Summary
Background
Prominent early features of COVID-19 include severe, often clinically silent, hypoxia and a pronounced reduction in B cells, the latter important in defence against SARS-CoV-2. This presentation resembles the phenotype of mice with VHL-deficient B cells, in which Hypoxia-Inducible Factors are constitutively active, suggesting hypoxia might drive B cell abnormalities in COVID-19.
Methods
Detailed B cell phenotyping was undertaken by flow-cytometry on longitudinal samples from patients with COVID-19 across a range of severities (NIHR Cambridge BioResource). The impact of hypoxia on the transcriptome was assessed by single-cell and whole blood RNA sequencing analysis. The direct effect of hypoxia on B cells was determined through immunisation studies in genetically modified and hypoxia-exposed mice.
Findings
We demonstrate the breadth of early and persistent defects in B cell subsets in moderate/severe COVID-19, including reduced marginal zone-like, memory and transitional B cells, changes also observed in B cell VHL-deficient mice. These findings were associated with hypoxia-related transcriptional changes in COVID-19 patient B cells, and similar B cell abnormalities were seen in mice kept in hypoxic conditions.
Interpretation
Hypoxia may contribute to the pronounced and persistent B cell pathology observed in acute COVID-19 pneumonia. Assessment of the impact of early oxygen therapy on these immune defects should be considered, as their correction could contribute to improved outcomes.
Funding
Evelyn Trust, Addenbrooke's Charitable Trust, UKRI/NIHR, Wellcome Trust
Keywords: COVID-19, Hypoxia, B cells, Lymphopenia
Research in context.
Evidence before this study
Hypoxemia is a prominent clinical feature of COVID-19. It often occurs silently, with many patients presenting to hospital in respiratory failure and with profoundly low blood oxygen saturations. This asymptomatic, “silent”, hypoxia is a particular trait of COVID-19 and is associated with a worse outcome. Another early feature of COVID-19 includes a pronounced reduction in B cells, which are important in defence against SARS-CoV-2. This was reminiscent of the phenotype of mice with VHL-deficient B cells, in which Hypoxia-Inducible Factors are constitutively active, suggesting hypoxia might drive B cell abnormalities in COVID-19.
Added value of this study
Through the NIHR Bioresource we undertook detailed B cell phenotyping linked with single-cell RNA-sequencing on a large cohort of samples from both acute COVID-19 and convalescence. We noted the similarity of B cell dysfunction in severe COVID-19 pneumonia in patients to unopposed activation of the Hypoxia Inducible Factor (HIF) pathway in animal models, suggesting that hypoxia may drive the persistent change in B cell function. We tested this model through studies in hypoxia-exposed mice, providing the first direct evidence of the profound impact oxygen availability has on the B cell immune response, and supporting the possibility that B cell dysfunction in COVID-19 could be driven by hypoxia.
Implications of all the available evidence
Our work could have important implications for our understanding of the biology of COVID-19 pneumonia. In particular, it suggests that appropriate and early use of oxygen therapy in COVID-19 may preserve the B cell immune response, something that could be tested in clinical trials. In addition, B cell dysfunction related to hypoxia may persist long after the initiation of infection and thus be implicated longer-term sequelae of COVID-19.
Alt-text: Unlabelled box
Introduction
The B cell response is a vital component of immune defence against SARS-CoV-2; neutralising antibodies contribute to protection from infection,1 monoclonal antibodies may be of benefit2 and antibody deficiency predisposes to viral persistence.3 Early and often persistent changes in B cell numbers are prominent in symptomatic COVID-19: increased plasmablasts and reduced memory B cells correlate with disease severity, and germinal centre (GC) responses, somatic hypermutation, and T follicular helper (TFH) cells may be reduced.4, 5, 6, 7
The reduction in B cell subsets in COVID-19 was reminiscent of the phenotype of mice with VHL-deficient B cells, which exhibit constitutive activation of Hypoxia-Inducible Factors (HIFs).8,9 These models had a Cre-mediated deletion of Vhl, targeted specifically to the B cell lineage, with Cre expression being driven by the B cell-specific promoter Mb-1, which deletes at the earliest Pro-B cell stage. A model of permanent Cre expression was used to assess developmental effects9 and a tamoxifen-inducible model used (Mb1-CreERT2)8 to assess the effect on B cell immune responses.
This raised the possibility that hypoxia might contribute to B cell dysregulation in COVID-19. Hypoxemia is prominent in COVID-19, often occurring “silently”, with many patients presenting to hospital with profoundly low blood oxygen saturations.10
The B cell abnormalities characteristic of COVID-19 might limit the efficiency of the anti-SARS-CoV-2 response,11 predispose to secondary infection,12 or contribute to sequelae such as autoimmunity.13 Understanding if and how hypoxia impacts upon them could therefore inform management strategies.
Methods
Participant recruitment and clinical data collection
This cohort has been previously described by Bergamaschi et al.6 Briefly, study participants were recruited between 31/3/2020 and 20/7/2020 from patients attending Addenbrooke's Hospital, Royal Papworth Hospital NHS Foundation Trust or Cambridge and Peterborough Foundation Trust with a confirmed diagnosis of COVID-19, together with Health Care Workers identified through staff screening as PCR positive for SARS-CoV-2.14 Controls were recruited among hospital staff attending Addenbrooke's for SARS-CoV-2 serology screening programme and having a negative serology result. All participants provided informed consent.
Inpatients were sampled at study entry, and then at regular intervals as long as they remained admitted to hospital (approximately weekly up to 4 weeks, and then every 2 weeks up to 12 weeks). Discharged patients were invited to provide a follow-up sample 4-8 weeks after study enrolment. Health care workers were sampled at study entry, and subsequently after approximately 2 and 4 weeks.
Clinical data were retrospectively collected by review of medical charts and extraction of data (laboratory test results, vital signs, medications) from Epic electronic health records (Addenbrooke's Hospital) and from MetaVision ICU (Royal Papworth Hospital).
Study volunteers were classified in 5 groups:
-
-
Group A: health care workers who were asymptomatic at the time of positive SARS-CoV-2 testing. This group included 10 volunteers who had possible COVID-19 symptoms before PCR testing (median time from symptoms to COVID-19 PCR test 26 days, range 9-42 days).
