Abstract
The clinical, social, and economic impacts of the coronavirus disease 2019 (COVID-19) pandemic, originated by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have motivated a massive search and investment to find treatments for this new disease. Repurposing drugs has been an appealing strategy for the rapid translation of in vitro and ex vivo drug discovery to the clinic. Several repurposed drugs have been assessed clinically, but no effective repurposed antiviral has been identified so far. Of note, no effective treatments for COVID-19 or for any other viral disease have been found by repurposing drugs identified through hypothesis-free screens. Here, I discuss whether drug repurposing is the best strategy for developing effective therapies to eradicate COVID-19 and other viral human infections.
Keywords: Antiviral, Repurposed drugs, Virus therapy, SARS-CoV-2
Introduction
Over the past few decades, screening of libraries of drugs approved for human use has been widespread to uncover new uses and targets for these medications.1 The aim of drug repurposing is to obtain a faster clinical benefit, because using already approved drugs would reduce the time and costs associated with conducting clinical studies. An additional benefit of repurposed drugs is that their safety in humans has already been assessed,2 and the lack of patent protection for many of these approved drugs also increases their appeal. Repurposing has occasionally proved successful, as with aspirin to treat coronary artery disease, erythromycin for impaired gastric motility, sildenafil for erectile dysfunction, thalidomide for multiple myeloma, and imatinib for gastrointestinal stromal tumors (3 and references within); however, these successes have been mostly serendipitous. Drug repurposing arising from such chance discoveries has promoted systematic searches for other drugs to reposition, with most efforts relying on high-throughput screening methods and computational modeling of extensive data.2 Several libraries of approved drugs are available that can be quickly screened for hits against the targeted disease.3, 4, 5 Unfortunately, systematic, hypothesis-free, and large-scale screening of these drug libraries has yet to yield effective treatments for most of the searched targets.1 It has been argued that, for numerous compounds, their pharmacology is probably different for a new indication that includes a different target, tissue, or dosing schedule.1 Cellular assays used for repurposing, including phenotypic and throughput screening assays, frequently are subject to assay interference or undesirable mechanisms of bioactivity.6
Nevertheless, drug repurposing has become an appealing approach as a fast way to recognize drugs against the spread of new infectious diseases, including those caused by viruses, such as Zika virus (ZIKV)7 and Ebola virus (EBOV).8 Indeed, repurposed drugs active against these two viruses have been identified in cellular assays or animal models, but nothing has yet reached the clinic. The emergence of the COVID-19 pandemic, caused by SARS-CoV-2, has likewise driven the search for numerous compounds that could be repurposed to quickly prevent the mortality, morbidity, and spread of this new virus disease.9 This search has been entirely justified because, despite the proven efficacy of current SARS-CoV-2 vaccines, it is crucial to identify different therapeutics, including effective antivirals. Vaccines are preventing disease but are not generating sterilizing immunity in many individuals, such that viral transmission is still possible even after vaccination.10, 11 Current vaccines induce mostly immunoglobulin (Ig) G production and not secretory IgA production.10 In addition, we cannot ignore problems associated with vaccine delivery delays, unsuccessful immunity, durability, vaccine denial, and the possible emergence of vaccine-resistant variants. Thus, under this scenario, it is not difficult to envisage the need for an effective antiviral against SARS-CoV-2 to reduce related morbidity and mortality and to control the spread of this virus.
In this review, I provide an overview of drugs that have been repurposed to treat SARS-CoV-2 infection and have reached the clinic, allowing for estimation of their overall efficacy (Table 1 ). I also discuss what past virus epidemics and pandemics have taught us about developing new effective antivirals against current and upcoming virus threats. The lessons learned with COVID-19 should offer a roadmap for approaching drug discovery against other current viral threats, such as influenza virus, coronaviruses, alphaviruses (e.g., Chikungunya), flaviviruses (e.g., dengue and ZIKV) and filoviruses (e.g., EBOV). The development of more systematic approaches to identifying repurposable drugs has resulted in the identification of several candidates to test against COVID-19. These methodologies have included hypothesis-free and mechanistic hypothesis approaches; however, when the drug candidates were tested in clinical studies, none were effective against SARS-CoV-2. In addition, some repurposed drugs that lack antiviral activity (e.g., corticosteroids) do show some effect in ameliorating COVID-19 symptoms and the mortality and morbidity of associated with severe COVID-19.9 These findings could offer clues regarding what should be searched for in terms of repurposed compounds.
Table 1.
