Abstract
Sclerotium rolfsii Sacc. the causative agent of white rot is one of the destructive pathogens of nightshade crops. In Côte d'Ivoire, this fungal pathogen constitutes a major constraint for the cultivation of tomato (Solanum lycopersicum) with 41.01% crop losses in humid forest areas. Controlling this fungus with synthetic chemicals can be effective, but harmful to human health and the environment. The use of biological control agents could be an alternative approach to control S. rolfsii. In this perspective, the objective of this work was to select fungi from the rhizosphere of tomato crops capable of inhibiting the growth of S. rolfsii. To do this, 153 fungi were isolated from the rhizosphere and from direct confrontation tests 10 fungi whose antagonistic power of S. rolfsii varied between 27 and 60% were selected. Molecular identification (ITS) of these antagonist fungi revealed that the isolates belonged to the genera Talaromyces sp. (n = 4), Trichoderma sp. (n = 3), Penicillium sp. (n = 2) and Clonostachys sp. (n = 1). Among these fungi, Talaromyces purpureogenus and Talaromyces assiutensis were able to diffuse compounds in agar capable of inhibiting the growth of S. rolfsii. The chemical study of these 2 fungi made it possible to identify mitorubrin and mitorubrinol produced by T. purpureogenus and spiculisporic acid produced by T. assiutensis. Mitorubrin and mitorubrinol had inhibitory activities of 100 and 70% at 10 mg/mL, respectively, whereas spiculisporic acid showed moderate inhibition of 38 at 20 mg/mL of the growth of S. rolfsii; however, its abundant production by the fungus could be an advantage in the control of this phytopathogen. Isolated from the same biotope as S. rolfsii, T. purpureogenus and T. assiutensis represent favorable candidates for the biological control against S. rolfsii.
Keywords: Antagonist fungi, Talaromyces sp, Antibiosis, Sclerotium rolfsii
Antagonist fungi, Talaromyces sp, Antibiosis, Sclerotium rolfsii.
1. Introduction
In agriculture, losses estimated at 20% of the annual yield are attributable to phytopathogenic fungi (Osman et al., 2017). The latter infect seeds, seedlings and mature or not mature plants causing many diseases such as damping-off, wilt (Nisha et al., 2009). Sclerotium rolfsii Sacc. causative agent of white rot is one of the destructive pathogens of plants worldwide (Le et al., 2012). This soil-borne fungus infects over 400 different plant species (Kim et al., 1999). In Côte d'Ivoire, this fungal pathogen constitutes a major constraint for the cultivation of tomatoes (Solanum lycopersicum) with 41.01% of crop losses in humid forest zones (Bolou et al., 2016a). The control of this fungal pathogen remains very difficult due to its survival structure (sclerotia) which remains viable in the soil for several years (Logan and Vos, 2011). The fight against this pathogenic fungus comes down to prophylactic measures and the use of synthetic pesticides (Bolou et al., 2015). However, the excessive use of synthetic fungicides leads to the development of resistant mutants and has harmful consequences on man and the environment. In Côte d'Ivoire, contamination of groundwater by organophosphate and organochlorine pesticides has been detected in agricultural regions where market gardening is practiced (Traoré et al., 2006). So many facts that require a sustainable and alternative management approach to fight against crop diseases.
Biological control by means of living microorganisms (biocontrol agent) could be a better approach to control S. rolfsii in order to reduce production losses and limit the use of synthetic fungicides. It consists of the use of microorganisms which either have a potential to inhibit the causal agent by different mechanisms of action, or an ability to increase the plant's defense mechanism. Microorganisms from the rhizosphere in areas where pest pressure is observed are the best candidate biocontrol agents. They are known for their ability to colonize the rhizosphere and the roots of plants, to co-evolve and sometimes control phytopathogenic microorganisms by various mechanisms including the production of bioactive metabolites. These bioactive metabolites are natural substances produced in response to interactions between protagonists (Galet, 2014; Belkadi and Koliai, 2016).
Songon, a town located in the south of Côte d'Ivoire with geographical coordinates 5° 18′59 ″ North latitude and 4° 14′27 ″ West longitude with an altitude of 10 m is a tomato production area where rife several pathogens including S. rolfsii (Bolou et al., 2016a; Kambiré et al., 2018). In addition to tomato plants, which suffer 83% of attacks from S. rolfsii, eggplant and pepper plants are also vulnerable. This showed an aggressiveness ranging from 52 to 90% against different varieties of pepper (Bolou et al., 2016a, 2016b). However, this proliferation of S. rolfsii can be inhibited by other microorganisms that coexist in the same habitat through different naturally developed mechanisms of action. The present work consists of the search for potential biocontrol agents against S. rolfsii isolated from soils under tomato crops.
