ABSTRACT
Background
Mammary serine protease inhibitor (maspin) is well known as a tumor suppressor gene in several types of cancers and its nuclear localization is essential for its tumor-suppressive function. We previously reported that the cytoplasmic-only localization of maspin is significantly correlated with unfavorable prognosis in patients with lung adenocarcinoma (LUAD). To clarify whether maspin in LUAD acts as a tumor promoter or suppressor, we examined the subcellular localization-dependent biological functions of maspin in human LUAD cell lines.
Methods
The expression levels and subcellular localization of maspin were investigated by performing immunoblotting and immunofluorescence in human LUAD cell lines (PC-9, A549, NCI-H23, RERF-LC-KJ) and human bronchial epithelial cell line (BEAS-2B). We then established stable cell lines overexpressing maspin (A549-maspin and RERF-LC-KJ-maspin) and investigated their subcellular localization. Cell invasion assays of these cell lines were performed to examine their invasiveness. Moreover, the mRNA expression levels between epithelial cell markers (E-cadherin) and mesenchymal cell markers (N-cadherin and vimentin) were compared.
Results
The expression of maspin in PC-9 cells was comparable to that in BEAS-2B cells, whereas its expression in A549, NCI-H23, and RERF-LC-KJ cells was decreased. The cell invasion capability of A549-maspin cells showing pancellular expression was significantly decreased compared with that of A549-control cells. By contrast, the cell invasion capability of RERF-LC-KJ-maspin cells showing cytoplasmic-only expression was significantly increased compared with that of RERF-LC-KJ-control cells. The mRNA expression levels of N-cadherin, but not E-cadherin and vimentin, in A549-maspin cells was significantly downregulated compared with that in A549-control cells. No significant differences in these markers were observed between RERF-LC-KJ-maspin and RERF-LC-KJ-control cells.
Conclusion
The invasive capability of LUAD cells is regulated by the intracellular localization of maspin. Clarification of the molecular mechanism underlying the subcellular localization-dependent function of maspin will promote a deeper understanding of LUAD development and progression.
Keywords: lung adenocarcinoma, maspin
Lung cancer is one of the leading causes of cancer-related death in both men and women worldwide.1 Non-small cell lung cancer (NSCLC) is a general term that includes various subtypes of epithelial lung cancers other than small cell lung cancer; lung adenocarcinoma (LUAD) is the most common subtype of NSCLC. Among NSCLC, especially LUAD, activating mutations in the epidermal growth factor receptor (EGFR) and rearrangements in the anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 receptor tyrosine kinase (ROS1) genes have been identified as oncogenic drivers and are currently a general target for molecular targeted therapy.2 Therefore, in patients with LUAD, targeted agents have demonstrated superior efficacy compared with chemotherapy. However, despite the advances in targeted therapies, some patients develop acquired resistance to small-molecule tyrosine kinase inhibitors.3 Hence, for patients who develop acquired resistance, empirical cytotoxic chemotherapy remains the treatment of choice. By contrast, mutation in the Kirsten rat sarcoma 2 viral oncogene homolog (K-RAS), one of the earliest known oncogenic drivers in NSCLC that is present in approximately 30% of LUAD patients, remains a challenging therapeutic target.2 Unfortunately, the driver mutations in 40% of patients with LUAD have not yet been identified, and classical cytotoxic chemotherapy is the only treatment option. As mentioned above, establishing a novel molecular targeted therapy for LUAD to increase the number of targeted treatment options is now the focus of research.
Human mammary serine protease inhibitor (maspin), which is located within a 10-gene serpin cluster on 18q21.13 and a protein encoded by the SERPINB5 gene, was originally identified in normal human mammary epithelial cells.4,5,6,7 The expression of maspin has been reported to correlate with induction of tumor cell apoptosis,8 suppression of cellular motility,9, 10 regulation of cell adhesion,11, 12 and regulation of drug sensitivity,13, 14 and has therefore been recognized as a tumor suppressor protein. Additionally, maspin expression correlates with better prognosis in various types of cancers such as breast, prostate, bladder, gastric, and lung cancers. However, maspin expression in pancreatic cancer is associated with a poor prognosis.15 As mentioned above, it remains controversial whether the expression of maspin suppresses or promotes tumor growth.16 We have previously reported that the cytoplasmic-only expression of maspin correlates with poor clinical prognosis in patients with breast cancer and NSCLC.17,18,19,20 In addition, the nuclear localization of maspin is essential for tumor suppression, because maspin expressed in the nucleus inhibits tumor growth and metastasis by binding to the promoter region of colony-stimulation factor-1.21 Based on these findings, we hypothesized that the function of maspin, which has tumor-promoting and tumor-suppressing properties, differs depending on its subcellular localization, and cytoplasmic maspin may contribute to the acquisition of aggressive phenotypes in LUAD cells. To the best of our knowledge, this is the first study to explore the association between LUAD cells and maspin subcellular localization.