Examples of antiviral drugs repurposed for COVID-19 that failed in the clinic.
| Repurposed drug | Original indication | Virus target | Refs |
|---|---|---|---|
| Favipiravir | Influenza virus | RNA polymerase | 18 |
| Remdesivir | HCV, Ebola, MERS-CoV | RNA polymerase | 23, 24, 25 |
| Lopinavir-ritonavir | HIV-1 | Protease | 38 |
| Darunavir/cobicistat | HIV-1 | Protease | 40 |
| Hydroxychloroquine | Malaria | Cell entry | 3, 25 |
| Azithromycin | Antibiotic | Not defined | 44 |
| Ivermectin | Intestinal strongyloidiasis and onchocerciasis | Not defined | 50 |
Failure of drugs repurposed against SARS-CoV-2
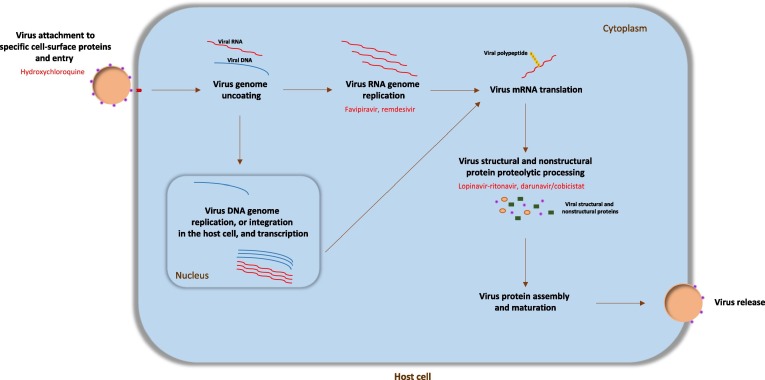
At the beginning of 2022, more than 280 million SARS-CoV-2 infections had been recorded and the death toll from the pandemic had reached 5.5 million deaths. The worldwide impact of the pandemic has encouraged the repurposing of several drugs. The repurposed drugs tested so far have been selected mostly because of their promising in vitro or ex vivo efficacy against SARS-CoV-2 or other human coronavirus diseases, such as SARS-CoV and Middle East respiratory syndrome (MERS-CoV).9 Protein structural analyses suggested that key drug-binding pockets in viral enzymes are conserved among different human coronaviruses.12 SARS-CoV-2, as described for many other viruses,13 can be targeted in different stages of its life cycle. Examples are the endocytic virus entry into host cells; RNA replication and transcription; translation and proteolytic processing of viral proteins; virion assembly; and release of new virus particles across exocytic mechanisms14 (Fig. 1 ). Different host cell factors, indispensable for viral replication, could also be targeted to inhibit SARS-CoV-2 replication. Nevertheless, it is difficult to find successful marketed antiviral compounds targeting host cell–virus cofactors.13, 15
Figure 1.
The life cycle of viruses and antiviral drug targets. The virus initially binds to the host cell-specific receptors through its surface proteins, to allow internalization of the virus particle. The internalized virus releases its genome into the cell cytoplasm to be replicated (RNA in cytosol and DNA in nucleus), transcribed, and translated to produce viral proteins. Viral components are assembled to produce progeny virions, which are released out of the cell by budding or lysis of the host cell. Targetable stages of the virus life cycle are the endocytic entry of the virus into host cells; RNA/DNA replication and transcription; translation and proteolytic processing of viral proteins; virion assembly; and release of new virus particles through the exocytic systems. In red are shown the possible viral targets of some repurposed coronavirus 2019 (COVID-19) drugs.
