Abstract
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins provide prokaryotes with nucleic acid–based adaptive immunity against infections of mobile genetic elements, including phages. To counteract this immune process, phages have evolved various anti-CRISPR (Acr) proteins which deactivate CRISPR-Cas–based immunity. However, the mechanisms of many of these Acr-mediated inhibitions are not clear. Here, we report the crystal structure of AcrIF13 and explore its inhibition mechanism. The structure of AcrIF13 is unique and displays a negatively charged surface. Additionally, biochemical studies identified that AcrIF13 interacts with the type I-F CRISPR-Cas surveillance complex (Csy complex) to block target DNA recognition and that the Cas5f-8f tail and Cas7.6f subunit of the Csy complex are specific binding targets of AcrIF13. Further mutational studies demonstrated that several negatively charged residues of AcrIF13 and positively charged residues of Cas8f and Cas7f of the Csy complex are involved in AcrIF13–Csy binding. Together, our findings provide mechanistic insights into the inhibition mechanism of AcrIF13 and further suggest the prevalence of the function of Acr proteins as DNA mimics.
Keywords: CRISPR/Cas, anti-CRISPR, crystal structure, protein–protein interaction, protein complex
Abbreviations: Acr, anti-CRISPR; Cas, CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeat; crRNA, CRISPR RNA; Csy, CRISPR-Cas surveillance complex; EMSA, electrophoretic mobility shift assay; MST, microscale thermophoresis
To fight against phage infection, prokaryotes have adopted multiple immune strategies, including Clustered regularly interspaced short palindromic repeat (CRISPR)-Cas systems, which belong to prokaryotic adaptive immune systems (1, 2, 3). CRISPR-Cas system, which copes with mobile genetic elements, is executed by three stages: adaptation, expression, and interference. At the adaptation stage, short fragments of the invading DNAs (protospacers) are captured by a scissor-like Cas enzyme (usually Cas1–Cas2 heterodimer) and then integrated into the CRISPR array to generate novel spacers (4). In the expression stage, the CRISPR array is transcribed to precursor transcripts (pre-crRNA), matured into small CRISPR RNA (crRNA), and assembled with Cas proteins to constitute a surveillance complex (5). During the interference stage, crRNA-guided surveillance complex directly cleaves later invasive DNA molecules or recruits a Cas nuclease to digest the DNA molecules, through recognizing protospacers with its crRNA (1, 2). CRISPR-Cas systems are divided into two distinct classes, class I and II, which are classified by multiprotein or single-protein Cas effector and further divided into six types (type I–VI) (6). The type I-F CRISPR-Cas surveillance complex, also called the Csy complex, consists of four types of Cas subunits including Cas8f, Cas5f, Cas7f, and Cas6f (Fig. S1A), in a stoichiometry of 1:1:6:1, and is guided by a ∼60-nt crRNA (Fig. S1B) (3, 7, 8). After the Csy complex binds the target DNA and forms an R-loop structure of the DNA, the Cas2/3 nuclease is recruited to the Csy–DNA complex and further degrades the DNA molecule (9, 10).
Nevertheless, phages in turn have developed anti-CRISPR (Acr) proteins during their long-term arms race with their prokaryotic hosts (11). Until now, more than 90 members of Acrs have been discovered through both bioinformatics and functional analysis (12, 13, 14). Out of these Acrs, the type I-F CRISPR–Cas system harbors the largest number of identified Acrs, ranging from AcrIF1 to AcrIF24 (15). These Acrs inhibit the CRISPR-Cas system through highly diverse mechanisms, resulting in inhibition of target DNA binding or the recruitment of Cas2/3 nuclease (3, 7, 8, 11, 16, 17, 18, 19, 20, 21). Among these 24 AcrIF proteins, until now, the atomic structures and inhibition mechanisms have been revealed for 11 Acrs, i.e., AcrIF1/2/3/4/6/7/8/9/10/11/14 (3, 7, 8, 16, 17, 18, 19, 20, 21). It still remains unknown what mechanisms are adopted by other Acrs.
AcrIF13 was an Acr discovered from Moraxella catarrhalis BC8 prophage and was reported to potently inhibit the type I-F CRISPR-Cas system (22); however, its structure and detailed mechanism of inhibition remains unknown. In this study, we have solved the crystal structure of AcrIF13, which displays a unique structure and a negatively charged surface. Biochemical analysis showed that AcrIF13 functions as a DNA mimic, targets the Cas5f-8f tail region of the Csy complex, and also needs the Cas7.6f subunit for stable binding with the Csy complex. Further mutational analysis identified that several negatively charged residues of AcrIF13 and specific regions of Cas8f and Cas7f subunits of the Csy complex are involved in AcrIF13–Csy binding. Taken together, our study provides structural and mechanistic insight into the function of AcrIF13 and facilitates its further application in modulating type I-F CRISPR system-based genome editing.
