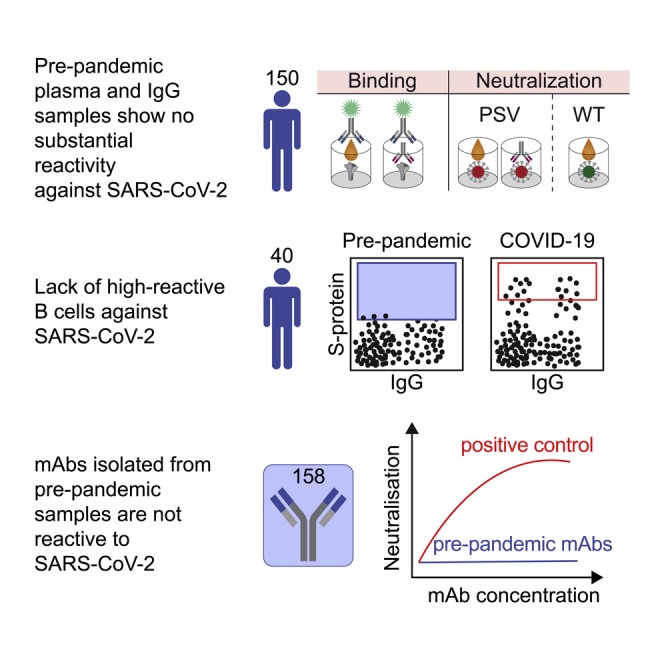
Summary
Preexisting immunity against SARS-CoV-2 may have critical implications for our understanding of COVID-19 susceptibility and severity. The presence and clinical relevance of a preexisting B cell immunity remain to be fully elucidated. Here, we provide a detailed analysis of the B cell immunity to SARS-CoV-2 in unexposed individuals. To this end, we extensively investigated SARS-CoV-2 humoral immunity in 150 adults sampled pre-pandemically. Comprehensive screening of donor plasma and purified IgG samples for binding and neutralization in various functional assays revealed no substantial activity against SARS-CoV-2 but broad reactivity to endemic betacoronaviruses. Moreover, we analyzed antibody sequences of 8,174 putatively SARS-CoV-2-reactive B cells at a single cell level and generated and tested 158 monoclonal antibodies. None of these antibodies displayed relevant binding or neutralizing activity against SARS-CoV-2. Taken together, our results show no evidence of competent preexisting antibody and B cell immunity against SARS-CoV-2 in unexposed adults.
Subject areas: Immunity, Immune response, Virology
Graphical abstract
Highlights
-
•
Comprehensive analysis of the B cell response to SARS-CoV-2 in pre-pandemic samples
-
•
No substantial plasma and IgG reactivity against SARS-CoV-2
-
•
MAbs isolated from pre-pandemic samples showed no SARS-CoV-2 neutralizing activity
-
•
No indication of competent preexisting B cell immunity against SARS-CoV-2
Immunity; Immune response; Virology ;
Introduction
The current pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represents a global health emergency that challenges health care systems throughout the world. SARS-CoV-2 infections appear with a broad spectrum of clinical manifestations ranging from asymptomatic infections to life-threatening acute respiratory distress syndrome (ARDS), multi organ failure, septic shock, and death. Although influenced by multiple contributing factors, disease severity is substantially shaped by innate and adaptive immune responses. Preexisting immunity against SARS-CoV-2 could represent a crucial determinant of disease severity and clinical outcome. For example, recognition of SARS-CoV-2 by a preexisting background immunity could limit disease severity by rapidly mounting specific immune responses. However, preexisting immunity may also be detrimental for the clinical course by mechanisms of antibody-dependent enhancement (ADE) (Khurana et al., 2013; de Alwis et al., 2014; Katzelnick et al., 2017; Arvin et al., 2020) or original antigenic sin (OAS) (Vatti et al., 2017), which have been previously described for other viral pathogens, such as dengue (de Alwis et al., 2014; Katzelnick et al., 2017; Mongkolsapaya et al., 2003; Midgley et al., 2011; Rothman, 2011) and influenza viruses (Linderman and Hensley, 2016; Zhang et al., 2019; Arevalo et al., 2020).
Preexisting T cell immune responses against SARS-CoV-2 have been observed in unexposed individuals (Mateus et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Braun et al., 2020; Bacher et al., 2020; Weiskopf et al., 2020; Echeverría et al., 2021). In these studies, T cell reactivity against the spike (S) and nucleocapsid (N) proteins as well the nonstructural proteins NSP7 and NSP13 was determined using antigen peptide pools (Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020). Importantly, pronounced T cell reactivity was detected against S protein peptides exhibiting a high degree of homology to endemic ‘common cold’ human coronaviruses (HCoV), including HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E. Therefore, preexisting T cell immunity against SARS-CoV-2 is hypothesized to originate from prior exposure to endemic HCoVs (Mateus et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Braun et al., 2020; Weiskopf et al., 2020).
Preexisting B cell immunity may be already germline-encoded in the naive B cell repertoire or originate from cross-reactive immune responses against related pathogens or variants. Many potent SARS-CoV-2 neutralizing antibodies (nAbs) exhibit binding by germline-encoded amino acid residues within the complementarity-determining regions 1 and 2 (CDRH1 and CDRH2) (Barnes et al., 2020a; Hurlburt et al., 2020; Shi et al., 2020; Wu et al., 2020; Yuan et al., 2020), are restricted to specific heavy chain V genes (Brouwer et al., 2020; Cao et al., 2020; Ju et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Wu et al., 2020; Zost et al., 2020), or exhibit a low degree of somatic mutations (Barnes et al., 2020b; Kreer et al., 2020a; Robbiani et al., 2020; Seydoux et al., 2020). This suggests that near-germline B cell receptor (BCR) sequences with close similarity to SARS-CoV-2-reactive antibodies might already be encoded in the naive B cell repertoire and can be readily selected to mount a potent B cell response without further affinity maturation. In line with this, we previously identified potential heavy and/or light-chain precursor sequences of SARS-CoV-2 binding as well as neutralizing antibodies by deep sequencing of naive B cell receptor repertoires sampled before the SARS-CoV-2 pandemic (Kreer et al., 2020a).
Cross-reactive immune responses to endemic HCoVs may also account for a potential preexisting humoral immunity against SARS-CoV-2. Recent studies provide controversial results regarding the frequency of cross-reactive antibodies in the sera of unexposed individuals (Anderson et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020; Poston et al., 2020; Shrock et al., 2020; Song et al., 2021) and their association with protection from disease severity or hospitalization (Anderson et al., 2021; Gombar et al., 2021; Sagar et al., 2021). Although one study found SARS-CoV-2 reactive antibodies in a considerable number of unexposed individuals - particularly among children, adolescents, and pregnant women (Ng et al., 2020), other studies did not find comparable evidence (Anderson et al., 2021; Nguyen-Contant et al., 2020; Poston et al., 2020; Song et al., 2021; Chau et al., 2021). Most studies to date depended on the investigation of SARS-CoV-2 reactivity in serum/plasma or affinity-enriched or secreted IgG fractions of unexposed individuals (Anderson et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020; Poston et al., 2020; Shrock et al., 2020; Song et al., 2021). These conflicting results call for more comprehensive and detailed investigations which beyond plasma and IgG fractions also involve BCR sequence analyses of SARS-CoV-2-reactive B cells and characterization of recombinant monoclonal antibodies.
To determine the presence of a relevant preexisting SARS-CoV-2 B cell immunity, we investigated plasma samples, single B cells, and monoclonal antibodies isolated from 150 SARS-CoV-2 unexposed individuals. We found no evidence of a competent preexisting B cell immunity that may account for the broad clinical spectrum of SARS-CoV-2 infections.
