SUMMARY
Activation of the innate immune system via pattern recognition receptors (PRRs) is key to generate lasting adaptive immunity. PRRs detect unique chemical patterns associated with invading microorganism, but if and how the physical properties of PRR ligands influence development of the immune response remains unknown. Through the study of fungal mannans we show that the physical form of PRR ligands dictates the immune response. Soluble mannans are immunosilent in the periphery but elicit a potent pro-inflammatory response in the draining lymph node (dLN). By modulating the physical form of mannans, we developed a formulation that targets both the periphery and dLN. When combined with viral glycoprotein antigens, this mannan formulation broadens epitope recognition, elicits potent antigen-specific neutralizing antibodies, and confers protection against viral infections of the lung. Thus, the physical properties of microbial ligands determine the outcome of the immune response and can be harnessed for vaccine development.
Graphical Abstract
In Brief
Modulating the physical form of fungal mannans improves lymph node targeting and improves the adjuvant properties of these immune stimuli when included in vaccine formulations against respiratory viruses.
INTRODUCTION
The dialogue between the innate and adaptive branches of the immune system is critical for protection against infections as well as the pathogenesis of autoimmune, allergic, and inflammatory diseases (Banchereau and Steinman, 1998; Iwasaki and Medzhitov, 2004; Janeway and Medzhitov, 2002; Matzinger, 1994). Peripheral tissue infection and/or damage leads to activation and migration of innate immune phagocytes to the draining lymph node (dLN), where they initiate an antigen-dependent adaptive immune response. Alternatively, innate stimuli or microbes with specific physical properties (e.g., diameter in the nanometer range) can directly drain to the dLN and activate LN-resident innate and adaptive immune cells (Bachmann and Jennings, 2010; Irvine et al., 2020). The dLN has been thoroughly scrutinized for its capacity to host adaptive immune responses, but recent reports indicate that the antigen-dependent adaptive immune response is preceded and supported by an antigen-independent LN innate response (Acton et al., 2014; Coccia et al., 2017; De Giovanni et al., 2020; Didierlaurent et al., 2014; Kastenmuller et al., 2012; Leal et al., 2021; Lian et al., 2020; Lynn et al., 2015; Martin-Fontecha et al., 2004; Soderberg et al., 2005; Wong et al., 2019; Wong et al., 2018; Xu et al., 2015b). The LN innate response allows antigen-independent LN expansion, establishment of a pro-inflammatory milieu and the development of an effective adaptive immune response (Acton and Reis e Sousa, 2016; Grant et al., 2020). It remains a mystery if, and how, the LN innate response may differ when it is driven by migration of phagocytes from the periphery as opposed to when it is governed by the direct targeting of LN-resident innate immune cells.
Innate immune cells recognize pathogen-associated molecular patterns (PAMPs) (Janeway and Medzhitov, 2002) via pattern recognition receptors (PRRs) (Brubaker et al., 2015). PRR activation is critical for triggering inflammation and for the ensuing development of adaptive immune responses. For this reason, targeting of PRRs has been harnessed for vaccine development (O’Hagan et al., 2020). Among PRRs, the biology of C-type lectin receptors (CLRs) and their potential as vaccine adjuvant targets has been less investigated.
CLRs control innate and adaptive immune responses to fungal infection through recognition of cell wall polysaccharides (Borriello et al., 2020; Brown et al., 2018). The CLRs Dectin-1 (Clec7a) and Dectin-2 (Clec4n) are activated by β-glucans and mannans, respectively. These fungal polysaccharides vary not only by chemical structure, but also by physical form (e.g., size and solubility). Dectin-1 and Dectin-2 bind fungal polysaccharides in soluble as well as insoluble forms, but only the latter induces efficient receptor clustering and activation (Goodridge et al., 2011; Zhu et al., 2013). Consequently, it is widely held that only particulate polysaccharides are immunostimulatory. In this study we re-examined this paradigm and show that the quality of LN innate and adaptive immune responses can be tuned by modulating the physical properties of fungal ligands, providing a promising approach for adjuvant design and vaccine development.
RESULTS
Mannans elicit LN-restricted IFN signatures that drive LN expansion
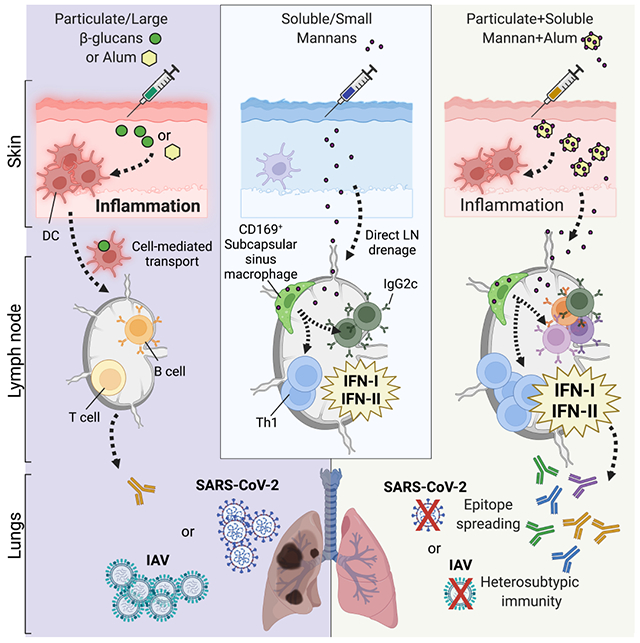
We employed preparations of β-glucans and mannans isolated from Candida albicans that exhibit distinct physical forms, being insoluble (with a diameter of ~500 nm) and soluble (with a diameter of ~20 nm) (Figure S1A). Particulate β-glucans, but not soluble mannans, elicited cytokine production and expression of co-stimulatory molecules by phagocytes in vitro (Figure S1B). As expected, signaling by β-glucans required Dectin-1 (Figure S1C). In contrast to soluble mannans, immobilization of mannans onto microbeads resulted in Dectin-2 and FcRγ-dependent activation of phagocytes (Figure S1C, lipopolysaccharide (LPS) and curdlan were used as controls). Accordingly, β-glucans elicited formation of skin abscesses and lesions upon in vivo intradermal injection, whereas mannans did not (Figure 1A). These results were confirmed by transcriptomic analysis of skin samples (Figure 1B and Table S1. Pathway analysis of the cluster of differentially expressed genes (DEGs) upregulated by β-glucans showed enrichment for pro-inflammatory and type II interferon (IFN) pathways, consistent with the response elicited by C. albicans skin infection (Figure S1D) (Santus et al., 2017). While soluble mannans did not induce skin inflammation, these fungal ligands induced dLN expansion and lymphocyte accrual as early as 6 hours post-injection (h.p.i.), which was sustained at 24 h.p.i. (Figure 1C and Figure S2A–C) and dependent on circulating leukocyte recruitment (Figure 1D). β-glucans also elicited LN expansion but only at 24 h.p.i. (Figure 1C and Figure S2A–C). Both fungal ligands increased the numbers of myeloid cells in the dLN (Figure S2D–G), with β-glucans preferentially increasing neutrophils, possibly reflecting drainage from the skin (Figure S2D). However, only mannans induced activation of myeloid cells, as measured by increased CD86 expression (Figure S2H). Considering the fast response and the diameter compatible with lymphatic drainage, we reasoned that mannans might activate LN-intrinsic circuits that eventually lead to dLN expansion. In keeping with this hypothesis, mannans rapidly accumulated in the dLN (Figure 1E) and induced dLN expansion even in Ccr7−/− mice (Figure 1F) in which migration of immune cells from the periphery (i.e. skin) to the dLNs is abolished (Ohl et al., 2004). A transcriptomic analysis of dLNs showed a completely opposite profile compared to the skin, with mannans eliciting an earlier and more pronounced response compared to β-glucans (Figure 1G, Table S2). Pathway analysis showed that mannans upregulated type I and II IFN pathways (Figure 1H, I).
Figure 1. Mannans elicit lymph node-restricted IFN signatures that drive lymph node expansion.
(A) Mice were injected intradermally with saline (Sal), mannans (Mann) or β-glucans (β-gluc). 24 hours later the injection site was assessed for the presence of an abscess with or without skin lesion. The graph depicts percentages of mice in each of the indicated categories. Representative pictures of skin appearance at injection sites of saline, mannans and β-glucans are also shown. N = 5 mice per group. (B) Transcriptomic analysis of skin samples collected 6 hours after injection of saline (Sal), β-glucans (β-gluc) or mannans (Mann). Heatmap of abundance (z-scored log2 normalized counts) of genes induced by β-glucans and/or mannans compared to saline control, ranked by abundance difference between β-glucans and mannans. The gap splits the genes into two clusters, one that is highly upregulated by β-glucans and one that is highly upregulated by mannans. N = 3 mice per group. (C) Mice were injected intradermally with saline, mannans (Mann) or β-glucans (β-gluc). 6 or 24 hours later dLNs were collected and analyzed for weight as well as absolute numbers of CD45+, B and T cells. Results are expressed as fold over contralateral, saline-injected LN. N = 5-9 mice per group. (D) Mice were injected intravenously with a blocking anti-CD62L antibody (αCD62L) or the same dose of an isotype control (Iso CTRL) one day before intradermal injections of saline or mannans. 24 hours later dLNs were collected and their weights were measured. Results are expressed as fold over contralateral, saline-injected LN. N = 4 mice per group. (E) WT and Ccr7−/− mice were intradermally injected with saline (Sal) or fluorescently labelled mannans (Mann). 1, 6 and 24 hours later dLNs were collected and homogenized to measure total fluorescence. Results are expressed as arbitrary units (A.U.) of fluorescence and shown as mean + SEM. N = 3 mice per timepoint. (F) Ccr7−/− mice were injected intradermally with saline or mannans. 24 hours later dLNs were collected and analyzed as indicated in C. N = 6 mice. (G) Transcriptomic analysis of dLNs collected 6 and 24 hours after intradermal injection of saline (Sal), β-glucans (β-gluc) or mannans (Mann). Heatmap of abundance (z-scored log2 normalized counts) of genes induced by β-glucans and/or mannans compared to saline control, ranked by abundance difference between β-glucans and mannans. The gap splits the genes into two clusters, one that is highly upregulated by β-glucans and one that is highly upregulated by mannans. N = 4-5 mice per group. (H) Pathway enrichment analysis of genes belonging to the cluster upregulated by mannans as depicted in G. (I) Heatmap representation of the average expression levels of the top 50 genes upregulated in mannan-treated dLNs 24 hours after the injection compared to the saline control. N = 4-5 mice per group. (J) WT and Ifnar−/− mice were intravenously injected with an anti-IFNγ blocking antibody (αIFNγ) or the same dose of an isotype control (Iso CTRL) on day −1 and 0. On day 0 mice were also intradermally injected with saline (Sal) or mannans (Mann). 24 hours later dLNs were collected, their weights were measured, and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected LN (weight) or as relative expression compared to Gapdh. N = 4 mice per group. (K) WT and Ifnar−/− Ifngr−/− mice were intradermally injected with saline (Sal) or Lipo-CpG. Samples were collected and analyzed as in J. N = 5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against the value 1 (which represent the contralateral control sample expressed as fold). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing among different experimental groups. See also Figure S1, S2, S3 and Table S1 and S2.
The presence of type II IFN (IFNγ)-producing cells in mannan-treated dLN was confirmed by flow cytometry (Figure S2I). The majority of IFNγ-producing cells were CD8+ T and NK cells (Figure S2J), with NK cells expressing IFNγ at higher levels (Figure S2K). When NK cells were depleted, expression of Ifng was impaired (Figure S2L). cDC1 produce cytokines that induce NK cell activation and IFNγ production (Cancel et al., 2019) and may therefore contribute to the response induced by mannans. Nevertheless, Batf3−/− mice that lack cDC1 showed no impairment of mannan-elicited Ifng expression (Figure S2M). Notably, NK cell depletion only partially affected LN expansion and cell accrual (Figure S2N. In keeping with this, IFNγ blockade only partially reduced mannan-induced expansion of LN (Figure 1J). We therefore tested whether the absence of both IFNγ and type I IFN signaling impacted the LN innate response. Simultaneous blockade of both IFN types prevented mannan-elicited LN expansion and induction of interferon-stimulated genes (ISGs) (Figure 1J). This mechanism was not restricted to mannans, since LN expansion and induction of ISGs elicited by Lipo-CpG, a well characterized LN-targeted Toll-like receptor (TLR)9 ligand (Liu et al., 2014), were also impaired in Ifnar−/− Ifngr−/− mice (Figure 1K). To further establish whether these differences are due to distinct physical forms of fungal polysaccharides, we evaluated the response to whole glucan particles (WGP) in dispersible (D) or soluble (S) forms, which have been characterized as Dectin-1 agonists and antagonists, respectively (Goodridge et al., 2011). Consistent with the pattern observed with mannans, WGP-S did not induce skin inflammation (Figure S3A) but elicited LN expansion and ISG expression (Figure S3B). Altogether, these results support a model in which the physical form of PAMPs drive expansion of the dLN and ISG induction.
Mannan-elicited LN innate response requires Dectin-2-expressing CD169+ sinus macrophages
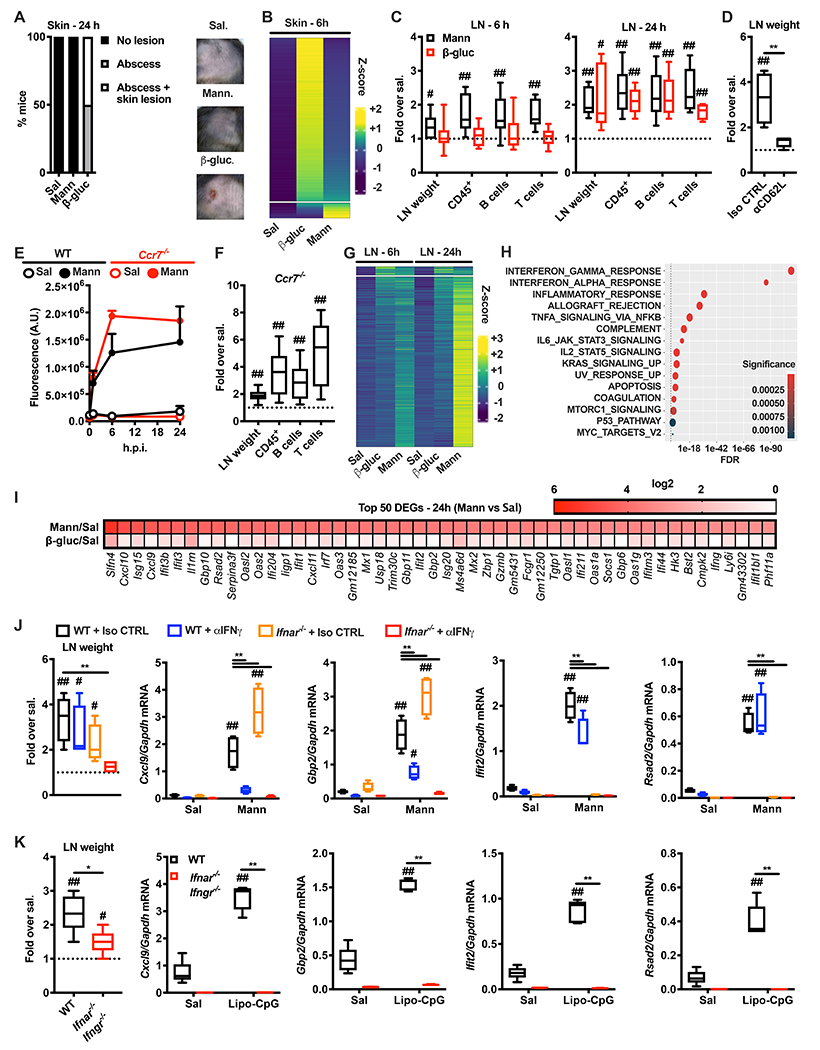
We found that Dectin-2, the major receptor for mannans (Borriello et al., 2020; Lionakis et al., 2017; Netea et al., 2008), and its co-receptor FcRγ were required for mannan-elicited LN expansion and ISG induction (Figure 2A). We employed fluorescently labeled mannans and found that mannan-laden cells were CD45+ cells (Figure 2B). Imaging cytometry analysis confirmed that these cells internalized mannans (Figure 2C) and confocal microscopy analysis showed colocalization of phospho-Syk and mannans (Figure 2D), indicative of Dectin-2 and FcRγ-mediated activation. Accordingly, mannan-laden cells showed the highest levels of expression of CD86 in an FcRγ-dependent manner (Figure 2E). We found that more than 50% of mannan-laden cells were Ly6G− (CD11b+ Ly6C+)− CD11c+ cells, while less abundant cell subsets were neutrophils (CD11b+ Ly6G+) and monocytes(-derived) cells (MoCs, Ly6G− CD11b+ Ly6C+) (Figure 2F). Depletion of CD11c+ cells, but not inhibition of monocyte egress from the bone marrow or depletion of neutrophils, abolished the mannan-elicited LN innate response (Figure 2G–I). CD11c+ cells can be further distinguished based on the expression of CD11b (Figure 2F). Since Dectin-2 is critical for mannan-elicited LN innate response, we assessed its expression on CD11b− CD11c+ and CD11b+ CD11c+ cells at steady state. Dectin-2 was expressed mainly by CD11b+ CD11c+ cells (Figure 2J). The majority of CD11b+ CD11c+ Dectin-2+ cells expressed the subcapsular and medullary sinus macrophage marker CD169 (Figure 2K). Confocal microscopy analysis confirmed colocalization of CD169 and Dectin-2 on cells lining the LN subcapsular sinus (Figure 2L). Furthermore, DT-mediated depletion of CD169+ cells in CD169-DTR mice phenocopied the results obtained with CD11c-DTR mice and completely abolished mannan-elicited LN expansion and ISG induction (Figure 2M). These results are consistent with a role for CD169+ sinus macrophages as sentinels of lymph-borne materials (Moran et al., 2019).