Results
AcrIF13 targets the Cas5f-Cas8f tail of the Csy complex
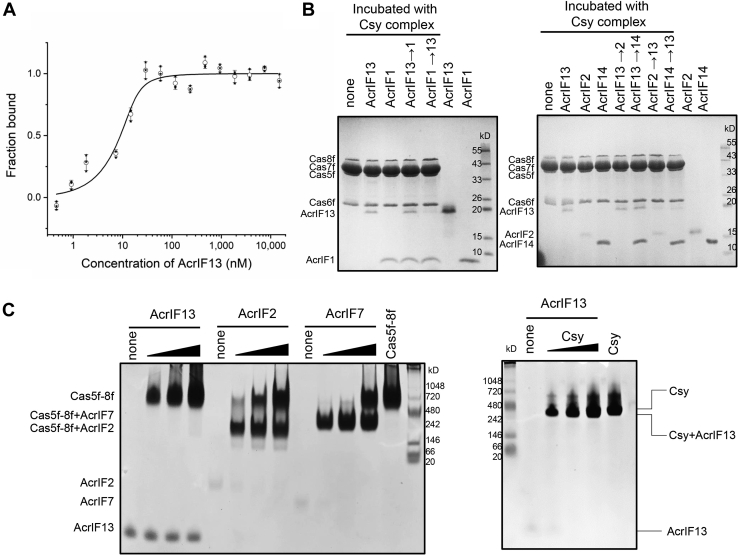
Our previous study has shown that AcrIF13 stably associates with the Csy complex in the gel filtration chromatography (8). First, we further confirmed their binding through microscale thermophoresis (MST) experiment, with a KD value of 1.51 ± 0.95 nM (Fig. 1A and Table 1), indicating a strong binding. Next, we moved on to investigate the binding mode of AcrIF13 on the Csy complex. Out of the known type I-F Acrs which stably bind the Csy complex, their binding modes can be divided into four types, i.e., Cas5f-8f tail targeting (AcrIF2, 6, 7, and 10) (3, 7, 16, 20), Cas7f targeting (AcrIF1, 9, and 14) (7, 18, 21, 23), mainly Cas8f targeting (AcrIF4) (19) and Cas8f-Cas7f targeting (AcrIF8) (16). Interestingly, while AcrIF1, 9, and 14 all engage the Cas7.6f and Cas7.4f subunits to prevent DNA binding, AcrIF9 and AcrIF14 could further induce non-sequence-specific DNA binding to the Csy complex (18, 21, 23). To investigate which binding mode is adopted by AcrIF13, we performed a competitive binding assay. The Csy complex with its Cas7f subunit tagged with 6xHis Tag was presaturated with untagged AcrIF1, AcrIF2, or AcrIF14, respectively. Then, untagged AcrIF13 was added into the above samples. After incubation, Acr-bound Csy complex was pulled down by Ni-NTA chromatography and detected by SDS-PAGE (Fig. 1B). We first incubated untagged AcrIF13 with the Csy complex, and then untagged AcrIF1, AcrIF2, or AcrIF14 was added. The competitive assay showed that only the binding of AcrIF2 but not AcrIF1 or AcrIF14 could be blocked by prior addition of AcrIF13. In turn, AcrIF13 binding could be partially blocked by prior addition of AcrIF2 or AcrIF14 and completely blocked by AcrIF1. In all, these results suggested that AcrIF13 and AcrIF2 bind the same binding site of the Csy complex, i.e., the Cas5f-8f tail, as AcrIF6, 7, and 10. Prior addition of AcrIF2 still allows AcrIF13 binding (lane 10 in Fig. 1B), suggesting that the binding between AcrIF2 and the Csy complex is weak, and a portion of the Csy complex are free for AcrIF13 binding. The reason why AcrIF13 binding is also blocked by prior addition of AcrIF1 and partially blocked by AcrIF14 remains to be investigated. Importantly, both AcrIF2 and AcrIF7 have been shown to directly interact with the purified Cas5f-8f subcomplex (20). Therefore, we moved on to test whether AcrIF13 also does. Native gel assay showed that unlike AcrIF2 and AcrIF7, AcrIF13 does not directly interact with the Cas5f-8f subcomplex (Fig. 1C, left panel). However, the binding between AcrIF13 and the whole Csy complex could be detected by the same native gel assay (Fig. 1C, right panel), indicating that AcrIF13 may also need other subunits of the Csy complex for binding. Taken together, biochemical assays indicated that AcrIF13 targets the Cas5f-8f tail but still needs other subunits of the Csy complex for binding.
Figure 1.
AcrIF13 targets the Cas5f-Cas8f tail of the Csy complex.A, MST assay of the binding between AcrIF13 and the Csy complex. AcrIF13 interacts with the Csy complex with a KD value equal to 1.51 ± 0.95 nM. B, untagged Acrs were mixed with the Csy complex containing 6xHis-tagged Cas7f. The samples were then affinity purified using Ni-NTA beads to remove unbound Acrs and proteins eluted from the beads and visualize through SDS-PAGE gel. In competitive binding experiments, one Acr was added after the first Acrs preincubated with Csy complex. An arrow means A replaced by B. C, native gel experiment for analyzing the interactions between Acrs (AcrIF2, AcrIF7, and AcrIF13) and the Cas5f-Cas8f heterodimer or the Csy complex. Acr protein (6 μM) (AcrIF2, AcrIF7, and AcrIF13) was incubated with different concentrations of Cas5f-Cas8f heterodimer of the Csy complex (3/6/12 μM). Lane 13 of the left panel was 12 μM Cas5f-Cas8f heterodimer alone. And then the mixture was visualized on 5% TBE native gels. Only AcrIF13 could not interact with Cas5f-Cas8f heterodimer. Acr, anti-CRISPR; Csy, CRISPR-Cas surveillance complex; MST, microscale thermophoresis.
