Abstract
As a result of a founder effect, a Leigh syndrome variant called Leigh syndrome, French-Canadian type (LSFC, MIM / 220,111) is more frequent in Saguenay–Lac-Saint-Jean (SLSJ), a geographically isolated region on northeastern Quebec, Canada. LSFC is a rare autosomal recessive mitochondrial neurodegenerative disorder due to damage in mitochondrial energy production. LSFC is caused by pathogenic variants in the nuclear gene leucine-rich pentatricopeptide repeat-containing (LRPPRC). Despite progress understanding the molecular mode of action of LRPPRC gene, there is no treatment for this disease.
The present study aims to identify the biological pathways altered in the LSFC disorder through microarray-based transcriptomic profile analysis of twelve LSFC cell lines compared to twelve healthy ones, followed by gene ontology (GO) and pathway analyses.
A set of 84 significantly differentially expressed genes were obtained (p ≥ 0.05; Fold change (Flc) ≥ 1.5). 45 genes were more expressed (53.57%) in LSFC cell lines compared to controls and 39 (46.43%) had lower expression levels. Gene ontology analysis highlighted altered expression of genes involved in the mitochondrial respiratory chain and energy production, glucose and lipids metabolism, oncogenesis, inflammation and immune response, cell growth and apoptosis, transcription, and signal transduction. Considering the metabolic nature of LSFC disease, genes included in the mitochondrial respiratory chain and energy production cluster stood out as the most important ones to be involved in LSFC mitochondrial disorder. In addition, the protein-protein interaction network indicated a strong interaction between the genes included in this cluster. The mitochondrial gene NDUFA4L2 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2), with higher expression in LSFC cells, represents a target for functional studies to explain the role of this gene in LSFC disease.
This work provides, for the first time, the LSFC gene expression profile in fibroblasts isolated from affected individuals. This represents a valuable resource to understand the pathogenic basis and consequences of LRPPRC dysfunction.
Keywords: Leigh syndrome French-Canadian type (LSFC), Gene expression, Microarrays, LRPPRC, NDUFA4L2, Mitochondrial chain respiration, Cytochrome c oxidase, rare diseases, Leigh syndrome
Abbreviations: LSFC, Leigh syndrome, French-Canadian type; SLSJ, Saguenay–Lac-Saint-Jean; LRPPRC, leucine-rich pentatricopeptide repeat-containing; GO, gene ontology; Flc, fold change; NDUFA4L2, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2; ATP, adénosine-5'-triphosphate; OXPHOS, oxidative phosphorylation; COX, cytochrome c-oxidase; DMEM, Dubelcco’s Modified Essential Medium; RMA, robust multi-array analysis; PPI, protein‐protein interaction; ND6, NADH dehydrogenase, subunit 6; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; HES1, hairy and enhancer of split 1; RPL13A, ribosomal protein L13a; qRT-PCR, Real-time PCR; SRA, steroid receptor RNA activator; SLIRP, stem-loop interacting protein; HIF-1, hypoxia inducible factor-1; ETC, electron transport chain; ROS, reactive oxygen species; NAFLD, non-alcoholic fatty liver disease; COPD, chronic obstructive pulmonary disease
1. Introduction
The principal function of the mitochondria is to carry out the oxidative energy metabolism [1], [2]. It produces adénosine-5′-triphosphate (ATP), by oxidative phosphorylation (OXPHOS), that is used by most mammalian cells for growth, survival and regular function [3]. The OXPHOS system is located in the inner mitochondrial membrane and comprises five enzyme complexes (complexes I-V) [4]. More than 150 distinct genetic mitochondrial syndromes have been defined [5]. Leigh syndrome, a metabolic disease affecting 1/40,000 newborn infants worldwide [6], is one of these disorders. It is characterized by a psychomotor regression, hypotonia, ataxia, lactic acidosis and by an estimated mean life expectancy of 3 to 5 years and can be caused by more than 30 genes [2]. A variant known as Leigh Syndrome French-Canadian type (LSFC, MIM / 220,111) was described in the founder population of Saguenay–Lac-Saint-Jean region (SLSJ) of Quebec, Canada where the largest cohort of LSFC patients was identified (56 patients in 2011) [7]. In SLSJ, around 1/2000 births are affected by LSFC and the carrier rate is 1/23 [8], [9]. LSFC is an autosomal recessive form of neurodegenerative congenital lactic acidosis that presents with developmental delay, hypotonia, ataxia, failure to thrive, and mild dysmorphic facial features [8], [10], [11]. It is biochemically characterized by tissue-specific defect in the respiratory chain complex IV (cytochrome c-oxidase, COX). In LSFC individuals, liver and brain are severely affected while fibroblasts and skeletal muscle are 50% affected, and kidney and heart have almost normal activities [10], [11], [12]. LSFC individuals presented also severe and often deadly neurological and/or acidotic crisis [13]. The responsible gene, LRPPRC, encoding for a pentatricopeptide repeat (PPR) family protein, was identified in 2003 [9]. Most SLSJ patients are homozygous for the founder missense mutation p.Ala354Val in exon 9 of this gene. Subsequently, significant advances in understanding the molecular mechanisms of LSFC were succeeded. A low steady state levels of the mutated LRPPRC protein was observed in all LSFC patient tissues [9] resulting in a defect in the translation of most mitochondrial messengers particularly those of the complex IV [14], [15]. Other evidences show implication of LRPPRC in various other diseases ranging from viral to tumour infections [16], [17].
All these recent findings and data illustrate the complexity of the LRPPRC function and the need to identify the downstream dysregulated pathways in LSFC patients and other caused diseases. This is why we conducted the present study on the gene expression profile of LSFC patients cell lines by microarray technology. It revealed several affected pathways and induction of cellular mechanism compensation and raised the possibility of designing novel therapeutic strategies for LSFC patients.
2. Patient and methods
2.1. Patients
Twelve unrelated French-Canadian LSFC patients were included in this study. Their samples were available in the LSFC Consortium Biobank (Université du Québec à Chicoutimi, Saguenay, QC, Canada) and clinical information was extracted from their medical reports (Table 1). Twelve healthy individuals were also recruited in this study and were paired to the LSFC patients according to their age (±3 years) and sex to perform comparative gene expression microarray analysis. The inclusion criteria for the control individuals included no health problems or being affected by diseases that did not involve nervous system degeneration or the mitochondrial respiratory chain. The ethic committee of the Centre intégré universitaire de santé et de services sociaux du SLSJ located in Saguenay, Quebec, Canada approved the study and all individuals (or their parents for affected children) gave informed consent.
Table 1.
Clinical and genetic characteristics of LSFC patients.
| LSFC Patients | Sex | Age | Number of acidosis crisis | Clinical presentation | Mutation in the LRPPRC gene |
|---|---|---|---|---|---|
| 1 | M | 25 | >10 | severe psychomotor delay, hypotonia, non autonomous | p.Ala354Val/p.Cys1,277Xdel8 |
| 2 | M | 25 | 0 | severe psychomotor delay, hypotonia, non autonomous | p.Ala 354Val |
| 3 | F | 23 | 1 | severe psychomotor delay, hypotonia, non autonomous | |
| 4 | M | 6 | 1 | mild psychomotor delay, autonomous | |
| 5 | F | 17 | 0 | mild psychomotor delay, autonomous | |
| 6 | F | 8 | 0 | mild psychomotor delay, autonomous | |
| 7 | F | 4 | 1 | moderate psychomotor delay, autonomous | |
| 8 | F | 21 | 3 | moderate psychomotor delay, hypotonia, semi autonomous | |
| 9 | M | 5 | 0 | mild psychomotor delay | |
| 10 | M | 2 months | 1 | NA | |
| 11 | M | 12 WA | – | NA | |
| 12 | F | 19 WA | – | NA |
WA: week of amenorrhea. NA: not applicable.