MATERIALS AND METHODS
Cell culture
The human LUAD cell lines, A549, RERF-LC-KJ, and PC-9, were provided by the RIKEN BioResource Research Center through the National BioResource Project of the Ministry for Education, Culture Sports, Science and Technology, Japan. The human LUAD, NCI-H23, and human bronchial epithelial (BEAS-2B) cell lines, were purchased from the American Type Culture Collection (ATCC; Manassas, VA). The A549 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Nissui Pharmaceutical, Tokyo, Japan, or Thermo Fisher Scientific, Waltham, MA) supplemented with 10% inactivated fetal bovine serum (FBS; Biological Industries, Cromwell, CT). The RERF-LC-KJ, PC-9, and NCI-H23 cells were cultured in Roswell Park Memorial Institute 1640 medium (RPMI1640; Thermo Fisher Scientific) supplemented with 10% inactivated FBS. The BEAS-2B cells were grown in Bronchial Epithelial Cell Growth Basal Medium (BEBM; Lonza, Walkersville, MD) supplemented with Bronchial Epithelial SingleQuots Kit (Lonza). The 293FT cell line was purchased from Thermo Fisher Scientific and maintained in DMEM (Nissui Pharmaceutical) supplemented with 10% inactivated FBS. The cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol Reagent (Thermo Fisher Scientific). cDNA was generated from isolated RNA using a High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The gene expression levels were determined using the TaqMan Gene Expression Master Mix (Thermo Fisher Scientific) on a LightCycler 96 system (Roche Diagnostics, Mannheim, Germany). The primer and probe sets used in this study were glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Human GAPDH Endogenous Control (Hs99999905_m1, Thermo Fisher Scientific); Maspin, TaqMan Gene Expression Assays (Hs00985285_m1, Thermo Fisher Scientific); E-cadherin, TaqMan Gene Expression Assays (Hs01023894_m1, Thermo Fisher Scientific); N-cadherin, TaqMan Gene Expression Assays (Hs00983056_m1, Thermo Fisher Scientific); and vimentin, TaqMan Gene Expression Assays (Hs00958111_m1, Thermo Fisher Scientific). The housekeeping gene, GAPDH, was used as the normalization standard.
Western blot
The total cellular proteins were extracted with a radioimmunoprecipitation assay (RIPA) buffer (containing 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with cOmpleteTM Protease Inhibitor Cocktail (Roche Diagnostics) and PhosSTOPTM (Roche Diagnostics). In the nuclear/cytoplasmic fractionation assay, the nuclear and cytoplasmic proteins were extracted using a subcellular protein fractionation kit for cultured cells (Thermo Fisher Scientific) according to the manufacturer’s instructions. The protein concentration was determined using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA). The protein concentration was adjusted with 5× sample buffer (60 mM Tris-HCl [pH6.8], 5% β-mercaptoethanol, 0.1% bromophenol blue, 2% SDS, 25% glycerol), and RIPA buffer, and the samples were boiled for 5 min. The samples were then subjected to SDS-polyacrylamide gel electrophoresis and transferred to a 0.45 μm PVDF membrane (Merck Millipore, Darmstadt, Germany). The membranes were blocked with ECL Prime blocking reagent (Cytiva/Global Life Sciences Solutions, Marlborough, MA) for 90 min at room temperature (RT), and then immunoblotting was performed at 4°C overnight. The primary antibodies used in this experiment were GAPDH (clone D16H11, #5174, Cell Signaling Technology, Danvers, MA), HSP90 (clone C45G5, #4877, Cell Signaling Technology), HDAC1 (clone 10E2, #5356, Cell Signaling Technology), and Maspin (clone EAW24, NCL-MASPIN, Leica Biosystems, Newcastle Ltd., UK). Immunoblotting was performed by further incubating the samples with secondary antibodies conjugated to horseradish peroxidase. Immunoreactivity was detected using ImageQuant LAS 4000 (Cytiva) using ECL Select Western Blotting Detection Reagent (Cytiva) according to the manufacturer’s instructions.
Immunofluorescence
The cells were cultured in a Nunc Lab-Tek II 8-well chamber slide (Thermo Fisher Scientific) for 24 h before the experiment. The cells were washed twice with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min at RT. The fixed cells were washed with PBS and permeabilized with 100% ice-cold methanol for 10 min at −20°C. The samples were treated with 0.2% Triton-X in PBS for 10 min at RT. After blocking with 10% goat serum in PBS for 60 min at RT, the samples were incubated with anti-maspin antibody (clone EAW24, NCL-MASPIN, Leica Biosystems) at 4°C overnight, followed by incubation with Alexa Fluor 488-conjugated or Alexa Fluor 647-conjugated secondary antibody (Thermo Fisher Scientific) in the dark. The slides were mounted using ProLong Diamond Antifade Mountant with 4’,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific). Confocal fluorescent images were obtained using a Zeiss LSM 780 confocal microscope (Carl Zeiss Microscopy, Oberkochen, Germany) and Zeiss ZEN2011 software (ver. 11.0.4.190; Carl Zeiss Microscopy).