Table 1.
Binding affinity between AcrIF13 and Csy mutants
| AcrIF13 | Csy complex | KD (nM) |
|---|---|---|
| WT | WT | 1.51 ± 0.95 |
| E10K | WT | 22.29 ± 7.59 |
| E18K | WT | 2.55 ± 1.03 |
| D21K/E22K | WT | 29.82 ± 9.50 |
| E40K | WT | 5.17 ± 1.84 |
| D43K/D44K | WT | 1.85 ± 1.24 |
| D45K/D46K | WT | 6.22 ± 2.44 |
| E50K | WT | 18.15 ± 5.73 |
| E57K | WT | 5.39 ± 1.80 |
| D63K | WT | 24.64 ± 5.30 |
| E82K/E83K/D84K | WT | 7.22 ± 2.68 |
| E97K | WT | 14.29 ± 3.97 |
| D102K | WT | 9.21 ± 2.50 |
| D113K | WT | 67.65 ± 18.97 |
| WT | Cas8f (K28E) | N.B. |
| WT | Cas8f (K247E) | N.B. |
| WT | Cas8f (N250D) | 647.23 ± 122.31 |
| WT | Cas7f (K58A/K60A) | 34.18 ± 14.78 |
Abbreviations: Acr, anti-CRISPR; Csy, CRISPR-Cas surveillance complex.
AcrIF13 displays a unique structure with a negatively charged surface
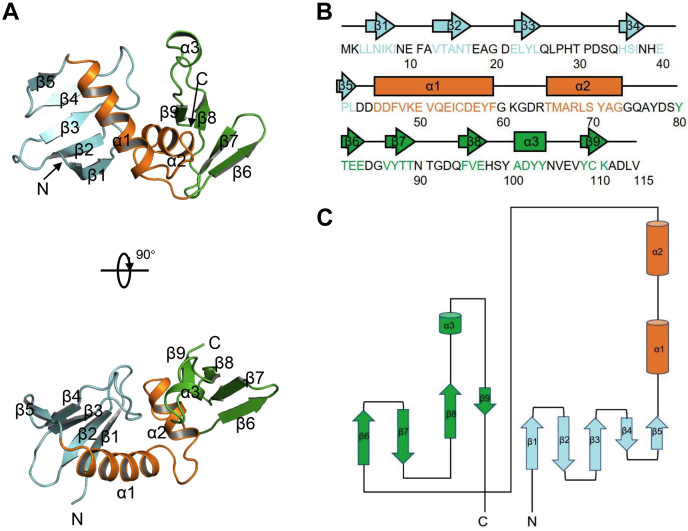
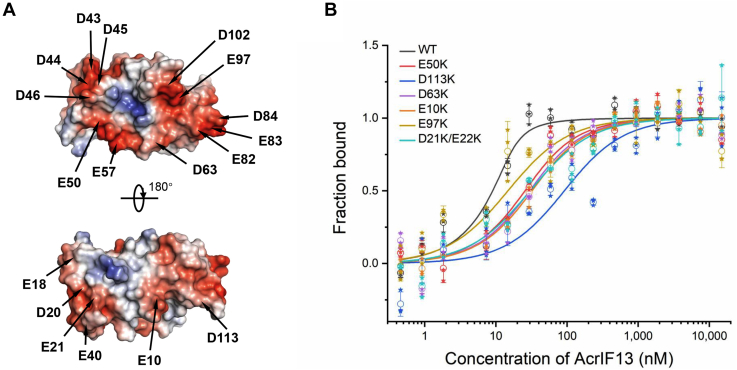
To further investigate the inhibitory mechanism of AcrIF13, we solved its crystal structure (full length, 115 residues) to a resolution of 2.9 Å. The structure of the selenomethionine-substituted L66M mutant of AcrIF13 was determined by single-wavelength anomalous dispersion phasing and was used as the template to solve the WT AcrIF13 structure (Table 2 and Fig. 2A). Each asymmetric unit of both WT and mutant structures contains eight protomers. To investigate the oligomeric state of AcrIF13, we performed a static light scattering assay of the protein, which showed that AcrIF13 is a monomer in solution (Fig. S2). The RMSD values of the Cα atoms between the WT and L66M structures were ∼0.19 Å, indicating that the mutation does not cause marked conformational changes to the structure (Fig. S3). Therefore, we use the WT structure hereafter for further analysis. Interestingly, the crystal structure of AcrIF13 reveals a typical three-layer structure, with a two-helix motif (44–78 residues) flanked by two antiparallel β sheets at both sides, acting like two halves of a shell wrapping the internal helix motif. The N-terminal β sheet (1–43 residues) is five stranded (β1-β5), and the C-terminal β sheet (79–115 residues) contains four strands (β6-β9) with a short α helix between β8 and β9 (Fig. 2, B and C). Dali search only returned entries with low Z scores, and no entries were aligned to the intact sequence of AcrIF13 (Table S1), suggesting that AcrIF13 displays an overall unique structure. Like other known type I-F Acrs that target the Cas5f-8f tail, AcrIF13 also displays a negatively charged surface (Fig. 3A), consistent with its low theoretical isoelectric point value of 4.17 (calculated by the ProtParam tool, https://web.expasy.org/protparam). Taken together, AcrIF13 displays a three-layer structure and a negatively charged surface.
