Abstract
Little is known about the genetic strain diversity and geographical range of Histoplasma capsulatum isolated in Rio de Janeiro State, Brazil. We characterized 13 environmental, 7 animal, and 28 clinical H. capsulatum isolates by using a PCR-based random amplified polymorphic DNA (RAPD) assay. DNA fingerprinting of these soil, animal, and clinical specimens was performed with four primers (1253, 1281, D-9355, and D-10513) and generated amplicons with considerable polymorphism. Although all of the isolates exhibited more than 80% genetic relatedness, they could be clustered into four to six genotypes for each primer. The RAPD profiles of H. capsulatum isolated from Rio de Janeiro State could be distinguished from those of the U.S. strains included in this study (Downs, G222B, G-186B, and FLS1) by showing less than 70% similarity to each primer. The genetic polymorphisms between H. capsulatum strains isolated from animals and soil obtained in the same geographic areas were 100% similar, suggesting that an environmental microniche could be acting as a source of infection for animals and the local human population.
Histoplasmosis, a systemic fungal disease caused by Histoplasma capsulatum, has a worldwide distribution and is a serious community-acquired endemic mycosis among immunocompetent hosts in some countries. Histoplasmosis has also become a serious problem in immunocompromised hosts, such as AIDS patients who develop a disseminated and often rapidly progressive clinical course, requiring prompt diagnosis and specific treatment (18). Before the advent of combination antiretroviral therapy, histoplasmosis accounted for 5 to 20% of the infections in AIDS patients in areas where it is endemic. Although incidence and prevalence data are lacking in Brazil, histoplasmosis may infect up to 5% of the individuals with AIDS in areas of the country where it is endemic.
Genome typing has the power to discriminate between clinical and environmental strains of microbes. In histoplasmosis, genome typing has achieved three levels of discrimination as follows. Broad groupings can be defined using hybridization with the yps3 probe (7), with additional discrimination provided by applying a mitochondrial DNA probe to the same Southern blots (15). Random amplification of polymorphic DNA (RAPD) is capable of revealing individual strain subtypes (8). More recently, another typing method has been described that is based on nucleotide sequence variation in the internal transcribed spacer regions of the rRNA gene, where Indianapolis isolates were classified into four types (4). A better understanding of how to identify clinical and environmental strains of H. capsulatum will help to estimate the contribution of endogenous versus exogenous sources of infection and aid in tracking the transmission of infection during outbreaks.
The molecular epidemiology of histoplasmosis has not been studied in Brazil. Brazilian isolates of H. capsulatum from patients with AIDS-related infections, immunologically intact hosts with community-acquired histoplasmosis, soil, and animals have not been compared by DNA fingerprinting, and little is known about the type and geographical range of H. capsulatum strains in Rio de Janeiro State. In this research, the genetic polymorphism of H. capsulatum strains from environmental and clinical sources was analyzed by using a RAPD assay.
MATERIALS AND METHODS
Cultures.
Forty-eight H. capsulatum strains were obtained from the Centro de Pesquisa Hospital Evandro Chagas—Fundação Oswaldo Cruz, the Hospital Universitário Pedro Ernesto—Universidade do Estado do Rio de Janeiro, and the Laboratório Lâmina, Rio de Janeiro, Brazil. Their sources and geographical origins are listed in Table 1. Fungal identification was done by conventional mycological methods, including morphology and the exoantigen test (16). H. capsulatum strains Downs (class I), G-222B (class II), G-186B (class III), and FLS-1 (class IV) from the United States were used in all studies as reference strains.
TABLE 1.
