ethanol for 20 min. Subsequently, cells were stained with PI (BD Biosciences, #556547) in the presence of ribonuclease (20 μg/ml; Thermo Fisher Scientific, #12091021). Cells were analyzed by flow cytometry.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (PCR) was performed as previously described (50). In brief, total RNA was extracted by using the RNeasy Mini Kit (Qiagen, Valencia, CA), and complementary DNA was synthesized by using the ProtoScript kit (New England BioLabs). Quantitative real-time PCR was conducted by using sequence-specific primers [CCRL2: 5′-CTTCGAGAAAAACGTCTC-3′ (forward) and 5′-CATCATATTCATCCTCTGGTG-3′ (reverse); DNMT1: 5′-CGTAAAGAAGAATTATCCGAGG-3′ (forward) and 5′-GTTTTCTAGACGTCCATTCAC-3′ (reverse); PIM1: 5′-CTCTTCAGAATGTCAGGATC-3′ (forward) and 5′-GGATGGTTCTGGATTTCTTC-3′ (reverse); BCL2: 5′-GATTGTGGCCTTCTTTGAG-3′ (forward) and 5′-GTTCCACAAAGGCATCC-3′ (reverse); MCL1: 5′-TAGTTAAACAAGAGGCTGG-3′ (forward) and 5′-ATAAACTGGTTTTGGTGGTG-3′ (reverse); MYC: 5′-TGAGGAGGAACAACAAGATG-3′ (forward) and 5′-ATCCAGACTCTGACCTTTTG-3′ (reverse); glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-CTTTTGCGTCGCCAG-3′ (forward) and 5′-TTGATGGCAACAATATCCA-3′ (reverse)] (Sigma-Aldrich), the Radian SYBR Green Lo-ROX qPCR Kit (Alkali Scientific, Fort Lauderdale, FL), and the CFX96 real-time PCR detection system (Bio-Rad). The RNA expression was normalized on the basis of GAPDH expression.
Western blotting
For Western blotting analysis, protein extraction was performed as previously described (52). Antibodies against P-JAK2 (Tyr1007/1008) (#3776), P-STAT3 (Tyr705) (#9131), STAT3 (#4904), P-AKT (Ser473) (#9271), AKT (#4691), P-ERK1/2 (Thr202/Tyr204) (#9101), ERK1/2 (#4695), DNMT1 (#5032), c-MYC (#5605), PIM1 (#3247), BCL2 (#4223), MCL-1 (#5453), and β-actin (#4970) were purchased from Cell Signaling Technology. Antibodies against JAK2 (sc-294), P-STAT5 (Tyr694) (sc11761), and STAT5 (sc-835) were purchased from Santa Cruz Biotechnology, and antibody against CCRL2 (LS-C382506) was purchased LSBio.
Coimmunoprecipitation
MDS-L cells were deprived of IL-3 for 48 hours. Cells were lysed before and after 30 min of IL-3 (20 ng/ml) treatment as described above (52). Cell lysates were incubated overnight with Sepharose bead conjugate JAK2 monoclonal antibody (Cell Signaling Technology, #4089) or Sepharose bead conjugate isotype control (Cell Signaling Technology, #3423). Beads were then washed extensively and boiled with 30 μl of loading buffer.
Colony formation assay
Clonogenic assays were performed as previously described (50). Briefly, cells following treatment were collected, counted, and resuspended at a density of 2000 cells/ml in methylcellulose-based media prepared as previously described (50). After 10 to 14 days of incubation at 37°C in 5% CO2, the recovery of colony-forming units was determined by colony counting under bright-field microscopy. A cell aggregate composed of >50 cells was defined as a colony.
MTT assay
MDS/AML cells were plated in 96-well plates (15,000 cells per well). After 72 hours of treatment with various doses of fedratinib, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (11465007001, Roche Diagnostics, Mannheim, Germany) was performed according to the manufacturer’s instructions. Absorbance was measured at 570 nm (reference, 750 nm). Cell viability was calculated by 100 × (absorbance of sample/average absorbance of untreated control) for the respective dose for each cell line.