2. Material and methods
2.1. Isolation of fungi from the soil
Rhizosphere soil samples were collected from six plots from different farmers. Sampling was carried out as described by Barillot et al. (2012). Briefly, using a sterile spatula, in each plot, a composite sample of 150 g of soil was made from 12 randomly selected subsamples in each plot. The composite samples were stored in sterile packaging bags (L x W: 200 × 145 mm). These were put into a cooler and then directly carried to the laboratory where they were stored at 4 °C. The various rhizospheric fungi were isolated by the suspension-dilution method (Rapilly 1968) on Potato Dextrose Agar (PDA) medium (Conda SA, Spain). For each soil sample, stock suspensions were prepared by mixing 1 g of soil into 9 mL of sterile distilled water. After vortexing for 5 min, a decimal dilution series is performed. The 10−3 to 10−5 dilutions were seeded on the PDA medium. Three tests are carried out for each of them. The dishes were incubated at 28 ± 1 °C for 7 days. The colonies obtained were subcultured on the PDA medium until pure cultures were obtained. All isolates were stored at -80 °C in 25% glycerol.
2.2. In vitro evaluation of the antifungal activity of soil fungi against S. rolfsii
The antifungal activity of fungi isolated from the soil against the phytopathogenic fungus S. rolfsii (provided by the Laboratory of Plant Physiology of the University Félix Houphouët-Boigny of Côte d'Ivoire) was studied by using the technique of opposite cultures, described by Hmouni et al. (1996). This test was carried out on the PDA medium. The fungal isolates and the phytopathogen were cultured on PDA medium and incubated for 7 days before the implementation of the antagonism test. After the 7 days of incubation, two explants including that of the pathogen and the antagonist agent were placed equidistant (3.5 cm) from the center of the Petri dish. The control consists of a subculture of the pathogen in the Petri dish. These dishes were incubated in a culture oven at 27 °C. The percentage of inhibition was calculated using Eq. (1):
| I (%) = (1 - Cn/Co) x 100 | (1) |
where:
Cn is the average diameter of the growth of the mycelium in the presence of the antagonist agent.
Co is the average diameter of the growth of the mycelium in the absence of the antagonist agent.
2.3. Morphological identification of isolated fungal isolates
Fungal isolates from soils under vegetable cultivation have been described using the fungal determination key from Barnett and Hunter (1972). More recent ones by Pitt and Hocking (1997); Houbraken et al. (2014) were used to define the genres to which they belong.
2.4. Molecular identification of antagonist fungi
2.4.1. DNA extraction
The extraction of genomic DNA from the fungi was carried out according to the method described by White et al. (1990) with some modifications. Briefly, the liquid cultures of the fungi carried out previously with Potato Dextrose Broth (PDB) medium were centrifuged at 10,000 g for 3 min. After removing the supernatant, the pellet was homogenized in 400 μL of TBS buffer (0.4 M NaCl, 10 mM Tris-HCl pH 8 and 2 mM EDTA pH 8) then vortexed for 1 min. Then 40 μL of 20% SDS and 8 μL of Proteinase K (20 mg/mL) were added, then the mixture was incubated at 60 °C for 3 h. After incubation, 300 μL of 6 M NaCl were introduced into the various tubes which were also vortexed for 30 s and then centrifuged at 10,000 g for 30 min at 4 °C. The supernatants were transferred to tubes of 2 mL to which was added an equal volume of 100% Isopropanol. These tubes were incubated at -20 °C for 1 h and then centrifuged at 10,000 g for 20 min at 4 °C. The pellet was washed with 70% ethanol and then centrifuged again for 10 min at maximum speed. The pellet from the centrifugation was subsequently recovered and dried at 37 °C for 5 min. The DNA thus obtained was re-suspended in 300 μL of TE buffer (Tris-EDTA) sterile and stored at -20 °C to be amplified. The quantity and quality of DNA were estimated visually under UV rays after migration on 1% agarose gel against the 100 bp molecular weight marker DNA Ladder (Solis biodyne).
