Highlights
-
•
Normal appearing brain matter (NABM) biomarkers in FLAIR MRI are related to cognition.
-
•
NABM texture in FLAIR MRI is correlated to mean diffusivity (MD) in dMRI.
-
•
Analysis conducted on large multicentre FLAIR MRI dataset: 1400 subjects, 87 centers.
-
•
NABM biomarkers vary differently across age and MoCA categories.
-
•
Biomarkers showed differences in patients with AD dementia and vascular disease.
Keywords: FLAIR, Texture analysis, Biomarkers, Normal-appearing brain matter (NABM), Alzheimer’s disease, Neurodegeneration
Abstract
A novel biomarker panel was proposed to quantify macro and microstructural biomarkers from the normal-appearing brain matter (NABM) in multicentre fluid-attenuation inversion recovery (FLAIR) MRI. The NABM is composed of the white and gray matter regions of the brain, with the lesions and cerebrospinal fluid removed. The primary hypothesis was that NABM biomarkers from FLAIR MRI are related to cognitive outcome as determined by MoCA score. There were three groups of features designed for this task based on 1) texture: microstructural integrity (MII), macrostructural damage (MAD), microstructural damage (MID), 2) intensity: median, skewness, kurtosis and 3) volume: NABM to ICV volume ratio. Biomarkers were extracted from over 1400 imaging volumes from more than 87 centres and unadjusted ANOVA analysis revealed significant differences in means of the MII, MAD, and NABM volume biomarkers across all cognitive groups. In an adjusted ANCOVA model, a significant relationship between MoCA categories was found that was dependent on subject age for MII, MAD, intensity, kurtosis and NABM volume biomarkers. These results demonstrate that structural brain changes in the NABM are related to cognitive outcome (with different relationships depending on the age of the subjects). Therefore these biomarkers have high potential for clinical translation. As a secondary hypothesis, we investigated whether texture features from FLAIR MRI can quantify microstructural changes related to how “structured” or “damaged” the tissue is. Based on correlation analysis with diffusion weighted MRI (dMRI), it was shown that FLAIR MRI texture biomarkers (MII and MAD) had strong correlations to mean diffusivity (MD) which is related to tissue degeneration in the GM and WM regions. As FLAIR MRI is routinely collected for clinical neurological examinations, novel biomarkers from FLAIR MRI could be used to supplement current clinical biomarkers and for monitoring disease progression. Biomarkers could also be used to stratify patients into homogeneous disease subgroups for clinical trials, or to learn more about mechanistic development of dementia disease.
1. Introduction
Half a million Canadians are living with dementia and 25,000 new cases are diagnosed every year. By 2031, this is expected to increase by 66% to more than 1 million Canadians, carrying a $16.6B cost per year to care for them (Chambers et al., 2016). Up until recently (FDA, 2021), there were numerous failed clinical trials targeting amyloid-beta plaques (Yiannopoulou et al., 2019). More research is needed to identify early precursors of cognitive decline and neurodegeneration, to detect disease early, monitor disease progression and to develop new therapies. Magnetic resonance imaging (MRI) shows brain damage accumulating in near-real time (Smith et al., 2019) and neuroimaging biomarkers from MRI are great candidates for disease monitoring and quantification.
Studies have investigated the relationship of macro- and micro-structural biomarkers in MRI of the brain to Alzheimer’s disease (AD) (first risk factor for dementia), and vascular disease (second dementia risk factor), including gray matter (GM) atrophy (Pini et al., 2016), ventricular enlargement (Nestor et al., 2008), white matter (WM) integrity (Ji et al., 2017) and white matter lesions (WML) (Frey, 2019, Oishi et al., 2008). In a meta-review (Lamar et al., 2020), the authors searched for commonalities between brain pathology in AD and cerebrovascular disease (CVD) risk factors. In total, 23 spatial regions were commonly associated with both CVD risk factors and Alzheimer’s dementia, including GM and subcortical structures, WM integrity and WML.
Gray matter atrophy has been studied for years for both dementia (Khan, 2016) and vascular diseases. One study controlled for vascular disease and examined both WM damage and GM pathology using diffusion MRI (dMRI) metrics fractional anisotropy (FA) and mean diffusivity (MD), and postulated that GM atrophy is secondary to WM damage (Pievani et al., 2010). For patients with CADASIL (genetic form of small vessel disease), more WM damage such as lacunes could induce more cortical thinning by impairing the microstructure of the WM tracts (Liu et al., 2021). White matter lesions, which are established CVD markers, predict cognitive decline, dementia, stroke, death, and WML progression increases these risks (Debette and Markus, 2010, Alber et al., 2019). WML represent increased and altered water content in hydrophobic white matter fibers and tracts. Changes in white matter vasculature likely contributes to WML pathogenesis (Gorelick et al., 2011). In Liu et al. (2021), WM tracts distal to WML were shown to have reduced integrity which is associated to a reduction of executive function and attention, which are both related to CVD. In Meng et al. (2017), the location of WML was found to play more of an important role on cognition, as compared to traditional total-lesion load measurements and implied that large-scale disconnection of networks could be the reason.
White matter disease spreads beyond the area of the visible FLAIR lesion and these subtle “pre-visible” alterations may be important vascular clues (Maniega et al., 2016). There is an increase in water diffusion in these regions which correlate to cognitive impairment (Smith et al., 2019). As shown in dMRI and FLAIR, WML that grow and develop de novo are preceded by changes in the normal-appearing white matter (NAWM). WM damage was seen as focal and cumulative effects of CVD and was a better predictor of cognition as compared to lesion load (Meng et al., 2017). In Zamboni et al. (2017), a lower FA in the WM correlated to a lower MoCA score indicating there is more widespread WM damage in patients with dementia. They also noted that FA and MD correlated more closely with cognitive deficits in vascular cognitive impairment (VCI). Dumont et al. (2019) computed a measure called free-water (FW) from dMRI which is the fraction of the diffusion signal described by isotropically unconstrained water. FW was able to differentiate normal controls (NC) from subjects with mild cognitive impairment (MCI) and AD and it is postulated that the differences are due to FW in the extracellular space around the axons or contamination from the CSF suggesting increased neuroinflammation.
Fluid Attenuation Inversion Recovery (FLAIR) MRI is preferred for WML analysis (Badji and Westman, 2020, Wardlaw et al., 2013, Ghafoorian et al., 2016, Chutinet and Rost, 2014), since the high signal from the cerebrospinal fluid (CSF) in T2 is suppressed, thus highlighting white matter disease (Lao et al., 2008). This is due to increased water content secondary to ischemia and demyelination are much more robustly seen in FLAIR than with T1/T2 (Gorelick et al., 2011). Although primarily used for lesion load and location analysis, studies are emerging that demonstrate that other measurements from FLAIR could be a valuable addition to biomarker pipelines. For example, in Maillard et al. (2013) FLAIR intensity in the regions around WML (the WML penumbra) were found to be an independent predictor for WML growth or de novo development, and the authors suggest that FLAIR should be considered as a continuous index of WM health and not be considered an inferior version of dMRI (Maillard et al., 2013). In De Groot et al. (2013), the authors show that dMRI and FLAIR signal intensity of the NAWM were associated with WML development independently (De Groot et al., 2013), which suggests they are capturing different pathological mechanisms. FLAIR contrast in WM is indirectly related to the attenuation of lipid protons within the myelin but there are many histopathological correlates suggesting more research into FLAIR MRI biomarkers is needed (Maillard et al., 2013). The authors stated that using FLAIR measurements from retrospectively collected FLAIR, with or without DTI, could “add substantial power to detecting treatment related differences” (Maillard et al., 2013). Although promising, the authors did not correlate FLAIR biomarkers to cognitive score.
To this end, we propose a novel biomarker panel that quantifies macro and microstructural biomarkers from the normal-appearing brain matter (NABM) in FLAIR MRI and investigate whether structural brain differences are associated with cognition. The NABM is composed of the normal white and gray matter regions of the brain, with the lesions and CSF stripped out. In Scola et al. (2010), analysis of GM and WM (NABM) regions for subjects with WML revealed increased water diffusion using DTI in the NABM which is suspected to indicate overall tissue loosening in GM and WM. In other works, both GM and WM were analyzed using dMRI and it was found that in regions with GM atrophy there is more diffusion and also less constrained flow in the WM for AD subjects (Scola et al., 2010). We opt for whole brain analysis where biomarkers are measured from the entire NABM region to quantify global disease burden as opposed to regions of interest or voxel-wise analysis since measurement variability in these methods can affect the ability (power) to detect change (Nave et al., 2007).
Three groups of biomarkers are proposed related to the intensity, texture and volume of the NABM region in FLAIR images. The texture biomarkers measure the structural integrity/damage of the NABM and three new features are proposed: macrostructural damage (MAD), microstructural damage (MID) and microstructural integrity (MII). The intensity features include the median intensity, the skewness and kurtosis of the intensity distribution of the NABM, and volume, measured as the ratio of the NABM volume normalized by the intracranial volume (ICV). Biomarkers are defined for a single sequence (FLAIR), without dependence on T1 or DWI. These were measured in over 1400 imaging volumes, from 87 international imaging centres from three datasets of patients with dementia, AD and vascular disease.
We hypothesize that FLAIR derived biomarkers in the NABM can differentiate between cognitively normal and cognitively impaired subjects. We further hypothesize that texture features from FLAIR MRI can be used to quantify microstructural changes related to how “structured” or “damaged” the tissue is. As FLAIR MRI is routinely collected for clinical neurological examinations, novel biomarkers from FLAIR MRI could supplement current clinical workflows for disease monitoring. Additionally, since FLAIR MRI contains CVD clues, this sequence could shed light into the relationship between CVD and dementia, for identifying early disease markers and determining optimal timing for therapeutic intervention.
2. Materials and methods
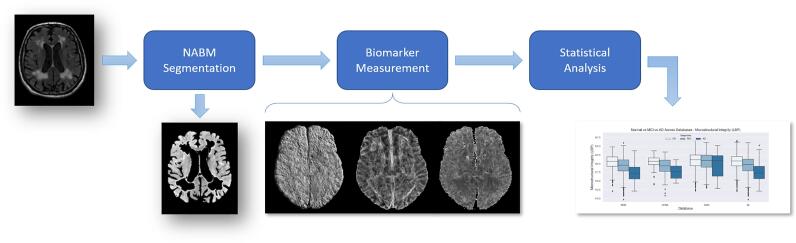
Three groups of NABM biomarkers for multicentre FLAIR MRI related to texture, intensity and volume were designed. In contrast to previous methods that mainly focus on volumetric biomarkers, the proposed biomarker panel quantifies both microstructural and macrostructural characteristics of the NABM region in one framework. The pipeline of the proposed work can be seen in Fig. 1.
Fig. 1.
Pipeline outlining major steps in this work; Original slice, NABM segmentation, resultant texture feature maps, along with statistical analysis.
2.1. Data
Three large, multicentre neuroimaging archives with axial FLAIR MRI and clinical data were used in this study. The summary of these three datasets are shown in Table 1. Data was acquired from over 80 international centres worldwide, and the imaging parameters are described in Table 2 The first dataset, ADNI (Alzheimer’s Disease Neuroimaging Initiative) (Aisen et al., 2021), includes Normal Controls (NC), Mild Cognitive Impairment (MCI) and Alzheimer’s Disease (AD) subjects from 1043 patients. The second data repository is CAIN (Canadian Atherosclerosis Imaging Network) (Tardif et al., 2013) which is a Canadian study investigating CVD on subjects with moderate to high stenosis and contains 383 patients. The last dataset, CCNA, is from the Canadian Consortium on Neurodegeneration in Aging (CCNA), a pan-Canadian study related to the common dementias (AD, Parkinson’s disease, Lewy Body, Frontotemporal Lobar Dementia and Subcortical Vascular Cognitive Impairment) (Mohaddes et al., 2021, Jessen et al., 2014). Only the 191 patients from CCNA with AD-type pathologies were analyzed. ADNI provides image quality scores where 3 and 4 indicates poor quality likely not optimal for automated analysis (Jack, 2011). In total 132 volumes were rated poor image quality and were excluded resulting in 1485 imaging volumes for ADNI, CCNA and CAIN combined.
Table 1.
Summary of three databases used in this study.
| Database | Disease | Centres | Patients | Volumes | Images | Scanners | Sex | Age |
|---|---|---|---|---|---|---|---|---|
| ADNI | AD | 58 | 911 | 911 | 32,796 | GE, Philips, Siemens | 479 M/ 432 F | 73.2 |
| CCNA | Dementia | 21 | 191 | 191 | 9168 | GE, Philips, Siemens | 96 M/ 95 F | 71.7 |
| CAIN | Vascular | 8 | 383 | 383 | 18,767 | GE, Philips, Siemens | 237 M/ 143 F | 70.3 |
| All | All | 87 | 1485 | 1485 | 60,731 | GE, Philips, Siemens | 812 M/ 670 F | 71.7 |
Table 2.
