Summary
Although there are numerous tissue clearing protocols, most are inadequate for clearing liver tissue. Here we present a flexible protocol for mouse liver tissue; we combine strategies from several previously published protocols for delipidation, decolorization, staining, and refractive index matching. LiverClear is sufficiently versatile to allow clearing of healthy and diseased mouse liver followed by immunofluorescence staining and imaging to visualize intact 3D structures such as bile ducts and hepatocyte canaliculi. We also adapted this protocol for clearing human livers.
For complete details on the use and execution of this protocol, please refer to Molina et al. (2021).
Subject areas: Developmental biology, Microscopy
Graphical abstract

Highlights
-
•
Step-by-step optimized protocol for tissue clearing of healthy or diseased liver
-
•
Visualization of livers cells in their 3-dimensional tissue organization
-
•
Adaptation of tissue clearing protocol for human livers
-
•
Immunostaining and ribbon-scanning confocal microscopy on cleared liver tissue
Although there are numerous tissue clearing protocols, most are inadequate for clearing liver tissue. Here we present a flexible protocol for mouse liver tissue; we combine strategies from several previously published protocols for delipidation, decolorization, staining, and refractive index matching. LiverClear is sufficiently versatile to allow clearing of healthy and diseased mouse liver followed by immunofluorescence staining and imaging to visualize intact 3D structures such as bile ducts and hepatocyte canaliculi. We also adapted this protocol for clearing human livers.
Before you begin
The previous 10 years have ushered in a revolution in methods that enable researchers to create optically transparent tissues. Initially, such protocols were developed for the brain, but they have progressively expanded towards multiple organ systems and even whole animals. Such tissue clearing methods have enabled the development of specialized optics, advanced light sheet and high-speed confocal microscopes that are capable of imaging very large volumes at micron and submicron scales. Such imaging strategies enable researchers to characterize whole organ systems allowing for simultaneous assessment at both macro- and microscopic scales. The liver however has proven to be a difficult organ to clear. The dense nature of the tissue prevents efficient removal of lipids and thus effective refractive index matching and abundant pigmentation absorbs light. Here we present LiverClear: a clearing strategy specifically tailored to address these gaps.
The cornerstone of all tissue clearing strategies is refractive index (RI) matching which mitigates scatter of photons as they move through tissue. Even under ideal RI matching conditions the liver remains opaque due to the presence of pigments like hemosiderin, bilirubin and hemoglobin which absorb light and counteract any benefits that come from RI matching. The LiverClear protocol presented here is a water-based method which uses techniques from various clearing/staining strategies to optimize RI matching, depigmentation and immunostaining – resulting in a tissue preparation suited for advanced whole-mount imaging.
LiverClear offers a simple and flexible approach to examine the tissue architecture of both normal and diseased liver of mice or similarly sized animal models as well as human liver. The protocol below describes how to process liver tissue for optimal clearing starting from the point of tissue harvest, fixation and hydrogel embedding to maintain the tissue architecture, active and passive delipidation, depigmentation, and immunostaining protocols for antigen visualization in healthy mouse liver as well as mouse hepatocellular carcinoma. LiverClear can be potentially applied to visualize diverse changes in the 3-dimensional organization of cells in healthy or diseased states and to correlate these with functional changes in metabolism, transport, proliferation, or fibrous deposition. For example, it has the potential to be used to examine liver zonation of metabolic processes and how zonated markers may be altered in regeneration or disease. It can also be used to visualize the biliary ductal system and how it integrates with hepatocytes (as shown in this protocol), as well as how the ductular reaction may or may not be connected with existing liver structure. Furthermore, in the setting of liver tumors this technique could be used to assess heterogeneity within tumors and visualize the relationship between tumors and the liver vasculature. We thus envision that this protocol can be adapted to the study of a variety of liver diseases and experiments examining liver injury and repair, making it a highly promising and versatile tool.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CK19 | Developmental Studies Hybridoma Bank (DSHB) | Cat.no.TROMAIII |
| CEACAM1 | LSBio | Cat.no. LS-C106710 |
| GS (Glutamine synthetase) | Sigma-Aldrich | Cat.no.G2781 |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium phosphate monobasic monohydrate | Sigma-Aldrich | Cat.no. S9638 |
| Sodium phosphate dibasic (anhydrous) | Sigma-Aldrich | Cat.no. S9763 |
| Sodium azide | Sigma-Aldrich | Cat.no. S2002 |
| 16% w/v Paraformaldehyde | Fisher Scientific | Cat.no. 50-980-487 |
| 2% bio-acrylamide | Bio-Rad | Cat.no. 161-0142 |
| 40% acrylamide | Bio-Rad | Cat.no. 161-0140 |
| VA-044 | Wako Chemicals, Fisher | Cat.no. NC0632395 |
| X-CLARITYTM Electrophoretic Tissue Clearing Solution | Logos Bio | Cat.no. C13001 |
| N,N,N′,N′ Tetrakis(2-Hydroxypropyl)ethylenediamine (Quadrol) | Sigma-Aldrich | Cat.no. 122262 |
| Boric acid | Fisher Scientific | Cat.no. A74-500 |
| Sodium sulfite | Sigma-Aldrich | Cat.no. S0505 |
| Bovine serum albumin (BSA) | Fisher Scientific | Cat.no. BP1605-100 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat.no. L3771 |
| Triton X-100 | Fisher Scientific | Cat.no. BP151-500 |
| Urea | Sigma-Aldrich | Cat.no. U5378 |
| Sucrose | Sigma-Aldrich | Cat.no. S9378 |
| Ammonium hydroxide 25% | Sigma-Aldrich | Cat.no. 105432 |
| 30% Hydrogen peroxide | Fisher Scientific | Cat.no. H325-500 |
| Experimental models: Organisms/strains | ||
| Wild type mice, C57BL6 | The Jackson Laboratory | N/A |
| Mouse: Foxa3-Cre YAP1 KO [Tg(Foxa3-Cre)1Khk, Yap1tm1.1Dupa, Gt(ROSA)26Sortm1(EYFP)Cos/J] | Molina et al., (2021) | N/A |
| Software and algorithms | ||
| Imaris 9.7.2 | Bitplane | N/A |
| Alternative image process and analysis software | ||
| Other | ||
| 50 mL Falcon conical tubes | Fisher Scientific | Cat.no. 05-539-9 |
| X-Clarity clearing system (this study) | LogosBio | Cat.no. XC-ETC 2 |
| Alternative clearing systems | See other resources section | N/A |
| Sample holder for X-Clarity clearing system (3D printed) | See other resources section | N/A |
| Ribbon Scanning Confocal (this study) | Caliber I.D. | Cat.no. RS-G4 |
| Alternative cleared tissue microscopy imaging platform | ||
Other resources
In this protocol we use a commercially available clearing system based on the ACT-PRESTO (Lee et al., 2016) and CLARITY (Chung and Deisseroth, 2013) protocols. Although the remainder of the protocol will discuss the use of the X-CLARITYTM system, numerous commercially available systems exist which we expect will yield similar results. In addition, resources are available for building a system from commonly available laboratory equipment described in the above citations as well as on this website: http://wiki.claritytechniques.org/index.php/Main_Page. Before designing an experiment, any system should be carefully tested under experimental conditions to pick the optimal clearing times and parameters.
