Abstract
Protein acetylation is conserved across phylogeny and has been recognized as one of the most prominent post-translational modifications since its discovery nearly sixty years ago. Histone acetylation is an active mark characteristic of open chromatin, but acetylation on specific lysine residues and histone variants occurs in different biological contexts and can confer varying outcomes. The significance of acetylation events is indicated by the implications of lysine acetyltransferases, deacetylases, and acetyl-lysine readers in developmental disorders and pathologies. Recent advances have uncovered new roles of acetylation regulators in chromatin-centric events, emphasizing the complexity of these functional networks. In this review, we discuss mechanisms and dynamics of acetylation in chromatin organization and DNA-templated processes, including gene transcription, DNA repair and replication.
Keywords: acetylation, acetyltransferase, deacetylase, chromatin organization, transcription, enhancer, DNA repair, DNA replication
eTOC Blurb
Chen et al. dive into the mechanisms and dynamics of protein acetylation in chromatin organization and DNA-templated processes, including gene transcription, DNA repair and replication. Newly discovered roles of acetylation regulators in chromatin-centric events have implications for human disease.
Introduction
Histone acetylation has long been associated with gene activity, providing one of the defining characteristics of “open” chromatin (Figure 1). Since acetylation neutralizes the charge of the epsilon amino group in lysine, it reduces electrostatic interactions between histones and DNA, thereby impacting both nucleosome structure and higher-order chromatin folding (Eberharter and Becker, 2002). The importance of specific highly conserved histone acetylation sites to gene activity and chromatin states was revealed by genetic studies in yeast (Megee et al., 1990) and by microscopic analyses using site-specific antibodies in flies and mammalian cells (Johnson et al., 1998; Munks et al., 1991). The identification of the first “writers” and “erasers” of acetylation, lysine acetyltransferases (KATs) and histone deacetylases (HDACs), in the mid-1990s further cemented the connections between this modification and gene regulation, as these enzymes had already been identified as transcriptional co-activators and repressors, respectively (Kuo et al., 1996; Taunton et al., 1996). Dozens of KATs have now been identified, along with several families of HDACs (Marmorstein and Zhou, 2014). In addition, specific acetyl-lysine “readers” have been discovered, reinforcing the regulatory role of this modification. Many acetylation modifiers and readers can catalyze and recognize, respectively, other acylations such as propionylation, butyrylation and crotonylation, which have been shown to serve cellular and physiological functions (Jiang et al., 2021; Sabari et al., 2017).
Figure 1. Association of histone acetylation with chromatin organization from local to higher-order scales.
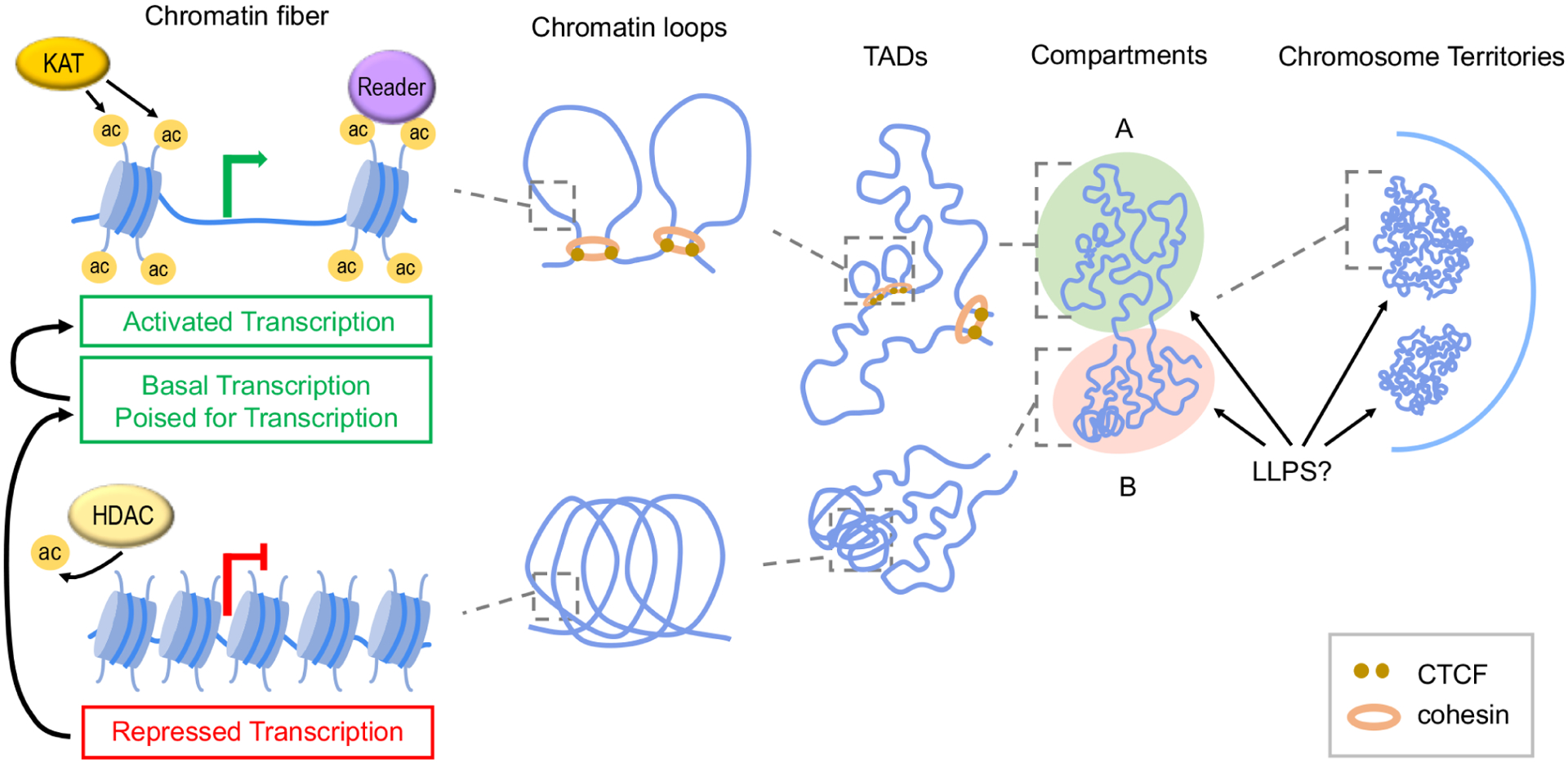
Histone acetylation (ac) conferred by KATs and recognized by reader proteins inhibits inter-nucleosome interactions and generates an open chromatin configuration, which allows gene transcription of various levels. On the contrary, deacetylation by HDACs leads to shorter inter-nucleosome distances on the chromatin fiber and repressed transcription. Acetylated chromatin is able to form enhancer–promoter chromatin loops, whereas lack of acetylation facilitates more compact conformations. Architectural proteins CTCF and cohesin mediate formation of chromatin loops and TAD borders. TADs segregate into broadly A and B chromatin compartments, and these compartments further form individual chromosome territories. LLPS may contribute to the segregation of chromatin compartments and territories.
Technologies have advanced beyond simple identification of open chromatin as nuclease sensitive regions, and these approaches now allow genome-scale resolution of acetylation patterns and levels of chromosome organization. Definition of the “when and where” of histone acetylation has provided new clues to the functions of individual acetylation events as well as global levels of acetylation in multiple histones. Here we review connections between histone acetylation states and specific steps in gene activation. We also discuss recent studies that reveal how acetylation impacts progressively higher-order chromatin and chromosome organization, from enhancer–promoter looping to formation of topologically associating domains (TADs) and chromosome territories. We additionally cover regulation of acetylation in other DNA-centered events, specifically DNA replication and repair. Throughout, we discuss how understanding the functions and regulation of protein acetylation informs the etiology of disease states, especially cancers.
Acetylation and Chromatin Organization
The “beads-on-a-string” view of nucleosomes separated by linker DNA provides the baseline for chromatin organization (Figure 1) (Woodcock and Ghosh, 2010). Interactions between nucleosomes create compacted higher-order structures, and further interactions between these secondary chromatin fibers form even more compact tertiary structures (Luger et al., 2012). Each level of organization is impacted by histone variants and by post-translational modifications. Acetylation decreases availability of histone tails to neighboring linker DNA and also disrupts inter-nucleosome interactions, leading to unfolding of chromatin fibers (Hansen, 2002)—a series of events more recently demonstrated by all-atom molecular dynamics simulations (Chang and Takada, 2016). These observations further support a role for histone acetylation in chromatin decompaction to expand chromatin access to transcription factors (TFs) and RNA polymerases, augmenting gene transcription (Eberharter and Becker, 2002).