Genotypes and characteristics of H. capsulatum strains isolated in Rio de Janeiro State, Brazil
| Strain | Source | Location | DNA fingerprinting pattern obtained by RAPD with primer:
|
|||
|---|---|---|---|---|---|---|
| 1253 | 1281 | D-9355 | D-10513 | |||
| IGS4/5 | Soil | Ilha Grande, RJd | Ia | Ia | Ia | Ia |
| IGS19 | Soil | Ilha Grande, RJ | Ia | Ia | Ia | Ia |
| RPS11 | Soil | Rio da Prata, RJ | Ia | Ia | Ia | Ia |
| RPS45 | Soil | Rio da Prata, RJ | Ia | Ia | Ia | Ia |
| RPS51 | Soil | Rio da Prata, RJ | Ia | Ia | Ia | Ia |
| RPS86 | Soil | Rio da Prata, RJ | Ia | Ia | Ia | Ia |
| EP2 | Soil | Itaipava, RJ | Ia | Ia | UKc | Ia |
| AC2 | Soil | Niteroi, RJ | Ia | Ia | Ia | Ia |
| AC5 | Soil | Niteroi, RJ | Ia | Ia | UK | Ia |
| TI1 | Soil | Tinguá, RJ | Ia | Ia | IIb | Ib |
| TI5 | Soil | Tinguá, RJ | Ib | Ia | IIb | Ib |
| TI14 | Soil | Tinguá, RJ | Ia | Ia | Ia | Ia |
| IT4 | Soil | Itaipava, RJ | Ib | II | Ib | II |
| RS1 | Rat | Ilha Grande, RJ | Ia | Ia | Ia | Ia |
| RS9 | Rat | Ilha Grande, RJ | Ia | Ia | Ia | Ia |
| RS36 | Rat | Ilha Grande, RJ | Ia | Ia | Ia | Ia |
| RP44 | Rat | Rio da Prata, RJ | Ia | Ia | Ib | Ia |
| RP93 | Opossum | Rio da Prata, RJ | Ia | Ia | Ia | Ia |
| CÃO4 | Dog | Itaipava, RJ | Ib | II | Ib | II |
| CADAM | Dog | Campo Grande, RJ | Ib | Ia | IIb | Ib |
| 2733 | HIV+a | Rio de Janeiro | IIb | IIIb | III | II |
| 2787 | HIV+ | Rio de Janeiro | Ia | Ia | Ia | Ia |
| 3612 | HIV+ | Rio de Janeiro | Ia | Ia | Ia | Ia |
| 3688 | HIV+ | Rio de Janeiro | Ia | Ia | Ia | Ia |
| 4334 | HIV+ | Rio de Janeiro | IIb | Ib | IIb | Ib |
| 4959 | HIV+ | Rio de Janeiro | Ib | IIIb | IIa | III |
| 5331 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 6404 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 6406 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 6503 | HIV+ | Rio de Janeiro | Ic | IV | Ib | II |
| 7098 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 7119 | HIV+ | Rio de Janeiro | Ib | IIIb | IIb | Ib |
| 8941 | HIV+ | Rio de Janeiro | Ic | IV | IIa | II |
| 9236 | HIV+ | Rio de Janeiro | Ia | IIIb | IIa | III |
| 9291 | HIV+ | Rio de Janeiro | IIa | IIIa | Ib | Ib |
| 9305 | HIV+ | Rio de Janeiro | IIa | IIIa | Ib | Ib |
| 9414 | HIV+ | Rio de Janeiro | Ic | IV | IIb | II |
| 84502 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 84544 | HIV+ | Rio de Janeiro | IIb | IIIb | III | Ib |
| 84564 | HIV+ | Rio de Janeiro | IIb | IIIb | III | II |
| 3237 | HIV−b | Rio de Janeiro | IIa | Ib | Ib | Ib |
| 3356 | HIV− | Rio de Janeiro | IIa | Ib | Ib | Ib |
| 3416 | HIV− | Rio de Janeiro | Ia | Ia | Ia | Ia |
| 3669 | HIV− | Rio de Janeiro | Ia | Ia | Ia | Ia |
| 4631 | HIV− | Rio de Janeiro | Ib | Ia | IIb | Ib |
| 75864 | HIV− | Rio de Janeiro | Ia | Ia | IIa | Ia |
| 78642 | HIV− | Rio de Janeiro | Ia | IIIb | IIa | III |
| B-670 | HIV− | Rio de Janeiro | IIb | Ib | Ib | II |
HIV+, histoplasmosis patient with AIDS.
HIV−, histoplasmosis patient who tested HIV negative.
UK, strain not amplified with primer D-9355.
RJ, Rio de Janeiro.
DNA isolation.
A single colony of yeast phase H. capsulatum was grown at 37°C in Pine's citrate broth (9) in a gyratory shaker at 120 rpm for 3 days. The yeast cells were harvested and washed three times in sterile distilled water by centrifugation at 2,000 × g. Genomic DNA was extracted from H. capsulatum yeast cells with a Puregene DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, Minn.). DNA quantification was done with a Gene Quant pro RNA/DNA Calculator (Amersham Pharmacia Biotech, Cambridge, United Kingdom).
PCR-RAPD.
