Abstract
Streptococcus pneumoniae (pneumococcus) is a leading cause of severe infections among children and adults. Interactions between commensal microbes in the upper respiratory tract and S. pneumoniae are poorly described. In this study, we sought to identify interspecies interactions that modify the risk of S. pneumoniae colonization during infancy and to describe development of the upper respiratory microbiome during infancy in a sub-Saharan African setting. We collected nasopharyngeal swabs monthly (0–6 months of age) or bimonthly (6–12 months of age) from 179 mother–infant dyads in Botswana. We used 16S ribosomal RNA gene sequencing to characterize the nasopharyngeal microbiome and identified S. pneumoniae colonization using a species-specific PCR assay. We detect S. pneumoniae colonization in 144 (80%) infants at a median age of 71 days and identify a strong negative association between the relative abundance of the bacterial genera Corynebacterium within the infant nasopharyngeal microbiome and the risk of S. pneumoniae colonization. Using in vitro cultivation experiments, we demonstrate growth inhibition of S. pneumoniae by secreted factors from strains of several Corynebacterium species isolated from these infants. Finally, we demonstrate that antibiotic exposures and the winter season are associated with a decline in the relative abundance of Corynebacterium within the nasopharyngeal microbiome, while breastfeeding is associated with an increase in the Corynebacterium relative abundance. Our findings provide novel insights into the interspecies interactions that contribute to colonization resistance to S. pneumoniae and suggest that the nasopharyngeal microbiome may be a previously unrecognized mechanism by which environmental factors influence the risk of pneumococcal infections during childhood. Moreover, this work lays the foundation for future studies seeking to use targeted manipulation of the nasopharyngeal microbiome to prevent infections caused by S. pneumoniae.
Subject terms: Bacterial infection, Microbial ecology
Introduction
Streptococcus pneumoniae (also known as pneumococcus) is a leading cause of severe infections among children and adults, with the highest incidence of pneumococcal disease occurring during infancy [1]. S. pneumoniae causes a broad range of infections ranging from mild respiratory illnesses, such as acute otitis media and acute sinusitis, to invasive pneumococcal disease (IPD), which includes serious illnesses such as bloodstream infection and meningitis [2]. Globally, S. pneumoniae is responsible for more than 300,000 child deaths each year, the overwhelming majority of which result from pneumonia [3] and occur in low- and middle-income countries [4]. Colonization of the nasopharynx precedes infections caused by S. pneumoniae and occurs in 25–65% of children and <15% of adults, with substantial variation in prevalence by geography and socioeconomic factors [5–9]. Pneumococcal conjugate vaccines effectively prevent IPD caused by vaccine serotypes [10, 11] but are less effective in preventing non-invasive infections, such as pneumonia [12, 13] and otitis media [14]. Moreover, the introduction of pneumococcal conjugate vaccines has been followed by the emergence of non-vaccine serotypes, some of which are highly virulent and multidrug resistant [15–17], and nonencapsulated pneumococci [18], which together threaten to compromise the long-term benefits of these vaccines [19, 20]. There is thus an urgent need to develop alternative approaches to preventing infections caused by S. pneumoniae.
A complex microbial community resides in the upper respiratory tract and has co-evolved with humans [21]. Over the last decade, accumulating evidence has emerged for this microbiome’s role in the pathogenesis of respiratory infections [22], with microbial communities at distinct anatomical sites within the upper respiratory tract contributing to resistance to colonization or infection by specific pathogens. Within the nasopharynx, resident microbes may resist colonization by S. pneumoniae through competition for nutrients and adhesion sites, secretion of antimicrobial factors, and modulation of host immune responses [23]. Most prior studies focused on associations between S. pneumoniae and colonization by other major respiratory pathobionts, demonstrating primarily positive associations with Haemophilus influenzae and Moraxella catarrhalis [24–26] and negative associations with Staphylococcus aureus [25, 27]. More recently, negative associations were reported between S. pneumoniae and other streptococcal species [28, 29], Lactobacillus species [30, 31], non-diphtheriae Corynebacterium species [6, 32, 33], and Dolosigranulum pigrum [32, 33]. The interspecies interactions that underlie these associations are often bidirectional, occur by diverse mechanisms, and may vary based on the local microenvironment. For instance, H. influenzae can inhibit S. pneumoniae by downregulating expression of pneumococcal adherence factors [34] or by stimulating complement-dependent phagocytosis of S. pneumoniae [35]. Still, these species often persist together in a multi-species biofilm that can confer protection to S. pneumoniae from antibiotics [36, 37]. Similarly, inhibition of pneumococcal growth by D. pigrum appears to require the presence of specific Corynebacterium species [38]. At the same time, Corynebacterium accolens inhibits S. pneumoniae through hydrolysis of free fatty acids from host skin surface triacylglycerols [33]. Although these laboratory studies have furthered our understanding of respiratory microbial community ecology, the extent to which these interspecies interactions contribute to population-level trends in S. pneumoniae colonization is unknown.
As described above, colonization of the nasopharynx is a necessary precursor to infections caused by S. pneumoniae, and is particularly prevalent among young children. Notably, the nasopharyngeal microbiome undergoes rapid shifts in composition during early childhood [39–41], primarily driven by environmental factors, such as delivery mode [42, 43], infant feeding practices [41, 44], contact with other children [39], season [40, 45], and antibiotic exposures [39]. Previous studies that evaluated associations between the nasopharyngeal microbiome and S. pneumoniae colonization in children were cross-sectional [6, 32, 33], and few data are available from low- and middle-income countries, where >80% of child deaths from S. pneumoniae occur [3] and household [46, 47] and environmental [48] exposures may differ from those in high-income countries. Longitudinal studies in these settings are necessary to determine the impact of environmental exposures on the developing infant nasopharyngeal microbiome and to identify microbiome features that facilitate or inhibit S. pneumoniae colonization.
In this study, we sought to identify interspecies interactions that modify the risk of pneumococcal colonization during infancy and to describe nasopharyngeal microbiome development during the first year of life in a sub-Saharan African setting. We studied the nasopharyngeal microbiomes of 179 mother–infant dyads in Botswana using 16S ribosomal RNA (rRNA) gene sequencing and identified S. pneumoniae colonization with a species-specific PCR assay. We describe changes in microbiome diversity and composition during infancy, evaluate associations between the nasopharyngeal microbiome and pneumococcal colonization risk, and identify environmental factors that influence nasopharyngeal microbiome composition during infancy.
Results
The nasopharyngeal microbiome is a low-diversity microbial community throughout infancy
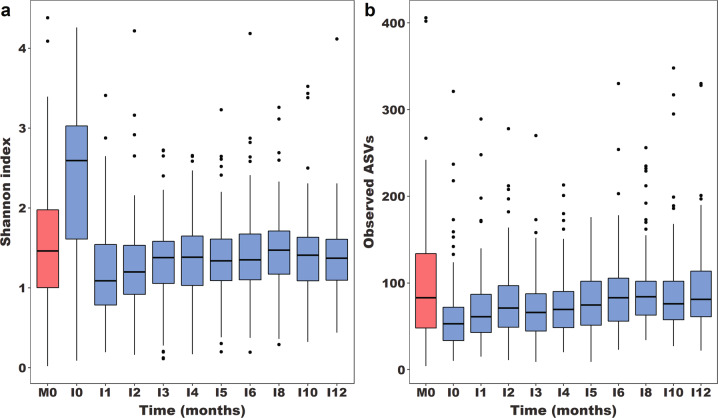
We collected nasopharyngeal swab samples monthly (0–6 months) or bimonthly (6–12 months) from 179 mother–infant dyads recruited at urban and rural study sites in southern Botswana. Infants were born vaginally, had a median [interquartile range (IQR)] birth weight of 3120 g (2855 g, 3408 g), and were predominantly breastfed (Table 1). Infants were followed in this study to a median (IQR) age of 12.0 (8.0, 12.1) months. Of 51 mothers with HIV, 44 (86%) received antiretroviral therapy during pregnancy for a median (IQR) duration of 9 [5, 9] months; median (IQR) CD4 count was 461 (312, 641) cells/µL. We performed 16S rRNA sequencing of infant nasopharyngeal samples from all study visits, and maternal nasopharyngeal samples from the delivery visit, resulting in 1368 infant and 172 maternal samples passing quality control procedures. The median (IQR) Shannon index and number of unique ASVs in these samples were 1.38 (1.03, 1.72) and 71 (50, 100), respectively. Infant nasopharyngeal microbiome diversity remained relatively stable during infancy and similar to maternal nasopharyngeal microbiome diversity (Fig. 1), except at birth (I0), when diversity was higher than later in infancy (Wilcoxon signed-rank tests, p < 0.0001) and compared to maternal samples (Wilcoxon signed-rank test, p < 0.0001). Nasopharyngeal microbiome richness increased with age during infancy (negative binomial regression, p < 0.0001) and did not differ from the richness of the maternal nasopharyngeal microbiome after five months of age (Wilcoxon signed-rank tests, p > 0.05).