Results
Pre-pandemic plasma samples and polyclonal IgG exhibit no significant reactivity to SARS-CoV-2
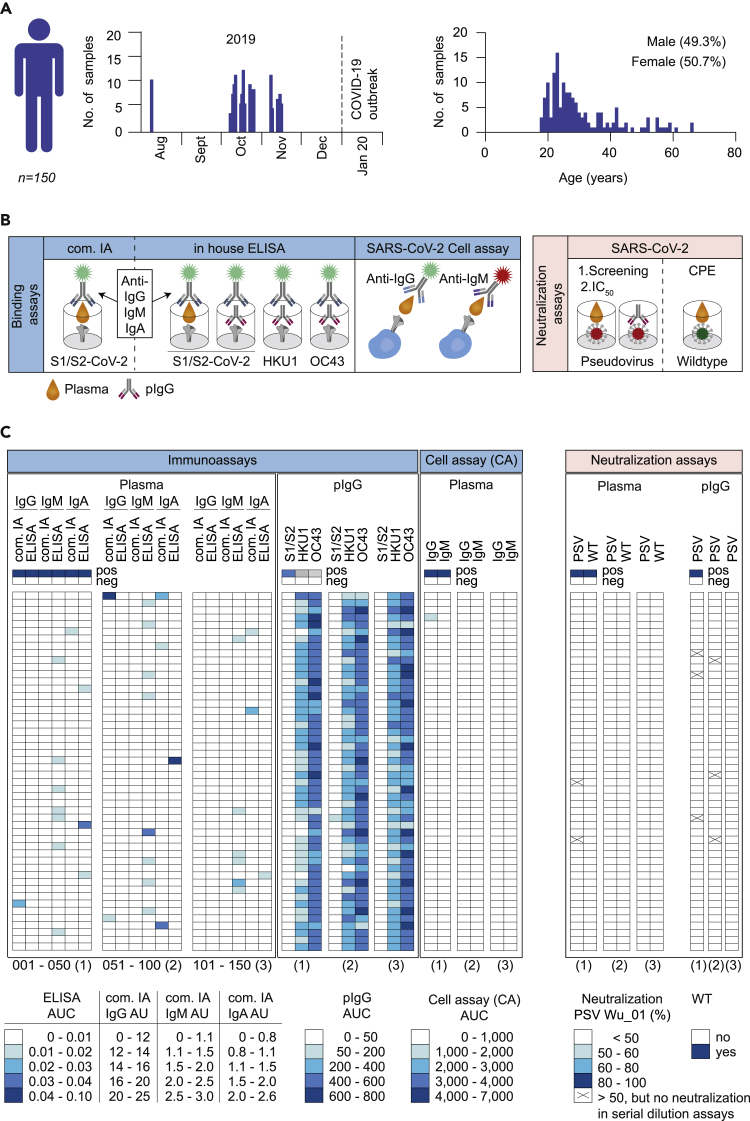
We previously identified unpaired rare heavy and light chain variable regions of pre-pandemic naive B cells that closely resembled near-germline SARS-CoV-2-reactive antibodies by performing NGS analyses on healthy individuals (Kreer et al., 2020a) (Figure S1). Some of these heavy and light chains derived from naive B cells can replace the original one of SARS-CoV-2-reactive antibodies without altering their functionality (Figure S1B). These findings complement previous observations that some SARS-CoV-2-reactive antibodies exhibit their activity by germline-encoded sequence features (Barnes et al., 2020b; Hurlburt et al., 2020; Robbiani et al., 2020; Seydoux et al., 2020). To investigate how these findings translate to plasma responses and whether SARS-CoV-2-reactive antibodies are already present in the plasma of unexposed individuals, we investigated pre-pandemic blood samples from 150 donors. Samples were collected between August and November 2019 and were studied for binding and neutralizing activity against SARS-CoV-2 (Figures 1A and 1B; Table S1). Donors were between 18 and 66 years of age (with a mean/median age of 30.6/27 years) (Figure 1A and Table S1). 49.3% of donors were male and 50.7% female (Figure 1A and Table S1). With regard to previously published conflicting results, we decided to use different assays in parallel as a stringent search for preexisting SARS-CoV-2-reactive antibodies. Initially, plasma samples of all 150 donors were tested for binding to the soluble full trimeric SARS-CoV-2 S ectodomain (S1/S2) or the S1 subunit (S1) by commercially available (com. IA) and in-house (ELISA) immunoassays, respectively (Figures 1B, 1C, and S2). Plasma samples showed mostly no or only minimal binding of IgG, IgM, and IgA to soluble SARS-CoV-2 S protein (Figures 1C and S2). In a few samples (Pre033 IgA, Pre051 IgG, Pre074 IgA, Pre084 IgM, and Pre097 IgA) notable reactivity was detected. However, in none of the investigated plasma samples binding was detected in both the commercial and in-house immunoassay. To substantiate this result, we assessed binding activity against cell surface expressed full-length SARS-CoV-2 S protein by flow cytometry (cell assay, CA; Figures 1B and 1C). Except for one donor (Pre004) that exhibited weak plasma IgG activity in this assay, all other samples did not reveal any binding activity (Figures 1C and S2). We attributed the occasional detection of binding activity in only one out of the three assays to the use of unpurified plasma samples that might yield nonspecific reactivity.
Figure 1.
Screening of pre-pandemic samples from 150 adults reveal no relevant reactivity to SARS-CoV-2
(A) Timeline of blood collections and demographic characteristics of 150 donors sampled before the SARS-CoV-2 pandemic. Pre-pandemic blood samples were collected as buffy coats between August and November of 2019. Gender and age distribution of donors are illustrated as bar plots.
(B) Pre-pandemic plasma samples and purified, polyclonal IgGs (pIgG) were tested for binding and neutralization using different experimental approaches. Plasma IgG, IgM, and IgA as well as pIgG were tested for binding to SARS-CoV-2, HKU1, and OC43 S proteins using in house (ELISA) and commercially available immunoassays (com. IA). Binding of plasma IgG and IgM to cell surface expressed SARS-CoV-2 S protein was also determined by FACS analysis (CA). Neutralizing activity was determined against SARS-CoV-2 pseudo- and authentic wild type viruses (PSV and WT).
(C) Heatmap visualization of binding (AUC or AU) and neutralizing activity (% or CPE) of pre-pandemic plasma samples and pIgG against SARS-CoV-2 and endemic HCoVs (HKU1 and OC43) S proteins or SARS-CoV-2 authentic wild type (WT) and pseudovirus (PSV Wu_01), respectively (see also Figures S2 and S3). Immunoassays were performed in duplicates and the AUCs are presented as geometric mean of duplicates. Neutralizing activity was first determined for single sample concentrations. Samples that displayed neutralization activity of ≥50% are indicated as (x) and were repeatedly investigated in serial dilutions (see also Figure S3). Samples were tested in duplicates. The average of neutralization is presented and each row represents one donor.
In order to reduce potential nonspecific signals, polyclonal IgGs (pIgG) were purified from pre-pandemic plasma samples and tested for binding to soluble full trimeric S proteins of SARS-CoV-2 (Figures 1C and S2). No notable S protein binding was detected in any of the samples. In contrast, all samples showed substantial activity against endemic human-pathogenic betacoronaviruses (HKU1 and OC43) (Figures 1C and S2). Therefore, results show no evidence for cross-reactivity of IgGs between HKU1 or OC43 and SARS-CoV-2 S proteins. Finally, neutralizing activity of plasma and pIgG against SARS-CoV-2 pseudotyped (PSV) and/or authentic wildtype virus (WT) was determined. Weak neutralization of 50–60% was found in two plasma and six pIgG samples, respectively. However, neutralizing activity of these samples could not be confirmed by testing serial dilutions (Figures 1C and S3).
In summary, none of the pre-pandemic samples obtained from 150 adults displayed reactivity against SARS-CoV-2 in more than one of the various assays applied. Thus, based on the parallel application of the various assays, we found no convincing evidence of SARS-CoV-2-reactive antibodies in the pre-pandemic blood samples examined.
No detection of SARS-CoV-2-reactive B cells in pre-pandemic samples
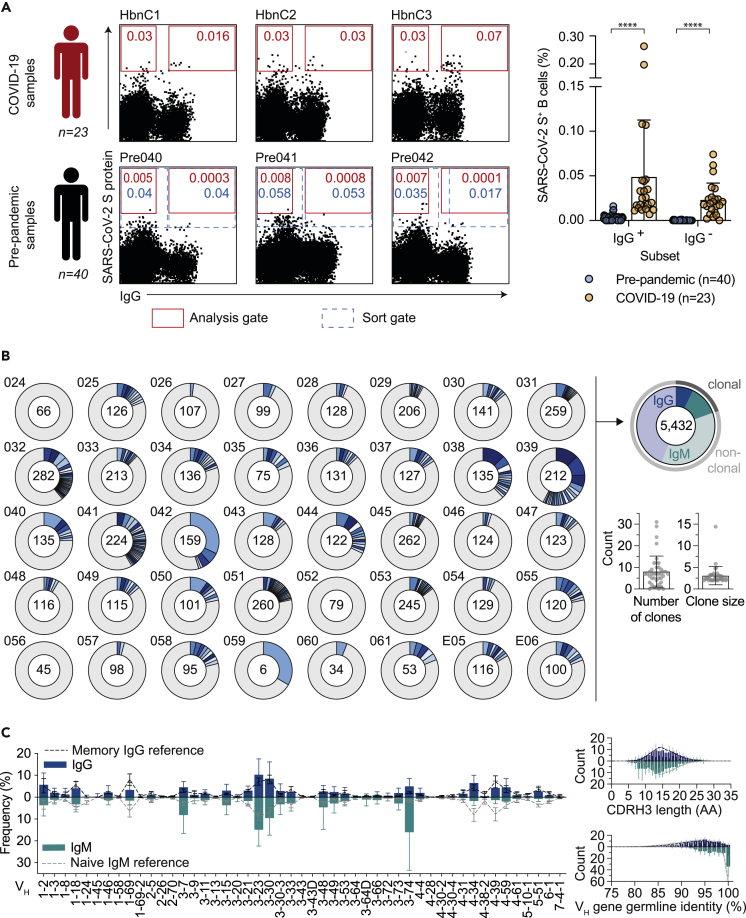
The lack of binding or neutralization activity against SARS-CoV-2 on plasma level does not exclude the existence of SARS-CoV-2-reactive B cells in unexposed individuals. To investigate the presence of low-frequency SARS-CoV-2 specific B cells, we aimed to identify single reactive B cells by cell sorting using the SARS-CoV-2 S protein as bait. In total, PBMCs of 40 donors sampled before the pandemic (Figures 2 and S4) were investigated. Using the same analysis gate as for COVID-19 convalescent donors, frequencies of SARS-CoV-2-reactive IgG+ and IgG− B cells isolated from pre-pandemic blood samples were significantly lower (p value < 0.0001). For COVID-19 convalescent donors, frequencies ranged from 0.002 to 0.065% for IgG+ (median 0.02%) and 0.007 to 0.39% for IgG− (median 0.031%) B cells (Figure 2A). Applying the same analysis gate of the COVID-19 convalescent samples to our pre-pandemic samples, frequencies ranged from 0 to 0.0013% for IgG+ (median 0.0001%) and from 0 to 0.016 for IgG− (median 0.003%) B cells (Figures 2A and S4). Therefore, we conclude that, if present at all, SARS-CoV-2-reactive B cells have a significantly lower frequency in pre-pandemic samples. We reasoned that gate settings applied for COVID-19 convalescent samples may exclude reactive B cells with low spike affinity which may be present in individuals unexposed to SARS-CoV-2. To assure that such cells do not remain undetected, we adjusted the actual sorting gate (Figure 2A) to isolate a total of 8,174 putatively SARS-CoV-2-reactive single B cells, of which 3,852 were IgG+ and 4,322 IgG− cells. Of those, we amplified a total of 5,432 productive heavy chain sequences, of which 2,789 sequences accounted for IgG and 2,643 for IgM heavy chains (Figure 2B). Sequence analyses showed that in each individual 0 to 58% of the sequences were clonally related with a mean clone number of 8.1 clones per individual and a mean clone size of 2.9 members per clone (Figure 2B). Heavy chain variable (VH) gene segment distribution, heavy chain complementarity-determining region 3 (CDRH3) length, and VH gene germline identities showed no notable divergence to a reference memory IgG and naive IgM repertoire dataset (Kreer et al., 2020a) (Figure 2C). We conclude that pre-pandemic samples of healthy adults usually lack high-reactive B cells against SARS-CoV-2.
