Abstract
Secondary metabolites profoundly affect microbial physiology, metabolism and stress responses. Increasing evidence suggests that these molecules can modulate microbial susceptibility to commonly used antibiotics; however, secondary metabolites are typically excluded from standard antimicrobial susceptibility assays. This may in part account for why infections by diverse opportunistic bacteria that produce secondary metabolites often exhibit discrepancies between clinical antimicrobial susceptibility testing results and clinical treatment outcomes. In this Review, we explore which types of secondary metabolites alter antimicrobial susceptibility, as well as how and why this phenomenon occurs. We discuss examples of molecules that opportunistic and enteric pathogens either generate themselves or are exposed to from their neighbors, and the nuanced impacts these molecules can have on tolerance and resistance to certain antibiotics.
Graphical Abstract
In this Review, Perry, Meirelles and Newman review the growing body of evidence that microbial secondary metabolites can modulate susceptibility to commonly used antibiotics, focusing on the mechanisms and why this phenomenon occurs, and they discuss the implications for the diagnosis of antibiotic resistance and therapeutic strategies.
Introduction
A vast number of organisms, many of which hail from the soil, produce a wide range of molecules classified as ‘secondary metabolites’1,2. They can be generated by Eukarya (for example, plants and fungi), Bacteria and Archaea, and are usually defined as organic compounds that do not directly support the producer’s growth or development3–5. In microbial planktonic cultures, they are typically produced during stationary phase, once doubling times have slowed6,7. As a result, these compounds were for many years assumed to be waste products of metabolism8. However, a more nuanced view of the biological functions of microbial secondary metabolites has emerged over the past two decades. Indeed, the moniker ‘secondary’ is something of a misnomer, as these molecules have been shown to have key roles in multiple physiological processes that are critical for microbial survival6,7, including but not limited to the acquisition of nutrients (such as iron or phosphate), cell–cell signaling and energy conservation in the absence of oxygen2,9–12.
In addition to conferring such pleiotropic benefits, many microbial secondary metabolites are also toxic, both to their producers and to neighboring organisms1,2,8. It is therefore not surprising that antibiotic development pipelines have driven the majority of secondary metabolite characterization and purification efforts, dating back to the discoveries of penicillin and streptomycin in the early 20th century. Most modern clinical antibiotics are derivatives of natural products that originated from soil microorganisms, and the soil-to-clinic axis continues to inspire natural product chemists in their search for and design of new drugs13. Yet, microbiologists have neglected to consider the potential for unintended consequences of this pipeline, particularly with respect to antibiotic efficacy against opportunistic pathogens that evolved in the same environment. Microorganisms are rarely, if ever, found in isolation, and therefore the presence of secondary metabolites in a microbial community exerts evolutionary pressure both on secondary metabolite-producing and non-producing members to develop means to withstand them. These defenses can in turn have collateral activity against clinical antibiotics.
In this Review, we highlight the growing body of evidence connecting bacterial secondary metabolites to the phenomena of antibiotic tolerance [G] (that is, the ability to survive transient antibiotic exposure) and resistance [G] (that is, the ability to grow in the presence of antibiotics at a given concentration)14–16. We also use the more generic term ‘antibiotic resilience’ [G] to refer to the ability of a bacterial population to be refractory to antibiotic treatment, which can arise from an increase in tolerance and/or resistance. Emphasizing examples of secondary metabolites produced by opportunistic or enteric pathogens (Table 1), we discuss common modes of action through which these molecules can alter antibiotic efficacy in both single-species and polymicrobial communities. Specifically, we draw attention to secondary metabolites that regulate multidrug resistance efflux systems, secondary metabolites that modulate the toxicity of antibiotics through interactions with reactive oxygen species, and the potential for secondary metabolite-induced antibiotic tolerance to provide an overlooked route for the evolution of antibiotic resistance. Although the impact of bacterial secondary metabolites on infection treatment outcomes has yet to be addressed in clinical studies, in vitro data indicating changes in antimicrobial susceptibility in the presence of diverse secondary metabolites strongly suggest that these molecules may represent an underappreciated factor in the recalcitrance of many opportunistic and chronic infections. We offer recommendations for future experiments to explore the breadth of relevance of these observations (Box 1) and discuss the implications that secondary metabolite production can have for the diagnosis of antibiotic resistance (Box 2). Finally, we consider how knowledge of interactions between secondary metabolites and antibiotic efficacy could be applied to optimize the use of existing antimicrobial drugs and generate targets for novel therapeutic strategies.
Table 1.
Secondary metabolites produced by opportunistic or enteric pathogens and their impacts on antibiotic efficacy.
| Metabolite | Producer | Antibiotic affected | Mechanism | Refs |
|---|---|---|---|---|
| PYO | Pseudomonas aeruginosa | Fluoroquinolones, aminoglycosides*, chloramphenicol, carbenicillin | Efflux induction, oxidative stress response induction | 36,44,45 |
| PCA, PCN | P. aeruginosa | Ciprofloxacin, tobramycin, carbenicillin | Metabolic changes | 44 |
| Paerucumarin | P. aeruginosa | Chloramphenicol, ciprofloxacin | Efflux induction | 50 |
| Indole | Escherichia coli | Fluoroquinolones, gentamicin, ampicillin, carbenicillin | Efflux induction, oxidative stress response induction | 27,31,77,111 |
| Salicylate | Burkholderia spp. | Chloramphenicol, trimethoprim, ciprofloxacin | Efflux induction | 55 |
| HQNO | P. aeruginosa | Meropenem | Increased extracellular DNA release and biofilm formation | 153 |
| Ergothioneine | Mycobacterium tuberculosis | Rifampicin, isoniazid, bedaquiline, clofazimine | Direct ROS detoxification, redox buffering | 89 |
| Polyamines (putrescine, spermidine) | E. coli, Burkholderia cenocepacia, P. aeruginosa | Levofloxacin, amikacin, cefotaxime, polymyxin B, norfloxacin, rifampicin, tobramycin | Direct ROS detoxification, decreased drug penetration | 99,101 |
| PQS | P. aeruginosa | Ciprofloxacin, oxofloxacin, imipenem, meropenem, gentamicin, colistin | Increased ROS generation | 103,105 |
| H2S | Diverse microorganisms | Gentamicin, amikacin, nalidixic acid, ciprofloxacin, ampicillin | Oxidative stress response induction, Fe2+ sequestration, redox buffering | 97,154 |
| Staphyloxanthin | Staphylococcus aureus | ND | Direct ROS detoxification | 90,91 |
| Carotenoids | Streptococcus spp. | ND | Direct ROS detoxification | 92,93 |
| 2,3-dihydroxybenzoate | E. coli | ND | Efflux induction | 25 |
| Toxoflavin | Burkholderia spp. | ND | Efflux induction**, oxidative stress response induction** | 65,68 |
| Phthiocol | M. tuberculosis | ND | Oxidative stress response induction** | 155,156 |
| D-alanylgriseoluteic acid | Pantoea agglomerans | ND | Oxidative stress response induction** | 157,158 |
| β−3H-indolydenopyruvate | Achromobacter sp. | ND | ND | 159 |
| Anthraquinones (for example, emodin, endocrocin) | Aspergillus spp. | ND | ND | 160,161 |
HQNO, 2-n-heptyl-4-hydroxyquinoline N oxide; PCA, phenazine 1-carboxylic acid; PCN, phenazine 1-carboxamide; PQS, Pseudomonas quinolone signal; PYO, pyocyanin, ROS, reactive oxygen species.
ND, not determined; these are molecules whose effects on antibiotics have not been directly tested.
For aminoglycosides, PYO has been shown to increase or decrease antibiotic resilience, depending on the studied conditions.
Hypothesized mechanisms by which the molecule might affect susceptibility.
Box 1: Guidelines for establishing causal links between secondary metabolite production and increased antibiotic tolerance or resistance.
We propose a few steps for the investigation of secondary metabolite-mediated changes in antibiotic susceptibility.
Identifying the candidate secondary metabolite.
This can be achieved in multiple ways, including based on molecular structure or its physiological effects on the cells. Genomic analysis could also reveal potentially relevant secondary metabolites produced by pathogens171. Researchers should investigate whether the putative secondary metabolite of interest shows any toxicity to the producer, as well as the molecular responses induced upon exposure to it. For example, the fact that pyocyanin (PYO) is toxic to producing cells35, shares structural similarities to fluoroquinolones (Fig. 1B), and induces efflux systems9,35, led to predictions about how this metabolite could decrease susceptibility to these drugs36. Transcriptomics approaches can be used to reveal responses caused by the specific secondary metabolite9,27,29,35,36. Importantly, it will often be necessary to construct a mutant strain lacking the biosynthetic genes for production of the secondary metabolite as a negative control.
Detecting the secondary metabolite.
It is essential to use an accurate and quantitative detection method because the secondary metabolite concentration might affect antibiotic susceptibility. Possible detection methods may include ultraviolet or visible light spectroscopy or mass spectrometry, both of which can be coupled to high-performance liquid chromatography in the case of analyzing complex samples (for example, extracts of microbial cultures or clinical samples). Investigators should attempt to detect the secondary metabolite in the relevant clinical context (for example, in infected sputum, wound exudates or stool samples)75,76,128,129. Importantly, concentrations of the secondary metabolite in vivo might vary greatly among patients, depending on a variety of infection parameters (for example, infection stage, the strain causing the infection, the number of bacterial cells present). For this reason, the guidelines provided will, at least initially, likely be most useful for cases of chronic infections, where longitudinal monitoring of patients coupled with repeated measurements of the in vivo concentrations of secondary metabolites can be used to constrain in vitro experiments (see below; and Box 2).
Considering the effects on nearby species.
It is also critical to account for the effect of secondary metabolites on the entire microbial community in the case of polymicrobial infections. Thus, investigators should consider which species are most commonly found together with the secondary metabolite producer in the type of infection being studied. Then, investigators should perform experiments in which the producer and non-producers are co-cultured, or the non-producers are separately exposed to controlled concentrations of the secondary metabolite. Understanding how the non-producers are affected, including the molecular mechanisms involved, is essential for predicting how the secondary metabolite might change the overall antibiotic susceptibility profile of a microbial community.
Testing the secondary metabolite-mediated effects on antibiotic susceptibility.