2.4.2. PCR amplification and sequencing
Fungal isolates were identified by sequencing the ITS (Internal Transcribed Spacer) regions of rDNA ITS1 (5′-GGA GTA AAA GTC GT A ACA GG-3 ′) and ITS4 (5′-TCC TCC GCT TATTGA TATGC-3 ′). 4 μL of 5x Master Mix buffer (Solis Biodyne FIREPOL), 0.75 μL of each fungal primer at 10 μM, 5 μL of fungal DNA were combined in a final volume of 25 μL. Amplification consisted of an initial denaturation step of 2 min at 94 °C followed by 30 cycles of 30 s at 94 °C (denaturation), 30 s at 52 °C (hybridization) and 1 min at 72 °C (elongation) with a final elongation step of 10 min at 72 °C. Analysis of the PCR products was carried out by electrophoresis on 1% Agarose gel in 1X TBE buffer stained with SYBR SAFE (1%). The PCR products obtained were packaged and sent to Eurofin Genomics for Sanger sequencing according to the service provider's recommendations. Quality of the FASTA sequences was assess and only good query sequences were used for the identification of fungi using the Megablast Algorithm of NCBI's Nucleotide BLAST Tool. The Fungi RefSeq ITS databases (Fungi FTP: ftp://ftp.ncbi.nlm.nih.gov/refseq/TargetedLoci/Fungi/) was targeted. It contains curated and re-annotated records of sequences from the ITS region in the nuclear ribosomal cistron, sourced from INSD records.
2.5. Production and extractions of secondary metabolites
The extraction of the secondary metabolites was carried out by first carrying out fermentation of the antagonist on a rice medium (40 g of rice for 80 mL of distilled water). To do this, a suspension of spores from a 7-day-old culture was produced in sterilized distilled water, then inoculated onto the previously sterilized rice medium. The rice media thus inoculated were incubated at 28 °C for 21 days. At the end of the incubation, the cultures were harvested in 1 L Erlenmeyer flasks. Then the extraction of the metabolites was carried out by adding 500 mL of ethyl acetate to each Erlenmeyer flask. The solution obtained after maceration was filtered three times through gas then through cotton wool. The collected filtrate was concentrated using a rotary evaporator to give the crude fungal extract.
2.6. Bioguided purification of bioactive secondary metabolites
The crude extracts were fractionated by chromatography on a silica gel column using an elution gradient of Cyclohexane/Ethyl Acetate (10-0/0–10). Fractions were pooled from their profile on TLC (Silica 60 F 254 Merck, Germany). The fractions obtained were evaluated against S. rolfsii. The fractions for were further purified in search of active molecules with the elution gradient indicated above. The 1D and 2D NMR spectra were recorded in DMSO-d6 and Acetone-d6 with Bruker 400 spectrometer using TMS as internal standard. The chemical shifts were given in δ (ppm). HR-ESI-MS spectra were obtained with an Applied Biosystem QSTAR Pulsar I mass spectrometer.
2.7. Evaluation of the biological activities of the fractions and of the purified compounds
For each fraction and purified compounds, a range of concentrations was achieved by the double dilution method, ranging from 5 to 20 mg/mL. Each concentration was spread beforehand on the PDA medium, then the different dishes were inoculated with a fungal disc of S. rolfsii 5 mm in diameter (obtained from a culture of 7 days of incubation on solid PDA medium). The control was obtained from a disc of S. rolfsii alone on PDA. The petri dishes were incubated at 28 °C for 4 days. The percentage of growth inhibition I (%) is expressed by the reduction in the diameter of the fungal colony relative to the control, calculated using Eq. (2):
| I (%) = [1 - (D test / Dcontrol)] x 100 | (2) |
with:
Dtest: diameter of the colony in the test in mm.
Dcontrol: control diameter of control colony in mm.
2.8. Statistical analysis
The means of the measurements of the diameters of inhibition carried out on the different days of collections were compared by ANOVA one way (GraphPad Prism 8.4.3).
3. Results
3.1. Antifungal activity of soil fungi against S. rolfsii
A total of 153 fungal isolates were collected, of which 10 (ie 15.3%) showed inhibitory activity against S. rolfsii. The antifungal activity was confirmed by measuring the diameter of inhibition of mycelial growth of S. rolfsii. The percent inhibition varied from 27.06 to 60.59% (Table 1). Alone isolates IIX2-13 and M2-10 exerted inhibition by diffusion of antifungal compounds into the agar with an observable zone of inhibition remained constant up to 11 days after incubation (Figure 1 and Figure 2). These antagonists were therefore selected for the identification of the compounds responsible for the antifungal activity.
Table 1.
S. rolfsii growth inhibition
| Isolate | Description | Inhibition rate (%) |
|---|---|---|
| TmF 13 | Talaromyces calidicanius | 60,59 |
| TmN 1 | Trichoderma yunnanense | 56,47 |
| M1-12 | Trichoderma ghanense | 42,94 |
| IIBio 5 | Trichoderma reesei | 40,59 |
| M4-7 | Penicillium citrinum | 37,06 |
| M4-8 | Penicillium citrinum | 35,29 |
| IX1-1 | Clonostachys swieteniae | 31,18 |
| IIX 1-2 | Talaromyces pratensis | 28,24 |
| IIX2-13 | Talaromyces purpureogenus | 27,65 |
| M2-10 | Talaromyces assiutensis | 27,06 |
Figure 1.