Table 1.
Characteristics of the 179 mother–infant dyads included in the study population.
| Subject characteristics | N | % |
|---|---|---|
| Infant sex | ||
| Female | 99 | 55% |
| Male | 80 | 45% |
| Median (IQR) infant birth weight, g | 3120 | (2855, 3408) |
| Infant HIV status | ||
| HIV-exposed, uninfected | 51 | 28% |
| HIV-unexposed, uninfected | 128 | 72% |
| Location of residence | ||
| Rural | 72 | 40% |
| Urban | 107 | 60% |
| Electricity in the home | 119 | 66% |
| Household use of solid fuels | 104 | 58% |
| Maternal educational level | ||
| None or primary | 13 | 7% |
| Secondary | 144 | 80% |
| Tertiary | 22 | 12% |
| Median (IQR) number of household members | ||
| <5 years | 1 | (0, 1) |
| 5–17 years | 1 | (0, 3) |
| ≥18 years | 3 | (2, 5) |
| Enrollment season | ||
| Summer (November to March) | 67 | 37% |
| Winter (April to October) | 112 | 63% |
| One or more PCV-13 doses | 153 | 85% |
| Infant breastfeeding, any | 153 | 85% |
| Infant antibiotic exposure, any | 114 | 64% |
| Amoxicillin | 83 | 46% |
| Metronidazole | 23 | 13% |
| Trimethoprim-sulfamethoxazole | 43 | 24% |
IQR interquartile range, PCV-13 13-valent pneumococcal conjugate vaccine.
Fig. 1. Alpha diversity of the nasopharyngeal microbiome among mother–infant dyads in Botswana.
Box plots depict nasopharyngeal microbiome diversity, as measured by the Shannon index (a), and richness, as measured by the number of unique ASVs (b). Maternal nasopharyngeal samples are from the birth visit only (M0), and are shown in red, while nasopharyngeal samples from infants were sequenced at ten time points during the first year of life (I0-I12), and are shown in blue. The Shannon index of the infant nasopharyngeal microbiome is higher at birth than at all later time points (Wilcoxon signed-rank tests, p < 0.0001) and compared to maternal samples (Wilcoxon signed-rank tests, p < 0.0001). The number of unique ASVs in infant nasopharyngeal samples is lower than in maternal samples from birth through five months of age (Wilcoxon signed-rank tests, p < 0.05). ASV amplicon sequence variants.
Rapid shifts in nasopharyngeal microbiome composition occur during early infancy
Six bacterial genera—Corynebacterium, Dolosigranulum, Haemophilus, Moraxella, Staphylococcus, and Streptococcus—accounted for more than 90% of the sequencing reads identified in infant nasopharyngeal samples collected at or after one month of age (Supplementary Table 1). Only at the birth visit were other bacterial genera abundant, including Acinetobacter, Gardnerella, Lactobacillus, and Sneathia, likely reflecting colonization of the infant upper respiratory tract by pioneering microbes from the maternal gut [49, 50] and vaginal microbiomes [50, 51]. Nasopharyngeal microbiome composition shifted dramatically during early infancy (Fig. 2); microbiome composition at each study visit differed from the preceding visit from birth through five months of age (PERMANOVA on Bray-Curtis dissimilarity, p < 0.05), after which no significant differences in overall microbiome composition were observed between consecutive study visits. While maternal nasopharyngeal microbiome composition at the birth visit differed from infant microbiome composition at all study visits (PERMANOVA on Bray-Curtis dissimilarity, p < 0.001), the dissimilarity of these microbiomes increased with age (linear mixed effects model, p < 0.0001), indicating progressive divergence of the infant nasopharyngeal microbiome from an adult microbiome profile during the first 12 months of life.
Fig. 2. Composition of the nasopharyngeal microbiome among mother–infant dyads in Botswana.
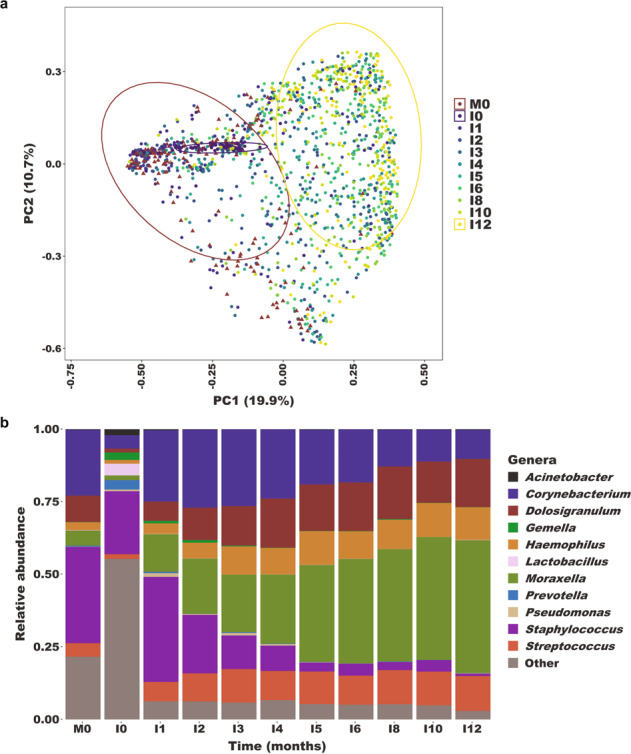
a Principal components (PCoA) plot based on Bray-Curtis distances showing nasopharyngeal microbiome composition among mothers at delivery (M0) and infants throughout the first year of life (I0-I12). During infancy, nasopharyngeal microbiome composition progressively diverges from the composition of the adult nasopharyngeal microbiome. Ellipses define the regions containing 80% of all samples that can be drawn from the underlying multivariate t distribution. Ellipses are shown for samples from mothers at delivery (M0), infants at birth (I0), and infants at 12 months of age (I12). b Relative abundances of highly abundant genera in nasopharyngeal samples from mothers (M0; n = 172 samples) and infants (I0-I12, n = 1368 samples) by month of visit.
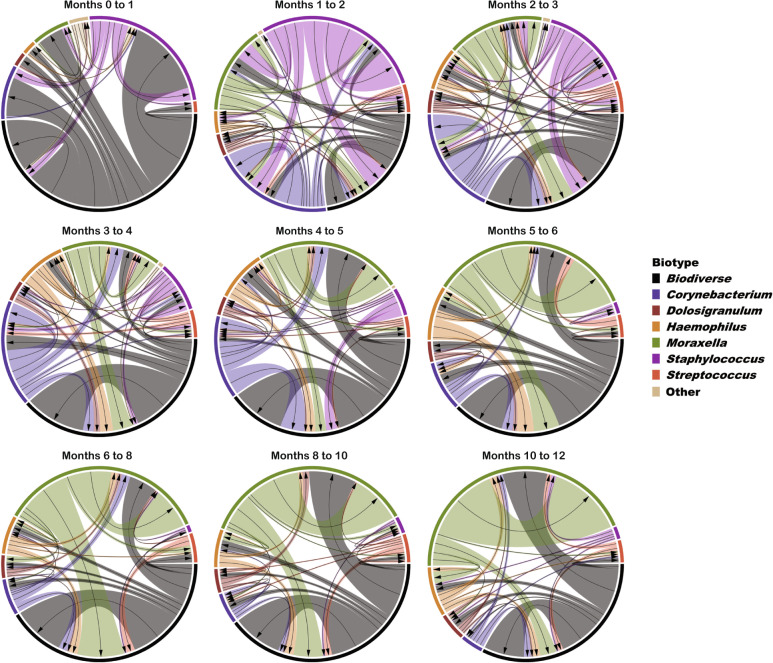
To further describe shifts in microbiome composition during infancy, we classified each infant nasopharyngeal sample based upon whether a single genus comprised 50% or more of the sequencing reads in that sample. Samples for which no genus met this relative abundance threshold were categorized as “biodiverse.” A single genus was dominant in 844 of 1368 (62%) infant samples with biodiverse (n = 524; 38%), Moraxella-dominant (n = 343; 25%), Corynebacterium-dominant (n = 153; 11%), and Staphylococcus-dominant (n = 142; 10%) being the most common microbiome “biotypes” (Supplementary Table 2). We considered a per sample microbiome profile to be “stable” if the next sample from that infant was classified as the same biotype. Based on this definition, within-infant stability of the nasopharyngeal microbiome varied by biotype (Fig. 3 and Supplementary Table 3). Compared to a biodiverse microbiome profile, a Moraxella-dominant biotype tended to be associated with higher stability (logistic regression model, p = 0.07), while lower stability was observed with Dolosigranulum-dominant (p = 0.005), Haemophilus-dominant (p = 0.001), and Streptococcus-dominant (p = 0.0004) biotypes.
