Abstract
Mutations in the RNA binding protein, Fused in Sarcoma (FUS), lead to amyotrophic lateral sclerosis (ALS), the most frequent form of motor neuron disease. Cytoplasmic aggregation and defective DNA repair machinery are etiologically linked to mutant FUS-associated ALS. Although FUS is involved in numerous aspects of RNA processing, little is understood about the pathophysiological mechanisms of mutant FUS. Here, we employed RNA-sequencing technology in Drosophila brains expressing FUS to identify significantly altered genes and pathways involved in FUS-mediated neurodegeneration. We observed the expression levels of DEAD-Box Helicase 17 (DDX17) to be significantly downregulated in response to mutant FUS in Drosophila and human cell lines. Mutant FUS recruits nuclear DDX17 into cytoplasmic stress granules and physically interacts with DDX17 through the RGG1 domain of FUS. Ectopic expression of DDX17 reduces cytoplasmic mislocalization and sequestration of mutant FUS into cytoplasmic stress granules. We identified DDX17 as a novel regulator of the DNA damage response pathway whose upregulation repairs defective DNA damage repair machinery caused by mutant neuronal FUS ALS. In addition, we show DDX17 is a novel modifier of FUS-mediated neurodegeneration in vivo. Our findings indicate DDX17 is downregulated in response to mutant FUS, and restoration of DDX17 levels suppresses FUS-mediated neuropathogenesis and toxicity in vivo.
Keywords: Neurodegeneration, ALS/FTD, Motor neuron disease, iPSC neurons, Drosophila, DNA-damage repair, FUS, DDX17, RGG-domain
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective loss of upper and lower motor neurons, resulting in progressive muscle atrophy [5, 14, 21, 39, 44, 59, 61, 78, 85]. Approximately 5–10% of ALS occurrences are inherited and referred to as familial ALS (fALS) [16], while the other 90–95% of cases are sporadic (sALS). Dominant mutations in the gene Fused in sarcoma (FUS) are the third most common monogenetic cause of ALS accounting for ~ 5% of fALS cases and ~ 1% of sALS cases [52, 55, 82, 96]. FUS is a member of the FET family of RNA binding proteins and harbors seven different domains critical for its protein function: An N-terminal transcriptional activation domain (QGSY-rich domain), three arginine-glycine-glycine repetitive regions (RGG1–RGG3), an RNA recognition motif, zinc finger (ZnF) motif, nuclear export signal (NES), and a C-terminus nuclear localization signal (NLS) [19, 27, 51, 53]. The most common disease-causing FUS mutations are located within the nuclear localization signal and include positions 521 (R converted to C, G, or H) and 525 (P mutated to L), which is also the most aggressive form of FUS-mediated ALS with juvenile onset [23]. The disease-causing NLS mutations of FUS result in nuclear FUS depletion and toxic cytoplasmic aggregation, which are key pathological hallmarks of ALS [6, 40].
FUS is a multifunctional RNA-binding protein involved in various aspects of RNA processing and metabolism (RNA transcription, alternative splicing, and RNA trafficking) [22, 24, 25, 31, 54, 74, 92, 105, 106]. Recent evidence has suggested FUS to be significantly involved in the DNA Damage Response (DDR) pathway in a PAR polymerase 1 (PARP1) dependent manner [65, 75, 86, 100]. Upon initial signs of DNA damage, PARP1 is recruited within seconds to DNA damage sites resulting in the recruitment of FUS [80]. In primary cortical neurons, FUS interacts with the DNA damage repair protein, histone deacetylase 1 (HDAC1), which is heavily involved in the DNA damage repair processes: homologous recombination (HR) and non-homologous end joining (NHEJ) [69, 99]. Interestingly, cells expressing mutant FUS resulted in a decreased interaction of FUS and HDAC1, suggesting their interaction to be crucial for effective DNA damage repair. This evidence is further supported by the observations of increased DNA damage in iPSC motor neurons and in the postmortem motor cortex of patients carrying FUS-R521C and P525L mutations [75, 99]. In addition, FUS-R521C transgenic mice exhibited evidence of DNA damage in cortical neurons and spinal motor neurons [81]. These findings suggest that DNA damage repair defects are central to the neuropathologies associated with mutant FUS expression [98]. Despite these advancements, molecular determinants or genetic modifiers to restore defective DNA damage repair machinery associated with FUS-mediated ALS have not been explored.
Here, we identified the RNA helicase, Dead-Box-Helicase 17 (DDX17), as a novel regulator of DNA damage repair and as a genetic modifier of FUS-mediated toxicity in mutant FUS iPSC neurons and Drosophila. Specifically, we found that disease-causing mutations in FUS significantly downregulate the levels of DDX17 in multiple models of mutant FUS, including iPSC neurons. Mutant FUS recruits nuclear DDX17 into cytoplasmic stress granules via interactions mediated by the FUS RGG1 domain. Ectopic expression of DDX17 significantly reduces insoluble mutant FUS aggregation and incorporation into stress granules in HEK cells. Furthermore, we observed that restoration of DDX17 levels strongly mitigates the DNA damage repair machinery defects exhibited in iPSC neurons expressing mutant FUS-P525L. Importantly, we observed DDX17 to be involved in the general DDR pathway, as shRNA-mediated knockdown of DDX17 is sufficient to cause DNA double strand breaks, whereas over expression of DDX17 is protective against DNA damage inducing agents in HEK cells. Furthermore, we observed the DDX17 Drosophila orthologue, Rm62, as a novel genetic modifier of FUS-associated ALS in Drosophila. These observations provide insight into the mechanisms of FUS toxicity and uncover the function of DDX17 in the DDR pathway, which may be useful in the development of potential therapeutic targets for ALS.
Methods
Drosophila lines
The FUS-WT, FUS-R518K, FUS-R521C lines were developed by site-specific insertion of the transgene (site-specific integration lines) at BestGene Inc. using the (attP2) insertion vector and were previously described [18, 58]. The information of the DDX fly lines can be found in Supplemental table 3 (Online resource 4).
Eye degeneration experiments in Drosophila
Utilizing the GAL4-UAS system, flies expressing the glass multiple reporter promoter element were crossed with flies expressing wild-type and mutant, exogenous, human FUS at 25 °C. Images of the left eyes of F1 generation, adult female Drosophila were taken at day 1 (or as indicated for aging experiments) using a Leica M205C dissection microscope equipped with a Leica DFC450 camera. External eye degeneration was quantified using a previously published scoring system (58, 18). Statistical analyses were performed using Prism 6 (GraphPad Software). Statistical comparisons between groups were performed using One-way ANOVA w/ Turkey’s multiple comparisons.
Drosophila motor function
ELAV-GS (neuron specific driver) was used to express the selected ALS-related genotypes, FUS-R521C, FUS- R518K, and FUS-WT with and without Rm62 over expression. Adult flies were transferred to RU486 drug food and placed in 25 °C for 20 days. 30–35 female Drosophila were transferred to empty vials and were knocked to the bottom of the vial by tapping each vial against the laboratory bench three times. A photo/video was taken 3 s after the third tap; three biological experimental replicates were performed for each group. The photos were analyzed using Kinovea video player and the ImageJ Cell Counter function was used to quantify how far the Drosophila climbed. The marked Drosophila were organized by genotype, vial number, and video number.
Sample preparation for RNA-seq of Drosophila brains
Adult Drosophila brains were dissected and placed in TRIzol (Ambion; 15596026) and RNA was isolated using a phenol–chloroform extraction method. 30–35 Adult Drosophila brains were dissected in triplicate for each genotype. In total, ~ 500 Adult Drosophila brains were dissected for sufficient RNA quantities for RNA-sequencing. Following lysis, chloroform was added, and the samples were centrifuged. The upper, aqueous layer was isolated, treated with isopropanol, and centrifuged. The resulting RNA pellets were washed with 75% ethanol, centrifuged, and dried by ambient air. RNA was suspended in RNase-free water. Quantity and purity (260/280 and 260/230 ratios) were determined using a NanoDrop ND-1000 spectrophotometer. RNA quality was assessed by 1% agarose gel electrophoresis using ethidium bromide.
RNA-seq and data analysis
RNA samples were subjected to analysis on an Agilent Bioanalyzer and spectrophotometric analysis to determine RNA quality. After passing this initial screening 500 ng of total RNA was used to prepare libraries for sequencing using the Lexogen SENSE mRNA-seq library kit for Ion Torrent. Libraries and amplified for 12 cycles as the final step of library preparation. Before sequencing, small aliquots of this material were quantified by qPCR utilizing the KAPA Library Quantification kit for the Ion Torrent. Quantification data from qPCR was used to balance the barcodes for final pooling before sequencing. Following this final pooling the library pools were sized to a target size of 300 bp on a Pippin Prep instrument. The sized libraries were examined on an Agilent High Sensitivity DNA chip, quantified using real-time PCR, and used for sequencing on the Ion Torrent Proton platform.
All fastq file were gathered from the sequencer. Quality assurance was performed using FASTQC. All reads were trimmed to remove any nucleotide that fails a phred score < Q20. The trimmed FASTQ files were aligned to the mm10 reference library using RNA STAR. Once aligned, the SAM files were collected and mined for read count information of each gene present in the reference file. Read counts were normalized using Counts per Million (CPM) method across the entire experiment. Principle component analysis and Pearson’s coefficient plots were performed on the normalized transcriptome profile. A Wilcoxon’s t test was used to determine significant between conditions. All genes that fail to yield a p-value greater than 0.05 were removed. Benjamini and Hochberg false discovery rate was performed on the trimmed gene list. All genes that fail to yield a false discovery rate of less than 0.05 were removed. The final significant differential gene lists were loaded into R to generate heatmaps. The targets were loaded into DAVID for pathway analysis, biological process identification, and disease association.
HEK Cell Data Set Comparison:
Quality controlled FASTQ files were aligned to the Ensembl Drosophila genome using the STAR aligner (version 2.5.1). Read summarization was done by feature Counts to produce a matrix of counts. Differential gene expression analysis between the different conditions was done using DESeq2 using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate, and differentially expressed genes were determined at the 5% threshold. Gene set enrichment analysis was used to assess the statistical enrichment of gene ontologies, and pathways. The FUS sequencing data from Drosophila were then compared to HEK cell FUS sequencing data to understand the transcriptome alteration of FUS mutants. The HEK sequencing data were processed using the same pipeline as Drosophila and the gene lists were compared. DIOPT-DRSC integrative ortholog prediction tool was used to find the human orthologs to Drosophila.
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Drosophila brains/tissues were lysed using TRIzol (Ambion; 15596026), and RNA was isolated using a phenol–chloroform extraction method. Following lysis, chloroform was added, and the samples were centrifuged. The upper, aqueous layer was isolated, treated with isopropanol, and centrifuged. The resulting RNA pellets were washed with 75% ethanol, centrifuged, and dried by ambient air. RNA was suspended in RNase-free water. Quantity and purity (260/280 and 260/230 ratios) were determined using a NanoDrop ND-1000 spectrophotometer. RNA quality was assessed by 1% agarose gel electrophoresis using ethidium bromide. The iScript Select cDNA Synthesis Kit (BioRad; 170-8897) was then used to produce cDNA from the RNA samples in an Omn-E PCR machine (Thermo Hybaid). Three RNA extractions were performed from each experimental group to produce cDNA; a sample lacking reverse transcriptase was used as a control to confirm the absence of genomic DNA. All cDNA samples were ran on 96-well plates (Applied Biosystems; 4306737) on a 7300 Real-Time PCR System (Applied Biosystems) using the Bio-Rad iQ Supermix (170-8862). α-tubulin was used as Drosophila and human housekeeping genes, respectively. A VIC-MGB TaqMan probe for actin beta served as normalizer (Thermo Fisher Scientific, Mm02619580_g1). Cycle threshold (CT) values were recorded and analyzed following the comparative (CT) method using Prism 6 (GraphPad Software) for statistical analyses.
All primers for qPCR were designed PrimerQuest primer design tool (Integrated DNA Technologies). The Primers were designed with the PrimerTime qPCR Assay tool (Integrated DNA Technologies). IDT PrimeTime qPCR Assays were used as the primer/probe solutions. The primers and probes used for qPCR assays are listed in Supplemental table 4 (Online resource 5).
Cell culture and transfections
HEK cells (ATCC® CRL-3216™) were cultured in Advanced DMEM supplemented with 10% FBS and 1% Glutamax and grown at 37 °C and 5% CO2. HEK cells were transiently transfected using Lipofectamine 3000 (Invitrogen L3000001) and used 24 h after transfection.
Plasmids
FLAG-FUS-WT, RS1, RS2, RS3, and RS4 were a gift from Dr. Jacob Schwartz [79]. HA-FUS-WT, and 4-FL constructs were previously generated in our lab [28]. The pLenti-C-mGFP-DDX17 plasmid was purchased from Origene (RC200599L4). The lentiviral vector used to overexpress HA-DDX17 in iPSC neuronal cells, pLV[Exp]-CMV > [HA-DDX17], was constructed and packaged by VectorBuilder, with the following ID: VB200909-1118dgf.
Immunoblotting
Nitrocellulose membranes were blocked in blocking buffer: 5% milk (BLOT- QuickBlocker™ EMD Millipore WB57) in TBST followed by overnight incubation with primary antibody at 4 °C. Membranes were washed with TBST and incubated with secondary antibody for 1 h at room temperature, followed by washed with TBST. The membranes were imaged on Odyssey® CLx (LI-COR Biosciences) and quantification of bands was performed using Image Studio™ (LI-COR Biosciences). Primary and secondary antibodies were prepared in blocking buffer.
