Abstract
Microbes can produce valuable natural products widely applied in medicine, food and other important fields. Nevertheless, it is usually challenging to achieve ideal industrial yields due to low production rate and poor toxicity tolerance. Evolution is a constant mutation and adaptation process used to improve strain performance. Generally speaking, the synthesis of natural products in microbes is often intricate, involving multiple enzymes or multiple pathways. Individual evolution of a certain enzyme often fails to achieve the desired results, and may lead to new rate-limiting nodes that affect the growth of microbes. Therefore, it is inevitable to evolve the biosynthetic pathways or the whole genome. Here, we reviewed the pathway-level evolution including multi-enzyme evolution, regulatory elements engineering, and computer-aided engineering, as well as the genome-level evolution based on several tools, such as genome shuffling and CRISPR/Cas systems. Finally, we also discussed the major challenges faced by in vivo evolution strategies and proposed some potential solutions.
Keywords: In vivo evolution, Natural products, Biosynthetic pathway evolution, Genome evolution, Microbes
1. Introduction
The complex metabolic network in microbes can produce a variety of valuable natural products, such as terpenes, flavonoids, and alkaloids [1]. To improve the yield of these microbial natural products, numerous evolution methods, including random mutagenesis, error-prone PCR, site-directed mutagenesis and site-saturation mutagenesis, have been implemented [2,3]. However, since most of these strategies can only be applied in the evolution of a single enzyme, optimized phenotypes may not be easily obtained [4]. Therefore, a great number of creative solutions have been inspired, including adaptive laboratory evolution (ALE), and synthetic biology mediated pathway and genome evolution (Fig. 1). ALE helps to screen beneficial mutants through long-term cultivation under special pressures, mostly physical or chemical factors [[5], [6], [7]]. It may generate mutations in the pathway or the whole genome randomly. However, ALE is usually time-consuming and laborious as several rounds of repetitive work are needed [8,9]. In recent years, with the development of synthetic biology, new tools have been exploited in pathway and genome evolution, including CRISPR-Cas9-and homology-directed-repair (HDR)-assisted genome-scale engineering (CHAnGE) [10], synthetic chromosome rearrangement and modification by LoxP-mediated evolution (SCRaMBLE), etc. [11]. In particular, synthetic biology mediated pathway and genome evolution are mostly rational designed engineering strategies, which makes it possible to rapidly evolve the target microbial biosynthetic pathways or the whole genomes [[12], [13], [14]].
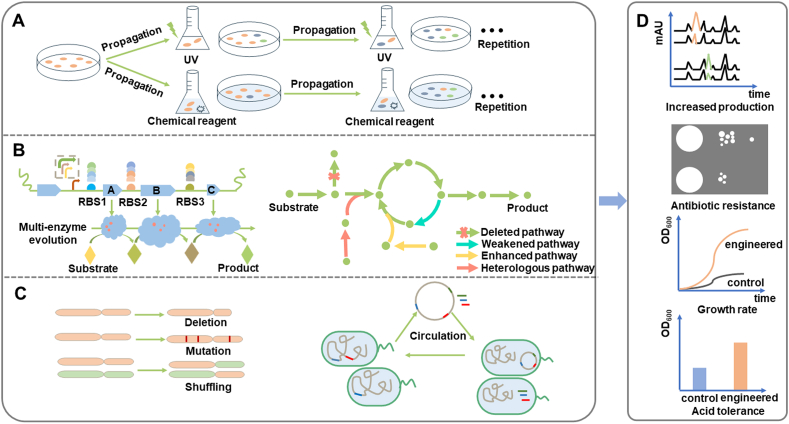
Fig. 1.
Overview of pathway and genome evolution strategies. A. Adaptive laboratory evolution: The strain grew under a certain environmental pressure through chemical and physical factors; B. Pathway-level evolution strategies: Pathway evolution was realized by multi-enzyme evolution, regulatory elements engineering and pathway reconstruction; C. Genome-level evolution strategies: By protoplast fusion or evolutionary tools that rely on synthetic biology, the evolution can be achieved on a genomic scale including deletion of large fragments, mutation of multiple sites, or genome shuffling; D. Evolutionary phenotype: Evolved strains could increase natural product yield, antibiotic resistance, acid tolerance, etc.
Here, we briefly summarized the latest advances in evolution strategies based on pathway-level evolution and genome-level evolution, as well as their applications within the recent five years. At the same time, the drawbacks and challenges of these evolutionary strategies in screening optimal strains are also discussed.
2. Pathway-level evolution strategies
The synthesis of natural or unnatural products in microbes usually requires a synergistic network of multiple metabolic pathways. Traditionally, biologists tend to deal with each step of the biosynthetic pathway in an isolated way [15]. Recently, assisted by synthetic biology, pathway-level evolution becomes popular (Fig. 2).
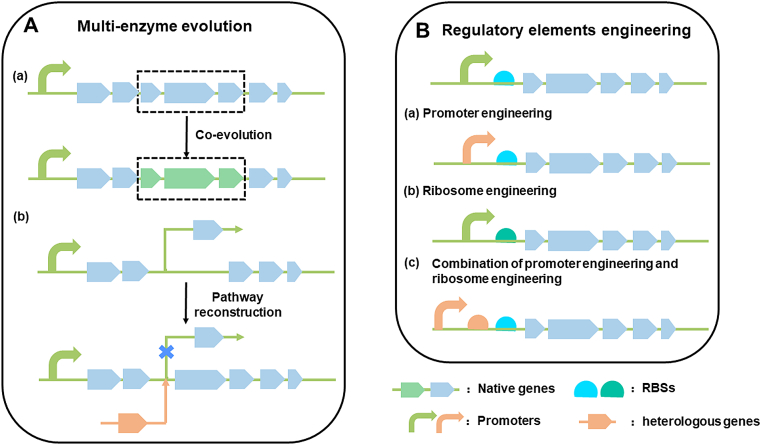
Fig. 2.
Pathway-level evolutionary strategies. A. Multi-enzyme evolution: (a) The co-evolution of multiple enzymes; (b) Pathway reconstruction. B. Regulatory elements engineering: (a) Promoter engineering; (b) Ribosome engineering; (c) Combination of promoter engineering and ribosome engineering.
2.1. Pathway-level evolution based on multi-enzyme evolution
In the vast majority of biosynthetic pathways, there are often more than one rate-limiting enzymes. Individual enzyme evolution may introduce new rate-limiting nodes, causing flux imbalance in the biosynthetic pathway [16]. Collaborative evolution of multiple enzymes may provide an effective solution to avoid this dilemma.
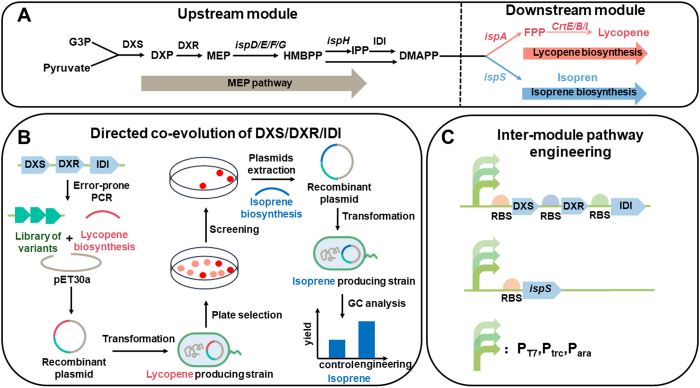
Traditional enzyme evolution mostly relies on DNA shuffling and error-prone PCR techniques, which are constantly used for multi-enzyme evolution [17]. For example, Lv et al. partitioned the isoprene biosynthetic pathway by using DMAPP as the connection node: the upstream module including the native methylerythritol-phosphate (MEP) pathway and the downstream module consisting of isoprene synthase (ISPS) (Fig. 3A). Since isoprene and lycopene biosynthetic pathways shared the same upstream module, lycopene was used as the colorimetric reporter for high-throughput screening. The optimization of upstream module, inculding the co-evolution of three rate-limiting enzymes DXS/DXR/IDI by error-prone PCR, increased the yield of isoprene by 60% (Fig. 3B) [18]. In another case, phenylpyruvate decarboxylase (ARO10) and phenylacetaldehyde dehydrogenase (FeaB) are important in 4-hydroxyphenylacetic acid (4-HPAA) biosynthesis. Error-prone PCR was also applied to generate mutants of ARO10 and FeaB simultaneously, and the yield of 4-HPAA increased 1.13-fold [19].
