Abstract
Corona Virus Disease 2019 (COVID-19) is a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This pandemic has brought the world to a standstill and threatened human lives. Many methods are known to date to detect this virus. Due to their relative sensitivity, polymerase chain reaction (PCR)-based assays are the most frequently applied and considered the gold standard. However, due to the rapid mutation rate of the viral genome and the emergence of new variants, existing protocols need to be updated and improved. Designing a fast and accurate PCR-based assay is of great importance for the early detection of this virus and more efficient control of the spread of this disease. This study describes a fast, reliable, easy-to-use, and high-throughput multiplex SARS-CoV-2 RT-PCR detection method. The assay was designed to detect two viral genes (N and RdRP) and a human gene (RP) simultaneously. The performance and the sensitivity of the assay were tested in 28 SARS-CoV-2 positive samples and compared with commercial kits, which showed 100% positive percent agreement with a limit of detection (LOD) value of 1.40 and 0.81 copies/µL or 35.13 and 20.31 copies/reaction for RdRP and N genes, respectively. The current assay is found accurate, reliable, simple, sensitive, and specific. It can be used as an optimized SARS-CoV-2 diagnostic assay in hospitals, medical centers, and diagnostic laboratories as well as for research purposes.
Subject terms: Biological techniques, Molecular medicine
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the subfamily Orthocoronavirinae of the family Coronaviridae of the order Nidovirales. As of December 18, 2021, about 273 million cases and almost 5.35 million deaths are related to SARS-CoV-21. The pandemic that started in late 2019 and continued to increase in 2020 has forced scientists and clinicians to develop new methods for diagnosis and treatment2. In addition to some known methods, studies to develop faster, easier, and more reliable methods for SARS-CoV-2 diagnosis continue intensively. These methods are mainly divided into nucleic acid-based amplification tests and serological tests. The nucleic acid-based amplification methods, such as rRT-PCR, are based on the amplification of viral RNA3. On the other hand, serological tests are based on detecting either the proteins of the COVID-19 virus or human antibodies generated in response to infection such as the immunoglobulin type M (IgM) or immunoglobulin type G (IgG). The most important disadvantage of serological tests is their limited sensitivity to early detection4. In general, the development of specific antibodies against the virus begins after the first week, and IgM and IgG production occurs mostly in the second week5. Therefore, the sensitivity of serological tests is limited in the acute stage of infection. Another drawback is the possibility of similar antibody responses to the viruses in the same or close families and the possibility of cross-reactivity. This possibility is a serious concern as most human coronaviruses are antigenically close related to each other. Therefore, the WHO and CDC do not recommend the use of point-of-care immunodiagnostic tests for clinical decision-making. Instead, serological tests can be used for research or clinical support purposes6–8.
On the other hand, the reverse transcription polymerase chain reaction (RT-PCR) is the most common and accepted as the gold standard method for the viral detection9. Although RT-PCR is a standard assay, it might produce false-negative and false-positive results due to lack of specificity and sensitivity9–11. According to Fang et al.12, RT-PCR was able to detect only 71% (36/51) of SARS-CoV-2 infections. This may be due to the low sensitivity of the test, low patient viral load, or inappropriate clinical sampling12. The probability of obtaining a true positive result decreases with time from symptom onset. Nasopharyngeal and oropharyngeal RT-PCR tests performed on the 10th day of the first symptom have a 25% and 47% chance of being false-negative, respectively13. Reports showed that patients with negative initial tests became positive after 3–5 repeated swabs14. The rate of false-negative result changes in between 1 and 30%15. Therefore, the sensitivity of this method should be improved. For this purpose, multiplex PCR protocol is proposed that targets multiple genes in the same reaction. By doing so, at least two viral genes are targeted that help to increase the probability to catch the virus especially in the patients having low viral load. In addition, a human housekeeping gene can be added as an internal control to prevent false-negative results that are due to inefficient sampling.
The genome of SARS-CoV-2 contains 14 ORFs encoding for 27 proteins16. The genome of this virus is composed of four major structural genes that are translated to spike protein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N)16, along with some accessory genes. The surface glycoproteins (S1 and S2) are responsible for binding to the ACE2 (Angiotensin-converting enzyme 2) receptors on the host cell allowing the virus to invade, where S1 bind to the ACE2 receptor and S2 fuses with the host cell membrane17,18. In addition, the genome harbors genes encoding non-structural proteins (Nsp) such as RdRP (Nsp12), the RNA-dependent RNA polymerase enzyme responsible for the replication of the viral genome19,20.
RT-PCR tests have been developed to target RdRP, E, N, or S genes10,21,22. Among these, those targeting RdRP gene were found to be the ones with the highest analytical sensitivity9,23. In addition, the human ribonuclease P (RNase P or RP) gene (responsible for the processing of tRNA molecules) is used as an internal control in multiplex RT-PCR protocols recommended by WHO and CDC24–26. There is also a need for studies to compare between recently developed assays based on their reproducibility, and sensitivity. Therefore, in addition to the experimental design, determining which genes to be targeted by the assay, are also extremely important goals to achieve when designing molecular testing protocols. Although multiplex RT-PCR method helps to improve the specificity, the design of multiplex primers and probe sets is very critical for the PCR efficiency. The formation of self-dimer or hetero-dimer structure reduce the target specificity and may lead to misinterpretation of the results. Therefore, the experimental design as well as the selection of the best primer and probe sets are crucial and needs standardization. Together with, real-time RT-PCR results may be affected by variations in viral RNA sequences27. A recent study estimated the nucleotide mutation rate of the SARS-CoV-2 genome as 6.677 × 10−4 substitution per site per year28, which lead loss of the assay sensitivity as shown by Peñarrubia et al.29. The emergence of new variants and the high mutation rate of the viral genome require updating existing diagnostic tests.