Primary antibodies
Rabbit HA-Tag (1:5000; Cell Signaling C29F4), Rabbit Anti-FLAG (1:3000; Sigma F7425), Rabbit Anti-FUS (1:3000; Bethyl A300), Mouse Anti-DDX17 (1:2000; Santa Cruz 271112), and Anti-Laminin B1 (1:1000, Abcam).
Secondary antibodies
Goat anti-mouse Dylight 680 (1:10000; LI-COR 925-68070); Goat anti-rabbit Dylight 680 (1:10000; Invitrogen 35568); Goat anti-mouse Dylight 800 (1:10000; Invitrogen SA5-10176); Goat anti-rabbit Dylight 800 (1:10000; Invitrogen SA5-35571).
Immunofluorescence
HEK or iPSC neuronal cells grown on coverslips were rinsed in PBS (Lonza 17-512F) and fixed in 4% paraform-aldehyde (Sigma P6148) for 20 min at room temperature. Following fixation, the samples were washed four times (× 10 min) in PBS and blocked with blocking buffer: 5% normal goat serum (NGS; Abcam AB7681) in PBS with 0.1% TritonX-100 (PBST). The samples were incubated with primary antibody overnight at 4 °C, washed four times (× 10 min) with 0.1% PBST, and incubated with secondary antibody for 2 h at room temperature followed by 0.1% PBST washes. Samples were mounted onto slides using Fluoroshield (Sigma F6057). Primary and secondary antibodies were prepared in blocking buffer.
Primary antibodies
Rabbit HA-Tag (1:1000; Cell Signaling C29F4), anti-G3BP1 (1:1000; Proteintech D5444), Rabbit Anti-CC3 (1:1000; Cell Signaling 9661S), Rabbit Anti-53BP1 (1:1000; Novus Bio NB100-304), Goat anti-MAP2 (1:1,000; Synaptic System 188-004), Chicken anti- beta-III Tubulin (1:1,000; NOVUS Biologicals NB100-1612).
Secondary antibodies
Alexa fluor-488, −568 and −647 secondary antibodies were used from Invitrogen.
Nuclei/cytoplasm fractionation
For cytosol/nuclear fractionation, cells were washed with ice-cold PBS and collected in PBS 1 × . Upon centrifugation, pellet was resuspended in solA (20 mM Tris–HCl pH8.0, 50 mM NaCl, 1% NP40, 1 mM DTT with protease inhibitors), incubated on ice for 10 min, and centrifuged at 4000 rpm for 5 min. The cytosolic fraction was collected (supernatant), whereas pellet was resuspended in solB (20 mM Tris–HCl pH 8.0, 0.4 M NaCL, 1% NP40, 1 mM DTT and protease inhibitor) and incubated on ice for 20 min. Supernatant was collected upon centrifugation at 12,000 rpm for 10 min (nuclear extract).
Soluble–insoluble fractionation
Cells were lysed and resuspended in NP40 lysis buffer: 0.5% NP40, 10 mM Tris HCl pH 7.8, 10 mM EDTA, 150 mM NaCl, 2.5 mM Na orthovanadate, 1 × protease inhibitor cocktail (Roche 11836170001). The lysate was sonicated in an ultrasonic bath and centrifuged at 21000xg for 25 min. The supernatant (soluble fraction) was collected and boiled in 1X NuPage™ LDS-Sample buffer (Invitrogen, NP0007) at 95 °C for 5 min. The pellet was washed by resuspending in Washing buffer: 50 mM Tris HCl pH 7.4, 150 mM NaCl, and centrifuged at 21000xg for 5 min. The pellet was then resuspended in resolubilization buffer: 50 mM Tris HCl pH 6.8, 5% SDS, 10% glycerol followed by sonication and centrifugation at 12000xg for 10 min. The supernatant (insoluble fraction) was collected and boiled in 1X NuPage™ LDS-Sample buffer (Invitrogen, NP0007) at 95 °C for 5 min.
Differentiation of FUS iPSCs into neuronal cells
The iPSCs were differentiated into neuronal cells by following the protocol. The iPSCs were cultured and maintained in mTeSR™ 1 media (STEMCELL technologies) on Matrigel coated plates. For differentiation, ~ 0.6 million cells were plated and let to grow for up to 80–90% confluency in mTeSR™ 1 for 2 days. For the first phase of differentiation, the confluent iPSC cells were grown for 9 days in N2B27 Neurobasal/DMEM-F12 medium (1:1 v/v) containing 1% N2 (Gibco, 17502-048), 2% B27 (Gibco, 17054-044), 1% Glutamax (Gibco), and non-essential amino acids (NEAA) (Gibco, 11140050) along with 10 μM SB431542 (STEMCELL technologies), 0.1 μM LDN (Sigma, SML0559), 1 μM retinoic acid (RA) (Sigma, R2625), 1 μM smoothened agonist (SAG, Cayman chemicals 11914). For days 9 to 15, cells were grown in N2B27 media supplemented with 1 μM RA, 1 μM SAG, 10 μM DAPT (Cayman, 13197), 16 μM SU5406 (Cayman, 131825). On day 15, cells were dissociated using TrypLE/ DNase I (Invitrogen) and cultured on poly-ornithine and laminin-coated coverslips or plates in neurobasal N2B27 media supplemented with 0.4 mg/ml ascorbic acid (Sigma, A4403), 10 μg/ml human brain-derived neurotrophic factor (BDNF) (Peprotech, 45002), 10 μg/ml glial cell derived neurotrophic factor (GDNF) (Peprotech, 45010), 10 μg/ml ciliary neurotrophic factor (CNTF) (Peprotech, 45013), 1% Glutamax and NEAA. The cells were differentiated into neurons for 28 days and processed for subsequent Immunofluorescence and Western blot analysis.
Lentiviral production and transduction
Lentiviral transfer vectors encoding DDX17 were co-transfected with Lenti packaging plasmids (OriGene) into HEK cells using the Turbofectin transfection reagent (OriGene) according to manufacturer’s instructions. Following an initial media change, lentiviral supernatant was collected at 24 and 48 h post-transfection prior to filtration and overnight incubation at 4 °C with 1X Lenti Concentrator Solution (Ori-Gene). The Lenti Particles were then centrifuged at 4000×g for 120 min at 4 °C. The resulting pellet was recentrifuged at 4000×g for 5 min prior to re-suspension in ice-cold, sterile PBS. Pellets were then allowed to dissolve for 1–2 days at 4 °C. Resuspended lentiviral particles were then aliquoted. Neuron transductions were performed by diluting lentiviral particles at an MOI of 5 in neuronal differentiation media. Media changes were performed after 48 h of incubation and all experiments were initiated at 72–96 h post-transduction.
Treatments and induced DNA damage
Etoposide (Sigma, E1383) was dissolved in DMSO to obtain a 500-μM stock. HEK cells were treated with a final 2 μM Etoposide for 1 h and then processed for experiments.
Data availability
RNA-sequencing data that support the findings of this study are available in the Gene Expression Omnibus (GEO) database under accession number GSE173838. Source data are provided with this paper.
Results
Disease-causing mutations in FUS lead to global transcriptomic alterations in Drosophila brains
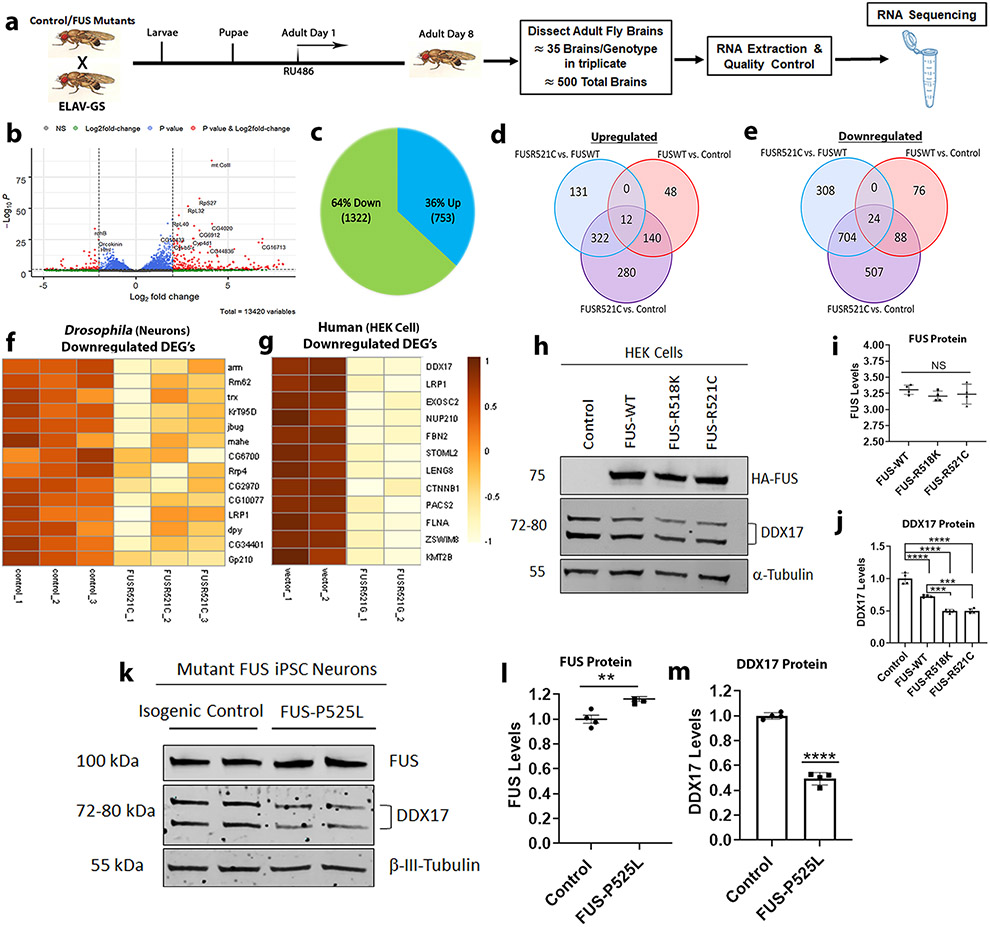
To investigate the global transcriptomic impact of wild-type and mutant FUS in an in vivo whole animal model, we utilized the inducible gene-switch GAL4 system with a conditional neuronal specific driver, ELAV-GS, to drive expression of wild-type and mutant FUS-R521C in Drosophila neurons (Fig. 1a). Using an in-silico approach, we identified the differentially expressed transcripts (DEGs) for each condition using a p-value threshold of ≤ 0.05 and log2 fold change of ≤ 1 and ≥ 1 [Supplemental Table 1 (Online Resource 2)]. We found a significant number of genes that were downregulated and upregulated in the FUS-R521C mutant compared to control (Fig. 1b) versus FUS-WT compared to control [Supplemental Fig. 1a (Online Resource 1)]. Specifically, we found 1322 (64%) downregulated and 753 (36%) upregulated genes in response to FUS-R521C (Fig. 1c), as opposed to only 200 (52%) downregulated and 188 (48%) upregulated genes in response to FUS-WT [Supplemental Fig. 1b (Online Resource 1)]. Interestingly, we found 280 upregulated genes to be specific to FUS-R521C apart from 48 genes specific to FUS-WT and found 131 shared genes that were upregulated between both FUS-WT and FUS-R521C (Fig. 1d). Furthermore, we identified most transcripts to be downregulated in response to both FUS-WT and FUS-R521C with 507 downregulated genes being specific to FUS-R521C apart from only 76 genes specific to FUS-WT (Fig. 1e).
Fig. 1.
Mutant FUS downregulates DDX17 in Drosophila brains and human cells. a Schematic showing the experimental setting used for sample preparation for RNA-sequencing. Wild-type and mutant FUS expression in Drosophila brains was achieved by a conditional neuronal driver, ELAV-GS, under the presence of the RU486 ligand. Drosophila were placed on treated RU486 drug food for 8 days, followed by brain dissection, RNA extraction, and RNA sequencing. b Volcano plot showing differentially expressed genes between FUS-R521C and control. Data for non-significant DEGs are plotted in black. Data for significant DEGs (FDR adjusted p value < 0.05) are plotted in blue. Data for significant DEGs (Bonferroni corrected p value < 0.05) and log2 fold changes less than or equal to – 1 or greater than or equal to 1 are plotted in red. c Pie chart showing the percentage and number of upregulated and downregulated genes in respect to FUS-R521C vs. control. d Venn diagram illustrating the number of upregulated DEGs (FDR adjusted p value < 0.05) between control, FUS-WT, and FUS-R521C. e Venn diagram illustrating the number of downregulated DEGs (FDR adjusted p value < 0.05) between control, FUS-WT, and FUS-R521C. f, g Heat maps of the 14-common downregulated DEGs of FUS-R521C mutation in Drosophila and FUS-R521G in HEK cells. Protein expression levels of endogenous DDX17 in (h) HEK cells transfected with FUS constructs and (k) FUS iPSC neurons harboring the FUS-P525L mutation. i, j Quantification of FUS and endogenous DDX17 protein levels from HEK cells (panel h) normalized to tubulin, indicating equivalent FUS expression in all groups and downregulated DDX17 protein levels in response to FUS constructs (n = 4 blots per condition, One-way ANOVA w/ Turkey’s multiple comparisons). l, m Quantification of endogenous FUS and DDX17 protein levels from mutant FUS iPSC neurons (panel k) normalized to the neuronal marker β-tubulin III (TUJ1). DDX17 protein levels are significantly downregulated in mutant FUS-P525L neurons compared to isogenic control (n = 4 blots, unpaired student t-test). Error bars indicate S.E.M. **p < 0.01; ***p < 0.001; ****p < 0.0001
To further assess the impact of mutant FUS-R521C, we conducted gene ontology and DAVID (v6.7) pathway analyses of the significantly altered DEGs. Interestingly, we identified ten downregulated pathways that were significantly enriched (p < 0.05) and nine upregulated pathways that were significantly enriched (p < 0.05) [Supplemental Fig. 2a (Online Resource 1)]. We found the most significantly enriched pathways to be loss of function or downregulated in response to mutant FUS and were associated with the integral membrane component (GO:0016021), nucleotide binding (GO:0000166), cellular ribonucleoprotein complex (GO:0030529), endoplasmic reticulum (GO:0005789), and ribosomal subunit/translation (GO:0002181).