Figure 2.
No substantial detection of SARS-CoV-2-reactive B cells
(A) Representative dot plots of SARS-CoV-2-reactive, CD19+CD20+, IgG+, and IgG- B cells of samples from COVID-19 convalescent individuals compared to before the pandemic (pre-pandemic). Depicted numbers indicate frequencies of S protein reactive B cells (see also Figure S4). Red colored gate indicates gating strategy for analysis and dotted gate indicates actual sorting gate. Dot plot bar graph displays the mean ± SD frequency of SARS-CoV-2-reactive, IgG+, and IgG- B cells in 40 pre-pandemic and 23 COVID-19 samples (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001; unpaired two-tailed t-test).
(B) Clonal relationship of heavy chain sequences amplified from single SARS-CoV-2-reactive IgG+ and IgG- B cells isolated from 40 donors sampled before the pandemic. Individual clones are colored in shades of blue, gray, and white. In the center of each pie chart, numbers of productive heavy chain sequences are illustrated. Presentation of clone sizes are proportional to the total number of productive heavy chain sequences per clone.
(C) VH gene distribution, VH gene germline identity, and CDRH3 length distribution in amino acids (AA) were separately determined for IgG and IgM. Distributions were calculated per individual. Bar and line plots show mean ± SD.
Monoclonal antibodies isolated from pre-pandemic samples are not reactive against SARS-CoV-2
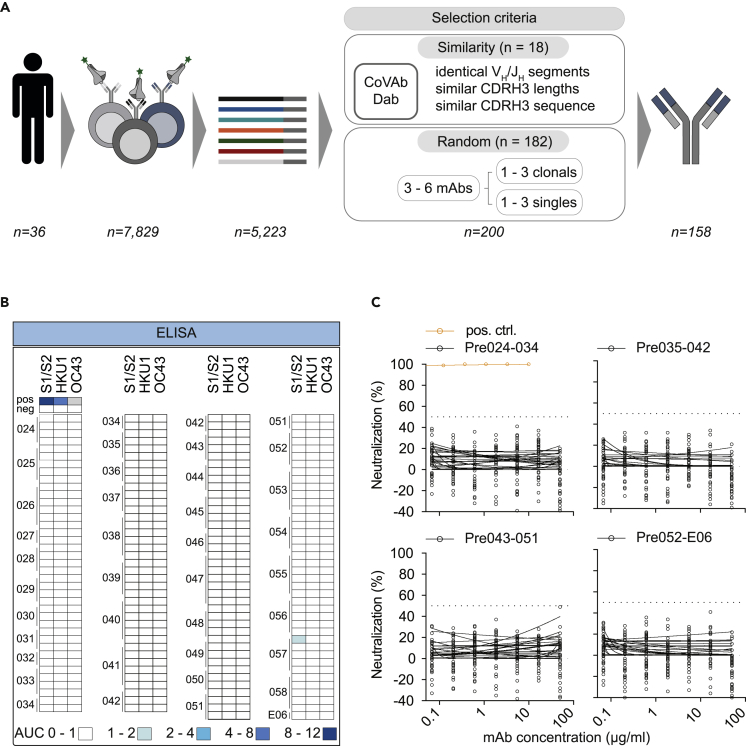
To confirm the absence of SARS-CoV-2-reactive B cells in pre-pandemic samples on a functional level, we selected 200 antibody candidates from 36 donors for production and functional testing (Figure 3A). Selection was conducted based on sequence similarity (see Methods section) to 920 SARS-CoV-2-reactive antibodies deposited in the CoVAbDab (n = 18) and on random sequence selection (n = 182) (Figure 3). Criteria for the similarity selection were identical VH/JH combinations, low level of CDRH3 length differences, and Levenshtein distances between isolated and deposited BCR sequences. The random selection included clonal as well as non-clonal sequences and was performed to ensure an equal selection of BCR sequences among different donors and clonotypes. In total, we successfully produced 158 monoclonal antibodies (81 IgM-, 77 IgG-derived) as IgG1 isotypes for functional testing. First, we determined binding activity of these antibodies to SARS-CoV-2 S protein and evaluated cross-reactivity to HKU1 and OC43 S proteins by ELISA (Figures 3B and S5). Monoclonal antibodies showed no relevant binding or cross-reactivity against SARS-CoV-2, HKU-1, or OC43 S proteins. Next, we tested all 158 monoclonal antibodies for neutralization activity against SARS-CoV-2 pseudovirus in single concentrations of 50 μg/mL and in serial dilutions (Figure 3C). In line with the lack of binding activity, none of the produced antibodies showed neutralizing activity up to concentrations of 50 μg/mL (Figure 3C). We conclude that putatively SARS-CoV-2-S+ B cells from pre-pandemic samples do not encode for SARS-CoV-2-reactive B cell receptors.
Figure 3.
Monoclonal antibodies isolated from pre-pandemic samples show no reactivity to SARS-CoV-2 and endemic HCoVs
(A) Illustration demonstrating the sequence selection for downstream antibody production. Sequences were selected for antibody production based on similarity to antibody sequences deposited in the CoVAbDab and on random selection. From 7,829 SARS-CoV-2 reactive IgG+ and IgG- B cells, 5,223 productive heavy chain sequences were amplified. For antibody production, 18 HC sequences were selected based on similarity selection and 182 HC sequences were selected based on random selection. In total, 158 antibodies were produced.
(B) Heatmap visualization of binding (AUC) and neutralizing activity (%) of monoclonal antibodies isolated from pre-pandemic blood samples against SARS-CoV-2 and endemic HCoVs (HKU1 and OC43) S proteins or SARS-CoV-2 pseudovirus (PSV Wu_01), respectively (see also Figure S5). Each row represents one monoclonal antibody. ELISAs were performed in duplicate experiments and the AUCs are presented as a geometric mean of duplicates. Neutralizing activity was determined for single sample concentrations. Samples were tested in duplicates.
(C) Neutralizing activity against SARS-CoV-2 pseudovirus (PSV Wu_01) was verified for all mAbs in serial dilutions.
Discussion
Adaptive immune responses against a pathogen are shaped by the naive immune repertoire and imprinted by previous encounters to the same pathogen or a related variant. Investigation of preexisting immunity to SARS-CoV-2 can advance our understanding of protective immunity, susceptibility to infection, disease severity, and guide the development of vaccination strategies (Imkeller and Wardemann, 2018; Niu et al., 2020; Schultheiβ et al., 2020). Although various studies have already provided evidence for preexisting T cell immunity against SARS-CoV-2, the presence of a preexisting B cell immunity remains to be elucidated (Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Sette and Crotty, 2021).