Fig. 3.
Combinatorial pathway optimization of isoprene production in E. coli. A. The isoprene biosynthetic pathway was divided into the upstream module and the downstream module using DMAPP as the connection node. Since the biosynthetic pathways of lycopene and isoprene shared the same upstream module, lycopene was used as the colorimetric reporter for high-throughput screening. B. The co-evolution of DXS/DXR/IDI. The mutant library was constructed by error-prone PCR. Recombinant strains with optimized upstream module were obtained by plate screening. The isoprene yield of the positive mutants was confirmed by GC analysis. C. Inter-module engineering of the isoprene biosynthetic pathway. The metabolic flux between the upstream and downstream modules was regulated by promoter replacement (PT7, PTrc, PAra) and inducer adjustment.
However, the evolutionary efficiency of error-prone PCR is usually low [17]. Recently, new enzyme evolution tools based on CRISPR techniques have also been developed [[20], [21], [22]]. Hao et al. developed a base editor by fusing nCas with a cytidine deaminase in Bacillus subtilis. This base editor could introduce approximately 100% of cytidine thymidine mutations across a 5 nt editable window, which was much higher than that of other base editors. Based on this base editor, co-evolution of secE and secG was realized and the transportation ability of SecE(V36I)/SecG(A62T/V63I) mutant was increased by 3.6 times [23]. Although CRISPR/Cas system has been widely used, it still has limitations, including the limited availability of PAM sites, and cytotoxicity [24]. Thus, Ho et al. optimized the pyrrolysine biosynthetic pathway by phage assisted non-continuous directed evolution (PANCE) in Escherichia coli, which can accommodate toxicity of Pyl biosynthetic genes by alternating mutagenic and selective phage growth. As a result, PylBCD genes were optimized simultaneously and the yield of pyrrolysine was increased by 32 times than the rationally engineered ancestors [25].
Pathway reconstruction is also a commonly used strategy to optimize the biosynthesis process. For instance, one-step fermentation of l-theanine was optimized in wild-type E. coli, through the introduction of a novel γ-glutamylmethylamide synthetase (GMAS) from Paracoccus aminovorans [26]. The glutamate dehydrogenase and the pyruvate carboxylase of Corynebacterium glutamicum were introduced, interrupting the tricarboxylic acid cycle to facilitate theanine production. In the meanwhile, the introduction of energy-saving phosphoenolpyruvate carboxykinase from Mannheimia succiniciproducens further increased the ATP yield for theanine synthesis. As a result, the theanine production of the recombinant strain was 70.6 g/L. Moreover, the complete biosynthesis of carminic acid from glucose was reconstructed in engineered E. coli. In this pathway, Type II PKS from Photorhabdus luminescens was introduced. A recombinant strain with high intermediate production was obtained through the co-expression of the Type II PKS and cyclase genes ZhuI and ZhuJ. Aklavinone 12-hydroxylase (DnrF) from Streptomyces peucetius and C-glucosyltransferase (GtCGT) from Gentiana triflora were also introduced. Then, in silico homology modeling and docking simulations were employed to enhance the activities of these two enzymes. In consequence, the yield of carminic acid from glucose was achieved 0.63 ± 0.02 mg/L in E. coli [27].
2.2. Pathway-level evolution based on regulatory elements engineering
When scientists study the biosynthesis process from the perspective of the whole pathway, not the individual genes, the evolutionary targets could be expanded. As the expression of targeted genes is often controlled by regulatory elements [28,29], engineering of these regulatory elements may drive the evolution of the whole pathway.
Precise control of gene expression through promoter engineering is the key to optimizing biosynthetic pathways [30]. The synthetic nar promoters (dissolved oxygen-dependent promoters) were divided into three groups based on their strength: strong, intermediate and weak. They were used to control the expression of enzymes in the d-lactate and 2,3-butanediol (BDO) biosynthetic pathways in E. coli [31]. The d-lactate and 2,3-BDO production of the recombinant strains were 34% and 72% higher than those using the native promoters, respectively. Except engineering individual promoters, promoter substitution is also a commonly applied strategy [30]. In the isoprene biosynthesis example mentioned in the previous section, the pathway was optimized through modular engineering in addition to the co-evolution of three rate-limiting enzymes of the MEP pathway. By tuning the metabolic flux between the upstream and downstream modules by promoter replacement (PT7, PTrc, PAra) and inducer adjustment, the final production of isoprene was improved by 4.7-fold (Fig. 3C) [18].
Since ribosomal binding sites (RBSs) are critical for enzyme activity regulation at the translational level, RBS engineering is also a commonly used evolution approach. Li et al. constructed a rationally designed RBS library in one-pot reaction through oligo linker mediated assembly (OLMA) and optimized the biosynthetic pathway of poly (3-hydroxybutyric acid) (PHB) in E. coli. Applying this method, strains accumulating 0%–92% PHB contents in cell dry weight (CDW) were obtained. PHB with various weight-average molecular weights (MW) of 2.7-6.8 × 106 were also efficiently produced in relatively high contents [32]. Besides, the Wood-Werkman cycle, which generated the propionate and 1-propanol in E. coli, was optimized through operon rearrangement, RBS adjustment, expression system optimization and adaptive laboratory evolution. As a result, nearly 30% of total carbon was redirected and the titers of propionic acid and propanol reached 9 mM and 5 mM, respectively [33].
In addition, there are cases of optimizing biosynthesis pathway through promoter-engineering and RBS-engineering in non-model bacteria as well. For instance, ecumicin biosynthesis was enhanced in the rare actinomycete Nonomuraea sp. MJM5123 by knocking into kasO∗p upstream of the ecuE gene. Since there were two potential start codons (ATG/GTG) among the initial region of ecuE, four different sites were engineered: (1) the one before the first start codon, (2) the RBS of the first start codon, (3) the one before the second start codon, and (4) the RBS of the second start codon. By comparison, engineered strain integrating the kasO∗p together with its own RBS before the first start codon had a higher ecumicin production. Importantly, production of a more active component EcuH16 was considerably increased in the double RBSs engineered strain, reaching 310 mg/L [34].
2.3. Pathway-level evolution based on computer-aided engineering
Although pathway-level evolution provides a good scheme for biosynthesis optimization, the metabolic engineering of target strains often requires a good understanding of the cell metabolism. However, sometimes the enzymes involved in the target biosynthetic pathways are still unknown, which poses a key challenge to their production. Constructing new biosynthetic pathways based on computer-aided engineering can partly overcome this challenge [35]. With the continuous increase of biological data in recent years, data-driven approaches have gradually become popular. For the design of metabolic pathways, previous studies used computer-aided inverse synthesis strategy to explore the huge chemical space that was difficult to navigate through manual inspection [36,37].