Database acquisition parameters.
| Database | Vendors | TR (ms) | TE (ms) | TI (ms) | Pixel Spacing | Slice Thickness |
|---|---|---|---|---|---|---|
| ADNI | GE, Philips, Siemens | 6000–11900 | 90–192.65 | 2000–2800 | 0.785–1.02 mm | 5 mm |
| CCNA | GE, Philips, Siemens | 9000–9840 | 120–146.44 | 2250–2500 | 0.9375 mm | 3 mm |
| CAIN | GE, Philips, Siemens | 8000–11000 | 117–150.24 | 2200–2800 | 0.42–1 mm | 3, 5 mm |
Clinical data included age and sex (all data sets), MoCA score (all data sets) and dementia diagnosis (ADNI, CCNA). MoCA was thresholded to find Normal Controls (NC) (MoCA 26), MCI (MoCA 19–25) and AD (MoCA 18) and the sample sizes are shown in Table 3. Diagnosis for both CCNA and ADNI were obtained through a battery of clinical instruments (Mohaddes et al., 2021, Alzheimer’s Disease Neuroimaging Initiative, 2008). The datasets were treated cross-sectionally and the baseline FLAIR volumes (first time point) of each subject was analyzed and used to determine cognitive status. Statistical analysis used all cohorts combined (ADNI, CCNA, CAIN) for a pooled analysis. Post-hoc exploratory analysis considered individual cohorts since the diagnostic labels were generated using different clinical instruments. There was appropriate consent and ethics approval to access the data.
Table 3.
MoCA datasplits and corresponding number of subjects per dataset.
| Database | Normal (26) | MCI (19–25) | AD (18) |
|---|---|---|---|
| ADNI | 256 | 464 | 98 |
| CCNA | 68 | 81 | 17 |
| CAIN | 216 | 127 | 14 |
| All | 540 | 672 | 129 |
For the same datasets, there were 52 CCNA imaging volumes and 46 CAIN imaging volumes with corresponding dMRI imaging data. CAIN dMRI data was acquired using a GE 3T Scanner, TR = 8800 ms, TE = 75.9–83.2 ms, along 25 directions using a b-value of 1000 s/mm2 and spatial resolutions of 1.48441.48443.6 mm. The CCNA data was acquired using a Siemens 3T Scanner, TR = 9400–13000 ms, TE = 64–101 ms along 31 directions using a b-value of 1000 s/mm2 with spatial resolutions of 222 mm.
2.2. NABM segmentation
To manage variability in multicentre FLAIR MRI data, the standardization pipeline in Reiche et al. (2019) was employed to reduce noise, bias field and intensity variability across scanners. The result is a more consistent intensity interval for tissues across different subjects. Following intensity standardization, whole volume brain extraction for FLAIR MRI (DiGregorio et al., 2021) is used to extract the intra-cranial volume (ICV) using UNET (DiGregorio et al., 2021). Spatial resampling was then performed to ensure local features are comparable across subjects that have different resolutions. The data is resampled to 1 mm 1 mm 1 mm using linear interpolation and cropping. The resultant resampled volumes have the same field of view, voxel size, and resolution. Using the intensity standardized, brain extracted and spatially resampled volumes, the same intensity thresholds were applied across the datasets to extract the NABM region by removing the CSF and hyperintense objects (white matter lesions) (Reiche et al., 2019, DiGregorio et al., 2021, Khademi et al., 2019). The WMH mask was dilated to avoid partial volume contamination. Several biomarkers are extracted from the NABM region as described below.
2.3. NABM biomarkers
Texture Biomarkers: Given an image slice with L number of gray levels in the volume, and spatial coordinates, features were extracted from local windows from each slice z. We investigated the finest window possible, which results in a physical width of 3 mm. The sliding window moves through the slice being processed , computes a texture feature at the spatial location to create a 2D feature map . The 2D feature maps were then averaged voxel-wise across slices from the 3D volume, to generate the mean feature map , that describes the global texture of the NABM per subject. Averaging helped to suppress noise, while amplifying the texture signal (as is common in synchronized averaging approaches (Michalski et al., 2011)). Based on the average feature maps , the median was extracted to result in a single metric (per imaging volume) that represents the global disease burden. In this work, three different texture features were explored.
Macrostructural Damage (MAD): The NABM macrostructural damage biomarker is based on a 2D version of Mantel’s test for spatial correlation as first proposed for images in Khademi (2009), which was later used to detect architectural distortion in mammograms (Rangayyan et al., 2012). The Mantel statistic (), quantifies the amount of spatial correlation between pixels by examining whether pixels that are close to one another are coupled with similar intensity values. The is large in regions with rapidly changing intensities, such as edges and textures, and low in homogeneous regions. Mantel’s statistic was calculated as a proxy for macrostructural damage by:
| (1) |
where, is the distance between pixels and and is the proximity of intensities and to one another. Window size for this analysis was fixed and as a result the distance between gray-levels is a driving factor in measuring texture properties with this metric. was computed on a per-slice and per-voxel basis to find the texture map for each slice, and the mean texture map was obtained through voxel-wise averaging of the 2D feature slices. Since high values can occur on boundaries of the brain, the brain mask was eroded with a kernel width equivalent to the analysis window size to remove them. In regions with rapidly changing intensities (textures, discontinuities, edges), is high, indicating more fluctuations in the local intensities. Since this both measures the local structural variations within the NABM tissue as well as larger scale features related to discontinuities (edges) between tissue classes we call this the macrostructural damage (MAD) marker.
Microstructural Damage (MID): To analyze microstructural texture, Mantel’s statistic was calculated per slice as per the macrostructural damage (MAD) marker, but instead, before performing voxel-wise averaging, the large values were automatically removed using Otsu’s thresholding to yield only small intensity differences. Although large features are clinically relevant and could be related to large intensity differences caused by edges or gross patterns (i.e. brains with more atrophy, or more lesions) the microstructural damage feature focuses more on the internal (small) variations within the NABM tissue. The microstructural damage feature map is denoted and voxel-wise averaging was completed to compute the global disease burden over the volume .
Microstructural Integrity (MII): A microstructural integrity (MII) marker based on local binary patterns (LBP) is proposed. LBP descriptors efficiently capture the local spatial patterns and the gray scale contrast in an image (Ojala et al., 1994, Ojala et al., 1996, Oppedal et al., 2015). Differences in intensity values in the local area are compared to the center pixel to define a texture pattern, and then LBP embeds this spatial structure into its descriptor. LBP descriptors can capture rapid changes in intensity, such as edge lines or ridges, since these regions have a higher intensity compared to their spatial neighborhood valleys. It has been used in many applications including facial recognition (Rahim et al., 2013) and MRI for brain tumor detection and dementia classification (Oppedal et al., 2015, Abbasi and Tajeripour, 2016). The LBP descriptor was used to quantify the overall microstructural integrity of the NABM in FLAIR MRI. Given a neighbourhood with a center pixel intensity and neighboring pixels , where P is the number of neighbors, the LBP descriptor is found by comparing the neighbouring pixels, , to the center pixel, , as in:
| (2) |
where s is the indicator function such that when and otherwise (Sairamya et al., 2019, Ojala et al., 2002). In this work, we employed the LBP operator on a 33 square neighbourhood (Tajeripour et al., 2007) to capture fine differences. The descriptor was used as a measure of structural integrity in this work since higher LBP descriptors generally describe patterns such as valleys and edges. In images with these similar “structured” patterns throughout the brain, there would be a high number of occurrences of these (larger) LBP descriptors (and more integrity).
Intensity Biomarkers: To measure the intensity of the NABM region, the median intensity was computed. In contrast to some recent works that examine FLAIR intensities only in regions surrounding WML, this biomarker investigates the changes in intensity of the NABM on a more global scale as both GM and WM are included in the analysis. In addition to median intensity, intensity skewness s and kurtosis were also investigated for the NABM of each volume. Skewness measures the (lack of) symmetry in the intensity distribution and gives indication whether the NABM region has more or less high intensities. Kurtosis measures how peaked a distribution is and could indicate more homogeneity in the NABM region for more peaked intensity distributions.
Volume Biomarkers: A macrostructural biomarker based on the NABM volume was included to complete the biomarker panel. Recall the NABM is the brain with hyperintensities and CSF stripped out. The volume in this region was computed by summing up the voxels in the NABM region and multiplying by the voxel resolution. To ensure differences in head size do not affect analysis, the NABM volume was normalized by the intracranial volume (ICV) to measure the NABM volume ratio:
| (3) |
2.4. Biomarker correlation analysis
To investigate characteristics of the FLAIR biomarkers, correlation analysis was performed. First, FLAIR MRI biomarkers were correlated to find relationships between biomarkers. Pearsons’s correlation was reported, along with p-value. The analysis investigated the relationship of FLAIR MRI biomarkers to quantitative metrics from the corresponding dMRI volumes. Fractional Anisotropy (FA) and Mean Diffusibility (MD) are common dMRI metrics used extensively in dementia and vascular disease research (Oishi et al., 2011, Alves et al., 2012). FA measures the degree of anisotropy of a water molecule (Morgan et al., 2013) and MD describes the total diffusion in each voxel related to disruption and degeneration of tissue microstructure in the WM (Chanraud et al., 2010) and demyelination and axonal loss (Lenglet, 2015) and gliosis, astrocytic alterations and other necrotic changes in the GM (Alexander et al., 2007). As shown in Scola et al. (2010), the MD of GM and WM both increase with progression of disease and neurodegeneration (Scola et al., 2010, Weston et al., 2015), indicating an increase of water diffusion is associated with both WM damage and GM loss (atrophy). However, FA histograms created from whole brain images containing GM and WM are the superimposition of two opposing trends (Della Nave et al., 2007). WM is a heavily anisotropic (higher FA values), while GM is much more isotropic (Lower FA values) (Della Nave et al., 2007, Hecke et al., 2016). The opposing values for GM and WM makes it difficult to elucidate changes in FA with both these two tissues combined. Therefore, only MD was explored for NABM analysis. MD is extracted using TractoFlow pipeline which was developed by the Sherbrooke Connectivity Imaging Lab (Theaud et al., 2020). FLAIR and MD were co-registered using ANTs SyN registration for the same subject and the FLAIR NABM mask was transformed to the dMRI space to avoid warping and to improve alignment, especially in ventricular regions. The median MD is extracted from each volume and compared to FLAIR MRI biomarkers using Pearson’s correlation analysis.
2.5. Analysis and statistics
To examine whether differences in micro- and macro-structure of the NABM in FLAIR MRI are related to cognition, ANOVA and ANCOVA analysis was used. As MoCA is available for all three datasets, MoCA was used to group patients into cognitive categories of NC, MCI and AD (see Table 3 for MoCA ranges and sample sizes). To gauge normality of the biomarker distributions, goodness of fit measures were performed and the data transformation was not necessary. Unadjusted univariable analyses (t-test, chi-square/ Fisher’s exact) were performed to assess relationships were appropriate. The Pearson correlation coefficient was used to compute the correlation between cross-sectional biomarkers and other continuous independent variables. First, unadjusted analysis is reported that examines the differences in biomarker means across cognitive groups. Results were reported as difference in means and p-value (Tukey adjustment for multiple comparisons) for all seven biomarkers. To examine the effect of age on the biomarkers, ANCOVA modeling with age and sex as covariates was performed to examine the interaction between age, sex and MOCA. For biomarkers that had a significant age*MoCA term, an additional two models were analyzed to investigate the two term interaction (sex*MoCA) and the three term interaction (age*sex*MoCA), respectively. If sex*MOCA term was significant in the first model, additional post hoc analysis is used to investigate differences in biomarkers between MoCA categories and sex. If biomarkers had a significant age*sex*MoCA term in the second model, an additional analysis was performed to estimate the slopes of the biomarkers for each MoCA-sex group as a function of age. The intent is to find biomarkers that stratify between disease groups that also have an age dependence, so biomarkers may be used to predict disease progression, trajectory and to determine optimal intervention times. Statistical significance was considered for p 0.05. All statistical analysis was performed using SAS 9.4 (The SAS Institute, Cary, NC, USA).
