Abstract
Modern linkages among magmatic, geochemical, and geobiological processes provide clues about the importance of thermophiles in the origin of biogeochemical cycles. The aim of this study was to identify the primary chemoautotrophs and host–virus interactions involved in microbial colonization and biogeochemical cycling at sublacustrine, vapor-dominated vents that represent the hottest measured ecosystems in Yellowstone National Park (~140 °C). Filamentous microbial communities exposed to extreme thermal and geochemical gradients were sampled using a remotely operated vehicle and subjected to random metagenome sequencing and microscopic analyses. Sulfurihydrogenibium (phylum Aquificae) was the predominant lineage (up to 84% relative abundance) detected at vents that discharged high levels of dissolved H2, H2S, and CO2. Metabolic analyses indicated carbon fixation by Sulfurihydrogenibium spp. was powered by the oxidation of reduced sulfur and H2, which provides organic carbon for heterotrophic community members. Highly variable Sulfurihydrogenibium genomes suggested the importance of intra-population diversity under extreme environmental and viral pressures. Numerous lytic viruses (primarily unclassified taxa) were associated with diverse archaea and bacteria in the vent community. Five circular dsDNA uncultivated virus genomes (UViGs) of ~40 kbp length were linked to the Sulfurihydrogenibium metagenome-assembled genome (MAG) by CRISPR spacer matches. Four UViGs contained consistent genome architecture and formed a monophyletic cluster with the recently proposed Pyrovirus genus within the Caudovirales. Sulfurihydrogenibium spp. also contained CRISPR arrays linked to plasmid DNA with genes for a novel type IV filament system and a highly expressed β-barrel porin. A diverse suite of transcribed secretion systems was consistent with direct microscopic analyses, which revealed an extensive extracellular matrix likely critical to community structure and function. We hypothesize these attributes are fundamental to the establishment and survival of microbial communities in highly turbulent, extreme-gradient environments.
Subject terms: Water microbiology, Biofilms, Metagenomics, Virus-host interactions
Introduction
The discovery of complex biological communities at marine hydrothermal vents [1] demonstrated that diverse ecosystems are supported by reduced chemical species such as sulfide, methane, and hydrogen. Past reports have shown that extreme chemosynthetic microbial habitats at deep thermal vents provide opportunities for determining relationships among microbial metabolism, host–virus interactions, and biogeochemical cycling [2–8]. Sulfur cycling has been intimately linked with the microbial metabolism of thermophiles since early in Earth’s evolutionary history [9–12], and recent studies have shown that microbial sulfur metabolisms may be more phylogenetically widespread than previously assumed [13]. Moreover, the discovery of viral auxiliary metabolic genes (AMGs) for sulfur assimilation and/or reduction demonstrated that viruses may also play major roles in sulfur cycling [4, 14–17]. Microbial metabolism, host–virus interactions, and geochemical cycles have coevolved [18, 19], but these relationships are difficult to isolate in complex microbial environments such as soils and/or natural waters with high species diversity [20–22]. High-temperature systems such as hydrothermal vents offer advantages for elucidating fundamental relationships among predominant primary producers and redox gradients because thermophilic communities have comparatively low diversity and are often associated with early evolved metabolic processes [23].
While most investigations of hydrothermal vent communities have focused on marine systems, sublacustrine hydrothermal vents offer important and often overlooked perspectives on the influence of chemosynthetic thermophiles on biogeochemical cycling. Yellowstone Lake (YLake) sits on top of the Yellowstone volcano, which accounts for two of the largest eruptions in the Holocene; unlike most large volcanos that are now dormant, Yellowstone remains active today [24–27]. Moreover, most modern large hot spots are found on the ocean crust (e.g., Hawaii, Galapagos), whereas the Yellowstone volcano is the only large, mid-continental hot spot [28, 29]. The highest heat flux and seismic activity from the Yellowstone volcano occur in the deepest region of YLake [30–32] where elevated pressure at thermal vents yields high-temperature fluids of up to 174 °C [33]. These sublacustrine vents are the hottest measured ecosystems in Yellowstone National Park (YNP) and offer unique opportunities for studying the genomic characteristics of deeply rooted, naturally occurring thermophilic microorganisms, which thrive at the interface between hot reduced fluids and cold (4 °C) [32] oxygenated lake water. Vapor-dominated thermal vents in YLake contain high concentrations of carbon dioxide, sulfide, hydrogen, and methane, which support thick filamentous biofilm communities growing across steep redox and temperature gradients [34]. Although logistically difficult to sample, hydrothermal vents in YLake are considerably more accessible using a remotely operated vehicle (ROV) than deep marine vents and provide analogous and relatively stable field laboratories for developing sensor technology and long term geochemical and/or microbiological monitoring [35]. Prior geochemical, microbiological, and geological inventories of YLake thermal vents revealed diverse vent geochemistry that was correlated with metabolic attributes of specific microbial lineages [34, 36, 37]. However, comprehensive metagenomic sequence datasets from the deepest and most extreme vents in YLake have not been available prior to this study.
The central goal of this research was to determine the function and host–virus interactions of predominant microbial community members thriving at the most extreme thermal and chemical gradient ecosystems in Yellowstone, and to determine their roles in mediating geochemical cycles important in Earth’s evolutionary history. Here we show that deep, vapor-dominated hydrothermal vents on the floor of Yellowstone Lake support unique thermophilic microbial communities rich in sulfur-oxidizing bacteria from the Aquificae, which are associated with a wide diversity of predominately lytic viruses. Integration of genomic and transcriptomic data coupled with microscopic and geochemical analyses showed that Sulfurihydrogenibium forms extensive filamentous structures, which are dominated by sulfur-active thermophiles and contain significant extracellular material. Highly expressed genes encoding novel type IV filament (TFF) systems and membrane porins were identified on Sulfurihydrogenibium plasmids, and a suite of diverse secretion and adhesion components were detected within the metagenome-assembled genome (MAG). These observations suggest that biofilm formation processes are crucial to the survival and growth of Sulfurihydrogenibium in situ and are consistent with the microscopic identification of extracellular material that dominates the biofilm matrix. Finally, we present evidence that predominant Sulfurihydrogenibium is a collection of several closely related sub-populations. Our report is part of a larger interdisciplinary effort to identify and predict multiscale processes driven by magmatic and tectonic forcing (Hydrothermal Dynamics of Yellowstone Lake) [38]. Geophysical processes responsible for changes in temperature and geochemistry are linked to rapid responses in local microbiota; consequently, microbiological signatures such as those identified here can be sensitive indicators of hydrothermal activity.
Results and discussion
Hydrothermal vent biofilm community structure
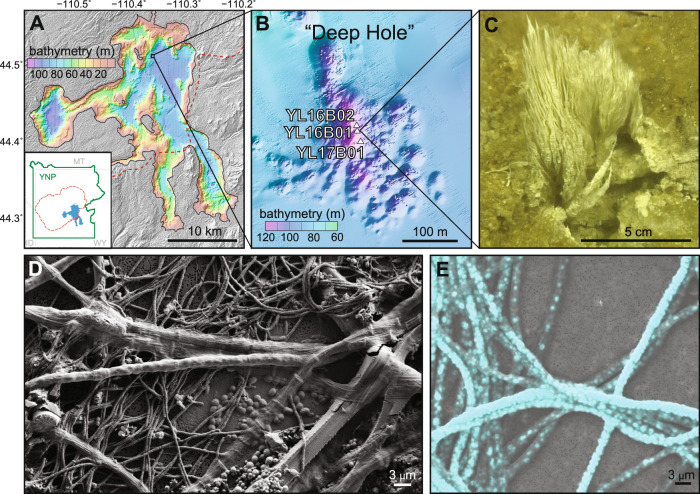
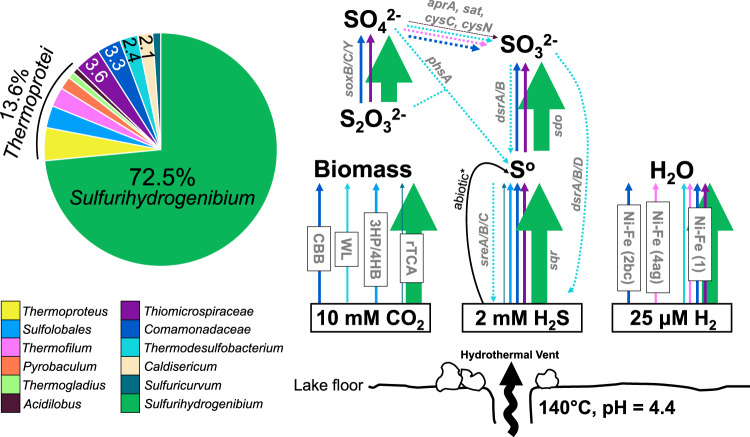
MAGs from 25 morphologically and phylogenetically diverse bacteria and archaea were recovered from sulfur-rich biofilms at three deep (110 m) hydrothermal vents in YLake (Fig. 1, Table 1). Vapor-dominated [33] vent fluids in the “Deep Hole” east of Stevenson Island [32, 38] discharge fluids at temperatures up to 174 °C and contain high concentrations of dissolved inorganic carbon (10 mM DIC), dissolved sulfide (1–2 mM), hydrogen (up to 25 µM), and methane (19–220 µM) [35, 39, 40], which provide abundant carbon and energy sources for microorganisms. Vent microbial communities reside within steep thermal and redox gradients created by fully oxygenated surrounding lake water at ca. 4 °C [32]. Vents sampled ranged from 95 to 140 °C during ROV collection. Predicted optimal growth temperatures (TOPT) calculated from total deduced protein sequences of each MAG [41] ranged from ca. 22.5 to 91.6 °C (Table 1), which indicated that the vent community was comprised of organisms that occupied different temperature niches. However, thermophilic populations with predicted TOPT >50 °C represented the vast majority (>95% relative abundance based on MAG sequence coverage) of community members when filamentous sulfur streamers were washed of surrounding sediments prior to DNA extraction (Sample 2016_B01_str; Supplementary Table 1). Sulfur-oxidizing Sulfurihydrogenibium spp. (phylum Aquificae) were the predominant thermophilic community members ranging from ~22 to 73% in unseparated samples and 84% when filaments were washed of surrounding sediments (Fig. 2; Supplementary Table 1; Supplementary Figure 1). This is consistent with previous observations of predominant Sulfurihydrogenibium detected by 16S rRNA gene sequencing at deep vents in YLake [37]. Diverse thermophilic archaea within the Thermoprotei together accounted for 11.6–13.1% relative abundance. Several important mesophiles were consistently associated with biofilm communities in lower abundance, including MAGs for Sulfuricurvum and Thiomicrospiraceae (Supplementary Discussion; Supplementary Table 1).
