Synopsis
Skeletal muscle is responsible for voluntary force generation across animals, and muscle architecture largely determines the parameters of mechanical output. The ability to analyze muscle performance through muscle architecture is thus a key step towards better understanding the ecology and evolution of movements and morphologies. In pennate skeletal muscle, volume, fiber lengths, and attachment angles to force transmitting structures comprise the most relevant parameters of muscle architecture. Measuring these features through tomographic techniques offers an alternative to tedious and destructive dissections, particularly as the availability of tomographic data is rapidly increasing. However, there is a need for streamlined computational methods to access this information efficiently. Here, we establish and compare workflows using partially automated image analysis for fast and accurate estimation of animal muscle architecture. After isolating a target muscle through segmentation, we evaluate freely available and proprietary fiber tracing algorithms to reconstruct muscle fibers. We then present a script using the Blender Python API to estimate attachment angles, fiber lengths, muscle volume, and physiological cross-sectional area. We apply these methods to insect and vertebrate muscle and provide guided workflows. Results from fiber tracing are consistent compared to manual measurements but much less time-consuming. Lastly, we emphasize the capabilities of the open-source three-dimensional software Blender as both a tool for visualization and a scriptable analytic tool to process digitized anatomical data. Across organisms, it is feasible to extract, analyze, and visualize muscle architecture from tomography data by exploiting the spatial features of scans and the geometric properties of muscle fibers. As digital libraries of anatomies continue to grow, the workflows and approach presented here can be part of the open-source future of digital comparative analysis.
Resumo [Brazilian Portuguese]
O músculo esquelético é responsável pela geração de força voluntária em animais, e a arquitetura muscular determina em grande parte os parâmetros de performance mecânica. A capacidade de analisar o desempenho muscular através da arquitetura muscular é, portanto, um passo fundamental para uma melhor compreensão da ecologia e evolução dos movimentos e morfologias. No músculo esquelético penado, o volume, o comprimento das fibras e os ângulos de fixação às estruturas de transmissão de força constituem os parâmetros mais relevantes da arquitetura muscular. A medição dessas propriedades por meio de técnicas tomográficas oferece uma alternativa às dissecções tediosas e destrutivas, especialmente pelo rápido aumento na disponibilidade de dados tomográficos. No entanto, há uma necessidade de métodos computacionais otimizados para acessar essas informações de forma eficiente. Aqui, estabelecemos e comparamos fluxos de trabalho usando análise parcialmente automatizada de imagem para estimativa rápida e precisa da arquitetura muscular em animais. Após isolar um músculo alvo por meio de segmentação, avaliamos os algoritmos disponíveis gratuitamente e pagos para o rastreamento de fibra (Fiber Tracing) para reconstruir as fibras musculares. Apresentamos um script que usa a API do Blender Python para estimar os ângulos de ligamento, o comprimento da fibra, o volume do músculo e a secção transversal fisiológica. Aplicamos esses métodos aos músculos de insetos e vertebrados e fornecemos instruções detalhadas de uso. Os resultados do rastreamento de fibra são consistentes em comparação com as medições manuais, mas requerem menos tempo. Finalmente, enfatizamos a capacidade do software de modelagem de código aberto Blender 3D tanto como uma ferramenta de visualização programável quanto uma ferramenta analítica para processar dados anatômicos digitalizados. Para muitos organismos, é possível extrair, analisar e visualizar a arquitetura muscular, explorando as características espaciais das tomografias e as propriedades geométricas das fibras musculares. À medida que as bibliotecas de anatomia digital continuam a crescer, os fluxos de trabalho e a abordagem apresentados aqui podem fazer parte do futuro da análise comparativa digital de código aberto.
Resumen [Spanish]
El músculo esquelético es responsable de la generación de fuerza voluntaria en los animales y la arquitectura muscular determina en gran medida los parámetros de producción mecánica. La capacidad de analizar el rendimiento muscular a través de la arquitectura muscular es, por tanto, un paso clave hacia una mejor comprensión de la ecología y evolución de los movimientos y morfologías. En el músculo esquelético pennado, el volumen, la longitud de las fibras y los ángulos de fijación a las estructuras de transmisión de fuerza comprenden los parámetros más relevantes de la arquitectura muscular. La medición de estas características mediante técnicas tomográficas ofrece una alternativa a las disecciones tediosas y destructivas, sobre todo porque la disponibilidad de datos tomográficos está aumentando rápidamente. Sin embargo, existe la necesidad de métodos computacionales optimizados para acceder a esta información de manera eficiente. Aquí, establecemos y comparamos flujos de trabajo utilizando análisis de imágenes parcialmente automatizados para una estimación rápida y precisa de la arquitectura del músculo animal. Después de aislar un músculo objetivo a través de la segmentación, evaluamos algoritmos de trazado de fibras (“Fiber Tracing”) patentados y gratuitos para reconstruir las fibras musculares. Presentamos un script que utiliza la API de Blender Python para estimar los ángulos de penación, la longitud de las fibras, el volumen muscular y el área de sección transversal fisiológica. Aplicamos estos métodos a los músculos de insectos y vertebrados y proporcionamos flujos de trabajo guiados. Los resultados del rastreo de fibras son consistentes en comparación con las mediciones manuales, pero requieren mucho menos tiempo. Por último, enfatizamos las capacidades del software 3D de código abierto Blender como herramienta de visualización y herramienta analítica programable para procesar datos anatómicos digitalizados. Por muchos organismos, es factible de extraer, analizar y visualizar la arquitectura muscular mediante la explotación de las características espaciales de las tomografías y las propiedades geométricas de las fibras musculares. A medida que las bibliotecas digitales de anatomías continúan creciendo, los flujos de trabajo y el enfoque presentados aquí pueden ser parte del futuro de código abierto del análisis comparativo digital.
Kurzfassung [German]
Die Skelettmuskulatur ist verantwortlich für die willentliche Krafterzeugung in Tieren und die Muskelarchitektur bestimmt weitgehend mechanisch relevante Parameter. Die Muskelleistung anhand der Muskelarchitektur analysieren zu können ist daher ein Schlüssel zum besseren Verständnis der Ökologie und Evolution von Bewegungen und Morphologien. In gefiederten Muskeln sind Volumen, Faserlängen, und Ansatzwinkel an kraftübertragenden Strukturen die wichtigsten Parameter der Muskelarchitektur. Diese Eigenschaften durch Verfahren in der Tomographie zu erfassen, bietet eine Alternative zur aufwendigen und irreversiblen Sektion, zumal die Verfügbarkeit tomographischer Daten rasch zunimmt. Es besteht jedoch ein Bedarf an standardisierten digitalen Analysemethoden, um effizient auf diese Informationen zuzugreifen. In dieser Arbeit entwickeln und vergleichen wir Abfolgen von Arbeitsschritten für eine genaue und schnelle Analyse der Muskelarchitektur. Nachdem ein Zielmuskel durch Segmentierung digital isoliert ist, werten wir sowohl frei verfügbare als auch kommerzielle Algorithmen der Faserverfolgung (“Fiber Tracing”) aus und rekonstruieren so Muskelfasern. Wir präsentieren ein Skript, welches die Blender-Python-API zur Berechnung von Ansatzwinkeln, Faserlängen und Physiologischer Querschnittsfläche verwendet. Wir wenden diese Methoden auf Insekten- und Vertebratenmuskel an und stellen dafür detaillierte Anleitungen zur Verfügung. Die Ergebnisse der Faserverfolgung stimmen mit manuellen Messungen überein, sind aber viel weniger zeitaufwändig. Zuletzt betonen wir hier die Möglichkeiten der Open-Source Software Blender für sowohl 3D Visualisierungen als die auch automatisierte Analyse in der Verarbeitung digitalisierter anatomischer Daten. Für verschiedenste Organismen besteht die Möglichkeit die Muskelarchitektur aus Tomographien zu extrahieren, analysieren und zu veranschaulichen, indem Bildeigenschaften von Scans und der gleiche Aufbau von Muskelfasern ausgenutzt werden. Bibliotheken, die die Anatomie digital katalogisieren, wachsen, und Arbeitsschritte und Anwendungsmöglichkeiten, die wir hier vorstellen, können Teil der offeneren und faireren Zukunft der digitalen vergleichenden Analyse sein.