Histone acetylation also impacts chromatin organization through ATP-dependent chromatin remodeling complexes, which often contain acetyl-lysine readers (Josling et al., 2012). For example, histone H4 lysine 16 acetylation (H4K16ac) directly inhibits secondary and tertiary higher-order structures, as well as nucleosome sliding mediated by the Drosophila ACF (ATP-utilizing chromatin assembly and remodeling factor) complex (Shogren-Knaak et al., 2006; Zhang et al., 2017). In vivo effects of acetylation on nucleosome occupancy have been visualized elegantly in recent years using three-dimensional, two-color super-resolution microscopy (Otterstrom et al., 2019). Previous work indicated that nucleosomes are non-uniformly grouped along the chromatin fiber, forming “nucleosome clutches” (Ricci et al., 2015). The numbers of nucleosomes per clutch were found to be heterogeneous within a given nucleus. Histone acetylation decreases nucleosome occupancy and thereby loosens DNA packing by nucleosome clutches, particularly in nucleosome clutch-rich regions (Otterstrom et al., 2019). Moreover, hyperacetylation of histone tails upon HDAC inhibition resulted in formation of smaller nucleosome clutches. These findings re-emphasize the impact of acetylation on nucleosome placement and density, which in turn impacts both primary and higher-order levels of chromatin organization, all of which influence gene activity.
Acetylation in Transcriptional Regulation
Levels of Gene Transcription
Understanding how transcription is regulated is complicated by the many states of gene expression, ranging from completely silent to highly induced (Figure 1) (Weake and Workman, 2010). Truly silent genes include cell- and lineage-specific genes whose expression is never needed in a different cellular context. Other genes are repressed under certain conditions, but are activated upon changes in the physiological or developmental state of the cell. Non-expressed genes may still be associated with paused RNA polymerases, poising these genes for rapid activation. Other genes are expressed at a basal level under normal conditions prior to high-level activation upon stimulation. Activation itself often occurs in bursts, rather than sustained high-level polymerase activity.
Each of the above gene expression states is associated with particular chromatin states, including different levels and compositions of histone acetylation. The many acetylation sites within each histone provide a wealth of regulatory potential and have been reviewed extensively (Choi and Howe, 2009; Latham and Dent, 2007). Here we will focus on the functions of a few specific acetylation events involved in particular structural features that control gene expression.
Histone Acetylation at Promoters and Gene Bodies
Chromatin structures at gene promoters are typified by a nucleosome free region (NFR) flanked by well positioned nucleosomes, referred to as −1 and +1 nucleosomes, reflecting their positions relative to the transcription start site (TSS) (Rando, 2007). Enrichment of a variant form of histone H2A, H2A.Z, is observed in these flanking nucleosomes, together with enrichment of activating modifications including histone H3 lysine 4 trimethylation (H3K4me3) and H3 lysine 9 acetylation (H3K9ac) (Jiang and Pugh, 2009). Altogether, these features are thought to increase chromatin accessibility and enable assembly of the RNA polymerase II (Pol II) preinitation complex (PIC). Acetylation of H3 and other histones at promoters and gene bodies can influence accessibility through effects on histone–DNA interactions, inter-nucleosome interactions, and recruitment of regulatory proteins through interactions with acetylation reader domains.
Although increased histone acetylation clearly correlates with gene activity, the causal relationship between promoter chromatin and transcription is still not entirely clear even after decades of study. Some findings, for example, suggest that modifications such as H3K4me3 and H3K9ac serve as markers for genes that have been transcribed rather than as drivers of transcription per se (Howe et al., 2017). A recent study revisited the long-standing “chicken and egg” relationship between histone acetylation and transcription by monitoring both RNA Pol II distributions and histone acetylation patterns following rapid inhibition of Pol II in yeast (Martin et al., 2021). Using immunoblots to monitor total levels of acetylation and chromatin immunoprecipitation followed by sequencing (ChIP-seq) to define genomic distributions, the authors found that acetylation at multiple sites in histones H3 and H4 decreased following acute Pol II inhibition. These decreases were not rescued by inhibition of HDAC activity by tricostatin A (TSA), indicating the decreases reflected loss of KAT activity, rather than increased HDAC activity. Consistent with this observation, the activity and recruitment of H4-specific KAT complexes, NuA4 (nucleosome acetyltransferase of H4) and Piccolo, were found to be dependent on association with Pol II. These results indicate strongly that maintenance of histone acetylation patterns at already active genes requires ongoing transcription. However, these studies do not address how histone acetylation at inactive, repressed genes enhances Pol II recruitment and transcription activation.
Significant clues to the importance of specific acetylation events have come from studies of the writers of these marks. H3K9ac can be deposited near promoters by a number of KAT complexes, but loss of GNAT (GCN5-related N-acetyltransferase) family members, GCN5 (general control non-repressed 5; KAT2A) and vertebrate-specific PCAF (p300/CBP-associated factor; KAT2B), uniquely causes a global depletion of this modification, indicating these KATs are major writers of H3K9ac in mammalian cells (Jin et al., 2011). GCN5 and PCAF function within multiprotein assemblies, including the SAGA (SPT-ADA-GCN5 acetyltransferase) and ATAC (ADA2A-containing) complexes (Helmlinger and Tora, 2017). The KAT modules in SAGA and ATAC are highly similar, except for unique isoforms of the ADA2 subunit (Kusch et al., 2003). SAGA contains ADA2B, whereas ATAC contains ADA2A. In both cases the ADA2 proteins enhance KAT activity (Kusch et al., 2003; Riss et al., 2015). Outside of the KAT module, SAGA and ATAC complexes are distinct in composition and in function (Chen and Dent, 2021; Helmlinger and Tora, 2017; Nagy et al., 2010). GCN5 and PCAF incorporation into these complexes is mutually exclusive, and an open question is whether the different catalytic subunits confer distinct functions. Although biochemically similar, loss of Gcn5 or PCAF conveys entirely different phenotypes in mice (Xu et al., 2000).
The ATAC complex houses a second, H4-specific KAT, ATAC2 (KAT14), as well as a reader of lysine acetylation, YEATS2 (YEATS domain containing 2). YEATS2 is amplified in non-small cell lung cancer (NSCLC), contributing to tumorigenic transcription programs (Mi et al., 2017). As a reader of H3K27ac, YEATS2 helps to recruit ATAC to gene promoters, enhancing H3K9ac. Interestingly, ATAC contains another H3 reader, the ZZ (ZZ-type zinc finger) domain in the ZZZ3 (zinc finger ZZ-type containing 3) protein, which also helps recruit ATAC to gene promoters for H3K9ac and gene activation (Mi et al., 2018). These interactions provide a nice example of the multi-functional nature of KAT complexes and more specifically, cooperative functions between KAT and reader proteins.
SAGA and ATAC impact specific gene expression programs in mammalian cells (Nagy et al., 2010). GCN5 is especially important as a co-activator for MYC, during both development and oncogenesis. However, the kinetics of SAGA and ATAC interactions with promoters or TFs is understudied. SAGA in particular has structural features similar to TFIID, a general TF required for TBP (TATA-binding protein) deposition (Antonova et al., 2019). Recent structures of SAGA in yeast indicate that it also can interact with and deposit TBP (Papai et al., 2020; Wang et al., 2020), but it is unclear whether the mammalian counterpart consistently serves this function. Understanding how and when SAGA, TFIID, ATAC and other factors interact with particular genes is important to understanding the division of labor among them. Vosnakis et al. addressed this important question using photobleaching approaches to monitor kinetics of ATAC, SAGA, TFIID, TFIIB and RNA Pol II in vivo (Vosnakis et al., 2017). Among these factors, only Pol II exhibited a long residence time. Both KAT complexes and the general TFs were transiently associated with chromatin. These findings may reflect a “hit and run” function for these factors that contribute to Pol II stability. Whether SAGA-mediated H3K9ac impacts Pol II stability on chromatin is not evident, but the residence time of Pol II was clearly influenced by transcription elongation and H3K4me levels.
Other KATs and acetylation events also influence gene activity at multiple levels. For example, Gaub et al. found that in both Drosophila and mouse embryonic stem cells (mESCs), acetylation of H4 by the NSL (non-specific lethal) complex is important for maintenance of activity of constitutively expressed genes through recruitment of the Brd4 (bromodomain containing 4) reader protein (Gaub et al., 2020). Consistent with these results, haploinsufficiency of the KANSL1 subunit (KAT8 regulatory NSL complex subunit 1) of the NSL complex is associated with Koolen-de Vries syndrome, and patient derived fibroblasts show altered expression of constitutive genes required for cellular homeostasis. More recently, Radzisheuskaya et al. also reported that the NSL complex is required for expression of housekeeping genes, and thereby cell survival (Radzisheuskaya et al., 2021). Altogether these studies illustrate nicely that acetylation impacts lower-level, constitutive transcription as well as high-level gene activation.