MDS xenograft studies
MDS-L cells were transduced with shControl and shCCRL2 lentiviral shRNA and selected under puromycin (2 μg/ml). Resistant cells were transduced with a retroviral vector carrying an enhanced GFP firefly luciferase fusion gene (53). Subsequently, GFP+ cells were selected by cell sorting and injected intravenously to 10-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ male mice (the Jackson Laboratory, stock no. 013062) (106 cells per mouse) 48 hours after intraperitoneal injection of Clophosome-A clodronate liposomes (100 μl per mouse), as previously described (26). Bioluminescence signal was measured by using IVIS spectrum in vivo imaging system at days 1, 12, 22, 34, 55, 69, and 77. Peripheral blood was collected from the mice at days 24, 55, and 78 after the injection. At day 78, the mice were euthanized, and the percentage of human CD45+ cells was assessed in peripheral blood, spleen, and bone marrow by flow cytometry. IACUC guidelines were followed throughout the experiment.
Publically available database
The analysis of mRNA levels of AR target genes in normal stem and progenitor cells and various types of AML and MDS was based on RNA sequencing data from the BloodSpot database (GSE42519, GSE13159, GSE15434, GSE61804, GSE14468, and TCGA) (20). Data from the Beat AML database (21) were derived to compare the expression of CCRL2 between healthy CD34+ cells, mononuclear cells, MDS, and AML.
Statistical analysis
Statistical analyses were performed by using GraphPad Prism (GraphPad Software, La Jolla, CA). R programming language was used to visualize the mRNA expression levels of AR target genes in healthy stem and progenitor cells and MDS and AML cells. Linear regression was performed to compare the mRNA and protein levels of CCRL2, correlated the CCRL2 expression with clinical and molecular characteristics, and to assess the correlation of the MFI of CCRL2 with the MFI of P-STAT3/STAT5 in CD34+ cells from patients with MDS and AML. Multivariable linear regression analysis was used to adjust for blast percentage and highest mutation allele frequency. Otherwise, an unpaired two-tailed Student t test was performed to evaluate statistical significance for comparisons between two groups. One-way analysis of variance (ANOVA) was performed for the comparisons between three or more groups. Dunnett’s test was used to account for multiple comparisons.
Acknowledgments
Funding: This study was supported by the National Cancer Institute/NIH (P01 CA225618-01A1, P30 CA06973, R01 HL156144, and K08 HL136894), NIH, National Heart, Lung, and Blood Institute (T32 HL007525), and the American Society of Hematology Research Training Award for Fellows (ASH RTAF).
Author contributions: T.K. and R.J.J. conceived and designed the study and wrote the manuscript. T.K. and B.P. performed the flow cytometry analysis. T.K., E.H., and W.B.D. performed the lentiviral transduction experiments. T.K., E.H., and T.R. performed the Western blot analysis. T.K., P.T., I.C., C.B., and G.G. performed the xenograft studies. T.K., B.P., B.C.P., and C.E. processed the primary samples. T.K. and C.E. performed the clonogenic assays. T.K. and P.T. performed the quantitative RT-PCR analysis. T.K. and A.R.M. performed the analysis of publicly available databases. T.K. and R.V. performed the statistical analysis. W.B.D., C.B., A.R.M., L.P.G., and M.J.L. interpreted the data and edited the manuscript.
