Abstract
Per- and Polyfluorinated alkyl substances (PFAS) are a broad class of synthetic compounds that have fluorine substituted for hydrogen in several or all locations and are globally categorized as PFCs (perfluorochemicals; commonly called fluorinated chemicals). These compounds have unique chemical and physical properties that enable their use in non-stick surfaces, fire-fighting efforts, and as slick coatings. However, recent concerns over the health effects of such compounds, specifically perfluorooctanoic acid and perfluorooctane sulfonic acid (PFOA, PFOS; PFOA/S), have led to increased attention and research by the global community into degradation methods. In this study, soil samples from PFAS-contamination sites were cultured and screened for microbes with PFOA/S degradation potential, which led to the identification of Delftia acidovorans. It was found that D. acidovorans isolated from PFAS-contaminated soils was capable of growth in minimal media with PFOA as a sole carbon resource, and an observable fluoride concentration increase was observed when cells were exposed to PFOA. This suggests potential activity of a dehalogenase enzyme that may be of use in PFOA or PFAS microbial remediation efforts. Several associated haloacid dehalogenases have been identified in the D. acidovorans genome and have been engineered for expression in Escherichia coli for rapid production and purification. These enzymes have shown potential for enzymatic defluorination, a significant step in biological degradation and removal of PFOA/S from the environment. We hypothesize that bioremediation of PFAS using naturally occurring microbial degradation pathways may represent a novel approach to remove PFAS contamination.
Keywords: iGEM, PFAS, Bioremediation, Bioengineering, Delftia acidovorans
Abbreviations: iGEM-international Genetic Engineered Machine, DeHa- Dehalogenase
1. Introduction
In 1930, per- and polyfluoroalkyl substances (PFAS) were invented and introduced into waterproof coatings, nonstick cookware, and a myriad of other uses for water repellency, heat resistance, and high durability needs [1,2]. PFAS were consistently used in firefighting aqueous film-forming foams (AFFFs) to combat liquid fuel fires [3]. They are set to be removed from use in 2024 [4,5]. The compounds are broadly defined as any organofluorine alkyl molecule. Often containing many fluorine atoms that shield the carbon chain, they exhibit high chemical and physical resistance to degradation [6].
Certain PFAS are also toxic to most forms of life as they are carcinogens, liver toxicants, immune system toxicants, and have been shown to affect hormone levels in the thyroid [[7], [8], [9], [10], [11], [12], [13]] [[7], [8], [9], [10], [11], [12], [13]] [[7], [8], [9], [10], [11], [12], [13]]. Other long-chain alkyl PFAS (typically >6 carbons) have potential for biomagnification which further complicates the issue of PFAS toxicity [14,15]. Due to their common use, they are often released into municipal water systems, spread into the runoff, and passed into groundwater, ultimately contaminating both environmental and human populations [14,16,17]. As materials made with PFAS compounds such as PFOA (perfluorooctanoic acid), enter the environment, they accumulate and are difficult to degrade with typical biological mechanisms [18]. There is no simple way for these highly stable compounds to be removed from the environment through natural or human-influenced processes.
The prevalence of PFAS in the natural environment means there are many highly contaminated sites in the U.S and other nations with a history with these compounds [19,20]. The current treatment for PFAS contamination typically involves expensive granular activated carbon filters and subsequent incineration, which may only serve to recycle PFAS back into the environment [21]. Known methods for chemical/physical degradation involve exotic materials, difficult workups, and are often time-consuming and expensive [22,23]. A biological method for PFAS degradation would thus serve as a cheaper, more effective bioremediation alternative to remove PFAS from the environment [[23], [24], [25]].
Recent studies have validated the potential for bioremediation in PFAS degradation using different microbes. Groups have uncovered aerobic and anaerobic bacteria surviving in PFAS-contaminated sites that can break down these fluorinated compounds [26,27]. However, these results are limited in capability by either enzymatic processes limited to the terminal, non-fluorinated carbon in aerobic species, or as in anaerobic species, the timeline for degradation is extremely long. These drawbacks represent significant hurdles in bioremediation of PFAS. Our work sought to find aerobic bacteria that can more quickly and efficiently degrade PFAS. Our group isolated the aerobic bacterium Delftia acidovorans from a PFAS-contaminated soil sample and sequenced the genome to identify that D. acidovorans possesses an enzymatic pathway for PFOA degradation [28]. The objectives of this work are to characterize the dehalogenase enzymatic activity and engineer it for expression in Escherichia coli (E. coli) or other species for rapid and efficient bioremediation and degradation of PFAS.