The sample holder chamber (Figure 1A) and lid (Figure 1B) were modeled/designed in Solidworks and 3D printed on a Formlabs Form2 printer using the Formlabs Tough1500 resin. This resin was chosen for its strength and resistance to absorbing fluids. After 3D printing is completed, the supports were removed. Since 3D printers operate within a certain size tolerance range, light sanding of the corners was necessary to ensure a proper fit into the clearing chamber. Sanding should allow the sample holder to fit precisely into the holder without any resistance. Files for 3D printing are included in the Online Supplements including clearing chamber lid (Methods S1) and clearing chamber (Methods S2) and can also be found at the following link: https://github.com/CBI-PITT/X-CLARITY-3D-Printed-Sample-Holder.
Figure 1.
Rendering of 3D printed sample holder
(A) Sample holder assembly.
(B) Sample holder lid.
(C) Sample holder chamber.
In this study, we used ribbon-scanning confocal microscopy (Watson et al., 2017) outfitted with a Nikon CFI90 20XC Glyc objective. Fluorescent imagery in all figures was generated in Bitplane Imaris. LiverClear samples can be imaged on any microscope platform adapted for whole organ cleared tissue; see more information here: http://wiki.claritytechniques.org/index.php/Main_Page
Materials and equipment
4× Phosphate buffered saline (4× PBS; adjust pH to 7.4)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium phosphate monobasic monohydrate | 25.8 mM | 3.1 g |
| Sodium phosphate dibasic (anhydrous) | 76.8 mM | 10.9 g |
| ddH2O | n/a | Adjust to 250 mL |
| Total | n/a | 250 mL |
Storage: Room temperature, indefinitely
1× Phosphate buffered saline (1× PBS)
| Reagent | Final concentration | Amount |
|---|---|---|
| 4× PBS | 1× | 250 mL |
| ddH2O | n/a | 750 mL |
| Total | n/a | 1 L |
Storage: Room temperature, indefinitely
Alternatives: You can use pre-mixed pouches from Fisher (Cat. no. PI28374)
PBSA (1× PBS/0.02% Sodium Azide)
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× PBS | n/a | 499 mL |
| 10% Sodium azide | 0.02 % w/v | 1.0 mL |
| Total | n/a | 500 mL |
Storage: Room temperature, indefinitely
4% w/v PFA in 1× PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| 4× PBS | 1× | 25 mL |
| 16% w/v Paraformaldehyde | 4% w/v | 25 mL |
| ddH2O | n/a | 50 mL |
| Total | n/a | 100 mL |
Storage: Keep at 4°C, 1–2 weeks
CRITICAL: Paraformaldehyde is a toxic carcinogen and irritant. Handle with gloves under a fume hood.
Hydrogel Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 2% bioacrylamide (cold) | 0.05% | 1.25 mL |
| 40% acrylamide (cold) | 4% | 5 mL |
| 1× PBS (cold) | n/a | 43.75 mL |
| VA-044 | 0.541 mM | 0.175 g |
| Total | n/a | 50 mL |
Storage: Best used fresh but can be kept at 4°C for a week.
CRITICAL: VA-044 may irritate the skin, eyes, and lungs if it comes in direct contact. Handle with care.
Electrophoresis Running Buffer (Alternative to Logos Bio C13001)
| Reagent | Final concentration | Amount |
|---|---|---|
| Boric acid | 200 mM | 12.37 g |
| SDS | 140 mM | 40 g |
| Sodium sulfite | 50 mM | 6.3 g |
| NaOH | 1N | As needed to adjust pH to 8.5 |
| ddH2O | n/a | Bring to 1 L |
| Total | n/a | 1 L |
Storage: Room temperature for up to 6 months.
CRITICAL: Boric acid and SDS may irritate the skin, eyes, and lungs if it comes in direct contact. Handle with care.
3% Hydrogen Peroxide Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 30% Hydrogen Peroxide | 3% | 10 mL |
| dH2O | n/a | 90 mL |
| Total | n/a | 100 mL |
Storage: Keep at 4°C, indefinitely.
Alternative: 3% Hydrogen Peroxide Solution for topical use can be purchased at stores providing medical supplies and used directly.
5% Ammonium Hydroxide Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 25% ammonium hydroxide | 5% | 100 mL |
| dH2O | n/a | 400 mL |
| Total | n/a | 500 mL |
Storage: Keep at room temperature, indefinitely.
Immunohistochemistry (IHC) Buffer/SWITCH ON
| Reagent | Final concentration | Amount |
|---|---|---|
| 10% Sodium azide | 0.02% w/v | 1.0 mL |
| Bovine serum albumin | 0.5% w/v | 2.5 g |
| Triton X-100 | 0.1 v/v | 0.5 mL |
| 1× PBS | n/a | Adjust to 500 mL |
| Total | n/a | 500 mL |
Storage: Keep at 4°C, up to 6 months
IHC Buffer with SDS/SWITCH OFF
| Reagent | Final concentration | Amount |
|---|---|---|
| 10% Sodium azide | 0.02% w/v | 1.0 mL |
| Bovine serum albumin | 0.5% w/v | 2.5 g |
| Triton X-100 | 0.1 v/v | 0.5 mL |
| SDS | 0.5 mM | 0.072 g |
| 1× PBS | n/a | Adjust to 500 mL |
| Total | n/a | 500 mL |
Storage: Keep at 4°C, up to 6 months
Note: The concentration of SDS can be up to 10 mM and should be tested for each antibody combination to ensure that it inhibits antibody binding.
CRITICAL: Direct contact with SDS may irritate the skin, eyes, and lungs. Handle with care.
CUBIC R2 Solution (without TEA)
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 25 wt% | 125 g |
| Sucrose | 50 wt% | 250 g |
| Triton X-100 | 0.1% v/v | 380 mL |
| dH2O | 15 wt% | 119.5 mL |
| Total | n/a | 500 mL |
Storage: Store up to 1 month at room temperature. Dispose of solution if it begins to smell strongly of ammonia.
CRITICAL: Mix 125 g urea (Sigma) and 119.5 mL dH2O in a glass beaker. Stir on a hot plate over low heat until the urea dissolves. Do not allow the solution to rise above 40°C or the urea will begin to decompose into ammonia. Slowly add 250 g sucrose (Sigma) and continue stirring on low heat until dissolved. Turn off heat and add 380 mL TritonX-100 (Fisher), stirring until well-mixed. The final solution should have no ammonia smell.
Step-by-step method details
Note: Unless otherwise stated, all storage, wash and incubation steps should be completed with a volume of solution that occupies at least 3 volumes of tissue. For example, if a tissue occupies approximately 2 mL of volume, all wash and incubation steps should be completed with >= 6 mL of solution.
Harvesting and fixing liver tissue
Timing: 2 days
-
1.
Mice should be housed, fed, and monitored in accordance with the protocols approved by the Institutional Animal Care and Use Committee. Mice should be euthanized humanely before the livers are removed. All mice used in this protocol were treated in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh School of Medicine.
Optional: Perfusion of the liver as described previously (Seglen, 1976; Gores et al., 1986) may be performed if desired, first with 1× PBS followed by 4% PFA in 1× PBS, prior to liver harvesting. This step, while optional, is strongly recommended, because it will remove any blood in circulation which facilitates tissue clearing by reducing the starting level of heme-containing compounds. Perfusion-fixation will result in rapid preservation and help maintain better tissue integrity over the course of clearing.
-
2.
Fix fresh tissue (whole liver or individual lobes, 5 mm thickness or less) in 4% PFA in 1× PBS for 24–48 h at 4°C.
Note: This step should be completed even following the above optional step of liver perfusion. Strong fixation is necessary to maintain tissue integrity during the clearing process.
Alternative: Tissues may also be fixed in 10% formalin. This is most applicable for human liver tissue which may be processed by a clinical pathology laboratory.
-
3.