Competing interests: C.B. has patents/patent applications in the fields of T cell and/or gene therapy for cancer. C.B. has received research funding from Merck, Sharpe, and Dohme; Kiadis Pharma; and Bristol Myers Squibb. The other authors declare that they have no disclosures or competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.Stevens B. M., Khan N., D’Alessandro A., Nemkov T., Winters A., Jones C. L., Zhang W., Pollyea D. A., Jordan C. T., Characterization and targeting of malignant stem cells in patients with advanced myelodysplastic syndromes. Nat. Commun. 9, 3694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shastri A., Will B., Steidl U., Verma A., Stem and progenitor cell alterations in myelodysplastic syndromes. Blood 129, 1586–1594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanumanthu V. S., Pirruccello S. J., GCSF-R expression in myelodysplastic and myeloproliferative disorders and blast dysmaturation in CML. Am. J. Clin. Pathol. 140, 155–164 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Lee H. J., Daver N., Kantarjian H. M., Verstovsek S., Ravandi F., The role of JAK pathway dysregulation in the pathogenesis and treatment of acute myeloid leukemia. Clin. Cancer Res. 19, 327–335 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Wang F., Ni J., Wu L., Wang Y., He B., Yu D., Gender disparity in the survival of patients with primary myelodysplastic syndrome. J. Cancer 10, 1325–1332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karantanos T., Chaturvedi S., Braunstein E. M., Spivak J., Resar L., Karanika S., Williams D. M., Rogers O., Gocke C. D., Moliterno A. R., Sex determines the presentation and outcomes in MPN and is related to sex-specific differences in the mutational burden. Blood Adv. 4, 2567–2576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karantanos T., Gondek L. P., Varadhan R., Moliterno A. R., DeZern A. E., Jones R. J., Jain T., Gender-related differences in the outcomes and genomic landscape of patients with myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes. Br. J. Haematol. 193, 1142–1150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mierzejewska K., Borkowska S., Suszynska E., Suszynska M., Poniewierska-Baran A., Maj M., Pedziwiatr D., Adamiak M., Abdel-Latif A., Kakar S. S., Ratajczak J., Kucia M., Ratajczak M. Z., Hematopoietic stem/progenitor cells express several functional sex hormone receptors-novel evidence for a potential developmental link between hematopoiesis and primordial germ cells. Stem Cells Dev. 24, 927–937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez-Aguilera A., Arranz L., Martín-Pérez D., García-García A., Stavropoulou V., Kubovcakova L., Isern J., Martín-Salamanca S., Langa X., Skoda R. C., Schwaller J., Méndez-Ferrer S., Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell 15, 791–804 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Chuang K. H., Altuwaijri S., Li G., Lai J. J., Chu C. Y., Lai K. P., Lin H. Y., Hsu J. W., Keng P., Wu M. C., Chang C., Neutropenia with impaired host defense against microbial infection in mice lacking androgen receptor. J. Exp. Med. 206, 1181–1199 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelbaset-Ismail A., Borkowska-Rzeszotek S., Kubis E., Bujko K., Brzeźniakiewicz-Janus K., Bolkun L., Kloczko J., Moniuszko M., Basak G. W., Wiktor-Jedrzejczak W., Ratajczak M. Z., Activation of the complement cascade enhances motility of leukemic cells by downregulating expression of HO-1. Leukemia 31, 446–458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S. Y., Fan J., Hong D., Zhou Q., Zheng D., Wu D., Li Z., Chen R. H., Zhao Y., Pan J., Qi X., Chen C. S., Hu S. Y., C3aR1 gene overexpressed at initial stage of acute myeloid leukemia-M2 predicting short-term survival. Leuk. Lymphoma 56, 2200–2202 (2015). [DOI] [PubMed] [Google Scholar]
- 13.P. R. Kumar R, Niemann J, Zanetti C, Azimpour A, Meister M, Godavarthy PS, Krause DS, paper presented at the ASH, Orlando, 2019.