The biosynthesis pathway with multiple-gene can be designed through computer-aided engineering. Ferreira et al. described the in silico driven design of butanol producing E. coli strains. Seven catalytic steps and nine heterologous genes from various sources were required. The recombinant strain was selected from an initial set of 105,954 different routes, which had the maximum butanol titer of 85 ± 1 mg/L [38]. Further, simulation tools can be used to optimize biosynthetic pathways as well. For example, genome scale metabolic model (GSM) simulation was used to design and develop metabolic pathways of B. subtilis. In a published GSM model of B. subtilis, iYO844, flux balance analysis (FBA) simulation was used to evaluate the effect of stepwise gene knockout to improve the 2,3-BDO yield in B. subtilis. As the result, Vikromvarasiri et al. found that lctE knockout led to a substantial increase in 2,3-BDO production and the improvement by mmgA knockout had never been investigated [39].
The establishment and enrichment of biosynthetic route databases are also a major application of computer-aided engineering. The SensiPath web server, which can screen multi-step enzymatic transformation of non-detectable compounds into detectable compounds, expands the potential applications of biosensors in synthetic biology [40]. Duigou et al. provided a complete set for RetroRules, which is a database of reaction rules for metabolic engineering, including more than 40,000 stereochemical perception reaction rules extracted from the public databases and expressed in the community standard SMARTS (SMIRKS) format [41].
The way to capture information from the databases can be optimized by computer-aided engineering as well. When computers design multi-step synthesis, they can rely on expert knowledge or information machines extracted from large reaction bases. However, both methods have insufficiencies to evaluate reaction choices: expert function is a heuristic based on chemical intuition, while machine learning relying on neural networks can only make meaningful predictions of popular reaction types [42,43]. Badowski et al. showed that the expert method and machine learning method could cooperate by training the neural network with the literature data matched to high-quality and expert coded reaction rules. After that, they can obtain higher synthesis accuracy. More importantly, they can also deal with rare and special reaction types [44].
3. Genome-level evolution strategies
Pathway-level evolution has succeeded in the generation of desired phenotypes. However, due to the complexity of cellular metabolic networks, beneficial modifications are non-intuitive and not limited to the pathway level. In this case, genome-level evolutionary strategies may offer a better outcome [45,46].
3.1. Genome-level evolution based on genome shuffling
In microbes, the complex interactions of metabolic networks are often poorly understood. Therefore, evolutionary strategies that don't consider strain genetic background and metabolic networks are urgently needed. Genome shuffling uses protoplast fusion to generate genomic diversity and screens the mutants under a special environmental stress, which is an effective way to evolve strains on the genome scale [47,48]. Importantly, genome shuffling breaks the restriction among species and has been widely applied in various fields, especially in improving desired product yield [13,49]. Genome shuffling was applied for the first time to improve tylosin yield based on recursive protoplast fusion. In 2002, Zhang et al. used the recursive protoplast to shuffle the genome of Streptomyces fradiae. As a result, the mutant strain exhibited 5 times higher tylosin yield [48]. The applications of surfactin, a promising natural product, was limited by its low yield. Recently, the recursive protoplast fusion was used to increase the production of surfactin in Bacillus velezensis LM3403. After three rounds of genome shuffling, the production of surfacin increased by about 679 mg/L [50]. Besides, genome shuffling has been widely used to improve strain tolerance, enhance substrates utilization, expand substrates scopes and other fields [[51], [52], [53], [54], [55], [56]].
However, genome shuffling through protoplast fusion tends to have low mutant diversity and low fusion rate [57]. A newly developed tool called SCRaMbLE could generate mutant diversity such as random deletion, duplication, and translocation at the genome level efficiently. It relies on the Cre recombinase that could recognize and cut the LoxP sites (Fig. 4) [11]. SCRaMbLE has been widely used in yeast [58,59]. Jia et al. used SCRaMbLE to evolve the haploid yeast genome, which increased the carotenoids production by 1.5 times. In order to quickly accumulate a large number of beneficial mutations that further improved carotenoids yield in yeast, the Multiple SCRaMbLE Iterative Cycle (MuSIC) was developed, which continuously increased the yield of carotenoids by 38.8 times after five iterative cycles of SCRaMbLE [60]. Chassis engineering and pathway optimization are usually carried out separately for expressing heterologous pathways, which is time-consuming and inefficient. SCRaMbLE-in was designed to rapidly evolve strains and improve the expression of heterologous pathways [61]. SCRaMbLE-in consists of two steps. First, regulatory elements were integrated into the targeted pathway by purified recombinant enzymes (Cre/VCre/Dre) in vitro. Then, the assembled pathway was integrated into the yeast chromosome, leading to genome shuffling. However, this approach might result in low integration rates due to re-excision of the integrated elements. In order to increase the integration rates, the mutated LoxP pairs (loxJT15 and loxJTZ17) were chosen. The strategy was applied to evolve violacein and β-carotene pathways with yields of 10 mg/L and 500 μg/L, respectively.
Fig. 4.
The experiment flow of SCRaMbLE. The LoxPsym locus needs to be inserted in yeast genomes in advance (Sc2.0). When the Cre enzyme was induced to express, it could recognize the LoxPsym locus and cut the yeast chromosome to achieve genetic diversity.
The SCRaMbLE system provides a powerful genomic evolution tool that could be applied to generate a variety of genomic diversities. In the meantime, SCRaMbLE also plays an important role in improving strain tolerance [62] and chromosome number variation [63]. However, there are still some limitations. For example, the expression of Cre recombinase is difficult to control. At present, strategies have been developed to strictly regulate the expression of Cre recombinase [64,65]. In addition, SCRaMbLE evolution strategy requires the construction of a genome sequence with LoxPsym locus in advance, thus the application of SCRaMbLE is mostly limited to synthetic yeasts.
3.2. Genome-level evolution based on CRISPR/Cas system
The CRISPR/Cas system, which is an adaptive immune system from bacteria or archaea, has been applied in genome editing [[66], [67], [68], [69], [70], [71]]. The evolutionary tools based on CRISPR/Cas system may overcome the blindness of genome shuffling and reduce the workload.
In recent years, researchers have made great progress in genome evolution using the CRISPR/Cas system. For example, CHAnGE could quickly generate tens of thousands of mutations at specific sites at the genome-level, with the assembly of gRNA and homology-directed-repair donors in the RNA expression cassette. CHAnGE improved the furfural tolerance of yeast to a high concentration (10 mM) [10]. Several other related genomic evolution tools have also been developed based on CRISPR/Cas system, including CRISPR-Enabled Trackable Genome Engineering (CREATE) [72], CRISPR/Cas9 (dCas9), the activation-induced cytidine deaminase (Target-AID) [[73], [74], [75]] and multiplexed accurate genome editing with short, trackable, integrated cellular barcodes (MAGESTIC) [76]. The common feature of the above methods is the requirement of RNA donors in vitro. In 2021, CRISPR- and RNA-assisted in vivo directed evolution (CRAIDE) strategy is the first example of an RNA-based directed evolution system in vivo (Fig. 5). Error-prone T7 RNA polymerase was used to continuously evolve the gRNA. CRAIDE was validated by evolving a new function of a nutrient deficiency marker gene and conferred with Saccharomyces cerevisiae. resistance for toxic amino acid analogues with the mutation rate >3,000 times [77].
Fig. 5.
The experiment flow of CRAIDE: CRISPR- and RNA-assisted in vivo directed evolution. This tool relies on the error-prone T7 RNA polymerase to evolve gRNA, which was introduced directly into genomic targets as RNA repair donors under the guidance of Cas9 or dCas9.