In this study, a multiplex real time RT-PCR (rRT-PCR) assay was designed and evaluated for the diagnosis of SARS-CoV-2 including the most recent variant of concerns (VOC). The study exhibits primer sets specific to SARS-CoV-2 and the accompanying optimized reaction conditions. The developed assay simultaneously detects viral N, RdRP and human RP genes in the same rRT-PCR reaction. The clinical performance of the test was screened with RNA samples from SARS-CoV-2 positive patients.
Materials and methods
SARS-CoV-2 genome sequences
In total, 344 SARS-CoV-2 genomes belongs to variants of concern (VOC) including alpha (B.1.1.7), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) were analyzed. The sequences were retrieved from GISAID database30 and chosen by covering all continents including Europa, North and South America, Asia, Africa, and Oceania. The sequences were aligned by using MAFFT software (https://mafft.cbrc.jp/alignment/server/) with default settings31. Then, the interested genome regions harboring the primer target sites for N and RdRP genes were selected and a possible substitution or single nucleotide polymorphism at the primer binding sites were screened by using JalView (v2.11.1.3) program32 (Supplementary Figs. S4 and S5). The consensus sequences (100% alignments) corresponding to the target genes were selected for primer design. The primers were synthesized by Molequle-On (Auckland, New Zealand) and purified by high-performance liquid chromatography.
Multiplex primer/probe design
To design the most suitable multiplex primers, PrimerPooler33, PrimerPlex (http://www.premierbiosoft.com/primerplex/index.html) and Primer334 programs were used with default settings. The possible secondary structures such as homo-dimer, hetero-dimer, and hairpin were checked by using OligoAnalyzer™ Tool of Integrated DNA Technologies (IDT) (https://eu.idtdna.com/pages/tools/oligoanalyzer). In the selection of SARS-CoV-2 primers, attention was paid to the selection of genome regions that differ from other SARS-CoV relatives (NC_004718.3, AY613947.1, AY502927.1, AY278491.2, AY502924.1, and AY559094.1) (Supplementary Figs. S5 and S6). Therefore, the primers are specific to SARS-CoV-2 virus only and are expected to be free from possible cross reactions with other SARS viruses. Each gene-specific probe was labeled with different fluorescent dye: fluorescein amidides (FAM) for the viral RdRp gene, hexachloro-fluorescein (HEX) for the viral N gene, and carboxyrodamine (ROX) for the human RP gene. The concentration, size, and sequence of each primer or probe are indicated in Table 1.
Table 1.
The sequence and concentration of primer and probe sets used in PCR reactions.
| Primer/probe | Sequence (5′–3′) | Working conc. (µM) | Final conc. (µM)a | Size (bp) | Reference |
|---|---|---|---|---|---|
| RdRp—F | CCTCACTTGTTCTTGCTCGC | 20 | 1 | 205 | This study |
| RdRp—R | GCCGTGACAGCTTGACAAAT | 20 | 1 | ||
| RdRp—Probe | FAM-GTGAAATGGTCATGTGTGGC-BHQ1 | 5 | 0.2 | ||
| N–F | TGAAACTCAAGCCTTACCGC | 20 | 1 | 160 | This study |
| N—R | TATAGCCCATCTGCCTTGTG | 20 | 1 | ||
| N—Probe | HEX-ATCCATGAGCAGTGCTGAC-BHQ | 5 | 0.2 | ||
| RP—F | AGATTTGGACCTGCGAGCG | 20 | 1 | 92 | Universal |
| RP—R | GATAGCAACAACTGAATAGCCAAGGT | 20 | 1 | ||
| RP—Probe | ROX-TTCTGACCTGAAGGCTCTGCGCG-BHQ2 | 5 | 0.2 |
aFinal concentration represents the concentration of each oligonucleotide in reaction mixture (20 µL).
Sample collection and RNA isolation
The RNA samples were extracted from nasopharyngeal swab or combined nasopharyngeal/oral swab collected from the patients at King Fahad Specialist Hospital (KFSH), Dammam, Saudi Arabia, between 01 and 30 November 2020. The swabs were collected in Virus Liquid Transport Medium-VTM per manufacture instruction for collecting and handling (Copan, U.S.A). Specimens were transported to the lab in cool box and kept refrigerated for not more than 8 h till time of nucleic acid extraction.
Prior to RNA extraction, collection tubes were vortexed; 200 or 400 µL of VTM were transferred to 2 mL tubes for RNA extraction using Magna Pure Compact (Roche, Germany) or ELITe InGenius (ELITechGroup, France) systems with elution set to 100 µL. Finally, 5 µL of extracted RNA was used as template for the rRT-PCR.
rRT-PCR conditions
The following reaction mixture was prepared in a micro-centrifuge tube: 2 µL of 10× Buffer (Procomcure Biotech, Austria), 0.25 µL of dNTPs (10 mM each; Procomcure Biotech, Austria), 0.2 µL of uracil-DNA glycosylase (UDG) (1 U/µL; New England BioLabs Inc., USA), 0.25 µL of molecular grade dimethyl sulfoxide (DMSO, Sigma-Aldrich), primer and probe mixture (Molequle-On, New Zealand), 0.4 µL of VitaTaq HS polymerase (2 U/µL; (Procomcure Biotech, Austria), 0.5 µL of M-MuLV Reverse Transcriptase (200 U/µL) (New England BioLabs Inc., USA), 5 µL of RNA template and RNase/DNase-free ddH2O up to 20 µL.