Comparative transcriptomic analysis of Drosophila and HEK cells reveals DDX17 as a downregulated target of mutant FUS
To further narrow down the large number of altered DEGs in Drosophila and identify pathways biologically relevant across multiple FUS-mediated ALS model systems, we used an unbiased approach and compared our sequencing data with a published mutant FUS RNA-seq data set in HEK cells [95]. We identified 14 common significantly downregulated genes of mutant FUS using a p-value threshold of ≤ 0.05 and log2 fold change of ≤ 1 and ≥ 1. [Supplemental table 2 (Online Resource 3)]. These 14 DEGs were not significantly altered in response to FUS-WT alone [Supplemental table 2 (Online Resource 3)]. Heat maps of these 14 DEGs represent a similar downregulated trend in FUS-R521C expressing flies (Fig. 1f), as well as FUS-R521G expressing human cells (Fig. 1g). For RNA-seq validation, we selected six common DEGs based on the most significantly enriched pathways [Supplemental Fig. 2 (Online Resource 1)], Supplemental table 2 (Online Resource 3)]. By qPCR of Drosophila brains, dmCTNNB1, DDX17 orthologue dmMahe, dmFBN2, dmLRP1, dmNUP210, and dmSTOML2 [Supplemental Fig. 3 (Online Resource 1)] were all significantly downregulated in mutant FUS-R521C compared to control. Out of the 14 common downregulated DEG’s between both FUS mutants, we identified 3 Drosophila orthologs of the human Dead Box Helicase 17 (DDX17) gene, dmRm62, dmCG10077, and dmMahe [Supplemental table 2 (Online Resource 3)]. These three homologs of DDX17 are components of the nucleotide binding pathway which was the second most significantly enriched pathway in mutant FUS-R521C fly brains.
Based on these results, we explored the link between FUS and DDX17 in HEK cells and mutant FUS ALS iPSC neurons. We transfected HEK cells with HA-tagged FUS (WT and mutant) and measured the protein levels by Western blot (Fig. 1h). We found that the expression levels of FUS were equivalent across FUS-expressing groups (Fig. 1i), while the levels of endogenous DDX17 were reduced by ~ 20–25% in response to FUS-WT and ~ 50–60% in mutant FUS (R518K and R521C) expressing HEK cells compared to control (Fig. 1j). To further test the levels of endogenous DDX17 in mutant FUS, we utilized FUS-EGFP iPSC lines harboring wild-type and mutant P525L [64] and differentiated them into neurons. We found DDX17 was reduced by ~ 60% in mutant FUS-P525L iPSC neurons as compared to the isogenic control (Fig. 1k-m). These observations suggest that DDX17 expression is significantly reduced in response to mutant FUS and prompted us to further explore the link between DDX17 and FUS in the context of FUS-associated ALS.
Mutant FUS recruits nuclear DDX17 into cytoplasmic stress granules
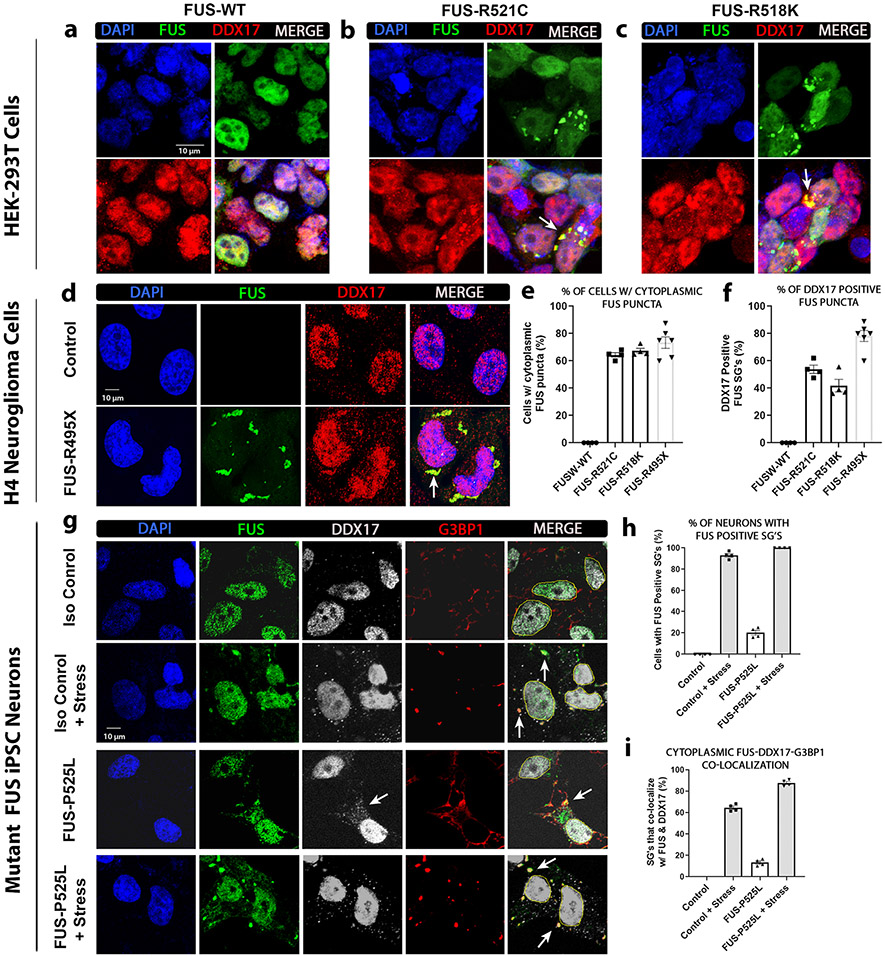
Mutant FUS cytoplasmic mislocalization and sequestration into stress granules (SG) are a well-known pathological hallmark of ALS [12, 33, 45, 52, 73, 96]. In addition, it has been shown that the incorporation of mutant FUS into SG’s alters SG dynamics by increased abundance and disrupted turnover [9]. These findings are supported by the observations that stress granules are formed under pathological or stress conditions resulting in trapped mRNAs and proteins leading to delays of global translation, as cells shift to translating essential proteins needed for survival [8, 15, 71]. To assess if DDX17 is being sequestered into mutant FUS-positive SGs in the cytoplasm, we utilized three different cellular models analyzing four unique disease-causing mutations in FUS. We transfected HEK cells with wild-type and mutant FUS (R518K and R521C) to assess the distribution pattern of endogenous DDX17. We have previously shown that ALS-associated mutations of FUS are sufficient for cytoplasmic SG formation in the absence of additional cellular stressors [18]. We found that DDX17 is predominantly nuclear in response to wild-type FUS expression (Fig. 2a), whereas under mutant FUS conditions, is recruited to the cytoplasm and colocalizes with mutant FUS-R521C (Fig. 2b) and FUS-R518K (Fig. 2c) positive stress granules. Furthermore, we found a similar disrupted localization pattern of DDX17 in response to mutant FUS-R495X expressing H4 neuroglioma cells. In response to disease-causing mutant FUS-R495X, DDX17 also colocalized with mutant FUS puncta in the cytoplasm while remaining mostly nuclear in control H4 cells not expressing mutant FUS (Fig. 2d). Compared to untransfected control and wild-type FUS expressing cells, all FUS mutants (R521C, R518K, and R495X) displayed a drastic increase in the number of cytoplasmic FUS puncta (Fig. 2e). In addition, all FUS mutants (R521C, R518K, and R495X) showed a drastic increase in the number of cytoplasmic FUS puncta that colocalized with DDX17 compared to wild-type FUS and untransfected control cells (Fig. 2f).
Fig. 2.
DDX17 is recruited into cytoplasmic stress granules in response to mutant FUS. HEK or H4 cells were transfected with the indicated FUS constructs (wild-type, R518K, R521C, or R495X). a–c For IF in HEK cells, the cells were transfected with HA tagged FUS-WT and mutants FUS-R521C and FUS-R518K. Representative confocal images showing the distributions of FUS and endogenous DDX17 in HEK cells. DAPI was used as a nuclear marker. White arrows indicate representative cytoplasmic FUS puncta in both FUS mutants that co-localize with DDX17. d For IF in H4 cells, the cells were transfected with EGFP tagged mutant FUS-R495X. Representative images showing the distributions of EGFP and endogenous DDX17 in H4 cells. DAPI was used as a nuclear marker. White arrows indicate representative cytoplasmic FUS puncta in mutant FUS-R495X that co-localize with DDX17. e Quantification of the percentage of cells with cytoplasmic FUS puncta per each FUS condition (n = 90–100 cells per FUS group). f Quantification of the percentage of DDX17 positive cytoplasmic FUS puncta. (n = 90–100 cells per FUS group). g Mutant FUS iPSC neurons expressing endogenous wild-type and mutant FUS with and without stress. Representative images showing the distributions of endogenous FUS, DDX17, and the stress granule marker G3BP1. White arrows in the 3rd column indicate cytoplasmic DDX17 in response to mutant FUS-P525L. White arrows in the last column indicate representative cytoplasmic colocalization of FUS, DDX17, and G3BP1. h Quantification of the percentage of neurons with cytoplasmic FUS puncta per each condition (n = 65–75 neurons per group). i Quantification of the percentage of G3BP1 positive SG’s that colocalize with both FUS and DDX17 (n = 65–75 neurons per group)
To rule out if the sequestration of DDX17 into FUS-positive cytoplasmic stress granules was a consequence of mutant FUS over expression, we again turned to human ALS iPSC neurons harboring wild-type and mutant FUS (P525L). Under native conditions in isogenic control neurons, both wild-type FUS and endogenous DDX17 are primarily distributed in the nucleus (Fig. 2g). However, when cells were challenged with sodium arsenite, we found that both FUS and DDX17 localized to the cytoplasm and sequestered into G3BP1-positve SGs, suggesting that FUS and DDX17 are components of SGs under stress conditions alone (Fig. 2g-h). We also examined the subcellular distribution of FUS and DDX17 in neurons harboring the disease-causing mutation FUS-P525L and found sequestration of mutant FUS and DDX17 into cytoplasmic G3BP1-positive SGs (Fig. 2g-h). These observations suggest that mutant FUS alters the subcellular distribution pattern of DDX17 by sequestering DDX17 into cytoplasmic stress granules, thus disturbing its normal nuclear biological and physiological processes.
DDX17 physically interacts with FUS and modifies FUS toxicity through the RGG domain in vivo
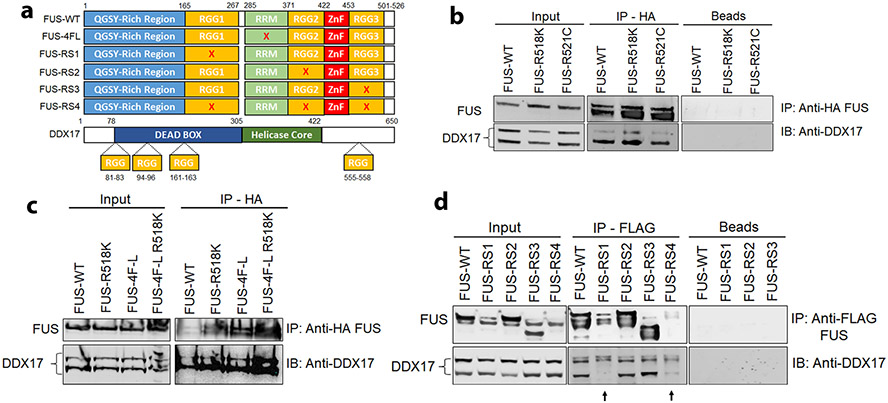
To identify the functional interaction between FUS and DDX17, we transfected HEK cells with FUS constructs each harboring a disruption of an individual functional domain of FUS protein (Fig. 3a). Interestingly, DDX17 and FUS both harbor RG/RGG motifs which are highly involved in DNA/RNA binding processes [93]. DDX17 is translated into two isoforms, p72 and p82, with each performing similar functions. p82 is larger and is translated via a non-AUG start site upstream from the normal canonical translation initiation site of p72 [48, 94]. Immunoprecipitation revealed a physical interaction between endogenous DDX17 and wild-type FUS as well as the FUS mutants R518K and R521C (Fig. 3b). To investigate if this interaction was mediated by the RNA Recognition Motif (RRM) of FUS, we transfected HEK cells with wild-type and RNA-binding incompetent FUS 4F-L (F305L, F341L, F359L, F368L) constructs that disrupt the conserved RRM domain of FUS [28]. Immunoprecipitation revealed a physical interaction between endogenous DDX17 and FUS in the RRM disrupted FUS 4F-L constructs (Fig. 3c). Interestingly, the FUS 4F-L mutations are sufficient to abolish RNA binding in both wild-type FUS 4F-L and FUS 4F-L R518K [28], which implies DDX17 and FUS are interacting through an RNA-independent manner. Furthermore, to investigate if this interaction was mediated by the glycine-arginine-rich domains (RGG) of FUS, we transfected HEK cells with FUS constructs harboring mutated arginine to serine amino acids of each separate RGG domain, thus disrupting the RGG domains of FUS [79]. Interestingly, disruption of the RGG1 domain of FUS abolished its interaction with p72, suggesting that the RGG1 domain is required for mediating interaction between FUS and DDX17 (Fig. 3d).