These applications are mainly focused on the class II CRISPR/Cas system that requires only a single Cas protein. However, the class II CRISPR/Cas system accounts for about only 10% of the bacteria and archaea CRISPR/Cas systems [78,79]. Thus, the class I CRISPR/Cas systems that widely present in microbes have been considered as well. Xu et al. developed a transferable system, which could stably integrate and express highly active I–F Cascade in a heterologous host [80]. The system relied on the mini-CTX-lacZ vector containing the integrated gene Int, the attachment site attP, strong promoter Ptat and λ-Red recombination system introduced to compensate for the defects of bacterial homologous recombination. By targeting the integrated regions Ps1, Ps2, Ps3 into genome, it was confirmed that the integration efficiency of I–F Cascade was about 6 times than Cas9. This work proves the application and potential of type I CRISPR/Cas system in genome evolution.
In addition, the CRISPR/Cas system could also be used to rapidly screen beneficial genes on the genome-level [81]. However, these methods have limitations, including specific microbes, or narrow mutant window. EvolvR combined an error-prone, Nick-translating DNA polymerase with nCas9 could enlarge the mutation window and increase the mutation rate by polymerase mutants [82]. The yEvolvR system was established in yeast. The polymerase mutant PolI-5M increased the mutation rate of the target sites by 12434 times [83]. Besides, new Cas proteins have gradually been discovered and may be used to develop genomic evolutionary strategies in the future [[84], [85], [86]].
3.3. Genome-level evolution based on other synthetic biology tools
With the development of genome sequencing and genome editing technology, more and more genome evolution tools have been developed and gradually improved.
Multiplex automated genome engineering (MAGE) is a multi-site evolution tool based on single-strand DNA (ssDNA) oligonucleotides, which is able to modify chromosomes at different positions [87]. The traditional MAGE tool requires optimization of model strains and genome modification in advance, resulting in unnecessary modifications and off-target accumulation. A plasmid-based evolutionary tool called pORTMAGE could effectively modify multiple sites without any off-target mutations and the optimization of strains [88]. In particular, pORTMAGE could be applied in Salmonella Enterica, expanding the host range of MAGE. To further improve the MAGE technology, CRISPR optimized MAGE (CRMAGE) was developed by combining MAGE with λ Red and CRISPR/Cas9. Single-site mutation and double-site mutation increased by 93% and 90% than using MAGE alone, respectively [89]. Since MAGE is limited to E. coli and some related enterobacteria [90]. A tool that could work in eukaryotes is urgently needed. The yeast oligo-mediated genome engineering (YOGE) was developed in S. cerevisiae [91]. Allele replacement efficiency was only 2% by YOGE, which was not enough for multi-site genome evolution. eMAGE relied on the annealing of ssDNA oligodeoxynucleotides (ssODNs) instead of the recombinant protein Rad51 during DNA replication [92]. The efficiency of chromosome modification was greater than 40%, including single-base mismatch, insertion, and deletion. In the meanwhile, eMAGE could achieve 105 genetic diversities through iterative transformation. However, when the number of target sites increases, the use of MAGE technology to carry out genome mutations will become difficult [93]. Bacterial retroelements (retron) has been used for genome editing and evolution. Using mutagenic T7 RNA polymerase-retron system to evolve antibiotic resistance genes, the mutation rate increased by 190 times. The tool has the potential to dynamically and continuously evolve in the selection of phenotypes. However, its mutational efficiency requires further improvement and is impossible to track the mutation sites except for the one-by-one amplification of the editing sites. The tool that could traceably, scarlessly introduce efficient mutations in E. coli are required. The CRISPR/retron system could introduce mutations into the E. coli genome with high throughput. The mutations could be tracked by analyzing reverse transcription plasmids [94].
Transposons are the DNA sequences that are able to move on the genome [95]. Therefore, transposons are also widely applied for genome perturbation [96]. A highly active transposon, Tn5, was used in Streptomyces coelicolor 51,443 transposition insertions were identified at the genome level. In the mutation library, 724 mutants changed the yield of tripyrrole antibiotic undecylprodigiosin (RED), including 17 genes in the RED biosynthetic gene cluster. The yield of RED was increased by more than 30 times [97]. In addition, transposons can be used to create expression libraries that are fine-tuned to take advantage of different transcriptional environments at each random integration site, resulting in library diversity. Hermes Transposon (HTn) together with the G-SA-U (GFP-SA-URA3) fragment, was introduced into a non-replicable circular DNA molecule. The strains that experienced completed transposition process could be screened by URA3. The biosynthetic pathway of (S)-norcoclaurine was divided into two modules to construct circular molecule, respectively, which were transferred into the strains Tyr-1 and Tyr-2. 73% and 46% positive colonies were obtained respectively. Finally, a strain capable of producing 97 μg/L (S)-norcoclaurine was obtained after fermentation for 120 h [98].
Transposons have also been applied to insert heterologous pathways into the genome in non-model microbes, achieving horizontal gene transfer. For example, chassis-independent recombinase-assisted genome engineering (CRAGE), which integrates biosynthetic gene clusters (BGCs) into the chromosomes randomly, consists of transposon Tn5 and Cre-lox site-specific recombination system [99]. Non-ribosomal peptide synthetase (NRPS) and NRPS-polyketide synthase (PKS) hybrid BGCs from Photorhabdus luminescens were integrated into 25 different γ-Proteobacteria species. 22 products were identified that their yields were higher than those in the native strains. However, some strains were resistant to the CRAGE technology, probably due to transposon inadequacy. A system with gene transfer and genome integration abilities was developed in Clostridium Ljungdahlii. The system was based on the conjugation and transfer of a donor plasmid and Himar transposons [100]. By regulating transposase in Clostridium Ljungdahlii, the production of acetone and isopropanol reached 0.6 mM and 2.4 mM respectively.
Those tools have opened up broad prospects for rapid genome editing and automation of genome-scale engineering. We summarized the research related to genome evolution tools in recent 5 years (Table 1). However, improvements are still needed. Most of them are limited to the evolution of specific strains. In the future, it is still necessary to rationally design and develop new methods to efficiently generate genetic diversity on a genome scale for different strains.
Table 1.
Genomic evolution tools developed within the last 5 years.
| Strategies | Strains | Advantages | Disadvantages | Effects | References |
|---|---|---|---|---|---|
| RAGE and CRISPR-Cas | Saccharomyces cerevisiae | accurate localization and optimization of phenotypes | high-quality complementary DNA library required | to improve acid tolerance | [101] |
| MAGIC | better control of gene expression levels; uncharacterized genetic factors identified |
low gene activation efficiency | to regulate gene expression levels at the genome-scale | [102] | |
| ICE | suitable for yeast containing long terminal repeat retrotransposon | the efficiency affected by homologous recombination | to generate mutant libraries up to 1.6 × 107|−1 per round by retrotransposon Ty1 in vivo | [103] | |
| Multiplexed CRISPRi | Kluyveromyces marxianus | multiple genes acted simultaneously | multiplexed sgRNAs required; low efficiency caused by the expression levels of Cas9 and sgRNA |
to increase the yield of ethyl acetate by 3.8-fold | [104] |
| CRISPR-Cas9-mediated HR | Yarrowia lipolytica | suitable for integrating targeted, markerless genes | low efficiency of multi-gene insertion | to produce 0.39 mg lycopene/g DCW | [105] |
| Transposase-mediated integration | Acidithiobacillus ferrooxidans | effective integration of exogenous genes into genome | site preference caused by Tn5 | to create strains that produce isobutyric acid | [106] |
| DIvERGE | Escherichia coli | effective mutation of multiple long genome fragments without off-target modification; suitable for different species |
target genes required to determine in advance | to increase the mutation rate of multiple sites by one million times | [107] |
| REXER | better insertion or replacement of synthetic DNA | time-consuming and high cost | to insert or replace genome with long synthetic DNA | [108] |
4. Summary and outlook
Although various evolutionary tools have been developed for pathway or genome evolution in recent years, high-throughput screening still seriously restricts the development of evolution strategies. Biosensors that could generate special signals provide a promising solution [[109], [110], [111], [112]]. Gwon et al. used ALE strategy assisted with tryptophan-responsive biosensor to improve the yield of violacein in E. coli [113]. The yield of violacein was 2.7 times higher than that of the native strain. In order to screen effectively, high sensitivity sensors are necessary. The biosensor of pentalactam and caprolactam from Pseudomonas putida KT2440 was 1,000 times more sensitive compared to previously reported biosensors [114]. In another study, a high-throughput screening method based on droplet microfluidic platform with a screening rate up to 10,000 strains/hour was proposed [115]. This technology has been used to screen constitutive promoter mutation libraries. The strength of ermE∗p and gapdh(EL)p was increased by 347.9% and 94.6%, respectively. Furthermore, the droplet microfluidic technology quickly screened out strains with a 69.2–111.4% increase in cellulase production. In recent years, with the development of automation and computer technology, the construction of automation platforms has provided a promising method for high-throughput screening [116].