Figure 2. Mannan-elicited lymph node innate response requires Dectin-2-expressing, CD169+ sinus macrophages.
(A) WT, Clec4n−/− and Fcer1g−/− mice were intradermally injected with saline or mannans. 24 hours later dLNs were collected, their weights were measured, and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected LN. N = 3-5 mice per genotype. (B) WT mice were intradermally injected with fluorescently labelled mannans (Mann-AF488). 6 hours later dLNs were collected and the absolute numbers of mannan-laden (Mann+) CD3/CD19/NK1.1− cells were quantified by flow cytometry. N = 6 mice. (C) Mice were treated as in B. Imaging cytometry analysis and quantification of mannan internalization was performed on CD3/CD19/NK1.1-depeted, CD45+ mannan-laden (Mann+) cells. N = 4 mice. (D) WT mice were intradermally injected with fluorescently labelled mannans (Mann). 1 hour later dLNs were collected for confocal microscopy analysis using antibodies against B220 and phospho-Syk (pSyk). DAPI was used for nuclear counterstaining. One representative image is shown. (E) WT and Fcer1g−/− mice were injected with saline or fluorescently labelled mannans. 6 hours later dLNs were collected and CD86 expression levels were assessed by flow cytometry on CD3/CD19/NK1.1− CD45+ mannan-laden (Mann+) cells, CD45+ cells that did not capture mannans (Mann−) and CD45+ cells from saline-injected dLNs (Sal). N = 6 mice per genotype. (F) WT mice were intradermally injected with fluorescently labelled mannans. 6 hours later dLNs were collected and the phenotype of CD3/CD19/NK1.1− CD45+ mannan-laden (Mann+) cells was assessed by flow cytometry. N = 6 mice. (G - I) Diphtheria toxin (DT)-treated CD11c-DT receptor (DTR), Ccr2−/− and isotype control (Iso CTRL)- or anti-Ly6G (αLy6G)-treated mice were treated and analyzed as in A. N = 4 mice per group. (J, K) LNs were isolated from untreated WT mice and the expression of Dectin-2 was evaluated by flow cytometry as percentage of expression in the indicated CD3/CD19/NK1.1− CD45+ cell subsets. N = 6 for J or 3 for K. (L) Confocal microscopy analysis of untreated LNs stained with antibodies against Dectin-2, B220 and CD169. DAPI was used for nuclear counterstaining. One representative image is shown. (M) DT-treated CD169-DTR mice were treated and analyzed as in A. N = 4 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against the value 1 (which represent the contralateral control sample expressed as fold) or saline control. * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing among different experimental groups.
Activation of the non-canonical NF-κB subunit RelB governs mannan-elicited LN innate responses
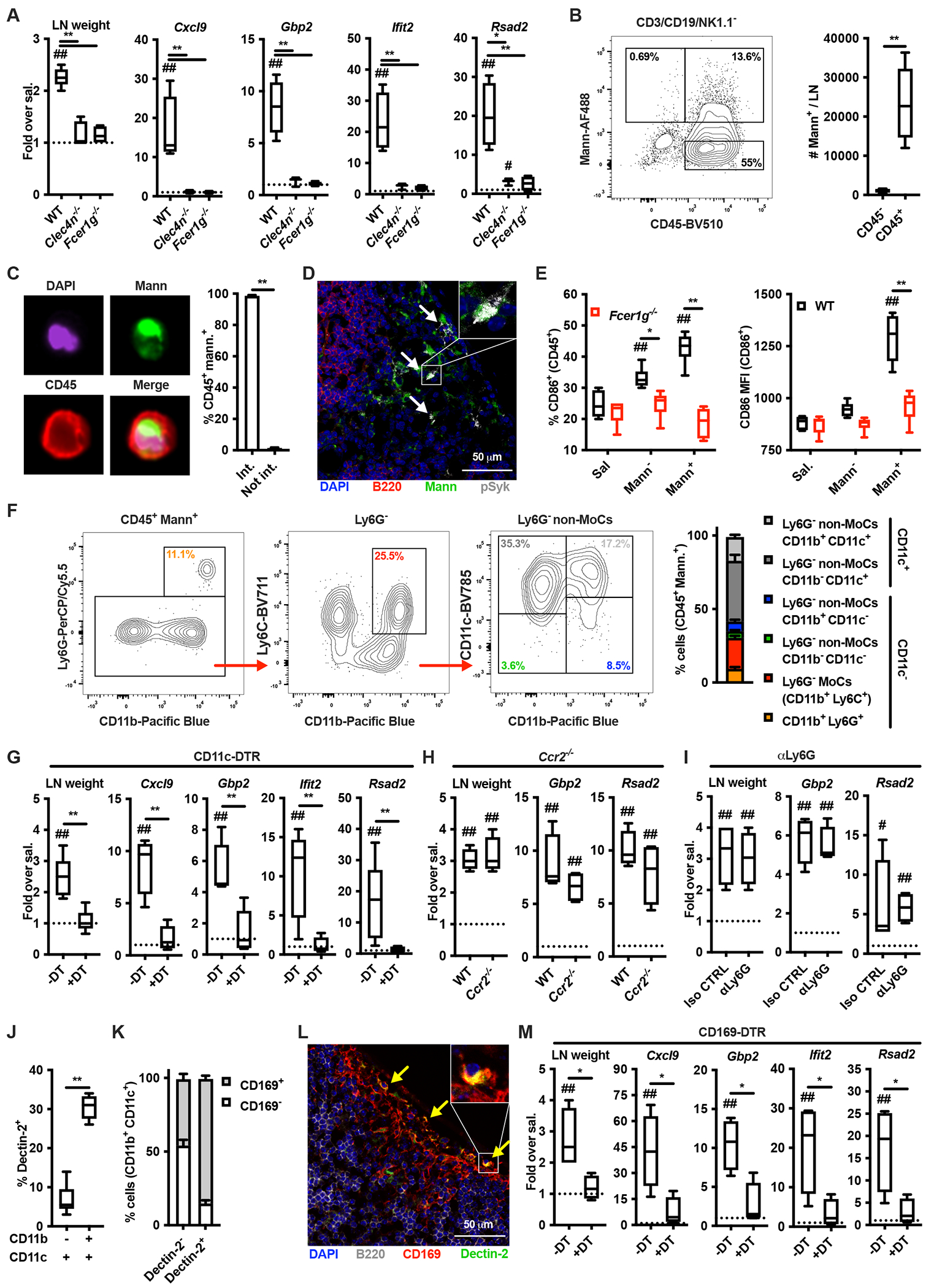
CARD9 is the major signaling adaptor of Dectins (Brubaker et al., 2015) but mannan-elicited LN expansion was comparable between wild type (WT) and Card9−/− mice (Figure 3A). In keeping with this, induction of ISGs was largely maintained in Card9−/− mice (Figure 3A). In particular, type I IFN-dependent genes were unchanged in Card9−/− compared to WT mice, while type II IFN-dependent ISGs, although significantly decreased compared to WT mice, were still partially induced (Figure 3A). Next, we performed a targeted transcriptomic analysis of mannan-laden CD11b+ CD11c+ cells isolated from WT, Fcer1g−/− and Card9−/− mice. While several genes were differentially expressed between WT (or Card9−/−) and Fcer1g−/− mice, cells isolated from WT and Card9−/− mice exhibited strikingly similar transcriptomes (Figure 3B, Table S3–S7). Pathway enrichment analysis showed that DEGs between cells isolated from WT and Fcer1g−/− are represented in TNF/NF-κB, type I and II IFN pathways (Figure S4). Downstream of Dectin-2, Syk activates the kinase NIK, which in turn leads to CARD9-independent activation of the non-canonical NF-κB transcription factor RelB (Gringhuis et al., 2009; Xu et al., 2018). We therefore generated mice in which RelB is conditionally deleted in the CD11c+ compartment (Cd11ccre Relbfl/fl). Mannan-induced LN expansion and expression of both type I and type II IFN-dependent ISGs were significantly reduced compared to control (Relbfl/fl mice (Figure 3C). Similar results were obtained when Lipo-CpG was employed (Figure 3D). These results support a model in which RelB regulates optimal IFN-dependent ISG expression elicited by LN-targeted stimuli to sustain LN expansion.
Figure 3. Activation of the non-canonical NF-kB subunit RelB governs the mannan-elicited lymph node innate response.
(A) WT and Card9−/− mice were treated and analyzed as in Figure 2A. N = 9 (for LN weight) or 4 (for gene expression analysis) mice per genotype. (B) CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ cells were sorted from dLNs of WT, Fcer1g−/− and Card9−/− mice 6 hours after AF488-mannan injection and transcriptional profiles were assessed by targeted transcriptome sequencing. Results are shown as heatmap of genes with an F-test FDR less than 0.05 and a log2 fold-change (FC) greater than 1 (or lower than −1) between a knockout mouse and WT control. (C, D) Relbfl/fl and Cd11ccre Relbfl/fl mice were treated with saline, mannans or Lipo-CpG, and analyzed as in Figure 2A. N = 4-13 mice per genotype. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against the value 1 (which represent the contralateral control sample expressed as fold) or saline control. * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing among different experimental groups. See also Figure S4 and Table S3, S4, S5, S6 and S7.
Molecular pathways required for mannan-elicited LN innate response regulate the magnitude of mannan adjuvant activity
We reasoned that lymphocyte accrual and IFN signatures induced by mannan may favor the encounter of T cells with their cognate antigen and its efficient presentation by the innate immune compartment, thereby improving the adaptive immune response. We employed a model of adoptive transfer of CFSE-labeled, OVA-specific OT-I CD8+ and OT-II CD4+ T cells to assess modulation of T cell proliferation (Figure 4A–H). Combining mannans with OVA resulted in a strong increase in the numbers of OT-I and OT-II cells in the dLN compared to mice injected with saline or OVA alone (Figure 4A, E). This increase was likely due to improved T cell recruitment to the dLN and more efficient antigen presentation/co-stimulation by LN-resident innate immune cells. Indeed, we detected a lower percentage of non-proliferating T cells as well as a higher percentage of T cells undergoing 6 or 7 divisions in mice treated with OVA and mannans compared to OVA alone (Figure 4B, F). Of note, the effect of mannans on T cell proliferation was abrogated in Fcer1g−/− mice, while it was only attenuated in Card9−/− mice (Figure 4C, D, G, H). These results show that pathways required for mannan-elicited LN innate response are also critical for the adjuvant activity of mannans on antigen-specific adaptive immune responses.
Figure 4. Molecular pathways required for mannan-elicited lymph node innate response regulate the magnitude of mannan adjuvant activity.
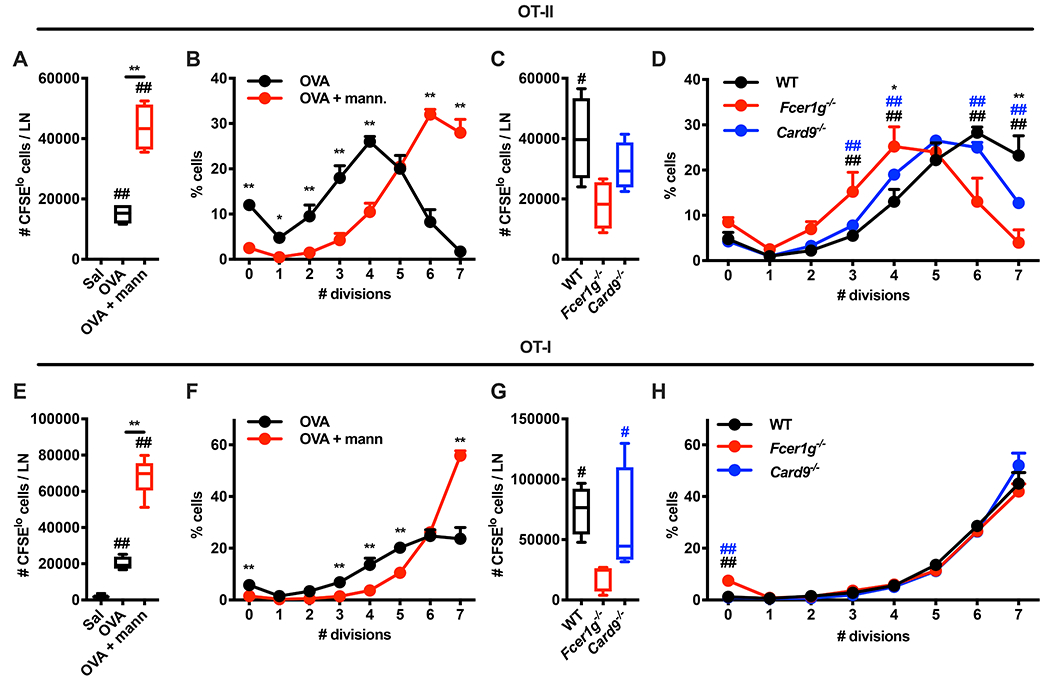
(A-H) CFSE-labelled OT-I CD8+ T or OT-II CD4+ T cells were injected intravenously in WT mice on day −1. On day 0 the mice were intradermally injected with saline, ovalbumin (OVA), or OVA combined with mannans (OVA + mann). 3 days later dLNs were isolated and the absolute numbers of CFSElo cells (i.e., cells that underwent at least one cycle of cell division) (A, E) or the percentages of cells in each division peak (B, F) were quantified by flow cytometry. N = 4 mice per group. ## indicates p ≤ 0.01 when comparing each group against saline control (A, E). * and ** respectively indicate p ≤ 0.05 and p ≤ 0.01 when comparing OVA vs OVA + mann. (A, B, E, F). (C, D, G, H) WT, Fcer1g−/− and Card9−/− mice were treated and analyzed as in A, B, E, F (with the exception that all mice received OVA combined with mannans). N = 4 mice per genotype. # and ## respectively indicate p ≤ 0.05 and p ≤ 0.01 when comparing WT vs Fcer1g−/− (black) or Card9−/− vs Fcer1g−/− (blue). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing WT vs Card9−/−. Results in B, F and D, H are shown as mean + SD.
Mannans formulated with aluminum hydroxide acquire physical properties that predict immunological functions
To further modulate the physical properties of mannans, we exploited the presence of phosphate groups of mannans to promote adsorption onto aluminum hydroxide (hereafter alum), as shown for other molecules (Morefield et al., 2005; Moyer et al., 2020). By 1H-NMR we found that alum adsorbed ~40% of mannans, with the remainder staying soluble (Figure S5A). We also quantified that alum bound mannans at approximately twice its mass in the formulation used in these experiments.
In vitro experiments showed that formulation of mannans with alum, but not alum or mannans alone, induced cytokine production in a Dectin-2- and CARD9-dependent manner (Figure S5B, C), while the expression of costimulatory molecules was Dectin-2-dependent but CARD9-independent (Figure S5D, E). Finally, mannans formulated or not with alum, but not alum alone, induced ISG expression in a Dectin-2-dependent but CARD9-independent manner, except for CXCL1 that was significantly upregulated only in response to mannans formulated with alum in WT cells (Figure S5F–I).