For RAPD analysis, each strain was tested with four primers described previously by Kersulyte et al. (8). The RAPD profiles were defined by bands that were present in different amplification reactions. Distinct RAPD profiles were designated by roman numerals; minor variations (at least one polymorphic band) were indicated by letters. The DNA amplification reaction was carried out in a 25-μl volume containing 20 ng of H. capsulatum DNA, 10× PCR buffer containing 500 mM KCl, 100 mM Tris-HCl (pH 8.3), 3 mM MgCl2, 200 μM deoxynucleoside triphosphate, 20 pmol of primer, and 1 U of Taq DNA polymerase (Roche Molecular Biochemicals). The PCR protocol used depended on the primer used in the reaction. For amplification with primers 1253 (5′-GTTTCCGCCC-3′) and 1281 (5′-AACGCGCAAC-3′), the thermal cycler (Perkin Elmer 2400) was programmed for 1 denaturation cycle at 94°C for 5 min, followed by 45 cycles of denaturation at 94°C for 1 min, annealing at 36°C for 1 min, and extension at 72°C for 2 min and a final extension period of 10 min at 72°C. For D-9355 (5′-CCGGATCCGTGATGCGGTGCG-3′) and D-10513 (5′-AACGTTCATGATAACTTCTGCTCTTCATCG-3′), the PCR program was as follows: (i) 4 cycles, each consisting of 5 min at 94°C, 5 min at 40°C, and 5 min at 72°C; (ii) 30 cycles, each consisting of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and (iii) incubation at 72°C for 10 min to complete the extension. The randomly amplified DNA fragments were analyzed by electrophoresis on a 1% agarose gel in TBE buffer (0.89 M Tris, 0.89 M boric acid, 0.02 M disodium EDTA, pH 8.4) stained with ethidium bromide (Roche Molecular Biochemicals) at a final concentration of 0.5 μg/ml. All of the bands visualized on the gel were counted, and data were scored for the presence or absence of amplification products. The reproducibility of this method was confirmed when identical electrophoretic profiles were observed in PCR assays repeated at least three times under the same conditions.
Computer-assisted data analysis.
The similarity coefficient or Dice index was determined for each isolate in a RAPD analysis by using the Molecular Analyst Fingerprinting Plus software, version 1.12 (Bio-Rad Laboratories, Richmond, Calif.). For clustering, the unweighted pair-group method with arithmetic means was used.
Discriminatory power.
To assess the discriminatory power of the method applied in this study, a single numerical index of discrimination (D) using Simpson's index of diversity (3) was calculated, based on the probability that two unrelated strains sampled from the test population will be placed into different typing groups.
RESULTS
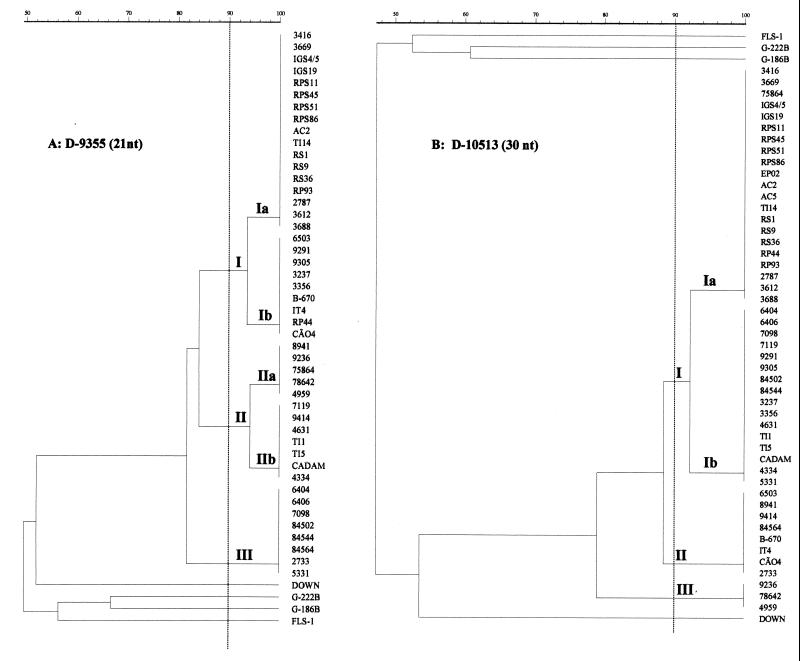
For representative gels showing the RAPD profiles of 48 H. capsulatum samples isolated from several geographic locations in Rio de Janeiro State (Table 1), see Fig. 1 and 3. Although all of the isolates exhibited more than 80% genetic relatedness, they could be clustered into four to six genotypes (see Fig. 1 and 3). Depending on the primer, 8 to 13 bands were separated, ranging in size from 0.3 to 4.4 kb. Common PCR fragments were seen among H. capsulatum isolates from Rio de Janeiro and from the United States when typed with all primers by this method: 1.5 kb (primer 1253), 0.8 kb (primer 1281), 0.9 kb (primer D-9355), and 0.5 kb (primer D-10513).
