Abstract
The mitochondrial unfolded protein response (UPRmt) is an evolutionarily conserved protective transcriptional response that maintains mitochondrial proteostasis by inducing the expression of mitochondrial chaperones and proteases in response to various stresses. The UPRmt-mediated transcriptional program requires the participation of various upstream signaling pathways and molecules. The factors regulating the UPRmt in Caenorhabditis elegans (C. elegans) and mammals are both similar and different. Cancer cells, as malignant cells with uncontrolled proliferation, are exposed to various challenges from endogenous and exogenous stresses. Therefore, in cancer cells, the UPRmt is hijacked and exploited for the repair of mitochondria and the promotion of tumor growth, invasion and metastasis. In this review, we systematically introduce the inducers of UPRmt, the biological processes in which UPRmt participates, the mechanisms regulating the UPRmt in C. elegans and mammals, cross-tissue signal transduction of the UPRmt and the roles of the UPRmt in promoting cancer initiation and progression. Disrupting proteostasis in cancer cells by targeting UPRmt constitutes a novel anticancer therapeutic strategy.
Keywords: Mitochondrial unfolded protein response, Cancer, Proteostasis, Mitochondrial heat shock protein, Mitochondrial protease
Background
Mitochondria are the power houses of cells, but their functions extend far beyond energy metabolism. Mitochondria play a vital role in the production of metabolic intermediates, calcium homeostasis, immune responses, cell differentiation, cell death, and the maintenance of proteostasis [1, 2]. Mitochondrial function requires the maintenance of mitochondrial homeostasis, which can be disrupted by many endogenous and exogenous stimuli. Therefore, a cytoprotective stress mechanism called the mitochondrial unfolded protein response (UPRmt) is activated upon mitochondrial damage and promotes mitochondrial recovery. UPRmt induces a transcriptional program involving numerous genes through retrograde mitochondrial-to-nucleus communication [3–5]. The UPRmt establishes a stable mitochondrial environment through the refolding of unfolded and misfolded proteins, degradation of damaged proteins through mitochondrial autophagy (mitophagy), regulation of mitochondrial biogenesis, and amelioration of oxidative stress [6–8].
The UPRmt, as an evolutionarily conserved mitochondrial stress response mechanism, can be induced in most organisms, such as Saccharomyces cerevisiae (S. cerevisiae), Caenorhabditis elegans (C. elegans), Drosophila, Homo sapiens, and Arabidopsis thaliana, to maintain mitochondrial proteostasis and function [9, 10]. Although different species may have unique signaling molecules upstream of UPRmt, the functional categories of UPRmt target genes show substantial overlap [10]. In summary, the UPRmt, a program for maintaining mitochondrial proteostasis, is indispensable in most organisms.
Inducers of the UPRmt
Inducers of the UPRmt in C. elegans
Numerous studies have shown that a large number of molecules or agents can activate the UPRmt (Table 1). Polyglutamine repeat protein (polyQ), an aggregation-prone protein, binds to mitochondria and triggers the UPRmt [11, 12]. Exposure of nematodes to nanoplastic particles induced the UPRmt in the intestine [13]. In addition, combined treatment with nanopolystyrene further enhanced the toxicity of microgravity stress to nematodes and induced UPRmt [14]. Statins, cholesterol-lowering drugs targeting the mevalonate pathway, have been shown to trigger the UPRmt [15]. Furthermore, a salicylic acid derivative, C8-SA, activates the UPRmt, and this response is associated with increased lifespan in C. elegans (Table 1) [16]. Through a screen of the C. elegans genome, a previous study showed that knockout of many mitochondrial process-related genes induced UPRmt, which explained the role of these genes in the maintenance of mitochondrial homeostasis (Table 2) [17]. These genes included those encoding mitochondrial large and small ribosomal subunits, transcription- and translation-related factors, tricarboxylic acid (TCA) cycle- and lipid metabolism-related proteins, mitochondrial chaperones and proteases, electron transport chain (ETC) components and ETC assembly factors (Table 2) [17–21]. The lack of respiratory chain components and assembly factors causes dysregulation of oxidative phosphorylation (OXPHOS), leading to accumulation of reactive oxygen species (ROS) and a decrease in the mitochondrial membrane potential. The decrease in the mitochondrial membrane potential acts as a signal to activate the UPRmt [17].
Table 1.