The reaction mixture was transferred to a 96-well plate (MicroAmp™ Fast Optical 96-well Reaction Plate 0.1 mL, Applied Biosystems) and sealed with transparent optical film (MicroAmp™ Optical Adhesive Film, Applied Biosystems). Pseudoviral RNA containing the viral RdRP and N genes and human RNaseP (RP) mRNA sequences were used as positive control material (EURM-019, single stranded SARS-CoV-2 RNA fragments https://crm.jrc.ec.europa.eu/p/EURM-019). In negative control samples, RNase/DNase-free ddH2O was added instead of the RNA template.
Applied Biosystems™, 7500 Fast Real-Time PCR system was used for rRT-PCR reactions. Prior to this procedure, the instrument was calibrated using the Applied Biosystems™ 7500 Rapid Real Time PCR Systems Spectral Calibration Kit. The following conditions were applied for the rRT-PCR reaction: (1) reverse transcription at 42 °C for 15 min, (2) pre-denaturation at 95 °C for 5 min. For the cyclic reactions (40×) (3) denaturation at 95 °C for 5 s and (4) amplification at 60 °C for 30 s. The fluorescence reading was performed at the amplification step. The reporter dye channel was determined as FAM for the RdRp gene, VIC for the N gene, and ROX for the RNAseP (RP) gene. For the Applied Biosystems™ real time PCR instrument (7500 and StepOne models), "passive reference" is set to "none".
Amplification efficiency and analytical sensitivity
The rRT-PCR amplification efficiency (E) was calculated for each viral gene. For this, dilution series of template RNA (Reference No: EURM-019, European Commission Joint Research Centre) was prepared and a standard curve was generated. Ct values were drawn for the logarithmic measurement of the template used. Amplification efficiency was calculated according to the following formula:
| 1 |
The RNA concentration was determined by Nanodrop 2000 (Thermo Fisher Scientific, USA) and the copy number was determined according to the following formula35,36:
| 2 |
Multiple units are used to display limit-of-detection (LOD), for example viral genomic RNA copies per milliliter of transfer medium (copies/mL), copies/µL, copies per reaction volume, or molarity of the test target, etc., which are sometimes confused37. To demonstrate the sensitivity of current assay, the LOD number was calculated and expressed in two different units: (1) the number of copies in one µL of the reaction mixture (copies/µL) and (2) the number of copies in the reaction (copies/reaction). For this, serial dilution of synthetic RNA (5 × 104, 5 × 103, 5 × 102, 5 × 101, and 5 × 100 copies/µL) was prepared. Since 5 µL of template solution was used in the PCR reaction, the lowest dilution (5 × 100 copies/µL) contains 25 copies of RNA. In total, 25 copies of RNA in 20 µL of reaction mixture corresponds to 1.25 copies/µL or 25 copies/reaction. In addition, the probit regression analysis was performed by using MedCalc program (version 19.2; MedCalc Software, Ostend, Belgium).
Diagnostic performance
To validate the new assay (called as mCoV-2), we performed rRT-PCR by using commercially available SARS-CoV-2 detection kits. For this purpose, the same RNA samples (n = 28) that were previously extracted from the COVID19 patients were used as the template. The reactions were run using either GeneFinderTMCOVID-19 Plus RealAmp Kit (GeneFinder, Korea) or RealStar SARS-CoV-2 RT-PCR Kit 1.0 (Altona, Germany).
Data analysis
Amplification curves of viral and human genes were considered to evaluate the results. After automatic adjustment of the cycle threshold (Ct or Cq) line, the representative Ct value of each gene was determined by using ABI 7500 software (v2.3). The positive cut-off value was set at cycle threshold number ≤ 37 with a sigmoidal curve. Any patient meets the criteria were accepted as positive.
Ethical approval
The study is approved by the Institutional Review Board (IRB) at Imam Abdulrahman bin Faisal University (IAU) with an IRB number of IRB-2020-13-406. All methods were carried out in accordance with relevant guidelines and regulations. The de-identified samples left over after completion of diagnostic tests were used; hence this study requires no consenting as per institutional ethics committee regulations and informed consent.