2. Materials and methods
2.1. Culturing microbes from soil samples
Soil was sampled from PFAS contamination sites around an Air Force Base in Colorado. This location was chosen due to availability and known long-term exposure to PFAS comounds [20,29]. Fire-fighting foams containing PFAS were used in training for several years at this site. From these soil samples, winogradsky columns were set up with varying amounts of PFAS. After establishment of the winogradsky column, aerobic bacteria were cultured in PFAS-containing tryptic soy broth (TSB), isolated and plated on tryptic soy agar (TSA) then identified by 16S sequence analysis. Individual colonies of each bacterial species were then taken from isolation plates and grown in TSB overnight to ensure cells were viable. Cells were pelleted at 4500×g for 5 min and suspended in phosphate buffered saline (PBS; pH 7.4) and pelleted again to wash the pellet. The pellet was then suspended in minimal media with no additional carbon source added. Following inoculation, samples were monitored for growth by measuring optical density at 600 nm.
2.2. Fluoride ion measurements
D. acidovorans, Escherichia coli, and DeHa1 transformed E.coli were grown in 1/10 Miller's LB Broth. 50 mL cultures of each microbe were grown overnight and spiked to 100 ppm PFOA at the 0hr time point. Measurements were performed by removing 5 mL aliquots from growth mixture and then removing cells by centrifugation. Following centrifugation, supernatant was mixed with 5 mL DI water and 5 mL TISAB II to make the probe mixture for each trial. This mixture was then tested directly for fluoride content using a F- ion probe (Mettler Toledo SG78-FK2 SevenGo Duo Pro Cat # 51302622) [27] sensitive down to a molarity of 1x10−6 mol/L.
2.3. Homology modeling of dehalogenases
It was of interest to initially develop a crude model of the identified Dehalogenases from D. acidovorans, thus the crystal structure of the dehalogenases were modeled using Phyre2. The peptide sequence was BLASTed against non-redundant proteins through the NCBI database and a multiple alignment from the top BLAST hits was used to generate a consensus sequence (>95% consensus). Highly-conserved amino acid residues in the original dehalogenase protein sequence were visualized with JMOL. Potential pockets were visually identified and binding affinity modeling was performed with 1 click docking software called MCule-1CD [30]. PFAS was visualized bound to the dehalogenases.
2.4. Molecular cloning and DeHa plasmid preparation
Two haloacid dehalogenase genes from D. acidovorans (PZP66635.1& WP_011137954.1) were designed to have restriction enzyme cut sites and ordered from IDT as gene blocks. These custom synthesized DNA fragments were then digested and re-ligated into the corresponding restriction enzyme opened plasmid backbone pSB1C3 containing chloramphenicol resistance genes. The resulting plasmids were sequence-verified that they contain the Dehalogenase genes in the correction location and orientation. The plasmids were transformed into chemically competent E. coli DH5α and verified and maintained on Chloramphenicol LB plates. The transformed E. coli containing the dehalogenase genes were then used in experimental design along with wild type E. coli and D. acidovorans.
2.5. Statistical analysis
The average was calculated for each of the replicate data and used to represent the mean value for the different replicates. The standard deviation was calculated for each set of data and the error bars are representing the standard error of the mean. A two tailed t-test was used to test for statistical significance.
3. Results
PFAS contaminated soil was collected and incubated to identify microbial species that can survive in highly PFAS contaminated regions. Several types of microbes were able to survive solely on minimal media and PFAS. After culturing candidate microbes, one species seemed to survive remarkably well on minimal PFAS media. This microbe was cultured and identified through 16S sequencing to be Delftia acidovorans and was used in further studies as a potential degrader of PFAS [31].