Fig. 3.
DDX17 and FUS physically interact via the RGG1 domain of FUS. a Schematic illustration of FUS constructs and DDX17 each harboring RGG domains. Mutated FUS 4F-L (F305L, F341L, F359L, F368L) disrupts the conserved RRM domain of FUS. Mutated FUS-RS1, RS2, and RS3 convert all arginine amino acids of each separate RGG domain to serine amino acids, thus disrupting individual RGG domains of FUS. In mutant FUS-RS4, arginine amino acids in all RGG domains were converted to serines, thus disrupting all RGG binding domains of FUS. b Immunoblot of co-immunoprecipitation of HA tagged FUS-WT, FUS-R518K, and FUS-R521C from HEK cells. Immunoprecipitation with HA antibody showed endogenous DDX17 (p72) in the input control samples and pulled down with HA-FUS-WT, FUS-R518K, and FUS-R521C, but absent from the negative control containing beads only. c Immunoblot of co-immunoprecipitation of HA-FUS-WT, FUS-R518K, FUS 4F-L, and FUS-R518K 4F-L from HEK cells expressing HA-FUS. Immunoprecipitation with HA antibody showed endogenous DDX17 (p72) in the input control samples and pulled down with HA-FUS-WT, FUS-R518K, FUS 4F-L, and FUS-R518K 4F-L. d Immunoblot of co-immunoprecipitation of FLAG-FUS-WT, FUS-RS1, FUS-RS2, FUS-RS3, and FUS-RS4 from HEK cells expressing FLAG-FUS. Immunoprecipitation with FLAG antibody showed endogenous DDX17 (p72) in the input control samples and pulled down with FLAG-FUS-WT, FUS-RS2, and FUS-RS3, but absent from FUS-RS1, FUS-RS4, and the negative control containing beads only. Interestingly, RGG1 disruption of FUS disrupts the interaction between FUS and DDX17
To further understand the functional consequences of the interaction between DDX17 and FUS, we performed a candidate genetic screen of other DDX family proteins. Using the glass multiple reporter (GMR), FUS-WT and FUS-R518K fly lines were crossed with EGFP control and the following RNAi-mediated DDX Drosophila orthologue fly lines: DDX3, DDX4, DDX9, DDX18, DDX19, DDX41, DDX49, DDX51 [Supplemental Fig. 4a (Online Resource 1)], Supplemental table 3 (Online Resource 4)]. Interestingly, only the Drosophila DDX proteins, DDX3, and DDX4, which harbor RGG motifs, similar to DDX17 [93], significantly modulated eye degeneration phenotypes associated with wild-type and mutant FUS [Supplemental Fig. 4b-c (Online Resource 1)]. Based on these results, we suggest the RGG motif to be crucial for regulating the physical and functional interactions between DDX17 and FUS.
Ectopic expression of DDX17 reduces insoluble FUS formation and sequestration into stress granules
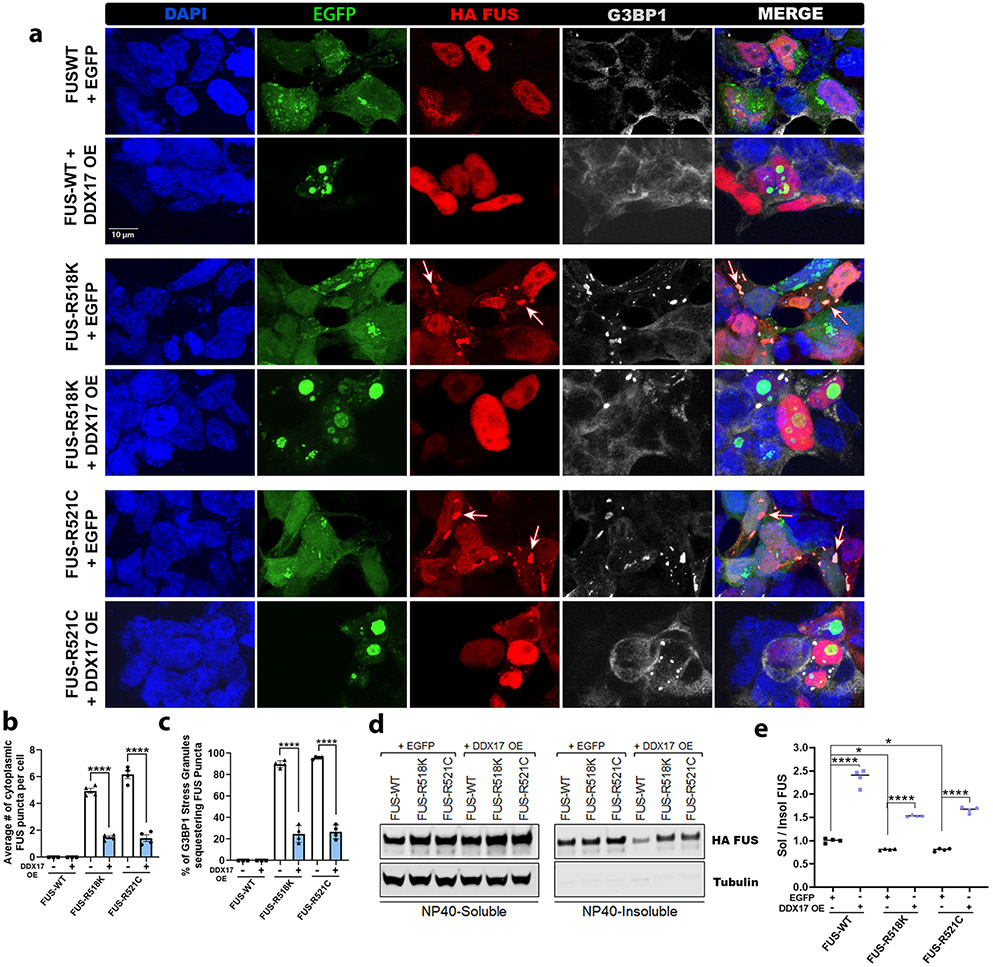
The accumulation of toxic insoluble cytoplasmic FUS aggregation has also been linked with protein toxicity and motor neuron death [4, 28, 68, 80, 102]. Since we assessed DDX17 as a downregulated target of mutant FUS, we hypothesized that restoring DDX17 protein levels via overexpression may reduce mutant FUS toxicity and incorporation into SGs. To determine the effect of DDX17 over expression on wild-type and mutant FUS localization and SG development in HEK cells, we co-transfected HEK cells with FUS (wild-type and mutant) and control EGFP, or EGFP tagged DDX17. The distribution of FUS and the stress granule marker, G3BP1, were visualized by fluorescent microscopy with and without over expression of DDX17 (Fig. 4a). In cells co-transfected with FUS and EGFP control, wild-type FUS localized to the nucleus, whereas FUS mutants (R518K and R521C) mislocalized to the cytoplasm and were positive for the stress granule marker G3BP1 (Fig. 4a). Compared to FUS-WT, both FUS mutants (R518K and R521C) showed a drastic increase in the number of cytoplasmic FUS puncta (Fig. 4b) and the number of G3BP1-positive SGs that sequestered cytoplasmic FUS (Fig. 4c). Interestingly, overexpression of DDX17 significantly reduced the number of cytoplasmic FUS puncta in both FUS mutants and drastically reduced the percentage of G3BP1-positve puncta sequestering mutant FUS (Fig. 4c). To assess if over expression of DDX17 had any effect on stress granule dynamics alone, we transfected control HEK cells with EGFP and EGFP tagged DDX17 and challenged the cells with sodium arsenate. We found that overexpression of DDX17 alone did not have any effect on the overall dynamics of stress granule formation compared to control cells [Supplemental Fig. 5 (Online Resource 1)]. These data provide evidence that restoration of DDX17 levels via over expression modifies FUS toxicity by reducing the recruitment of mutant FUS into cytoplasmic stress granules.
Fig. 4.
Ectopic DDX17 expression reduces insoluble cytoplasmic FUS formation and sequestration into stress granules. HEK cells were co-transfected with the indicated HA tagged FUS constructs (wild-type, R518K, or R521C) and either control EGFP or DDX17-EGFP over expression. a Representative confocal images showing FUS, DDX17 OE, and stress granule marker G3BP1 distributions in HEK cells. For IF, cells were co-transfected with HA tagged FUS (wild-type and mutant) and control EGFP, or EGFP tagged DDX17 OE at equal concentrations. White arrows indicate representative cytoplasmic FUS puncta in both FUS mutants that also co-localize with the stress granule marker, G3BP1. (b) Quantification of the average number of cytoplasmic FUS puncta per cell confirms that over expression of DDX17 significantly reduced the number of cytoplasmic FUS puncta in the R518K and R521C FUS mutants. (n = 55–70 cells per FUS group, One-way ANOVA w/ Turkey’s multiple comparisons). c Quantification of the percentage of G3BP1 positive cytoplasmic stress granules sequestering mutant FUS. Interestingly, overexpression of DDX17 significantly reduced mutant FUS integration into SGs (n = 55–70 cells per FUS group, One-way ANOVA w/ Turkey’s multiple comparisons). d Representative blots of soluble–insoluble fractionation of FUS protein for FUS-WT, FUS-R518K, and FUS-R521C plus EGFP control and FUS-WT, FUS-R518K, FUS-R521C plus DDX17 OE. Tubulin was used as a soluble loading control. e Quantification of soluble/insoluble (Sol/Insol) FUS formation from representative blots shown in panel (d) (n = 4 blots per condition, One-way ANOVA w/ Turkey’s multiple comparisons). Over expression of DDX17 significantly increases the sol/insol fraction of FUS expressing groups. Error bars indicate S.E.M. *p < 0.05, ****p < 0.0001
DEAD-box proteins were also identified as global regulators of phase separated organelles [42]. Therefore, we reasoned that ectopic expression of DDX17 might be reducing the recruitment of mutant FUS into SGs by mitigating toxic protein insolubility. To test this possibility, we co-transfected HEK cells with FUS (wild-type and mutant) and control EGFP, or EGFP tagged DDX17 [Supplemental Fig. 6 (Online Resource 1)] and analyzed if overexpression of DDX17 had any effect on the solubility of FUS via NP40 soluble/insoluble fractionation (Fig. 4d). In cells co-transfected with FUS and EGFP, mutant FUS (R518K and R521C) proteins were significantly less soluble, with more FUS protein accumulating in the insoluble fraction (Fig. 4e). Interestingly, over expression of DDX17 significantly increased the solubility of FUS (wild-type and mutant), while drastically decreasing the formation of toxic insoluble species (Fig. 4e). In addition, to assess if the reduced sequestration of mutant FUS into cytoplasmic SGs upon over expression of DDX17 had any effect on the overall levels of FUS, we measured the levels of FUS via qPCR and Western blot [Supplemental Fig.7a-c (Online Resource 1)]. We found that over expression of DDX17 significantly reduced the mRNA and protein levels of FUS. Furthermore, we measured the nuclear and cytoplasmic protein levels of FUS (wild-type and mutant R518K) in cellular fractions, with and without DDX17 overexpression. In cells co-transfected with FUS and EGFP, the nuclear/cytoplasmic ratio (N/C ratio) of FUS-R518K was significantly reduced compared to wild-type FUS. In cells co-transfected with FUS and EGFP tagged DDX17, the N/C ratio of mutant FUS-R518K was drastically increased [Supplemental Fig. 7d-f (Online Resource 1)]. These data suggest that the reduction of FUS-positive stress granules in the cytoplasm following over expression of DDX17 may be due to the reduction of toxic insoluble cytoplasmic mutant FUS formation.