Dual culture test A: Isolate M2-10 and S. rolfsii after 5 days of incubation B: Isolate M2-10 and S. rolfsii after 11 days of incubation.
Figure 2.
Dual culture test A: Isolate IIX2-13 and S. rolfsii after 5 days of incubation B: Isolate IIX2-13 and S. rolfsii after 11 days of incubation.
3.2. Morphological identification of selected antagonist fungi
Microscopic identification of the antagonist fungi revealed the genera Penicillium (5 isolates) and Trichoderma (3 isolates).
3.3. Molecular identification of selected antagonist fungi
The request sequence, a DNA fragment corresponding to the different antagonists, was used to perform a BLASTn with the Megablast program. The data bank indicated was RNA types trains/ITS_RefSeq_Fungi. Table 2 lists the species identified by molecular biology. Analysis of the sequencing data revealed that the isolates fall into 4 genera with 4 species (Isolates TmF13, IIX 1–2, IIX2-13 and M2-10) for the genus Talaromyces; 3 species for the genus Trichoderma (Isolates TmN 1, M1-12, and IIBio 5); one species for the genus Penicillium (Isolates M4-7, and M4-8); a species for the genus Clonostachys (Isolate IX1-1).
Table 2.
BLASTn results of 18S rRNA sequences of antagonist fungi.
| Isolate | Description | % Identity | Accession nb |
|---|---|---|---|
| TmF 13 | Talaromyces calidicanius | 97,79 | NR_103665.2 |
| TmN 1 | Trichoderma yunnanense | 100 | NR_134419.1 |
| M1-12 | Trichoderma ghanense | 99,33 | NR_120299.2 |
| IIBio 5 | Trichoderma reesei | 99,83 | NR_120297 |
| M4-7 | Penicillium citrinum | 100 | NR_121224 |
| M4-8 | Penicillium citrinum | 99,43 | NR_121224 |
| IX1-1 | Clonostachys swieteniae | 96,89 | NR_171105 |
| IIX 1-2 | Talaromyces pratensis | 98,78 | NR_165529 |
| IIX2-13 | Talaromyces purpureogenus | 98,99 | NR_121529 |
| M2-10 | Talaromyces assiutensis | 96,74 | NR_172040 |
3.4. Biological activities of secondary metabolites
After extracting the cultures of the fungi, a crude extract of dark red color with a mass of 4 g was obtained for T. purpureogenus and a crude extract of brown color with a mass of 510 mg was obtained for T. assiutensis. Bioguided purification of crude extracts of T. purpureogenus led to compounds 1 (153 mg) and 2 (284 mg). The crude extract of T. assiutensis yielded compound 3 (168 mg) (Table 3).
Table 3.
Growth inhibition rate of S. rolfsii by crude extracts.
| Crude extracts | Concentration (mg/mL) | Inhibition rate |
|---|---|---|
| T. purpureogenus IIX2-13 | 50 | 81% |
| 25 | 65% | |
| 12,5 | 55% | |
| T. assiutensis M2-10 | 50 | 68% |
| 25 | 58% | |
| 12,5 | 46% |
Compound 1 with a concentration of 10 mg/mL completely inhibited the growth of S. rolfsii. On the other hand, compound 2 with the same concentration (10 mg/mL) exerted an inhibition of 70%. Compound 3 inhibited the mycelial growth of S. rolfsii by 38% (Table 4).
Table 4.
Growth inhibition rate of S. rolfsii by compounds.
| Compounds | 20 mg/mL | 10 mg/mL | 5 mg/mL |
|---|---|---|---|
| 1 | 100 % | 100 % | 40 % |
| 2 | 90 % | 70 % | 32 % |
| 3 | 38 % | 04 % | 00 % |
3.5. Purified antifungal compounds
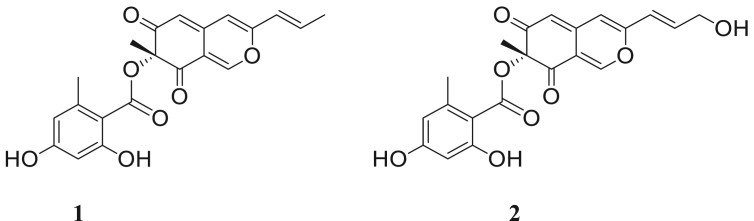
Analysis of the mass spectrum of compound 1 made it possible to demonstrate a peak of molecular ion HR-ESI-MS m/z [M + H]+ 383.111 (calc. for C21 H18 O7 + H+: 383.112), and m/z [M + Na]+ 405.094 (calc. for C21 H18 O7 + Na+: 405.115). This analysis combined with the 1D and 2D NMR data made it possible to determine the chemical structure of 1 in accordance with the data in the literature corresponding to Mitorubrin (Figure 3). Analysis of the mass spectrum of compound 2 made it possible to demonstrate an HR-ESI-MS m/z [M + H]+ 399.1074 (calc. for C21 H18 O8 + H+: 399.10711) and m/z [2M + H]+ 797.21 (calc. for C42 H36 O16 + H+). This analysis allowed to determine the chemical structure of 2 in agreement with the data of the literature, which is the hydroxylated form of 1, which corresponds to Mitorubrinol (Figure 3).
