Abstract
Claviceps purpurea produces many pharmacologically important ergot alkaloids (EAS), which are widely used to treat migraine and hypertension and to aid childbirth. Although an EAS biosynthetic cluster of C. purpurea has been discovered more than 20 years ago, the complete biosynthetic pathway of EAS has not been fully characterized until now. The main obstacle to elucidating this pathway and strain modification is the lack of efficient genome-editing tools for C. purpurea. The conventional gene manipulation method for C. purpurea relies on homologous recombination (HR), although the efficiency of HR in C. purpurea is very low (∼1–5%). Consequently, the disruption of target genes is laborious and time-consuming. Although CRISPR/Cas9 genome-editing methods based on in vivo Cas9 expression and gRNA transcription have been reported recently, their gene-disruption efficiency is still very low. Here, we developed an efficient genome-editing system in C. purpurea based on in vitro assembled CRISPR/Cas9 gRNA ribonucleoprotein complexes. As proof of principle, three target genes were efficiently knocked out using this CRISPR/Cas9 ribonucleoprotein complex-mediated HR system, with editing efficiencies ranging from 50% to 100%. Inactivation of the three genes, which are closely related to uridine biosynthesis (ura5), hypha morphology (rac), and EAS production (easA), resulted in a uridine auxotrophic mutant, a mutant with a drastically different phenotype in axenic culture, and a mutant that did not produce EAS, respectively. Our ribonucleoprotein-based genome-editing system has a great advantage over conventional and in vivo CRISPR/Cas9 methods for genome editing in C. purpurea, which will greatly facilitate elucidation of the EAS biosynthetic pathway and other future basic and applied research on C. purpurea.
Keywords: Ribonucleoprotein, Genome editing, Ergot alkaloids, Biosynthetic pathway, Homologous recombination
1. Introduction
Claviceps purpurea is a phytopathogenic fungus that grows on cereals and forage grasses. It produces pharmacologically important ergot alkaloids (EAS), such as ergosine, ergocristine, ergotamine, and ergonovine, which have been used to treat migraine, hypertension, or to aid childbirth [1]. An EAS biosynthetic cluster of C. purpurea was first discovered in 1999 [2], and several genes in the cluster have been characterized functionally by gene disruption and analysis of the intermediates [[3], [4], [5], [6], [7]]. However, the P450 enzyme responsible for the formation of key precursor elymoclavine and the non-ribosomal peptide synthases (NRPS) for the biosyntheses of downstream ergosine and ergocristine remained elusive [5]. Candidate genes for the above miss enzymes have been proposed either within or beyond the EAS cluster, efficient gene disruption tools would largely benefit the characterization and validation of these candidates.
The conventional gene-manipulation method in C. purpurea incorporates selection markers into the genome via homologous recombination (HR); however, the efficiency of HR in C. purpurea is very low (1–5%) [[5], [8]]. The disruption of target genes is expensive and time-consuming. Therefore, it is necessary to develop an efficient, versatile genome editing tool for C. purpurea. NHEJ (non-homologous end joining)-deficient background (such as Δku70, ΔkusA and ΔligD) strains are generally used to improve the HR efficiency [9]. Operation of key HR-relating genes (such as Rad51, Rad52, and Mph1) is an alternative strategy [10]. Moreover, longer flanking sequences of a selection marker also facilitate the higher HR efficiency [11]. Recently, the CRISPR/Cas9 system has been successfully engineered in both prokaryotes and eukaryotes [[12], [13], [14]] and has proved to be a feasible, efficient, versatile, genome-engineering tool. This system consists of a Cas9 nuclease and a functional guide RNA (gRNA) [15]. The gRNA recognizes the target sequence in the genome, and the Cas9 nuclease cleaves the sequence to generate double-strand DNA breaks (DSBs) [16]. Then, the cells repair these breaks via nonhomologous end-joining or HR using exogenous DNA (donor DNA), resulting in insertion or deletion mutagenesis at targeted genomic loci [17,18].