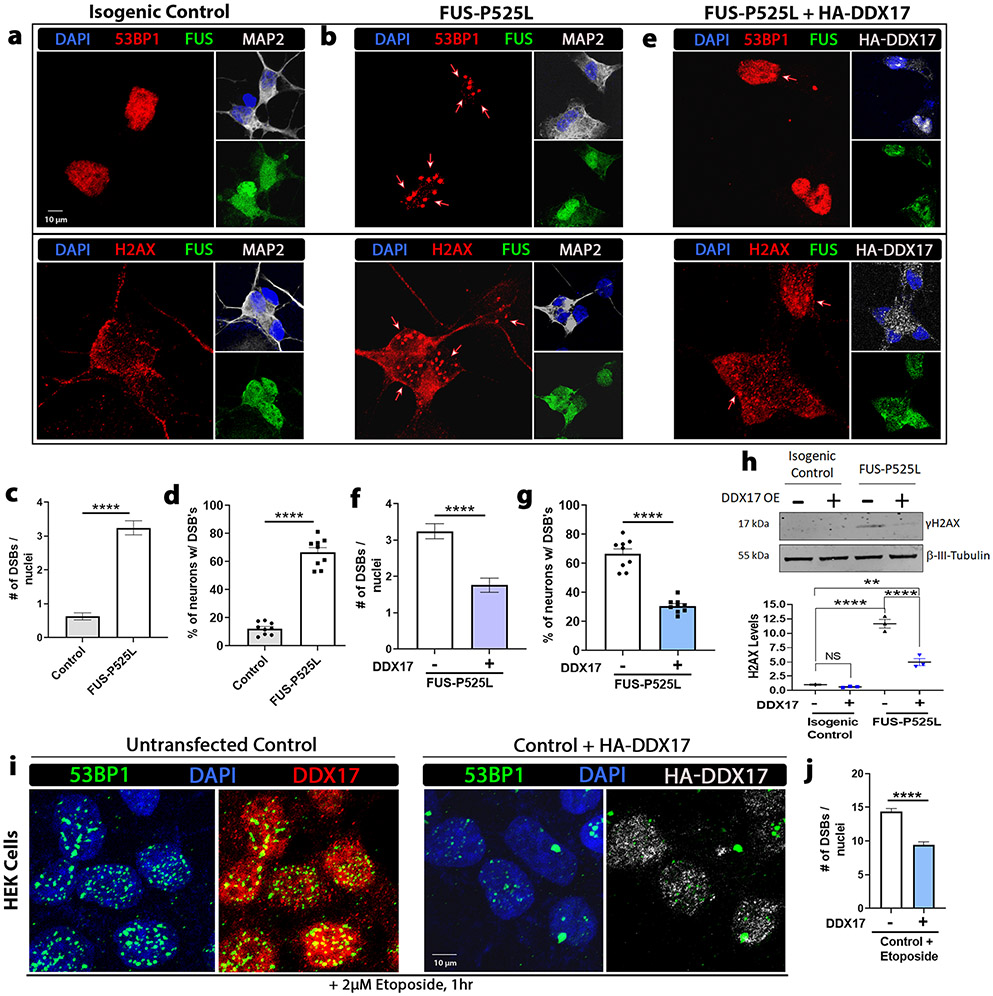
DDX17 suppresses DNA damage in mutant FUS iPSC neurons and is a regulator of DNA damage repair mechanisms
The increased occurrence of DNA damage and double-strand break (DSB) formation has been linked in patients with mutant FUS-associated ALS [75, 99] as well as animal models of FUS [81]. DDX17 has also been implicated as an epigenetic regulator and found to be recruited to DNA damage sites by microirradiation, suggesting a possible role for DDX17’s involvement in regulating DNA damage repair [3, 38]. In addition, several key DNA damage response proteins contain RGG/RG motifs with many of these proteins being involved in DSB repair [2, 93]. To assess the role of DDX17 in the DNA damage response pathway in the context of mutant FUS pathology, we analyzed if restoration of DDX17 levels has any effect on the defective DNA damage repair machinery exhibited in mutant FUS iPSC neurons. Therefore, we assessed the occurrence of DSBs in human ALS iPSC neurons harboring wild-type and mutant FUS (P525L). By immunofluorescence analysis, we analyzed two markers of DNA damage, γH2AX and tumor suppressor p53-binding protein 1 (53BP1), in control and mutant FUS-P525L derived neuronal cells (Fig. 5a, b). We observed significantly more DSBs in the mutant P525L line compared to the isogenic control line (Fig. 5c). We also observed approximately 65–70% of mutant FUS neurons exhibited DSBs compared to only 15% of isogenic control neurons (Fig. 5d). Next, we utilized a lentiviral expression system to selectively express HA tagged DDX17 [Supplemental Fig. 6 (Online Resource 1)] in the mutant FUS-P525L neuronal cells and assessed the occurrence of DSBs via γH2AX and 53BP1 (Fig. 5b, e). Interestingly, over expression of DDX17 significantly reduced the number of DSBs and percentage of mutant FUS neuronal cells expressing DSBs compared to mutant FUS neuronal cells without DDX17 over expression (Fig. 5f, g). Furthermore, Western blot confirmed that DDX17 OE prevents the induction of γH2AX caused by mutant FUS-P525L (Fig. 5h).
Fig. 5.
Upregulation of DDX17 mitigates DNA damage exhibited in mutant FUS iPSC neurons. Representative confocal images of a isogenic control and b, e mutant FUS iPSC neurons with and without overexpression of HA-DDX17 from the same set of neuronal differentiations. Neurons were probed for FUS, the DNA damage and double-strand break (DSB) markers γH2AX and p53-binding protein 1 (53BP1), HA-DDX17, and the neuronal marker microtubule-associated protein 2 (MAP2). Nuclei were stained with DAPI. c, d Quantification of the number of DNA DSBs and the percentage of neurons with DSBs between control (panel a) and mutant FUS-P525L (panel b) (n = 60–70 neurons, unpaired students t-test). Mutant FUS-P525L neurons show a significant increase in the number of DNA DSBs per neuron compared to control. f, g Quantification of the number of DNA DSBs and the percentage of neurons with DSBs between mutant FUS-P525L (panel b) and mutant FUS-P525L with overexpression of HA-DDX17 (panel e) (n = 60–70 neurons, unpaired students t-test). Mutant FUS-P525L neurons expressing HA-DDX17 show a significant decrease in the number of DNA DSBs per neuron compared to mutant FUS-P525L alone. h Representative blots of γH2AX protein in isogenic control and mutant FUS-P525L neuronal cells with and without overexpression of DDX17. β-tubulin III was used as a loading control (n = 3 blots, One-way ANOVA w/ Turkey’s multiple comparisons). FUS-P525L neurons showed a significant increase in γH2AX protein levels compared to FUS-P525L neurons expressing HA-DDX17. i Confocal images of HEK cells exposed to the DNA damage-inducing agent, etoposide (2 μM), for 1 h after being transduced with and without HA-DDX17 for 72 h. The cellular distributions of 53BP1, HA-DDX17, and endogenous DDX17 were assessed. j Quantification of the number of DNA DSBs between HEK cells exposed to etoposide with and without over expression of HA-DDX17 (n = 70–80 cells, unpaired students t-test). Cells over expressing DDX17 show a significant decrease in the number of DNA double-strand breaks per cell compared to control after etoposide exposure. Error bars indicate S.E.M. **p < 0.01, ****p < 0.0001
Next, we assessed the involvement of DDX17 in the DNA damage response pathway under non pathological mutant FUS conditions. Under mutant FUS-associated conditions, we identified a 50–60% significant reduction of DDX17 protein levels (Fig. 1h-m). Mimicking these conditions, we knocked down DDX17 in control HEK cells [Supplemental Fig. 8a, b (Online Resource 1)] and tested the occurrence of DNA DSBs via immunofluorescence. In HEK cells expressing a~ 50% reduction of endogenous DDX17, we found significantly more DSBs per cell nuclei than scramble control [Supplemental Fig. 8c (Online Resource 1)]. Cells expressing DDX17 shRNA resulted in an average of 5 DNA DSBs per nuclei, compared to ~ 2.5 DSBs per nuclei for the scrambled control [Supplemental Fig. 8d (Online Resource 1)]. We were then interested in determining if DDX17 over expression was also protective against DNA damage-inducing agents. We transduced HEK cells with and without DDX17 for 48–72 h and treated the cells with etoposide for 1 h (Fig. 5i). Interestingly, cells treated with etoposide that over expressed DDX17 exhibited a significant reduction in DNA DSBs per nuclei compared to control cells also treated with etoposide (Fig. 5j). In addition, DDX17 was also recruited into 53BP1 puncta in both conditions, suggesting its recruitment to DNA damage sites. These data suggest that the reduction of DDX17 levels is sufficient to cause DSBs, whereas over expression of DDX17 repairs defective DNA damage repair machinery in induced DNA damage and pathological mutant FUS-associated ALS conditions.
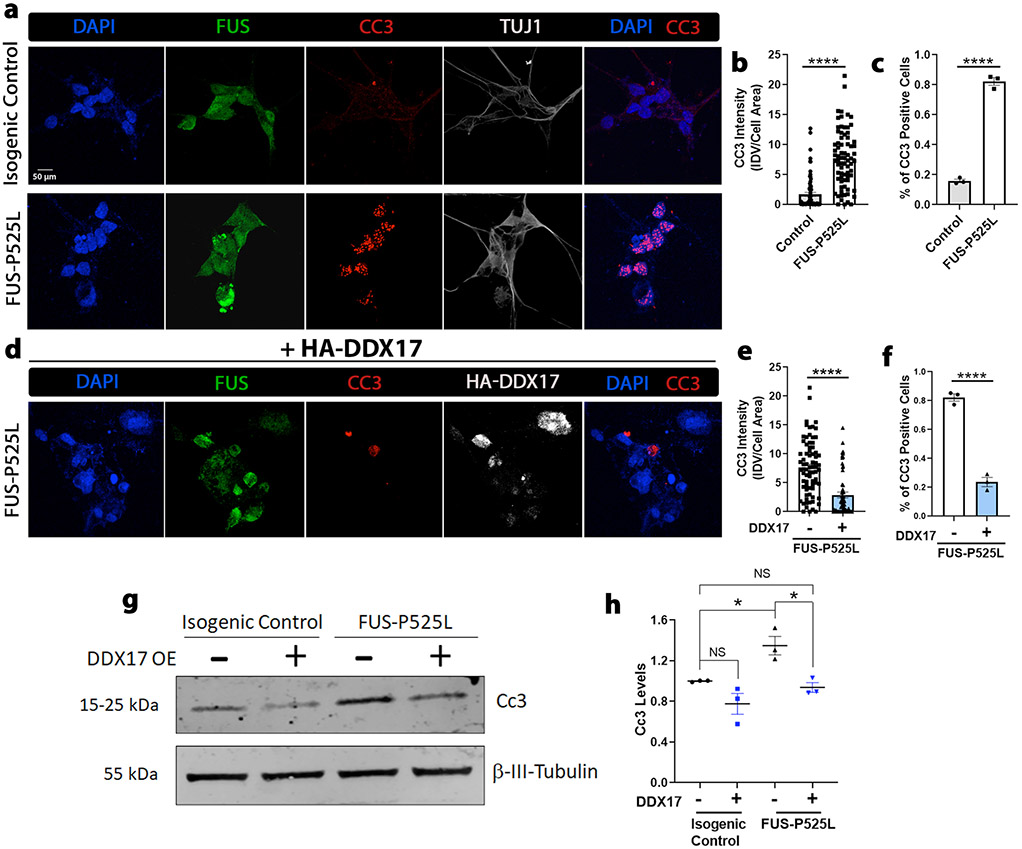
Ectopic expression of DDX17 protects against induced cell death of mutant FUS iPSC neurons
Since we identified that over expression of DDX17 restored defective DNA damage repair machinery in mutant FUS iPSC neurons, we hypothesized that DDX17 over expression could also be neuroprotective in mutant FUS iPSC neurons. Increased apoptotic signaling is often correlated with defects in the DNA damage repair system, as apoptosis is a secondary response to DNA damage [32, 62]. In addition, mutant FUS patient iPSC neurons have been shown to exhibit positive markers of apoptotic cell death [64, 75]. Thus, we analyzed if over expression of DDX17 had any effect on the induced cell death of mutant FUS iPSC neurons by immunostaining for the apoptotic cell marker, cleaved-CASPASE3 (CC3) in isogenic control and mutant FUS-P525L iPSC differentiated neurons. In all neurons labeled with the neuronal marker β-tubulin III (TUJ1), we found a significant increase in the intensity of CC3 for mutant FUS-P525L neurons compared to isogenic control neurons (Fig. 6a, b). In addition, we found the percentage of neurons that exhibited apoptotic activity was also significantly increased in neurons harboring the FUS-P25L mutation (Fig. 6c). Next, we overexpressed DDX17 in the same set of mutant FUS-P525L differentiated neurons using lentiviral transduction (Fig. 6d) Interestingly, we found that over expression of DDX17 significantly reduced the overall intensity of CC3 and drastically reduced the percentage of mutant FUS-P525L neurons that exhibited apoptotic activity (Fig. 6e, f). In addition to immunofluorescence, we also assessed the levels of CC3 via Western blot and observed a significant increase in CC3 protein levels in mutant FUS-P525L neurons compared to the isogenic control. Upregulation of DDX17 significantly reduced CC3 protein levels in mutant FUS-P525L neurons, similar to the CC3 levels observed in isogenic control neurons (Fig. 6g, h). These results suggest that overexpression of DDX17 in mutant FUS iPSC neurons is neuroprotective by reducing mutant FUS apoptotic activity.
Fig. 6.