Inducers of the UPRmt
| Inducers | Functional role | Impact | References |
|---|---|---|---|
| Abnormal respiratory chain | Dysregulation of OXPHOS | ROS accumulation, decrease in the mitochondrial membrane potential | [17] |
| C8-SA | Activation of DAF-16 | Antioxidant stress | [16] |
| FUS proteinopathies | Interference of ATP synthase complex formation | Inhibition of mitochondrial ATP synthesis | [26] |
| Heat shock | Activation of HSF1, upregulation of mitochondrial chaperones | Maintenance of mitochondrial proteostasis | [64, 65] |
| Inflammation | Involvement of TNF-α | ROS accumulation, overload of damaged proteins | [24] |
| Microgravity stress | Not mentioned | ROS accumulation, decrease in locomotion behavior | [14] |
| Mitochondrial-related genes knockout | Disorder of mitochondrial function | Imbalance of mitochondrial homeostasis | [17–21] |
| Nanopolystyrene | Activation of ELT-2 signaling, Wnt signaling, and insulin signaling | ROS accumulation, decrease in locomotion behavior | [13] |
| Nicotinamide riboside | Activation of SIRT3 | Antioxidant stress | [22, 23, 58] |
| PolyQ | Direct interaction with the outer membrane of the mitochondria | ROS accumulation, decrease in the mitochondrial membrane potential | [11, 12] |
| Statins | Inhibition of HMG-CoA reductase in the mevalonate pathway | Interference of mitochondrial electron carriers | [15] |
| TDP-43 proteinopathies | Inhibition of ATP synthesis | abnormal cristae and a loss of cristae, ROS accumulation | [27] |
Table 2.
List of mitochondrial process-related genes associated with UPRmt
| Group | Gene name | Gene description | References |
|---|---|---|---|
| Mitochondrial large ribosomal subunits | mrpl-1 | Mitochondrial ribosomal protein L1 | [17, 20, 21] |
| mrpl-2 | Ribosomal L2 C domain-containing protein | [17, 20, 21] | |
| mrpl-9 | 39S ribosomal protein L9, mitochondrial | [17] | |
| mrpl-11 | Putative 39S ribosomal protein L11, mitochondrial | [17] | |
| mrpl-50 | 39S ribosomal protein L50, mitochondrial | [17, 21] | |
| Mitochondrial small ribosomal subunits | mrps-2 | Mitochondrial ribosomal protein, S2 | [17, 20, 21] |
| mrps-5 | Putative 28S ribosomal protein S5, mitochondrial | [17, 20, 21] | |
| mrps-18b | Mitochondrial ribosomal protein, S18b | [17, 20] | |
| mrps-24 | 28S ribosomal protein S24, mitochondrial | [17] | |
| mrps-35 | MRP-S28 domain-containing protein | [17, 20] | |
| Complex I ETC | nuo-3 | NADH ubiquinone oxidoreductase | [17, 21] |
| gas-1 | Putative NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 | [17, 20] | |
| lpd-5 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | [17, 21] | |
| Complex V ETC | atp-2 | ATP synthase subunit beta, mitochondrial | [17, 20, 21] |
| hpo-18 | Mitochondrial F1F0-ATP synthase, subunit epsilon/ATP15 | [17, 20] | |
| asb-1 | ATP synthase subunit b | [17] | |
| Mitochondrial ETC assembly factors | sco-1 | Putative cytochrome C oxidase assembly protein | [17, 20] |
| cox-18 | Cytochrome oxidase assembly protein | [17] | |
| Mitochondrial import | dnj-21 | Mitochondrial import inner membrane translocase subunit TIM14 | [17, 20, 21] |
| gop-3 | SAM50-like protein gop-3 | [17] | |
| Lipid metabolism | acdh-13 | Acyl CoA dehydrogenase | [17] |
| mecr-1 | Enoyl-[acyl-carrier-protein] reductase | [17, 21] | |
| Translation elongation factors | tufm-1 | Elongation factor Tu, mitochondrial | [17, 20] |
| gfm-1 | Elongation factor G, mitochondrial | [17] | |
| Mitochondrial proteases | spg-7 | AFG3-like protein spg-7 | [17, 21] |
| atad-3 | ATPase family AAA domain-containing protein 3 | [17] | |
| Mitochondrial chaperones | hsp-60 | Chaperonin homolog Hsp-60, mitochondrial | [17, 20, 21] |
| phb-2 | Mitochondrial prohibitin complex protein 2 | [17, 20, 21] | |
| Mitochondrial transcription | hoe-1 | Ribonuclease Z | [17] |
Inducers of the UPRmt in mammals and other animals
Mitochondrial proteostasis impairment is one of the most common inducers of the UPRmt (Table 1). Inhibition of mitochondrial chaperones, proteases or electron transfer complexes strongly triggers initiation of the UPRmt. In addition, nicotinamide riboside, a precursor of NAD+, can activate UPRmt, accompanied by a significant increase in the level of UPRmt-related proteins [22, 23]. In airway smooth muscle, inflammation-induced accumulation of ROS and overload of damaged proteins activate the UPRmt, and the proinflammatory cytokine TNF-α plays a crucial role in this event [24]. Moreover, caloric restriction can activate the UPRmt to improve mitochondrial function, and miRNAs are involved as key mediators [25]. Heat shock elicits the production of various molecular chaperones, including mitochondrial heat shock proteins (HSPs) (Table 1). As a master inducer of a class of neurodegenerative diseases, fused in sarcoma (FUS) proteinopathies, FUS interacts with the catalytic subunit of mitochondrial ATP synthase, which interferes with the formation of ATP synthase complexes and hinders the production of energy, thus triggering UPRmt activation in Drosophila (Table 1) [26]. Additionally, mutations or dysregulation of TDP-43 can cause TDP-43 proteinopathies. Mitochondrial impairment is one of the characteristics of TDP-43 proteinopathies; morphologically, mitochondrial cristae exhibit reduced numbers and abnormal phenotypes in animal experiments and patient brain samples. In mammalian cell and Drosophila models, increased TDP-43 expression inhibits ATP synthesis and accelerates ROS production, thus activating the UPRmt (Table 1) [27].