Table 2.
Data collection and refinement statistics
| Statistics | AcrIF13 (L66M) | AcrIF13 |
|---|---|---|
| Data collection | ||
| Space group | F23 | F23 |
| Cell dimensions | ||
| a, b, c (Å) | 263.847, 263.847, 263.847 | 262.771, 262.771, 262.771 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50–3.03 (3.14–3.03) | 50–2.95 (3.06–2.95)a |
| Rsym or Rmerge (%) | 16.6 (133.7) | 15.7 (169.2) |
| I/σ(I) | 21.2 (2.3) | 30.7 (3.2) |
| Completeness (%) | 99.6 (100.0) | 99.2 (99.9) |
| Redundancy | 31.1 (29.8) | 40.4 (41.3) |
| Refinement | ||
| Resolution (Å) | 50–3.03 | 50–2.95 |
| No. reflections | 29,467 | 31,488 |
| Rwork/Rfree# | 0.2438/0.2843 | 0.2363/0.2705 |
| No. atoms | 7431 | 7425 |
| Protein | 7431 | 7425 |
| Ligand/ion | 0 | 0 |
| Water | 0 | 0 |
| B factors | 72.72 | 54.26 |
| Protein | 72.72 | 54.26 |
| Ligand/ion | ||
| Water | ||
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 0.85 | 1.25 |
| Ramachandran plot (%) | ||
| Favored | 97.17 | 96.84 |
| Allowed | 2.83 | 2.83 |
| Outliers | 0 | 0.33 |
For each structure one crystal was used. Values in parentheses are for highest-resolution shell.
Figure 2.
Crystal structure of AcrIF13.A, the crystal structure of AcrIF13 shown in cartoon model. The N-terminal β-sheet, middle α-helices, and C-terminal β-sheet are shown in cyan, yellow, and green, respectively. Two perpendicular views are shown with secondary structures labeled. B, secondary structural elements of AcrIF13 are shown above the sequences, colored as in A. C, topological diagram of AcrIF13. Acr, anti-CRISPR; Csy, CRISPR-Cas surveillance complex.
Figure 3.
The negatively charged surface of AcrIF13 is involved in Csy binding.A, electrostatic surface representation of AcrIF13. The negatively charged residues which are mutated in mutagenesis assays are marked. B, MST assay of the binding between the Csy complex and AcrIF13 or its mutants. The binding curves of the six AcrIF13 mutants with binding KD values of over 10-fold higher than that of WT AcrIF13 are displayed. Error bars represent SD; n = 3. Acr, anti-CRISPR; Csy, CRISPR-Cas surveillance complex; MST, microscale thermophoresis.
AcrIF13 blocks the target DNA binding by the Csy complex
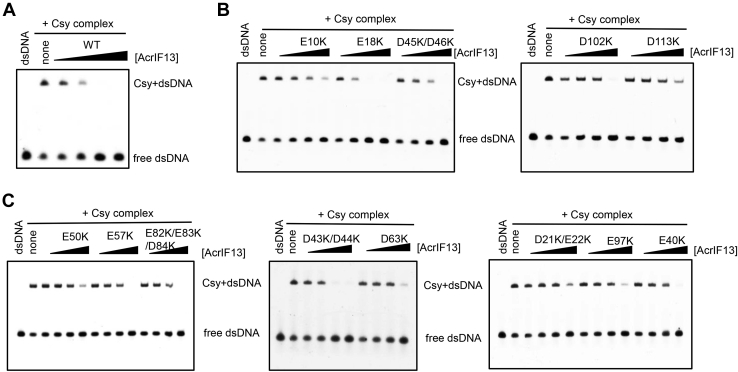
The similar binding site of AcrIF13 as that of AcrIF2 and its negatively charged surface suggest that AcrIF13 may also act as a DNA mimic protein to block the recognition of target DNA by the Csy complex. To test this, we performed an electrophoretic mobility shift assay (EMSA) with the Csy complex, target double-stranded DNA, and AcrIF13. The result showed that binding of target DNA by the Csy complex was potently inhibited by AcrIF13 when incubated at a molar ratio as low as 1:1 (AcrIF13: Csy, Fig. 4A). Therefore, AcrIF13 acts as a DNA mimic and prevents target DNA binding.
Figure 4.
EMSA results of AcrIF13 and its mutants.A–C, EMSA experiments using Csy complex, target DNA, and AcrIF13 or its mutants. DNA binding by Csy complex produced a shift of the target DNA. Addition of gradually increasing concentrations of AcrIF13 or its mutants would decrease the shifted DNA to different extents. Acr, anti-CRISPR; Csy, CRISPR-Cas surveillance complex; EMSA, electrophoretic mobility shift assay.
