Abstract
Uranium is a naturally occurring radionuclide. Its redistribution, primarily due to human activities, can have adverse effects on human and non-human biota, which poses environmental concerns. The molecular mechanisms of uranium tolerance and the cellular response induced by uranium exposure in bacteria are not yet fully understood. Here, we carried out a comparative analysis of four actinobacterial strains isolated from metal and radionuclide-rich soils that display contrasted uranium tolerance phenotypes. Comparative proteogenomics showed that uranyl exposure affects 39–47% of the total proteins, with an impact on phosphate and iron metabolisms and membrane proteins. This approach highlighted a protein of unknown function, named UipA, that is specific to the uranium-tolerant strains and that had the highest positive fold-change upon uranium exposure. UipA is a single-pass transmembrane protein and its large C-terminal soluble domain displayed a specific, nanomolar binding affinity for UO22+ and Fe3+. ATR-FTIR and XAS-spectroscopy showed that mono and bidentate carboxylate groups of the protein coordinated both metals. The crystal structure of UipA, solved in its apo state and bound to uranium, revealed a tandem of PepSY domains in a swapped dimer, with a negatively charged face where uranium is bound through a set of conserved residues. This work reveals the importance of UipA and its PepSY domains in metal binding and radionuclide tolerance.
Subject terms: Bacteria, Proteomics, Structural biology
Introduction
Uranium is a long-lived radionuclide naturally found in the earth’s crust. Its redistribution in the environment is mainly due to anthropogenic activities raising environmental and human health concerns [1]. Uranium toxicity stems from chemical toxicity rather than radiotoxicity [2]. In the environment, uranium has two major oxidation states, U(IV) and U(VI). The U(IV) form is found under anoxic conditions and has a limited toxicity due to its low solubility. The U(VI) form is prevalent under oxic conditions and its speciation is pH-dependent. The water-soluble uranyl cation UO22+ is the most toxic form of U(VI) and dominates at pH<5 [3]. As a hard cation, U(VI) in the uranyl molecule forms complexes with oxygen ligands such as hydroxyl, amide, phosphate and carboxylate groups and can therefore interact with many biological molecules. For instance, uranyl can replace physiological cations such as Ca2+ and Fe3+ and impair functional binding sites on proteins [4].
Soil bacteria play a crucial role in the functioning of soils and ecosystems. It has been shown that uranium can affect the structure and activity of soil bacterial communities [5–7]. At the same time, high bacterial diversity has been reported in uranium-rich environments, from both natural and anthropogenic origins [8–11], and these environments are reservoirs of uranium- and metal-tolerant bacteria [11–17].
Bacteria interact with uranium and can modulate its speciation, playing a major role in its mobility and transfer [18]. Bacteria can immobilize uranium by different mechanisms such as reduction, sorption, precipitation and mineralization. Under anoxic conditions, some bacterial species mediate reductive precipitation of uranium [19]. Under oxic conditions, the biosorption of uranyl on chelating groups occurs on bacterial surfaces [20, 21]. Uranium mobility can also be reduced by a variety of bacterial strains via bioprecipitation and biomineralization with inorganic phosphate [22]. Both processes can take place intra- or extracellularly and involve phosphatase activity [23–26]. Metabolites such as siderophores may also be involved in uranium sequestration [27, 28]. All of these mechanisms have been proposed to participate in uranium detoxification [26, 29]. In addition, transporters such as PIB-ATPases and cation diffusion facilitator (CDF) may participate in uranium tolerance [30–33]. The ability of some bacterial species to sequester uranium makes them promising candidates to develop bioremediation processes [34].
Although a large body of work has been dedicated to the mechanisms of uranium-bacteria interaction, less is known about the cellular response induced by uranium exposure. Transcriptome and proteome profiling has revealed various cellular responses, including general and membrane stress responses, DNA repair, protection against oxidative stress, two-component signal transduction systems, efflux proteins, and iron transport systems [27, 29, 35–40]. In all of these studies, a large proportion of the identified genes/proteins have no known function, indicating that there is still much to discover.
In an effort to uncover the molecular mechanisms of uranium stress response and tolerance, we have isolated bacteria from metal- and radionuclide-rich soils [10, 41–43]. One of the most uranium-tolerant strains in our collection, Microbacterium oleivorans A9, has been subjected to a detailed analysis with a multidisciplinary approach. Using tightly controlled exposure conditions, a three-step process involving uranyl sorption, release and biomineralization was evidenced [44]. Using proteogenomics, we further showed the high impact of uranyl on the proteome of this strain [45, 46]. In the present work, we have examined three additional Microbacterium strains for which we obtained the whole-genome [47], and subjected them to comparative analysis (Fig. 1a). We first showed that these four strains differ in their uranium tolerance. Key proteins involved in the uranium stress response were identified by comparative proteomics, which led to the discovery of a protein named UipA specific to uranium-tolerant strains and highly up-produced in response to uranyl. Next, we established that UipA is a single-pass membrane protein showing high affinity for UO22+ and Fe3+ with dissociation constants in the nanomolar range. FTIR- and XAS-spectroscopy showed that uranyl coordination involves carboxylate groups of Asp and Glu residues. The three-dimensional structure of UipA, solved in its apo and uranium bound states, finally revealed two PepSY domains with uranium binding sites located on a highly negatively charged face, and stabilized by a set of conserved residues.
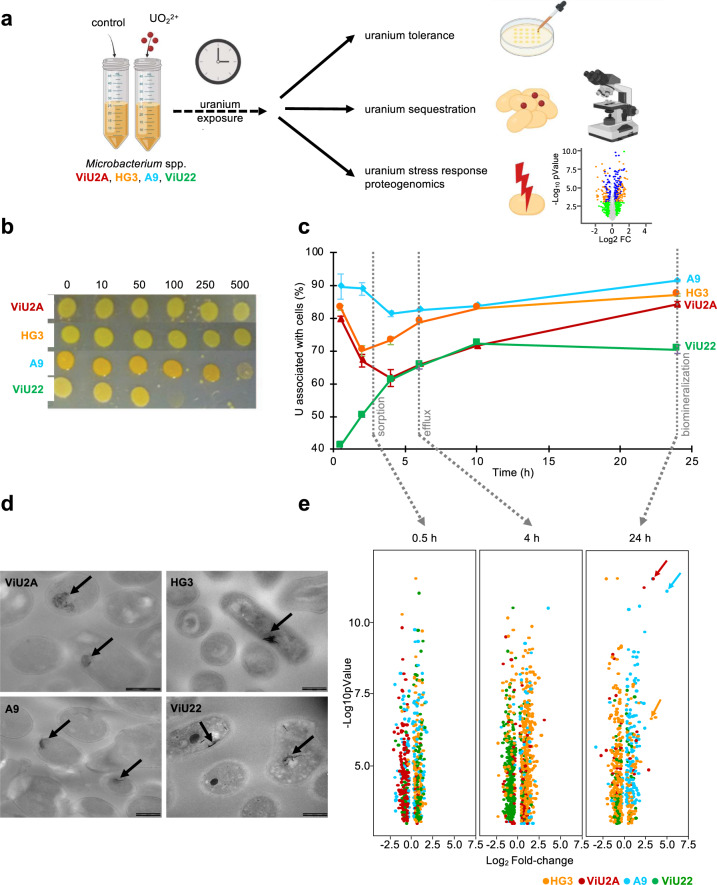
Fig. 1. Interactions of Microbacterium strains ViU2A, HG3, A9 and ViU22 with uranium.
a Schematic workflow to monitor uranium tolerance, sequestration and stress response of strains ViU2A, HG3, A9 and ViU22. b Uranium tolerance assay: cells of strains ViU2A, HG3, A9 and ViU22 were exposed to 0–500 µM uranium for 24 h and drops of cell suspension were spotted onto LB agar plates. Cell growth was observed after 2 days at 25 °C. c Time evolution of the uranium (% of the initial concentration) associated with the cells of strains ViU2A (red), HG3 (orange), A9 (blue) and ViU22 (green). The sampling times for proteomics analysis are indicated by the dotted lines. Error bars denote the standard deviations of triplicate measurements. When not visible, error bars are smaller than the symbols. d TEM micrograph of cells exposed to 10 µM U(VI) for 24 h at 25 °C. The needle-like structures indicated by the arrows were previously shown to contain U, Ca, K and P in strain A9 [44]. Scale bars correspond to 300 nm for ViU2A and HG3 and to 200 nm for A9 and ViU22. e Volcano plot showing proteomic data at 0.5, 4 and 24 h of uranium exposure for ViU2A (red dots), HG3 (orange dots), A9 (blue dots) and ViU22 (green dots). The position of UipA protein plots at 24 h is indicated by arrows.
Materials and methods
A detailed protocol is available in Supplementary Information.
Strains and culture conditions
Microbacterium sp. ViU2A and Microbacterium lemovicicum ViU22T strains were isolated from a natural uranium-rich soil collected in Bessines, France [10, 42]. The Microbacterium oleivorans sp. A9 strain was isolated from a contaminated waste storage site in Chernobyl [43]. The Microbacterium sp. HG3 strain was isolated from black sand collected in Vik, Iceland [41]. The bacteria were routinely cultivated in Luria Broth (LB, Difco Laboratories) at 32 °C with shaking.
Uranium tolerance assay
Bacteria were cultured until the exponential growth phase and exposed to uranyl following a previously described procedure [44]. Briefly, cells were suspended in 0.1 M NaCl (pH 5.0) supplemented with 0, 10, 50, 100, 250 or 500 µM uranyl nitrate and incubated 24 h at 25 °C. Subsequently, cells were washed, deposited on LB agar plates and incubated for 48 h at 32 °C to estimate cell survival.
Sequestration of uranyl by Microbacterium spp. and sample preparation for proteomics
Bacteria were cultured until the exponential growth phase and exposed to 0 or 10 µM uranyl nitrate in 0.1 M NaCl (pH 5.0) as previously described [44]. Samples were taken after 0.5, 2, 4, 6, 10 and 24 h exposure. Cell pellets and supernatant were separated by centrifugation, and uranium concentration in the supernatant was measured by inductively coupled plasma-atomic emission spectrometry. Controls without bacteria demonstrated that uranium remains soluble and that no loss of uranium due to adsorption on the vial wall occurred within the 24 h exposure experiment. The fraction of uranium associated with the cells was calculated as follows: Ucells = (Ublank – Usolution)*100/Ublank. Ultra-thin sections of bacterial cells exposed to uranium for 24 h were analyzed by transmission electron microscopy as previously described [44]. To estimate cell viability, aliquots of cell suspensions taken at 0.5 and 24 h were diluted in LB and spread on LB agar plates. Colony forming units were counted after 24 h at 30 °C.
For the proteomic analysis, ~2 mg of cell pellets exposed or not to uranyl were collected at 0.5, 4 and 24 h by centrifugation and were immediately frozen in liquid nitrogen and kept at −80 °C until further processing. Four independent biological replicates were made.