3. Results
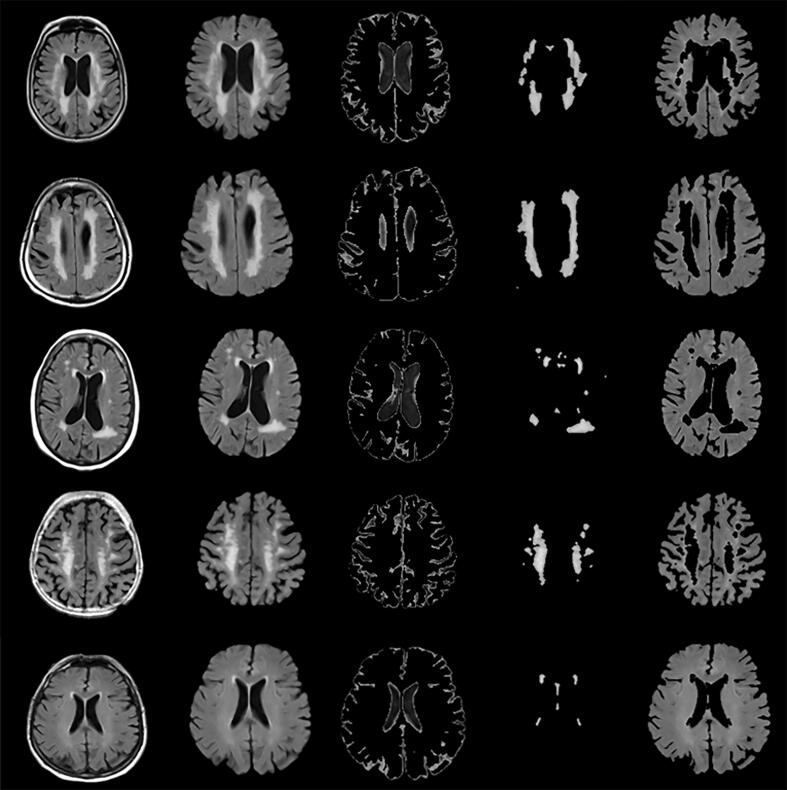
Seven NABM biomarkers were extracted across ADNI, CAIN and CCNA cohorts and their relationship to cognition is analyzed in this section. Intensity standardization was first employed to normalize the intensity interval of the brain region as shown in Fig. 12. The NABM region was then found by thresholding all images with the same intensity thresholds for CSF and WML across patients (Reiche et al., 2019). Example NABM segmentations are shown in Fig. 2. For each imaging volume, the following NABM biomarkers were then extracted: microstructural integrity (MII), microstructural damage (MID), macrostructural damage (MAD), intensity, kurtosis, skewness and NABM to ICV volume ratio.
Fig. 12.
Original intensity histograms (left) per subject which shows intensity variability across brain regions in multicentre data. The intensity histograms of the intensity standardized images (right) show that intensity intervals for tissues across subjects are normalized for fair biomarker comparisons.
Fig. 2.
NABM Segmentation in FLAIR MRI. Left to Right: Intensity standardized FLAIR MRI, skull-stripped, CSF, WML and final NABM segmentation.
3.1. Biomarker extraction
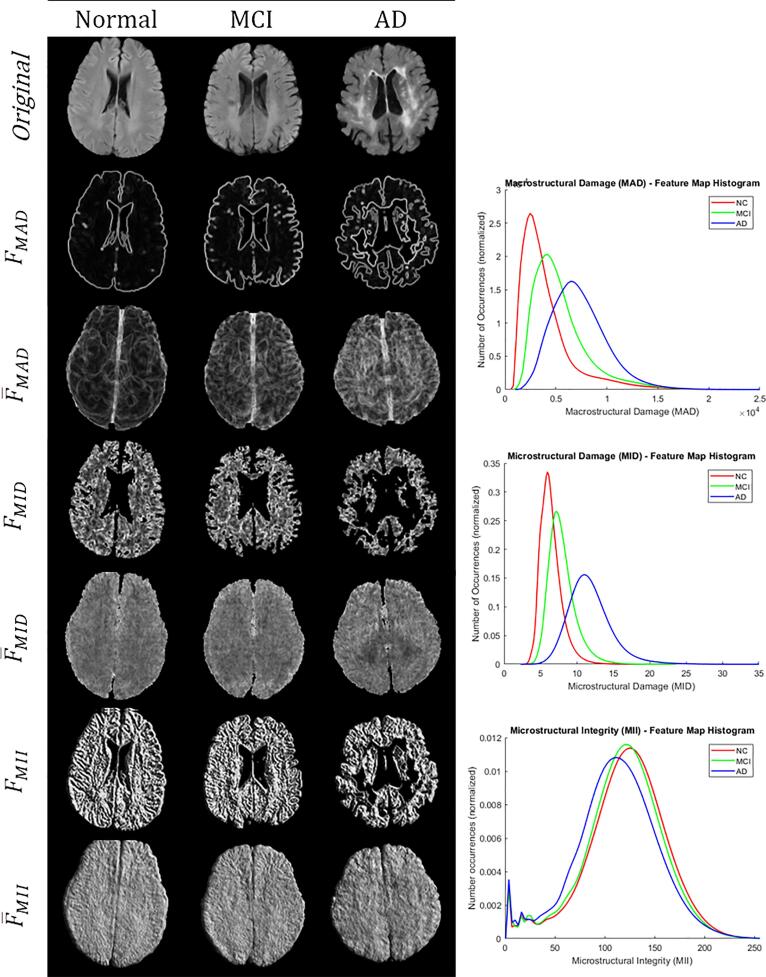
Intensity and volume features were computed directly from the extracted NABM region. For texture features, the 2D feature maps were first computed for every slice: macrostructural damage , microstructural damage and microstructural integrity . The mean volume feature maps were computed through voxel-wise averaging of each of the respective feature maps to quantify global texture trends in the NABM. Biomarkers were measured as the median of for each volume. Fig. 3 has visual representations of the slice-based and volume-based texture features for normal, mild cognitive impairment (MCI) and Alzheimer’s disease (AD) determined from MoCA scores (see Data). For the macrostructural damage marker, boundaries have large feature values in the slice-based analysis along with finer variations within the NABM. In the feature map , there are more rapidly changing intensities within the NABM (less homogeneity) with decreasing cognitive performance. The microstructural integrity (MII) marker measures intensity patterns within the cerebral tissue related to linear structures, ridges and valleys. As shown in Fig. 3 there is more structure (or integrity) in the cognitively normal subject. The histograms for each of the texture biomarkers in Fig. 3 exhibit differences between normal, MCI and AD disease groups, as well. There are smaller differences and more overlapping values for the MII biomarker across cognitive groups. This could be due to some regions of the brain having heterogeneity (normal and abnormal tissue).
Fig. 3.
Left: microstructural feature extraction exhibiting 2D feature slices and the corresponding averaged feature map, for normal (MoCA 26), MCI (MoCA 19–25) and AD (MoCA 18). Right: corresponding histograms for the mean feature maps.
For each biomarker, outlier analysis was conducted to exclude extreme values caused by possible remaining image quality issues or measurement error. Outliers were detected as 1.5 interquartile distance above the upper quartile or below the lower quartile. Outlier detected volumes were visually inspected and removed if the volumes displayed evidence of ringing artifact, bias field, or brain mask issues. The remainder were included in the final dataset. In CCNA, an entire center had 100% of scans rejected with outlier analysis and upon inspection, this center had an extreme bias field issues. In total there were 14 CAIN (3.7%), 14 ADNI (1.5%) and 25 CCNA (13.1%) volumes, that were excluded resulting in 53 volumes in total (3.6%).
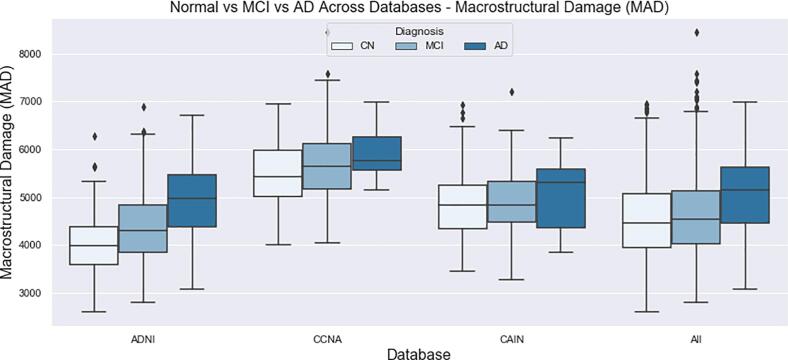
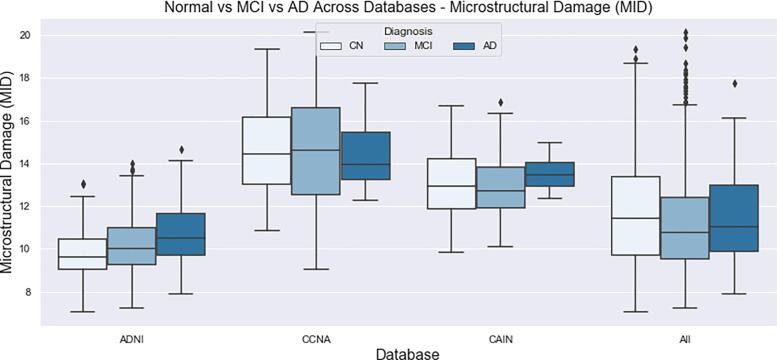
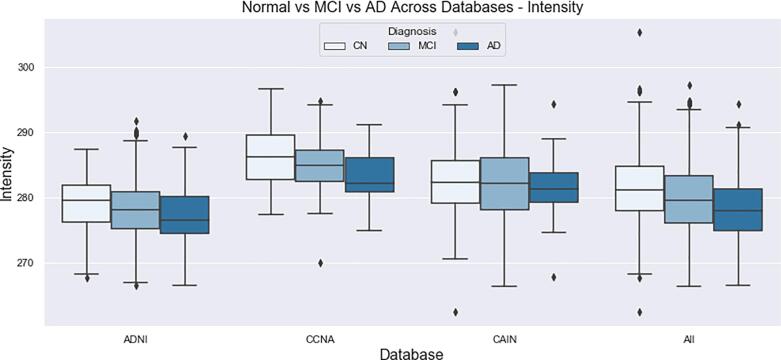
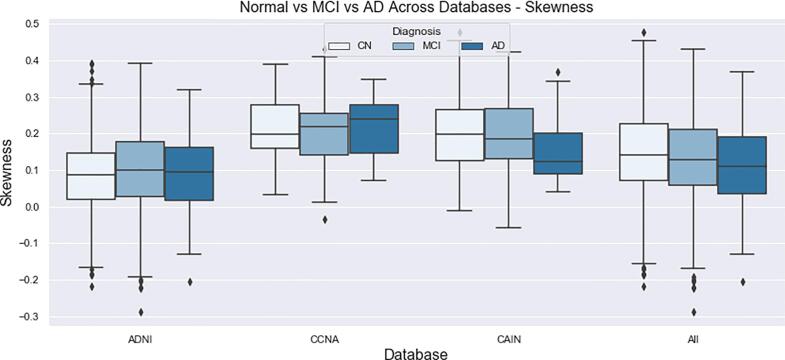
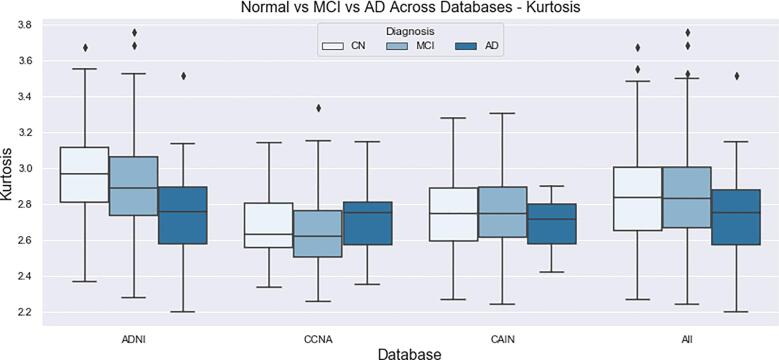
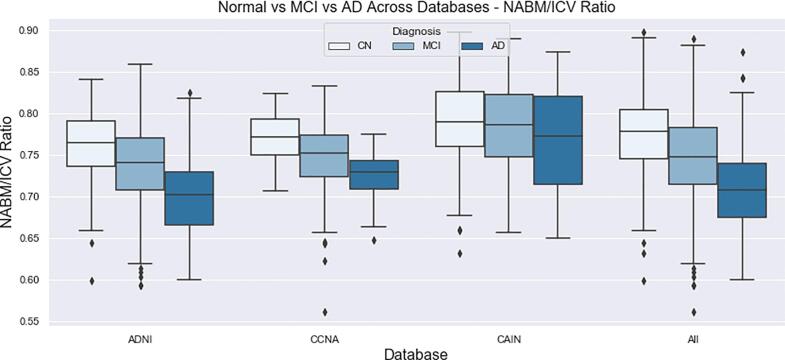
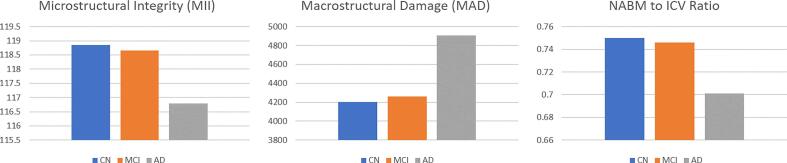
The final distributions of biomarkers over all cohorts, for Normal, MCI and AD groups are shown in Fig. 13, Fig. 14, Fig. 15, Fig. 16, Fig. 17, Fig. 18, Fig. 19. Integrity quantifies the amount of structure that is related to edge-based patterns in the data. Over all the datasets, there is a trend of decreasing integrity for worse cognitive condition, indicating there is less structure in the brain. In diseased brains, the NABM contains more random patterns due to neurodegeneration. The macro (MAD) and microstructural damage (MID) markers quantify NABM texture roughness by measuring local differences in intensity values. Lower values were found for better cognitive outcome (which increased for MCI and AD groups). A low value indicates smooth or homogeneous tissue (less damage) whereas coarse textures and regions with rapid intensity changes imply more neurodegeneration. In the interior of the NABM region, MAD and MID measure the fine changes in intensities which can be associated with WM tract degeneration and GM atrophy. The MAD biomarker also captures more global intensity differences related to NABM tissue loss as there are more edges due to atrophy, enlarged ventricles, and lesions. A decrease in median NABM intensity values for worse cognitive outcome was found. This may seem counter-intuitive as some recent studies found higher intensities in regions surrounding WML in FLAIR MRI (Maillard et al., 2013), which suggests there is pre-visible WM damage surrounding WML that could be earlier signs of disease (ischemia/demylination). However, the NABM contains both WM and GM and changes in both tissue are reflected in the median FLAIR intensity measurement. As shown in Siemonsen et al. (2008), the intensity of the GM may decrease in T2 weighted images for atrophy and neuronal loss, and therefore, the decrease in the GM intensities may be overpowering the subtle changes in the WM. Higher kurtosis and skewness values were found across MoCA groups for better cognitive condition, indicating more tissue homogeneity and higher intensities overall. The NABM to ICV volume ratio measures the normalized volume of the GM and WM together with CSF and WML stripped out. Over all datasets, NABM to ICV volume ratio decreased for lower MoCA scores (worse cognitive condition), indicating that NABM tissue loss is related to cognitive decline. For healthy individuals, there can be little or no lesions present and the CSF spaces should be small, which results in a higher NABM volume in these subjects. For subjects with neurodegenerative disease, there is an increase in lesion load, ventricle size, CSF spaces and a decrease of GM and WM and these differences were robustly detected using the NABM to ICV volume ratio biomarker.