Viral enzymes are the main targets of antiviral agents,13, 15 and inhibitors of these enzymes represent more than two-thirds of all antivirals approved for human use. Of these antivirals, polymerase inhibitors are the largest class of agent and represent two groups: nucleoside/nucleotide analogs and non-nucleoside/nucleotide inhibitors. Nucleoside/nucleotide analogs are chain terminators that, after being incorporated into RNA/DNA strands, stop viral polymerases. Non-nucleoside/nucleotide inhibitors serve as noncompetitive inhibitors by binding viral polymerases and impairing their catalytic activity. Several nucleoside/nucleotide analog compounds have been marketed to target diverse viruses, such as HIV, hepatitis B virus (HBV), hepatitis C virus (HCV), influenza virus, human cytomegalovirus, herpes simplex virus, and varicella zoster virus.13
Favipiravir is a purine nucleoside analog broad-spectrum inhibitor of viral RNA-dependent RNA polymerase (RdRp) that has been approved for treatment of influenza virus infection in Japan.16 This compound was chosen to treat COVID-19 because it displayed ex vivo activity against SARS-CoV-2. Although its inhibitory capacity is poor, within the high micromolar range,17 favipiravir was chosen as an experimental drug to treat COVID-19 in humans. An open-label nonrandomized trial of 80 patients with COVID-19 showed a faster SARS-CoV-2 viral clearance in patients treated with favipiravir compared with controls treated with lopinavir-ritonavir, along with a significantly lower rate of adverse events (11.4% versus 55.6%; P < 0.01).18 However, all study patients were co-treated with interferon-α1b until viral clearance and, therefore, it is difficult to estimate the true clinical efficacy of favipiravir. A recent prospective, randomized, open-label trial showed no improvement in viral clearance with favipiravir.19
Remdesivir (GS-5734) is an adenine nucleoside analog monophosphoramidate prodrug approved by the US Food and Drug Administration (FDA) for COVID-19. This compound, originally designed to treat HCV, is converted in the cell to its active nucleoside triphosphate (GS-443902).20, 21 Remdesivir and its parent molecules have shown potential for targeting emerging viruses, such as SARS-CoV, MERS-CoV, ZIKV, dengue virus, and EBOV.22 However, despite impressive preclinical and animal model data obtained with use of remdesivir against EBOV,26 similar results have not been seen in clinical studies.27
During the early stages of the COVID-19 pandemic, remdesivir was considered the most promising antiviral compound against SARS-CoV-2. Initial interest in the drug was based on its potency in cell culture models of SARS-CoV-2, in which low micromolar potency was observed in several cell lines, including primary human airway epithelial cells.28 Another aspect that promoted excitement about remdesivir is its specificity in targeting coronavirus RdRp. After 23 cell culture virus passages with the drug, two mutations (F276L and V553L) were found in the SARS-CoV viral RdRp gene, which conferred viral resistance to remdesivir.29 These cell culture data also were supported by experiments in mouse models of SARS-CoV30 and MERS-CoV,31 and in a rhesus macaque model of MERS-CoV infection.32 Overall, these results highlighted remdesivir as a promising candidate for repurposing against COVID-19. Although preclinical and animal model data were encouraging, the clinical efficacy of remdesivir for COVID-19 remains unclear. Several clinical trials have found no significant differences in time to clinical recovery or mortality between remdesivir-treated and control groups.23, 24, 25
Pioneer clinical studies exploring the efficacy of remdesivir against COVID-19 involved compassionate use of this compound in 61 patients with severe disease. In 36 of 53 (68%) patients assessed, clinical improvement in oxygen support status was observed, and overall mortality was 13% during a median follow-up of 18 days. Given that no control group was included in this study, it is challenging to estimate the efficacy of remdesivir.33 Double-blind randomized controlled trials have shown no statistical difference in clinical improvement between remdesivir- and placebo-treated patients.34 Moreover, a reduction in mortality has been not reported.35 The observed differences between the promising remdesivir preclinical data and its questionable clinical activity suggest that some assumptions made at the preclinical level do not reflect real-world clinics.24 In addition, to search for low effective concentration (EC)50 or inhibitory concentration (IC)50 values, more attention should be paid to other aspects, such as drug exposure, drug target specificity, tissue-specific localization, and cellular toxicity [i.e., cytotoxicity concentration (CC)50 values]. The use of new or different animal models also might be considered.
Viral polyprotein processing is an essential step in most virus life cycles. Protease inhibitors have been highly successful as therapies for HIV-1 and HCV. The HIV-1 protease inhibitor lopinavir-ritonavir showed cell culture inhibitory activity against the main protease (3CLpro or Mpro) of SARS-CoV.36 Lopinavir-ritonavir also improved clinical parameters, disease progression, and virus titers in MERS-CoV-infected marmosets.37 These results encouraged the repurposing of lopinavir-ritonavir as a possible antiviral against COVID-19. Regrettably, a randomized, controlled, open-label clinical trial in patients hospitalized with COVID-19 showed no differences between lopinavir-ritonavir treatment and standard-of-care.38 Mortality at 28 days and detectable viral RNA at different time points were similar between treated and nontreated patients. These results were confirmed in studies showing that lopinavir-ritonavir has little or no effect on patients hospitalized with COVID-19.39 Darunavir/cobicistat, another HIV-1 protease inhibitor, also has been tested against COVID-19, with no sign of efficacy in hospitalized patients, thus not supporting its use in COVID-19.40