Recently published studies that investigated a preexisting B cell immunity in unexposed individuals provide partially conflicting results. For instance, some studies based on the examination of serum samples reported detection of SARS-CoV-2 cross-reactive antibodies and preexisting humoral immunity in uninfected individuals (Ng et al., 2020; Majdoubi et al., 2021; Sagar et al., 2021; Woudenberg et al., 2021). Cross-reactive humoral immune responses were mainly observed against S protein structures within the S2 subunit, the N protein, or against ORF polypeptides such as nonstructural proteins protein 2 (NSP2) and 15 (NSP15) that are relatively conserved among endemic HCoVs and SARS-CoV-2. However, the frequency of cross-reactive serum responses varied greatly between the individual studies. For instance, although some of these studies report an overall seroprevalence of cross-reactive serum responses in 10%–20% in unexposed subjects (Ng et al., 2020; Nguyen-Contant et al., 2020; Woudenberg et al., 2021), another study determined antibody responses in more than 90% of all samples analyzed (Majdoubi et al., 2021). Varying frequency of cross-reactive serum responses between these studies are attributed to differences in cohort composition and heterogeneous sensitivities of applied diagnostic assays. Furthermore, in the described studies the extent to which prior immunity against HCoVs may modify COVID-19 disease severity and susceptibility as well as seasonal and geographical transmission patterns is controversially discussed (Ng et al., 2020; Sagar et al., 2021; Woudenberg et al., 2021). In contrast, other studies did not or only rarely detect cross-reactive serum and B cell responses against SARS-CoV-2 in pre-pandemic samples and argued against a broad and clinically relevant preexisting B cell immunity (Anderson et al., 2021; Nguyen-Contant et al., 2020; Poston et al., 2020; Shrock et al., 2020; Song et al., 2021). Of note, most studies to date only investigated plasma/serum or enriched IgG fractions of pre-pandemic samples (Anderson et al., 2021; Majdoubi et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020; Poston et al., 2020; Sagar et al., 2021; Shrock et al., 2020; Song et al., 2021; Woudenberg et al., 2021). They did not involve the analysis of naive B cell receptor repertoires and putatively SARS-CoV-2-reactive B cells or the characterization of recombinant monoclonal antibodies derived from these cells. Therefore, such plasma-based studies may miss preexisting potent naive B cell precursors and low frequency memory B cells. In our study, we performed an extensive examination of plasma and IgG fractions as well as reached out further to investigate the presence of SARS-CoV-2-reactive B cell precursors, memory B cells, and the respective monoclonal antibodies.
We recently isolated SARS-CoV-2-reactive antibodies from convalescent individuals and identified highly similar heavy and/or light chain sequences in naive B cell receptor repertoires from pre-pandemic samples (Kreer et al., 2020a). Here, we show that some of these chains can be components of SARS-CoV-2-reactive antibodies without the need for further affinity maturation. This finding supports previous observations that the readily detected antibody response in some individuals might use existing antibody heavy or light chains with distinct CDR3 recombination patterns or germline encoded sequence features (Barnes et al., 2020a, 2020b; Hurlburt et al., 2020; Kreer et al., 2020a; Robbiani et al., 2020). In line with this, SARS-CoV-2 neutralizing antibodies were successfully isolated from the human naive B cell compartment using antigen-specific single B cell sorts or phage display (Bertoglio et al., 2021; Feldman et al., 2021). Feldman et al. sequentially applied diverse SARS-CoV-2 spike protein subdomains as antigen probes for the isolation of single SARS-CoV-2 and sarbecovirus-reactive naive B cells. Following this approach, they report a median frequency of 0.0025% for RBM-reactive naive B cells. BCR sequence analyses revealed a diverse and polyclonal gene usage for heavy and light chains but an increase in the mean repertoire frequency of 20% for the heavy chain v gene IGHV3-9 (Feldman et al., 2021). Finally, monoclonal antibodies were isolated from naive B cell precursors that exhibited binding and neutralization activity against circulating SARS-CoV-2 variants of concern and bat-derived coronaviruses. Bertoglio et al. applied phage display to isolate antibody candidate STE73-2E9 from naive B cell libraries that targets the ACE2-RBD interface without cross-reactivity to other coronaviruses and neutralizes authentic SARS-CoV-2 wildtype virus with an IC50 of 0.43 nM. However, because phage display relies on random sequence recombination, this study does not provide evidence that SARS-CoV-2-reactive antibodies naturally occur in the naive B cell repertoire.
Using various immunological and functional assays assessing SARS-CoV-2 binding and neutralization activity as well as cross-reactivity to endemic betacoronaviruses, we found no evidence of significant plasma or IgG reactivity against SARS-CoV-2 in pre-pandemic samples. We comprehensively investigated plasma binding activity of IgG, IgM, and IgA immunoglobulin isotypes against diverse beta coronavirus S proteins (SARS-CoV-2 S1 and S1/S2, HCoV-HKU1, and HCoV-OC43 S) by in house and commercially available ELISAs. Moreover, we validated our ELISA results against cell-surface-expressed S protein using flow cytometry and by applying SARS-CoV-2 pseudo- as well as wildtype neutralization assays. Our results are consistent with recently published studies that disagreed on a broad preexisting B cell background immunity (Anderson et al., 2021; Chau et al., 2021; Nguyen-Contant et al., 2020; Poston et al., 2020; Shrock et al., 2020; Song et al., 2021). These studies report an overall prevalence of cross-reactive serum responses against the SARS-CoV-2 S protein or the S1 subdomain below 5%. In line with this, only 3% of analyzed plasma samples (5/150) in our study displayed some binding activity against the SARS-CoV-2 S protein. Furthermore, published studies from Anderson et al. and Poston et al. report a lack of SARS-CoV-2 neutralization activity in pre-pandemic serum sample and no protection from disease severity by hCoV-reactive antibodies (Anderson et al., 2021; Poston et al., 2020). Consistent with these results, we could also not detect any notable neutralizing activity in pre-pandemic serum samples. However, the findings of our plasma screening may not be directly comparable to the recent studies that indeed reported preexisting humoral immunity because of cohort composition or differences in experimental assays. For example, Ng et al. detected serum neutralization activity in unexposed children, adolescents, and pregnant women who are not included in our study cohort (Ng et al., 2020).
To investigate potential preexisting immunity on a molecular level, we applied antigen-specific single cell sorts to 40 donors and isolated a total of 8,174 putatively SARS-CoV-2-reactive B cells. Consistent with our findings from binding and neutralization screening of plasma samples and polyclonal IgG, frequencies of putatively SARS-CoV-2-reactive B cells in pre-pandemic samples were very low especially when compared to COVID-19 convalescent samples. The choice of an appropriate bait protein for single cell sorting critically determines the isolation of antigen-specific B cells. Immunogenic structures that are not displayed by the chosen bait protein are excluded from isolation. To ensure the comprehensive isolation of antigen-specific B cells, we therefore chose to apply the native, full trimeric SARS-CoV-2 S protein and to adjust the sorting gate. With regard to the low frequencies of SARS-CoV-2-reactive B cells in our study, it can be argued that application of alternative bait proteins, e.g., the S2 subunit would have been favorable (Ng et al., 2020; Nguyen-Contant et al., 2020; Shrock et al., 2020; Song et al., 2021). Antibody sequence analyses of isolated putatively SARS-CoV-2-reactive B cells revealed diverse heavy chain V gene usage, normally distributed CDRH3 lengths, and VH gene germline identities similar to the naive BCR receptor repertoire of healthy individuals sampled before the pandemic. In particular, we found no evidence of enrichment of specific V regions which were preferentially used in antibodies of SARS-CoV-2 convalescent individuals such as IGHV3-30, IGHV3-53, or IGHV3-63 (Barnes et al., 2020a, 2020b; Robbiani et al., 2020). Furthermore, we detected no relevant reactivity of 158 recombinantly produced monoclonal antibodies against SARS-CoV-2 and endemic HCoV S proteins.
Limitations of the study
In the presented study, we could not detect cross-reactivity of pre-pandemic serum samples between endemic HCoVs HKU1 or OC43 and SARS-CoV-2 S proteins. However, individuals involved in the cohort were not investigated for recent HCoV infections or a documented history of infection prior to blood sampling. Therefore, this study cannot rule out that recent HCoV infections may be important determinants for the detection of serum cross-reactivity to SARS-CoV-2. In this light, recent HCoV infections may, for example, elicit transient serum cross-reactivity against SARS-CoV-2 that is not detected in our study. Moreover, HCoV infections appear with higher prevalence in children and young adolescents who were not included in the investigated study cohort (Chiu et al., 2005; Esper et al., 2005; Woudenberg et al., 2021).