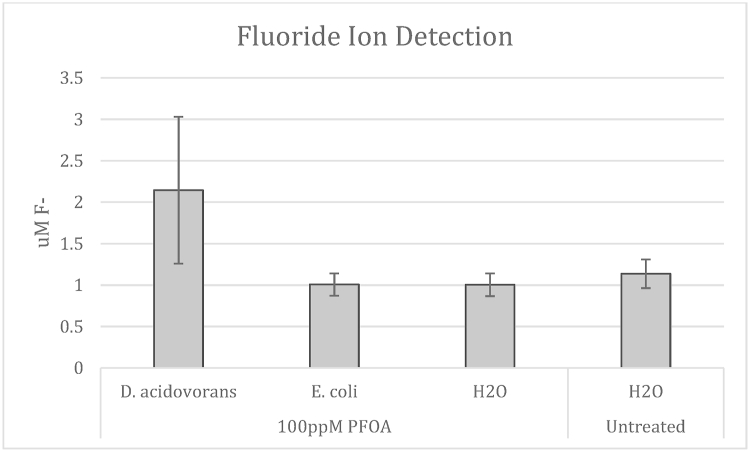
D. acidovorans was analyzed for potential PFAS degradation by looking at fluoride ion release into the growth media throughout incubation and exposure to PFOA. F- ion release from PFOA by D. acidovorans was compared to that of E. coli, both exposed to 100 ppm PFOA, and two control samples: DI water and 100 ppm PFOA DI water. The D. acidovorans cultures showed an increase in F- ion release at 4 h of exposure compared to control samples. A T-test comparing F- ion release between D. acidovorans and control samples gave a p-value of 0.23 (see Fig. 1).
Fig. 1.
D. acidovorans releases F- when exposed to PFAS. Fluoride measurement of D. acidovorans when exposed to 100PPM PFOA. F- specific ion probe was used at times indicated to measure free F- ions. D. acidovorans shows an increase trend in F- release compared to E. coli (100 ppm PFOA) (two tailed t-test. p = 0.23), DI water (100 ppm PFOA), and un-spiked DI water (0 ppm PFOA) samples. Error bars represent the Standard Error of the Mean. (N = 7).
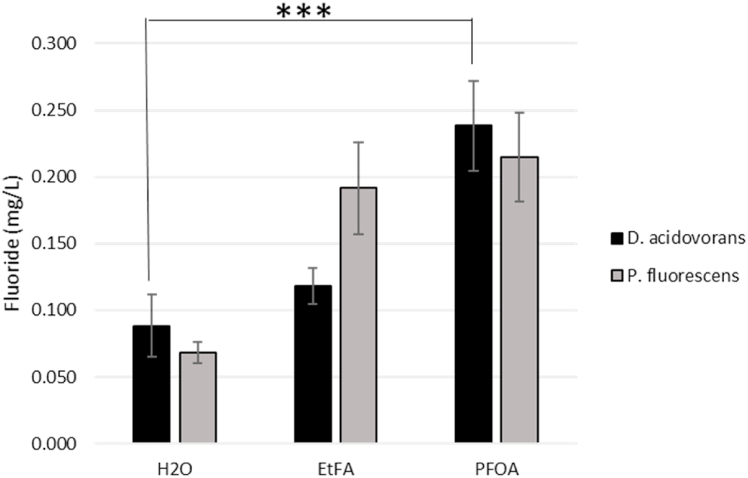
To validate the selective ion probe and F- release, Pseudomonas fluorescens, a known degrader of the fluorinated compound ethylfluoroacetate (EtFA) [32] was grown in similar conditions and exposed to either EtFA or PFOA. As expected, P. fluorescens showed F- ion release when given EtFA, while D. acidovorans didn't show much F- ion release with EtFA. Interestingly, both P. fluorescens and D. acidovorans showed an increase in F- ion release after PFOA treatment (Fig. 2).
Fig. 2.
D. acidovorans and P. fluorescens F- ion release with EtFA and PFOA. Fluoride measurement of D. acidovorans (black) and P. fluorescens (gray) when exposed to water (H20), ethylfluoroacetate (EtFA) 1000PPM, and Perfluorooctanoic acid (PFOA) 100PPM. Data represents average F- ion release over exposure time of 58 h (N = 6) Error bars represent the Standard Error of the Mean. (Two-tailed t-test. ***p < 0.005).