Results
Standardization of the multiplex rRT-PCR
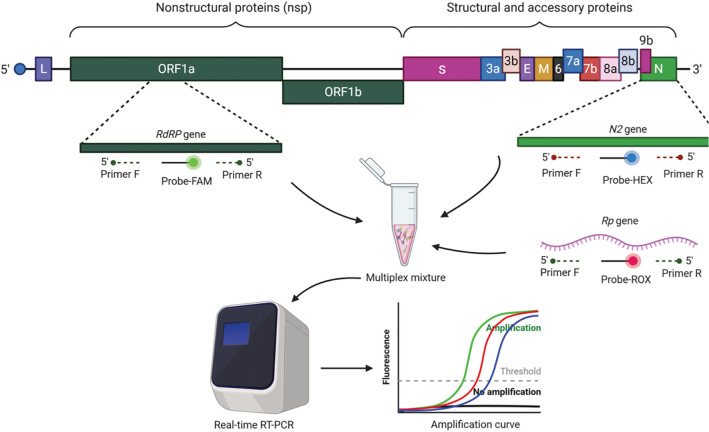
A multiplex rRT-PCR assay was optimized for the diagnosis of SARS-CoV-2. The assay simultaneously targets two viral genes (RdRP and N) and one human gene (RP) as internal control (Fig. 1). The assay tested in 28 RNA samples collected from COVID-19 positive individuals. Supplementary Figure S1 exhibits the rRT-PCR outputs belonging to COVID-19 positive and negative individuals. In COVID-19 positive samples, the RP, RdRP, and N genes amplified simultaneously, forming S-shaped sigmoidal curves (Supplementary Fig. S1a). In the COVID-19 negative specimen, the internal control gene (RP) was the only gene amplified with a sigmoidal amplification curve (Supplementary Fig. S1b). In the positive control reactions, pseudoviral RNA including N and RdRP regions and a human RP mRNA was used as template. The amplification curves were obtained for all targeted genes (Supplementary Fig. S1c). In the negative control reactions, ddH2O was used as the template, which led no amplification line without primer dimer formation (homo-dimer or hetero-dimer). The results showed that the multiplex primer and probe design successfully amplify all targeted genes both in SARS-CoV-2 positive specimen and synthetic positive control samples without forming primer dimer or self-amplification.
Figure 1.
Genome structure of SARS-CoV-2 and the targeted genes in multiplex rRT-PCR assay.
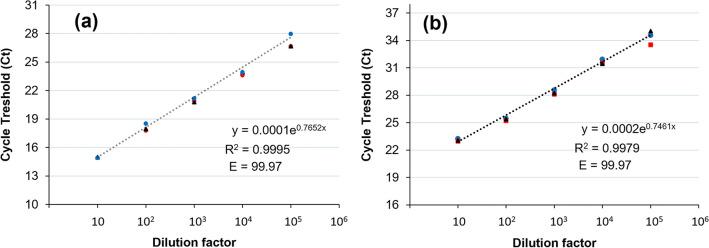
In addition, the standard curve analysis was performed to test the sensitivity of the assay. For this purpose, a dilution series of clinical RNA was prepared with a dilution factor range of 105 to 101 (Fig. 2; Supplementary Fig. S2). Triplicate rRT-PCR analysis revealed that the results are consistent across technical replicates. The test ran successfully even on samples diluted 105 times. The rRT-PCR efficiency for both RdRP and N genes is 99.7 and the R2 value is > 0.997, which shows the consistency and reliability of the assay.
Figure 2.
Standard curve analysis for multiplex rRT-PCR of (a) RdRP and (b) N primers. The template RNA was serially diluted with a range of 105 to 101. The reactions were carried out in triplicate. The amplification efficiencies (E) were shown on each graph. The error bars represent the standard deviation between the replicates. The error bars represent the standard deviation between the replicates. The amplification plots are shown in Supplementary Fig. S2.
Validation of the assay
The validation of the results has been performed by using two different commercially available kits (GeneFinderTMCOVID-19 Plus RealAmp Kit (GeneFinder, Korea) and RealStar SARS-CoV-2 RT-PCR Kit 1.0 (Altona, Germany)) that are targeting different genes such as RdRP, N, S, and E. Among 28 clinically ‘confirmed’ SARS-CoV-2 positive samples, the current assay found 25 positives and three negatives (Supplementary Table S1). Accordingly, the Ct value equals and lower than 37 is accepted as positive. Besides, in both assays, the Ct score of those negative samples was higher than 37, which is out of the CDC and WHO recommendations38,39. Therefore, the samples having a Ct score of ≥ 37.01 are accepted as SARS-CoV-2 negative. In this case, the assay exhibited 100% positive percent agreement with those commercial assays. The distribution of Ct value obtained from both commercial methods and this mCoV-2 assay are displayed in Fig. 3. Since these kits target different genes, the Ct scores of those genes were combined. Accordingly, it is obvious that the average Ct value of the current assay is lower than those of the genes targeted in the comparative commercial kits. This result demonstrates the high sensitivity of the current assay.
Figure 3.
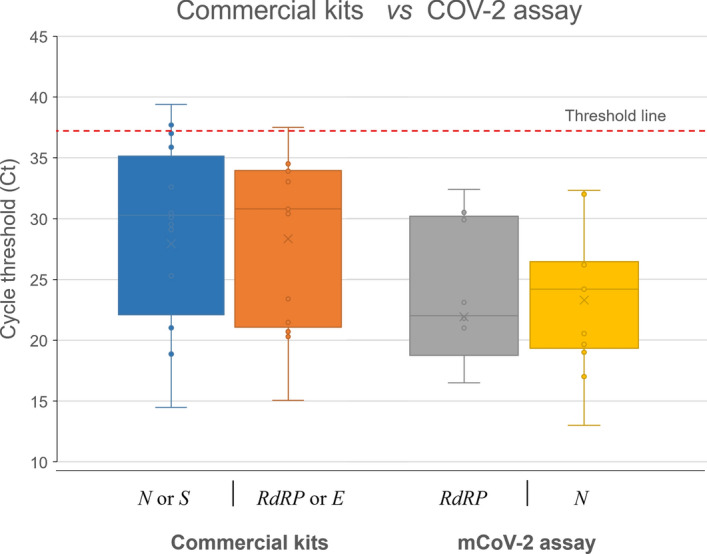
The cycle threshold (Ct) scores of the same clinical samples tested either the current mCoV-2 assay or commercial kits. Each bar represents different genes, which are RdRP (gray) and N (yellow) for mCoV-2 assay; and N or S (blue) and RdRP or E (orange) for commercial kits. Dashed line shows the positivity cut-off level equals to Ct 37.