Establishment of A549-maspin and RERF-LC-KJ-maspin cell lines
pLenti/maspin-ZsGreen and pLenti/ZsGreen vectors created in the previous study were co-transfected with ViraPower Packaging Mix (Thermo Fisher Scientific) into 293FT cells,22 and lentivirus stocks were collected according to the manufacturer’s instructions. The lentivirus stocks were concentrated using a Lenti-X Concentrator (Takara Bio, Shiga, Japan) and then titrated using qPCR Lentivirus Titration Kit (Applied Biological Materials, Richmond, BC, Canada) according to the manufacturer’s instructions. To establish the A549 and RERF-LC-KJ stable cell lines that overexpress either maspin-ZsGreen or ZsGreen alone (control), the cells were infected with a lentivirus-based vector at a multiplicity of infection of 25 and were pooled with 10 μg/mL blasticidin (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan). The surviving cells of the A549-maspin, A549-control, RERF-LC-KJ-maspin, and RERF-LC-KJ-control cell lines were used in subsequent experiments.
Cell invasion assay
The established cell lines were starved in a serum-free medium for 24 h. The cells (8 × 105 cells/mL) were added in the upper chamber of the transwells with 8.0 μm pore size membranes in 24-well plates; then, the lower chambers were loaded with 10% inactivated FBS in cell culture medium. The invasion capability of the cells was evaluated by performing a QCM ECMatrix Cell Invasion Assay using a 24-well (8 μm) colorimetric kit (Merck Millipore, Billerica, MA) according to the manufacturer’s instructions. After 24 h of incubation for A549 stable cell lines or 72 h incubation for RERF-LC-KJ stable cell lines, the cells were fixed with 4% paraformaldehyde for 15 min at RT and then stained with crystal violet for 10 min. The number of invasive cells was counted using ImageJ/Fiji software (ver. 2.1.0/1.53c).
Statistical analysis
Statistical analyses were performed using Excel and SPSS Statistics 25 (IBM Corp., Armonk, NY). The data were presented as mean ± standard deviation (SD). The differences between means were evaluated using Student’s t test or Dunnett’s test. A P value of < 0.05 was considered significant.
RESULTS
Expression level and subcellular localization of maspin in BEAS-2B and LUAD cell lines
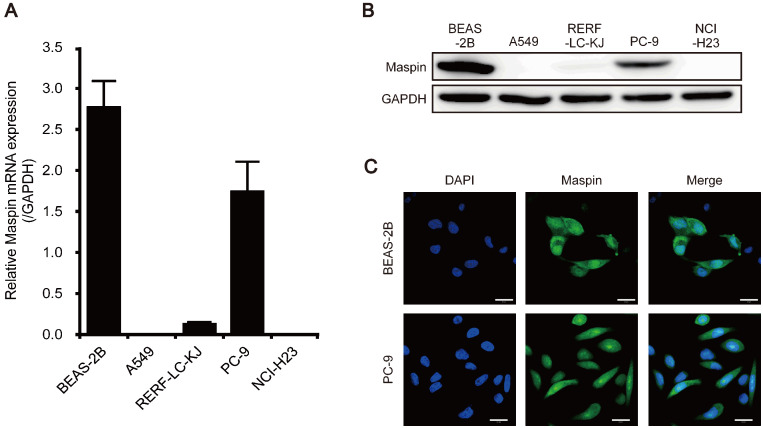
Our previous studies showed that maspin exhibits differential subcellular localization, such as cytoplasmic-only, pancellular (combined nuclear and cytoplasmic), and no expression, in tumor tissues of patients with LUAD, and found that cytoplasmic localization of maspin was associated with poor prognosis.18 The expression level of maspin in a human bronchial epithelial cell line (BEAS-2B) and four LUAD cell lines (A549, RERF-LC-KJ, PC-9, and NCI-H23) was investigated. In BEAS-2B cells, the mRNA expression of maspin was relatively high (Fig. 1A). Among the LUAD cells, maspin mRNA expression was the highest in the PC-9 cells (Fig. 1A). By contrast, this expression was slightly lower in RERF-LC-KJ cells and was not detected in A549 and NCI-H23 cells (Fig. 1A). The protein expression of maspin was investigated by western blotting, and results showed that maspin was highly expressed in BEAS-2B and PC-9 cells, as with mRNA expression levels (Fig. 1B). In addition, a small amount of maspin protein was detected in RERF-LC-KJ cells, but not in A549 and NCI-H23 cells (Fig. 1B). To investigate the subcellular localization of maspin, immunofluorescence was performed using two cell lines with high maspin expression levels. In BEAS-2B and PC-9 cells, maspin protein was detected in the nucleus and cytoplasm (Fig. 1C). These results suggest that although the LUAD cell lines show different maspin expression profiles, the mechanisms underlying the maspin nuclear transport in PC-9 cells, which exhibit pancellular maspin expression, were similar to those in BEAS-2B cells.
Fig. 1.