In summary, pathway and genome level evolution have effectively promoted the improvement of strain phenotypes. Different evolution strategies exhibit their own advantages and disadvantages. To overcome difficulties, a combination of different evolutionary strategies may be able to compensate for their limitations. In the meantime, evolution strategies used in humans or mammals may also be applied in microbes, like transposon piggyBac [117]. In addition, developing evolutionary tools for non-model microbes may overcome the problem that could not be solved in the mode microbes. However, it may be difficult to develop evolutionary tools in non-model strains. On the one hand, the genomic information of non-model strains and metabolic pathway are not thoroughly studied. The techniques such as plasmid transformation and base editing are difficult to apply. On the other hand, most of non-model strains are resistant to be engineered [118]. Destroying or inhibiting the strain's own repair mechanism may break this limitation. In the meantime, a transient suppression system may be needed. Besides, CRISPR/Cas9 system that has been widely applied in genome editing of non-model strains, like Halomonas [119], Lactobacillus plantarum [120], has the potential to be used in genome evolution. With the development of synthetic biology, evolutionary strategies and screening methods that can be applied in multiple strains simultaneously will also be updated.
Credit Author Statement
Huang Chaoqun: Writing-Original draft preparation. Wang Chang: Writing-Original draft preparation. Luo Yunzi: Writing-Reviewing and Editing.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0903300), the National Natural Science Foundation of China (32071426), the Natural Science Foundation of Tianjin Province (19JCYBJC24200), and the Key Area Research and Development Program of Guangdong Province (2020B0303070002).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Kumar A., Jaitak V. Natural products as multidrug resistance modulators in cancer. Eur J Med Chem. 2019;176:268–291. doi: 10.1016/j.ejmech.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Tan Z.L., Zheng X., Wu Y., Jian X., Xing X., Zhang C. In vivo continuous evolution of metabolic pathways for chemical production. Microb Cell Factories. 2019;18(1):82. doi: 10.1186/s12934-019-1132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markel U., Essani K.D., Besirlioglu V., Schiffels J., Streit W.R., Schwaneberg U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem Soc Rev. 2020;49(1):233–262. doi: 10.1039/c8cs00981c. [DOI] [PubMed] [Google Scholar]
- 4.Currin A., Parker S., Robinson C.J., Takano E., Scrutton N.S., Breitling R. The evolving art of creating genetic diversity: from directed evolution to synthetic biology. Biotechnol Adv. 2021;50:107762. doi: 10.1016/j.biotechadv.2021.107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker E.P., Hittinger C.T. Evolution of a novel chimeric maltotriose transporter in Saccharomyces eubayanus from parent proteins unable to perform this function. PLoS Genet. 2019;15(4) doi: 10.1371/journal.pgen.1007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo H., Yang L., Kim S.H., Wulff T., Feist A.M., Herrgard M., et al. Directed metabolic pathway evolution enables functional pterin-dependent aromatic-amino-acid hydroxylation in Escherichia coli. ACS Synth Biol. 2020;9(3):494–499. doi: 10.1021/acssynbio.9b00488. [DOI] [PubMed] [Google Scholar]
- 7.Sandberg T.E., Salazar M.J., Weng L.L., Palsson B.O., Feist A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng. 2019;56:1–16. doi: 10.1016/j.ymben.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biot-Pelletier D., Martin V.J. Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol. 2014;98(9):3877–3887. doi: 10.1007/s00253-014-5616-8. [DOI] [PubMed] [Google Scholar]
- 9.Pál C., Papp B., Pósfai G. The dawn of evolutionary genome engineering. Nat Rev Genet. 2014;15(7):504–512. doi: 10.1038/nrg3746. [DOI] [PubMed] [Google Scholar]
- 10.Bao Z., HamediRad M., Xue P., Xiao H., Tasan I., Chao R., et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat Biotechnol. 2018;36(6):505–508. doi: 10.1038/nbt.4132. [DOI] [PubMed] [Google Scholar]
- 11.Szymanski E., Calvert J. Designing with living systems in the synthetic yeast project. Nat Commun. 2018;9(1):2950. doi: 10.1038/s41467-018-05332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chae T.U., Choi S.Y., Kim J.W., Ko Y.S., Lee S.Y. Recent advances in systems metabolic engineering tools and strategies. Curr Opin Biotechnol. 2017;47:67–82. doi: 10.1016/j.copbio.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Chen L., Xin Q.H., Ma L.M., Li R.F., Bian K. Applications and research advance of genome shuffling for industrial microbial strains improvement. World J Microbiol Biotechnol. 2020;36(10):158. doi: 10.1007/s11274-020-02936-w. [DOI] [PubMed] [Google Scholar]
- 14.Ye L., Yang C., Yu H. From molecular engineering to process engineering: development of high-throughput screening methods in enzyme directed evolution. Appl Microbiol Biotechnol. 2018;102(2):559–567. doi: 10.1007/s00253-017-8568-y. [DOI] [PubMed] [Google Scholar]
- 15.Cobb R.E., Si T., Zhao H. Directed evolution: an evolving and enabling synthetic biology tool. Curr Opin Chem Biol. 2012;16(3–4):285–291. doi: 10.1016/j.cbpa.2012.05.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivy T.L., Fall R., Rosenstiel T.N. Evidence of isoprenoid precursor toxicity in Bacillus subtilis. Biosci Biotechnol Biochem. 2011;75(12):2376–2383. doi: 10.1271/bbb.110572. [DOI] [PubMed] [Google Scholar]
- 17.Tiwari V. In vitro Engineering of novel bioactivity in the natural enzymes. Front Chem. 2016;4:39. doi: 10.3389/fchem.2016.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv X., Gu J., Wang F., Xie W., Liu M., Ye L., et al. Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering. Biotechnol Bioeng. 2016;113(12):2661–2669. doi: 10.1002/bit.26034. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y.P., Fong L.S., Yan Z.B., Liu J.Z. Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli. Biotechnol Biofuels. 2019;12:94. doi: 10.1186/s13068-019-1438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch-Guiteras N., Uroda T., Guillen-Ramirez H.A., Riedo R., Gazdhar A., Esposito R., et al. Enhancing CRISPR deletion via pharmacological delay of DNA-PKcs. Genome Res. 2021;31(3):461–471. doi: 10.1101/gr.265736.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S., Shin J., Cho B.K. Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci. 2018;19(4) doi: 10.3390/ijms19041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Wang L., Luo Y. Blossom of CRISPR technologies and applications in disease treatment. Synth Syst Biotechnol. 2018;3(4):217–228. doi: 10.1016/j.synbio.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao W., Cui W., Cheng Z., Han L., Suo F., Liu Z., et al. Development of a base editor for protein evolution via in situ mutation in vivo. Nucleic Acids Res. 2021;49(16):9594–9605. doi: 10.1093/nar/gkab673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Wang H., Liu H., Zhao Q., Liu B., Wang L., et al. Improved CRISPR-Cas12a-assisted one-pot DNA editing method enables seamless DNA editing. Biotechnol Bioeng. 2019;116(6):1463–1474. doi: 10.1002/bit.26938. [DOI] [PubMed] [Google Scholar]