When injected into mice, mannans formulated with alum elicited skin inflammation (Figure 5A), but also drained to the LN in a CCR7-independent manner (Figure 5B). Mannans formulated with alum induced over time a higher LN expansion compared to alum, mannans or β-glucans (Figure 5C). Moreover, mannans alone or in combination with alum elicited comparable ISG expression in the dLN (Figure 5D). As expected, β-glucans induced LN expansion but were largely excluded from the LN and did not induce expression of ISG (Figure 5B–D). LN expansion induced by mannans formulated with alum, but not β-glucans, was impaired in Ifnar−/− mice treated with an anti-IFNγ blocking antibody (Figure 5E). Similar results were obtained when IFN production was transiently blocked by treating WT mice with anti-IFNAR and anti-IFNγ blocking antibodies (Figure 5F). Overall, these results are compatible with a model in which the particulate fraction (mannans adsorbed onto alum) promotes skin inflammation, while the soluble fraction (unbound mannans) drains to the LN and induces the ISG expression. Side by side injections of mice with either particulate or soluble fractions of mannans formulated with alum validated this model (Figure 5G, H). Finally, we assessed whether the LN innate response induced by mannans formulated with alum exploits the same cellular and molecular mechanisms of mannans or if the immunomodulatory functions of alum (Eisenbarth et al., 2008) completely rewired these requirements. We found that LN expansion and ISG induction were impaired in Clec4n−/− and Fcer1g−/− mice (Fig. 5I). In addition, Ifng expression in the dLN was reduced when NK cells were depleted (Fig. 5J) and preserved in Batf3−/− mice lacking cDC1 (Fig. 5K). Overall, formulation with alum endows mannans with enhanced immunological functions that can be predicted based on their physical properties (i.e., particulate vs soluble) and reflect triggering of mannan-dependent Dectin-2-activated pathways.
Figure 5. Formulation of mannans with aluminum hydroxide confers physical properties that predict immunological functions.
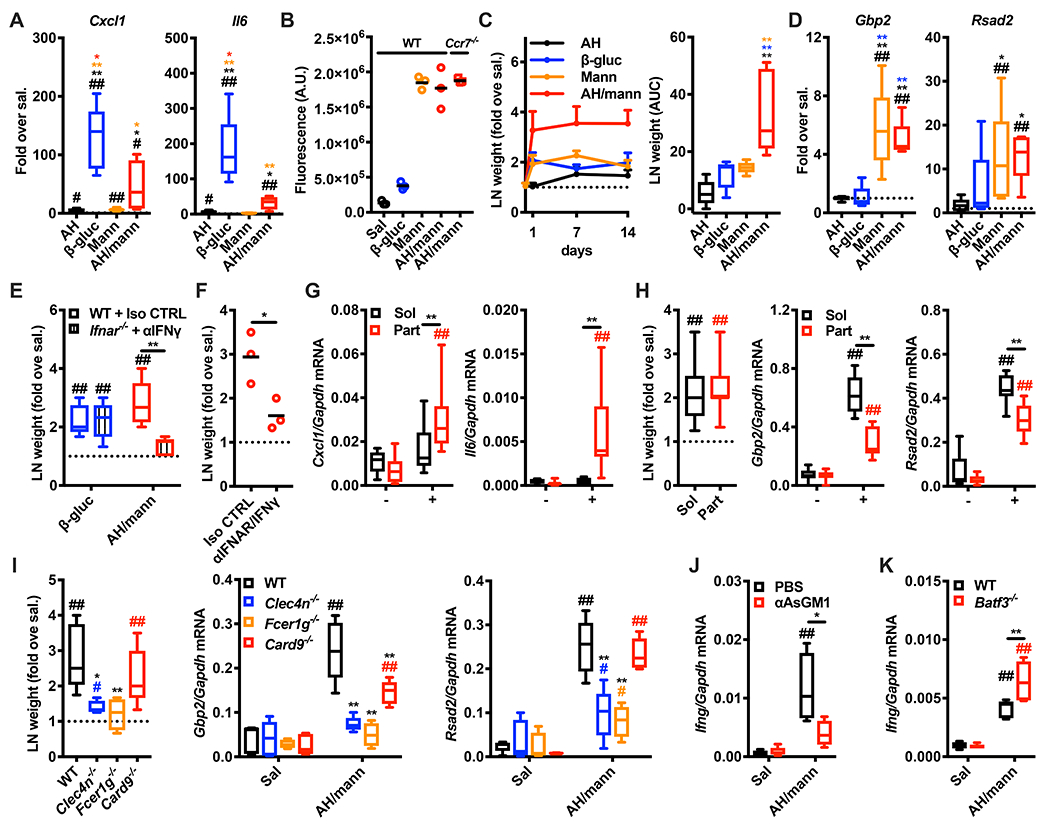
(A) Mice were intradermally injected with saline (sal.), alum (AH), β-glucans (β-gluc), mannans (Mann) or AH/mannans (AH/mann). 24 hours later skin samples were collected, and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected skin sample. N = 4-5 mice per group. (B) WT mice were intradermally injected with saline (Sal), fluorescently labelled β-glucans (β-gluc), fluorescently labelled mannans (Mann) or their formulation with AH (AH/mann). In addition, Ccr7−/− mice were intradermally injected with AH/mann. 24 hours later dLNs were collected and homogenized to measure total fluorescence. Results are expressed as arbitrary units (A.U.) of fluorescence and shown as individual data points (horizontal bars represent means). N = 3 mice. (C) Mice were treated as in A. 1, 7 and 14 days later dLNs were collected, their weights were measured and expressed as fold over contralateral, saline-injected LN. Results are represented as mean + SEM (left panel) or area under the curve (AUC, right panel). (D) Mice were treated as in A. 24 hours later dLNs were collected, and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected LN. N = 5 per group. (E) Ifnar−/− and WT mice were respectively treated with a blocking anti-IFNγ antibody (αIFNγ) or the same dose of an isotype control (Iso CTRL) on day −1 and 0, and on day 0 mice were intradermally injected with saline (sal.), β-glucans (β-gluc), or AH/mannans (AH/mann). 24 hours later dLNs were collected and their weights were measured. Results are expressed as fold over contralateral, saline-injected LN. N = 5 mice per group. (F), WT mice treated with blocking anti-IFNAR plus anti-IFNγ (αIFNAR/IFNγ) antibodies or the same doses of isotype controls (Iso CTRL) on day −1 and 0, and on day 0 mice were intradermally injected with saline (sal.) or AH/mannans (AH/mann). 24 hours later dLNs were collected and their weights were measured. Results are expressed as fold over contralateral, saline-injected LN. N = 5 mice per group. (G, H) Mice were treated with the soluble (Sol) or particulate (Part) fractions of AH/mannans. 24 hours later skin samples (G) and dLNs (H) were collected, dLN weights were measured and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected skin sample or LN. N = 5 mice per group. (I) Mice of the indicated backgrounds were injected with saline (sal.) or AH/mannans (AH/mann). 24 hours later dLNs were collected, their weights were measured, and RNA was extracted for gene expression analysis. Results are expressed as fold over contralateral, saline-injected sample or as relative expression compared to Gapdh. (J, K) WT mice injected on day −1 and 0 with the same volumes of PBS or a depleting anti-Asialo GM1 antibody (αAsGM1) (J), or WT and Batf3−/− mice (K) were injected intradermally on day 0 with saline (Sal) or AH/mannans (AH/mann). 24 hours later dLNs were collected, and RNA was extracted for gene expression analysis. Results are reported as relative expression compared to Gapdh N = 5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against its untreated control (CTRL) or the value 1 (which represent the contralateral control sample expressed as fold). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing among different experimental groups. See also Figure S5.
Immunization with SARS-CoV-2 Spike protein and mannans formulated with alum generates anti-Spike type 1 immunity and neutralizing antibodies with broad epitope specificity
To investigate adjuvant activities of mannans in an immunization model of translational relevance, we used the pre-fusion stabilized SARS-CoV-2 Spike trimer (hereafter Spike) (Corbett et al., 2020; Jackson et al., 2020; Keech et al., 2020; Mercado et al., 2020; Walls et al., 2020; Walsh et al., 2020; Wrapp et al., 2020). We immunized mice with Spike alone or with Spike admixed with alum, β-glucans, or mannans formulated or not with alum, with a prime - boost schedule. Mannans formulated with alum induced the highest levels of anti-Spike or anti-receptor binding domain (RBD) antibodies (Figure 6A, B). Type-1 immunity has been associated with reduced risk of vaccine-associated enhanced respiratory disease upon viral infection (Graham, 2020), and mannans formulated with alum promoted anti-Spike type-1 immunity by inducing anti-Spike IgG2c and antigen-specific T cells skewed toward IFNγ production (Figure 6A–C). Elevated levels of anti-Spike IgG1 and IgG2c were maintained for up to 98 days post immunization (Figure S6A, B. Next, we performed a surrogate virus neutralization test and an actual SARS-CoV-2 neutralization test and found that mice immunized with Spike and mannans formulated with alum showed the highest degree of neutralization (Figure 6D, E). We also assessed whether there were spatial and temporal constraints to the adjuvant effect of mannans formulated with alum. Mice were injected with Spike and mannans formulated with alum at the same site, or with Spike formulated with alum at one injection site and with mannans at an adjacent site, either on the same day or on consecutive days. An enhanced antibody response was observed only when Spike was admixed with mannans formulated with alum (Fig. 6F, G. In contrast to B cell responses, IFNγ production by antigen-specific T cells was observed in mice immunized with alum and mannans, regardless of when and where mannans were injected (Fig. 6H).
Figure 6. Immunization with SARS-CoV-2 Spike protein and mannans formulated with alum generates anti-Spike type 1 immunity and neutralizing antibodies.
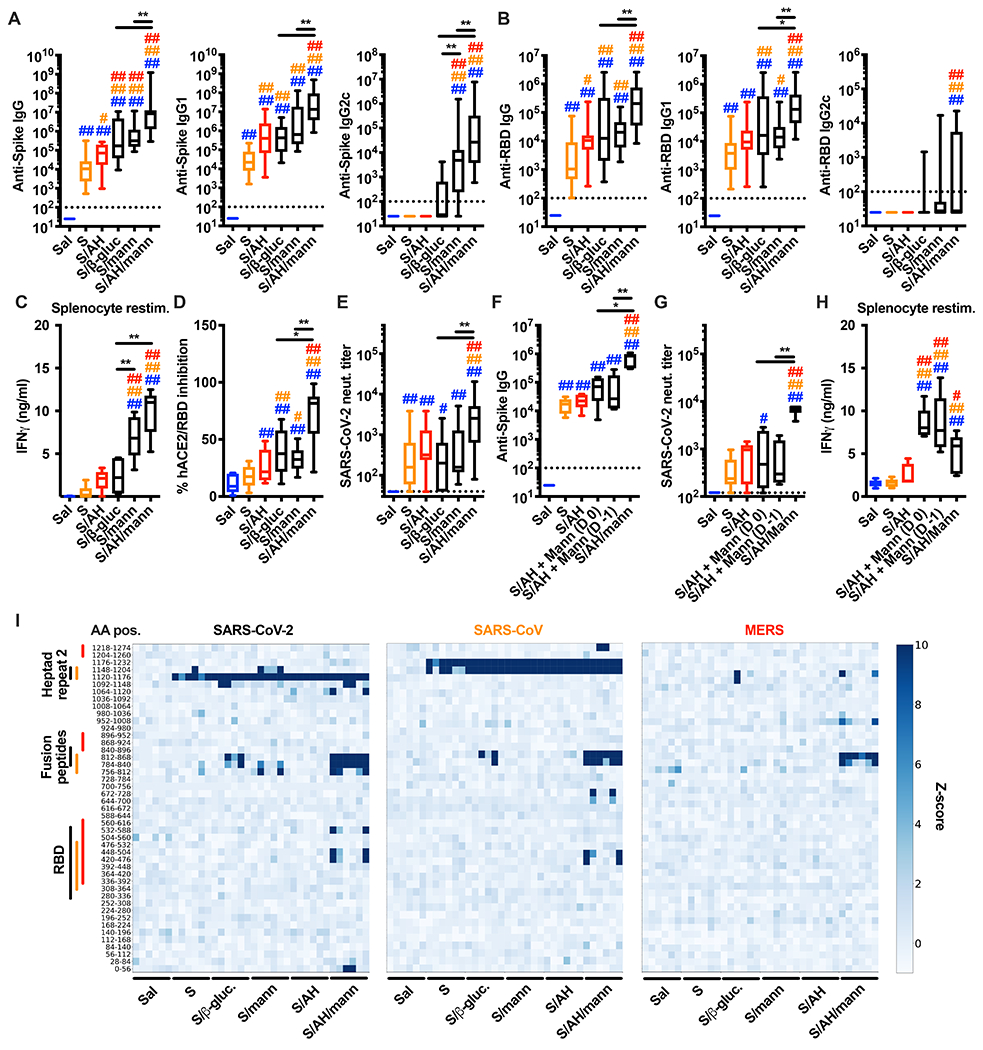
(A-E) Mice were injected intradermally with saline (Sal), pre-fusion stabilized SARS-CoV-2 trimer alone (S) or combined with alum (AH) (S/AH), β-glucans (S/β-gluc.), mannans (S/mann) or AH/mannans (S/AH/mann) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess anti-Spike (A) and anti-RBD (B) antibody levels, SARS-CoV-2 surrogate virus neutralization test (D) and neutralization titer (E). In selected experiments (C), mice were sacrificed on day 35 to collect spleens and isolate splenocytes for in vitro restimulation with Spike peptides. After 96 hours supernatants were collected and IFNγ protein levels were measured by ELISA. N = 16-18 (A, B), 10 (C), 8-10 (D) or 13-15 (E) mice per group. (F-H) Mice were injected intradermally with saline (Sal), pre-fusion stabilized SARS-CoV-2 trimer alone (S), or combined with AH (S/AH). Mannans (Mann) were injected separately on the same side of the S/AH injection in a proximal site, either the same day (S/AH + Mann (D 0)) or the day before (S/AH + Mann (D −1)). As a control, SARS-CoV-2 trimer combined with AH and mannans (S/AH/Mann) was also injected. Formulations were injected on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess anti-Spike antibody levels (F) and SARS-CoV-2 neutralization titer (G). In selected experiments (H), mice were sacrificed on day 35 to collect spleens and isolate splenocytes for in vitro restimulation as in C. N = 6-8 mice per group. #, * and ##, ** respectively indicate p ≤ 0.05 and 0.01 when comparing among different experimental groups. Comparisons are indicated by the color code. (I) Mice were immunized as in A-E. VirScan analysis was performed on serum samples collected on day 28. Each column represents a single serum sample collected from an individual mouse and each row represents a peptide tile. Tiles are labeled by amino acid start and end position. Color intensity represents the degree of enrichment (z-score) of each peptide. Color-coded lines indicate the approximate aminoacidic positions (AA pos.) of RBD, Fusion peptides and Heptad repeat 2 of each virus. N = 6 mice per group. See also Figure S6.
B and T cell responses induced by mannans formulated with alum were abrogated in Clec4n−/− mice (Fig. S6C, D) but only partially impaired in Card9−/− mice (Fig. S6E, F). Immunization of Cd11ccre Relbfl/fl or Ifnar−/− Ifngr−/− mice, as well as of WT mice in which IFN signaling was transiently abrogated during the immunization phase by administration of anti-IFNAR and anti-IFNγ antibodies, all showed impairment in the anti-Spike IgG response (Fig. S6G–I). Finally, we found that cDC1 were also required, as assessed by impaired IgG production in Batf3−/− mice (Fig. S6J).
To probe the extent of the Spike epitopes targeted by antibodies elicited by the different adjuvant formulations tested above, we performed a VirScan analysis (Shrock et al., 2020; Xu et al., 2015a). All formulations induced antibodies against the Heptad repeat 2 region, but only mannans formulated with alum induced antibodies directed toward the Fusion peptide and RBD (Figure 6I). This epitope targeting profile was comparable for SARS-CoV-2 and SARS-CoV Spike proteins. Notably, although fewer epitopes were recognized, the MERS Spike was also detected by antibodies elicited in mice immunized with SARS-CoV-2 spike and mannans formulated with alum (Figure 6I). Altogether, our results show that mannan formulations enhance anti-Spike antibody levels and promote anti-Spike type 1 immunity, and that mannans formulated with alum are particularly effective at inducing anti-Spike neutralizing antibodies with broad epitope specificity.
The formulation of mannans and alum confers protection against viral infections of the lung.
Next, we benchmarked mannans formulated with alum against FDA-approved adjuvants: squalene-based oil-in-water nano-emulsions (AS03-like AddaS03 or MF59-like AddaVax) and the AS04-like formulation prepared by simple admixture of alum and PHAD, a synthetic structural analog of the monophosphoryl lipid A (alum/PHAD).
Immunization of mice with Spike and mannans formulated with alum, AddaS03 or alum/PHAD elicited comparable levels of anti-Spike and anti-RBD IgG antibodies (Figure 7A). Increased anti-Spike and anti-RBD antibodies correlated with increased neutralization capacity compared to alum alone (Figure S7A, B). When mice were infected with the mouse-adapted SARS-CoV-2 MA10 strain (Leist et al., 2020), we found markedly reduced viral lung titers in mice immunized with mannans formulated with alum, AddaS03 or alum/PHAD compared to saline-treated or alum-immunized mice (Figure 7B).
Figure 7. The adjuvant formulation of mannans and alum confers protection against lung viral infections.