CRISPR/Cas9 systems can be established by either in vivo or in vitro strategies. In the in vivo strategy, cassettes expressing Cas9 and gRNA with appropriate promoters are introduced into cells to produce a functional Cas9/gRNA complex in vivo [19]. Recently, an approach for CRISPR/Cas9-mediated mutagenesis in C. purpurea using in vivo expression of Cas9 and gRNA was described [20]. However, this strategy does not work well, since the editing efficiency of CRISPR/Cas9-mediated homology-directed repair was only slightly better than HR-mediated knock-out of the TrpE gene (4/116 and 6/384, respectively). In the in vitro strategy, the purified Cas9 protein and in vitro transcript gRNA are assembled in vitro to generate a functional Cas9/gRNA ribonucleoprotein (RNP) and then transformed into cells for genome editing. This strategy has obvious advantages over the in vivo strategy, as assembly of the Cas9/gRNA complex is not limited by the amount or rate of Cas9 translation or gRNA transcription in vivo. The editing efficiency reached 100% in the filamentous fungi Trichoderma reesei [11,21,22] and Cordyceps militaris [23,24] after optimizing the RNP-based method.
Here, we developed an efficient CRISPR/Cas9-mediated genome-editing method for C. purpurea using the in vitro strategy. Three target genes, which are closely related to uridine biosynthesis (ura5, encoding orotate phosphoribosyl transferase catalyzing the transformation of orotate to OMP in the pyrimidine pathway; accession No. KAG6317199) [25], hypha morphology (rac, encoding small GTPase involved in polarity, sporulation and hyphal growth of C. purpurea; accession No. CAO82105) [26], and EAS production (easA, encoding chanoclavine-I aldehyde oxidoreductase catalyzing chanoclavine-I-aldehyde to form more-complex ergot alkaloids; accession No. Q6ZXC1) [27], were selected to verify the efficiency of this system. In the experiments, all three genes were efficiently knocked out by the CRISPR/Cas9-mediated HR.
2. Materials and methods
2.1. Strains and culture media
The strain C. purpurea 3.1003 was purchased from China General Microbiological Culture Collection Center (CGMCC, Beijing, China), which was grown on potato dextrose agar medium (200 g potato infusion, 20 g dextrose, 20 g agar to 1 L water). For the fermentation of C. purpurea strain, the mycelia were cultivated for 5 days in the seed medium TS (100 g/L sucrose, 10 g/L asparagine, 0.5 g/L KH2PO4, 0.3 g/L MgSO4·7H2O, 0.007 g/L FeSO4·7H2O, 0.006 g/L ZnSO4·7H2O, 0.1 g/L yeast extract, at pH 5.2), then transferred to the fermentation medium B (100 g/L sucrose, 10 g/L citric acid, 1 g/L Ca(NO3)2, 0.5 g/L KH2PO4, 0.25 g/L MgSO4·7H2O, 0.007 g/L FeSO4·7H2O, 0.006 g/L ZnSO4·7H2O, 0.12 g/L KCl, at pH 5.2) and cultivated in the dark at 24 °C on a rotary shaker (200 rpm) for 4 weeks. C. purpurea was grown at 28 °C either for 3 days in CD medium [11] for protoplast preparation, or for 2 days in SDB medium (40 g/L dextrose, 10 g/L yeast extract, 10 g/L tryptone) for genomic DNA isolation. Protoplast regeneration medium was used as described by Zou et al. (2020) [11]. Escherichia coli Top10 used for gene cloning was cultured at 37 °C in Luria Bertani (LB) medium with ampicillin (100 μg/mL). Primers used in this study are listed in Table S2.
2.2. Preparation of target gene, Cas9 protein and gRNAs
Cas9 protein tagged with a nuclear localization signal was purchased from Novoprotein, Inc. (Shanghai, China). Since the genomic information of C. purpurea 3.1003 has not been reported yet, primers were designed to amplify the target genes using C. purpurea 20.1 as a reference. In vitro transcription of gRNA was carried out as previously described by Zou et al. [11]. The in vitro RNA product was incubated with Cas9 at 37 °C for 15 min to form a Cas9-gRNA complex before transformation into protoplasts. The assembling reaction system contained 6 μL Cas9 (1 μg/μL), 30 μL 10 × Cas9 activity buffer (0.2 M HEPES, 1.5 M KCl, 5.0 mM dithiothreitol, 1.0 mM EDTA, 0.1 M MgCl2, pH 7.5), 5 μL gRNA (∼3 μg/μL) and 46 μL 2 × S2 solution (2 M sorbitol, 100 mM CaCl2, 20 mM Tris-HCl, pH 7.5).