-
-
Group B: health care workers who had possible COVID-19 symptoms at the time of PCR testing.
-
-
Group C: patients in hospital who did not receive any supplemental oxygen for COVID-19. Five patients were discharged soon after initial diagnosis and assessment but followed up as part of the study.
-
-
Group D: patients in hospital who received supplemental oxygen using low flow nasal prongs, simple face mask, Venturi mask or non re-breather face mask
-
-
Group E: patients in hospital who received any of non-invasive ventilation (NIV), mechanical ventilation or ECMO. Patients who received supplemental oxygen (but no ventilation) and deceased in hospital were also assigned to group E.
Study results were analysed according to time since onset of COVID-19 symptoms, or otherwise time since positive SARS-CoV-2 testing (in group A and in 4 asymptomatic patients in group C).
Peripheral blood mononuclear cell preparation and flow cytometry immunophenotyping
For direct enumeration of T, B and NK cells, an aliquot of whole blood EDTA (50μl) was added to BD TruCount࣪ tubes with 20μl BD Multitest 6-colour TBNK reagent (BD Biosciences) and processed as per the manufacturer's instructions.
Peripheral venous blood (up to 27 ml per sample) for isolation of Peripheral Blood Mononuclear Cells (PBMCs) was collected into 10% sodium citrate tubes. PBMCs were isolated using Leucosep tubes (Greiner Bio-One) with Histopaque 1077 (Sigma) by centrifugation at 800x g for 15 minutes at room temperature. PBMCs at the interface were collected, rinsed twice with autoMACS running buffer (Miltenyi Biotech) and cryopreserved in FBS with 10% DMSO. All samples were processed within 4 hours of collection.
Approximately 106 cells have been stained with: anti-human IgM (clone: G20-127, BD), CD19 (clone: SJ25C1, BD), CD38 (clone: HIT2, BD), IgD (clone: IA6-2, BD), CD20 (clone: 2H7, BD), CD3 (clone: UCHT1, BioLegend), CD14 (clone: 63D3, BioLegend), CD15 (clone: W6D3, BioLegend), CD193 (clone: 5E8, BioLegend), CD27 (clone: O323, BioLegend), CD56 (clone: MEM188, Thermo), CD24 (clone: ML5, BD), IgA (polyclonal goat IgG, Jackson), IgG (clone: G18-145, BD), and Zombie Yellow (BioLegend) as described in detail by Bergamaschi et al,6 Samples were stored at 4°C and acquired within 4 hours using a 5-laser BD Symphony X-50 flow cytometer. Single colour compensation tubes (BD CompBeads) or cells were prepared for each of the fluorophores used and acquired at the start of each flow cytometer run.
Samples were gated in FlowJo v10.2 and number of cells falling within each gate was recorded. For analysis, these were expressed as an absolute concentration of cells per μl using the BD TruCount™ system. Some previously reported data, detailed by Bergamaschi et al is represented for comparison.
CRP, complement components and cytokines
As detailed in Bergamaschi et al,6 concentrations of complement components were measured in EDTA plasma using commercially available enzyme-linked immunosorbent assays (ELISA) kits. High sensitivity CRP and cytokines (IL-6, IL-10, IL-1β, TNFα and IFNγ) were assayed in serum using standard laboratory assays.
Total Immunoglobulin levels
Serum immunoglobulin levels were measured for 186 COVID-19 patients and 45 healthy controls at the time of enrolment using the standard assay by the Immunology Department at Peterborough City hospital.
Whole blood bulk RNA-Seq
Whole blood RNA was extracted from PAXgene Blood RNA tubes (BD Biosciences) of 188 COVID-19 patients at up to 2 time points and 42 healthy volunteers. RNA-Sequencing libraries were generated using the SMARTer® Stranded Total RNA-Seq v2 - Pico Input Mammalian kit (Takara) using 10ng RNA as input following the manufacturer's protocol. Libraries were pooled together (n = 96) and sequenced using 75bp paired-end chemistry across 4 lanes of a Hiseq4000 instrument (Illumina) to achieve 10 million reads per sample. Read quality was assessed using FastQC v.0.11.8 (Babraham Bioinformatics, UK), and SMARTer adaptors trimmed and residual rRNA reads depleted in silico using Trim galore v.0.6.4 (Babraham Bioinformatics, UK) and BBSplit (BBMap v.38.67(BBMap - Bushnell B. - sourceforge.net/projects/bbmap/)), respectively. Alignment was performed using HISAT2 v.2.1.0 (Kim et al., 2019) against the GRCh38 genome achieving a greater than 95% alignment rate. Count matrices were generated using featureCounts (Rsubreads package)15 and stored as a DGEList object (EdgeR package)16 for further analysis.
All downstream data handling was performed in R (R Core Team, 2015). Counts were filtered using filterByExpr (EdgeR) with a gene count threshold of 10 CPM and the minimum number of samples set at the size of the smallest disease group. Library counts were normalised using calcNormFactors (EdgeR) using the method ‘weighted trimmed mean of M-values’. The function ‘voom’17 was applied to the data to estimate the mean-variance relationship, allowing adjustment for heteroscedasticity.
The analyses were carried out splitting the samples in 12 days bins post screening (group A) or symptom onset (groups B-E).
Single cell RNA-seq
CITE-seq data were generated from frozen PBMCs of 36 COVID-19 patients and 11 healthy controls as described by Stephenson et al.7 Briefly, after thawing, pools of 4 samples were generated by combined 500,000 viable cells per individual (total of 2 million cells per pool). TotalSeq-C™ antibody cocktail (BioLegend 99813) was used to perform cell surface marker staining on 500,000 cells per pool. 50,000 live cells (up to a maximum of 60,000 total cells) for each pool were processed using Single Cell V(D)J 5’ version 1.1 (1000020) together with Single Cell 5’ Feature Barcode library kit (1000080), Single Cell V(D)J Enrichment Kit, Human B Cells (1000016) and Single Cell V(D)J Enrichment Kit, Human T Cells (1000005) (10xGenomics) according to the manufacturer's protocols. Samples were sequenced on NovaSeq 6000 (Illumina) using S1 flowcells. Droplet libraries were processed using Cellranger v4.0. Reads were aligned to the GRCh38 human genome concatenated to the SARS-Cov-2 genome (NCBI SARS-CoV-2 isolate Wuhan-Hu-1) using STAR18 and unique molecular identifiers (UMIs) deduplicated. CITE-seq UMIs were counted for GEX and ADT libraries simultaneously to generate feature X droplet UMI count matrices.