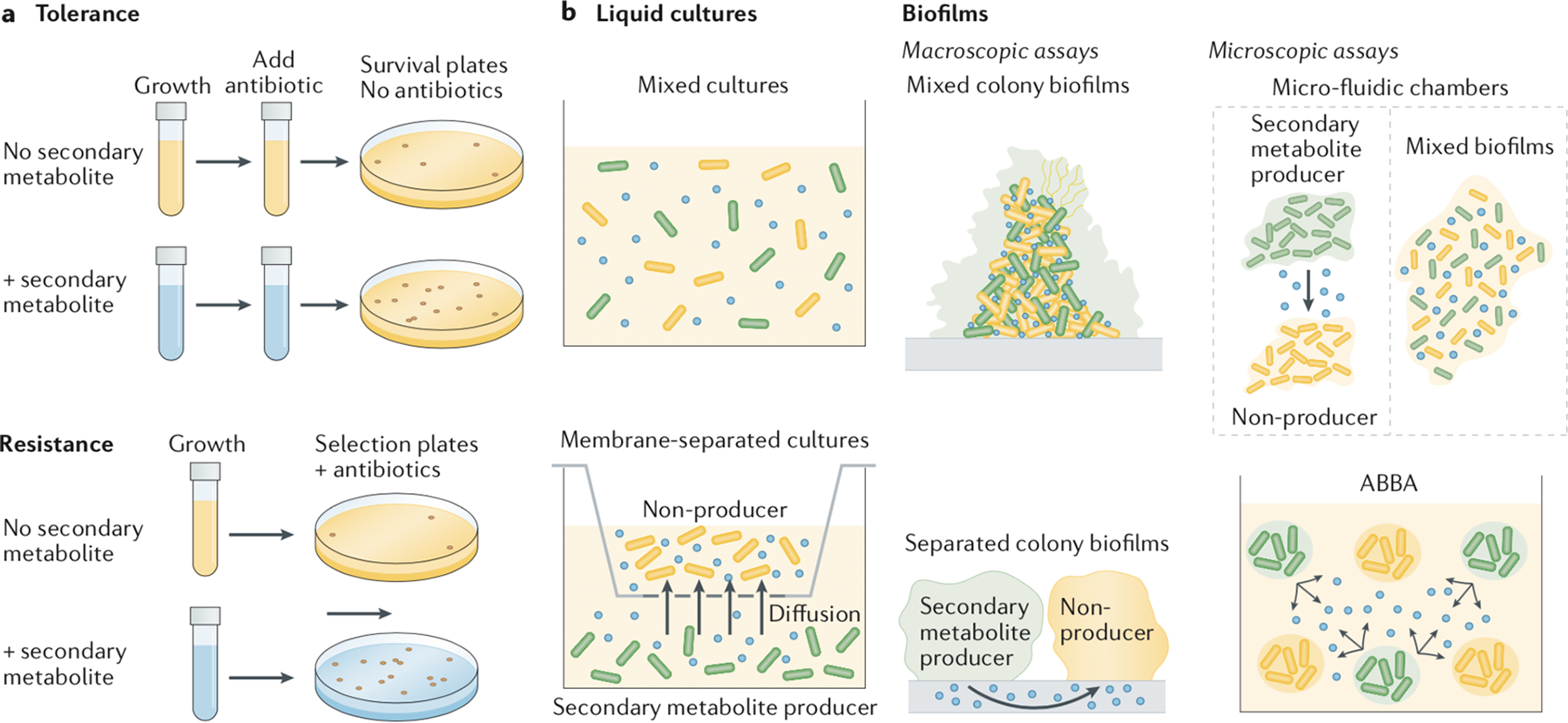
Tolerance and resistance are two different modes of antibiotic resilience that should be tested separately14–16 (see the figure, part a); the former can be measured by determining the percentage of surviving cells following a temporary exposure to the antibiotic, whereas the latter calls for assessing the ability to grow in the presence of the antibiotic. There are also various assays available for testing secondary metabolite-mediated effects on drug susceptibility in multispecies communities (see the figure, part b). In liquid co-cultures, the species can be grown with or without separation by a permeable membrane that restricts interactions to those mediated by diffusible small molecules. For biofilms, experiments can be performed in macroscopic assays (for example, colony biofilms, with species mixed or separated), or microscopic assays (for example, microfluidics172,173 or alternative assays like the agar block biofilm assay (ABBA)148,174, to which concomitant measurements of microenvironment variables such as pH or O2 levels have been applied (see the figure, part b)). Importantly, the decision to perform experiments on liquid cultures versus biofilms can influence the results, as some secondary metabolite-mediated tolerance mechanisms are specific to biofilms. For example, the increased release of extracellular DNA mediated by secondary metabolites can stimulate biofilm formation, which results in increased tolerance levels153. In addition, redox-active secondary metabolites, such as phenazines, greatly affect metabolism within biofilms44,175, which in turn modulates antimicrobial drug efficacy44,74. By contrast, planktonic cultures provide better control for testing specific secondary metabolite-mediated responses and are amenable to more sophisticated methods when evaluating resistance, such as fluctuation tests, in which a large number of parallel cultures are plated on a selective medium and the number of spontaneous resistant mutants from each culture serves as the input into a mathematical formula for inferring mutation rates176,177. The investigators should therefore decide what is more appropriate for their particular questions.
[Figure] The blue color in tubes or plates (in part a) or the blue dots (in part b) represent the secondary metabolite.
Box 2: Accounting for secondary metabolite production during AST.
As the traditional basis for determining appropriate courses of treatment for infections, antimicrobial susceptibility testing (AST) is a cornerstone of clinical microbiology. However, in many clinical contexts, there is little to no correlation between AST results and treatment outcomes178,179, which may stem in some cases from the fact that standard AST conditions generally preclude detection of interactions between secondary metabolites and antimicrobial drugs. Clinical AST relies on a readout of absence of growth in cultures that are inoculated at low cell densities180, whereas secondary metabolites are typically not produced until at least late log-phase or early stationary phase7. Moreover, AST is routinely performed on single-species cultures even in the case of polymicrobial infections, eliminating the possibility of detecting secondary metabolite-mediated interspecies interactions that could affect antimicrobial efficacy.
We suggest a few speculative but testable modifications to clinical AST protocols that could help account for the effects of microbial secondary metabolites produced during infections. Our intent is to inspire concrete discussion of how a better understanding of interactions between secondary metabolites and clinical antibiotic efficacy could eventually be applied to improve treatment outcomes, while also recognizing the probable complexity of such endeavors. Given that secondary metabolites can substantially affect antimicrobial efficacy, the proposed modifications might improve the empirical correlation between in vitro AST results and the success of clinical treatments. However, many factors beyond secondary metabolite production (for example, biofilm formation, nutritional conditions and/or host-derived molecules) are likely to also affect antibiotic susceptibility in situ during infections. The approaches proposed below would need to be refined and validated in animal models of infection, and ultimately tested in clinical trials comparing patient outcomes against the use of traditional AST for antimicrobial drug selection. In addition, if studies indicate that secondary metabolite-related modifications improve the predictive value of AST, scalability in terms of cost and throughput will need to be further optimized before widespread implementation becomes possible in clinical microbiology laboratories.
Use filtered supernatants from overnight cultures of the infective microorganism or microorganisms).
By setting up AST assays using a mixture of fresh growth media and filtered supernatant from an overnight culture of the infective agent, the effects of any secondary metabolites produced during late log phase or stationary phase could be accounted for without needing to modify other aspects of standard AST protocols (for example, visible growth as a readout). A key advantage of this approach is that it is agnostic to which secondary metabolite is produced, avoiding the need for prior knowledge of the biosynthetic capabilities of the infective agent, as well as accounting for the combined effects of multiple secondary metabolites. However, this approach would significantly increase the time to result. In addition, mixing spent supernatant with fresh growth media might dilute the concentrations of the secondary metabolites below clinically relevant levels. The concentrations of different nutrients in the media would also be affected, possibly in unpredictable ways across different strains, which in turn could also impact antibiotic susceptibility; indeed, the choice of growth medium in general can significantly influence MIC measurements181.
Add purified secondary metabolites exogenously to cultures.
If it is known that the infective species produces specific secondary metabolites that have been validated as clinically relevant (for example, pyocyanin (PYO) in Pseudomonas aeruginosa or indole in Escherichia coli), purified forms of the secondary metabolites could be added to traditional AST assays. An advantage of this approach is the ability to control the level of exposure to the secondary metabolites, which would make it possible to ensure that the concentrations in the AST assay are similar to those detected in clinical samples. However, this approach requires that the secondary metabolites be commercially available or otherwise economically viable to synthesize or purify from cultures. In addition, prior knowledge of the biosynthetic capabilities of the infective agent would be necessary. An important caveat in this regard is that microbial secondary metabolite production can vary greatly from strain to strain within the same species. Thus, basing the working concentration of a secondary metabolite on an average across strains or patient samples may lead to over- or underestimation of the effects of the secondary metabolite in individual cases.
Adjust antibiotic ‘breakpoint’ guidelines according to in situ levels of secondary metabolites.
For secondary metabolites that are commonly found in infections and can be procured in purified form, in vitro testing could lead to the development of mathematical models that quantitatively relate the concentration of the secondary metabolite to the change in the producing species’ resistance to different antimicrobial drugs. Once validated across a range of different strains, such models could potentially enable secondary metabolite-based adjustments to the standard ‘breakpoint’ antibiotic concentrations used by clinicians to classify microorganisms as susceptible or resistant. Assays for the quantification of the secondary metabolite in patient samples could then be combined with traditional AST testing. Such an approach would enable taking into account strain variability in secondary metabolite production and avoid necessitating modifications to existing AST protocols. However, this approach might also lead to underestimation of secondary metabolite effects in some cases, as bulk measurements of a secondary metabolite in a patient sample may obscure heterogeneity at the micron scale.
Grow multiple species together if they are co-isolated from a polymicrobial infection.
Secreted secondary metabolites produced by one species can affect antimicrobial efficacy in neighboring species. Thus, in the case of polymicrobial infections, inoculating multiple species together in AST assays may improve prediction of the overall community response to antimicrobial drugs. To account for the production of secondary metabolites, this approach would still need to be combined with other modifications, such as those proposed above (points 1–3). In addition, the optimal ratios at which to inoculate different species would need to be investigated and standardized. Alternatively, it may be possible to perform AST using mixed cultures derived directly from patient samples, either bypassing or in parallel to the step in which individual isolates are obtained and identified.
Induction of efflux systems
Activation of efflux pumps [G] that export toxins out of the cell is one mechanism used by diverse bacteria to thrive during clinical antibiotic treatment17,18. However, efflux pumps long predate human use of synthetic antibiotics, and therefore are presumed to have originally evolved to transport other, naturally occurring substrates, such as secondary metabolites19,20. The types and components of efflux pumps have been extensively reviewed21,22. In this Review, we focus on how the induction of efflux systems in response to self-produced secondary metabolites can affect antibiotic tolerance and resistance in pathogenic bacteria (Fig. 1A). The examples we discuss fall mostly within the resistance-nodulation-division (RND) efflux systems, but the same principles could, in theory, be applicable to other types of efflux systems that are regulated by secondary metabolites. Importantly, efflux pumps vary in their specificity, with regard to both their regulation and their substrate affinity22. Therefore, to predict whether a secondary metabolite will increase antibiotic resilience in its producer through the induction of a particular efflux system, it is essential to understand how the secondary metabolite interacts with the transcriptional regulation of the system, as well as which classes of drugs the system can transport. Many known efflux-regulating secondary metabolites have at least one aromatic or heterocyclic ring (Fig. 1B), possibly suggesting that secondary metabolites with this structural motif are particularly likely to affect antibiotic resilience through the induction of multidrug efflux systems.
Fig 1. Secondary metabolite-mediated regulation of multidrug resistance efflux pumps.