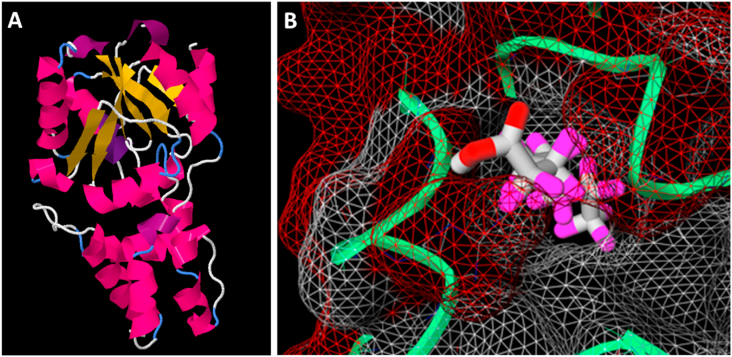
In an attempt to identify possible dehalogenases from D. acidovorans, a search was performed on a previously published genome. From this, two haloacid dehalogenases were identified (PZP66635.1& WP_011137954.1), further referred to as DeHa type I and type II. These two enzymes were crudely modeled for PFOA binding capability through the use of Phyre2 to model a crystal structure, and MCule-1CD to model PFOA binding centered on a region of highly conserved residues. No crystal structure of either DeHa I or II has been published at the time of writing. However, both dehalogenase's sequences are similar to α/β hydrolases, of which there are many known crystal structures. This allows for the use of the Phyre2 server to create an approximation of DeHa I & II's tertiary structure. The output PDB file for both enzymes was used for further modeling.
Using the PFAS contaminated soil, Winogradsky columns were created and spiked with different concentrations of PFAS contaminated water. In the presence of PFAS, certain genera of bacteria increased in abundance compared to non-PFAS-spiked samples. All genera that greatly increased in abundance with PFAS exposure were loaded into a protein BLAST with the query of DeHa I & II. From this, a list of similar proteins to the identified dehalogenases were compiled. A consensus sequence (>95%) was generated from the protein sequence file and visualized on DeHa I & II. The result was a central region in both dehalogenases of highly conserved residues, which coincided with an interior pocket between the α and β domains typical of these types of enzymes [33].
To further model PFOA's binding capability to DeHa I & II, the software MCule-1CD was utilized. PFOA was initially centered on the interior pocket, and the ligand binding energy was maximized through the software. PFOA modeled to bind to a highly conserved region on modeled DeHa type I in MCule 1CD. A binding score of −6.7 was calculated. As a point of comparison, the known peptide, A2A adenosine receptor complexed to caffeine (PDB: 5MZP) was modeled to bind caffeine in MCule 1CD after ligand removal, with an output score of [−5.7] (data not shown). PFOA was effectively modeled to bind well into the interior pocket of DeHa I & II, although in a follow-up analysis using a more robust, global affinity software, there was not a significantly dominant binding mode inside the pocket, likely owing to a crude crystal structure and the very flexible nature of PFOA. Overall, these findings confirmed our hypothesis that microbes that survive in high PFAS contamination areas may have enzymes that allow their degradation. Many of the microbes identified have putative proteins and enzymatic domains that may be able to bind PFAS (see Fig. 3).
Fig. 3.
Protein modeling of D. acidovorans dehalogenase. A) DeHa Type I estimated crystal structure from Phyre2 server. This dehalogenase has a strong match to α/β hydrolases. B) PFOA modeled to bind to a highly conserved region on modeled DeHa type I in MCule 1CD. A binding score of −6.7 was calculated. As a point of comparison, the known peptide, A2A adenosine receptor complexed to caffeine (PDB: 5MZP) was modeled to bind caffeine in MCule 1CD after ligand removal, with an output score of −5.7 (data not shown).