Fig. 13.
Microstructural Integrity (MII) NABM biomarkers for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 14.
Macrostructural Damage (MAD) NABM biomarkers for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 15.
Microstructural Damage (MID) NABM biomarkers for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 16.
Intensity of the NABM for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 17.
Intensity skewness of the NABM for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 18.
Intensity kurtosis of the NABM for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
Fig. 19.
NABM/ICV volume ratios for normal (MoCA 26), MCI (MoCA 19–25), AD (MoCA 18).
3.2. Biomarker correlation analysis
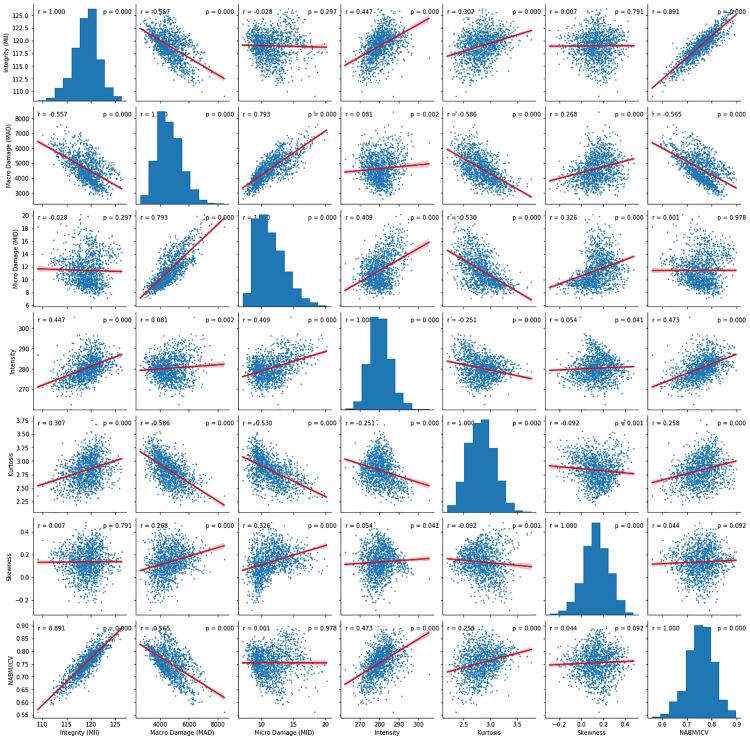
The relationships among FLAIR NABM biomarkers pairs shown in Fig. 4 and corresponding r and p-values are listed in Table 4. Correlations that are higher than 0.5 are underlined in the table. Microstructural integrity (MII) was negatively correlated with macro (MAD) and microstructural damage (MID), with a higher overall correlation between integrity and macrostructural damage (r = −0.56) as compared with microstructural damage (r = −0.03), although statistical significance was only seen between MII and MAD (p 0.01). Other significant relationships show lower tissue integrity (MII) for decreased intensity (r = 0.45) and higher MII for higher NABM volumes (r = 0.89) respectively while demonstrating statistically significant relationships as well (p 0.01). Macrostructural damage (MAD) was strongly negatively correlated to the NABM to ICV volume ratio (r = −0.57, p 0.01), indicating that subjects with more damage, there is less NABM volume. The highest correlations were found between NABM to ICV volume ratio and microstructural integrity (r = 0.89, p 0.01) as well as microstructural damage vs macrostructural damage (r = 0.79, p 0.01).
Fig. 4.
All FLAIR biomarkers versus each other. This figure shows the relationships between the biomarkers.
Table 4.
Correlation between FLAIR biomarkers. Underline is and bold means significant.
| Features | MII |
MAD |
MID |
Intensity |
Kurtosis |
Skewness |
NABM/ICV |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | |
| MII | – | – | -0.56 | 0.01 | -0.03 | 0.30 | 0.45 | 0.01 | 0.31 | 0.01 | 0.01 | 0.79 | 0.89 | 0.01 |
| MAD | −0.56 | 0.01 | – | – | 0.79 | 0.01 | 0.08 | 0.01 | −0.59 | 0.01 | 0.27 | 0.01 | −0.57 | 0.01 |
| MID | −0.03 | 0.30 | 0.79 | 0.01 | – | – | 0.41 | 0.01 | −0.53 | 0.01 | 0.33 | 0.01 | 0 | 0.98 |
| Intensity | 0.45 | 0.01 | 0.08 | 0.01 | 0.41 | 0.01 | – | – | −0.25 | 0.01 | 0.05 | 0.04 | 0.47 | 0.01 |
| Kurtosis | 0.31 | 0.01 | −0.59 | 0.01 | 0.53 | 0.01 | −0.25 | 0.01 | – | – | −0.09 | 0.01 | 0.26 | 0.01 |
| Skewness | 0.01 | 0.79 | 0.27 | 0.01 | 0.33 | 0.01 | 0.05 | 0.04 | −0.09 | 0.01 | – | – | 0.04 | 0.09 |
| NABM/ICV | 0.89 | 0.01 | −0.57 | 0.01 | 0 | 0.98 | 0.47 | 0.01 | 0.26 | 0.01 | 0.04 | 0.09 | – | – |
To investigate how FLAIR NABM biomarkers are related to water diffusion in the brain, FLAIR NABM biomarkers were correlated to the corresponding dMRI biomarkers. As previously discussed, since the NABM contains both WM and GM, only MD is explored for NABM analysis. In total, there were 52 CCNA dMRI volumes with 31 directions, and 46 dMRI volumes with 25 directions for CAIN. The MD was extracted and the registered NABM mask was used to mask out CSF and lesions. Median MD is extracted for the NABM and correlated to the corresponding FLAIR biomarkers for the same imaging volume.
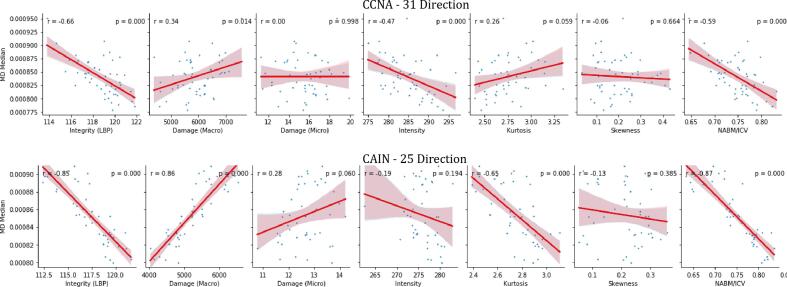
Fig. 5 shows FLAIR biomarkers plotted against median MD. As MD increases, integrity (MII), intensity, kurtosis and NABM to ICV volume ratio decreased while micro (MID) and macrostructural damage (MAD) increased. The strongest (significant) correlations were found between MD and: microstructural integrity (CCNA: r = −0.66, CAIN: r = −0.85, p 0.01 respectively), macrostructural damage (CAIN: r = −0.86, p 0.01) and NABM to ICV volume ratio (CCNA r = −0.59, CAIN: r = −0.87, p 0.01) the remaining correlation values can be seen in Table 5. Microstructural integrity relates to how structured the NABM tissue is, with less structure being attributed to neurodegeneration (higher MD). Conversely, subjects with higher MII values (more integrity) also have lower MD values which are indicative of constrained water flow in a healthier brain. Micro and macrostructural damage biomarkers measure rapid changes in intensity, and quantify tissue roughness and loss. For increased micro and macrostructural damage, there was a similar increase in MD, indicative of more damage to the brain. Higher MD values can be attributed to less integrity of the WM and atrophy of the GM which creates less obstacles for water diffusion and as a result an increase in MD. With intensity, there is a negative relationship with MD which shows that for healthy subjects (low MD), there was higher intensities. While there may be more localized (increased) intensity changes in the WM (around WML for example), the NABM as a whole was found to be of lower intensity, which correlated to a reduction in MD compared to normal subjects (although the correlations were moderate, CCNA:r = −0.47, p 0.01 and CAIN: r = −0.19, p = 0.19). The NABM to ICV volume ratio was also highly correlated to MD in both datasets, indicating that as NABM volumes decreasd, MD increased. As NABM volume is related to tissue loss (i.e. reduced GM volume, increased CSF spaces, more lesions) the correlations suggest a relationship to water diffusion in the brain. It is worth noting that CAIN and CCNA are comprised of 25 and 31 directions, respectively, which can account for minor differences in the MD values between these datasets. Also, the sample size is small and in CAIN there were only two subjects classified as AD by their MoCA score so it may be hard to compare cohorts directly. Although there are minor differences in the level of correlations between dMRI and FLAIR across datasets (CCNA vs. CAIN), the trends are mostly the same. The main difference is between CAIN and CCNA in the kurtosis and MID biomarkers and this may be due to a bias field artifact in CCNA (see discussion), which spreads out the intensity distribution. Due to the small sample size this affect may be more noticeable in this analysis.
Fig. 5.
Comparison of DTI to FLAIR Biomarkers on the CCNA and CAIN Datasets – MD Median.
Table 5.