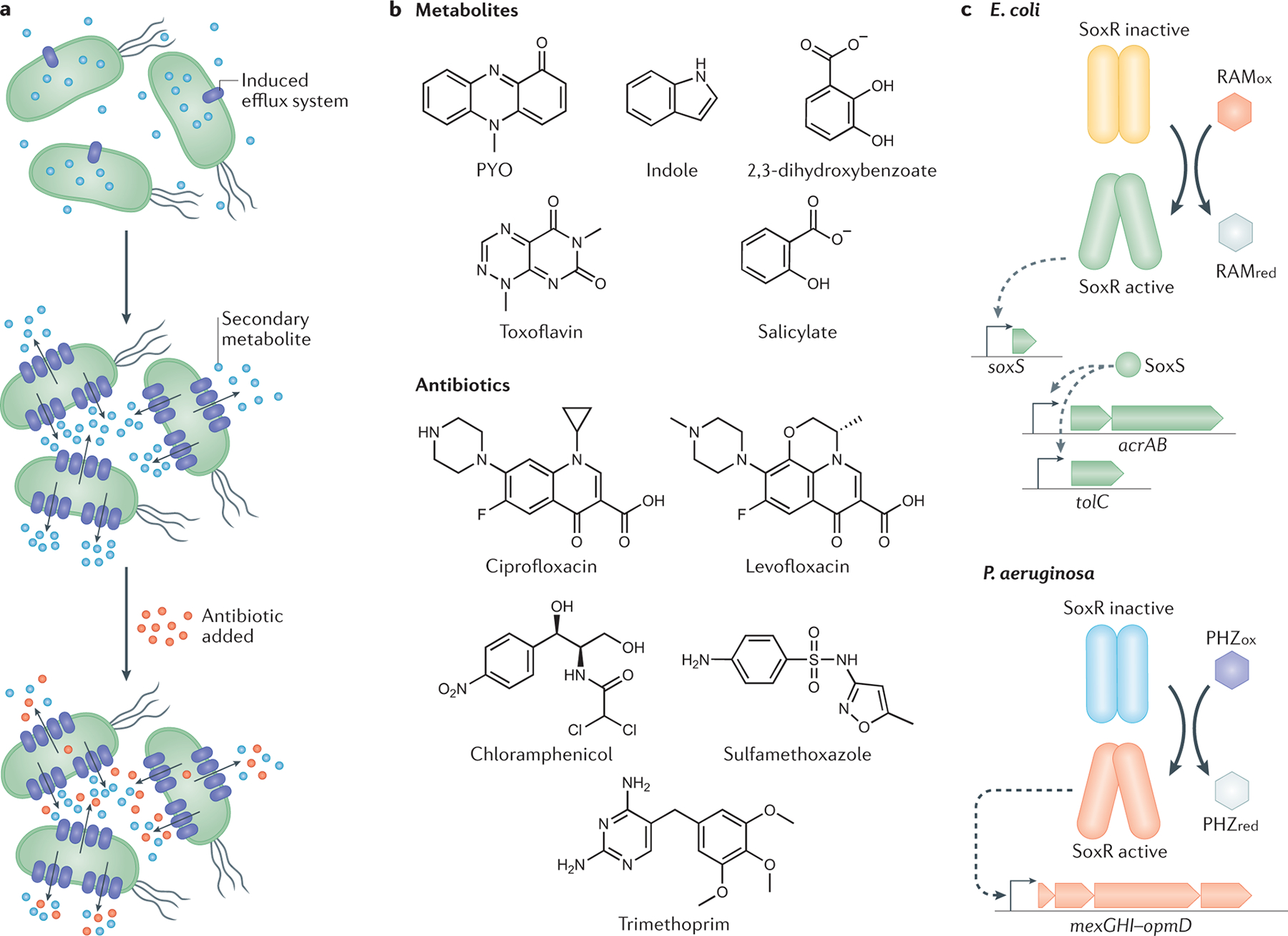
a. Secondary metabolites induce the expression of efflux systems, which export the metabolite. The increased expression of efflux pumps can provide collateral resilience to antibiotics used in the clinic by expelling the drugs out of the microbial cells. b. Structures of known efflux pump-regulating secondary metabolites and selected clinical antibiotics, showing the shared prevalence of aromatic and/or heterocyclic ring motifs. c. SoxR-regulated efflux systems in Escherichia coli and Pseudomonas aeruginosa. Each SoxR monomer contains a Fe–S cluster that can be directly oxidized by redox-active molecules, which leads to its activation and transcriptional induction of efflux systems162,163. In E. coli (top), several molecules can induce transcription of the efflux system AcrAB–TolC through the activation of SoxR162,164,165. Note that acrAB and tolC are transcribed separately, and this is a simplified version of a very complex regulatory system22. Importantly, TolC can assemble with efflux systems from different classes, such as AcrAB–TolC and several others from the resistance-nodulation-division (RND) efflux systems superfamily, EmrAB–TolC (major facilitator superfamily), and MacAB–TolC (ABC superfamily)23,166,167. When studying how additional secondary metabolites might affect E. coli susceptibility to antibiotics, it will be important to determine which specific efflux system TolC is part of, the regulation involved in the induction of the system, and its substrate-specificity. In P. aeruginosa (bottom), the activation of SoxR is mediated by two endogenous phenazines, 5-Me-PCA (not shown) and PYO, and leads to the induction of the efflux system MexGHI–OpmD9,34,36,163. PYO, pyocyanin; PHZox, phenazine in the oxidized state; PHZred, phenazine in the reduced state; RAMox, redox-active molecule in oxidized state, RAMred, redox-active molecule in reduced state.
In the enteric bacterium Escherichia coli, one of the best-studied multidrug resistance efflux systems is AcrAB–TolC, which has a complex regulatory system and an extensive substrate range, being part of a general stress response22,23. Although AcrAB–TolC is generally expressed at high intrinsic levels22,24, numerous molecules have been shown to further upregulate its transcription, including self-produced secondary metabolites such as the compound 2,3-dihydroxybenzoate, an intermediate in the biosynthesis of the siderophore enterobactin25. In fact, 2,3-dihydroxybenzoate directly binds to MarR26, a transcriptional repressor that modulates the expression of AcrAB–-TolC alongside the redox-sensing SoxRS regulatory system22 (Fig. 1C). Although it has not been directly tested whether the production of enterobactin or 2,3-dihydroxybenzoate per se increases antibiotic resilience, it is well established that AcrAB–TolC provides protection against many classes of clinical antibiotics23. The signaling molecule indole is another example of a self-produced secondary metabolite in E. coli with an efflux-mediated effect on antibiotic susceptibility. Indole triggers the expression of certain multidrug efflux pumps in enteric bacteria27–30. Indeed, the production of high levels of indole by a subpopulation of mutants has been characterized as a ‘charity’ mechanism that induces population-level resistance against norfloxacin and gentamicin in E. coli, with the MdtEF–TolC efflux system being upregulated by this secondary metabolite31. Intriguingly, a tolC mutant of Shewanella oneidensis, an environmental isolate related to opportunistic pathogens of fish32, was sensitive to anthraquinone-2,6-disulfonate (an analog of naturally occurring redox-active humic substances in soils and sediments that resembles quinone-containing synthetic antibiotics)33, further suggesting that functional relationships between structurally similar natural and synthetic molecules may be common.
Efflux pumps can also provide protection against secondary metabolites that are toxic to their producers, an effect that might confer resilience to clinical antibiotics. For example, in the opportunistic pathogen Pseudomonas aeruginosa, the redox-sensing transcription factor SoxR (Fig. 1C) activates expression of the MexGHI–OpmD efflux system in response to phenazines, which are toxic, redox-active self-produced secondary metabolites9,34–36. Phenazines have important roles both in natural environments (for example, by protecting plants against fungal pathogens) as well as in infections (for example, by increasing P. aeruginosa virulence in the lungs of patients with cystic fibrosis)37,38. The phenazine pyocyanin (PYO) also induces a second efflux operon in P. aeruginosa, mexEF–oprN35,36, which is clinically relevant39,40. Experiments with a broad-spectrum efflux pump inhibitor that reduces the activity of various RND efflux systems41,42, as well as a knockout mutant lacking the mexGHI–opmD operon, have confirmed that efflux is a major mechanism underlying the tolerance and resistance of P. aeruginosa to its own phenazines34,36,43. Importantly, both phenazine-regulated efflux pumps are also known to transport fluoroquinolones, and multiple studies have reported a strong antagonistic effect of phenazines on fluoroquinolone efficacy36,43–45. For example, P. aeruginosa cells exposed to PYO (either self-produced or exogenously added) display increased tolerance against the fluoroquinolones ciprofloxacin and levofloxacin36. This phenotype was recapitulated in one study by artificially overexpressing MexGHI–OpmD in a phenazine-null mutant to a level similar to that achieved in the presence of PYO, which suggests that drug efflux drives PYO-mediated increases in fluoroquinolone tolerance36. Interestingly, besides fluoroquinolones and chloramphenicol, MexEF–OprN is also thought to transport trimethoprim and sulfamethoxazole, to which P. aeruginosa is intrinsically resistant46, but not aminoglycosides46, against which phenazines have demonstrated mixed effects on tolerance36,44,45. Like phenazines, but unlike aminoglycosides, all of the known antibiotic substrates for MexEF–OprN have at least one aromatic ring (Fig. 1B), which suggests that analysis of shared structural motifs may enable prediction of which clinical antibiotics will be most affected. Such structural comparisons would only be appropriate for efflux systems that are relatively specific in substrate recognition, and would not apply to efflux systems that export a wide range of antibiotics46.
In addition to MexGHI–OpmD and MexEF–OprN, P. aeruginosa possesses at least nine other efflux systems that belong to the RND family, many of which have been studied in detail with respect to their structures and biochemistry22,46–48. Whether any P. aeruginosa-produced secondary metabolites regulate these other efflux systems under clinically relevant circumstances remains to be determined. Notably, however, phenazines are not the only secondary metabolites produced by P. aeruginosa that promote increased resilience against antibiotics by inducing efflux. Recent work showed that production of the secondary metabolite paerucumarin also stimulates transcription of the MexEF–OprN efflux system in P. aeruginosa, with consequent increases in resistance to both chloramphenicol and ciprofloxacin49,50. These findings underscore the potential for self-produced secondary metabolites to promote resilience to clinical antibiotics in opportunistic pathogens by triggering the upregulation of efflux pumps.
Other bacterial opportunistic pathogens inhabiting soils or plant roots also produce secondary metabolites that promote efflux and decrease antibiotic susceptibility. For example, several strains of ‘Burkholderia cepacia complex’ species isolated from patients with cystic fibrosis can produce salicylate51,52. Salicylate has Fe-chelating properties and is used as a siderophore by the producing cells53,54. It also induces specific efflux systems in Burkholderia species (for example, CeoAB–OpcM in B. cenocepacia), which leads to increased antibiotic resistance55. The salicylate-derived antibiotic resistance effect is not limited to the Burkholderia genus; for example, in enterobacteria, salicylate binds to and inactivates MarR, which leads to upregulation of efflux pump expression and increased resistance to multiple clinical antibiotics56,57. Burkholderia species also produce several other secondary metabolites that could affect antibiotic resilience, including many natural antibiotics with strong inhibitory capacities relevant during plant host colonization58–61. One intriguing example is toxoflavin, which is produced by several Burkholderia species, including Burkholderia gladioli, a common species found in patients with cystic fibrosis62,63. Although it is still unknown whether toxoflavin poisons producing cells, it is redox-active64–66, presumably causing oxidative stress through the generation of H2O2 (Ref. 65), and it is toxic to other bacteria and fungi65,67. More importantly, like PYO, toxoflavin induces a specific RND efflux system, ToxFGHI, which is used for its export68. It is not yet known if B. gladioli or other opportunistic pathogens within the Burkholderia genus produce toxoflavin during infections, or if the toxoflavin-induced RND efflux system ToxFGHI can transport any of the currently used clinical antibiotics; however, this is a possibility, given that toxoflavin, like PYO, bears structural similarity to fluoroquinolones.