2.2. Mutation screening
Genomic DNA was extracted from peripheral blood lymphocytes using the QIAamp DNA Blood Midi kit (Qiagen, ON, Canada) according to the manufacturer's instructions. Total DNA of the participants and their parents when available was used as a template for amplification of the genomic sequences of LRPPRC. LRRPRC segments (including 38 exons and all exon–intron borders) were amplified as previously described [9]. Sequence analyses were performed using Big Dye terminator technology (ABI 3730xl) (Applied Biosystems, ON, Canada) and were analyzed using variant reporter software 2 (Applied Biosystems).
2.3. Cell culture
Skin fibroblasts were obtain from LSFC patients and controls as this tissue is easier to obtain than brain, liver or lung cells. Moreover, the respiratory chain complex IV in LSFC skin fibroblasts is decreased of 50% compared to control cell lines. It was therefore considered a good model for this study. Briefly, primary skin fibroblasts of LSFC participants and age matched control individuals were isolated from cutaneous biopsies and were grown in Dubelcco's Modified Essential Medium (DMEM) rich in glucose and enriched with 10% fetal bovine serum and 100 μl/ml penicillin and streptomycin. Cultures were maintained at 37 °C in a 5% humidified CO2 atmosphere.
2.4. Microarray screening
RNA was isolated from 3 × 106 fibroblasts using RNeasy plus mini kit (Qiagen, Valencia, CA). Microarray analysis was performed with Affymetrix Genechip HG-U133plus2 microarrays containing 54,675 probe sets (Affymetrix, Santa Clara, CA). This chip offers coverage of nearly the entire mitochondrial and nuclear transcriptome, defined by over 47,000 transcripts which, in turn, represent approximately 39,000 genes (www.thermofisher.com). Hybridization and scanning of images were performed at the McGill University and Genome Quebec Innovation Centre (www.genomequebec.mcgill.ca). RNA processing steps (RNA extraction, probe labeling and chip hybridization) were performed in parallel for each pair of control and LSFC samples to minimize technical variability. Nevertheless, the microarrays were performed in two different sets spaced out by 5 years because of the recruitment of new LSFC participants to increase the statistical power of the study. The raw image files (CEL format) generated from the analysis of the scanned image were used for the statistical analysis. The analysis was performed using several packages available in Bioconductor (http://www.bioconductor.org) which uses R language (http://www.R-project.org). We used Affy package to assess artifacts and variability among microarrays and we normalized the probe intensities with robust multi-array analysis (RMA), which includes background correction, quantile normalization, and median polish steps. As batch effects was a parameter to take into account in the microarrays, due to different hybridization dates, we used the inSilicoMerging package with the Empirical Bayes method (COMBAT) to adjust the variance through the microarrays. Finally, Smyth's moderated t-test in Limma package was used to identify the genes that were differentially expressed between the LSFC participants and the control individuals with a cut off of 1.5 for fold change (Flc) and 0.05 for p values.
2.5. Gene ontology and construction of protein-protein interaction network
To understand the functional alterations behind the gene changes in LSFC cells, we performed gene ontology (GO) analyses based on two bioinformatic tools: DAVID (http://david.abcc.ncifcrf.gov) and Panther gene list analysis (http://www.pantherdb.org/). Then, we used STRING database (https://string-db.org) to identify the protein-protein interaction (PPI) networks for both the higher- and under-expressed genes using a combined interaction score of > 0.4 for significant interaction [18] and visualized the results using the network visualization software Cytoscape [19].
2.6. Real-time PCR (qRT-PCR)
To validate differences in gene expression levels observed in microarrays, qRT-PCR was performed on a selected set of genes according to their known functions in mitochondrial activities or glucose metabolism. Four genes were selected: NADH dehydrogenase, subunit 6 (complex I) (ND6), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (NDUFA4L2), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4), and hairy and enhancer of split 1, (Drosophila) (HES1). Reverse transcription of RNA was performed using the qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, Maryland). TaqMan qRT-PCR reaction were performed in 100-wells discs using the Rotor-Gene 6000 (Qiagen/Corbett, Valencia, CA) with the Perfecta qPCR ToughMix (Quanta Biosciences) in a final volume of 20 μl. Each sample was run in triplicate with a negative control. Each gene expression measure was repeated twice. A standard curve was done with three serial dilutions in triplicate for each selected gene and for ribosomal protein L13a (RPL13A), which was selected as housekeeping gene [20]. Quantification obtained from standard curves of each gene was normalized to the relative amount of RPL13A according to the two standard curves method (Rotor-Gene 6 software (version 6.0)). Expression level of each selected gene was measured in the two groups. Data were expressed as mean ± standard error of the mean (SEM) and was compared by Student's t-test. A p value <0.05 was considered significant.
3. Results
3.1. Clinical and mutational diagnosis
Twelve LSFC affected individuals and twelve control participants cell lines were included in this study. LSFC participants were six females and six males aged from 12 weeks of amenorrhea to 25 years. Most participants, among the patients who were born, presented hypotonia, developmental delay, mild facial dysmorphism, and chronic well-compensated metabolic acidosis. Six patients (6/12, 50%) developed one or more acidosis crisis and have survived, exept one who died before the age of five years.
Mutational analysis of LRPPRC gene identified the homozygous founder mutation of missense type c.1,061 C > T transition in exon 9 predicting a missense p.Ala354Val in eleven LSFC individuals. One patient was compound heterozygous; he was heterozygous for the p.Ala354Val amino acid change and for n 8-nt deletion in exon 35 resulting in a premature stop at amino acid 1277 (Table 1).
3.2. Gene expression analysis
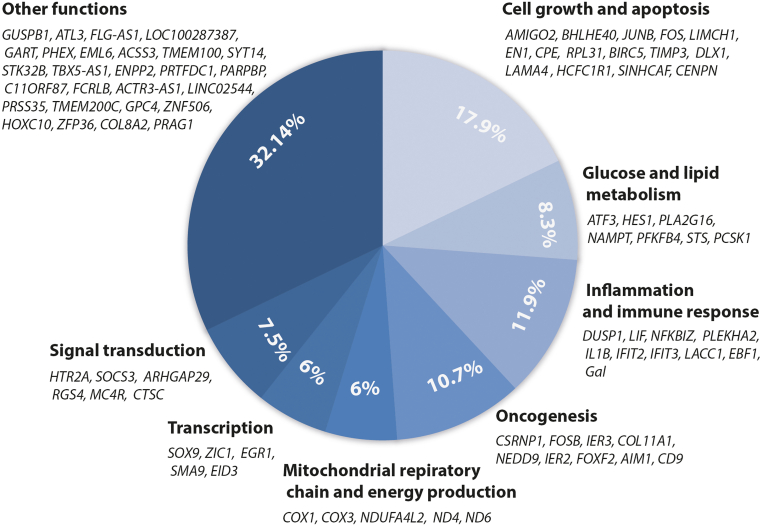
Transcriptional profiling of twelve LSFC affected individuals and twelve control participants fibroblasts was performed using the Affymetrix Genechip HG-U133plus2 chip platform. Microarray gene expression analysis showed significant differences in the expression of 84 genes between LSFC and control fibroblasts (p < 0.05 and Flc > 1.5). Four genes were mitochondrial and the others were nuclear. These differentially expressed genes were classified into eight clusters based on their main function: mitochondrial respiratory chain and energy production (5) , glucose and lipids metabolism (7) , oncogenesis (9) , immune response (10) , cell growth and apoptosis (15) , transcription (5) , signal transduction (6) , and 27 genes with other or not yet known function. Table 2 and Fig. 1 summarize the results of the microarray profiling. Table 3 shows the more and less expressed genes in each cluster. In total, 45 genes were higher expressed and 39 genes were under expressed.