- 14.Orr S. J., Morgan N. M., Elliott J., Burrows J. F., Scott C. J., McVicar D. W., Johnston J. A., CD33 responses are blocked by SOCS3 through accelerated proteasomal-mediated turnover. Blood 109, 1061–1068 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Bonecchi R., Graham G. J., Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front. Immunol. 7, 224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schioppa T., Sozio F., Barbazza I., Scutera S., Bosisio D., Sozzani S., Del Prete A., Molecular basis for CCRL2 regulation of leukocyte migration. Front Cell Dev. Biol. 8, 615031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regan-Komito D., Valaris S., Kapellos T. S., Recio C., Taylor L., Greaves D. R., Iqbal A. J., Absence of the non-signalling chemerin receptor CCRL2 exacerbates acute inflammatory responses in vivo. Front. Immunol. 8, 1621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Prete A., Martínez-Muñoz L., Mazzon C., Toffali L., Sozio F., Za L., Bosisio D., Gazzurelli L., Salvi V., Tiberio L., Liberati C., Scanziani E., Vecchi A., Laudanna C., Mellado M., Mantovani A., Sozzani S., The atypical receptor CCRL2 is required for CXCR2-dependent neutrophil recruitment and tissue damage. Blood 130, 1223–1234 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Schinke C., Giricz O., Li W., Shastri A., Gordon S., Barreyro L., Bhagat T., Bhattacharyya S., Ramachandra N., Bartenstein M., Pellagatti A., Boultwood J., Wickrema A., Yu Y., Will B., Wei S., Steidl U., Verma A., IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood 125, 3144–3152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagger F. O., Kinalis S., Rapin N., BloodSpot: A database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 47, D881–D885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyner J. W., Tognon C. E., Bottomly D., Wilmot B., Kurtz S. E., Savage S. L., Long N., Schultz A. R., Traer E., Abel M., Agarwal A., Blucher A., Borate U., Bryant J., Burke R., Carlos A., Carpenter R., Carroll J., Chang B. H., Coblentz C., d’Almeida A., Cook R., Danilov A., Dao K. T., Degnin M., Devine D., Dibb J., Edwards D. K. t., Eide C. A., English I., Glover J., Henson R., Ho H., Jemal A., Johnson K., Johnson R., Junio B., Kaempf A., Leonard J., Lin C., Liu S. Q., Lo P., Loriaux M. M., Luty S., Macey T., MacManiman J., Martinez J., Mori M., Nelson D., Nichols C., Peters J., Ramsdill J., Rofelty A., Schuff R., Searles R., Segerdell E., Smith R. L., Spurgeon S. E., Sweeney T., Thapa A., Visser C., Wagner J., Watanabe-Smith K., Werth K., Wolf J., White L., Yates A., Zhang H., Cogle C. R., Collins R. H., Connolly D. C., Deininger M. W., Drusbosky L., Hourigan C. S., Jordan C. T., Kropf P., Lin T. L., Martinez M. E., Medeiros B. C., Pallapati R. R., Pollyea D. A., Swords R. T., Watts J. M., Weir S. J., Wiest D. L., Winters R. M., McWeeney S. K., Druker B. J., Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tohyama K., Tsutani H., Ueda T., Nakamura T., Yoshida Y., Establishment and characterization of a novel myeloid cell line from the bone marrow of a patient with the myelodysplastic syndrome. Br. J. Haematol. 87, 235–242 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka A., Tochigi A., Kishimoto M., Nakahara T., Kondo T., Tsujioka T., Tasaka T., Tohyama Y., Tohyama K., Lenalidomide induces cell death in an MDS-derived cell line with deletion of chromosome 5q by inhibition of cytokinesis. Leukemia 24, 748–755 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Bento L. C., Correia R. P., Pitangueiras Mangueira C. L., De Souza Barroso R., Rocha F. A., Bacal N. S., Marti L. C., The use of flow cytometry in myelodysplastic syndromes: A review. Front. Oncol. 7, 270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhyasen G. W., Wunderlich M., Tohyama K., Garcia-Manero G., Mulloy J. C., Starczynowski D. T., An MDS xenograft model utilizing a patient-derived cell line. Leukemia 28, 1142–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeisberger S. M., Odermatt B., Marty C., Zehnder-Fjällman A. H., Ballmer-Hofer K., Schwendener R. A., Clodronate-liposome-mediated depletion of tumour-associated macrophages: A new and highly effective antiangiogenic therapy approach. Br. J. Cancer 95, 272–281 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura T., Tange T., Terasawa T., Chiba S., Kuwaki T., Miyagawa K., Piao Y. F., Miyazono K., Urabe A., Takaku F., Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J. Cell. Physiol. 140, 323–334 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Handin R. I., DAMI cell line. Blood 89, 4238 (1997). [PubMed] [Google Scholar]