Statistics
All statistical analyses were conducted using custom scripts in R (R Core Team, 2015). Appropriately age matched healthy controls were included in all analyses. Absolute cell counts (cells/uL) were offset by +1 to allow subsequent log2 transformation of zero counts. Unless otherwise specified, longitudinally collected data was grouped by bins of 12 days from symptom onset or first positive SARS-CoV2 swab. Pairwise statistical comparisons of absolute cell counts and proportions and immunoglobulin levels between individuals in a given severity group at a given time bin and HCs, or between severity groups, was conducted by Wilcoxon test unless otherwise specified. For analyses involving repeated measures, false discovery rate corrected (Benjamini & Hochberg) p values were reported. For individuals sampled more than once within a given time bin, data from the earliest blood collection was used.
Gene set enrichment analysis (GSEA)19 was used to identify biological pathways enriched in COVID-19 severity groups relative to healthy controls. Briefly, a list of ranked genes, determined by Signal-To-Noise ratio was generated. An enrichment score was calculated, determined by how often genes from the geneset of interest appeared at the top or the bottom of the pre-ranked set of genes with the enrichment score representing the maximum deviation from zero. To assess statistical significance, an empirical phenotype- based permutation test was run, where a collection of enrichment scores was generated from the random assignment of phenotype to samples and used to generate a null distribution. To account for multiple testing, an FDR rate q < 0.20 was deemed significant. HALLMARK gene sets from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb) were used in analysis.
The relationships between immunological parameters and transcriptional data in the form of gene expression modules were assessed using Pearson's correlation (Hmisc package) and visualized with corrplot.
B cell receptor repertoire
Library preparation
B cell receptor repertoire libraries have been generated for 119 COVID-19 patients and 71 healthy controls using the protocol describe by Bashford-Rogers et al.20 Briefly, 200ng of total RNA from PAXgenes (14ul volume) was combined with 1uL 10mM dNTP and 10uM reverse primer mix (2uL) and incubated for 5 min at 70°C. The mixture was immediately placed on ice for 1 minute and then subsequently combined with 1uL DTT (0.1 M), 1uL SuperScriptIV (Thermo Fisher Scientific), 4ul SSIV Buffer (Thermo Fisher Scientific) and 1uL RNAse inhibitor. The solution was incubated at 50°C for 60 min followed by 15 min inactivation at 70°C. cDNA was cleaned with AMPure XP beads and PCR-amplified with a 5′ V-gene multiplex primer mix and 3′ universal reverse primer using the KAPA protocol and the following thermal cycling conditions: 1cycle (95°C, 5min); 5cycles (98°C, 20s; 72°C, 30s); 5cycles (98°C, 15s; 65°C, 30s; 72°C, 30s); 19cycles (98°C, 15s; 60°C, 30s; 72°C, 30s); 1 step (72°C, 5 min). Sequencing libraries were prepared using Illumina protocols and sequenced using 300-bp paired-end sequencing on a MiSeq.
Sequence analysis
Raw reads were filtered for base quality using a median Phred score of ≥32 (http://sourceforge.net/projects/quasr/). Forward and reverse reads were merged where a minimum 20bp identical overlapping region was present. Sequences were retained where over 80% base sequence similarity was present between all sequences with the same barcode. The constant-region allele with highest sequence similarity was identified by 10-mer matching to the reference constant-region genes from the IMGT database. Sequences without complete reading frames and non-immunoglobulin sequences were removed and only reads with significant similarity to reference IGHV and J genes from the IMGT database using BLAST were retained. Immunoglobulin gene use and sequence annotation were performed in IMGT V-QUEST, and repertoire differences were performed by custom scripts in Python.
Murine models
Vhl−/− mice21 were crossed with Cd79a-cre (Mb1-cre)22 or Cd19-cre (JAX, stock no. 004126) to delete Vhl in the B cell lineage. All mice with loxP-flanked alleles were hemizygous for Cre. Deletion efficiency was determined via real-time PCR of genomic DNA. The degree of excision was calculated by comparison of Vhl intact DNA relative to an unexcised gene Actb. The primers and probes used were Vhl forward 5′-GCTTGCGAATCCGAGGG, Vhl reverse 5′-TCCTCTGGACTGGCTGCC, Vhl Probe 5′-E6-FAM−CCCGTTCCAATAATGCCCCGG (Life Technologies) and Actb (mouse assay ID: Mm00607939_s1; Life Technologies). The deletion efficiency for mature B cells was 52% (95% CI 30-75%) in Vhl−/−Cd19-cre mice and 98% (95% CI, 97–99%) in Vhl−/−Mb1-cre mice.9 The mice were backcrossed for at least eight generations and maintained on a C57BL/6J background. These mice, along with C57BL/6J mice (JAX, stock no. 000664) were housed in specific pathogen-free animal facilities (at 20–23 °C, with 40–60% humidity, 12-h light:12-h dark cycle). All experiments included age- and litter-matched mice that were not selected for gender. Where possible, the resource equation was used to determine sample size for experiments. Randomization was genetic and, where possible, investigators were blinded to the genetic status. For hypoxic exposure studies, a randomization algorithm was used (Excel) to allocate mice into experimental groups. Mice were immunised with 100μg 4-hydroxy-3-nitrophenylaceyl-keyhole limpet hemocyanine (NP-KLH, loading 31-33) (Biosearch Technologies) adjuvanted with Alum (Thermo Scientific) via intraperitoneal injection. C57BL/6J mice were exposed to 10% O2 in a hypoxic chamber for 1 day, then immunised. Mice remained in the hypoxic chamber for 10-, 14- or 20-days post immunisation. Normoxic (21% O2) mice were treated the same way and were kept in standard conditions. The reoxygenation groups were removed from the hypoxic chamber on day 10 post immunisation to standard conditions for 4 or 10 days.