Table 2.
List of genes differentially expressed in LSFC fibroblasts in comparison with healthy controls.
| Clusters | Probe set | ACCNUM | Gene Symbola | Gene name | Cytobandb | p | Flcc | Functiond |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial respiratory chain and energy production | 1553538_s_at | – | COX1 | cytochrome c oxidase subunit I | M | 4.73E-07 | −2.15 | complex IV subunit1 |
| 238199_x_at | – | COX3 | cytochrome c oxidase III | M | 1.96E-13 | −3.22 | complex IV subunit2 | |
| 224372_at | NC_012920.1 | ND4 | NADH Dehydrogenase Subunit 4 | M | 1.69E-06 | −1.57 | complex I subunit3 | |
| 1553575_at | – | *ND6 | NADH dehydrogenase, subunit 6 (complex I) | M | 1.81E-05 | 1.75 | complex I subunit4 | |
| 218484_at | NM_020142 | *NDUFA4L2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | 12q13.3 | 0.04 | 2.02 | complex I inhibition in hypoxia5 | |
| Glucose and lipid metabolism | 202672_s_at | NM_001030287 | ATF3 | activating transcription factor 3 | 1q32.3 | 0.042 | 1.94 | regulation of metabolic homeostasis 6 |
| 203394_s_at | NM_005524 | *HES1 | hairy and enhancer of split 1, (Drosophila) | 3q28-q29 | 0.032 | 2.14 | alpha-glucosidase activator7 | |
| 209581_at | NM_0011282 | PLA2G16 | phospholipase A2, group XVI | 11q12.3 | 0.016 | 1.79 | phospholipase 8 | |
| 243296_at | NAMPT | nicotinamide phosphoribosyltransferase | 7q22.3 | 0.041 | 1.53 | regulation/reprogramming of cellular metabolism9 | ||
| 228499_at | NM_004567 | *PFKFB4 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | 3p22-p21 | 0.03 | 1.55 | activator of glycolysis enzyme 10 | |
| 203767_s_at | AI122754 | STS | steroid sulfatase | Xp22.31 | 0.036 | −1.61 | steroid metabolism11 | |
| 205825_at | NM_000439 | PCSK1 | proprotein convertase subtilisin/kexin type 1 | 5q15 | 0.045 | −2.16 | regulation of glucose homeostasis and food intake12 | |
| Oncogenesis | 225557_at | NM_033027 | CSRNP1 | cysteine-serine-rich nuclear protein 1 | 3p22 | 0.007 | 1.60 | tumor suppressor13 |
| 202768_at | NM_0011141 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 19q13.32 | 0.015 | 2.76 | reduction of Fos and Jun proteins14 | |
| 201631_s_at | NM_003897 | IER3 | immediate early response 3 | 6p21.3 | 0.011 | 1.69 | immune regulation and tumorigenesis15 | |
| 206377_at | NM_001452 | FOXF2 | forkhead box F2 | 6p25.3 | 0.015 | −1.58 | regulation of gene expression in embryonic development, tumoreginicity16 | |
| 212543_at | NM_001624 | AIM1 | absent in melanoma 1 | 6q21 | 0.015 | −1.65 | melanoma suppression17 | |
| 204320_at | NM_0011907 | COL11A1 | collagen, type XI, alpha 1 | 1p21 | 0.019 | 3.79 | stimulation of cancer progression18 | |
| 201005_at | NM_001769 | CD9 | CD9 molecule | 12p13.3 | 0.022 | −1.76 | tumor cell motility and adhesion19 | |
| 202149_at | NM_0011423 | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 | 6p25-p24 | 0.007 | 2.2 | support of oncogenic signaling20 | |
| 202081_at | NM_004907 | IER2 | immediate early response 2 | 19p13.2 | 0.001 | 1.56 | may be involved in the regulation of tumor progression and metastasis21 | |
| Inflammation and immune response | 229487_at | NM_024007 | EBF1 | early B-cell factor 1 | 5q34 | 0.024 | −1.61 | activation of the B cell lineage program22 |
| 201044_x_at | NM_004417 | DUSP1 | dual specificity phosphatase 1 | 5q34 | 0.021 | 1.6 | regulation of anti-inflammatory genes23 | |
| 214240_at | NM015973 | GAL | galanin prepropeptide | 11q13.3 | 0.024 | −1.55 | skin immunity24 | |
| 205266_at | NM_002309 | LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 22q12.2 | 0.029 | 1.53 | anti-inflammatory and pro-gestational activities25 | |
| 223217_s_at | NM_001005474 | NFKBIZ | kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | 3p12-q12 | 0.021 | 1.57 | inflammatory and immune response26 | |
| 238013_at | NM_021623 | PLEKHA2 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 | 8p11.22 | 0.024 | 1.57 | B-cell activation27 | |
| 39402_at | M15330 | IL1B | interleukin 1 beta | 2q14.1 | 0.034 | 1.53 | key mediator of the inflammatory response28 | |
| 226757_at | AA131041 | IFIT2 | interferon induced protein with tetratricopeptide repeats 2 | 10q23.31 | 0.047 | −1.61 | antiviral immune response and innate immunity29 | |
| 229450_at | AI075407 | IFIT3 | interferon induced protein with tetratricopeptide repeats 3 | 10q23.31 | 0.029 | −1.54 | antiviral immune response and innate immunity29 | |
| 1553142_at | NM_153218 | LACC1 | laccase domain containing 1 | 13q14.11 | 0.041 | −1.63 | cytokine secretion and bacterial clearance30 | |
| Cell growth and apoptosis | 222108_at | NM_181847 | AMIGO2 | adhesion molecule with Ig-like domain 2 | 12q13.11 | 0.046 | 1.56 | apoptosis inhibition 31 |
| 202094_at | NM_001012270 | BIRC5 | baculoviral IAP repeat containing 5 | 17q25 | 0.033 | −1.74 | apoptosis inhibition32 | |
| 201147_s_at | NM_000362 | TIMP3 | TIMP metallopeptidase inhibitor 3 | 22q12.3 | 0.036 | −1.54 | apoptosis regulation33 | |
| 201170_s_at | NM_003670 | BHLHE40 | basic helix-loop-helix family, member e40 | 3p26 | 0.002 | 1.57 | chondrocytes differentiation34 | |
| 201473_at | NM_002229 | JUNB | jun B proto-oncogene | 19p13.2 | < 0.001 | 1.92 | control of cell growth and differentiation35 | |
| 209189_at | NM_005252 | FOS | FBJ murine osteosarcoma viral oncogene homolog | 14q24.3 | 0.024 | 2.36 | bone growth36–37 | |
| 242138_at | NM_001038493 | DLX1 | distal-less homeobox 1 | 2q32 | 0.016 | −1.83 | production of forebrain GABAergic interneurons38 | |
| 212327_at | NM_0011127 | LIMCH1 | LIM and calponin homology domains 1 | 4p13 | 0.027 | 2.14 | non muscle myosin-II regulation and cell migration supression39 | |