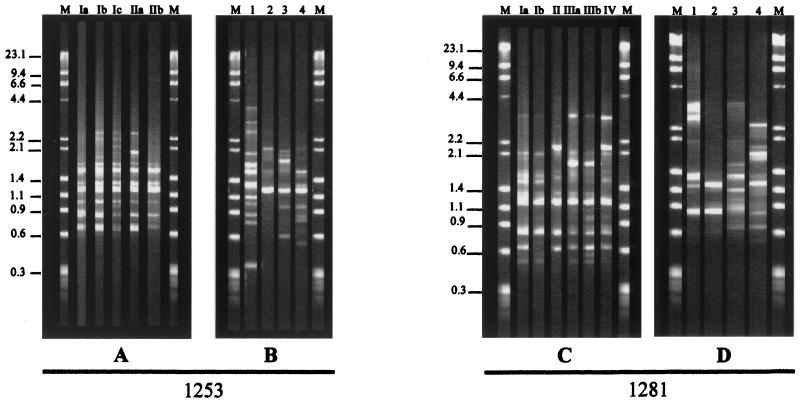
FIG. 1.
Representative RAPD profiles of H. capsulatum isolates from Rio de Janeiro State, Brazil (A and C), and U.S. strains in classes 1, 2, 3, and 4 (B and D) with primers 1253 and 1281, respectively. Lanes M, DNA molecular size marker (Roche Biochemicals). The values on the left are molecular sizes in kilobases.
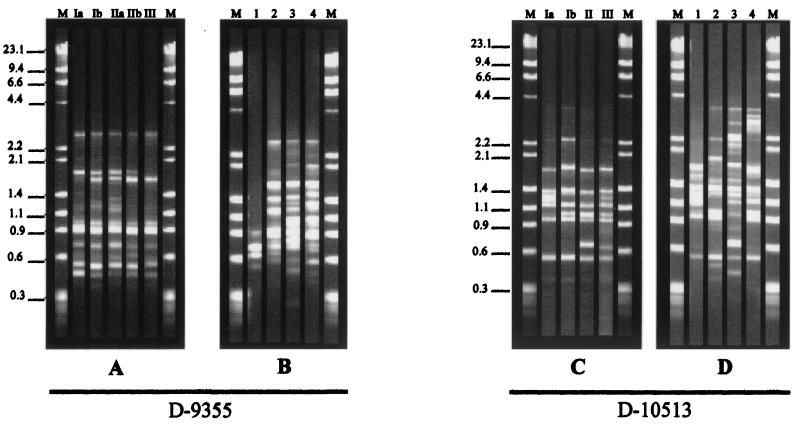
FIG. 3.
Representative RAPD profiles of H. capsulatum isolates from Rio de Janeiro State, Brazil (A and C), and U.S. strains in classes 1, 2, 3, and 4 (B and D) with primers D-9355 and D-10513, respectively. Lanes M: DNA molecular size marker (Roche Biochemicals). The values on the left are molecular sizes in kilobases.
The RAPD profiles of H. capsulatum isolates from Rio de Janeiro State were distinguished from those of the U.S. strains included in this study (Downs, G222B, G-186B, and FLS1) by showing less than 70% similarity to all of the primers.
Primer 1253.