Finally, we note that the above examples of efflux system induction due to the presence of secondary metabolites are all from Gram-negative bacteria, which are traditionally the organisms in which drug efflux has been studied in more detail. However, a related phenomenon has been demonstrated in the non-pathogenic Gram-positive bacterium Streptomyces coelicolor, which produces a natural antibiotic, actinorhodin, that stimulates expression of a transporter similar to those that export tetracycline69,70. Future work should investigate the extent to which secondary metabolite-mediated induction of multidrug efflux pumps might occur in pathogenic Gram-positive bacteria and its consequences for antibiotic resilience. Taken together, the examples discussed highlight the rich diversity of bacterial secondary metabolites that induce efflux activity in their producers, potentially compromising the efficacy of clinical antibiotics.
Modulation of oxidative stress
More than a decade ago, it was proposed that bactericidal antibiotics exert their lethal effects in part by inducing oxidative stress, regardless of the specific cellular targets of different antibiotic classes71. Although this hypothesis has engendered controversy72,73, evidence reviewed elsewhere74 suggests that bactericidal antibiotics have an impact on cellular redox states, and that the resulting increases in reactive oxygen species (ROS) and oxidative stress can contribute to cell death. Importantly, many secondary metabolites also interface with cellular redox homeostasis and oxidative stress responses. In this section, we discuss three different modes of action by which these metabolites can potentially antagonize or potentiate the toxicity of clinical antibiotics (Fig. 2A): upregulation of oxidative stress response genes; direct detoxification of ROS; and increased endogenous ROS generation.
Fig. 2. Secondary metabolite interactions with oxidative stress.
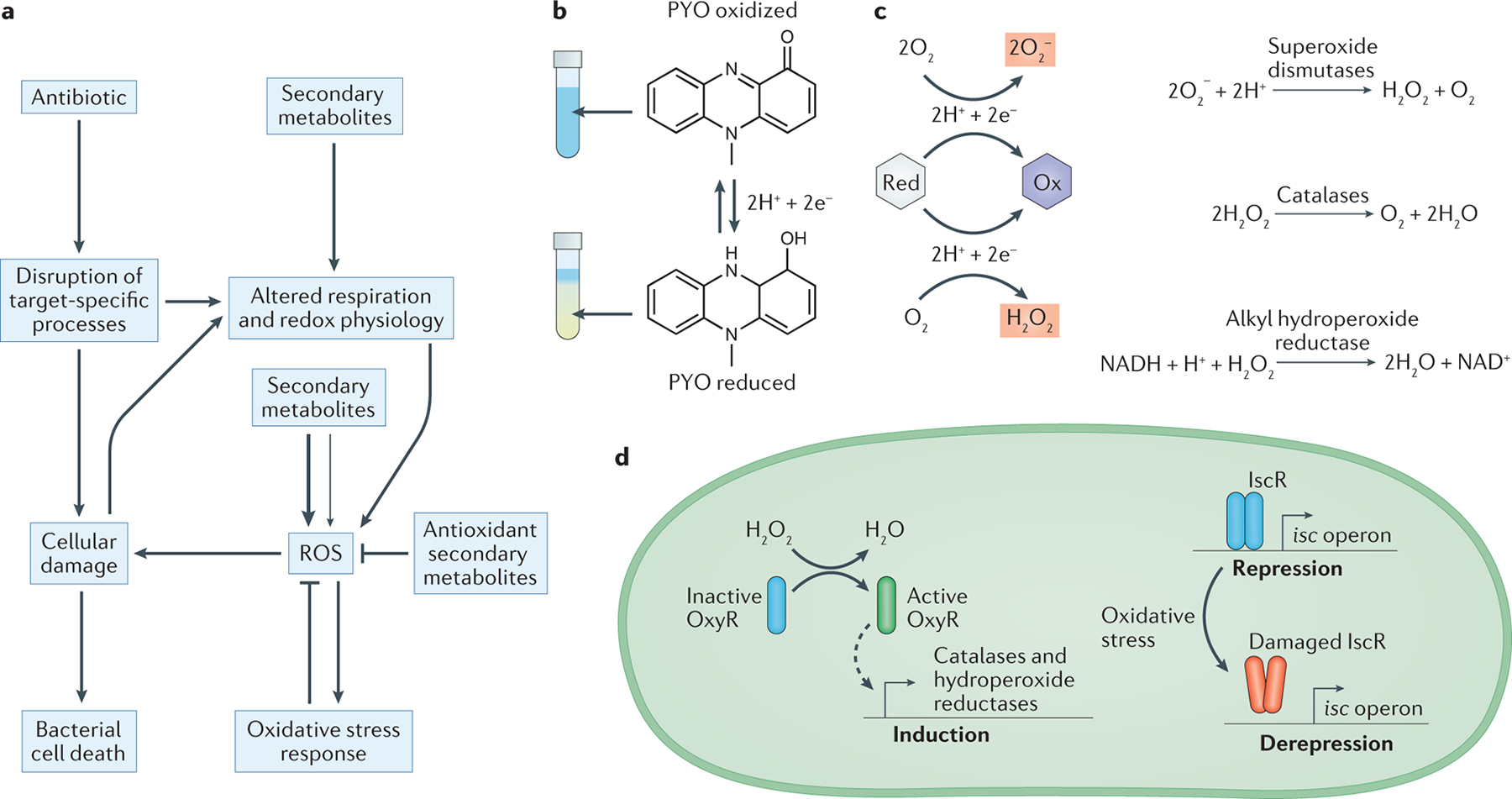
a. Schematic depicting how bactericidal antibiotics can cause cell death both by directly disrupting target-specific processes, and by indirectly promoting the formation of reactive oxygen species (ROS) as a consequence of altered respiration and cellular damage87. Secondary metabolites can interface with these pathways at multiple points, including by interfering with respiration and redox homeostasis, directly generating ROS through redox-cycling, and detoxifying ROS via one-electron reactions. Secondary metabolites that promote oxidative stress can either antagonize or potentiate antibiotic toxicity, which is likely to depend on whether the resulting increases in ROS are moderate (thin arrow) or severe (thick arrow). Moderate increases in ROS may induce protective oxidative stress responses that can counteract antibiotic toxicity, whereas severe increases in ROS may overwhelm the defenses of the cell, which leads to synergistic effects with bactericidal antibiotics. b. The redox-active nature of pyocyanin (PYO) is visually apparent as it undergoes a color change from blue (oxidized) to colorless (reduced) upon gaining two electrons and two protons from cellular reductants. This reaction is reversible under physiological conditions. c. Many redox-active secondary metabolites can donate electrons to molecular oxygen in the process of cycling from a reduced (Red) to an oxidized (Ox) state, which leads to the formation of superoxide or hydrogen peroxide. Cells can detoxify these forms of ROS through enzymatic reactions catalyzed by superoxide dismutase, catalase, and alkyl hydroperoxide reductase162. d. Bacterial oxidative stress responses are typically regulated through multiple pathways. For example, in Pseudomonas aeruginosa, the H2O2-sensing transcription factor OxyR controls the expression of catalases and alkyl hydroperoxide reductases (AHPs)168,169 (left). In addition, the transcription factor iron-sulfur cluster regulator (IscR) upregulates the biosynthesis of iron–sulfur cluster (isc) operons in response to oxidative stress caused by ROS such as superoxide or H2O2, which can directly damage the iron–sulfur cluster in IscR itself and thereby lead to depression of its regulon170.
Upregulation of defenses against oxidative stress.
Secondary metabolites that upregulate the expression of oxidative stress responses can prime bacterial cells for tolerance and/or resistance to clinical antibiotics, analogous to the protective effects of exposure to sub-lethal concentrations of oxidants like H2O2. Among this class of metabolites, indole is perhaps the best-studied. As mentioned above, indole can also influence antibiotic susceptibility by upregulating efflux pump expression. However, this effect is thought to happen primarily at high concentrations of indole (>1 mM)27,29; indole concentrations in human feces tend to be lower75, though measurements up to the equivalent of 1100 μM have been recorded76. At these lower concentrations, indole is non-toxic to its producer, E. coli, but still induces oxidative stress response genes regulated by OxyR, including alkyl hydroperoxide reductases, thioredoxin reductase, and the DNA-binding protein Dps77. Exposure to indole increases the frequency of E. coli persisters [G] to antibiotics that belong to three different classes (fluoroquinolones, aminoglycosides and β-lactams) by at least an order of magnitude, and deletion of oxyR substantially diminishes this effect, which demonstrates that upregulation of oxidative stress responses by a secondary metabolite can contribute to bacterial persistence77. The molecular pathway through which indole activates OxyR remains unclear, but indole readily undergoes one-electron reduction to a radical form in the presence of hydroxyl radicals or other strong oxidants78, which suggests that it might interact with and potentially amplify endogenous ROS generated as a byproduct of respiration. Indole has also been proposed to disrupt the arrangement of membrane lipids, which would enable the direct interaction of respiratory quinones with dioxygen and thereby lead to the generation of superoxide79.
PYO is another example of a bacterial secondary metabolite that induces oxidative stress responses. As a redox-active metabolite that can gain and lose electrons reversibly under physiological conditions (Fig. 2B), PYO can generate ROS under aerobic conditions through direct reduction of oxygen to superoxide (Fig. 2C), in addition to interfering with respiration80,81. In its producer, P. aeruginosa, PYO increases superoxide dismutase activity82 and upregulates the transcription of several other oxidative stress response genes, including those encoding alkyl hydroperoxide reductases, thioredoxin reductase, catalase and iron-sulfur cluster biogenesis machinery35 (Fig. 2C–D). Intriguingly, PYO has been shown to increase the frequency of gentamicin-resistant mutants in P. aeruginosa cultures in a manner that is independent of drug efflux, as PYO does not upregulate aminoglycoside-transporting efflux pumps36. Given that gentamicin is known to promote increased intracellular ROS levels through the formation of complexes with iron83,84, and that pre-treating cells with oxidants can prime them to tolerate antibiotics85, a plausible explanation for this phenomenon is that PYO-induced oxidative stress responses counteract ROS-related gentamicin toxicity. This in turn could decrease the rate at which spontaneous mutants are stochastically lost from the population86, which would lead to the observed increase in the frequency of resistant mutants. PYO can also promote the growth of P. aeruginosa in the presence of other aminoglycosides (kanamycin, streptomycin and tobramycin) and a β-lactam antibiotic (carbenicillin)45. Like gentamicin, these antibiotics are not known to be substrates for PYO-regulated efflux systems36,46, but they belong to classes of drugs that have been shown to perturb cellular redox states87,88, which again suggests that the observed decreases in antibiotic efficacy could be related to PYO-induced oxidative stress responses.
Detoxification of ROS.