Correlation between FLAIR and dMRI (MD) biomarkers in CAIN and CCNA. Underline is and bold means significant.
| Dataset | MII |
MAD |
MID |
Intensity |
Kurtosis |
Skewness |
NABM/ICV |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p-value | r | p-value | r | p-value | r | p-value | r | p-value | r | p-value | r | p-value | |
| CCNA | −0.66 | 0.01 | 0.34 | 0.014 | 0.00 | 1.00 | −0.47 | 0.01 | 0.26 | 0.06 | −0.06 | 0.66 | −0.59 | 0.01 |
| CAIN | −0.85 | 0.01 | 0.86 | 0.01 | 0.28 | 0.06 | −0.19 | 0.19 | −0.65 | 0.01 | −0.13 | 0.39 | −0.87 | 0.01 |
3.3. Group analysis
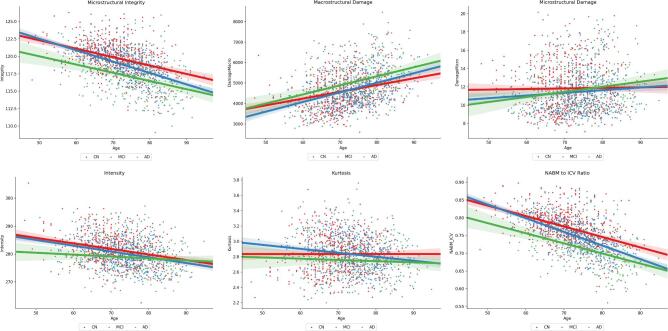
Differences in biomarker means across different cognitive groups were investigated using ANOVA and ANCOVA as detailed in the Analysis and Statistics section. The imaging volumes were grouped into NC, MCI and AD categories according to MoCA ranges across CCNA, CAIN and ADNI (see Data for ranges and sample sizes) and biomarkers for each imaging volume were computed. The biomarker distributions (boxplots) as a function MoCA category are shown in Fig. 13, Fig. 14, Fig. 15, Fig. 16, Fig. 17, Fig. 18, Fig. 19 for all datasets. With increasing disease progression (worse cognitive outcome), microstructural integrity (MII), intensity, kurtosis and NABM to ICV volume ratio biomarkers decreased while, micro (MID) and macrostructural damage (MAD) biomarkers increased.
Unadjusted ANOVA analysis for MoCA category alone is reported for each biomarker in Table 6, and post hoc analysis showed significant differences across most biomarkers and groups, with the exception of MID (Normal vs. AD, Normal vs. MCI), intensity (MCI vs. AD), kurtosis (Normal vs. MCI) and skewness (Normal vs. AD, MCI vs. AD) comparisons. ANCOVA analysis for each biomarker across MoCA groups with age as a covariate is reported in Table 7 for the age*MoCA interaction term. Biomarkers with significant differences for the age*MOCA interaction term were MII, MAD, intensity, kurtosis and NABM to ICV volume ratio, indicating these biomarkers change with age differently for each MoCA category. Fig. 7 plots each biomarker with respect to age and MoCA category. Earlier in life, the MII, MAD, intensity, skewness and NABM volume biomarkers of MCI subjects are similar to the Normal subjects, whereas later in life the biomarkers of MCI subjects are more similar to those of AD subjects. The slopes of several biomarkers as a function of age are shown in Table 10 and Fig. 6. While there is a general trajectory with age and disease progression for each biomarker (i.e. increasing or decreasing with age), the MCI group demonstrated a more significant slope compared to the other two categories (normal and AD), which indicates that degeneration is happening fastest in this group. As this group is comprised of subjects that deteriorate rapidly, identifying them early and providing treatment could possibly improve outcomes.
Fig. 7.
FLAIR NABM biomarkers vs Age and MoCA Groups.
Fig. 6.
Slope of FLAIR NABM biomarkers vs age, for MoCA category and sex. NC: MoCA 26, MCI: MoCA (19–25) and AD: MoCA 18.
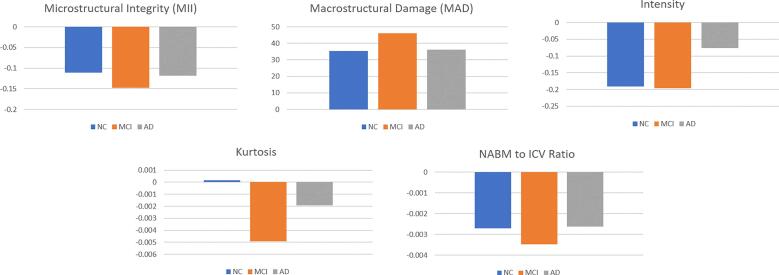
An additional model was considered to investigate any two-term or possible three-term interactions between age, sex and MoCA. While many multicentre data issues have been corrected through preprocessing, there are other influencers in this relationship such as age and sex, as well as residual nuisance parameters related to scanner manufacturer and database. To ensure the relationship between MoCA and biomarker values were not confounded by other factors, in this analysis we adjusted for age, database, scanner and sex in the final model. The adjusted means over all data are listed in Table 9 and plotted in Fig. 8, demonstrating similar trends to the unadjusted analysis.
Fig. 8.
Adjusted biomarker means over all datasets for MoCA categories (NC/MCI/AD) determined using MoCA (see Data). As indicated in Table 8, there were significant differences between Normal vs. MCI (p 0.01) for MII, MAD and NABM to ICV ratio biomarkers. Significant differences were also seen between MCI vs. AD (p 0.01) for all biomarkers except for MID and skewness. Only the skewness biomarker did not show any significant difference (p 0.01) between the Normal vs. AD groups.
Post-hoc analysis in Table 8 revealed similar trends as unadjusted analysis in that adjusted means were significant across most groups and biomarkers, with few that showed no differences across groups: skewness (across all groups), MID (Normal vs. MCI and MCI vs. AD), kurtosis (Normal vs. MCI) and intensity (Normal vs. MCI). No statistical significance was found for sex*age*MoCA and sex*MoCA interaction terms over all biomarkers. It was observed that over all biomarkers with the exception of intensity and skewness (believed to be as a result of CCNA bias field issues - see Discussion), females exhibited generally “healthier” biomarker values for MII, MAD, MID, kurtosis and NABM to ICV volume ratio as per Table 11. The means of sex by MoCA category differed, however, did not provide significance.
3.4. Post-hoc exploratory analysis
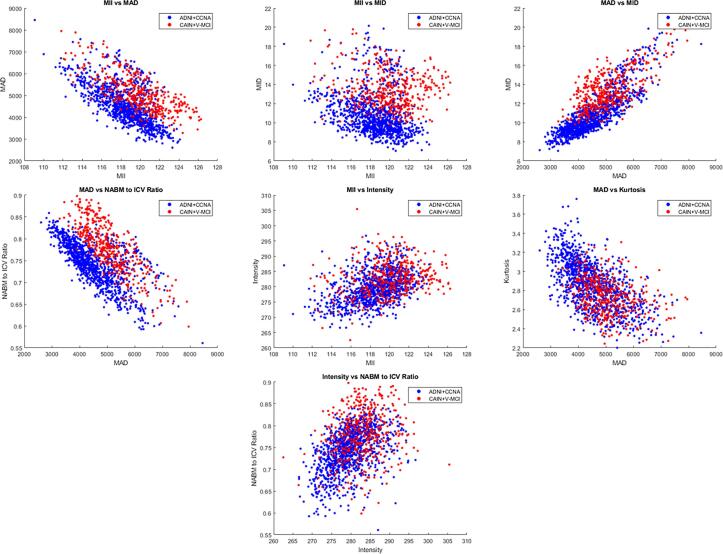
ADNI and CCNA are AD pathology datasets and CAIN is a vascular disease group. In this analysis, differences across datasets and diagnostic labels were examined through post hoc exploratory analysis. Using CCNA diagnostic labels there were 43 SCI, 88 MCI, 59 V-MCI (vascular MCI) and 35 AD subjects. In ADNI, there were 199 normal (CN) and 97 subjective memory complaint subjects (merged into one CN group), 283 early and 181 late MCI subjects (merged into an MCI group) and 137 AD subjects. To examine differences across disease types, a few of biomarkers that had high correlations with one another were investigated in Fig. 9. There are clusters in the biomarkers that seem to differentiate between dementia (ADNI + CCNA) and CVD (CAIN + V-MCI), but also overlapping regions suggesting there are similarities in neuroanatomical profiles between diseases. That being said, there are some subtle differences that can be mentioned. Firstly, when observing MAD vs. MID, subjects with vascular disease related pathology appeared to have higher microstructural and macrostructural damage as compared to the AD set. AD subjects are likely experiencing increased atrophy, ventricle sizes and decreased WM integrity that are reflected in these metrics. CVD is often associated with vascular lesions which increased the MAD biomarker (more tissue removed from stripping out the lesions as well as WM degeneration which caused rapid intensity changes) but the changes seem to be noticeable in the vascular group. Similarly, in the NABM volume vs. MAD comparison, the vascular group had higher NABM volumes over all, along with higher MAD compared to the AD group. With the MII vs. MID scatter plot, it can be seen that CVD clusters are on the higher end of the integrity range coupled with higher values of micro damage. There are slight differences between CAIN + vMCI and ADNI + CCNA, which may be due to differences in pathology (i.e. AD vs. vascular disease) but it also can be due to the fact that the CAIN cohort is comprised largely of cognitively in tact individuals (mean MoCA = 26). Differences between AD pathology and vascular disease will be investigated further in the future using these biomarkers.
Fig. 9.
Relationship between two FLAIR NABM biomarkers computed for dementia (ADNI + CCNA) and vascular disease patients (CAIN + vMCI).
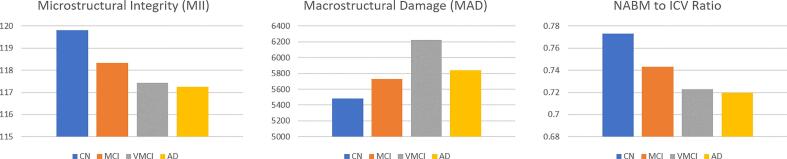
A secondary group analysis was investigated here where biomarker differences were examined according to the diagnostic labels that were available in ADNI and CCNA. The mean values for the MII, MAD and NABM to ICV volume ratio biomarkers for CCNA and ADNI are shown in Fig. 10 and Fig. 11. The overall trends in biomarkers over diagnostic labels follow those seen in the MoCA group analysis. For increasing cognitive impairment there was decline in MII (reduction in integrity or structure in the WM and GM) and NABM to ICV volume ratio (more tissue loss related to atrophy, large ventricles and lesions), along with an increase in MAD (increased ventricle sizes and pathology removed). The mean MII and NABM to ICV volume ratio biomarkers were similar between V-MCI and AD patients, whereas MAD was much higher for the V-MCI group compared to all. In ADNI there were similarities in mean biomarkers for the cognitively intact and MCI groups. The unadjusted ANOVA analysis is reported in Table 12, Table 13, Table 14, Table 15. There was statistical significance between the differences of biomarkers of cognitively normal (or SCI) and AD groups in ADNI and CCNA (p 0.01) for MII, intensity and NABM to ICV volume ratio. In terms of V-MCI and MCI, there were significant differences in biomarker means for MII, MAD and NABM to ICV volume ratio, indicating that subjects with V-MCI and MCI had different structural integrity, volumes and macrostructural damage. The intensity (related to GM loss) and MID (local tissue homogeneity) were significantly different between V-MCI and MCI. When comparing to AD, V-MCI and AD were similar over all biomarkers except for MID which could suggest microstructural changes were different between these two diseases (whereas there were similarities in integrity, intensity and volume). In contrast, MCI and AD in CCNA were similar over all biomarkers except for MII, intensity and NABM to ICV volume ratio, indicating that structural brain changes between MCI and AD are different (higher intensities, integrity and NABM tissue for better cognitive state).
Fig. 10.
Mean biomarker values over the CCNA dataset. Datasplits for SCI/MCI/vMCI/AD determined using diagnostic label. As shown in Table 13, there were significant differences between MCI vs. V-MCI and SCI vs. V-MCI (p 0.01) for MII, MAD and NABM to ICV ratio markers. Significant differences were also seen between V-MCI vs. AD for MID (p 0.01), MCI vs. AD (p 0.01) for MII and NABM to ICV ratio. Lastly, significant differences were found between SCI vs. AD for the MII, intensity and NABM to ICV ratio markers.
Fig. 11.
Mean biomarker values in ADNI with groups determined by diagnosis label. As described in Table 15, MII, MAD, MID, intensity and NABM ratio had significant differences (p 0.01) for CN and AD, as well as MCI and AD. There were no significant differences in biomarkers found between CN and MCI.
4. Discussion
Compared to majority of works investigating biomarkers from T1 or DWI, this work defines biomarkers purely from FLAIR MRI which is a major novelty of the proposed approach. Other studies have begun investigating FLAIR features for neurodegenerative disease analysis and we believe this is an emerging field strongly supported by our work. In Maillard et al. (2013) the authors show FLAIR features in non-lesion regions uniquely predicted voxels that would convert into lesions, suggesting FLAIR contains subtle signs of microstructural damage and neurodegeneration. In Maillard et al. (2013), FLAIR intensity provided information on the underlying tissue integrity of the WM and is also related to demyelination and ischemia. In De Groot et al. (2013), dMRI and FLAIR signal intensity of the NAWM were associated with WML development independently (De Groot et al., 2013), which suggests these sequences are capturing different pathological mechanisms. The authors stated that using FLAIR measurements from retrospectively collected FLAIR, with or without DTI, could “add substantial power to detecting treatment related differences” (Maillard et al., 2013). This is largely due to the fact that FLAIR is routinely acquired and widely available. Therefore, biomarkers from FLAIR can be used in clinical trials to improve study power, stratify patients and to determine optimal intervention times. Clinically, such biomarkers can be easily integrated into routine workflows.