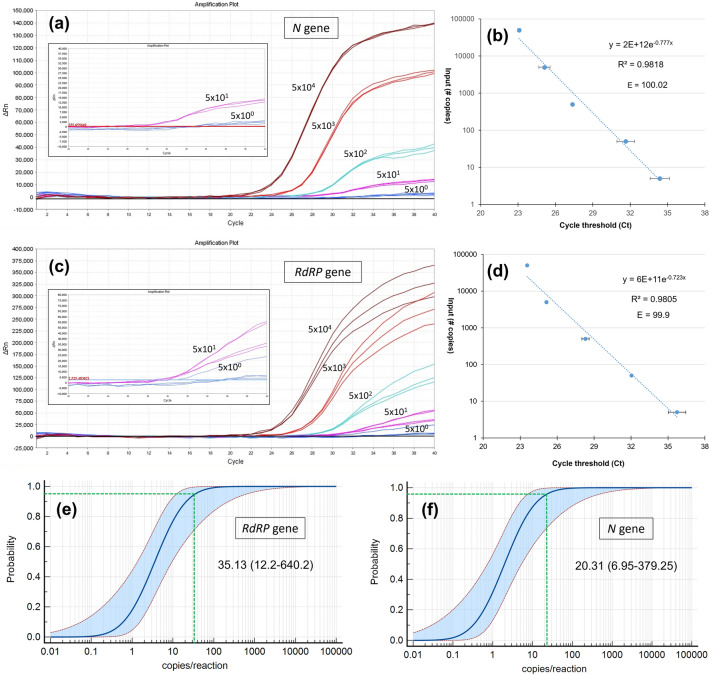
Limit-of-detection (LOD) and rRT-PCR efficiency
A serial dilution of synthetic RNA (5 × 104, 5 × 103, 5 × 102, 5 × 101, and 5 × 100 copies/µL) was prepared to find out the limit-of-detection (LOD) for RdRP and N genes. The amplification plots, the amplification efficiencies (E), and R2 score are represented in Fig. 4. The probit regression analysis of the serially diluted synthetic RNAs (at least 10 replicates) determined the LOD of N gene as 0.81 copy/µL or 20.31 copy/reaction (Fig. 4e). Together with, the LOD of RdRP gene was computed as 1.40 copy/µL or 35.13 copy/reaction (Fig. 4f). The estimated 95% confidence intervals (CI) for the N and RdRP genes are between 6.95 and 379.25 and 12.2 and 640.2 copies/reaction, respectively. The standard curve analysis revealed that the E value of N and RdRP genes are 100.2 and 99.9, respectively. The R2 values are 0.9818 for the N and 0.9805 for the RdRP gene.
Figure 4.
Determination of the limit of detection (LOD) for RdRP and N primers. The 5 × 104 copy/µl pseudoviral RNA was serially diluted. The amplification plots (a,c) and the amplification efficiencies (E) (b,d) for N and RdRP genes were represented, respectively. The R2 value of the trendline and the efficiency (E) of the standard curve were displayed on each graph. The error bars represent the standard deviation between the replicates. The probit regression analysis of the LODs based on 10 technical replicates of serially diluted synthetic RNA: (e) RdRP and (f) N genes. Green dotted lines indicate LOD and values are denoted with 95% confidence intervals (CI) in parenthesis.
Discussion
Since rRT-PCR method is considered the gold standard in the diagnosis of SARS-CoV-2, WHO and CDC recommend it as the diagnostic test for asymptomatic and mildly symptomatic patients6,40. However, rRT-PCR methods also have some drawbacks such as possible false-negative or false-positive results, the cost, etc41. In order to eliminate or minimize those drawbacks, multiplex rRT-PCR methods have been developed that target more than one gene at the same time. By doing this, it is aimed to improve rRT-PCR efficiency and sensitivity. Until now, many studies have been conducted to find the method that can detect the SARS-CoV-2 RNA with the highest sensitivity. For this, different combinations of targeted viral genes were tested in multiplex. According to the WHO recommendations, four viral genes (RdRp, E, N and S genes) can be used in multiplex rRT-PCR reactions in different combinations42. Along with these genes, primer and probe sequences of the human internal/positive control RNase P (RP) gene have been published and their use has been recommended by US CDC24. The studies to improve these protocols are still ongoing. Designing primers with the highest sensitivity towards the target gene, eliminating their cross reactivity, minimizing possible false negative and positive results, optimizing rRT-PCR conditions are examples of what can be improved by those studies. It is worth mention here that recently, Dekker et al.43 demonstrated a faulty design in RP primer sets defined by the CDC, which is another example of the importance of those improvement studies.
Together with, it has been shown by many studies that the SARS-CoV-2 genome evolve very fast as a consequence of the lack of proofreading activity of polymerases29,44. In a recent study, the nucleotide mutation rate of the SARS-CoV-2 genome was found as 6.677 × 10−4 substitution per site per year28. Due to the high susceptibility to mutation, it has been shown that more than one variant can be present in a patient at the same time45. For instance, a meta-transcriptome analyses of 110 SARS-CoV-2 sequences obtained from BAF samples (Bronchoalveolar fluid) from eight patients revealed the evolution of SARS-CoV-2 in the patient and that many variants can be found simultaneously in the individual45. Emergence of variations by new mutations, in particular when they placed on the RT-PCR primer binding sites, and active viral recombination46 can cause loss of the assay sensitivity29 and false-negative results27. Therefore, the selection and update of primer target regions and targeting more than one gene are of great importance. The RT-PCR protocols in particular the target regions/sequences should be updated by considering the most recent mutations and variants.