Shotgun proteomic analysis
The cell samples were analyzed by shotgun proteomics according to the protocol described for strain A9 in [46]. Peptides obtained after in-gel proteolysis of proteins with trypsin were characterized using a Q-Exactive HF mass spectrometer (ThermoFisher) coupled to an UltiMate 3000 LC system (Dionex-LCPackings) operated according to a Top20 data-dependent acquisition method, as previously described [48]. Data were interpreted as described in [46] against the genome-annotated protein sequence databases of the four Microbacterium strains [47]. The statistical protein abundance variations between the exposure conditions were compared using the T-Fold option of the PatternLab 2.0 software [49], taking into account the four replicates. The full proteomics data for each strain are available in SI Tables S1–S4. The mass spectrometry proteomics data were deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [50] with the dataset identifiers: PXD020778 and 10.6019/PXD020778 for Microbacterium sp. HG3, PXD020767 and 10.6019/PXD020767 for Microbacterium lemovicicum ViU22, PXD020737 and 10.6019/PXD020737 for Microbacterium sp. ViU2A, and PXD020998 and 10.6019/PXD020998 for Microbacterium oleivorans sp. A9.
Genomic context and sequence analysis
Genomic analyses were performed with Genobrowser, an in-house bioinformatics tool for data management. Sequence alignment of UipA proteins was calculated with Clustal W and visualized in Jalview [51]. A phylogenetic tree was constructed using 16S rRNA gene sequences from 482 Microbacteriaceae genomes available at GTDB (Genome Taxonomy Database) [52], identified only as species representatives. A subtree of Microbacterium strains containing particular features was extracted and analyzed with the ETE3 Python package [53]. For this, alignment was made using MAFFT [54], overhangs were trimmed with TRIMAL [55], and the tree was built with Fasttree [56]. The final phylogenic tree visualization with the Uip operon was made by Interactive tree of life (iTOL) tools [57]. Synteny analyses were performed with the MicroScope platform [58].
Topology of UipA in vivo
To determine in vivo the orientation of the UipA protein in the membrane, the pKtop plasmid encoding a dual PhoA-LacZα reporter was used [59]. Three pKtop derivatives were constructed, in which phoA-lac was fused in frame after the uipA-ViU2A codons R69, G96 and D281 (see SI). The constructions were made by overlap extension PCR cloning according to the method described in [60]. In a first step of PCR, the inserts corresponding to the targeted regions of uipA and the overlapping regions of the pKtop plasmid were amplified. The PCR products were purified and used as megaprimers to perform a second PCR with the plasmid pKtop as matrix DNA. PCR products were digested by DpnI and transformed in E. coli DH5α. Transformants were selected on LB agar plates supplemented with 50 µg mL−1 kanamycin and 0.2% glucose. For protein topology assays in vivo, freshly transformed colonies were streaked on LB agar plates supplemented with 50 µg mL−1 kanamycin, 1 mM IPTG, 80 µg mL−1 Red-Gal, and 100 µg mL−1 BCIP. Plates were incubated 24 h at 30 °C and 14 h at room temperature.
Production and purification of the extracellular domain of UipA recombinant proteins
The DNA fragments coding for the soluble domain of UipA proteins were amplified by PCR with primers that introduce the TEV protease recognition site upstream of the start codon (see Supplementary Information). The PCR products were ligated into the pQE30 plasmid to obtain the pQE-UipA recombinant plasmids for each of the three homologs. The recombinant proteins were expressed overnight at 17 °C after addition of 0.1 mM IPTG in E. coli M15Rep4. Cells were lysed and the soluble extract was loaded on a HiTrap HP chelation column. The recombinant proteins were eluted from the resin with imidazole, desalted, and incubated 48 h with TEV protease to remove the His tags. The samples were then loaded on a HiTrap HP column, and recombinant proteins without the His tag were collected in the eluate fraction. A final purification step was performed on an exclusion chromatography column to increase the purity. Protein purity and integrity were checked by SDS-PAGE and mass spectrometry in denaturing conditions.
Non-covalent mass spectrometry
Non-covalent mass spectrometry analyses were performed on a MicrOTOF-Q (Bruker, Wissembourg, France) with an electrospray ionization source. Protein-uranyl complexes were prepared, and the samples were continuously infused at a flow rate of 7 µL min−1. The mass spectra were recorded in the 50–4000 mass-to-charge (m/z) range. Data were acquired in the positive mode, and the calibration was performed using a calibrating solution of ESI Tune Mix (Agilent) in CH3CN/H2O (95/5-v/v). The system was controlled using the MicrOTOF Control 2.2 software package, and data were processed with DataAnalysis 3.4.
Metal-binding affinities of the UipA soluble domain measured by fluorescence titration
The metal binding affinities of the three UipAext peptides for different metal cations (UO22+, Fe3+, Cu2+, Ca2+, Ni2+ and Zn2+) were examined by monitoring the fluorescence intensity of tryptophan and tyrosine. Fe3+ and Ca2+ were selected because they can be replaced by UO22+ on protein. The other metals were selected as representatives of physiological metals with no known competition with UO22+. To remove all traces of metal, each protein solution was incubated with 3 mM EDTA and then washed. For fluorescence titrations in the presence of uranyl or iron, iminodiacetate (IDA) was added to the peptide solution at a IDA:peptide ratio of 10:1 to avoid the formation of hydroxo complexes, as previously described [61]; this was also done to control iron and uranyl speciation. Competition experiments between the UipAext proteins and IDA were performed to determine the conditional dissociation constants of the protein-cation complexes (uranyl or iron) at pH 6. IDA binds uranyl and iron with moderate affinity and forms three and two complexes, respectively: UO2IDA, [UO2(IDA)2]2−, [(UO2)2(IDA)2(OH)2]2− and [FeIDA]+, [Fe(IDA)2]−. The stability constants at 25 °C and 0.1 M ionic strength were obtained from [62]. For iron-IDA complexes, the stability constants were obtained from [63]. The experiments were carried out with 10 µM protein in 20 mM MES buffer (100 mM KCl; pH 6). Fluorescence was measured at 25 °C using a 275 nm excitation wavelength. Emission was recorded from 300 to 380 nm. The excitation and emission slits were 10 nm. The reported dissociation constants are averages of three experimental values, including standard deviation. The analysis of the multiwavelength dataset was performed using the commercially available ReactLab software [64]. The goodness of fit was assessed by the convergence value σ and visual inspection of the residuals.
Fourier-transformed infrared spectroscopy
For Fourier-transformed infrared spectroscopy (FTIR), proteins were concentrated to about 500 µM in 20 mM MES buffer (pH 6). Uranyl or iron from stock solutions of uranyl nitrate and iron chloride at 50 mM in 2% HCl were added to reach a stoichiometric peptide:metal ratio of 1:1. In parallel, control samples were prepared in 20 mM MES buffer (pH 6) by adding an equivalent volume of 2% HCl solution. Samples with proteins were then deposited onto the ATR-FTIR crystal. For spectra acquisition, samples were first dried for half an hour after deposition. The spectra were recorded on an IFS66 SX FTIR spectrometer (Bruker) equipped with a KBr beam splitter, a DTGS-KBR detector, and an attenuated total reflection device (ATR; SensIR Technologies, CT) equipped with a 4.3 mm diameter diamond 9-bounce microprism and ZnSe optics. Each single spectrum corresponded to 50 co-added scans at a resolution of 4 cm−1. All reported frequencies have an accuracy of ±1 cm−1. The spectra correspond to the average of data recorded with 2–4 replicates in the same conditions. To calculate the difference spectra between uranyl or iron-containing samples and control samples, interactive subtraction was used to minimize the effect of small differences in absorption between samples. These differences in total absorption have consequences on bands associated with the buffer in the difference spectra (noted as * in the spectra). Interpretation of the FTIR data was limited to bands that were reproductively observed in spectra obtained with different samples. IR bands of MES buffer and nitrate were identified according to [65] and [66] respectively.
Synchrotron-based analysis
UipA-uranyl complexes were prepared by mixing UipAext proteins at 120 µM in 20 mM MES buffer 100 mM KCl pH 6 with 100 µM uranyl nitrate. Next, UipA-uranyl complexes were loaded in a kapton-teflon liquid cell for synchrotron analysis. EXAFS experiments were performed on the MARS beam line at the SOLEIL synchrotron facility [67]. Energy calibration was performed at the yttrium K edge at 17038 eV, and EXAFS experiments at the U LIII edge. All measurements were conducted in fluorescence mode using a 13-element high purity germanium detector (ORTEC) at room temperature. EXAFS data processing was performed using the ATHENA code [68]. The E0 energy was identified at the maximum of the absorption edge. Fourier transformation (FT) with k2 weighting was performed between 2.8 and 11.8 Å−1 with a Hanning window. The fits were performed using the DEMETER code (Demeter version 0.9.25) in R space between 1 and 5 Å. For EXAFS data fitting, one global amplitude factor S0² and one energy threshold correction factor E0 were used for every path of the fits. The agreement factor r (%) and the quality factor (QF = reduced χ²) of the fits were provided directly by DEMETER. Phases and amplitudes were calculated using the FEFF6 simulation code integrated in DEMETER based on the structural model of the uranyl-acetate complex (UO2(CH3COO)2) [69]. During the fitting procedure, the relative number of mono and bidentate carboxylates was allowed to fluctuate, but the total number of Oeq was fixed to 5.
Crystal structure of UipA
UipAext proteins were concentrated to ~15 mg ml−1. We tested the crystallization of all three UipAext proteins by vapor diffusion using a TECAN robot and different commercial crystallization kits (JCSG, Pact Premier and Stura (Molecular Dimensions)). Only crystals of UipAext-ViU2A could be obtained, which diffracted poorly (to 15–20 Å at synchrotron sources). Different strategies were therefore examined to improve diffraction. The screening of additives proved to be the most efficient, as formamide improved the diffraction quality by up to 3.2 Å when using a rotating anode. The final crystallization conditions consisted in mixing 0.7 mL of UipAext-ViU2A with an equal volume of a reservoir solution (100 mM MES pH6, PEG6K 22%, formamide 6% and ZnCl2 5–15 mM). For phasing, one crystal was soaked in a mother liquor directly supplemented with gadolinium powder (Gd-HPDO3A). Another crystal was soaked in a mother liquor supplemented with 5 mM uranium nitrate (U02(NO3)2). All crystals were briefly soaked with a mother liquor containing 5% glycerol before flash-cooling directly in liquid nitrogen before data collection.
Diffraction data were collected on a PROXIMA-1 beam line at the SOLEIL synchrotron (France). Data were processed with the XDS program [70] using XDSME scripts [71]. The structure of UipAext-ViU2A was solved by the SAD method using the Gd derivative, and the gadolinium positions were found using the SHELX C/D/E suite of programs [72]. Structure was built semi-automatically using the ARP/wARP program [73] and completed by rounds of model building using COOT [74], with refinements using Refmac [75]. Details of the crystallographic analysis are presented in Table S5.