| 220559_at | NM_001426 | EN1 | engrailed homeobox 1 | 12q23.3 | 0.025 | 1.64 | regulation in early development40 | |
| 202202_s_at | NM_0011052 | LAMA4 | laminin, alpha 4 | 6q21 | 0.01 | −1.53 | constituent of basement membranes41 | |
| 201116_s_at | NM_001873 | CPE | carboxypeptidase E | 4q32.3 | 0.032 | 2.18 | involved in the processing of the majority of neuropeptides and peptide hormones42 | |
| 200962_at | NM_001098577 | RPL31 | ribosomal protein L31 | 2q11.2 | 0.010 | 1.82 | component of the 60S subunit43 | |
| 45714_at | AA436930 | HCFC1R1 | host cell factor C1 regulator 1 | 16p13.3 | 0.015 | 1.5 | cell cycle regulation44 | |
| 222118_at | AK023669 | CENPN | centromere protein N | 16q23.2 | 0.038 | −1.56 | cell cycle regulation45 | |
| 223038_s_at | BG479856 | SINHCAF | SIN3-HDAC complex associated factor | 12p11.21 | 0.046 | 1.52 | cell cycle regulation46 | |
| Transcription | 228531_at | NM_001193307 | SMAD9 | sterile alpha motif domain containing 9 | 7q21.2 | 0.015 | −1.60 | transcriptional regulation in BMP signaling47 |
| 231292_at | NM_0010083 | EID3 | EP300 interacting inhibitor of differentiation 3 | 12q23.3 | 0.026 | −1.62 | transcriptional control of testicular tissue48 | |
| 202935_s_at | NM_000346 | SOX9 | SRY (sex determining region Y)-box 9 | 17q23 | 0.031 | 2.59 | transcription factor49 | |
| 206373_at | NM_003412 | ZIC1 | Zic family member 1 (odd-paired homolog, Drosophila) | 3q24 | 0.014 | 3.03 | transcription factor, differentiation and growth 50 | |
| 201693_s_at | NM_001964 | EGR1 | early growth response 1 | 5q31.1 | 0.001 | 2.31 | regulation of gene transcription51 | |
| Signal transduction | 1558280_s_at | NM_004815 | ARHGAP29 | Rho GTPase activating protein 29 | 1p22.1-p21.3 | 0.014 | −1.62 | regulation of the RhoA-LIMK-cofilin pathway52 |
| 207135_at | NM_000621 | HTR2A | 5-hydroxytryptamine (serotonin) receptor 2A | 13q14-q21 | 0.039 | 1.8 | serotonin receptor53 | |
| 221467_at | NM_005912 | MC4R | melanocortin 4 receptor | 18q22 | 0.004 | −1.98 | key regulator of energy homeostasis, food intake and body weight54 | |
| 225647_s_at | NM_0011141 | CTSC | cathepsin C | 11q14.2 | 0.002 | −3.13 | activation of granule serine proteases55 | |
| 227697_at | NM_003955 | SOCS3 | suppressor of cytokine signaling 3 | 17q25.3 | 0.018 | 1.57 | suppressor of cytokine signaling56 | |
| 204338_s_at | NM_005613 | RGS4 | regulator of G protein signaling 4 | 1q23.3 | 0.043 | 1.89 | cell Signaling57 | |
| Other functions | 236532_at | NM_207645 | C11orf87 | chromosome 11 open reading frame 87 | 11q22.3 | 0.025 | −2.09 | not known |
| 235888_at | NR027026 | GUSPB1 | glucuronidase, beta pseudogene 1 | 5p14.3 | 0.004 | 1.51 | not known | |
| 238452_at | NM_001002901 | FCRLB | Fc receptor-like B | 1q23.3 | 0.021 | −1.72 | not known | |
| 237075_at | AI191591 | ACTR3-AS1 | ACTR3 antisense RNA 1 | 2q14.1 | 0.001 | 1.67 | not known | |
| 223453_s_at | BC005096 | ATL3 | atlastin GTPase 3 | 11q13.1 | 0.005 | −1.6 | GTPase58 | |
| 1561141_at | AF086258 | LINC02544 | long intergenic non-protein coding RNA 2544 | 6q27 | 0.015 | 1.94 | not known | |
| 235874_at | AL574912 | PRSS35 | serine protease 35 | 6q14.2 | 0.016 | 1.7 | not known | |
| 241014_at | H09620 | FLG-AS1 | FLG antisense RNA 1 | 1q21.3 | 0.017 | −1.56 | not known | |
| 229523_at | N66694 | TMEM200C | transmembrane protein 200C | 18p11.31 | 0.017 | 1.53 | not known | |
| 217220_at | AL050153 | LOC100287387 | uncharacterized LOC100287387 | 2q37.3 | 0.019 | −1.59 | not known | |
| 230097_at | AI207338 | GART | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 21q22.11 | 0.02 | −1.52 | purine synthesis59 | |
| 239229_at | AI342246 | PHEX | phosphate regulating endopeptidase homolog X-linked | Xp22.11 | 0.03 | −1.69 | not known | |
| 229656_s_at | AA236463 | EML6 | EMAP like 6 | 2p16.1 | 0.03 | −1.62 | not known | |
| 229222_at | AI123815 | ACSS3 | acyl-CoA synthetase short chain family member 3 | 12q21.31 | 0.031 | −1.62 | not known | |
| 204984_at | NM_001448 | GPC4 | glypican 4 | Xq26.2 | 0.032 | 1.51 | not known | |
| 1568720_at | BC018100 | ZNF506 | zinc finger protein 506 | 19p13.11 | 0.033 | 1.58 | not known | |
| 218959_at | NM_017409 | HOXC10 | homeobox C10 | 2q13.13 | 0.034 | 2.12 | not known | |
| 219230_at | NM_018286 | TMEM100 | transmembrane protein 100 | 17q22 | 0.039 | −1.64 | not known | |
| 1553654_at | NM_153262 | SYT14 | synaptotagmin 14 | 1q32.2 | 0.041 | −1.57 | not known | |
| 201531_at | NM_003407 | ZFP36 | ZFP36 ring finger protein | 19q13.2 | 0.042 | 1.55 | not known | |
| 219686_at | NM_018401 | STK32B | serine/threonine kinase 32B | 4p16.2 | 0.046 | −1.61 | not known | |
| 233947_s_at | U47671 | TBX5-AS1 | TBX5 antisense RNA 1 | 12q24.21 | 0.047 | −2.16 | not known | |
| 221900_at | AI806793 | COL8A2 | collagen type VIII alpha 2 chain | 1p34.3 | 0.047 | 1.62 | Not known | |
| 210839_s_at | D45421 | ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 8q24.12 | 0.048 | −1.57 | not known | |
| 222803_at | AI871620 | PRTFDC1 | phosphoribosyl transferase domain containing 1 | 10p12.1 | 0.048 | −1.5 | not known | |
| 227928_at | AI224977 | PARPBP | PARP1 binding protein | 12q23.2 | 0.049 | −1.5 | not known | |
| 235085_at | BF739767 | PRAG1 | PEAK1 related, kinase-activating pseudokinase 1 | 8p23.1 | 0.036 | 1.59 | not known |