Wash tissue with PBSA 3 times for 15 min each at room temperature (25°C–37°C) on a rotating nutator, or longer if desired. Fixed tissues can be stored at 4°C in PBSA for up to a year. It is imperative to store tissue in PBSA which includes sodium azide as a preservative to prevent growth of fungus or bacteria in the tissues (see problem 1).
Hydrogel embedding of liver tissue
Timing: 2 days
In this step, the tissue is embedded in an acrylamide-based hydrogel which is crosslinked to proteins within the tissue and holds them in place to preserve the overall tissue architecture. This is essential to ensure that structures are visualized in their native spatial relationships and to minimize structural disruption caused by the next steps of the clearing process.
-
4.
Incubate the tissue in hydrogel solution overnight (for at least 12 h and up to 24 h) at 4°C in a 50 mL Falcon container, making sure the tissue is fully submerged. This solution should be made fresh each time and can be stored at 4°C for about a week.
-
5.
Polymerize the hydrogel by placing the 50 mL container with tissue and hydrogel solution into a water bath at 37°C for 2.5 h. The hydrogel should be neither firm nor runny, with a gelatin-like texture.
-
6.
Remove polymerized hydrogel containing the tissue from the container and rub off excess hydrogel surrounding the tissue. Wash the tissue with 1× PBS.
Pause point: If necessary, tissues may be kept at 4°C in polymerized hydrogel solution overnight.
Electrophoresis-based tissue clearing and depigmentation
Note: This protocol refers to the X-CLARITYTM clearing system. See the other resources section for information on alternative clearing systems.
Equipment setup
Timing: 3–4 h
-
7.Set up X-CLARITYTM Electrophoretic Tissue Clearing System.
-
a.Run distilled water through the system 2 times to ensure that it is clean. Ensure there are no leaks in any of the tubing connecting the main system, the sample holder, and the buffer reservoir.
-
b.Place 1–1.5 L of Electrophoretic Clearing SolutionTM (or see alternative electrophoresis clearing buffer in the materials and equipment section) into the reservoir and run the system at temperature 37°C, voltage 70 V, pump speed 100 rpm, and current ∼1.2 Amps for 20 min to ensure that the temperature is maintained at or below 37°C.
-
a.
Electrophoresis-based tissue clearing and depigmentation
Timing: 5–9 days depending on tissue size and composition
During this stage, the tissue is submerged in a buffered solution containing the detergent SDS. An electrophoretic gradient is applied to facilitate removal of lipids from the tissue. At the same time, we add N,N,N′,N′-tetrakis(2-hydroxypropyl)ethylenediamine (Quadrol) to the solution, which contributes to the depigmentation of heme-containing macromolecules within the liver.(Susaki et al., 2014)
-
8.Begin active tissue clearing process.
-
a.Turn on the X-CLARITYTM Tissue Clearing System (set up as per manufacturer specifications).
-
b.Add 1.5 L of X-CLARITYTM Electrophoretic Tissue Clearing Solution to the reservoir. Then add 10–20 mL of Quadrol to the reservoir. Quadrol is very viscous. Do not try to dissolve the Quadrol manually, it will gradually dissolve over time when the buffer begins to circulate.
-
c.Place the tissue into the desired sample holder (see 3D-printing instructions in the other resources section) and place it into the ETC chamber of the X-CLARITYTM system.
 CRITICAL: Make sure the tissue is secured and cannot float out of the chamber as this could block the buffer from circulating freely and raise the pressure inside the chamber which may cause it to crack and leak. Also make sure the chamber lid and reservoir lids are sealed securely to prevent leaks.
CRITICAL: Make sure the tissue is secured and cannot float out of the chamber as this could block the buffer from circulating freely and raise the pressure inside the chamber which may cause it to crack and leak. Also make sure the chamber lid and reservoir lids are sealed securely to prevent leaks. -
d.Adjust the settings on the X-CLARITYTM system to set the temperature at 37°C, voltage at 70 V, pump speed at 100 rpm, and current at ∼1.2 Amps (both voltage and Amps will vary as the buffer is consumed). Set the machine to constant current.
-
e.Begin the run by starting the X-CLARITYTM system and selecting ETC Active Clearing.
-
f.Replace the tissue clearing solution every 24–48 h.
 CRITICAL: How long the tissue will take to clear depends on the dimensions of the tissue as well as the composition of the tissue which may be influenced by genetic, environmental and experimental factors. A small chunk of liver 2 mm in thickness may clear in 48–72 h, while a whole median lobe, left lobe, or right lobe of murine liver may clear in 1–2 weeks, after which it will reach a steady state and is unlikely to clear any further. The Quadrol will initially turn the tissue into a brown/green color due to dissolved heme, but as the clearing proceeds, the color will be released into solution and the liver will begin to turn pale. Under most circumstances, the color will never be fully removed from the liver at this stage. You can expect the tissue to have a light red/brown hue. The liver will not appear clear at this stage. (See problem 2).
CRITICAL: How long the tissue will take to clear depends on the dimensions of the tissue as well as the composition of the tissue which may be influenced by genetic, environmental and experimental factors. A small chunk of liver 2 mm in thickness may clear in 48–72 h, while a whole median lobe, left lobe, or right lobe of murine liver may clear in 1–2 weeks, after which it will reach a steady state and is unlikely to clear any further. The Quadrol will initially turn the tissue into a brown/green color due to dissolved heme, but as the clearing proceeds, the color will be released into solution and the liver will begin to turn pale. Under most circumstances, the color will never be fully removed from the liver at this stage. You can expect the tissue to have a light red/brown hue. The liver will not appear clear at this stage. (See problem 2). CRITICAL: The tissue clearing solution will turn yellow over time and should be replaced every 24–48 h to encourage continuous dissolution and removal of lipids from the tissue into circulation. The tissue can be cleared for up to 2 weeks if needed with regular changes of clearing solution. You may pause the run at any time to visually check on the state of the sample. If clearing multiple tissues at a time, you may need to change the buffer more frequently. Additionally, when clearing multiple samples simultaneously, it may be helpful to rotate samples into different compartments within the clearing chamber to ensure uniformity of clearing among the samples. An opportune time to rotate samples is during changes of clearing solution.
CRITICAL: The tissue clearing solution will turn yellow over time and should be replaced every 24–48 h to encourage continuous dissolution and removal of lipids from the tissue into circulation. The tissue can be cleared for up to 2 weeks if needed with regular changes of clearing solution. You may pause the run at any time to visually check on the state of the sample. If clearing multiple tissues at a time, you may need to change the buffer more frequently. Additionally, when clearing multiple samples simultaneously, it may be helpful to rotate samples into different compartments within the clearing chamber to ensure uniformity of clearing among the samples. An opportune time to rotate samples is during changes of clearing solution.
-
a.
-
9.
When the color of the liver has reached a steady state (see the note above), stop the run, drain the sample chamber and turn off the X-CLARITYTM system. Remove the sample from the sample holder and wash the tissue in PBSA.
Pause point: The tissue can be stored at this point in PBSA at 4°C for up to 1 year.
Ammonium hydroxide treatment (optional for mouse liver; recommended for human liver)
Timing: 24–72 h, depending on tissue size and composition
Next, the tissue is treated with ammonium hydroxide to further dissolve heme and further decolorize the tissue (Jing et al., 2018).
-
10.
Wash the tissue in PBSA for 2 h on a shaker.
-
11.
Place the tissue in a solution of 5% ammonium hydroxide for 24–72 h on a gently rotating shaker at 30°C–37°C. Change the solution every 12–24 h.