Statistical analyses
All measurements were performed at least in triplicate. The symbol ± corresponds to standard deviation.
Results
Microbacterium spp. strains show contrasted U tolerance phenotypes
Bacterial culture collections were previously established from uranium-rich soil samples collected in France [10], radionuclide-contaminated soil samples collected at Chernobyl [43], and volcanic black sand collected in Iceland [41]. A preliminary broad plate screening of these bacterial strain collections indicated that four actinobacterial strains, namely Microbacterium sp. HG3, Microbacterium sp. ViU2A, Microbacterium oleivorans A9 and Microbacterium lemovicicum ViU22, had contrasted uranium tolerance capabilities (data not shown). To refine this result, the tolerance of these four strains against uranium was evaluated by a drop test. As shown in Fig. 1b, the survival of strains HG3 and ViU2A was not affected by uranium at 500 µM, and strain A9 could tolerate up to 250 µM of uranium. In contrast, the survival of strain ViU22 was severely affected by 100 µM of uranium.
Interactions of Microbacterium spp. strains with uranyl
Bacteria remained viable after 24 h in the controls without uranyl and with 10 µM uranyl (Fig. S1). All strains were able to deplete an important fraction of the metal from the solution (Fig. 1c) and to biomineralize it within 24 h (Fig. 1d). The time evolution of uranium removal by the uranium-tolerant strains ViU2A, HG3 and A9 had similar overall profiles, comprising three steps: a rapid removal of 80–90% of uranium within the first 30 min, likely by biosorption; a release of uranium into the solution between 0.5 and 4 h; and a final slow uranium removal step leading to removal of more than 80% of the initial uranium. The release of uranium by strain A9 was less efficient compared to the other two uranium-tolerant strains (ViU2A and HG3). Regarding the uranium-sensitive strain ViU22, only 40% of uranium was associated with the cells after 30 min, no uranium release step was observed, and uranium removal after 24 h was ~60% of the initial amount of uranium.
U-stress molecular response of Microbacterium spp. strains
Given the different behaviors of the Microbacterium spp. strains toward uranium, a comparative proteogenomics approach was used to decipher the molecular changes occurring in these strains upon uranyl exposure, and to highlight the key proteins possibly involved in uranium tolerance. We recorded large shotgun proteomics datasets for each strain at three times after uranyl addition, corresponding to the sorption, release and biomineralization phases. We interpreted these data with proteomes constructed from the corresponding complete genome sequences (GenBank accession numbers CP031421, CP031422, CP031338, and CP031423). Overall, a total of 2038, 1156, 1523 and 1312 proteins were validated by at least two peptides for strains HG3, ViU2A, A9 and ViU22, respectively (Tables S1–S4). These proteins represented between 29.7 and 53.4% of the proteins predicted by the genome, reflecting a good coverage of the theoretical proteome. The functional classification of all proteins showed a distribution between all clusters of orthologous genes (COG) similar to the distribution of Coding DNA Sequences predicted from the genomes (Table S6). As expected for the overlooked Microbacterium genus, the “poorly characterized” proteins represented a large fraction, ranging from 38.9 to 48.4% of the total proteins.
In response to uranium, the abundance of proteins in strains HG3 (966 proteins), ViU2A (452 proteins), A9 (596 proteins) and ViU22 (535 proteins) was significantly modulated, with fold-change (FC) values varying from −10.3 to +32.3 (log2FC varying from −3.36 to 5.01; Fig. 1e). This represents 39.1–47.4% of the total proteins detected, showing that uranium has a strong impact on the metabolism of the four strains, regardless of their tolerance phenotype. The proteomics data shows that uranyl has an impact on phosphate and iron metabolism in all strains (Table S7). In particular, several proteins were modulated by uranyl, including proteins of the phosphate (Pho) regulon and the high-affinity phosphate transport system (Pst) (Fig. S2), and proteins involved in siderophore transport (FepC, YclQ) and in iron-sulfur cluster assembly (SufB, SufD). Several predicted transporters including ABC transporters were also modulated. Noteworthy, the six proteins with the highest FC values are all predicted to be membrane proteins (Table 1).
Table 1.
Top10 proteins with the highest FC values and their homologs in the other strains after 0.5, 4 and 24 h of uranium exposure.
| ViU2A | HG3 | A9 | ViU22 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 h | 4 h | 24 h | 0.5 h | 4 h | 24 h | 0.5 h | 4 h | 24 h | 0.5 h | 4 h | 24 h | Functional annotation and subcellular localization |
| 1.4 | ns | 10.5 | ns | ns | 8.6 | −1.3 | 11.5 | 32.3 | abs | abs | abs | Unknown function (UipA)—membrane |
| 1.4 | 2 | ns | ns | 6.8 | 12 | 2.2 | 1.7 | 4.8 | ns | ns | ns | Unknown function (UipB)—membrane |
| nd | nd | nd | ns | ns | 2 | ns | 3.2 | 9.9 | nd | nd | nd | ABC transporter ATP-binding protein YxdL—membrane |
| ns | 8.3 | 7.1 | ns | 2.2 | 2.1 | 1.4 | 1.6 | 2.9 | nd | nd | nd | Putative ABC transporter solute-binding prot YclQ—membrane |
| ns | ns | 1.8 | 1.3 | 5.4 | 3 | abs | abs | abs | abs | abs | abs | 4-hydroxyacetophenone monooxygenase—membrane |
| ns | 1.7 | 4.7 | ns | 2.0 | 3.1 | ns | 1.5 | 5.3 | ns | −1.3 | ns | Periplasmic serine endoprotease DegP —membrane |
| nd | nd | nd | ns | ns | ns | ns | 2.8 | 5.2 | nd | nd | nd | Unknown function |
| ns | ns | ns | ns | 4.7 | −1.4 | 2 | ns | ns | nd | nd | nd | Unknown function |
| nd | nd | nd | ns | 4.6 | −2.0 | ns | ns | 1.7 | ns | ns | ns | Putative SOS response-associated peptidase YedK |
| −1.7 | ns | ns | ns | 4.5 | 1.8 | ns | −1.6 | ns | ns | −1.7 | ns | Protein RecA— cytoplasm |
The data are sorted in descending order of maximum FC value. FC (below 1.5) or p value (above 0.05): not significant (ns); not detected in the proteome (nd); absent gene (abs). Proteins specific to the uranium-tolerant strains are shown in italics.
To search for proteins potentially involved in uranium tolerance, we performed a comparative proteogenomic analysis of the four strains. We identified only four proteins modulated by uranium that were common to the tolerant strains, and which were absent or not modulated in the sensitive strain (in italics in Table 1). One protein is annotated as YclQ. In response to uranium, its abundance increased in the three tolerant strains (FC values: 8.4 and 7.1 in strain ViU2A; 2.2 and 2.1 in strain HG3 at 4 and 24 h, respectively; and 1.4, 1.6 and 2.9 in strain A9 at 0.5, 4 and 24 h, respectively). In contrast, it was not detected in the proteome of the uranium-sensitive strain ViU22, although the corresponding gene is present in the genome. A second protein is annotated as DegP. This protein was more abundant in the uranium-tolerant strains under uranium stress (FC values: 1.7 and 4.7 in strain ViU2A; 2.0 and 3.1 in strain HG3; and 1.5 and 5.3 at 4 and 24 h in strain A9), and was either not modulated or less abundant in ViU22 exposed to uranium (FC = −1.3 at 4 h).
The two other proteins had no functional annotation and were named UipA and UipB for Uranium-Induced Protein A and B. The FC values of UipB at 4 h were 2, 6.8 and 1.7 in strains ViU2A, HG3 and A9, respectively (Table 1). The protein was detected in the proteome of strain ViU22, but its abundance was not significantly modulated by uranyl exposure. UipB is predicted to be a membrane protein of 210 kDa with most of it located outside of the cell. It displayed several conserved domains consisting of a cadherin repeat (cd11304) and cadherin-like domains (pfam 17803), bacterial immunoglobulin domains (Big-9, pfam17963), and fibronectin type III domains (Fn3, pfam00041). Finally, UipA was the most modulated protein at 24 h in strain ViU2A (FC = 10.5), and at 4 and 24 h in strain A9 (FC = 11.5 and 32.3 respectively), and was the second-most modulated protein at 24 h in strain HG3 (FC = 8.62) (Table 1). The corresponding gene was absent from the genome of the uranium-sensitive strain ViU22. Thus, the UipA protein is specific to uranium-tolerant strains and is highly accumulated in cells in response to uranyl exposure. This unique profile, which is expected for a protein involved in uranium tolerance, clearly sets UipA apart from the other proteins. We therefore focused on this protein for a detailed characterization.
Genomic context of uipA
Upstream of the uipAViU2A (CVS53_03692), uipAHG3 (CVS54_03678) and uipAA9 (BWL13_02796) genes, we found two genes homologous to the two-component czcRS system involved in metal tolerance in Pseudomonas aeruginosa and Cupriavidus metallidurans [76, 77]. Based on this homology, these genes were named uipR and uipS (Fig. S3A). The uipA genes are orphans with no homologs in any other lineages than Microbacterium. A deeper analysis showed that the genomic context of uipRSA is conserved in almost all Microbacterium strains with the uipA gene (Fig. S4). As was the case for the uipA gene, the uipR and uipS genes were also absent from the genome of the U-sensitive strain ViU22.
Sequence analysis and topology of UipA proteins
The genes uipAViU2A, uipAHG3 and uipAA9 respectively encode proteins of 281, 285 and 247 aa in length (Fig. S3B, C), and share 36.6–74.6% sequence identity with each other. Two conserved PepSY domains (Peptidase pro-peptide and YPEB domain; pfam03413) were predicted at their C-terminal part by HHpred [78]. UipA proteins are predicted to be single-pass transmembrane proteins with a cytoplasmic N-terminal domain (38–75 aa in length), and an extracellular C-terminal domain of 185–186 residues according to CCTOP [79]. We obtained experimental validation of this predicted topology for UipAViU2A using a PhoA-LacZα reporter system [59] (Fig. S5).
Remarkably, these proteins have low pI values (3.86–3.99) due to their richness in negatively charged residues (aspartate and glutamate), accounting for 18.6–20.7% of the total amino acids, as compared to 12.0% on average in the proteomes (Fig. S3B). Since carboxyl groups of aspartate and glutamate can provide oxygen atoms for uranyl coordination [80], we speculated that the UipA extracellular domain could bind uranyl with a high affinity. Thus, we investigated the ability of the recombinant extracellular domain of UipA to bind uranyl using a combination of mass spectrometry in the native mode and spectroscopic approaches.