The aminoquinoline drug chloroquine and its hydroxyl analog hydroxychloroquine are antimalarial generic drugs that are also used to treat amoebic liver abscess and rheumatic disease. Again, initial reported results with chloroquine and hydroxychloroquine described ex vivo micromolar antiviral activity against SARS-CoV-2. It was suggested that chloroquine and hydroxychloroquine prevent virus entry into the cell through the reduction of autophagosome–lysosome fusion and the inhibition of autophagic flux.41 The therapeutic usage of chloroquine and hydroxychloroquine to treat COVID-19 has become an archetypal example of how preliminary and wrongly analyzed results from small clinical observational studies led to misguided repurposing of several drugs at the beginning of the pandemic.23, 42 These misinterpreted studies fostered public interest and widespread off-label use of hydroxychloroquine as an intended treatment of, and prophylactic against, COVID-19. The FDA even issued an emergency use authorization for off-label use of chloroquine phosphate and hydroxychloroquine sulfate for COVID-19, which was later revoked.43 Significantly, randomized and observational clinical trials that included a larger number of participants have shown no effect of these aminoquinolines in reducing COVID-19 mortality and/or morbidity.25, 35, 44 Additionally, treatment with chloroquine or hydroxychloroquine displayed in some hospitalized patients a more severe infection course45 and an increased overall mortality.46 Cell culture experiments have demonstrated that, in human lung cells, chloroquine does not block SARS-CoV-2 infection.47 The anti-SARS-CoV-2 efficacy of chloroquine and hydroxychloroquine in tissue culture, mentioned above, was determined using African green monkey kidney-derived Vero cells. Remarkably, the inhibitory capacity of chloroquine was prevented when Vero cells were transfected with a cellular protease, TMPRSS2, which promotes SARS-CoV-2 entry into susceptible cells.47 Moreover, chloroquine does not inhibit SARS-CoV-2 infection in human lung Calu-3 cells that express TMPRSS2, suggesting that chloroquine targets a viral activation pathway that is not present in lung cells, and, consequently, it is improbable that this drug will stop the spread of SARS-CoV-2. These results highlight once again why early preclinical data should be viewed with caution.
Ivermectin has been also prescribed to treat and prevent COVID-19. Similar to chloroquine or hydroxychloroquine, ivermectin is another cheap antiparasitic drug that also displayed anti-SARS-CoV-2 activity in Vero cells.48 At specific doses, ivermectin tablets are approved as a treatment for some parasitic worms, and there are topical formulations for head lice and skin conditions, such as rosacea.49 However, currently available data show no effectiveness of ivermectin against COVID-19.50, 51 The results of the several clinical trials that have been performed are inconsistent. Different doses and dosing schedules among the studies have also confounded findings and conclusions, which are further complicated by several retractions.51 Intriguingly, many countries have recommended the use of ivermectin, despite advice to the contrary from the WHO. The FDA has not authorized ivermectin for use in preventing or treating COVID-19 in humans or animals.
In addition to antiviral drugs targeting SARS-CoV-2 replication, compounds that modulate COVID-19-related inflammation and the cytokine storm also have been explored. In contrast to repurposed direct antivirals, some repurposed anti-inflammatory drugs have proved more effective against COVID-19 morbidity and mortality. Examples are monoclonal antibodies, such as tocilizumab and sarilumab, which target interleukin (IL)-6 receptors. The accentuated inflammatory response of COVID-19-associated acute hypoxemic respiratory failure is linked to elevated IL-6 concentrations. Patients with COVID-19 who developed severe forms of COVID-19 respiratory failure had high levels of IL-6. Thus, it was hypothesized that blocking IL-6 might be beneficial for severe infection. Randomized controlled trials with tocilizumab and sarilumab yielded conflicting results. Patients needing high-flow nasal oxygen, non-invasive ventilation, or invasive mechanical ventilation appear to benefit from treatment with tocilizumab.52 By contrast, these compounds could harm patients with mild COVID-19. Corticosteroids also appear to work against COVID-19 inflammation with appropriate timing of administration. Randomized clinical trials of dexamethasone have shown an association of this steroid with significantly increased COVID-19 survival and reduced morbidity.53, 54 Compounds that have been successfully used in the clinical treatment of several rheumatologic and inflammatory diseases, such as Janus kinase inhibitors, also have been repurposed as a COVID-19 therapeutic alternative. A Phase III randomized placebo-controlled trial with one such inhibitor, baricitinib, has shown promising results in patients hospitalized with COVID-19.55
A confounding example in the search for repurposed drugs against SARS-CoV-2
A recent study has raised important concerns about many repurposed drugs for SARS-CoV-2 that displayed antiviral activity in hypothesis-free cellular screens.56 That study demonstrated that phospholipidosis is a shared mechanism underlying the anti-SARS-CoV-2 activity of many tested compounds. Phospholipidosis interferes with lysosomal lipid catabolism and trafficking and, in doing so, disrupts the virus-generated double-membrane vesicles that are essential for virus propagation. Cationic amphiphilic drugs can provoke cell phospholipidosis.57 Tummino and colleagues showed that, with the cationic amphiphilic drugs they tested (e.g., hydroxychloroquine and azithromycin), phospholipidosis was repeatedly correlated with their antiviral efficacy. By contrast, these authors demonstrated that drugs that did not induce phospholipidosis did not display antiviral activity. Their work confirms that ex vivo cationic amphiphilic drug phospholipidosis correlates with the absence of success in vivo, suggesting that this apparently general mechanism explains the lack of clinical efficacy of most drugs repurposed to date for SARS-CoV-2. Eliminating this confounding effect could accelerate the identification of genuinely potent antivirals against SARS-CoV-2 and other viruses. Of note, phospholipidosis is a confounder that only affects repurposed antiviral drugs and is irrelevant for drugs, such as dexamethasone, that target COVID-19 immunomodulation. As has been suggested,58 the main lesson from this study is not the process of phospholipidosis itself but the potential for many such confounders in drug screens. Antiviral screening hits derived from biochemical or cell culture assays must be treated with skepticism, whether they come from newly synthesized drug libraries or from approved repurposed drugs.