The catalytic subunit of NSL complex, the MYST (MOZ-YBF2/SAS3-SAS2-TIP60) family member Mof (males absent on the first; Kat8), also drives H4 acetylation in the context of the MSL (male specific lethal) complex, which was first discovered as a factor required to increase chromatin accessibility and amplify gene expression across broad regions of the male X chromosome in Drosophila to achieve dosage compensation (Gelbart et al., 2009). Radzisheuskaya et al. found that Mof exhibits different substrate specificity in the context of NSL (acetylating H4K5 and K8) than as part of MSL (acetylating K16) (Radzisheuskaya et al., 2021). These studies reveal that the same KAT can orchestrate different chromatin environments in different contexts, leading to sharp domains of H4K5ac and K8ac at individual promoters as part of NSL and large-scale domains that affect expression of multiple genes as part of MSL. Importantly, Mof-mediated H4K16ac also influences heritability of chromatin states. Samata et al. found that H4K16ac distributions in oocytes are maintained in fertilized embryos in both flies and mammals, where it drives formation of large accessible domains for gene promoters prior to zygotic gene activation (Samata et al., 2020). Furthermore, H4K16ac can be inherited from mothers through oocytes to influence gene expression in offsprings. Other KATs and modifications may also reinforce these states before and after fertilization. For example, p300 (E1A binding protein p300; EP300) and Brd4 are required for the first round of zygotic transcription in zebrafish (Chan et al., 2019).
Interestingly, mammalian BRD4 is reported to have intrinsic KAT activity, indicating it is both a writer and a reader of acetylation (Devaiah et al., 2016). H3K122ac by BRD4 results in histone eviction, nucleosome clearance, chromatin decondensation and increased transcription at target genes, including oncogenic cell cycle regulators MYC, FOXS and AURKB.
Another modification associated with gene activation, particularly at promoters, is H3K14ac (Karmodiya et al., 2012). Regadas et al. recently investigated the role of this modification in cell type-specific gene expression in both Drosophila and human cells (Regadas et al., 2021). ChIP-seq analyses of stage 15 Drosophila embryos revealed H3K14ac is enriched not just at promoters, but also at introns and intergenic regions. In addition, almost one third of H3K14ac was found within gene bodies, at exons that were devoid of other activating modifications, such as H3K9ac, H3K27ac and H3K4me3. Further analyses revealed that H3K14ac is particularly important for expression of genes that direct wing and gut development, and this modification is read by Brm (brahma), the catalytic subunit of the chromatin remodeling SWI/SNF (switch/sucrose non-fermentable) complex. This work not only shows the unique importance of an individual acetylation event, but also how acetylation can direct chromatin organization by other factors. These findings are reminiscent of previous findings in yeast that revealed functional connections between SAGA, NuA4 and SWI/SNF in gene activation (Hassan et al., 2001).
HDACs as erasers of acetylation are also crucial in creating chromatin environments that regulate promoter activity. Traditionally, HDACs are linked to creation of inactive, closed states, but they also impact gene expression at other levels. For example, Etchegaray et al. discovered that the SIRT6 (sirtuin 6) deacetylase interacts with Pol II and inhibits release of the negative elongation factor (NELF), thereby stabilizing Pol II pausing near promoters in mESCs (Etchegaray et al., 2019). SIRT6-mediated deacetylation of H3K9ac and K56ac directly impacts NELF release, providing new insights as to how these modifications regulate gene activity.
Transcriptional Bursting
The advent of single-cell sequencing technologies is revolutionizing our view of how transcription is regulated. Studies of populations of cells provide a view of the average state of transcription, giving the impression that genes are continuously transcribed once they are activated. However, single-cell RNA analyses have revealed that expression of most genes occurs in bursts, with intervals of inactivity (Suter et al., 2011). The frequency and duration of bursts determines the overall expression of a given gene, and these parameters are tuned by specific chromatin states at promoters and enhancers.
Several recent papers indicate that p300/CBP and H3K27ac at enhancers are key to regulation of transcriptional bursting in various cellular contexts. Using single-molecule RNA fluorescence in situ hybridization (FISH), Nicolas et al. examined bursting at the promoter of Bmal1, a circadian gene, in transgenes in fibroblasts and found that histone acetylation by p300 regulates burst frequency (Nicolas et al., 2018). Another study used single-cell RNA sequencing to address the roles of CBP (CREB binding protein) recruitment and H3K27ac at enhancers of two neuronal activity inducible genes, Fos and Npas4 (Chen et al., 2019). Membrane depolarization induces activation of these genes by increasing the frequency of transcriptional bursts. Comparing the effects of targeting Cas9 fusions with either CBP or HDAC8 to enhancers that regulate these genes revealed that targeted increases in CBP-mediated histone acetylation lengthened burst duration, whereas targeted loss of acetylation upon HDAC8 recruitment decreased burst frequency. These results illustrate the importance of H3K27ac to regulation of both duration and frequency aspects of bursts.
Enhancer–Promoter Interactions and Chromatin Looping
Inducible gene promoters are regulated by enhancer elements, which are often located distally from the promoter. Enhancer–promoter interactions (EPIs) are facilitated by formation of chromatin loops, which brings stretches of genomic sequence on the same chromosome to closer physical proximity to each other than to intervening sequences. Looping occurs between enhancers and promoters and also between insulator elements (Kadauke and Blobel, 2009). Enhancer-bound transcriptional activators physically interact with Pol II and other factors at cognate promoters through these chromatin loops to promote transcription (Figure 2).
Figure 2. Acetylation events in enhancer–promoter interactions.
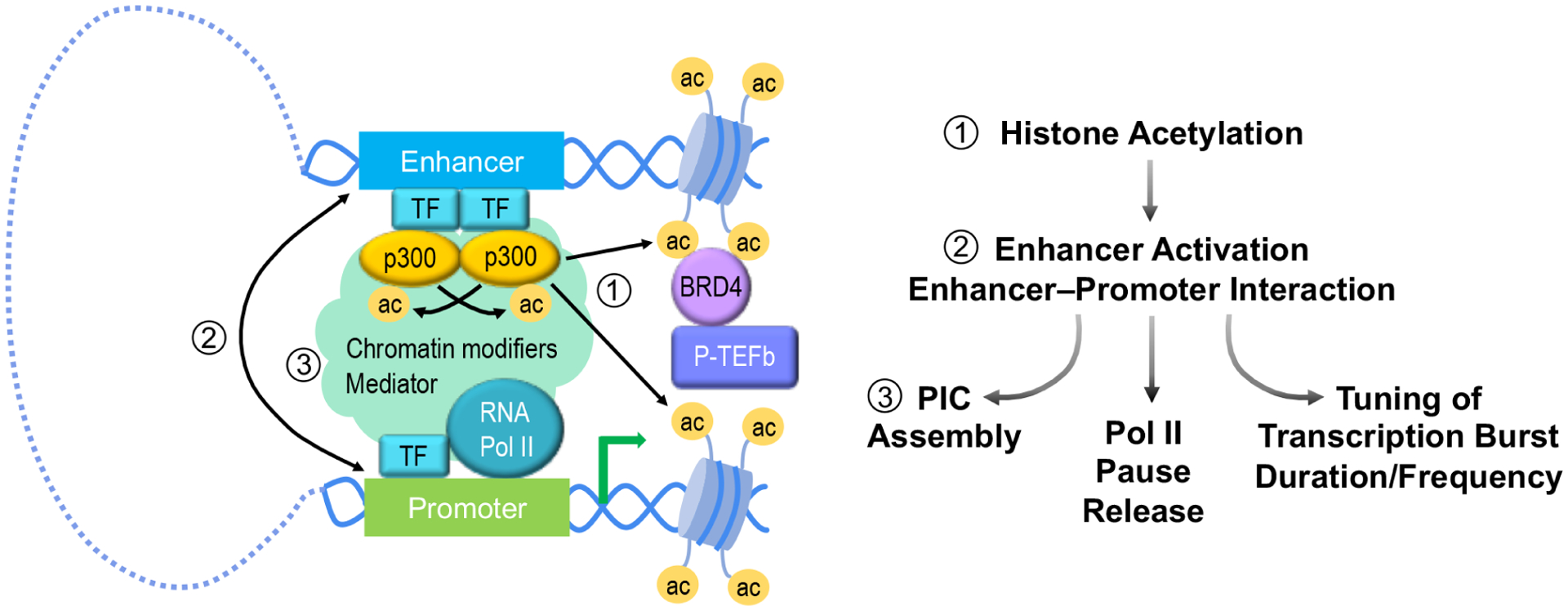
Dimerization of TFs recruits two p300 molecules that trans-acetylate each other, activating their KAT activities. p300 acetylates H3K27 at both enhancer and promoter regions. BRD4 binds to acetylated histones and further recruits or activates P-TEFb to induce release of paused Pol II. Histone acetylation also promotes chromatin looping, allowing enhancer-bound TFs to interact with chromatin regulators and transcription machinery at cognate promoters. Collectively, histone acetylation is crucial for enhancer activation and EPIs, which enhance PIC assembly, release of paused Pol II, and/or tuning of transcriptional bursting.