Stoichiometry of UipAext protein-(UO22+) complexes
The extracellular domains of UipA proteins (named UipAext) were produced in Escherichia coli and purified. The formation of UipAext protein-(UO22+) complexes was first analyzed by native mass spectrometry. This method enables detecting the formation of protein-ligand complexes and assessing the stoichiometry of these complexes. The spectra displayed peaks compatible with 1:1 and 1:2 UipAext–(UO22+) complexes (Fig. S6).
Binding affinities of UipA proteins for uranyl and other metal cations
The affinity of UipAext recombinant proteins for UO22+ was measured by fluorescence titration, taking advantage of the presence of a conserved Trp residue on each of the proteins as well as an additional Tyr on UipAext-HG3 and UipAext-ViU2A (Figs. S7–S12). In line with native mass spectrometry results, the best fitting curves were obtained with the assumption of two uranyl-binding sites, and Kd values were calculated accordingly (Figs. S7, S9 and S11). As shown in Table 2, the conditional dissociation constants of the 1:1 and 1:2 UipAext – UO22+ complexes were within the nanomolar range, with Kd values ranging from 1.3 × 10–9 to 5.0 × 10–8 M. This indicates a high binding affinity of UipAext domains for the two uranyl molecules.
Table 2.
Macroscopic dissociation constants for 1:1 and 1:2 protein-metal complexes of UipAext proteins and selected metals.
| Metal | UipAext-ViU2A | UipAext-HG3 | UipAext-A9 | |||
|---|---|---|---|---|---|---|
| Prot-Me | Prot-Me2 | Prot-Me | Prot-Me2 | Prot-Me | Prot-Me2 | |
| Uranyl (UO22+) | (4.2 ± 1.8) × 10–8 | (5.0 ± 1.1) × 10–8 | (3.1 ± 2.5) × 10–8 | (3.7 ± 1.5) × 10–8 | (1.3 ± 0.6) × 10–9 | (3.6 ± 0.2) × 10–8 |
| Iron (Fe3+) | (3.3 ± 1.1) × 10–9 | (2.3 ± 0.1) × 10–8 | (2.9 ± 0.2) × 10–8 | (2.6 ± 0.4) × 10–5 | (3.2 ± 0.3) × 10–8 | (4.4 ± 0.1) × 10–5 |
| Calcium (Ca2+) | (1.1 ± 0.7) × 10–4 | (6.9 ± 0.7) × 10–4 | (1.2 ± 0.5) × 10–4 | (2.5 ± 0.2) × 10–3 | (2.9 ± 1.6) × 10–4 | – |
| Nickel (Ni2+) | (1.7 ± 0.3) × 10–5 | (2.2 ± 0.1) × 10–5 | (4.0 ± 0.9) × 10–5 | (2.8 ± 0.2) × 10–5 | (1.8 ± 0.8) × 10–4 | – |
| Zinc (Zn2+) | (8.9 ± 3.2) × 10–5 | (1.3 ± 0.1) × 10–4 | (3.3 ± 0.9) × 10–4 | (6.9 ± 0.4) × 10–5 | (5.5 ± 4.4) × 10–5 | – |
Values are given in M ± corresponds to the standard deviation, calculated from three independent experiments.
To determine whether UipAext proteins have a high affinity for other metals, the Kd values of complexes between UipAext proteins and Ca2+, Fe3+, Ni2+ and Zn2+ were measured (Figs. S13–S33). The best fitting curves of the three UipAext proteins and Fe3+ were obtained with the assumption of two metal binding sites (Figs. S13, S15 and S17). The dissociation constants of the 1:1 protein - Fe3+ complexes were in the nanomolar range, with Kd values ranging from 3.3 × 10−9 to 3.2 × 10–8 M. The dissociation constants of the 1:2 protein - Fe3+ complexes were more contrasted, with Kd values ranging from 2.3 × 10−8 to 4.4 × 10–5 M (Table 2).
By contrast, the Kd values measured for Ca2+, Ni2+and Zn2+ were at least three orders of magnitude higher than the dissociation constants measured for UO22+ and Fe3+ (ranging from 1.7 × 10–5 to 2.5 × 10–3 M), reflecting a much lower affinity of UipAext proteins for these divalent metal cations, as compared to UO22+ and Fe3+. It should be noted that for the three metal cations, two binding sites were inferred from the fluorescence titrations for the HG3 and ViU2A strains. The 1:2 protein:metal complex was not detected on the UipAext-A9 protein, unlike the other two proteins (Figs. S23, S28 and S33). This is probably due to the lack of the Tyr residue in this protein, which could not be used as a spectroscopic probe.
Characterization of uranyl and iron coordination
The coordination of UO22+ and Fe3+ by the UipAext domains was further studied by ATR-FTIR spectroscopy. ATR-FTIR spectra were recorded for each UipAext protein with or without metal, and the difference spectra were calculated (Figs. 2a and S34). In these spectra, positive and negative bands represent vibration modes of functional groups that are subject to changes due to the coordination of uranyl or iron by the protein. The FTIR spectra are dominated by contributions from the carboxylate groups of Asp or Glu (νas and νs(COO−) IR modes), which are highly sensitive to the coordination mode of the carboxyl group to metal cations or uranyl [81–84]. The spectra indicate uranyl coordination by monodentate and bidentate carboxylate groups (Fig. 2a). The νas(COO−) and νs(COO−) IR modes of the bidentate carboxylate(s) were assigned at 1510 and 1454 cm−1 for UipAext-A9, and at 1534 and 1456 cm−1 for UipAext-HG3 and UipAext-ViU2A. For the monodentate carboxylate groups, the νas(COO−) modes were observed at 1627 cm−1 for UipAext-A9 and at 1606–1610 cm−1 for UipAext-HG3 and UipAext-ViU2A. The frequency of the νas(UO22+) stretching mode also differed slightly for UipAext-A9 at 912 cm−1, and for the other strains at 916 cm−1. A similar involvement of mainly carboxylate groups was observed upon iron binding to the UipAext proteins (Fig. S34). As compared to uranyl, a greater number of bidentate ligands appear to be present in the iron binding site, with a larger relative intensity of bands assigned to the νas(COO−) and νs(COO−) IR modes of bidentate carboxylates at 1513–1526 and 1441–1441 cm−1 (Fig. S34). Note that for Fe3+, coordination is expected to involve 6–7 ligands in all directions, whereas for uranyl, 5–6 ligands are expected to contribute in the equatorial plane.
Fig. 2. Characterization of uranyl coordination by FTIR and EXAFS spectroscopy.
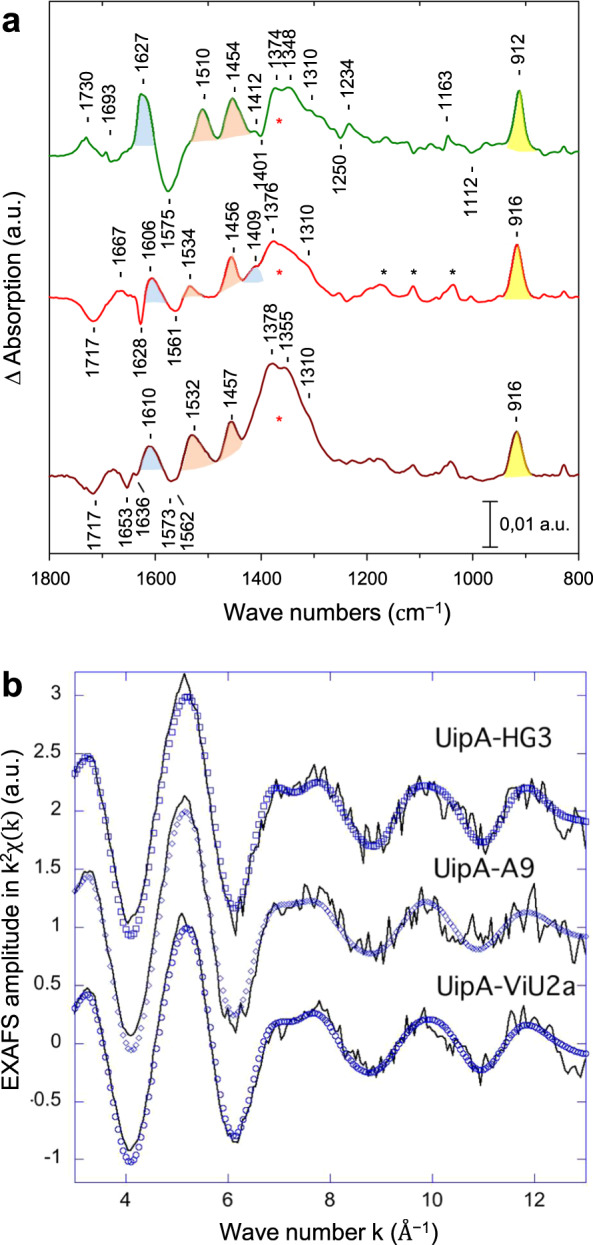
a FTIR difference spectra recorded with UipAext with UO22+ – minus – without UO22+. Green line: UipAext-A9; red line: UipAext-HG3; brown line: UipAext-ViU2A. Black * correspond to IR bands of the MES buffer and red * to a large band that is essentially due to the ν3as(NO3−) IR mode. The bands assigned to the modes of monodentate carboxylates are colored blue, and those assigned to bidentate carboxylates are colored orange. The band assigned to the νas(UO2) mode is colored yellow. b EXAFS spectra at the U LIII edge of UipAext-HG3, UipAext-A9 and UipAext-ViU2a. Experimental spectra = straight line, fit = blue forms.
EXAFS spectroscopy was performed as a complementary method to probe the uranyl coordination environment within UipAext proteins. It can be observed that all spectra exhibit similar waves (Fig. 2b), suggesting that the uranyl environment is very similar in all three proteins. Metrical parameters associated with the fit of the EXAFS data are summarized in Table S8. All uranyl cations are made of 2 (fixed) oxo bonds at 1.79 Å, in very good agreement with molecular uranyl in general [85] and close to solid state uranyl acetate for instance [69]. In all three proteins, the distance associated with equatorial oxygen atoms is very similar, between 2.36 and 2.38 Å. Although the signal of the second coordination sphere is relatively weak due to the presence of backscatters of low atomic number, best fits have been obtained with a shell of C atoms (see Materials and Methods section), indicating that carboxylate functions (mono and/or bidentate) are most likely involved in the uranyl coordination. Accordingly, a U-C single scattering path accounting for the C atoms of bidentate functions and a U-O-C triple scattering path accounting for the monodentate functions have been fitted. The values obtained for the U-Oeq average distance are indicative of the presence of mostly monodentate carboxylate groups [84–87], although only average values are discussed here. Regarding the carbon atoms, the U-C average distances reported in Table 1 in the range of 2.9–3.0 Å also support the presence of bidentate carboxylate functions [82, 86]. Therefore, we propose the presence of approximately two monodentate functions (Table S8) and complementary bidentate functions. It should be noted that the fit of the second coordination sphere in UipAext-A9 is associated with very high uncertainties, and the values must be considered carefully. Therefore, we propose the presence of at least one bidentate function (Table S8).