要約 [Japanese]
骨格筋は、動物の随意的な力の生成を担っており、骨格筋の構造は、力学的出力パラメータを大きく左右する。そのため骨格筋の構造学的要素を通して筋パフォーマンスを解析することは、運動や形態の生態や進化をよりよく理解するための重要なステップとなる。羽状筋では、体積、線維長、力伝達構造への付着角度といったものが、筋構造を構成する最も重要なパラメータとなる。これらの特徴を断層撮影技術によって測定することは、複雑で不可逆的な解剖の代替法であり、とりわけ断層撮影データの利便性は急速に高まっている。 ただし、情報に効率的にアクセスするための正確な計算方法が必要である。本稿では、動物の筋肉構造を高速かつ正確に推定するため、部分的自動画像解析を用いたワークフローを確立・比較する。セグメンテーションによってターゲットの筋を分離した後、筋線維を再構築するために、自由に利用できる線維トレーシング・アルゴリズムと独自のアルゴリズムを評価する。次にBlender Python APIを使用し、付着角度、線維長、筋体積、生理学的断面積を推定するスクリプトを示す。これらの方法を昆虫と脊椎動物の筋肉に適用し、ガイド付きのワークフローを提供する。線維トレーシングからの結果は、手動測定と比較して一貫性があるとともに、はるかに時間が節約できる。最後に、デジタル化された解剖学的データ処理用の可視化ツール兼スクリプト可能な分析ツールとして、オープンソースの3DソフトウェアBlenderの機能に焦点を当てる。生物全体において、スキャンの空間的特徴と筋線維の幾何学的特性を利用することで、断層撮影データから筋構造を抽出、分析、可視化できる。解剖学のデジタルライブラリが増え続ける中、ここで紹介したワークフローとアプローチは、デジタル比較解析のオープンソースの未来の一部となり得る。
Introduction
The rise of X-ray computed micro-tomography (XCT) techniques in organismal research has led to an increase in quantifiable, digital morphological data (Friedrich et al. 2014; Buser et al. 2020). Now, the rate of processing the large amounts of raw images is limiting research more than their acquisition (Muñoz and Price 2019). Skeletal muscle is relevant to analyze for various questions in organismal biology, it can be resolved three-dimensionally (3D), but it is difficult to quantify effectively. Skeletal muscle as a hierarchically organized fibrous tissue that contracts to generate force is present across the animal kingdom (Huxley and Niedergerke 1954; Huxley and Hanson 1954; Hodge 1956; Pringle 1967), and the quantitative analysis of muscle can link form with function. Muscles are resolved in XCT when contrast-enhancing agents are used to stain specimens, and protocols have been developed for various animals (Metscher 2009a; Faulwetter et al. 2013). Using controlled drying methods or synchrotron XCT, staining is not even necessary (van de Kamp et al. 2015). So far, however, only a few studies tackled a more thorough quantitative analysis of muscle regarding its fibrous structure using XCT (Kupczik et al. 2015; Dickinson et al. 2018; Nyakatura et al. 2019; Sullivan et al. 2019; Peeters et al. 2020; Püffel et al. 2021). The hierarchical fibrous nature of muscle offers possibilities to apply standardized computational methods to quantify this functionally relevant tissue across taxa.
Muscle architecture
Skeletal muscle is characterized by a hierarchical organization of fibrous, contractile units on different scales, and a highly variable composition of these units. The explanatory power of muscle anatomy for performance is degraded when muscle is quantified purely based on volume or weight (Sacks and Roy 1982). A reliable, anatomy-based approach to estimate function and performance of skeletal muscle is to quantify muscle architecture (Sacks and Roy 1982; Powell et al. 1984; Lieber and Friden 2000; Lieber and Ward 2011). Muscle architecture is defined as the arrangement of muscle fibers and fascicles, which are bundles of fibers (Gans and Bock 1965; Lieber and Friden 2000). Muscle volume, varying fiber length and, if the muscle is pennate, attachment angle are the most encountered metrics of muscle architecture. Small changes in pennation angle and fiber lengths result in significant differences in force output (Stark et al. 2008). Comparing pennation angles alone is useful due to an inherent trade-off of more parallel attaching fibers transmitting force more efficiently with obliquely attaching fibers utilizing space constraints better (Gans and Bock 1965; Sacks and Roy 1982; Paul and Gronenberg 1999). Physiological cross-sectional area (PCSA) is a summary value calculated from muscle architecture parameters to estimate performance as it was found to be proportional to force generation (Powell et al. 1984; Lieber and Ward 2011). Total muscle force can be estimated from PCSA by multiplication with muscle-specific tension, which is the muscle's force generation capability per unit area (Sacks and Roy 1982; Powell et al. 1984). A single value for specific tension has been suggested for all mammalian muscle, and benchmarks exist for the variation found in muscles of other animal groups (Lieber and Ward 2011; Weihmann et al. 2015). However, a unified model for skeletal muscle has not been achieved and it might be impossible to do so, emphasizing the importance of effectively gathering various muscle architecture parameters (Winkel 2020). Skeletal muscle is heterogeneously arranged at all levels, but the principles of hierarchical muscle structure can be exploited to estimate function from anatomy.
Muscle architecture is usually estimated based on dissections, but several imaging-based methods have been popularized. The reconstruction of muscle architecture is most advanced in the biomedical field, where dissection techniques can be substituted with sonomicrometry or diffusion-based imaging to allow for in vivo measurements of human muscle (Franchi et al. 2018; Bilston et al. 2019). The most common method to reconstruct 3D muscle architecture in biomedical applications is through muscle diffusion tensor imaging (mDTI) whose key strength is that orientation information is stored in each image increment during acquisition (Damon et al. 2017). However, mDTI is expensive and exclusively tailored towards humans. Measuring muscle architecture through dissections has been extended to invertebrates. For example, by cutting tissue in the position of apodemes, endoskeletal cuticle serving similar functions to tendons, the pennation angle of mandible muscles was compared across ants (Paul and Gronenberg 1999; Paul 2001). As dissections are highly destructive and in vivo techniques are difficult to realize in animals, ultimately, non-invasive XCT imaging techniques harbor the potential to analyze muscle architecture more completely across animal taxa.
Digitally analyzing muscle architecture using XCT
Across animals, the hierarchical structure, and the homogeneity in voxel colors along fibers paired with a contrast to the surrounding tissues depicts fibrous tissue as such in XCT images. Vertebrate muscle fibers are bundled into fascicles that are sheathed by perimysium. Using iodine staining and sufficient resolution, these fascicles and perimysium show different contrasts and the fibrous structure of muscle is revealed with XCT (Jeffery et al. 2011). Successful attempts to resolve muscle fascicles have been made for diverse vertebrate taxa, such as rats and mice, squirrels, dogs, pigs, monkeys, and birds (Jeffery et al. 2011; Kupczik et al. 2015; Dickinson et al. 2018; Nyakatura et al. 2019; Sullivan et al. 2019). Using the right XCT setup, it is even possible to resolve contrast between myofibers and their very fine surrounding sheaths of endomysium below resolutions of 10 µm per voxel (Jeffery et al. 2011). Resolving individual muscle fibers for whole muscles is easier when the organism is smaller. Scans of small-bodied invertebrates routinely resolve muscle fibers as resolution is often below 5 µm. Effective staining is easy to implement in small-bodied animals and exoskeletons provide a natural protection from deformations (Metscher 2009a). Following best practices to resolve fibrous tissues in tomograms in addition to publishing the data on open-source platforms, the availability of XCT scans that resolve muscle fibers or fascicles will continuously increase to allow broader comparative scales.
To process these large, volumetric images and quantify fibrous tissue, several automatic techniques have been developed. A wider array of solutions exists for two-dimensional (2D) images, for example, implemented in the freely available image analysis software ImageJ (Encarnacion-Rivera et al. 2020). The only program of our knowledge developed for muscle architecture analysis in 3D is ImageXd (Kupczik et al. 2015). Sullivan et al. (2019) used a fiber tracking approach within the software Avizo to trace fascicles in several muscles around the pectoral girdle of a starling. This approach was applied to thoracic muscles in ants to compare workers and queens in the sister software Amira (Peeters et al. 2020). The algorithm was developed focusing on microtubuli (Redemann et al. 2014; Westenberger and Blanc 2016). Amira/Avizo fiber tracing requires a template, which corresponds to the fraction of fiber that is then algorithmically placed across the tomogram constrained by several input parameters. ImageXd in contrast directly estimates possible directionality in voxels based on the texture and stores the resulting values as vectors (Kupczik et al. 2015; Dickinson et al. 2018). Püffel et al. (2021) designed a custom tracing method for near linear fibers that requires nothing but segmentations of highly resolved target anatomy. Automated fiber tracing techniques using XCT data give the advantages of reproducible and extensive digital measurements and access to the 3D structure of specimens without the necessity to destroy specimens. If these automated techniques produce reliable results, they hold the potential to enable analysis of muscle architecture across the tree of life.
Quantifications of anatomy and biomechanics using XCT data are still scarce, but comparative studies benefit from muscle architecture as data to correlate with other traits. Muscles between closely related, yet ecologically divergent squirrels differ corresponding to their life histories (Nyakatura et al. 2019). Intra-colony analysis of muscle architecture in leafcutter ants showed that these polymorphic ants tend to capitalize on muscle volume to generate stronger individuals (Püffel et al. 2021). Architectural parameters of thoracic muscles of ponerine ant workers indicate adaptations for strength as compared to their queens (Peeters et al. 2020). Extending such analyses to more comparative datasets can give important insights into the evolution of behaviors and ecological adaptations.