Fig. 1. Microbial streamer communities collected from sulfidic hydrothermal vents (Stevenson Island Deep Hole, Yellowstone Lake, WY) contain morphologically diverse cells and accumulate elemental sulfur.
A Bathymetric map of Yellowstone Lake relative to the boundary (red dashed line) of the most recent caldera-forming eruption. B Bathymetric map of three sampling sites within the Deep Hole east of Stevenson Island. C A thermophilic sulfur-rich biofilm sampled at one hydrothermal vent (Sample 2016_B02). D Scanning electron microscopy demonstrates morphological diversity of different microorganisms. E Elemental analysis of sulfur (blue) within the largest (~3 µm diameter) filamentous cells of the vent biofilm (more detail in Supplementary Figure 5).
Table 1.
Predominant MAGsa identified in sulfidic hydrothermal vent streamers (Stevenson Island Deep Hole, Yellowstone Lake, WY) sorted as a function of predicted optimal growth temperature.
| Genome populationa | Phylumb | TOPTc | Len.d | Comp.e | Red.f | G + C %g | Coverageh | Cont.i | N50 (kb)j | Viral cont.k | SNV Dens.l | Trans.m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pyrobaculum DHV | A: Crenarchaeota | 91.6 | 1.18 | 75.9 | 3.7 | 56.7 (4.2) | 20.3 (3.2) | 197 | 6.84 | 13 | 40.7 | 1 |
| Thermogladius DHV | A: Crenarchaeota | 85.3 | 1.15 | 95.1 | 3.1 | 52.1 (2.4) | 56.3 (12.6) | 64 | 31.07 | 0 | 71.8 | 0 |
| Acidilobus DHV | A: Crenarchaeota | 82.7 | 1.26 | 95.1 | 3.1 | 59.3 (2.5) | 21.1 (5.9) | 39 | 56.84 | 0 | 9.6 | 2 |
| Thermoproteus DHV | A: Crenarchaeota | 81.9 | 1.44 | 93.8 | 3.7 | 61.2 (3) | 104.1 (24.1) | 87 | 30.03 | 1 | 30.5 | 1 |
| Thermodesulfobacterium DHV | B: Thermodesulfobacteria | 78.8 | 1.73 | 89.9 | 12.9 | 42.6 (2.4) | 47.1 (16.4) | 158 | 18.31 | 1 | 44.1 | 2 |
| Thermofilum DHV | A: Crenarchaeota | 78.7 | 2.16 | 90.7 | 2.5 | 45.1 (4.5) | 41.3 (11.0) | 225 | 16.54 | 37 | 36.3 | 4 |
| Thermocrinis DHV | B: Aquificae | 78.0 | 1.23 | 87.8 | 0.0 | 42.8 (2.9) | 118.4 (353.4) | 177 | 8.57 | 1 | 58.6 | 5 |
| Sulfolobus DHV | A: Crenarchaeota | 76.6 | 1.21 | 99.4 | 2.5 | 52.1 (4.6) | 86.5 (18.0) | 81 | 27.15 | 2 | 71.9 | 1 |
| Desulfurococcaceae DHV | A: Crenarchaeota | 71.8 | 1.46 | 97.5 | 3.1 | 36.4 (1.2) | 16.0 (5.4) | 80 | 30.03 | 0 | 7.8 | 3 |
| Thermoprotei DHV | A: Crenarchaeota | 70.9 | 1.22 | 87.7 | 2.5 | 43.5 (2.6) | 12.3 (4.7) | 138 | 10.97 | 1 | 3.4 | 7 |
| — | ||||||||||||
| Thermoplasmata DHV | A: Thermoplasmata | 67.8 | 1.30 | 97.5 | 6.2 | 35.6 (2.4) | 39.1 (11.3) | 80 | 26.53 | 0 | 17.1 | 4 |
| Archaea DHV | A: Thaum./Aig. | 67.3 | 1.28 | 87.7 | 7.4 | 36.1 (1.3) | 7.4 (3.2) | 176 | 8.56 | 0 | 0.5 | 2 |
| Caldisericum DHV | B: Caldiserica | 66.1 | 1.62 | 97.1 | 0.7 | 36.8 (2) | 42.8 (11.1) | 161 | 19.60 | 4 | 30.8 | 3 |
| Thaumarchaeota DHV | A: Thaumarchaeota | 63.1 | 0.71 | 69.1 | 1.2 | 67.3 (2) | 8.6 (3.8) | 132 | 5.90 | 0 | 1.4 | 4 |
| Sulfurihydrogenibium DHV | B: Aquificae | 59.0 | 2.96 | 94.2 | 54.0** | 31.7 (2.5) | 1414.1 (460.5) | 908 | 4.24 | 35 | 58.5 | 37 |
| Ignisphaera DHV | A: Crenarchaeota | 50.5 | 2.56 | 84.0 | 4.3 | 38.9 (4.1) | 12.7 (4.9) | 405 | 6.64 | 63 | 12.0 | 1 |
| Aminicenantes DHV | B: Aminicenantes | 50.1 | 2.21 | 90.6 | 6.5 | 42.4 (2.1) | 5.5* (1.8) | 134 | 28.98 | 0 | 0.9 | 5 |
| — | ||||||||||||
| Acidobacteria DHV | B: Acidobacteria | 37.7 | 1.91 | 89.2 | 4.3 | 60.1 (1.4) | 11 (4.3) | 275 | 7.66 | 0 | 3.7 | 2 |
| Tibeticola DHV | B: Betaproteobacteria | 36.7 | 2.38 | 96.4 | 0.7 | 66.5 (1.9) | 6.9* (3.3) | 183 | 16.72 | 0 | 1.7 | 12 |
| Campylobacteraceae DHV | B: Epsilonproteobacteria | 34.2 | 1.43 | 95.0 | 2.9 | 42.8 (1.8) | 10.5 (4.2) | 145 | 13.42 | 0 | 3.3 | 3 |
| Sulfuricurvum DHV | B: Epsilonproteobacteria | 29.8 | 1.74 | 95.7 | 7.9 | 44.5 (3.2) | 12.2 (4.7) | 158 | 15.43 | 1 | 5.2 | 8 |
| Nitrosopumilus DHV | A: Thaumarchaeota | 26.4 | 1.61 | 84.0 | 8.0 | 33.2 (1.9) | 4.6* (2.8) | 205 | 8.62 | 2 | 2.2 | 1 |
| Burkholderiales DHV | B: Betaproteobacteria | 25.9 | 4.62 | 94.2 | 7.2 | 65.9 (1.7) | 6.9* (3.3) | 422 | 13.81 | 2 | 2.2 | 35 |
| Thiomicrospiraceae DHV | B: Gammaproteobacteria | 25.3 | 2.25 | 100.0 | 5.8 | 43.8 (2.8) | 53.8 (11.6) | 174 | 28.70 | 15 | 12.0 | 8 |
| Lentimicrobium DHV | B: Bacteriodetes | 22.5 | 2.66 | 92.8 | 3.6 | 34.5 (1.4) | 9.9 (4.1) | 305 | 11.33 | 2 | 2.8 | 10 |
Bold: Sulfurihydrogenibium was the predominant MAG in all four metagenomes and is represented by varying micro-populations that could not be subdivided by tetranucleotide frequency or coverage-based clustering analyses. Empty rows separate groups based on predicted optimal growth temperature, TOPT(upper = hyperthermophiles; middle = thermophiles; lower = mesophiles).
*Co-assembly coverage is mean coverage between both assemblies.
**High redundancy for this MAG was examined carefully and determined to be related to Sulfurihydrogenibium (Supplementary Figure 2; Fig. 4; Supplementary Table 8; Supplementary Table 6).
aMAGs with >1% relative abundance (within their respective metagenome assembly) and greater than ~70% estimated completeness were named by the highest resolution taxonomic rank determined by robust phylogenomic analysis (Supplementary Figure 1), with the suffix “DHV” for Deep Hole Vent. Comprehensive lists of all MAGs recovered are provided in Supplemental Table 2.
bThe corresponding microbial phylum is listed for each MAG, preceded by A for archaea or B for bacteria.
cTOPT = predicted optimal growth temperature based on 2-mer amino acid compositions [41].
dLength (Mb) = cumulative sequence length of all contigs within a MAG.
eComp. = estimated completeness based on single-copy gene detection.
fRed. = estimated redundancy based on single-copy gene detection.
gG + C% = mean percentage of G and C nucleotides determined for each contig within a MAG. Standard deviation from the mean is listed in parentheses.
hCoverage = mean depth of short read recruitment across all contigs within a MAG. For relative abundance see individual assembly information (Supplementary Tables – individual assemblies). Standard deviation from the mean is listed in parentheses. Asterisk indicates average of coverage values between both samples in the co-assembly.
iCont. = the number of contigs within each MAG.
jN50 = minimum contig length required to constitute 50% of the MAG (in kilobase).
kViral contigs listed here identified based on VirSorter and mapping CRISPR variable spacer regions (see “Materials and methods”).
lSNV Dens. = single nucleotide variants per kilobase per genome.
mNumber of transposase sequences detected in each MAG.
Fig. 2. Community composition and key pathways of Sulfurihydrogenibium-dominated hydrothermal streamers.
Relative abundance is shown for biofilm metagenome from vent sample 2016_B01, which was the cleanest unmanipulated sample retrieved (the relative abundance of other samples is provided in Supplementary Table 1). Concentrations of H2, H2S, and CO2 were previously determined for YLake vents in the Deep Hole [33, 35, 39]. Arrows either represent (i) autotrophic pathways, (ii) enzymatic conversions between sulfur species, or (iii) hydrogenases. Arrow color corresponds to the microbial lineage encoding that particular enzyme; arrow thickness is weighted by mean coverage for the contig containing the relevant gene/s. [apr = adenylylsulfate reductase; sat = sulfate adenylyltransferase; cys = adenylylsulfate kinase; phs = polysulfide reductase; dsr = dissimilatory sulfite reductase; sdo = sulfur dioxygenase; sre = sulfur reductase; sqr = sulfide:quinone oxidoreductase].