Note: Step 11 can be repeated several times. The tissue should appear yellowish-white after this step. However, once no obvious improvement in color is seen after repeating step 11, then continue to step 12.
Pause point: The tissue can be stored at this point in PBSA at 4°C for up to 1 year.
Hydrogen peroxide bleaching
Timing: 24–72 h, depending on tissue size and composition
Next, the tissue is bleached with hydrogen peroxide which will further eliminate light absorption. Treatment with substances like hydrogen peroxide can contribute to bleaching of pigments through oxidation and structural degradation (Tainaka et al., 2016). The duration of treatment can be adjusted based on the size of the tissue.
-
12.
Wash the tissue in PBSA for 2 h on a shaker.
-
13.Place the tissue in a solution of 3% hydrogen peroxide for 24–72 h. Change the solution every 6–12 h.Note: Step 13 can be repeated several times. The tissue should appear yellow/white after this step. However, once no obvious improvement in color is seen after repeating step 13, then continue to step 14. (See problem 2).
-
14.
Wash the tissue in PBSA for 2 h on a shaker.
Pause point: The tissue can be stored at this point in PBSA at 4°C for up to 1 year.
Immunofluorescence staining
Timing: 2–3 weeks
At this stage, the cleared tissue can be used for immunofluorescent labeling. We have based our method on the SWITCH protocol (Murray et al., 2015), which has been shown to improve the distribution of antibody throughout thick tissue samples while reducing antibody-antibody aggregates which may contribute to artifacts during imaging. This method uses SDS to prevent antibody binding while the antibodies are diffusing through the sample to achieve an even distribution of stain throughout the tissue. Upon removal of the SDS the antibodies can then bind to their targets. The antibodies used here are the same as those used in conventional immunolabeling, and protocols should be optimized accordingly (see problem 3, problem 4, and problem 5). Concentrations of antibody in LiverClear should remain the same or be increased relative to conventional immunofluorescence staining protocols to account for the size of the sample that is being stained and the commensurate increase in antigen. In general, the total amount of antibody used is generally much greater than for conventional staining. We suggest optimizing on smaller fragments of cleared tissues prior to scaling up. After staining, the samples are post-fixed in paraformaldehyde to lock the antibodies in place long-term which enables prolonged imaging sessions and storage of the sample.
Note: Since hydrogen peroxide is reactive, it is suggested that each antibody be tested on tissue sections to determine whether it still performs as expected after hydrogen peroxide treatment. Hydrogen peroxide did not affect the performance of antibodies used in our studies.
Primary antibody stain:
-
15.
Acclimate the tissue in IHC buffer with SDS / SWITCH-OFF for 1 h – overnight. See note included with the SWITCH-OFF solution recipe regarding the flexibility of SDS concentration. SDS concentration in SWITCH-OFF should be tested for each antibody combination.
-
16.
Incubate tissue in SWITCH-OFF along with primary antibodies of choice for 4–7 days depending on the size of the tissue. The tissue should be kept on a shaker rotating gently at a temperature ranging from 25°C–37°C. Multiple primary antibodies can be added at the same time, as with conventional immunofluorescence staining protocols.
CRITICAL: Tissues should be incubated in wide-bottom tubes (such as 50 mL Falcon containers) to allow for free circulation of the solution containing antibodies all around the tissue. The tissue should be fully submerged. The volume should be only enough to cover the whole tissue to minimize antibody waste and maximize antibody concentration.
-
17.
Wash the tissue in IHC buffer / SWITCH-ON without antibodies for 24 h, on a shaker at a temperature ranging from 25°C–37°C.
-
18.
Wash the tissue in SWITCH-ON solution 3 times for 2 h each, on a shaker at a temperature ranging from 25°C–37°C.
Secondary antibody stain:
-
19.
Acclimate the tissue in SWITCH-OFF buffer for 1 h – overnight.
-
20.
Incubate tissue in SWITCH-OFF solution as well as secondary antibodies conjugated to fluorophores of choice for 4–7 days depending on the size of the tissue. The tissue should be kept on a shaker rotating gently at a temperature ranging from 25°C–37°C. Keep the tissue shielded from light by covering the container in aluminum foil.
Note: At this stage nuclear stains like ToPro3 and DRAQ5/7 can be added. These stains are compatible with SWITCH and it is suggested that you use SWITCH when using these dyes in large cleared samples. Avoid DAPI for the reasons discussed in the quantification and statistical analysis section.
-
21.
Incubate tissue in SWITCH-ON for 24 h, on a shaker at a temperature ranging from 25°C–30°C.
-
22.
Wash the tissue in SWITCH-ON or PBSA 3 times for 2 h each, on a shaker rotating gently at a temperature ranging from 25°C–37°C.
-
23.
Incubate the tissue in 4% PFA for 2 h on a shaker rotating gently at a temperature ranging from 25°C–30°C. This step fixes the antibody stain in place and enable the sample to undergo prolonged imaging sessions and storage.
-
24.
Wash tissue in PBSA for 1 h on a shaker rotating gently at a temperature ranging from 25°C–30°C.
Pause point: It is strongly suggested to continue to the next steps to get the best imaging results. However, the tissue can be stored at this point in PBSA at 4°C for up to 1 year. Prolonged storage is likely to result in signal degradation.
CUBIC R2 clearing
Timing: 24 h–2 weeks (or storage for 1–2 years)
The final stage of clearing involves submerging the tissue in CUBIC R2 solution, which has a refractive index similar to liver tissue. The tissue will appear more transparent the longer it is left in the CUBIC R2 solution. Clearing is usually complete within a few days but can require up to 2 weeks for large samples.
Note: CUBIC R2 solution contains urea which will cause the liver tissue to expand over time. Expansion of the tissue will improve clearing, but because the tissue is physically larger it may be less amenable to imaging on some microscope platforms.
-
25.
Place tissue in CUBIC R2 solution at room temperature for at least 24 h. Change the solution every 24–48 h until the desired clarity is achieved.
Pause point: It is strongly suggested to continue to the next steps to get the best imaging results; however, the tissue can be stored at this point in CUBIC R2 at room temperature in the dark for up to 1–2 years.
Microscopy and image processing
Timing: 1 week
In this study, we used ribbon-scanning confocal microscopy (Watson et al., 2017) outfitted with a Nikon CFI90 20XC Glyc objective. Fluorescent imagery in all figures was generated in Bitplane Imaris. LiverClear samples can be imaged on any microscope platform designed for large cleared tissue.
Expected outcomes
LiverClear results in rapid, simple, and effective clearing of mouse liver tissue
The original CLARITY protocol, first published in 2013, combines sodium dodecyl sulfate (SDS)-based solubilizing of lipids with an electrophoretic gradient to drive the SDS-lipid micelles out of the tissue. This results in rapid delipidation within a few days, which is faster than protocols relying on passive diffusion (Epp et al., 2015; Yang et al., 2014; Lee et al., 2014; Chung et al., 2013). Furthermore, the use of an acrylamide-based hydrogel helps preserve tissue architecture and protein content during the delipidation process, improving the quality of subsequent protein detection and tissue imaging (Chung et al., 2013). Even though the protocol allowed for effective clearing for many organs, it only resulted in moderate clearing of the liver, largely due to the lack of specific depigmentation strategies (Yang et al., 2014; Lee et al., 2014; Epp et al., 2015; Chung and Deisseroth, 2013).