Aims of this paper
Here, we present and compare methods to analyze and visualize muscle architecture from XCT. As a case study, we estimate muscle architecture of the pharaoh ant Monomorium pharaonis, an insect that only measures about 2.5 mm in total. We trace muscle fibers using ImageXd as a freely available software and Amira XTracing as a proprietary software. We further include the recently published methods from Püffel et al. (2021). We aim to establish that soft tissue tomographic data will yield consistent results when subjected to different architecture reconstruction techniques as we compare the three approaches to manually measured validation data. Additionally, we use the published results from Sullivan et al. (2019) to extend analysis from insect to vertebrate muscle and compare our findings to theirs. We cover both an invertebrate and a 50.000 times more voluminous vertebrate muscle to test the workflows on phylogenetically distant and physiologically different specimens. We then provide detailed summaries of the fiber tracing workflows to promote the future use of muscle architecture as a collection of traits in comparative analyses.
Materials and methods
To compare and validate fiber tracing and muscle architecture estimation, we chose a specimen of the ant M. pharaonis and focused on the mandible closer muscle (M. craniomandibularis internus M.11/0md1). Due to the small size of M. pharaonis, it has a tractable number of muscle fibers to manually count and measure attachment angles relative to the direction of force production. Specimen CASENT0709255 is part of the Global Ant Genomics Alliance (GAGA) project (Boomsma et al. 2017). The specimen resided in 99% ethanol until it was transferred to iodine staining solution (2%) for several weeks. Prior to scanning, we washed the specimen in 99% ethanol and chose a plastic pipette tip filled with 99% ethanol as a specimen holder. We scanned M. pharaonis using a Zeiss Xradia 510 Versa 3D X-ray microscope equipped with a 20x magnification lens. As the model for testing various techniques of muscle architecture reconstruction, we cropped the scan towards the volume of the left closer muscle and rotated the scan so that the y-axis is in line with the apodeme to manually estimate muscle architecture parameters (Fig. 1). The tomogram is characterized by isometric voxel resolution of 0.67 µm³ with acquisition settings of 13 s exposure time, 50 kV at 4 W, and 18 mm distance between X-ray source and microscope. To save computation time for the free software fiber tracing approach, we resampled the image stack and reduced resolution down to 2 µm³ voxel size in 3D Slicer (URL: https://slicer.org; Fedorov et al. 2012; Kikinis et al. 2014). Prior to resampling, we enhanced contrast using an unsharp mask in Fiji (sigma 5 px, weight 0.75; Schindelin et al. 2012).
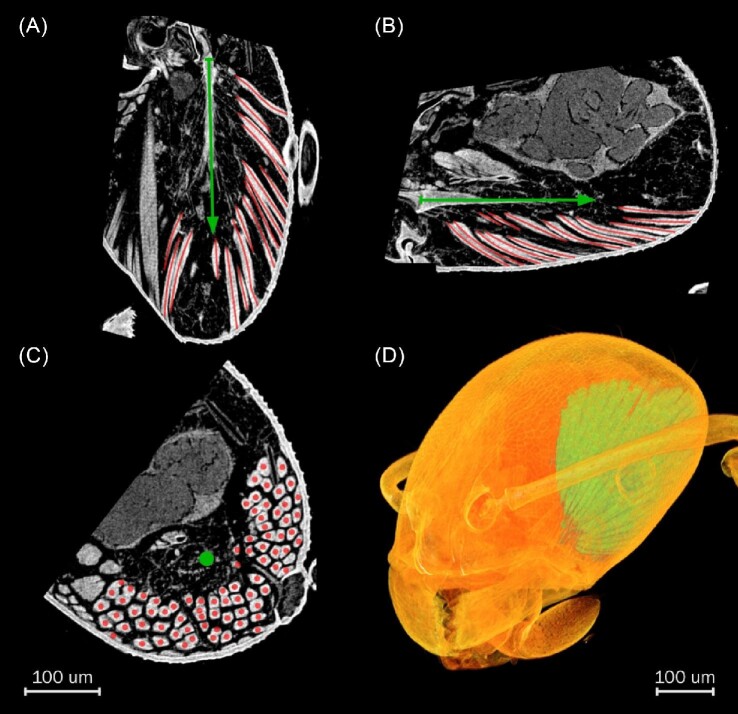
Fig. 1.
Contrast-enhanced XCT resolves individual muscle fibers in the M. pharaonis head. (A, B, C) Axial, sagittal, and coronal slices through the M. pharaonis left closer muscle. Apodeme highlighted as green arrow pointing in direction of force generation. Red lines and dots indicate muscle fibers. (D) Volume rendering of the M. pharaonis head visualizing the closer muscle that occupies much space in the left hemisphere.
We further accessed a tomogram of the starling Sturnus vulgaris (URL: https://morphosource.org, Specimen ID: 000S30577) to estimate the bird's M. supracoracoideus muscle architecture. Details of specimen preparation and scanning are elaborated in Sullivan et al. (2019). Briefly, soft tissue of S. vulgaris was stained with iodine and the resolution of this scan is isometric with 43.86 µm3 per voxel, which resolves muscle fascicles.
All workflows we present here require segmentation, the digital dissection of tissues. To save time and improve results, we used Biomedisa to drastically reduce segmentation effort and improve accuracy (URL: https://biomedisa.de; Lösel et al. 2020). Here, only every nth slice (in this case, every fifth) requires presegmentation and after uploading the segmentation to the platform, the full segmentation is interpolated. Masking is a crucial step for fiber tracing where segmentation is used to extract only the areas of the tomogram that show the target tissue. The principle of masking is to preserve the gray values of segmented voxels and replace the values of all non-segmented voxels with 0 (resulting in the color black), and to save the result as a new, cleaner image stack. Optionally, image filters can be applied to the image before or after masking to enhance contrast, sharpness, or reduce artifacts and noise.
Voxel-transformation-based fiber tracing methods
Fascicles or individual muscle fibers are traced according to patterns in the texture of XCT images. Both ImageXd and Amira XTracing process the grayscale voxel data to instead produce new 3D images, where regions corresponding to fibrous structures are highlighted. The algorithms exploit the assumption that in a digital image, contrast between neighboring voxels along fibers is lower than in perpendicular directions. Fibers are tracked within the newly generated images and are stored as connected points in a 3D space of the same dimensionality as the original image stack.
The Supplements feature detailed walkthroughs on how to estimate muscle architecture by voxel-based fiber tracing using proprietary or free software. At the initial step of both workflows, we expect that X-ray projections have already been reconstructed into an XCT image stack. The last steps to retrieve muscle architecture parameters from fiber files are identical using our custom Blender-Python script.
ImageXd version 4.1.3 (Heiko Stark, Jena, Germany, URL: https://starkrats.de)
ImageXd is a freely available software to process volumetric data. By writing out a script (imagexd.macro), the fiber tracking analysis can be performed based on the approach presented in Kupczik et al. (2015). The basic principle of ImageXd fiber tracing is to form streamlines from a field of orientation vectors. The orientation vectors derive from individual voxels, where a sphere with a given voxel radius is computed to find the direction of least variation in contrast. Streamlines are the most plausible connections of vectors. The only data input ImageXd requires to trace fibers from XCT data is an image stack in NIfTI-format showing the fibrous tissue. After loading the stack, a vector field is calculated in a computationally demanding step. For large files and depending on the computer available, it may be advisable to resample the image stack to reduce computation time. However, the number of vectors relates to voxels. Hence, higher resolution results in a more detailed vector field. Upon processing the vector field, it should be smoothed to facilitate the tracing of streamlines. Streamlines are traced from seed points set all throughout the masked vector field. To make streamline calculation more biologically adequate and easier to handle for visualization, streamlines can be pruned based on various parameters and number and location of seeds can be constrained.
Workflow using Fiji, 3D Slicer, and ImageXd (see Supplements)
For this workflow, Fiji as an extended ImageJ is most important for changing file types and to process the image stack to adjust contrast. Here, we applied unsharp masking and Gaussian blurring to emphasize the fibrous texture. We recommend using 3D Slicer to further adjust the image stack, perform muscle segmentations, and trace the direction of force production. In 3D Slicer, we produced pre-segmentations limited to voxel intensity thresholds that we chose to interpolate with Biomedisa. Alternatively, 3D Slicer on its own possesses an extensive set of segmentation tools. From complete muscle segmentation, we used the “Mask Volume” tool (“SegmentEditorExtraEffects” extension) to obtain an isolated version of the muscle within the image stack. This masked stack is then exported to NIfTI file format. For S. vulgaris, we further segmented the tendon, used the “Margin” segmentation tool to grow the segmentation, and exported the resulting region to NIfTI to constrain seed points in ImageXd. To trace the orientation of the tendon/apodeme, we use the “Create and Place – Fiducial” tool and export the four points into FCSV-format that our custom script can read in.
ImageXd is best executed via command line and works with adjusting an “imagexd.macro” script. Within ImageXd, there are several parameters to limit the final number of streamlines and restrict the fiber tracing based on a priori information. We set input and output to the desired file names and set muscle-specific parameters. To compute the transformation of greyscale-voxels into vectors without border artifacts, the regular image stack or a generously masked muscle can be used. The vector field is then smoothed and subsequently masked with the muscle segmentation to constrain seeds and to reduce border artifacts. We sampled references lengths from the image stack and set those values to constrain the streamlines to avoid unrealistically short or long fibers. We further constrained the tracing of streamlines by allowing each path to only be occupied by one streamline and limited segment angles. In the case of S. vulgaris, we restricted the seeds to the area surrounding the tendon and we found it necessary to allow streamlines to be traced more easily. Otherwise, resulting fibers were consistently too short. ImageXd can produce various file formats to represent fibers. For our custom script, we set the output to SWC (Cannon et al. 1998).