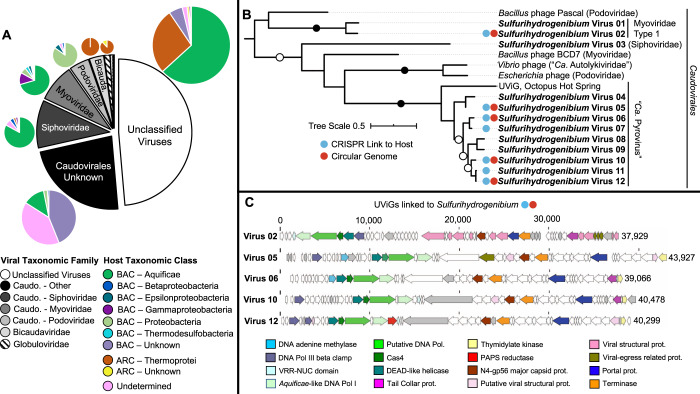
Thermal vent biofilms in YLake also host a wide diversity of viruses (Fig. 3, Table 2; Supplementary Table 2). The highest abundance of viral contigs was associated with the predominant archaea and bacteria within the vent biofilm community based on overlapping tetranucleotide signatures between putative viral sequence and host MAGs [42, 43]: Thermoprotei (n = 63), Thermofilum spp. (n = 37), Sulfurihydrogenibium spp. (n = 35), and Thiomicrospiraceae (n = 15) (Table 1; Fig. 2). The viral sequence dataset was dominated by members of the Caudovirales, within the Siphoviridae, Myoviridae, Podoviridae, and miscellaneous unknown families (Fig. 3A). Nearly half (49%) of the putative viral sequences could not be assigned to known relatives, which indicates a large proportion of uncharacterized viruses within these communities. Phylum Aquificae (which includes Sulfurihydrogenibium) was targeted by viruses from all viral families of the Caudovirales as well as the unclassified group based on (i) phylogenetic signatures of overall TNF clusters and/or (ii) taxonomic hits of viral protein sequences to microbial entries in public databases [44] (Fig. 3A, in green). Moreover, 87% (99/114) of all CRISPR repeat sequences with perfect sequence matches to metagenome sequence were taxonomically identified as Aquificae, and 81% (309/382) of variable spacer sequences within CRISPR arrays matched to putative viral sequences within Sulfurihydrogenibium MAGs (Supplementary Table 3; Supplementary Discussion). The Myoviridae and Podoviridae families were also associated with members of the γ- and ε-proteobacteria, which contain Thiomicrospiraceae and Sulfuricurvum, respectively. A unique, 30-bp CRISPR repeat sequence was detected for Sulfuricurvum and associated with 1893 spacers on separate arrays. Less abundant Bicaudaviridae and Globuloviridae families were associated exclusively with archaeal hosts (especially Thermoprotei), which is consistent with previously established archaeal host taxonomy for these viral families [45, 46]; 27% of uncharacterized viruses were also associated with Thermoprotei hosts. Finally, 15% (56/382) of CRISPR spacer sequences and 7% (8/114) of direct repeats matched contigs within Thermoprotei clusters. These observations demonstrate that the predominant bacterial and archaeal members of the microbial community (i.e., Sulfurihydrogenibium and various Thermoprotei) experience significant and diverse viral selective pressures.
Fig. 3. Virus–host diversity and genomic characteristics of Sulfurihydrogenibium viruses.
A Family-level community structure of recovered viral sequences in the black and white chart is related to microbial host at the class level in colored pie charts. B Maximum-likelihood phylogenetic tree of deduced capsid protein sequences from Sulfurihydrogenibium viruses showing a monophyletic relationship with the newly proposed “Ca. Pyrovirus” genus within the Caudovirales [66]. Entries with red circles represent circularized viral genomes; entries with blue circles had CRISPR-resolved links to a Sulfurihydrogenibium host. Vibrio phage corresponds to the 1.232.O._10N.261.51.E11 identifier; Escherichia phage corresponds to the vB_EcoP_PTXU04 identifier. Contig IDs corresponding to Sulfurihydrogenibium viruses are listed in Supplementary Table 16. C Genome architecture and annotations of circular UViGs that had CRISPR links to a Sulfurihydrogenibium host.
Table 2.
Gene content of putative viral (or plasmid) scaffolds (>20 kbp) associated with Aquificales and Thermoprotei in sulfur-rich filamentous biofilms.
| Putative virus/plasmid | Length (bp) | Genes | CRISPR links to hosta | UViG (circular) | VirSorter rank | Phage hallmark | Nucleotide metabolism | Genetic information processing |
|---|---|---|---|---|---|---|---|---|
| Sulfurihydrogenibium virus 13 | 117,954 | 190 | 0 | N | 2 | 3 | spoT, comEB, thyX | dnaB, polB, rfcS, lig1, recA |
| Sulfurihydrogenibium virus 14 | 48,861 | 58 | 0 | N | 1 | 5 | – | – |
| Sulfurihydrogenibium virus 05 | 44,006 | 52 | 2 | Y | 2 | 1 | tmk, thyX | polA, dnaN, dam |
| Sulfurihydrogenibium virus 10 | 40,586 | 51 | 5 | Y | 2 | 1 | tmk, thyX | polA, dnaN |
| Sulfurihydrogenibium virus 12 | 40,337 | 47 | 5 | Y | 2 | 1 | tmk | polA, dnaN |
| Sulfurihydrogenibium virus 06 | 39,121 | 46 | 3 | Y | 2 | 1 | tmk | polA, dam |
| Sulfurihydrogenibium virus 02 | 38,070 | 55 | 66 | Y | 2 | 4 | – | polA, dnaN |
| Sulfurihydrogenibium virus 07 | 33,189 | 45 | 5 | N | 2 | 1 | tmk, thyX | polA, dnaN |
| Sulfurihydrogenibium plasmid | 30,807 | 40 | 4 | N | 3 | nd | tmk | ssb |
| Sulfurihydrogenibium virus 15 | 29,694 | 42 | 61 | N | 2 | 5 | – | polA |
| Sulfurihydrogenibium virus 16 | 27,080 | 18 | 1 | N | 6 | nd | thyX | dam, ccrM |
| Sulfurihydrogenibium virus 17 | 25,469 | 29 | 4 | N | 2 | 1 | – | polA |
| Thermofilum virus 01 | 52,269 | 62 | 1 | N | 3 | nd | – | – |
| Thermofilum virus 02 | 31,158 | 56 | 1 | N | 3 | nd | pcnB | dam |
| Thermofilum virus 03 | 27,347 | 38 | 1 | N | 2 | 1 | – | dcm |
| Thermofilum virus 04 | 25,126 | 42 | 10 | N | 2 | nd | – | tfb |
| Amphipod virus 01 | 105,300 | 98 | 0 | N | 3 | 0 | – | – |
Contig IDs corresponding to Sulfurihydrogenibium viruses are listed in Supplementary Table 16.
UViG Uncultivated Virus Genome, tmk dTMP kinase, thyX thymidylate synthase (FAD), pcnB polyA polymerase, polA DNA polymerase I, dnaN DNA polymerase III subunit beta, dam DNA adenine methylase, ssb single-stranded DNA binding protein, dcm DNA (cytosine-5) methyltransferase I, tfb transcription initiation factor TFIIB, ccrM modification methylase, spoT GTP diphosphokinase, comEB dCMP daminase, dnaB replicative DNA helicase, polB DNA polymerase, rfcS replication factor C, small subunit, lig1 DNA ligase I, recA recombination protein RecA. Membrane transport gene content was detected in Thermofilum virus 04 (transitional endoplasmic reticulum ATPase VCP) and the Sulfurihydrogenibium plasmid (gspG, gspE, gspDsecretin, and a beta-barrel porin).
aTotal CRISPR hits via MinCED and CRASS (may include duplicate entries where both analyses detected the same CRISPR).
Metabolic reconstruction and biogeochemical cycling
Functional gene analysis showed that all predominant microbial community members are dependent on different species of sulfur (Fig. 2; Supplementary Table 4; Supplementary Table 1). The oxidation of reduced sulfur compounds (sulfide, elemental S, thiosulfate) is an important energy source for the highly abundant Sulfurihydrogenibium, which is consistent with well-established metabolic functions of this genus [47–50]. Sulfurihydrogenibium has been detected in high abundance at other sulfidic vent sites in shallower regions of YLake, including Inflated Plain and West Thumb [34], as well as terrestrial geothermal springs elsewhere in YNP [48, 49, 51, 52]. The biogeography of Sulfurihydrogenibium in YNP typically corresponds to high concentrations of dissolved sulfide and carbon dioxide, and a slightly sub-neutral pH (5–7) [34, 48]. pH is an important environmental control on Sulfurihydrogenibium biogeography because thiosulfate concentrations peak within a pH range of 5.5–6.8 [53], and thiosulfate is a key energy source for Sulfurihydrogenibium growth [49, 50]. Indeed, the Sulfurihydrogenibium MAG from the YLake Deep Hole encodes the SoxY carrier protein (Supplementary Table 4) for covalent binding of thiosulfate during thiosulfate oxidation [54], as well as genes required for the oxidation of sulfide to elemental sulfur (i.e., sqr: sulfide-quinone oxidoreductase, 3614X coverage) and elemental sulfur to sulfite (i.e., sdo: sulfur dioxygenase, 3580X coverage) (Supplementary Table 5). Group I Ni-Fe hydrogenases were also present at high coverage (3749–4165X), which confirmed previous reports that Sulfurihydrogenibium at hydrothermal vents in YLake utilize H2 as an energy source in addition to reduced sulfur compounds [34]. In contrast, other isolates of Sulfurihydrogenibium from terrestrial sites in YNP (e.g., S. yellowstonense and sp. Y03AOP1) do not encode hydrogenases [49, 55]; this is correlated with lower environmental concentrations of dissolved H2 measured in terrestrial sites. The concentrations of dissolved H2 at deep YLake vents (up to 25 µM) [40] are three orders of magnitude higher than values from terrestrial hot springs in YNP colonized by Sulfurihydrogenibium, such as Mammoth or Calcite Hot Springs (14–30 nM) [51, 52]. Sublacustrine hydrothermal vents in YLake thus expand the biogeography and functional diversity of Sulfurihydrogenibium to include hydrogen metabolism. Group I Ni-Fe hydrogenases have also been observed in isolates of Sulfurihydrogenibium obtained from terrestrial hot springs in the Azores, Portugal [47] (S. azorense) and Iceland (S. kristjanssonii) [56].
Three putative viral contigs associated with Sulfurihydrogenibium contained auxiliary metabolic genes that encode the phosphoadenosine phosphosulfate (PAPS) reductase protein (i.e., cysH). PAPS reductase is involved in the synthesis of sulfite from sulfate during assimilatory sulfate reduction. Viral cysH has also been observed in diverse environments globally, including marine cold seep sediments [14], bovine rumen [57], stratified sulfidic mine tailings [17], and deep freshwater lakes [16]. However, cysH homologs have recently been associated with CRISPR-Cas systems in viruses and thus in some cases may be viral CRISPR accessory genes [58] (discussion below). Sulfur oxidation genes have been detected in viral genomes from deep-sea hydrothermal plumes of the Eastern Lau Spreading Center (Western Pacific Ocean) and Guaymas Basin (Gulf of California) [4], but viral rdsr genes were not observed here. Genes for sulfur oxidation were also detected in other members of the YLake vent biofilm community including Sulfolobales, Thiomicrospiraceae, Comamonadaceae, and Sulfuricurvum, though in much lower relative abundance (Fig. 2). Elemental analysis demonstrated that the largest filaments (3–5 µm diameter, >100 µm length) accumulated elemental sulfur within the biofilm (Fig. 1E), which has been shown previously for members of the Thiomicrospiraceae [59]. However, this lineage is typically composed of smaller, single-cell sulfur oxidizers; the identity of the large, sulfur-accumulating filaments in YLake biofilms remains to be determined (Supplementary Discussion).