Upregulation of DDX17 protects against apoptotic activity of mutant FUS iPSC neurons. a Representative confocal images of isogenic control and mutant FUS iPSC neurons showing FUS, the apoptotic marker cleaved caspase 3 (CC3), HA-DDX17, and the neuronal marker β-tubulin III (TUJ1). Nuclei were stained with DAPI. b, c Quantification of integrated CC3 intensity per cell normalized to the area of each neuron (panel a) and the percentage of neurons that expressed CC3 signal (panel c) between FUS-P525L and control (n = 80 neurons, unpaired students t-test). FUS-P525L neurons showed a drastic increase in CC3 signal compared to control. d Representative confocal images of the same set of differentiated ALS mutant FUS-P525L iPSC neurons from (panel a) overexpressing HA-DDX17. The neurons were probed for the apoptotic marker CC3, the neuronal marker TUJ1, and DAPI. e, f Quantification of integrated CC3 intensity per cell normalized to the area of each neuron (panel e) and the percentage of neurons that expressed CC3 signal (panel f) between FUS-P525L and FUS-P525L with HA-DDX17 expression (n = 80 neurons, unpaired students t-test). g Representative blots of CC3 protein in isogenic control and mutant FUS-P525L neuronal cells with and without overexpression of DDX17. β-tubulin III was used as a loading control (n = 3 blots, One-way ANOVA w/ Turkey’s multiple comparisons). h FUS-P525L neurons showed a drastic increase in CC3 protein levels compared to FUS-P525L neurons overexpressing HA-DDX17. Error bars indicate S.E.M. *p < 0.05, ****p < 0.0001
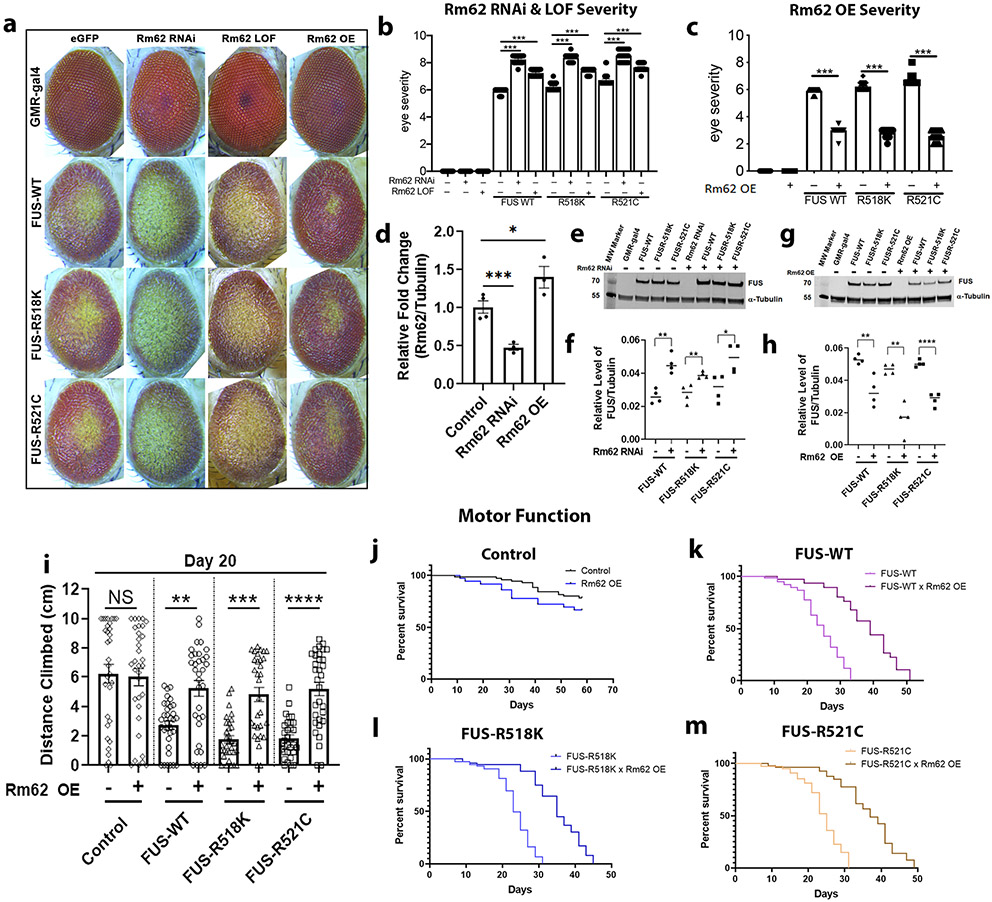
DDX17 Drosophila ortholog, Rm62, is a novel modifier of FUS toxicity in vivo
Our data suggest that genetic upregulation of DDX17 levels suppresses mutant FUS toxicity defects in vitro. To investigate this further, we tested if downregulation of DDX17 or restoration of DDX17 levels via overexpression had any effect on the toxicity of FUS in vivo. Thus, DDX17 was further analyzed as a modifier of FUS toxicity in Drosophila using well-established site-specific wild-type and mutant FUS fly lines [18]. Using the GMR promoter, wild-type and two ALS-linked mutant FUS (R518K and R521C) fly lines were crossed with EGFP control (to rule out gal4 dilution) and Rm62 RNAi, LOF, and over expression lines, which were also generated using site-specific integration (Fig. 7a). Further reduction of Rm62 levels around 50% (Fig. 7d) significantly enhanced external eye degeneration in all FUS-expressing Drosophila compared to control (Fig. 7b). In contrast, over expression of Rm62 levels increased by around 40% compared to control (Fig. 7d) significantly suppressed external eye degeneration in all FUS-expressing Drosophila (Fig. 7c). Interestingly, knockdown of endogenous Rm62 increased FUS protein levels, indicating that this effect could be due to an increase in the amount of toxic FUS protein (Fig. 7e, f). On the other hand, over expression of endogenous Rm62 decreased FUS protein levels, indicating that this effect could be due to a decrease in the amount of toxic FUS protein (Fig. 7g, h). More importantly, knockdown and overexpression of Rm62 alone in control flies was well-tolerated and did not cause any external eye degeneration itself (Fig. 7a). To further assess if DDX17 was a modifier of FUS toxicity in vivo, we crossed an additional DDX17 Drosophila ortholog, CG10077, with wild-type and mutant FUS-R518K and FUS-R521C fly lines using GMR-gal4 [Supplemental Fig. 9a (Online Resource 1)]. Similar to Rm62, we found that knockdown of endogenous CG10077 resulted in significantly enhanced external eye degeneration in all FUS-expressing groups, while knockdown of CG10077 alone had no effect on external eye degeneration itself [Supplemental Fig. 9b (Online Resource 1)].
Fig. 7.
DDX17 Drosophila orthologue, Rm62, is a novel modifier of FUS toxicity in vivo. a Representative images of Drosophila eyes of wild-type and ALS-linked mutant FUS protein alone (left column), in combination with RNAi-mediated knockdown of Rm62 (middle left column), LOF-mediated Rm62 (middle right column), or with overexpression of Rm62OE (right column). b Quantification of eye degeneration severity from panel (a) confirms significant enhancement of FUS toxicity following depletion of Rm62, and c significant suppression of FUS toxicity following overexpression of Rm62 (n = 15–25 Drosophila per group, One-way ANOVA w/ Turkey’s multiple comparisons). d qPCR of RNA from (n = 4 biological replicates of 6 Drosophila heads per group) confirms significant knockdown of endogenous Rm62 in the RNAi line and significant overexpression in the Rm62OE line (One-way ANOVA w/ Turkey’s multiple comparisons). e Western blot analysis showing FUS and tubulin (housekeeping control) protein levels in Drosophila eyes expressing wild-type and mutant FUS, with and without Rm62 knockdown (n = 4 biological replicates of 5 Drosophila heads per group). f Quantification of FUS from panel e normalized to tubulin, indicating equivalent FUS expression in all FUS alone-expressing groups. Interestingly, knockdown of Rm62 further enhanced FUS protein levels (n = 4 blots per condition, One-way ANOVA w/ Turkey’s multiple comparisons). g Western blot analysis showing FUS and tubulin (housekeeping control) protein levels in Drosophila eyes expressing wild-type and mutant FUS, with and without Rm62 overexpression (n = 4 biological replicates of 5 Drosophila heads per group). h Quantification of FUS from panel g normalized to tubulin, indicating equivalent FUS expression in all FUS alone-expressing groups. Interestingly, overexpression of Rm62 significantly reduced FUS protein levels (n = 4 blots per condition, One-way ANOVA w/ Turkey’s multiple comparisons). i Quantification of the total height climbed (cm) of adult flies in the first 3 s of RING climbing assay. Over expression of Rm62 significantly improved the climbing ability of neuronal expressing FUS-WT, FUS-R518K, and FUS-R521C flies. (n = 30–35 adult flies, One-way ANOVA w/ Turkey’s multiple comparisons). j–m Kaplan–Meier survival curve of adult flies conditionally expressing j control, k FUS-WT, l FUS-R518K, and m FUS-R521C under the neuronal specific driver, ELAV-GS, with and without Rm62 over expression. Over expression of Rm62 drastically improved longevity of FUS-WT, FUS-R518K, and FUS-R521C expressing flies. (n = 75 adult flies). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
In addition, it has been shown that degenerative effects associated with FUS toxicity in Drosophila increase in an age-dependent manner [18, 57, 58]. We tested whether modulation of Rm62 levels had any effect on FUS-induced external eye degeneration over time [Supplemental Fig. 10 (Online Resource 1)]. We found that knockdown of Rm62 resulted in further enhanced external eye degeneration in FUS-expressing flies aged to day 15 compared to day 1. In contrast, over expression of Rm62 over time resulted in almost complete suppression of external eye degeneration in FUS-expressing groups [Supplemental Fig. 10 (Online Resource 1)].
To determine if Rm62 is a novel modifier of other ALS-linked proteins or specific to FUS-associated ALS, Rm62 fly lines were crossed with fly lines expressing wild-type or mutant forms of two other ALS linked proteins (TDP-43 and C9orf72). Modulation of Rm62 via knockdown and overexpression did not suppress or enhance the toxicity of TDP-43 and C9orf72 expressing Drosophila [Supplemental Fig. 11 (Online Resource 1)]. Therefore, the enhanced and suppressed phenotypes observed with DDX17 appear to be specific to FUS associated ALS.
As ALS primarily affects motor neurons of the brain, we tested whether restoration of Rm62 levels via over expression could ameliorate FUS-associated toxicity in Drosophila neurons. Wild-type and mutant FUS were expressed in fly neurons using ELAV-GS-gal4 and crossed with control and Rm62 over expressing flies. We assessed the motor climbing ability using the RING assay [77] and assessed the life span of FUS-expressing flies alone and FUS-expressing flies that also over expressed Rm62. Ectopic expression of Rm62 alone in control flies was well-tolerated and did not cause any motor deficits or changes in the adult survival (Fig. 7i, j). We found that over expression of Rm62 significantly increased the motor ability of flies across each FUS-expressing group (Fig. 7i), thereby rescuing motor function. We also found that over expression of Rm62 drastically increased the adult survival of each neuronal FUS-expressing group (Fig. 7k-m). Based on these findings, we propose Drosophila DDX17 (Rm62 and CG10077) as a novel modifier of ALS-associated FUS toxicity in vivo.
Discussion
Disease-causing FUS mutations are associated with the most aggressive forms of ALS involving juvenile onset and rapid disease progression [14, 44, 59, 85, 97]. To comprehensively evaluate the detrimental outcomes of disease-causing FUS mutations in a whole animal Drosophila model system, we performed RNA-sequencing on Drosophila brains exhibiting targeted expression of wild-type and mutant FUS. To narrow down the significant list of innumerable altered DEGs in Drosophila and identify consistent gene expression changes across multiple FUS-mediated ALS model systems with high confidence, we performed comparative transcriptomic analysis on FUS-expressing Drosophila brains and human cells. Nevertheless, the comparative analysis revealed to be highly impactful, as we identified many common downregulated targets of mutant FUS across both species [Supplemental table 2 (Online Resource 3)]. Specifically, we identified DDX17 as one of the most common and highly significant downregulated targets of mutant FUS in Drosophila, human cells, and iPSC neurons (Fig. 1). While DDX17 has never been linked directly to driving ALS pathogenesis, proteomic studies have identified DDX17 as an interactor of the ALS-causing genes, FUS [20, 90] and C9orf72 (Poly-PR) [91], suggesting a possible role of DDX17 in ALS pathogenesis.
DDX17 is a multifunctional protein of the DEAD box RNA Helicase family and is involved in many aspects of RNA processing [36, 38] which include: alternative splicing [30, 56], chromatin modification [30, 36], and transcriptional regulation [17, 30, 36, 56]. There are two isoforms of DDX17: p72 and p82, with each isoform arising from the DDX17 gene through different in-frame translation initiation codons. p72 is the main canonical isoform of DDX17 which is translated downstream of p82. Biochemical studies suggest that p72 and p82 have nearly identical properties with equivalent protein expression [36, 41, 48, 94]. DDX17 is predominantly nuclear, but under osmotic and stress conditions, recent studies have implicated DDX17 as a novel component of cytoplasmic stress granules (SGs) [41, 47, 102]. The response of RNA metabolism to various stressors such as infection, temperature, and oxidative stress, typically results in the formation of cytoplasmic SGs [7, 15, 83]. In the presence of stress, SGs form through the process of liquid–liquid phase separation (LLPS) and quickly disassemble through reversible phase transitions once stress is removed [11, 49]. SG formation is a key pathological hallmark of many neurodegenerative diseases including FUS-mediated ALS [60, 64, 67, 88, 102]. Under mutant FUS pathological conditions, we found that DDX17 also becomes trapped in cytoplasmic SGs, thus likely resulting in inhibition of its normal biological functions (Fig. 2). Although the underlying mechanism for the recruitment of DDX17 into cytoplasmic FUS positive SGs is not clear, we hypothesize this recruitment is mediated in an RGG-domain dependent manner, as the RGG1 motif of FUS is crucial for mutant-FUS mislocalization into SGs [9, 63, 93], and we found the FUS-DDX17 interaction to be mediated in an RGG-domain dependent manner [Fig. 3, Supplemental Fig. 4 (Online Resource 1)]. These findings suggest that mutant FUS impedes the normal processes of DDX17 by its sequestration into FUS-positive cytoplasmic SGs in an RGG domain dependent manner.