Specific histone modification signatures determine enhancer functionality: active enhancers are enriched with both H3K27ac and H3K4me1, whereas primed enhancers are marked solely by H3K4me1, and poised or repressed enhancers are distinguished by the presence of H3K27me3 (Calo and Wysocka, 2013). The importance of H3K27ac at enhancers is depicted by a recent study in normal and lymphoma B cells, which demonstrated that changes in the local abundance of H3K27ac modulate the frequency and spreading of EPIs (Sungalee et al., 2021). H3K27ac is mediated by p300/CBP KAT activity, and treatment of cells with the p300/CBP small-molecular inhibitor A-485 reduced genome-wide H3K27ac levels, which led to weakened EPIs as well as loss of larger-scale chromatin interactions (in the A compartment, described below). Less frequent EPIs upon p300/CBP inhibition were associated with lower expression of several oncogenes, including BCL6, BCL11A and MYC. A role for H3K27ac in looping is additionally supported by the finding that this modification is required for formation of new EPIs upon oncogenic t(3:8) chromosomal translocations that bring a large enhancer region on chromosome 3 in close proximity to the MYC locus on chromosome 8. A-485 treatment reduced interactions between these regions, resulting in decreased MYC expression.
Multiple studies have linked p300/CBP to human cancers, and translocations targeting these KATs are common in acute leukemias (Iyer et al., 2004). The catalytic activity of p300/CBP also appears to be particularly important in multiple myeloma and non-Hodgkin’s lymphoma, as indicated by a recent analysis of the Cancer Dependency Map (DepMap) dataset together with A-485 inhibitor survival screens across cancer types (Hogg et al., 2021). Inhibition of p300/CBP depleted acetylation at enhancer elements throughout the genome and resulted in impaired expression of key oncogenic TFs (Hogg et al., 2021; Raisner et al., 2018). The dynamic regulation of acetylation events is indicated by the finding that HDAC3, as part of NCOR/SMRT (nuclear receptor corepressor/silencing mediator for retinoid and thyroid receptor) co-repressor complexes, antagonizes p300/CBP function in multiple myeloma cell lines (Hogg et al., 2021). Removal of H3K27ac subsequently allowed methylation at this site, reinforcing the repressed state. Opposition of p300/CBP functions at enhancers by HDACs, as reflected by changes in expression of enhancer RNAs (eRNAs), has also been observed in mESCs and fibroblasts, indicating enhancers are dynamically regulated in both normal and disease settings (Narita et al., 2021; Weinert et al., 2018).
In addition to H3K27, p300/CBP acetylate multiple lysines in H3 and other histones as well as non-histone proteins (Narita et al., 2019). Acetylation of oncogenic MITF (microphthalmia-associated TF) by p300/CBP reduces the DNA-binding affinity of the TF, increasing its activity and leading to its genome-wide redistribution, ultimately driving tumorigenesis and melanocyte development (Louphrasitthiphol et al., 2020). Interestingly, activation of p300/CBP activity often involves interactions between TFs. Structural studies revealed that dimerization of IRF3 (interferon regulatory factor 3) or STAT1 (signal transducer and activator of transcription 1) results in trans-acetylation of associated p300 molecules in an auto-inhibitory lysine rich loop, thereby activating p300 catalytic activity (Ortega et al., 2018). A separate study found that interactions between TF activation domains and the disordered region of p300 creates “co-condensates” that activate the KAT and facilitate BRD4 recruitment through H3K27ac (Ma et al., 2021). Narita et al. recently reported the unexpected finding that inhibition of p300/CBP KAT activity does not disrupt p300/CBP or TF binding, but does severely attenuate Pol II recruitment and transcription initiation at enhancers and enhancer-regulated genes (Figure 2) (Narita et al., 2021). Protein acetylation conferred via p300/CBP promotes BRD4 binding to induce release of paused Pol II through recruiting or activating P-TEFb (positive transcription elongation factor b). Thus, p300/CBP affects different steps in transcription by diverse mechanisms. Interestingly, these effects appear to be tied to changes in chromatin accessibility in mESCs, but not in malignant hematopoietic cells (Hogg et al., 2021; Narita et al., 2021).
Enhancers often occur in clusters, sometimes achieving “super enhancer” (SE) status. Cell type-specific SEs regulate identity-determining core regulatory (CR) TFs and the overexpression of key CR TFs drives oncogenesis in disease settings. For example, BRD4 interactions at a SE drive MYC oncogene expression, re-emphasizing the potential of BET (bromodomain and extra-terminal) domain inhibitors as possible cancer therapies (Filippakopoulos et al., 2010; Loven et al., 2013; Zuber et al., 2011). Surprisingly, a small-molecule screen for drugs capable of selective inhibition of CR circuitry identified HDACs to be important for CR TF transcription as well (Gryder et al., 2019). Specifically, HDAC1/2/3 co-inhibition disrupted chromatin looping between SEs and CR TFs to suppress CR transcription. Similarly, HDAC7 binds to SEs and regions near TSS to upregulate H3K27ac and SE-associated oncogenes such as MYC in breast cancer stem cells (Caslini et al., 2019). These studies indicate a critical balance between KAT and HDAC activities at SEs appears to be important to regulating transcription dynamics.
Importantly, other histone acetylation events also impact enhancer functions. Nucleosomes containing H2A.Zac are commonly redistributed to newly formed enhancers in cancer, in concert with increased chromatin accessibility and activation of gene expression (Jin et al., 2009; Valdes-Mora et al., 2017). In prostate cancer, upregulated H2A.Zac and redistribution of H2A.Zac to promoters and neo-enhancers facilitate androgen response by generating nucleosome-free regions that allow binding of the androgen receptor, which may then promote EPIs (Valdes-Mora et al., 2017).
Acetylation in TADs and Beyond
Chromatin loops and EPIs occur within limited chromatin domains separated by insulators or boundary elements to prevent inappropriate activation of genes (Brasset and Vaury, 2005). Binding of architectural proteins such as cohesin and CTCF (CCCTC-binding factor) in a convergent orientation to these elements creates higher-order TADs, likely via dynamic extrusion processes (Gu et al., 2020). Hyperacetylated chromatin is also critical for the demarcation of TADs, as TAD boundaries contain highly acetylated nucleosomes (Ulianov et al., 2016). Each TAD contains smaller, discrete chromatin nanodomains (CNDs) of various sizes. Condensed inactive chromatin regions fold into fewer CNDs than active regions, suggesting that CND formation may rely on interactions between nucleosomes and thus may be regulated by histone acetylation (Shogren-Knaak et al., 2006). Indeed, histone hyperacetylation induced by TSA treatment disrupts CND organization in mESCs (Szabo et al., 2020). CND architecture is additionally governed by master regulators such as MYC (Kieffer-Kwon et al., 2017). During B cell activation, histone acetylation is amplified to induce rapid release of nucleosomes from the nuclear matrix into the nucleoplasm, while MYC functions to decompact CND clusters into mononucleosome fibers, doubling the number of chromatin loops and contact domains, ultimately resulting in reduced residence times and fewer non-specific collisions of TFs.
TADs further aggregate into chromatin compartments and finally chromosome territories (Szabo et al., 2019). Histone modifications contribute to the segregation of chromatin into compartments, termed A and B, which are broadly distinguished by their enrichment in active and repressive histone marks, respectively (Lieberman-Aiden et al., 2009). Acetylation reader proteins therefore also play roles in chromatin compartmentalization. For example, the oncoprotein BRD4-NUT (nuclear protein in testis) that drives aggressive NUT carcinoma, was recently found to initiate long-range interactions between hyperacetylated “megadomains” that are separated by tens to hundreds of megabases (Mb) in cancer cells (Rosencrance et al., 2020). These interactions occur within and between chromosomes and form a specific nuclear subcompartment with elevated transcription activity compared to other subcompartments, but do not disturb local loop and TAD structures. Modulating chromosome structure via pharmacological degradation of master regulators such as BRD4-NUT thus may present an option for treating human disease.
Formation of chromatin subcompartments may also be regulated by liquid-liquid phase separation (LLPS), which could enhance gene expression by increasing local concentrations of transcription components, or conversely, inhibit transcription by excluding these factors (Erdel and Rippe, 2018). In vitro reconstituted chromatin undergoes LLPS under physiologic salt conditions, and these states are preserved when microinjected into nuclei (Gibson et al., 2019). Mutations of the histone H4 basic amino acid patch, alterations in linker DNA lengths, and the presence of linker histone H1 all impact chromatin LLPS. Histone acetylation by p300 inhibits droplet formation, but subsequent binding of an artificial multi-BRD (bromodomain) protein restores LLPS of acetylated chromatin in vitro. These findings suggest that acetylation and BRD proteins may regulate LLPS in vivo, but the presence of additional histone modifications may further modulate such functions, and single BRD proteins are not sufficient to induce LLPS. Indeed, repressive marks H3K9me2/3 and their regulators including reader HP1 (heterochromatin protein 1), writer SUV39H1 (suppressor of variegation 3–9 homolog 1) and scaffolding protein TRIM28 (tripartite motif containing 28), also drive LLPS, forming highly dense droplets reminiscent of heterochromatin that exclude TFIIB (Wang et al., 2019).