Sulfurihydrogenibium spp. at sublacustrine thermal vents in YLake [33–35] exploit high concentrations of CO2, reduced sulfur and hydrogen (Fig. 2). The oxidation of sulfur and/or hydrogen provides reducing equivalents for carbon fixation via the reverse tricarboxylic acid (rTCA) cycle. Marker genes for the rTCA cycle (i.e., ATP citrate lyase alpha and beta subunits) were detected in the Sulfurihydrogenibium MAG with extremely high sequence coverages of 3828X and 3379X, respectively (Supplementary Table 5), which demonstrates that Sulfurihydrogenibium spp. are likely the dominant primary producers in the vent biofilm. Indeed, previous work at deep YLake vents demonstrated that 16S rRNA gene sequences from Sulfurihydrogenibium coincided with maximum CO2 fixation rates of 9 µM C/h [37]. This observation was made at temperatures between 50 and 60 °C, which is consistent with the predicted temperature optimum of 59 °C for the Sulfurihydrogenibium MAG (Table 1). Other members within the community have putative abilities to fix carbon via the Calvin–Benson–Bassham (Comamonadaceae), Wood–Ljungdahl (Thermodesulfobacterium), and 3-Hydroxypropionate/4-Hydroxybutyrate (Sulfolobales) pathways. However, the much lower abundance of these community members indicates that Sulfurihydrogenibium spp. provides most of the organic carbon generated in the vent biofilm.
Genomic variability in Sulfurihydrogenibium
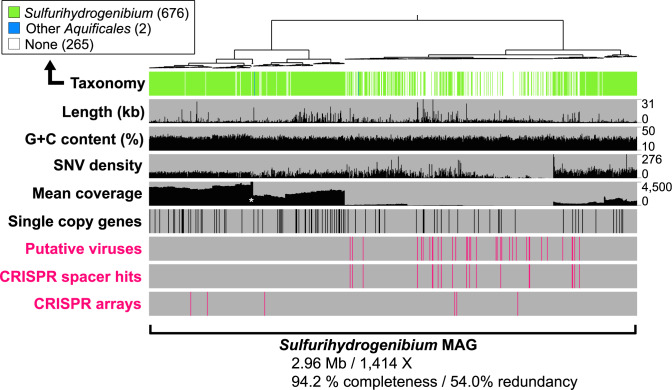
Sulfurihydrogenibium spp. in YLake thermal vents exhibited significant genomic variability as indicated by (i) a wide range of contig coverage values (~50–5000X) (Fig. 4; Supplementary Table 6), (ii) high single nucleotide and single amino acid variants per kilobase (i.e., SNV and SAAV “density”) (Supplementary Table 7), and (iii) multiple Sulfurihydrogenibium single-copy gene (SCG) variants (Supplementary Table 8; Supplementary Figure 2). High mean sequence coverage (1414X) of this MAG revealed micro-diversity among closely related sub-populations (Fig. 4). A nearly full suite of SCGs [60] were detected, resulting in an estimated genome completeness of 94.2%; however, the estimated redundancy was 54.0% (Table 1) due to homologous sequence variants of SCGs that assembled on separate contigs (discussed below). Sequence down-sampling protocols (prior to assembly) reduced MAG redundancy but resulted in lower genome completeness and a loss of sequence content related to the Sulfurihydrogenibium (Fig. 4; Supplementary Table 9; Supplementary Discussion). Removal of lower coverage contigs also decreased redundancy but decreased completeness to below 50%. Genomic redundancy in high-abundance microbial MAGs demonstrates the importance of reporting pangenomes [61, 62], which more comprehensively describe the genetic repertoire of microorganisms in nature. However, due to potential impacts of high coverage on assembly quality, we also include a low-redundancy (only 7%) Sulfurihydrogenibium MAG obtained using randomly subsampled reads (10% of reads) (Supplementary Genome File).
Fig. 4. Taxonomic identity and sequence character of contigs within the Sulfurihydrogenibium MAG.
Contig sequences were clustered based on mean coverage values (shown as nX) and tetranucleotide frequencies. Sulfurihydrogenibium contigs (n = 680) exhibited consistent G + C content (%) with a wide range of coverage values from 32 to 4500X (white asterisk indicates a group of five contigs with coverage values >4500X). 35 putative viruses contained within the same tetranucleotide cluster are indicated, and line up with positive BLASTn hits to CRISPR spacer sequences (i.e., “CRISPR Spacer Hits”; Supplementary Table 3). Six complete CRISPR arrays detected in the assembled metagenome (Supplementary Table 3) are also indicated [SNV density = single nucleotide variant density, per kilobase].
Examination of redundant SCG variants showed that they were closely related to Sulfurihydrogenibium spp. (Supplementary Table 8), which also confirmed that the sequence redundancy did not represent contamination from unrelated taxa. For example, deduced protein sequences from nine SCG variants of GidB (i.e., 16S rRNA guanine(537)-N(7)-methyltransferase) all had a top amino acid sequence identity (from 96 to 99%) to Sulfurihydrogenibium sp. Y03AOP1 [55]. GidB sequence variants also formed a closely related monophyletic cluster with S. yellowstonense (Supplementary Figure 2A). Nine variant copies of DNA polymerase III (subunit beta II) were also detected in the MAG and seven of them exhibited highest sequence similarities to various Sulfurihydrogenibium spp., ranging in similarity from 30 to 77% (expect value <1 × 10−10; Supplementary Table 8). The remaining two sequences had no significant similarities to the sequence database, but all nine copies formed a monophyletic clade related to Sulfurihydrogenibium spp. (Supplementary Figure 2B). Sequence variants of phylogenetically related SCGs indicate genomic variability that could arise from closely related populations of Sulfurihydrogenibium.
SNV and SAAV densities (Supplementary Table 7) and transposase occurrences (Table 1) provided further evidence of genomic variability in the Sulfurihydrogenibium MAG. A high SNV density of 58.5 SNVs per kilobase (SNVs kb−1) was observed for the Sulfurihydrogenibium MAG compared to an average of 20.8 SNV kb−1 for other vent community members. SNV density remained high (52.3 SNV kb−1) after down-sampling of metagenomic reads at 10%, which resulted in a lower-redundancy (7%) Sulfurihydrogenibium MAG (Supplementary Genome File; Supplementary Table 9), demonstrating that SNV density is not an artifact of high redundancy. Amino acid variants were also examined to determine whether the observed genomic variability translated to changes in amino acid content. The Sulfurihydrogenibium MAG (subsampled at 10%) represented the second-highest SAAV density at 38.9 SAAV kb−1, while the other MAGs from the same metagenome had a lower average SAAV density of 13.0 SAAV kb−1. Finally, the Sulfurihydrogenibium MAG contained a significant number of transposases (n = 37) (Table 1), which suggests the potential for gene movement among related sub-populations.
Host–virus interactions
Differences in viral replication and dispersal strategies in natural environments have significant effects on the ecology of microbial communities. Numerous contigs (840) containing predicted virus sequence were detected among the hydrothermal vent metagenomes and nearly all (98.7%) were ranked as free-living and/or actively replicating viral particles (VirSorter [63] categories 1–3; Supplementary Table 10). Lytic viruses kill their host on a short time scale, whereas lysogenic viruses integrate into host genomes and have a period of latency prior to induction of a lytic cycle. While lytic viruses act mainly as an antagonistic force on their microbial hosts, lysogenic viruses can act as a positive force by providing ecological benefits such as horizontally acquired genes and protection from super-infection (see review [64]). The predominance of sequences predicted as free-living viral particles in YLake biofilms suggests that lytic viruses contribute significantly to selective pressures experienced by the vent community.
Tetranucleotide frequency (TNF) analysis [42, 43] and CRISPR [65] sequence matches can be used to link viruses with specific hosts. Viral contigs >20 kbp were associated with the primary thermophilic organisms in the biofilm community (i.e., Sulfurihydrogenibium, Thermofilum) based on nucleotide signatures and CRISPR spacer matches (Table 2). Most variable spacers (95.1%, n = 288) from microbial CRISPR arrays had high-identity matches (0–1 mismatches) to putative viral sequences within the same TNF cluster, which established a direct linkage among viruses and hosts (Supplementary Table 3). Together, these observations demonstrate that thermophilic archaea and bacteria within the vent community are targeted by a variety of mostly lytic viruses against which numerous CRISPR systems have been developed.
Putative Sulfurihydrogenibium viruses were examined extensively to investigate potential effects of viral selective pressures and/or gene augmentation related to the predominant member of the biofilm. Phylogenetic analysis of viral capsid proteins from nine different Sulfurihydrogenibium contigs revealed a monophyletic cluster that was closely related to an Uncultivated Virus Genome (UViG) from Octopus Hot Spring (YNP, Wyoming) [66] (Fig. 3B), another high-temperature habitat containing filamentous Aquificales biofilms [51]. Five putative Sulfurihydrogenibium viruses (Sulfurihydrogenibium viruses 02, 05, 06, 10, 12) were identified as circular UViGs and linked to the Sulfurihydrogenibium host by CRISPR spacer matches (Table 2; Supplementary Table 3). These Sulfurihydrogenibium UViGs exhibit a highly syntenic set of core genes very similar to the proposed “Pyrovirus” genus [66], including several structural (capsid, terminase, portal protein, and tail collar protein), and functional (Aquificae-like DNA polymerase, helicase, and a Cas4 homolog) proteins (Fig. 3C, Supplementary Table 11). One of the viral genomes (Sulfurihydrogenibium virus 02) showed less gene synteny and formed a separate branch outside of the primary Sulfurihydrogenibium virus cluster (Fig. 3B). The 38-kbp genome of Virus 02 contained 66 loci that matched to spacer sequences within Sulfurihydrogenibium CRISPR arrays (Table 2, Supplementary Table 3), which may signify relatively recent or frequent infection [67] and establishes that this virus was targeted extensively by the host immune system.