2.3. Protoplast preparation, transformation and selection
Mature conidia of C. purpurea were collected with 0.85% NaCl-0.02% Tween 80 and inoculated in 100 mL CD liquid medium for 3 days at 28 °C. The mycelia were then collected by filtration, and washed with solution 1 (1.2 M sorbitol, 0.1 M KH2PO4, pH 5.5). After that, the mycelia were suspended in 20 mL solution 1 with 12.5 mg/L lysing enzymes from Trichoderma harzianum (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 28 °C for 2 h by mild agitation (50 rpm). The protoplasts were then separated from mycelia by filtration through sterilized Miracloth (Sigma-Aldrich). Then they were gently precipitated by centrifugation (2000 × g, 10 min, 4 °C) and washed once again with 4 mL solution 2 (1 M sorbitol, 50 mM CaCl2, 10 mM Tris-HCl, pH 7.5). Finally, the protoplasts were resuspended in solution 2 to a concentration of 107/mL for subsequent transformation. Protoplasts release was checked by microscopic observation. The Cas9-gRNA complex and donor DNA were added into 200 μL of the above protoplast suspension. The aliquot was incubated on ice for 20 min and then solution 3 (2 mL) was added to the aliquot. After 30 min of incubation at 25 °C, solution 2 (4 mL) was added to the mixture. The resulting mixture was then poured into 50 mL melted regeneration medium containing 1.5 mg/mL hygromycin B and divided into three plates. After 7 days cultivation at 28 °C, transformants were inoculated onto a 24-well PDA plate (with 0.5 mg/mL hygromycin B). Colonies were picked and cultured in SDB for genomic DNA extraction. Gene disruption was validated by PCR amplification with primer pairs flanking the target site. All the sequence analyses of transformants were carried out by Sangon Biotech (Shanghai, China).
2.4. Construction of donor DNAs
The donor DNAs containing the 5′ and 3′ flanking sequences of target gene and the selectable marker cassette (Ptrpc-hph-Ttrpc) were generated by overlapping PCR using I5 DNA polymerase (Tsingke, Beijing, China) and ligated into the pMD-18T vector (Takara, Tokyo, Japan). The generated vector was propagated in E. coli Top10 and purified using the Plasmid Midi Kit (Qiagen, Hilden, Germany).
2.5. Analysis of ergot alkaloids
For alkaloid extraction and determination, cultures were extracted twice with ethyl acetate, and concentrated for further alkaloid analysis by HPLC. An Agilent 1100 HPLC system with a RP18-column (5 μm particle diameter, 250 × 4.6 mm) was used to separate and detect the alkaloids. The flow rate was 0.8 mL/min with a gradient elution of 15–95% (v/v) acetonitrile in 10 mM ammonium carbonate for 50 min. Detection was at 320 nm. Authentic ergocristine was purchased from Sigma–Aldrich Analyticals (Taufkirchen, Germany).
3. Results and discussion
3.1. Establishing genetic transformation and in vitro assembled Cas9/gRNA complex for C. purpurea
The PEG-mediated protoplast transformation method was used to establish the genetic transformation system (Fig. 1). C. purpurea protoplasts were collected based on the preparation method used in Aspergillus oryzae with some modifications [11]. As shown in Fig. S1, approximately 107 protoplasts/mL were obtained after incubating mycelia with 12.5 mg/mL lysing enzyme for 2 h, which is sufficient for a transformation experiment. The efficiency of this genetic transformation system was confirmed by transferring the Ptrpc-ble-Ttrpc fragment into the prepared protoplasts. After culturing at 28 °C for 7 days, monophyletic colonies were observed in the regeneration medium (containing phleomycin 30, 60, and 120 μg/mL) (Fig. S2), indicating that the Ptrpc-ble-Ttrpc fragment was successfully inserted into the C. purpurea genome, as confirmed by diagnostic PCR (Fig. S3).
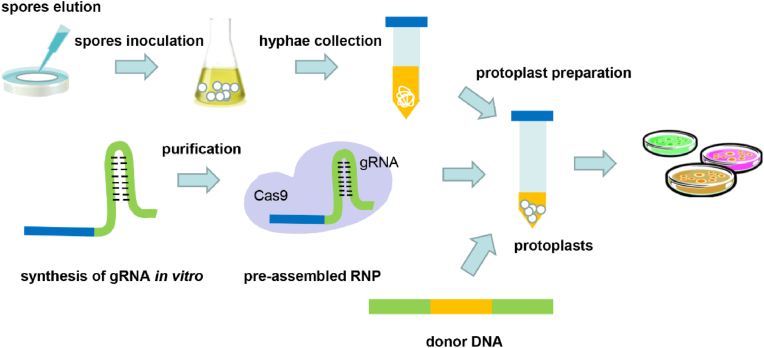
Fig. 1.