Tissue processing and immunophenotyping of murine cells by flow cytometry was performed as described.9 B cells were gated as total B cells (B220+), FO (B220+CD93–CD23+CD21+), MZ (B220+CD93−CD23−CD21+), GC (B220+CD95highGL-7high), PCs (B220−CD138+), early memory (B220+IgDneg/loCD95+GL7−CD38+CD73+), T cells (CD3+) and Tfh cells (CD3+CD4+PD-1highCXCR5highFoxP3−). Antibodies are listed in Supplementary table 1.
Murine total immunoglobulin (Ig) and NP-specific ELISAs
Detection of total IgM and NP-specific IgG1 was performed as described.23
Confocal microscopy
10µm sections were mounted on Superfrost Plus slides and air dried at RT for 1h. Samples were then fixed in -20°C acetone for 10 minutes and air dried again at RT for 1h before blocking in 0.1M Tris containing 1% BSA, 1% normal mouse serum and 1% normal rat serum. Samples were stained in a wet chamber at RT for 1h30 with the appropriate antibodies, washed 3 times in PBS and mounted in Fluoromount-G. Images were acquired using a TCS SP8 inverted confocal microscope on a 40x oil immersion objective. Raw imaging data were processed using Imaris.
Murine BCR amplification and sequencing
BCR amplification and sequencing was performed as described in Burrows et al.9 Data are available at the Sequence Research Archive (SRA) database (BioProject accession nos. PRJNA574931, PRJNA574906 and PRJNA574628). Briefly, total RNA was extracted from isolated plasma cells (B220−CD138+). Reverse transcription (RT) was performed using constant region-specific primers (including unique molecular identifiers (UMIs)), followed by cDNA cleanup and PCR amplification using V gene specific primers.
Sequencing libraries were prepared using Illumina protocols and sequenced using 300bp paired-ended MiSeq (Illumina). Raw reads were filtered as Burrows et al.9 Ig gene sequence annotations were performed in IMGT V-QUEST, where somatic hypermutation repertoire and isotype usage differences were performed by custom scripts in python, and statistics were performed in R using Wilcoxon tests for significance (non-parametric test of differences between distributions).
Role of funding source
Financial support from CVC Capital Partners, the Evelyn Trust (20/75), Addenbrooke's Charitable Trust (12/20A), the UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC) and NIHR Cambridge BioResource centre (Grant codes: RG85445 and RG94028) funded sample collection and processing. The Wellcome Trust (no. 19710) for supporting murine studies. Funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Ethics
Ethical approval was obtained from the East of England – Cambridge Central Research Ethics Committee (“NIHR BioResource” REC ref 17/EE/0025, and “Genetic variation AND Altered Leucocyte Function in health and disease - GANDALF” REC ref 08/H0308/176).
All procedures were ethically approved by the University of Cambridge Animal Welfare and Ethical Review Body and complied with the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012, under the authority of a UK Home Office Licence. The ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines (https://arriveguidelines.org/arrive-guidelines) were used for planning, conducting and reporting experiments.
Results
SARS-CoV-2 PCR-positive subjects were recruited between March and July 2020 and categorized by peak clinical severity6 (Figure 1a and Supp Fig 1a):
-
A)
asymptomatic healthcare workers (HCWs) recruited from routine screening (n=18).
-
B)
HCWs either still working with mild symptoms, or symptomatic and self-isolating (n=40).
-
C)
patients who presented to hospital but never required oxygen supplementation (n=46).
-
D)
admitted patients whose maximal respiratory support was supplemental oxygen (n=37).
-
E)
patients who required assisted ventilation (57 of 60) or died without ventilation (3 of 60).
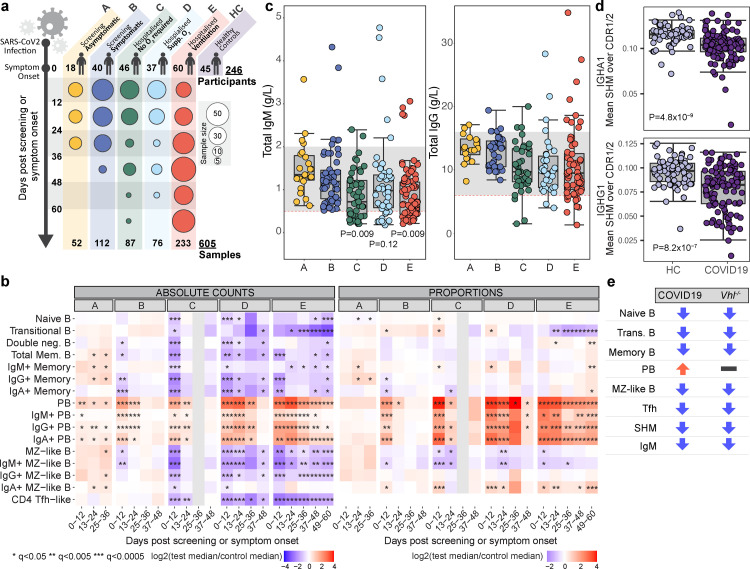
Figure 1.
B cells in COVID-19 and VHL-deficient mice. a, Cohort details. Time post positive swab (group A) or symptom onset (groups B-E). b, Median absolute cell counts (left) or proportions relative to total B cells (right) (log2 fold change relative to healthy controls). (Wilcoxon test FDR adjusted p-value (q)): *<0.05, **<0.005, ***<0.0005. c, Serum IgG and IgM (g/L) at enrolment. Grey band: 5-95th centiles of healthy reference range (see methods). Significant P values listed. d, Somatic hypermutation frequency in IgA and IgG within 0-12 days post symptom onset, calculated over the CDR1/CDR2 regions using BCR sequencing of whole blood. (Wilcoxon test). c,d, Circles represent individual donors. e, Phenotype comparison: COVID-19 patients versus mice with Vhl-deficient B cells.