Regulation of the UPRmt
Mitochondrial retrograde signaling pathways in Saccharomyces cerevisiae
Retrograde signaling in S. cerevisiae is the first retrograde pathway and has been elucidated in great detail [28, 29]. The most important event in retrograde signaling in S. cerevisiae is the nuclear translocation of retrograde transcription factors from the cytoplasm, including Rtg1p, Rtg2p, and Rtg3p [28–30]. Rtg1p and Rtg3p are basic helix-loop-helix/leucine zipper proteins and form a heterodimer to bind to the DNA binding site R box (GTCAC), thus regulating the expression of various genes that encode mitochondrial proteins. The translocation of this heterodimeric transcription factor is partially regulated by the dephosphorylation of Rtg3p, which is mediated by Mks1p when it binds to the 14-3-3 protein Bmh1p or Bmh2p [28–30]. Rtg2p is also an activator of this retrograde signaling pathway. Rtg2p facilitates the dephosphorylation of Rtg3p by binding to Mks1p and inhibiting Mks1p to form a complex with Bmh1p/Bmh2p, a complex maintaining Rtg3p in a hyperphosphorylated state [31, 32]. Interestingly, there are currently no reports strictly related to UPRmt in yeast. Although there is no signaling pathway strictly classified as UPRmt in yeast, very similar mechanisms exist that repair mitochondrial dysfunction by activating the expression of nuclear genes.
The UPRmt in C. elegans
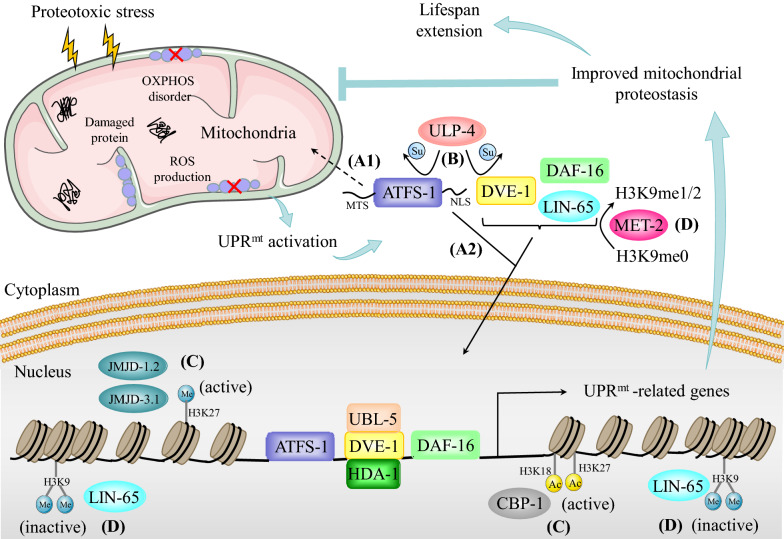
A multicellular eukaryote, C. elegans has been widely used as a model organism in genetics, developmental biology, neurobiology, and molecular biology. Studies using C. elegans as a model organism have gradually deepened our understanding of the UPRmt and have revealed its components and regulatory pathways. The most well-known regulator of the UPRmt in C. elegans is activating transcription factor associated with stress-1 (ATFS-1) (Fig. 1). The amino terminus of ATFS-1 contains the mitochondrial targeting sequence (MTS). Under normal conditions, ATFS-1 enters the mitochondrial matrix in an MTS-dependent manner through channel proteins located on the mitochondrial membrane and is degraded by the protease Lon peptidase 1 (LONP1) [4, 33]. Under mitochondrial stress conditions, a decrease in the mitochondrial transport efficiency of ATFS-1 causes it to be retained in the cytoplasm. The nuclear localization sequence (NLS) located at the carboxyl terminus of ATFS-1 mediates its nuclear transport, and after nuclear translocation, ATFS-1 functions as a transcription factor to drive the UPRmt transcriptional program (Fig. 1) [17, 33]. When the ETC is disrupted, a large proportion of ATFS-1 eventually enters the nucleus. Subsequently, ATFS-1 fine-tunes the expression of OXPHOS-related genes (such as atp-3, nuo-4 and nduf-6) and TCA cycle-related genes (such as aco-2, idh-1 and pyc-1) in both the nucleus and mitochondria to adapt the metabolic capacity to the limited protein folding capacity, thereby reducing the load of misfolded proteins and promoting the recovery of OXPHOS [4]. In addition, when proteostasis is destroyed, DVE-1 and UBL-5 form a complex; DVE-1 then binds to the promoters of genes encoding mitochondrial chaperones, including heat shock protein 60 (HSP60), activating their transcription to maintain proteostasis (Fig. 1) [34]. Moreover, a genome-wide RNAi screen identified ULP-4 as a regulator of UPRmt [35]. Knockdown of ULP-4 greatly impaired the activation of UPRmt, but not ER stress or the heat shock response (HSR), in C. elegans. Yeast two-hybrid screening identified two key substrates