The nuclear lamina plays a critical role in chromatin organization, providing an anchoring framework for chromatin domains. MOF, the major metazoan KAT targeting H4K16ac, additionally acetylates lamin A/C (Karoutas et al., 2019). This function of MOF is mediated through the NSL complex, and loss of NSL components results in nuclear envelope rupture and severe genome instability including chromothripsis. An altered epigenetic landscape characterized by reduced H3K27ac and enriched facultative heterochromatin mark H3K27me3 was concomitantly observed. These combined changes constituted a state termed “heterochromatin enrichment in nuclear abnormalities” (HENA).
Fully condensed, metaphase chromosomes represent the most compact organization of chromatin. Classically defined, visible banding patterns in metaphase chromosomes reflect gene expression potential at other cell cycle states. To define the roles of specific histone modifications in these states, Halsall et al. used immunofluorescence and ChIP-seq to monitor H3K9ac, H3K4me3 and H3K27me3 levels and distributions across the cell cycle in HeLa and lymphoblastoid cells (Halsall et al., 2021). Remarkably, they found that 10–50 Mb regions that are visible as bands in mitosis persist in G1 and G2, and are made up of 1–5 Mb sub-bands. The sub-bands differed between the two cell types which may reflect cell type-specific transcription programs, but remained constant throughout the cell cycle. Modifications associated with gene activation, H3K9ac and H3K4me3, co-localized in the same sub-bands. Altogether, these findings indicate that histone modification patterns provide a cell type-specific “bar code” that may enable rapid re-establishment of gene expression programs upon exit from mitosis.
Acetylation in DNA Repair and Genome Stability
All DNA-templated processes are tightly linked to chromatin organization in eukaryotes. In response to DNA damage (DDR) and during DNA repair, chromatin structures undergo extensive reorganization and remodeling, both locally at the site of damage and genome-wide (Hauer and Gasser, 2017). Production of gamma-H2A.X in DDR is a classic example of how chromatin modulations contribute to DNA repair pathways (Mah et al., 2010). Other modifications, including acetylation, are also triggered by DNA damage and important for repair mechanisms (Ramanathan and Smerdon, 1986). For example, acetylation of histone H4 is critical for chromatin decondensation, enabling DNA repair factors to access damaged DNA (Bird et al., 2002; Shogren-Knaak et al., 2006). DNA break-induced H4 acetylation is catalyzed by TIP60 and recognized by several readers including the BRD of PCAF (Kim et al., 2019; Tang et al., 2013). Recruitment of PCAF as part of the SAGA complex triggers deubiquitination at H2BK120 by the deubiquitinase (DUB) module of SAGA, allowing PCAF to then acetylate H2BK120 (Clouaire et al., 2018; Kim et al., 2019). These events facilitate subsequent repair of DNA double-strand breaks (DSBs) by homologous recombination (HR).
In contrast to the positive role of H4 acetylation in promoting DNA repair, H1K85ac is reduced by HDAC1 in DDR, and subsequently restored by PCAF after completion of DNA repair (Li et al., 2018). The dynamics of H1K85ac impact chromatin folding as well, but in reversed effects as those of H4 acetylation. An H1K85ac-mimic mutation (H1K85Q) results in increased nucleosome binding of H1 and enhanced association between H1 and core histones. Moreover, H1K85ac recruits HP1 to promote chromatin compaction, therefore deacetylation of H1K85ac is required for chromatin relaxation and DNA repair. These findings demonstrate that acetylation events on different histones can lead to varying effects on chromatin organization.
Global changes in histone acetylation levels that are associated with DDR have been observed. Unbiased quantitative mass spectrometry analyses indicate that acetylation of all core histones is dramatically reduced in response to UV-induced DNA damage in mammalian cells (Mandemaker et al., 2018). The degradation of acetylated histones is mediated by proteasome activator 200 (PA200) and is independent of ubiquitylation. A similar loss of acetylated histones was detected upon induced replication stress and in non-replicating cells but not upon transcription inhibition, indicating that depletion of acetylated histones is the direct consequence of replication stress which could be triggered by DNA damage. The means by which acetylated nucleosomes are targeted for destruction, or the purpose of their degradation, is not yet clear.
Acetylation dynamics of non-histone proteins contribute to various pathways of DNA repair (reviewed in (Li et al., 2020). The mammalian E2F1 TF is acetylated by multiple KATs, such as p300/CBP, in DDR (Galbiati et al., 2005). This modification allows binding of the BRDs of p300/CBP to E2F1, which in turn recruits p300/CBP to DSBs to induce histone acetylation at damage sites (Manickavinayaham et al., 2019). E2F1 also recruits GCN5 to sites of UV damage to deposit H3K9ac, which likely increases chromatin access for the nucleotide excision repair (NER) machinery (Guo et al., 2011). Another example is ATM (ataxia telangiectasia mutated), an apical kinase that drives DDR signaling. ATM is activated upon DNA damage via various mechanisms including acetylation by TIP60 (Sun et al., 2005). SIRT7 was recently identified as the HDAC responsible for deacetylating and deactivating ATM (Tang et al., 2019). Deactivation of ATM is crucial at the late stages of DDR, and SIRT7-deficient cells show compromised DSB repair. ATM also triggers acetylation of other proteins, such as RAD52 (radiation sensitive 52). p300/CBP-mediated acetylation sustains co-localization of RAD52 and its interacting partner RAD51 at DSBs (Yasuda et al., 2018). RAD52 acetylation is reversed by HDACs SIRT2 and SIRT3, causing dissociation of both RAD51 and RAD52 from DSB sites and disrupting HR pathway of DNA repair.
DNA damage is not only induced by exogenous agents, it also occurs in the process of normal cellular events that create vulnerable structures such as R-loops. These three-stranded structures are formed during transcription and consist of DNA–RNA hybrids plus the displaced single-strand DNA (ssDNA) (Niehrs and Luke, 2020). Failed resolution of R-loops hinders transcription as well as genome stability. Histone deacetylation is important to prevent and to resolve accumulation of deleterious R-loops by creating compact chromatin conformations that are less prone to DNA–RNA hybridization (Figure 3). The human RNA-binding THO/TREX (transcription-export) complex interacts with the SIN3A (suppressor-interacting 3A) HDAC complex to remove histone acetylation, suppressing formation of R-loops and protecting against DNA damage and replication fork stalling (Salas-Armenteros et al., 2017). The histone acetylation readers BRD2 and BRD4 also participate in blocking R-loop accumulation (Kim et al., 2019; Lam et al., 2020). BRD2 interacts with and enhances activity of TOP1 (topoisomerase I) to restrain R-loops, and BRD4 indirectly promotes TOP1 activity via phosphorylation of CTD (carboxy-terminal domain) of the largest subunit of Pol II (Baranello et al., 2016; Kim et al., 2019).
Figure 3. Regulation of acetylation in prevention of R-loops and DNA damage.