Mutational studies reveal key residues in AcrIF13–Csy binding
To investigate the detailed binding mechanism of AcrIF13 with Csy, we constructed 13 reverse charge mutants of AcrIF13 by replacing its Glu or Asp with Lys residues. We then tested the affinity of all the mutants for the Csy complex through both MST and EMSA experiments. Out of the 13 mutants, six of them displayed a binding KD value of over 10-fold higher than that of WT AcrIF13 (Table 1 and Fig. 3B). Particularly, the D113K mutant of AcrIF13 displayed the weakest binding with a binding KD value of 67.65 nM, 40-fold as that of WT AcrIF13. Besides, the E97K, D63K, E50K, D21K/E22K, and E10K mutants of AcrIF13 also showed weaker binding to different extents with binding KD values of 10- to 15-fold higher than that of WT AcrIF13. Meanwhile, two mutations, E18K and E43/44K, did not cause marked binding defects with the Csy complex (Fig. S4). The EMSA results were also consistent with the binding affinity obtained by the MST assay, showing weaker inhibition of target DNA–Csy binding by the mutants with higher binding KD values than WT AcrIF13 (Fig. 4B). The fact that none of the AcrIF13 mutants completely abolished the Csy binding suggested that AcrIF13 may explore multiple negatively charged residues to bind the Csy complex. In turn, we also constructed several mutants of the Csy complex. We first focused on K28, K247, and N250 of Cas8f due to their important roles in Acr binding in previous studies (7, 8, 20). The results showed that binding cannot be detected between AcrIF13 and K247ECas8f or K28ECas8f mutants of Csy, and the N250DCas8f mutant of Csy also displays much weaker binding to AcrIF13 (Table 1 and Fig. S5). This indicated that these important sites of Cas8f are also engaged by AcrIF13, suggesting a common binding strategy shared by these Cas5f-8f targeting Acrs. Since the purified Cas5f-8f subcomplex does not maintain binding with AcrIF13, we further tested which subunits of the Csy complex are also involved in AcrIF13 binding. Due to the close position and validation of involvement in the binding of AcrIF2 (7), Cas7.6f was chosen for further mutagenesis experiments by mutating the Cas7f genes in the vector expressing the Csy complex. This time, we focus on K58/K60 of the thumb domain of Cas7f because the two residues have been verified to be involved in AcrIF2 binding (7). MST assay showed that the K58A/K60ACas7f mutant of Csy also displayed a weaker binding to AcrIF13 than WT Csy (Table 1 and Fig. S5), suggesting the involvement of this region in AcrIF13 binding. Taken together, mutational studies showed that several negatively charged regions of AcrIF13 and K28, K247, N250 of Cas8f, and K58/K60 of Cas7f of the Csy complex are involved in the AcrIF13–Csy binding.
Discussion
Acrs display highly variable amino acid sequences and structures. Prevention of target DNA hybridization with the crRNA has been identified as a common inhibition strategy utilized by multiple Acrs. For type I–F Acrs, this strategy has been adopted by AcrIF1, 2, 6, 7, 8, 9, 10, 11, and 14, although the detailed mechanisms differ among them. In this study, the biochemical and structural analysis showed that AcrIF13 is another Acr, acting as a DNA mimic and targeting the Cas5f-8f tail, like AcrIF2, 6, 7, and 10. Interestingly, while AcrIF2 and AcrIF7 can interact with the purified Cas5f-8f subcomplex, AcrIF13 does not show this binding. This suggests that the binding affinity of AcrIF13 to the Cas5f-8f subcomplex may be not high enough to maintain this binding, and the binding from Cas7.6f is also essential for the binding of AcrIF13.
We think that the necessity of Cas7.6f in AcrIF13 binding can also help explain the competitive binding assay results regarding AcrIF1 and AcrIF14. In the assay, we found that AcrIF13 binding is partially blocked by prior addition of AcrIF14 and completely blocked by AcrIF1. In turn, only the binding of AcrIF2, but not AcrIF1 or AcrIF14, can be blocked by prior addition of AcrIF13 (Fig. 1B). Both AcrIF1 and AcrIF14 target the thumb domains of Cas7.6f and Cas7.4f subunits with two copies of protein (3, 7, 19, 21). Structures of both the Csy–AcrIF1 and Csy–AcrIF14 complexes show that the thumb domain of Cas7.6f, where K58 and K60 are localized (Fig. S6), is engaged by one AcrIF1 or AcrIF14 molecule. Therefore, prior addition of AcrIF1 or AcrIF14 may hinder the recognition of K58/K60 residue by AcrIF13. However, while prior addition of AcrIF13 may prevent the binding of one AcrIF1/14 molecule to the Cas7.6f subunit, the Cas7.4f subunit can still be engaged by another AcrIF1/14 molecule. Therefore, prior addition of AcrIF1/14 can hinder the binding of AcrIF13 to different extents, but in turn, prior addition of AcrIF13 still allows the binding of AcrIF1/14.