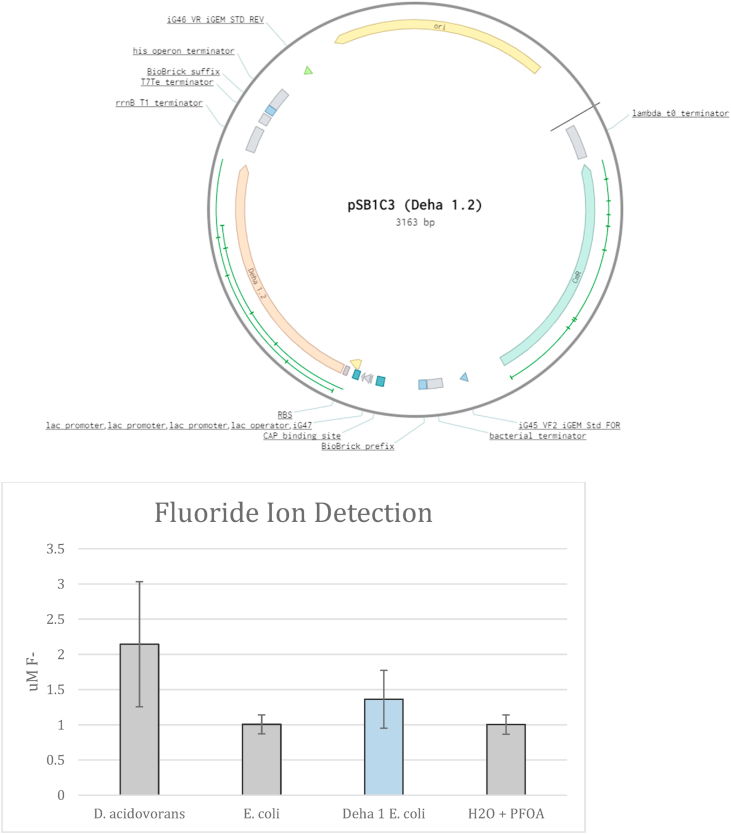
The first haloacid dehalogenase gene (DeHa 1) from D. acidovorans was genetically engineered and inserted into an iGEM plasmid (pSB1C3) for expression in E. coli. These transformed E. coli were then grown and exposed to 100 ppm PFOA. Over an incubation period of 4 h, F- ion release was detected from transformed E. coli cultures containing the engineered plasmid and Dehalogenase gene and compared to F- ion release from D. acidovorans and negative controls of DI water and 100 ppm PFOA DI water (Fig. 4). A t-test comparing F- ion release between transformed E. coli and control samples did not result in a significant p-value.
Fig. 4.
Engineered Dehalogenase plasmid and effect in transformed E. coli A) Plasmid map of pSB1C3 containing the Dehalogenase gene. B) Fluoride measurement of E. coli containing the plasmid when grown in 100 ppm PFOA spiked media. F- specific ion probe was used at times indicated to measure free F- ions. (N = 7).
4. Discussion
The research presented here was from a university level, international genetic engineering machine (iGEM) competition. The team identified an emerging global problem and used biological systems and synthetic biology to find a solution. Through intentional screening of PFAS contaminated soil, a large variety of microorganisms were identified that had capability to degrade or utilize PFAS in some fashion, or at least survive high levels of PFAS contamination. From this, D. acidovorans was identified as a likely candidate for dehalogenases with activity against PFOA. Growth of D. acidovorans in minimal media with only PFOA as a carbon source, indicated that the microbe could utilize PFAS, potentially for an energy source. Further exploration into how D. acidovorans utilizes PFOA was undertaken to determine if there was evidence for enzymatic defluorination. PFOA contains a terminal carboxylic group that can be enzymatically removed without a strict requirement of defluorination. This posed the potential for use of PFOA as a carbon resource without performing the desired function of removing fluorine. However, initial determination of fluoride ion increase in solution with actively growing cultures of D. acidovorans supported the idea that D. acidovorans exposed to PFOA results in an increase in free fluoride. While not conclusive, this is encouraging evidence of enzymatic activity against PFOA.