Examples from other human virus pandemics
The previous achievements of antiviral therapies developed against new pandemic viruses, such as HIV-1 and HCV, should guide us in the development of effective SARS-CoV-2 antivirals. HIV-1 was identified in 1983, but effective antiretroviral therapies were not available until 1996. Between 1983 and 1996, it was only possible to use prophylactic therapies against common opportunistic viruses that helped to manage AIDS-related illnesses. During the mid-1990 s, HIV-1 reverse transcriptase and protease inhibitors reached the clinic, and the combination regimens of these drugs transformed the clinical approach to HIV-1 and changed HIV-1 infection from a life-threatening disease to a controllable chronic condition. HIV-1 therapy is probably one of the most successful treatments of any human disease, and its success has been founded on the development of different antiretroviral agents that target diverse and specific steps of the HIV-1 life cycle.59 The FDA has approved 23 compounds for the treatment of HIV-1 infection, all directed against most of the targetable stages of HIV-1 life cycle. Considering the molecular mechanism and virus targets, antiretrovirals are classified in eight different groups: nonnucleoside reverse transcriptase inhibitors; nonnucleoside reverse transcriptase inhibitors; protease inhibitors; integrase inhibitors; CCR5 antagonists that block the CCR5 co-receptor, which is needed for HIV-1 to enter susceptible cells; fusion inhibitors, which prevent HIV-1 entering the cell; attachment inhibitors that bind HIV-1 glycoprotein 120; and post-attachment inhibitors that block the cellular CD4 receptor. A crucial difference between antiretrovirals and antiviral repurposed drugs against SARS-CoV-2 is the high specificity of approved antiretrovirals. For instance, in addition to other considerations (e.g., structure–activity relationships), the in vitro or in vivo selection of HIV-1-resistant variants has been considered essential to confirm the specificity of an antiretroviral. Indeed, every approved antiretroviral drug has demonstrated its target specificity.
Recent development of effective antivirals against other virus infections has been based on antiretroviral success. One of the best examples is another human, life-threatening virus, HCV, which was first characterized in 1989.60 Following its identification that year, the identification of effective HCV therapies was delayed by the impossibility of propagating this virus in tissue culture. In 1997, interferon alfacon-1 became the first drug approved by the FDA against HCV infections. The arsenal for combatting HCV grew to include ribavirin (a guanosine nucleoside analog) in 1998. Although interferon and ribavirin showed low clinical efficacy, they were the standard-of-care therapies until 2011. Interferon-based therapies involved long treatment duration (48 weeks) and caused serious adverse effects. Again, neither interferon nor ribavirin target any specific viral protein, and they can be considered repurposed drugs that previously showed some efficacy against other viruses. The development in 1999 of an ex vivo HCV replicon system was crucial to elucidate the HCV life cycle and for the unbiased identification of new classes of HCV direct-acting antiviral (DAA) targeting different HCV enzymes.61 Approved HCV DDAs are NS3/4A protease inhibitors, NS5A phosphoprotein inhibitors, NS5B RdRp nonnucleoside polymerase inhibitors, and nucleoside and nucleotide NS5B RdRp inhibitors, which block viral RNA synthesis.60 Use of HCV DAAs allows a shorter treatment duration of 6–8 weeks and yields an increased cure rate of almost 100%. Current HCV therapy is an outstanding example of success in the antiviral field. DAAs have completely changed the management of HCV infection and are offering the possibility of HCV eradication.