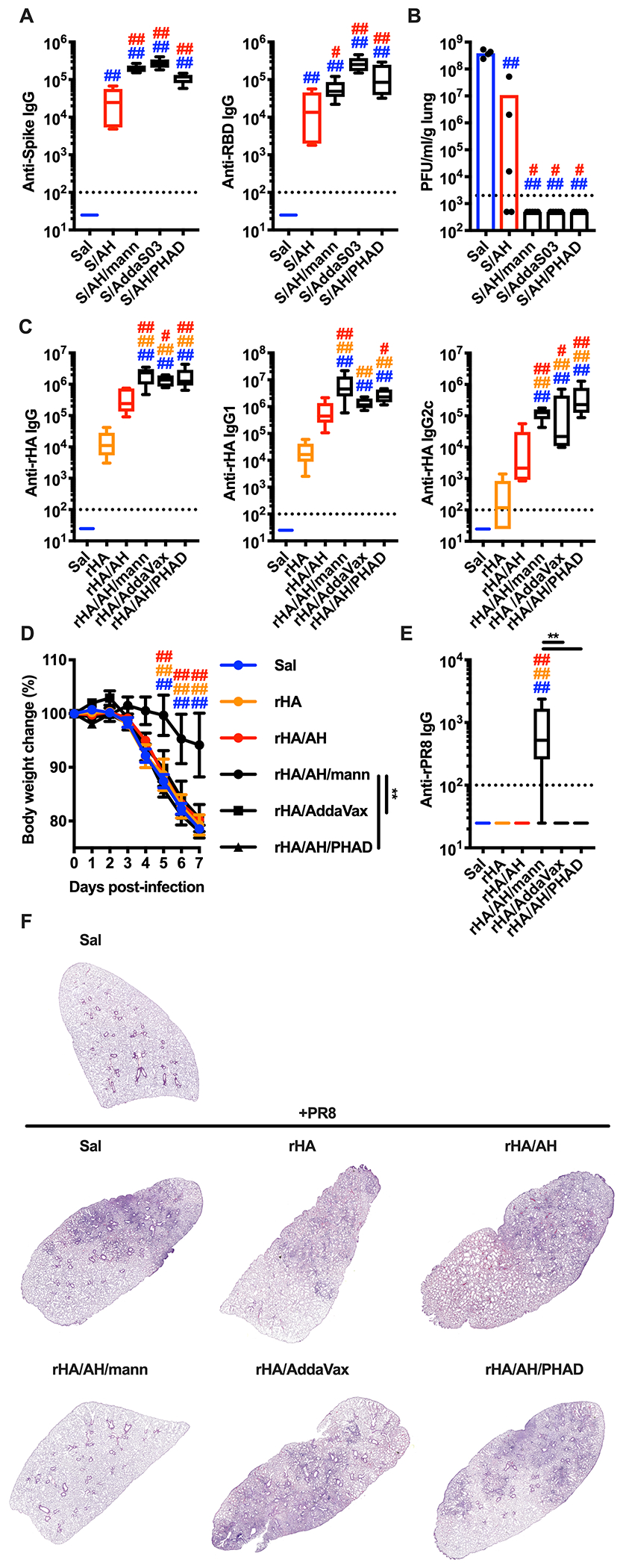
(A, B) Mice were injected intradermally with saline (Sal), pre-fusion stabilized SARS-CoV-2 trimer alone (S) or combined with alum (AH) (S/AH), AH/mannans (S/AH/mann), AddaS03 (S/AddaS03), or AH/PHAD (S/AH/PHAD) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess anti-Spike and anti-RBD antibody levels (A). On day 35 mice were intranasally infected with SARS-CoV-2 MA10 on day 35 and 2 days later numbers of plaque forming units (PFU) were quantified in the lungs (B). N = 4-5 mice per group. (C - F) Mice were injected intradermally with saline (Sal), Flublok alone (rHA) or combined with AH (rHA/AH), AH/mannans (rHA/AH/mann), AddaVax (rHA/AddaVax), or AH/PHAD (rHA/AH/PHAD) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess antibodies against rHA (anti-rHA, C) or IAV A/PR/8/1934 recombinant hemagglutinin (anti-rPR8, E). On day 35 mice were intranasally infected with IAV A/PR/8/1934 and body weights were recorded for 7 days (D). N = 5 (C, E) or 8 (D) mice per group. On day 7 post-infection mice were sacrificed and lungs were collected for histological analysis (hematoxylin eosin staining, F). One representative image per group is shown. #, * and ##, ** respectively indicate p ≤ 0.05 and 0.01. Comparisons are indicated by the color code. See also Figure S7.
The enhanced magnitude and breadth of the antigen-specific antibody response elicited by mannans formulated with alum might be relevant for additional viral glycoproteins of high antigenic variability, such as influenza A virus (IAV) hemagglutinin (HA). IAV is characterized by many strains within multiple serotypes, generating high subtypic diversity (Sangesland and Lingwood, 2021). We reasoned that the mannan formulation might not only promote a robust antibody response against target antigens but heterosubtypic immunity upon influenza vaccine immunization. We employed the clinically relevant recombinant HA (rHA) vaccine Flublok, and immunized mice with Flublok alone or formulated with alum, mannans formulated with alum, Addavax, or alum/PHAD. Anti-rHA antibodies were significantly increased in mice immunized with rHA and mannans formulated with alum, AddaVax or alum/PHAD (Figure 7C). We then challenged the mice intranasally with the IAV strain A/PR/8/1934, whose HA is not part of the Flublok vaccine, and found that only mice previously immunized with Flublok and mannans formulated with alum were significantly protected. These results correlated with high IgG levels against recombinant HA derived from A/PR/8/1934 (rPR8) in mice immunized with Flublok and mannans formulated with alum, but not the other adjuvant formulations (Figure 7D–F, S7D).
Overall, these data show that the mannans formulated with alum enhances both magnitude and breadth of the antibody response against multiple viral glycoproteins in clinically relevant immunization models.
DISCUSSION
Activation of innate immune cells by PRR ligands is a critical step to initiate an adaptive immune response (Banchereau and Steinman, 1998; Iwasaki and Medzhitov, 2004; Janeway and Medzhitov, 2002; Matzinger, 1994). The study of cellular and molecular events triggered by PRRs led to the identification of signaling organelles, metabolic pathways and gene expression profiles that shape the innate immune response (Brubaker et al., 2015). However, cell-intrinsic features of PRR activation and signaling alone do not explain the complexity of the in vivo inflammatory response elicited by innate stimuli. Indeed, their localization at cellular and organismal levels plays a key role in determining the activation status of innate immune cells (Evavold and Kagan, 2019).
The study of CLRs, and specifically of Dectin-1 and -2, is of particular interest since only the particulate form of their ligands induces efficient receptor clustering and activation (Goodridge et al., 2011; Zhu et al., 2013). In our study, we show that soluble mannans, while largely inactive in vitro and in vivo at the injection site, traffic to the LN due to their size and elicit a potent innate response, characterized by LN expansion and expression of type I and II IFN transcriptional programs. Remarkably, the responses we observe bypass the need for dendritic cell migration from the periphery to the dLN and soluble mannans directly target CD169+ sinus macrophages. We further modulated the physical properties of mannans by adsorbing them onto alum to obtain formulations in which mannans are present in both soluble and particulate forms. We show that this formulation results in enhanced immunological properties, such as full activation of innate immune cells in vitro and simultaneous targeting of the periphery and the LN. When tested as an adjuvant system formulated with viral glycoprotein antigens (IAV HA or SARS-CoV-2 Spike), mannans enabled induction of neutralizing antibodies with broad epitope specificity. Mannans formulated with alum protects against a SARS-CoV-2 MA10 challenge to the same extent as adjuvants that resemble those included in licensed vaccines. Immunization of mice with rHA-based Flublok vaccine, formulated with alum and mannans, elicited heterosubtypic immunity. Overall, our work sheds light on the molecular pathways activated by mannan formulations that trigger LN innate and adaptive responses and result in antibody responses against viral glycoproteins of enhanced magnitude and breadth.
A key aspect of our work is the requirement for both type I and II IFNs to sustain mannan-induced lymphocyte accrual and LN expansion. Also, IFNs are necessary to potentiate the induction of anti-glycoprotein antibodies. IFNs act on both myeloid cells and LN stromal cells where they modulate a range of functions, including chemokine expression and vascular permeability (Barrat et al., 2019; Ivashkiv, 2018). Hence, our results raise the possibility that mannan-elicited IFN signatures affect LN-resident myeloid and stromal compartments, eventually leading to potentiated adaptive immune responses. Our data on the augmented production and epitope specificity of anti-Spike antibody and data on the induction of heterosubtypic anti-A/PR/8/1934 HA-antibody suggest that the IFN signature driven by mannans formulated with alum broadens epitope recognition and potentiates protection against viruses. Nevertheless, it will be important in the future to assess a possible ‘innate training’ effect promoted by a more potent IFN response that may yield to cross-protective innate immunity.
Mannans formulated with alum acquire remarkable new immunological properties. Although we cannot exclude synergy between AH and mannans, our results are more readily explained by the concurrent presence in the formulation of mannans and alum of unbound and alum-adsorbed mannans. The former is responsible for ISG expression in the dLN, while the latter mediates skin inflammation. Evaluation of mannans formulated with alum in our immunization model with SARS-CoV-2 Spike supports this model. Direct synergy between alum and mannan inflammatory activity was excluded when alum and mannans were injected separately but in close physical proximity, an approach that led to a loss of the capacity of these adjuvants to enhance anti-Spike IgG. Such loss did not apply to T cell-mediated responses, which were boosted by the presence of mannans, independently of their coformulation with alum. Analysis of soluble mannans formulated or not with alum and admixed to Spike also showed that both formulations induce humoral and cellular type 1 immunity. Early induction of type I and II IFN signatures in the dLN is therefore sufficient to explain polarization of the adaptive immune response. Transient disruption of type I/II IFN signaling by administration of blocking antibodies significantly decreased levels of anti-Spike IgG, suggesting that early IFN signatures in the dLN translate into long-term potentiation of the immune response.
A remarkable property of the formulation of mannans and alum is its ability to induce neutralizing anti-Spike antibodies with broad epitope specificity. These antibodies cross-react with SARS-CoV Spike and, to a lesser extent, with MERS Spike. The epitope specificity profile observed in mice immunized with alum and mannans is comparable to that observed in COVID-19 patients, highlighting the translational relevance of our results (Shrock et al., 2020). Production of these antibodies has the same molecular and cellular requirements as the LN innate reaction induced by mannans formulated with alum, with the sole exception of cDC1 that play important roles in driving the IgG response, but not the LN innate response. Neutralizing antibodies are important for protection against SARS-CoV-2 infection in animal models (Cao et al., 2020; Hassan et al., 2020; Lv et al., 2020; McMahan et al., 2020; Rogers et al., 2020; Schafer et al., 2021; Shi et al., 2020; Tortorici et al., 2020; Zost et al., 2020). Accordingly, mice immunized with Spike and mannans formulated with alum show undetectable lung viral titers after infection with SARS-CoV-2 MA10, similar to mice immunized with clinically relevant adjuvant formulations. We then employed a model of influenza vaccination, using the clinically relevant Flublok vaccine. While all adjuvants tested enhanced the antibody response to Flublok, only mannans formulated with alum induced heterosubtypic immunity, that was paralleled by detection of antibodies against HA of A/PR/8/1934. It will be important in the future to clarify how the formulation of mannans and alum modulates germinal center dynamics and B cell repertoire selection. From a translational perspective, the broadening of epitope specificity suggests that mannans formulated with alum in combination with appropriate antigens might be a promising candidate for the development of vaccines that target multiple coronavirus or influenza A strains.
Overall, our study provides mechanistic and translational insights into how modulation of the physical form of mannans enables Dectin-2 targeting to enhance vaccine immunogenicity and protection, thereby illuminating the relationship between the physical form of innate stimuli and LN innate and adaptive immune responses.
Limitations of the study:
Our study provides a detailed mechanistic analysis of mannan-induced LN innate response at the molecular and cellular level. We demonstrated that mannans alone, or admixed with alum, require Dectin-2-expressing, CD169+ sinus macrophages, but not cDC1, neutrophils or monocytes for LN expansion, while cDC1 are required to potentiate antibody production. It remains to be elucidated whether other immune cells, such as cDC2, may be involved either in the LN innate or adaptive response. Also, it will be important to unravel the possible relationship between NK cells and DCs. Another unexpected finding is the only partial requirement of CARD9, the key signaling adaptor downstream of Dectins (Borriello et al., 2020; Brubaker et al., 2015). CARD9-independent pathways downstream of Dectins have also been described, including NIK-dependent activation of the non-canonical NFκB subunits p52 and RelB (Gringhuis et al., 2009; Xu et al., 2018). Mice that lack NIK, p52 or RelB have profound defects in secondary lymphoid organ development (Sun, 2017). Our results show that RelB activation in CD11c-expressing cells regulates mannan-induced LN expansion and expression of type I ISGs and cooperates with CARD9 in modulating the expression of type II ISGs. Although the role of RelB in modulating IFN responses and ISG expression is ambiguous (Le Bon et al., 2006; Saha et al., 2020). and more studies will be required to decipher its role upon mannan encounter, our findings create an intriguing parallelism between LN development and the innate response, setting the stage for RelB as a central regulator of the biology of secondary lymphoid organs.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ivan Zanoni (ivan.zanoni@childrens.harvard.edu; @Lo_Zanzi)
Materials Availability
All experimental models and reagents will be made available upon installment of a material transfer agreement.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
RNA sequencing data accession number: GSE193419.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice:
C57BL/6J (Jax 00664) (wild type), CB6F1 (Jax 100007), B6.129P2(C)-Ccr7tm1Rfor/J (Ccr7−/−, Jax 006621), B6.129S2-Ifnar1tm1Agt/Mmjax (Ifnar−/−, Jax 32045-JAX), B6.Cg-Ifngr1tm1Agt Ifnar1tm1.2Ees/J (Ifnar−/− Ifngr−/−, Jax 029098), B6.FVB-1700016L21RikTg(Itgax-DTR/EGFP)57Lan/J (CD11c-DTR, Jax 004509), B6.Cg-Tg(Itgax-cre)1-1Reiz/J (Cd11ccre, Jax 008068), B6.Cg-Relbtm1Ukl/J (Relbfl/fl, Jax 028719), B6.129S4-Ccr2tm1Ifc/J (Ccr2−/−, Jax 004999), B6.129S(C)-Batf3tm1Kmm/J (Batf3−/−, Jax 013755), B6.129-Card9tm1Xlin/J (Card9−/−, Jax 028652), B6.129S6-Clec7atm1Gdb/J (Clec7a−/−, Jax 012337) and C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I, Jax 003831) were purchased from Jackson Labs. B6.129P2-Fcer1gtm1Rav N12 (Fcer1g−/−, Model 583) were purchased from Taconic. Clec4n−/− mice were kindly provided by Drs. Nora A. Barrett and Yoichiro Iwakura. B6;129-Siglec1<tm1(HBEGF)Mtka> (CD169-DTR) mice were kindly provided by Dr. F. Pucci and are from the Riken Institute (No. RBRC04395), deposited by Drs. Kenji Kohno and Masato Tanaka (Miyake et al., 2007; Saito et al., 2001). B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II, Jax 004194) were kindly provided by Juan Manuel Leyva-Castillo. 6-8 week-old female mice were used for all the experiments. Mice were housed under specific pathogen-free conditions at Boston Children’s Hospital, and all the procedures were approved under the Institutional Animal Care and Use Committee (IACUC) and operated under the supervision of the department of Animal Resources at Children’s Hospital (ARCH).
Cell lines:
VeroE6 cells (ATCC, CRL-1586) were cultured in DMEM (Quality Biological, 112-014-101) supplemented with 10% FBS and 1% pen/strep. Expi293 cells were grown in Expi293 Expression Medium (ThermoFisher Scientific). Cells were maintained at 37°C (5% CO2) and passaged when confluent.
Viruses:
Influenza virus A/Puerto Rico/8/1934 (H1N1) (NR-348) was obtained through BEI Resources, NIAID, NIH, and propagated in the allantoic cavity of 9-11 days old specific-pathogen-free chicken (SPF) eggs (Charles River Laboratories, Wilmington, MA). Samples of SARS-CoV-2 were obtained from the CDC following isolation from a patient in Washington State (WA-1 strain - BEI #NR-52281). SARS-CoV-2 MA10 was generated in mice infected with the SARS-CoV-2 MA stock virus for the first passage and with lung homogenates of the previous passage for all following passages (passage 2 – 10). Clonal isolate from P10 was plaque purified to obtain SARS-CoV-2 MA10. All virus stocks were propagated on Vero E6 cells.
Yeasts:
Candida albicans strain SC5314 was maintained on blood agar (Remel) plates grown at 37°C.