In contrast to ROS-generating redox-active secondary metabolites that induce enzymatic oxidative stress responses, secondary metabolites that possess antioxidant activity [G] can protect against antibiotic assaults by directly detoxifying antibiotic-derived ROS. One example is ergothioneine, which is one of two major sulfur-containing redox buffers in mycobacteria, along with mycothiol. Loss of ergothioneine biosynthesis genes in Mycobacterium tuberculosis decreases minimum inhibitory concentrations (MICs) for rifampicin, isoniazid, bedaquiline and clofazimine, in addition to decreasing survival under treatment at the wildtype MICs by at least 30–60% (Ref. 89). Other secondary metabolites with antioxidant activity have been shown to be important for resistance to ROS generated by host immune cells, including macrophages and neutrophils. One such metabolite is staphyloxanthin, a membrane-embedded carotenoid pigment that protects Staphylococcus aureus against ROS90,91 and consequently decreases killing by neutrophils91. Likewise, carotenoids produced by group B Streptococcus92 and the dental pathogen Streptococcus mutans93 have been implicated in resistance to ROS. The antioxidant capacity of these pigments has been attributed to their highly conjugated polyene backbones94, though the exact mechanisms by which different carotenoids scavenge ROS remain unclear95. Importantly, antioxidant agents need not necessarily be located in the cytoplasm to protect against antibiotic-induced ROS accumulation, as exogenous catalase has been shown to increase bacterial survival following exposure to trimethoprim96. Thus, although most studies on membrane-embedded carotenoids have focused on interactions with extracellular sources of ROS, these pigments might also be able to dampen oxidative stress that originates inside the cell during treatment with bactericidal antibiotics. Considering the growing body of evidence that redox imbalance and oxidative stress are downstream effects of many antimicrobial drugs74, the possibility that staphyloxanthin or other membrane-associated pigments promote resilience to clinical antibiotics is worthy of further investigation.
In addition to the above examples, certain microbially produced compounds that are not classic secondary metabolites, either because they are inorganic or are not always dispensable for normal growth, also contribute to antibiotic tolerance or resistance by enhancing the antioxidant capacity of bacterial cells. For example, endogenously generated H2S protects a diverse range of bacteria, including E. coli, P. aeruginosa and S. aureus, against the toxicity of antibiotics known to exert oxidative stress, such as gentamicin97. This phenomenon has been proposed to stem from a dual-action mechanism whereby H2S both inhibits the Fenton reaction and stimulates the activities of catalase and superoxide dismutase97. Polyamines have also been shown to protect bacteria from antibiotic toxicity by counteracting oxidative stress. This is thought to be due in part to their capacity to neutralize free radicals98,99, though physical protection of cellular components and indirect upregulation of other oxidative stress responses may also be involved98,100. E. coli upregulates production of putrescine and spermidine upon exposure to antibiotics under aerobic conditions, and these polyamines in turn statistically significantly increase viability under antibiotic treatment by decreasing ROS production and oxidative damage101. Similarly, putrescine secreted by Burkholderia cenocepacia protects its producer against oxidative stress arising from treatment with polymyxin B, norfloxacin, or rifampicin99. Finally, in P. aeruginosa, the BqsRS regulatory system drives increased polyamine production upon sensing ferrous iron, which is prevalent in the lungs of patients with cystic fibrosis, thereby promoting survival in the presence of cationic antibiotics such as polymyxins and aminoglycosides102. Together, these examples demonstrate how interactions between diverse bacterial metabolites and oxidative stress can lead to increased resilience against clinical antibiotics.
Synergistic interactions between secondary metabolites and antibiotics.
Besides the examples in which secondary metabolites decrease antibiotic efficacy by attenuating oxidative stress, it is also important to note that in some cases, secondary metabolites that increase ROS generation can amplify the toxicity of clinical antibiotics. One example is 2-heptyl-3-hydroxy-4-quinolone, also known as the Pseudomonas quinolone signal (PQS), which is produced by P. aeruginosa. PQS is a redox-active molecule that can reduce not only free radicals but also metal ions, and consequently possesses both antioxidant properties and pro-oxidant activity [G], as reduction of iron promotes ROS formation through the Fenton reaction103. The pro-oxidant activity seems to dominate in cells, as PQS induces oxidative stress responses and increases sensitivity to hydrogen peroxide and ciprofloxacin103,104. The pro-oxidant activity of PQS is also evidenced by the fact that overproduction of PQS acts synergistically with impairment of superoxide dismutase and catalase activity to increase endogenous oxidative stress and antibiotic susceptibility105. Notably, abolishment of PQS production increases tolerance to ciprofloxacin, imipenem and gentamicin103. Another redox-active secondary metabolite that increases antibiotic susceptibility under certain conditions is PYO. Although pre-exposure of P. aeruginosa to PYO increases tolerance to fluoroquinolones and promotes the establishment of gentamicin-resistant mutants, PYO and other phenazines have also been shown to increase the sensitivity of P. aeruginosa to cationic antimicrobial peptides, including colistin36,44 and polymyxin B45. The underlying mechanism of this synergistic interaction has yet to be determined, but it is notable that polymyxin B precipitates severe oxidative stress in P. aeruginosa and other Gram-negative opportunistic pathogens106,107. Moreover, unlike fluoroquinolones or aminoglycosides, cationic antimicrobial peptides also permeabilize the outer membrane108 and consequently might increase phenazine uptake, which would accelerate ROS generation even further. Thus, the synergy between phenazines and cationic antimicrobial peptides may ultimately be driven by an overwhelming cascade of oxidative stress. Given that ROS-generating secondary metabolites can both potentiate and diminish antibiotic efficacy depending on the circumstances, future studies focused on revealing which of these effects take precedence during infections will be critical to better understand how such secondary metabolites may affect clinical treatment outcomes.
Interspecies antibiotic resilience
So far, we have discussed examples of secondary metabolites that either are known to affect their producer’s susceptibility to clinical antibiotics, or have the potential to do so based on their interactions with established mechanisms of antibiotic tolerance and resistance. However, equally important is the fact that many secondary metabolites, in particular those that are secreted, can also modulate interspecies antibiotic resilience (Fig. 3). Indeed, how interactions among members of a polymicrobial infection [G] might affect antibiotic treatment outcomes has recently received much attention109,110.
Fig. 3. Secondary metabolites as interspecies modulators of antibiotic resilience.
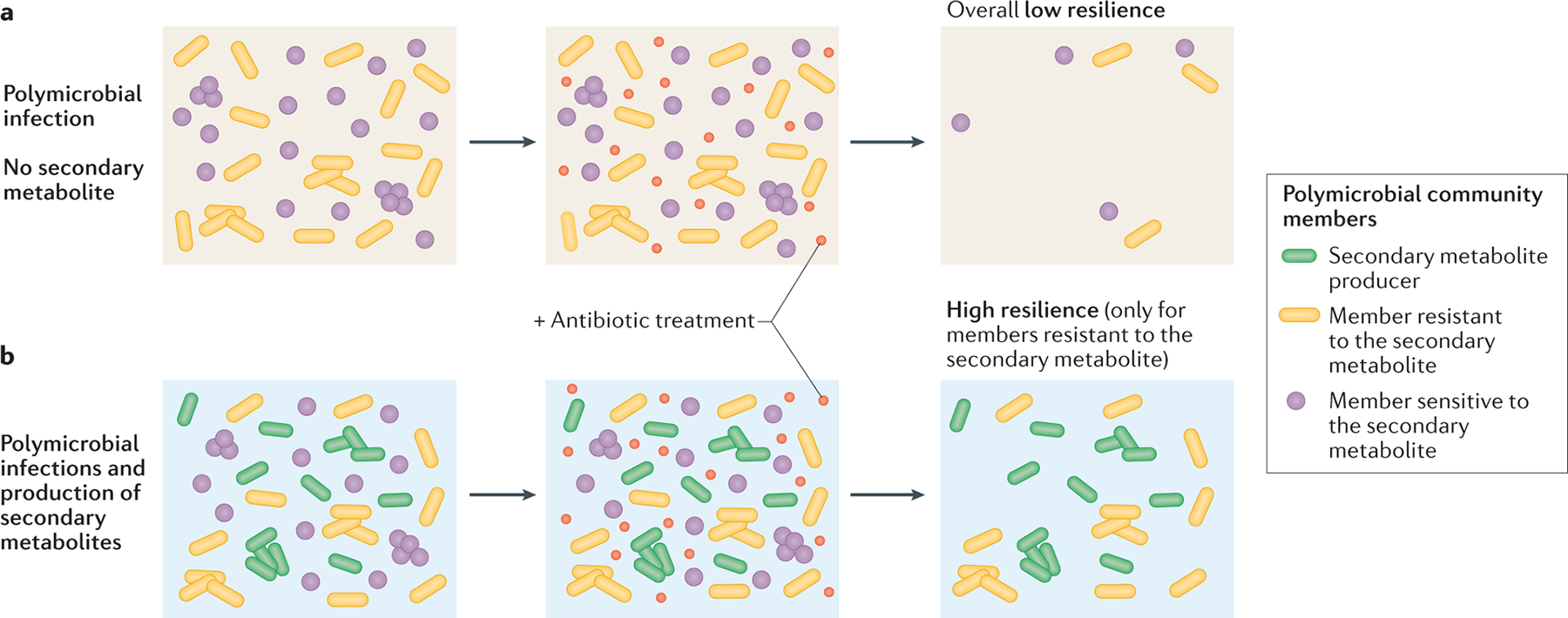
The presence of secondary metabolite producers in polymicrobial infections can alter the community susceptibility profile to antibiotic treatment. When the producer is not present (part a), overall resilience levels upon antibiotic treatment are low. However, through the secretion of the secondary metabolite, the producer’s presence (part b; green cells) can have distinct effects on different community members. For members intrinsically resistant to the secondary metabolite (yellow cells), the molecule’s presence can increase resilience to antibiotic treatment. However, if a member is sensitive to the secondary metabolite (purple cells), the added toxicity can overwhelm cellular defenses, potentiating the killing by the clinical drug.
Although our understanding of how exchangeable secondary metabolites affect community-level antibiotic resilience is still in its infancy, there have been several reports on this subject. For example, indole has been identified as an interspecies modulator of antibiotic tolerance between E. coli and Salmonella enterica subsp. enterica serovar Typhimurium29,111. S. Typhimurium, which is likely to interact with commensal E. coli during infection, does not produce indole, yet its tolerance to ciprofloxacin increased by greater than threefold in the presence of exogenously added indole, as well as in co-cultures with indole-producing E. coli111. Similar to its effect in E. coli, indole induces OxyR-regulated oxidative stress responses in S. Typhimurium, and deletion of oxyR abolished the indole-mediated increase in ciprofloxacin tolerance111. Interestingly, indole has also been reported to increase resistance to ampicillin in the indole non-producer P. aeruginosa, not by inducing oxidative stress responses but rather by stimulating the expression of efflux pumps and a chromosomal β-lactamase112. These examples highlight the potential for secreted secondary metabolites to have both conserved and mechanistically divergent effects on antibiotic resilience in neighboring species.
Other secondary metabolites besides indole have shown potential as interspecies modulators of antibiotic resilience. For example, the protective effect against polymyxin B of putrescine secreted by B. cenocepacia extends not only to the producer, but also to neighboring species in co-cultures, including E. coli and P. aeruginosa100. Similarly, PYO produced by P. aeruginosa considerably increases the tolerance of multiple clinically relevant Burkholderia species to ciprofloxacin in co-cultures36. In yet another opportunistic pathogen, Acinetobacter baumannii, exposure to PYO increases the frequency of persisters to amikacin and carbenicillin by three- to four-fold113, possibly through the upregulation of superoxide dismutase and catalase113,114. Finally, quorum sensing signals produced by P. aeruginosa induce fluconazole resistance in the yeast Candida albicans115, which indicates that bacterial secondary metabolites can mediate not only interspecies but also interkingdom effects on antimicrobial efficacy. So far, these interactions have only been demonstrated in vitro. However, given that Burkholderia species, Acinetobacter species and fungal pathogens can all be found together with P. aeruginosa in chronic infections62,116,117, these findings suggest that valuable insights could be gained from future studies focused on the in vivo relevance of secondary metabolite-mediated interspecies induction of antibiotic resilience.