It was hypothesized that biomarkers from the normal-appearing brain matter (NABM) in FLAIR MRI can differentiate between cognitively normal and cognitively impaired subjects. There were three groups of features defined for this task: texture (microstructural integrity (MII), macrostructural damage (MAD), microstructural damage (MII)), intensity (median, skewness, kurtosis) and volume biomarkers (NABM to ICV volume ratio). Intensity standardization and spatial normalization allowed for fair comparison across biomarkers in a diverse dataset acquired with varying acquisition parameters and scanner vendors. Through statistical analysis of NABM biomarkers extracted in over 1400 imaging volumes from 87 centres worldwide, we established a relationship between structural brain changes in the NABM and cognition. There was a decrease in MII, intensity, kurtosis, skewness and NABM to ICV volume ratio for worse cognitive outcome, compared to MAD and MID that increased. In the adjusted ANCOVA model, there were significant differences in biomarker means across all MoCA categories (normal, MCI and AD) for the MII, MAD and NABM to ICV volume ratio biomarkers, demonstrating NABM biomarkers from FLAIR are related to cognition.
We further hypothesized that texture features from FLAIR MRI can be used to quantify microstructural changes related to how “structured” or “damaged” the tissue is. Texture features measure subtle intensity variations related to how rapid the intensities are changing in local regions, or how smooth the tissue is. FLAIR contrast in the WM is related to attenuation of lipid protons within the myelin and thus carries important information regarding the microstructure of the tissue (Maillard et al., 2013). Additionally, FLAIR highlights white matter lesions exceptionally well and as shown by pathological examination of post-mortem tissue, these lesions, which appear bright in FLAIR, exhibited venous collagenosis, vessel tortuosity, edema and demyelination (Black, 2009, Keith et al., 2017) – indicating that FLAIR is capturing microstructural changes. Since subtle changes in FLAIR contrast are related to microstructural changes, texture measures from FLAIR may be related to neurodegeneration, ischemia, edema, vessel collagenosis, white matter tract degeneration, and/or GM tissue loss/loosening. When comparing to dMRI metrics, mean diffusivity (MD) was used and lower integrity (MII), and higher MAD and MID, were correlated to higher MD. As MD measures the total (magnitude of) diffusion in each voxel and higher MD values are associated to tissue degeneration, trends in these features further support the hypothesis that NABM texture is related to structure and damage of the GM and WM. As a result, the novel FLAIR-only texture biomarkers have some relationship with tissue microstructure and open up the possibility for new clinical applications and avenues for measuring neurodegeneration in the brain without dependence on other sequences such as dMRI. In the future, more analysis into the histopathological correlates of the FLAIR-only texture features will be considered.
A major innovation of the paper is the identification of age-dependent trends in the MII, MAD, intensity, kurtosis and NABM to ICV volume ratio biomarkers that vary differently across MoCA categories. These were discovered, we believe, due to the novel nature of the biomarkers, the rigorous preprocessing strategies to reduce measurement variability and increase sensitivity, and the size of the cohort that was analyzed. This relationship exists even after adjusting for confounding effects of database and manufacturer. Age-dependent biomarker curves have utility in tracking disease progression, in an analogous manner as growth curves or charts for children. Clinically, it could be possible to measure a patient’s NABM biomarkers and compare them to population curves for the NC, MCI and AD groups to monitor neurodegeneration and determine optimal treatment points. For drug development, these biomarkers can be used to stratify patients into homogeneous groups for clinical trials. MCI is a promising group to target since 1) they degenerate the fastest and 2) permanent brain damage has not occurred yet. The proposed biomarkers, which also were shown to be different among the sexes (although not significant), could be used to stratify and monitor patients. In spite of the fact that a trend was seen between sex and MoCA level, more data would be needed to confirm.
Interestingly, the MCI group progressed the fastest (largest slope), which was followed by NC and then AD (had the lowest slope). The change in the normal group, which is less than the MCI group could be attributed to normal aging processes. However, for the AD group, as shown by Fig. 7 (age slope regression graphs), younger subjects in the AD group usually start off with a lower or higher “baseline” biomarker value earlier in life (more neurodegeneration) indicating these subjects could be presenting with underlying disease even early in life. As a result, the rate of degeneration may not be as high as the MCI group since damage had already been accruing.
In post hoc exploratory analysis, group analysis was completed using the biomarkers over diagnostic labels in CCNA and ADNI. Unadjusted analysis found differences between biomarkers across many groups, including MCI, V-MCI and AD groups. V-MCI patients are diagnosed according to MoCA (13–24 inclusive), CERAD word list recall 6, Lawton-Brody IADL scale score 14 and global CDR 0.5 criteria (which is the same for the MCI group) but to determine the vascular nature, the T2 FLAIR was used, and Age-Related White Matter Change (ARWMC) was assessed. Compared to MCI, there are significant differences in the structural integrity (MII), intensity (GM loss) and NABM volume biomarkers compared to V-MCI. The MII biomarker was lower in patients with V-MCI than MCI, and was on the same order as subjects with AD indicating the structural integrity of V-MCI and AD patients are similar. Similar trends were seen with NABM to ICV volume ratio, in that the NABM volumes of V-MCI subjects were similar to subjects with AD. While the NABM ratio takes into account tissue loss in the GM/WM and enlarged CSF spaces which are associated with AD, larger lesion loads are typical in V-MCI and could reduce the total NABM volume. Recent evidence is pointing towards a “two–hit” vascular hypothesis for AD and vascular disease, where hit one includes CVD risk factors that lead to blood brain barrier dysfunction and reduced cerebral blood flow that precedes dementia, followed by hit two caused by an increase in beta-amyloid amplifying neuronal dysfunction, neurodegeneration and disease (Lamar et al., 2020). In Meng et al. (2017), the authors suggest that vascular and neurodegenerative pathological processes have a supra-additive effect on cognitive performance. Perhaps these biomarkers are demonstrating this effect. In future works, further investigation into the differences across biomarker profiles for vascular disease and dementia will be conducted. Understanding the overlap in neuroanatomical profiles associated with CVD risk factors and dementia can lead to more optimized therapies. Cerebrovascular disease (CVD) is the second most common contributor to dementia risk (Smith et al., 2017) and early CVD may represent a pivotal stage to intervene before irreversible brain injury and disability occurs (Smith et al., 2017, Black et al., 2011). CVD risk factors are no longer seen as more relevant to vascular dementia but share a common etiology for all the dementias (Lamar et al., 2020). Targeting modifiable vascular risk factors that contribute to cognitive impairment and neurodegeneration would be paradigm changing (Badji and Westman, 2020).
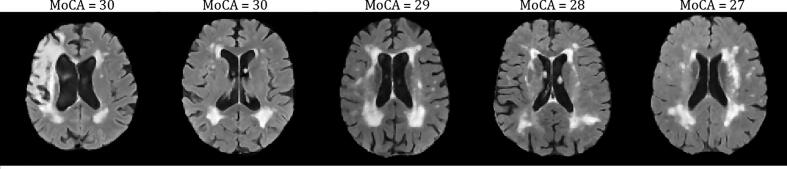
Limitations and challenges were noted with this work. First, only MD (and not FA) was investigated here, since as was shown in Nave et al. (2007) the changes in fractional anisotropy (FA) for GM and WM were opposing, and any small differences are a superposition of these opposing trends which makes it difficult to elucidate any changes. In the future we will further investigate both MD and FA dMRI metrics for GM and WM separately to further understand neurodegeneration as described by the proposed FLAIR NABM biomarkers. If possible, histopathological studies related to FLAIR intensity and texture could be considered. In the CCNA dataset, despite intensity standardization and bias field reduction, slightly higher intensities were noticed across patients compared to ADNI and CAIN. This could have an impact on the intensity biomarkers, as well as microstructural damage feature which is found by Otsu thresholding the macrostructural damage (MAD) feature maps. In cases with severe bias field artifact, they were detected as outliers and removed from the analysis but some volumes still remained. In the future, we will investigate a more robust bias field reduction technique and/or better outlier reduction techniques. This intensity bias likely affected correlation between the MD metrics and the FLAIR-based MID and intensity biomarkers, such as kurtosis. Sample size in the DTI analysis also likely contributed to these issues, although some prominent trends were found, especially between MII, MAD and NABM/ICV biomarkers. Another challenge found was in some CAIN cases that had biomarkers that were close to the interquartile ranges but were not rejected. Upon inspection, some of these subjects have moderate to severe WML patterns (vascular damage) - but have high MoCA scores indicating they are cognitively intact (see example slices from these subjects and their respective MoCA in Fig. 20). The NABM biomarker panel for these patients show that the biomarkers fall into the range of MCI and AD indicating that the tool is picking up neurodegeneration. It is possible that these patients while not suffering noticeable cognitive deficits, may suddenly suffer decline if more damage accumulates (tip the balance). There are other factors that can come into play here, as well, such as education and socio-economic status (Russ et al., 2013, Wilson et al., 2009) that have known to be protective against dementia. Therefore, these subjects may be on the verge of developing cognitive deficits. Lastly, analysis was performed cross-sectionally and without cross validation studies, which may be considered as a major limitation of the work. However, the main goal was to find associates between NABM features and cognition in the baseline cohort and believe we have satisfied those goals. In the future, we will examine the biomarkers in a longitudinal study and investigate clinically relevant cut-offs for the biomarkers, in the hopes of quantifying disease progression robustly and to determine optimal treatment points for patients suffering cognitive decline.
Fig. 20.
Sample slices from varying CAIN volumes with normal MoCA scores.
Although many proof of concept biomarkers exist (tested in single centres with limited variability), there are only a few that are technically validated in large multicentre datasets to establish “proof of effectiveness” which is a barrier to translation. Technical validation includes biomarker tests related to feasibility, accuracy, reproducibility and repeatability (Smith et al., 2019, Sullivan et al., 2015, Obuchowski et al., 2015), and should precede clinical validation, otherwise it is difficult to determine if biomarker changes are from the biological process or technical variability of the biomarkers (Smith et al., 2019). Biomarkers should be investigated on smaller, more controlled datasets; then scaled to large multi-centre sets to prove effectiveness. Once effectiveness is shown, clinical utility can be investigated. We believe this work makes headway on many of those goals. All preprocessing and segmentation tools have been extensively validated on multicenter datasets, and clinical utility is established by demonstrating that structural brain differences in the normal-appearing brain matter (NABM) in FLAIR MRI are associated with cognition.
5. Conclusions
In this work, a novel biomarker panel from the normal-appearing brain matter (NABM) in FLAIR MRI was designed, which included intensity, texture and volume-based features. The texture-based NABM biomarkers had high correlation with diffusion MRI metrics, suggesting that FLAIR MRI can be used to characterize subtle details related to tissue microstructure. On a dataset of more than1400 subjects from 87 international centers, statistical analysis revealed that the new NABM biomarkers were significantly associated with cognition. Age-based analysis also demonstrated that NABM biomarkers vary significantly as a function of age and cognitive group, with the mild cognitive impairment (MCI) category often displaying the most rapid decline over age, as compared to normal controls and patients with advanced dementia. On post hoc exploratory analysis, the biomarkers differentiated between subjects with vascular disease and dementia, which creates possibilities of using these biomarkers to learn more about disease etiology and for patient stratification.
Acknowledgement
-
•
Natural Sciences and Engineering Research Council of Canada (NSERC) and the NSERC Discovery Grant program.
-
•
Canadian Atherosclerosis Imaging Network (CAIN) which was established through funding from a Canadian Institutes of Health Research Team Grant for Clinical Research Initiatives (CIHR-CRI 88057). Funding for the infrastructure was received from the Canada Foundation for Innovation (CFI-CAIN 20099), with matching funds provided by the governments of [H]ta, Ontario, and Quebec.
-
•
Data collection and sharing for this project was partially funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defence award number W81XWH-12–2-0012).
-
•
Canadian Consortium on Neurodegeneration and Aging (CCNA) for data, data storage, management and standardized MRI protocols. See Chertkow et al. (2019) about the CCNA data from the COMPASS-ND study, Mohaddes et al. (2021) for CCNA data storage and management and Duchesne et al. (2019) for developing the MRI protocols.
-
•
We would like to thank and acknowledge, Dr. Sandra E. Black, Neurologist and Professor of Medicine at the University to Toronto, for her contributions to this research by providing feedback on the manuscript and guidance on the work and data, as well as Principle Investigator (PI) of CAIN.