In this study, a multiplex rRT-PCR method has been developed that simultaneously targets the viral N and RdRP genes and the human RP gene. The primer and probe sets were designed to obtain the best PCR efficiency and target specificity. Due to the rapid evolution of SARS-CoV-2 genome47, attention was paid to designing the primers that are covering the most recent variant of concerns (VOC) such as alpha (B.1.1.7), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta). For this purpose, 344 SARS-CoV-2 genomes from different lineages and locations (including all continents) were screened and the primer binding sites were defined as the most conserved regions (Supplementary Figs. S3 and S4). In addition, to improve the specificity, the oligos were set to the variable region between SARS-CoV-2 and other SARS viruses (Supplementary Figs. S5 and S6). A recent genome analysis of 31,000 SARS-CoV-2 sequences from nasopharyngeal samples of 30 patients showed that 99% of the genome regions targeted by the RT-PCR primers were identical48. On the other hand, 1% of heterogeneous sequences presented inconsistencies, particularly a mismatch between the SARS-CoV-2 genome and the commercial primer, including genes selected in WHO-recommended RT-PCR detection tests. For example, two regions of genetic variability were identified in the sequence of RdRP gene primers recommended by the US CDC48,49. This variation can have a critical impact on the reliability and sensitivity of the assay.
According to the results, the rRT-PCR efficiencies of tenfold dilutions series of the standards were > 99 for both N and RdRP genes (Fig. 2; Supplementary Fig. S2), which matches the criteria for an efficient RT-qPCR assay50. Besides, the current protocol allows the diagnosis of SARS-CoV-2 RNA with a limit of detection (LOD) value of 0.81 copies/µL or 20.31 copies/reaction for the N gene, and 1.40 copies/µL or 35.13 copies/reaction for the RdRP gene (Fig. 4). The estimated 95% confidence intervals (CI) for the LODs of N and RdRP genes are 6.95–379.25 and 12.2–640.2 copies/reaction, respectively. The corresponding copy number of each gene per µL (copies/µL) varied between 0.27 and 15.17 copies/µL for the N gene, and 0.48 and 25.60 for the RdRP gene. The LOD of the CDC’s 2019-nCoV Real-Time RT-PCR Diagnostic Panel is 10 copies/μL. According to Vogels et al.51, all SARS-CoV-2 primer and probe sets that has capacity to detect 500 copies/reaction can be used to diagnose SARS-CoV-2. Pfefferle et al.52 demonstrated that the LOD of RT-PCR assay was 689.3 copies/mL with 275.72 copies per reaction. In this study, GeneFinderTMCOVID-19 Plus RealAmp Kit (GeneFinder, Korea) and RealStar SARS-CoV-2 RT-PCR Kit 1.0 (Altona, Germany) has been used to verify the assay control. The LOD of the GeneFinder kit is 0.5 copies/µL for both upper respiratory and sputum specimen as measured on the ABI 7500, ABI 7500 Fast, and CFX96 instruments53. Another study using a digital PCR (dPCR) assay found it as 0.149 and 0.163 copies/µL for N1 and N2 genes, respectively54. The LOD of the RealStar kit was 1.2 copies/µL in nasopharyngeal swabs (NP) specimens; but 12 copies/µL in bronchoalveolar lavage (BAL) specimens55. The LODs of three SARS-CoV-2 RT-PCR kits, Allplex (Seegene, Korea), PowerChek (KogeneBiotech, Korea), and Real-Q (BioSewoom, Korea), were found to be 153.94, 84.12, and 80.60 copies/reaction, respectively56. Overall, it can be said that the current assay has at least as high sensitivity as the recommended and known tests, or even more than some.
The validation of the assay was tested by using viral RNA samples extracted from the swabs of SARS-CoV-2 positive individuals (n = 28). It is revealed that three samples out of 28 did not match with the results of the commercial kits (Supplementary Table S1). In both assays, either commercial or the current one, it is estimated that the Ct score of those negative samples was higher than > 37.01 which is out of the WHO recommendation36. Therefore, in the current mCoV-2 assay, they are called as negative. Accordingly, the Ct value equals and lower than 37 is accepted as positive.
For COVID19, RT-PCR detection kits are commonly destined to amplify the genes S, E, N, RdRP, and ORF1a/b, but ORF1a/b and E were mostly applied57,58. In China, Orf1ab and N genes are regularly used, while N1, N2 and N3 genes were utilized in US CDC and E, N, and RdRP genes in Europe59. The importance of N1 and N2 primer-probes is for providing a less conservative but more sensitive than the RdRP primer-probes especially in samples that have low viral titers60,61. In a recent study, where swabs from confirmed cases were taken from nasopharynx and pharynx targeting ORF1ab and N genes yielded the best sensitivity when compared to positive confirmed samples61. Chu et al.62 have reported two assays that had the capability to achieve a large dynamic range and recommended targeting N gene for screening and the ORF1b gene to confirm the result. Together with, a case report found that after 9 and 10 days from the onset of disease, an RT-PCR kit detected a single positive gene which is nucleocapsid (N), but not the orf1ab gene. On day 16, both genes resulted in positive amplification which suggests that the N gene is more sensitive to amplify than the orf1ab gene63. The abovementioned studies confirmed the capability of targeting N gene to be utilized in the detection of COVID-19 with other genes. While Li et al.64 concluded that the more stable E gene is the target for the standardization of coronavirus tests, but N and RdRP genes are mostly targeted to confirm the results. In our study, it seems that targeting RdRP and N genes will make the test more sensitive. However, more studies are needed for fast and accurate COVID19 detection.