Crystal structure of UipA and the uranyl-binding site
We tested the crystallization of all three UipAext proteins and obtained crystals of UipAext-ViU2A (hereafter abbreviated as UipA) that diffracted up to 2.3 Å using a synchrotron source. The three-dimensional structure was solved using the SAD method with a crystal soaked with Gd-HPDO3A (Table S5). Overall, the three-dimensional structure of UipA (built from residues 143–280, the first 48 residues connecting the membrane helix to residue 143 being disordered) displays a domain-swapped dimer, with each monomer folding as two distinct domains (Fig. 3a).
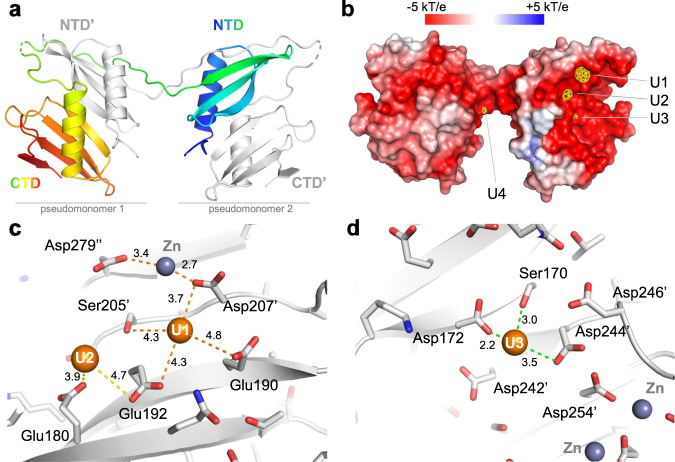
Fig. 3. Crystallographic structure of UipAext-ViU2A.
a An illustration of the dimer structure, with one monomer colored from the N-Terminus (blue) to the C-Terminus (red) and the second monomer colored in gray. b Molecular surface representation of the UipAext-ViU2A dimer (in the same orientation as in a) colored according to its electrostatic potential using the APBS plugin in pymol. The surface is superimposed with the anomalous map (yellow) collected at the uranium edge and contoured at 4σ all around the dimer. At this contour level, there are four uranium binding sites, three of which are localized on the negatively charged face of UipA (labeled U1-U3 as a function of peak height in the anomalous electron density map). c View of the two uranium binding sites U1 and U2. Ser205’ and Asp207’ indicate residues that are contributed by the second monomer of the domain-swapped dimer. d View of the third uranium binding site (U3). In (c and d), all of the residues are shown in stick form, and all distances from U atoms to residues located in a 5 Å radius are shown. The 2Fobs-Fcalc refined electron densities around the three uranium binding sites are shown in Fig. S37.
Each domain adopts the same fold, a central α-helix packed on a four-stranded antiparallel β-sheet (see Fig. S35B for a superimposition of all individual domains) reminiscent of the PepSY (pfam03413) domain. In a lipoprotein from Clostridium difficile (pdb code 4EXR; with 13% sequence identity), a distant 3D-homologue found by threading, these two domains appear close together in a protein apparently forming a monomer, while in UipA they are swapped in a dimer related by a crystallographic axis. However, the two pseudo-monomers of UipA (the NTer domain of one monomer plus the CTer domain of the other monomer and vice versa) superimpose very well with the lipoprotein, suggesting that the folding unit of UipA is this pseudomonomer (Fig. S35A). Although dimerization might be a crystallization artifact, it also suggests that dimerization of UipA may be functional and could occur at high concentration, such as within its membrane context, in which local protein concentration could be high due to 2D-limited diffusion. Since the two α-helices of both domains of the pseudomonomer are on the same face, this arrangement creates a six-stranded β-sheet concave surface which appears to be highly negatively charged (Fig. 3b). In the native crystal structure of UipA, five zinc ions arise from the crystallization conditions, each of which were located from an anomalous map collected at the Zn edge. Two of these zinc binding sites are on the α-helical side and the other three are on the opposite concave face (Fig. S36).
To decipher the uranium binding site on UipA, we soaked a crystal in a mother liquor supplemented with 5 mM uranyl nitrate. Since we collected data at a wavelength where U contributes to the anomalous signal but not Zn, we were able to unambiguously determine the position of four uranyl-binding sites, three of which were located on the β-sheet concave face (Figs. 3b–d, S37). The highest peak (labeled U1) is made of residues Glu190 and Glu192 from one monomer, and Ser205’ and Asp207’ from an adjacent monomer (Fig. 3c). However, the side chains of these residues are not well-resolved (see Fig. S37 for the refined 2Fobs-Fcalc map), the distance to the uranium atom is quite long (3.7–4.8 Å), and the Asp207’ side chain is also involved in binding to a Zn ion. The second uranium binding site (U2) is contributed by Glu192 and Glu180. The side chains of these residues are not well defined, and the distances to the uranium atom are quite long (3.9 Å and 4.7 Å). The third uranium peak (U3) is surrounded by the side chains of Ser170, Asp172, Asp244’ and possibly Asp242’. This site is well resolved in the electron density, and it was even possible to tentatively model one or two additional water molecules (although this could be an over-interpretation, due to the medium resolution of 2.9 Å achieved by this crystal; see Fig. S37). The distances of the oxygen atoms to the uranium atom ranged from 2.2–3.5 Å. Both carboxylates of these aspartate residues are monodentate, as observed on all of the other sites, although the moderate resolution obtained might not be sufficient to precisely determine their binding mode. The fourth uranium site (U4) is contributed by the side chains of Glu152 and Glu197, with the uranyl to oxygen distances ranging from 2.8–3.9 Å (Fig. S37). Sites U1 and U3 are remarkably similar, as they are made by four residues distributed on two adjacent β-strands (Fig. S38). Finally, we found that within the crystal packing, the dimers of UipA and the U and Zn binding sites create large tunnels of approximately 35 Å along the P43 crystallographic axis that are lined with the concave acidic face of UipA, and where almost all of the zinc and uranium ions bind (Fig. S39).
Discussion
Microbacterium is a widespread genus of actinobacteria that remains largely understudied. In this work, we confirmed the ability of Microbacterium spp. A9, ViU2A and HG3 to tolerate uranium. This phenotype has been reported for several other Microbacterium strains of diverse origin [11, 16, 17, 88]. Our work has shown that this trait is not strictly conserved across the Microbacterium genus, as M. lemovicicum ViU22 was shown to be sensitive to uranium.
When exposed to a non-toxic concentration of bioavailable uranyl, all strains were able to sequester the metal by biosorption and intracellular biomineralization, regardless of their uranium tolerance phenotype. Therefore, biomineralization, which has been proposed to contribute to uranium tolerance [26, 89], does not appear to be sufficient on its own. Phosphatases, in particular acid and alkaline phosphatase, play a major role in the biomineralization of uranium. Here, alkaline phosphatase was not detected in the proteome in any of the four stains, while acid phosphatase was detected in the proteome of strains A9, HG3 and ViU22, albeit with a very low abundance (0.006%, 0.005% and 0.016% of the total proteins, respectively; Tables S2, S3 and S4). The proteins were not modulated by uranyl, except in strain HG3 at 4 h (FC = 3.7). In strain ViU2A, one gene annotated as acid phosphatase was detected in the genome but the corresponding protein was not detected in the proteome, which can be due to the low abundance of the protein. The contribution of constitutive phosphatases to uranium biomineralization has been observed in Serratia sp. strain OT II 7 [90] and Caulobacter crescentus [89].
Interestingly, we only observed a transitory step of uranyl release in the uranium-tolerant strains. A uranium release step has been observed in other microorganisms [26, 37, 91, 92]. Although the mechanism of uranyl release is unexplained, it could involve an efflux transporter. Efflux is a conserved mechanism that can mitigate the intoxication of a variety of physiological and toxic metals [93]. Up-regulation of genes encoding metal efflux pumps of the RND, CDF and PIB-ATPase families during uranium exposure has been reported [31, 40, 94, 95], and horizontal gene transfer of PIB-ATPases has been observed in bacteria isolated from uranium-rich soils [30, 32]. However, even though multidrug resistance pumps have been induced in C. crescentus by uranyl exposure, none of them were found to play a role in uranium tolerance [96]. In fact, except for C. crescentus, in which the membrane transporter RsaFa (involved in S-layers export) is proposed to be involved in uranium efflux, the molecular mechanisms of uranyl trafficking across membranes remain largely unknown. We have shown that several proteins of the ABC transporter family (YxdL, YclQ, GlnP, NatA, YbiT, PstA and PstC) were up-produced by uranyl in at least one uranium-tolerant strain. YxdL is the only one of these proteins predicted to be an efflux transporter, mediating the efflux of cationic antimicrobial peptides and drugs in B. subtilis [97] and Clostridium hathewayi [98], respectively. Its potential role in uranyl efflux needs to be further examined. Regarding the other ABC transporters, YclQ is a petrobactin-binding protein that is part of an ABC transporter involved in iron transport in B. subtilis [99]; GlnP is a permease responsible for glutamate uptake in bacteria [100]; and PstA and PstC form a membrane channel responsible for phosphate internalization. We speculate that all of these transporters could mitigate uranyl intoxication by importing a variety of ligands (siderophores, phosphate and glutamate), which can chelate the metal and thereby reduce its bioavailability.
Among the proteins that are positively modulated by uranyl, only four were specific to the uranium-tolerant strains: YclQ and DegP, and two proteins with no functional annotation, UipA and UipB. Interestingly, these four proteins were among those with the highest FC values in at least one strain. The corresponding genes have no relationship at the genomic level. All of these proteins are predicted to be located at the cell membrane, showing that uranyl causes membrane damages, which is consistent with the literature (reviewed in [2]). DegP, a serine protease, is involved in the degradation of misfolded and aggregated proteins in the periplasm of Gram-negative bacteria [101]. In Gram-positive bacteria, DegP is located at the membrane and could also be involved in the envelope stress response, as recently suggested for Streptococcus gordonii [102]. In Microbacterium, DegP could alleviate the toxic effect of uranyl on membrane proteins and participate in uranyl tolerance.
The protein UipB displayed several conserved domains such as a cadherin repeat, and cadherin-like, Fn3 and Big-9 domains. Cadherin repeats and cadherin-like domains are found in Ca2+-binding proteins involved in cell-cell interactions and adhesion. In bacteria, Fn3 domains could be involved in the anchoring of extracellular glycohydrolases to their substrate [103]. Big domains are proposed to be involved in various functions such as adhesion and biofilm formation [104], and have been described as Ca2+-binding domains in a surface adhesin [105]. Big-9 domains are located at the N-terminal part of MtrF, a membrane cytochrome involved in the utilization of extracellular mineral forms of iron and manganese as respiratory electron acceptors in Shewanella oneidensis [106]. In addition, immunoglobulins have been identified as uranyl-binding proteins in the human serum [107]. These data suggest that UipB could be a modular protein with adhesion and uranyl- and/or metal cation-binding properties.