We compared absolute B cell subset numbers in COVID-19 patients to 45 healthy controls (Figure 1b).6 In more severe groups C-E, profound reductions in TFH-like cells and many B cell subsets were seen at the first bleed, including memory and marginal zone like (MZL) B cells. Most then showed some recovery. Changes were far less pronounced in groups A and B (Figure 1b). Single-cell RNA-sequencing coupled with analysis of surface proteins on a subset of patients confirmed proportional differences (Figure S1b). We also explored cell kinetics in groups C–E, assigning patients to two categories based on whether their CRP concentrations remained elevated above 10 mg/L (“persisting CRP”) or fell below 10 mg/L (“resolving CRP”) by their final bleed within 3 months post symptom onset. The latter group included both individuals with early high CRP that then fell, together with those for which CRP remained low (10 mg/L) throughout (see Figure 6, Bergamaschi et al.6). B cell derangements including low transitional B cells and elevated plasmablasts persisted regardless of CRP, while MZ, memory and naïve B cell reductions recovered as the CRP did (Figure S1).
Total serum IgM fell as disease severity increased, with many patients in groups C-E having IgM levels below the normal range, while IgG and IgA were less impacted (Figures 1c and S1d). Anti- SARS-CoV-2 spike antibodies rose over time in all severity groups, reaching highest titres in the more severe groups.6 BCR sequencing showed reduced somatic hypermutation (SHM) in COVID-19 patients, most prominent in IgA and IgG1/2 (Figures 1d and S1e), consistent with previous reports.5
Having noted a similarity in B cell phenotype in COVID-19 and mice with constitutively active HIF(Figure 1e),8,9 we hypothesised that B cell loss might relate to hypoxia in vivo. The effects of acute hypoxia on immune responses in mice and humans have not been assessed. Immunised Vhl−/−Cd19-cre mice (in which VHL is deleted at the pre-B cell stage) were studied to allow more granular comparison with the B cell pathology in COVID-19: they showed reductions in follicular (FO), MZ and GC B cells and increased plasma cell (PC) to B cell ratio (Figure S2a). Findings were confirmed in Vhl−/−Mb1-cre mice (VHL deleted in pro-B cells) immunised with NP-KLH, in which reduced NP-specific GC and memory B cells, and TFH cells were also observed (Figure S2b-c). Serum IgM, but not IgG and IgA, was reduced, as was affinity maturation and SHM in some isotypes (Figure S2d-f). Reduced GC and memory B cells, along with defects in affinity maturation, were similar to those observed in an inducible model of B cell specific Vhl deletion.8 Thus, HIF activation in multiple mouse models produces similar changes to those in patients with moderate to severe COVID-19 (Figure 1e), supporting the possibility that hypoxia could be implicated in COVID-19 B cell pathology.
Hypoxia in COVID-19 patients, as determined by monitoring peripheral oxygen saturation (SpO2), was common early in disease and tended to improve with recovery, or with ventilation or ECMO in intensive care (group E) (Figure S1f), but was hard to correlate directly with immune changes, as recorded SpO2 is usually taken on oxygen replacement, which is commonly administered in the ambulance or immediately on arrival in hospital. Thus these data will underestimate the real hypoxia on admission, which is likely to have been sustained for hours or days before the patient presented. Furthermore, SpO2 is not reliably reflective of tissue hypoxia, which may persist in severe pneumonia and acute lung injury. We therefore instead measured the impact of hypoxia on the transcriptome in COVID-19 blood samples. An eigengene representative of the curated Hallmark hypoxia signature in whole blood was associated with disease severity (Figure 2a). Hallmark hypoxia gene set enrichment in single-cell RNA-sequencing data7 showed enrichment in almost all B cell subsets in groups C, D and E but in only plasma cells in A and B (Figure 2b).
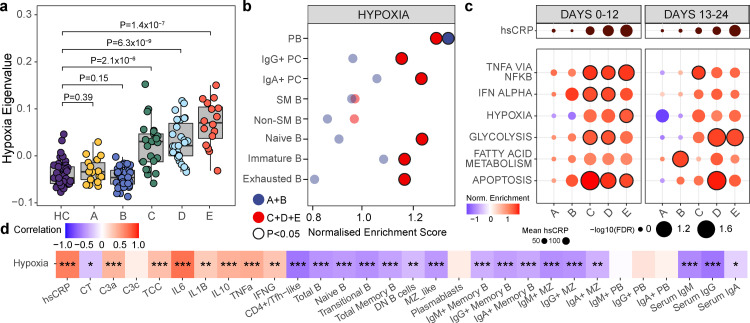
Figure 2.
Hypoxia-related transcription signatures in COVID-19. a, Eigenvalues of Hallmark Hypoxia geneset grouped by severity at 0-12 days post symptom onset, (unpaired, two-sided Student's t-test). Circles represent individual donors. b, B cell subpopulations identified using CITEseq with Gene set enrichment analysis (GSEA) of Hallmark hypoxia geneset assessed on a single cell level comparing HC to COVID-19, grouped by severity (A/B n=8, C/D/E n =20), within 24 days of symptom onset (B-E)/positive swab(A). c, GSEA of Hallmark genesets in COVID-19 versus HC grouped by severity and time. Outlined circles: nominal P value <0.05 and FDR adjusted P <0.2. Mean hsCRP represented. d, Correlation between Hallmark hypoxia geneset eigengenes and parameters shown at 0-12 days post symptom onset in COVID-19 patients. Boxes coloured by strength of correlation, Pearson correlation.
The “Hallmark hypoxia” signature was enriched for genes regulated by HIF, and could therefore be activated by reduced oxygen-tension and/or inflammatory stimuli. We therefore compared Hallmark signatures of hypoxia with inflammation in whole blood - the hypoxia signature was prominent in early severe disease (groups C-E) before declining, perhaps due to recovering disease and effective oxygen supplementation, but was not enriched in mild disease (A and B). In contrast inflammation-related signatures were often seen in these mild groups (Figure 2c). The differential enrichment of hypoxic and inflammatory signatures in asymptomatic and mild disease suggests a specific role for hypoxia, but an additional role for inflammation cannot be excluded. Finally the hypoxia eigengene correlated inversely with B cell number across most subsets, with the exception of plasma cells (see discussion) (Figure 2d).