Importantly, although the above examples suggest that certain secreted secondary metabolites have the potential to raise the community-wide level of antibiotic resilience in polymicrobial infections, a secondary metabolite that promotes antibiotic resilience in one species will not always do so in others. One key consideration is that in order for a secondary metabolite to trigger cross-species induction of resilience to clinical antibiotics, the non-producing species must be able to tolerate any stress caused by the secondary metabolite. If toxicity outweighs benefits from defense induction or antibiotic detoxification, the non-producing species would not gain a benefit. In fact, the secondary metabolite might even act synergistically with the clinical antibiotic118. Examples of this type of interaction have been found between S. aureus and P. aeruginosa, two species that often co-occur in patients with cystic fibrosis119–121. S. aureus is sensitive to several secondary metabolites secreted by P. aeruginosa122,123, and some of these can increase the susceptibility of S. aureus to clinical antibiotics. For example, P. aeruginosa-produced rhamnolipids potentiate tobramycin toxicity in S. aureus by increasing membrane permeability122. Another P. aeruginosa-produced secondary metabolite, 2-n-heptyl-4-hydroxyquinoline N oxide (HQNO), was recently shown to increase the sensitivity of S. aureus biofilms to fluoroquinolones and membrane-targeting antibiotics via a similar mechanism124. However, the toxic effects of P. aeruginosa secondary metabolites on S. aureus are not always synergistic with clinical antibiotics. By inhibiting growth, HQNO promotes tolerance in S. aureus biofilms specifically to antibiotics targeting cell wall synthesis and protein synthesis125, contrary to its effect on other classes of antibiotics. By contrast, in planktonic cultures of S. aureus, HQNO can induce multidrug tolerance by inhibiting respiration and depleting intracellular ATP122. The interference of PYO with respiration in S. aureus similarly selects for non-respiring small colony variants123,126, which are often resistant to antibiotic treatment127. In such cases where different secondary metabolites produced by one species seem to have conflicting and condition-dependent effects on a neighboring species, in vivo studies and co-culture experiments are particularly necessary to determine the overall impact on clinical antibiotic efficacy.
The potential for P. aeruginosa-produced secondary metabolites to have complex effects on antibiotic resilience has also been observed in another opportunistic pathogen, Stenotrophomonas maltophilia. Compared to members of the Burkholderia cepacia complex, which strongly benefit from exposure to PYO during treatment with ciprofloxacin, S. maltophilia is more sensitive to PYO toxicity, exhibiting growth inhibition at concentrations as low as 50 μM36. At a low but still lethal dose of ciprofloxacin, PYO at concentrations up to 50 μM statistically significantly increased survival of S. maltophilia, which suggests that defenses against fluoroquinolones are indeed induced by PYO in this species36. Yet at a ten-fold higher dose of ciprofloxacin, even 10 μM PYO was detrimental36. Notably, although in situ levels of PYO can vary greatly across patients infected with P. aeruginosa, PYO has been detected in infected sputum and wound exudates at concentrations up to 130 μM or 0.31 mg/g, respectively128,129. Thus, the example of PYO and S. maltophilia suggests that to predict how a secondary metabolite will affect community-wide levels of resilience to clinical antibiotics during a polymicrobial infection, it is imperative to characterize the directionality and magnitude of interspecies effects over a range of clinically relevant concentrations of both the secondary metabolite and the clinical antibiotic.
Implications for resistance evolution
In recent years, it has increasingly become appreciated that antibiotic tolerance cannot only directly contribute to infection treatment failure, but also promote the establishment of heritable resistance mutations130–134. Exploration of the connection between antibiotic tolerance and the evolution of resistance has largely focused on tolerance resulting from spontaneous mutations that are selected by treatment with clinical antibiotics130,131,133,134. However, effects on the establishment of heritable resistance mutations have also been demonstrated for at least one secondary metabolite produced by an opportunistic pathogen, namely, PYO. Experiments based on classic fluctuation tests revealed that PYO increases the rate of mutation to antibiotic resistance not only in the producing species, P. aeruginosa, but also in a clinical isolate of a co-occurring opportunist, Burkholderia multivorans36. Notably, the impact of PYO on the acquisition of heritable resistance varied across different classes of antibiotics. In particular, strong effects were observed for drugs against which PYO-induced defenses confer increased tolerance, which suggests that this phenomenon is indeed driven by tolerance as opposed to a mutagenic effect of PYO. Remarkably, in B. multivorans, treatment with PYO increased mutation rates for ciprofloxacin resistance to a level rivaling clinical hypermutator strains36,135. In addition, in both P. aeruginosa and B. multivorans, pre-treatment with PYO prior to antibiotic treatment was sufficient to increase the rate at which resistant mutants became established, even without continued exposure to high levels of PYO36. These findings collectively reveal the potential for tolerance-inducing secondary metabolites to have a substantial impact on the evolution of antibiotic resistance. However, future experiments will be necessary to investigate the clinical relevance of these observations, and whether the link between tolerance and resistance triggered by PYO exposure can be generalized to other secondary metabolites remains to be determined.
Concluding remarks and future directions
Although direct connections between bacterial secondary metabolite production or exposure and resilience to clinical antibiotics have only been pursued in a small number of studies, many more secondary metabolites are known to interface with cellular functions that are relevant to antibiotic tolerance and resistance, especially drug efflux and oxidative stress responses. We hope the examples discussed in this Review stimulate further investigation into the conditions under which these and related secondary metabolites alter the efficacy of clinical antibiotics, particularly during treatment of infections. Currently, PYO represents the secondary metabolite for which the most detailed evidence on this topic is available, likely due to the extensive research attention that its producer, P. aeruginosa, has received. However, we expect that deliberately searching for such molecules across a broad range of opportunistic pathogens (Box 1) will reveal additional as-yet-uncharacterized secondary metabolites that have the potential to affect antibiotic treatment outcomes. Soil-borne opportunistic pathogens in particular often possess the biosynthetic capacity to produce a great variety of secondary metabolites60,136–138, perhaps because they have to cope with the extraordinary complexity and heterogeneity that typifies the soil environment139. In most cases, the biological functions of these secondary metabolites, as well as whether they are produced during infections, remain unknown. However, biosynthetic gene clusters for secondary metabolites are often located near or co-transcribed with genes encoding efflux pumps140–142, and numerous microbial secondary metabolites are redox-active and therefore have the potential to generate or detoxify ROS12,143, which suggests that interactions with clinical antibiotic efficacy are likely to be far more common than is currently appreciated.
Importantly, understanding the molecular mechanisms involved in the cellular responses triggered by a secondary metabolite, as well as the chemical properties of the secondary metabolite itself, can provide practical insights regarding which clinical antibiotics are likely to be affected. With this knowledge in hand, combined with an understanding of other environmental and physiological factors that affect antibiotic susceptibility144,145, we posit that it will be possible to optimize the use of existing antibiotics so as to better minimize the risk of treatment failure and prevent the evolution of resistance in vivo. For example, if a pathogen produces a secondary metabolite that upregulates efflux pumps specific to fluoroquinolones and other aromatic molecules, the chances of successful treatment would likely be higher with another class of antibiotics that is not susceptible to this defense, such as aminoglycosides. The biosynthetic pathways for these secondary metabolites, or even the molecules themselves, could also be targets for the development of new adjuvants for antimicrobial drugs. Such efforts are already underway for secondary metabolites such as PYO and staphyloxanthin146–149, which are also known to act as virulence factors150. For example, enzymatic removal of PYO increased antibiotic killing of P. aeruginosa biofilms in vitro149. In addition, inhibiting a bacterial enzyme that generates H2S increased the potency of bactericidal antibiotics both in vitro and in mouse infection models151. These results encourage further exploration of whether targeting the production or presence of specific bacterial metabolites could optimize the clinical efficacy of treatments for infections. Finally, retooling clinical antimicrobial susceptibility testing protocols to account for the impact of secondary metabolites on antibiotic efficacy (Box 2) could potentially improve the predictive value of these assays, especially in the case of secondary metabolite-producing opportunistic pathogens, such as members of the Burkholderia cepacia complex, that often exhibit discrepancies between in vitro minimum inhibitory concentration measurements and clinical treatment outcomes152.
In conclusion, our ability to address the vexing challenges posed by antibiotic tolerance and resistance in the future has much to gain by reflecting on the past. The evolutionary and ecological history of natural antibiotics intersects directly with the history of clinical antibiotic discovery. While the soil has continued to provide a rich reservoir for natural product mining efforts, what has gotten lost is the fact that alongside the evolution of pathways that synthesize these molecules, other pathways have co-evolved that respond to them. Remembering this shared historical context is important for predicting how secondary metabolites might affect the response of polymicrobial communities to conventional antibiotics, and compels creative thinking about novel ways to manage such responses.
Acknowledgements
Work in the corresponding author’s laboratory was supported by grants to D.K.N. from the NIH (1R01AI127850-01A1, 1R01HL152190-01) and the Doren Family Foundation. E.K.P. was supported by a National Science Foundation Graduate Research Fellowship under Grant No. DGE-1745301.
Glossary
- Tolerance
The ability to survive transient antibiotic exposure
- Resistance
The ability to grow in the presence of antibiotics at a given concentration
- Antibiotic resilience
The ability of a bacterial population to be refractory to antibiotic treatment via tolerance and/or resistance
- Efflux pumps
Membrane-associated transport proteins that are responsible for the extrusion of various compounds out of the cell
- Persisters
A subpopulation of bacteria that is killed by a given antibiotic at a much slower rate than the rest of the population, in a manner that is non-heritable
- Antioxidant activity
The ability to neutralize highly reactive free radicals
- Pro-oxidant activity
The ability to induce oxidative stress
- Polymicrobial infection
An infection that is caused by more than one species of microorganism
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Maplestone RA, Stone MJ & Williams DH The evolutionary role of secondary metabolites – a review. Gene 115, 151–157 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Demain AL & Fang A The natural functions of secondary metabolites. Adv Biochem Eng Biotechnol 69, 1–39 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Keller NP, Turner G & Bennett JW Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol 3, 937–947 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Tyc O, Song C, Dickschat JS, Vos M & Garbeva P The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol 25, 280–292 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Wang S & Lu Z in Biocommunication of Archaea (ed. Witzany G) 235–239 (Springer International Publishing, 2017). doi: 10.1007/978-3-319-65536-9_14 [DOI] [Google Scholar]
- 6.Price-Whelan A, Dietrich LEP & Newman DK Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol 2, 71–78 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Davies J Specialized microbial metabolites: functions and origins. J Antibiot 66, 361–364 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Haslam E Secondary metabolism – fact and fiction. Nat. Prod. Rep 3, 217 (1986). [Google Scholar]
- 9.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M & Newman DK The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol 61, 1308–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Glasser NR, Kern SE & Newman DK Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol 92, 399–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol 193, 3606–3617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McRose DL & Newman DK Redox-active antibiotics enhance phosphorus bioavailability. Science 371, 1033–1037 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling LL et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kester JC & Fortune SM Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit Rev Biochem Mol Biol 49, 91–101 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Brauner A, Fridman O, Gefen O & Balaban NQ Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Balaban NQ et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piddock LJV Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev 19, 382–402 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X-Z, Plésiat P & Nikaido H The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev 28, 337–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piddock LJV Multidrug-resistance efflux pumps – not just for resistance. Nat. Rev. Microbiol 4, 629–636 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Martinez JL et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev 33, 430–449 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Mousa JJ & Bruner SD Structural and mechanistic diversity of multidrug transporters. Nat Prod Rep 33, 1255–1267 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Du D et al. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol 16, 523–539 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Anes J, McCusker MP, Fanning S & Martins M The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol 6, 587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X-Z & Nikaido H in Efflux-Mediated Antimicrobial Resistance in Bacteria (eds. Li X-Z, Elkins CA & Zgurskaya HI) 219–259 (Springer International Publishing, 2016). doi: 10.1007/978-3-319-39658-3_9 [DOI] [Google Scholar]
- 25.Ruiz C & Levy SB Regulation of acrAB expression by cellular metabolites in Escherichia coli. J. Antimicrob. Chemother 69, 390–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identifies endogenous cellular metabolites that induce expression of a major multidrug efflux pump in E. coli.