Viral taxonomy cannot be resolved from capsid phylogeny alone [68], therefore Sulfurihydrogenibium viral sequences were analyzed for differential protein occurrences associated with characterized viral lineages [69] (Supplementary Figure 3). Virus 02 contained 8 deduced structural head/neck proteins, including a sheath protein, which provides basis for assignment to Myoviridae type I (Fig. 3B), and Sulfurihydrogenibium Virus 03 was related to the Siphoviridae based on analysis with BLAST (Basic Local Alignment Search Tool [70]) (Supplementary Table 2). These results indicate that at least three taxonomically distinct viruses (Myoviridae Type I, Siphoviridae, and Ca. Pyrovirus) are likely capable of lysing Sulfurihydrogenibium cells. Further evidence for an antagonistic, “Kill-the-Winner” relationship [71] between Sulfurihydrogenibium and its viruses included the presence of putative viral anti-CRISPR genes such as PAPS reductases, Cas4 nuclease homologs, and neighboring DEAD-like helicases. PAPS reductases are thought to be involved in sulfur assimilation but may also serve as CRISPR-Cas accessory proteins in some viruses [58]. Helicase-nuclease fusions are also hypothesized to serve defense functions [72] (Supplementary Discussion). The presence of a highly conserved gene neighborhood in complete genomes of Sulfurihydrogenibium viruses that is likely dedicated to defense mechanisms suggests that antagonistic relationships including lytic infection are likely a dominant host–virus relationship in YLake vent biofilms. This is further supported by the low percentage (1.3%) of prophage sequences detected by VirSorter [63] (Supplementary Table 10) and the lack of integrase genes present in Sulfurihydrogenibium viruses (Supplementary Table 11). The predominance of lytic viruses at YLake advective-flow vents contrasts with the prevalence of lysogenic viruses at diffuse-flow seafloor hydrothermal vents [7, 73], and may be more similar to increased phage to bacteria ratios observed in host-associated mucus layers [74, 75]. Identification of UViGs associated with Sulfurihydrogenibium (this study) and other Aquificales [66] using metagenomic approaches provides impetus for future isolation and characterization of the life history strategies of thermophilic viruses in YLake vent biofilms.
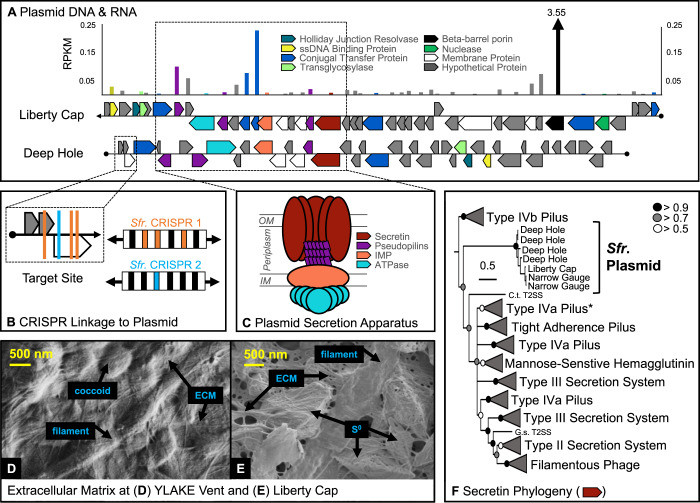
CRISPR-targeted plasmid and biofilm processes in Sulfurihydrogenibium
The Sulfurihydrogenibium MAG also has two CRISPR systems that target plasmid DNA, which contains genes for a TFF system [76] (Fig. 5; Table 2; Supplementary Table 3; Supplementary Table 12). Mobile genetic elements including plasmids are sometimes targeted by host CRISPR arrays for modulating DNA exchange [77–79]. Four CRISPR spacer sequences from two separate CRISPR arrays linked this plasmid with the Sulfurihydrogenibium MAG (Fig. 5). The plasmid is 31 kbp long and contains 40 genes. Highly similar sequences to this plasmid were also detected in several other geothermal habitats where Sulfurihydrogenibium is the predominant organism, including other vent sites in YLake, as well as Mammoth and Liberty Cap Hot Springs (YNP) [34, 48]. A highly syntenic 40-kbp plasmid containing 51 genes was recovered from the Liberty Cap filamentous community, and a metatranscriptome from this site showed high transcript levels of several genes on this plasmid.
Fig. 5. Early evolved filament system in Sulfurihydrogenibium spp. is encoded and transcribed from CRISPR-targeted plasmid.
A Plasmid sequences from the Deep Hole and Liberty Cap are compared and RNA expression is reported as reads per kilobase per megabase (RPKM) for the Liberty Cap sequence. Hash marks in the beta-barrel porin bar indicate RNA expression of 3.6, beyond the limits of the y-axis. B Four CRISPR target sites are indicated along the plasmid sequence from the Deep Hole. C A conserved gene neighborhood in both plasmid sequences encodes a complete TFF system [OM outer membrane, IM inner membrane, IMP inner membrane platform]. D, E Scanning electron micrographs of biofilm matrices at (D) the Deep Hole and (E) Liberty Cap. F Bayesian phylogeny of deduced secretin protein sequences from Sulfurihydrogenibium plasmids detected in three regions of YNP. Posterior probabilities are indicated by circles at nodes. Asterisk indicates the clade containing secretin genes from the genomic (non-plasmid) DNA of Sulfurihydrogenibium.
TFF systems contain homologous components (secretin proteins, pseudopilins, inner membrane platform, and ATPase components) and serve diverse functions, including protein secretion, adhesion, motility, and conjugation for DNA exchange [76, 80]. Phylogenetic analysis of the deduced secretin protein from the Sulfurihydrogenibium plasmid revealed a new, deeply rooted clade, which split the previous phylogenetic root (Type IVb Pilus) from all other entries (Fig. 5F). Sulfurihydrogenibium plasmid genes also encode a ssDNA binding protein, Holliday junction resolvase, multiple conjugal transfer proteins, and a nuclease (Fig. 5A), which suggests that the TFF system may be involved in conjugation and DNA exchange (Supplementary Discussion). Transcriptomic data from Liberty Cap (Mammoth Hot Springs) showed that all components of the plasmid-conferred Sulfurihydrogenibium TFF system were expressed, as well as an outer-membrane beta-barrel porin (3.55 RPKM) (not part of the TFF system, Fig. 5A), that may promote the diffusion of smaller substrates through the outer membrane [81].
Protein secretion, adhesion, motility, and the production of a thick and resilient extracellular matrix (ECM, Fig. 5D, E) are likely crucial processes for microbial colonization in highly turbulent environments (Supplementary Video). Several filament systems involved in key biofilm processes were identified (TXSScan) [82] in the Sulfurihydrogenibium MAG (in addition to the plasmid, above) (Supplementary Table 13). Multiple copies of 11 genes encoding all necessary components of a complete flagellum structure were identified, and a near-complete type IVa pilus for twitching motility [83] was detected and confirmed by phylogenetic analysis of the secretin protein sequence (see asterisk in Fig. 5F). Components of this type IVa pilus were also highly transcribed in a recent study of Sulfurihydrogenibium at Mammoth Hot Springs and likely interact with ECM for stronger adhesion [48]. Multiple copies of genes encoding a complete type I secretion system were also found, which suggests that protein secretion can occur in a single step across the inner and outer membranes [84]. Incomplete gene sets were also detected for the “Tight-adherence” complex and the type II, type III, and type VI secretion systems. Together, these secretion systems perform functions involved in pili production and “tenacious biofilm formation” (Tad) [85], secretion of periplasmic proteins across the outer membrane (T2SS) [86], and delivery of effector proteins directly into neighboring cells (T3SS and T6SS) [87, 88]. Nine out of 14 accessory genes of the type IV secretion system (F-type) were observed, and may be involved in conjugation or protein secretion [89]. Homologous components of varying secretion systems may be interchangeable; consequently, incomplete systems detected in silico may interact to perform a unified function in vivo [90, 91].
Summary
Members of the deeply rooted bacterial phylum Aquificae colonize high temperature, vapor-dominated sublacustrine thermal vents in YLake and form extensive filamentous structures intermixed with elemental sulfur. These highly cohesive streamer biofilms experience steep temperature and geochemical gradients, and they provide habitats for other archaeal and bacterial hyperthermophiles as well as some mesophiles with optimum growth temperatures as low as 22 °C. The composition of microbial communities that thrive in these types of thermal vents (high carbon dioxide, hydrogen, and sulfide) are highly consistent across different vent sites not only within the Deep Hole near Stevenson Island, but also those sampled in a prior study at the Northern Dome (also known as Inflated Plain, YLake) [34]. Geochemical forcing due to high amounts of different sulfur species results in community functions that are intimately linked with sulfur cycling (both oxidation and reduction reactions). Moreover, evidence of viral interactions in the highly abundant thermophiles (Aquificales, Thermoprotei) indicates that numerous lytic viruses play a direct role in modulating carbon fixation and sulfur cycling in both archaeal and bacterial domains, even if due primarily to effects on host turnover. Finally, the presence of multiple secretion systems in the Sulfurihydrogenibium MAG and plasmid is consistent with direct microscopic observations of an extensive extracellular matrix, which is critical to the survival and development of filamentous biofilms common in the extremely hot, turbulent vent environments.
Materials and methods
Site selection and hydrothermal vent sampling
A remotely operated vehicle (ROV) Yogi and research vessel (R/V) Annie (operated by the Global Foundation for Ocean Exploration; https://www.engineeringfordiscovery.org/) were used to locate and sample microbial biofilm colonies at the deepest and hottest hydrothermal vents in YLake near Stevenson Island (referred to as the Deep Hole). Filamentous biofilm communities were sampled in 2016 and 2017 from three hydrothermal vents (2016_B01, 2016_B02, 2017_B01) using a manipulator arm and the ROV Yogi suction sampler (Supplementary Table 14). Biofilm “streamer” material was captured in a canister and samples retrieved aseptically upon ROV return to R/V Annie. Temperatures of hydrothermal vent fluid and ambient lake water were measured for several minutes with a ROV temperature probe and detailed chemical analyses of these vent fluids were performed in collaborative work [33, 35].
Elemental analysis and electron microscopy
Subsamples of microbial streamer material were fixed in 2% glutaraldehyde on site for microscopic and elemental analysis in the Imaging and Chemical Analysis Laboratory (Montana State University). Fixed biofilm samples were mounted and washed with sterile DI H2O on 13-mm diameter 0.2 µm polycarbonate filters (Millipore), then powder-coated with Iridium prior to imaging using a Zeiss SUPRA 55VP field-emission scanning electron microscope. Sulfur and nitrogen were identified using energy-dispersive X-ray spectroscopy (Bruker X-Flash detector) of Ir-coated samples with an integrated Auger nanoprobe field emission electron microscope (PHI 710).