METHOD DETAILS
Reagents and antibodies:
for flow cytometry, imaging cytometry, fluorescence-activated cell sorting (FACS) and confocal microscopy experiments the following reagents and antibodies were used: anti-CD45 BV510 (30-F11), anti-CD45 Alexa Fluor 700 (30-F11), anti-CD45 APC (30-F11), anti-CD45 PerCP/Cy5.5 (30-F11), anti-CD3 PE/Dazzle 594 (17A2), anti-CD3 BV510 (17A2), anti-TCRβ Alexa Fluor 400 (H57-597), anti-CD4 PE/Cy5 (GK1.5), anti-CD19 PE/Dazzle 594 (6D5), anti-CD19 BV650 (6D5), anti-NK1.1 PE/Dazzle 594 (PK136), anti-NK1.1 BV421 (PK136), anti-CD49b Alexa Fluor 488 (DX5), anti-Ter119 PE/Dazzle 594 (TER-119), anti-I-A/I-E PE/Cy7 (M5/114.15.2), anti-Ly6G PerCP/Cy5.5 (1A8), anti-CD11b Pacific Blue (M1/70), anti-Ly6C BV711 (HK1.4), anti-CD11c BV785 (N418), anti-CD11c APC (N418), anti-CD86 APC/Cy7 (GL-1), anti-CD86 APC (GL-1), anti-OX40L PE (RM134L), anti-CD169 APC (3D6.112), anti-CD169 Alexa Fluor 647 (3D6.112), anti-CD4 APC/Fire 750 (GK1.5), anti-CD8 PE/Cy7 (53-6.7), anti-CD45R/B220 Alexa Fluor 594 (RA3-6B2), anti-IFNγ PE (XMG1.2), anti-CD3 Biotin (145-2C11), anti-CD19 Biotin (6D5), anti-NK1.1 Biotin (PK136), anti-Ter119 Biotin (TER-119), TrueStain FcX (93), True-Stain Monocyte Blocker and Zombie Red Fixable Viability Kit were purchased from BioLegend; anti-Dectin-2 PE (REA1001) was purchased from Miltenyi Biotec; rat anti-Dectin-2 (D2.11E4) was purchased from GeneTex; anti-phopsho-Syk (Tyr525/526) (C87C1) was purchased from Cell Signaling Technology; CellTrace CFSE Cell Proliferation Kit, Alexa Fluor 488 NHS Ester (Succinimidyl Ester) and DAPI were purchased from Thermo Fisher Scientific.
For in vitro and in vivo experiments the following reagents were used: Iscove’s Modified Dubecco’s Medium (IMDM), Phosphate Buffer Saline (PBS), penicillin/streptomycin (pen/strep) and L-Glutamine (L-Gln) were purchased from Lonza; Fetal Bovine Serum (FBS) was purchased from Thermo Fisher Scientific; collagenase from Clostridium histolyticum, deoxyribonuclease (DNase) I from bovine pancreas and dispase II were purchased from MilliporeSigma; TLRGrade Escherichia coli LPS (Serotype O555: B5, 1 μg/ml) was purchased from Enzo Life Sciences; curdlan (10 μg/ml) was purchased from Wako Chemicals; mannans, β-glucans and their Alexa Fluor 488-conjugates (10 μg/ml for in vitro experiments, 500 μg/mouse for in vivo experiments) were provided by Michael D Kruppa, Zuchao Ma and David L Williams (East Tennessee State University); carboxyl latex beads 3 μm were purchased from Thermo Fisher Scientific and used directly (cell:bead ratio 1:10 for in vitro experiments) or after coating with diaminopropane derivatized mannans provided by Michael D Kruppa, Zuchao Ma and David L Wiliams (East Tennessee State University); WGP-S and WGP-D (500 μg/mouse for in vivo experiments) were purchased from InvivoGen; Lipo-CpG was provided by Darrell J. Irvine (Koch Institute for Integrative Cancer Research at MIT); diphtheria toxin (unnicked) from Corynebacterium diphtheriae (200 ng/mouse for CD11-DTR mice, 500 ng/mouse for CD169-DTR mice) was purchased form Cayman Chemical; ovalbumin (OVA) EndoFit (5 μg/mouse), Alhydrogel adjuvant 2% (AH, 2 μg/ml for in vitro experiments, 100 μg/mouse for in vivo experiments), AddaVax (25 μl/mouse) and AddaS03 (25 μl/mouse) were purchased from InvivoGen; PHAD (synthetic monophosphoryl lipid A, 50 μg/mouse) was purchased from Avanti Polar Lipids; recombinant pre-fusion stabilized SARS-CoV-2 Spike trimer (1 μg/mouse) and RBD were expressed and purified from plasmids generously provided by Drs. Berney S. Graham (NIH Vaccine Research Center) and Aaron G. Schmidt (Ragon Institute), respectively; SARS-CoV-2 Spike peptide pools (PepTivator SARS-CoV-2 Prot_S) were purchased from Miltenyi Biotec; Flublok vaccine (quadrivalent formula 2020 – 2021, composed of HAs from IAV A/Guangdong-Maonan/SWL1536/2019 [H1N1], IAV A/HongKong/2671/2019 [H3N2], influenza B virus B/Washington/02/2019 and influenza B virus B/Phuket/3073/2013) was purchased from the Boston Children’s Hospital Pharmacy; Influenza A H1N1 (A/Puerto Rico/8/1934) recombinant hemagglutinin (rPR8) was purchased from Sino Biological; anti-Asialo-GM1 (Poly21460, 25 μl/mouse) was purchased from BioLegend; anti-CD62L (Mel-14, 100 μg/mouse), anti-IFNAR1 (MAR1-5A3, 500 μg/mouse), anti-IFNγ (XMG1.2, 200 μg/mouse), anti-Ly6G (1A8, 50 μg/mouse) and their isotype controls rat IgG2a (2A3), rat IgG1 (HRPN) and mouse IgG1 (MOPC-21) were purchased from Bio X Cell.
The formulation of alum and mannans (AH/mann) was obtained by admixture of alum (100 μg/10 μl), mannans (500 μg/25 μl) and saline (15 μl). When formulated with an antigen (e.g., SARS-CoV-2 Spike trimer or Flublok) the volume of saline was reduced accordingly in order to keep the total volume constant. This formulation is further described in the NIH/NIAID Vaccine Adjuvant Compendium (https://vac.niaid.nih.gov).
Isolation of mannan from C. albicans.
For mannan isolation, C. albicans was inoculated into 15 l of YPD (1% yeast extract, 2% peptone, 2% dextrose) and grown for 20 hours at 37°C. Cells were harvested by centrifugation at 5000 g for 5 minutes. This resulted in a 100 g pellet from 15 l of media. We used a standard protocol for isolation and NMR characterization of the mannan (Kruppa et al., 2011; Lowman et al., 2011). In brief, the cell pellets were suspended in 200 ml of acetone to delipidate the cells for 20 minutes prior to centrifugation at 5000 g for 5 minutes, removal of acetone and drying of the pellet for an hour. Dried pellets were broken up and transferred to a tissue homogenizer. An equivalent volume of acid-washed glass beads was added and 200 ml of dH2O was added to the mixture. The cells were subjected to bead beating for three 30 second pulses before the entire mixture was transferred to a 1 l flask. The material was autoclaved for 2 hours, allowed to cool and then centrifuged for 5 minutes at 5000 g. The supernatant was retained and the cell pellet discarded. Pronase (500 mg in 20 ml dH2O), which had been filter sterilized and heat treated for 20 minutes at 65°C (to remove any glycosidic activity) was added to the supernatant along with sodium azide to a concentration of 1 mM. The mixture was then incubated overnight (20 hours) at 37°C to allow for degradation of any proteins in the solution. Mannans were extracted by addition of an equal volume of Fehling’s solution to the protease treated mannan solution and allowed to mix for one hour at room temperature. After mixing the solution was allowed to stand for 20 minutes to facilitate mannan precipitation. The supernatant was decanted and the precipitate was dissolved in 10 ml of 3M HCl, to enable release of copper from the reducing ends of the mannans. To the dissolved mannan solution 500 ml of an 8:1 mixture of methanol:acetic acid was added, and the mixture stirred to allow the mannan to precipitate overnight. After the material had settled, the supernatant was decanted, washed again with 500 ml of methanol, allowing six hours for the mannans to settle. The supernatant was decanted and the remaining precipitate was dissolved in 200 ml dH2O. The mannans were dialyzed against a 200-fold change of dH2O over 48 hours using a 2000 MW cutoff membrane to remove residual acid, methanol and other compounds from the extraction process. The dialysate was then subjected to lyophilization and stored at −20°C until needed. A small sample (10 mg) of the material was subjected to NMR to confirm for the purity of the N-linked mannans (Lowman et al., 2011) and for assessment of molecular weight (Kruppa et al., 2011). Prior to in vitro or in vivo use the mannan is depyrogenated to remove any residual endotoxin and filter sterilized.
Preparation of the diaminopropane (DAP) derivatized mannan.
Mannan (100 mg) was dissolved in 6 ml of water, followed by addition of 1,3-diaminopropane (0.6 ml). The reaction mixture was stirred at ambient temperature for 1 hour. Sodium cyanoborohydride (300 mg) was added and the pH value of the reaction mixture was adjusted to ~4.8 by slowly adding acetic acid (~1.1 ml). The reaction mixture was stirred at room temperature for 48 hours, then dialyzed with a 2000 MWCO RC membrane against ultrapure water (1000 ml x 4). The retentate was harvested and lyophilized to yield the DAP attached mannan. The recovery was 88.5 mg, ~88%. The mannan-DAP was characterized by 1H-NMR to confirm the identity of the compound.
For conjugation with Alexa Fluor 488 NHS Ester (Succinimidyl Ester), ~15 mg of mannan-DAP were resuspended in 1 ml of sodium borate conjugation buffer (100 mM, pH 8.5) and allowed to solvate for at least 24 hours. Then, 1 mg of Alexa Fluor 488 NHS Ester resuspended in 35 μl of DMSO was added to the solution and incubated overnight in the dark at room temperature with gentle agitation. The reaction mixture was dialyzed with a 6000-8000 MWCO RC membrane against saline (1000 ml x 4) and the retentate was filter sterilized.
For conjugation with carboxyl latex beads 3 μm, mannan-DAP was resuspended at a concentration of 10 mg/1 ml of BupH MES conjugation buffer pH 4.5 (Thermo Fisher Scientific) and allowed to solvate for at least 24 hours. 1 ml of mannan-DAP was added to 50 x 106 beads and then mixed with 4 mg/1 ml of EDC (Thermo Fisher Scientific) resuspended in pure water. The reaction mixture was incubated for 4 hours in the dark at room temperature with gentle agitation. Then, the beads were washed twice (4000 g for 10 minutes) with saline and resuspended in saline at a concentration of 108 beads/ml.
Preparation of Candida albicans β-glucan particles.
β-glucan particles were isolated from Candida albicans SC5314 as previously described by our laboratory (Lowman et al., 2014). Briefly, glucan was isolated from C.albicans using a base/acid extraction approach with provides water insoluble glucan particles that are > 95% pure. The structure and purity of the glucan was determined by 1H-NMR in DMSO-d6 (Lowman et al., 2014). Prior to in vitro or in vivo use the β-glucan particles are depyrogenated and sterilized.
Preparation of the diaminopropane (DAP) derivatized β-glucan.
β-glucan particles (20 mg) were dissolved in 1 ml of dimethyl sulfoxide (DMSO) in 4 ml vial after one hour of stirring. 1,3-Diaminopropane (100 μL) was added and stirred at ambient temperature for 3 hours. Sodium cyanoborohydride (100 mg) was added and the reaction mixture was stirred for 48 hours, followed by addition of sodium borohydride (50 mg) and stirring for 24 hours. Acetic acid (200 μl) was added dropwise at 0°C to quench the reaction and the reaction mixture was stirred at ambient temperature for 3 hours. The β-glucan particles were harvested and washed five times in water by centrifugation (862 g). The recovery was >95%. The glucan-DAP was characterized by 1H-NMR to confirm the derivatized glucan was still intact. The glucan-DAP was lyophilized to dryness and stored at −20°C in the dark in a desiccator until needed.
For conjugation with Alexa Fluor 488 NHS Ester (Succinimidyl Ester), 20 mg of glucan-DAP were suspended in 1 ml of sodium borate conjugation buffer (100 mM, pH 8.5) and allowed to hydrate for at least 24 hours at 4°C. 1 mg of Alexa Fluor 488 NHS Ester resuspended in 35 μl of DMSO was then added to the solution which was incubated overnight in the dark at room temperature with gentle agitation. The reaction mixture was centrifuged, washed five times in water with centrifugation (862 g) and the 488 labeled glucan particles were harvested.
Quantification of mannans in the alum/mannans formulation.
Supernatants were harvested from the alum/mannans mixture and were lyophilized. Lyophilized supernatants and a standard mannan sample (4.0 mg) that was not mixed with AH were respectively dissolved in 500 μl of deuterium oxide (99.9% D) with 0.01% (W/V) internal standard TMSP-2,2,3,3-D4 (98.0% D). 1H-NMR data were collected on a 400MHz Bruker Avance Ultra Shield NMR spectrometer at 295 K with the same acquisition parameters for all the samples. NMR spectra were processed using TOPSPIN 2.1 running on the Avance 400MHz NMR. The ring proton resonances (3.25 - 4.50 ppm) were integrated referencing to the integral of internal standard (−0.02 - 0.02 ppm) calibrated as 1.0. Based on the ratio between the mass of standard mannan (4.0 mg) and its ring proton integral (39.12), the mass of mannan in the supernatant was calculated using the detected ring proton integral multiplied by 4.0/39.2. Thus, the amount of mannan absorbed by the AH was determined by difference between the mass of total mannan added and the mass of mannan remaining in the supernatant after formulation.
Analysis of skin and LN responses.
To assess skin and LN innate responses, mice were injected intradermally on day 0 with the indicated compounds in a volume of 50 μl on each side of the back (one side for the compound and the contralateral side for saline of vehicle control). 6 or 24 hours post-injection skin samples at the injection sites and draining (brachial) LNs were collected for subsequent analysis. In selected experiment LNs were also collected 7 and 14 days post-injection.
Skin samples were transferred to a tissue homogenizer and disrupted with beads in 1 ml of TRI Reagent (Zymo Research). Then, samples were centrifuged 12000 g for 10 minutes and 800 μl of cleared supernatant were transferred to a new tube for subsequent RNA isolation.
LNs were weighted on an analytical scale prior to transfer to a tissue homogenizer and disrupted with beads in TRI Reagent as indicated for skin samples or processed to generate a LN cell suspension by modification of a previously published protocol (Fletcher et al., 2011). Briefly, individual LNs were incubated at 37°C for 20 minutes in 400 μl of digestion mix (IMDM + pen/strep + FBS 2% + collagenase 100 mg/ml + dispase II 100 mg/ml + DNase 10 mg/ml). Then, LNs were ground by pipetting with a 1000 μl tip, supernatants were transferred to new tubes and kept at 4°C while 200 μl of digestion mix were added to the pellets and incubated at 37°C for 10 minutes. This cycle was repeated one more time, then pooled supernatants of individual LNs were divided into two aliquots: one for flow cytometry analysis, another one was centrifuged at 300 g for 5 minutes and the cell pellet was resuspended in 800 μl of TRI Reagent for subsequent RNA isolation. In selected experiment LN suspensions were cultured in the presence of Brefeldin A (BioLegend) for 4 hours and then processed for flow cytometry analysis (both surface staining and intracellular cytokine staining).
For specific experiments mice were treated with: anti-CD62L blocking antibody or isotype control, intravenous injections on day −1; anti-IFNγ and anti-IFNAR1 blocking antibody or isotype controls, intravenous injections on day −1 and 0; anti-Asialo-GM1 or PBS, intravenous injections on day −1 and 0; anti-Ly6G depleting antibody or isotype control, intraperitoneal injections on day −1 and 0; diphtheria toxin, intravenous injections on day −1 and intradermal injections (co-injected with mannans) on day 0 for CD11c-DTR mice, intraperitoneal injection on day −2 for CD169-DTR mice.
In vitro stimulation of GM-CSF-differentiated, bone marrow-derived phagocytes.
Bone marrow-derived phagocytes were differentiated from murine bone marrow in IMDM + 10% B16-GM-CSF derived supernatant + 10% FBS + pen/strep + L-Gln and used after 7 days of culture. Then, cells were harvested, plated in flat bottom 96 well plates at a density of 105 cells/200 μl/well in IMDM + 10% FBS + pen/strep + L-Gln and stimulated with the indicated compounds for 18-21 hours. At the end of stimulation, supernatants were harvested, and TNF and IL-2 concentrations were measured by ELISA (BioLegend) according to the manufacturer’s protocol. Cells were detached with PBS + EDTA 2 mM and transferred to a round bottom 96 well plate for subsequent flow cytometry staining and analysis. Alternatively, cells were stimulated for 6 hours, lysed in TRI Reagent and RNA was extracted for gene expression analysis.
In vivo quantification of fluorescently labelled β-glucans and mannans.