1.Dennerlein, S et al. J Cell Sci 2015, 128 (5), 833–7. 2.Remacle, C et al. Plant Mol Biol 2010, 74 (3), 223–33. 3.Alharbi, M. A et al. Biomed Rep 2019, 11 (6), 257–268. 4.Bai, Y et al. EMBO 1998, 17 (16), 4848–58. 5.Tello, D et al. Cell Metab 2011, 14 (6), 768–79. 6.Ku, H. C et al. Front Endocrinol (Lausanne) 2020, 11, 556. 7.Yan, B et al. J Biol Chem 2001, 276 (3), 1789–93. 8.Xiong, S et al. Proc Natl Acad Sci U S A 2014, 111 (30), 11,145–50. 9.Audrito, V et al. Front Oncol 2020, 10, 358. 10.Wang, G et al. Biochem Biophys Res Commun 2020, 526 (4), 978–985. 11.Reed, M. J et al. Endocr Rev 2005, 26 (2), 171–202. 12.Muhsin, N. I. A et al. Mamm Genome 2020, 31 (1-2), 17–29. 13.Ishiguro, H et al. Oncogene 2001, 20 (36), 5062–6. 14.Nakabeppu, Y et al. Cell 1991, 64 (4), 751–9. 15.Arlt, A et al. Eur J Cell Biol 2011, 90 (6-7), 545–52. 16.He, W et al. Cell Death Dis 2020, 11 (6), 424. 17.Trent, J. M et al. Science 1990, 247 (4942), 568–71. 18.Patra, R et al. Front Genet 2021, 12, 608,313. 19.Maecker, H. T et al. FASEB 1997, 11 (6), 428–42. 20.Gabbasov, R et al. Oncogene 2018, 37 (35), 4854–4870. 21.Wu, W et al. Int J Mol Med 2015, 36 (4), 1104–10. 22.Nechanitzky, R et al. Nat Immunol 2013, 14 (8), 867–75. 23.Hoppstadter, J et al. Front Immunol 2019, 10, 1446. 24.Bauer, J. W et al. Cell Mol Life Sci 2008, 65 (12), 1820–5. 25.Hamelin-Morrissette, J et al. Mol Immunol 2020, 120, 32–42. 26.Yamamoto, M et al. Nature 2004, 430 (6996), 218–22. 27.Costantini, J. L et al. Blood 2009, 114 (21), 4703–12. 28.Lopez-Castejon, G et al. Cytokine & growth factor reviews 2011, 22 (4), 189–95. 29.Pidugu, V. K et al. Front Mol Biosci 2019, 6, 148. 30.Lahiri, A et al. Nat Commun 2017, 8, 15,614. 31.Ono, T et al. J Neurosci 2003, 23 (13), 5887–96. 32.Ambrosini, G et al. Nat Med 1997, 3 (8), 917–21. 33.Hojilla, C. V et al. PloS one 2011, 6 (10), e26718. 34.Shen, M et al. Biochem Biophys Res Commun 1997, 236 (2), 294–8. 35.Yang, M. Y et al. Blood 2003, 101 (8), 3205–11. 36.Dony, C et al. Nature 1987, 328 (6132), 711–4. 37.Amary, F et al. Am J Surg Pathol 2019, 43 (12), 1661–1667. 38.Cobos, I et al. Nat Neurosci 2005, 8 (8), 1059–68. 39.Lin, Y. H et al. Mol Biol Cell 2017, 28 (8), 1054–1065. 40.Logan, C et al. Dev Genet 1992, 13 (5), 345–58. 41.Richards, A. J et al. Genomics 1994, 22 (1), 237–9. 42.Alsters, S. I et al. PloS one 2015, 10 (6), e0131417. 43.Fabijanski, S et al. Mol Gen Genet 1981, 184 (3), 551–6. 44.Mahajan, S. S et al. J Biol Chem 2002, 277 (46), 44,292–9. 45.Oka, N et al. J Cancer 2019, 10 (16), 3728–3734. 46.Munoz, I. M et al. J Biol Chem 2012, 287 (39), 32,346–53. 47.Tsukamoto, S et al. Sci Rep 2014, 4, 7596. 48.Bavner, A et al. Nucleic Acids Res 2005, 33 (11), 3561–9. 49.Sudbeck, P et al. Nat Genet 1996, 13 (2), 230–2. 50.Salero, E et al. J Biol Chem 2001, 276 (3), 1881–8. 51.Havis, E et al. . Int J Mol Sci 2020, 21 (5). 52.Qiao, Y et al. Cell Rep 2017, 19 (8), 1495–1502. 53.Choi, J. R et al. BMC Med Genet 2020, 21 (1), 5. 54.Doulla, M et al. Paediatr Child Health 2014, 19 (10), 515–8. 55.Romero-Quintana, J. G et al. BMC Med Genet 2013, 14, 7. 56.Hill, G. R et al. Blood 2010, 116 (2), 287–96. 57.Stratinaki, M et al. Proc Natl Acad Sci U S A 2013, 110 (20), 8254–9. 58.Steiner, B et al. Commun Integr Biol 2018, 11 (2), 1–5. 59.Li, T et al. J Cell Mol Med 2020, 24 (12), 6704–6715.
Genes marked by an asterisk were selected to be tested by real-time PCR (qRT-PCR).
Gene location obtained from National Center for Biotechnology Information public database (http://www.ncbi.nlm.nih.gov).
Fold-changes (Flc) are indicated for each probe set significantly more or less expressed between LSFC and control fibroblasts (p < 0.05; absolute Flc > 1.5). Positive data indicate that the genes are more expressed by LSFC fibroblasts; negative data indicate that the genes are less expressed by LSFC fibroblasts.
References that allow classification of differentially expressed genes in function categories:
Fig. 1.
Differentially expressed genes clusters according to their molecular function Comparison of gene expression profile of twelve paired LSFC and controls cell lines (fibroblasts) by microarrays showed a set of 84 significant differentially expressed genes (Flc ≥ 1.5 and p ≤ 0.05). Based on the molecular function of these genes, they were classified on seven clusters: mitochondrial respiratory chain and energy production (5), glucose and lipids metabolism (7), oncogenesis (9), immune response (10), cell growth and apoptosis (15), transcription (5), signal transduction (6), and 27 genes with other not yet known function.
Table 3.
List of higher and under expressed genes in LSFC patients.
| Clusters | Higher expressed genes | Under expressed genes |
|---|---|---|
| Mitochondrial respiratory chain and energy production | ND6, NDUFA4L2 | COX1, COX3, ND4 |
| Glucose and lipid metabolism | ATF3, HES1, PLA2G16, NAMPT, PFKFB4 | STS, PCSK1 |
| Oncogenesis | CSRNP1, FOSB, IER3, COL11A1, NEDD9, IER2 | FOXF2, AIM1, CD9 |
| Inflammation and immune response | DUSP1, LIF, NFKBIZ, PLEKHA2, IL1B | EBF1, Gal, IFIT2, IFIT3, LACC1 |
| Cell growth and apoptosis | AMIGO2, BHLHE40, JUNB, FOS, LIMCH1, EN1, CPE, RPL31, HCFC1R11, SINHCAF | BIRC5, TIMP3, DLX1, LAMA4, CENPN |
| Transcription | SOX9, ZIC1, EGR1 | SMA9, EID3 |
| Signal transduction | HTR2A, SOCS3, RGS4 | ARHGAP29, MC4R, CTSC |
| Other functions | GUSPB1, ACTR3-AS1, LINC02544, PRSS35, TMEM200C, GPC4, ZNF506, HOXC10, ZFP36, COL8A2, PRAG1 | C11orf87, FCRLB, ATL3, FLG-AS1, LOC100287387, GART, PHEX, EML6, ACSS3, TMEM100, SYT14, STK32B, TBX5-AS1, ENPP2, PRTFDC1, PARPBP |
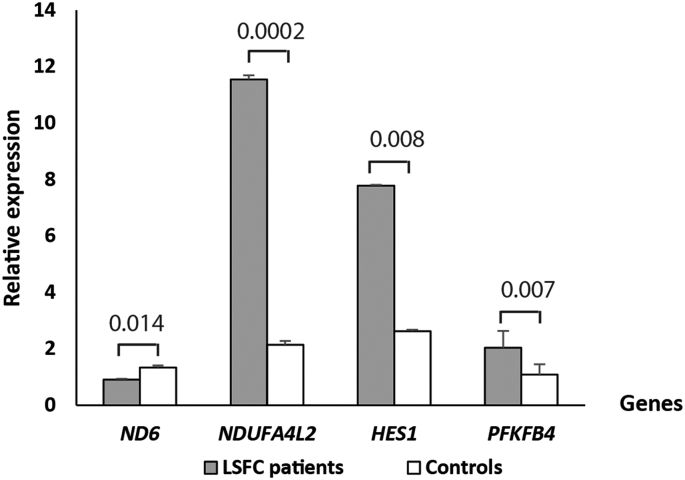
Four genes were analyzed by qRT-PCR to confirm the gene expression data obtained from microarray analysis (ND6, NDUFA4L2, PFKFB4 and HES1) on the twelve LSFC and twelve control fibroblasts. The qRT-PCR results were agreeing with the microarray data except for ND6 which was found to be higher expressed in microarrays but under expressed in qRT-PCR results (Fig. 2).