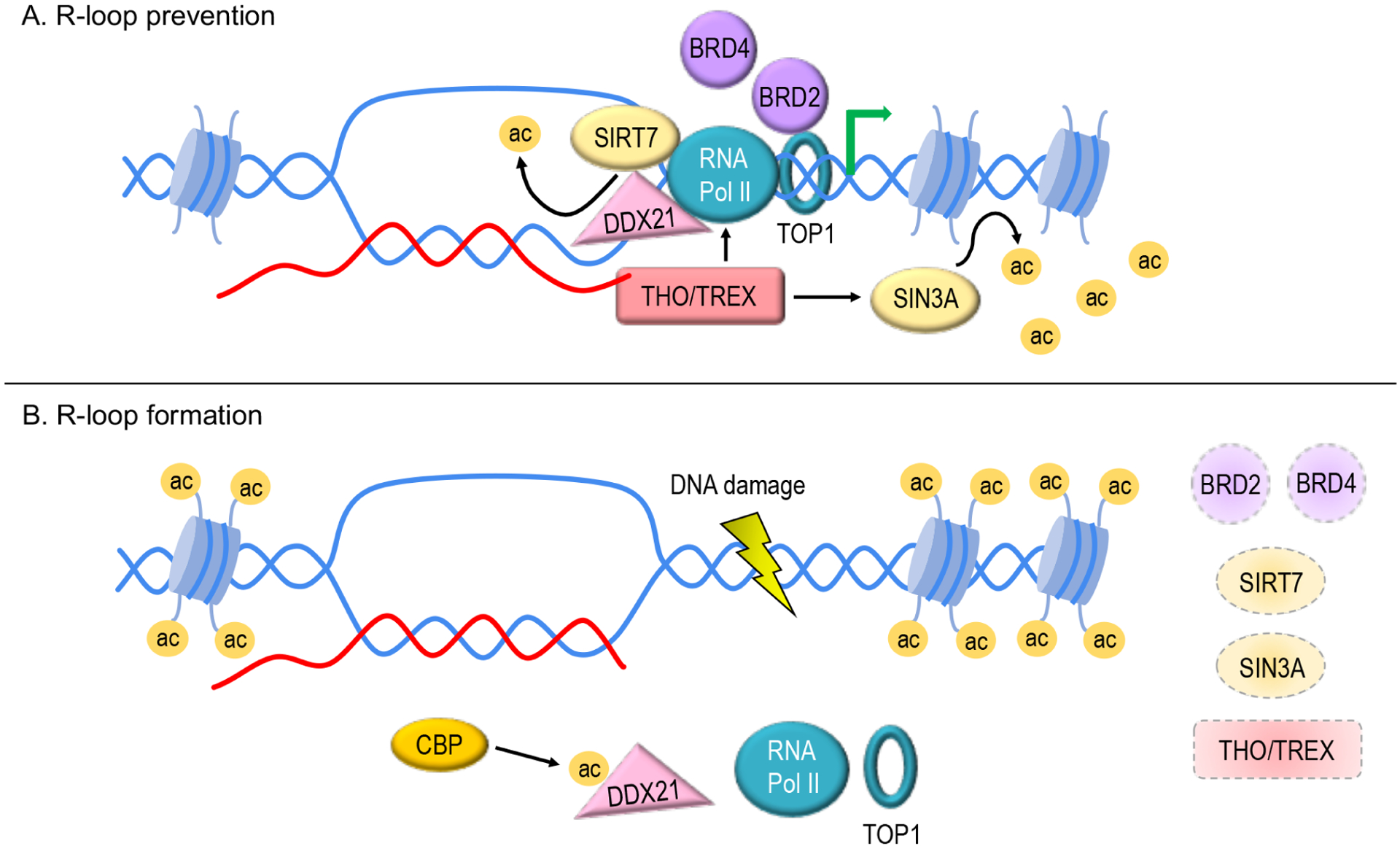
A. Under normal conditions, the RNA-binding THO/TREX complex promotes Pol II activity and SIN3A-mediated histone deacetylation, which inhibits R-loop formation. BRD2 directly interacts with and enhances TOP1 activity to restrain R-loops. BRD4 also participates in R-loop prevention, but the direct mechanism of action is yet to be determined. SIRT7 deacetylates and activates the helicase activity of DDX21, and DDX21 unwinds the DNA–RNA hybrid strands to resolve R-loops.
B. Upon inhibition or degradation of BRD2, BRD4, SIRT7, SIN3A and/or THO/TREX complex, R-loop accumulates and ultimately leads to DNA damage. Acetylation of DDX21 by CBP suppresses the activity of DDX21. Histone acetylation enhances transcription and thus increases the potential of R-loop formation.
An example of acetylation-mediated regulation of a non-histone protein involved in the prevention of R-loops is the DEAD (Asp–Glu–Ala–Asp)-box RNA helicase DDX21 (Figure 3). DDX21 efficiently unwinds the hybrid nucleic acid strands, and its activity is adversely modulated by acetylation in human cells (Song et al., 2017). CBP-conferred acetylation of DDX21 inhibits its function whereas deacetylation by SIRT7 augments its helicase activity. DDX21 functions are crucial in estrogen receptor positive breast cancer cells, in which estrogen treatment rapidly escalates global transcription and thus potential R-loop formation. DDX21 is required to resolve R-loops on estrogen-responsive genes to prevent stalling of elongating Pol II and to limit vulnerability to DNA damage.
Acetylation in DNA Replication
During DNA replication, a crucial interplay occurs between the replication machinery and chromatin dynamics that involves histone eviction and recycling at the fork, specific post-transcriptional modifications, and exchange of canonical histones with histone variants via histone chaperones (Cortez, 2019). When replication forks encounter obstacles that induce replication stress by blocking replicative DNA polymerase progression, chromatin components including acetylation-regulating complexes contribute to checkpoint machineries that rescue potential replication fork collapse. BRD protein function in replication stress was recently reviewed in (Lee et al., 2021).
The KAT HBO1 (histone acetyltransferase binding to origin recognition complex 1) coordinates multiple acetylation events at replication forks. HBO1 activity in the JADE1 (jade family PHD finger 1) complex is required for MCM2–7 (minichromosome maintenance protein complex 2–7) replication factor loading during replication licensing, and HBO1 acetylates histone H4 at K5, K8, and K12 at mammalian replication origins (Iizuka et al., 2006; Miotto and Struhl, 2010). A recent study identified a new role for HBO1 activity in complex with BRPF3 (BRD and PHD finger containing 3) that targets H3K14 but not H4K5/K8/K12 for acetylation, and that is required for the activation of pre-replication complexes during S phase, the step subsequent to licensing (Feng et al., 2016). Loss-of-function mutations of HBO1 and BRPF3 show a clear role for these proteins in replication fork regulation, however, the mechanism of how particular histone acetylation states impact replication origin specification and efficiency remains unclear.
Histone H4 acetylation is also required for replication fork recovery after DNA damage. A recent study performed in fission yeast shows a critical role for the NuA4 complex specifically in the repair of broken DNA at replication forks. Recruitment of NuA4 to replication forks is mediated by the Swi1-Swi3 replication fork protection complex and replication fork recovery requires acetylation of H4 at K5, K8 and K12 (Noguchi et al., 2019). Despite the fact that the NuA4 complex also targets H4K16ac, levels of H4K16ac during DNA replication are mostly regulated by the SAS-I (something about silencing I) complex in yeast, which acetylates H4K16 immediately upon H4 deposition likely via its interaction with CAF-I (chromatin assembly factor 1) and histone chaperone Asf1 (anti-silencing factor 1) (Boltengagen et al., 2021).
In mammalian cells, H4K8ac catalyzed by PCAF promotes degradation of stalled replication forks (Kim et al., 2020). This activity is important in BRCA1/2-deficient cells, where H4K8ac directly recruits MRE11 (meiotic recombination 11) and EXO1 (exonuclease 1) nucleases to stalled forks, inducing nascent DNA degradation. Reduced PCAF levels or activity stabilize stalled replication forks, decreasing the efficacy of PARP (poly (ADP-ribose) polymerase) inhibitors as therapies for BRCA1/2-deficient cancer cells. These results highlight the importance of understanding the functions of specific histone acetylation events in DNA replication, as these functions can influence response to replication/repair-based therapies.
Key components of the DNA replication machinery, such as PCNA, are also regulated by acetylation. PCNA is acetylated at several lysines in the inner surface of the trimer ring-like structure that is important for its sliding-clamp function (Billon et al., 2017). Acetylation of a specific lysine, K20, in PCNA by the cohesin KAT Eco1 (establishment of cohesion 1) is triggered by DNA damage to stimulate HR and suppresses translesion synthesis, thereby promoting DNA damage resistance in yeast. Structural studies indicate that acetylation alters the interfaces between PCNA molecules in the trimer. Additionally, BRD4 inhibits PCNA unloading on nascent DNA in part through binding acetylated histones in mammalian cells (Kang et al., 2019). These studies again highlight the diverse regulatory functions of lysine acetylation in DNA replication and repair pathways.
Concluding Remarks and Prospects
Past and recent advances have unraveled the fundamental impact of histone acetylation across the spectrum of chromatin structures. However, the roles of individual acetylation sites in histones and non-histone proteins are still not completely defined, although the many studies described here indicate evident progress in addressing the missing pieces. In part this lack of clarity may reflect functional redundancy among acetylation events. Unlike different lysine methylation marks in histones, such as H3K4me3 and H3K27me3, which are associated with very distinct active or repressed transcription states respectively, most histone acetylation modifications are associated with gene activation. Whether or how H3K9ac or K14ac, for example, uniquely impacts transcription initiation remains unclear, especially in mammalian cells. Genetic approaches to address this question have so far been impossible in multicellular organisms due to the multiple copies of each of the histone genes, but perhaps current state-of-the-art gene editing strategies will provide new opportunities to determine unique or shared phenotypes caused by mutations of specific acetylation sites in individual histones. Combinations of mutations might then allow identification of functional synergies and crosstalk between modifications, where the presence or absence of a modification at one site influences occurrence of modifications at other sites (Latham and Dent, 2007). Another outstanding question is whether additional reader domains for specific acetylation events exist. Currently only a few domains are known to read acetyl-lysine, including BRD, YEATS and some PHD (plant homeodomain) fingers (Gong et al., 2016), and the interactions of the most commonly studied BRDs with acetylated histones are somewhat promiscuous (Filippakopoulos et al., 2012). The relative lack of specificity may again reflect partial redundancy in functions of acetylation sites. Given the past and emerging connections between KATs and HDACs in human cancers (Goutas et al., 2021; Waddell et al., 2021), neurodegenerative diseases (Cappelletti et al., 2021), and memory and learning (Burns and Graff, 2021), understanding the molecular impacts of specific acetylation events is imperative. HDAC inhibitors were the second epigenetic drugs to be approved for cancer treatment (Ganesan et al., 2019). The development of additional KAT, HDAC and BRD inhibitors and degraders (Brand et al., 2021; Ramaiah et al., 2021; Xiang et al., 2021) as possible cancer therapies likely foreshadows the development of new agents to tailor acetylation states to impact other pathological conditions.