The protein UipA stood out from the other proteins modulated by uranyl because it was specific to the uranium-tolerant strains and had the highest FC values. Here, we established that UipA is a single-pass transmembrane protein with a large extracellular C-terminal part, displaying two high-affinity uranyl-binding sites with conditional Kd values in the nanomolar range. By comparison, a calmodulin-derived peptide considered to be a strong chelator of uranyl has conditional Kd value for UO22+ in the vicinity of 30 nM [61]. ATR-FTIR and EXAFS spectroscopy established that uranyl coordination involves monodentate and bidentate carboxylate groups, in line with what is described in the literature [80]. Crystallographic analysis showed that the extracellular part of UipA is organized in two PepSY domains, forming a domain-swapped dimer with a highly negatively charged concave surface. Four uranyl-binding sites were detected by crystallography, three of which are located on the negatively charged surface of UipA formed by 2 PepSY domains. However, in two of these sites (U2 and U4), the uranyl coordination only involves two ligands. This is unexpected for uranyl coordination in proteins [80], and could be an artefact due to the high concentration of the uranyl solution used to soak the crystal. In contrast, the U1 and U3 binding sites, which involve conserved D and E residues, are highly similar (Figs. 3c, d, S38) and may correspond to the two binding sites identified by mass spectrometry and fluorescence titration.
PepSY domains (pfam 03413) are found in the N-terminal pro-peptide of the M4 peptidase family, where they inhibit protease activity [108]. However, PepSY domains have also been detected in several non-peptidase proteins classified in different families, where they play an unknown function. Interestingly, membrane proteins with PepSY domains have been reported as iron-regulated [109–111]. Here, we assigned for the first time a function to the PepSY domain in a non-peptidase protein by showing that it can bind iron and uranyl with a very high affinity. Regarding uranium, UipA could participate in uranium tolerance through metal capture by its PepSY domains and sequestration at the cell surface, consequently limiting uranium entry into the cytoplasm. However, this sequestration would not completely prevent metal internalization in our experimental conditions since biomineralization occurred in all strains. It should be mentioned that uranium was not detected by TEM at the cell membrane. This indicates that if UipA-uranyl complexes formed at the membrane, they were not detectable by microscopy, possibly because non-precipitated forms of uranium are under the detection limit.
The high binding affinity of UipA for iron raises many questions about its physiological role and its potential functions in iron metabolism. Upstream of the uipA gene we found uipR and uipS, two genes predicted to encode a two-component system. One attractive hypothesis is that UipRSA constitutes a novel system involved in iron/uranium sensing (Fig. 4). UipR and UipS were either not detected (strain ViU2A) or not significantly modulated by uranyl (strains A9 and HG3) in the proteomic analysis, but this may be due to their very low abundance. Hence, the role of UipR and S as an iron/uranium sensing system is a working hypothesis that will need to be further examined.
Fig. 4. Working model showing the genomic organization of uipRS and A genes, and the proposed mechanism of U sensing and the function of UipA.
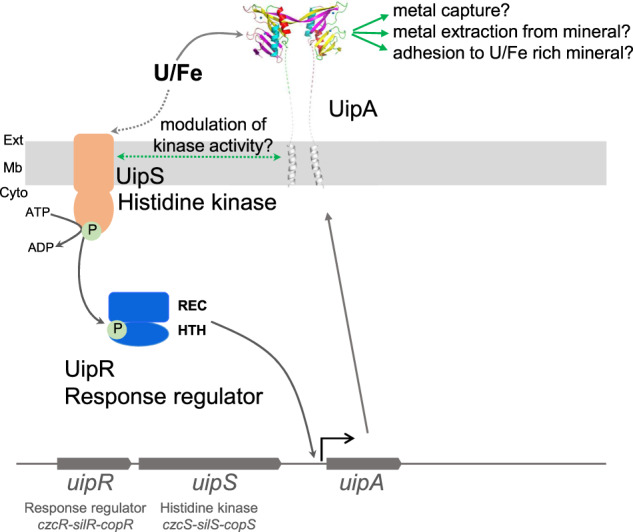
The topology of UipS was predicted using the CCTOP program. UipS is predicted to be a histidine kinase, which could be involved in Fe and U sensing and activation of its cognate regulator, UipR (REC = receiver domain; HTH = Helix Turn Helix domain), via phosphate transfer. The activated UipR would then induce uipA gene expression, leading to overproduction of the protein UipA, formation of UipA dimers at the cell surface, and metal binding. Hypothetical functions of UipA are proposed.
A striking comparison can be made between uipRSA and the uranium-induced operon CCNA_01361-urpR-urpS in C. crescentus [112] and the ferrous iron-responsive operon bqsPQRS in P. aeruginosa [111]. In these systems, the expression of PepSY membrane protein genes (CCNA_01361 as well as BqsP and Q) is mediated by a two-component system (RS proteins) in response to metal exposure. In B. subtilis, YycI (a PepSY-containing membrane protein) has been shown to modulate the kinase activity of the YycFG two-component system through transmembrane domain interactions [113], and the same role has been proposed for BqsP and BqsQ in P. aeruginosa [111]. A similar role can thus be suggested for UipA in modulating the UipRS two-component system in response to iron concentration. In addition, our results suggest that UipA, with its nanomolar binding affinity for iron, captures the metal outside of the cell and may participate in iron homeostasis. Because iron is a critical resource, one hypothesis is that UipA could constitute an iron reservoir for the cell. In this scenario, UipA would likely interact with another protein for iron transport. The crystallographic structure shows that UipA has a flat, negatively charged surface. Based on this observation, we speculate that another possible function of UipA could be to promote interaction with solid surfaces such as iron-containing minerals. More generally, the interplay between uranium and iron in these environmental bacteria will have to be explored in more detail.
Supplementary information
Acknowledgements
This work was supported by the Toxicology program of the CEA (BEnUr project), the CNRS/CEA/AREVA NEEDS-Ressources Program (SURE project) and the CNRS/IRSN GDR TRASSE program. The PhD grants of Nicolas Gallois and Abbas Mohamad Ali were funded by the PhD program of the CEA. The PhD grant of Nicolas Theodorakopoulos was funded by the IRSN/PACA regional council. We thank the AFMB lab (Marseille, France) for the use of the rotating anode. This work has benefitted from the facilities and expertize of the PROXIMA-1 beam line for XRD and MARS beam line for EXAFS at the SOLEIL synchrotron, Saint Aubin, France. We warmly thank Séverine Zirah for providing us with the HG3 strain used in this study, and Badreddine Douzi for the pKtop plasmid.
Author contributions
VC designed and supervised the study. VC and LP performed the uranium exposure experiments and prepared the samples for proteomics. NT, MF and LF performed microscopic analysis and ICP-AES measurements. BAB acquired the proteomics data and NG, BA-B and JA performed the proteomic analyses. PO and MB performed genomic and phylogenetic analyses and constructed ORF databases for proteomics. AMA established UipA topology in vivo. NG purified the recombinant proteins and performed fluorescence titration experiments, assisted by NB, NG and DL performed native mass spectrometry experiments. NG acquired FTIR data under CB supervision. NG prepared the samples for synchrotron-based analysis and participated in EXAFS data acquisition. CDA performed EXAFS data acquisition and processing. NB and NG performed crystallization tests and obtained the protein crystals under supervision of PA. PA and PL resolved the UipA structure. NG, BA-B, PO, MB, NB, AMA, DL, CDA, PA, CB, JA and VC analyzed the data. NG and VC wrote the paper and PO, MB, LF, DL, CDA, PA, CB. BA-B and JA edited it.
Competing interests
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential competing interests.
Footnotes
The original online version of this article was revised due to a mix-up in the table captions.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/3/2022
A Correction to this paper has been published: 10.1038/s41396-021-01164-w
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01113-7.