- 26.Chubiz LM & Rao CV Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J. Bacteriol 192, 4786–4789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirakawa H, Inazumi Y, Masaki T, Hirata T & Yamaguchi A Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol 55, 1113–1126 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Nishino K, Honda T & Yamaguchi A Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol 187, 1763–1772 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido E et al. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog 4, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishino K, Nikaido E & Yamaguchi A Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol 189, 9066–9075 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HH, Molla MN, Cantor CR & Collins JJ Bacterial charity work leads to population-wide resistance. Nature 467, 82–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that the production of indole by highly antibiotic-resistant mutants of E. coli increases the antibiotic tolerance and resistance of less-resistant strains, thus establishing a precedent for the role of a secondary metabolite in mediating the overall antibiotic susceptibility of a bacterial population.
- 32.Paździor E, Pękala-Safińska A & Wasyl D Phenotypic diversity and potential virulence factors of the Shewanella putrefaciens group isolated from freshwater fish. J. Vet. Res 63, 321–332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shyu JBH, Lies DP & Newman DK Protective role of tolC in efflux of the electron shuttle anthraquinone-2,6-disulfonate. J. Bacteriol 184, 1806–1810 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakhtah H et al. The Pseudomonas aeruginosa efflux pump MexGHI–OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Natl. Acad. Sci. USA 113, E3538–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meirelles LA & Newman DK Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol 110, 995–1010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meirelles LA, Perry EK, Bergkessel M & Newman DK Bacterial defenses against a natural antibiotic promote collateral resilience to clinical antibiotics. PLoS Biol 19, e3001093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows that a toxic secondary metabolite can increase tolerance to fluoroquinolones in strains of Pseudomonas aeruginosa and other opportunistic pathogens, and can also promote the establishment of spontaneous antibiotic-resistant mutants in populations of these bacteria.
- 37.Lau GW, Hassett DJ, Ran H & Kong F The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med 10, 599–606 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Mavrodi DV, Blankenfeldt W & Thomashow LS Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol 44, 417–445 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Llanes C et al. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother 55, 5676–5684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardot C et al. Amino acid substitutions account for most MexS alterations in clinical nfxC mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother 60, 2302–2310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomovskaya O & Bostian KA Practical applications and feasibility of efflux pump inhibitors in the clinic – a vision for applied use. Biochem. Pharmacol 71, 910–918 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Jamshidi S, Sutton JM & Rahman KM Computational study reveals the molecular mechanism of the interaction between the efflux inhibitor PAβN and the AdeB transporter from Acinetobacter baumannii. ACS Omega 2, 3002–3016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolloscheck D, Krishnamoorthy G, Nguyen J & Zgurskaya HI Kinetic control of quorum sensing in Pseudomonas aeruginosa by multidrug efflux pumps. ACS Infect. Dis 4, 185–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiessl KT et al. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun 10, 762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals that phenazine production alters both the metabolic profile of biofilms and their tolerance to different classes of clinical antibiotics, suggesting that beyond induction of specific cellular defenses, secondary metabolites can also impact antibiotic susceptibility via indirect mechanisms.
- 45.Zhu K, Chen S, Sysoeva TA & You L Universal antibiotic tolerance arising from antibiotic-triggered accumulation of pyocyanin in Pseudomonas aeruginosa. PLoS Biol 17, e3000573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors report that sublethal antibiotic treatment can trigger pyocyanin production in P. aeruginosa, and that pyocyanin enables multiple bacterial species to grow to higher cell densities in the presence of diverse clinical antibiotics.
- 46.Lister PD, Wolter DJ & Hanson ND Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev 22, 582–610 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yonehara R, Yamashita E & Nakagawa A Crystal structures of OprN and OprJ, outer membrane factors of multidrug tripartite efflux pumps of Pseudomonas aeruginosa. Proteins 84, 759–769 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Glavier M et al. Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat. Commun 11, 4948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke-Pearson MF & Brady SF Paerucumarin, a new metabolite produced by the pvc gene cluster from Pseudomonas aeruginosa. J. Bacteriol 190, 6927–6930 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iftikhar A et al. Mutation in pvcABCD operon of Pseudomonas aeruginosa modulates MexEF–OprN efflux system and hence resistance to chloramphenicol and ciprofloxacin. Microb. Pathog 104491 (2020). doi: 10.1016/j.micpath.2020.104491 [DOI] [PubMed] [Google Scholar]
- 51.Sokol PA, Lewis CJ & Dennis JJ Isolation of a novel siderophore from Pseudomonas cepacia. J. Med. Microbiol 36, 184–189 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Darling P, Chan M, Cox AD & Sokol PA Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun 66, 874–877 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visca P, Ciervo A, Sanfilippo V & Orsi N Iron-regulated salicylate synthesis by Pseudomonas spp. J. Gen. Microbiol 139, 1995–2001 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Bakker PAHM, Ran L & Mercado-Blanco J Rhizobacterial salicylate production provokes headaches! Plant Soil 382, 1–16 (2014). [Google Scholar]
- 55.Nair BM, Cheung K-J, Griffith A & Burns JL Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia). J. Clin. Invest 113, 464–473 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen SP, Levy SB, Foulds J & Rosner JL Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol 175, 7856–7862 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brochado AR et al. Species-specific activity of antibacterial drug combinations. Nature 559, 259–263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burkhead KD, Schisler DA & Slininger PJ Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol 60, 2031–2039 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong Y et al. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis 87, 890–895 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Depoorter E et al. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol 100, 5215–5229 (2016). [DOI] [PubMed] [Google Scholar]; This review catalogues the toxic secondary metabolites known to be produced by Burkholderia species and describes what is known about their regulation, thus serving as a useful resource for identifying endogenous compounds that might affect antibiotic susceptibility in this family of opportunistic pathogens.
- 61.Depoorter E, De Canck E, Coenye T & Vandamme P Burkholderia bacteria produce multiple potentially novel molecules that inhibit carbapenem-resistant Gram-Negative Bacterial Pathogens. Antibiotics (Basel) 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipuma JJ The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev 23, 299–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones C et al. Kill and cure: genomic phylogeny and bioactivity of Burkholderia gladioli bacteria capable of pathogenic and beneficial lifestyles. Microb. Genom 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stern KG Oxidation-reduction potentials of toxoflavin. Biochem. J 29, 500–508 (1935). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Latuasan HE & Berends W On the origin of the toxicity of toxoflavin. Biochim. Biophys. Acta 52, 502–508 (1961). [DOI] [PubMed] [Google Scholar]
- 66.Gencheva R, Cheng Q & Arnér ESJ Efficient selenocysteine-dependent reduction of toxoflavin by mammalian thioredoxin reductase. Biochim. Biophys. Acta Gen. Subj (2018). doi: 10.1016/j.bbagen.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 67.Li X, Li Y, Wang R, Wang Q & Lu L Toxoflavin produced by Burkholderia gladioli from Lycoris aurea is a new broad-spectrum fungicide. Appl. Environ. Microbiol 85, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J et al. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol 54, 921–934 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Tahlan K et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol 63, 951–961 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Willems AR et al. Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J. Mol. Biol 376, 1377–1387 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA & Collins JJ A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Keren I, Wu Y, Inocencio J, Mulcahy LR & Lewis K Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Liu Y & Imlay JA Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dwyer DJ, Collins JJ & Walker GC Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol 55, 313–332 (2015). [DOI] [PubMed] [Google Scholar]; This work comprehensively discusses the evidence that oxidative stress contributes to antibiotic lethality.
- 75.Zuccato E et al. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig. Dis. Sci 38, 514–519 (1993). [DOI] [PubMed] [Google Scholar]
- 76.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR & Lorentz O Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J. Cancer Res. Clin. Oncol 109, 135–141 (1985). [DOI] [PubMed] [Google Scholar]
- 77.Vega NM, Allison KR, Khalil AS & Collins JJ Signaling-mediated bacterial persister formation. Nat. Chem. Biol 8, 431–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen X, Lind J & Merenyi G One-electron oxidation of indoles and acid-base properties of the indolyl radicals. J. Phys. Chem 91, 4403–4406 (1987). [Google Scholar]
- 79.Garbe TR, Kobayashi M & Yukawa H Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch. Microbiol 173, 78–82 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Perry EK & Newman DK The transcription factors ActR and SoxR differentially affect the phenazine tolerance of Agrobacterium tumefaciens. Mol. Microbiol 112, 199–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voggu L et al. Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J. Bacteriol 188, 8079–8086 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hassett DJ, Charniga L, Bean K, Ohman DE & Cohen MS Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect. Immun 60, 328–336 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Priuska EM & Schacht J Formation of free radicals by gentamicin and iron and evidence for an iron/gentamicin complex. Biochem. Pharmacol 50, 1749–1752 (1995). [DOI] [PubMed] [Google Scholar]
- 84.Prayle A, Watson A, Fortnum H & Smyth A Side effects of aminoglycosides on the kidney, ear and balance in cystic fibrosis. Thorax 65, 654–658 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mosel M, Li L, Drlica K & Zhao X Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob. Agents Chemother 57, 5755–5759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alexander HK & MacLean RC Stochastic bacterial population dynamics restrict the establishment of antibiotic resistance from single cells. Proc. Natl. Acad. Sci. USA 117, 19455–19464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dwyer DJ et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 111, E2100–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belenky P et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13, 968–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saini V et al. Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep 14, 572–585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work indicates that a metabolite involved in redox homeostasis plays a key role in mediating antibiotic resistance and tolerance in M. tuberculosis, suggesting that such metabolites might also contribute to these phenotypes in other bacteria.