Overview of the RNP-based genome-editing system in C. purpurea in this study. Mature C. purpurea conidia were collected using 0.85% NaCl-0.02% Tween 80 and inoculated into 100 mL CD liquid medium. After 3 days, mycelia were collected and then suspended in solution 1 containing lysing enzymes to prepare protoplasts. Protoplasts were resuspended in solution 2 to a concentration of 107/mL for subsequent transformation. RNA prepared in vitro was incubated with Cas9 at 37 °C for 15 min to form a Cas9/gRNA RNP complex. This complex was transformed into protoplasts with donor DNA. Finally, the resulting mixture was poured into 50 mL melted regeneration medium containing 1.5 mg/mL hygromycin B and divided into three plates.
To identify a resistance marker for genetic manipulation, phleomycin was primarily used as a selection agent based on a previous study in C. purpurea [5,8]. The resistance of C. purpurea to phleomycin increased with the pH of the culture medium. On pH 6.0 plates, phleomycin at 40–200 μg/mL lacked toxic effects, while phleomycin had strong restraining effects on a pH 8.0 plate containing 40 μg/mL phleomycin (Fig. S4). However, a high pH was not beneficial for the regeneration of C. purpurea protoplasts. Therefore, hygromycin, which is commonly used for genetic manipulation of fungi, was tested. After culturing at 28 °C for 27 days, mycelia growth was completely inhibited at a minimum inhibitory concentration of 0.5 mg/mL hygromycin on PDA plates (pH 6.0), which was used in the subsequent experiments (Fig. S5).
To develop an in vitro strategy for efficient genome editing in C. purpurea based on CRISPR/Cas9 technology, gRNAs for the target genes were transcribed in vitro via runoff reactions using T7 RNA polymerase. To improve the editing efficiency, gRNAs were designed for each target gene. Then, the in vitro RNA products were incubated with Cas9 protein to generate Cas9/gRNA complexes, according to our previously established CRISPR/Cas9 genome editing approaches for several fungi [11]. Next, the complexes were transformed into C. purpurea via PEG-mediated protoplast transformation (Fig. 1).
3.2. Gene disruption using the RNP-based CRISPR/Cas9 system in C. purpurea
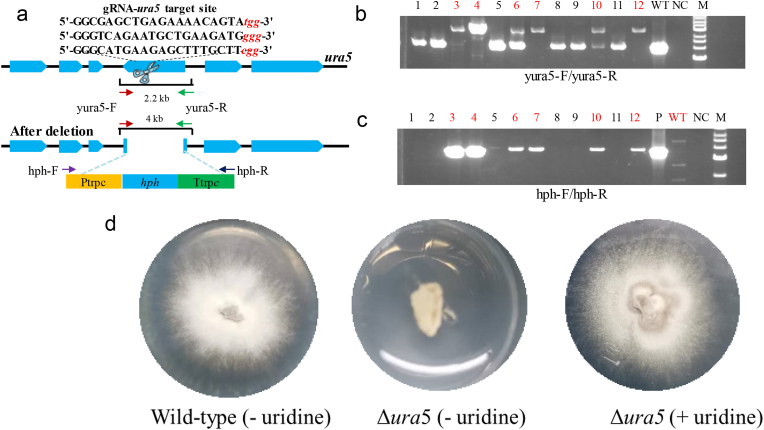
A previous study used an HR strategy for genome editing in C. purpurea. However, HR was inefficient (9 positive transformants of 181 selected) [5]. Here, we first tested the efficiency of the ribonucleoprotein-based CRISPR/Cas9 system to stimulate HR using exogenous DNA (donor DNA) in C. purpurea by targeting the ura5 gene encoding orotate phosphoribosyl transferase; this enzyme converts 5-fluoroorotic acid (5-FOA) into a toxic compound that severely inhibits the growth of wild fungal strains on medium. By contrast, the ura5-disruption mutant survives on plates with additional 5-FOA and uridine in the medium [11]. Three gRNAs were designed to target ura5 (Fig. 2a) and were then incubated with Cas9 protein to generate Cas9/gRNA complexes. After co-transformation of the Cas9/gRNA complexes and donor DNA (containing the hph expression cassette) into C. purpurea, 12 transformants were obtained by hygromycin screening. Successful disruption of the ura5 gene was indicated by a 4 kb PCR fragment produced using primers yUra5-F and yUra5-R and confirmed by sequencing the PCR product. Of the 12 colonies checked, 6 ones showed correct homologous integration (Fig. 2b), representing an editing efficiency of 50%. The PCR sequence from the remaining 6 obtained colonies demonstrated that the hygromycin selection marker was not incorporated into these colonies (Fig. 2c) as well as no mutations were found in all of the three targeted regions of their ura5 genes (Fig. S6). These results indicated that we obtained half true colonies with the expected homologous recombination and half false positive colonies. The phenotypic study showed that the ura5-disruption mutant cannot survive on CD plates without uridine (Fig. 2d). Moreover, the uridine auxotrophic mutant could survive on PDA plates supplemented with 0.3 mg/mL 5-FOA and 10 mM uridine, while the wild-type showed no 5-FOA resistance (Fig. S7).