References
- 1.Cothern CR, Lappenbusch WL, Cotruvo JA. Health effects guidance for uranium in drinking water. Health Phys. 1983;44:377–84. doi: 10.1097/00004032-198306001-00035. [DOI] [PubMed] [Google Scholar]
- 2.Gao N, Huang ZH, Liu HQ, Hou J, Liu XH. Advances on the toxicity of uranium to different organisms. Chemosphere. 2019;237:124548. doi: 10.1016/j.chemosphere.2019.124548. [DOI] [PubMed] [Google Scholar]
- 3.Markich SJ. Uranium speciation and bioavailability in aquatic systems: an overview. ScientificWorldJournal. 2002;2:707–29. doi: 10.1100/tsw.2002.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidaud C, Gourion-Arsiquaud S, Rollin-Genetet F, Torne-Celer C, Plantevin S, Pible O, et al. Structural consequences of binding of UO22+ to apotransferrin: can this protein account for entry of uranium into human cells? Biochemistry. 2007;46:2215–26. doi: 10.1021/bi061945h. [DOI] [PubMed] [Google Scholar]
- 5.Suriya J, Shekar MC, Nathani NM, Suganya T, Bharathiraja S, Krishnan M. Assessment of bacterial community composition in response to uranium levels in sediment samples of sacred Cauvery River. Appl Microbiol Biotechnol. 2017;101:831–41. doi: 10.1007/s00253-016-7945-2. [DOI] [PubMed] [Google Scholar]
- 6.Antunes SC, Pereira R, Marques SM, Castro BB, Goncalves F. Impaired microbial activity caused by metal pollution: a field study in a deactivated uranium mining area. Sci Total Environ. 2011;410:87–95. doi: 10.1016/j.scitotenv.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Yan X, Luo XG. Radionuclides distribution, properties, and microbial diversity of soils in uranium mill tailings from southeastern China. J Environ Radioactivity. 2015;139:85–90. doi: 10.1016/j.jenvrad.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Radeva G, Kenarova A, Bachvarova V, Flemming K, Popov I, Vassilev D, et al. Bacterial diversity at abandoned uranium mining and milling sites in Bulgaria as revealed by 16S rRNA genetic diversity study. Water Air Soil Poll. 2013;224:1748. [Google Scholar]
- 9.Islam E, Paul D, Sar P. Microbial diversity in uranium deposits from Jaduguda and Bagjata uranium mines, India as revealed by clone library and denaturing gradient gel electrophoresis analyses. Geomicrobiol J. 2014;31:862–74. [Google Scholar]
- 10.Mondani L, Benzerara K, Carriere M, Christen R, Mamindy-Pajany Y, Fevrier L, et al. Influence of uranium on bacterial communities: a comparison of natural uranium-rich soils with controls. Plos ONE. 2011;6:e25771. doi: 10.1371/journal.pone.0025771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam E, Sar P. Diversity, metal resistance and uranium sequestration abilities of bacteria from uranium ore deposit in deep earth stratum. Ecotoxicol Environ Saf. 2016;127:12–21. doi: 10.1016/j.ecoenv.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Jaswal R, Pathak A, Edwards B, Lewis R, Seaman JC, Stothard P, et al. Metagenomics-guided survey, isolation, and characterization of uranium resistant microbiota from the Savannah River Site, USA. Genes-Basel. 2019;10:325. doi: 10.3390/genes10050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, Nongkhlaw M, Acharya C, Joshi SR. Uranium (U)-tolerant bacterial diversity from U ore deposit of Domiasiat in North-East India and its prospective utilisation in bioremediation. Microbes Environ. 2013;28:33–41. doi: 10.1264/jsme2.ME12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki YB, Resistance JF. to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol J. 2004;21:113–21. [Google Scholar]
- 15.Martinez RJ, Beazley MJ, Taillefert M, Arakaki AK, Skolnick J, Sobecky PA. Aerobic uranium (VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ Microbiol. 2007;9:3122–33. doi: 10.1111/j.1462-2920.2007.01422.x. [DOI] [PubMed] [Google Scholar]
- 16.Nedelkova M, Merroun ML, Rossberg A, Hennig C, Selenska-Pobell S. Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. Fems Microbiol Ecol. 2007;59:694–705. doi: 10.1111/j.1574-6941.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Castro I, Arnador-Garcia A, Moreno-Romero C, Lopez-Fernandez M, Phrommavanh V, Nos J, et al. Screening of bacterial strains isolated from uranium mill tailings porewaters for bioremediation purposes. J Environ Radioactivity. 2017;166:130–41. doi: 10.1016/j.jenvrad.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Andres Y, Abdelouas A, Grambow B. Microorganisms effects on radionuclides migration. Radioprotection. 2002;37:C1-3 C1–9. [Google Scholar]
- 19.Wufuer R, Wei YY, Lin QH, Wang HW, Song WJ, Liu W, et al. Uranium bioreduction and biomineralization. Adv Appl Microbiol. 2017;101:137–68. doi: 10.1016/bs.aambs.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Fowle DAF JB, Martin AM. Experimental study of uranyl adsorption onto Bacillus subtilis. Environ Sci Technol. 2000;34:3737–41. [Google Scholar]
- 21.Merroun ML, Raff J, Rossberg A, Hennig C, Reich T, Selenska-Pobell S. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl Environ Microbiol. 2005;71:5532–43. doi: 10.1128/AEM.71.9.5532-5543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadd GM. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology. 2010;156:609–43. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 23.Macaskie L, Bonthrone K, Rouch D. Phosphatase-mediated heavy metal accumulation by a Citrobacter sp. and related enterobacteria. FEMS Microbiol Lett. 1994;121:141–6. doi: 10.1111/j.1574-6968.1994.tb07090.x. [DOI] [PubMed] [Google Scholar]
- 24.Beazley MJ, Martinez RJ, Sobecky PA, Webb SM, Taillefert M. Nonreductive biomineralization of uranium(VI) phosphate via microbial phosphatase activity in anaerobic conditions. Geomicrobiol J. 2009;26:431–41. [Google Scholar]
- 25.Sousa T, Chung AP, Pereira A, Piedade AP, Morais PV. Aerobic uranium immobilization by Rhodanobacter A2-61 through formation of intracellular uranium-phosphate complexes. Metallomics. 2013;5:390–7. doi: 10.1039/c3mt00052d. [DOI] [PubMed] [Google Scholar]
- 26.Acharya C, Chandwadkar P, Nayak C. Unusual versatility of the filamentous, diazotrophic cyanobacterium Anabaena torulosa revealed for its survival during prolonged uranium exposure. Appl Environ Microbiol. 2017;83:e03356–16. doi: 10.1128/AEM.03356-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee A, Wheaton GH, Blum PH, Kelly RM. Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proc Natl Acad Sci USA. 2012;109:16702–7. doi: 10.1073/pnas.1210904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashmi V, Shylajanaciyar M, Rajalakshmi R, D’Souza SF, Prabaharan D, Uma L. Siderophore mediated uranium sequestration by marine cyanobacterium Synechococcus elongatus BDU 130911. Bioresour Technol. 2013;130:204–10. doi: 10.1016/j.biortech.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Yung MC, Ma J, Salemi MR, Phinney BS, Bowman GR, Jiao Y. Shotgun proteomic analysis unveils survival and detoxification strategies by Caulobacter crescentus during exposure to uranium, chromium, and cadmium. J Proteome Res. 2014;13:1833–47. doi: 10.1021/pr400880s. [DOI] [PubMed] [Google Scholar]
- 30.Martinez RJ, Wang YL, Raimondo MA, Coombs JM, Barkay T, Sobecky PA. Horizontal gene transfer of P-IB-type ATPases among bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Appl Environ Microbiol. 2006;72:3111–8. doi: 10.1128/AEM.72.5.3111-3118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nongkhlaw M, Kumar R, Acharya C, Joshi SR. Occurrence of horizontal gene transfer of P-IB-type ATPase genes among bacteria isolated from an uranium rich deposit of Domiasiat in North East India. Plos ONE. 2012;7:e48199. doi: 10.1371/journal.pone.0048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nongkhlaw M, Joshi SR. Molecular insight into the expression of metal transporter genes in Chryseobacterium sp. PMSZPI isolated from uranium deposit. Plos ONE. 2019;14:e0216995. doi: 10.1371/journal.pone.0216995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khare D, Kumar R, Acharya C. Genomic and functional insights into the adaptation and survival of Chryseobacterium sp. strain PMSZPI in uranium enriched environment. Ecotoxicol Environ Saf. 2020;191:110217. doi: 10.1016/j.ecoenv.2020.110217. [DOI] [PubMed] [Google Scholar]
- 34.Newsome L, Morris K, Lloyd JR. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem Geol. 2014;363:164–84. [Google Scholar]
- 35.Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol. 2005;187:8437–49. doi: 10.1128/JB.187.24.8437-8449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khemiri A, Carriere M, Bremond N, Ben Mlouka MA, Coquet L, Llorens I, et al. Escherichia coli response to uranyl exposure at low pH and associated protein regulations. Plos ONE. 2014;9:e89863. doi: 10.1371/journal.pone.0089863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal M, Pathak A, Rathore R, Prakash O, Singh R, Jaswal R, et al. Proteogenomic analysis of Burkholderia species strains 25 and 46 isolated from uraniferous soils reveals multiple mechanisms to cope with uranium stress. Cells. 2018;7:269. doi: 10.3390/cells7120269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panda B, Basu B, Acharya C, Rajaram H, Apte SK. Proteomic analysis reveals contrasting stress response to uranium in two nitrogen-fixing Anabaena strains, differentially tolerant to uranium. Aquat Toxicol. 2017;182:205–13. doi: 10.1016/j.aquatox.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Orellana R, Hixson KK, Murphy S, Mester T, Sharma ML, Lipton MS, et al. Proteome of Geobacter sulfurreducens in the presence of U(VI) Microbiology. 2014;160:2607–17. doi: 10.1099/mic.0.081398-0. [DOI] [PubMed] [Google Scholar]
- 40.Pinel-Cabello M, Jroundi F, Lopez-Fernandez M, Geffers R, Jarek M, Jauregui R, et al. Multisystem combined uranium resistance mechanisms and bioremediation potential of Stenotrophomonas bentonitica BII-R7: Transcriptomics and microscopic study. J Hazard Mater. 2021;403:123858. doi: 10.1016/j.jhazmat.2020.123858. [DOI] [PubMed] [Google Scholar]
- 41.Francois F, Lombard C, Guigner JM, Soreau P, Brian-Jaisson F, Martino G, et al. Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl Environ Microbiol. 2012;78:1097–106. doi: 10.1128/AEM.06522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mondani L, Piette L, Christen R, Bachar D, Berthomieu C, Chapon V. Microbacterium lemovicicum sp nov., a bacterium isolated from a natural uranium-rich soil. Int J Syst Evolut Microbiol. 2013;63:2600–6. doi: 10.1099/ijs.0.048454-0. [DOI] [PubMed] [Google Scholar]
- 43.Chapon V, Piette L, Vesvres M-H, Coppin F, Marrec CL, Christen R, et al. Microbial diversity in contaminated soils along the T22 trench of the Chernobyl experimental platform. Appl Geochem. 2012;27:1375–83. [Google Scholar]
- 44.Theodorakopoulos N, Chapon V, Coppin F, Floriani M, Vercouter T, Sergeant C, et al. Use of combined microscopic and spectroscopic techniques to reveal interactions between uranium and Microbacterium sp A9, a strain isolated from the Chernobyl exclusion zone. J Hazard Mater. 2015;285:285–93. doi: 10.1016/j.jhazmat.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Ortet P, Gallois N, Long J, Barakat M, Chapon V. Draft genome sequence of Microbacterium oleivorans strain A9, a bacterium isolated from Chernobyl radionuclide-contaminated soil. Genome Announc. 2017;999:e00092–17. doi: 10.1128/genomeA.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallois N, Alpha-Bazin B, Ortet P, Barakat M, Piette L, Long J, et al. Proteogenomic insights into uranium tolerance of a Chernobyl’s Microbacterium bacterial isolate. J Proteom. 2018;177:148–57. doi: 10.1016/j.jprot.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Ortet P, Gallois N, Long J, Zirah S, Berthomieu C, Armengaud J, et al. Complete genome sequences of four Microbacterium strains isolated from metal- and radionuclide-rich soils. Microbiol Resour Announc. 2019;8:e00846–19. doi: 10.1128/MRA.00846-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein G, Mathe C, Biola-Clier M, Devineau S, Drouineau E, Hatem E, et al. RNA-binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology. 2016;10:1555–64. doi: 10.1080/17435390.2016.1244299. [DOI] [PubMed] [Google Scholar]
- 49.Carvalho PC, Hewel J, Barbosa VC, Yates Iii JR. Identifying differences in protein expression levels by spectral counting and feature selection. Genet Mol Res. 2008;7:342–56. doi: 10.4238/vol7-2gmr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D50. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;38:1079–86. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 53.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–8. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, et al. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020;48:D579–D89. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karimova G, Ladant D. Defining membrane protein topology using pho-lac reporter fusions. Methods Mol Biol. 2017;1615:129–42. doi: 10.1007/978-1-4939-7033-9_10. [DOI] [PubMed] [Google Scholar]
- 60.Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. BioTechniques. 2010;48:463–5. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardoux R, Sauge-Merle S, Lemaire D, Delangle P, Guilloreau L, Adriano JM, et al. Modulating uranium binding affinity in engineered calmodulin EF-hand peptides: effect of phosphorylation. PLoS ONE. 2012;7:e41922. doi: 10.1371/journal.pone.0041922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang J, Renshaw JC, Sarsfield MJ, Livens FR, Collison D, Charnock JM, et al. Solution chemistry of uranyl ion with iminodiacetate and oxydiacetate: a combined NMR/EXAFS and potentiometry/calorimetry study. Inorg Chem. 2003;42:1233–40. doi: 10.1021/ic020460o. [DOI] [PubMed] [Google Scholar]
- 63.Smith RM, Martell AE. Critical stability-constants, enthalpies and entropies for the formation of metal-complexes of aminopolycarboxylic acids and carboxylic-acids. Sci Total Environ. 1987;64:125–47. [Google Scholar]
- 64.Maeder M, King P. ReactLab. Jplus Consulting Pty Ltd East Fremantle, West Australia, Australia; 2009.