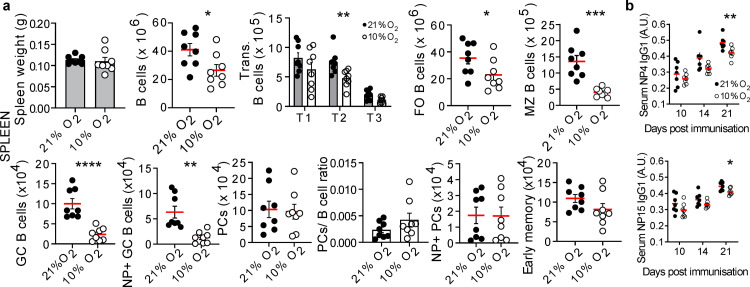
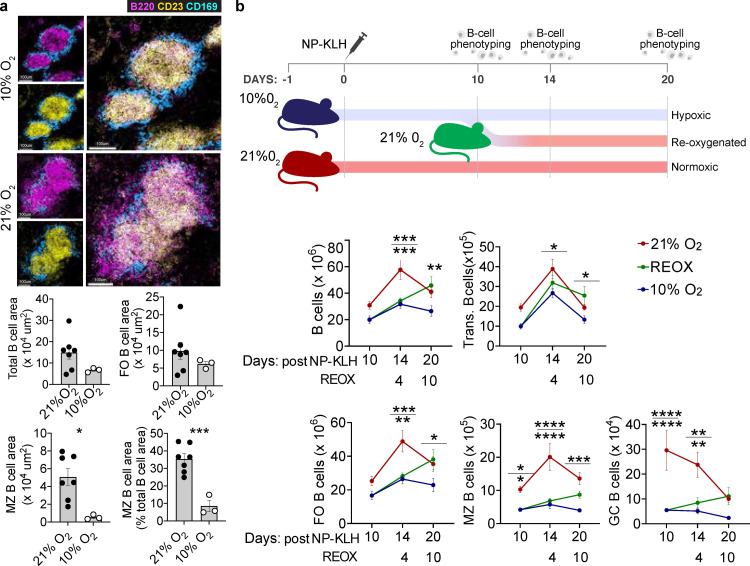
To differentiate inflammatory-driven from hypoxia-driven HIF-mediated effects, we studied mice in hypoxic conditions (10% O2) after immunisation with NP-KLH. After 11 days of hypoxia we observed reduced transitional, FO, MZ and GC B cells, whilst PCs were normal (Figure S3a). These defects persisted when hypoxia was prolonged (20 days; Figure 3a and S3b). Little effect was observed on B cell development in the bone marrow (Figure S3c) and as FO and MZ B cells turn over every 7-8 weeks,24,25 it is unlikely these early reductions are due to a developmental defect. In hypoxic mice there was a tendency to reduced early memory B cells (Figure 3a and S3d) and serum Ig was normal but antigen-specific IgG1 was reduced (Figure 3b and S3e). Histological analysis revealed that, in hypoxic conditions, B cells were almost absent from the MZ, which appeared otherwise structurally intact (Figure 4a and S3f). Some mice were removed from the hypoxic chamber after 11 days: B cell subsets generally recovered following this reoxygenation. MZ and GC B cells were most prominently affected by hypoxia, continuing to decline under hypoxic conditions, and recovering more slowly following reoxygenation, than other subsets (Figure 4b). Hypoxia induced only minor reductions in T cells and NK cells, and no changes in macrophage numbers, indicating that B cells seem particularly sensitivity to perturbations in oxygenation and HIF (Figure S4 and data not shown). Thus while it is likely that hypoxia will have other effects that will warrant more detailed examination, B cells seem particularly sensitivity to perturbations in oxygenation and HIF activity.
Figure 3.
The response of mouse B cells to hypoxia in vivo. a, WT mice exposed to 21% or 10% O2, were immunized with NP-KLH at day 1, then absolute spleen B cells enumerated at day 21. (Unpaired, two-sided Student's t-test). FO, (Unpaired, two-sided Mann-Whitney U-test). Gating in methods, mean ± s.e.m, circles represent individual mice (n=8 per group), data pooled from two experiments, results confirmed in a third. b, Serum NP-specific antibodies after NP-KLH immunization, (unpaired, two-sided Student's t-test). mean ± s.e.m, circles represent individual mice, (n=8 per group), data pooled from two experiments, confirmed in a third. *P < 0.05,**P < 0.01,***P < 0.001,**** P < 0.0001.
Figure 4.
The response of mouse B cells to re-oxygenation in vivo. a, Spleen confocal images from immunised mice (Figure 3a), MZ B cells (magenta, B220+CD23−), FO B cells (yellow, B220+CD23+) and MZ metallophilic macrophages (blue, CD169+). B cell area, circles represent individual follicles from one spleen per condition, mean ± s.e.m. b, Experiment outline and absolute spleen B cells, (two-way ANOVA with Tukey's multiple comparisons test). Gating in methods. Mean ± s.e.m, data pooled from two experiments. *P < 0.05,**P < 0.01,***P < 0.001,**** P < 0.0001.