MST and EMSA experiments showed that none of the mutations of AcrIF13 tested in this study can completely abolish the AcrIF13–Csy binding, indicating that multiple regions of AcrIF13 might be required for Csy binding. In contrast, the single-residue mutations of the Cas8f subunit of Csy have much severer effect on the binding. Interestingly, the residues of Cas8f and Cas7f which are engaged by AcrIF2 and AcrIF7 are also engaged by AcrIF13. This suggests that Acrs of divergent origins evolve not only their specificity for a common protein target (Cas5f-8f tail of the Csy complex) through convergent evolution of their surface charges but also the specificity for the specific binding residues of the Csy complex with their completely distinct three-dimensional structures. It appears very unclever for phages that they use Acrs of different origins to target the same sites of the Csy complex, since prokaryotes could overcome this by mutation of the targeted sites. However, K28/K247/N250 residues of Cas8f have been shown to be rather important for target DNA binding by the Csy complex, which therefore restricts mutations of the residues. In all, our study characterizes the inhibition mechanism of AcrIF13, highlighting the prevalence of mechanisms as DNA mimics utilized by Acrs and facilitating its further application as a modulating tool in genome editing by CRISPR-Cas technology.
Experimental procedures
Cloning, expression, and purification
The synthetic gene of AcrIF13 was cloned into pGEX-6P-1 with an N-terminal glutathione S-transferase (GST) tag and a cleavage site for PreScission protease. The mutant AcrIF13 genes were generated by two-step PCR and were subcloned, overexpressed, and purified in the same way as the WT protein. The WT and mutant constructs were transformed into Escherichia coli BL21(DE3) cells, and the cells were cultured in LB medium at 37 °C until the absorbance at A600 reached 1.0. Protein expression was induced by the addition of 0.2 mM isopropyl β-D-1-thiogalactopyranoside followed by incubation at 18 °C for 12 h. The E. coli cells were harvested by centrifugation and resuspended in the lysis/wash buffer (1 × PBS, 2 mM DTT, and 1 mM PMSF). After high pressure homogenization and centrifugation, the supernatant was loaded onto a glutathione resin column (Genscript) pre-equilibrated with the lysis/wash buffer (1 × PBS with 2 mM DTT). The column was washed with the same buffer. The N-terminal GST tag was cleaved by PreScission protease in cleavage buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [Hepes], pH 7.5, 200 mM NaCl, 2 mM DTT) at 19 °C for 2 to 4 h, and the AcrIF13 without GST tag was eluted by the same buffer. The eluant was concentrated and further purified using a Superdex-200 increase 10/300 GL (GE Healthcare) column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. The purified protein was analyzed by SDS-PAGE. Selenomethionine (Se-Met)-labeled AcrIF13 L66M was expressed in E. coli BL21(DE3) cells grown in M9 minimal medium supplemented with 35 mg/L Se-Met (Acros) and specific amino acids: Ile, Leu, and Val at 50 mg/L and Lys, Phe, and Thr at 100 mg/L. The Se-Met protein and the mutants were purified as described above.
The Csy complex from Pseudomonas aeruginosa was purified as follows. The Cas8f/Cas5f and Cas7f/Cas6f genes were cloned into pETDuet-1 and pACYCDuet-1, respectively. The crRNA fragment was cloned into pRSFDuet-1 vector. After co-expression of the three plasmids in E. coli strain BL21(DE3), the Csy complex (6 × His tag on Cas7f) was purified through Ni-column, anion exchange chromatography, and gel filtration. The mutations of Cas8f and Cas7f were made as above, and the Csy complex with Cas8f and Cas7f mutations were purified similarly as the WT complex. The Cas8f/Cas5f complex (6 × His tag on Cas8f) was purified similarly as the Csy complex.
Crystallization, data collection, and structure determination
The WT and L66M AcrIF13 was concentrated to 12 mg/ml in 10 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. Crystals were grown using the hanging-drop vapor diffusion method. Crystals of AcrIF13 were grown at 18 °C by mixing an equal volume of the protein (12 mg/ml) with reservoir solution containing 2.4 M sodium malonate monohydrate. The crystals appeared overnight and grew to full size in about 4 to 5 days. The crystals were cryoprotected in the reservoir solution containing 15% hexanediol before its transferring to liquid nitrogen. Se-Met–labeled L66M AcrIF13 was crystallized in the same buffer. After crystal diffraction tests at home and beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility, a crystal of the Se-Met–labeled protein suitable for structure determination was finally obtained. All the data were collected at the Shanghai Synchrotron Radiation Facility beamlines BL17U1 (24) and BL19U1 (25), integrated and scaled using the HKL2000 package (26). The initial model was solved by Autosol in PHENIX (27) and refined manually using COOT (28). The structure was further refined with PHENIX (27) against the native data using noncrystallographic symmetry and stereochemistry information as restraints. The final structure was obtained through several rounds of refinement. Data collection and structure refinement statistics are summarized in Table 2. All of the structural illustrations were generated using the software PyMOL.