Alternative protocols, such as Scale and CUBIC, rely on passive delipidation combined with hyperhydration of the sample using concentrated solutions of urea and sucrose (Susaki et al., 2014; Tainaka et al., 2014, 2018; Hama et al., 2011). Hyperhydration lowers the refractive index of the tissues, reducing light scattering and improving the transparency of the tissue in aqueous solutions (Susaki et al., 2014; Tainaka et al., 2014, 2018; Hama et al., 2011). The development of CUBIC in particular resulted in the discovery that certain polyhydric aminoalcohols facilitated lipid solvation when combined with traditionally used detergents (Susaki et al., 2014). Interestingly, aminoalcohols such as N,N,N′,N′-Tetrakis(2-Hydroxypropyl)ethylenediamine (Quadrol), which is a main component of one of the CUBIC clearing solutions, also contributes to decoloration of heme-based substances, possibly by promoting heme dissociation from cellular proteins(Susaki et al., 2014). In addition, it was shown that treatment with substances like hydrogen peroxide can contribute to bleaching of pigments through oxidation and structural degradation(Susaki et al., 2014; Tainaka et al., 2016). However, the advancement of depigmentation strategies alone, described in both CUBIC and Scale, did not allow for substantial progress in clearing the liver.
LiverClear combines features of both CLARITY and CUBIC to efficiently clear liver tissue using aqueous-based methods, which are less toxic and result in only minimal distortion of the tissue. The protocol combines rapid delipidation by electrophoresis and adds several steps to promote depigmentation. As a result, significant clearing of liver tissue is achieved in a matter of 1–2 weeks.
Figure 2 shows the step-by-step progression of clearing of a single lobe of liver tissue after each step of the clearing protocol. Each square in the grid beneath the tissue is 5 mm × 5 mm. Figure 2B shows fresh fixed liver tissue consisting of one large lobe of mouse liver, of about 4 mm thickness. Figure 2C shows the same tissue after 6 days of electrophoretic clearing with Quadrol added to the circulating buffer. This process alone produces decolorization of the liver tissue and minor translucency around the edges of the tissue, but for a large lobe is not sufficient to achieve translucency. Figure 2D shows improved decolorization evident by the lighter color tone after 2 days of incubation in 3% hydrogen peroxide. Figure 2E shows the appearance of the liver after 1 day of incubation in CUBIC R2 solution, and we can see that the grid lines are visible beneath a large part of the liver tissue except for the very middle which is the thickest part. Finally, Figure 2F shows further clearing of the liver tissue after incubation in CUBIC R2 for 12 days. The tissue can be left in CUBIC R2 solution indefinitely at room temperature and continues clearing over time. However, satisfactory immunostaining and imaging can be achieved without extended incubation times, as demonstrated by Figures 3, 4, and 5.
Figure 2.
Diagram of tissue clearing and staining protocol with example images
(A) Timeline of mouse liver tissue clearing protocol including immunostaining and image processing.
(B–F) Liver tissue at various stages of the clearing process showing the gradual increase in translucency of the tissue over time, from (B) fresh fixed tissue, (C) post-electrophoretic clearing, (D) after hydrogen peroxide treatment, (E) after 1 day incubation in CUBIC R2, and (F) after 12 days incubation in CUBIC R2 solution. Grid cubes are 5 mm × 5 mm.
Figure 3.
3D imaging of biliary transport structures in healthy and diseased livers with a genetic absence of bile ducts from birth
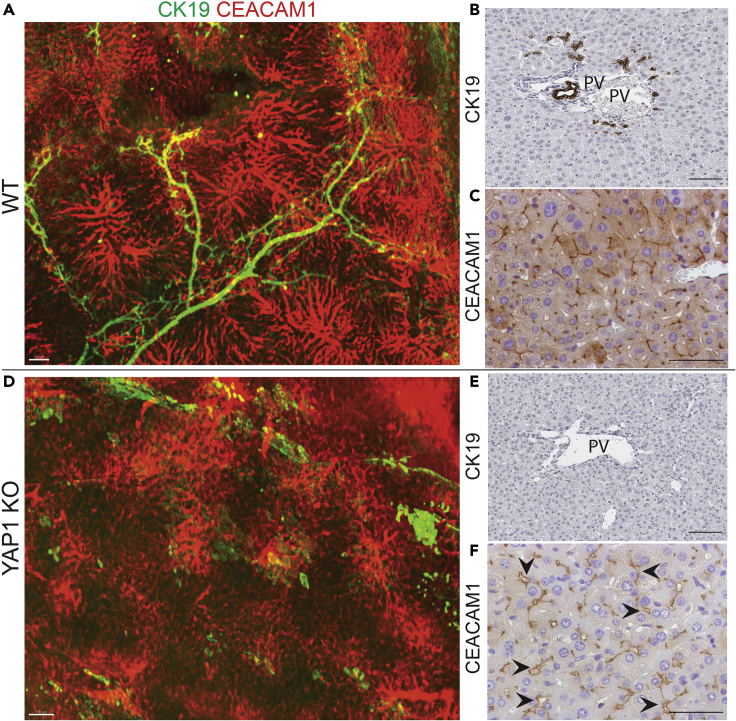
(A and B) 3D imaging of normal (WT) liver and diseased liver lacking in bile ducts (YAP1 KO), stained for CK19 which marks bile ducts and CEACAM1 which marks hepatocyte canaliculi.
(C and D) Whole slide scans of slides from WT and KO liver stained for CK19 showing the 2D histological correlates to the 3D imaging. Arrows point to bile ducts. Scale bars: A. 1500 μm, B. 1000 μm, C. 500 μm, D. 1000 μm.
Figure 4.
Higher magnification of 3D imaging of biliary transport structures in healthy and diseased livers along with histological correlates
(A and D) Zoomed view of 3D reconstructions from Figure 3, showing WT (A) and KO (D) livers stained for CK19 and CEACAM1, highlighting the loss of bile ducts and disorganization of canaliculi in KO livers vs WT.
(B and C, E and F) Immunofluorescence showing CK19 and CEACAM1 staining for WT (B and C) and KO (E and F) livers showing the histological correlates to A and D. Arrows point to dilated canaliculi. Scale bars: A. 100 μm B. 100 μm C. 50 μm D. 100 μm E. 100 μm F. 50 μm
Figure 5.
3D imaging of hepatocellular carcinoma liver cancer model
(A and B) 3D imaging of mouse liver with hepatocellular carcinoma stained for glutamine synthetase (A) and inset in (B) highlighting irregular sheets and clusters of tumor cells.
(C) Histology of the same cancer model showing that tumor cells are strongly positive for glutamine synthetase and are irregularly spread out through the parenchyma in nodular aggregates. Scale bars: A. 5000 μm B. 500 μm C. 200 μm
Immunostaining and ribbon-scanning confocal microscopy on cleared liver tissue can produce high-resolution images of micron-level tissue structures
Figures 3 and 4 demonstrate the results of immunofluorescent staining to visualize the biliary transport structures, namely the bile ducts labeled with cytokeratin 19 (CK19) and the hepatocyte canaliculi labeled with CEACAM1. In healthy adult liver (Figures 3A and 4A–4C), bile is produced by hepatocytes and secreted into the canaliculi, which are small channels between hepatocytes that push the bile towards the larger bile ducts, which form a branching network that dumps bile into the common bile duct exiting the liver. After clearing and immunostaining, we were able to visualize the biliary tree throughout a whole liver lobe of 3–4 mm thickness (CK19) and also visualize the fine network of canaliculi which are normally 1–2 mm wide (CEACAM1). Figure 4 offers a zoomed view of samples from Figure 3 which illustrate the level of detail, specificity, and precision that can be achieved with this immunostaining method. Figures 3C, 3D and 4B, 4C also show the 2-dimensional histological correlates which are the standard imaging techniques used in the field. Traditional immunostaining allows CK19-labeled bile ducts to be seen mostly in cross-section, whereas 3D-imaging allows us to appreciate the precise branching structure and interconnectedness of the ducts. This is critical to our understanding of bile duct formation, how structure changes in the setting of injury, and how duct repair takes place at an organ level, not just a cellular level.