-
•
We also would like to acknowledge the help of Dr. Maxime Descoteaux from the Computer Science Department, University of Sherbrooke, for help with processing the dMRI data.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Appendix
Table 6.
Unadjusted ANOVA analysis of difference in biomarker means for all datasets using MoCA groups: Normal (26) vs MCI (19–25) vs AD (18). Reported as difference in means (effect) and p-value. P-values 0.01 indicate significant differences (*).
| Features | Normal vs MCI |
MCI vs AD |
Normal vs AD |
|||
|---|---|---|---|---|---|---|
| Effect | p | Effect | p | Effect | p | |
| Integrity (MII) | 1.21 | 0.01* | 1.63 | 0.01* | 2.84 | 0.01* |
| Macrostructural Damage (MAD) | 150.40 | 0.01* | 516.47 | 0.01* | 666.90 | 0.01* |
| Microstructural Damage (MID) | 0.31 | 0.06 | 0.62 | 0.01* | 0.31 | 0.30 |
| Intensity | 1.52 | 0.01* | 0.97 | 0.09 | 2.49 | 0.01* |
| Kurtosis | 0 | 0.93 | 0.09 | 0.01* | 0.095 | 0.01* |
| Skewness | 0.02 | 0.01* | 0 | 0.94 | 0.02 | 0.25 |
| NABM/ICV | 0.02 | 0.01* | 0.04 | 0.01* | 0.06 | 0.01* |
Table 7.
Interaction terms. Age*MoCA is from the ANCOVA model with only age as a covariate. Reported as F and p-values.
| Features | Age*MoCA | |
|---|---|---|
| F | PrF | |
| Integrity (MII) | 3.95 | 0.0191 |
| Macrostructural Damage (MAD) | 3.86 | 0.0213 |
| Microstructural Damage (MID) | – | – |
| Intensity | 3.70 | 0.0248 |
| Kurtosis | 6.95 | 0.0010 |
| Skewness | – | – |
| NABM/ICV | 6.00 | 0.0025 |
Table 8.
ANCOVA analysis of difference in biomarker means over MoCA groups: Normal (26) vs MCI (19–25) vs AD (18) adjusted for sex, age, scanner and database. Effect reported as difference in means. P-values 0.01 indicate significant differences (*).
| Features | Normal vs MCI | MCI vs AD | Normal vs AD | |||
|---|---|---|---|---|---|---|
| Effect | p | Effect | p | Effect | p | |
| Integrity (MII) | 0.48 | 0.01* | 1.28 | 0.01* | 1.76 | 0.01* |
| Macrostructural Damage (MAD) | 145.51 | 0.01* | 320.07 | 0.01* | 467.58 | 0.01* |
| Microstructural Damage (MID) | 0.13 | 0.32 | 0.20 | 0.24 | 0.33 | 0.04* |
| Intensity | 0.28 | 0.51 | 1.44 | 0.01* | 1.72 | 0.01* |
| Kurtosis | 0.02 | 0.11 | 0.07 | 0.01* | 0.09 | 0.01* |
| Skewness | 0 | 0.99 | 0 | 0.94 | 0 | 0.92 |
| NABM/ICV | 0.02 | 0.01* | 0.02 | 0.01* | 0.04 | 0.01* |
Table 9.
Adjusted biomarker means for all datasets using MoCA groups: Normal (26) vs MCI (19–25) vs AD (18).
| Features | NC | MCI | AD |
|---|---|---|---|
| Microstructural Integrity (MII) | 119.27 | 118.79 | 117.51 |
| Macrostructural Damage (MAD) | 4897.93 | 5045.44 | 5365.51 |
| Microstructural Damage (MID) | 12.60 | 12.73 | 12.93 |
| Intensity | 281.67 | 281.39 | 279.95 |
| Kurtosis | 2.75 | 2.73 | 2.66 |
| Skewness | 0.171 | 0.170 | 0.167 |
| NABM/ICV Ratio | 0.77 | 0.75 | 0.73 |
Table 10.
Slopes for each MoCA category when age*MoCA interaction was significant.
| Features | All |
|||
|---|---|---|---|---|
| MoCA 0 | MoCA 1 | MoCA 2 | ||
| Slope | −0.1113 | −0.1474 | −0.1180 | |
| MII | Standard Error | 0.0106 | 0.0098 | 0.0233 |
| p-value | 0.01* | 0.01* | 0.01* | |
| Slope | 35.3789 | 45.9693 | 36.2244 | |
| MAD | Standard Error | 3.3555 | 3.0301 | 6.4972 |
| p-value | 0.01* | 0.01* | 0.01* | |
| Slope | −0.1907 | −0.1962 | −0.0753 | |
| Intensity | Standard Error | 0.0248 | 0.0210 | 0.0448 |
| p-value | 0.01* | 0.01* | 0.0949 | |
| Slope | 0.0001 | −0.0049 | −0.0019 | |
| Kurtosis | Standard Error | 0.0012 | 0.0010 | 0.0018 |
| p-value | 0.9015 | 0.01* | 0.2776 | |
| Slope | −0.0027 | −0.0035 | −0.0026 | |
| NABM/ICV Ratio | Standard Error | 0.0002 | 0.0002 | 0.0004 |
| p-value | 0.01* | 0.01* | 0.01* | |
Table 11.
Average unadjusted biomarker values by sex.
| Features | Female | Male |
|---|---|---|
| Integrity (MII) | 119.43 2.50 | 118.59 2.54 |
| Macrostructural Damage (MAD) | 4453.93 826.67 | 4775.58 834.12 |
| Microstructural Damage (MID) | 11.03 2.26 | 11.72 2.24 |
| Intensity | 280.22 5.35 | 280.45 5.31 |
| Kurtosis | 2.86 0.24 | 2.80 0.24 |
| Skewness | 0.11 0.12 | 0.15 0.11 |
| NABM/ICV | 0.76 0.05 | 0.75 0.05 |
Table 12.
Unadjusted Means for CCNA using diagnostic labels.
| CCNA | SCI | MCI | V-MCI | AD |
|---|---|---|---|---|
| Microstructural Integrity (MII) | 119.80 1.32 | 118.33 2.07 | 117.43 2.52 | 117.26 1.89 |
| Macrostructural Damage (MAD) | 5482.39 685.92 | 5727.30 802.31 | 6220.15 809.60 | 5842.65 748.46 |
| Microstructural Damage (MID) | 14.83 2.04 | 14.76 2.31 | 15.65 2.27 | 14.24 2.14 |
| Intensity | 286.65 4.36 | 285.63 4.46 | 285.20 3.80 | 283.36 4.42 |
| NABM/ICV Ratio | 0.773 0.03 | 0.743 0.04 | 0.723 0.05 | 0.720 0.04 |
Table 13.
P-Values for CCNA using diagnostic labels. P-values 0.01 indicate significant differences (*).
| CCNA | MCI vs V-MCI | V-MCI vs AD | MCI vs AD | SCI vs V-MCI | SCI vs MCI | SCI vs AD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microstructural Integrity (MII) | 0.01* | 0.98 | 0.01* | 0.01* | 0.21 | 0.01* | ||||||
| Macrostructural Damage (MAD) | 0.01* | 0.11 | 0.46 | 0.01* | 0.80 | 0.18 | ||||||
| Microstructural Damage (MID) | 0.08 | 0.01* | 0.65 | 0.26 | 1.00 | 0.65 | ||||||
| Intensity | 0.93 | 0.18 | 0.04 | 0.33 | 0.58 | 0.01* | ||||||
| NABM/ICV Ratio | 0.01* | 0.99 | 0.01* | 0.01* | 0.15 | 0.01* |
Table 14.
Unadjusted Means for ADNI using diagnostic labels.
| ADNI | CN | MCI | AD |
|---|---|---|---|
| Microstructural Integrity (MII) | 118.85 2.17 | 118.65 2.28 | 116.79 2.53 |
| Macrostructural Damage (MAD) | 4203.64 641.61 | 4262.00 737.37 | 4906.86 776.63 |
| Microstructural Damage (MID) | 9.95 1.21 | 10.05 1.26 | 10.82 1.43 |
| Intensity | 278.97 4.46 | 278.56 4.12 | 277.18 4.21 |
| NABM/ICV Ratio | 0.750 0.04 | 0.746 0.05 | 0.701 0.05 |
Table 15.
P-Values for ADNI using diagnostic labels. P-values 0.01 indicate significant differences (*).
| ADNI | CN vs MCI | CN vs AD | MCI vs AD |
|---|---|---|---|
| Microstructural Integrity (MII) | 0.48 | 0.01* | 0.01* |
| Macrostructural Damage (MAD) | 0.51 | 0.01* | 0.01* |
| Microstructural Damage (MID) | 0.49 | 0.01* | 0.01* |
| Intensity | 0.40 | 0.01* | 0.01* |
| NABM/ICV Ratio | 0.64 | 0.01* | 0.01* |
References
- Abbasi, S., Tajeripour, F., 2016. Detection of Brain Tumor in 3D MRI Images using Local Binary Patterns and Histogram Orientation Gradient. Neurocomputing 219 (2016). doi: 10.1016/j.neucom.2016.09.051.
- Aisen P.S., et al. Alzheimer’s disease neuroimaging initiative 2 clinical core: progress and plans. Alzheimer’s Dementia. 2021;11(6):734–739. doi: 10.1016/j.jalz.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber J., et al. White matter hyperintensities in vascular contributions to cognitive impairment and dementia (VCID): Knowledge gaps and opportunities. Alzheimer’s and Dementia: Translational Research & Clinical Interventions. 2019;5:107–117. doi: 10.1016/j.trci.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander A.L., et al. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G.S., et al. Diffusion tensor imaging studies in vascular disease: A review of the literature. Dement Neuropsychol. 2012;6(3):158–163. doi: 10.1590/S1980-57642012DN06030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Disease Neuroimaging Initiative (ADNI2), 2008. Defining Alzheimer’s Disease, Procedures Manual.
- Badji A., Westman E. Cerebrovascular pathology in Alzheimer’s disease: Hopes and gaps. Psychiatry Res. Neuroimag. 2020;111184:1–6. doi: 10.1016/j.pscychresns.2020.111184. [DOI] [PubMed] [Google Scholar]
- Black S., Gao F., Bilbao J. Understanding white matter disease: imaging-pathological correlations in vascular cognitive impairment. Stroke. 2009;40(3):S48–52. doi: 10.1161/STROKEAHA.108.537704. [DOI] [PubMed] [Google Scholar]
- Black S.E., DeCarli C., Greenberg S.M., et al. AHA/ASA Scientific Statement Vascular Contributions to Cognitive Impairment and Dementia. A Statement for Healthcare Professionals From the American Heart Association/American Stroke Association Council on Epidemiology and Prevention, Council on Cardiovas. Stroke. 2011;42:2672–2713. doi: 10.1161/STR.0b013e3182299496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, L.W., Bancej, C., McDowell, I., 2016. Prevalence and Monetary Costs of Dementia in Canada: Population Health Expert Panel. In: Alzheimer Society of Canada in collaboration with the Public Health Agency of Canada.
- Chanraud S., et al. MR Diffusion Tensor Imaging: A Window into White Matter Integrity of the Working Brain. Neuropsychol. Rev. 2010;20(2):209–225. doi: 10.1007/s11065-010-9129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertkow H., Borrie M., Black S.E., et al. The Comprehensive Assessment of Neurodegeneration and Dementia: Canadian Cohort Study. Can. J. Neurol. Sci. 2019;46(5):499–511. doi: 10.1017/cjn.2019.27. [DOI] [PubMed] [Google Scholar]
- Chutinet A., Rost N.S. White Matter Disease as a Biomarker for Long-Term Cerebrovascular Disease and Dementia. Curr. Treatment Options Cardiovasc. Med. 2014;16(3) doi: 10.1007/s11936-013-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debette S., Markus H.S. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ. 2010;341(c3666):1–9. doi: 10.1136/bmj.c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot M., et al. Changes in Normal-Appearing White Matter Precede Development of White Matter Lesions. Stroke. 2013;44(4):1037–1042. doi: 10.1161/STROKEAHA.112.680223. [DOI] [PubMed] [Google Scholar]
- Della Nave R., Foresti S., Pratesi A., et al. Whole-brain histogram and voxel-based analyses of diffusion tensor imaging in patients with leukoaraiosis: correlation with motor and cognitive impairment. Am. J. Neuroradiol. 2007;28(7):1313–1319. doi: 10.3174/ajnr.A0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio J., Tyrrell P.N., Khademi A., et al. Intracranial volume segmentation for neurodegenerative populations using multicentre flair mri. Neuroimage: Reports. 2021;1(1) doi: 10.1016/j.ynirp.2021.10000. [DOI] [Google Scholar]
- Duchesne S., Chouinard I., Potvin O., et al. The Canadian Dementia Imaging Protocol: Harmonizing National Cohorts. J. Magn. Resonance Imag. 2019;49(2):456–465. doi: 10.1002/jmri.26197. [DOI] [PubMed] [Google Scholar]
- Dumont M., et al. Free Water in White Matter Differentiates MCI and AD From Control Subjects. Front. Aging Neurosci. 2019;11:270. doi: 10.3389/fnagi.2019.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA’s Decision to Approve New Treatment for Alzheimer’s Disease FDA. https://www.fda.gov/drugs/news-events-human-drugs/fdas-decision-approve-new-treatment-alzheimers-disease.