Figure 3.
Structures of mitorubrin (1) and mitorubrinol (2).
Analysis of the mass spectrum of compound 3 made it possible to demonstrate a high resolution molecular ion peak ESI [2M + H]+ m/z 657.3849; [M + H]+ m/z 329.1962 (calc. for C17 H28 O6 + H+: 329.1986). This analysis, combined with the 1D and 2D NMR data, made it possible to determine the chemical structure of the compound in accordance with the data in the literature. It was Spiculisporic acid (Figure 4).
Figure 4.
Structure of spiculisporic acid (3).
4. Discussion
The tomato (Solanum lycopersicum) is subject to many fungal constraints in relation to the infectious potential of the soils. The city of Songon, located in the south of Côte d'Ivoire is a market gardening area, particularly tomatoes. This area is also described in the literature as being favorable to the proliferation of S. rolfsii (Bolou et al., 2015), thus devastating the tomato crops. However, the growth of S. rolfsii could be inhibited by other microorganisms that coexist in this same habitat through different naturally developed mechanisms of action. With a view to implementing biological control, soil investigations in this area were carried out. These investigations led to the isolation of 153 rhizospheric fungi.
The screening carried out from the 153 soil-based fungal isolates by means of a direct confrontation test on the PDA medium made it possible to select 10 fungi which inhibit the growth of S. rolfsii. The percentages of inhibition ranged from 27.06 to 60.59%. These results are statistically identical (ANOVA test, F = 0.8042; p = 0.3932). The mechanisms of action observed during this inhibition were competition and antibiosis.
Rapid growth of some fungi (percent inhibition greater than 50%) inhibited mycelial growth of the pathogen. This phenomenon reminds us of the phenomenon of antagonism by competition of space and nutrients. This is because the depletion of nutrients caused by the rapid growth of the antagonist will slow down or inhibit the growth of the pathogen. Competition is particularly intense in the soil, which is an oligotrophic medium where the microorganisms are essentially at rest in the form of preservation. The rhizo-deposits released by the root system (supply of organic matter), will allow the activation of microorganisms in the rhizosphere of plants. Only the most competitive microorganisms will be able to take advantage of this nutrient supply and attempt to colonize the roots of plants (Aouar et al., 2019).
The second mechanism observed was antibiosis. This is based on the release by a microorganism of soluble or volatile secondary metabolites responsible for inhibiting the growth of a second microorganism. This inhibitory activity was observed by the appearance of a zone of inhibition followed by growth arrest of S. rolfsii.
Microscopic identification of the antagonist fungi revealed the genera Penicillium and Trichoderma. However, analysis of the ITS sequences confirmed one Penicillium in all 5 isolates and assigned 3 isolates to the genus Talaromyces and 1 isolate to the genus Clonostachys. This difference in result could be explained by the fact that the genera Talaromyces and Penicillium are anamorphs and therefore difficult to differentiate microscopically (Benjamin, 1955). In fact, the microscopic observation of Talaromyces is identical to that of the subgenus Penicillium biverticillium, so the subgenus Penicillium biverticillium has been transferred to the genus Talaromyces (Yilmaz et al., 2014). It is also difficult to differentiate on microscopy the genus Penicillium from the genus Clonostachys because both present a monoverticulate Conidiophore.
All isolates belonging to the genus Trichoderma (n = 3) exerted inhibition on the growth of S. rolfsii. Trichoderma species are rapid, invasive, filamentous colonizers, exhibiting a symbiotic relationship with plants. They exhibit antagonistic behavior against several phytopathogens such as bacteria, nematodes and in particular fungi by inhibiting their growth either by direct interaction (hyper parasitism, competition for nutrients and space, and antibiosis) or indirectly by improving plant growth and vigor and improving stress tolerance (Zhang et al., 2017). These different properties explain why approximately 90% of biological fungal control agents against pathogenic microorganisms to date belong to different strains of Trichoderma (Sood et al., 2020). In addition to the Trichoderma species, 40% (n = 4) of the antagonists identified belong to the genus Talaromyces. Talaromyces species are cosmopolitan filamentous fungi that play various roles in natural ecosystems and in biotechnology. Several species of Talaromyces isolated from soil have the capacity to exert antagonistic activities against phytopathogenic fungi and to produce secondary metabolites unique to this genus (Manoch et al., 2013). As a result, several species of this genus including T. flavus are used in the biological control of fungal phytopathogens such as Verticillium dahliae and Rhizoctonia solani (Kakvan et al., 2013).