- 25.Ho J.M.L., Miller C.A., Smith K.A., Mattia J.R., Bennett M.R. Improved pyrrolysine biosynthesis through phage assisted non-continuous directed evolution of the complete pathway. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-24183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan X., Zhang T., Ji Y., Li J., Long K., Yuan Y., et al. Pathway engineering of Escherichia coli for one-step fermentative production of L-theanine from sugars and ethylamine. Metab Eng Commun. 2020;11 doi: 10.1016/j.mec.2020.e00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang D., Jang W.D., Lee S.Y. Production of carminic acid by metabolically engineered Escherichia coli. J Am Chem Soc. 2021;143(14):5364–5377. doi: 10.1021/jacs.0c12406. [DOI] [PubMed] [Google Scholar]
- 28.Ren J., Lee J., Na D. Recent advances in genetic engineering tools based on synthetic biology. J Microbiol. 2020;58(1):1–10. doi: 10.1007/s12275-020-9334-x. [DOI] [PubMed] [Google Scholar]
- 29.Sterk M., Romilly C., Wagner E.G.H. Unstructured 5'-tails act through ribosome standby to override inhibitory structure at ribosome binding sites. Nucleic Acids Res. 2018;46(8):4188–4199. doi: 10.1093/nar/gky073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu N., Wei L., Liu J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J Microbiol Biotechnol. 2019;35(2) doi: 10.1007/s11274-019-2606-0. [DOI] [PubMed] [Google Scholar]
- 31.Hwang H.J., Lee S.Y., Lee P.C. Engineering and application of synthetic nar promoter for fine-tuning the expression of metabolic pathway genes in Escherichia coli. Biotechnol Biofuels. 2018;11:103. doi: 10.1186/s13068-018-1104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T., Ye J., Shen R., Zong Y., Zhao X., Lou C., et al. Semirational approach for ultrahigh poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening. ACS Synth Biol. 2016;5(11):1308–1317. doi: 10.1021/acssynbio.6b00083. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Garcia R.A., McCubbin T., Turner M.S., Nielsen L.K., Marcellin E. Engineering Escherichia coli for propionic acid production through the Wood-Werkman cycle. Biotechnol Bioeng. 2020;117(1):167–183. doi: 10.1002/bit.27182. [DOI] [PubMed] [Google Scholar]
- 34.Su C., Tuan N.Q., Lee M.J., Zhang X.Y., Cheng J.H., Jin Y.Y., et al. Enhanced production of active ecumicin component with higher antituberculosis activity by the rare actinomycete Nonomuraea sp. MJM5123 using a novel promoter-engineering strategy. ACS Synth Biol. 2020;9(11):3019–3029. doi: 10.1021/acssynbio.0c00248. [DOI] [PubMed] [Google Scholar]
- 35.Jang W.D., Kim G.B., Kim Y., Lee S.Y. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr Opin Biotechnol. 2021;73:101–107. doi: 10.1016/j.copbio.2021.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Hatzimanikatis V., Li C., Ionita J.A., Henry C.S., Jankowski M.D., Broadbelt L.J. Exploring the diversity of complex metabolic networks. Bioinformatics. 2005;21(8):1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- 37.Kumar A., Wang L., Ng C.Y., Maranas C.D. Pathway design using de novo steps through uncharted biochemical spaces. Nat Commun. 2018;9(1):184. doi: 10.1038/s41467-017-02362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira S., Pereira R., Liu F., Vilaca P., Rocha I. Discovery and implementation of a novel pathway for n-butanol production via 2-oxoglutarate. Biotechnol Biofuels. 2019;12:230. doi: 10.1186/s13068-019-1565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vikromvarasiri N., Shirai T., Kondo A. Metabolic engineering design to enhance (R,R)-2,3-butanediol production from glycerol in Bacillus subtilis based on flux balance analysis. Microb Cell Factories. 2021;20(1) doi: 10.1186/s12934-021-01688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delepine B., Libis V., Carbonell P., Faulon J.L. SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucleic Acids Res. 2016;44(W1):W226–W231. doi: 10.1093/nar/gkw305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duigou T., du Lac M., Carbonell P., Faulon J.L. RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Res. 2019;47(D1):D1229–D1235. doi: 10.1093/nar/gky940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segler M.H.S., Preuss M., Waller M.P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555(7698):604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 43.Szymkuc S., Gajewska E.P., Klucznik T., Molga K., Dittwald P., Startek M., et al. Computer-assisted synthetic planning: the end of the beginning. Angew Chem Int Ed Engl. 2016;55(20):5904–5937. doi: 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- 44.Badowski T., Gajewska E.P., Molga K., Grzybowski B.A. Synergy between expert and machine-learning approaches allows for improved retrosynthetic planning. Angew Chem Int Ed Engl. 2020;59(2):725–730. doi: 10.1002/anie.201912083. [DOI] [PubMed] [Google Scholar]
- 45.Jiang S., Si T., Dai J. Whole-genome regulation for yeast metabolic engineering. Small Methods. 2020;4(2) doi: 10.1002/smtd.201900640. [DOI] [Google Scholar]
- 46.Lian J., Mishra S., Zhao H. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab Eng. 2018;50:85–108. doi: 10.1016/j.ymben.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Johansen E. Use of natural selection and evolution to develop new starter cultures for fermented foods. Annu Rev Food Sci Technol. 2018;9:411–428. doi: 10.1146/annurev-food-030117-012450. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y.X., Perry K., Vinci V.A., Powell K., Stemmer W.P., del Cardayré S.B. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415(6872):644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 49.Gong J., Zheng H., Wu Z., Chen T., Zhao X. Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv. 2009;27(6):996–1005. doi: 10.1016/j.biotechadv.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Chen L., Chong X.Y., Zhang Y.Y., Lv Y.Y., Hu Y.S. Genome shuffling of Bacillus velezensis for enhanced surfactin production and variation analysis. Curr Microbiol. 2020;77(1):71–78. doi: 10.1007/s00284-019-01807-4. [DOI] [PubMed] [Google Scholar]
- 51.Gu C., Wang G., Mai S., Wu P., Wu J., Wang G., et al. ARTP mutation and genome shuffling of ABE fermentation symbiotic system for improvement of butanol production. Appl Microbiol Biotechnol. 2017;101(5):2189–2199. doi: 10.1007/s00253-017-8093-z. [DOI] [PubMed] [Google Scholar]
- 52.Han G.G., Song A.A., Kim E.B., Yoon S.H., Bok J.D., Cho C.S., et al. Improved antimicrobial activity of Pediococcus acidilactici against Salmonella Gallinarum by UV mutagenesis and genome shuffling. Appl Microbiol Biotechnol. 2017;101(13):5353–5363. doi: 10.1007/s00253-017-8293-6. [DOI] [PubMed] [Google Scholar]
- 53.Hu S., You Y., Xia F., Liu J., Dai W., Liu J., et al. Genome shuffling improved acid-tolerance and succinic acid production of Actinobacillus succinogenes. Food Sci Biotechnol. 2019;28(3):817–822. doi: 10.1007/s10068-018-0505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jetti K.D., Gns R.R., Garlapati D., Nammi S.K. Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis. Int Microbiol. 2019;22(2):247–254. doi: 10.1007/s10123-018-00044-2. [DOI] [PubMed] [Google Scholar]