For single cell sorts, we only expressed and functionally characterized a small proportion (n = 158) of amplified BCR sequences (n = 5,223). Although we applied defined criteria for selection of BCR sequences for downstream antibody production, we cannot rule out that SARS-CoV-2-reactive BCR sequences were indeed present among isolated sequences amplified but omitted by the bioinformatics approach.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human IgG-APC (Clone G18-145) | BD Biosciences | Cat#550931; RRID: AB_398478 |
| Anti-Human CD20-Alexa Fluor 700 (Clone 2H7) | BD Biosciences | Cat#560631; RRID: AB_1727447 |
| Anti-Human CD27-PE (Clone M-T271) | BD Biosciences | Cat#560985; RRID: AB_10563213 |
| Brilliant Violet 421(TM) Anti-Human IgG Fc (Clone HP6017) | BioLegend | Cat#409318; RRID: AB_2562176 |
| APC Anti-Human IgM (Clone MHM-88) | BioLegend | Cat#314510; RRID: AB_493011 |
| Anti-6X His tag antibody | Abcam | Cat#ab9108; RRID: AB_307016 |
| Goat Anti-Human IgG-HRP | SouthernBiotech | Cat#2040-05; RRID: AB_2795644 |
| Goat Anti-Human IgM-HRP | ThermoFisher Scientific | Cat#A18835; RRID: AB_2535612 |
| Goat Anti-Human IgA-HRP | ThermoFisher Scientific | Cat#A18781; RRID: AB_2535558 |
| Bacterial and virus strains | ||
| E. coli DH5α | Thermo Fisher Scientific | Cat#18263012 |
| SARS-CoV-2 CoV2-P3 authentic virus | Vanshylla et al., 2021 | N/A |
| SARS-CoV-2 pseudovirus Wu01 | Crawford et al., 2020 | N/A |
| Biological samples | ||
| PBMCs, Plasma, and IgGs of 150 donors sampled pre-pandemically | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Thermo Fisher | Cat#D1306; CAS: 581-88-4 |
| RNasin | Promega | Cat#N2515 |
| RNaseOUT | Thermo Fisher | Cat#10777019 |
| SuperScript IV Reverse Transcriptase | Thermo Fisher | Cat#18090050 |
| Platinum Taq DNA Polymerase | Thermo Fisher | Cat#10966034 |
| Platinum Taq Green Hot Start | Thermo Fisher | Cat#11966034 |
| Q5 Hot Start High Fidelity DNA Polymerase | NEB | Cat#M0493L |
| T4 DNA Polymerase | New England Biolabs | Cat#M0203L |
| Branched Polyethylenimine, 25 kDa | Sigma-Aldrich | Cat#408727; CAS: 9002-98-6 |
| FreeStyle Expression Medium | Thermo Fisher | Cat#12338001 |
| Protein G Sepharose 4 Fast Flow | GE Life Sciences | Cat#17061805 |
| ABTS solution | Thermo Fisher | Cat#002024 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F9665 |
| Sodium Pyruvate | Thermo Fisher | Cat#11360-070 |
| L-Glutamine | Thermo Fisher | Cat#25030024 |
| HEPES | Thermo Fisher | Cat#15630-080 |
| GlutaMAX Supplement | Thermo Fisher | Cat#35050-061 |
| 0.05% Trypsin-EDTA (1x) | Gibco | Cat#25300-096 |
| Pen Strep | Gibco | Cat#15070-063 |
| L-Glutamine 200 mM (100x) | Gibco | Cat#250030-123 |
| Ni-NTA Agarose | Macherey-Nagel | Cat#745400.25 |
| Strep-Tactin®XT Superflow® 50% suspension | IBA lifesciences | Cat#2-4010-010 |
| 10x Buffer BXT; Strep-Tactin®XT Elution Buffer | IBA | Cat#2-1042-025 |
| 10x Buffer W; Strep-Tactin®/Strep-Tactin®XT Wash Buffer | IBA | Cat#2-1003-100 |
| Critical commercial assays | ||
| CD19-Microbeads | Miltenyi Biotec | Cat#130-050-301 |
| Microscale Antibody Kit (DyLight 488) | Thermo Fisher Scientific | Cat#53025 |
| Molecular Probes™ Alexa Fluor™ 488 NHS Ester (Succinimidyl Ester) | Thermo Fisher Scientific | Cat#10266262 |
| Anti-SARS-CoV-2 ELISA (IgG) | Euroimmun | Cat#2606-9601 G |
| LIAISON SARS-CoV-2 S1/S2 IgG serology test | DiaSorin | Cat#311450 |
| LIAISON SARS-CoV-2 S1/S2 IgM serology test | DiaSorin | Cat#311470 |
| Anti-SARS-CoV-2 ELISA (IgA) | Euroimmun | Cat#2606-9601 A |
| Deposited Data | ||
| Cloned and tested monoclonal antibodies | This paper | GenBank: OM631046 - OM631381 |
| Cloned and tested chimeric antibodies generated from NGS data | This paper | GenBank: OM631382 - OM631443 |
| Experimental models: Cell lines | ||
| 293-6E cells | NRC | NRC file 11565 |
| HEK293T cells | ATCC | Cat#CRL-11268 |
| VeroE6 cells | ATCC | Cat#CRL-1586; RRID: CVCL_0574 |
| Oligonucleotides | ||
| Optimized PCR primers for single cell PCR | Kreer et al., 2020a | N/A |
| Random Hexamer Primer | Thermo Fisher | Cat#SO142 |
| SLIC heavy chain reverse primer (GGGTGCCAGGGGGAAGAC CGATGGGCCCTTGGTCGAGGC) |
This paper | N/A |
| SLIC kappa chain reverse primer (CTCATCAGATGGCGGGAAG ATGAAGACAGATGGTGCAGCCACCGTACG) |
This paper | N/A |
| SLIC lambda chain reverse primer (GAAGCTCCTCACTCGAGG GYGGGAACAGAGTG) |
This paper | N/A |
| Recombinant DNA | ||
| Human antibody expression vectors (IgG1, Igλ, Igκ) | Tiller et al., 2008 | N/A |
| Plasmid encoding SARS-CoV-2 S ectodomain (amino acids 1−1208 of SARS-CoV-2 S; GenBank: MN908947) | Wrapp et al., 2020 | GenBank: MN908947 |
| Plasmid encoding cell surface expressed SARS-CoV-2 full trimeric S protein | This paper | N/A |
| pCAGGS-EBOV GPΔTM-GCN4-HIS-Avi, encoding the EBOV Makona glycoprotein (GP) ectodomain (GenBank: KJ660347; aa:1-651) | Ehrhardt et al., 2019 | (GenBank: KJ660347; aa:1-651) |
| Plasmid encoding HCoV-HKU1 S protein (amino acids 1 - 1295; GenBank: YP_173238.1) | Woo et al., 2005 | GenBank: YP_173238.1) |
| Plasmid encoding HCoV-OC43 (amino acids 1 - 1300; GenBank: AY585229) | St-Jean et al., 2004 | GenBank: AY585229) |
| Software and algorithms | ||
| Geneious R10 and Geneious Prime | Geneious | RRID: SCR_010519 |
| Python 3.6.8 | Python Software Foundation, https://www.python.org/ | RRID: SCR_008394 |
| IgBLAST 1.13.0 | Ye et al., 2013 | RRID: SCR_002873 |
| FlowJo 10.5.3 | FlowJo, LLC | https://www.www.flowjo.com |
| Prism 7 | GraphPad | https://www.graphpad.com/; RRID: SCR_002798 |
| MacVector 16.0.9 | MacVector | https://www.macvector.com |
| Adobe Illustrator CC 2021 | Adobe | https://www.adobe.com |
| Other | ||
| Amicon MWCO 30 kDa/10 kDa | Merck Millipore | Cat#Z677108 |
| Leica DMI-Mikroskop | Leica Biosystem | N/A |
| BD FACSAria Fusion Cell Sorter | BD Bioscience | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Florian Klein (florian.klein@uk-koeln.de).
Materials availability
There are restrictions to the availability of SARS-CoV-2-reactive antibodies due to limited production capacities and ongoing consumption. Reasonable amounts of antibodies will be made available by the Lead Contact upon request under a Material Transfer Agreement (MTA) for non-commercial usage. Nucleotide sequences and expression plasmids will be shared upon request.
Experimental model and subject details
Study participants and collection of clinical samples
Study participants were recruited at the University Hospital Cologne. Buffy coats from study participants were collected according to a study protocol approved by the Institutional Review Board of the University of Cologne (study protocol 16–054). Buffy coats were provided as residual products in the context of regular blood donations from the Institute for Transfusion Medicine at the University Hospital of Cologne. All study participants provided informed consent. The study cohort comprised 150 individuals with 50.7% female and 49.3% male adults. The influence of gender on the results of the study was not explicitly measured. All study participants were ≥18 years old. Blood samples were collected between August and November 2019.
Cell lines
HEK293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 1 x antibiotic-antimycotic (Thermo Fisher), 1 mM sodium pyruvate (Gibco) and 2 mM L-glutamine (Gibco) at 37°C and 5% CO2. HEK293-6E cells were maintained in FreeStyle 293 Expression Medium (Life Technologies) supplemented with 0.2% penicillin/streptomycin under constant shaking at 90–120 rpm, 37°C and 6% CO2. VeroE6 and HEK293T-ACE2 cells were maintained in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 1 x penicillin-streptomycin (Gibco), 1 mM sodium pyruvate (Gibco) and 2 mM L-glutamine (Gibco) at 37°C and 5% CO2. The sex of HEK293T, HEK293-6E and VeroE6 cell lines is female. Cell lines were not specifically authenticated.