Expression and subcellular localization of maspin in human bronchial epithelial cell line (BEAS-2B) and lung adenocarcinoma (LUAD) cell lines. (A) Relative mRNA expression levels of maspin in BEAS-2B cell line and LUAD cell lines. mRNA expression was normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are shown as mean ± SD (n = 3). (B) Protein expression levels of maspin in BEAS-2B cell line and LUAD cell lines. GAPDH is shown for loading control. (C) Representative examples of subcellular localization of maspin in BEAS-2B and PC-9 cells. The expression of maspin was detected using anti-maspin antibody. The nucleus was counterstained with 4’,6-diamidino-2-phenylindole (DAPI). Scale bars represent 20 μm. Confocal fluorescent images were obtained using Zeiss LSM 780 confocal microscope and Zeiss ZEN2011 software (ver. 11.0.4.190).
Subcellular localization of exogenous maspin in stable cell lines
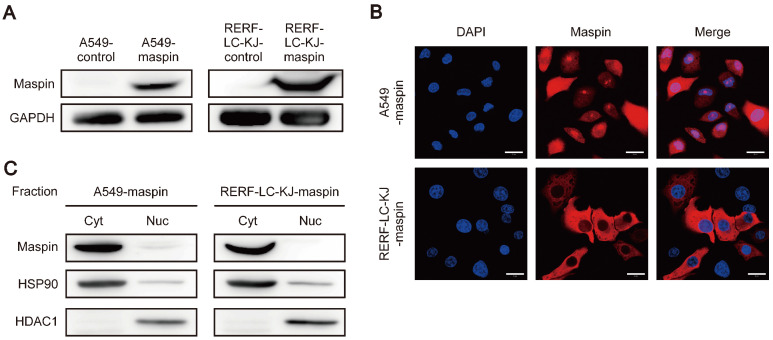
To confirm the subcellular localization of overexpressed maspin in LUAD cell lines with low maspin expression, maspin stably expressing cell lines were established using two different LUAD cell lines, the A549 and RERF-LC-KJ. The two established cell lines (A549-maspin and RERF-LC-KJ-maspin) showed increased maspin expression compared to the control cell lines (A549-control and RERF-LC-KJ-control) (Fig. 2A). Immunofluorescence staining of these cell lines showed that A549-maspin cells expressed maspin in both the nucleus and cytoplasm, whereas the RERF-LC-KJ-maspin cells expressed maspin only in the cytoplasm (Fig. 2B). Additionally, the different subcellular localizations of maspin in these cell lines, as with the results of immunofluorescence staining, were also confirmed by nuclear/cytoplasmic fractionation assay (Fig. 2C).
Fig. 2.
Maspin expression and subcellular localization in maspin stable cell lines. (A) Protein expression levels of maspin in A549-control, A549-maspin, RERF-LC-KJ-control, and RERF-LC-KJ-maspin cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown for loading control. (B) Representative examples of subcellular localization of maspin in A549-maspin and RERF-LC-KJ-maspin cells. Maspin expression was detected using anti-maspin antibody. The nucleus was counterstained with 4’,6-diamidino-2-phenylindole (DAPI). Scale bars represent 20 μm. Confocal fluorescent images were obtained using Zeiss LSM 780 confocal microscope and Zeiss ZEN2011 software (ver. 11.0.4.190). (C) The expression levels of maspin in the cytoplasm (Cyt) and nucleus (Nuc) fractions. The expression levels of HSP90 and HDAC1 were used as protein loading control in the cytoplasm and nucleus fractions, respectively.
Subcellular localization-dependent effect of maspin on the invasion capability of LUAD cells
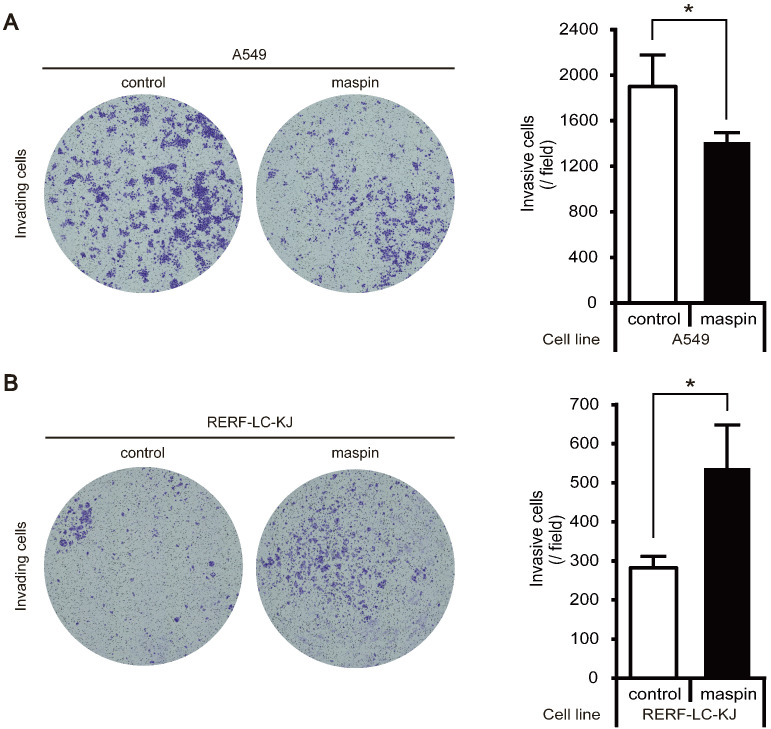
To determine whether the role of maspin in the biological properties of LUAD cells is altered by its subcellular localization, cell invasion assay was performed using two established stable cell lines (pancellular-maspin model: A549-maspin; cytoplasmic-maspin model: RERF-LC-KJ-maspin). The cell invasion capability of A549-maspin cells was significantly decreased compared with that of A549-control cells (Fig. 3A). By contrast, the increased expression of maspin in the cytoplasm of RERF-LC-KJ-maspin cells significantly promoted cell invasiveness when compared with RERF-LC-KJ-control cells (Fig. 3B).