Amira 2019.2 XTracing (FEI, Thermo Fisher Scientific)
Template-based fiber tracking in Amira searches for as many fibers as possible matching the given constraints of a cylindrical template and expected contrast (Redemann et al. 2014; Westenberger and Blanc 2016). Presegmentation is carried out in the “Segmentation Editor,” data are masked either with the “Mask” or “Arithmetic” tools, and various image filters are available if necessary. Fibers are tracked based on contrast within the masked correlation result, the orientation from template fitting and other input parameters, such as expected curvature, length of fibers, and distance between fibers.
Amira workflow (see Supplements)
As with 3D Slicer, we interpolate from a voxel-intensity-limited presegmentation using Biomedisa. Alternatively, Amira itself offers several segmentation tools. We attach a “Cylinder Correlation” module from the Amira XTracing extension to the image stack and sample a template either by adjusting a visual example or using previously measured parameters. To generate the outputs from the “Cylinder Correlation” module, we recommend using a grown mask or the full image stack to not generate border artifacts. We recommend masking after running the cylinder correlation because masking after cylinder correlation reduces border artifacts. For fiber tracing, we set necessary values, particularly minimum fiber length and distance from fiber to fiber. The last step is to export the fibers to SWC. The tendon/apodeme is sampled in Amira using the “Landmark Editor” and exported to ASCII format. All modules used here should also exist in Avizo under slightly different names (Sullivan et al. 2019). In both programs, however, purchasing an extension to the base program is necessary.
Fibers projected from seeds; Püffel et al. (2021)
For M. pharaonis, we include the fiber tracing method recently published in Püffel et al. (2021), which was applied to mandible muscles of the polymorphic ant Atta vollenweideri. Here, the muscle segmentation is intersected with a surrounding, dilated cuticle segmentation. When the muscle fibers are highly resolved, the intersecting area represents muscle fibers as discs. The center points of these discs are then used as seeds to project straight lines towards the apodeme within the muscle segmentation to optimize fiber length and pennation angle.
Complete fiber segmentation to validate automated fiber tracing
To verify the automated fiber tracing methods, we sampled the muscle fibers of M. pharaonis manually. First, we individually segmented each fiber. We created several copies of the original XCT scan rotated around the y-axis to sample pennation angles manually. Slices showing the apodeme allowed us to estimate the direction of force production as a simple 2D projection. We manually measured pennation angles in fibers summing up to 305 measurements from 171 different fibers. We then averaged the resulting angles from multiple measurements of the same fiber. We further used the complete segmentation of muscle fibers to estimate individual fiber lengths in Blender (https://www.blender.org). We converted the segmentations into meshes and as the muscle was previously oriented along the y-axis, we measured the distance between those vertices having the most extreme values in y for each fiber. As muscle fibers shrink individually in ethanol-stored insects and XCT methods can resolve the space in between muscle fibers in good contrast, calculating muscle volume from segmenting fibers alone does not present a reliable value. Therefore, in Amira, we used the “Grow” operation on the fiber segmentation until the space in between fibers was filled. We then subtracted a segmentation of the head cuticle from this dilation to remove excess material and measured volume as number of segmented voxels times voxel volume.
Blender Python script to estimate muscle architecture
With a custom script, we use the point data from automated approaches to estimate, summarize, and visualize muscle architecture using the Python API of the open-source 3D software Blender. Traced fibers are stored and read into Blender first as arrays of individual points that make up the fiber. We included several steps into the script to improve the quality of the fiber dataset (listed and explained in the Supplemental Data Sheet). The summed distances of all subsequent points per fiber constitute fiber length. To estimate pennation angles, we use the orientation of fibers in 3D space and define the direction of force production through locating the tendon/apodeme. We estimate muscle volume with an input parameter referring to fiber thickness. We convert the fiber curves to a mesh, remesh it according to fiber diameter, and then extend the resulting volume by fiber diameter. By using a loose unification of fibers, we include space in between fibers and account for tissue shrinkage. Remeshing is carried out with the “Remesh Modifier.” Subsequently, the mesh is copied, smoothed (“Subsurface Modifier”), and grown by fiber diameter (“Shrinkwrap Modifier”). Lastly, as a measure of performance, we calculate PCSA2 based on Sacks & Roy (1982) as:
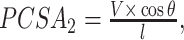
where V is muscle volume, Θ is mean pennation angle, and l is mean fiber length. Running the script results in a Blender scene and several .csv files that contain individual fiber parameters to calculate statistics and a summary including PCSA2 (Table 1).
Table 1.
Commonly encountered metrics for muscle architecture are calculated and written out by our custom Blender-Python script.
| Output file | Data | Explanation |
|---|---|---|
| out_angles.csv | Pennation angles [deg] | Calculated pennation angle for each fiber as angle towards tendon/apodeme |
| out_lengths.csv | Fiber lengths [mm] | Calculated fiber length for each fiber as sum of point-to-point distances |
| out_summary.csv | Volume [mm3] | Muscle volume from muscle fibers recalculated as single mesh |
| Average length [mm] | Sum of fiber lengths by n fibers | |
| Average angle [deg] | Sum of pennation angles by n fibers | |
| N fibers | Total number of fibers | |
| PCSA2 [mm2] | PCSA as PCSA2 = volume*cos(angle)/avg length |
Visualization
The Blender scene initially shows fibers and tendon in three versions: normalized to length 1 at the scene origin, as connected points, and straight from start- to endpoint. The curves are shown as “NURBS,” which allows smooth visualization. In the script, we included code that sets two panels in the “Scene Properties” tab in Blender to change the colorization of straightened fibers from gold to a gradient based on pennation angle or fiber length in “Render View” (see Fig. 4). The “Color Ramp” defining the gradient colors can be modified in the “Shader Editor.” The tendon/apodeme is visualized in default color green at scene origin and where the points were sampled. The blender scene can be customized further with additional data input, such as meshes from segmentations. The usage of Blender for visualizations is then subject to user preference and skills in Blender.
Fig. 4.
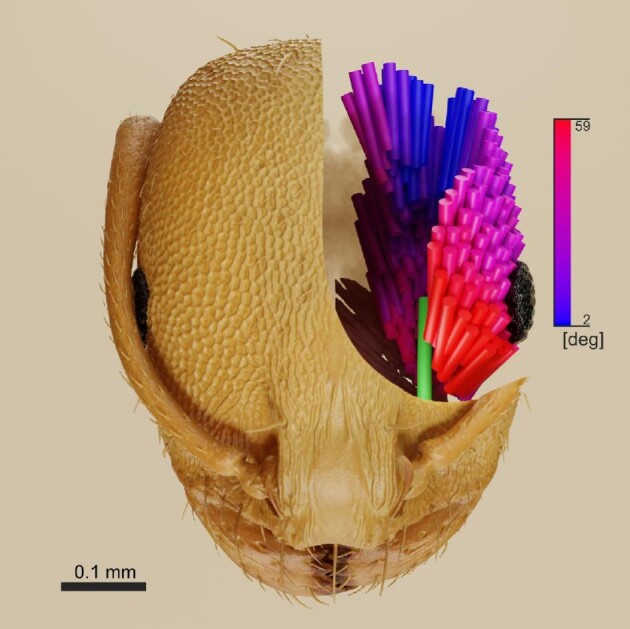
Monomorium pharaonis mandible closer muscle fibers colored according to attachment angle embedded in a rendering of the head cuticle. Fiber attachment angle: high = red, low = blue. Apodeme = green. Gradient extremes are set between minimum and maximum pennation angle. All head colors and material properties are artificial based on microscope photographs of M. pharaonis.
Statistical methods to compare the three fiber tracing approaches
We used statistical tests to compare the distributions of pennation angles and fiber lengths (see Supplemental Data R Code). We used the Shapiro–Wilks statistic to test for non-normality in the distribution (R Core Team 2019). We then chose t-tests, Kolmogorov–Smirnov, and Mann–Whitney tests to compare the resulting distributions of angles and lengths against the references and against each other in R (R Core Team 2019). For M. pharaonis, we further computed the Kullback–Leibler divergence (R package “philentropy”; Drost 2018) of random samples from fiber tracking results against the manual measurements to estimate what approaches yielded results closer to the ground-truthed distribution of pennation and fiber lengths. In addition, we plotted the distributions to visually inspect the differences in results from the fiber tracing approaches.
Results
As a case study, we report below the results from digitally reconstructing muscle architecture parameters for the ant M. pharaonis against measurements from manual segmentation of individual fibers. We further report results for the starling S. vulgaris using the same automated approaches against published results that incorporate dissection and the template-based approach that we reproduced here.