Method details
Isolation of PBMCs, plasma and total IgG
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats and whole blood by density gradient separation using Histopaque separation medium (Sigma-Aldrich) and Leucosep cell tubes (Greiner Bio-one) according to the manufacturer’s instructions. Isolated PBMCs were cryopreserved at 150°C in 90% FBS supplemented with 10% DMSO until further use. Plasma was collected and stored at −80°C. Plasma samples were heat-inactivated at 56°C for 40 min prior to further use. For IgG isolation, 1 ml of heat-inactivated plasma was incubated with Protein G Sepharose (GE Life Sciences) overnight at 4°C (GE Life Sciences) under constant rotation. Protein G beads were transferred to chromatography columns and washed twice with sterile PBS. IgGs were eluted from columns using 0.1 M glycine (pH = 3.0) and buffered in 0.1 M Tris (pH = 8.0). Buffer exchange to PBS and concentration of IgGs was performed by centrifugation using 30 kDa Amicon spin membranes (Millipore). The final concentration of purified IgGs was determined by UV/Vis spectroscopy using a Nanodrop (A280). Subsequently, purified IgGs were stored at 4°C.
Expression and purification of viral surface proteins
Constructs encoding the stabilized spike protein ectodomain of the endemic HCoV-HKU1 (amino acids 1–1295, GenBank ID.: ABD75497) and HCoV-OC43 (amino acids 1–1300, GenBank ID.: AAX84792) were kindly provided by Raiees Andrabi (California, USA) and Victor Corman (Berlin, Germany) and described previously (St-Jean et al., 2004; Woo et al., 2005; Corman et al., 2012; Kreye et al., 2020; Song et al., 2021). Constructs encoding the prefusion stabilized SARS-CoV-2 S ectodomain (amino acids 1–1207; GenBank ID.: MN908947) were kindly provided by Jason McLellan (Texas, USA) and Florian Krammer (New York, USA) and previously described (Stadlbauer et al., 2020; Wrapp et al., 2020). All three constructs contain the following mutations: two proline substitutions for prefusion state stabilization (SARS-CoV-2: residues 986 and 987; OC43: residues 1078 and 1079; HKU1: residues 1071 and 1072) and the furin cleavage sites were mutated (SARS-CoV-2: ‘‘GGGG’’ substitution at residues 682–685; OC43: ‘‘GSAS’’ substitution at residues 762–766; HKU1‘‘GSAS’’ substitution at residues 756–760). Ebola surface glycoprotein (EBOV Makona, GenBank ID.: KJ660347; amino acids 1–651) was used as a negative control for HCoV S protein ELISAs. The Ebola surface glycoprotein was stabilized with GCN4 trimerization domain and expressed without the transmembrane domain as previously described (Ehrhardt et al., 2019). The different coronavirus ectodomains were amplified from the synthetic gene plasmids by PCR and subsequently cloned into a modified sleeping beauty transposon expression vector containing a C-terminal T4 fibritin trimerization motif (foldon) followed by a Twin-Strep-Tag purification tag. For the recombinant protein productions, stable HEK293 EBNA cell lines were generated employing the sleeping beauty transposon system (Kowarz et al., 2015). Briefly, expression constructs were transfected into the HEK293 EBNA cells using FuGENE HD transfection reagent (Promega). After selection with puromycin, cells were induced with doxycycline. Cell supernatants were filtered and the recombinant proteins purified via Strep-Tactin XT (IBA Lifescience) resin. Proteins were then eluted by biotin-containing TBS-buffer (IBA Lifescience), and dialyzed against TBS-buffer.
Isolation of single SARS-CoV-2-reactive B cells
CD19+ B cells were enriched from PBMCs by immunomagnetic cell separation using CD19 microbeads (Miltenyi Biotec) according to the manufacturer’s protocol. Enriched B cells were labeled for 20 min on ice with 4’,6-Diamidin-2-phenlindol (DAPI; Thermo Fisher Scientific), anti-human CD20-Alexa Fluor 700 (BD), anti-human IgG-APC (BD), anti-human CD27-PE (BD) and DyLight488-labeled SARS-CoV-2 S protein (10 μg/ml). DAPI−, CD20+, SARS-CoV S protein+, IgG+/IgG− single cells were sorted into 96 well plates using a FACSAria Fusion (Becton Dickinson). Each well of the 96 well plate was prefilled with 4 μl of sorting buffer consisting of 0.5 x PBS, 0.5 U/μl RNAsin (Promega), 0.5 U/μl RNaseOut (Thermo Fisher Scientific), and 10 mM DTT (Thermo Fisher Scientific). Plates were cryopreserved at −80°C immediately after sorting.
B cell receptor amplification and sequence analysis
Generation of cDNA and amplification of antibody heavy and light chain genes from sorted single cells was performed as previously described (Gieselmann et al., 2021; Kreer et al., 2020a; Schommers et al., 2020). For reverse transcription, sorted cells were incubated with 7 μl of a random-hexamer-primer master mix (RHP mix) consisting of 0.75 μl Random Hexamer Primer (Thermo Fisher Scientific), 0.5 μl NP-40 (Thermo Fisher Scientific), 0.15 μl RNaseOut (Thermo Fisher Scientific), and 5.6 μl of nuclease-free H2O at 65°C for 1 min. Subsequently, samples were incubated with a reverse-transcription master mix (RT mix) consisting of 3 μl of 5 x Superscript IV RT buffer (Thermofisher Scientific), 0.5 μl dNTPs (Thermo Fisher Scientific), 1 μl DTT (Thermo Fisher Scientific), 0.1 μl of RNasin (Promega), 0.1 μl of RNaseOut (Thermo Fisher Scientific), 2.05 μl of nuclease-free H2O and 0.25 μl of Superscript IV (Thermofisher Scientific) per well and incubated at 42°C for 10 min, 25°C for 10 min, 50 °C for 10 min and 94°C for 5 min. Heavy and light chains were amplified from cDNA by semi-nested, single cell PCRs using optimized V gene-specific primer mixes(Kreer et al., 2020b) and Platinum Taq DNA Polymerase or Platinum Taq Green Hot Start Polymerase (Thermo Fisher Scientific) as previously described (Kreer et al., 2020a, 2020b; Schommers et al., 2020; Gieselmann et al., 2021). PCR products were analyzed by agarose gel electrophoresis for correct product size and subsequently send for Sanger sequencing. Only chromatograms with a mean Phred score of 28 and sequences with a minimal length of 240 nucleotides were selected for downstream sequence analyses. Filtered sequences were annotated with IgBlast (Ye et al., 2013) according to the IMGT system (Lefranc et al., 2009) and only the variable region from FWR1 to the end of the J gene was extracted. Base calls within the variable region with a Phred score below 16 were masked and sequences with more than 15 masked nucleotides, stop codons, or frameshifts were excluded from further analyses. Sequence analyses to inform on sequence clonality were performed separately for each study participant. All productive heavy chain sequences were grouped by identical VH/JH gene pairs and the pairwise Levenshtein distance for their CDRH3s was determined. Starting from a random sequence, clone groups were assigned to sequences with a minimal CDRH3 amino acid identity of at least 75% (with respect to the shortest CDRH3). 100 rounds of input sequence randomization and clonal assignment were performed and the result with the lowest number of remaining unassigned (non-clonal) sequences was selected for downstream analyses. All clones were cross-validated by the investigators taking shared mutations and light chain information into account.
Sequence selection for antibody production
Antibody selection for cloning was was performed with an in-house python script (Python v3.6.8) by two approaches. First, a similarity search was performed against 868 and 52 SARS-CoV-2-binding antibodies from the CoVAbDab (Raybould et al., 2020) (retrieved on 21.08.20) and Kreer et al., 2020 (Kreer et al., 2020a), respectively. To this end, SARS-CoV-2 antibodies were annotated with IgBLAST (Ye et al., 2013). Annotations and sequence information from published antibodies and sequences from this study were loaded with the pandas module (McKinney, 2010) into data frames. Both data frames were grouped by their V/J gene combination (ignoring any allele information). For all V/J groups that were found in both data frames, a pairwise comparison was performed by looping over all CDRH3s. For CDRH3 pairs, where the amino acid length difference was ≤2 AA, the Levenshtein distance was computed with the python-Levenshtein package. Sequences from this study, where the Levenshtein distance was ≤3 AA in comparison to at least one SARS-CoV-2-binding antibody, were selected as similar and produced (18 antibodies in total).
Second, a random selection was performed with the python random module (Python standard library) on clonal and non-clonal sequences from each individual (determined as described in “B cell receptor amplification and sequence analysis”) to select at least 3 different clones and at least 3 non-clonal sequences. If less than 3 clones were available for an individual, random non-clonal sequences were used to fill up the selection to yield at least 6 antibodies per individual.
Cloning and production of monoclonal antibodies
Heavy and light chain variable regions of selected antibodies were cloned into expression vectors by sequence and ligation independent cloning (SLIC) (von Boehmer et al., 2016) as previously described (Tiller et al., 2008; Schommers et al., 2020; Kreer et al., 2020a; Gieselmann et al., 2021). 1st PCR product was amplified using Q5 Hot Start High Fidelity DNA Polymerase (New England Biolabs) and specific forward and reverse primers including adaptor sequences which are homologous to the restriction sites of the antibody expression vector (IgG1, IgL, IgK (Tiller et al., 2008). Forward primers were designed according to 2nd PCR primers (Kreer et al., 2020b) and encode for the complete native leader sequence of all heavy and light chain V genes, whereas reverse primers bind to the conserved sequence motifs at the 5’ end of heavy and light chain immunoglobulin constant regions. PCR was run at 98°C for 30 s; 35 cycles of 98°C for 10 s, 72°C for 45 s; and 72°C for 2 min. Subsequently, PCR products were purified with 96-well format silica membranes, cloned by SLIC (von Boehmer et al., 2016) into linearized expression vectors with T4 DNA polymerase (NEB) and transformed into chemically competent Escherichia coli (DH5α). Heavy chain variable regions of IgG− B cells were cloned into IgG1 expression vectors. Correct insertion of the V region sequence into the expression vector was examined by colony PCR and Sanger sequencing. Positive colonies were propagated in midi cultures and plasmids purified.