Acknowledgements
This work was supported by NIH R35GM131678-01 and R01HD094400 grants to SYRD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
SYRD is a member of the Advisory Board for Molecular Cell.
References
- Baranello L, Wojtowicz D, Cui K, Devaiah Ballachanda N., Chung H-J, Chan-Salis Ka Y., Guha R, Wilson K, Zhang X, Zhang H, et al. (2016). RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell 165, 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Li J, Lambert JP, Chen Y, Tremblay V, Brunzelle JS, Gingras AC, Verreault A, Sugiyama T, Couture JF, et al. (2017). Acetylation of PCNA Sliding Surface by Eco1 Promotes Genome Stability through Homologous Recombination. Mol Cell 65, 78–90. [DOI] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, and Christman MF (2002). Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415. [DOI] [PubMed] [Google Scholar]
- Boltengagen M, Samel-Pommerencke A, Fechtig D, and Ehrenhofer-Murray AE (2021). Dynamics of SAS-I mediated H4 K16 acetylation during DNA replication in yeast. PLoS One 16, e0251660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Clayton J, Moroglu M, Schiedel M, Picaud S, Bluck JP, Skwarska A, Bolland H, Chan AKN, Laurin CMC, et al. (2021). Controlling Intramolecular Interactions in the Design of Selective, High-Affinity Ligands for the CREBBP Bromodomain. J Med Chem 64, 10102–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasset E, and Vaury C (2005). Insulators are fundamental components of the eukaryotic genomes. Heredity (Edinb) 94, 571–576. [DOI] [PubMed] [Google Scholar]
- Burns AM, and Graff J (2021). Cognitive epigenetic priming: leveraging histone acetylation for memory amelioration. Curr Opin Neurobiol 67, 75–84. [DOI] [PubMed] [Google Scholar]
- Calo E, and Wysocka J (2013). Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti G, Calogero AM, and Rolando C (2021). Microtubule acetylation: A reading key to neural physiology and degeneration. Neurosci Lett 755, 135900. [DOI] [PubMed] [Google Scholar]
- Caslini C, Hong S, Ban YJ, Chen XS, and Ince TA (2019). HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells. Oncogene 38, 6599–6614. [DOI] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich-Codore H, Vejnar CE, Beaudoin JD, Musaev D, Fernandez JP, Benitez MDJ, Bazzini AA, et al. (2019). Brd4 and P300 Confer Transcriptional Competency during Zygotic Genome Activation. Dev Cell 49, 867–881 e868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, and Takada S (2016). Histone acetylation dependent energy landscapes in trinucleosome revealed by residue-resolved molecular simulations. Sci Rep 6, 34441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Lin YT, Gallegos DA, Hazlett MF, Gomez-Schiavon M, Yang MG, Kalmeta B, Zhou AS, Holtzman L, Gersbach CA, et al. (2019). Enhancer Histone Acetylation Modulates Transcriptional Bursting Dynamics of Neuronal Activity-Inducible Genes. Cell Rep 26, 1174–1188 e1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, and Dent SYR (2021). Conservation and diversity of the eukaryotic SAGA coactivator complex across kingdoms. Epigenetics Chromatin 14, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, and Howe LJ (2009). Histone acetylation: truth of consequences? Biochem Cell Biol 87, 139–150. [DOI] [PubMed] [Google Scholar]
- Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, et al. (2018). Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol Cell 72, 250–262.e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D (2019). Replication-Coupled DNA Repair. Mol Cell 74, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, and Singer DS (2016). BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 23, 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, and Becker PB (2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, and Rippe K (2018). Formation of Chromatin Subcompartments by Phase Separation. Biophys J 114, 2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Zhong L, Li C, Henriques T, Ablondi E, Nakadai T, Van Rechem C, Ferrer C, Ross KN, Choi JE, et al. (2019). The Histone Deacetylase SIRT6 Restrains Transcription Elongation via Promoter-Proximal Pausing. Mol Cell 75, 683–699 e687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Vlassis A, Roques C, Lalonde ME, Gonzalez-Aguilera C, Lambert JP, Lee SB, Zhao X, Alabert C, Johansen JV, et al. (2016). BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J 35, 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. (2012). Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati L, Mendoza-Maldonado R, Gutierrez MI, and Giacca M (2005). Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle 4, 930939. [DOI] [PubMed] [Google Scholar]
- Ganesan A, Arimondo PB, Rots MG, Jeronimo C, and Berdasco M (2019). The timeline of epigenetic drug discovery: from reality to dreams. Clinical Epigenetics 11, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub A, Sheikh BN, Basilicata MF, Vincent M, Nizon M, Colson C, Bird MJ, Bradner JE, Thevenon J, Boutros M, et al. (2020). Evolutionary conserved NSL complex/BRD4 axis controls transcription activation via histone acetylation. Nat Commun 11, 2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, and Kuroda MI (2009). Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, and Rosen MK (2019). Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484 e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Chiu LY, and Miller KM (2016). Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLoS Genet 12, e1006272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutas D, Theocharis S, and Tsourouflis G (2021). Unraveling the Epigenetic Role and Clinical Impact of Histone Deacetylases in Neoplasia. Diagnostics (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Wu L, Woldemichael GM, Pomella S, Quinn TR, Park PMC, Cleveland A, Stanton BZ, Song Y, Rota R, et al. (2019). Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun 10, 3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, and Wysocka J (2020). Opposing Effects of Cohesin and Transcription on CTCF Organization Revealed by Super-resolution Imaging. Mol Cell 80, 699–711 e697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, and Johnson DG (2011). GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 39, 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsall JA, Andrews S, Krueger F, Rutledge CE, Ficz G, Reik W, and Turner BM (2021). Histone modifications form a cell-type-specific chromosomal bar code that persists through the cell cycle. Sci Rep 11, 3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC (2002). Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct 31, 361–392. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, and Workman JL (2001). Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Hauer MH, and Gasser SM (2017). Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev 31, 2204–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, and Tora L (2017). Sharing the SAGA. Trends Biochem Sci 42, 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg SJ, Motorna O, Cluse LA, Johanson TM, Coughlan HD, Raviram R, Myers RM, Costacurta M, Todorovski I, Pijpers L, et al. (2021). Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Mol Cell 81, 2183–2200 e2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe FS, Fischl H, Murray SC, and Mellor J (2017). Is H3K4me3 instructive for transcription activation? Bioessays 39, 1–12. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, and Smith MM (2006). Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, and Caldas C (2004). p300/CBP and cancer. Oncogene 23, 4225–4231. [DOI] [PubMed] [Google Scholar]
- Jiang C, and Pugh BF (2009). Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Li C, Lu M, Lu K, and Li H (2021). Protein lysine crotonylation: past, present, perspective. Cell Death & Disease 12, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, and Felsenfeld G (2009). H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, and Ge K (2011). Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CA, O’Neill LP, Mitchell A, and Turner BM (1998). Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res 26, 9941001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josling GA, Selvarajah SA, Petter M, and Duffy MF (2012). The Role of Bromodomain Proteins in Regulating Gene Expression. Genes 3, 320–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, and Blobel GA (2009). Chromatin loops in gene regulation. Biochim Biophys Acta 1789, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Kim J, Ryu E, Ha NY, Hwang S, Kim BG, Ra JS, Kim YJ, Hwang JM, Myung K, et al. (2019). PCNA Unloading Is Negatively Regulated by BET Proteins. Cell Rep 29, 4632–4645.e4635. [DOI] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, and Tora L (2012). H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoutas A, Szymanski W, Rausch T, Guhathakurta S, Rog-Zielinska EA, Peyronnet R, Seyfferth J, Chen HR, de Leeuw R, Herquel B, et al. (2019). The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat Cell Biol 21, 1248–1260. [DOI] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. (2017). Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation. Mol Cell 67, 566–578 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee SY, Choi JH, Woo HG, Xhemalce B, and Miller KM (2020). PCAF-Mediated Histone Acetylation Promotes Replication Fork Degradation by MRE11 and EXO1 in BRCA-Deficient Cells. Mol Cell 80, 327–344 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang CM, Xhemalce B, Paull TT, et al. (2019). Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev 33, 1751–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, and Allis CD (1996). Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272. [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, and Workman JL (2003). Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol 23, 3305–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FC, Kong YW, Huang Q, Vu Han TL, Maffa AD, Kasper EM, and Yaffe MB (2020). BRD4 prevents the accumulation of R-loops and protects against transcriptionreplication collision events and DNA damage. Nat Commun 11, 4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, and Dent SY (2007). Cross-regulation of histone modifications. Nat Struct Mol Biol 14, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Lee SY, Kim JJ, and Miller KM (2021). Bromodomain proteins: protectors against endogenous DNA damage and facilitators of genome integrity. Exp Mol Med 53, 1268–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shi B, Liu X, and An H-X (2020). Acetylation and Deacetylation of DNA Repair Proteins in Cancers. Frontiers in Oncology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Z, Dong L, Tang M, Zhang P, Zhang C, Cao Z, Zhu Q, Chen Y, Wang H, et al. (2018). Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res 46, 7716–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louphrasitthiphol P, Siddaway R, Loffreda A, Pogenberg V, Friedrichsen H, Schepsky A, Zeng Z, Lu M, Strub T, Freter R, et al. (2020). Tuning Transcription Factor Availability through Acetylation-Mediated Genomic Redistribution. Mol Cell 79, 472–487.e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, and Tremethick DJ (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, and Lin Y (2021). Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol Cell 81, 1682–1697.e1687. [DOI] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, and Karagiannis TC (2010). gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24, 679–686. [DOI] [PubMed] [Google Scholar]