DNA isolation, metagenome sequencing, QC, and assembly
Three hydrothermal vent streamer samples (2016_B01, 2016_B02, and 2017_B01) were split into five fractions for DNA isolation. Streamer material from the 2016_B02 sample was cleaned by passing through soft 0.1% sterile agar followed by sterile 1X PBS, splitting the sample into two separate subsamples: 2016_B02_str, which represented agar-cleaned streamer filaments and 2016_B01_sed, which represented smaller filamentous materials and surrounding sediments left behind after cleaning. 2017_B01 was divided into two biological replicates, 2017_B01a and 2017_B01b. DNA was isolated from five subsamples originating from three vents (2016_B01, 2016_B02_str, 2016_B02_sed, 2017_B01a, 2017_B01b) with the MP Biomedicals FastDNA Spin Kit for Soil according to the manufacturer’s protocol. DNA concentrations were measured with a Qubit fluorometer and sent to the Census of Deep Life (at Marine Biological Laboratory, Woods Hole, MA) for shotgun metagenomic sequencing (Illumina NextSeq) in a 2 × 150 paired-end run with dedicated read indexing and demultiplexed with bcl2fastq.
Raw fastq files were filtered for quality with the “Illumina-utils” toolkit with default parameters [92]. Spades (v3.11.1) was used with the “--meta” option to separately assemble 2016_B01, 2016_B02_str, and 2016_B02_sed, while MEGAHIT (v1.1.2) was used to co-assemble the biological replicates, 2017_B01a and 2017_B01b, resulting in four total metagenome assemblies [93, 94]. Bowtie2 (v2.2.6) was used to map quality-filtered short reads back to assembled contigs [95]. We used t-stochastic neighbor-embedding to cluster metagenome contigs based on tetranucleotide frequencies (TNF) [23, 96] (Supplementary Figure 4) and we refined each cluster by examination of mean coverage values and sequence composition in anvi’o (v5.3) [97] to achieve estimated redundancy values <10% based on detection of single-copy genes for Archaea [98] and Bacteria [60].
Curation of the Sulfurihydrogenibium MAG
Regardless of assembly or binning methodology, cases existed in which microbial MAGs for Sulfurihydrogenibium were recovered with very high coverage (>1000X) but very low completeness (<50%). To resolve this, we identified short contigs (1000–3000 bp) that were taxonomically related to Sulfurihydrogenibium using Basic Local Alignment Search Tool for nucleotide sequences (BLASTn) [70] by comparing them to four reference genomes for Sulfurihydrogenibium: S. yellowstonense [49], S. azorense [47], S. Y03AOP1 [55], and S. subterraneum [50]. We identified positive hits with e values <1E–20 and nucleotide similarities >90%, re-uploaded these shorter contig sequences with the original bin to our tSNE clustering analysis, and redefined a new cluster with overlapping Sulfurihydrogenibium-related short sequences >1000 bp combined with the original longer sequences >3000 bp. This process resulted in a new MAG with 989 contigs and an estimated completeness of 94.2%. We decontaminated this Sulfurihydrogenibium MAG by removing 81 contigs that were either (i) related by BLASTn to a different taxonomic lineage at >80% identity with an alignment length of >500 bp, (ii) related by BLASTn to a different member of the Aquificales at >90% identity with an alignment length of >500 bp, (iii) related to a different taxonomic lineage by Centrifuge taxonomy software [99], or (iv) contained a single-copy gene with BLASTp relatedness to a different taxonomy lineage. We also performed four random subsampling [92, 100] and two coverage-based normalization techniques (bbnorm function within the BBMap program [101]) to achieve high completeness and low redundancy. Subsampling quality-filtered reads at 10% was the only case that resulted in redundancy <10% (Supplementary Table 9; Supplementary Genome File) but we confirmed the loss of Sulfurihydrogenibium-related sequence during the process.
Identification of related MAGs from different assemblies
All genomic clusters from the four assemblies were then compared for average nucleotide identity (ANI) with PyANI [102] to determine overlapping MAGs among assemblies. Replicate MAGs were designated by >95% ANI [103], and the replicate MAG with the highest estimated genome completeness was selected as the representative MAG for downstream sequence analyses (Table 1). We used anvi’o [97] to calculate genomic characteristics, including G + C content and standard deviation, length, N50, completeness, redundancy, and mean coverage and standard deviation. Three methods were used to determine taxonomy. Initial taxonomic identification was determined with the BLASTn [70] of every contig within every MAG to the nucleotide sequence database from the National Center for Biotechnology Information (NCBI). Majority consensus of contig hits was used to formulate taxonomic hypotheses for each MAG, which were tested with robust phylogenomic analyses (below). GTDB-Tk [104] was then used to confirm taxonomy (Supplementary Table 15). All three methods (BLASTn of binned contigs, phylogenomics of concatenated ribosomal proteins, and GTDB-Tk) agreed on taxonomic identity of the MAGs discussed in this report. Temperature optima were predicted from deduced amino acid content using previously established methods [41, 105].
Phylogenomic analyses of archaea and bacteria
Hidden Markov Models (HMM) were downloaded from the protein families (PFAM) database [106] for the following 16 ribosomal proteins: L27a, S10, L2, L3, L4, L18p, L6, S8, L5, L24, L14, S17, S3c, L22, S19, L16. Custom HMMs were used for archaeal ribosomal proteins. HMMer was used to scan MAGs for HMM hits and ribosomal protein sequences were extracted from each MAG. Individual ribosomal proteins were aligned with MAFFT (--maxiterate 1000 --localpair --nomemsave) and positions with >50% gaps were trimmed with trimAl. Following the concatenation of 16 alignments, MrBayes [107] (version 3.2.7a; MPI) was used for Bayesian inference analysis (2 million generations, 0.25 burn-in fraction, 4 parallel chains, 4 rate categories, and invariable gamma models for rate variation, LG model with empirical amino acid frequencies and heating factors set to 0.125).
Phylogenetic analyses single-copy gene variants
Nine variant copies of two single-copy genes—GidB and DNA polymerase III beta II subunit—were detected by HMM in the Sulfurihydrogenibium MAG. Deduced amino acid sequences were extracted for each, aligned with MAFFT (separately for GidB and DNApoly3beta2), trimmed with trimAl [108] v1.4.rev22, and phylogenetically compared to reference sequences from other Aquificae, Epsilonproteobacteria, and Thermotogales with MrBayes [107]. Both analyses ran for 600,000 generations until standard deviation of split frequencies reached 0.0059 for GidB and 0.0020 for DNApoly3beta2 (0.25 burn-in fraction, 8 parallel chains, 8 rate categories, and invariable gamma models for rate variation, LG model with empirical amino acid frequencies). Both trees were viewed and edited in iTOL [109].
Identification of viral contigs and host–virus interactions
VirSorter [63] was used to scan all assembled contigs against the VIROME database [110], and to identify viral gene content and putative viral contigs. Putative viral contigs that clustered with primary MAGs based on tetranucleotide frequency signatures, as well as other viral contigs with lengths >20 kbp, were compared by BLAST analysis against four databases: NCBI nt (non-redundant), NCBI aa (non-redundant), UniProt Viral nt, UniProt Viral aa (Supplementary Table 2). We assigned viral taxonomy at the family level based on BLAST results to the viral subset of the nr database with an e-value cutoff of 10−3, selecting the most common occurrence of a viral family as the representative taxonomy for each contig. In the event of a tie, i.e., the same number of hits to two or three different viral families, lowest e values were used to determine viral taxonomy. Relative abundance of viral families among all four assemblies was determined by normalizing mean contig coverage values to assembly read depth; normalized coverage values were subtotaled for each viral family for cumulative normalized coverage and presented in Fig. 3. Relationships between viruses and hosts were assigned by (i) overlapping tetranucleotide signatures between viral and host sequences [42, 43], (ii) taxonomic hits of viral protein sequences to microbial entries in public databases [44], and/or (iii) sequence identity matches between host CRISPR spacers and viral genomes.
CRISPR arrays were identified using assembled and unassembled metagenomic sequences with MinCED (v0.4.2) [111] and Crass [112] (v0.3.12), respectively, with default parameters. Thirty-four CRISPR arrays were detected by the MinCED analysis and we used BLASTn to identify CRISPR spacer sequences that match putative viral sequences (with <2 bp mismatches) in the four metagenome assemblies. We also searched for CRISPR arrays by running Crass on quality-filtered metagenomic reads (unassembled) and used the associated “crisprtools” to extract repeat and spacer sequences. BLAST was used to create a custom nucleotide database for all putative viral contigs identified by VirSorter (above) and the ca. 29,000 spacer sequences from the Crass analysis were compared by BLASTn to this database (allowing <2 bp mismatches) revealing 382 matches.
Metabolic potential of archaea, bacteria, and viruses
We used anvi’o (v5.3) [97] to scan for open reading frames with Prodigal [113] and to compare deduced protein sequences with NCBI Clusters of Orthologous Groups (COGs) [114]. After binning metagenomic sequences into MAGs, METABOLIC software (v2) was used to identify enzymes involved in metabolic pathways with specific influences on biogeochemical cycles [115]. METABOLIC output files were examined and specific genes for sulfur, hydrogen, and carbon metabolism were reported. Anvi’o was further used to extract mean coverage values for functional genes detected within metagenomic sequence.
Examination of Sulfurihydrogenibium viruses
A curated group of putative Sulfurihydrogenibium-infecting viruses was created using the initial list identified via tetranucleotide frequency from each of the four individual metagenomes. This list was refined by removing viral contigs with <10 genes (or <10 kbp), putative viruses that were determined to be plasmids, and viral contigs that received a low-confidence score of three or six from VirSorter [63]. The list for the four individual metagenomes was then condensed by aligning using MAFFT [116] v7.407. Contigs with >95% identity were considered identical for analysis purposes. Individual viral contigs were annotated by identifying putative ORFs using MetaGeneAnnotator [117]. The predicted ORFs were translated and used to search NCBI non-redundant database, the Conserved Domain Database (CDD), TIGRFam [118], Pfam [119], SMART [120], PRK [121], COG [114], and InterPro [122] databases. Functional annotations were made if e-values to databases were <10e−5. In addition, the HHpred/HHsearch algorithm [123] was used to perform profile-profile comparisons of the head-neck region of viral contigs on the VIRFAM platform [69]. Phylogenetic analysis of the major capsid protein (cl22542) was performed by aligning viral capsid proteins from to nearest neighbors via BLASTp database searching. An alignment was built using MAFFT and a phylogenetic tree was created using RAxML [124] and evaluated with 1000 bootstrap replicates.
Detecting homologous plasmid DNA and RNA at other sites
The deduced amino acid sequence for secretin (i.e., Fig. 5F) was compared by BLASTp analysis to deduced proteins from publicly available metagenomes from Liberty Cap and Narrow Gauge hot springs in YNP [48]. This method identified two additional plasmid contigs, one from each hot spring metagenome, that were similarly annotated with Holliday Junction Resolvases, ssDNA binding proteins, and conjugation proteins. This observation was further confirmed by a matching spacer sequence (1 mismatch) from the Sulfurihydrogenibium CRISPR array that targeted the same plasmid at the Liberty Cap site, as indicated by BLASTn comparison to the hot spring metagenome. The associated Liberty Cap metatranscriptome (JGI GOLD ID Gp0055116) was used to calculate reads per kilobase per megabase (RPKM) values for genes along the plasmid contig.