- 90.Hall JW, Yang J, Guo H & Ji Y The Staphylococcus aureus AirSR two-component system mediates reactive oxygen species resistance via transcriptional regulation of staphyloxanthin production. Infect. Immun 85, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clauditz A, Resch A, Wieland K-P, Peschel A & Götz F Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun 74, 4950–4953 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu GY et al. Sword and shield: linked group B streptococcal beta-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc. Natl. Acad. Sci. USA 101, 14491–14496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu C et al. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol 76, 5815–5826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edge R & Truscott TG Singlet oxygen and free radical reactions of retinoids and carotenoids – a review. Antioxidants (Basel) 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young AJ & Lowe GL Carotenoids – antioxidant Properties. Antioxidants (Basel) 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hong Y, Zeng J, Wang X, Drlica K & Zhao X Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. USA 116, 10064–10071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shatalin K, Shatalina E, Mironov A & Nudler E H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 (2011). [DOI] [PubMed] [Google Scholar]
- 98.Tkachenko AG & Fedotova MV Dependence of protective functions of Escherichia coli polyamines on strength of stress caused by superoxide radicals. Biochemistry. (Mosc) 72, 109–116 (2007). [DOI] [PubMed] [Google Scholar]
- 99.El-Halfawy OM & Valvano MA Putrescine reduces antibiotic-induced oxidative stress as a mechanism of modulation of antibiotic resistance in Burkholderia cenocepacia. Antimicrob. Agents Chemother 58, 4162–4171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.El-Halfawy OM & Valvano MA Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One 8, e68874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tkachenko AG, Akhova AV, Shumkov MS & Nesterova LY Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res. Microbiol 163, 83–91 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Kreamer NN, Costa F & Newman DK The ferrous iron-responsive BqsRS two-component system activates genes that promote cationic stress tolerance. MBio 6, e02549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Häussler S & Becker T The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4, e1000166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bredenbruch F, Geffers R, Nimtz M, Buer J & Häussler S The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol 8, 1318–1329 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Nguyen D et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Han M-L et al. Comparative metabolomics and transcriptomics reveal multiple pathways associated with polymyxin killing in Pseudomonas aeruginosa. mSystems 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sampson TR et al. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother 56, 5642–5649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trimble MJ, Mlynárčik P, Kolář M & Hancock REW Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bottery MJ, Pitchford JW & Friman V-P Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J 15, 939–948 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Welp AL & Bomberger JM Bacterial community interactions during chronic respiratory disease. Front. Cell Infect. Microbiol 10, 213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vega NM, Allison KR, Samuels AN, Klempner MS & Collins JJ Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 110, 14420–14425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work reveals how a secondary metabolite that induces antibiotic tolerance can work across bacterial species.
- 112.Kim J, Shin B, Park C & Park W Indole-induced activities of β-lactamase and efflux pump confer ampicillin resistance in Pseudomonas putida KT2440. Front. Microbiol 8, 433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhargava N, Sharma P & Capalash N Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect. Immun 82, 3417–3425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heindorf M, Kadari M, Heider C, Skiebe E & Wilharm G Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One 9, e101033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bandara HMHN et al. Fluconazole resistance in Candida albicans is induced by Pseudomonas aeruginosa quorum sensing. Sci. Rep 10, 7769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chmiel JF et al. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, Gram-negative bacteria, and multiple infections. Annals of the American Thoracic Society 11, 1120–1129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwab U et al. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect. Immun 82, 4729–4745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wood KB & Cluzel P Trade-offs between drug toxicity and benefit in the multi-antibiotic resistance system underlie optimal growth of E. coli. BMC Syst. Biol 6, 48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yung DBY, Sircombe KJ & Pletzer D Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol (2021). doi: 10.1111/mmi.14699 [DOI] [PubMed] [Google Scholar]
- 120.Camus L, Briaud P, Vandenesch F & Moreau K How bacterial adaptation to cystic fibrosis environment shapes interactions between Pseudomonas aeruginosa and Staphylococcus aureus. Front. Microbiol 12, 617784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Briaud P et al. Impact of coexistence phenotype between Staphylococcus aureus and Pseudomonas aeruginosa isolates on clinical outcomes among cystic fibrosis patients. Front. Cell Infect. Microbiol 10, 266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Radlinski L et al. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15, e2003981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors screen clinical isolates of P. aeruginosa for effects on the antibiotic susceptibility of S. aureus and find that the interactions are strain-specific and complex, highlighting the challenges of understanding how secondary metabolites impact polymicrobial infections.
- 123.Noto MJ, Burns WJ, Beavers WN & Skaar EP Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J. Bacteriol 199, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Orazi G, Ruoff KL & O’Toole GA Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orazi G & O’Toole GA Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biswas L, Biswas R, Schlag M, Bertram R & Götz F Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl. Environ. Microbiol 75, 6910–6912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McNamara PJ & Proctor RA Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 14, 117–122 (2000). [DOI] [PubMed] [Google Scholar]
- 128.Wilson R et al. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun 56, 2515–2517 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cruickshank CN & Lowbury EJ The effect of pyocyanin on human skin cells and leucocytes. Br J Exp Pathol 34, 583–587 (1953). [PMC free article] [PubMed] [Google Scholar]
- 130.Levin-Reisman I, Brauner A, Ronin I & Balaban NQ Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. USA 116, 14734–14739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Santi I, Manfredi P, Maffei E, Egli A & Jenal U Evolution of antibiotic tolerance shapes resistance development in chronic Pseudomonas aeruginosa infections. mBio 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors analyze the microevolution of Pseudomonas aeruginosa within chronically infected patients and find that antibiotic tolerance promotes the evolution of antibiotic resistance, validating earlier in vitro studies that showed a link between tolerance and resistance.
- 132.Windels EM et al. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13, 1239–1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Levin-Reisman I et al. Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 134.Liu J, Gefen O, Ronin I, Bar-Meir M & Balaban NQ Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 135.Martina P et al. Hypermutation in Burkholderia cepacia complex is mediated by DNA mismatch repair inactivation and is highly prevalent in cystic fibrosis chronic respiratory infection. Int. J. Med. Microbiol 304, 1182–1191 (2014). [DOI] [PubMed] [Google Scholar]
- 136.Ryan RP et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol 7, 514–525 (2009). [DOI] [PubMed] [Google Scholar]
- 137.Walterson AM & Stavrinides J Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev 39, 968–984 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Smith DDN, Kirzinger MWB & Stavrinides J Draft genome sequence of the antibiotic-producing cystic fibrosis isolate Pantoea agglomerans Tx10. Genome Announc 1, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.König S, Vogel H-J, Harms H & Worrich A Physical, chemical and biological effects on soil bacterial dynamics in microscale models. Front. Ecol. Evol 8, (2020). [Google Scholar]
- 140.Severi E & Thomas GH Antibiotic export: transporters involved in the final step of natural product production. Microbiology (Reading, Engl.) 165, 805–818 (2019). [DOI] [PubMed] [Google Scholar]
- 141.Martín JF, Casqueiro J & Liras P Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Opin. Microbiol 8, 282–293 (2005). [DOI] [PubMed] [Google Scholar]
- 142.Crits-Christoph A, Bhattacharya N, Olm MR, Song YS & Banfield JF Transporter genes in biosynthetic gene clusters predict metabolite characteristics and siderophore activity. Genome Res (2020). doi: 10.1101/gr.268169.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Glasser NR, Saunders SH & Newman DK The colorful world of extracellular electron shuttles. Annu. Rev. Microbiol 71, 731–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yan J & Bassler BL Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ersoy SC et al. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20, 173–181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Song Y et al. Inhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic results. J. Med. Chem 52, 3869–3880 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu C-I et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319, 1391–1394 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Costa KC, Glasser NR, Conway SJ & Newman DK Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science 355, 170–173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.VanDrisse CM, Lipsh-Sokolik R, Khersonsky O, Fleishman SJ & Newman DK Computationally designed pyocyanin demethylase acts synergistically with tobramycin to kill recalcitrant Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This works reveals that degradation of a secondary metabolite that promotes antibiotic tolerance in biofilms can increase antibiotic lethality, suggesting that targeting such secondary metabolites may be a viable approach to potentiating exisiting clinical antibiotics.
- 150.Liu GY & Nizet V Color me bad: microbial pigments as virulence factors. Trends Microbiol 17, 406–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shatalin K et al. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science 372, 1169–1175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors demonstrate that antibiotic efficacy is increased both in vitro and in a mouse infection model by inhibiting the production of a bacterial metabolite previously implicated in intrinsic antibiotic tolerance and resistance.
- 152.Abbott IJ & Peleg AY Stenotrophomonas, Achromobacter, and nonmelioid Burkholderia species: antimicrobial resistance and therapeutic strategies. Semin Respir Crit Care Med 36, 99–110 (2015). [DOI] [PubMed] [Google Scholar]
- 153.Hazan R et al. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol 26, 195–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shukla P et al. “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem. Sci 8, 4967–4972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Anderson RJ & Newman MS The chemistry of the lipids of tubercle bacilli. J. Bio. Chem 103, 405–412 (1933). [Google Scholar]
- 156.Gardner PR Superoxide production by the mycobacterial and pseudomonad quinoid pigments phthiocol and pyocyanine in human lung cells. Arch. Biochem. Biophys 333, 267–274 (1996). [DOI] [PubMed] [Google Scholar]
- 157.Giddens SR, Feng Y & Mahanty HK Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol 45, 769–783 (2002). [DOI] [PubMed] [Google Scholar]
- 158.Giddens SR & Bean DC Investigations into the in vitro antimicrobial activity and mode of action of the phenazine antibiotic D-alanylgriseoluteic acid. Int. J. Antimicrob. Agents 29, 93–97 (2007). [DOI] [PubMed] [Google Scholar]
- 159.Krishnamurthi VS, Buckley PJ & Duerre JA Pigment formation from L-tryptophan by a particulate fraction from an Achromobacter species. Arch. Biochem. Biophys 130, 636–645 (1969). [DOI] [PubMed] [Google Scholar]
- 160.Wells JM, Cole RJ & Kirksey JW Emodin, a toxic metabolite of Aspergillus wentii isolated from weevil-damaged chestnuts. Appl Microbiol 30, 26–28 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lim FY et al. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl. Environ. Microbiol 78, 4117–4125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Imlay JA The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dietrich LEP, Teal TK, Price-Whelan A & Newman DK Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321, 1203–1206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gu M & Imlay JA The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol 79, 1136–1150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Singh AK, Shin J-H, Lee K-L, Imlay JA & Roe J-H Comparative study of SoxR activation by redox-active compounds. Mol. Microbiol 90, 983–996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tanabe M et al. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem. Biophys. Res. Commun 380, 338–342 (2009). [DOI] [PubMed] [Google Scholar]
- 167.Lu S & Zgurskaya HI Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter. Mol. Microbiol 86, 1132–1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wei Q et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40, 4320–4333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K & Hassett DJ Role of the Pseudomonas aeruginosa oxyR–recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB–ankB, ahpB, and ahpC–ahpF. J. Bacteriol 182, 4533–4544 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Romsang A, Dubbs JM & Mongkolsuk S in Stress and environmental regulation of gene expression and adaptation in bacteria (ed. de Bruijn FJ) 1090–1102 (John Wiley & Sons, Inc., 2016). doi: 10.1002/9781119004813.ch106 [DOI] [Google Scholar]
- 171.Blin K et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu J et al. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liu J et al. Coupling between distant biofilms and emergence of nutrient time-sharing. Science 356, 638–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Spero MA & Newman DK Chlorate specifically targets oxidant-starved, antibiotic-tolerant populations of Pseudomonas aeruginosa biofilms. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Saunders SH et al. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 182, 919–932.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rosche WA & Foster PL Determining mutation rates in bacterial populations. Methods 20, 4–17 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zheng Q A new practical guide to the Luria-Delbrück protocol. Mutat. Res 781, 7–13 (2015). [DOI] [PubMed] [Google Scholar]
- 178.Somayaji R et al. Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: a systematic review. J Cyst Fibros 18, 236–243 (2019). [DOI] [PubMed] [Google Scholar]
- 179.Hurley MN, Ariff AHA, Bertenshaw C, Bhatt J & Smyth AR Results of antibiotic susceptibility testing do not influence clinical outcome in children with cystic fibrosis. J Cyst Fibros 11, 288–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jorgensen JH & Ferraro MJ Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis 49, 1749–1755 (2009). [DOI] [PubMed] [Google Scholar]
- 181.Brown MR Nutrient depletion and antibiotic susceptibility. J. Antimicrob. Chemother 3, 198–201 (1977). [DOI] [PubMed] [Google Scholar]