- 29.Dutta R., Zhang T. Y., Köhnke T., Thomas D., Linde M., Gars E., Stafford M., Kaur S., Nakauchi Y., Yin R., Azizi A., Narla A., Majeti R., Enasidenib drives human erythroid differentiation independently of isocitrate dehydrogenase 2. J. Clin. Invest. 130, 1843–1849 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa U., Grignani F., Hassan H. J., Rogaia D., Masciulli R., Gelmetti V., Guerriero R., Macioce G., Liberatore C., Barberi T., Mariani G., Pelicci P. G., Peschle C., Terminal megakaryocytic differentiation of TF-1 cells is induced by phorbol esters and thrombopoietin and is blocked by expression of PML/RARalpha fusion protein. Leukemia 12, 563–570 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Tvorogov D., Thomas D., Liau N. P. D., Dottore M., Barry E. F., Lathi M., Kan W. L., Hercus T. R., Stomski F., Hughes T. P., Tergaonkar V., Parker M. W., Ross D. M., Majeti R., Babon J. J., Lopez A. F., Accumulation of JAK activation loop phosphorylation is linked to type I JAK inhibitor withdrawal syndrome in myelofibrosis. Sci. Adv. 4, eaat3834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palandri F., Palumbo G. A., Elli E. M., Polverelli N., Benevolo G., Martino B., Abruzzese E., Tiribelli M., Tieghi A., Latagliata R., Cavazzini F., Bergamaschi M., Binotto G., Crugnola M., Isidori A., Caocci G., Heidel F., Pugliese N., Bosi C., Bartoletti D., Auteri G., Cattaneo D., Scaffidi L., Trawinska M. M., Stella R., Ciantia F., Pane F., Cuneo A., Krampera M., Semenzato G., Lemoli R. M., Iurlo A., Vianelli N., Cavo M., Breccia M., Bonifacio M., Ruxolitinib discontinuation syndrome: Incidence, risk factors, and management in 251 patients with myelofibrosis. Blood Cancer J. 11, 4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kesarwani M., Huber E., Kincaid Z., Evelyn C. R., Biesiada J., Rance M., Thapa M. B., Shah N. P., Meller J., Zheng Y., Azam M., Targeting substrate-site in Jak2 kinase prevents emergence of genetic resistance. Sci. Rep. 5, 14538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talpaz M., Kiladjian J. J., Fedratinib, a newly approved treatment for patients with myeloproliferative neoplasm-associated myelofibrosis. Leukemia 35, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karantanos T., Jain T., Moliterno A. R., Jones R. J., DeZern A. E., Sex-related differences in chronic myeloid neoplasms: From the clinical observation to the underlying biology. Int. J. Mol. Sci. 22, 2595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes N., Benedetti I., Rebollo J., Correa O., Geliebter J., Atypical chemokine receptor CCRL2 is overexpressed in prostate cancer cells. J. Biomed. Res. 33, 17–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akram I. G., Georges R., Hielscher T., Adwan H., Berger M. R., The chemokines CCR1 and CCRL2 have a role in colorectal cancer liver metastasis. Tumour Biol. 37, 2461–2471 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Del Prete A., Sozio F., Schioppa T., Ponzetta A., Vermi W., Calza S., Bugatti M., Salvi V., Bernardini G., Benvenuti F., Vecchi A., Bottazzi B., Mantovani A., Sozzani S., The atypical receptor CCRL2 is essential for lung cancer immune surveillance. Cancer Immunol. Res. 7, 1775–1788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Habbel J., Arnold L., Chen Y., Möllmann M., Bruderek K., Brandau S., Dührsen U., Hanoun M., Inflammation-driven activation of JAK/STAT signaling reversibly accelerates