Determining putative functions of plasmid-based genes
Initial gene annotations were performed in anvi’o [97] with the NCBI COG database (Supplementary Table 5), and each deduced protein sequence from the plasmid genome was also compared by BLASTp [70] to the NCBI non-redundant database [125] (Supplementary Table 12). Components related to a TFF system were further scanned with ConjScan [126] and TXSScan [127] for functional identification with inconclusive results. The deduced secretin protein sequence was aligned by MAFFT [116] and phylogenetically compared with MrBayes [107] to previously published sequences [80]. This formed a deeply rooted cluster distantly related to all other sequences and precluded any functional information for the Sulfurihydrogenibium plasmid secretin. MAFFT alignment was performed with default parameters and MrBayes phylogenetic analysis was run for 3,000,000 generations with a fixed amino acid model and burn-in fraction of 0.25.
Supplementary information
Acknowledgements
The authors appreciate support from the National Science Foundation (Subaward A101357, WHOI Project 81636100 (LJM and WPI); DEB 1950770 (WPI and MD) and EPSCoR1736255 (LJM and MWF)), the Montana Agricultural Experiment Station (MAES 911300; WPI), the National Institutes of Health IDeA Program (COBRE grant GM110732; MD), and by the US Department of Energy—Ecosystems and Networks Integrated with Genes and Molecular Assemblies (DE-AC02–05CH11231; MWF). Metagenome sequencing was performed by the Census of Deep Life (Deep Carbon Observatory). Computations were performed on the Hyalite High-Performance Computing System (MSU Information Technology). Electron microscopy and elemental analyses were performed at the Montana Nanotechnology Facility, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the NSF (ECCS-2025391). We also appreciate significant synergistic contributions from the HD-YLake project and support for KML (NSF EAR 1514865) in sampling and data sharing efforts. We thank the Global Foundation for Ocean Exploration and Capt. Dave Lovalvo for crucial shipboard support and ROV expertise, which made it possible to sample this extreme environment. Finally, we are grateful for helpful discussions with Drs. S. Abby, R. Denise, E. Rocha, Z. Jay, M. Myers, and A. Segall. Research in Yellowstone Lake (YNP, Wyoming, USA) was conducted under permit YELL-2016/17-SCI-7018. The sample from Liberty Cap (Mammoth Hot Springs, YNP) was collected in collaboration with Dr. B. Fouke under YNP research permit to WPI (YELL-2011-SCI-5686).
Author contributions
LJM and WPI designed the study. LJM, ODN, and WPI wrote the manuscript. WPI acquired grant funding to support the investigation. LJM and DBR performed metagenome binning. LJM analyzed MAGs for genomic characteristics and metabolic potential. ODN scanned metagenomes for viral content, analyzed putative viral sequence, and annotated UViGs. LJM and ODN scanned and analyzed metagenomes for CRISPR information. MD constructed phylogenomic trees from concatenated protein alignments. MD and LJM calculated predicted optimal growth temperatures and performed metagenomic subsampling. LJM constructed phylogenetic trees of SCGs and annotated plasmid DNA. WPI performed scanning electron microscopy and LJM assisted in elemental analyses. KML assisted in sampling YLake biofilms, recovered ROV data and videos, and created maps of YLake. MWF assisted in metabolic analyses. All authors reviewed and edited the manuscript.
Data availability
Genomic sequence information for this investigation can be found in the NCBI database under BioProject PRJNA669531 and BioSamples SAMN16454225, SAMN16454373, SAMN16454375, SAMN16454377, and SAMN16454376. Quality-filtered short reads used for single metagenomic assemblies are available in the NCBI Sequence Read Archive as SRR12852619 (2016_B01), SRR12852620 (2016_B02_str), and SRR12852618 (2016_B02_sed). Quality-filtered short reads used for coassembled metagenomes are available in the NCBI Sequence Read Archive as SRR12852621 (2017_B01_S1) and SRR12852622 (2017_B01_S2).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luke J. McKay, Email: mcgucas@gmail.com
William P. Inskeep, Email: binskeep@montana.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01132-4.
References
- 1.Corliss JB, Dymond J, Gordon LI, Edmond JM, von Herzen RP, Ballard RD, et al. Submarine thermal springs on the galápagos rift. Science. 1979;203:1073–83. doi: 10.1126/science.203.4385.1073. [DOI] [PubMed] [Google Scholar]
- 2.Jannasch HW, Mottl MJ. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–25. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 3.Dick GJ. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol. 2019;17:271–83. doi: 10.1038/s41579-019-0160-2. [DOI] [PubMed] [Google Scholar]
- 4.Anantharaman K, Duhaime MB, Breier JA, Wendt KA, Toner BM, Dick GJ. Sulfur oxidation genes in diverse deep-sea viruses. Science. 2014;344:757–60. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 5.Teske A, Reysenbach A-L. Editorial: Hydrothermal microbial ecosystems. Front Microbiol. 2015;6:884. doi: 10.3389/fmicb.2015.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He T, Li H, Zhang X. Deep-sea hydrothermal vent viruses compensate for microbial metabolism in virus-host interactions. mBio. 2017;8:e00893-17. doi: 10.1128/mBio.00893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson SJ, Cary SC, Williamson KE, Helton RR, Bench SR, Winget D, et al. Lysogenic virus–host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2008;2:1112–21. doi: 10.1038/ismej.2008.73. [DOI] [PubMed] [Google Scholar]
- 8.Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 2018;9:4999. doi: 10.1038/s41467-018-07418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhillon A, Goswami S, Riley M, Teske A, Sogin M. Domain evolution and functional diversification of sulfite reductases. Astrobiology. 2005;5:18–29. doi: 10.1089/ast.2005.5.18. [DOI] [PubMed] [Google Scholar]
- 10.Heinen W, Lauwers AM. Organic sulfur compounds resulting from the interaction of iron sulfide, hydrogen sulfide and carbon dioxide in an anaerobic aqueous environment. Orig Life Evol Biosph. 1996;26:131–50. doi: 10.1007/BF01809852. [DOI] [PubMed] [Google Scholar]
- 11.Nisbet EG, Sleep NH. The habitat and nature of early life. Nature. 2001;409:1083–91. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Buick R, Canfield DE. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature. 2001;410:77–81. doi: 10.1038/35065071. [DOI] [PubMed] [Google Scholar]
- 13.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–28. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Pan D, Wei G, Pi W, Zhang C, Wang J-H, et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15:2366–78. doi: 10.1038/s41396-021-00932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mara P, Vik D, Pachiadaki MG, Suter EA, Poulos B, Taylor GT, et al. Viral elements and their potential influence on microbial processes along the permanently stratified Cariaco Basin redoxcline. ISME J. 2020;14:3079–92. doi: 10.1038/s41396-020-00739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki Y, Nishimura Y, Yoshida T, Ogata H, Nakano S-I. Genome-resolved viral and cellular metagenomes revealed potential key virus-host interactions in a deep freshwater lake. Environ Microbiol. 2019;21:4740–54. doi: 10.1111/1462-2920.14816. [DOI] [PubMed] [Google Scholar]
- 17.Gao S-M, Schippers A, Chen N, Yuan Y, Zhang M-M, Li Q, et al. Depth-related variability in viral communities in highly stratified sulfidic mine tailings. Microbiome. 2020;8:89. doi: 10.1186/s40168-020-00848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 19.Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2020;18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 20.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 21.Weitz JS, Stock CA, Wilhelm SW, Bourouiba L, Coleman ML, Buchan A, et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015;9:1352–64. doi: 10.1038/ismej.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weitz JS, Wilhelm SW. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep. 2012;4:17. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay LJ, Dlakić M, Fields MW, Delmont TO, Eren AM, Jay ZJ, et al. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat Microbiol. 2019;4:614–22. doi: 10.1038/s41564-019-0362-4. [DOI] [PubMed] [Google Scholar]
- 24.Huang H-H, Lin F-C, Schmandt B, Farrell J, Smith RB, Tsai VC. Volcanology. The Yellowstone magmatic system from the mantle plume to the upper crust. Science. 2015;348:773–6. doi: 10.1126/science.aaa5648. [DOI] [PubMed] [Google Scholar]
- 25.Farrell J, Smith RB, Husen S, Diehl T. Tomography from 26 years of seismicity revealing that the spatial extent of the Yellowstone crustal magma reservoir extends well beyond the Yellowstone caldera. Geophys Res Lett. 2014;41:3068–73. [Google Scholar]
- 26.Mason BG, Pyle DM, Oppenheimer C. The size and frequency of the largest explosive eruptions on Earth. Bull Volcano. 2004;66:735–48. [Google Scholar]
- 27.Christiansen RL. The quaternary and pliocene Yellowstone Plateau volcanic field of Wyoming, Idaho, and Montana. U.S. Department of the Interior, U.S. Geological Survey. 2001.
- 28.Foulger GR, Natland JH. Is “hotspot” volcanism a consequence of plate tectonics? Science. 2003;300:921–2. doi: 10.1126/science.1083376. [DOI] [PubMed] [Google Scholar]
- 29.Christiansen RL, Foulger GR, Evans JR. Upper-mantle origin of the Yellowstone hotspot. GSA Bull. 2002;114:1245–56. [Google Scholar]
- 30.Morgan P, Blackwell DD, Spafford RE, Smith RB. Heat flow measurements in Yellowstone Lake and the thermal structure of the Yellowstone Caldera. J Geophys Res. 1977;82:3719–32. [Google Scholar]
- 31.Morgan LA, Shanks WC, Lovalvo DA, Johnson SY, Stephenson WJ, Pierce KL, et al. Exploration and discovery in Yellowstone Lake: results from high-resolution sonar imaging, seismic reflection profiling, and submersible studies. J Volcano Geotherm Res. 2003;122:221–42. [Google Scholar]
- 32.Sohn RA, Luttrell K, Shroyer E, Stranne C, Harris RN, Favorito JE. Observations and modeling of a hydrothermal plume in Yellowstone lake. Geophys Res Lett. 2019;46:6435–42. [Google Scholar]
- 33.Fowler APG, Tan C, Cino C, Scheuermann P, Volk MWR, Pat Shanks WC, et al. Vapor-driven sublacustrine vents in Yellowstone Lake, Wyoming, USA. Geology. 2019;47:223–6. [Google Scholar]
- 34.Inskeep WP, Jay ZJ, Macur RE, Clingenpeel S, Tenney A, Lovalvo D, et al. Geomicrobiology of sublacustrine thermal vents in Yellowstone Lake: geochemical controls on microbial community structure and function. Front Microbiol. 2015;6:1044. doi: 10.3389/fmicb.2015.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan C, Cino CD, Ding K, Seyfried WE. High temperature hydrothermal vent fluids in Yellowstone Lake: observations and insights from in-situ pH and redox measurements. J Volcano Geotherm Res. 2017;343:263–70. [Google Scholar]
- 36.Clingenpeel S, Macur RE, Kan J, Inskeep WP, Lovalvo D, Varley J, et al. Yellowstone Lake: high-energy geochemistry and rich bacterial diversity. Environ Microbiol. 2011;13:2172–85. doi: 10.1111/j.1462-2920.2011.02466.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang T, Lyons S, Aguilar C, Cuhel R, Teske A. Microbial communities and chemosynthesis in Yellowstone lake sublacustrine hydrothermal vent waters. Front Microbiol. 2011;2:130. doi: 10.3389/fmicb.2011.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohn R, Harris R, Linder C, Luttrell K, Lovalvo D, Morgan L, et al. Exploring the restless floor of Yellowstone lake. Eos 2017;98. 10.1029/2017EO087035.