Monomorium pharaonis closer muscle architecture
All muscle fibers are filament-attached, which is one of three fiber types in ant mandible muscle theorized to capitalize on strength per volume (Paul and Gronenberg 1999). Configurations including other fiber types were previously reported for small myrmicine ants, such as Wasmannia affinis (Richter et al. 2019). The muscle generates force via attaching to a simple apodeme and is held in place by attaching to the dorsal and lateral left hemisphere of the head. Muscle fibers even attach to the cuticular carina surrounding the eye. They are separated where the optic nerve connects brain and eye. Further, a dorsomedial portion of the muscle appears disconnected from the main muscle body, but that might be an artifact of tissue shrinkage.
Number of fibers
Through semiautomatic segmentation, we established that our specimen of M. pharaonis has exactly 223 fibers in its left closer muscle. Using template matching in Amira XTracing and running our custom script on the results, fiber tracking results in 223 fibers as well. Using ImageXd, we overestimate the number of fibers recovered from using the original-sized dataset with 301 fibers. However, using the sharpened and resampled image stack, we retrieve 220 fibers. Using the intersection of fibers and head for sampling fiber seeds, we recover 190 fibers.
Muscle volume
We inflated the segmentation of the muscle fibers to cover all the space occupied by the closer muscle to account for shrinkage (manual in Table 2). Compared to this measurement, the script recovers a slightly larger volume for all approaches. The difference between manual and automatic calculation is less than 10%. However, as we have no data for a fresh M. pharaonis, all data are subject to error resulting from tissue shrinkage.
Table 2.
Estimated muscle architecture values for M. pharaonis and S. vulgaris from different approaches. Column ‘Script Fiber diam’ relates to an estimated value of individual fiber/fascicle diameter from sampling fiber width in image slices plus minimally accounting for tissue shrinkage. Other columns relate to output from the Blender-Python script. Rows S. vulgaris ‘Dissection’ and ‘Amira published’ from Sullivan et al. (2019). ‘*’ indicates volume estimation from segmentation in Amira.
| Specimen | Method | Pennation angle [deg] | Muscle volume [mm3] | Fiber lengths [mm] | PCSA2 [mm2] | Script fiber diameter [mm] | N fibers |
|---|---|---|---|---|---|---|---|
| Monomorium pharaonis | Manual | 27.3 (SD 11.8, min 3.0, max 59.0) | 0.0083(*) | 0.114 (SD 0.014, min 0.064, max 0.155) | 0.067 | - | 223 |
| Amira | 27.3 (SD 12.0, min 2.1, max 59.2) | 0.0089 | 0.106 (SD 0.021, min 0.050, max 0.177) | 0.074 | 0.018 | 223 | |
| ImageXd | 24.7 (SD 11.0, min 2.5, max 57.9) | 0.0089 | 0.106 (SD 0.023, min 0.065, max 0.167) | 0.076 | 0.018 | 301 | |
| ImageXd-Resampled | 23.2 (SD 10.4, min 1.0, max 55.1) | 0.0086 | 0.111 (SD 0.026, min 0.065, max 0.172) | 0.071 | 0.018 | 220 | |
| Puffel et al. (2021) | 28.01 (SD 12.66, min 3.78, max 72.56) | 0.0083(*) | 0.117 (SD 0.023, min 0.049, max 0.186) | 0.063 | - | 190 | |
| Sturnus vulgaris | Dissection | 18.7 (SD 8.79) | 559 | 12.7 (SD 3.67) | 41.6 | - | - |
| Amira published | 23.5 (SD 12.46) | 690 | 10.3 (SD 3.60) | 61.4 | - | 218 | |
| Amira reproduced | 19.76 (SD 9.71, min 1.80, max 43.03) | 610.09 (*628.5) | 10.81 (SD 3.96, min 5.47, max 29.16) | 53.11 | 0.35 | 226 | |
| ImageXd | 19.72 (SD 11.06, min 0.63, max 52.22) | 616.36 | 10.23 (SD 3.5, min 4.92, max 19.99) | 56.71 | 0.35 | 1331 |
Pennation angle
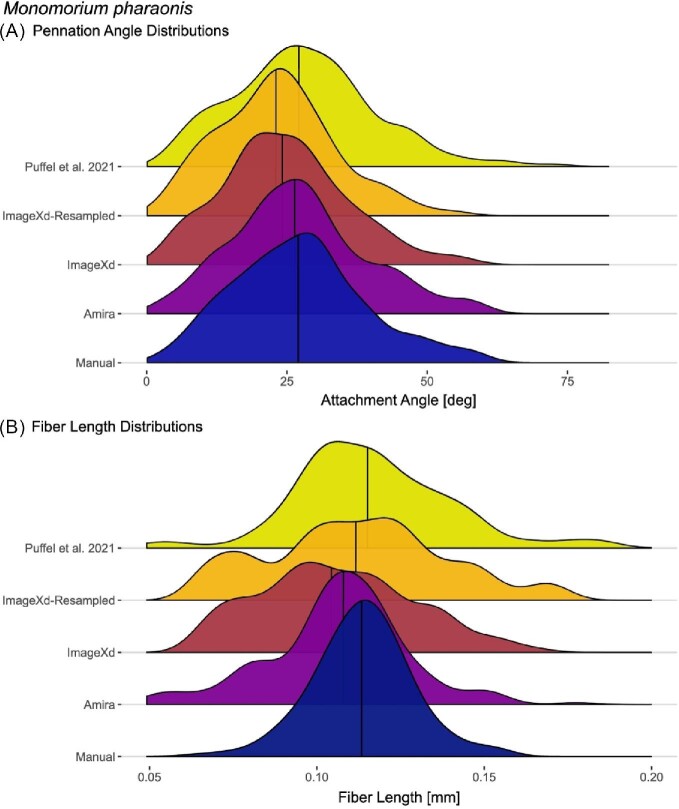
We calculated an average angle of 27.3° (SE = 0.69°) through manually sampling fibers in axial and sagittal sections with the apodeme fully visible (Table 2). The mean of differences in angle between fibers sampled more than once is 1.89°. The Amira and Püffel et al. (2021) approaches are nearly identical to this ground-truthed average pennation angle. Tracing from ImageXd led to a slightly lower mean of 24.7°. Using the resampled stack with the same parameters in ImageXd, the average angle remained lower still at 23.65°. Out of the implemented statistical methods, only the ImageXd-Resampled approach differs significantly from the manual measurements and other approaches. None of the four different approaches can outcompete the others consistently when calculating the Kullback–Leibler divergence with all of them falling around 0.3. Plotting a distribution of pennation angles from the traced fibers reveals the congruence of the chosen methods in more detail, as they all recover heterogeneity in pennation angles (Fig. 2A). Many fibers attach at angles around the total mean around 27°. However, there are other slight second peak at above 40° that is recovered in all methods and another at 15°.
Fig. 2.
The distribution of pennation angles for the left M. pharaonis mandible closer muscle is consistent across different approaches whereas fiber lengths recovered from automated methods tend to have a broader distribution. (A) For attachment, next to the peak around 25°, all automated methods show bumps at above 40° and 10° angle also present in the manual measurements. (B) Fiber lengths estimated from individual fiber segmentation (manual) are uniformly distributed whereas all automated methods show local peaks. Horizontal lines show mean values.
Fiber lengths
Using the segmentation of each fiber, average length is at 0.114 mm (SE = 0.001 mm), which agrees with ImageXd and Amira each at 0.106 mm. Using ImageXd with the resampled image data and the method from Püffel et al. (2021), the results are even closer at 0.111 mm and 0.117 mm, respectively. In contrast to automated fiber tracing methods, the distribution of lengths from manual segmentation is just normal (p = 0.05). From the chosen statistical tests, only the ImageXd-Resampled approach consistently fails to be significantly different from the manual measurements. However, according to Kullback–Leibler divergence, the Amira and Püffel et al. (2021) approaches are closer to the manual measurements. Plotting the fiber length distribution of all approaches reveals the differences among the approaches further (Fig. 2B). In both ImageXd approaches, the curves are flattened, and the resulting values are more spread than the manual measurements. The total peak of fiber lengths is close to the mean in all methods. However, fibers are always present close to the chosen cutoff lengths and the voxel-based tracking methods recover a few fibers longer than reasonable.
Physiological cross-sectional area
We approximate force generation capabilities by estimating PCSA. PCSA2 calculation, which incorporates volume, fiber length, and pennation angle resulted in 0.067 mm² using manual measurements and individual fiber segmentation. In general, the ImageXd and Amira approaches differ from this value by less than 15%. ImageXd and Amira result in higher values with 0.076 mm² and 0.074 mm² from the full-sized image-stack, respectively. Using the resampled image stack with ImageXd and the Püffel et al. (2021) method yielded the closest PCSA2 of 0.071 mm2 and 0.063 mm2 due to slightly less estimated volumes and longer fibers.