Fig. 3.
Cell invasion capability of maspin stable cell lines. (A) Cell invasion assay in A549 stable cell lines. Representative examples of invading cells stained with crystal violet and total cell count passed through the basement membrane matrix. The invasion of A549 stable cell lines was assessed after 24 h. Data are shown as mean ± SD (n = 3). *P < 0.05; Student’s t-test. (B) Cell invasion assay of RERF-LC-KJ stable cell lines. Representative examples of invading cells stained with crystal violet and total cell count passed through the basement membrane matrix. Invasion of RERF-LC-KJ stable cell lines was assessed after 72 h. Data are shown as mean ± SD (n = 3). *P < 0.05; Student’s t-test
mRNA expression of epithelial and mesenchymal cell markers in stable cell lines
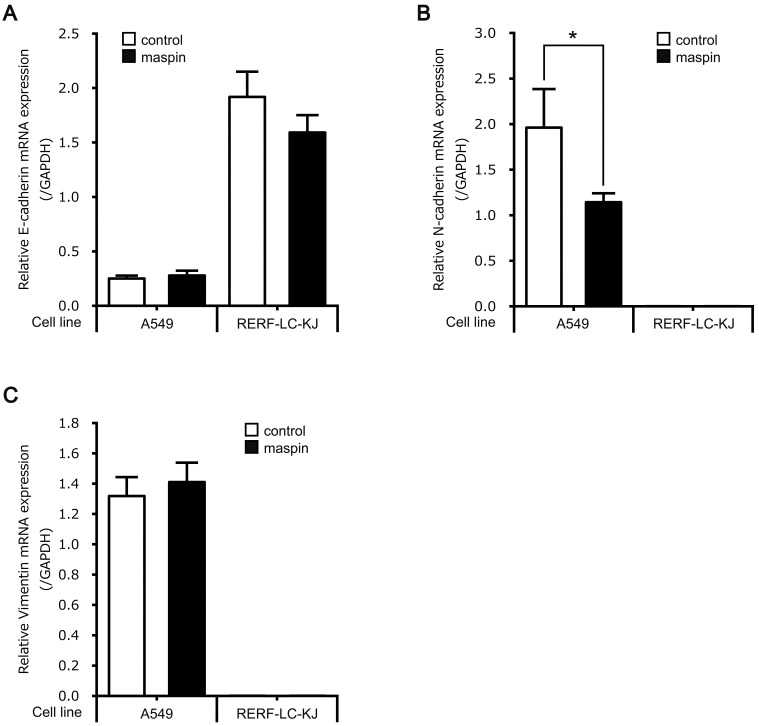
To investigate in detail the subcellular localization-dependent functions of maspin on cell invasiveness, the mRNA expression levels of an epithelial cell marker (E-cadherin) and mesenchymal cell markers (N-cadherin and vimentin) were compared using two different maspin subcellular localization models. The mRNA expression levels of E-cadherin were not changed by the maspin overexpression in either A549 or RERF-LC-KJ cell lines (Fig. 4A). In A549 cells, the mRNA expression level of N-cadherin, but not vimentin, was significantly downregulated in A549-maspin cells when compared with A549-control cells (Figs. 4B and C). In contrast to the A549 cell line, the mRNA expression levels of N-cadherin and vimentin in RERF-LC-KJ cells were not detectable, and no significant difference was observed between RERF-LC-KJ-maspin and RERF-LC-KJ-control cells (Figs. 4B and C). These results indicate that the overexpression of pancellular maspin in A549 cells suppresses cell invasion by inhibiting the transformation of these cells to mesenchymal cells, which is also known as epithelial mesenchymal transition (EMT).
Fig. 4.
mRNA levels of EMT markers of A549-control, A549-maspin, RERF-LC-KJ-control, and RERF-LC-KJ-maspin. (A) Relative mRNA expression level of E-cadherin. mRNA expression was normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are shown as mean ± SD (n = 3). (B) The relative mRNA expression level of N-cadherin. mRNA expression was normalized to the level of GAPDH. Data are shown as mean ± SD (n = 3). *P < 0.05; Student’s t-test. (C) The relative mRNA expression level of vimentin. The mRNA expression level was normalized to the level of GAPDH. Data are shown as mean ± SD (n = 3).