The RGG domain is one of the most evolutionary conserved nucleic acid binding motifs that has been shown to be involved in telomere maintenance, transcription, translation of G-quadruplex-containing regions, and suppression of virus replication [1, 10, 35]. Recently studies have demonstrated that FUS recognizes G-quadruplex RNA sequences within neuronal mRNAs [46, 66]. G-quadruplex folding and unfolding are regulated by G-quadruplex binding proteins that include several RNA-binding proteins containing RGG domains such as FUS, heterogenous ribonucleoproteins (RNPs), and others [34, 89]. G-quadruplex’s have been shown to regulate various physiological pathways including transcription, translation, DNA replication, and histone modifications [13, 66, 72]. Helicase activity of DDX17 is required for the regulation of both exon inclusion and skipping [30]. Furthermore, DDX17 cooperates on G-quadruplex structures to regulate splicing events [30]. Importantly, a recent study showed a specific binding of FUS to RNA G-quadruplex structures formed by the post-synaptic density protein 95 (PSD-95) and Shank1 mRNAs [46]. The FUS R495X mutation blocks binding to Shank1 and PSD-95 with high affinity as compared to wild-type FUS [46]. It is possible that DDX17 overexpression exerts protective effects through the regulation of G-quadruplex structure and function. Alternatively, since FUS and DDX17 both contain RGG domains, DDX17 over expression may have a compensatory function which is sufficient to modulate FUS toxicity. However, these possibilities remain to be experimentally tested in the future.
Components of cytoplasmic SGs have been shown to modulate phenotypes associated with various neurodegenerative diseases [18, 26, 29, 50]. In addition, the DEAD-box family of proteins were shown to be global regulators of RNA-containing-phase-separated organelles [42]. Thus, we hypothesized that restoration of DDX17 in the background of mutant FUS may reduce the accumulation of toxic insoluble FUS formation in the cytoplasm. We found that ectopic expression of DDX17 significantly reduced the incorporation of mutant FUS into cytoplasmic SGs. More importantly, we see the biochemical properties of FUS are significantly changed by the mutations, as mutant FUS drastically decreases the overall protein solubility. Upregulation of DDX17 significantly reduces the formation of insoluble FUS species and increases the overall protein solubility (Fig. 4). These findings indicate that upregulation of DDX17 may be reducing the sequestration of mutant FUS into cytoplasmic stress granules through reduction of toxic insoluble FUS formation.
In Drosophila and humans, DDX17 is essential for orchestrating transcription and regulating the splicing profile of cells in early cellular differentiation, as downregulation of DDX17 leads to cell-specific alterations in the splicing profile creating various phenotypic changes [30, 38]. In addition, several mutations in DDX17 (missense and frameshift) have been associated with various cancers involving multiple tissue types. These mutations are thought to lead to disruptions in DDX17 protein function by causing a drastic decrease in the ability of DDX17 to hydrolyze ATP and interact with other RNAs [37, 76, 84]. This suggests possible implications of reduced DDX17 protein function contributing to disease and supports our findings of a loss of function mechanism of DDX17 in FUS-mediated ALS, as upregulation of DDX17 leads to suppression of FUS toxicity. Since DDX17 is a multifunctional protein similar to FUS, it is possible DDX17 and FUS regulate similar cellular pathways, as the complete role of DDX17 in RNA processing is not completely understood. A more mechanistic understanding of the functions of DDX17 and other related RNA-binding proteins like FUS is another area of investigation for future studies.
Recent evidence has brought to light the significant involvement of FUS in the DDR pathway, as multiple documentations of increased DNA damage have been identified in mutant FUS-associated ALS patient tissues and mutant FUS-expressing transgenic mice [75, 81, 99]. Recent work highlighting the impairment of proper DNA damage response signaling caused by mutations in FUS suggests the dire need for possible development of novel therapies involving DNA damage pathways in FUS-mediated ALS [75, 98]. We hypothesized that upregulation of the epigenetic regulator, DDX17, might play a significant role in restoration of the defective DNA damage repair machinery exhibited in FUS-mediated ALS, as DDX17 has been shown to be recruited to DNA damage sites and strongly interacts with DNA damage repair protein HDAC1 [101]. The close interaction of DDX17 and HDAC1 further suggests a role for DDX17 in DDR processes, as FUS was also shown to interact with HDAC1 [99]. Here, we report the first evidence of proof-of-concept studies showing DDX17s involvement in the DDR pathway, as upregulation of DDX17 restores defective DNA damage repair machinery seen in mutant FUS iPSC neurons (Fig. 5). In addition, we found that when DDX17 was upregulated in the background of mutant FUS iPSC neurons, there was a drastic reduction in apoptotic activity, suggesting repairment of DNA damage machinery may be neuroprotective (Fig. 6). To gain further insights into the involvement of DDX17s role in the DDR pathway, we shifted to HEK cells under normal physiological conditions. We downregulated the levels of endogenous DDX17 in control cells, comparable to the levels seen in mutant FUS conditions, and found a significant increase in the number of DSBs in cells expressing shRNA mediated knockdown of DDX17 compared to scrambled control [Supplemental Fig. 8 (Online Resource 1)]. This may imply that increased evidence of DNA damage in FUS-mediated ALS could also be a result of significant downregulation of DDX17 protein. Furthermore, upregulation of DDX17 was also protective against the DNA damage-inducing agent, etoposide, which further supports the role of DDX17s involvement in the DDR pathway. These findings provide novel insights into the functions of DDX17 in the DDR pathway and further supports the proper development of DNA damage repair therapies for FUS-mediated ALS patients.
Collectively, our data support the notion that DDX17 is a novel modifier of FUS toxicity in FUS-expressing human cells. Our in vivo data confirm this claim as genetic modulation of the DDX17 Drosophila orthologues, Rm62 and CG10077, significantly modified the external eye degeneration caused by expression of wild-type and mutant FUS. In addition, upregulation of Rm62 significantly suppressed the motor impairments of FUS-expressing (wild-type and mutant) flies (Fig. 7). Overexpression of numerous wild-type ALS-linked RNA binding proteins including FUS are sufficient to recapitulate pathogenetic pathways in cell and animal models [18, 43, 70, 103, 104]. In addition, upregulation of endogenous FUS protein in human ALS patients has been shown to cause neurodegenerative symptoms [87]. As wild-type FUS also leads to mild toxicity in Drosophila and slight downregulation of endogenous DDX17 in HEK cells, our data suggest restoration of DDX17 via overexpression may be reducing wild-type FUS toxicity through reducing the levels of wild-type FUS overexpression. Notably, the genetic modulation of Rm62 had no effect on the other ALS-causing genes, TDP-43 and C9orf72, suggesting the specificity to FUS-mediated ALS.
In summary, our results are the first to demonstrate that DDX17 may play a significant role in the pathogenesis of FUS-mediated ALS, as it becomes significantly downregulated and trapped in cytoplasmic stress granules. In addition, these results highlight the novel involvements of DDX17 in DNA damage repair and provide a suitable therapeutic target for restoring defective DNA damage repair machinery found in FUS-mediated ALS. Our results are the first to show that DDX17 modifies FUS-associated phenotypes in vivo. In conclusion, our data demonstrate that restoration of DDX17 levels in mutant FUS ALS patients could be an effective therapeutic strategy for reducing toxic insoluble FUS formation and repairing the defective DNA damage repair machinery exhibited by mutant FUS.
Supplementary Material
Acknowledgements
This work was supported by the National Institute on Neurological Disorders and Stroke (NINDS) and National Institute on Aging (NIA) R01 NS081303 (U.B.P.), R21 NS094921, R21 NS101661, R21 NS111768, R21 AG064940, R21 NS100055 (U.B.P. and C.J.D.), R01NS105756 (C.J.D) and NIGMS R35GM119790 (F.S.), Muscular Dystrophy Association (U.B.P), the ALS Association (U.B.P.), and the Robert Packard Center for ALS at Johns Hopkins (U.B.P.). AH is supported by the NOMIS foundation and the Hermann und Lilly Schilling-Stiftung für medizinische Forschung im Stifterverband. RNA Sequencing was performed by The Molecular Resource Center (MRC) at the University of Tennessee Health Sciences Center (UTHSC). We are thankful to Dr. William Taylor, Dr. Lawrence Reiter, and Dr. Daniel Johnson for their assistance with the RNA-sequencing. We are also thankful to Dr. Jacob Schwartz for sharing FUS plasmids for this study. The computational analysis was performed using the high performance cluster hosted by the Center for Research Computing, University of Pittsburgh.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00401-021-02333-z.
References
- 1.Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM et al. (2011) Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res 39(19):8513–8530. 10.1093/nar/gkr488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MM, Wang B, Xia Z, Morales JC, Lu X, Donehower LA et al. (2005) 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 4(12):1854–1861. 10.4161/cc.4.12.2282 [DOI] [PubMed] [Google Scholar]
- 3.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ (2012) A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14(3):318–328. 10.1038/ncb2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti S, Hyman AA (2016) Are aberrant phase transitions a driver of cellular aging? BioEssays 38(10):959–968. 10.1002/bies.201600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso A, Logroscino G, Jick SS, Hernan MA (2009) Incidence and lifetime risk of motor neuron disease in the United Kingdom: a population-based study. Eur J Neurol 16(6):745–751. 10.1111/j.1468-1331.2009.02586.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An H, Skelt L, Notaro A, Highley JR, Fox AH, La Bella V et al. (2019) ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol Commun 7(1):7. 10.1186/s40478-019-0658-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson P, Kedersha N, Ivanov P (2015) Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849(7):861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baradaran-Heravi Y, Van Broeckhoven C, van der Zee J (2020) Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol Dis 134:104639. 10.1016/j.nbd.2019.104639 [DOI] [PubMed] [Google Scholar]
- 9.Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ et al. (2013) Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener 8:30. 10.1186/1750-1326-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian WX, Xie Y, Wang XN, Xu GH, Fu BS, Li S et al. (2019) Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res 47(1):56–68. 10.1093/nar/gky1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F et al. (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol 28(6):420–435. 10.1016/j.tcb.2018.02.00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ Jr et al. (2010) Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19(21):4160–4175. 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brazda V, Haronikova L, Liao JC, Fojta M (2014) DNA and RNA quadruplex-binding proteins. Int J Mol Sci 15(10):17493–17517. 10.3390/ijms151017493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown RH, Al-Chalabi A (2017) Amyotrophic lateral sclerosis. N Engl J Med 377(2):162–172. 10.1056/NEJMra1603471 [DOI] [PubMed] [Google Scholar]
- 15.Buchan JR (2014) MRNP granules. Assembly, function, and connections with disease. RNA Biol 11(8):1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne S, Walsh C, Lynch C, Bede P, Elamin M, Kenna K et al. (2011) Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 82(6):623–627. 10.1136/jnnp.2010.224501 [DOI] [PubMed] [Google Scholar]
- 17.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V et al. (2006) The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 11(4):547–560. 10.1016/j.devcel.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 18.Casci I, Krishnamurthy K, Kour S, Tripathy V, Ramesh N, Anderson EN et al. (2019) Muscleblind acts as a modifier of FUS toxicity by modulating stress granule dynamics and SMN localization. Nat Commun 10(1):5583. 10.1038/s41467-019-13383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Ding X, Akram N, Xue S, Luo SZ (2019) Fused in sarcoma: properties, self-assembly and correlation with neurodegenerative diseases. Molecules. 10.3390/molecules24081622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi B, O’Connell JD, Yamazaki T, Gangopadhyay J, Gygi SP, Reed R (2018) Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II machinery and contains multiple ALS/SMA-causative proteins. Sci Rep 8(1):8755. 10.1038/s41598-018-27136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chio A, Mora G, Calvo A, Mazzini L, Bottacchi E, Mutani R et al. (2009) Epidemiology of ALS in Italy: a 10-year prospective population-based study. Neurology 72(8):725–731. 10.1212/01.wnl.0000343008.26874.d1 [DOI] [PubMed] [Google Scholar]
- 22.Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E et al. (2012) TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287(19):15635–15647. 10.1074/jbc.M111.333450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conte A, Lattante S, Zollino M, Marangi G, Luigetti M, Del Grande A et al. (2012) P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord 22(1):73–75. 10.1016/j.nmd.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 24.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA 108(52):20881–20890. 10.1073/pnas.110943410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T et al. (2012) Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 21(13):2899–2911. 10.1093/hmg/dds116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cziko AM, McCann CT, Howlett IC, Barbee SA, Duncan RP, Luedemann R et al. (2009) Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics 182(4):1051–1060. 10.1534/genetics.109.103234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Da Cruz S, Cleveland DW (2011) Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol 21(6):904–919. 10.1016/j.conb.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daigle JG, Lanson NA Jr, Smith RB, Casci I, Maltare A, Monaghan J et al. (2013) RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet 22(6):1193–1205. 10.1093/hmg/dds526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigle JG, Krishnamurthy K, Ramesh N, Casci I, Monaghan J, McAvoy K et al. (2016) Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity. Acta Neuropathol 131(4):605–620. 10.1007/s00401-015-1530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H et al. (2014) RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep 7(6):1900–1913. 10.1016/j.celrep.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 31.De Santis R, Santini L, Colantoni A, Peruzzi G, de Turris V, Alfano V et al. (2017) FUS mutant human motoneurons display altered transcriptome and microrna pathways with implications for ALS pathogenesis. Stem Cell Rep 9(5):1450–1462. 10.1016/j.stemcr.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Zio D, Cianfanelli V, Cecconi F (2013) New insights into the link between DNA damage and apoptosis. Antioxid Redox Signal 19(6):559–571. 10.1089/ars.2012.4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A et al. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J 29(16):2841–2857. 10.1038/emboj.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fay MM, Lyons SM, Ivanov P (2017) RNA G-quadruplexes in biology: principles and molecular mechanisms. J Mol Biol 429(14):2127–2147. 10.1016/j.jmb.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z et al. (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471(7339):532–536. 10.1038/nature09772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller-Pace FV (2013) The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 1829(8):756–763. 10.1016/j.bba-grm.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 37.Fuller-Pace FV, Moore HC (2011) RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol 7(2):239–251. 10.2217/fon.11.1 [DOI] [PubMed] [Google Scholar]