Compared with SARS-CoV-2, HIV-1 and HCV involve an extended course of infection that is presently measured in years. It can be argued that, during the asymptomatic or mildly symptomatic phase of HIV-1 and HCV infection, there are many opportunities to administrate an antiviral drug. Given that HIV-1 or HCV infection generate a persistent, prolonged, and steady viremia, an infected individual can serve as their own control for gauging drug efficacy. A different scenario is displayed during SARS-CoV-2 infection, in which a rapid progressive disease is observed. In these patients, obtaining a detectable benefit from antivirals might be more difficult because, in to make the antiviral treatment effective, the disease has to be diagnosed at the beginning of the infection. Some have noted that the philosophy of ‘one drug, one bug’ that has served the industry in developing new drugs against HIV-1 or HCV might be inefficient in terms of rapidly addressing epidemics or pandemics.62 Also debated is whether it is preferable to target generic cell pathways used by a broad array of different viruses. As mentioned above, targeting host cell–virus cofactors13 and disrupting host cell targets might be more toxic than targeting virus-specific processes.9, 25 We must not forget that the examples of HIV-1 and HCV can give us a point of reference for further development of SARS-CoV-2 antivirals and for more distantly related pathogens that pose a pandemic risk.
Concluding remarks
The early rapid spread of SARS-CoV-2 and lack of specific therapies against COVID-19 encouraged off-label testing of repurposed approved drugs. Despite the interest that this approach has generated and the considerable resources invested in it, it has had limited success in stopping COVID-19.1, 25 These failures emphasized the need for new and more specific drugs. Over the past few decades, successful approved antivirals have saved millions of lives, and the challenge is to obtain effective and well-tolerated compounds for treating COVID-19 and stopping SARS-CoV-2 propagation.
SARS-CoV-2, similar to other respiratory virus infections (e.g., influenza virus), generates an acute disease that is best combatted during the first few days after the onset of symptoms63 and when the virus is actively replicating. This treatment window is probably limited in most patients to the first week of symptoms.64 Thus, antiviral drugs that require a healthcare facility for administration should be avoided. Orally administered antivirals are preferable for widespread use in patients with mild-to-moderate disease.
Early administration of influenza virus neuraminidase inhibitors is recommended for preventing the development of severe disease. By contrast, they might not produce a detectable effect in patients hospitalized with severe influenza. A goal with antiviral therapy might be to avoid severe disease and to prevent hospitalization rather than to cure severe disease. This early treatment strategy could also prevent person-to-person transmission. Thus, SARS-CoV-2 should preferably be stopped in the first days of infection. In contrast to the lifelong administration of HIV-1 antiretrovirals, SARS-CoV-2 antivirals would have to be administered for a narrow period of time and, consequently, the associated drug toxicity of SARS-CoV-2 therapies might be considered less important and higher drug doses could be used.
An important lesson from the experience with repurposing several drugs as antivirals for SARS-CoV-2 infection is to exclude nonspecific mechanisms during drug development.65 Viruses are obligatory parasites and need the cellular machinery to carry out their replication; it is not unusual that a screened drug affects a cell pathway and leads to false-positive hits. Target specificity should be examined before treatments are initiated in affected patients. It has been previously suggested that the study of structure–activity relationships is an interesting approach to explore drug specificity.58 Nevertheless, the efficient correlation of structural changes of the study drugs with their activity against the target, in vitro and ex vivo, is not immediate and might require time and the synthesis of multiple compounds. It also has been suggested that virus target specificity can be explored and determined by the isolation of drug-resistant viruses, as previously demonstrated with other virus infection models.23 Counter-assays should be used to rule out nonspecific mechanisms. The emergence of SARS-CoV-2 in 2019 is not a unique or improbable event, given that other zoonotic coronaviruses, SARS-CoV and MERS-CoV, emerged in 2002 and 2012, respectively.66 Consequently, the development of pancoronavirus compounds directed against highly conserved features within the coronavirus family would appear an attractive strategy.67 As discussed in the previous section on compounds targeting virus–host cofactors, a broad spectrum antiviral remains to be discovered and marketed.
Drug repurposing is perhaps an effective strategy for identifying antivirals, but there are no shortcuts for drug development.1 Development of drugs, whether approved or not, requires time and funding to know and understand the drug target and toxicity and how to use it appropriately. The rational design and development of new DAAs against SARS-CoV-2 appears an informed approach to reach an effective COVID-19 therapy. The strategy could take more time but could be useful not only to stop SARS-CoV-2, but also to develop drugs for newly emerging viruses and future pandemics. New antiviral drugs targeting different SARS-CoV-2 enzymes and proteins are approaching the clinic (e.g., molnupiravir and nirmatrelvir)68, 69 and we soon will see the impact of these newly developed drugs on COVID-19. Experience with previous virus pandemics has shown us that developing an ample repertoire of antiviral drugs with different targets will be essential to improve antiviral therapy.
Acknowledgment
This work was supported by the Spanish Ministry of Science and Innovation (PID2019-103955RB-100).