Fig. 2.
Knock-out of ura5 in C. purpurea by CRISPR/Cas9-mediated homologous recombination (HR). (a) The knock-out strategy for the ura5 gene. (b) Diagnostic PCR using primers yUra5-F and yUra5-R. (c) Diagnostic PCR using primers hph-F and hph-R. (d) The phenotypes of C. purpurea and the ura5-deficient mutant on CD plates with or without uridine cultured for 4 weeks at 28 °C. WT, wild-type; NC, negative control (using water as the amplification template); P: plasmid containing hygromycin selectable marker; M, marker.
The ura5 gene has been used as an effective selection marker for genome editing in fungi [11,28]. As there are few selection markers for genome editing in C. purpurea, the ura5 gene may serve as an additional selectable marker for genome engineering in C. purpurea.
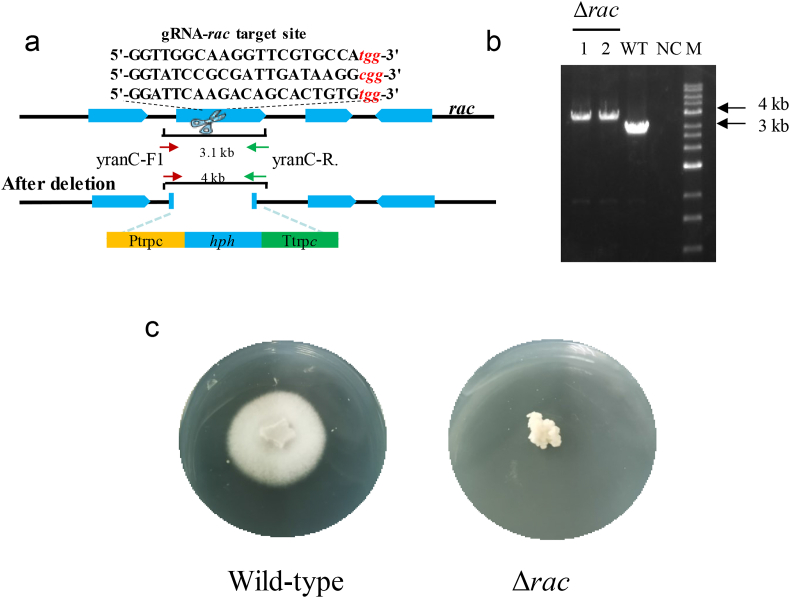
We subsequently tested the efficiency of the ribonucleoprotein-based CRISPR/Cas9 system at stimulating HR in C. purpurea by targeting the rac gene, which strongly influences the morphology of C. purpurea hyphae [26]. Three gRNAs were also designed to target rac (Fig. 3a). After co-transformation of the Cas9/gRNA complexes and donor DNA (containing the hph expression cassette) into C. purpurea, only two transformants were obtained by hygromycin screening. Fortunately, both were positive mutants, as confirmed by diagnostic PCR and Sanger sequencing (Fig. 3b). The rac-disruption mutants had a drastically different phenotype in axenic culture compared with the wild-type; they had a convoluted three-dimensional coralline form (without invading the agar), while the wild-type had a normal flat, two-dimensional form (Fig. 3c), as reported previously [26].
Fig. 3.
Knock-out of rac in C. purpurea by CRISPR/Cas9-mediated homologous recombination (HR). (a) The knock-out strategy for the rac gene. (b) Diagnostic PCR using primers yranC-F1 and yranC-R. (c) The phenotypes of C. purpurea and the Δrac mutant on PDA plates. WT, wild-type; NC, negative control (using water as the amplification template); M, marker.