Fig. 2.
Expression of the four selected genes using real-time PCR (qRT-PCR). NADH dehydrogenase, subunit 6 (complex I) (ND6), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (NDUFA4L2), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4), and hairy and enhancer of split 1, (Drosophila) (HES1) mRNA was extracted from skin fibroblasts of LSFC (gray bars) and paired controls (white bars) individuals. Measure of the mRNA expression by real-time RT-PCR was done twice in triplicate with negative control and normalized to RPL13A expression using two-standard curves method. Data are expressed as mean + SEM values. NDUFA4L2, PFKFB4, and HES1 mRNA level are significantly (p < 0.05) higher in LSFC skin fibroblasts participants compared with controls.
3.3. Protein-protein interactions network of the differentially expressed genes
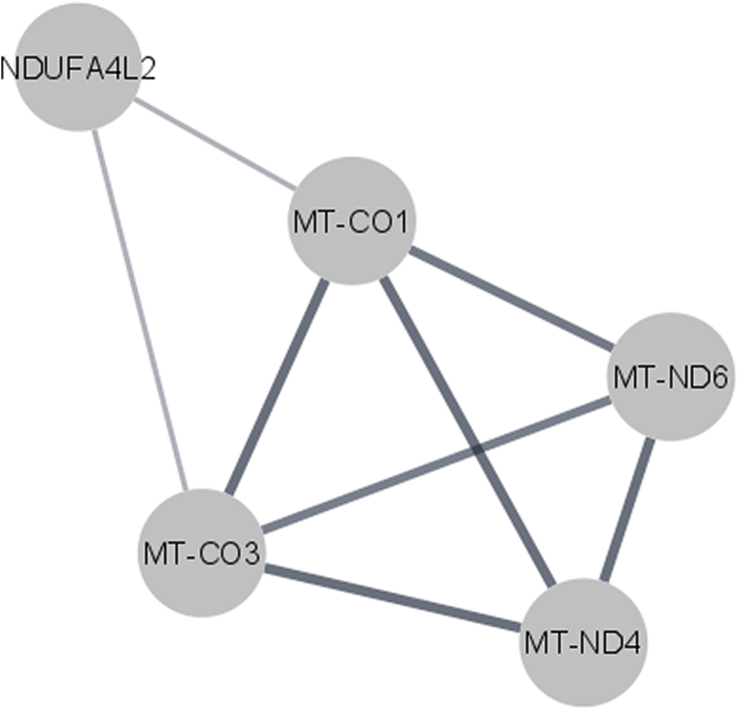
Functional annotation and pathway profiling of the differentially regulated genes, using DAVID, Panther and STRING online database, provided an overview of the molecular function of each gene and its potential involvement in biological and cellular processes. STRING PPI network showed a strong interaction between the protein NDUFA4L2 and proteins of the other dysregulated mitochondrial respiratory chain ND6, ND4, COX1, and COX3 (Fig. 3).
Fig. 3.
Protein protein interactions network. Network analysis of dysregulated genes was performed using STRING database, considering a combined interaction score > 0.4 cut off for significant interaction. A strong interaction between the differentially expressed genes of the mitochondrial and energy production cluster was observed.
4. Discussion
Currently, the pathogenic mechanisms underlying LSFC disease remain unclear and no cure exists. The unique available option to reduce the high-energy demands of digestion is eating several small meals throughout the day. The responsible gene for LSFC disorder, LRPPRC, was discovered in 2003 [9]. The encoded protein LRPPRC belongs to the family of pentatricopeptide repeat proteins that is involved in post transcriptional mitochondrial gene expression. LRPPRC regulates the stability and handling of mature messenger RNAs. In mitochondria, LRPPRC forms a mitochondrial ribonucleoprotein complex with steroid receptor RNA activator (SRA) stem-loop interacting protein (SLIRP) [10]. This complex controls polyadenylated mRNAs and is required for mitochondrial mRNA stability [21]. As shown in Table 1, two LRPPRC mutations have been identified in the studied LSFC individuals: the transition c.1061C > T (p.Ala354Val) and p.Cys1277Xdel8. The missense variation p.Ala354Val is identified in 95% of the cases of LSFC in SLSJ. The carrier rate of this is variant in the SLSJ region is 1/23. A carrier-screening test for this founder mutation has become routinely offered to couples with SLSJ ancestry. It was shown that LSFC fibroblasts present several mitochondrial functional abnormalities including reduced mitochondrial membrane potential, fragmentation of the mitochondrial network, and impaired OXPHOS capacity [13]. In mice harboring an hepatocyte-specific inactivation of Lrpprc, it was observed an alteration of the mitochondrial pemeability transition pore and of the lipid composition of mitochondrial membranes [22].
Little is currently known about the sequences of biological pathways altered in LSFC patients. We conducted a microarray gene expression of twelve LSFC patient primary fibroblasts compared to twelve control ones paired for age and sex in order to better understand the functional impact of LRPPRC gene mutations and the molecular mechanisms linking the LRPPRC mutations to the LSFC disorder.
The microarray gene expression analysis showed 84 significant differentially expressed genes (p value<0.05 and Flc > 1.5) between the LSFC and control cells lines. These genes are implicated in several cellular deregulated processes including the mitochondrial respiratory chain and energy production, glucose and lipid metabolism, oncogenesis, cell growth and apoptosis, inflammation and immune response, signaling transduction and transcription. Considering the mitochondrial type of LSFC disorder and the known LRPPRC role in mitochondrial mRNA stability, we think that the potential altered genes, related to the LSFC disorder, are those implicated in the mitochondrial function. This cluster includes two genes encoding for complex IV subunits (COX1 and COX3), two genes encoding for complex I subunits (ND4 and ND6) and NDUFA4L2 gene whose function and association with respiratory chain complexes remains obscure. COX1, COX3, ND6 and ND4 were under expressed, whereas NDUFA4L2 is higher expressed in LSFC cells.
These results are partially in agreement with the previous study of Xu et al. (2004) showing that LRPPRC is required for the expression of COX1 and COX3 [14].
NDUFA4L2 is expressed two times more in LSFC fibroblasts compared to control fibroblasts (p = 0.04; Flc = 2.02). NDUFA4L2 protein is the target of the hypoxia inducible factor-1 (HIF-1) gene, which is activated in low oxygen conditions. It has been shown that NDUFA4L2, in hypoxic conditions, inhibits electron transport chain (ETC) activity and this reduces mitochondria oxygen consumption, which limits intracellular reactive oxygen species production and plays an important role in the control of glycolysis and glucose oxidation [23] [24]. Consequently, NDUFA4L2 can mediate the function of oxidative phosphorylation and reactive oxygen species (ROS) production in mitochondria. In the case of LSFC patients for which we observed an increase of NDUFA4L2 expression, we hypothesize that COX deficiency could lead to relative hypoxia similar to the one induced by HIF-1. Consequently, NDUFA4L2 expression is induced which could counterbalance the oxygen decrease by preventing the overloading of the respiratory chain, thus resulting in metabolic acidosis.