DISCUSSION
The downregulation of maspin expression, and the involvement of transcriptional regulation, miRNAs, and aberrant promoter methylation in this silencing, has been reported in several types of cancers.23,24,25 Consistent with the results of these studies, results of the maspin expression analysis in LUAD cell lines demonstrated that maspin protein was absent in A549 and NCI-H23 cells, and its expression was downregulated in RERF-LC-KJ cells (Figs. 1A and B). Since the re-expression of maspin in cancer cells has tumor-suppressive effects,8,9,10, 13, 14 we developed A549 and RERF-LC-KJ cell lines in which maspin was overexpressed. Additionally, to establish stable cell lines, a lentiviral vector incorporating maspin cDNA cloned from MCF10A, in which maspin expression was detected in both cytoplasm and nucleus in previous studies,22 was used. Exogenous maspin expression was detected in both the nucleus and cytoplasm of A549 cells, whereas it was detected only in the cytoplasm of RERF-LC-KJ cells (Fig. 2). These findings suggest that maspin expression in LUAD cells is regulated by both expression levels and subcellular localization, and that the regulatory mechanism of subcellular localization, such as nuclear transport, rather than genetic alterations in maspin gene, is disrupted in RERF-LC-KJ cells. Dzinic SH et al. speculate that the subcellular localization of maspin may be indirectly regulated in a manner dependent on the localization of an unknown protein that is translocated out of the nucleus by the nuclear export protein CRM1, because treatment with leptomycin B, an inhibitor of CRM1, reduced the nuclear accumulation of maspin.26 Furthermore, Reina et al. reported that the transport of maspin from the cytoplasm to the nucleus involves a 28-amino acid sequence within the maspin protein and three-dimensional regulation of protein structure.27 These findings may help explain our observations, which indicated that the mechanism regulating the subcellular localization of maspin is disrupted in RERF-LC-KJ cells. However, since the detailed molecular mechanisms of nuclear translocation of maspin have not yet been elucidated, further investigations are required to clarify this finding.
The antitumor mechanisms of maspin have been actively investigated for a long time. Li et al. reported that the re-expression of maspin in prostate cancer cell lines inhibited histone deacetylase 1 (HDAC1) activity by directly binding to HDAC1, and increased the expression of tumor suppressor genes such as p21WAF1/CIP1, Bax, and CK18, which are downstream of HDAC1.28 Maspin also reduced the release of active urokinase-type plasminogen activator (uPA) and uPA receptor, which promote tumor metastasis by mediating pericellular plasminogen activation in prostate cancer cell lines.29 Additionally, an investigation of the relationship between the nuclear localization of maspin and its tumor-suppressive effect revealed that nuclear localization of maspin is essential for tumor suppression.21 These findings are consistent with our results; that is, A549-maspin cells, which showed nuclear and cytoplasmic localization of maspin, suppressed cell invasion (Fig. 3A), suggesting that the nuclear localization of maspin also suppresses tumor progression in LUAD. On the contrary, the relationship between cytoplasmic maspin and tumor progression remain unclear; however, we have previously reported that cytoplasmic maspin increases the invasiveness of MDA-MB-231 cells by promoting EMT in breast cancer cells.22 In the present study, we found that cytoplasmic maspin promoted the invasiveness of RERF-LC-KJ cells (Fig. 3B). These findings indicate that the role of maspin in LUAD cells is dependent on its subcellular localization and that cytoplasmic maspin promotes invasion in LUAD cells. Our results suggest that the role of maspin in promoting or inhibiting the invasive capability of LUAD cells is regulated by its subcellular localization. These findings support our previous observations, that is, increased cytoplasmic maspin expression correlates with poor prognosis in patients with LUAD.18, 20
EMT, the process by which differentiated epithelial cells lose cell-cell adhesion structures and acquire the characteristics of invasive mesenchymal cells, is strongly associated with cancer invasion and metastasis.30 The expression of epithelial markers, such as E-cadherin, and mesenchymal markers, such as N-cadherin and vimentin, are used as specific markers of the EMT process.31 The downregulation of E-cadherin, also known as the cell adhesion molecule, may enhance the metastatic potential of cancer cells.32 N-cadherin, a single-chain transmembrane glycoprotein, is a calcium-dependent homophilic cell-cell adhesion molecule.33 The aberrant expression of N-cadherin promotes tumorigenesis and invasion in multiple epithelial cancer cell types.34,35,36,37,38 A previous study reported that the upregulation of vimentin, an intermediate filament, also promoted the invasion of prostate cancer and lung cancer cells.39, 40 Based on these findings, evaluating the expression of these EMT markers can help determine the extent of EMT in cancer cells. N-cadherin expression was significantly decreased in A549-maspin cells, which showed pancellular expression, when compared with A549-control cells (Fig. 4B). Previous studies and our findings suggest that the nuclear expression of maspin is important for the tumor-suppressive capability of maspin in LUAD, and that the presence of maspin in the nucleus of A549-maspin cells suppresses the acquisition of mesenchymal phenotypes, such as EMT. By contrast, RERF-LC-KJ-maspin cells, which show cytoplasmic expression, did not show significant differences in the expression of the three markers (Fig. 4). These results suggested that enhancement of the invasive capability of RERF-LC-KJ cells by cytoplasmic maspin involves different molecular mechanisms rather than the promotion of EMT, and prompted us to investigate the detailed molecular mechanisms underlying the cytoplasmic maspin-dependent enhancement of cell invasion in LUAD.