- 38.Giraud G, Terrone S, Bourgeois CF (2018) Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep 51(12):613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon PH (2013) Amyotrophic lateral sclerosis: an update for 2013 clinical features, pathophysiology, management and therapeutic trials. Aging Dis 4(5):295–310. 10.14336/AD.2013.0400295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerrero EN, Wang H, Mitra J, Hegde PM, Stowell SE, Liachko NF et al. (2016) TDP-43/FUS in motor neuron disease: complexity and challenges. Prog Neurobiol 145–146:78–97. 10.1016/j.pneurobio.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirai Y, Domae E, Yoshikawa Y, Tomonaga K (2020) Differential roles of two DDX17 isoforms in the formation of membraneless organelles. J Biochem 168(1):33–40. 10.1093/jb/mvaa023 [DOI] [PubMed] [Google Scholar]
- 42.Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA et al. (2019) DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573(7772):144–148. 10.1038/s41586-019-1502-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C, Zhou H, Tong J, Chen H, Liu YJ, Wang D et al. (2011) FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet 7(3):e1002011. 10.1371/journal.pgen.1002011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huisman MH, de Jong SW, van Doormaal PT, Weinreich SS, Schelhaas HJ, van der Kooi AJ et al. (2011) Population based epidemiology of amyotrophic lateral sclerosis using capture-recapture methodology. J Neurol Neurosurg Psychiatry 82(10): 1165–1170. 10.1136/jnnp.2011.244939 [DOI] [PubMed] [Google Scholar]
- 45.Humphrey J, Birsa N, Milioto C, McLaughlin M, Ule AM, Robaldo D et al. (2020) FUS ALS-causative mutations impair FUS autoregulation and splicing factor networks through intron retention. Nucleic Acids Res 48(12):6889–6905. 10.1093/nar/gkaa410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imperatore JA, McAninch DS, Valdez-Sinon AN, Bassell GJ, Mihailescu MR (2020) FUS recognizes G quadruplex structures within neuronal mRNAs. Front Mol Biosci 7:6. 10.3389/fmolb.2020.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164(3):487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jalal C, Uhlmann-Schiffler H, Stahl H (2007) Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res 35(11):3590–3601. 10.1093/nar/gkm058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kedersha N, Ivanov P, Anderson P (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 38(10):494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ et al. (2014) Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 46(2):152–160. 10.1038/ng.2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King OD, Gitler AD, Shorter J (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462:61–80. 10.1016/j.brainres.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323(5918):1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- 53.Lagier-Tourenne C, Polymenidou M, Cleveland DW (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19(R1):R46–64. 10.1093/hmg/ddq137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC et al. (2012) Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 15(11):1488–1497. 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, Dillman A et al. (2011) FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging 32(3):550 e1–4. 10.1016/j.neurobiolaging.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lambert MP, Terrone S, Giraud G, Benoit-Pilven C, Cluet D, Combaret V et al. (2018) The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res 46(15):7686–7700. 10.1093/nar/gky545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lanson NA Jr, Pandey UB (2012) FUS-related proteinopathies: lessons from animal models. Brain Res 1462:44–60. 10.1016/j.brainres.2012.01.039 [DOI] [PubMed] [Google Scholar]
- 58.Lanson NA Jr, Maltare A, King H, Smith R, Kim JH, Taylor JP et al. (2011) A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet 20(13):2510–2523. 10.1093/hmg/ddr150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee CT, Chiu YW, Wang KC, Hwang CS, Lin KH, Lee IT et al. (2013) Riluzole and prognostic factors in amyotrophic lateral sclerosis long-term and short-term survival: a population-based study of 1149 cases in Taiwan. J Epidemiol 23(1):35–40. 10.2188/jea.je20120119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lenzi J, De Santis R, de Turris V, Morlando M, Laneve P, Calvo A et al. (2015) ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech 8(7):755–766. 10.1242/dmm.020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Logroscino G, Traynor BJ, Hardiman O, Chio A, Mitchell D, Swingler RJ et al. (2010) Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry 81(4):385–390. 10.1136/jnnp.2009.183525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madabhushi R, Pan L, Tsai LH (2014) DNA damage and its links to neurodegeneration. Neuron 83(2):266–282. 10.1016/j.neuron.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marko M, Vlassis A, Guialis A, Leichter M (2012) Domains involved in TAF15 subcellular localisation: dependence on cell type and ongoing transcription. Gene 506(2):331–338. 10.1016/j.gene.2012.06.088 [DOI] [PubMed] [Google Scholar]
- 64.Marrone L, Poser I, Casci I, Japtok J, Reinhardt P, Janosch A et al. (2018) Isogenic FUS-EGFP iPSC reporter lines enable quantification of FUS stress granule pathology that is rescued by drugs inducing autophagy. Stem Cell Rep 10(2):375–389. 10.1016/j.stemcr.2017.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS (2013) The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 288(34):24731–24741. 10.1074/jbc.M113.497974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masuzawa T, Oyoshi T (2020) Roles of the RGG domain and RNA recognition motif of nucleolin in G-quadruplex stabiliza-tion. ACS Omega 5(10):5202–5208. 10.1021/acsomega.9b04221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matus S, Bosco DA, Hetz C (2014) Autophagy meets fused in sarcoma-positive stress granules. Neurobiol Aging 35(12):2832–2835. 10.1016/j.neurobiolaging.2014.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miguel L, Avequin T, Delarue M, Feuillette S, Frebourg T, Campion D et al. (2012) Accumulation of insoluble forms of FUS protein correlates with toxicity in Drosophila. Neurobiol Aging 33(5):1008 e1–15. 10.1016/j.neurobiolaging.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 69.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S et al. (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17(9):1144–1151. 10.1038/nsmb.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell JC, McGoldrick P, Vance C, Hortobagyi T, Sreedharan J, Rogelj B et al. (2013) Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol 125(2):273–288. 10.1007/s00401-012-1043-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163(1):123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukherjee AK, Sharma S, Chowdhury S (2019) Non-duplex G-quadruplex structures emerge as mediators of epigenetic modifications. Trends Genet 35(2):129–144. 10.1016/j.tig.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakaya T, Maragkakis M (2018) Amyotrophic lateral sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci Rep 8(1):15575. 10.1038/s41598-018-33964-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakaya T, Alexiou P, Maragkakis M, Chang A, Mourelatos Z (2013) FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. RNA 19(4):498–509. 10.1261/rna.037804.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A et al. (2018) Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun 9(1):335. 10.1038/s41467-017-02299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ngo TD, Partin AC, Nam Y (2019) RNA specificity and autoregulation of DDX17, a modulator of microRNA biogenesis. Cell Rep 29(12):4024–4035 e5. 10.1016/j.celrep.2019.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nichols CD, Becnel J, Pandey UB (2012) Methods to assay Drosophila behavior. J Vis Exp. 10.3791/3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Toole O, Traynor BJ, Brennan P, Sheehan C, Frost E, Corr B et al. (2008) Epidemiology and clinical features of amyotrophic lateral sclerosis in Ireland between 1995 and 2004. J Neurol Neurosurg Psychiatry 79(1):30–32. 10.1136/jnnp.2007.117788 [DOI] [PubMed] [Google Scholar]
- 79.Ozdilek BA, Thompson VF, Ahmed NS, White CI, Batey RT, Schwartz JC (2017) Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res 45(13):7984–7996. 10.1093/nar/gkx460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162(5):1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 81.Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S et al. (2014) ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest 124(3):981–999. 10.1172/JCI72723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Renton AE, Chio A, Traynor BJ (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17(1):17–23. 10.1038/nn.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riggs CL, Kedersha N, Ivanov P, Anderson P (2020) Mammalian stress granules and P bodies at a glance. J Cell Sci. 10.1242/jcs.242487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robert F, Pelletier J (2013) Perturbations of RNA helicases in cancer. Wiley Interdiscip Rev RNA 4(4):333–349. 10.1002/wrna.1163 [DOI] [PubMed] [Google Scholar]
- 85.Rowland LP, Shneider NA (2001) Amyotrophic lateral sclerosis. N Engl J Med 344(22):1688–1700. 10.1056/NEJM200105313442207 [DOI] [PubMed] [Google Scholar]
- 86.Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F et al. (2014) PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res 42(1):307–314. 10.1093/nar/gkt835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sabatelli M, Moncada A, Conte A, Lattante S, Marangi G, Luigetti M et al. (2013) Mutations in the 3’ untranslated region of FUS causing FUS overexpression are associated with amyotrophic lateral sclerosis. Hum Mol Genet 22(23):4748–4755. 10.1093/hmg/ddt328 [DOI] [PubMed] [Google Scholar]
- 88.Sama RR, Ward CL, Kaushansky LJ, Lemay N, Ishigaki S, Urano F et al. (2013) FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol 228(11):2222–2231. 10.1002/jcp.24395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spiegel J, Adhikari S, Balasubramanian S (2020) The structure and function of DNA G-quadruplexes. Trends Chem 2(2):123–136. 10.1016/j.trechm.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun S, Ling SC, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S et al. (2015) ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun 6:6171. 10.1038/ncomms7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suzuki H, Shibagaki Y, Hattori S, Matsuoka M (2018) The proline-arginine repeat protein linked to C9-ALS/FTD causes neuronal toxicity by inhibiting the DEAD-box RNA helicase-mediated ribosome biogenesis. Cell Death Dis 9(10):975. 10.1038/s41419-018-1028-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan AY, Manley JL (2010) TLS inhibits RNA polymerase III transcription. Mol Cell Biol 30(1):186–196. 10.1128/MCB.00884-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thandapani P, O’Connor TR, Bailey TL, Richard S (2013) Defining the RGG/RG motif. Mol Cell 50(5):613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 94.Uhlmann-Schiffler H, Rossler OG, Stahl H (2002) The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J Biol Chem 277(2):1066–1075. 10.1074/jbc.M107535200 [DOI] [PubMed] [Google Scholar]
- 95.van Blitterswijk M, Wang ET, Friedman BA, Keagle PJ, Lowe P, Leclerc AL et al. (2013) Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq. PLoS ONE 8(4):e60788. 10.1371/journal.pone.0060788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323(5918):1208–1211. 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waibel S, Neumann M, Rosenbohm A, Birve A, Volk AE, Weishaupt JH et al. (2013) Truncating mutations in FUS/TLS give rise to a more aggressive ALS-phenotype than missense mutations: a clinico-genetic study in Germany. Eur J Neurol 20(3):540–546. 10.1111/ene.12031 [DOI] [PubMed] [Google Scholar]
- 98.Wang H, Hegde ML (2019) New mechanisms of DNA repair defects in fused in sarcoma-associated neurodegeneration: stage set for DNA repair-based therapeutics? J Exp Neurosci 13:1179069519856358. 10.1177/1179069519856358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC et al. (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16(10):1383–1391. 10.1038/nn.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang H, Guo W, Mitra J, Hegde PM, Vandoorne T, Eckelmann BJ et al. (2018) Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in amyotrophic lateral sclerosis. Nat Commun 9(1):3683. 10.1038/s41467-018-06111-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV (2004) The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol 5:11. 10.1186/1471-2199-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolozin B, Ivanov P (2019) Stress granules and neurodegeneration. Nat Rev Neurosci 20(11):649–666. 10.1038/s41583-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia R, Liu Y, Yang L, Gal J, Zhu H, Jia J (2012) Motor neuron apoptosis and neuromuscular junction perturbation are prominent features in a Drosophila model of Fus-mediated ALS. Mol Neurodegener 7:10. 10.1186/1750-1326-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu YF, Gendron TF, Zhang YJ, Lin WL, D’Alton S, Sheng H et al. (2010) Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci 30(32):10851–10859. 10.1523/JNEUR0SCI.1630-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang L, Embree LJ, Tsai S, Hickstein DD (1998) Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem 273(43):27761–27764. 10.1074/jbc.273.43.27761 [DOI] [PubMed] [Google Scholar]
- 106.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D (1997) TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci 110(Pt 15):1741–1750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data that support the findings of this study are available in the Gene Expression Omnibus (GEO) database under accession number GSE173838. Source data are provided with this paper.