Moreover, other researchers showed that loss of LRPPRC function in LSFC fibroblasts displayed primarily a COX deficiency and a global reduction in the steady-state levels of all mitochondrial mRNAs except ND3 and ND6 [10], [11]. Indeed, ND6 mRNA lacks poly A tail that is why its steady-state level was shown to not be changed in the absence of LRPPRC in the mouse heart [21].The present microarrays expression results showed a variable expression of ND6 gene in LSFC fibroblasts compared to control ones. We think that ND6 expression may be variable between heart and fibroblasts specially that it was shown that usually heart is less affected in LSFC patients [10], [12]. Nevertheless, ND6 gene was higher expressed in microarrays (p = 3.34E-05; Flc = 1.72) and under expressed in qRT-PCR (p = 0.014; Flc = 1.46). This contradiction could be explained by the fact that the microarrays were performed in two different sets spaced out by 5 years and were not all carried out at the same time nor by the same manipulator.
STRING showed strong interactions between COX1, COX3, ND4, ND6 and NDUFA4L2, such interaction is crucial to induce an adaptive response of mitochondria in LSFC cells (Fig. 3). This is in agreement with a previously study showing that LSFC fibroblasts preserved ATP levels in basal conditions, suggesting the activation of a compensatory mechanism [13].
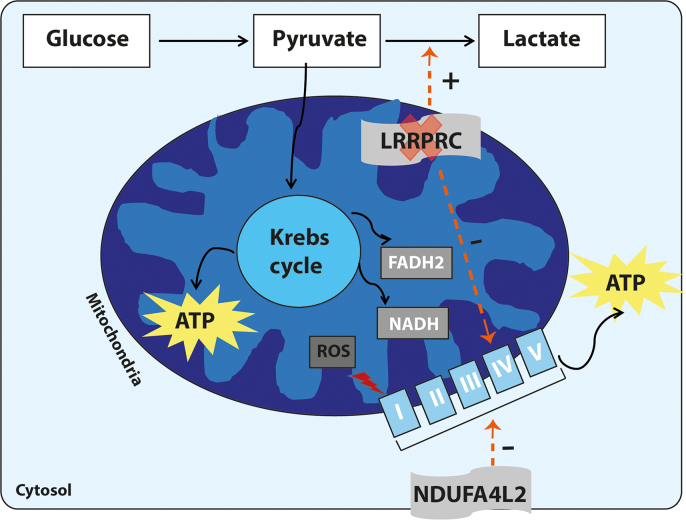
Based on the present microarray results analysis, we hypothesize that in LSFC fibroblasts, LRPPRC loss causes a COX deficiency by decreasing its two subunits COX1 and COX3. This could lead to relative hypoxia that induced the expression of NDUFA4L2. NDUFA4L2 attenuates mitochondrial oxygen consumption by ETC inhibition via the decreasing expression of ND4 subunit. This reduces function of the transcription/translation mitochondrial machinery, and limits the intracellular reactive oxygen species production under low-oxygen conditions [23] (Fig. 4).
Fig. 4.
Depiction of the respiratory chain defects in LSFC patients. The five mitochondrial complexes are shown embedded in the inner mitochondrial membrane and called I, II, III, IV, and V. Loss of LRPPRC decreases the activity of the mitochondrial complex IV that results in accumulation of reactive oxygen species (ROS) in the mitochondria. As an adaptative mechanism, cells switch away from mitochondrial ATP production toward glycolysis, a necessary adaptation to the loss of mitochondrial respiratory capacity in LSFC cells leading to increasing level of blood lactic acid. This will cause hypoxia condition that increases the expression of the NDUFA4L2 gene. NDUFA4L2 decreases oxygen consumption by inhibiting the electron transport chain activity.
Mitochondrial respiration is crucial for cellular metabolic function. In normal cells, LRPPRC promotes fatty acid uptake and oxidation of hepatocytes by increasing oxidative phosphorylation activity, which limit blood lipid level and interdicts non-alcoholic fatty liver disease (NAFLD) in mice [25]. In LSFC disorder, many perturbations were observed in fatty acid metabolism in mitochondria [26] as well as a lipid dyshomeostasis [27]. Indeed, loss of LRPPRC caused oxidative phosphorylation deficiency and decreased the capacity to oxidize fatty acids. In the present work, we observed higher expression of several genes involved in lipid and glucose metabolism as PFKFB4 gene encoding for an activator of glycolysis enzyme and PLA2G16, a phospholipase. The increased expression of glycolytic and lipidic genes may in part represent a biochemical adaptation to compensate for the loss of mitochondrial ATP production by enhancing glycolytic ATP production. Previous studies have shown increased expression of genes involved in glycolysis in mitochondrial DNA mutant cells [28], [29]. The higher expression of these genes may in part lead to a metabolic switch away from mitochondria toward glycolysis, a necessary adaptation to the loss of mitochondrial respiratory capacity in LSFC cells.
Non-targeted lipidomic analysis was also performed and thirty-three distinct lipids were shown to be altered in H-Lrpprc−/− mice mitochondria indicating that LRPPRC deficiency leads to changes in the lipid composition of mitochondrial membranes [22].
The present LSFC gene expression profile analysis showed also a dysregulation of the expression of several genes involved in tumor progression and cancer. This is not surprising as recent studies have shown that LRPPRC expression increases in various cancer tissues and tumor cell lines, including prostate cancer [30], [31], [32], gastric cancer [16], and lung adenocarcinoma [16], [33]. Further experiments are needed to explore the eventual implication of these oncogenesis genes in LSFC disorder.
We also observed the altered expression of genes involved in cell growth and apoptosis, inflammation and immune response, transcription and transduction signaling. These pathways are in majority a result of the mitochondrial respiratory chain defect. It was reported that the reactive oxygen species are a major activator of apoptosis that has been linked with oxidative stress in acute respiratory distress syndrome, chronic obstructive pulmonary disease (COPD and lung fibrosis [34], [35], [36], [37], [38]. Interestingly, a close link was observed between oxidative stress and inflammatory responses [38].
5. Conclusion
In summary, the present study used global high-throughput microarray analysis together with bioinformatics-assisted functional clustering to identify the expression profile in LSFC patients cell lines. Our data demonstrates that LSFC fibroblasts present a series of adaptations to potentially overcome the decrease in mitochondrial respiration. A set of interesting differentially expressed genes in LSFC patients was identified. Specifically, genes involved in the mitochondrial chain respiratory, seem to be directly involved in the LSFC disease. The present work provides a better understanding of the biological pathways altered in LSFC disorder. Nevertheless, the downregulation of LRPPRC expression is tissue specific, that is why, these data cannot be extrapolated to other tissues such as brain and liver, which have different energetic metabolism. Further functional gene expression studies in these tissue cells are required to strengthen the significance of our findings in the biology of LSFC disorder.
Author contributions
CL build and manage the LSFC biobank, design the study and reach the financial support, supervise trainee and research staff, edit paper and approval the final version. CM is a pediatrician involved in patient recruitment and sampling and revised the paper. JT performed experiments and participated in data analysis. MB participated in data analysis and interpretation. MB and JT wrote the first draft of the manuscript.
Declaration of Competing Interest
The authors declare that there is no conflict of interest.
Acknowledgements
The LSFC Consortium supported by CIHR grants dedicated to investigating the causes of and developing treatments for LSFC. The authors thank the participants and their families for their valuable participation in this study. Catherine Laprise and Mbarka Bchetnia are part of the Quebec ThéCell Network (http://www.reseauthecell.qc.ca). The authors would like to thank Vanessa Tremblay-Vaillancourt for her technical work in biobanking, Anne-Marie Madore and Anne-Marie Boucher-Lafleur for their involvement respectively as research professional and research technician, and Dr. Grant Mitchell for the recruitment, evaluation and sampling of the LSFC patients from the Centre hospitalier universitaire (CHU) Sainte-Justine.