Analysis of the D. acidovorans genome identified at least 2 dehalogenase enzymes that have conserved binding sites and potential activity against PFAS. An additional 3–5 dehalogenases have been recently identified through active collaboration with the Air Force Research Lab and they are currently being investigated for PFAS degradation abilities as well. Interestingly, when the gene for a D. acidovorans dehalogenase is transformed into E. coli, the E. coli also shows a release of F- ions when exposed to PFOA. The exact degradation pathways and molecular intermediates that are produced have yet to be resolved, but the data indicates that the D. acidovorans dehalogenase enzyme may have biodegradation activity against PFOA. The next steps will be to determine exactly where the F- ions are coming from. Samples are currently being analyzed through LC-MS to detect specific perfluorinated compounds to quantitatively measure the input of PFOA and determine what breakdown products (if any) exist after exposure to a D. acidovorans dehalogenase. Additionally, it is important to determine if the enzyme can function outside of the cell and how the activity of the enzyme is affected through various purification processes and to see if the enzyme works in isolation, or what additional factors are needed to see enzymatic activity. It will also be critical to determine at what concentration the enzymes work to remove fluoride ions from PFAS. Perhaps enzymatic activity is only noticeable when exposed to high PFAS concentrations, such as the environments used in our experiments. Understanding the range of activity and enzymatic activity would be critical in identifying potential uses for these enzymes in future bioremediation efforts.
There were several microbes that were identified from the PFAS contaminated soil that could survive high PFAS concentrations and are currently being tested and identified for future bioremediation efforts. It is also possible that P. fluorescens has undocumented activity against PFOA as well. P. fluorescens was utilized in our experiments as a positive control for F- release, but it also showed intriguing ability to release F- when exposed to PFOA. This will need to be evaluated further. It is reasonable to assume that a species which is known to defluorinate EtFA has cellular mechanisms to perform this defluorination on other fluorinated compounds.
5. Conclusion
Even a single defluorination step and break in the encompassing shell of fluorine atoms constituting PFAS's stable structure could potentially allow for significant improvement in the ability (chemical or biological) to reduce toxicity, lower cost of removal/destruction, and ensure these synthetic compounds are able to be removed from the environment. If the D. acidovorans dehalogenase enzyme that is identified and described here can be better characterized and implemented in bioremediation, it may have critical impacts and provide novel options for dealing with the global PFAS contamination. The results presented are very encouraging and justify the need to look for more naturally occurring bioremediation pathways. Enzymes, such as the D. acidovorans dehalogenases may already and can be identified, engineered, and utilized to solve the PFAS issue.
IRB/human subject/animal experiments
Not Applicable.
Public release
PA#: USAFA-DF-2021-108.
Submission declaration
This has not been submitted to any other publisher.
CRediT authorship contribution statement
Jackson D. Harris: Conceptualization, Methodology, Investigation, Experimental design and setup, Writing, Supervision. Collin M. Coon: Conceptualization, Methodology, Investigation, Experimental design and setup, Writing, Supervision. Megan E. Doherty: Conceptualization, Methodology, Investigation, Experimental design and setup, Writing, Supervision. Eamon A. McHugh: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. Margaret C. Warner: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. Conley L. Walters: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. Olivia M. Orahood: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. Abigail E. Loesch: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. David C. Hatfield: Investigation, Experimental design and setup, Formal analysis, Visualization, Writing. John C. Sitko: Conceptualization, Methodology, Investigation, Experimental design and setup, Funding acquisition, Management and Supervision. Erin A. Almand: Conceptualization, Methodology, Investigation, Experimental design and setup, Funding acquisition, Management and Supervision. J. Jordan Steel: Conceptualization, Methodology, Investigation, Experimental design and setup, Writing, Supervision.
Declaration of competing interest
None.
Acknowledgements
We would like to thank Col Steven Hasstedt, Basic Science Division chair, and Gen Linell Letendre, Dean of the US Air Force Academy, for their continual support for the USAFA iGEM team. Dr. Don Veverka, Life Science Research Center Director, for his support in acquiring and managing funding. Dr. Nereyda Sevilla and the Defense Health Agency (DHA), for the financial support in this critical research and educational endeavor. Dr. Anthony Arment for helping advice the iGEM team and acquiring the original soil samples. We also acknowledge the other iGEM team members from past and present USAFA iGEM teams and our collaborators at the Air Force Research Labs that helped with training, advice, and general guidance with the research project.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Wilder L., Worley R., Community Breysse P. Exposures to per- and polyfluoroalkyl substances in drinking water: a national issue. J Environ Health. 2017;80(2):38–50. [Google Scholar]
- 2.Darlington R., Barth E., McKernan J. The challenges of PFAS remediation EPA public access. Ecosystem Services. 2018;23(1994):58–60. [PMC free article] [PubMed] [Google Scholar]
- 3.Moody C.A., Martin J.W., Kwan W.C., Muir D.C.G., Mabury S.A. Monitoring perfluorinated surfactants in biota and surface water samples following an accidental release of fire-fighting foam into Etobicoke Creek. Environ Sci Technol. 2002;36(4):545–551. doi: 10.1021/es011001+. [DOI] [PubMed] [Google Scholar]
- 4.US EPA. Laboratory-scale thermal degradation of perfluorooctanyl sulfo- nate and related substances.