of ULP-4: DVE-1 and ATFS-1 [35]. Surprisingly, SUMOylation influences DVE-1 and ATFS-1 through two different molecular mechanisms. Normally, DVE-1 is SUMOylated on the K327 residue. Knockdown of ULP-4 significantly increased the SUMOylation level of DVE-1, thus impeding the nuclear translocation of DVE-1 and suppressing the activation of UPRmt during mitochondrial stress [35]. ATFS-1 is another substrate of ULP-4. ULP-4 interacted with and deSUMOylated ATFS-1 at residue K326 following mitochondrial stress. Although knockdown of ULP-4 did not affect the nuclear translocation of ATFS-1, the protein stability and transcriptional activity of ATFS-1 were greatly decreased (Fig. 1) [35]. These posttranslational modifications enhance innate immunity and prolong the lifespan of nematodes. The NAD+-dependent protein deacetylase sir-2.1 leads to activation of the FOXO transcription factor DAF-16 and promotes DAF-16 to induce the expression of antioxidant genes when proteostasis is dysregulated. Activation of UPRmt prolongs the lifespan of nematodes after exogenous addition of the NAD+ precursor increases the NAD+ level [36].
Fig. 1.
Mechanism of UPRmt in C. elegans. When mitochondrial proteostasis is disturbed, signaling molecules of UPRmt mediate mitochondria-to-nucleus retrograde communication. Transcription factors, including ATFS-1, DVE-1, and DAF-16, are activated by upstream signals and bind to the promoters of target genes, thus inducing the transcription of UPRmt-related genes. A1 Under nonstress conditions, ATFS-1 enters the mitochondria through the MTS and is subsequently degraded. A2 Under mitochondrial stress conditions, ATFS-1 is transported to the nucleus in an NLS-dependent manner. B ULP-4-mediated deSUMOylation of ATFS-1 and DVE-1 enhances ATFS-1- and DVE-1-dependent transcription programs. C JMJD-1.2-, JMJD-3.1- and CBP-1-dependent epigenetic modifications facilitate the formation of a nucleosome conformation that is conducive to transcription. D Additionally, MET-2- and LIN-65-mediated chromatin silencing of non-UPRmt gene regions is crucial for triggering UPRmt. Consequently, the activation of UPRmt promotes the recovery of mitochondria from damage, enhances mitochondrial function, and prolongs the lifespan of C. elegans
Changes in chromatin structure also play a substantial role in the UPRmt via a mechanism related to epigenetic modification [37]. H3K9 dimethylation (H3K9me2) mediated by the histone methyltransferase MET-2 and its cofactor LIN-65 silences the expression of some genes, while ATFS-1 and DVE-1 synergistically promote the transcription of UPRmt genes in nonsilenced regions (Fig. 1). Interestingly, the nuclear enrichment of DVE-1 and LIN-65 is interdependent [38]. The nematode HDA-1, a homologous protein of mammalian HDAC, interacts with DVE-1 to activate the UPRmt [39]. The histone demethylases JMJD-1.2 and JMJD-3.1 cooperate to promote the epigenetic transition from H3K27me3 to H3K27me1, inducing the transcription of UPRmt effectors and delaying senescence in nematodes (Fig. 1) [40]. A recent study indicated that the acetyltransferase CBP-1 mediates the acetylation of histone H3 at K18 and K27, which facilitates the binding of transcription factors to the promoters of UPRmt genes (Fig. 1) [41]. Although great progress has been made in understanding the regulation of UPRmt in nematodes, the more precise regulatory mechanism is still unclear. The scientific question of how ATFS-1, DVE-1 and DAF-16 coordinate their functions to drive transcription programs needs to be further explored.