Discussion
We demonstrate profound B cell abnormalities in severe COVID-19 and provide evidence they may be the result of hypoxia. B cell lymphopenia extends across all subsets, is present soon after symptom onset, and is often persistent. There is an associated reduction in total serum IgM and in SHM in switched B cells. Despite this, patients in all groups develop neutralising anti-SARS-CoV-2 antibodies,6 known not to require affinity maturation.26 It nonetheless seems likely that these profound B cell deficits could have an impact on disease. As we have suggested for bystander CD8 T cell responses,6 early B cell defects may increase COVID-19 severity through non-antigen-specific mechanisms, perhaps reducing early antigen localisation, transport or presentation, or cytokine or “natural” antibody production.27 Later, B cell defects may predispose to problematic secondary bacterial infection.12 MZ B cells are key players in early defence against blood-borne bacterial infection,28 and MZL cells are profoundly reduced in COVID-19, and hypoxia almost ablates MZ B cells in mice. This may also predispose to re-infection by new variants, as affinity maturation might generate a broader spectrum of neutralizing antibodies and B cell memory.26 B cell dysregulation might also play a part in driving COVID-19-associated autoimmunity.13
Three HIF-α isoforms are known, with HIF-1α and HIF-2α providing the main transcriptional response to oxygen gradients. HIF-1α is ubiquitously expressed but HIF-2α is restricted to specific cell types, including B cells.9 Both HIF-1α and HIF-2α are regulated by prolyl hydroxylation and VHL-mediated protein degradation, although the kinetics of their stabilisation in oxygen gradients differ, with a more prolonged HIF-2 response compared to HIF-1. HIF-1 and HIF-2 also share a number of target genes and are sometimes expressed in the same cells.29 How these distinct HIF isoforms regulate gene expression is an important area of ongoing study, but the hypoxic signature we observe is entirely consistent with activation of HIF target genes. These findings are also supported by studies in mice with B cell-specific VHL deletion, and thus constitutively active HIF, which show abnormal B cell development and reduced GC B cells, antibody class-switching and affinity-maturation. Deleting HIF-1 or both HIF-1 and HIF-2 in these models rescued the defects, confirming a HIF-1/-2-dependent effect8,9 and suggesting that hypoxia-induced HIF stabilization might be physiologically important in B cell biology.
Hypoxia might drive transcriptional and cellular changes independently of HIFs via epigenetic modifications involving DNA/histone demethylation. These occur through the oxygen-dependence of several lysine demethylases (KDMs)30,31 or through impaired DNA methylcytosine hydroxylation via the oxygen-sensitive TET enzymes.32 The oxygen affinity of KDMs and TETs are higher than the HIF prolyl hydroxylases. Therefore HIF transcriptional responses will occur first, and epigenetic changes may only be observed in severe tissue hypoxia. The role of KDMs and TETs in hypoxic immune regulation have yet to be studied, but they could contribute to the longer-term defects observed on B cells once hypoxia has resolved.
While inflammation can contribute to activate HIF-1α, our demonstration that hypoxia alone can induce profound, reversible B cell abnormalities supports a major role for hypoxia in driving the B cell abnormalities in COVID-19. Hypoxia exerts its major effect on HIF-1α by preventing its degradation, while the main inflammatory impact is through enhancing HIF transcription, making a synergistic impact of hypoxia and inflammation on HIF function plausible.33,34 While it is impossible to conclusively separate the two impacts in humans with COVID-19, the fact that a tighter correlation is seen between disease severity and hypoxia than with inflammatory signatures, together with the demonstration that hypoxia alone can induce profound, reversible B cell abnormalities in mice, and supports a role for hypoxia in driving B cell abnormalities in COVID-19. Only plasmablasts showed HIF activation in the absence of hypoxia and these were the only B cell subset in which cell numbers do not correlate with the hypoxia signature, nor fall in mice subject to hypoxia, raising the possibility that HIF is constitutively active in these cells. This is consistent with a growing literature demonstrating that HIF-1α is active in multiple myeloma.35 Given this aspect of COVID-19 B cell pathology does not appear impacted by hypoxia, therapeutic approaches to plasma cell control may need to involve pharmacological antagonism of HIF rather than increased oxygenation.
Supplemental oxygen in established COVID-19 may not correct localised areas of hypoxia following acute lung injury/ARDS and could account for a persistent hypoxic transcriptional signal. Inflammation will exacerbate these transcriptional changes, and it is not possible to distinguish the relative contribution of hypoxia versus inflammation in severe disease. However, our corroboratory observations in mice clearly demonstrate that hypoxia is sufficient to drive these B cell changes, irrespective of inflammation, and the observation that hypoxia perturbs B cell immunity has implications in a wide range of clinical settings. In COVID-19, appropriate early oxygen therapy may lead to improved immune responsiveness, impacting on both the short-term and long-term outcomes of the disease, and this could be tested in clinical studies.
Contributors
Conceptualization: C.H., J.R.B., P.H.M., M.R.C, J.A.N., P.A.L., N.B. and K.G.C.S.
Data curation: P.K., F.M., L.T., L.B., A.P., Z.K.T., N.R., S.D.M., B.M.O., M.D.M., and N.B.
Data Analysis: P.K., F.M., A.H., L.T., L.B., M.D.M., P.A.L. and N.B.
Project Administration: F.M., L.T., L.B. and B.J.D.
Funding Acquisition: P.H.M, C.H., J.R.B., P.A.L., and K.G.C.S.
Writing – Original Draft: P.A.L., P.K., N.B. and K.G.C.S.
Writing – Review & Editing: P.K., F.M., A.H., L.T., L.B., B.J.D., B.G., M.T., C.H., J.R.B, P.H.M., M.R.C., J.A.N., P.A.L., N.B. and K.G.C.S.
All authors have read and approved the final version of the manuscript.
Declaration of interests
P.H.M. declares consultancy fees from Mission Therapeutics and AstraZeneca, has received speaker honoraria from AstraZeneca, Dana Farber Cancer Institute and Astellas, participates on an advisory board for Mission Therapeutics and has a fiduciary role on committees for the Academy of Medical Sciences, Medical Schools Council and Cambridge Enterprise. J.N. declares an ITEN grant (Pfizer) to explore role of deubiquitinating enzymes in hypoxia. K.G.C.S is a co-founder and/or consultant with PredictImmune, Rheos Medicines, GSK and Kymab. M.T is on the Scientific Advisory Board of MorphogenIX. C.H. is co-founder and CSO of Hornet Therapeutics, has recently received speaker honoraria from GSK and the NIH (USA), and is board member of the Novartis Foundation for Medical-Biological Research. J.R.B has received a speaker honorarium from AstraZeneca. M.C declares an academia-industry collaborative grant from Sanofi iAward Europe to study kidney immunity, has received a speaker honorarium from Novartis and is a board member of the Medical Research Council for Population and Systems Medicine. P.A.L has received a grant from EU H2020, has received royalties and/or consulting fees from PredictImmune and Ducentis, has received a speaker honorarium from GSK and is on the committee for UKIVAS. Declaration of interests listed by these authors are outside and not related to the submitted work. All other authors have no declarations.