Fig. 3. State transitions of the nasopharyngeal microbiome during infancy.
Each nasopharyngeal sample was classified based on the dominant bacterial genus in that sample or, if no genus occupied ≥50% of the sequencing reads, the sample was classified as biodiverse. Each diagram depicts changes in infant nasopharyngeal microbiome biotype between two consecutive study visits. Arrows point to the directionality of microbiome transitions while ribbon widths represent the frequency of these transitions. The colors correspond to specific microbiome biotypes.
Environmental exposures influence nasopharyngeal microbiome composition during infancy
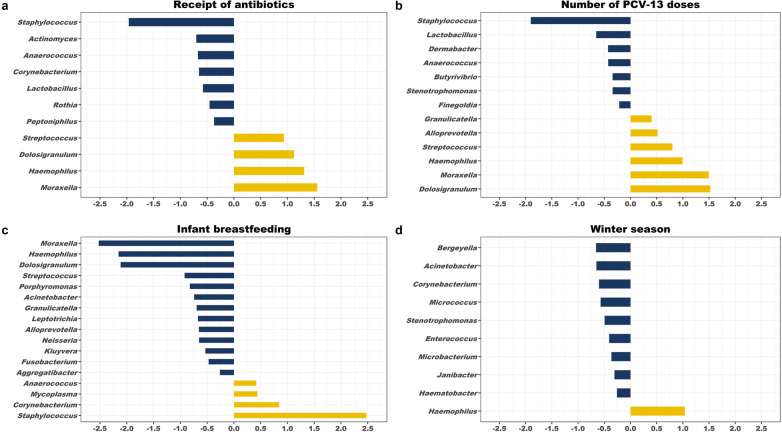
To identify early life factors that influence the nasopharyngeal microbiome, we used MaAsLin2 [52] to fit generalized linear mixed models evaluating associations between sociodemographic factors and environmental exposures and the abundances of specific bacterial genera within the nasopharyngeal microbiome (Supplementary Table 4). The most substantial microbiome composition changes were associated with recent antibiotic exposures, 13-valent pneumococcal conjugate vaccine (PCV-13) doses, breastfeeding, and the winter season (Fig. 4). Antibiotic exposures were associated with decreases in the relative abundances of several bacterial genera generally associated with respiratory health, including Corynebacterium and Lactobacillus [31, 53–55], and increases in the relative abundances of genera containing common respiratory pathobionts (Haemophilus, Moraxella, Streptococcus). Similarly, during winter months, the relative abundance of Corynebacterium declined and was accompanied by an increase in the relative abundance of Haemophilus. In contrast, breastfeeding was associated with an increase in the relative abundance of Corynebacterium and decreases in the relative abundances of Haemophilus, Moraxella, and Streptococcus. No significant differences in nasopharyngeal microbiome composition were observed by sex, low birth weight status, and urban residence.
Fig. 4. Associations between environmental exposures and the composition of the nasopharyngeal microbiome during infancy.
MaAsLin2 was used to fit log-transformed generalized linear mixed models evaluating associations between sociodemographic factors and environmental exposures and the relative abundances of bacterial genera within the infant nasopharyngeal microbiome. The coefficients from these models, which correspond to the relative effect sizes of associations, are shown for significant associations (q < 0.20) identified for (a) antibiotic exposures, (b) number of PCV-13 doses, (c) breastfeeding, and (d) the winter season. The number of PCV-13 doses was modeled as an ordinal variable such that the coefficients represent the relative effect sizes associated with each successive vaccine dose. Bacterial genera that increase in relative abundance with the exposure are shown as yellow bars. Bacterial genera that decrease in relative abundance with the exposure are shown as blue bars.
Corynebacterium species are associated with a lower risk of S. pneumoniae colonization
S. pneumoniae colonization was identified in 144 of 179 (80%) infants at a median (IQR) age of 71 (39, 126) days (Supplementary Fig. 1). To identify nasopharyngeal microbiome features that precede S. pneumoniae colonization, we first fit a Cox proportional hazards model to evaluate associations between nasopharyngeal microbiome biotypes and acquisition of S. pneumoniae at the next study visit (Table 2). Compared to a biodiverse microbiome profile, a Corynebacterium-dominant biotype was associated with a lower hazard of S. pneumoniae colonization [hazard ratio (HR): 0.43, 95% confidence interval (CI): 0.23–0.80]. In addition, the risk of S. pneumoniae colonization increased during winter months (HR: 1.48, 95% CI: 1.07–2.04) and with each additional child household member (HR: 1.21, 95% CI: 1.09–1.34), and declined with increasing number of PCV-13 doses (HR: 0.10, 95% CI: 0.06–0.16). To further explore the negative association between Corynebacterium species and S. pneumoniae, we classified samples into quartiles based on Corynebacterium relative abundance and evaluated the association between these sample quartiles and S. pneumoniae acquisition. The hazard ratio for pneumococcal colonization declined with each successive quartile increase in Corynebacterium relative abundance (Supplementary Table 5); compared to the lowest quartile, the highest quartile of Corynebacterium relative abundance was associated with a 69% lower hazard of S. pneumoniae colonization (HR: 0.31, 95% CI: 0.18–0.53).
Table 2.
Cox proportional hazards model analyses evaluating associations between microbiome “biotypes” identified in infant nasopharyngeal samples and the risk of acquisition of S. pneumoniae prior to the next study visit.
| Hazard ratio | 95% CI | p | |
|---|---|---|---|
| Microbiome biotype† | |||
| Biodiverse | 1.00 | Ref. | – |
| Dominated by a single genus | |||
| Corynebacterium | 0.43 | (0.23–0.80) | 0.008 |
| Dolosigranulum | 1.40 | (0.79–2.48) | 0.25 |
| Haemophilus | 1.32 | (0.49–3.54) | 0.58 |
| Moraxella | 1.15 | (0.73–1.82) | 0.55 |
| Staphylococcus | 0.70 | (0.39–1.25) | 0.23 |
| Streptococcus | 1.24 | (0.23–6.68) | 0.81 |
| Other genera | 3.34 | (1.28–8.70) | 0.01 |
| Male infant sex | 0.97 | (0.71–1.31) | 0.82 |
| Infant low birth weight (<2500 g) | 0.82 | (0.45–1.50) | 0.52 |
| HIV-exposed, uninfected infant | 1.11 | (0.70–1.76) | 0.65 |
| Urban residence | 1.35 | (0.98–1.86) | 0.07 |
| Household use of solid fuels | 1.12 | (0.80–1.57) | 0.51 |
| Number of child household members | 1.21 | (1.09–1.34) | 0.0003 |
| Winter season† | 1.48 | (1.07–2.04) | 0.02 |
| Number of PCV-13 doses† | 0.10 | (0.06–0.16) | <0.0001 |
| Infant breastfeeding† | 0.99 | (0.63–1.55) | 0.97 |
| Receipt of antibiotics† | 0.79 | (0.54–1.17) | 0.24 |
CI confidence interval, PCV-13 13-valent pneumococcal conjugate vaccine.
†Variable was modeled as a time-dependent covariate.
Identification of Corynebacterium species negatively associated with S. pneumoniae colonization
Given the diversity of Corynebacterium species isolated from the human upper respiratory tract [56–58], we sought to determine if specific species accounted for the negative association with S. pneumoniae colonization. We used BLAST searches to identify the species or group of closely related species (supraspecies) corresponding to each of the 279 Corynebacterium amplicon sequence variants (ASVs) identified in infant nasopharyngeal samples [59]. We operationally classified those ASVs into 45 unique Corynebacterium species or supraspecies (Supplementary Table 6), the most abundant of which were C. pseudodiphtheriticum/propinquum (90.9% sample prevalence, 10.4% mean relative abundance), C. accolens/macginleyi (72.1% sample prevalence, 5.8% mean relative abundance), and C. tuberculostearicum (54.5% sample prevalence, 1.2% mean relative abundance). Subsequent analyses demonstrated that the relative abundances of each of these three Corynebacterium species or supraspecies were inversely associated with the risk of S. pneumoniae colonization (Supplementary Table 7). These findings illustrate the enormous diversity of Corynebacterium species that colonize the human upper respiratory tract and indicate that multiple Corynebacterium species likely contribute to colonization resistance to S. pneumoniae.