Mice were intradermally injected with 500 μg/mouse of Alexa Fluor 488-conjugated β-glucans and mannans (in selected experiments mannans were formulated with 100 μg/mouse of AH before injection). At the indicated timepoints dLNs were collected, transferred to a tissue homogenizer and disrupted with beads in 400 μl of deionized water. Then, samples were centrifuged (12000 g for 10 minutes) and cleared supernatants were transferred to a 96 well clear bottom black plate. Fluorescence values were measured with SpectraMax i3x microplate reader (Molecular Devices) and expressed as arbitrary units after background (deionized water) subtraction.
Flow cytometry, fluorescence-activated cell sorting (FACS), imaging cytometry and confocal microscopy.
For flow cytometry analysis, cells were first stained with Zombie Red Fixable Viability in PBS for 5 minutes at 4°C, washed once with PBS + BSA 0.2% + NaN3 0.05% (300 g for 5 minutes) and then stained with antibodies against surface antigens diluted in PBS + BSA 0.2% + NaN3 0.05% for 20 minutes at 4°C. Cells were then washed, fixed with 2% paraformaldehyde for 10 minutes at room temperature, washed again and resuspended in PBS + BSA 0.2% + NaN3 0.05%. Samples were acquired on a BD LSRFortessa flow cytometer and data were analyzed using FlowJo v.10 software (BD Biosciences). CountBright Absolute Counting Beads were used to quantify absolute cell numbers. In selected experiments, after fixation with 2% paraformaldehyde cells were permeabilized by incubation with a saponin-based permeabilization buffer (BioLegend) for 10 minutes at 4°C and stained with antibodies against intracellular cytokines diluted in permeabilization buffer for 20 minutes at 4°C. Then, cells were washed with permeabilization buffer, resuspended in PBS + BSA 0.2% + NaN3 0.05% and acquired as indicated before.
For FACS and imaging cytometry, mice were intradermally injected with AF488-mannans and 6 hours later dLNs were harvested to obtain LN cell suspensions. For FACS, cells were stained with antibodies against surface antigens diluted in PBS + BSA 0.2% for 20 minutes at 4°C. Cells were then washed once, resuspended in 1 ml of PBS + BSA 0.2%, filtered through 70 μm cell strainers (Fisher Scientific) and sorted with a Sony MA900 cell sorter directly into 1 ml of TRI Reagent. The following cell subset was sorted: CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+. For imaging cytometry, cells were depleted of lymphoid and erythroid cells by sequential staining with biotinylated antibodies against anti-CD3, anti-CD19, anti-NK1.1, anti-Ter119 and Streptavidin Microbeads (Miltenyi Biotec) according to the manufacturer’s protocol. The remaining cells were stained with anti-CD45 APC, fixed with 2% paraformaldehyde, washed once and resuspended in 60 μl of PBS + DAPI (0.2 μg/ml). Samples were then acquired on an Amnis ImageStream X Mark II (Luminex Corporation). Mannan internalization was analyzed with Amnis Ideas Software and calculated with Internalization Feature as AF488 signal within the APC mask.
For confocal microscopy, dLNs were isolated at steady state or 1 hour post-injection of AF488-mannans and fixed with 4% paraformaldehyde overnight. Tissue slides were prepared from frozen LN samples at the Beth Israel Deaconess Medical Center (BIDMC) Histology Core Facility and stained at the BIDMC Confocal Imaging Core Facility. Briefly, frozen sections were air-dried for 30 minutes and rehydrated. The sections were permeabilized using 0.05% Triton X-100 for 10 minutes at room temperature and washed three times with TBS. The sections were then incubated with 5% normal donkey serum (Jackson ImmunoResearch Lab) for 1 hour at room temperature. For Dectin-2 staining of steady state LNs, slides were incubated with rat anti-Dectin-2 overnight at 4°C. The slides were washed three times and incubated with: Alexa Fluor 488-conjugated Donkey anti-rat secondary antibody (Jackson ImmunoResearch Lab) for 90 minutes at room temperature and washed four times. Slides were then incubated with Alexa Fluor 647-conjugated rat anti-CD169 primary antibody and Alexa Fluor 594-conjugated rat anti-CD45R/B220 primary antibody for 90 minutes at room temperature and then washed with TBS. For phospho-Syk staining of AF488-mannan-treated LNs, slides were incubated with rabbit anti-phospho-Syk (Cell Signaling Technology) overnight at 4°C. The slides were washed three times and incubated with Alexa Fluor 647-conjugated Donkey anti-rabbit secondary antibody (Jackson ImmunoResearch Lab) for 90 minutes at room temperature and washed four times. Slides were then incubated with Alexa Fluor 594-conjugated rat anti-CD45R/B220 primary antibody for 90 minutes at room temperature and then washed with TBS. rabbit anti-phospho-Syk (Cell Signaling Technology). Samples were counterstained with Hoechst 33342 (Thermo Fisher Scientific) and washed three times with TBS. Slides were mounted with Prolong Gold anti-fade mounting media (Thermo Fisher Scientific) and imaged on a Zeiss 880 laser scanning confocal microscope at the Boston Children’s Hospital Harvard Digestive Disease Center.
RNA isolation, qPCR, transcriptomic and pathway analyses.
RNA was isolated from TRI Reagent samples using phenol-chloroform extraction or column-based extraction systems (Direct-zol RNA Microprep and Miniprep, Zymo Research) according to the manufacturer’s protocol. RNA concentration and purity (260/280 and 260/230 ratios) were measured by NanoDrop (Thermo Fisher Scientific).
Purified RNA was analyzed for gene expression by qPCR on a CFX384 real time cycler (Biorad) using pre-designed KiCqStart SYBR Green Primers (MilliporeSigma) specific for Cxcl9 (M_Cxcl9_1), Gbp2 (M_Gpb2_1), Ifit2 (M_Ifit2_1), Rsad2 (M_Rsad2_1), Il6 (M_Il6_1), Cxcl1 (M_Cxcl1_1), Rpl13a (M_Rpl13a_1) or pre-designed PrimeTime qPCR Primers (Integrated DNA Technologies) specific for Gapdh (Mm.PT.39a.1).
For bulk RNAseq analysis, RNA isolated from skin or LN samples was submitted to Genewiz. RNA samples were quantified using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and RNA integrity was checked with RNA Screen Tape on Agilent 2200 TapeStation (Agilent Technologies). RNA sequencing library preparation was prepared using TruSeq Stranded mRNA library Prep kit following manufacturer’s protocol (Illumina, Cat# RS-122-2101). Briefly, mRNAs were first enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 8 minutes at 94°C. First strand and second strand cDNA were subsequently synthesized. The second strand of cDNA was marked by incorporating dUTP during the synthesis. cDNA fragments were adenylated at 3’ends, and indexed adapter was ligated to cDNA fragments. Limited cycle PCR was used for library enrichment. The incorporated dUTP in second strand cDNA quenched the amplification of second strand, which helped to preserve the strand specificity. Sequencing libraries were validated using DNA Analysis Screen Tape on the Agilent 2200 TapeStation (Agilent Technologies), and quantified by using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) as well as by quantitative PCR (Applied Biosystems). The sequencing libraries were multiplexed and clustered on 1 lane of flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2x150 Pair-End (PE) High Output configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS) on the HiSeq instrument. Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and demultiplexed using Illumina bcl2fastq program version 2.17. One mismatch was allowed for index sequence identification. Reads were quality-controlled using FastQC. Illumina adapters were removed using cutadapt. Trimmed reads were mapped to the mouse transcriptome (GRCm38) based on Ensembl annotations using Kallisto (Bray et al., 2016). Transcript counts were imported and aggregated to gene counts using tximport (Soneson et al., 2015). Gene counts were analyzed using the R package DESeq2 (Love et al., 2014). When applicable, batch was used as a blocking factor in the statistical model. Differentially expressed genes (DEGs) were identified as those passing a threshold of FDR significance threshold (0.05 for skin; 0.01 for lymph nodes, a more stringent threshold thanks to the greater power due to higher number of replicates) where the alternate hypothesis was that the absolute log2 FC was greater than 0. Genes induced by mannan or glucan treatment over saline were plotted in heatmaps using the R package ComplexHeatmap, using Z-scored log2 normalized abundance. Genes were arranged by abundance delta between glucan and mannan (aggregated from multiple time points when appropriate), with a gap delimiting two clusters: genes more highly expressed upon mannan stimulation vs genes more highly expressed upon glucan stimulation. Pathway analysis was performed with the R package hypeR (Federico and Monti, 2020), using hypergeometric enrichment tests of genes belonging to a cluster of interest and the Hallmark gene set collection from the Broad Institute’s MSigDB collection.
For targeted transcriptome sequencing, 25 ng of RNA isolated from sorted cells was retrotranscribed to cDNA using Superscript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). Barcoded libraries were prepared using the Ion AmpliSeq Transcriptome Mouse Gene Expression Kit as per the manufacturer’s protocol and sequenced using an Ion S5 system (Thermo Fisher Scientific). Differential gene expression analysis was performed using the ampliSeqRNA plugin (Thermo Fisher Scientific). To quantify the number of DEGs, gene-level fold change < −1.5 or > 1.5 and gene-level p value < 0.05 (ANOVA) were considered. For heatmap representation, DEGs were defined with an F-test FDR less than 0.05 and a log2 fold-change (FC) greater than 1 (or lower than −1) between a mutant and WT control. Hierarchical clustering was performed with Pearson correlation and average linkage. Pathway analysis was performed with the R package hypeR, using Kolmogorov Smirnov Test on genes ranked according to their log2FC.
In vivo CD4+ and CD8+ T cell proliferation assay.
Spleens were isolated from OT-II or OT-I mice and meshed with the plunger end of a syringe. Then, splenocyte cell suspensions were treated with ACK lysis buffer (2 ml for 2 minutes at room temperature), washed with PBS (300 g for 5 minutes) and filtered through 70 μm cell strainers. CD4+ and CD8+ T cells were respectively purified using CD4 (L3T4) or CD8a (Ly-2) MicroBeads (Miltenyi Biotec) according to the manufacturer’s protocol and stained with CellTrace CFSE (5 μM in PBS + FBS 2.5% for 20 minutes in the dark). At the end of incubation, cells were washed twice with PBS, resuspended at a concentration of 5 x 105 cells/100 μl saline and 100 μl of cell suspension was intravenously injected into each mouse. 24 hours later (day 0) mice were intradermally injected with OVA (5 μg/mouse) alone or combined with mannans (500 μg/mouse). Saline-injected mice were used as control. On day +3 dLNs were harvested and LN cell suspension were stained with anti-CD19, anti-Ter119, anti-CD3 and anti-CD4 or anti-CD8 antibodies. Adoptively transferred, CFSE-labelled OT-II and OT-I cells were respectively detected in the CD19− Ter119− CD3+ CD4+ and CD19− Ter119− CD3+ CD8+ gates. Results are expressed as absolute number of CD19− Ter119− CD3+ CD4+ CFSElo or CD19− Ter119− CD3+ CD8+ CFSElo cells (i.e., cells undergoing at least one division cycle) or percentage of each division peak within the CD19− Ter119− CD3+ CD4+ or CD19− Ter119− CD3+ CD8+ gates.
SARS-CoV-2 Spike and RBD expression and purification.
Full length SARS-CoV-2 spike glycoprotein and RBD constructs (amino acid residues R319-K529), both with an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag at C-terminus, were obtained from Drs. Barney S. Graham (NIH Vaccine Research Center) and Aaron G. Schmidt (Ragon Institute), respectively. These expression vectors were used to transiently transfect Expi293 cells (Thermo Fisher Scientific) using polyethylenimine (Polysciences). Protein was purified from filtered cell supernatants using either StrepTactin resin (IBA) or Cobalt-TALON resin (Takara). Affinity tags were cleaved off from eluted protein samples by HRV3C protease and tag removed proteins were subjected to additional purification by size-exclusion chromatography using either a Superose 6 10/300 column (GE Healthcare) or a Superdex 75 10/300 Increase column (GE Healthcare) in PBS (pH 7.4) buffer.
Immunization and antibody quantification.
CB6F1 mice were immunized by intradermal injection of Spike (1 μg/mouse) alone or formulated with AH (100 μg/mouse), β-glucans (500 μg/mouse), mannans (500 μg/mouse), alum (AH)/mannans (AH/mann), AddaS03 or AH/PHAD on day 0 and day +14. Alternatively, C57BL/6 mice were immunized by intradermal injection Flublok vaccine (1 μg/mouse) alone or formulated with AH, AH/mann, AddaVax or AH/PHAD on day 0 and day +14. Saline-injected mice were used as control. Blood samples were collected by retroorbital bleeding on day +14 (pre-boost) and day +28, and serum samples were isolated after centrifugation of blood samples twice at 1500 g for 10 minutes. In selected experiments blood samples were collected on day +98 or 7 days post-challenge. Spike-, RBD-, Flublok- and rPR8-specific IgG, IgG1, IgG2c antibody levels were quantified in serum samples by ELISA by modification of a previously described protocol (Borriello et al., 2017). Briefly, high binding flat bottom 96-well plates were coated with 0.5 μg/ml Spike, 1 μg/ml RBD, 1 μg/ml Flublok or 1 μg/ml rPR8 in PBS, incubated overnight at 4°C, washed once with PBS + 0.05% Tween-20 (PBST) and blocked with PBS + BSA 1% for 1 hour at room temperature. Then, serum samples were added with an initial dilution of 1:100 and 1:4 serial dilutions in PBS + BSA 1% to generate 11-point curves and incubated for 2 hours at room temperature. Plates were then washed three times with PBST and incubated for 1 hour at room temperature with HRP-conjugated anti-mouse IgG, IgG1 or IgG2c (Southern Biotech) antibodies. At the end of the incubation, plates were washed five times with PBST and developed with tetramethylbenzidine (BD OptEIA Substrate Solution for Spike, rHA, and rPR8; Thermo Fisher Scientific 1-Step Ultra TMB-ELISA Substrate Solution for RBD ELISA) for 5 min, then stopped with 2 N H2SO4. Optical densities (ODs) were read at 450 nm with SpectraMax iD3x microplate reader (Molecular Devices) and endpoint titers were calculated using as cutoff three times the optical density of the background. Values < 100 were reported as 25.
Splenocyte restimulation assay.
Immunized mice were sacrificed on day 35 and their spleens were collected. To isolate splenocytes, spleens were mashed through a 70 μm cell strainer and the resulting cell suspensions were washed with PBS and incubated with ACK lysis buffer (2 ml for 2 minutes at room temperature) to lyse erythrocytes. Splenocytes were re-washed with PBS and plated in flat bottom 96-well plates (2 x 106 cells per well). Then, SARS-CoV-2 Spike peptides (PepTivator SARS-CoV-2 Prot_S, Miltenyi Biotec) were added at a final concentration of 0.6 nmol/ml (total cell culture volume, 200 μl per well). After 96 hours, supernatants were harvested and IFNγ levels were measured by ELISA (Thermo Fisher Scientific) according to the manufacturer’s protocol.
SARS-CoV-2 surrogate virus neutralization tests.
The surrogate virus neutralization test was performed by modification of a previously published protocol (Tan et al., 2020). Briefly, high binding flat bottom 96-well plates were coated with 2 μg/ml recombinant human ACE2 (hACE2, MilliporeSigma) in PBS, incubated overnight at 4°C, washed three times with PBST and blocked with PBS + BSA 1% for 1 hour at room temperature. In the meantime, each serum sample (final dilution 1:160) was pre-incubated with 3 ng of RBD-Fc (R&D Systems) in PBS + BSA 1% for 1 hour at room temperature and then transferred to the hACE2-coated plate. As positive control, RBD-Fc was also added to hACE2-coated wells without pre-incubation with serum samples. After 1 hour at room temperature, plates were washed three times with PBST and incubated with an HRP-conjugated anti-human IgG Fc antibody (Southern Biotech) for 1 hour at room temperature. At the end of the incubation, plates were washed five times with PBST and developed with tetramethylbenzidine (BD Biosciences) for 5 min, then stopped with 2 N H2SO4. The optical density was read at 450 nm with SpectraMax iD3x microplate reader (Molecular Devices). Percentage inhibition of RBD binding to hACE2 was calculated with the following formula: Inhibition (%) = [1 - (Sample OD value - Background OD value) / (Control OD value - Background OD value)] x 100.
SARS-CoV-2 Neutralization Titer Determination.