- 5.US EPA O. Drinking water health advisories for PFOA and PFOS. [DOI] [PubMed]
- 6.Schultz M.M., Higgins C.P., Huset C.A., Luthy R.G., Barofsky D.F., Field J.A. Fluorochemical mass flows in a municipal wastewater treatment facility. Environ Sci Technol. 2006;40(23):7350–7357. doi: 10.1021/es061025m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau C., Anitole K., Hodes C., Lai D., Pfahles-Hutchens A., Seed J. Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicol Sci. 2007;99(2):366–394. doi: 10.1093/toxsci/kfm128. [DOI] [PubMed] [Google Scholar]
- 8.Kudo N., Kawashima Y. Toxicity and toxicokinetics of perfluorooctanoic acid in humans and animals. J Toxicol Sci. 2003;28(2):49–57. doi: 10.2131/jts.28.49. [DOI] [PubMed] [Google Scholar]
- 9.Pelch K.E., Reade A., Wolffe T.A.M., Kwiatkowski C.F. Environment International130; 2019. PFAS health effects database: protocol for a systematic evidence map; p. 104851. [DOI] [PubMed] [Google Scholar]
- 10.Rodea-Palomares I., Makowski M., Gonzalo S., González-Pleiter M., Leganés F., Fernández-Piñas F. Effect of PFOA/PFOS pre-exposure on the toxicity of the herbicides 2,4-D, Atrazine, Diuron and Paraquat to a model aquatic photosynthetic microorganism. Chemosphere. 2015;139:65–72. doi: 10.1016/j.chemosphere.2015.05.078. [DOI] [PubMed] [Google Scholar]
- 11.Chaparro-Ortega A., Betancourt M., Rosas P., et al. Endocrine disruptor effect of perfluorooctane sulfonic acid (PFOS) and perfluorooctanoic acid (PFOA) on porcine ovarian cell steroidogenesis. Toxicol Vitro. 2018;46:86–93. doi: 10.1016/j.tiv.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Zhao G., Wang J., Wang X., et al. Mutagenicity of PFOA in mammalian cells: role of mitochondria-dependent reactive oxygen species. Environ Sci Technol. 2011;45(4):1638–1644. doi: 10.1021/es1026129. [DOI] [PubMed] [Google Scholar]
- 13.Melzer D., Rice N., Depledge M.H., Henley W.E., Galloway T.S. Association between serum perfluorooctanoic acid (PFOA) and thyroid disease in the U.S. National Health and Nutrition Examination Survey. Environ Health Perspect. 2010;118(5):686–692. doi: 10.1289/ehp.0901584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van De Vijver K.I., Hoff P.T., Das K., et al. Perfluorinated chemicals infiltrate ocean waters: link between exposure levels and stable isotope ratios in marine mammals. Environ Sci Technol. 2003;37(24):5545–5550. doi: 10.1021/es0345975. [DOI] [PubMed] [Google Scholar]
- 15.Higgins C.P., Mcleod P.B., Macmanus-Spencer L.A., Luthy R.G. Bioaccumulation of perfluorochemicals in sediments by the aquatic oligochaete Lumbriculus variegatus. Environ Sci Technol. 2007;41(13):4600–4606. doi: 10.1021/es062792o. [DOI] [PubMed] [Google Scholar]
- 16.Darlington R, Barth E, Mckernan J. The challenges of PFAS remediation the problem with PFAS EPA public access. [PMC free article] [PubMed]
- 17.Parsons J.R., Sáez M., Dolfing J., de Voogt P. Biodegradation of perfluorinated compounds. Rev Environ Contam Toxicol. 2008;196:53–71. doi: 10.1007/978-0-387-78444-1_2. [DOI] [PubMed] [Google Scholar]
- 18.Glüge J., Scheringer M., Cousins I.T., et al. An overview of the uses of per- and polyfluoroalkyl substances (PFAS) Environ Sci: Processes and Impacts. 2020;22(12):2345–2373. doi: 10.1039/d0em00291g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contamination W., Wr U., Wkh H., et al. 2019. These chemicals are forever. February. [Google Scholar]