To highlight the utility of this technique, we show results of imaging with the same markers on murine liver in which YAP1 was deleted in early development, resulting in a congenital absence of bile ducts (YAP1 KO)(Molina et al., 2021). Figure 3B grossly shows the absence of CK19-labeled ductal structures throughout the liver (consistent with the absence of CK19-positive ducts illustrated by histology in Figure 4E). Figure 3B also shows that the hepatocyte canaliculi are present but much more disorganized than in normal liver. Figure 4D highlights this structural change, showing dropout of some canaliculi and swelling of others, which is consistent with a loss of an outflow tract and thus impaired fluid flow through these structures. Histology (Figures 4E and 4F) shows the presence of canaliculi marked by CEACAM1 and also reveals some structural dilatation (arrows), but it does not convey the level of disorganization that can be clearly understood from Figure 4D.
LiverClear protocol allows for the clearing and imaging of murine hepatocellular carcinoma tumors
Figures 5A and 5B shows an example of a liver tissue from a murine model of hepatocellular carcinoma driven by constitutively activated forms of β-catenin and Met receptor.(Zhan et al., 2018; Puliga et al., 2017; Tao et al., 2016) In this case, we have labeled the tumors using glutamine synthetase (GS), a canonical target of β-catenin signaling which is strongly expressed in β-catenin-activated HCC tumors, as can be seen by immunohistochemical staining of tumors from the same mouse model in Figure 5C. We can visualize clumps of GS-positive tumor cells which resemble sheets and aggregates matching the clusters seen by IHC, and we can even resolve individual cells within the tumor areas as seen in Figure 5B. This is a promising start and shows the potential of visualizing complex disease structures.
LiverClear protocol is effective at clearing human liver
Deidentified human liver tissue was collected under an approved University of Pittsburgh IRB protocol (#PRO08010372) and obtained from the Clinical Biospecimen Repository and Processing Core (CBRPC) of the Pittsburgh Liver Research Center. We collected non-tumor liver tissue adjacent to hepatocellular carcinoma tumors from two different patients undergoing surgical resection of tumor tissue. Patient Hu-1 had high levels of fibrosis particularly around the hepatocytes (pericellular fibrosis), while patient Hu-2 had predominantly fatty liver, as shown through H&E and Sirius Red staining in Figures 6A and 6B. Tissues were fixed in either 4% PFA (as with the mouse tissue) or 10% formalin, in which most pathology specimens are fixed clinically. Figure 6C shows the time course of clearing human liver tissue. In this case, we added an additional step, incubating the tissues in 5% ammonium hydroxide to further encourage dissociation of heme-containing compounds. Treatment with hydrogen peroxide and ammonium hydroxide resulted in dramatic discoloration of the fatty liver tissue, while the fibrotic liver was less affected by this treatment. After 3 weeks of incubation in CUBIC R2 solution, both samples are mostly translucent. The fatty liver fixed in 4% PFA showed no difference to that cleared in 10% formalin, and indeed the fibrotic liver fixed in 10% formalin looks slightly clearer than that fixed in 4% PFA, showing that 10% formalin is a viable fixative for this process and opening the door to using most clinical pathologic liver specimens. Clearing human liver tissue will allow for the direct observation of tissue architecture in human disease, providing a new investigative method that can be correlated to traditional imaging studies such as ultrasound, CT, and MRI for the tracking of disease progression or response to treatment.
Figure 6.
Tissue clearing of human liver tissue
(A and B) H&E (A) and Sirius Red (B) staining of formalin fixed liver tissue. Yellow arrows point to fibrosis, black arrows point to fatty tissue.
(C and D) Timeline of tissue clearing for human tissue fixed in either 4% PFA or 10% Formalin. Grid cubes are 5 mm × 5 mm. Scale bars: A-B. 200 μm
Quantification and statistical analysis
Prior to designing an experiment that will utilize large cleared tissues, one should carefully consider the way that imaging data will be analyzed. There are a few key factors to consider before embarking on a cleared tissue experiment. These include 1) the types of quantitative data analysis to be performed and 2) the size of the dataset to be acquired and/or the time required to image a large tissue.
Quantitative data analysis
Even under an ideal clearing scenario where LiverClear renders the tissue optically indistinguishable from the refractive index matching solution, commonly used wavelengths for fluorescent imaging ∼400–800nM continue to be absorbed and scattered within a tissue. Generally, the proportions of scattering and absorption are higher towards the blue end of the spectrum and decrease as wavelengths become redder. (Jacques, 2013) This phenomenon is most apparent in thick tissues where blue light detection will attenuate very quickly often reducing imaging depths to a fraction of 1 mm (we suggest avoiding blue-fluorescent reporters such as DAPI, Hoechst, AlexaFluor 405, etc. in most scenarios) whereas red wavelengths may appear to penetrate unimpeded for many millimeters into the sample experiencing relatively minimal attenuation in fluorescence intensity. For these reasons, quantitative analysis of protein expression levels, which depend on precise fluorescent intensity measurements, are generally not an appropriate analysis for thick cleared tissues. If these types of analyses are desired, it is suggested that more traditional immunofluorescent assays be performed on thin sections. Generally, sections should be no thicker than 50 μm when imaging by confocal and 5 μm when imaging by widefield.
Cleared tissue studies like those using LiverClear are well suited for outcomes that will quantify numbers or the morphologies of structures – for example the presence of cell populations, the proportion of neuronal innervation or the length and number of branches in a biliary tree. While it is true that the fluorescent intensity of these structures is important during object detection analysis, the final metrics used to quantify the structures are based on object properties and not on the absolute fluorescent intensity of the underlying data.
Size of datasets and time required for acquisition
The predictable consequence of imaging large tissues is that the user spends considerably more time acquiring data, and the data generated is proportionately greater. This should be considered carefully when designing an experiment. The acquisition of two colors in a lobe of mouse liver at cellular resolution can require 24 or more hours of continuous acquisition on a high-speed imaging system like the ribbon scanning confocal microscopes used in this study or a modern light-sheet instrument. On less specialized instrumentation, acquisition at a similar scale can take weeks or months.
A good rule of thumb is that a lobe of liver will generate a minimum of 1 terabyte of data per channel. When designing an experiment, it is critical to success that these factors are considered. It is critical to prepare the proper software, hardware and storage space to deal with the datasets that will be generated. (Watson and Watkins, 2019) Consider whether these factors affect the feasibility of imaging the samples. Critically determine the sample size required to produce the appropriate statistical power. It may not be feasible to image enough samples to produce the necessary statistical power, thus it is important to consider whether more conventional imaging approaches or alternative methods can be used to replace or support the observations made with LiverClear.
Limitations
Sample thickness
We have tested LiverClear with tissue up to 5mm thick. At these sizes, the tissue is noticeably less clear towards the center but sufficient to enable imaging of the whole tissue. We expect that thicker tissues may be increasingly more difficult to clear; however, we have not tested the protocol under these conditions. Since tissues become progressively clearer the longer they incubate in the CUBIC R2 RI matching solution, we expect that LiverClear has the ability to clear much thicker specimens given sufficient time. This can be seen by looking at the human tissues, where prolonged (3+ week) incubation in CUBIC R2 rendered them completely transparent (Figure 6C).