Conclusion
Due to the spread of COVID-19 all over the world and emergence of new SARS-CoV-2 variants, there is an urgent need to develop more reliable and sensitive methods and to improve existing methods. Establishing sensitive primers and PCR conditions are extremely important to detect SARS-CoV-2 early and to control the spread of the disease. Current study describes a multiplex rRT-PCR assay that simultaneously targets two viral (RdRP and N) and one human internal control gene (RP). In addition, the experimental design is free from background (self- or hetero-dimer formations) with high sensitivity. Thanks to this strategy, fast, reliable, and easy-to-use rRT-PCR method is obtained to detect SARS-CoV-2. The sensitivity of the method should be evaluated in different RT-PCR devices.
Supplementary Information
Acknowledgements
This study is funded by Institute for Research and Medical Consultations (IRMC) under the project number 2020-IRMC-S-3 and the Deanship of Scientific Research (DSR) of Imam Abdulrahman Bin Faisal University (IAU) Fast track fund of COVID-19 (COVID19-2020-026-IRMC). We would like to thank Mr. Geoffrey James T. Moro for technical assistance in conducting RT-PCR experiments. The schema is created with BioRender.com.
Author contributions
H.T. designed the experiments. H.T., J.K., and M.A. carried out the experiments. H.A., N.A., and J.K. provided the clinical samples. E.A., and H.A. analyzed and interpreted the data. H.T., H.S. and E.A. wrote the manuscript. E.A., H.A., and N.A. revised the manuscript. All authors read and commented on the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-06977-z.
References
- 1.Worldometer. Covid-19 Coronavirus Pandemic. (2021). https://www.worldometers.info/coronavirus (accessed 05 Sep 2021).
- 2.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 3.Caruana G, Croxatto A, Coste AT, Opota O, Lamoth F, Jaton K, Greub G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 2020;26:1178–1182. doi: 10.1016/j.cmi.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubina R, Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6):434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. What we know about the COVID-19 immune response. (2020). https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update-34-immunity-2nd.pdf?sfvrsn=8a488cb6_2 (accessed 10 Jan 2021).
- 6.WHO (2020). Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases.https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 (accessed 10 Jan 2021).
- 7.WHO. Advice on the Use of Point-Of-Care Immunodiagnostic Tests for COVID-19. (2020). https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 (accessed 10 Jan 2021).
- 8.CDC. Overview of Testing for SARS-CoV-2. (2020). https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html#:~:text=Authorized%20assays%20for%20viral%20testing,COVID%2D19%2C%20is%20present. (accessed 14 Jan 2021).
- 9.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JFW, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e0031020. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infect. 2020;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikramaratna PS, Paton RS, Ghafari M, Lourenço J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eurosurveillance. 2020;25(50):2000568. doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long DR, Gombar S, Hogan CA, Greninger AL, Shah VOR, Bryson-Cahn C, et al. Occurrence and timing of subsequent SARS-CoV-2 RT-PCR positivity among initially negative patients. Clin. Infect. Dis. 2021;72(2):323–326. doi: 10.1093/cid/ciaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Petitjean SJL, Koehler M, et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Yang C, Xu X, et al. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S, Tombuloglu H, Hassanein SE, Rehman S, Bozkurt A, Cevik E, et al. Coronavirus diseases 2019: Current biological situation and potential therapeutic perspective. Eur. J. Pharmacol. 2020;886:173447. doi: 10.1016/j.ejphar.2020.173447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konrad R, Eberle U, Dangel A, Treis B, Berger A, Bengs K, et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance. 2020;25(9):2000173. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tombuloglu H, Sabit H, Al-Suhaimi E, Al Jindan R, Alkharsah KR. Development of multiplex real-time RT-PCR assay for the detection of SARS-CoV-2. PLoS ONE. 2021;16(4):e0250942. doi: 10.1371/journal.pone.0250942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Böger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021;49(1):21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CDC. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. (2020). https://www.fda.gov/media/134922/download (accessed 14 Jan 2021). [DOI] [PMC free article] [PubMed]
- 25.Dahdouh E, Lázaro-Perona F, Romero-Gómez MP, Mingorance J, García-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J. Infect. 2020;82:414–451. doi: 10.1016/j.jinf.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhen W, Berry GJ. Development of a new multiplex real-time RT-PCR assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. J. Mol. Diagn. 2020;22(12):1367–1372. doi: 10.1016/j.jmoldx.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Xu X, Wei C, Li S, Zhao J, Zheng Y, et al. Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J. Med. Virol. 2021;94:310–317. doi: 10.1002/jmv.27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peñarrubia L, Ruiz M, Porco R, Rao SN, Juanola-Falgarona M, Manissero D, et al. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int. J. Infect. Dis. 2020;97:225–229. doi: 10.1016/j.ijid.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The GISAID (Global Initiative on Sharing All Influenza Data. https://www.gisaid.org (accessed 05 May 2010).