- Mandemaker IK, Geijer ME, Kik I, Bezstarosti K, Rijkers E, Raams A, Janssens RC, Lans H, Hoeijmakers JH, Demmers JA, et al. (2018). DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickavinayaham S, Velez-Cruz R, Biswas AK, Bedford E, Klein BJ, Kutateladze TG, Liu B, Bedford MT, and Johnson DG (2019). E2F1 acetylation directs p300/CBP-mediated histone acetylation at DNA double-strand breaks to facilitate repair. Nat Commun 10, 4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, and Zhou MM (2014). Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6, a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BJE, Brind’Amour J, Kuzmin A, Jensen KN, Liu ZC, Lorincz M, and Howe LJ (2021). Transcription shapes genome-wide histone acetylation patterns. Nat Commun 12, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, and Smith MM (1990). Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247, 841–845. [DOI] [PubMed] [Google Scholar]
- Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, et al. (2017). YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun 8, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Zhang Y, Lyu J, Wang X, Tong Q, Peng D, Xue Y, Tencer AH, Wen H, Li W, et al. (2018). The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat Commun 9, 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, and Struhl K (2010). HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munks RJ, Moore J, O’Neill LP, and Turner BM (1991). Histone H4 acetylation in Drosophila. Frequency of acetylation at different sites defined by immunolabelling with site-specific antibodies. FEBS Lett 284, 245–248. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, and Tora L (2010). The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci 67, 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Ito S, Higashijima Y, Chu WK, Neumann K, Walter J, Satpathy S, Liebner T, Hamilton WB, Maskey E, et al. (2021). Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol Cell 81, 2166–2182 e2166. [DOI] [PubMed] [Google Scholar]
- Narita T, Weinert BT, and Choudhary C (2019). Functions and mechanisms of non-histone protein acetylation. Nature Reviews Molecular Cell Biology 20, 156–174. [DOI] [PubMed] [Google Scholar]
- Nicolas D, Zoller B, Suter DM, and Naef F (2018). Modulation of transcriptional burst frequency by histone acetylation. Proc Natl Acad Sci U S A 115, 7153–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, and Luke B (2020). Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol 21, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, Singh T, Ziegler MA, Peake JD, Khair L, Aza A, Nakamura TM, and Noguchi E (2019). The NuA4 acetyltransferase and histone H4 acetylation promote replication recovery after topoisomerase I-poisoning. Epigenetics Chromatin 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterstrom J, Castells-Garcia A, Vicario C, Gomez-Garcia PA, Cosma MP, and Lakadamyali M (2019). Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo. Nucleic Acids Res 47, 8470–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, and Ben-Shem A (2020). Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577, 711–716. [DOI] [PubMed] [Google Scholar]
- Radzisheuskaya A, Shliaha PV, Grinev VV, Shlyueva D, Damhofer H, Koche R, Gorshkov V, Kovalchuk S, Zhan Y, Rodriguez KL, et al. (2021). Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. Mol Cell 81, 1749–1765 e1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, et al. (2018). Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep 24, 1722–1729. [DOI] [PubMed] [Google Scholar]
- Ramaiah MJ, Tangutur AD, and Manyam RR (2021). Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci 277, 119504. [DOI] [PubMed] [Google Scholar]
- Ramanathan B, and Smerdon MJ (1986). Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis 7, 1087–1094. [DOI] [PubMed] [Google Scholar]
- Rando OJ (2007). Chromatin structure in the genomics era. Trends Genet 23, 67–73. [DOI] [PubMed] [Google Scholar]
- Regadas I, Dahlberg O, Vaid R, Ho O, Belikov S, Dixit G, Deindl S, Wen J, and Mannervik M (2021). A unique histone 3 lysine 14 chromatin signature underlies tissue-specific gene regulation. Mol Cell 81, 1766–1780 e1710. [DOI] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, Garcia-Parajo MF, Lakadamyali M, and Cosma MP (2015). Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158. [DOI] [PubMed] [Google Scholar]
- Riss A, Scheer E, Joint M, Trowitzsch S, Berger I, and Tora L (2015). Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J Biol Chem 290, 28997–29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosencrance CD, Ammouri HN, Yu Q, Ge T, Rendleman EJ, Marshall SA, and Eagen KP (2020). Chromatin Hyperacetylation Impacts Chromosome Folding by Forming a Nuclear Subcompartment. Mol Cell 78, 112–126 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Zhang D, Allis CD, and Zhao Y (2017). Metabolic regulation of gene expression through histone acylations. Nature Reviews Molecular Cell Biology 18, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Armenteros I, Perez-Calero C, Bayona-Feliu A, Tumini E, Luna R, and Aguilera A (2017). Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J 36, 3532–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samata M, Alexiadis A, Richard G, Georgiev P, Nuebler J, Kulkarni T, Renschler G, Basilicata MF, Zenk FL, Shvedunova M, et al. (2020). Intergenerationally Maintained Histone H4 Lysine 16 Acetylation Is Instructive for Future Gene Activation. Cell 182, 127–144 e123. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, and Peterson CL (2006). Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- Song C, Hotz-Wagenblatt A, Voit R, and Grummt I (2017). SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev 31, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, and Price BD (2005). A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 102, 13182–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungalee S, Liu Y, Lambuta RA, Katanayeva N, Donaldson Collier M, Tavernari D, Roulland S, Ciriello G, and Oricchio E (2021). Histone acetylation dynamics modulates chromatin conformation and allele-specific interactions at oncogenic loci. Nat Genet 53, 650–662. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, and Naef F (2011). Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474. [DOI] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, and Cavalli G (2019). Principles of genome folding into topologically associating domains. Sci Adv 5, eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Donjon A, Jerkovic I, Papadopoulos GL, Cheutin T, Bonev B, Nora EP, Bruneau BG, Bantignies F, and Cavalli G (2020). Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat Genet 52, 1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, and Greenberg RA (2013). Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature Structural & Molecular Biology 20, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Li Z, Zhang C, Lu X, Tu B, Cao Z, Li Y, Chen Y, Jiang L, Wang H, et al. (2019). SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci Adv 5, eaav1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, and Schreiber SL (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Ulianov SV, Khrameeva EE, Gavrilov AA, Flyamer IM, Kos P, Mikhaleva EA, Penin AA, Logacheva MD, Imakaev MV, Chertovich A, et al. (2016). Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res 26, 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Mora F, Gould CM, Colino-Sanguino Y, Qu W, Song JZ, Taylor KM, Buske FA, Statham AL, Nair SS, Armstrong NJ, et al. (2017). Acetylated histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer. Nat Commun 8, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosnakis N, Koch M, Scheer E, Kessler P, Mely Y, Didier P, and Tora L (2017). Coactivators and general transcription factors have two distinct dynamic populations dependent on transcription. EMBO J 36, 2710–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell AR, Huang H, and Liao D (2021). CBP/p300: Critical Co-Activators for Nuclear Steroid Hormone Receptors and Emerging Therapeutic Targets in Prostate and Breast Cancers. Cancers (Basel) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dienemann C, Stutzer A, Urlaub H, Cheung ACM, and Cramer P (2020). Structure of the transcription coactivator SAGA. Nature 577, 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, et al. (2019). Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol Cell 76, 646–659 e646. [DOI] [PubMed] [Google Scholar]
- Weake VM, and Workman JL (2010). Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11, 426–437. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Narita T, Satpathy S, Srinivasan B, Hansen BK, Scholz C, Hamilton WB, Zucconi BE, Wang WW, Liu WR, et al. (2018). Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 174, 231–244 e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, and Ghosh RP (2010). Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2, a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Wang Q, Ran K, Ren J, Shi Y, and Yu L (2021). Structure-guided discovery of novel potent and efficacious proteolysis targeting chimera (PROTAC) degrader of BRD4. Bioorg Chem 115, 105238. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, and Roth SY (2000). Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 26, 229–232. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Kagawa W, Ogi T, Kato TA, Suzuki T, Dohmae N, Takizawa K, Nakazawa Y, Genet MD, Saotome M, et al. (2018). Novel function of HATs and HDACs in homologous recombination through acetylation of human RAD52 at double-strand break sites. PLoS Genet 14, e1007277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Erler J, and Langowski J (2017). Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys J 112, 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]