- Frey B.M. Characterization of White Matter Hyperintensities in Large-Scale MRI-Studies. Front. Neurol. 2019 doi: 10.3389/fneur.2019.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., van Uden I.W.M., et al. Automated detection of white matter hyperintensities of all sizes in cerebral small vessel disease. Med. Phys. 2016;43(12):6246–6258. doi: 10.1118/1.4966029. [DOI] [PubMed] [Google Scholar]
- Gorelick P.B., Scuteri A., Black S.E., et al. Vascular contributions to cognitive impairment and dementia: A statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2011;42(9):2672–2713. doi: 10.1161/STR.0b013e3182299496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecke, W.V., Emsell, L., Sunaert, S., (Eds.), 2016. Diffusion Tensor Imaging: A Practical Handbook. Springer-Verlag, New York. doi: 10.1007/978-1-4939-3118-7.
- Jack, C.R., Jr, 2011. ADNI GO/2 MRI QC Procedures in Aging and Dementia Imaging Research Laboratory. p. 3.
- Jessen F., Wolfsgruber S., et al. Ad dementia risk in late mci, in early mci, and in subjective memory impairment. Alzheimer’s Dementia. 2014;10(1):76–83. doi: 10.1016/j.jalz.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Ji F., Pasternak O., Liu S., et al. Distinct white matter microstructural abnormalities and extracellular water increases relate to cognitive impairment in Alzheimer’s disease with and without cerebrovascular disease. Alzheimer Res. Therapy. 2017;9(1) doi: 10.1186/s13195-017-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith J., et al. Collagenosis of the Deep Medullary Veins: An Underrecognized Pathologic Correlate of White Matter Hyperintensities and Periventricular Infarction. J. Neuropathol. Exp. Neurol. 2017;76(4):299–312. doi: 10.1093/jnen/nlx009. [DOI] [PubMed] [Google Scholar]
- Khademi A., et al. 2009 16th International Conference on Digital Signal Processing. 2009. Nonparametric statistical tests for exploration of correlation and nonstationarity in images. [DOI] [Google Scholar]
- Khademi A., Reiche B., DiGregorio J., et al. Whole Volume Brain Extraction for Multi-Centre, Multi-Disease FLAIR MRI Datasets. Magn. Reson. Imaging. 2019;66:116–130. doi: 10.1016/j.mri.2019.08.022. [DOI] [PubMed] [Google Scholar]
- Khan T.K. Neuroimaging Biomarkers in Alzheimer’s Disease. Biomarkers Alzheimer’s Dis. 2016:51–100. doi: 10.1016/b978-0-12-804832-0.00003-1. [DOI] [Google Scholar]
- Lamar M., et al. Common Brain Structural Alterations Associated with Cardiovascular Disease Risk Factors and Alzheimer’s Dementia: Future Directions and Implications. Neuropsychol. Rev. 2020;30(4):546–557. doi: 10.1007/s11065-020-09460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Z., Shen D., Liu D., et al. Computer-Assisted Segmentation of White Matter Lesions in 3D MR Images Using Support Vector Machine. Acad Radiol. 2008;15(3):300–313. doi: 10.1016/j.acra.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenglet C. Diffusion Tensor Imaging. Brain Mapping. 2015:245–251. doi: 10.1016/b978-0-12-397025-1.00291-8. [DOI] [Google Scholar]
- Liu Y., et al. White matter hyperintensities induce distal deficits in the connected fibers. Hum. Brain Mapp. 2021 doi: 10.1002/hbm.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P., et al. FLAIR and diffusion MRI signals are independent predictors of white matter hyperintensities. Am. J. Neuroradiol. 2013;34(1):54–61. doi: 10.3174/ajnr.A3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniega S.M., et al. Integrity of normal-appearing white matter: Influence of age, visible lesion burden and hypertension in patients with small-vessel disease. J. Cerebral Blood Flow Metabolism. 2016;37(2):644–656. doi: 10.1177/0271678X16635657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D., et al. Lesion Topography and Microscopic White Matter Tract Damage Contribute to Cognitive Impairment in Symptomatic Carotid Artery Disease. Radiology. 2017;282(2):505–515. doi: 10.1148/radiol.2016152685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski, D., et al. Synchronized-averaging of 4D CT Data Set For Improved Quality Treatment Planning CT Scan. Int. J. Rad. Oncol.- Biol.- Phys. 81(2) (2011) S818. doi: 10.1016/j.ijrobp.2011.06.1440.
- Mohaddes, Z., et al. National Neuroinformatics Framework for Canadian Consortium on Neurodegeneration in Aging (CCNA). Front. Neuroinf. 12, 85. doi: 10.3389/fninf.2018.00085. [DOI] [PMC free article] [PubMed]
- Morgan, J.T., Nordahl, C.W., Schumann, C.M., 2013. Chapter 3.5 – The Amygdala in Autism Spectrum Disorders. In: The Neuroscience of Autism Spectrum Disorders. pp. 297–312. doi: 10.1016/B978-0-12-391924-3.00021-1.
- Nave R.D., et al. Whole-Brain Histogram and Voxel-Based Analysis of Diffusion Tensor Imaging in Patients with Leukoaraiosis: Correlation with Motor and Cognitive Impairment. Am. J. Neuroradiol. 2007;28(7):1313–1319. doi: 10.3174/ajnr.a0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor S.M., et al. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131(9):2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuchowski N.A., Reeves A.P., Huang E.P., et al. Quantitative imaging biomarkers: A review of statistical methods for computer algorithm comparisons. Stat. Methods Med. Res. 2015;24(1):68–106. doi: 10.1177/0962280214537390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., et al. Human Brain White Matter Atlas: Identification and Assignment of Common Anatomical Structures in Superficial White Matter. NeuroImage. 2008;43(3):447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., et al. DTI Analyses and Clinical Applications in Alzheimer’s Disease. J. Alzheimer’s Dis. 2011;26(s3):287–296. doi: 10.3233/jad-2011-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala, T., Pietikainen, M., Harwood, D., 1994. Performance evaluation of texture measures with classification based on kullback discrimination of distributions. In: Proceedings of the 12th International Conference on Pattern Recognition, vol. 1. pp. 582–585, Jerusalem, Israel. doi: 10.1109/ICPR.1994.576366.
- Ojala T., Pietikainen M., Harwood D. A comparative study of texture measures with classification based on feature distributions. Pattern Recogn. 1996;29(1):51–59. doi: 10.1016/0031-3203(95)00067-4. [DOI] [Google Scholar]
- Ojala T., Pietikainen M., Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24(7):971–987. doi: 10.1109/TPAMI.2002.1017623. [DOI] [Google Scholar]
- Oppedal, et al., 2015. Classifying Dementia Using Local Binary Patterns from Different Regions in Magnetic Resonance Images. Int. J. Biomed. Imaging 15. doi: 10.1155/2015/572567. [DOI] [PMC free article] [PubMed]
- Oppedal, K., et al., 2015. Classifying Dementia Using Local Binary Patterns from Different Regions in Magnetic Resonance Images. Int. J. Biomed. Imag. 1–14. doi:10.1155/2015/572567. [DOI] [PMC free article] [PubMed]
- Pievani M., et al. Assessment of white matter tract damage in mild cognitive impairment and Alzheimer’s disease. Hum. Brain Mapp. 2010;31(12):1862–1875. doi: 10.1002/hbm.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pini L., et al. Brain atrophy in Alzheimer’s Disease and aging. Ageing Res. Rev. 2016;30:25–48. doi: 10.1016/j.arr.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Rahim A., Hossain N., Wahid T., Azam S. Face Recognition using Local Binary Patterns (LBP), in Global Journal of Computer Science and Technology. Graphics Vis. 2013;13(4) [Google Scholar]
- Rangayyan R.M., Banik S., Leo Desautels J.E. 2012 IEEE International Symposium on Medical Measurements and Applications Proceedings. 2012. Detection of architectural distortion in prior mammograms using measures of angular dispersion. [DOI] [Google Scholar]
- Reiche B., Moody A.R., Khademi A. Pathology-preserving intensity standardization framework for multi-institutional FLAIR MRI datasets. Magn. Resonance Imag. 2019;62:59–69. doi: 10.1016/j.mri.2019.05.001. [DOI] [PubMed] [Google Scholar]
- Russ T.C., Stamatakis E., Hamer M., et al. Socioeconomic status as a risk factor for dementia death: individual participant meta-analysis of 86 508 men and women from the UK. Br. J. Psychiatry. 2013;203(01):10–17. doi: 10.1192/bjp.bp.112.119479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairamya N.J., Susmitha L., Thomas George S., Subathra M.S.P. Intelligent Data Analysis for Biomedical Applications. Academic Press; 2019. Chapter 12 - Hybrid Approach for Classification of Electroencephalographic Signals Using Time-Frequency Images With Wavelets and Texture Features; pp. 253–273. [DOI] [Google Scholar]
- Scola E., et al. A diffusion tensor MRI study of patients with MCI and AD with a 2-year clinical follow-up. J. Neurol. Neurosurg. Psychiatry. 2010;81(7):798–805. doi: 10.1136/jnnp.2009.189639. [DOI] [PubMed] [Google Scholar]
- Siemonsen S., Finsterbusch J., Matschkeat J. Age-Dependent Normal Values of T2* and T2 in Brain Parenchyma. Am. J. Neuroradiol. 2008;29(5):950–955. doi: 10.3174/ajnr.a0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.E., Cieslak A., Barber P., et al. Therapeutic Strategies and Drug Development for Vascular Cognitive Impairment. J. Am. Heart Assoc. 2017;6(5):1–12. doi: 10.1161/JAHA.117.005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.E., Biessels G.J., De Guio F., et al. Harmonizing brain magnetic resonance imaging methods for vascular contributions to neurodegeneration. in Alzheimer’s Dementia Diagnosis. Assessment Disease Monit. 2019;11:191–204. doi: 10.1016/j.dadm.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.C., Obuchowski N.A., Kessler L.G., et al. Metrology standards for quantitative imaging biomarkers. Radiology. 2015;277(3):813–825. doi: 10.1148/radiol.2015142202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajeripour, F., Kabir, E., Sheikhi, A., 2007. Defect Detection in Patterned Fabrics Using Modified Local Binary Patterns. In: Conference on Computational Intelligence and Multimedia Applications, vol. 2. doi: 10.1109/ICCIMA.2007.50.
- Tardif J.C., et al. Atherosclerosis imaging and the canadian atherosclerosis imaging network in Canadian. J. Cardiol. 2013;29(3):297–303. doi: 10.1016/j.cjca.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Theaud, G., Houde, J.-C., Boré, A., Rheault, F., Morency, F., Descoteaux, M. TractoFlow: A robust, efficient and reproducible diffusion MRI pipeline leveraging Nextflow & Singularity. NeuroImage. doi: 10.1016/j.neuroimage.2020.116889. [DOI] [PubMed]
- Wardlaw J.M., Smith E.E., Biessels G.J., et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12(8):822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston P.S.J., et al. Diffusion imaging changes in grey matter in Alzheimer’s disease: a potential marker of early neurodegeneration. Alzheimer’s Res. Ther. 2015;7(1) doi: 10.1186/s13195-015-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.S., Hebert L.E., Scherr P.A., et al. Educational attainment and cognitive decline in old age. Neurology. 2009;72(5):460–465. doi: 10.1212/01.wnl.0000341782.71418.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiannopoulou K.G., Anastasiou A.I., Zachariou V., Pelidou S. Reasons for Failed Trials of Disease-Modifying Treatments for Alzheimer Disease and Their Contribution in Recent Research. MDPI Biomed. 2019;7(97):1–16. doi: 10.3390/biomedicines7040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni G., et al. White Matter Imaging Correlates of Early Cognitive Impairment Detected by the Montreal Cognitive Assessment After Transient Ischemic Attack and Minor Stroke. Stroke. 2017;48(6):1539–1547. doi: 10.1161/STROKEAHA.116.016044. [DOI] [PubMed] [Google Scholar]