- 55.Liu H., Jiang C., Lin J., Zhuang Z., Kong W., Liu L., et al. Genome shuffling based on different types of ribosome engineering mutants for enhanced production of 10-membered enediyne tiancimycin-A. Appl Microbiol Biotechnol. 2020;104(10):4359–4369. doi: 10.1007/s00253-020-10583-2. [DOI] [PubMed] [Google Scholar]
- 56.Monerawela C., Bond U. Recombination sites on hybrid chromosomes in Saccharomyces pastorianus share common sequence motifs and define a complex evolutionary relationship between group I and II lager yeasts. FEMS Yeast Res. 2017;17(5) doi: 10.1093/femsyr/fox047. [DOI] [PubMed] [Google Scholar]
- 57.Wang M., Zhang W., Xu W., Shen Y., Du L. Optimization of genome shuffling for high-yield production of the antitumor deacetylmycoepoxydiene in an endophytic fungus of mangrove plants. Appl Microbiol Biotechnol. 2016;100(17):7491–7498. doi: 10.1007/s00253-016-7457-0. [DOI] [PubMed] [Google Scholar]
- 58.Chen S., Xie Z.X., Yuan Y.J. Discovering and genotyping genomic structural variations by yeast genome synthesis and inducible evolution. FEMS Yeast Res. 2020;20(2) doi: 10.1093/femsyr/foaa012. [DOI] [PubMed] [Google Scholar]
- 59.Xie Z.X., Liu D., Li B.Z., Zhao M., Zeng B.X., Wu Y., et al. Design and chemical synthesis of eukaryotic chromosomes. Chem Soc Rev. 2017;46(23):7191–7207. doi: 10.1039/c7cs00208d. [DOI] [PubMed] [Google Scholar]
- 60.Jia B., Wu Y., Li B.Z., Mitchell L.A., Liu H., Pan S., et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast. Nat Commun. 2018;9(1):1933. doi: 10.1038/s41467-018-03084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W., Luo Z., Wang Y., Pham N.T., Tuck L., Pérez-Pi I., et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat Commun. 2018;9(1):1936. doi: 10.1038/s41467-018-04254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma L., Li Y., Chen X., Ding M., Wu Y., Yuan Y.J. SCRaMbLE generates evolved yeasts with increased alkali tolerance. Microb Cell Factories. 2019;18(1):52. doi: 10.1186/s12934-019-1102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J., Xie Z.X., Ma Y., Chen X.R., Huang Y.Q., He B., et al. Ring synthetic chromosome V SCRaMbLE. Nat Commun. 2018;9(1):3783. doi: 10.1038/s41467-018-06216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hochrein L., Mitchell L.A., Schulz K., Messerschmidt K., Mueller-Roeber B. L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast. Nat Commun. 2018;9(1):1931. doi: 10.1038/s41467-017-02208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steensels J., Gorkovskiy A., Verstrepen K.J. SCRaMbLEing to understand and exploit structural variation in genomes. Nat Commun. 2018;9(1):1937. doi: 10.1038/s41467-018-04308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 67.Cao M., Gao M., Ploessl D., Song C., Shao Z. CRISPR-Mediated genome editing and gene repression in Scheffersomyces stipitis. Biotechnol J. 2018;13(9) doi: 10.1002/biot.201700598. [DOI] [PubMed] [Google Scholar]
- 68.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526(7571):55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 70.Tong Y., Charusanti P., Zhang L., Weber T., Lee S.Y. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol. 2015;4(9):1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J., Zhang D., Zhu J., Liu H., Liang S., Luo Y. Efficient multiplex genome editing in Streptomyces via engineered CRISPR-Cas12a systems. Front Bioeng Biotechnol. 2020;8:726. doi: 10.3389/fbioe.2020.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garst A.D., Bassalo M.C., Pines G., Lynch S.A., Halweg-Edwards A.L., Liu R., et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol. 2017;35(1):48–55. doi: 10.1038/nbt.3718. [DOI] [PubMed] [Google Scholar]
- 73.Després P.C., Dubé A.K., Nielly-Thibault L., Yachie N., Landry C.R. Double selection enhances the efficiency of Target-AID and Cas9-based genome editing in yeast. G3 (Bethesda) 2018;8(10):3163–3171. doi: 10.1534/g3.118.200461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Després P.C., Dubé A.K., Seki M., Yachie N., Landry C.R. Perturbing proteomes at single residue resolution using base editing. Nat Commun. 2020;11(1):1871. doi: 10.1038/s41467-020-15796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305) doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 76.Roy K.R., Smith J.D., Vonesch S.C., Lin G., Tu C.S., Lederer A.R., et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol. 2018;36(6):512–520. doi: 10.1038/nbt.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jensen E.D., Laloux M., Lehka B.J., Pedersen L.E., Jakočiūnas T., Jensen M.K., et al. A synthetic RNA-mediated evolution system in yeast. Nucleic Acids Res. 2021;49(15):e88. doi: 10.1093/nar/gkab472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crawley A.B., Henriksen J.R., Barrangou R. CRISPRdisco: an automated pipeline for the discovery and analysis of CRISPR-Cas systems. Crispr j. 2018;1(2):171–181. doi: 10.1089/crispr.2017.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vento J.M., Crook N., Beisel C.L. Barriers to genome editing with CRISPR in bacteria. J Ind Microbiol Biotechnol. 2019;46(9–10):1327–1341. doi: 10.1007/s10295-019-02195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Z., Li Y., Cao H., Si M., Zhang G., Woo P.C.Y., et al. A transferrable and integrative type I-F Cascade for heterologous genome editing and transcription modulation. Nucleic Acids Res. 2021;49(16):e94. doi: 10.1093/nar/gkab521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang L., Fan J., Luo S., Chen Y., Wang C., Cao Y., et al. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids. Nat Commun. 2021;12(1):4976. doi: 10.1038/s41467-021-25243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Halperin S.O., Tou C.J., Wong E.B., Modavi C., Schaffer D.V., Dueber J.E. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 2018;560(7717):248–252. doi: 10.1038/s41586-018-0384-8. [DOI] [PubMed] [Google Scholar]
- 83.Tou C.J., Schaffer D.V., Dueber J.E. Targeted diversification in the S. cerevisiae genome with CRISPR-Guided DNA polymerase I. ACS Synth Biol. 2020;9(7):1911–1916. doi: 10.1021/acssynbio.0c00149. [DOI] [PubMed] [Google Scholar]
- 84.Hu Z., Wang S., Zhang C., Gao N., Li M., Wang D., et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020;18(3) doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pausch P., Al-Shayeb B., Bisom-Rapp E., Tsuchida C.A., Li Z., Cress B.F., et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–337. doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pausch P., Soczek K.M., Herbst D.A., Tsuchida C.A., Al-Shayeb B., Banfield J.F., et al. DNA interference states of the hypercompact CRISPR-CasΦ effector. Nat Struct Mol Biol. 2021;28(8):652–661. doi: 10.1038/s41594-021-00632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R., et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nyerges A., Csorgo B., Nagy I., Balint B., Bihari P., Lazar V., et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc Natl Acad Sci U S A. 2016;113(9):2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ronda C., Pedersen L.E., Sommer M.O.A., Nielsen A.T. CRMAGE: CRISPR optimized MAGE recombineering. Sci Rep. 2016;6:19452. doi: 10.1038/srep19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wannier T.M., Nyerges A., Kuchwara H.M., Czikkely M., Balogh D., Filsinger G.T., et al. Improved bacterial recombineering by parallelized protein discovery. Proc Natl Acad Sci U S A. 2020;117(24):13689–13698. doi: 10.1073/pnas.2001588117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DiCarlo J.E., Conley A.J., Penttilä M., Jäntti J., Wang H.H., Church G.M. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013;2(12):741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barbieri E.M., Muir P., Akhuetie-Oni B.O., Yellman C.M., Isaacs F.J. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell. 2017;171(6):1453–1467. doi: 10.1016/j.cell.2017.10.034. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simon A.J., Morrow B.R., Ellington A.D. Retroelement-based genome editing and evolution. ACS Synth Biol. 2018;7(11):2600–2611. doi: 10.1021/acssynbio.8b00273. [DOI] [PubMed] [Google Scholar]