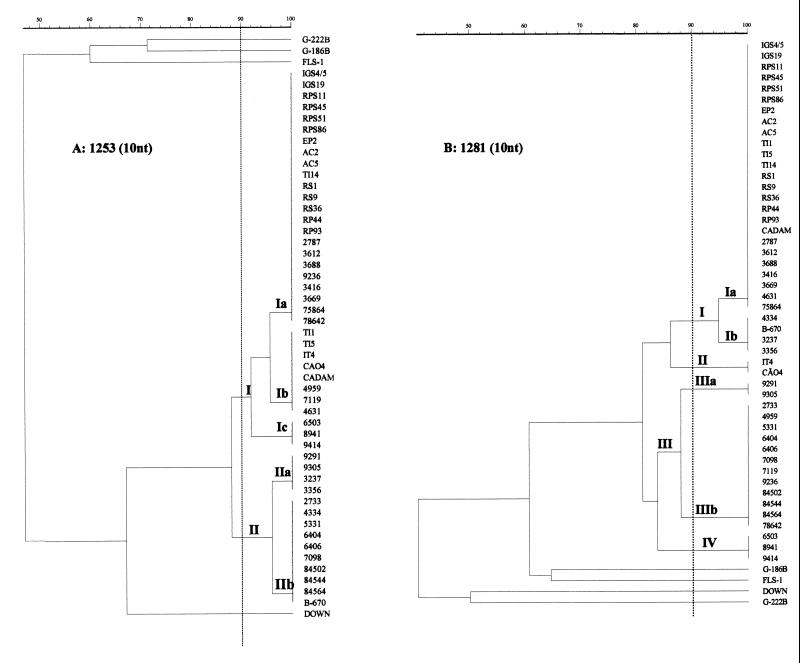
Amplifications performed with primer 1253 resulted in the identification of five distinct genotypes among the 48 H. capsulatum strains (Table 1; Fig. 1A and 2A), permitting the discrimination of two distinct clusters (groups I and II) exhibiting 87.2% relatedness and five subtypes (Ia, Ib, Ic, IIa, and IIb) with a high degree of similarity among them. Twenty-three isolates were included in subtype Ia, with 10 strains isolated from soil, 5 from animals, and 8 from patients with histoplasmosis. Subtype Ib accounted for three isolates from soil, two from animals, and three from patients with confirmed histoplasmosis. Three strains, all isolated from patients, made up subtype Ic. Subtypes IIa and IIb, with 95.9% similarity, also contained H. capsulatum strains isolated only from histoplasmosis patients.
FIG. 2.
Dendrogram derived from analysis of the RAPD profiles of genomic DNAs of H. capsulatum strains using Molecular Analyst Fingerprinting Plus, version 1.12 (Bio-Rad). Panels: A, primer 1253; B, primer 1281. The numbers on the right are strain designations.
Primer 1281.
With primer 1281, Rio de Janeiro isolates were classified into groups I (subtypes Ia and Ib), II, III (subtypes IIIa and IIIb), and IV, showing greater DNA polymorphism among them (81.2% similarity). Group I contained a majority of soil and animal isolates, with subtype Ia predominating (12 isolates from soil and 6 from animals). Three of the 20 AIDS patient isolates and four of the eight strains from human immunodeficiency virus (HIV)-negative patients were also included in this subtype, presenting 94.6% relatedness to subtype Ib, which is composed of 4 human isolates (Table 1; Fig. 1C and 2B). Group II contained a cluster of two other isolates from soil (IT4) and animals (CÃO4), respectively. Most of the AIDS-associated isolates were found in group III, although subtype IIIb also contained one isolate from an HIV-negative patient (78642). Group IV comprised H. capsulatum strains 8941 and 9414, which were isolated from HIV-positive patients.
Primer D-9355.
When primer D-9355 was used for typing of the H. capsulatum isolates, five genotypes divided into three groups (I, II, and III) (Fig. 3A and 4A) were also observed. Most of the isolates were clustered into group I, subtype Ia, which presented 93.5% relatedness with subtype Ib, which is composed of one strain from soil, two from animals, three from AIDS patients, and three from HIV-negative patients. Group II, also divided into subtypes IIa and IIb, included 12 isolates of different origins (six AIDS patient strains, three strains from HIV-negative patients, two strains from soil, and one animal isolate). Group III, which presented 81.4% similarity to the other groups, was made up of only isolates from AIDS patients.
FIG. 4.
Dendrogram derived from analysis of the RAPD profiles of genomic DNAs of H. capsulatum strains using Molecular Analyst Fingerprinting Plus, version 1.12 (Bio-Rad). Panels: A, primer D-9355; B, primer D-10513. The numbers on the right are strain designations.
Primer 10513.
Genetic differences were also expressed in three clusters by RAPD with primer D-10513 (Fig. 3C and 4B). Group I contained 77% of the strains from different sources and was divided into subtypes Ia and Ib, with a high degree of relatedness between them (92.3%). Groups II and III did not show any subtype division and comprised isolates from several origins: IT4 (soil), CÃO4 (animal), 2733, 6305, 8941, 9414, 84564, 4959, 9236 (AIDS patients), B-670, and 78642 (HIV-negative patients).