Sturnus vulgaris M. supracoracoideus muscle architecture
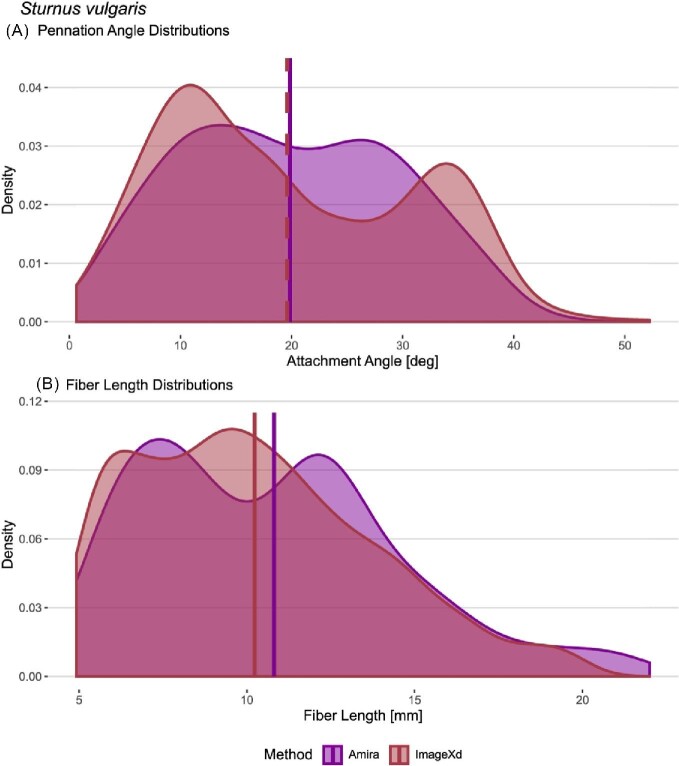
Similar patterns as in M. pharaonis emerge when comparing different approaches of estimating muscle architecture parameters for the starling specimen. Overall, calculating the parameters with our script reproduces the results from Sullivan et al. (2019) when tracing fascicles with Amira and ImageXd (Table 2). None of the results are normally distributed in accordance with the bipennate muscle arrangement. Statistical tests fail to find significant differences between the pennation angle results (P > 0.05). The same goes for a Mann–Whitney test on fascicle lengths (P = 0.06). Only the Kolmogorov–Smirnov test is significant for lengths (P = 0.005). However, the distance metric remains low between the distributions (D = 0.13). Looking at the distributions of pennation angles and fascicle lengths, the different automated approaches produce almost identical results in the means with slight differences in distribution shapes (Fig. 3).
Fig. 3.
Automated fiber tracing approaches yield consistent results in pennation angles and fiber length distributions for the M. supracoracoideus of S. vulgaris. Both ImageXd and Amira recover a bimodal distribution of pennation angles and lengths, which fits the bipennate architecture of the muscle. Amira XTracing does not support a cutoff for maximum length. However, unrealistically long fibers (>20 mm) had a negligible effect on the results. Horizontal lines show mean values.
Discussion
We compared methods of fiber tracing to estimate muscle architecture parameters. Our evaluation shows that ImageXd, Amira, and the methods published in Püffel et al. (2021) produce results comparable to manual measurements, previously published data using the same methods, and dissection-based measurements. The overall consistency between automated approaches and validations provides evidence that digital muscle architecture analysis from XCT scans is a reliable tool to generate comparative data. For S. vulgaris, reproduced results are comparable with those from Sullivan et al. (2019) with both ImageXd and Amira XTracing showing a tendency to underestimate fiber lengths given their validation through dissection. Our method to track the direction of force production shows pennation angles more in agreement with the dissection than the Amira XTracing results from the original publication (see Table 2). In contrast to our general underestimation, previous fascicle lengths in mammalian muscles were overestimated using ImageXd, but that may have been due to tissue shrinkage in the time between scanning and dissection (Kupczik et al. 2015; Dickinson et al. 2018). For M. pharaonis, the two workflows yielded accurate results in muscle architecture compared to our manual measurements. The Amira XTracing template matching and ImageXd streamline approaches resulted in less than 15% deviation in PCSA2. In Püffel et al. (2021), the approach was validated by sampling fibers across Atta ants and by estimating the expected error rates. As they found high accuracy in the method, we also find high agreement compared to manual measurements in M. pharaonis with only 6% difference in PCSA2. As we recover them for bird and ant, pennation angles are often reported to be heterogeneous within muscles (Paul and Gronenberg 1999; Lieber and Friden 2000). XCT image-based analysis of muscle in situ is not only less destructive, but also offers more measurements than a sample of dissected fibers/fascicles. Furthermore, automated approaches are much faster and less labor intensive than sampling angles manually. As the same structural principles of muscle apply at different scales, fiber tracing approaches are versatile enough to study heterogeneous muscle architecture across organisms. Especially when the methods from Püffel et al. (2021) or ImageXd are applied to trace muscle fibers, only freely available software is necessary and the only limitations are available scans, scan quality, and computing power.
However, there are caveats for comparative digital muscle architecture reconstruction that represent areas for future work. First, even when the same animal could be fixed multiple times, results would likely vary because of artifacts and the state of muscle contraction. Stark and Schilling (2010) showed that muscle architecture in mammals varies considerably in all parameters measured here depending on contraction. Further, all measurements after chemical preservation may be affected by tissue shrinkage and the amount of shrinkage has been reported to increase over time in mammals (Hedrick et al. 2018). The extensive methods-oriented literature on contrast-enhanced XCT targets different tissues and taxa (e.g., Metscher 2009a, 2009b; Gignac et al. 2016; Smith et al. 2016; Davies et al. 2017). To better understand preservation artifacts, future studies could quantify shrinkage and then improve fixation and staining protocols. However, it is possible to select specimens with muscles fixed in a similar state of action using the same chemical agents to hold a potential error constant in a comparative approach. Until shrinkage issues are clarified and accounted for, we address shrinkage in highly resolved fibers by interpolating over empty space between muscle fibers within our Blender-Python script.
Second, processing all voxels into directional information is computationally demanding, so one might face a trade-off of accuracy with computation time. To address this, it is possible to mask regions of interest, but border artifacts are a threat to accuracy (Kupczik et al. 2015). For ImageXd, resampling the M. pharaonis image stack led to slightly worse results as we traded off contrast against computation time. However, computation time decreased dramatically from two days to less than an hour. Sensitivity analysis for Amira/Avizo template matching showed that fiber tracking was improved by investing in heavier computation efforts through reducing the “Angular Sampling” parameter (Sullivan et al. 2019). Higher image quality and more computation time improve results using the automated approaches and future work could specifically optimize the trade-off between computation time and data size.
Third, all automated approaches require manual adjustments, which introduces potential error sources. Chiverton et al. (2018) focused on an algorithmic technique to estimate fiber diameters from XCT data, which might be useful to implement into fiber tracing tools. Fiber diameters within a muscle are variable, which is especially problematic for Amira template-based tracking (Redemann et al. 2014; Sullivan et al. 2019). However, sensitivity analysis showed overall invariance for the various set parameters in Amira XTracing (Sullivan et al. 2019). ImageXd fiber tracing is initially agnostic calculating vectors and streamlines purely based on image texture, but therefore, streamline numbers need not represent fiber number. Sensitivity analysis for ImageXd suggested that a random sample of seed-points does not considerably affect results in fiber lengths (Dickinson et al. 2018). However, we initially recovered results biased to regions of the image where fibers are better resolved affecting the means. Therefore, we first use as many seeds as there are voxels and then reduce streamlines based on fiber diameter. To control for excessively long or short fibers, Kupczik et al. (2015) used the distribution of lengths of all streamlines to identify unrealistic accumulations to set cutoffs in fascicle length. We instead sampled those values directly from the images. For the masseter muscle analyzed in Kupczik et al. (2015), a grid in the central part of the muscle was defined to equal number of seeds to fascicles. This was not feasible here due to the radial arrangement of fibers in both muscles. Instead, for the S. vulgaris muscle, we chose to constrain the location of seeds by using the tendon. To find an accurate number of seeds, Püffel et al. (2021) chose the intersections between muscle fibers and cuticle to produce geometrically similar objects whose number represents the number of fibers and whose centers depict the centers of muscle fibers. This approach implemented for M. pharaonis shows accurate results in number and should be easily extended to other arthropod taxa and muscles. For biomechanics, accurate numbers of fibers could then extend estimations from muscle fiber mechanical analysis (Shi et al. 2021) as an alternative to muscle specific tension and PCSA. Despite the necessary extra-tasks that come with the different approaches, they are significantly faster than segmenting fibers individually or dissecting but perform equally well in muscle architecture reconstruction according to our results.
Other tools for fiber tracing
We attempted to implement two other software for fiber tracing rooted in the material sciences. We tried to trace fibers using the Python-based quanfima (Shkarin et al. 2019) and the program FeatureScout (Weissenbock et al. 2014) in the standalone software package Open_iA (Fröhler et al. 2019), which draws from the “Insight Toolkit” iTk (Yoo et al. 2002; McCormick et al. 2014). However, both require more intensive pre-processing, such as individual fiber segmentation or complete skeletonization. The segmentation of individual fibers introduces a serious bottleneck that has been approached by multiple methods in the field of material sciences (Salaberger et al. 2011; Heinzl et al. 2018). In our specimens, however, we found fibers to be too heterogenous, so sophisticated preprocessing algorithms would be necessary to separate fibers at areas of little contrast. Therefore, we only report here results from methods that require quick segmentations for masking.