Strain-specific secretion of antipneumococcal factors by Corynebacterium species
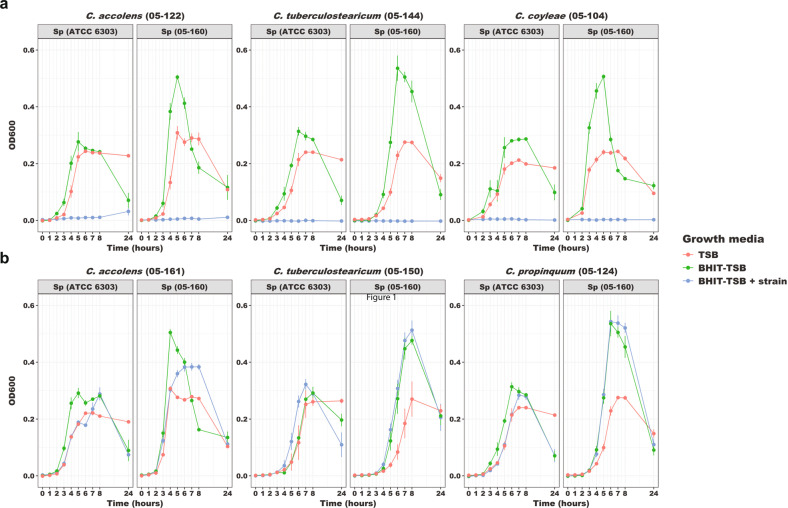
To further investigate the inhibition of S. pneumoniae by Corynebacterium species, we cultured infant nasopharyngeal samples on selective media and identified culture isolates to the species level using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). We isolated a total of 35 morphologically distinct Corynebacterium strains from 21 infants, including strains of C. accolens (n = 21), C. tuberculostearicum (n = 5), C. pseudodiphtheriticum (n = 4), C. coyleae (n = 2), C. propinquum (n = 2), and C. striatum (n = 1). We then screened supernatants from cell cultures of each of these Corynebacterium strains for inhibition of growth of reference (ATCC 6303; serotype 3) and contemporary infant nasopharyngeal strains (05-160; serotype 11 A) of S. pneumoniae. We observed inhibition of pneumococcal growth by cell-free supernatants from cultures of seven (20%) strains (Fig. 5), including species accolens (1 of 21; 5%), tuberculostearicum (3 of 5; 60%), coyleae (2 of 2; 100%), and striatum (1 of 1; 100%). While the characteristics and mechanisms underlying the inhibition require further investigation, these assays support a causal basis for the inverse relationship between Corynebacterium spp. and S. pneumoniae observed in microbiome data analyses. Furthermore, these data indicate that specific species and strains have the capacity to inhibit S. pneumoniae.
Fig. 5. Strain-specific inhibition of pneumococcal growth by Corynebacterium.
Two strains of S. pneumoniae, one reference strain (ATCC 6303; serotype 3) and one strain isolated from an infant nasopharyngeal sample (05-160; serotype 11 A) were separately added to sterile, cell-free media from overnight cultures of Corynebacterium strains. Growth of these strains of S. pneumoniae was determined by OD600 readings that were normalized to blank media controls (TSB and BHIT-TSB). a Growth curves of S. pneumoniae strains in different media, including cell-free media from inhibitory strains of C. accolens (05-122), C. tuberculostearicum (05-144), and C. coyleae (05-104). b Growth curves of S. pneumoniae strains in different media, including cell-free media from strains of C. accolens (05-161), C. tuberculostearicum (05-150), and C. propinquum (05-124) that do not demonstrate pneumococcal growth inhibition. The decline in OD600 observed in these experiments after the peak density is reached reflects a transition of S. pneumoniae from the exponential growth phase to the autolysis phase. TSB tryptic soy broth, BHIT-TSB brain heart infusion medium with 0.2% Tween80 diluted 50% with tryptic soy broth.
Discussion
In this study, we described the nasopharyngeal microbiome dynamics during the first year of life and identified environmental factors that influence infant nasopharyngeal microbiome composition. We also found that higher abundances of Corynebacterium species in the nasopharyngeal microbiome are associated with a lower risk of S. pneumoniae colonization during infancy. Finally, we demonstrated inhibition of pneumococcal growth by secreted factors from strains of several Corynebacterium species.
We found that the composition of the nasopharyngeal microbiome of infants in Botswana bears similarities to that reported in previous studies of infants and young children in high-income countries [39, 41]. Specifically, independent of the resource setting, this low-diversity microbial community is typically comprised mostly of bacteria from six genera, with Corynebacterium and Staphylococcus predominating during the first several months of life and Dolosigranulum and Moraxella becoming more abundant later in infancy [39, 40]. These trends in nasopharyngeal microbiome composition during infancy appear to be highly conserved despite substantial differences in host characteristics, household exposures, and climate. However, some caution must be taken in these statements given that most studies of the infant nasopharyngeal microbiome have used 16S rRNA gene sequencing. Substantial functional or genomic differences may exist between the bacterial strains that colonize the upper respiratory tracts of children in geographically distinct human populations that would only be revealed with alternative methodologies. We also found that the nasopharyngeal microbiome of infants in Botswana is highly dynamic, although microbiome stability varies markedly based on the dominant genus. In particular, higher microbiome stability was observed with nasopharyngeal microbiome profiles that were dominated by Moraxella, while lower stability was seen with profiles that were dominated by Dolosigranulum, Haemophilus, or Streptococcus. Findings from studies conducted in high-income countries were broadly similar, although notably nasopharyngeal microbiome profiles with high abundance of Dolosigranulum were reported to be highly stable in these settings [39, 41]. Further research is needed to investigate associations between the presence and abundance of Dolosigranulum in the nasopharyngeal microbiome and child respiratory health in low- and middle-income countries.
Our findings also demonstrate the impact of early life environmental exposures on the nasopharyngeal microbiome. In particular, the composition of the nasopharyngeal microbiome of infants varied with season, as reported in several prior studies [40, 45], with winter months associated with a declining relative abundance of Corynebacterium. The association between season and the abundance of Corynebacterium in the nasopharyngeal microbiome could contribute to the higher incidence of S. pneumoniae colonization during the winter season observed in our cohort and reported in several prior studies [60–63]. In contrast, we found few changes in the infant nasopharyngeal microbiome associated with maternal HIV infection, suggesting that other factors may account for the increased risks of pneumococcal colonization and disease observed in HIV-exposed, uninfected infants [64–66]. Alternatively, the mild immunosuppression of mothers with HIV in our study may have limited the effect of HIV infection on the maternal upper respiratory microbiome; indeed, mothers with and without HIV in our cohort had similar nasopharyngeal microbiome composition (PERMANOVA on Bray-Curtis dissimilarity, p = 0.99). Feeding practices also influenced the composition of the nasopharyngeal microbiome of infants in this study, with breastfeeding promoting the enrichment of the nasopharyngeal microbiome with Corynebacterium species. Interestingly, previous studies have not reported an association between breastfeeding and pneumococcal colonization during infancy [6, 67], which could reflect the complexity of the effect of breastfeeding on the infant upper respiratory microbiome. Taken together, our findings indicate that the nasopharyngeal microbiome may be a previously unrecognized and potentially modifiable mechanism by which environmental factors influence the risk of pneumococcal infections during childhood.
We found that higher abundances of Corynebacterium species within the nasopharyngeal microbiome are associated with a lower risk of S. pneumoniae colonization during infancy. Moreover, we identified strains of multiple Corynebacterium species that secrete factors that inhibit pneumococcal growth in laboratory experiments. Corynebacterium is a diverse bacterial genus that includes common residents of the upper respiratory tracts, skin, and gastrointestinal tracts of humans and animals. Although >150 species of Corynebacterium have been identified to date [68], the most common species isolated from the human respiratory tract are C. accolens, C. pseudodiphtheriticum, C. propinquum, C. striatum, and C. tuberculostearicum [56–58]. With the exception of Corynebacterium diphtheriae, the etiological agent of diphtheria [69], Corynebacterium species only rarely cause human disease, often in the setting of compromised host immunity [70, 71], indwelling prosthetic material [71–73], or chronic respiratory diseases [74, 75]. This low pathogenicity of non-diphtheriae Corynebacterium species is a key characteristic that supports their further evaluation for use as biotherapeutics. Despite the commonality of these species in the human microbiome, surprisingly little is known about the mechanisms by which Corynebacterium species adhere to human mucosal surfaces, interact with co-occurring microbial species and the host immune system, and rarely cause invasive infection.