acute myeloid leukemia in vitro. Blood Adv. 4, 3000–3010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shastri A., Choudhary G., Teixeira M., Gordon-Mitchell S., Ramachandra N., Bernard L., Bhattacharyya S., Lopez R., Pradhan K., Giricz O., Ravipati G., Wong L. F., Cole S., Bhagat T. D., Feld J., Dhar Y., Bartenstein M., Thiruthuvanathan V. J., Wickrema A., Ye B. H., Frank D. A., Pellagatti A., Boultwood J., Zhou T., Kim Y., MacLeod A. R., Epling-Burnette P. K., Ye M., McCoon P., Woessner R., Steidl U., Will B., Verma A., Antisense STAT3 inhibitor decreases viability of myelodysplastic and leukemic stem cells. J. Clin. Invest. 128, 5479–5488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris R., Kershaw N. J., Babon J. J., The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27, 1984–2009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hubbard S. R., Mechanistic insights into regulation of JAK2 tyrosine kinase. Front. Endocrinol. 8, 361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas C., Moraga I., Levin D., Krutzik P. O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S. E., Glenn J. S., Nolan G. P., Piehler J., Schreiber G., Garcia K. C., Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greiser J. S., Stross C., Heinrich P. C., Behrmann I., Hermanns H. M., Orientational constraints of the gp130 intracellular juxtamembrane domain for signaling. J. Biol. Chem. 277, 26959–26965 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Gerber J. M., Zeidner J. F., Morse S., Blackford A. L., Perkins B., Yanagisawa B., Zhang H., Morsberger L., Karp J., Ning Y., Gocke C. D., Rosner G. L., Smith B. D., Jones R. J., Association of acute myeloid leukemia’s most immature phenotype with risk groups and outcomes. Haematologica 101, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranganathan P., Yu X., Na C., Santhanam R., Shacham S., Kauffman M., Walker A., Klisovic R., Blum W., Caligiuri M., Croce C. M., Marcucci G., Garzon R., Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 120, 1765–1773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moliterno A. R., Spivak J. L., Posttranslational processing of the thrombopoietin receptor is impaired in polycythemia vera. Blood 94, 2555–2561 (1999). [PubMed] [Google Scholar]
- 48.Liu Z., Tian M., Ding K., Liu H., Wang Y., Fu R., High expression of PIM2 induces HSC proliferation in myelodysplastic syndromes via the IDH1/HIF1-α signaling pathway. Oncol. Lett. 17, 5395–5402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghiaur G., Yegnasubramanian S., Perkins B., Gucwa J. L., Gerber J. M., Jones R. J., Regulation of human hematopoietic stem cell self-renewal by the microenvironment’s control of retinoic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 16121–16126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang Y. T., Hernandez D., Alonso S., Gao M., Su M., Ghiaur G., Levis M. J., Jones R. J., Role of CYP3A4 in bone marrow microenvironment-mediated protection of FLT3/ITD AML from tyrosine kinase inhibitors. Blood Adv. 3, 908–916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Engeland M., Ramaekers F. C., Schutte B., Reutelingsperger C. P., A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry 24, 131–139 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Karantanos T., Karanika S., Wang J., Yang G., Dobashi M., Park S., Ren C., Li L., Basourakos S. P., Hoang A., Efstathiou E., Wang X., Troncoso P., Titus M., Broom B., Kim J., Corn P. G., Logothetis C. J., Thompson T. C., Caveolin-1 regulates hormone resistance through lipid synthesis, creating novel therapeutic opportunities for castration-resistant prostate cancer. Oncotarget 7, 46321–46334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vera J., Savoldo B., Vigouroux S., Biagi E., Pule M., Rossig C., Wu J., Heslop H. E., Rooney C. M., Brenner M. K., Dotti G., T lymphocytes redirected against the kappa light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood 108, 3890–3897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3