The invasive capability of LUAD cells is regulated by the subcellular localization of maspin. Pancellular maspin, especially nuclear maspin, suppressed the cell invasion in LUAD cell lines, whereas cytoplasmic maspin promoted it. In conclusion, these findings suggest that cytoplasmic maspin and the regulation of its nuclear translocation may be potential therapeutic targets for LUAD.
Acknowledgments
Acknowledgments: This work was supported by JSPS KAKENHI (grant number: JP19K09304). This research was partly performed at the Tottori Bio Frontier, managed by Tottori prefecture.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Sung H,Ferlay J,Siegel RL,Laversanne M,Soerjomataram I,Jemal A,et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Chan BA,Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu W,Du Y,Wen R,Yang M,Xu J. Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Pharmacol Ther. 2020;206:107438. 10.1016/j.pharmthera.2019.107438 [DOI] [PubMed] [Google Scholar]
- 4.Smith SL,Watson SG,Ratschiller D,Gugger M,Betticher DC,Heighway J. Maspin – the most commonly-expressed gene of the 18q21.3 serpin cluster in lung cancer – is strongly expressed in preneoplastic bronchial lesions. Oncogene. 2003;22:8677-87. 10.1038/sj.onc.1207127 [DOI] [PubMed] [Google Scholar]
- 5.Zou Z,Anisowicz A,Hendrix M,Thor A,Neveu M,Sheng S,et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526-9. 10.1126/science.8290962 [DOI] [PubMed] [Google Scholar]
- 6.Sheng S,Pemberton PA,Sager R. Production, purification, and characterization of recombinant maspin proteins. J Biol Chem. 1994;269:30988-93. 10.1016/S0021-9258(18)47379-6 [DOI] [PubMed] [Google Scholar]
- 7.Schneider SS,Schick C,Fish KE,Miller E,Pena JC,Treter SD,et al. A serine proteinase inhibitor locus at 18q21.3 contains a tandem duplication of the human squamous cell carcinoma antigen gene. Proc Natl Acad Sci USA. 1995;92:3147-51. 10.1073/pnas.92.8.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latha K,Zhang W,Cella N,Shi HY,Zhang M. Maspin mediates increased tumor cell apoptosis upon induction of the mitochondrial permeability transition. Mol Cell Biol. 2005;25:1737-48. 10.1128/MCB.25.5.1737-1748.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng S,Carey J,Seftor EA,Dias L,Hendrix MJ,Sager R. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA. 1996;93:11669-74. 10.1073/pnas.93.21.11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi HY,Zhang W,Liang R,Abraham S,Kittrell FS,Medina D,et al. Blocking tumor growth, invasion, and metastasis by maspin in a syngeneic breast cancer model. Cancer Res. 2001;61:6945-51. [PubMed] [Google Scholar]
- 11.Odero-Marah VA,Khalkhali-Ellis Z,Chunthapong J,Amir S,Seftor REB,Seftor EA,et al. Maspin regulates different signaling pathways for motility and adhesion in aggressive breast cancer cells. Cancer Biol Ther. 2003;2:398-403. 10.4161/cbt.2.4.471 [DOI] [PubMed] [Google Scholar]
- 12.Cella N,Contreras A,Latha K,Rosen JM,Zhang M,Cella N,et al. Maspin is physically associated with β1 integrin regulating cell adhesion in mammary epithelial cells. FASEB J. 2006;20:1510-2. 10.1096/fj.05-5500fje [DOI] [PubMed] [Google Scholar]
- 13.Nam E,Park C. Maspin suppresses survival of lung cancer cells through modulation of Akt pathway. Cancer Res Treat. 2010;42:42-7. 10.4143/crt.2010.42.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben Shachar B,Feldstein O,Hacohen D,Ginsberg D. The tumor suppressor maspin mediates E2F1-induced sensitivity of cancer cells to chemotherapy. Mol Cancer Res. 2010;8:363-72. 10.1158/1541-7786.MCR-09-0137 [DOI] [PubMed] [Google Scholar]
- 15.Liu H,Shi J,Anandan V,Wang HL,Diehl D,Blansfield J,et al. Reevaluation and identification of the best immunohistochemical panel (pVHL, Maspin, S100P, IMP-3) for ductal adenocarcinoma of the pancreas. Arch Pathol Lab Med. 2012;136:601-9. 10.5858/arpa.2011-0326-OA [DOI] [PubMed] [Google Scholar]
- 16.Berardi R,Morgese F,Onofri A,Mazzanti P,Pistelli M,Ballatore Z,et al. Role of maspin in cancer. Clin Transl Med. 2013;2:8. 10.1186/2001-1326-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umekita Y,Ohi Y,Sagara Y,Yoshida H. Expression of maspin predicts poor prognosis in breast-cancer patients. Int J Cancer. 2002;100:452-5. 10.1002/ijc.10500 [DOI] [PubMed] [Google Scholar]
- 18.Takagi Y,Matsuoka Y,Shiomi T,Nosaka K,Takeda C,Haruki T,et al. Cytoplasmic maspin expression is a predictor of poor prognosis in patients with lung adenocarcinoma measuring <3 cm. Histopathology. 2015;66:732-9. 10.1111/his.12586 [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka Y,Takagi Y,Nosaka K,Sakabe T,Haruki T,Araki K,et al. Cytoplasmic expression of maspin predicts unfavourable prognosis in patients with squamous cell carcinoma of the lung. Histopathology. 2016;69:114-20. 10.1111/his.12921 [DOI] [PubMed] [Google Scholar]