Prior studies demonstrated antagonistic relationships between Corynebacterium species and several important bacterial pathobionts. C. propinquum was recently recognized to produce siderophores that inhibit the growth of coagulase-negative staphylococci through iron restriction [76], a strategy that this species may use to colonize the upper respiratory tract. Intranasal administration of a C. pseudodiphtheriticum strain effectively eradicated S. aureus nasal carriage in adults [77], demonstrating the potential use of Corynebacterium species as respiratory probiotics. Strains of C. pseudodiphtheriticum also inhibited growth of M. catarrhalis in co-cultivation experiments [78]. Lower relative abundances of Corynebacterium species were previously observed in the upper respiratory tracts of children with S. pneumoniae colonization [6, 32, 33]. Moreover, Bomar et al. identified a C. accolens strain that inhibited S. pneumoniae through the production of a lipase that releases antipneumococcal free fatty acids from human skin triacylglycerols [33]. Our findings demonstrate the substantial strain-level heterogeneity that characterizes interactions between Corynebacterium spp. and S. pneumoniae and point towards the need to study large collections of Corynebacterium strains to understand the breadth and distribution of phenotypes exhibiting anti-pneumococcal activity.
Our study has several limitations. First, this study was conducted at urban and rural sites in southern Botswana, and may not be generalizable to infants living in other settings. We excluded infants who were born by caesarian delivery, and were thus unable to evaluate the effects of delivery mode on the nasopharyngeal microbiome or the risk of S. pneumoniae colonization. Interestingly, in a study of >2000 Fijian infants 5–8 weeks of age, the prevalence and density of pneumococcal colonization were higher among infants born by vaginal delivery than among infants delivered by caesarian birth [79]. Comparative analyses of microbiome composition were based on sample relative abundances, and we are thus unable to determine if the observed associations represent differences in the absolute amounts of bacterial genera or species within the nasopharynx across samples. Our study employed 16S rRNA gene sequencing to characterize the nasopharyngeal microbiome of infants and mothers; future studies incorporating shotgun metagenomic sequencing may provide a more detailed understanding of interspecies interactions within the upper respiratory tract. Although the analyses adjusted for a large number of potential confounders in evaluating associations between the nasopharyngeal microbiome and S. pneumoniae colonization, residual confounding remains possible. Finally, we have not yet identified the secreted factors by which specific Corynebacterium strains inhibited S. pneumoniae in growth inhibition assays, nor did we assess Corynebacterium strains for pneumococcal inhibition through non-secretome-based mechanisms.
Conclusions
In summary, we identify environmental exposures that shape the developing infant nasopharyngeal microbiome and may influence colonization resistance to S. pneumoniae. Moreover, we demonstrate an inverse relationship between the abundances of Corynebacterium species within the nasopharyngeal microbiome and the risk of S. pneumoniae colonization during infancy. Future studies to define the molecular mechanisms of these bacterial interactions could lead to development of the first rationally-designed biotherapeutics for the prevention of infections caused by S. pneumoniae.
Materials and methods
Setting
Botswana is a landlocked country in southern Africa with a semi-arid climate and a rainy summer season that typically occurs from November to March. The country’s under-five child mortality rate was estimated to be 41.6 per 1000 live births in 2019 [80]. Haemophilus influenzae type B (Hib) and 13-valent pneumococcal conjugate (PCV-13) vaccinations were included in the national immunization program in November 2010 and July 2012, respectively. Complete vaccine series coverage rates in 2019 were estimated to be 95% for Hib and 92% for PCV-13 [81]. Botswana’s capital and largest city, Gaborone, is located in the country’s South-East district and has a population of 231,626 based on a census conducted in 2011 [82]. The HIV prevalence among adults 15 to 49 years of age in Botswana was 20.7% in 2019 [83]. More than 95% of pregnant women with HIV in Botswana receive antiretroviral therapy, and the mother-to-child HIV transmission rate is estimated to be <2% [83].
Data and biospecimen collection
Mother–infant dyads (n = 179) were enrolled within 72 h of delivery between February 2016 and December 2018 at three sites: a referral hospital in Gaborone, a public clinic in a low-income urban neighborhood in Gaborone, and a public clinic in a rural village located ~15 km outside of Gaborone. Exclusion criteria included maternal age <18 years, infant birth weight <2000 g, multiple gestation pregnancy, and caesarian delivery. Participants were seen for monthly study visits until the infant was six months of age and every other month thereafter until the infant was 12 months of age. At all study visits, a caregiver questionnaire was administered and nasopharyngeal swabs were collected from mothers and infants by trained study personnel. Nasopharyngeal samples were placed directly into MSwab medium (Copan Italia), transported to the National Health Laboratory in Gaborone, and frozen within 4 h of collection to −80 °C. Testing for S. pneumoniae was performed using a quantitative PCR assay targeting the autolysin gene (lytA), as previously described [6, 84]. A dried blood spot was collected from HIV-exposed infants by heel prick at two months of age and, for infants who were breastfed for any duration, again at 12 months of age. These samples were tested for HIV-1 DNA using the Cobas AmpliPrep/Cobas TaqMan HIV-1 Qualitative Assay, version 2.0 (Roche) [85]. All study participants or their legal guardians provided written informed consent to participate in this study. The study protocol was approved by the Botswana Ministry of Health, the Princess Marina Hospital ethics committee, and institutional review boards at the University of Pennsylvania, Children’s Hospital of Philadelphia, McMaster University, and Duke University.
Processing of nasopharyngeal samples for 16S ribosomal RNA gene sequencing
The Duke Microbiome Core Facility extracted DNA from nasopharyngeal samples using Powersoil Pro extraction kits (Qiagen) following the manufacturer’s instructions. DNA concentrations were determined using Qubit dsDNA high-sensitivity assay kits (Thermo Fisher Scientific). Negative extraction and PCR controls were amplified with all four batches of samples included in analyses to evaluate for background contamination. For the first two sample batches, these negative controls were verified to not have visible bands on gel electrophoresis. For the final two sample batches, sequencing was performed on these negative control samples. Bacterial community composition was characterized by PCR amplification of the V4 variable region of the 16S rRNA gene using the forward primer 515 and the reverse primer 806 following the Earth Microbiome Project protocol [86]. These primers carry unique barcodes that allow for multiplexed sequencing. Equimolar 16S rRNA PCR products from all samples were quantified and pooled prior to sequencing. Sequencing was performed by the Duke Sequencing and Genomic Technologies Core Facility on a MiSeq instrument (Illumina, Inc.) configured for 250 base-pair paired-end sequencing. Raw sequences were trimmed using Trimmomatic version 0.36 [87], demultiplexed using QIIME2 tools [88], and analyzed through a pipeline that used DADA2 version 1.16 [89]. ASVs were given taxonomic assignments based on alignment to the expanded Human Oral Microbiome Database version 15.1 [90]. The most abundant genera in negative control samples were Delftia, Brevundimonas, Bacteroides, and Leptothrix. Presumed reagent contaminant ASVs (n = 248) were identified and removed based on presence in negative control samples or negative correlation with DNA concentration using the frequency method (threshold = 0.10) implemented in the decontam R package version 1.12 [91]. Samples with <1000 sequencing reads after quality filtering were excluded. Sequencing reads were classified into 7167 ASVs representing 200 genera from 8 phyla. For each of the 279 ASVs assigned to the genus Corynebacterium, we performed a standard nucleotide REFSEQ BLAST search using the National Center for Biotechnology Information’s Bacteria and Archaea 16S ribosomal RNA project database [59]. We assigned species information to ASVs using a best-hit approach based on the E value with a minimum percent identity of 97%.