Tissue variability
We anticipate that LiverClear will be effective at clearing most well fixed liver samples. We demonstrate acceptable results in human and mouse as well as fibrotic and fatty liver. However, it is expected that tissues will require differences in electrophoresis or incubation times, or application of decoloring reagents based on factors that may include species, genetics, age, diet, disease, environmental factors or specific experimental conditions. It is critical for success that the conditions required to clear various tissues are determined empirically before designing an experiment.
Choice of markers
A strength of LiverClear is its ability to enable RI matching in an aqueous environment. Key to this process is the removal of lipids during the electrophoresis phase of clearing. This process does preserve proteins and nucleic acids which can be targets for visualization; however, lipids and lipophilic dyes will not work under these conditions. Furthermore, it is important to validate the preservation of endogenous fluorescent proteins by comparing the expected distribution after conventional tissue processing and LiverClear. We have successfully used anti-GFP antibody to detect endogenous EYFP both by conventional means and following LiverClear (images not shown). Our animal modes favor antibody enhancement of GFP signal, because endogenous signal is weak and may be further quenched during tissue preparation. We have not systematically analyzed the preservation of endogenous fluorescent proteins; however, we expect that LiverClear is compatible with such markers similar to the CLARITY and CUBIC protocols. Compatibility should be assessed for each experimental condition, and signal will depend on the abundance of protein, method of expression, distribution and quantity of cells expressing the fluorescent protein, and any association between the fluorescent protein and specific cell compartments.
Porphyrin plugs in liver tissue
This protocol cannot remove precipitates of porphyrins, such as those modeled in animals treated with DDC diet to induce porphyrin accumulation and severe cholestasis (Gayathri and Padmanaban, 1974).
Fibrotic tissue
Tissue clearing on a sample of human liver tissue with high levels of fibrosis was less effective than other tissue types, although partial clearing was still achieved. Smaller size of the tissue and longer treatment times at each step may improve clearing for fibrotic tissues.
Tissue distortion
Tissue distortion is common among most clearing protocols because they often cause the inflation or shrinkage of the tissue. LiverClear will cause the tissue to swell during the clearing process: first during the electrophoretic phase (Chung et al., 2013) and finally during the RI matching phase (Susaki et al., 2014). Although the hydrogel components of LiverClear are designed to provide integrity to the tissue and are expected to promote an even expansion of tissue, one should practice caution before using LiverClear (or any clearing protocol) to make conclusions about the size or absolute relationships between molecular targets. We suggest the use of photographs to track tissue changes to ensure that LiverClear does not cause excessive distortion of the tissue. Where possible, fluorescent images before and after expansion of fiducial markers inside the tissue can be used to verify that clearing preserved the geometry of the tissue.
Troubleshooting
Problem 1
Growth of fungus or bacteria in the tissue
Since tissues are processed for weeks and stored for months during this process, there is risk of colonization by fungus or bacteria. This may be visible as cloudiness in the buffer solution containing the tissue and/or whitish-yellow growths attached to the tissue. A putrid smell is also an indication of microbial growth.
Potential solution
The affected tissue must be discarded. To prevent this from occurring, it is imperative to maintain a high concentration of sodium azide (0.02%) in all solutions during tissue incubations, treatment and storage. Sodium azide is extremely toxic. One should take necessary precautions to ensure that it does not come into contact with the skin or ingested into the body.
Problem 2
The center of the tissue remains darker than the edges after clearing
Many of the techniques used in this protocol rely on passive or active diffusion of lipids and other substances through the tissue, and thus it is easier to clear the edges of a tissue rather than the center, particularly when using a large piece of liver tissue (greater than 5 mm thickness, 1 cm length, or 1 cm width).
Potential solution
There are several options to resolve this problem. First, consider if you can use one or more smaller pieces of tissue for your analysis. Reducing all three dimensions of the tissue sample will reduce the amount of time needed to clear the tissue and will generally facilitate more even clearing throughout the whole sample. Otherwise, tissues may be actively clearing with CLARITY/Quadrol for one to two weeks and incubated for longer times in H2O2 and CUBIC R2 solutions as needed to fully penetrate a large piece of tissue. Furthermore, periodically replacing the active clearing buffer once it turns yellow (every 1-2 days during active clearing) improves the efficiency of active clearing and makes better use of clearing time particularly for larger tissue samples that may require over a week to clear.
Problem 3
Antibody validation
The selection and validation of antibodies for staining large tissues can be challenging.
Potential solution
Choosing antibodies against any marker can be a frustrating process. We recommend first validating antibodies by conventional immunostaining of thin tissue sections using the same combinations of primary and secondary antibodies and/or the conjugated fluorophores you would like to use for LiverClear. In our experience, antibodies that work under conventional conditions continue to work after LiverClear processing, saving considerable time compared to testing directly on cleared tissue. 50 μm frozen sections work well. Tissue used for validations should have been fixed and stored in the same manner as the tissue to be used for LiverClear. Additionally, we recommend validating your antibodies on the types of liver tissue (healthy vs diseased) that you intend to apply to LiverClear. The goal is to maximize the sensitivity and specificity of the antibody for your target of interest, while reducing artifact and background noise. Properly validating antibodies will lead to savings of time, tissue, and cost by optimizing the concentrations of antibodies required for staining much larger samples. This will also help you understand what you expect to see in the 3D reconstruction, including the expression levels of your target and their distribution pattern in the tissue. Prior to staining a large tissue, we strongly recommend further validating your antibody on a small slice of tissue processed by LiverClear. Slices (0.1–1 mm) can be removed using a vibratome, microtome or razor blade then tested as described above. This additional step will provide the best results for assessing compatibility with LiverClear.
Problem 4
Antibody signal from immunostaining does not penetrate to the middle of the tissue
The tissue may appear clear, but after immunostaining and imaging the center of the tissue may have decreased or absent fluorescence intensity as compared to the edges of the tissue, resulting in a dark region without detectable signal in the center of the image.
Potential solution
There are several contributing factors that can affect antibody penetration. First, ensure that the tissue is free-floating in the solution containing antibodies to allow for free circulation of the buffer at all times while on a rotating shaker. For this we recommend incubating tissues in 50 mL wide-bottom plastic tubes (such as 50 mL Falcon tubes). Next, it may be necessary to increase the concentration of primary antibody to ensure there is enough to penetrate the tissue without being depleted by the antigen it encounters around the periphery of the tissue. It may also be necessary to increase the incubation time for antibodies to fully penetrate the tissue. This needs troubleshooting for individual antibodies. This also depends on the thickness of the tissue, with less time needed for smaller tissues. Finally, you can increase the concentration of SDS in SWITCH OFF solutions.
Note: Avoid blue wavelength fluorescent reporters where possible. One reason that antibodies appear absent in the cleared tissue during imaging may be due to absorption and scattering of light in thick cleared tissues. The use of fluorescent reporters closer to the red end of the spectrum will increase the success of detecting signal in deep tissues. See the discussion in the quantification and statistical analysis section for more detail.
Problem 5
Fluorescent artifact seen outside the tissue boundaries after imaging
Sometimes fluorescent signal is observed outside the boundaries of the tissue sample after imaging. This may be due to antibody clumping in the hydrogel surrounding the tissue.
Potential solution
Carefully remove excess hydrogel surrounding the tissue sample prior to incubating the tissue in antibody solutions for immunostaining. This can be done by gently rubbing off the hydrogel while keeping the tissue moist with PBSA. This will help to prevent antibody clumping outside the tissue, reduce fluorescent artifacts visible after imaging, and improve antibody penetrance during immunostaining.