T. purpureogenus and T. assiutensis exerted the antibiosis mechanism. The low inhibitory powers of IIX2-13 and M2-10 are related to their slow growth, compared to that of S. rolfsii. In confrontation, their slow growth was compensated by the biosynthesis and the diffusion of antifungal compounds completely preventing the growth of S. rolfsii. This inhibitory activity was observed by the appearance of a zone of inhibition followed by stopping the growth of S. rolfsii. The zone of inhibition could be explained by the presence of toxic substances secreted by the antagonist agent. Indeed, the presence and size of the zone of inhibition have been used as evidence of the production of antibiotics by strains of Trichoderma (Barari, 2016). Some authors including Abouzeid (2014) and Gnancadja et al. (2015) used the mechanism of antibiosis to select different isolates likely to be biocontrol agents.
Mitorubrin and mitorubinol obtained by bioguided fractionation of the crude extract of T. purpureogenus belong to the azaphilone family: a set of fungal secondary metabolites with a pyrone-quinone structure containing a highly oxygenated bicyclic nucleus and a chiral nucleus (Venkatachalam et al., 2018). They group together compounds exhibiting a broad spectrum of biological activity such as antimicrobial, antiviral, antioxidant, anti-inflammatory, cytotoxic and nematicidal activities. A synergistic effect of these two compounds could be at the origin of the inhibitory activity of T. purpureogenus against S. rolfsii. Pakora et al. (2018) have indeed shown a synergistic effect of gliovirin and viridin isolated from Trichoderma sp against Phytophthora sp., The agent of brown rot in cocoa pods.
Spiculisporic acid resulting from the purification of the crude extract produced by T. assiutensis is a bioactive γ- butenolide, a mycotoxin found in certain species of Talaromyces such as T. trachyspermus (Yaguchi et al., 1994) and T. purpureogenus (Yilmaz et al., 2012). Spiculisporic acid is used commercially as a biosurfactant (Ishigami et al., 2000). This is the first time that spiculisporic acid has been reported to have been isolated from T. assiutensis. Hamza et al. (2017) showed nematophagous activity of T. assiutensis against juvenile strains of Meloidogyne javanica. The soluble n-hexane and chloroform fractions of T. assiutensis cultured filtrates showed strong antimicrobial activity against root rot fungi (Farhat et al., 2021).
5. Conclusion
This study made it possible to highlight the diversity of antagonistic fungi originating from the rhizosphere of tomato crops. Molecular identification made it possible to highlight the antagonist fungi T. purpureogenus and T. assiutensis among the fungi exerting the mechanism of antibiosis. Bio-guided purification and NMR identified mitorubrin, mitorubrinol and spiculisporic acid as molecules responsible for inhibiting S. rolfsii. These molecules, already known but used in biotechnology, this study revealed another application of them, in particular in agriculture. Isolated from the same biotope as S. rolfsii, T. purpureogenus and T. assiutensis represent favorable candidates for biological control against S. rolfsii.
Declarations
Author contribution statement
Anne-Edwige Coulibaly: Performed the experiments; Wrote the paper.
Gilles Alex Pakora: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Aristide Berenger Ako: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Georges Elisée Ler-N’Ogn Dadé Amari, Carine Aya N'Guessan, Abo Kouabenan, Daouda Kone & Joseph Allico Djaman: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Interprofessional Fund for Agricultural Research and Council (FIRCA) of Côte d’Ivoire, with the project “BIOCONTRÔLE”.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Prof. Kacou N'douba Adèle who granted us access to the Bacteriology-Virology laboratory of the Félix Houphouët-Boigny University in Cocody for carrying out this work. We also thank Abinan Kouman Brice and Brou Sygnoh Eve Pristile for their assistance during the bioguided purification of bioactive secondary metabolites.
The authors wish to thank L. Dubost for mass spectra, A. Deville for NMR spectra (Laboratoire Molécules de Communication et Adaptation des Micro-organismes (MCAM), Muséum national d’Histoire naturelle, CNRS; CP54, 57 Rue Cuvier, 75005 Paris, France).