Alternative workflows to reconstruct muscle architecture
We present a custom Blender-Python script to specifically retrieve muscle architecture parameters directly including volume without a necessity to rearrange the entire image stack. However, it is possible to retrieve muscle architecture parameters using other programs. The Graphical User Interface software Cloud2 (Heiko Stark, Jena, Germany, URL: https://starkrats.de) is scriptable and can process data generated from ImageXd or other applications. Within Cloud2, it is possible to calculate further properties of the fibers and estimate muscle architecture. For calculations regarding pennation, this must be done towards the axis planes of the image stack coordinate system (Dickinson et al. 2018). To align muscle and tendon effectively towards these planes, image stacks can either be readjusted before or the whole coordinate system can be recalculated in Cloud2. In Amira/Avizo, when the image stack is aligned correctly towards the direction of force production, tensors that represent pennation angle can be recovered (Sullivan et al. 2019). For a fiber object in Amira, errors can be cleaned by manipulating the object and a spreadsheet can be displayed with information for each tracked fiber. We here provide an alternative to rearranging a whole image stack by tracking the direction of force production directly as a vector and provide a script for Blender that is dedicated to calculating muscle architecture parameters including volume from fibers.
Visualizing muscle architecture
When working with 3D organismal data, it is often important to produce figures that effectively visualize the data and convey further information to the reader immediately. Alternative to Blender, Amira can dynamically change the thickness of fibers, and color can be set according to fiber properties (Sullivan et al. 2019). Cloud2 not only offers to visualize fibers as streamlines but can further convey information on the vector field (Kupczik et al. 2015). Despite an initial steep learning curve, Blender is an excellent tool to consider because only few settings will generate high-quality renders with the options to employ more realistic materials and lighting. In Blender, one has a variety of options to modify the data into formats for 2D figures, simple animations or animations for movement simulation also considering physical properties, interactive 3D models, and meshes for 3D printing. Our final image rendering of M. pharaonis shows data directly calculated within the script. It has the goal to be aesthetically pleasing but also effectively convey the information of heterogenous pennation angles within the closer muscle through a color gradient (Fig. 4).
Conclusions
Access to data after publishing in online repositories, the rise of high-throughput XCT, and large-scale digitization efforts are offering researchers the opportunities to analyze soft tissue anatomy for more taxa and larger numbers of datasets. Based on our results, digital fiber tracing is a reliable tool to make use of these datasets for which we lay out analysis workflows that allow estimating muscle architecture and function comparatively at intra- and interspecific levels or within specimens for different muscles. Existing fiber tracing tools offer opportunities to gain deeper knowledge about muscle architecture in a digital image-based approach. We hope here to promote the usage of those and provide an additional tool to analyze and visualize digitally traced muscle fibers or fascicles for comparative biology. Image-based analysis of skeletal muscle can become a powerful addition to the repertoire of techniques within biomechanics and comparative biology. Especially when researchers aim to gain insight into the evolution of musculature, muscle architecture is a key variable to be quantified. Digital muscle architecture reconstruction not only improves accuracy but also amplifies the possibilities of measuring muscle architecture on larger scales across the tree of life.
Supplementary Material
Acknowledgements
We thank the Scientific Computing and Data Analysis section and Imaging Section of the Research Support Division at Okinawa Institute of Science and Technology Graduate University (OIST) for supporting different aspects of this work and Mari Takenouchi of the Communication and Public Relations Division at OIST for help in translating the abstract to Japanese. DeepL (https://deepl.com) and Google Translate (https://translate.google.com) were used to generate preliminary abstract translations. We thank Kosmas Deligkaris for aid in setting up ImageXd on computing servers, Alexandre Casadei-Ferreira for advice on 3D Slicer and Brazilian abstract translation, and Lazzat Aibekova for testing the workflows. We thank Francisco Hita Garcia for help on the Spanish abstract translation. We greatly appreciate the support of Frederik Püffel in applying his recently published fiber tracing methods to our ant. Special thanks to Samuel “Spiro” P. Sullivan for providing data, giving insights into his Avizo workflow, and helping to reproduce originally published results. We want to express our gratitude for the work done by the reviewers.
Contributor Information
Julian Katzke, Biodiversity and Biocomplexity Unit, Okinawa Institute of Science and Technology Graduate University, Onna, 904-0495 Okinawa, Japan.
Pavel Puchenkov, Scientific Computing and Data Analysis Section, Research Support Division, Okinawa Institute of Science and Technology Graduate University, Onna, 904-0495 Okinawa, Japan.
Heiko Stark, Institute of Zoology and Evolutionary Research, Friedrich Schiller University Jena, Fürstengraben 1,07743 Jena, Germany.
Evan P Economo, Biodiversity and Biocomplexity Unit, Okinawa Institute of Science and Technology Graduate University, Onna, 904-0495 Okinawa, Japan.
Funding
This work was supported by funding from the Okinawa Institute of Science and Technology Graduate University. The funders had no role in study design.
Declaration of Competing Interest
The authors declare no competing interests.
Data Availability
A run of the script at the state of publication can be done using supplemental data. Script versions will be updated under: https://github.com/julesforfools/get_fibers. Full data available at: doi:10.5061/dryad.4mw6m90c3.
References
- Bilston LE, Bolsterlee B, Nordez A, Sinha S. 2019. Contemporary image-based methods for measuring passive mechanical properties of skeletal muscles in vivo. J Appl Physiol 126:1454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma JJ, Brady SG, Dunn RR, Gadau J, Heinze J, Keller L, Moreau CS, Sanders NJ, Schrader L, Schultz TRet al. 2017. The Global Ant Genomics Alliance (GAGA). Myrmecol News 25:61–6. [Google Scholar]
- Buser TJ, Boyd OF, Cortés Á, Donatelli CM, Kolmann MA, Luparell JL, Pfeiffenberger JA, Sidlauskas BL, Summers AP.. 2020. The Natural historian's guide to the CT galaxy: step-by-step instructions for preparing and analyzing computed tomographic (CT) data using cross-platform, open access software. Integr Org Biol 2:obaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV.. 1998. An on-line archive of reconstructed hippocampal neurons. J. Neurosci. Methods 84:49–54. [DOI] [PubMed] [Google Scholar]
- Chiverton JP, Kao A, Roldo M, Tozzi G.. 2018. Automatic diameter and orientation distribution determination of fibrous materials in micro X-ray CT imaging data. J Microsc 272:180–95. [DOI] [PubMed] [Google Scholar]
- Damon BM, Froeling M, Buck AK, Oudeman J, Ding Z, Nederveen AJ, Bush EC, Strijkers GJ.. 2017. Skeletal muscle diffusion tensor-MRI fiber tracking: rationale, data acquisition and analysis methods, applications and future directions. NMR Biomed 30:e3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TG, Rahman IA, Lautenschlager S, Cunningham JA, Asher RJ, Barrett PM, Bates KT, Bengtson S, Benson RB, Boyer DMet al. 2017. Open data and digital morphology. Proc Biol Sci 284:20170194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson E, Stark H, Kupczik K.. 2018. Non-destructive determination of muscle architectural variables through the use of diceCT. Anat Rec (Hoboken) 301:363–77. [DOI] [PubMed] [Google Scholar]
- Drost HG. 2018. Philentropy: information theory and distance quantification with R. J Open Source Softw 3:765. [Google Scholar]
- Encarnacion-Rivera L, Foltz S, Hartzell HC, Choo H.. 2020. Myosoft: an automated muscle histology analysis tool using machine learning algorithm utilizing FIJI/ImageJ software. PLoS One 15:e0229041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulwetter S, Dailianis T, Vasileiadou A, Arvanitidis C.. 2013. Contrast enhancing techniques for the application of micro-CT in marine biodiversity studies. Microsc Microanal 27:S4–7. [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka Met al. 2012. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi MV, Raiteri BJ, Longo S, Sinha S, Narici MV, Csapo R.. 2018. Muscle Architecture assessment: strengths, shortcomings and new frontiers of in vivo imaging techniques. Ultrasound Med Biol 44:2492–504. [DOI] [PubMed] [Google Scholar]
- Friedrich F, Matsumura Y, Pohl H, Bai M, Hörnschemeyer T, Beutel RG.. 2014. Insect morphology in the age of phylogenomics: innovative techniques and its future role in systematics. Entomol Sci 17:1–24. [Google Scholar]