The cell-nonautonomous UPRmt
The nervous system plays a vital role in the maintenance of organism homeostasis in C. elegans [42]. Previous studies have revealed that mitochondrial stress in nematode neurons can be transmitted to distal tissues and activate the UPRmt in a nonautonomous manner [18, 43]. The substances released from neurons, collectively known as mitokines, are involved in activating the UPRmt in distal tissue [44, 45]. Downregulation of cco-1 in C. elegans neurons triggers the secretion of Wnt, subsequently leading to activation of the Wnt signaling-dependent UPRmt in peripheral tissues, which regulates the organism’s mitochondrial homeostasis [18]. In C. elegans, neurons with abnormal respiratory chain function and ROS accumulation release serotonin, which requires UNC-31-mediated neurosecretion. Immediately after its release, serotonin acts on the distal intestine, activates the UPRmt and triggers adaptive metabolic changes in response to proteotoxic stress [11]. Activation of FSHR-1 in neurons promotes the function of SPHK-1 in the intestine and consequently triggers UPRmt activation in response to stress [46]. Under stimulation by neuroendocrine signals, SPHK-1 localizes to the mitochondrial membrane and catalyzes the conversion of SPH to S1P, which participates in the activation of the cell-nonautonomous UPRmt as an early event. SPHK-1 mutants lacking kinase activity or mitochondrial localization ability cannot effectively induce UPRmt [47]. In addition, neural circuits mediated by three types of sensory neurons (ASK, AWA and AWC) and an interneuron (AIA) are involved in the sensing and communication of neuronal mitochondrial dysfunction, in which the neuropeptide FLP-2 plays a crucial role [43]. In summary, the nervous system of C. elegans systematically coordinates the cellular nonautonomous UPRmt through the release of endocrine signals called mitokines, which facilitate the communication of information across cells and tissues to regulate metabolism, maintain homeostasis and prolong lifespan. Moreover, communication between germline and intestinal cells can also be conducted through endocrine signals, and translational repression of CYC-2.1 mediated by the RNA-binding protein GLD-1 in germline cells is involved in this process [48].
The UPRmt in Drosophila
Research on UPRmt with Drosophila as the experimental material has made a significant contribution to our current understanding of UPRmt. A previous study showed that mild muscle mitochondrial damage maintains mitochondrial function, inhibits the deterioration of muscle structure and function, and extends the lifespan of Drosophila [49]. The mechanism underlying this phenomenon involves redox-dependent induction of UPRmt-related genes and systemic repression of insulin signaling via the Drosophila ortholog of insulin-like growth factor-binding protein 7 (IGFBP7) [49]. Phosphoglycerate mutase 5 (PGAM5), a mitochondrial phosphatase, is cleaved by the rhomboid protease PARL and released from membranes during mitochondrial stress. Numerous studies have indicated that PGAM5 is involved in the regulation of mitochondrial homeostasis not only by activating mitochondrial biogenesis and mitophagy but also by inducing excessive mitochondrial fission and different types of cell death, such as apoptosis and necroptosis [50–52]. In Drosophila, the mitochondrial membrane protein PGAM5 senses mitochondrial stress and activates the transcription factor FoxO through ASK1 and JNK [53]. Persistent activation of FoxO upregulates the expression of multiple chaperones, thereby promoting the recovery of mitochondria and extending the lifespan of organisms [53].
The UPRmt in mammals
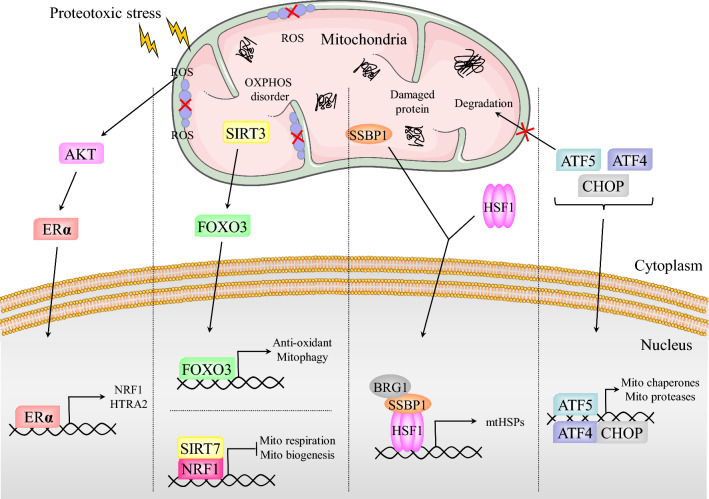
The factors and mechanisms that regulate UPRmt in mammals are more complex than those in C. elegans. However, our understanding of the mammalian UPRmt is far less advanced than our understanding of the C. elegans UPRmt. Proteostasis is essential for cell survival, and the ClpXP complex protease plays a key role in maintaining proteostasis. The ClpXP complex is composed of two subunits: ClpX and the caseinolytic mitochondrial matrix peptidase proteolytic subunit (ClpP). Overexpression of ClpX leads to upregulation of some genes associated with UPRmt, suggesting that ClpX is involved in the initiation of UPRmt [54]. The mitochondrial matrix peptidase ClpP not only maintains mitochondrial proteostasis by degrading unfolded and misfolded proteins but also mediates UPRmt induction. A decrease