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abouzeid N. Effect of volatile and non-volatile compounds of Trichoderma spp. on Botrytis fabae the causative agent of faba bean chocolate spot. Am. J. Life Sci. 2014;2(6-2):11–18. [Google Scholar]
- Aouar L., Inas A.B., Benadjila H., Medjoudj Z., Mourad Streptomyces griseus Lac1: biocontrôle et propriétés promotrices de la croissance des plantes. Revue des BioRessources. 2019;9(1):27–37. [Google Scholar]
- Barari H. Biocontrol of tomato Fusarium wilt by Trichoderma species under in vitro and in vivo conditions. Cercet. Agronomice Moldova. 2016;49(1):91–98. [Google Scholar]
- Barillot C.D.C., Sarde C.-O., Bert V., Tarnaud E., Cochet N. A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 2012;63:471–476. [Google Scholar]
- Barnett H.L., Hunter B.B. third ed. Burgess Publishing Co.; Minneapolis: 1972. Illustrated Genera of Imperfect Fungi; p. 241. [Google Scholar]
- Belkadi Z., Yasmina K. University Mouloud Mammeri; 2016. Isolation of Rhizospheric Bacteria with Antagonistic Activity and Testing in the Biocontrol of Botrytis Cinerea on Tomato.https://dl.ummto.dz/handle/ummto/4869 Thesis. [Google Scholar]
- Benjamin C.R. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47:669–687. [Google Scholar]
- Bolou Bi B.A., Kouakou T.H., Kouame G.K., Kassi F. Inhibition of Sclerotium rolfsii sacc. (Corticiaceae), the causal agent of tomato stem collar rot (Solanaceae), by Xylopia aethiopica (dunal) a. Rich (Annonaceae) and Trichoderma sp. Euro. Sci. J. ESJ. 2015;11(12) [Google Scholar]
- Bolou Bi B.A., Abo K., Doumbouya M., Kouamé K.G., Kassi K.F., Tuo S., Nahoua K., Kouassi N.P., Koné D. Analysis of sclerotinia expression due to Sclerotium rolfsii fungus in market gardening crops in the different agroecological zones of Côte d’Ivoire. J. Agri. Ecol. Res. Int. 2016;6(3):1–12. [Google Scholar]
- Bolou Bi Bolou Antoine, Kouabenan Abo, Gaston Kouamé Koffi, Daouda Kone. Evaluation of resistance of seven pepper varieties to sclerotinia caused by Sclerotium rolfsii, in the locality of Songon-Te, in the south of Cote d’Ivoire. Int. J. Agricul. Innov. Res. 2016;4(4):703–707. [Google Scholar]
- Farhat Hafiza, Urooj Faizah, Muhammed Irfan, Nida Sohall, Salma Majeed, Shafique Hafiza, Hameedi Sidra, Ehteshamul-Haque Syed. Biocontrol potential of endophytic fungus Talaromyces trachyspermus against root rot pathogens of sunflower. Res. Square. 2021 [Google Scholar]
- Galet J. University of Lorraine; 2014. Towards the Understanding of Microbial Dialogues in Soil Ecosystems: Study of the Interaction between Streptomyces and Pseudomonas.https://hal.univ-lorraine.fr/tel-01751378 Thesis. [Google Scholar]
- Gnancadja L., Tonon D., Faton E., Douro K., Dannon E., Akoegninou A. Efficacy of the antagonistic agent Trichoderma harzianum on Fusarium oxysporum f. sp. lycopersici pathogen of tomato. Int. J. Brain Cognit. Sci. 2015;9(2):770. [Google Scholar]
- Hamza Mohamed Aït, Lakhtar Hicham, Tazi Hafssa, Moukhli Abdelmajid, Fossati-Gaschignard OOdile, Miché Lucie, Roussos Sebastianos, Ferji Zahra, El Mousadik Abdelhamid, Mateille Thierry, Hassan Boubaker. Diversity of nematophagous fungi in Moroccan olive nurseries : highlighting prey-predator interactions and efficient strains against root-knot nematodes. Biol. Contr. 2017 [Google Scholar]
- Hmouni A., Hajlaoui M.R., Mlaiki A. Resistance of Botrytis cinerea to benzimidazoles and dicarboximides in sheltered tomato crops in Tunisia. OEPP/EPPO Bull. 1996;26(3-4):697–705. [Google Scholar]
- Houbraken J., Vries RP de, Samson R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv. Appl. Microbiol. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Ishigami Y., Zhang Y.J., Ji F.X. Spiculisporic acid. Functional development as biosurfactant. Chim. Oggi – Chem. Today. 2000;18:32–34. ISSN :0392-839X. [Google Scholar]
- Kakvan Nikoo, Heydari Asghar, Zamanizadeh Hamid Reza, Rezaee Saeed, Naraghi Laleh. Development of new bioformulations using Trichoderma and Talaromyces fungal antagonists for biological control of sugar beet damping-off disease. Crop Protect. 2013;53:80–84. [Google Scholar]