- 31.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown SS, Chen YW, Wang M, Clipson A, Ochoa E, Du MQ. PrimerPooler: Automated primer pooling to prepare library for targeted sequencing. Biol. Methods Protoc. 2017;2(1):bpx006. doi: 10.1093/biomethods/bpx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y., Zhou, Y., Ma, Y., Xu, R., Jin, X., & Zhang, C. A mismatch-tolerant RT-LAMP method for molecular diagnosis of highly variable viruses. Bio-protocol. 9(21) (2019). [DOI] [PMC free article] [PubMed]
- 36.Dong Y, Wu X, Li S, Lu R, Li Y, Wan Z, et al. Comparative evaluation of 19 reverse transcription loop-mediated isothermal amplification assays for detection of SARS-CoV-2. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-020-80314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnaout, R., Lee, R. A., Lee, G. R., Callahan, C., Yen, C. F., Smith, K. P., et al. SARS-CoV2 testing: the limit of detection matters. bioRxiv. (2020). [DOI] [PMC free article] [PubMed]
- 38.CDC. FAQs about the CDC DENV-1–4 Real-Time RT-PCR Multiplex Assay. (2020). https://www.cdc.gov/dengue/healthcare-providers/testing/molecular-tests/faq_rt-pcr.html (accessed 14 Jan 2021).
- 39.WHO. WHO Emergency Use Assessment Coronavirus disease (COVID-19) IVDs PUBLIC REPORT. October 2020, version 1.0. EUL-0517-204-00. (2020). https://extranet.who.int/pqweb/sites/default/files/documents/201005_final_pqpr_eul_0517_204_00_sars_cov2_nucleic_acid_detection%20%281%29.pdf. (accessed 24 Feb 2021).
- 40.CDC. Testing for COVID-19. (2020). https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/testing.html (accessed 14 Jan 2021).
- 41.Shen M, Zhou Y, Ye J, Al-Maskri AAA, Kang Y, Zeng S, Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WHO. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020 (No. WHO/COVID-19/laboratory/2020.4). (2020). World Health Organization. (accessed 10 Jan 2021).
- 43.Dekker, R. J., Ensink, W. A., van Leeuwen, S., Rauwerda, H., & Breit, T. M. Overhauling a faulty control in the CDC-recommended SARS-CoV-2 RT-PCR test. bioRxiv. (2020).
- 44.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi H. 2019 novel coronavirus is undergoing active recombination. Clin. Infect. Dis. 2020;71(15):884–887. doi: 10.1093/cid/ciaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal A, Dobhal S, Dey KK, Sharma AK, Savani V, Negi VS. Polymorphic landscape of SARS-CoV-2 genomes isolated from Indian population in 2020 demonstrates rapid evolution in ORF3a, ORF8, nucleocapsid phosphoprotein and spike glycoprotein. Comput. Biol. Chem. 2021;95:107594. doi: 10.1016/j.compbiolchem.2021.107594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Álvarez-Díaz DA, Franco-Muñoz C, Laiton-Donato K, Usme-Ciro JA, Franco-Sierra ND, Flórez-Sánchez AC, Mercado-Reyes M. Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia. Infect. Genet. Evol. 2020;84:104390. doi: 10.1016/j.meegid.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bezier C, Anthoine G, Charki A. Reliability of real-time RT-PCR tests to detect SARS-Cov-2: A literature review. Int. J. Metrol. Qual. Eng. 2020;11:13. [Google Scholar]
- 50.Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance. 2020;25(9):2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.GeneFinder™ COVID-19 Plus RealAmp Kit https://www.fda.gov/media/137116/download (accessed 15 Dec 2021).
- 54.Poggio P, Songia P, Vavassori C, Ricci V, Banfi C, Barbieri SS, et al. Digital PCR for high sensitivity viral detection in false-negative SARS-CoV-2 patients. Sci. Rep. 2021;11(1):1–7. doi: 10.1038/s41598-021-83723-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhteg K, Jarrett J, Richards M, Howard C, Morehead E, Geahr M, et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hur KH, Park K, Lim Y, Jeong YS, Sung H, Kim MN. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front. Med. 2020;7:521. doi: 10.3389/fmed.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui F, Zhou HS. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;95:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheikhzadeh E, Eissa S, Ismail A, Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220:121392. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bai, H., Cai, X. & Zhang, X. Landscape coronavirus disease 2019 test (COVID-19 test) in vitro—A comparison of PCR vs immunoassay vs Crispr-based test. 10.31219/osf.io/6eagn (2020).
- 60.Lista, M. J., Page, R., Sertkaya, H., Matos, P., Ortiz-Zapater, E., Maguire, T. J., et al. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation. medRxiv. (2020). [DOI] [PMC free article] [PubMed]
- 61.Zhou Y, Pei F, Ji M, Wang L, Zhao H, Li H, et al. Sensitivity evaluation of 2019 novel coronavirus (SARS-CoV-2) RT-PCR detection kits and strategy to reduce false negative. PLoS One. 2020;15(11):e0241469. doi: 10.1371/journal.pone.0241469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv DF, Ying QM, Weng YS, Shen CB, Chu JG, Kong JP, et al. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clin. Chim. Acta. 2020;506:172–175. doi: 10.1016/j.cca.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D, Zhang J, Li J. Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2. Theranostics. 2020;10(16):7150. doi: 10.7150/thno.47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.