All serum samples were heat inactivated at 56°C for 30 minutes to deactivate complement and allowed to equilibrate to room temperature prior to processing for neutralization titer. Samples were diluted in duplicate to an initial dilution of 1:5 or 1:10 followed by 1:2 serial dilutions, resulting in a 12-dilution series with each well containing 100 μl. All dilutions were performed in DMEM (Quality Biological), supplemented with 10% (v/v) fetal bovine serum (heat inactivated, MilliporeSigma), 1% (v/v) penicillin/streptomycin (Gemini Bio-products) and 1% (v/v) L-glutamine (2 mM final concentration, Thermo Fisher Scientific). Dilution plates were then transported into the BSL-3 laboratory and 100 μl of diluted SARS-CoV-2 (WA-1, courtesy of Dr. Natalie Thornburg/CDC) inoculum was added to each well to result in a multiplicity of infection (MOI) of 0.01 upon transfer to titering plates. A non-treated, virus-only control and a mock infection control were included on every plate. The sample/virus mixture was then incubated at 37°C (5.0% CO2) for 1 hour before transferring to 96-well titer plates with confluent VeroE6 cells. Titer plates were incubated at 37°C (5.0% CO2) for 72 hours, followed by cytopathic effect (CPE) determination for each well in the plate. The first sample dilution to show CPE was reported as the minimum sample dilution required to neutralize >99% of the concentration of SARS-CoV-2 tested (neut99).
VirScan.
We performed phage IP and sequencing as described previously (Xu et al., 2015a) with slight modifications. A sublibrary encoding a 56-mer peptide library tiling every 28 amino acids through the proteomes of the six HCoVs and three bat coronaviruses most closely related to SARS-CoV-2 (Shrock et al., 2020) was mixed with the original VirScan library to enable mapping of SARS-CoV-2 epitopes. 0.6 μl mouse sera, or approximately 2 μg of IgG, was included in each VirScan reaction. Immunoprecipitations were performed using magnetic protein A and protein G Dynabeads (Thermo Fisher Scientific) as previously described (Xu et al., 2015a).
SARS-CoV-2 challenge and lung viral titer determination.
Mice were anesthetized with intraperitoneal injection of ketamine/xylazine prior to intranasal inoculation with 1x104 pfu MA-10 (Leist et al., 2020) per mouse in 50μL PBS. Weights were measured daily starting from the day of inoculation (Day 0) to the day of euthanization (Day 2). Two days post-infection, mice were euthanized with isoflurane and dissected. The entire left lung was fixed in 4% paraformaldehyde (PFA) for histology and the right lung was halved and stored in either TRIzol for RNA isolation or PBS for titering by plaque assay.
SARS-CoV-2 Plaque Assay.
The day prior to infection, 2x105 VeroE6 cells (ATCC, CRL-1586) were seeded per well in 12-well plates. Lung samples were homogenized and then 25μL of each sample was serially diluted 1:10 in 225μL DMEM. Media was removed from the cells and 200μL of each dilution was added to each well. The plates were incubated at 37°C and 5% CO2 for one hour, rocking every 15 minutes. The cells were then overlaid with 2mL of a semi-solid agar overlay (DMEM+2%FBS+0.05% w/v Ultra Pure Agarose (Invitrogen, A9539). Plates were incubated at 37°C and 5% CO2 for 72 hours before being fixed with 4% PFA and stained with a 0.25% w/v crystal violet + 20% Ethanol solution. Plaques were counted and titers calculated.
Influenza A virus challenge and lung pathology.
C57BL/6 mice were anesthetized by isoflurane before intranasally inoculated with 50 μl of 4 x 103 plaque-forming units (PFU) of influenza virus A/Puerto Rico/8/1934 (H1N1). Mice were monitored daily and the body weights were recorded for 7 days. On day 7 post-infection mice were sacrificed and lungs were collected for histological analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS.
When necessary, data were Log-transformed before statistical analysis to approximate normal distributions. One-sample t test was used to compare each group against the value 1 (or 0 after Log-transformation, which represent the contralateral control sample expressed as fold). Statistical differences between groups in datasets with one categorical variable were evaluated by two sample t test (2 groups) or one-way ANOVA (more than 2 groups) corrected for multiple comparisons. Statistical differences between groups in datasets with two categorical variables were evaluated by two-way ANOVA corrected for multiple comparisons. # or * and ** or ## respectively indicate p ≤ 0.05 and 0.01.
Supplementary Material
Table S1. Related to Figure 1. List of genes detected by bulk RNAseq analysis that are expressed in the skin 6 hours after the injection of saline, β-glucans or mannans.
Table S2. Related to Figure 1. List of genes detected by bulk RNAseq analysis that are expressed in the dLN 6 and 24 hours after the injection of saline, β-glucans or mannans.
Table S3. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT and Fcer1g−/− mice 6 hours after the injection of mannans.
Table S4. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from Card9−/− and Fcer1g−/− mice 6 hours after the injection of mannans.
Table S5. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT and Card9−/− mice 6 hours after the injection of mannans.
Table S6. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT, Fcer1g−/− and Card9−/− mice 6 hours after the injection of mannans that are upregulated in Fcer1g−/− compared to WT and Card9−/− mice.
Table S7. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT, Fcer1g−/− and Card9−/− mice 6 hours after the injection of mannans that are upregulated in WT and Card9−/− compared to Fcer1g−/− mice.
Figure S3. Related to Figure 1. Soluble whole glucan particles elicit LN expansion and ISG expression. Mice were injected intradermally with saline (Sal), dispersible (WGP-D) or soluble (WGP-S) whole glucan particles. 24 hours later injection sites were assessed as indicated in Figure 1A (A), skin samples and dLNs were collected, LN weights were measured, and RNA extracted for gene expression analysis (B). Results are expressed as fold over the median value of saline-injected skin samples or LNs. N = 4 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against the value 1 (which represent the saline control samples expressed as fold). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing between different experimental groups.
Figure S2. Related to Figure 1. Mannans and β-glucans induce unique patterns of immune cell recruitment and activation in the draining LNs. (A-H) Mice were injected intradermally with saline (black plots), mannans (Mann, red plots) or β-glucans (β-gluc, red plots). 6 or 24 hours later dLNs were collected and analyzed by flow cytometry for absolute numbers (A-G) or CD86 expression (H) of the indicated populations. N = 5-6 mice per group. (I - K) Mice were injected intradermally with saline (Sal) or mannans (Mann). 24 hours later dLNs were collected and analyzed for absolute numbers of total IFNγ+ cells (I) or the indicated cell populations among CD45+ cells (J). Levels of IFNγ production in CD8+ T cells or NK cells were evaluated by measuring the MFI of IFNγ among IFNγ-expressing cells (K). (L - N) WT mice treated on day −1 and 0 with the same volumes of PBS or a depleting anti-Asialo GM1 antibody (αAsGM1) (L, N), or WT and Batf3−/− mice (M) were injected intradermally on day 0 with saline or mannans. 24 hours later dLNs were collected and RNA was extracted for gene expression analysis (L, M), or dLNs were weighted and analyzed by flow cytometry for absolute numbers of the indicated populations (N). Results are expressed as fold over contralateral, saline-injected LN. N = 4-5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing between treatments (Mann vs β-gluc, A - H) or each group against the value 1 (which represent the contralateral control sample expressed as fold, L and M). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing between treatments (Mann, β-gluc) and their respective saline controls (A - H) or between the indicated experimental groups (K, L).
Figure S1. Related to Figure 1. Mannans and β-glucans exhibit different diameters and inflammatory properties. (A) Hydrodynamic diameters of mannan (Mann) and β-glucan (β-gluc) preparations were measured by dynamic light scattering. Results from 1 representative experiment are shown. (B, C) GM-CSF differentiated, bone marrow-derived phagocytes were generated from WT (B, C), Clec7a−/−, Clec4n−/− and Fcer1g−/− mice (C) and stimulated with LPS, curdlan, β-glucans (β-gluc), soluble mannans (Mann) (B, C), uncoated microbeads (B only) and microbeads covalently linked to mannans (B:mann) (C). After 18-21 hours supernatants were collected, and TNF and IL-2 concentrations measured by ELISA while cells were harvested and expression levels of CD86 and OX40L were measured by flow cytometry. N = 4-6 independent experiments. ## indicate p ≤ 0.01 when comparing each group against its untreated control (CTRL). Results are shown as mean +SEM. (D) Pathway enrichment analysis of genes belonging to the cluster upregulated by β-glucans as depicted in Figure 1B.
Figure S4. Related to Figure 3. Mannans elicit CARD-9-independent responses in the dLN. Pathway analysis of genes significantly induced in WT compared to Fcer1g−/− mice as depicted inFigure 3B.
Figure S6. Related to Figure 6. B and T cell responses activated by immunization with SARS-CoV-2 Spike formulated with alum/mannans have the same cellular and molecular requirements of the LN innate response elicited by alum/mannans. (A - I) WT mice, transgenic mice or WT mice treated on day −1, 0, 13 and 14 with blocking anti-IFNAR plus anti-IFNγ (αIFNAR/IFNγ) antibodies or the same doses of isotype control antibodies (Iso CTRL), were injected intradermally with pre-fusion stabilized SARS-CoV-2 trimer combined with alum (AH) (S/AH) or AH/mannans (S/AH/mann) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 (B-I) or on day 98 (A, WT mice) to assess anti-Spike antibody levels. In selected experiments (C, E), mice were sacrificed on day 35 to collect spleens and isolate splenocytes for in vitro restimulation with Spike peptides. After 96 hours supernatants were collected and IFNγ protein levels were measured by ELISA (F). N = 4-7 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing S/AH and S/AH/mann vs Sal (A, B) or S/AH/mann vs S/AH (C - I). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing S/AH/mann vs S/AH (A, B) or S/AH/mann across treatments or genotypes (C - I). Comparisons are indicated by the color code.
Figure S7. Related to Figure 7. The adjuvant formulation of mannans and alum confers protection against lung viral infections. (A, B) Mice were injected intradermally with saline (Sal), pre-fusion stabilized SARS-CoV-2 trimer alone (S) or combined with alum (AH) (S/AH), AH/mannans (S/AH/mann), AddaS03 (S/AddaS03), or AH/PHAD (S/AH/PHAD) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess SARS-CoV-2 surrogate virus neutralization test (A) and neutralization titer (B). N = 4-5 mice per group. (C, D) Mice were injected intradermally with saline (Sal), Flublok alone (rHA) or combined with AH (rHA/AH), AH/mannans (rHA/AH/mann), AddaVax (rHA/AddaVax), or AH/PHAD (rHA/AH/PHAD) on day 0 (prime) and day 14 (boost). On day 35 mice were intranasally infected with IAV A/PR/8/1934. On day 42 serum samples were collected to assess anti-IAV A/PR/8/1934 recombinant hemagglutinin (Anti-rPR8) levels (C). Histology score of lung images collected as in in Figure 7F (D). N = 5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01. Comparisons are indicated by the color code.
Figure S5. Related to Figure 5. Mannans formulated with aluminum hydroxide acquire physical properties that predict immunological functions. (A) Soluble mannans (Mann), AH and the formulation of alum (AH) and mannans (AH/mann) were incubated at room temperature for 30 minutes, then spun down and the supernatants were collected for 1H-nuclear magnetic resonance quantification of unbound mannans. The reaction contained an excess of mannan. The results indicate that the mannan absorption capacity of AH is approximately two times its mass in this formulation strategy. Results are expressed as percentage of soluble mannans and shown as mean + SD. (B - I) GM-CSF differentiated, bone marrow-derived phagocytes were generated from WT or the indicated knock out mice and stimulated with alum (AH), mannans (Mann), AH/mannans (AH/mann). After 18-21 hours supernatants were collected, and TNF and IL-2 protein concentrations were measured by ELISA (B, C). Cells were harvested and expression levels of CD86 and OX40L were measured by flow cytometry (D, E). Alternatively, cells were stimulated for 6 hours, and RNA was extracted for gene expression analysis. Results are reported as relative expression compared to Rpl13a (F-I). N = 3 (A) or 4 (B, I) independent experiments. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing the same genotype across treatments. * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing the same treatment across genotypes. Comparisons are indicated by the color code.
Highlights:
Size and solubility of microbial ligands determine the immune response
Fungal mannans are directly drained to the LN to drive cell accrual via interferon release
Tuning mannan properties widens epitope specificity of SARS-CoV-2 anti-spike antibodies
An adjuvant formulation of mannan/alum induces heterosubtypic immunity against IAV
ACKNOWLEDGEMENTS
IZ is supported by NIH grant 1R01AI121066, 1R01DK115217, 1R01AI165505 and contract no. 75N93019C00044, Lloyd J. Old STAR Program CRI3888, and holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. DJD is supported by NIH grant 1R21AI137932-01A1 and contract nos. 75N93019C00044 and 75N93020C00038. SJE is funded by grants from the Value of Vaccine Research Network and the MassCPR. MDK is supported by NIH grant R21 1R21AI159877. DLW is supported by NIH grants 1R01GM119197, 1R01GM083016 and C06RR0306551. ES is funded by the NSF Graduate Research Fellowship Program. SJE is an Investigator with the Howard Hughes Medical Institute. MF is supported by NIH grants R01 AI148166, R21AI153480, DARPA HR0011-20-2-0040, and DHS/BARDA ASPR-20-01495. We thank Drs. Barney S. Graham (NIH Vaccine Research Center) and Aaron G. Schmidt (Ragon Institute) for providing plasmids for pre-fusion stabilized SARS-CoV-2 Spike trimer and RBD, respectively. RS thanks the UCLA Institute for Quantitative and Computational Biosciences (QCBio) Collaboratory community directed by Matteo Pellegrini.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
FB, EN, TRO’M, IZ, DJD, and OL are named inventors on invention disclosures and patents involving vaccine adjuvants. SJE is a founder of TSCAN Therapeutics, ImmuneID, MAZE Therapeutics and Mirimus. SJE serves on the scientific advisory board of Homology Medicines, TSCAN Therapeutics, MAZE Therapetics, XChem, and is an advisor for MPM, none of which impact this work. SJE is an inventor on a patent application issued to the Brigham and Women’s Hospital (US20160320406A) that covers the use of the VirScan library to identify pathogen antibodies in blood. The other authors declare no commercial or financial conflict of interest.
Data and materials availability:
all data is available in the manuscript or the supplementary materials.