- Kambire B., Ymba M., Ouattara K. Liquid waste management and population vulnerability to disease: the case of Songon-Agban, district d’Abidjan. Tropicultura. 2018;36(2):407–416. [Google Scholar]
- Kim W.-G., Cho W.-D., Jee H.-J. Occurrence of sclerotinia rot on cucurbitaceous vegetable crops in greenhouses. Korean J. Mycol. 1999;27(3):198–205. [Google Scholar]
- Le C.N., Kruijt M., Raaijmakers J.M. Involvement of phenazines and lipopeptides in interactions between Pseudomonas species and Sclerotium rolfsii, causal agent of stem rot disease on groundnut. J. Appl. Microbiol. 2012;112(2):390–403. doi: 10.1111/j.1365-2672.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- Logan N.A., Vos P.D. Springer Science & Business Media; 2011. Endospore-forming Soil Bacteria. [Google Scholar]
- Manoch Leka, Dethoup Tida, Yilmaz Neriman, Houbraken Jos, Robert A., Samson Two new Talaromyces species from soil in Thailand. Mycoscience. 2013;54:335–342. [Google Scholar]
- Nisha Aggarwal, Kumar Rajesh, Prem Dureja, Rawat Diwan. Schiff bases as potential fungicides and nitrification. J. Agric. Food Chem. 2009;57(18):8520–8525. doi: 10.1021/jf902035w. [DOI] [PubMed] [Google Scholar]
- Osman Mohamed Ali Eisa, Shakil Najam Akhtar, Rana Virendra Singh, Sarkar Dhruba Jyoti, Majumder Sujan, Kaushik Parshant, Singh Braj Bhushan, Kumar Jitendra. Antifungal activity of nano emulsions of neem and citronella oils against phytopathogenic fungi, Rhizoctonia solani and Sclerotium rolfsii. Ind. Crop. Prod. 2017;108:379–387. [Google Scholar]
- Pakora G.-A., Mpika J., Kone D., Ducamp M., KebeI, Nay B., Buisson D. Inhibition of Phytophthora species, agents of cocoa black pod disease, by secondary metabolites of Trichoderma species. Environ. Sci. Pollut. Contr. Ser. 2018;25(30):29901–29909. doi: 10.1007/s11356-017-0283-9. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Hocking A.D. second ed. Blackie Academic and Professional; London: 1997. Fungi and Food Spoilage. [Google Scholar]
- Rapilly F. Les techniques de mycologie en pathologie végétale. Ann. Epiphytes. 1968;19 https://www.sudoc.fr/060599316 [Google Scholar]
- Sood Monika, Kapoor Dhriti, Kumar Vipul, Sheteiwy Mohamed S., Ramakrishnan Muthusamy, Landi Marco, Araniti Fabrizio, Sharma Anket. Trichoderma: the “secrets” of a multitalented biocontrol agent. Plants. 2020;2020(9):762. doi: 10.3390/plants9060762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traore Karim S., Mamadou K., Dembélé A., Lafrance P., Mazellier P., Houénou P. Contamination of groundwater by pesticides in agricultural regions in Côte d'Ivoire (centre, south and south-west) SOACHIM J. 2006;1:1–9. [Google Scholar]
- Venkatachalam Ekala, Zelena Miroslava, Cacciola Francesco, Ceslova Lenka, Girard Valenciennes Emmanuelle. Partial characterization of the pigments produced by the marine-derived fungus Talaromyces albobiverticillius 30548. Towards a new fungal red colorant for the food industry. J. Food Compos. Anal. 2018;67:38–47. Elsevier. [Google Scholar]
- White T.B., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. PCR Protoc.: A Guide Method Appl. 1990:315–322. https://www.researchgate.net/publication/223397588 [Google Scholar]
- Yaguchi Takashi, Someya Ayako, Miyadoh Shinji, Udagawa Shun-ichi. New variety of Talaromyces wortmannii and some observation on Talaromyces assiutensis. Mycoscience. 1994;35:63–68. [Google Scholar]
- Yilmaz N., Houbraken J., Hoekstra E.S., Frisvad J.C., Visagie C.M., Samson R.A. Delimitation and characterisation of Talaromyces purpurogenus and related species. Persoonia. 2012;29(2012):39–54. doi: 10.3767/003158512X659500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen G.-Y., Li X.-Z., Hu M., Wang B.-Y., Ruan B.-H., Zhou H., Zhao L.-X., Zhou J., Ding Z.-T. Phytotoxic, antibacterial, and antioxidant activities of mycotoxins and other metabolites from Trichoderma sp. Nat. Prod. Res. 2017;2017(31):2745–2752. doi: 10.1080/14786419.2017.1295235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.