in the ClpP level weakens the UPRmt in mouse cells, resulting in mitochondrial dysregulation [55]. The results of previous studies indicate that CHOP is activated by UPRmt. In turn, CHOP transcriptionally upregulates the molecular chaperones HSP10 and HSP60 to increase the protein folding ability of mitochondria (Fig. 2) [56]. Notably, CHOP is also involved in regulating the endoplasmic reticulum unfolded protein response (UPRER). However, disruption of mitochondrial proteostasis does not induce the UPRER, indicating that CHOP triggers the expression of only mitochondria-localized stress proteins. The selective and specific induction of CHOP during the UPRmt may be caused by the activation of AP-1 [56]. Furthermore, ATF4 is reported to be involved in the UPRmt by resetting cellular metabolism [5]. More importantly, as a mammalian homolog of ATFS-1, activating transcription factor 5 (ATF5) is similar to ATFS-1 in its regulatory mechanisms and transcriptional programs induced in the UPRmt process (Fig. 2) [3]. ATF5 also possesses an MTS and an NLS. ATF5 is transported to mitochondria via the TIM-TOM complex under nonstress conditions by recognition of its MTS and is subsequently degraded. Under overload of misfolded and unfolded proteins as well as protein aggregation in mitochondria, ATF5 translocates to the nucleus by recognition of its NLS and upregulates a variety of molecular chaperones and proteases to promote mitochondrial recovery [3].
Fig. 2.
Mechanism of UPRmt in mammals. Upon ROS accumulation in IMS, the AKT-ERα axis is activated to trigger the transcription of NRF1 and HTRA2 in response to IMS damage. When ATF5 senses mitochondrial disorders, it translocates to the nucleus together with ATF4 and CHOP to synergistically promote the expression of mitochondrial chaperones and proteases. HSF1 forms a complex with SSBP1 and recruits the chromatin remodeling factor BRG1, consequently inducing the expression of mtHSPs. The SIRT3-FOXO3 axis is involved in antioxidant stress and mitophagy. The interaction of NRF1 and SIRT7 inhibits mitochondrial respiration and biogenesis, thereby reducing the load of damaged proteins. In summary, the mammalian UPRmt promotes mitochondrial recovery and maintains proteostasis through a variety of pathways
Previous studies revealed that the deacetylase SIRT3 protects cells from mitochondrial damage through upregulation of antioxidant activity and mitophagy (Fig. 2) [57]. Mechanistically, SIRT3 deacetylates FOXO3 at K271 and K290 in response to the accumulation of ROS and a decrease in the mitochondrial membrane potential, and the resulting activated FOXO3 is then redistributed into the nucleus and transcriptionally upregulates antioxidant- and mitophagy-related genes (Fig. 2). Among these events, FOXO3-mediated upregulation of PGC-1α and SOD2 plays a critical role [58, 59]. As an important mediator of antioxidant stress in the UPRmt, SOD2 plays an indispensable role in reducing the level of mitochondrial ROS [60]. Additionally, activation of the SIRT1/UPRmt/SOD signaling axis is involved in the elimination of excess mitochondrial ROS [36]. In breast cancer cells, the accumulation of misfolding-prone proteins in the mitochondrial intermembrane space (IMS) interferes with ETC homeostasis, leading to an increase in the level of ROS in the IMS and subsequently triggering the activation of estrogen receptor α (ERα or ESR1) (Fig. 2). ERα regulates the expression of its target genes NRF1 and HTRA2. The protease HTRA2 degrades misfolded proteins in the IMS to ameliorate IMS stress [61]. Upon protein folding stress, NRF1, a major mitochondrial regulator, binds to the histone deacetylase SIRT7 (Fig. 2). NRF1 transports SIRT7 to the promoter region of target genes and suppresses their transcription by compressing the local region of chromatin [62]. The NRF1-SIRT7 complex regulates energy metabolism by inhibiting mitochondrial respiration and balances suboptimal protein folding and degradation capabilities by inhibiting mitochondrial biogenesis (Fig. 2) [62]. Inhibition of protein translation is an important component of the UPRmt. In response to the disruption of proteostasis, cells rapidly express high levels of mitochondrial chaperones, while UPRmt reduces the biogenesis of mitochondria-localized proteins by impeding pre-RNA processing and translation. Through this mechanism, a balance between the enhanced protein folding capacity and the reduced protein folding load is achieved, enabling accelerated repair of cellular injury [63].