Electrophoretic mobility shift assay
Target DNA strand with 5′ end FAM (6-carboxyfluorescein) labeling was purchased from Sangon. Target and nontarget DNA strands were hybridized with a molar ratio of 1:1.5 to obtain a dsDNA. Incubation were performed in a 20 μl buffer system containing 4 μM Csy complex, 0.2 μM dsDNA, and 1/2/3/4 μM AcrIF13 or its mutants. The Csy complex and AcrIF13 were incubated at 4 °C for 30 min, and then the dsDNA was added and was incubated at 37 °C for another 30 min in the buffer containing 20 mM Hepes pH 7.5, 150 mM NaCl, and 5% glycerol. The mixture was separated using 5% native polyacrylamide gels and visualized by fluorescence imaging.
Target DNA sequence (54 bp, 5′-FAM fluorescein labeled):
GGAAGCCATCCAGGTAGACGCGGACATCAAGCCCGCCGTGAAGGTGCAGCTGCT
Multiangle light scattering analysis
Multiangle light scattering experiment was performed in 10 mM Tris pH 8.0 and 200 mM NaCl using a Superdex-75 10/300 Gl size-exclusion column from GE Healthcare. 1 mg/ml AcrIF13 was used. The chromatography system was connected to a Wyatt DAWN HELEOS Laser photometer and a Wyatt Optilab T-rEX differential refractometer. Wyatt ASTRA 7.3.2 software was used for data analysis.
Protein binding assays
Purified AcrIF13 (6 μM) was incubated with a concentration gradient of Cas5f-Cas8f heterodimer (3/6/12 μM) or the Csy complex for 1 h at 4 °C in binding buffer (20 mM Hepes pH 7.5, 150 mM NaCl, and 5% glycerol), as well as AcrIF2 and AcrIF7. The mixture was separated using 5% native polyacrylamide gel in 4 °C and visualized by Coomassie blue R250 staining.
Acrs competitive binding to the Csy complex
Purified 6xHis-tagged Csy complex (3 μM) was incubated with excess Acrs (15 μM) for 30 min at 4 °C in binding buffer (20 mM Hepes pH 7.5, 150 mM NaCl, and 5% glycerol). Competitor Acr (75 μM) was added and incubated for 30 min at 4 °C. The mixture was then loaded onto Ni-NTA beads, and it was incubated for 30 min at 4 °C. Bound Csy complex and Acr were collected through centrifugation at 12,000 rpm for 1 min to remove unbound Acr. The reaction was then washed three times with wash buffer (50 mM Tris pH 8.0, 200 mM NaCl, 30 mM imidazole) with a centrifugation step after each wash. The sample was then eluted in elution buffer (50 mM Tris pH 8.0, 200 mM NaCl, 300 mM imidazole). The samples were visualized on Coomassie blue R250 stained SDS-PAGE gels. Each experiment was conducted at least three times, and the same result was obtained each time.
MST measurements
All MSTs were performed on a NanoTemper Monolith NT.115 instrument (NanoTemper Technologies) using the Standard Treated Capillaries K002 of the supplier. Each of the 16 solutions of one titration series was filled into a capillary, which were measured successively to create the respective data points in the experiment. General settings were applied for all MST experiments as follows: manual temperature control: 25 °C; LED laser: RED, fluorescence measurement before MST: 5 s; MST (IR laser) on: 45 s; fluorescence after MST: 15 s; and delay: 25 s. LED power is set at 100%, and MST power is set at medium.
For MST assay, all the proteins were exchanged into the MST buffer (20 mM Hepes pH7.5, 150 mM KCl, 5% glycerol). The Csy complex and its mutants were fluorescence-labeled using the Protein Labeling Kit RED-NHS second Generation (NanoTemper Technologies) at 15 μM. The 15 nM fluorescence-labeled Csy complex and its mutants were added in a 1:1 ratio to a 1:2 dilution series with a final concentration of 15 μM down to 0.458 nM for AcrIF13 and its mutants. Each experiment was conducted at least three times, and the similar result was obtained each time. Each protein KD value was obtained with a signal-to-noise ratio higher than 7. Datasets were processed with the MO (Monolith). Affinity Analysis v2.3 software, using the signal from the initial fluorescence. The analysis of dose–response curves was carried out with Origin 2018.
Data availability
The accession numbers for the coordinate and structure factor of WT and L66M mutant of AcrIF13 are PDB: 7VEH and 7FI4, respectively.
Supporting information
This article contains supporting information
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank the staff at beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility for their assistance with data collection. We would like to thank the Tsinghua University Branch of China National Center for Protein Sciences Beijing and Shilong Fan for providing facility support for X-ray diffraction of the crystal samples. The authors would like to thank Mrs Wu Yao (State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences) for assistance in the microscale thermophoresis experiment. This work was supported by National Natural Science Foundation of China (32171274, 31822012, and 32000901), the National Key Research and Development Program of China (2017YFA0506500, 2019YFC1200500, and 2019YFC1200502), the Beijing Natural Science Foundation (5204038), and the Beijing Nova Program.
Author contributions
H. W., T. G., and Y. F. methodology; H. W., T. G., J. R., and J. G. investigation; Yu Zhou, J. Z., and Y. X. software; Yu Zhou, J. Z., and Y. X. data analysis; Yi Zhang supervision; Y. F. conceptualization; Y. F. writing—review & editing.