Statistical methods for analyzing infant nasopharyngeal microbiome diversity and stability
We calculated alpha (Shannon index and observed ASVs) and beta diversity (Bray-Curtis dissimilarity) using the phyloseq R package version 1.36 [92]. We used Wilcoxon signed-rank tests to compare microbiome alpha diversity across infant ages and between paired infant and maternal samples. To evaluate associations between infant age and alpha diversity measures, we used negative binomial mixed effect models with subject as a random effect to account for repeated sampling of individuals. We compared beta diversity with PERMANOVA using the adonis function within the vegan R package version 2.5.7 [93]; for comparisons of infant samples collected at different ages and between maternal and infant samples, we included a unique identifier for each mother–infant pair as a blocking variable [94]. For initial analyses of specific microbiome features, we aggregated ASVs at the genus level and classified a sample as dominated if 50% or more of the sequencing reads generated from this sample were assigned to a single genus. Samples dominated by a genus other than the six most highly abundant genera were classified in a single “other” category. Samples for which no single genus accounted for the majority of the sequencing reads were classified as “biodiverse.” We considered a sample’s microbiome profile to be “stable” if the next visit’s sample from that infant was classified as the same biotype. We then used logistic regression to evaluate associations between specific nasopharyneal biotypes and microbiome stability, adjusting for infant age in days.
Statistical analyses for analyzing infant nasopharyngeal microbiome composition
We used MaAsLin2 version 1.6 [52] to fit log-transformed linear mixed models evaluating associations between sociodemographic factors and environmental exposures and the relative abundances of bacterial genera within the infant nasopharyngeal microbiome. These analyses considered the following variables identified based on a literature review: sex, low birth weight (<2500 g), HIV exposure status, location of residence (urban vs. rural), household use of solid fuels, number of other child household members (<5 years of age), season (summer vs. winter), breastfeeding, number of PCV-13 doses, and systemic antibiotic exposures [39–41, 44, 45, 95–97]. MaAsLin2 analyses were limited to bacterial genera present in at least 10% of nasopharyngeal samples. The comparisons were corrected for the false discovery rate using the Benjamini–Hochberg procedure and a q value threshold for significance of 0.20. The models included subject as a random effect. To identify compositional features of the nasopharyngeal microbiome that influence the risk of pneumococcal colonization, we first fit a Cox proportional hazards model evaluating the association between nasopharyngeal microbiome biotype and S. pneumoniae colonization detected at the subsequent study visit. Given the observed negative association between a Corynebacterium-dominant biotype and pneumococcal colonization in this model, additional Cox proportional hazards models were fit to evaluate the association between the relative abundance of Corynebacterium, modeled on the genus level and separately for specific highly abundant species, and S. pneumoniae colonization. Two (1%) infants who were colonized with S. pneumoniae at the birth visit were excluded from these analyses. In addition, infants who were not colonized with S. pneumoniae at any study visit were censored at the last visit for which S. pneumoniae colonization data were available. Time was modeled as a continuous variable corresponding to infant age in days. All models were implemented using the survival R package version 3.2.11 [98] and were adjusted for all previously specified sociodemographic factors and environmental exposures, with the season, breastfeeding, PCV-13 doses, and systemic antibiotic exposures modeled as time-dependent covariates.
Laboratory experiments evaluating for pneumococcal growth inhibition by Corynebacterium strains
To isolate Corynebacterium strains from infant nasopharyngeal samples, we streaked 10 µL of sample on plates containing brain heart infusion (BHI) medium (Sigma–Aldrich) supplemented with 50 µg/mL of fosfomycin disodium (Fisher Scientific) and 1% Tween 80 (VWR). We subcultured single bacterial colonies to 5% sheep blood agar plates (Fisher Scientific) and identified bacterial species using a VITEK MS automated mass spectrometry microbial identification system (bioMérieux). We then used cell-free growth inhibition assays to screen Corynebacterium strains for the secretion of anti-pneumococcal factors. Corynebacterium strains were grown in 20 mL of BHI medium supplemented with 0.2% Tween 80 for 12–18 h at 37 °C and 5% CO2. These cultures were centrifuged at 3000 rpm for 10 min to generate cell pellets, and the supernatants were sterile-filtered with a 0.22-µM filter. The resulting cell-free media was diluted 50% in tryptic soy broth (TSB; Fisher Scientific) and glycerol stocks of two strains of S. pneumoniae, one a reference strain (ATCC 6303; serotype 3) and the second strain isolated from an infant nasopharyngeal sample (05-160; serotype 11 A), were separately diluted 1:50 into the diluted cell-free media. Growth was assessed by OD600 readings relative to blank media controls for each growth medium over 24 h.
Supplementary information
Acknowledgements
We would like to thank Copan Italia (Brescia, Italy) for the donation of the MSwab media and flocked swabs used in the collection of nasopharyngeal specimens. We also thank the Duke University School of Medicine for the use of the Microbiome Core Facility, which performed the DNA extractions for this research. We offer sincere gratitude to the children and families who participated in this research.
Author contributions
MSK, APS, TA, KAF, JFR SSS, CKC and PCS contributed to the study concept and design. MSK, CP, YY, JNA, SMP, JHH, RRY, MS, SB, TL, TM, MZP, JJ, CRP collected the data or assisted with data analysis or interpretation. MSK drafted the paper and all other authors revised it critically for important intellectual content. All authors approved of the final version of the paper.
Funding
This research was supported by a Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene Postdoctoral Fellowship in Tropical Infectious Diseases, by Children’s Hospital of Philadelphia and the Pincus Family Foundation, and through core services from the Penn Center for AIDS Research, a National Institutes of Health (NIH)-funded program (P30-AI045008). MSK and CKC received financial support from the NIH through the Duke Center for AIDS Research (P30-AI064518). MSK was supported by a NIH Career Development Award (K23-AI135090) and a research grant from the Society for Pediatric Research. PCS received funding from the NIH through a Research Project Grant (7R01-GM108494). SMP was supported by a VECD Global Health Fellowship, funded by the Office of AIDS Research and the Fogarty International Center of the NIH (D43-TW009337). APS and TAM received financial support from the NIH through the Penn Center for AIDS Research (P30-AI045008).
Availability of data and materials
The sequencing dataset supporting the conclusions of this study is available in the Sequence Read Archive (PRJNA698366). The statistical files and script used for data analyses are also publicly available (https://github.com/mskelly7/Coryne_Sp_manuscript).
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Botswana Ministry of Health, the Princess Marina Hospital ethics committee, and institutional review boards at the University of Pennsylvania, Duke University, and McMaster University. Written informed consent was obtained from all participants or their legal guardians.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01108-4.
References
- 1.Backhaus E, Berg S, Andersson R, Ockborn G, Malmström P, Dahl M, et al. Epidemiology of invasive pneumococcal infections: manifestations, incidence and case fatality rate correlated to age, gender and risk factors. BMC Infect Dis. 2016;16:367. doi: 10.1186/s12879-016-1648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogaert D, de Groot R, Hermans P. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–54. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 3.Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health. 2018;6:e744–57. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAllister DA, Liu L, Shi T, Chu Y, Reed C, Burrows J, et al. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob Health. 2019;7:e47–57. doi: 10.1016/S2214-109X(18)30408-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdullahi O, Karani A, Tigoi CC, Mugo D, Kungu S, Wanjiru E, et al. The prevalence and risk factors for pneumococcal colonization of the nasopharynx among children in Kilifi District, Kenya. PloS ONE. 2012;7:e30787. doi: 10.1371/journal.pone.0030787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly MS, Surette MG, Smieja M, Rossi L, Luinstra K, Steenhoff AP, et al. Pneumococcal colonization and the nasopharyngeal microbiota of children in Botswana. Pediatr Infect Dis J. 2018;37:1176–83. doi: 10.1097/INF.0000000000002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, Pelton SI, et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009;124:e1–e11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoek AJ, Sheppard CL, Andrews NJ, Waight PA, Slack MP, Harrison TG, et al. Pneumococcal carriage in children and adults two years after introduction of the thirteen valent pneumococcal conjugate vaccine in England. Vaccine. 2014;32:4349–55. doi: 10.1016/j.vaccine.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Almeida ST, Nunes S, Paulo ACS, Valadares I, Martins S, Breia F, et al. Low prevalence of pneumococcal carriage and high serotype and genotype diversity among adults over 60 years of age living in Portugal. PloS ONE. 2014;9:e90974. doi: 10.1371/journal.pone.0090974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan SL, Mason EO, Wald ER, Schutze GE, Bradley JS, Tan TQ, et al. Decrease of invasive pneumococcal infections in children among 8 children’s hospitals in the United States after the introduction of the 7-valent pneumococcal conjugate vaccine. Pediatrics. 2004;113:443–9. doi: 10.1542/peds.113.3.443. [DOI] [PubMed] [Google Scholar]