- 39.Cino C. An analysis of the hydrothermal fluid chemistry and isotopic data of Yellowstone Lake vents. Retrieved from the University of Minnesota Digital Conservancy. 2018. https://hdl.handle.net/11299/198977.
- 40.Fowler APG, Tan C, Luttrell K, Tudor A, Scheuermann P, Pat Shanks WC, et al. Geochemical heterogeneity of sublacustrine hydrothermal vents in Yellowstone Lake, Wyoming. J Volcano Geotherm Res. 2019;386:106677. [Google Scholar]
- 41.Li G, Rabe KS, Nielsen J, Engqvist MK. Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth Biol. 2019;8:1411–20. doi: 10.1021/acssynbio.9b00099. [DOI] [PubMed] [Google Scholar]
- 42.Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017;45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol Rev. 2015;40:258–72. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Shayeb B, Sachdeva R, Chen L-X, Ward F, Munk P, Devoto A, et al. Clades of huge phages from across Earth’s ecosystems. Nature. 2020;578:425–31. doi: 10.1038/s41586-020-2007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krupovic M, Quemin ERJ, Bamford DH, Forterre P, Prangishvili D. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol. 2014;88:2354–8. doi: 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prangishvili D, Krupovic M. ICTV Report Consortium. ICTV virus taxonomy profile: globuloviridae. J Gen Virol. 2018;99:1357–8. doi: 10.1099/jgv.0.001123. [DOI] [PubMed] [Google Scholar]
- 47.Aguiar P, Beveridge TJ, Reysenbach A-L. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int J Syst Evol Microbiol. 2004;54:33–39. doi: 10.1099/ijs.0.02790-0. [DOI] [PubMed] [Google Scholar]
- 48.Dong Y, Sanford RA, Inskeep WP, Srivastava V, Bulone V, Fields CJ, et al. Physiology, metabolism, and fossilization of hot-spring filamentous microbial mats. Astrobiology. 2019;19:1442–58. doi: 10.1089/ast.2018.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa S, Shtaih Z, Banta A, Beveridge TJ, Sako Y, Reysenbach A-L. Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfur-oxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int J Syst Evol Microbiol. 2005;55:2263–8. doi: 10.1099/ijs.0.63708-0. [DOI] [PubMed] [Google Scholar]
- 50.Takai K, Kobayashi H, Nealson KH, Horikoshi K. Sulfurihydrogenibium subterraneum gen. nov., sp. nov., from a subsurface hot aquifer. Int J Syst Evol Microbiol. 2003;53:823–7. doi: 10.1099/ijs.0.02506-0. [DOI] [PubMed] [Google Scholar]
- 51.Takacs-Vesbach C, Inskeep WP, Jay ZJ, Herrgard MJ, Rusch DB, Tringe SG, et al. Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three Aquificales lineages. Front Microbiol. 2013;4:84. doi: 10.3389/fmicb.2013.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, Schoonen MAA, Nordstrom DK, Cunningham KM, Ball JW. Sulfur geochemistry of hydrothermal waters in Yellowstone National Park: I. the origin of thiosulfate in hot spring waters. Geochim Cosmochim Acta. 1998;62:3729–43. [Google Scholar]
- 54.Grabarczyk DB, Berks BC. Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS ONE. 2017;12:e0173395. doi: 10.1371/journal.pone.0173395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reysenbach A-L, Hamamura N, Podar M, Griffiths E, Ferreira S, Hochstein R, et al. Complete and draft genome sequences of six members of the Aquificales. J Bacteriol. 2009;191:1992–3. doi: 10.1128/JB.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flores GE, Liu Y, Ferrera I, Beveridge TJ, Reysenbach A-L. Sulfurihydrogenibium kristjanssonii sp. nov., a hydrogen- and sulfur-oxidizing thermophile isolated from a terrestrial Icelandic hot spring. Int J Syst Evol Microbiol. 2008;58:1153–8. doi: 10.1099/ijs.0.65570-0. [DOI] [PubMed] [Google Scholar]
- 57.Anderson CL, Sullivan MB, Fernando SC. Dietary energy drives the dynamic response of bovine rumen viral communities. Microbiome. 2017;5:155. doi: 10.1186/s40168-017-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shmakov SA, Makarova KS, Wolf YI, Severinov KV, Koonin EV. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci USA. 2018;115:E5307–16. doi: 10.1073/pnas.1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Javor BJ, Wilmot DB, Vetter RD. pH-Dependent metabolism of thiosulfate and sulfur globules in the chemolithotrophic marine bacterium Thiomicrospira crunogena. Arch Microbiol. 1990;154:231–8. [Google Scholar]
- 60.Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA. 2013;110:5540–5. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–94. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018;6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Correa AMS, Howard-Varona C, Coy SR, Buchan A, Sullivan MB, Weitz JS. Revisiting the rules of life for viruses of microorganisms. Nat Rev Microbiol. 2021;19:501–13. doi: 10.1038/s41579-021-00530-x. [DOI] [PubMed] [Google Scholar]
- 65.Anderson RE, Brazelton WJ, Baross JA. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol. 2011;77:120–33. doi: 10.1111/j.1574-6941.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 66.Palmer M, Hedlund BP, Roux S, Tsourkas PK, Doss RK, Stamereilers C, et al. Diversity and distribution of a novel genus of hyperthermophilic aquificae viruses encoding a proof-reading family—a DNA polymerase. Front Microbiol. 2020;11:583361. doi: 10.3389/fmicb.2020.583361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shipman SL, Nivala J, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353:aaf1175. doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopes A, Tavares P, Petit M-A, Guérois R, Zinn-Justin S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genomics. 2014;15:1027. doi: 10.1186/1471-2164-15-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 71.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. [Google Scholar]
- 72.Hudaiberdiev S, Shmakov S, Wolf YI, Terns MP, Makarova KS, Koonin EV. Phylogenomics of Cas4 family nucleases. BMC Evol Biol. 2017;17:232. doi: 10.1186/s12862-017-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labonté JM, Pachiadaki M, Fergusson E, McNichol J, Grosche A, Gulmann LK, et al. Single cell genomics-based analysis of gene content and expression of prophages in a diffuse-flow deep-sea hydrothermal system. Front Microbiol. 2019;10:1262. doi: 10.3389/fmicb.2019.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silveira CB, Rohwer FL. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes. 2016;2:16010. doi: 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denise R, Abby SS, Rocha EPC. Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol. 2019;17:e3000390. doi: 10.1371/journal.pbio.3000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 78.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 2013;9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denise R, Abby SS, Rocha EPC. The evolution of protein secretion systems by co-option and tinkering of cellular machineries. Trends Microbiol. 2020;28:372–86. doi: 10.1016/j.tim.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Koebnik R, Locher KP, Van, Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–53. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 82.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha EPC. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–4. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas S, Holland IB, Schmitt L. The type 1 secretion pathway—the hemolysin system and beyond. Biochimica et Biophysica Acta (BBA)-Mol Cell Res. 2014;1843:1629–41. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 85.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–75. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 86.Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–51. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–36. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wallden K, Rivera-Calzada A, Waksman G. Microreview: type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010;12:1203–12. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peabody CR, Chung YJ, Yen M-R, Vidal-Ingigliardi D, Pugsley AP, Saier MH. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–72. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 91.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci USA. 2001;98:2503–8. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eren AM, Vineis JH, Morrison HG, Sogin ML. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One. 2013;8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 95.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–605. [Google Scholar]
- 97.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for’omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 99.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–9. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hug LA. Subsampled assemblies and hybrid nucleotide composition/differential coverage binning for genome-resolved metagenomics. Methods Mol Biol. 2018;1849:215–25. doi: 10.1007/978-1-4939-8728-3_14. [DOI] [PubMed] [Google Scholar]
- 101.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Lab; 2014.
- 102.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. [Google Scholar]
- 103.Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL. Microbial genomic taxonomy. BMC Genomics. 2013;14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–27. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–32. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S, et al. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genom Sci. 2012;6:427–39. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinforma. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Skennerton CT, Imelfort M, Tyson GW. Crass: identification and reconstruction of CRISPR from unassembled metagenomic data. Nucleic Acids Res. 2013;41:e105. doi: 10.1093/nar/gkt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Z, Tran P, Liu Y, Kieft K, Anantharaman K. METABOLIC: a scalable high-throughput metabolic and biogeochemical functional trait profiler based on microbial genomes. bioRxiv [preprint] 2020. Available from: 10.1101/761643.
- 116.Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–96. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–3. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–80. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–6. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009;37:D216–23. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cury J, Abby SS, Doppelt-Azeroual O, Néron B, Rocha EPC. Identifying conjugative plasmids and integrative conjugative elements with CONJscan. Methods Mol Biol. 2020;2075:265–83. doi: 10.1007/978-1-4939-9877-7_19. [DOI] [PubMed] [Google Scholar]
- 127.Abby SS, Rocha EPC. Identification of protein secretion systems in bacterial genomes using MacSyFinder. Methods Mol Biol. 2017;1615:1–21. doi: 10.1007/978-1-4939-7033-9_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequence information for this investigation can be found in the NCBI database under BioProject PRJNA669531 and BioSamples SAMN16454225, SAMN16454373, SAMN16454375, SAMN16454377, and SAMN16454376. Quality-filtered short reads used for single metagenomic assemblies are available in the NCBI Sequence Read Archive as SRR12852619 (2016_B01), SRR12852620 (2016_B02_str), and SRR12852618 (2016_B02_sed). Quality-filtered short reads used for coassembled metagenomes are available in the NCBI Sequence Read Archive as SRR12852621 (2017_B01_S1) and SRR12852622 (2017_B01_S2).