For production of recombinant monoclonal antibodies, HEK293-6E suspension cells were co-transfected chemically with human heavy chain and corresponding light chain antibody expression vectors using 25 kDa branched polyethylenimine (PEI; Sigma-Aldrich). Transfected cells were propagated in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) supplemented with 0.2% penicillin/streptomycin (Thermo Fisher Scientific) at 37°C and 6% CO2 under constant shaking at 90–120 rpm for seven days. Cell supernatants were harvested by centrifugation, filtered through PES filters and incubated overnight at 4°C under constant rotation with Protein-G-coupled Sepharose beads (GE Life Sciences). The suspension was transferred to chromatography columns, washed twice with sterile PBS and antibodies were eluted with 0.1 M glycine (pH = 3) and buffered using 1 M Tris (pH = 8). Buffer exchange to PBS and concentration of antibodies was performed by centrifugation using 30kDa Amicon spin membranes (Millipore). Antibodies were filtered through Ultra-free-MC 0.22 μm membranes (Millipore) and stored at 4°C. The final concentration of purified antibodies was determined by UV/Vis spectroscopy using a Nanodrop (A280). Subsequently, purified antibodies were stored at 4°C.
Cell-surface-expressed S protein immunoassay
HEK293T cells were transfected with plasmids encoding the full-length SARS-CoV-2 S protein (GenBank ID.: MN908947) using TurboFect™TM transfection reagent (Thermo Fisher Scientific). After incubation at 37°C and 5% CO2 for 48 h, adherent cells were detached with PBS supplemented with 1 mM EDTA (pH = 7.4) and resuspended in FACS buffer (1x PBS supplemented with 2% FCS and 2 mM EDTA). 3 × 104 cells were distributed in 50 μl of FACS buffer to each well of a V-bottom-shaped 96 well plate. Starting with an initial dilution of 1:25, plasma samples were prepared in a 4-fold serial dilution for a total of 6 dilutions. Cells were incubated with 50 μl/well of diluted plasma samples and incubated for 30 min on ice. After incubation, cells were washed once with 100 μl/well of FACS buffer and stained in 50 μl/well of 1:160 dilution of BV421 IgG and 1:100 dilution of APC IgM on ice for 30 min. After staining, cells were washed once with 100 μl/well of FACS buffer and analyzed on a FACS BD Aria Fusion. Evaluations were performed using FlowJo10 software. Geometric Mean values of all cells/single cells/mCherry positive (transfected cells) in APC-A channel (for IgM) and BV421-A channel (for IgG) were determined and graphically displayed using GraphPad Prism.
SARS-CoV-2 and HCoV S protein immunoassays
ELISA plates (Greiner Bio-One 655092) were coated with 2 μg/ml protein (spike ectodomains of SARS-CoV-2, HKU1, OC43) for IgG measurement or 5 μg/ml (spike ectodomain of SARS-CoV-2) for IgM and IgA measurements in PBS at 4°C overnight. Plates were blocked with blocking buffer (BB) consisting of PBS supplemented with 5% nonfat dried milk powder (Carl Roth T145.2) for 60 min at RT. Monoclonal antibodies were tested with a starting concentration of 10 μg/ml in PBS, polyclonal IgGs with 500 μg/ml in BB and plasma samples with a starting dilution of 1:20 for IgG detection and 1:10 for IgM and IgA detection in BB. Monoclonal antibodies were serially diluted 1:5. Polyclonal IgGs and plasma were diluted in 1:4 serial dilutions. After incubation with samples for 90 min at RT, plates were incubated with anti-human IgG-HRP (Southern Biotech 2040-05) diluted 1:2500 in BB, anti-human IgM-HRP (Thermo Fisher Scientific A18835) diluted 1:2000 in BB or anti-human IgA-HRP (Thermo Fisher Scientific A18781) 1:2000 in BB. Plates were developed using ABTS solution (Thermo Fisher Scientific 002024) and absorbance was measured at 415 and 695 nm by a Microplate Reader (Tecan).
Anti-SARS-CoV-2 S1/S2 IgG and IgM antibody titers of plasma samples were also assessed using the automated DiaSorin’s LIAISON® SARS-CoV-2 S1/S2 protein ELISA kit according to the manufacturer’s instructions. IgG and IgM result values were interpreted with the following cut-off values: negative <12.0 AU/ml, equivocal ≥12.0 - < 15.0 AU/ml, and positive ≥15.0 AU/ml. Anti-SARS-CoV-2 S1 IgA antibody titers of plasma samples were also measured using the Euroimmun anti-SARS-CoV-2 ELISA on the automated system Euroimmun Analyzer I and S/CO values interpreted with following cut-off values: negative S/CO < 0.8, equivocal S/CO ≥ 0.8 - < 1.1, positive S/CO ≥ 1.1.
SARS-CoV-2 pseudovirus neutralization assays
SARS-CoV-2 pseudovirus expressing the Wu01 spike (EPI_ISL_406716) was generated by co-transfection of individual plasmids encoding HIV Tat, HIV Gag/Pol, HIV Rev, luciferase followed by an IRES and ZsGreen, and the SARS-CoV-2 spike protein into HEK 293T cells using the FuGENE 6 Transfection Reagent (Promega). Cell culture supernatants containing pseudovirus particles were harvested and stored at −80°C till use. The pseudovirus was titrated by infecting HEK293T cells expressing human ACE2 (Crawford et al., 2020). Following a 48-h incubation at 37 °C and 5% CO2, luciferase activity was determined by addition of luciferin/lysis buffer (10 mM MgCl2, 0.3 mM ATP, 0.5 mM Coenzyme A, 17 mM IGEPAL (all Sigma-Aldrich), and 1 mM D-Luciferin (GoldBio) in Tris-HCL) using a microplate reader (Berthold). For neutralization assays, a virus dilution with a relative luminescence unit (RLU) of at least at least1000-fold in infected cells versus non-infected cells was selected.
For testing neutralization at a single dilution, polyclonal IgG samples at a concentration of 1000 μg/ml, plasma samples at a dilution of 1:10, or mAbs at a concentration of 50 μg/ml, were co-incubated with pseudovirus supernatants for 1 h at 37°C, following which 293T-ACE-2 cells were added. After a 48 h incubation at 37°C and 5% CO2, luciferase activity was determined using the luciferin/lysis buffer. After subtracting background RLUs of non-infected cells, % of neutralization was calculated and the mean value was used for reporting. Each sample was tested in duplicates.
To determine IC50 values for mAbs a 3-fold dilution series of the antibody was performed starting with 50 μg/ml. IC50 values were calculated as the antibody concentration causing a 50% reduction in signal compared the virus-only controls using a dose-response curve in GraphPad Prism.
Authentic virus neutralization assays
Authentic virus neutralization was tested using a virus previously grown out from an oro-/naso-pharyngeal swab using VeroE6 cells (Vanshylla et al., 2021). For testing neutralization, plasma samples at a single dilution of 1:10 were co-incubated with the 2000 TCID50 virus for 1 h at 37°C, following which VeroE6 cells were added. After 4 days, cytopathic effects (CPE) were analysed under a bright-field microscope and neutralization was determined as the absence of CPE. Cells without any virus served as reference for lack of CPE and cells with virus only served as reference for positive CPE.
Quantification and statistical analysis
Flow cytometry analyses and quantification were performed using FlowJo10 software. Statistical tests and analyses were done with GraphPad Prism (v7 and v8), Python (v3.6.8), R (v4.0.0) and Mircosoft Excel for Mac (v14.7.3 and 16.4.8). CDRH3 lengths, V gene usage and germline identity distributions for clonal sequences (Figure 2C) were assessed for all input sequences without further collapsing. A two-tailed unpaired t test (Prism, GraphPad) (significance level, p < 0.05) was performed to test for the frequency differences of SARS-CoV-2-reactive IgG+ and IgG− B cells between pre-pandemic and convalescent blood samples (Figure 2A). Normality of data was evaluated using GraphPad Prism (v7 and v8). Data representation, dispersion and precision measures can be viewed in the figure legends. The absolute number of donors, samples, monoclonal antibodies or antibody sequences included (exact n value) in each experiment can be viewed in the figures.