Edited by Karen Fleming
Supporting information
References
- 1.Barrangou R., Marraffini L.A. CRISPR-cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The biology of CRISPR-Cas: Backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 3.Guo T.W., Bartesaghi A., Yang H., Falconieri V., Rao P., Merk A., Eng E.T., Raczkowski A.M., Fox T., Earl L.A. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell. 2017;171:414–426.e12. doi: 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., Van Der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin E.V., Makarova K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374:20180087. doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury S., Carter J., Rollins M.F., Golden S.M., Jackson R.N., Hoffmann C., Bondy-Denomy J., Maxwell K.L., Davidson A.R., Fischer E.R. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169:47–57.e11. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu Y., Yang L., Gao T., Dong C., Zhang B., Yin P., Hopp A.-K., Li D., Gan R., Wang H. A type IF anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-ribosylation. Mol. Cell. 2020;80:512–524.e5. doi: 10.1016/j.molcel.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Rollins M.F., Chowdhury S., Carter J., Golden S.M., Miettinen H.M., Santiago-Frangos A., Faith D., Lawrence C.M., Lander G.C., Wiedenheft B. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol. Cell. 2019;74:132–142.e5. doi: 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuminauskaite D., Norkunaite D., Fiodorovaite M., Tumas S., Songailiene I., Tamulaitiene G., Sinkunas T. DNA interference is controlled by R-loop length in a type I-F1 CRISPR-Cas system. BMC Biol. 2020;18:1–16. doi: 10.1186/s12915-020-00799-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondy-Denomy J., Davidson A.R., Doudna J.A., Fineran P.C., Maxwell K.L., Moineau S., Peng X., Sontheimer E.J., Wiedenheft B. A unified resource for tracking anti-CRISPR names. CRISPR J. 2018;1:304–305. doi: 10.1089/crispr.2018.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C., Hao G.-F., Hua H.-L., Liu S., Labena A.A., Chai G., Huang J., Rao N., Guo F.-B. Anti-CRISPRdb: A comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 2018;46:D393–D398. doi: 10.1093/nar/gkx835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson A.R., Lu W.-T., Stanley S.Y., Wang J., Mejdani M., Trost C.N., Hicks B.T., Lee J., Sontheimer E.J. Anti-CRISPRs: Protein inhibitors of CRISPR-Cas systems. Annu. Rev. Biochem. 2020;89:309–332. doi: 10.1146/annurev-biochem-011420-111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinilla-Redondo R., Shehreen S., Marino N.D., Fagerlund R.D., Brown C.M., Sørensen S.J., Fineran P.C., Bondy-Denomy J. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K., Wang S., Li S., Zhu Y., Pintilie G.D., Mou T.-C., Schmid M.F., Huang Z., Chiu W. Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type IF CRISPR–Cas complex revealed by cryo-EM. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7176–7182. doi: 10.1073/pnas.1922638117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Yao D., Xu J.-G., Li A.-R., Xu J., Fu P., Zhou Y., Zhu Y. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 2016;23:868–870. doi: 10.1038/nsmb.3269. [DOI] [PubMed] [Google Scholar]
- 18.Hirschi M., Lu W.-T., Santiago-Frangos A., Wilkinson R., Golden S.M., Davidson A.R., Lander G.C., Wiedenheft B. AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabel C., Li Z., Zhang H., Chang L. Structural basis for inhibition of the type I-F CRISPR–Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14. Nucleic Acids Res. 2020;49:584–594. doi: 10.1093/nar/gkaa1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I., Koo J., An S.Y., Hong S., Ka D., Kim E.-H., Bae E., Suh J.-Y. Structural and mechanistic insights into the CRISPR inhibition of AcrIF7. Nucleic Acids Res. 2020;48:9959–9968. doi: 10.1093/nar/gkaa690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Zhang L., Xiu Y., Gao T., Huang L., Xie Y., Yang L., Wang W., Wang P., Zhang Y., Yang M., Feng Y. Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex. Nucleic Acids Res. 2021;49:10178–10191. doi: 10.1093/nar/gkab738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marino N.D., Zhang J.Y., Borges A.L., Sousa A.A., Leon L.M., Rauch B.J., Walton R.T., Berry J.D., Joung J.K., Kleinstiver B.P. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362:240–242. doi: 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu W.-T., Trost C.N., Müller-Esparza H., Randau L., Davidson A.R. Anti-CRISPR AcrIF9 functions by inducing the CRISPR–Cas complex to bind DNA non-specifically. Nucleic Acids Res. 2021;49:3381–3393. doi: 10.1093/nar/gkab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q., Xu C.-Y., Yu F., He J.-H. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018;29:68. [Google Scholar]
- 25.Zhang W.Z., Tang J.C., Wang S.S., Wang Z.J., He J.H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 2019;30:170. [Google Scholar]
- 26.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Adams P.D., Grosse-Kunstleve R.W., Li W.H., Ioerger T.R., Terwilliger T.C. Phenix: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Lohkamp B., Scott W., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the coordinate and structure factor of WT and L66M mutant of AcrIF13 are PDB: 7VEH and 7FI4, respectively.