The mammalian histone demethylases PHF8 and JMJD3, homologs of C. elegans JMJD-1.2 and JMJD-3.1, respectively, induce the expression of UPRmt-related genes by altering epigenetic patterns [40]. Mammalian CBP/p300-dependent H3K18Ac and H3K27Ac participate in the formation of an active chromatin state, thereby promoting the transcription of UPRmt genes [41]. In addition, inhibition of HDAC1/2 suppresses the transcriptional program mediated by UPRmt in mammalian cells [39].
Our previous studies have shown that heat shock factor 1 (HSF1), a master regulator of the heat shock response (HSR), is also involved in UPRmt [64]. Activated HSF1 recruits mitochondrial single-stranded DNA binding protein 1 (SSBP1) to the promoters of cytoplasmic and mitochondrial chaperone genes (Fig. 2). Subsequently, the HSF1-SSBP1 complex recruits the chromatin remodeling factor BRG1, thereby promoting the formation of the chromatin remodeling complex [64]. The upregulation of the mitochondrial chaperones HSP10, HSP60 and mitochondrial heat shock protein 70 (mtHSP70) induced by HSF1 participates in UPRmt to combat proteotoxicity stress (Fig. 2) [64–66]. RNA-seq analysis identified that interferon-α inducible protein 6 (IFI6) confers radioprotection in skin cells. IFI6 translocates into nucleus in response to radiation and interacts with SSBP1 to increase the transcriptional activity of HSF1 [67]. Recently, a mitochondrial stress-specific variant of HSF1, dephosphorylated by the PP2A complex, was found in C. elegans upon ETC impairment and exhibited a protective role in age-related dysfunction of proteostasis by selectively upregulating the expression of small HSPs [68]. Therefore, it will be interesting to explore whether mitochondrial localization of HSF1 exists in mammalian cells. Overall, although increased research has gradually improved the framework of UPRmt regulation in mammals, the details of the regulation of each branch of UPRmt are still unclear. With the increasing depth of research, more proteins involved in UPRmt will be discovered, and the potential relationships among branches will be clarified.
Relationships between Ca2+ regulation, mitophagy, ISR and UPRmt
The UPRmt and Ca2+ regulation
Calcium, as a key intracellular second messenger, plays a pivotal role in many physiological and pathological processes. Ca2+ transfer from the endoplasmic reticulum (ER) to mitochondria is the main source of mitochondrial Ca2+, which is mediated by mitochondria-associated membranes (MAMs). Ca2+ regulation in mitochondria is involved in various biological processes, including ATP synthesis and metabolism [69, 70]. Previous studies have shown that the decline in brain metabolic activity during aging is associated with mitochondrial dysfunction. Disorders of OXPHOS, imbalanced Ca2+ buffering, and dysregulation of UPRmt-related proteins lead to neuronal decline during aging [71]. Under ER stress, Ca2+ is transported to mitochondria through the MAM, which ultimately leads to increased expression of the mitochondrial protease LONP1 [72, 73]. Recent studies have demonstrated that LONP1 is required for the maintenance of mitochondrial proteostasis and gene expression. LONP1 depletion leads to loss of mitochondrial DNA (mtDNA), impaired ribosome biogenesis, accumulation of insoluble protein aggregates in the matrix, stabilization of PINK1, and activation of the integrated stress response (ISR) [74]. However, the exact relationship between mitochondrial Ca2+ signaling and UPRmt has not been elucidated. The potential crosstalk between Ca2+ regulation in mitochondria and UPRmt urgently needs to be explored.