Acknowledgments
We thank all study participants for supporting our research by blood donation; members of the Klein and Becker laboratories for their support and inspiring discussions; Raiees Andrabi, Victor Corman, and Jason McLellan for sharing and providing the SARS-CoV-2 and endemic HCoVs S ectodomain plasmids; Daniela Weiland and Nadine Henn for lab management and assistance; Sabine Adam of the Institute of Transfusion Medicine of the University Hospital of Cologne for organizing and providing blood donations of study participants; technical assistants of the serology department at the Institute of Virology of the University Hospital of Cologne for assistance with ELISA testing of plasma samples; as well as Verena Kraehling for sharing VeroE6 cells, and Susanne Berghöfer for excellent technical assistance with neutralization assays. This work was funded by grants from the German Center for Infection Research (DZIF) to F.K. and S.B., the German Research Foundation (DFG) CRC1310, European Research Council (ERC) ERC-stG639961, and COVIM: NaFoUniMed-Covid19 to F. Klein.
Author contributions
Conceptualization, F.K., M.S.E., L.G., C.K., and M.Z.; Methodology, M.S.E., L.G., S. Dähling, N.P., S. Detmer, M. Korenkov, K.V., V.D.C., S.H., Michael Klüver, and C.K.; Formal analysis, M.S.E., L.G., C.K., V.D.C, S.H., V.K., and S. Dähling; Investigation, M.S.E., L.G., C.K., V.D.C., S.H., V.K., S. Dähling, S. Detmer, M. Korenkov, and N.P.; Resources, B.G.; Writing – Original Draft, M.S.E., L.G., C.K., and F.K.; Writing – Review & Editing, all authors; Supervision, F.K. and C.K.; Funding acquisition, F.K. These authors contributed equally to the work: M.S.E. and L.G.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103951.
Supplemental information
Data and code availability
-
•
Nucleotide sequences of produced monoclonal and chimeric antibodies have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Further antibody sequences and NGS data will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo C.P., Le Sage V., Bolton M.J., Eilola T., Jones J.E., Kormuth K.A., Nturibi E., Balmaseda A., Gordon A., Lakdawala S.S., et al. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. Proc. Natl. Acad. Sci. 2020;117:17221–17227. doi: 10.1073/pnas.1920321117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoglio F., Meier D., Langreder N., Steinke S., Rand U., Simonelli L., Heine P.A., Ballmann R., Schneider K.-T., Roth K.D.R., et al. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat. Commun. 2021;12:1577. doi: 10.1038/s41467-021-21609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau N.V.V., Nhan L.N.T., Nguyet L.A., Tu N.T.K., Hong N.T.T., Man D.N.H., Ty D.T.B., Nhu L.N.T., Yen L.M., Khanh T.H., et al. Absence of SARS-CoV-2 antibodies in pre-pandemic plasma from children and adults in Vietnam. Int. J. Infect. Dis. 2021;111:127–129. doi: 10.1016/j.ijid.2021.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.S., Hung Chan K., Wing Chu K., Kwan S.W., Guan Y., Man Poon L.L., Peiris J.S.M. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M., Nitsche A., et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R., Williams K.L., Schmid M.A., Lai C.-Y., Patel B., Smith S.A., Crowe J.E., Wang W.-K., Harris E., de Silva A.M. Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. Plos Pathog. 2014;10:e1004386. doi: 10.1371/journal.ppat.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverría G., Guevara Á., Coloma J., Ruiz A.M., Vasquez M.M., Tejera E., de Waard J.H. Pre-existing T-cell immunity to SARS-CoV-2 in unexposed healthy controls in Ecuador, as detected with a COVID-19 Interferon-Gamma Release Assay. Int. J. Infect. Dis. 2021;105:21–25. doi: 10.1016/j.ijid.2021.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt S.A., Zehner M., Krähling V., Cohen-Dvashi H., Kreer C., Elad N., Gruell H., Ercanoglu M.S., Schommers P., Gieselmann L., et al. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 2019;25:1589–1600. doi: 10.1038/s41591-019-0602-4. [DOI] [PubMed] [Google Scholar]
- Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 2005;191:492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J., Bals J., Altomare C.G., St. Denis K., Lam E.C., Hauser B.M., Ronsard L., Sangesland M., Moreno T.B., Okonkwo V., et al. Naive human B cells engage the receptor binding domain of SARS-CoV-2, variants of concern, and related sarbecoviruses. Sci. Immunol. 2021;6:eabl5842. doi: 10.1126/sciimmunol.abl5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann L., Kreer C., Ercanoglu M.S., Lehnen N., Zehner M., Schommers P., Potthoff J., Gruell H., Klein F. Effective high-throughput isolation of fully human antibodies targeting infectious pathogens. Nat. Protoc. 2021;16:3639–3671. doi: 10.1038/s41596-021-00554-w. [DOI] [PubMed] [Google Scholar]
- Gombar S., Bergquist T., Pejaver V., Hammarlund N.E., Murugesan K., Mooney S., Shah N., Pinsky B.A., Banaei N. SARS-CoV-2 infection and COVID-19 severity in individuals with prior seasonal coronavirus infection. Diagn. Microbiol. Infect. Dis. 2021;100:115338. doi: 10.1016/j.diagmicrobio.2021.115338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Seydoux E., Wan Y.-H., Edara V.V., Stuart A.B., Feng J., Suthar M.S., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11:5413. doi: 10.1038/s41467-020-19231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkeller K., Wardemann H. Assessing human B cell repertoire diversity and convergence. Immunol. Rev. 2018;284:51–66. doi: 10.1111/imr.12670. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Loving C.L., Manischewitz J., King L.R., Gauger P.C., Henningson J., Vincent A.L., Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- Kowarz E., Löscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Döring M., Lehnen N., Ercanoglu M.S., Gieselmann L., Luca D., Jain K., Schommers P., Pfeifer N., Klein F. openPrimeR for multiplex amplification of highly diverse templates. J. Immunol. Methods. 2020;480:112752. doi: 10.1016/j.jim.2020.112752. [DOI] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.-C.D., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lefranc M.-P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman S.L., Hensley S.E. Antibodies with “original antigenic sin” properties are valuable components of secondary immune responses to influenza viruses. Plos Pathog. 2016;12:e1005806. doi: 10.1371/journal.ppat.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdoubi A., Michalski C., O’Connell S.E., Dada S., Narpala S., Gelinas J., Mehta D., Cheung C., Winkler D.F.H., Basappa M., et al. A majority of uninfected adults show preexisting antibody reactivity against SARS-CoV-2. JCI Insight. 2021;6:e146316. doi: 10.1172/jci.insight.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. 2010. Data Structures for Statistical Computing in Python. (Austin, Texas) pp. 56–61. [Google Scholar]
- Midgley C.M., Bajwa-Joseph M., Vasanawathana S., Limpitikul W., Wills B., Flanagan A., Waiyaiya E., Tran H.B., Cowper A.E., Chotiyarnwon P., et al. An in-depth analysis of original antigenic sin in dengue virus infection. J. Virol. 2011;85:410–421. doi: 10.1128/JVI.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsapaya J., Dejnirattisai W., Xu X., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S., Duangchinda T., Dong T., Rowland-Jones S., et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. MBio. 2020;11 doi: 10.1128/mBio.01991-20. e01991–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Li S., Li P., Pan W., Wang Q., Feng Y., Mo X., Yan Q., Ye X., Luo J., et al. Longitudinal analysis of T and B cell receptor repertoire transcripts reveal dynamic immune response in COVID-19 patients. Front. Immunol. 2020;11:582010. doi: 10.3389/fimmu.2020.582010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston D., Weisblum Y., Wise H., Templeton K., Jenks S., Hatziioannou T., Bieniasz P. Absence of severe acute respiratory syndrome coronavirus 2 neutralizing activity in prepandemic sera from individuals with recent seasonal coronavirus infection. Clin. Infect. Dis. 2020;73:e1208–e1211. doi: 10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinforma. 2020;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A.L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011;11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommers P., Gruell H., Abernathy M.E., Tran M.-K., Dingens A.S., Gristick H.B., Barnes C.O., Schoofs T., Schlotz M., Vanshylla K., et al. Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell. 2020;180:471–489.e22. doi: 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., et al. Next-Generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53:442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K., Di Cristanziano V., Kleipass F., Dewald F., Schommers P., Gieselmann L., Gruell H., Schlotz M., Ercanoglu M.S., Stumpf R., et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe. 2021;29:917–929.e4. doi: 10.1016/j.chom.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatti A., Monsalve D.M., Pacheco Y., Chang C., Anaya J.-M., Gershwin M.E. Original antigenic sin: a comprehensive review. J. Autoimmun. 2017;83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- von Boehmer L., Liu C., Ackerman S., Gitlin A.D., Wang Q., Gazumyan A., Nussenzweig M.C. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C., Chan K., Tsoi H., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg T., Pelleau S., Anna F., Attia M., Donnadieu F., Gravet A., Lohmann C., Seraphin H., Guiheneuf R., Delamare C., et al. Humoral immunity to SARS-CoV-2 and seasonal coronaviruses in children and adults in north-eastern France. EBioMedicine. 2021;70:103495. doi: 10.1016/j.ebiom.2021.103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Stacey H.D., Mullarkey C.E., Miller M.S. Original antigenic sin: how first exposure shapes lifelong anti–influenza virus immune responses. J. Immunol. 2019;202:335–340. doi: 10.4049/jimmunol.1801149. [DOI] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Nucleotide sequences of produced monoclonal and chimeric antibodies have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Further antibody sequences and NGS data will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.