- 11.Hammitt LL, Etyang AO, Morpeth SC, Ojal J, Mutuku A, Mturi N, et al. Effect of ten-valent pneumococcal conjugate vaccine on invasive pneumococcal disease and nasopharyngeal carriage in Kenya: a longitudinal surveillance study. Lancet. 2019;393:2146–54. doi: 10.1016/S0140-6736(18)33005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutts F, Zaman S, Enwere GY, Jaffar S, Levine O, Okoko J, et al. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet. 2005;365:1139–46. doi: 10.1016/S0140-6736(05)71876-6. [DOI] [PubMed] [Google Scholar]
- 13.Congdon M, Hong H, Young RR, Cunningham CK, Enane LA, Arscott-Mills T, et al. Effect of Haemophilus influenzae type b and 13-valent pneumococcal conjugate vaccines on childhood pneumonia hospitalizations and deaths in Botswana. Clin Infect Dis. 2020; e-pub ahead of print 8 July 2020; 10.1093/cid/ciaa919. [DOI] [PMC free article] [PubMed]
- 14.Eskola J, Kilpi T, Palmu A, Jokinen J, Eerola M, Haapakoski J, et al. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl J Med. 2001;344:403–9. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- 15.Pelton SI, Huot H, Finkelstein JA, Bishop CJ, Hsu KK, Kellenberg J, et al. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2007;26:468–72. doi: 10.1097/INF.0b013e31803df9ca. [DOI] [PubMed] [Google Scholar]
- 16.Pichichero ME, Casey JR. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. JAMA. 2007;298:1772–8. doi: 10.1001/jama.298.15.1772. [DOI] [PubMed] [Google Scholar]
- 17.Neves FP, Cardoso NT, Snyder RE, Marlow MA, Cardoso CA, Teixeira LM, et al. Pneumococcal carriage among children after four years of routine 10-valent pneumococcal conjugate vaccine use in Brazil: the emergence of multidrug resistant serotype 6C. Vaccine. 2017;35:2794–800. doi: 10.1016/j.vaccine.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Bradshaw JL, McDaniel LS. Selective pressure: rise of the nonencapsulated pneumococcus. PLoS Pathog. 2019;15:e1007911. doi: 10.1371/journal.ppat.1007911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladhani SN, Collins S, Djennad A, Sheppard CL, Borrow R, Fry NK, et al. Rapid increase in non-vaccine serotypes causing invasive pneumococcal disease in England and Wales, 2000–17: a prospective national observational cohort study. Lancet Infect Dis. 2018;18:441–51. doi: 10.1016/S1473-3099(18)30052-5. [DOI] [PubMed] [Google Scholar]
- 20.Ouldali N, Levy C, Varon E, Bonacorsi S, Béchet S, Cohen R, et al. Incidence of paediatric pneumococcal meningitis and emergence of new serotypes: a time-series analysis of a 16-year French national survey. Lancet Infect Dis. 2018;18:983–91. doi: 10.1016/S1473-3099(18)30349-9. [DOI] [PubMed] [Google Scholar]
- 21.Zaneveld J, Turnbaugh PJ, Lozupone C, Ley RE, Hamady M, Gordon JI, et al. Host-bacterial coevolution and the search for new drug targets. Curr Opin Chem Biol. 2008;12:109–14. doi: 10.1016/j.cbpa.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Steenhuijsen Piters WA, Binkowska J, Bogaert D. Early life microbiota and respiratory tract infections. Cell Host Microbe. 2020;28:223–32. doi: 10.1016/j.chom.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke H, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–2. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 24.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14:1584. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiri T, Nunes MC, Adrian PV, Van Niekerk N, Klugman KP, Madhi SA. Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother–child dyads. BMC Infect Dis. 2013;13:483. doi: 10.1186/1471-2334-13-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby P, Watson K, Bowman J, Taylor A, Riley TV, Smith DW, et al. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25:2458–64. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nzenze S, Shiri T, Nunes M, Klugman K, Kahn K, Twine R, et al. Temporal association of infant immunisation with pneumococcal conjugate vaccine on the ecology of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus nasopharyngeal colonisation in a rural South African community. Vaccine. 2014;32:5520–30. doi: 10.1016/j.vaccine.2014.06.091. [DOI] [PubMed] [Google Scholar]
- 28.Faden H, Stanievich J, Brodsky L, Bernstein J, Ogra PL. Changes in nasopharyngeal flora during otitis media of childhood. Pediatr Infect Dis J. 1990;9:623–6. [PubMed] [Google Scholar]
- 29.Shekhar S, Khan R, Schenck K, Petersen FC. Intranasal Immunization with the commensal Streptococcus mitis confers protective immunity against pneumococcal lung infection. Appl Environ Microbiol. 2019;85:e02235–18. doi: 10.1128/AEM.02235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cangemi de Gutierrez R, Santos V, Nader-Macias ME. Protective effect of intranasally inoculated Lactobacillus fermentum against Streptococcus pneumoniae challenge on the mouse respiratory tract. FEMS Immunol Med Microbiol. 2001;31:187–95. doi: 10.1111/j.1574-695X.2001.tb00519.x. [DOI] [PubMed] [Google Scholar]
- 31.Wong SS, Quan Toh Z, Dunne EM, Mulholland EK, Tang ML, Robins-Browne RM, et al. Inhibition of Streptococcus pneumoniae adherence to human epithelial cells in vitro by the probiotic Lactobacillus rhamnosus GG. BMC Res Notes. 2013;6:135. doi: 10.1186/1756-0500-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011;2:e00245–10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio. 2016;7:e01725–15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cope EK, Goldstein-Daruech N, Kofonow JM, Christensen L, McDermott B, Monroy F, et al. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PloS ONE. 2011;6:e28523. doi: 10.1371/journal.pone.0028523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PloS Pathog. 2005;1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weimer KE, Juneau RA, Murrah KA, Pang B, Armbruster CE, Richardson SH, et al. Divergent mechanisms for passive pneumococcal resistance to β-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis. 2011;203:549–55. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tikhomirova A, Kidd SP. Haemophilus influenzae and Streptococcus pneumoniae: living together in a biofilm. Pathog Dis. 2013;69:114–26. doi: 10.1111/2049-632X.12073. [DOI] [PubMed] [Google Scholar]
- 38.Brugger SD, Eslami SM, Pettigrew MM, Escapa IF, Henke MT, Kong Y, et al. Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere. 2020;5. [DOI] [PMC free article] [PubMed]
- 39.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–15. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol. 2015;135:905–12. doi: 10.1016/j.jaci.2014.12.1909. [DOI] [PubMed] [Google Scholar]
- 41.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–92. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 42.Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine. 2016;9:336–45. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 45.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, Van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PloS ONE. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruce N, Perez-Padilla R, Albalak R. Indoor air pollution in developing countries: a major environmental and public health challenge. Bull World Health Organ. 2000;78:1078–92. [PMC free article] [PubMed] [Google Scholar]
- 47.Pelissari DM, Diaz-Quijano FA. Household crowding as a potential mediator of socioeconomic determinants of tuberculosis incidence in Brazil. PloS ONE. 2017;12:e0176116. doi: 10.1371/journal.pone.0176116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mannucci PM, Franchini M. Health effects of ambient air pollution in developing countries. Int J Environ Res Public Health. 2017;14:1048. doi: 10.3390/ijerph14091048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. bioRxiv. 2021. 10.1101/2021.01.20.427420. [DOI] [PMC free article] [PubMed]