- 20.Hu X.C., Andrews D.Q., Lindstrom A.B., et al. Detection of poly- and perfluoroalkyl substances (PFASs) in U.S. Drinking water linked to industrial sites, military fire training areas, and wastewater treatment plants. Environ Sci Technol Lett. 2016;3(10) doi: 10.1021/acs.estlett.6b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoiber T., Evans S., Naidenko O.V. Disposal of products and materials containing per- and polyfluoroalkyl substances (PFAS): a cyclical problem. Chemosphere. 2020:127659. doi: 10.1016/j.chemosphere.2020.127659. [DOI] [PubMed] [Google Scholar]
- 22.Fischl W., Bartenschlager R. Exploitation of cellular pathways by Dengue virus. Curr Opin Microbiol. 2011:470–475. doi: 10.1016/j.mib.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 23.Kucharzyk K.H., Darlington R., Benotti M., Deeb R., Hawley E. Novel treatment technologies for PFAS compounds: a critical review. J Environ Manag. 2017;204:757–764. doi: 10.1016/j.jenvman.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Mejia Avendaño S. Microbial degradation of polyfluoroalkyl chemicals in the environment: a review. Environ Int. 2013;61:98–114. doi: 10.1016/j.envint.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 25.Kwon B.G., Lim H.-J., Na S.-H., Choi B.-I., Shin D.-S., Chung S.-Y. Biodegradation of perfluorooctanesulfonate (PFOS) as an emerging contaminant. Chemosphere. 2014;109:221–225. doi: 10.1016/j.chemosphere.2014.01.072. [DOI] [PubMed] [Google Scholar]
- 26.Yi L.B., Chai L.Y., Xie Y., Peng Q.J., Peng Q.Z. Isolation, identification, and degradation performance of a PFOA-degrading strain. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15028043. [DOI] [PubMed] [Google Scholar]
- 27.Huang S., Jaffé P.R. Defluorination of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) by acidimicrobium sp. strain A6. Environ Sci Technol. 2019;53(19):11410–11419. doi: 10.1021/acs.est.9b04047. [DOI] [PubMed] [Google Scholar]
- 28.Harris J., Gross M., Kemball J., et al. Draft genome sequence of the bacterium Delftia acidovorans strain D4B, isolated from soil. Microbiol Res Ann. 2021;10(44) doi: 10.1128/MRA.00635-21. Available from: /pmc/articles/PMC8567785/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke B.O., Anumol T., Barlaz M., Snyder S.A. Investigating landfill leachate as a source of trace organic pollutants. Chemosphere. 2015;127:269–275. doi: 10.1016/j.chemosphere.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 30.Babkova P., Dunajova Z., Chaloupkova R., Damborsky J., Bednar D., Marek M. Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics. Comput Struct Biotechnol J. 2020;18:1497–1508. doi: 10.1016/j.csbj.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braña V., Cagide C., Morel M.A. Microbial models: from environmental to industrial sustainability. Springer Singapore; 2016. The sustainable use of Delftia in agriculture, bioremediation, and bioproducts synthesis; pp. 227–247. [Google Scholar]
- 32.Camboim E.K.A., Almeida A.P., Tadra-Sfeir M.Z., et al. Isolation and identification of sodium fluoroacetate degrading bacteria from caprine rumen in Brazil. Sci World J. 2012:2012. doi: 10.1100/2012/178254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmquist M. Alpha beta-hydrolase fold enzymes structures, functions and mechanisms. Curr Protein Pept Sci. 2005;1(2):209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]