- Fröhler B, Weissenböck J, Schiwarth M, Kastner J, Heinzl C.. 2019. open_iA: a tool for processing and visual analysis of industrial computed tomography datasets. J Open Source Software 4:1185. [Google Scholar]
- Gans C, Bock WJ.. 1965. The functional significance of muscle architecture: a theoretical analysis. Adv Anat Embryol Cell Biol 38:115–42. [PubMed] [Google Scholar]
- Gignac PM, Kley NJ, Clarke JA, Colbert MW, Morhardt AC, Cerio D, Cost IN, Cox PG, Daza JD, Early CMet al. 2016. Diffusible iodine-based contrast-enhanced computed tomography (diceCT): an emerging tool for rapid, high-resolution, 3-D imaging of metazoan soft tissues. J Anat 228:889–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick BP, Yohe L, Vander Linden A, Davalos LM, Sears K, Sadier A, Rossiter SJ, Davies KTJ, Dumont E.. 2018. Assessing soft-tissue shrinkage estimates in museum specimens imaged with diffusible iodine-based contrast-enhanced computed tomography (diceCT). Microsc Microanal 24:284–91. [DOI] [PubMed] [Google Scholar]
- Heinzl C, Amirkhanov A, Kastner J. 2018. Processing, analysis and visualization of CT data. In: Carmignato S, Dewulf W, Leach R, editors. Industrial X-Ray Computed Tomography. New York, NY: Springer Cham. p. 99–142. [Google Scholar]
- Hodge AJ. 1956. The fine structure of striated muscle; a comparison of insect flight muscle with vertebrate and invertebrate skeletal muscle. J Biophys Biochem Cytol 2:131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R.. 1954. Structural changes in muscle during contraction. Nature 174:971–3. [DOI] [PubMed] [Google Scholar]
- Huxley H, Hanson J.. 1954. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 173:973–6. [DOI] [PubMed] [Google Scholar]
- Jeffery NS, Stephenson RS, Gallagher JA, Jarvis JC, Cox PG. 2011. Micro-computed tomography with iodine staining resolves the arrangement of muscle fibres. J Biomech 44:189–92. [DOI] [PubMed] [Google Scholar]
- Kikinis R, Pieper SD, Vosburgh KG.. 2014. 3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support. In: Jolesz FA, editor. Intraoperative Imaging and Image-Guided Therapy. New York, NY: Springer New York. p. 277–89. [Google Scholar]
- Kupczik K, Stark H, Mundry R, Neininger FT, Heidlauf T, Rohrle O.. 2015. Reconstruction of muscle fascicle architecture from iodine-enhanced microCT images: a combined texture mapping and streamline approach. J Theor Biol 382:34–43. [DOI] [PubMed] [Google Scholar]
- Lieber RL, Friden J.. 2000. Functional and clinical significance of skeletal muscle architecture. Muscle Nerve 23:1647–66. [DOI] [PubMed] [Google Scholar]
- Lieber RL, Ward SR.. 2011. Skeletal muscle design to meet functional demands. Philos Trans R Soc Lond B Biol Sci 366:1466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lösel PD, van de Kamp T, Jayme A, Ershov A, Farago T, Pichler O, Tan Jerome N, Aadepu N, Bremer S, Chilingaryan SAet al. 2020. Introducing Biomedisa as an open-source online platform for biomedical image segmentation. Nat Commun 11:5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M, Liu X, Jomier J, Marion C, Ibanez L.. 2014. ITK: enabling reproducible research and open science. Front Neuroinform 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD. 2009a. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD. 2009b. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn 238:632–40. [DOI] [PubMed] [Google Scholar]
- Muñoz MM, Price SA.. 2019. The future is bright for evolutionary morphology and biomechanics in the era of big data. Integr Comp Biol 59:599–603. [DOI] [PubMed] [Google Scholar]
- Nyakatura JA, Baumgarten R, Baum D, Stark H, Youlatos D.. 2019. Muscle internal structure revealed by contrast-enhanced μCT and fibre recognition: the hindlimb extensors of an arboreal and a fossorial squirrel. Mamm Biol 99:71–80. [Google Scholar]
- Paul J. 2001. Mandible movements in ants. Comp Biochem Physiol A: Mol Integr Physiol 131:7–20. [DOI] [PubMed] [Google Scholar]
- Paul J, Gronenberg W. 1999. Optimizing force and velocity: mandible muscle fibre attachments in ants. J Exp Biol 202:797–808. [DOI] [PubMed] [Google Scholar]
- Peeters C, Keller RA, Khalife A, Fischer G, Katzke J, Blanke A, Economo EP.. 2020. The loss of flight in ant workers enabled an evolutionary redesign of the thorax for ground labour. Front Zool 17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell PL, Roy RR, Kanim P, Bello MA, Edgerton VR.. 1984. Predictability of skeletal muscle tension from architectural determinations in guinea pig hindlimbs. J Appl Physiol Respir Environ Exerc Physiol 57:1715–21. [DOI] [PubMed] [Google Scholar]
- Pringle JW. 1967. The contractile mechanism of insect fibrillar muscle. Prog Biophys Mol Biol 17:1–12. [DOI] [PubMed] [Google Scholar]
- Püffel F, Pouget A, Liu X, Zuber M, van de Kamp T, Roces F, Labonte D. 2021. Morphological determinants of bite force capacity in insects: a biomechanical analysis of polymorphic leaf-cutter ants. J R Soc Interface 18:20210424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Redemann S, Weber B, Möller M, Verbavatz J-M, Hyman AA, Baum D, Prohaska S, Müller-Reichert T.. 2014. The segmentation of microtubules in electron tomograms using Amira. In: Sharp DJ, editor. Mitosis: Methods and Protocols. New York, NY: Springer New York. p. 261–278. [DOI] [PubMed] [Google Scholar]
- Richter A, Keller RA, Baumgarten Rosumek F, Economo EP, Hita Garcia F, Beutel RG. 2019. The cephalic anatomy of workers of the ant species Wasmannia affinis (Formicidae, Hymenoptera, Insecta) and its evolutionary implications. Arthropod Struct Dev 49:26–49. [DOI] [PubMed] [Google Scholar]
- Sacks RD, Roy RR.. 1982. Architecture of the hind limb muscles of cats: functional significance. J Morphol 173:185–95. [DOI] [PubMed] [Google Scholar]
- Salaberger D, Kannappan KA, Kastner J, Reussner J, Auinger T. 2011. Evaluation of computed tomography data from fibre reinforced polymers to determine fibre length distribution. Int Polym Proc 26:283–91. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid Bet al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Bethea JP, Hetzel-Ebben HL, Landim-Vieira M, Mayper RJ, Williams RL, Kessler LE, Ruiz AM, Gargiulo K, Rose JSMet al. 2021. Mandibular muscle troponin of the Florida carpenter ant Camponotus floridanus: extending our insights into invertebrate Ca(2+) regulation. J Muscle Res Cell Motil 42:399–417. [DOI] [PubMed] [Google Scholar]
- Shkarin R, Shkarin A, Shkarina S, Cecilia A, Surmenev RA, Surmeneva MA, Weinhardt V, Baumbach T, Mikut R. 2019. Quanfima: an open source Python package for automated fiber analysis of biomaterials. PLoS One 14:e0215137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Bernhardt G, Raine NE, Abel RL, Sykes D, Ahmed F, Pedroso I, Gill RJ.. 2016. Exploring miniature insect brains using micro-CT scanning techniques. Sci Rep 6:21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Fischer MS, Schilling N.. 2008. Die 3D-Architektur der Muskelfaszikel in ausgewählten Muskeln und ihre Relevanz zur Kraftentwicklung [Ph.D. thesis]. Friedrich-Schiller-Universität Jena. [Google Scholar]
- Stark H, Schilling N.. 2010. A novel method of studying fascicle architecture in relaxed and contracted muscles. J Biomech 43:2897–903. [DOI] [PubMed] [Google Scholar]
- Sullivan SP, McGechie FR, Middleton KM, Holliday CM.. 2019. 3D muscle architecture of the pectoral muscles of European starling (Sturnus vulgaris). Integr Org Biol 1:oby010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kamp T, Cecilia A, dos Santos Rolo T, Vagovic P, Baumbach T, Riedel A. 2015. Comparative thorax morphology of death-feigning flightless cryptorhynchine weevils (Coleoptera: Curculionidae) based on 3D reconstructions. Arthropod Struct Dev 44:509–23. [DOI] [PubMed] [Google Scholar]
- Weihmann T, Reinhardt L, Weissing K, Siebert T, Wipfler B.. 2015. Fast and powerful: biomechanics and bite forces of the mandibles in the American cockroach Periplaneta americana. PLoS One 10:e0141226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbock J, Amirkhanov A, Weimin L, Reh A, Amirkhanov A, Groller E, Kastner J, Heinzl C.. 2014. FiberScout: an interactive tool for exploring and analyzing fiber reinforced polymers. Proceedings of the 2014 IEEE Pacific Visualization Symposium. Yokohama, Japan. p. 153–60. [Google Scholar]
- Westenberger P, Blanc R.. 2016. Advanced fiber evaluation workflows. Proceedings of the 6th Conference on Industrial Computed Tomography. Wels, Austria. [Google Scholar]
- Winkel B. 2020. A three-dimensional model of skeletal muscle for physiological, pathological and experimental mechanical simulations [doctoral thesis]. Bauhaus-Universität Weimar. [Google Scholar]
- Yoo TS, Ackerman MJ, Lorensen WE, Schroeder W, Chalana V, Aylward S, Metaxas D, Whitaker R. 2002. Engineering and algorithm design for an image processing API: a technical report on ITK–the Insight toolkit. Stud Health Technol Inform 85:586–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A run of the script at the state of publication can be done using supplemental data. Script versions will be updated under: https://github.com/julesforfools/get_fibers. Full data available at: doi:10.5061/dryad.4mw6m90c3.