The majority of the H. capsulatum strains isolated from soil and animals, independent of which primer was used in the RAPD assay, were assembled into group I, subtype Ia (Table 1). Table 2 lists the DNA fingerprinting patterns achieved among H. capsulatum isolates from animals and soil obtained from the same geographic area. All of the H. capsulatum strains isolated from Ilha Grande presented the same genotype, with 100% similarity, and were clustered into group I, subtype Ia. Similar results were observed with the isolates from Rio da Prata. Most of these isolates were clustered into group I, subtype Ia (83% similarity). However, H. capsulatum isolate RP44, from a wild rodent, was classified into group I, subtype Ib.
TABLE 2.
Genetic relationship among H. capsulatum strains isolated from soils and animals in the same geographic area in Rio de Janeiro State
| Location | Isolate
|
Genotype(s)
|
||||
|---|---|---|---|---|---|---|
| Soil | Animal | 1253 | 1281 | D-9355 | D-10513 | |
| Ilha Grande, RJa | IGS4/5 | RS1 | Ia | Ia | Ia | Ia |
| IGS19 | RS9 | Ia | Ia | Ia | Ia | |
| RS36 | Ia | Ia | Ia | Ia | ||
| Rio da Prata, RJb | RPS11 | Ia | Ia | Ia | Ia | |
| RPS45 | RP44 | Ia | Ia | Ia/Ib | Ia | |
| RPS51 | RP93 | Ia | Ia | Ia | Ia | |
| RPS86 | Ia | Ia | Ia | Ia | ||
| Tinguá, RJb | TI1 | None | Ib | Ia | IIb | Ib |
| TI5 | Ib | Ia | IIb | Ib | ||
| TI14 | Ia | Ia | Ia | Ia | ||
| Niterói, RJc | AC2 | None | Ia | Ia | Ia | Ia |
| AC5 | Ia | Ia | NTd | Ia | ||
| Itaipava, RJd | IT4 | CÃO4 | Ib | II | Ib | II |
| EP2 | Ia | Ia | NT | Ia | ||
| CADAM | Ib | Ia | IIb | Ib | ||
County region of Angra dos Reis municipality.
County region of Rio de Janeiro (RJ) municipality.
City of Rio de Janeiro State.
NT, not typed.
The DNA fingerprinting patterns obtained from clinical samples of patients with histoplasmosis, whether or not they were isolated from patients with AIDS, showed a high degree of genetic strain diversity (Table 1). H. capsulatum strains were isolated on different dates from the same patients, two HIV-positive patients (9291 and 9305 and 84502 and 84544) and two HIV-negative patients (3237 and 3356 and 3416 and 3669). The DNA profiles of strains from the same patient isolated at different times showed 100% similarity, independent of the primer used in the test (Table 3).
TABLE 3.
Genetic relatedness between H. capsulatum isolates from the same histoplasmosis patients on different dates
| Patient no. | HIV status | Date of isolation | Culture no. | Genotype
|
|||
|---|---|---|---|---|---|---|---|
| 1253 | 1281 | D-9533 | D-10513 | ||||
| 1 | Negative | 10/27/88 | 3237 | IIa | Ib | Ib | Ib |
| Negative | 12/07/88 | 3356 | IIa | Ib | Ib | Ib | |
| 2 | Negative | 01/10/89 | 3416 | Ia | Ia | Ia | Ia |
| Negative | 05/12/89 | 3669 | Ia | Ia | Ia | Ia | |
| 3 | Positive | 05/28/93 | 9291 | IIa | III | Ib | Ib |
| Positive | 06/03/93 | 9305 | IIa | III | Ib | Ib | |
| 4 | Positive | 01/22/99 | 84502 | IIb | IV | III | Ib |
| Positive | 01/22/99 | 84544 | IIb | IV | III | Ib | |
DISCUSSION
The molecular epidemiology of histoplasmosis has not previously been studied in Brazil. The present study establishes the genetic relatedness among Brazilian (from Rio de Janeiro State) H. capsulatum samples isolated from patients with AIDS-related infections, immunologically intact hosts with community-acquired histoplasmosis, soil, and animals by using RAPD assays. Analysis of our findings showed that the H. capsulatum isolates included in this study had 80 to 100% similarity and that despite this genetic similarity, they could be separated into four to six different genotypes. The primers used for RAPD analysis were chosen based on a previous study (8). Although the choice of other primers with higher discriminatory power could be applied to this study, many arbitrary oligonucleotides can serve as informative primers. It has been demonstrated that 50% G+C primers generally give good amplification with the larger and more complex genomes of eukaryotes (13) such as H. capsulatum. The G+C contents of our primers are 40 to 70%. Also, the usefulness of primers with higher G+C contents probably stems from their generally stronger binding and thus their ability to utilize a larger number of partially mismatched template DNA sites (8). The RAPD profiles of H. capsulatum isolated from Rio de Janeiro State could be distinguished from those of the U.S. strains included in this study (Downs, G222B, G-186B, and FLS1) with each of the primers tested. Similar studies with H. capsulatum isolates from Thailand showed identical results. Those isolates were also genetically homogeneous and clustered into two or three groups with DNA profiles distinct from those of American strains (10), suggesting that they could be environmentally and/or geographically distinct strains. Our data corroborate this hypothesis and provide the beginning of a population genetic analysis of this species. These data also suggest geographical specificity, as previously demonstrated for Cryptococcus neoformans serotypes (1, 14).