REFERENCES
- Acton SE, Farrugia AJ, Astarita JL, Mourao-Sa D, Jenkins RP, Nye E, Hooper S, van Blijswijk J, Rogers NC, Snelgrove KJ, et al. (2014). Dendritic cells control fibroblastic reticular network tension and lymph node expansion. Nature 514, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton SE, and Reis e Sousa C (2016). Dendritic cells in remodeling of lymph nodes during immune responses. Immunol Rev 271, 221–229. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, and Jennings GT (2010). Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol 10, 787–796. [DOI] [PubMed] [Google Scholar]
- Banchereau J, and Steinman RM (1998). Dendritic cells and the control of immunity. Nature 392, 245–252. [DOI] [PubMed] [Google Scholar]
- Barrat FJ, Crow MK, and Ivashkiv LB (2019). Interferon target-gene expression and epigenomic signatures in health and disease. Nature immunology 20, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello F, Pietrasanta C, Lai JCY, Walsh LM, Sharma P, O’Driscoll DN, Ramirez J, Brightman S, Pugni L, Mosca F, et al. (2017). Identification and Characterization of Stimulator of Interferon Genes As a Robust Adjuvant Target for Early Life Immunization. Front Immunol 8, 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello F, Zanoni I, and Granucci F (2020). Cellular and molecular mechanisms of antifungal innate immunity at epithelial barriers: The role of C-type lectin receptors. Eur J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brown GD, Willment JA, and Whitehead L (2018). C-type lectins in immunity and homeostasis. Nat Rev Immunol 18, 374–389. [DOI] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni I, and Kagan JC (2015). Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33, 257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JC, Crozat K, Dalod M, and Mattiuz R (2019). Are Conventional Type 1 Dendritic Cells Critical for Protective Antitumor Immunity and How? Front Immunol 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, et al. (2020). Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 73–84 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M, Collignon C, Herve C, Chalon A, Welsby I, Detienne S, van Helden MJ, Dutta S, Genito CJ, Waters NC, et al. (2017). Cellular and molecular synergy in AS01-adjuvanted vaccines results in an early IFNgamma response promoting vaccine immunogenicity. NPJ Vaccines 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, et al. (2020). Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med 383, 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giovanni M, Cutillo V, Giladi A, Sala E, Maganuco CG, Medaglia C, Di Lucia P, Bono E , Cristofani C, Consolo E, et al. (2020). Spatiotemporal regulation of type I interferon expression determines the antiviral polarization of CD4(+) T cells. Nature immunology 21, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierlaurent AM, Collignon C, Bourguignon P, Wouters S, Fierens K, Fochesato M, Dendouga N, Langlet C, Malissen B, Lambrecht BN, et al. (2014). Enhancement of adaptive immunity by the human vaccine adjuvant AS01 depends on activated dendritic cells. J Immunol 193, 1920–1930. [DOI] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, and Flavell RA (2008). Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold CL, and Kagan JC (2019). Inflammasomes: Threat-Assessment Organelles of the Innate Immune System. Immunity 51, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A, and Monti S (2020). hypeR: an R package for geneset enrichment workflows. Bioinformatics 36, 1307–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AL, Malhotra D, Acton SE, Lukacs-Kornek V, Bellemare-Pelletier A, Curry M, Armant M, and Turley SJ (2011). Reproducible isolation of lymph node stromal cells reveals site-dependent differences in fibroblastic reticular cells. Front Immunol 2, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, et al. (2011). Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 472, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BS (2020). Rapid COVID-19 vaccine development. Science 368, 945–946. [DOI] [PubMed] [Google Scholar]
- Grant SM, Lou M, Yao L, Germain RN, and Radtke AJ (2020). The lymph node at a glance - how spatial organization optimizes the immune response. J Cell Sci 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Wevers B, Bruijns SC, and Geijtenbeek TB (2009). Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nature immunology 10, 203–213. [DOI] [PubMed] [Google Scholar]
- Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, et al. (2020). A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 182, 744–753 e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DJ, Aung A, and Silva M (2020). Controlling timing and location in vaccines. Adv Drug Deliv Rev 158, 91–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB (2018). IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol 18, 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, and Medzhitov R (2004). Toll-like receptor control of the adaptive immune responses. Nature immunology 5, 987–995. [DOI] [PubMed] [Google Scholar]
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. (2020). An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med 383, 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA Jr., and Medzhitov R (2002). Innate immune recognition. Annu Rev Immunol 20, 197–216. [DOI] [PubMed] [Google Scholar]
- Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, and Germain RN (2012). A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell 150, 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, Plested JS, Zhu M, Cloney-Clark S, Zhou H, et al. (2020). Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med 383, 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa M, Greene RR, Noss I, Lowman DW, and Williams DL (2011). C. albicans increases cell wall mannoprotein, but not mannan, in response to blood, serum and cultivation at physiological temperature. Glycobiology 21, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Montoya M, Edwards MJ, Thompson C, Burke SA, Ashton M, Lo D, Tough DF, and Borrow P (2006). A role for the transcription factor RelB in IFN-alpha production and in IFN-alpha-stimulated cross-priming. Eur J Immunol 36, 2085–2093. [DOI] [PubMed] [Google Scholar]
- Leal JM, Huang JY, Kohli K, Stoltzfus C, Lyons-Cohen MR, Olin BE, Gale M Jr., and Gerner MY (2021). Innate cell microenvironments in lymph nodes shape the generation of T cell responses during type I inflammation. Sci Immunol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist SR, Dinnon KH 3rd, Schafer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, et al. (2020). A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 183, 1070–1085 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Ozga AJ, Sokol CL, and Luster AD (2020). Targeting Lymph Node Niches Enhances Type 1 Immune Responses to Immunization. Cell Rep 31, 107679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Iliev ID, and Hohl TM (2017). Immunity against fungi. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Moynihan KD, Zheng Y, Szeto GL, Li AV, Huang B, Van Egeren DS, Park C, and Irvine DJ (2014). Structure-based programming of lymph-node targeting in molecular vaccines. Nature 507, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman DW, Ensley HE, Greene RR, Knagge KJ, Williams DL, and Kruppa MD (2011). Mannan structural complexity is decreased when Candida albicans is cultivated in blood or serum at physiological temperature. Carbohydr Res 346, 2752–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman DW, Greene RR, Bearden DW, Kruppa MD, Pottier M, Monteiro MA, Soldatov DV, Ensley HE, Cheng SC, Netea MG, et al. (2014). Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J Biol Chem 289, 3432–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z, Deng YQ, Ye Q, Cao L, Sun CY, Fan C, Huang W, Sun S, Sun Y, Zhu L, et al. (2020). Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369, 1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn GM, Laga R, Darrah PA, Ishizuka AS, Balaci AJ, Dulcey AE, Pechar M, Pola R, Gerner MY, Yamamoto A, et al. (2015). In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat Biotechnol 33, 1201–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, and Sallusto F (2004). Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nature immunology 5, 1260–1265. [DOI] [PubMed] [Google Scholar]
- Matzinger P (1994). Tolerance, danger, and the extended family. Annu Rev Immunol 12, 991–1045. [DOI] [PubMed] [Google Scholar]
- McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, et al. (2020). Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, et al. (2020). Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, and Tanaka M (2007). Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest 117, 2268–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran I, Grootveld AK, Nguyen A, and Phan TG (2019). Subcapsular Sinus Macrophages: The Seat of Innate and Adaptive Memory in Murine Lymph Nodes. Trends Immunol 40, 35–48. [DOI] [PubMed] [Google Scholar]
- Morefield GL, Jiang D, Romero-Mendez IZ, Geahlen RL, Hogenesch H, and Hem SL (2005). Effect of phosphorylation of ovalbumin on adsorption by aluminum-containing adjuvants and elution upon exposure to interstitial fluid. Vaccine 23, 1502–1506. [DOI] [PubMed] [Google Scholar]
- Moyer TJ, Kato Y, Abraham W, Chang JYH, Kulp DW, Watson N, Turner HL, Menis S, Abbott RK, Bhiman JN, et al. (2020). Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat Med 26, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, and Gow NA (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 6, 67–78. [DOI] [PubMed] [Google Scholar]
- O’Hagan DT, Lodaya RN, and Lofano G (2020). The continued advance of vaccine adjuvants - ‘we can work it out’. Semin Immunol 50, 101426. [DOI] [PubMed] [Google Scholar]
- Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G , and Forster R (2004). CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 21, 279–288. [DOI] [PubMed] [Google Scholar]
- Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, et al. (2020). Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha I, Jaiswal H, Mishra R, Nel HJ, Schreuder J, Kaushik M, Singh Chauhan K, Singh Rawat B, Thomas R, Naik S, et al. (2020). RelB suppresses type I Interferon signaling in dendritic cells. Cell Immunol 349, 104043. [DOI] [PubMed] [Google Scholar]
- Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, Mekada E, Kimata Y, Tsuru A, and Kohno K (2001). Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol 19, 746–750. [DOI] [PubMed] [Google Scholar]
- Sangesland M, and Lingwood D (2021). Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santus W, Barresi S, Mingozzi F, Broggi A, Orlandi I, Stamerra G, Vai M, Martorana AM, Polissi A, Kohler JR, et al. (2017). Skin infections are eliminated by cooperation of the fibrinolytic and innate immune systems. Sci Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Muecksch F, Lorenzi JCC, Leist SR, Cipolla M, Bournazos S, Schmidt F, Maison RM, Gazumyan A, Martinez DR, et al. (2021). Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. (2020). A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124. [DOI] [PubMed] [Google Scholar]
- Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y, Robinson ML, Sie BM, Li MZ, Chen Y, et al. (2020). Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg KA, Payne GW, Sato A, Medzhitov R, Segal SS, and Iwasaki A (2005). Innate control of adaptive immunity via remodeling of lymph node feed arteriole. Proc Natl Acad Sci U S A 102, 16315–16320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, and Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC (2017). The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol 17, 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, Hu Z, Chen VC, Young BE, Sia WR, et al. (2020). A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 38, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Tortorici MA, Beltramello M, Lempp FA, Pinto D, Dang HV, Rosen LE, McCallum M, Bowen J, Minola A, Jaconi S, et al. (2020). Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, and Veesler D (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EE, Frenck RW Jr., Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, et al. (2020). Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N Engl J Med 383, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Montoya B, Stotesbury C, Ferez M, Xu RH, and Sigal LJ (2019). Langerhans Cells Orchestrate the Protective Antiviral Innate Immune Response in the Lymph Node. Cell Rep 29, 3047–3059 e3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Xu RH, Rubio D, Lev A, Stotesbury C, Fang M, and Sigal LJ (2018). Migratory Dendritic Cells, Group 1 Innate Lymphoid Cells, and Inflammatory Monocytes Collaborate to Recruit NK Cells to the Virus-Infected Lymph Node. Cell Rep 24, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, and McLellan JS (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, et al. (2015a). Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348, aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Wong EB, Rubio D, Roscoe F, Ma X, Nair S, Remakus S, Schwendener R, John S, Shlomchik M, et al. (2015b). Sequential Activation of Two Pathogen-Sensing Pathways Required for Type I Interferon Expression and Resistance to an Acute DNA Virus Infection. Immunity 43, 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu JF, Zheng G, Lu HW, Duan JL, Rui W, Guan JH, Cheng LQ, Yang DD, Wang MC, et al. (2018). CARD9(S12N) facilitates the production of IL-5 by alveolar macrophages for the induction of type 2 immune responses. Nature immunology 19, 547–560. [DOI] [PubMed] [Google Scholar]
- Zhu LL, Zhao XQ, Jiang C, You Y, Chen XP, Jiang YY, Jia XM, and Lin X (2013). C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity 39, 324–334. [DOI] [PubMed] [Google Scholar]
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, et al. (2020). Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 1. List of genes detected by bulk RNAseq analysis that are expressed in the skin 6 hours after the injection of saline, β-glucans or mannans.
Table S2. Related to Figure 1. List of genes detected by bulk RNAseq analysis that are expressed in the dLN 6 and 24 hours after the injection of saline, β-glucans or mannans.
Table S3. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT and Fcer1g−/− mice 6 hours after the injection of mannans.
Table S4. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from Card9−/− and Fcer1g−/− mice 6 hours after the injection of mannans.
Table S5. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT and Card9−/− mice 6 hours after the injection of mannans.
Table S6. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT, Fcer1g−/− and Card9−/− mice 6 hours after the injection of mannans that are upregulated in Fcer1g−/− compared to WT and Card9−/− mice.
Table S7. Related to Figure 3. List of genes detected by targeted transcriptomic analysis of CD3− CD19− NK1.1− Ter119− CD45+ AF488-mannan+ Ly6G− (CD11b+ Ly6C+)− CD11b+ CD11c+ isolated from WT, Fcer1g−/− and Card9−/− mice 6 hours after the injection of mannans that are upregulated in WT and Card9−/− compared to Fcer1g−/− mice.
Figure S3. Related to Figure 1. Soluble whole glucan particles elicit LN expansion and ISG expression. Mice were injected intradermally with saline (Sal), dispersible (WGP-D) or soluble (WGP-S) whole glucan particles. 24 hours later injection sites were assessed as indicated in Figure 1A (A), skin samples and dLNs were collected, LN weights were measured, and RNA extracted for gene expression analysis (B). Results are expressed as fold over the median value of saline-injected skin samples or LNs. N = 4 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing each group against the value 1 (which represent the saline control samples expressed as fold). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing between different experimental groups.
Figure S2. Related to Figure 1. Mannans and β-glucans induce unique patterns of immune cell recruitment and activation in the draining LNs. (A-H) Mice were injected intradermally with saline (black plots), mannans (Mann, red plots) or β-glucans (β-gluc, red plots). 6 or 24 hours later dLNs were collected and analyzed by flow cytometry for absolute numbers (A-G) or CD86 expression (H) of the indicated populations. N = 5-6 mice per group. (I - K) Mice were injected intradermally with saline (Sal) or mannans (Mann). 24 hours later dLNs were collected and analyzed for absolute numbers of total IFNγ+ cells (I) or the indicated cell populations among CD45+ cells (J). Levels of IFNγ production in CD8+ T cells or NK cells were evaluated by measuring the MFI of IFNγ among IFNγ-expressing cells (K). (L - N) WT mice treated on day −1 and 0 with the same volumes of PBS or a depleting anti-Asialo GM1 antibody (αAsGM1) (L, N), or WT and Batf3−/− mice (M) were injected intradermally on day 0 with saline or mannans. 24 hours later dLNs were collected and RNA was extracted for gene expression analysis (L, M), or dLNs were weighted and analyzed by flow cytometry for absolute numbers of the indicated populations (N). Results are expressed as fold over contralateral, saline-injected LN. N = 4-5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing between treatments (Mann vs β-gluc, A - H) or each group against the value 1 (which represent the contralateral control sample expressed as fold, L and M). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing between treatments (Mann, β-gluc) and their respective saline controls (A - H) or between the indicated experimental groups (K, L).
Figure S1. Related to Figure 1. Mannans and β-glucans exhibit different diameters and inflammatory properties. (A) Hydrodynamic diameters of mannan (Mann) and β-glucan (β-gluc) preparations were measured by dynamic light scattering. Results from 1 representative experiment are shown. (B, C) GM-CSF differentiated, bone marrow-derived phagocytes were generated from WT (B, C), Clec7a−/−, Clec4n−/− and Fcer1g−/− mice (C) and stimulated with LPS, curdlan, β-glucans (β-gluc), soluble mannans (Mann) (B, C), uncoated microbeads (B only) and microbeads covalently linked to mannans (B:mann) (C). After 18-21 hours supernatants were collected, and TNF and IL-2 concentrations measured by ELISA while cells were harvested and expression levels of CD86 and OX40L were measured by flow cytometry. N = 4-6 independent experiments. ## indicate p ≤ 0.01 when comparing each group against its untreated control (CTRL). Results are shown as mean +SEM. (D) Pathway enrichment analysis of genes belonging to the cluster upregulated by β-glucans as depicted in Figure 1B.
Figure S4. Related to Figure 3. Mannans elicit CARD-9-independent responses in the dLN. Pathway analysis of genes significantly induced in WT compared to Fcer1g−/− mice as depicted inFigure 3B.
Figure S6. Related to Figure 6. B and T cell responses activated by immunization with SARS-CoV-2 Spike formulated with alum/mannans have the same cellular and molecular requirements of the LN innate response elicited by alum/mannans. (A - I) WT mice, transgenic mice or WT mice treated on day −1, 0, 13 and 14 with blocking anti-IFNAR plus anti-IFNγ (αIFNAR/IFNγ) antibodies or the same doses of isotype control antibodies (Iso CTRL), were injected intradermally with pre-fusion stabilized SARS-CoV-2 trimer combined with alum (AH) (S/AH) or AH/mannans (S/AH/mann) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 (B-I) or on day 98 (A, WT mice) to assess anti-Spike antibody levels. In selected experiments (C, E), mice were sacrificed on day 35 to collect spleens and isolate splenocytes for in vitro restimulation with Spike peptides. After 96 hours supernatants were collected and IFNγ protein levels were measured by ELISA (F). N = 4-7 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing S/AH and S/AH/mann vs Sal (A, B) or S/AH/mann vs S/AH (C - I). * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing S/AH/mann vs S/AH (A, B) or S/AH/mann across treatments or genotypes (C - I). Comparisons are indicated by the color code.
Figure S7. Related to Figure 7. The adjuvant formulation of mannans and alum confers protection against lung viral infections. (A, B) Mice were injected intradermally with saline (Sal), pre-fusion stabilized SARS-CoV-2 trimer alone (S) or combined with alum (AH) (S/AH), AH/mannans (S/AH/mann), AddaS03 (S/AddaS03), or AH/PHAD (S/AH/PHAD) on day 0 (prime) and day 14 (boost). Serum samples were collected on day 28 to assess SARS-CoV-2 surrogate virus neutralization test (A) and neutralization titer (B). N = 4-5 mice per group. (C, D) Mice were injected intradermally with saline (Sal), Flublok alone (rHA) or combined with AH (rHA/AH), AH/mannans (rHA/AH/mann), AddaVax (rHA/AddaVax), or AH/PHAD (rHA/AH/PHAD) on day 0 (prime) and day 14 (boost). On day 35 mice were intranasally infected with IAV A/PR/8/1934. On day 42 serum samples were collected to assess anti-IAV A/PR/8/1934 recombinant hemagglutinin (Anti-rPR8) levels (C). Histology score of lung images collected as in in Figure 7F (D). N = 5 mice per group. # and ## respectively indicate p ≤ 0.05 and 0.01. Comparisons are indicated by the color code.
Figure S5. Related to Figure 5. Mannans formulated with aluminum hydroxide acquire physical properties that predict immunological functions. (A) Soluble mannans (Mann), AH and the formulation of alum (AH) and mannans (AH/mann) were incubated at room temperature for 30 minutes, then spun down and the supernatants were collected for 1H-nuclear magnetic resonance quantification of unbound mannans. The reaction contained an excess of mannan. The results indicate that the mannan absorption capacity of AH is approximately two times its mass in this formulation strategy. Results are expressed as percentage of soluble mannans and shown as mean + SD. (B - I) GM-CSF differentiated, bone marrow-derived phagocytes were generated from WT or the indicated knock out mice and stimulated with alum (AH), mannans (Mann), AH/mannans (AH/mann). After 18-21 hours supernatants were collected, and TNF and IL-2 protein concentrations were measured by ELISA (B, C). Cells were harvested and expression levels of CD86 and OX40L were measured by flow cytometry (D, E). Alternatively, cells were stimulated for 6 hours, and RNA was extracted for gene expression analysis. Results are reported as relative expression compared to Rpl13a (F-I). N = 3 (A) or 4 (B, I) independent experiments. # and ## respectively indicate p ≤ 0.05 and 0.01 when comparing the same genotype across treatments. * and ** respectively indicate p ≤ 0.05 and 0.01 when comparing the same treatment across genotypes. Comparisons are indicated by the color code.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
RNA sequencing data accession number: GSE193419.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
all data is available in the manuscript or the supplementary materials.