- 20.Ohno T,Kubouchi Y,Wakahara M,Nosaka K,Sakabe T,Haruki T,et al. Clinical significance of subcellular localization of maspin in patients with pathological stage ia lung adenocarcinoma. Anticancer Res. 2018;38:3001-7. [DOI] [PubMed] [Google Scholar]
- 21.Goulet B,Kennette W,Ablack A,Postenka CO,Hague MN,Mymryk JS,et al. Nuclear localization of maspin is essential for its inhibition of tumor growth and metastasis. Lab Invest. 2011;91:1181-7. 10.1038/labinvest.2011.66 [DOI] [PubMed] [Google Scholar]
- 22.Sakabe T,Wakahara M,Shiota G,Umekita Y. Role of cytoplasmic localization of maspin in promoting cell invasion in breast cancer with aggressive phenotype. Sci Rep. 2021;11:11321. 10.1038/s41598-021-90887-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettstetter M,Woenckhaus M,Wild PJ,Rümmele P,Blaszyk H,Hartmann A,et al. Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J Pathol. 2005;205:606-14. 10.1002/path.1732 [DOI] [PubMed] [Google Scholar]
- 24.Zhu S,Wu H,Wu F,Nie D,Sheng S,Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350-9. 10.1038/cr.2008.24 [DOI] [PubMed] [Google Scholar]
- 25.Villares GJ,Zigler M,Dobroff AS,Wang H,Song R,Melnikova VO,et al. Protease activated receptor-1 inhibits the Maspin tumor-suppressor gene to determine the melanoma metastatic phenotype. Proc Natl Acad Sci USA. 2011;108:626-31. 10.1073/pnas.1006886108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzinic SH,Kaplun A,Li X,Bernardo M,Meng Y,Dean I,et al. Identification of an intrinsic determinant critical for maspin subcellular localization and function. PLoS One. 2013;8:e74502. 10.1371/journal.pone.0074502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reina J,Zhou L,Fontes MRM,Panté N,Cella N. Identification of a putative nuclear localization signal in the tumor suppressor maspin sheds light on its nuclear import regulation. FEBS Open Bio. 2019;9:1174-83. 10.1002/2211-5463.12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X,Yin S,Meng Y,Sakr W,Sheng S. Endogenous inhibition of histone deacetylase 1 by tumor-suppressive maspin. Cancer Res. 2006;66:9323-9. 10.1158/0008-5472.CAN-06-1578 [DOI] [PubMed] [Google Scholar]
- 29.Biliran H Jr,Sheng S. Pleiotrophic inhibition of pericellular urokinase-type plasminogen activator system by endogenous tumor suppressive maspin. Cancer Res. 2001;61:8676-82. [PubMed] [Google Scholar]
- 30.Son HJ,Moon A. Epithelial-mesenchymal transition and cell invasion. Toxicol Res. 2010;26:245-52. 10.5487/TR.2010.26.4.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dongre A,Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69-84. 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- 32.Pećina-Šlaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003;3:17. 10.1186/1475-2867-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vunnam N,Pedigo S. Calcium-induced strain in the monomer promotes dimerization in neural cadherin. Biochemistry. 2011;50:8437-44. 10.1021/bi200902s [DOI] [PubMed] [Google Scholar]
- 34.Nieman MT,Prudoff RS,Johnson KR,Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631-44. 10.1083/jcb.147.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazan RB,Phillips GR,Qiao RF,Norton L,Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol. 2000;148:779-90. 10.1083/jcb.148.4.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G,Satyamoorthy K,Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819-25. [PubMed] [Google Scholar]
- 37.Nakajima S,Doi R,Toyoda E,Tsuji S,Wada M,Koizumi M,et al. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin Cancer Res. 2004;10:4125-33. 10.1158/1078-0432.CCR-0578-03 [DOI] [PubMed] [Google Scholar]
- 38.Da C,Wu K,Yue C,Bai P,Wang R,Wang G,et al. N-cadherin promotes thyroid tumorigenesis through modulating major signaling pathways. Oncotarget. 2017;8:8131-42. 10.18632/oncotarget.14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J,Xu G,Wu M,Zhang Y,Li Q,Liu P,et al. Overexpression of vimentin contributes to prostate cancer invasion and metastasis via src regulation. Anticancer Res. 2008;28:327-34. [PubMed] [Google Scholar]
- 40.Tadokoro A,Kanaji N,Liu D,Yokomise H,Haba R,Ishii T,et al. Vimentin regulates invasiveness and is a poor prognostic marker in non-small cell lung cancer. Anticancer Res. 2016;36:1545-51. [PubMed] [Google Scholar]