3.3. Using the RNP-based genome-editing system to confirm that a key enzyme participates in EAS synthesis in C. purpurea
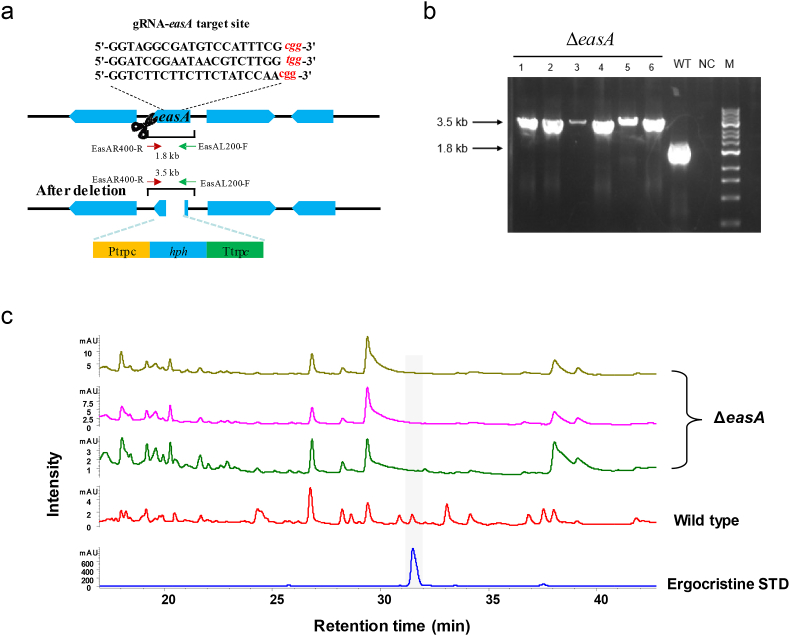
The synthesis of clavines and lysergic acid derivatives starts with the formation of dimethylallyltryptophan (DMAT) from tryptophan and dimethylallylpyrophosphate (DMAPP) catalyzed by the first specific enzyme, i.e., dimethylallyltryptophan synthase (DMATS, coded by dmaW). Then, EasF methylates DMAT into MeDMAT. The generated MeDMAT is catalyzed by chanoclavine synthases (easC and easE) to form tetracylic ergolene, chanoclavine-I. In the presence of EasD, the chanoclavine-I aldehyde is generated. The easA gene was identified to encode chanoclavine-I aldehyde oxidoreductase. Through the catalysis of EasG, agroclavine is produced. However, the P450 for the production of elymoclavine has yet been defined in the EAS pathway. Another P450 (CloA) is responsible for paspalic acid producing. The following process is controversial. Some researchers believe that paspalic acid spontaneously isomerizes to d-lysergic acid. However, some researchers believe that it is catalyzed by an unknown enzyme [27]. In ergopeptine-producing ergot fungi, d-lysergic acid is assembled into the corresponding D-lysergyl peptides by the action of NRPS called lysergyl peptide synthetase 1 and 2 (LPS1 and 2). Among the EAS biosynthetic cluster, easA is a confirmed essential gene for EAS synthesis. Using the knock-out strategy shown in Fig. 4a, the partial easA ORF was replaced by an integrated hph expression cassette. After protoplast transformation, six transformants were obtained on PDA plates with hygromycin B. These transformants were tested by PCR using the primers easA-F/R, and all six transformants produced a 3.5 kb fragment (Fig. 4b), demonstrating that the hph cassette replaced parts of the easA gene. DNA sequencing of the PCR products confirmed that the targeted partial ORF of easA had been knocked out. The editing efficiency of HR and CRISPR/Cas9-mediated HR was compared by introducing dDNA-easA (containing the Ptrpc-hph-Ttrpc cassette and the 3ʹ and 5ʹ flanking regions of easA) into C. purpurea protoplasts. No transformants were obtained by the direct HR approach, indicating that our CRISPR/Cas9 system is an efficient, versatile genome-editing tool for C. purpurea. The easA gene encodes a key enzyme that is thought to take part in the EAS biosynthetic pathway in C. purpurea (Fig. S8) [5]. Disruption of this gene may lead to biosynthetic interruption of the downstream EAS, of which ergocristine is an end product (in red in Fig. S8). Thus, we checked whether the ΔeasA mutants and wild-type produced ergocristine. The wild-type and easA mutant C. purpurea strains were fermented for 4 weeks, and the fermentation broths were extracted with ethyl acetate to analyze the EAS products. HPLC analysis detected no ergocristine in the fermentation extracts of the ΔeasA mutants, while it was detected in the fermentation extracts of the wild-type strain (Figs. 4c and S9). These results indicate that inactivation of easA abolished EAS production in C. purpurea.