References
- 1.Edwards A. What are the odds of finding a COVID-19 drug from a lab repurposing screen? J Chem Inf Model. 2020;60:5727–5729. doi: 10.1021/acs.jcim.0c00861. [DOI] [PubMed] [Google Scholar]
- 2.Huang R, ZHu H, Shinn P, Ngan D, Ye L, Thakur A, et al. The NCATS Pharmaceutical Collection: a 10-year update. Drug Discov Today 2019; 24: 2341–2349. [DOI] [PubMed]
- 3.Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med 2017 234 2017; 23: 405–408. [DOI] [PMC free article] [PubMed]
- 4.Jan J.-T., Cheng T.-J.-R., Juang Y.-P., Ma H.-H., Wu Y.-T., Yang W.-B., et al. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2021579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang W.D., Jeon S., Kim S., Lee S.Y. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2024302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlin J.L., Auld D.S., Rothenaigner I., Haney S., Sexton J.Z., Nissink J.W.M., et al. Nuisance compounds in cellular assays. Cell Chem Biol. 2021;28:356–370. doi: 10.1016/j.chembiol.2021.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saiz J.C., Martín-Acebes M.A. The race to find antivirals for zika virus. Antimicrob Agents Chemother. 2017;61:e00411–e417. doi: 10.1128/AAC.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bixler SL, Duplantier AJ, Bavari S. Discovering drugs for the treatment of Ebola virus. Curr Treat Options Infect Dis 2017 93 2017; 9: 299–317. [DOI] [PMC free article] [PubMed]
- 9.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob Agents Chemother. 2020;64:e00399–e420. doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 11.Cromer D., Juno J.A., Khoury D., Reynaldi A., Wheatley A.K., Kent S.J., et al. Prospects for durable immune control of SARS-CoV-2 and prevention of reinfection. Nat Rev Immunol. 2021;21:395–404. doi: 10.1038/s41577-021-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 2021 193 2020; 19: 149–150. [DOI] [PubMed]
- 15.Tompa D.R., Immanuel A., Srikanth S., Kadhirvel S. Trends and strategies to combat viral infections: a review on FDA approved antiviral drugs. Int J Biol Macromol. 2021;172:524–541. doi: 10.1016/j.ijbiomac.2021.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020 303 2020; 30: 269–271. [DOI] [PMC free article] [PubMed]
- 18.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi Y., Hibino M., Hase R., Yamamoto M., Kasamatsu Y., Hirose M., et al. A prospective, randomized, open-label trial of early versus late favipiravir therapy in hospitalized patients with COVID-19. Antimicrob Agents Chemother. 2020;64:e01897-20. doi: 10.1128/AAC.01897-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho A., Zhang L., Xu J., Lee R., Butler T., Metobo S., et al. Discovery of the first C-nucleoside HCV polymerase inhibitor (GS-6620) with demonstrated antiviral response in HCV infected patients. J Med Chem. 2014;57:1812–1825. doi: 10.1021/jm400201a. [DOI] [PubMed] [Google Scholar]
- 21.Malin J.J., Suárez I., Priesner V., Fätkenheuer G., Rybniker J. Remdesivir against COVID-19 and other viral diseases. Clin Microbiol Rev. 2020;34:1–21. doi: 10.1128/CMR.00162-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel D, Hui HC, Doeffler E, CLarke MO, Chun K, Zhang L, et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo2,1-ftriazin-4-amino adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem 2017; 60: 1648–1661. [DOI] [PubMed]
- 23.Martinez M.A. Lack of Effectiveness of repurposed drugs for COVID-19 treatment. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.635371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan V.C., Muller F.L. Why remdesivir failed: preclinical assumptions overestimate the clinical efficacy of remdesivir for COVID-19 and Ebola. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.01117-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez M.A. Clinical trials of repurposed antivirals for SARS-CoV-2. Antimicrob Agents Chemother. 2020;64:e01101–e1120. doi: 10.1128/AAC.01101-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulangu S., Dodd L.E., Davey R.T., Tshiani Mbaya O., Proschan M., Mukadi D., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9:e00221–e318. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinksi L.E., Case J.B., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wit W, Feldmann F, Cronin J, Jordan R, Okumura A, homas T, et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A 2020; 117: 6771–6776. [DOI] [PMC free article] [PubMed]
- 33.Grein J, Ohomagari N, Shin D, Diaz G, Asperges E, A Castagna A, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020; 382: 2327–2336. [DOI] [PMC free article] [PubMed]
- 34.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., et al. Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci U S A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan J.-F.-W., Yao Y., Yeung M.-L., Deng W., Bao L., Jia L., et al. Treatment With lopinavir/ritonavir or Interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A Trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO Solidarity Trial Consortium Repurposed antiviral drugs for Covid-19 — Interim WHO Solidarity Trial Results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Xia L., Liu L., Xu Q., Ling Y., Huang D., et al. Antiviral activity and safety of darunavir/cobicistat for the treatment of COVID-19. Open Forum. Infect Dis. 2020;7:ofaa241. doi: 10.1093/ofid/ofaa241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.-J., et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.FDA. EUA Hydroxychloroquine sulfate Health Care Provider Fact Sheet, version date 4/27/2020. Bethesda; FDA: 2020.