- 65.Hienerwadel R, Gourion-Arsiquaud S, Ballottari M, Bassi R, Diner BA, Berthomieu C. Formate binding near the redox-active Tyrosine(D) in Photosystem II: consequences on the properties of Tyr(D) Photosynth Res. 2005;84:139–44. doi: 10.1007/s11120-005-0637-x. [DOI] [PubMed] [Google Scholar]
- 66.Goebbert DJ, Garand E, Wende T, Bergmann R, Meijer G, Asmis KR, et al. Infrared spectroscopy of the microhydrated nitrate ions NO(3)(-)(H2O)(1-6) J Phys Chem A. 2009;113:7584–92. doi: 10.1021/jp9017103. [DOI] [PubMed] [Google Scholar]
- 67.LLorens I, Solari PL, Sitaud B, Bes R, Cammelli S, Hermange H, et al. X-ray absorption spectroscopy investigations on radioactive matter using MARS beamline at SOLEIL synchrotron. Radiochimica Acta. 2014;102:957–72. [Google Scholar]
- 68.Ravel B, Newville M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J Synchrotron Radiat. 2005;12:537–41. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 69.Howatson J, Grev DM, Morosin B. Crystal and molecular-structure of uranyl acetate dihydrate. J Inorg Nucl Chem. 1975;37:1933–5. [Google Scholar]
- 70.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr Sect D, Biol Crystallogr. 2010;66:133–44. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Legrand P. XDSME: XDS Made Easier. GitHub repository. 2017.
- 72.Sheldrick GM. A short history of SHELX. Acta Crystallogr Sect A, Found Crystallogr. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 73.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D, Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D, Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grosse C, Grass G, Anton A, Franke S, Santos AN, Lawley B, et al. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–93. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caille O, Rossier C, Perron K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol. 2007;189:4561–8. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, et al. A Completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2018;430:2237–43. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 79.Dobson L, Remenyi I, Tusnady GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–12. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin YW. Uranyl binding to proteins and structural-functional impacts. Biomolecules. 2020;10:457. doi: 10.3390/biom10030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kakihana M, Nagumo T, Okamoto M, Kakihana H. Coordination structures for uranyl carboxylate complexes in aqueous-solution studied by Ir and C-13 nmr-spectra. J Phys Chem-Us. 1987;91:6128–36. [Google Scholar]
- 82.Sauge-Merle S, Brulfert F, Pardoux R, Solari PL, Lemaire D, Safi S, et al. Structural analysis of uranyl complexation by the EF-Hand motif of calmodulin: effect of phosphorylation. Chem-Eur J. 2017;23:15505–17. doi: 10.1002/chem.201703484. [DOI] [PubMed] [Google Scholar]
- 83.Deacon G, Phillips R. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord Chem Rev. 1980;33:227–50. [Google Scholar]
- 84.Groenewold GS, de Jong WA, Oomens J, Van, Stipdonk MJ. Variable denticity in carboxylate binding to the uranyl coordination complexes. J Am Soc Mass Spectr. 2010;21:719–27. doi: 10.1016/j.jasms.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 85.Perez-Conesa S, Torrico F, Martinez JM, Pappalardo RR, Marcos ES. A general study of actinyl hydration by molecular dynamics simulations using ab initio force fields. J Chem Phys. 2019;150:104504. doi: 10.1063/1.5083216. [DOI] [PubMed] [Google Scholar]
- 86.Lahrouch F, Chamayou AC, Creff G, Duvail M, Hennig C, Rodriguez MJL, et al. A combined spectroscopic/molecular dynamic study for investigating a methyl-carboxylated PEI as a potential uranium decorporation agent. Inorg Chem. 2017;56:1300–8. doi: 10.1021/acs.inorgchem.6b02408. [DOI] [PubMed] [Google Scholar]
- 87.Denecke MA, Reich T, Bubner M, Pompe S, Heise KH, Nitsche H, et al. Determination of structural parameters of uranyl ions complexed with organic acids using EXAFS. J Alloy Compd. 1998;271:123–7. [Google Scholar]
- 88.Brown SD, Palumbo AV, Panikov N, Ariyawansa T, Klingeman DM, Johnson CM, et al. Draft genome sequence for Microbacterium laevaniformans strain OR221, a bacterium tolerant to metals, nitrate, and low pH. J Bacteriol. 2012;194:3279–80. doi: 10.1128/JB.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yung MC, Jiao YQ. Biomineralization of uranium by PhoY phosphatase activity aids cell survival in Caulobacter crescentus. Appl Environ Microbiol. 2014;80:4795–804. doi: 10.1128/AEM.01050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chandwadkar P, Misra HS, Acharya C. Uranium biomineralization induced by a metal tolerant Serratia strain under acid, alkaline and irradiated conditions. Metallomics. 2018;10:1078–88. doi: 10.1039/c8mt00061a. [DOI] [PubMed] [Google Scholar]
- 91.Bader M, Muller K, Foerstendorf H, Drobot B, Schmidt M, Musat N, et al. Multistage bioassociation of uranium onto an extremely halophilic archaeon revealed by a unique combination of spectroscopic and microscopic techniques. J Hazard Mater. 2017;327:225–32. doi: 10.1016/j.jhazmat.2016.12.053. [DOI] [PubMed] [Google Scholar]
- 92.Kolhe N, Zinjarde S, Acharya C. Impact of uranium exposure on marine yeast, Yarrowia lipolytica: Insights into the yeast strategies to withstand uranium stress. J Hazard Mater. 2020;381:121226. doi: 10.1016/j.jhazmat.2019.121226. [DOI] [PubMed] [Google Scholar]
- 93.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15:338–50. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Junier P, Dalla Vecchia E, Bernier-Latmani R. The response of Desulfotomaculum reducens MI-1 to U(VI) exposure: a transcriptomic study. Geomicrobiol J. 2011;28:483–96. [Google Scholar]
- 95.Sutcliffe B, Chariton AA, Harford AJ, Hose GC, Stephenson S, Greenfield P, et al. Insights from the genomes of microbes thriving in uranium-enriched sediments. Micro Ecol. 2018;75:970–84. doi: 10.1007/s00248-017-1102-z. [DOI] [PubMed] [Google Scholar]
- 96.Yung MC, Park DM, Overton KW, Blow MJ, Hoover CA, Smit J, et al. Transposon mutagenesis paired with deep sequencing of Caulobacter crescentus under uranium stress reveals genes essential for detoxification and stress tolerance. J Bacteriol. 2015;197:3160–72. doi: 10.1128/JB.00382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Staron A, Finkeisen DE, Mascher T. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrobial Agents Chemother. 2011;55:515–25. doi: 10.1128/AAC.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rafii F, Park M. Detection and characterization of an ABC transporter in Clostridium hathewayi. Arch Microbiol. 2008;190:417–26. doi: 10.1007/s00203-008-0385-3. [DOI] [PubMed] [Google Scholar]
- 99.Zawadzka AM, Kim Y, Maltseva N, Nichiporuk R, Fan Y, Joachimiak A, et al. Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc Natl Acad Sci USA. 2009;106:21854–9. doi: 10.1073/pnas.0904793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kronemeyer W, Peekhaus N, Kramer R, Sahm H, Eggeling L. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J Bacteriol. 1995;177:1152–8. doi: 10.1128/jb.177.5.1152-1158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–28. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 102.Lima BP, Kho K, Nairn BL, Davies JR, Svensater G, Chen R, et al. Streptococcus gordonii type I lipoteichoic acid contributes to surface protein biogenesis. mSphere. 2019;4:e00814–19. doi: 10.1128/mSphere.00814-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kataeva IA, Seidel RD, 3rd, Shah A, West LT, Li XL, Ljungdahl LG. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl Environ Microbiol. 2002;68:4292–300. doi: 10.1128/AEM.68.9.4292-4300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huttener M, Prieto A, Aznar S, Bernabeu M, Glaria E, Valledor AF, et al. Expression of a novel class of bacterial Ig-like proteins is required for IncHI plasmid conjugation. PLoS Genet. 2019;15:e1008399. doi: 10.1371/journal.pgen.1008399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raman R, Rajanikanth V, Palaniappan RU, Lin YP, He H, McDonough SP, et al. Big domains are novel Ca(2)+-binding modules: evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS ONE. 2010;5:e14377. doi: 10.1371/journal.pone.0014377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clarke TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J, et al. Structure of a bacterial cell surface decaheme electron conduit. Proc Natl Acad Sci USA. 2011;108:9384–9. doi: 10.1073/pnas.1017200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vidaud C, Dedieu A, Basset C, Plantevin S, Dany I, Pible O, et al. Screening of human serum proteins for uranium binding. Chem Res Toxicol. 2005;18:946–53. doi: 10.1021/tx050038v. [DOI] [PubMed] [Google Scholar]
- 108.Yeats C, Rawlings ND, Bateman A. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem Sci. 2004;29:169–72. doi: 10.1016/j.tibs.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 109.Manck LE, Espinoza JL, Dupont CL, Barbeau KA. Transcriptomic study of substrate-specific transport mechanisms for iron and carbon in the marine copiotroph Alteromonas macleodii. Msystems. 2020;5:e00070–20. doi: 10.1128/mSystems.00070-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lim CK, Hassan KA, Tetu SG, Loper JE, Paulsen IT. The effect of iron limitation on the transcriptome and proteome of Pseudomonas fluorescens Pf-5. PLoS ONE. 2012;7:e39139. doi: 10.1371/journal.pone.0039139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kreamer NN, Wilks JC, Marlow JJ, Coleman ML, Newman DK. BqsR/BqsS constitute a two-component system that senses extracellular Fe(II) in Pseudomonas aeruginosa. J Bacteriol. 2012;194:1195–204. doi: 10.1128/JB.05634-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park DM, Taffet MJ. Combinatorial sensor design in Caulobacter crescentus for selective environmental uranium detection. ACS Synth Biol. 2019;8:807–17. doi: 10.1021/acssynbio.8b00484. [DOI] [PubMed] [Google Scholar]
- 113.Szurmant H, Bu L, Brooks CL, 3rd, Hoch JA. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc Natl Acad Sci USA. 2008;105:5891–6. doi: 10.1073/pnas.0800247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.