Fig. 4.
Knock-out of easA in C. purpurea by CRISPR/Cas9-mediated homologous recombination (HR). (a) The knock-out strategy for the easA gene. (b) Diagnostic PCR using primers EasAL200-F and EasAR400-R. (c) Analysis of the fermentation products of C. purpurea and the ΔeasA mutant. Three ΔeasA transformants were tested. WT, wild-type; NC, negative control (using water as the amplification template); M, marker.
In summary, our CRISPR/Cas9 system based on in vitro assembled Cas9/gRNA RNP effectively stimulates HR in C. purpurea. Compared with the in vivo strategy in C. purpurea, this system has obvious advantages in terms of the process and editing efficiency (Table 1). Moreover, for those strains lacking genomic information, such as C. purpurea 3.1003 used here, it is difficult to establish a CRISPR/Cas9 system via an in vivo strategy, since neither codon-optimized Cas9 genes nor RNA polymerase III promoters can be produced without genomic information. The RNP-based method established here overcomes these difficulties via in vitro transcription of gRNA and purification of Cas9 protein. Because of the transient existence of the RNP complex in cells, in vitro assembled and delivered Cas9/gRNA-based genome editing reduces the risk of unnecessary gene targeting and avoids the integrated mutations that may result from gene-delivery strategies [[29], [30], [31]].
Table 1.
Comparison of in vivo and in vitro CRISPR/Cas9 strategies.
| Strategy | Strain | Genome information | Target gene | HR vs NHEJ | Expression of Cas9 and gRNA | Reference |
|---|---|---|---|---|---|---|
| Plasmids | C. purpurea 20.1 | Reported | Trpe | 4/116 (3.4%) | Expression of Cas9 and gRNA based on transforming plasmids with gRNA and codon-optimized Cas9 gene in vivo | [15] |
| RNPs | C. purpurea 3.1003 | None | easA | 6/6 (100%) | Unrestricted synthesis/expression of Cas9 and gRNA for pre-assembling RNPs in vitro | This study |
In conclusion, we developed a simple, efficient CRISPR/Cas9 system in C. purpurea based on gRNA in vitro transcription. Target genes were efficiently disrupted by CRISPR/Cas9-mediated HR. Based on genomic information, the candidate genes involved in EAS pathway could be deleted efficiently using our developed method. With the advantage of counter selection using 5-FOA, ura5 could be utilized as the bidirectional selection marker for recyclable genome editing. These strategies rapidly achieved simultaneous or stepwise deletion of multiple candidate genes. This work will be useful for metabolic engineering and gene regulation of C. purpurea, as well as elucidating the EAS pathway.
CRediT authorship contribution statement
Lu Yu: Conceptualization, Investigation, Writing – original draft. Meili Xiao: Conceptualization, Investigation, Writing – original draft. Zhihua Zhu: Investigation. Yinmei Wang: Investigation. Zhihua Zhou: Project administration, Conceptualization, Supervision. Pingping Wang: Conceptualization, Writing – review & editing, Supervision. Gen Zou: Conceptualization, Writing – review & editing, Supervision.
Declaration of competing interest
The authors have no interests to declare.
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2018YFA0900500), the National Natural Science Foundation of China (Nos. 31921006, 31470201, and, 31741003), and the Strategic Biological Resources Service Network Plan of the Chinese Academy of Sciences (Grant No. KFJ-BRP-009).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.02.002.
Contributor Information
Lu Yu, Email: yulu31081@126.com.
Meili Xiao, Email: xiaomeili@sippe.ac.cn.
Zhihua Zhu, Email: zhzhu2016@cemps.ac.cn.
Yinmei Wang, Email: wangyinmei@cemps.ac.cn.
Zhihua Zhou, Email: zhouzhihua@sippe.ac.cn.
Pingping Wang, Email: ppwang@cemps.ac.cn.