Problem 6
Bubbles present in cleared tissue and/or hydrogel matrix
Sometimes after clearing you may observe bubbles on or in the tissue. This may distort tissue architecture and interfere with imaging.
Potential solution
First determine whether the bubbles may be in the solution and not the tissue itself. CUBIC R2 may be prone to bubble formation due to its high viscosity. These bubbles can be removed by allowing sufficient time for the bubbles to surface and dissipate or removing the bubbles by pushing them away from the tissue. The bubbles may be inside the tissue itself which may form due to a number of factors including 1) gasses dissolved in the hydrogel during polymerization, 2) strong/prolonged electrophoretic currents, and 3) tissue-specific factors. If bubbles occur, consider degassing the hydrogel in a vacuum for 30 min prior to polymerization to remove excess dissolved gasses. Polymerizing under gentle movement, like a nutator, can also disrupt the formation of large bubbles inside the tissue, but may require a prolonged polymerization time. If you suspect that electrophoresis is causing bubbles to accumulate in the tissue, the voltages can be reduced, but this will increase clearing time. Also, consider increasing the rate at which buffer circulates within the electrophoretic clearing chamber to hasten the removal of bubbles formed by electrolysis. Finally, tissue-specific factors may result in gasses being trapped in the tissue. This can result when tissues are exposed for prolonged periods to air during tissue harvest and prior to fixation. Where possible, reduce exposure to air by more quickly moving tissues into fixative following harvest. We have also observed that livers with heavy tumor burden particularly cystic or bloody tumors often have an increase in bubbles. Unfortunately, it is often not possible to know that air bubbles are present in the tissue until the LiverClear protocol is complete. We have not identified a way to remove bubbles once they have been formed without damaging the tissue. In rare circumstances, a needle and syringe can be used to remove air pockets by suction, but use caution as this is likely to damage the tissue.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Satdarshan P. Monga (smonga@pitt.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
Funding was provided by 2T32EB001026 and 1F30DK121393 to L.M.M., and 5R01CA204586, 1R01DK62277, and Endowed Chair for Experimental Pathology to S.P.M. Funding was also provided by NIH grant 1P30DK120531 to the Pittsburgh Liver Research Center, for services provided by the Advanced Cell and Tissue Imaging Core and the Clinical Biospecimen Repository and Processing Core. The study was also in part funded by 2P30CA047904-32 to S.W. and the Center for Biological Imaging.
Author contributions
Conceptualization, L.M.M., A.M.W., and S.P.M.; methodology, L.M.M. and A.M.W.; investigation, L.M.M., Y.K., N.J., M.S., and A.M.W.; resources, J.T. and T.W.; data curation, N.J., M.S., and A.M.W.; writing – original draft, L.M.M. and A.M.W.; writing – review and editing, L.M.M., A.M.W., and S.P.M.; visualization, L.M.M., Y.K., and A.M.W.; supervision, A.M.W., S.W., and S.P.M.; funding acquisition, L.M.M., S.W., A.M.W., and S.P.M.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101178.
Contributor Information
Alan M. Watson, Email: alan.watson@pitt.edu.
Satdarshan P. Monga, Email: smonga@pitt.edu.
Supplemental information
Data and code availability
The raw data files supporting the current study have not been deposited in a public repository, but they are available from the corresponding authors on request.
References
- Chung K., Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K., et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp J.R., Niibori Y., Liz Hsiang H.L., Mercaldo V., Deisseroth K., Josselyn S.A., Frankland P.W. Optimization of CLARITY for clearing whole-brain and other intact organs. eNeuro. 2015;2 doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri A.K., Padmanaban G. Biochemical effects of 3,5-diethoxycarbonyl-1,4-dihydrocollidine in mouse liver. Biochem. Pharmacol. 1974;23:2713–2725. doi: 10.1016/0006-2952(74)90042-2. [DOI] [PubMed] [Google Scholar]
- Gores G.J., Kost L.J., Larusso N.F. The isolated perfused rat liver: conceptual and practical considerations. Hepatology. 1986;6:511–517. doi: 10.1002/hep.1840060331. [DOI] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Jacques S.L. Optical properties of biological tissues: a review. Phys. Med. Biol. 2013;58:R37–R61. doi: 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- Jing D., Zhang S., Luo W., Gao X., Men Y., Ma C., Liu X., Yi Y., Bugde A., Zhou B.O., et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 2018;28:803–818. doi: 10.1038/s41422-018-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Choi J., Jo Y., Kim J.Y., Jang Y.J., Lee H.M., Kim S.Y., Lee H.J., Cho K., Jung N., et al. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Park J.H., Seo I., Park S.H., Kim S. Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev. Biol. 2014;14:48. doi: 10.1186/s12861-014-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L.M., Zhu J., LI Q., Pradhan-Sundd T., Krutsenko Y., Sayed K., Jenkins N., Vats R., Bhushan B., Ko S., et al. Compensatory hepatic adaptation accompanies permanent absence of intrahepatic biliary network due to YAP1 loss in liver progenitors. Cell Rep. 2021;36:109310. doi: 10.1016/j.celrep.2021.109310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E., Cho J.H., Goodwin D., Ku T., Swaney J., Kim S.Y., Choi H., Park Y.G., Park J.Y., Hubbert A., et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puliga E., Min Q., Tao J., Zhang R., Pradhan-Sundd T., Poddar M., Singh S., Columbano A., Yu J., Monga S.P. Thyroid hormone receptor-beta agonist GC-1 inhibits met-beta-catenin-driven hepatocellular cancer. Am. J. Pathol. 2017;187:2473–2485. doi: 10.1016/j.ajpath.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T.M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S., et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kubota S.I., Suyama T.Q., Susaki E.A., Perrin D., Ukai-Tadenuma M., Ukai H., Ueda H.R. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kuno A., Kubota S.I., Murakami T., Ueda H.R. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev Biol. 2016;32:713–741. doi: 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Murakami T.C., Susaki E.A., Shimizu C., Saito R., Takahashi K., Hayashi-Takagi A., Sekiya H., Arima Y., Nojima S., et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 2018;24:2196–2210.e9. doi: 10.1016/j.celrep.2018.07.056. [DOI] [PubMed] [Google Scholar]
- Tao J., Xu E., Zhao Y., Singh S., Li X., Couchy G., Chen X., Zucman-Rossi J., Chikina M., Monga S.P.S. Modeling a human hepatocellular carcinoma subset in mice through coexpression of met and point-mutant β-catenin. Hepatology. 2016;64:1587–1605. doi: 10.1002/hep.28601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A.M., Rose A.H., Gibson G.A., Gardner C.L., Sun C., Reed D.S., Lam L.K.M., St Croix C.M., Strick P.L., Klimstra W.B., Watkins S.C. Ribbon scanning confocal for high-speed high-resolution volume imaging of brain. PLoS ONE. 2017;12:e0180486. doi: 10.1371/journal.pone.0180486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A.M., Watkins S.C. Massive volumetric imaging of cleared tissue: the necessary tools to be successful. Int. J. Biochem. Cell Biol. 2019;112:76–78. doi: 10.1016/j.biocel.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Yang B., Treweek J.B., Kulkarni R.P., Deverman B.E., Chen C.K., Lubeck E., Shah S., Cai L., Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan N., Adebayo Michael A., Wu K., Zeng G., Bell A., Tao J., Monga S.P. The effect of selective c-met inhibitor on hcc in the met-active, β-catenin mutated mouse model. Gene Expr. 2018;18:135. doi: 10.3727/105221618X15174108894682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data files supporting the current study have not been deposited in a public repository, but they are available from the corresponding authors on request.