References
- 1.Shoubridge E.A. Nuclear gene defects in respiratory chain disorders. Semin. Neurol. 2001;21(3):261–267. doi: 10.1055/s-2001-17943. [DOI] [PubMed] [Google Scholar]
- 2.Lee N., Daly M.J., Delmonte T., Lander E.S., Xu F., Hudson T.J., et al. A genomewide linkage-disequilibrium scan localizes the Saguenay-lac-saint-Jean cytochrome oxidase deficiency to 2p16. Am. J. Hum. Genet. 2001;68(2):397–409. doi: 10.1086/318197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duchen M.R. Mitochondria and calcium: from cell signalling to cell death. J. Physiol. 2000;529(Pt 1):57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alston C.L., Rocha M.C., Lax N.Z., Turnbull D.M., Taylor R.W. The genetics and pathology of mitochondrial disease. J. Pathol. 2017;241(2):236–250. doi: 10.1002/path.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vafai S.B., Mootha V.K. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 6.Rahman S., Blok R.B., Dahl H.H., Danks D.M., Kirby D.M., Chow C.W., et al. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann. Neurol. 1996;39(3):343–351. doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- 7.Debray F.G., Morin C., Janvier A., Villeneuve J., Maranda B., Laframboise R., et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 2011;48(3):183–189. doi: 10.1136/jmg.2010.081976. [DOI] [PubMed] [Google Scholar]
- 8.Morin C., Mitchell G., Larochelle J., Lambert M., Ogier H., Robinson B.H., et al. Clinical, metabolic, and genetic aspects of cytochrome C oxidase deficiency in Saguenay-lac-saint-Jean. Am. J. Hum. Genet. 1993;53(2):488–496. [PMC free article] [PubMed] [Google Scholar]
- 9.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U. S. A. 2003;100(2):605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E.A. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21(8):1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasarman F., Nishimura T., Antonicka H., Weraarpachai W., Shoubridge E.A. Tissue-specific responses to the LRPPRC founder mutation in french Canadian Leigh syndrome. Hum. Mol. Genet. 2015;24(2):480–491. doi: 10.1093/hmg/ddu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merante F., Petrova-Benedict R., MacKay N., Mitchell G., Lambert M., Morin C., et al. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-lac-saint-Jean region of Quebec. Am. J. Hum. Genet. 1993;53(2):481–487. [PMC free article] [PubMed] [Google Scholar]
- 13.Burelle Y., Bemeur C., Rivard M.E., Thompson Legault J., Boucher G., Morin C., et al. Mitochondrial vulnerability and increased susceptibility to nutrient-induced cytotoxicity in fibroblasts from leigh syndrome french Canadian patients. PloS one. 2015;10(3) doi: 10.1371/journal.pone.0120767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F., Morin C., Mitchell G., Ackerley C., Robinson B.H. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. The Biochemical journal. 2004;382(Pt 1):331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gohil V.M., Nilsson R., Belcher-Timme C.A., Luo B., Root D.E., Mootha V.K. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285(18):13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian T., Ikeda J., Wang Y., Mamat S., Luo W., Aozasa K., et al. Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur. J. Cancer. 2012;48(15):2462–2473. doi: 10.1016/j.ejca.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Cui J., Wang L., Ren X., Zhang Y., Zhang H. LRPPRC: a multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 2019;10:595. doi: 10.3389/fphys.2019.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Camara Y., et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31(2):443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuillerier A., Honarmand S., Cadete V.J.J., Ruiz M., Forest A., Deschenes S., et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Hum. Mol. Genet. 2017;26(16):3186–3201. doi: 10.1093/hmg/ddx202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tello D., Balsa E., Acosta-Iborra B., Fuertes-Yebra E., Elorza A., Ordonez A., et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting complex I activity. Cell Metab. 2011;14(6):768–779. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Mukaneza Y., Cohen A., Rivard M.E., Tardif J., Deschenes S., Ruiz M., et al. mTORC1 is required for expression of LRPPRC and cytochrome-c oxidase but not HIF-1alpha in Leigh syndrome french Canadian type patient fibroblasts. Am. J. Physiol. Cell Physiol. 2019;317(1):C58–C67. doi: 10.1152/ajpcell.00160.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akie T.E., Liu L., Nam M., Lei S., Cooper M.P. OXPHOS-mediated induction of NAD+ promotes complete oxidation of fatty acids and interdicts non-alcoholic fatty liver disease. PloS one. 2015;10(5) doi: 10.1371/journal.pone.0125617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson Legault J., Strittmatter L., Tardif J., Sharma R., Tremblay-Vaillancourt V., Aubut C., et al. A metabolic signature of mitochondrial dysfunction revealed through a monogenic form of Leigh syndrome. Cell Rep. 2015;13(5):981–989. doi: 10.1016/j.celrep.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz M., Cuillerier A., Daneault C., Deschenes S., Frayne I.R., Bouchard B., et al. Lipidomics unveils lipid dyshomeostasis and low circulating plasmalogens as biomarkers in a monogenic mitochondrial disorder. JCI insight. 2019;4(14) doi: 10.1172/jci.insight.123231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heddi A., Stepien G., Benke P.J., Wallace D.C. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J. Biol. Chem. 1999;274(33):22968–22976. doi: 10.1074/jbc.274.33.22968. [DOI] [PubMed] [Google Scholar]
- 29.Behan A., Doyle S., Farrell M. Adaptive responses to mitochondrial dysfunction in the rho degrees Namalwa cell. Mitochondrion. 2005;5(3):173–193. doi: 10.1016/j.mito.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X., Li X., Huang H., Jiang F., Lin Z., He H., et al. Elevated levels of mitochondrion-associated autophagy inhibitor LRPPRC are associated with poor prognosis in patients with prostate cancer. Cancer. 2014;120(8):1228–1236. doi: 10.1002/cncr.28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X., Zhong W., Huang H., He H., Jiang F., Chen Y., et al. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients. Mol. Carcinog. 2015;54(10):1194–1204. doi: 10.1002/mc.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H.Y., Ma Y.D., Zhang Y., Cui J., Wang Z.M. Elevated levels of autophagy-related marker ULK1 and mitochondrion-associated autophagy inhibitor LRPPRC are associated with biochemical progression and overall survival after androgen deprivation therapy in patients with metastatic prostate cancer. J. Clin. Pathol. 2017;70(5):383–389. doi: 10.1136/jclinpath-2016-203926. [DOI] [PubMed] [Google Scholar]
- 33.Fahrmann J.F., Grapov D., Phinney B.S., Stroble C., DeFelice B.C., Rom W., et al. Proteomic profiling of lung adenocarcinoma indicates heightened DNA repair, antioxidant mechanisms and identifies LASP1 as a potential negative predictor of survival. Clin. Proteomics. 2016;13:31. doi: 10.1186/s12014-016-9132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beers M.F., Morrisey E.E. The three R's of lung health and disease: repair, remodeling, and regeneration. J. Clin. Invest. 2011;121(6):2065–2073. doi: 10.1172/JCI45961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey C.J., Thimmulappa R.K., Singh A., Blake D.J., Ling G., Wakabayashi N., et al. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009;46(4):443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangasamy T., Cho C.Y., Thimmulappa R.K., Zhen L., Srisuma S.S., Kensler T.W., et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Invest. 2004;114(9):1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida T., Mett I., Bhunia A.K., Bowman J., Perez M., Zhang L., et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat. Med. 2010;16(7):767–773. doi: 10.1038/nm.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thimmulappa R.K., Scollick C., Traore K., Yates M., Trush M.A., Liby K.T., et al. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-imidazolide. Biochem. Biophys. Res. Commun. 2006;351(4):883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]