Gen Zou, Email: zougen@sibs.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Silberstein S.D., Shrewsbury S.B., Hoekman J. Dihydroergotamine (DHE) - then and now: a narrative review. Headache. 2020;60(1):40–57. doi: 10.1111/head.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tudzynski P., Holter K., Correia T., Arntz C., Grammel N., Keller U. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol Gen Genet. 1999;261(1):133–141. doi: 10.1007/s004380050950. [DOI] [PubMed] [Google Scholar]
- 3.Correia T., Grammel N., Ortel I., Keller U., Tudzynski P. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem Biol. 2003;10(12):1281–1292. doi: 10.1016/j.chembiol.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Haarmann T., Machado C., Lubbe Y., et al. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry. 2005;66(11):1312–1320. doi: 10.1016/j.phytochem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Haarmann T., Ortel I., Tudzynski P., Keller U. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. Chembiochem. 2006;7(4):645–652. doi: 10.1002/cbic.200500487. [DOI] [PubMed] [Google Scholar]
- 6.Coyle C.M., Cheng J.Z., O'Connor S.E., Panaccione D.G. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol. 2010;76(12):3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenz N., Olsovska J., Sulc M., Tudzynski P. Alkaloid cluster gene ccsA of the ergot fungus Claviceps purpurea encodes chanoclavine I synthase, a flavin adenine dinucleotide-containing oxidoreductase mediating the transformation of N-methyl-dimethylallyltryptophan to chanoclavine I. Appl Environ Microbiol. 2010;76(6):1822–1830. doi: 10.1128/AEM.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oeser B., Heidrich P.M., Muller U., Tudzynski P., Tenberge K.B. Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet Biol. 2002;36(3):176–186. doi: 10.1016/s1087-1845(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 9.van Leeuwe T.M., Arentshorst M., Ernst T., Alazi E., Punt P.J., Ram A.F.J. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol Biotechnol. 2019;6:13. doi: 10.1186/s40694-019-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai P., Duan X., Wu X., Gao L., Ye M., Zhou Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021;49(13):7791–7805. doi: 10.1093/nar/gkab535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou G., Xiao M., Chai S., Zhu Z., Wang Y., Zhou Z. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb Biotechnol. 2021;14(6):2343–2355. doi: 10.1111/1751-7915.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong L., Ran F.A., Cox D., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 14.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Y., Wei Y., Jiao S., Yu H. A CRISPR/Cas9-based single-stranded DNA recombineering system for genome editing of Rhodococcus opacus PD630. Synthetic Syst. Biotechnol. 2021;6(3):200–208. doi: 10.1016/j.synbio.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Fang H., Zhang D. Expanding application of CRISPR-Cas9 system in microorganisms. Synthetic Syst. Biotechnol. 2020;5(4):269–276. doi: 10.1016/j.synbio.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kralova M., Bergougnoux V., Frebort I. CRISPR/Cas9 genome editing in ergot fungus Claviceps purpurea. J Biotechnol. 2021;325:341–354. doi: 10.1016/j.jbiotec.2020.09.028. [DOI] [PubMed] [Google Scholar]
- 21.Chai S., Zhu Z., Tian E., et al. Building a versatile protein production platform using engineered Trichoderma reesei. ACS Synth Biol. 2021 doi: 10.1021/acssynbio.1c00570. [DOI] [PubMed] [Google Scholar]
- 22.Zou G., Zhou Z. CRISPR/Cas9-Mediated genome editing of Trichoderma reesei. Methods Mol Biol. 2021;2234:87–98. doi: 10.1007/978-1-0716-1048-0_8. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Yang Z., Bao D., et al. Improving hypoxia adaption causes distinct effects on growth and bioactive compounds synthesis in an entomopathogenic fungus Cordyceps militaris. Front Microbiol. 2021;12:698436. doi: 10.3389/fmicb.2021.698436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou G., Li B., Wang Y., et al. Efficient conversion of spent mushroom substrate into a high value-added anticancer drug pentostatin with engineered Cordyceps militaris. Green Chem. 2021;23(24):10030–10038. doi: 10.1039/D1GC03594K. [DOI] [Google Scholar]
- 25.Wyka S.A., Mondo S.J., Liu M., Dettman J., Nalam V., Broders K.D. Whole-genome comparisons of ergot fungi reveals the divergence and evolution of species within the genus claviceps are the result of varying mechanisms driving genome evolution and host range expansion. Genome Biol Evol. 2021;13(2) doi: 10.1093/gbe/evaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolke Y., Tudzynski P. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol Microbiol. 2008;68(2):405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- 27.Young C.A., Schardl C.L., Panaccione D.G., et al. Genetics, genomics and evolution of ergot alkaloid diversity. Toxins. 2015;7(4):1273–1302. doi: 10.3390/toxins7041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R., Chen L., Jiang Y., Zhou Z., Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3 doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24(6):1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuris J.A., Thompson D.B., Shu Y., et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.