- 53.Hojsak I, Snovak N, Abdović S, Szajewska H, Mišak Z, Kolaček S. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin Nutr. 2010;29:312–6. doi: 10.1016/j.clnu.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Gluck U, Gebbers JO. Ingested probiotics reduce nasal colonization with pathogenic bacteria (Staphylococcus aureus, Streptococcus pneumoniae, and beta-hemolytic streptococci) Am J Clin Nutr. 2003;77:517–20. doi: 10.1093/ajcn/77.2.517. [DOI] [PubMed] [Google Scholar]
- 55.Feleszko W, Jaworska J, Rha RD, Steinhausen S, Avagyan A, Jaudszus A, et al. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin Exp Allergy. 2007;37:498–505. doi: 10.1111/j.1365-2222.2006.02629.x. [DOI] [PubMed] [Google Scholar]
- 56.Nhan T-X, Parienti J-J, Badiou G, Leclercq R, Cattoir V. Microbiological investigation and clinical significance of Corynebacterium spp. in respiratory specimens. Diagn Microbiol Infect Dis. 2012;74:236–41. doi: 10.1016/j.diagmicrobio.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Díez-Aguilar M, Ruiz-Garbajosa P, Fernández-Olmos A, Guisado P, Del Campo R, Quereda C, et al. Non-diphtheriae Corynebacterium species: an emerging respiratory pathogen. Eur J Clin Microbiol Infect Dis. 2013;32:769–72. doi: 10.1007/s10096-012-1805-5. [DOI] [PubMed] [Google Scholar]
- 58.Teutsch B, Berger A, Marosevic D, Schönberger K, Lâm T-T, Hubert K, et al. Corynebacterium species nasopharyngeal carriage in asymptomatic individuals aged ≥ 65 years in Germany. Infection. 2017;45:607–11. doi: 10.1007/s15010-017-1017-0. [DOI] [PubMed] [Google Scholar]
- 59.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner P, Turner C, Green N, Ashton L, Lwe E, Jankhot A, et al. Serum antibody responses to pneumococcal colonization in the first 2 years of life: results from an SE Asian longitudinal cohort study. Clin Microbiol Infect. 2013;19:e551–8. doi: 10.1111/1469-0691.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Numminen E, Chewapreecha C, Turner C, Goldblatt D, Nosten F, Bentley SD, et al. Climate induces seasonality in pneumococcal transmission. Sci Rep. 2015;5:11344. doi: 10.1038/srep11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelly MS, Smieja M, Luinstra K, Wirth KE, Goldfarb DM, Steenhoff AP, et al. Association of respiratory viruses with outcomes of severe childhood pneumonia in Botswana. PloS ONE. 2015;10:e0126593. doi: 10.1371/journal.pone.0126593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.le Roux DM, Myer L, Nicol MP, Zar HJ. Incidence and severity of childhood pneumonia in the first year of life in a South African birth cohort: the Drakenstein Child Health Study. Lancet Glob Health. 2015;3:e95–103. doi: 10.1016/S2214-109X(14)70360-2. [DOI] [PubMed] [Google Scholar]
- 64.von Mollendorf C, von Gottberg A, Tempia S, Meiring S, de Gouveia L, Quan V, et al. Increased risk and mortality of invasive pneumococcal disease in HIV-exposed-uninfected infants <1 year of age in South Africa, 2009-2013. Clin Infect Dis. 2015;60:1346–56. doi: 10.1093/cid/civ059. [DOI] [PubMed] [Google Scholar]
- 65.Farley JJ, King JC, Jr., Nair P, Hines SE, Tressler RL, Vink PE. Invasive pneumococcal disease among infected and uninfected children of mothers with human immunodeficiency virus infection. J Pediatr. 1994;124:853–8. doi: 10.1016/s0022-3476(05)83170-1. [DOI] [PubMed] [Google Scholar]
- 66.Kinabo GD, van der Ven A, Msuya LJ, Shayo AM, Schimana W, Ndaro A, et al. Dynamics of nasopharyngeal bacterial colonisation in HIV-exposed young infants in Tanzania. Trop Med Int Health. 2013;18:286–95. doi: 10.1111/tmi.12057. [DOI] [PubMed] [Google Scholar]
- 67.Koliou MG, Andreou K, Lamnisos D, Lavranos G, Iakovides P, Economou C, et al. Risk factors for carriage of Streptococcus pneumoniae in children. BMC Pediatr. 2018;18:1–8. doi: 10.1186/s12887-018-1119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.List of prokaryotic names with standing in nomenclature. Available at: http://www.bacterio.net/corynebacterium.html. Accessed 4 February 2021.
- 69.Efstratiou A, George R. Microbiology and epidemiology of diphtheria. Rev Med Microbiol. 1996;7:31–42. [Google Scholar]
- 70.Spach DH, Opp DR, Gabre-Kidan T. Bacteremia due to Corynebacterium jeikeium in a patient with AIDS. Rev Infect Dis. 1991;13:342–3. doi: 10.1093/clinids/13.2.342. [DOI] [PubMed] [Google Scholar]
- 71.Wang C, Mattson D, Wald A. Corynebacterium jeikeium bacteremia in bone marrow transplant patients with Hickman catheters. Bone Marrow Transplant. 2001;27:445. doi: 10.1038/sj.bmt.1702808. [DOI] [PubMed] [Google Scholar]
- 72.Morris A, Guild I. Endocarditis due to Corynebacterium pseudodiphtheriticum: five case reports, review, and antibiotic susceptibilities of nine strains. Rev Infect Dis. 1991;13:887–92. doi: 10.1093/clinids/13.5.887. [DOI] [PubMed] [Google Scholar]
- 73.Bookani KR, Marcus R, Cheikh E, Parish M, Salahuddin U. Corynebacterium jeikeium endocarditis: a case report and comprehensive review of an underestimated infection. IDCases. 2018;11:26–30. doi: 10.1016/j.idcr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Renom F, Gomila M, Garau M, Gallegos M, Guerrero D, Lalucat J, et al. Respiratory infection by Corynebacterium striatum: epidemiological and clinical determinants. N. Microbes N. Infect. 2014;2:106–14. doi: 10.1002/nmi2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bittar F, Cassagne C, Bosdure E, Stremler N, Dubus J-C, Sarles J, et al. Outbreak of Corynebacterium pseudodiphtheriticum infection in cystic fibrosis patients, France. Emerg Inf Dis. 2010;16:1231. doi: 10.3201/eid1608.100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stubbendieck RM, May DS, Chevrette MG, Temkin MI, Wendt-Pienkowski E, Cagnazzo J, et al. Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl Environ Microbiol. 2019;85:e02406–18. doi: 10.1128/AEM.02406-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiryukhina N, Melnikov V, Suvorov A, Morozova YA, Ilyin V. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiotics Antimicrob Proteins. 2013;5:233–8. doi: 10.1007/s12602-013-9147-x. [DOI] [PubMed] [Google Scholar]
- 78.Lappan R, Peacock CS. Corynebacterium and Dolosigranulum: future probiotic candidates for upper respiratory tract infections. Microbiol Aust. 2019;40:172–7. [Google Scholar]
- 79.Neal EFG, Nguyen C, Ratu FT, Matanitobua S, Dunne EM, Reyburn R, et al. A comparison of pneumococcal nasopharyngeal carriage in very young Fijian infants born by vaginal or cesarean delivery. JAMA Netw Open. 2019;2:e1913650. doi: 10.1001/jamanetworkopen.2019.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.UN Interagency Group for Child Mortality Estimation. Levels & trends in child mortality, report 2020. Available at: https://childmortality.org/wp-content/uploads/2020/09/UNICEF-2020-Child-Mortality-Report.pdf. Accessed 3 Nov 2020.
- 81.United Nations Children’s Fund, World Health Organization. Botswana: WHO and UNICEF estimates of immunization coverage, 2019 revision. Available at: https://www.who.int/immunization/monitoring_surveillance/data/bwa.pdf. Accessed 18 Jan 2021.
- 82.Statistics Botswana. 2011 Population and Housing Census Analytical Report. Available at: http://www.cso.gov.bw/images/analytical_report.pdf. Accessed 24 July 2016.
- 83.Joint United Nations Programme on HIV/AIDS. UNAIDS estimates 2019: Botswana. Available at: http://www.unaids.org/en/regionscountries/countries/botswana. Accessed 3 Feb 2021.
- 84.McAvin JC, Reilly PA, Roudabush RM, Barnes WJ, Salmen A, Jackson GW, et al. Sensitive and specific method for rapid identification of Streptococcus pneumoniae using real-time fluorescence PCR. J Clin Microbiol. 2001;39:3446–51. doi: 10.1128/JCM.39.10.3446-3451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gueye SB, Diop-Ndiaye H, Diallo MM, Ly O, Sow-Ndoye A, Diagne-Gueye ND, et al. Performance of Roche CAP/CTM HIV-1 qualitative test version 2.0 using dried blood spots for early infant diagnosis. J Virol Methods. 2016;229:12–15. doi: 10.1016/j.jviromet.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 86.Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Brown CT, et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genom Sci. 2010;3:243–8. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Escapa IF, Huang Y, Chen T, Lin M, Kokaras A, Dewhirst FE, et al. Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets. Microbiome. 2020;8:1–16. doi: 10.1186/s40168-020-00841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:719. [Google Scholar]
- 94.Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 95.Biesbroek G, Wang X, Keijser BJ, Eijkemans RM, Trzcinski K, Rots NY, et al. Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerg Infect Dis. 2014;20:201–10. doi: 10.3201/eid2002.131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med. 2016;8:349ra100. doi: 10.1126/scitranslmed.aaf5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214:1924–8. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Therneau T. A package for survival analysis in R. R package version 3.2.11, https://CRAN.R-project.org/package=survival.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing dataset supporting the conclusions of this study is available in the Sequence Read Archive (PRJNA698366). The statistical files and script used for data analyses are also publicly available (https://github.com/mskelly7/Coryne_Sp_manuscript).