- 94.Lim H., Jun S., Park M., Lim J., Jeong J., Lee J.H., et al. Multiplex generation, tracking, and functional screening of substitution mutants using a CRISPR/Retron system. ACS Synth Biol. 2020;9(5):1003–1009. doi: 10.1021/acssynbio.0c00002. [DOI] [PubMed] [Google Scholar]
- 95.O'Neill K., Brocks D., Hammell M.G. Mobile genomics: tools and techniques for tackling transposons. Philos Trans R Soc Lond B Biol Sci. 2020;375:20190345. doi: 10.1098/rstb.2019.0345. 1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu J., Zhu Q., Gong R., Xu Q., Cai M., Jiang T., et al. PiggyBac transposon-mediated mutagenesis and application in yeast Komagataella phaffii. Biotechnol Lett. 2018;40(9–10):1365–1376. doi: 10.1007/s10529-018-2592-6. [DOI] [PubMed] [Google Scholar]
- 97.Xu Z., Wang Y., Chater K.F., Ou H.Y., Xu H.H., Deng Z., et al. Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl Environ Microbiol. 2017;83(6) doi: 10.1128/aem.02889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Y., Yao Z., Ploessl D., Ghosh S., Monti M., Schindler D., et al. Leveraging the hermes transposon to accelerate the development of nonconventional yeast-based microbial cell factories. ACS Synth Biol. 2020;9(7):1736–1752. doi: 10.1021/acssynbio.0c00123. [DOI] [PubMed] [Google Scholar]
- 99.Wang G., Zhao Z., Ke J., Engel Y., Shi Y.M., Robinson D., et al. CRAGE enables rapid activation of biosynthetic gene clusters in undomesticated bacteria. Nat Microbiol. 2019;4(12):2498–2510. doi: 10.1038/s41564-019-0573-8. [DOI] [PubMed] [Google Scholar]
- 100.Philipps G., de Vries S., Jennewein S. Development of a metabolic pathway transfer and genomic integration system for the syngas-fermenting bacterium Clostridium ljungdahlii. Biotechnol Biofuels. 2019;12:112. doi: 10.1186/s13068-019-1448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Si T., Chao R., Min Y., Wu Y., Ren W., Zhao H. Automated multiplex genome-scale engineering in yeast. Nat Commun. 2017;8:15187. doi: 10.1038/ncomms15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lian J., Schultz C., Cao M., HamediRad M., Zhao H. Multi-functional genome-wide CRISPR system for high throughput genotype-phenotype mapping. Nat Commun. 2019;10(1):5794. doi: 10.1038/s41467-019-13621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crook N., Abatemarco J., Sun J., Wagner J.M., Schmitz A., Alper H.S. In vivo continuous evolution of genes and pathways in yeast. Nat Commun. 2016;7:13051. doi: 10.1038/ncomms13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lobs A.K., Schwartz C., Thorwall S., Wheeldon I. Highly multiplexed CRISPRi repression of respiratory functions enhances mitochondrial localized ethyl acetate biosynthesis in Kluyveromyces marxianus. ACS Synth Biol. 2018;7(11):2647–2655. doi: 10.1021/acssynbio.8b00331. [DOI] [PubMed] [Google Scholar]
- 105.Schwartz C., Shabbir-Hussain M., Frogue K., Blenner M., Wheeldon I. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synth Biol. 2017;6(3):402–409. doi: 10.1021/acssynbio.6b00285. [DOI] [PubMed] [Google Scholar]
- 106.Inaba Y., Banerjee I., Kernan T., Banta S. Transposase-mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2018;84(21) doi: 10.1128/AEM.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nyerges Á., Csörgő B., Draskovits G., Kintses B., Szili P., Ferenc G., et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc Natl Acad Sci U S A. 2018;115(25) doi: 10.1073/pnas.1801646115. E5726-e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang K., Fredens J., Brunner S.F., Kim S.H., Chia T., Chin J.W. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539(7627):59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Armetta J., Berthome R., Cros A., Pophillat C., Colombo B.M., Pandi A., et al. Biosensor-based enzyme engineering approach applied to psicose biosynthesis. Synth Biol (Oxf) 2019;4(1):ysz028. doi: 10.1093/synbio/ysz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leavitt J.M., Wagner J.M., Tu C.C., Tong A., Liu Y., Alper H.S. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae. Biotechnol J. 2017;12(10) doi: 10.1002/biot.201600687. [DOI] [PubMed] [Google Scholar]
- 111.Liu Y., Zhuang Y., Ding D., Xu Y., Sun J., Zhang D. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synth Biol. 2017;6(5):837–848. doi: 10.1021/acssynbio.6b00328. [DOI] [PubMed] [Google Scholar]
- 112.Mahr R., Gatgens C., Gatgens J., Polen T., Kalinowski J., Frunzke J. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum. Metab Eng. 2015;32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 113.Gwon D.A., Seok J.Y., Jung G.Y., Lee J.W. Biosensor-assisted adaptive laboratory evolution for violacein production. Int J Mol Sci. 2021;22(12) doi: 10.3390/ijms22126594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thompson M.G., Pearson A.N., Barajas J.F., Cruz-Morales P., Sedaghatian N., Costello Z., et al. Identification, characterization, and application of a highly sensitive lactam biosensor from Pseudomonas putida. ACS Synth Biol. 2020;9(1):53–62. doi: 10.1021/acssynbio.9b00292. [DOI] [PubMed] [Google Scholar]
- 115.Tu R., Zhang Y., Hua E.B., Bai L.K., Huang H.M., Yun K.Y., et al. Droplet-based microfluidic platform for high-throughput screening of Streptomyces. Commun Biol. 2021;4(1) doi: 10.1038/s42003-021-02186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dörr M., Fibinger M.P., Last D., Schmidt S., Santos-Aberturas J., Böttcher D., et al. Fully automatized high-throughput enzyme library screening using a robotic platform. Biotechnol Bioeng. 2016;113(7):1421–1432. doi: 10.1002/bit.25925. [DOI] [PubMed] [Google Scholar]
- 117.Beckermann T.M., Luo W., Wilson C.M., Veach R.A., Wilson M.H. Cognate restriction of transposition by piggyBac-like proteins. Nucleic Acids Res. 2021;49(14):8135–8144. doi: 10.1093/nar/gkab578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thorwall S., Schwartz C., Chartron J.W., Wheeldon I. Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nat Chem Biol. 2020;16(2):113–121. doi: 10.1038/s41589-019-0452-x. [DOI] [PubMed] [Google Scholar]
- 119.Qin Q., Ling C., Zhao Y.Q., Yang T., Yin J., Guo Y.Y., et al. CRISPR/Cas9 editing genome of extremophile. Halomonas spp. Metab Eng. 2018;47:219–229. doi: 10.1016/j.ymben.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 120.Zhou D., Jiang Z., Pang Q., Zhu Y., Wang Q., Qi Q. CRISPR/Cas9-assisted seamless genome editing in Lactobacillus plantarum and its application in N-acetylglucosamine production. Appl Environ Microbiol. 2019;85(21) doi: 10.1128/aem.01367-19. [DOI] [PMC free article] [PubMed] [Google Scholar]