The ecology of H. capsulatum in Rio de Janeiro State has been the subject of several investigations (2, 19). The fungus has been isolated from soil in different geographic areas, suggesting that H. capsulatum microniches exist at these localities and could be acting as the source of infection for animals and the local human population (19). The high similarity observed among the H. capsulatum strains isolated from animals and soil at the same locality favors that hypothesis. H. capsulatum isolates from Ilha Grande were clustered into group I, subtype la, presenting 100% identity. Similar results were obtained with isolates from Rio da Prata. Five isolates from soil and one from an animal showed identical DNA profiles, being classified into group I, subtype Ia. Although the genotype profiles of the RP44 sample (isolated from a rodent) were clustered into group I, subtype Ib (Table 2), when it was typed with the D-9355 primer, it showed 93% similarity to other isolates from the same locality, showing a close genetic relationship. This could be explained by the existence of one or more genetic events, such as an insertion or deletion of nucleotides in the genomic DNA region that hybridizes with this primer (17). However, we cannot rule out the possibility that the animal (Rattus rattus) was infected at a different ecological site. The IT4 strain was isolated from the soil of a cave in Itaipava, where a human histoplasmosis outbreak occurred (B. Wanke, M. F. Ferreira da Cruz, M. S. Lazera, and G. Bretas, Ann. 19th Congr. Soc. Bras. Med. Trop. abstr. 26, 1983), and the CÃO-4 strain was isolated from a dog that died after being exposed to fungal propagules in the same place to induce an experimental infection (12). The high percentage of genetic similarity demonstrated between these isolates suggests the possibility that only one virulent genetic population was present in this microniche. Although it has been demonstrated that H. capsulatum reproduction could be recombinant (17) and hybrid genotypes could be found in nature, this event has not been demonstrated, even with molecular techniques such as sequencing of several genes encoding four H. capsulatum proteins (5).
H. capsulatum soil isolates from a trench under a bridge in Pendotiba and from a chicken house in Tinguá were suspected to be the source of infection in two outbreaks of histoplasmosis that were investigated in our laboratory. Samples AC2 and AC5, obtained in the first outbreak, presented the same genotype, with 100% similarity between them. Different results were observed when three H. capsulatum isolates from the chicken house (TI1, TI5, and TI14) were RAPD typed. They were included in two different groups and subtypes (Table 2). This fact suggested the presence of a unique genetic population in microniches less exposed to climatic variations. However, the possibility of a genetic alteration in nature cannot be ruled out.
Low-virulence strains (Downs strain) are more prevalent in AIDS patients (6). Our results were not able to show such an association, and this view is consistent with the observation made by Reyes-Montes et al. (11) in typing H. capsulatum strains isolated in Mexico. The strains from HIV-positive and HIV-negative patients showed genetic diversity, being included in all of the groups and subtypes discriminated by the RAPD technique. Moreover, when this genotype profile was compared with those of strain Downs and classes II, III, and IV (7, 15), less than 70% similarity was observed among them.
All of the H. capsulatum strains isolated from four patients on different dates presented the same genotype pattern (Table 3). Possibly, these patients harbored only one genetic population. This approach will be useful in determining whether infection is the result of endogenous reactivation of a quiescent focus, an event already demonstrated by Keath et al. (6).
Poonwan et al. (10) analyzed the genetic diversity of strains from Thailand and observed a 700-bp genomic DNA fragment in all of the strains included in their study. Those authors considered this fragment to be a species-specific marker. In our study, some species-specific markers were observed and further study will be done in order to obtain the complete nucleotide sequence of these fragments for standardization of a specific diagnosis of histoplasmosis.