- 44.Butler C.C., Dorward J., Yu L.-M., Gbinigie O., Hayward G., Saville B.R., et al. Azithromycin for community treatment of suspected COVID-19 in people at increased risk of an adverse clinical course in the UK (PRINCIPLE): a randomised, controlled, open-label, adaptive platform trial. Lancet. 2021;397:1063–1074. doi: 10.1016/S0140-6736(21)00461-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., et al. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Medicine. 2020;1:114–127. doi: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., et al. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Medicine. 2020;1:114–127. doi: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., et al. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 48.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.FDA. www.fda.gov/consumers/consumer-updates/why-you-should-not-use-ivermectin-treat-or-prevent-covid-19. [Accessed February 15, 2022].
- 50.Popp M., Stegemann M., Metzendorf M.I., Gould S., Kranke P., Meybohm P., et al. Ivermectin for preventing and treating COVID-19. Cochrane Database Syst Rev. 2021;7:CD015017. doi: 10.1002/14651858.CD015017.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker N., Coronapod R.S. Ivermectin, what the science says. Nature. Published online August 6. 2021 doi: 10.1038/d41586-021-02178-2. [DOI] [PubMed] [Google Scholar]
- 52.Angriman F., Ferreyro B.L., Burry L., Fan E., Ferguson N.D., Husain S., et al. Interleukin-6 receptor blockade in patients with COVID-19: placing clinical trials into context. Lancet Respir Med. 2021;9:655–664. doi: 10.1016/S2213-2600(21)00139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021; 384: 693–704. [DOI] [PMC free article] [PubMed]
- 54.BM omazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. JAMA 2020; 324: 1307–1316. [DOI] [PMC free article] [PubMed]
- 55.Florescu D.F., Kalil A.C. Janus Kinase inhibitors for the treatment of hospitalized patients with COVID-19. Curr Opin Crit Care. 2021;27:493–496. doi: 10.1097/MCC.0000000000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O’Meara M.J., Monel B., et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breiden B., Sandhoff K. Emerging mechanisms of drug-induced phospholipidosis. Biol Chem. 2019;401:31–46. doi: 10.1515/hsz-2019-0270. [DOI] [PubMed] [Google Scholar]
- 58.Edwards A., Hartung I.V. No shortcuts to SARS-CoV-2 antivirals. Science. 2021;373:488–489. doi: 10.1126/science.abj9488. [DOI] [PubMed] [Google Scholar]
- 59.Arts E.J., Hazuda D.J. HIV-1 Antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez M.A., Franco S. Discovery and development of antiviral therapies for chronic hepatitis C virus infection. Adv Exp Med Biol. 2021;1322:139–157. doi: 10.1007/978-981-16-0267-2_6. [DOI] [PubMed] [Google Scholar]
- 61.Lohmann V., Körner F., Koch J., Herian U., Theilmamm L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 62.Dolgin E. The race for antiviral drugs to beat COVID - and the next pandemic. Nature. 2021;592:340–343. doi: 10.1038/d41586-021-00958-4. [DOI] [PubMed] [Google Scholar]
- 63.Kumar A. Early versus late oseltamivir treatment in severely ill patients with 2009 pandemic influenza A (H1N1): speed is life. J Antimicrob Chemother. 2011;66:959–963. doi: 10.1093/jac/dkr090. [DOI] [PubMed] [Google Scholar]
- 64.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 65.Quancard J., Cox B., Finsinger D., Guéret S.M., Hartung I.V., Koolman H.F., et al. The European Federation for Medicinal Chemistry (EFMC) Best Practice Initiative: validating chemical probes. ChemMedChem. 2020;15:2388–2390. doi: 10.1002/cmdc.202000597. [DOI] [PubMed] [Google Scholar]
- 66.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vries M., Mohamed A.S., Prescott R.A., Valero-Jimenez A.M., Desvignes L., O’Connor R., et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CL pro inhibitor PF-00835231 as a potential new treatment for COVID-19. J Virol. 2021;95:e01819–e1820. doi: 10.1128/JVI.01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clinical Trials Register. Clinicaltrials.gov [Accessed February 15, 2022].
- 69.Fischer W, Eron JJ, Holman W, Cohen MS, Fang L, Szewczyk LJ, et al. Molnupiravir, an oral antiviral treatment for COVID-19. MedRxiv. 2021: 2021.06.17.21258639.