Acknowledgments
Acknowledgements
We thank the patients and Health Care Workers who took part in this study. We are grateful for financial support from CVC Capital Partners, the Evelyn Trust (20/75), Addenbrooke's Charitable Trust (12/20A) and the UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC). We acknowledge NIHR Cambridge BioResource Centre (Grant codes: RG85445 and RG94028), NIHR Cambridge Clinic Research Facility outreach team, relevant NHS Trusts and their staff, and the Wellcome Trust (no. 19710) for supporting murine studies. K.G.C.S. is the recipient of a Wellcome Investigator Award (200871/Z/16/Z); C.H. was funded by a Wellcome COVID-19 Rapid Response DCF and the Fondation Botnar. J.A.N is the recipient of a Wellcome Senior Clinical Research Fellowship (215477/Z/19/Z) and a Lister Institute Research Fellowship. NB and AP are supported by the Wellcome Trust, Senior Investigator Award to P.H.M, and the Rosetrees Trust. NR is supported by the NIHR Cambridge Biomedical Research Centre. Z.K.T. and M.R.C. are supported by a Medical Research Council Human Cell Atlas Research Grant (MR/S035842/1). M.R.C is supported by an NIHR Research Professorship (RP-2017-08-ST2-002). PK is the recipient of a Jacquot Research Entry Scholarship of the Royal Australasian College of Physicians Foundation.
We thank the NIHR, NHS Blood and Transplant, and Health Data Research UK as part of the Digital Innovation Hub Programme, and the NIHR Cambridge Biomedical Research Centre Cell Phenotyping Hub, the CRUK Cambridge Institute cytometry core and the Cambridge NIHR BRC Stratified Medicine Core Laboratory NGS Hub.
Data sharing statement
The dataset from our study can be explored interactively through a web portal: https://covid19cellatlas.org.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103878.
Contributor Information
Natalie Burrows, Email: nb470@cam.ac.uk.
Kenneth G.C. Smith, Email: kgcs2@cam.ac.uk.
Appendix. Supplementary materials
References
- 1.Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz LM. (A little) clarity on convalescent plasma for Covid-19. N Engl J Med. 2021;384:666–668. doi: 10.1056/NEJMe2035678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckland MS, Galloway JB, Fhogartaigh CN, et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneko N, Kuo HH, Boucau J, et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157. doi: 10.1016/j.cell.2020.08.025. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen SCA, Yang F, Jackson KJL, et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525. doi: 10.1016/j.chom.2020.09.002. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergamaschi L, Mescia F, Turner L, Bradley JR, Lyons PA, Smith KGC. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–1275. doi: 10.1016/j.immuni.2021.05.010. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephenson E, Reynolds G, Botting RA, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nature Med 2021 27:5. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SH, Raybuck AL, Stengel K, et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537:234–238. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrows N, Bashford-Rogers RJM, Bhute VJ, et al. Dynamic regulation of hypoxia-inducible factor-1α activity is essential for normal B cell development. Nat Immunol. 2020;21:1408–1420. doi: 10.1038/s41590-020-0772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couzin-Frankel J. The mystery of the pandemic's ‘happy hypoxia. Science. 2020;368:455–456. doi: 10.1126/science.368.6490.455. [DOI] [PubMed] [Google Scholar]
- 11.Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021:1–10. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripa M, Galli L, Poli A, et al. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clin Microbiol Infect. 2021;27:451–457. doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang EY, Mao T, Klein J, et al. Diverse functional autoantibodies in patients with COVID-19. medRxiv. 2020 doi: 10.1038/s41586-021-03631-y. : 2020.12.10.20247205. [DOI] [PubMed] [Google Scholar]
- 14.Rivett L, Sridhar S, Sparkes D, et al. Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. eLife. 2020;9 doi: 10.7554/eLife.58728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law CW, Chen Y, Shi W, Smyth GK. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashford-Rogers RJM, Bergamaschi L, McKinney EF, et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. 2019;574:122–126. doi: 10.1038/s41586-019-1595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Nat Acad Sci USA. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobeika E, Thiemann S, Storch B, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Nat Acad Sci USA. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownlie RJ, Lawlor KE, Niederer HA, et al. Distinct cell-specific control of autoimmunity and infection by FcγRIIb. J Exp Med. 2008;205:883–895. doi: 10.1084/jem.20072565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprent J, Basten A. Circulating T and B lymphocytes of the mouse: II. Lifespan. Cell Immunol. 1973;7:40–59. doi: 10.1016/0008-8749(73)90181-0. [DOI] [PubMed] [Google Scholar]
- 25.B S, WJ Q, K H, J E, D A. Characterization of marginal zone B cell precursors. J Exp Med. 2005;202:1225–1234. doi: 10.1084/jem.20051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark SA, Clark LE, Pan J, et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021 doi: 10.1016/j.cell.2021.03.027. Published online March 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez AM, Holodick NE. Editorial: Natural Antibodies in Health and Disease. Front Immunol. 2017;8:1795. doi: 10.3389/fimmu.2017.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemazee D. Natural history of MZ B cells. J Exp Med. 2021;218 doi: 10.1084/JEM.20202700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia? J Clin Invest. 2007;117:862–865. doi: 10.1172/JCI31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty AA, Laukka T, Myllykoski M, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363:1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbarash RA, Toll L, Sahn SA. Alpha-difluoromethylornithine infusion and cardiac arrest. Ann Intern Med. 1986;105:141–142. doi: 10.7326/0003-4819-105-1-141_2. [DOI] [PubMed] [Google Scholar]
- 32.Thienpont B, Steinbacher J, Zhao H, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrows N, Maxwell PH. Hypoxia and B cells. Exp Cell Res. 2017;356:197–203. doi: 10.1016/j.yexcr.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Watts ER, Walmsley SR. Inflammation and hypoxia: HIF and PHD isoform selectivity. Trends Mol Med. 2019;25:33–46. doi: 10.1016/j.molmed.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Martin SK, Diamond P, Gronthos S, Peet DJ, Zannettino A. The emerging role of hypoxia, HIF-1 and HIF-2 in multiple myeloma. Leukemia. 2011;25:1533–1542. doi: 10.1038/leu.2011.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.