This is the first study of histoplasmosis done by molecular typing methods in Brazil. The data obtained suggest that the application of RAPD methodology could be a useful tool for H. capsulatum typing. This method of strain identification and analyses of genome diversity is sensitive, reliable, and fast; requires much less DNA than is needed for Southern blotting; and avoids the need for restriction endonuclease cleavage, which is often difficult with DNA extracted from H. capsulatum (8). New comparative studies using RAPD and other molecular typing methods are in development to delineate the polymorphism present in H. capsulatum isolates from Rio de Janeiro in comparison with those from other Brazilian states in order to provide more information and new insight into the population genetics of H. capsulatum and the epidemiology of histoplasmosis.
ACKNOWLEDGMENTS
The research was supported in part by grant Proc. 400175/98-3 from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and grant Proc. E-26/150.130/99 from the Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ) Brazil.
We thank Andréa Pussenti Derossi and Rosane Orofino Costa for providing the H. capsulatum isolates from the Hospital Universitário Pedro Ernesto and the Laboratório Lâmina and Mark F. Schinsky for suggestions and review.
REFERENCES
- 1.Bennett J E, Kwon-Chung K J, Howard D H. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 2.Capone D, Wanke B, Monteiro P C F, Lazera M S, Andrade G N, Valle A C F, Moreno A M H, Londero A T. Chronic pulmonary histoplasmosis in the State of Rio de Janeiro, Brazil. Mycopathologia. 1999;145:75–79. doi: 10.1023/a:1007016414833. [DOI] [PubMed] [Google Scholar]
- 3.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1998;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang B, Bartlett M S, Allen S D, Smith J W, Wheat L J, Connolly P A, Lee C. Typing Histoplasma capsulatum isolates based on nucleotide sequence variation in the internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 2000;38:241–245. doi: 10.1128/jcm.38.1.241-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasuga T, Taylor J W, White T J. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J Clin Microbiol. 1999;37:653–663. doi: 10.1128/jcm.37.3.653-663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphism in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keath E J, Spitzer E D, Painter A A, Travis S J, Kobayashi G S, Medoff G. DNA probe for the identification of Histoplasma capsulatum. J Clin Microbiol. 1989;27:2369–2372. doi: 10.1128/jcm.27.10.2369-2372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pine L. Growth of Histoplasma capsulatum. VI. Maintenance of the mycelial phase. Appl Microbiol. 1970;19:413–420. doi: 10.1128/am.19.3.413-420.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poonwan N, Imai T, Mekha N, Yazawa K, Mikami Y, Ando A, Nagata Y. Genetic analysis of Histoplasma capsulatum strains isolated from clinical specimens in Thailand by a PCR-based random amplified polymorphic DNA method. J Clin Microbiol. 1998;36:3073–3076. doi: 10.1128/jcm.36.10.3073-3076.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Montes M R, Bobadilla-Del Valle M, Martínes-Rivera M A, Rodrígues-Arellanes G, Maravilla E, Sifuentes-Osornio J, Taylor M L. Relatedness analyses of Histoplasma capsulatum isolates from Mexican patients with AIDS-associated histoplasmosis by using histoplasmin electrophoretic profiles and randomly amplified polymorphic DNA patterns. J Clin Microbiol. 1999;35:1404–1408. doi: 10.1128/jcm.37.5.1404-1408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro V L S. Histoplasmose canina no Rio de Janeiro. M.S. thesis. Rio de Janeiro, Brazil: Fundação Oswaldo Cruz; 1985. [Google Scholar]
- 13.Serikawa T, Montagutelli X, Simon-Chazottes D, Guenet J L. Polymorphisms revealed by PCR with single, short-sized, arbitrary primers are reliable markers for mouse and rat gene mapping. Mamm Genome. 1992;3:65–72. doi: 10.1007/BF00431248. [DOI] [PubMed] [Google Scholar]
- 14.Soll D R. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–370. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitzer E D, Lasker B A, Travis S J, Kobayashi G S, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Standard P G, Kaufman L. Specific immunological test for the rapid identification of members of the genus Histoplasma. J Clin Microbiol. 1976;3:191–199. doi: 10.1128/jcm.3.2.191-199.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheat L J, Connolly-Stringfield P, Blair R, Connolly K, Garringer T, Katz B P. Histoplasmosis relapse in patients with AIDS: detection using Histoplasma capsulatum variety capsulatum antigens levels. Ann Intern Med. 1991;115:936–941. doi: 10.7326/0003-4819-115-12-936. [DOI] [PubMed] [Google Scholar]
- 19.Zancopé-Oliveira R M, Wanke B. Isolamento do Histoplasma capsulatum de animais silvestres no município do Rio de Janeiro. Cad Saúde Pública. 1987;2:42–52. [Google Scholar]