The UPRmt and mitophagy
Mitochondria are the first-line defense of cells under various stresses. Dysfunctional mitochondria can be repaired by a complex set of adaptive responses, such as mitochondrial biogenesis, mitochondrial fission/fusion, UPRmt and mitophagy. UPRmt and mitophagy are two major axes that maintain mitochondrial proteostasis [75]. As discussed above, UPRmt facilitates the recovery and survival of cells by inducing the expression of mitochondrial chaperones, proteases, antioxidant genes and protein import and assembly factors. When damaged mitochondria cannot be accurately repaired by a small-scale stress response, such as UPRmt, dysfunctional mitochondria can be removed by mitophagy. Mitophagy targets damaged mitochondria through autophagosome phagocytosis and lysosomal degradation to ensure that they are cleared before they become toxic to cells [76–78]. The molecular mechanisms of PINK1/Parkin- and BNIP3/NIX-regulated mitochondrial clearance have previously been reviewed in detail [76–78]. Therefore, it is speculated that UPRmt appears to be activated prior to mitophagy. Inconsistent with this conclusion, a recent study revealed that UPRmt was a downstream pathway of mitophagy [79, 80]. Both UPRmt and mitophagy were slightly activated in LPS-treated cardiomyocytes to sustain mitochondrial function. Treatment with urolithin A, an inducer of mitophagy, significantly reduces sepsis-mediated heart injury by restoring mitochondrial function without influencing the UPRmt [80]. However, deletion of FUN14 domain containing 1 (FUNDC1), a mammalian mitophagy receptor localized on the outer mitochondrial membrane, greatly increased the expression of UPRmt-related genes in LPS-treated mouse hearts [80]. In contrast, the enhancement of UPRmt by oligomycin effectively alleviated sepsis-induced mitochondrial damage and myocardial dysfunction. This cardioprotective effect was not evident in FUNDC1 CKO mice [80]. Moreover, when UPRmt was suppressed, mitophagy-mediated cardiac protection was partially attenuated [80]. In summary, UPRmt and mitophagy are important quality control mechanisms. However, the precise molecular mechanism by which mitophagy modulates UPRmt has not been clarified. Whether other classic mitophagy signaling pathways and regulatory proteins mediate UPRmt also needs to be further verified by in vivo and in vitro experiments.
The UPRmt and integrated stress response (ISR)
The ISR is also an evolutionarily conserved cellular signaling pathway that facilitates cells and tissues to cope with various stresses, such as ER stress, heme deficiency, amino acid starvation, viral infection and hypoxia [81–83]. A large amount of evidence from mammals highlights that the ISR is the core element of UPRmt. In mammalian cells, ISR assists in the regulation of UPRmt, and its molecular mechanisms depend on the phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α), thereby suppressing the ability of eIF2 to transmit methionylated initiator transfer RNA (Met-tRNAi) to ribosomes [84]. The ancestral kinase general control nonderepressible 2 (GCN2) mediates phosphorylation of eIF2α during nutrient deprivation in S. cerevisiae. GCN-2-dependent phosphorylation of eIF2α is necessary for the development and extension of lifespan in C. elegans. Depletion of GCN-2 could significantly upregulate the expression of mitochondrial chaperones to activate UPRmt, suggesting that GCN-2-dependent translation attenuation is a parallel signaling pathway to maintain mitochondrial proteostasis during mitochondrial stress. In mammalian cells, four kinases have been identified to phosphorylate eIF2α, including GCN2, PKR-like endoplasmic reticulum kinase (PERK), protein kinase R (PKR), and heme-regulated inhibitor (HRI), in response to different forms of cellular stress [84, 85]. GCN2 is associated with ribosomes and activated by amino acid depletion. PERK, an ER membrane protein, is stimulated by the presence of unfolded or misfolded proteins in the ER. PKR is activated by the accumulation of double-stranded RNA derived from mitochondria in the cytoplasm, thus promoting eIF2α phosphorylation-mediated ISR [86]. HRI activity is regulated by the depletion of heme [84, 85, 87]. Recently, two outstanding studies revealed that mitochondrial stress induced HRI activation to promote eIF2α phosphorylation even in the presence of full heme [88, 89]. In addition, they found that OMA1, a protease localized on the inner mitochondrial membrane, cleaved the DELE1 protein. A fragment of DELE1 accumulated in the cytosol and then interacted with and phosphorylated eIF2α. Inhibition of the OMA1-DELE1-HRI signaling pathway induced the expression of specific molecular chaperones [88, 89]. These studies further established crosstalk between mitochondrial dysfunction and ISR. Phosphorylation of eIF2α leads to a reduction in the overall translation of protein synthesis to adapt to environmental changes. Paradoxically, several mRNAs containing upstream open reading frames (uORFs) can be selectively translated during ISR [81, 82]. The mRNA sequences of the transcription factors CHOP, ATF4 and ATF5 contain uORFs and require eIF2α phosphorylation to initiate their translation. These transcription factors are involved not only in the ER stress response but also in the regulation of UPRmt. The accumulation of unfolded proteins in the mitochondrial matrix of mammalian cells leads to CHOP-dependent transcriptional upregulation of mitochondrial chaperones and proteases but not ER stress proteins [90]. CHOP binding elements were identified in some gene promoters, such as HSP60, HSP10, mtDNAJ, ClpP, YME1L1, MPPβ, TIM17A, NDUFB2, endonuclease G and thioredoxin 2 [90, 91]. Moreover, four different mitochondrial stressors, doxycycline, actinonin, FCCP and MitoBloCK, significantly activated ATF4 to modulate ISR activation, reduce mitochondrial ribosomal proteins and suppress mitochondrial translation [5]. A recent study revealed that the noncanonical initiation factors eIF2D and DENR also participated in the translational induction of ATF4 during ISR [92, 93]. In contrast to ATF4, ATF5 seems to directly participate in the UPRmt process [3, 6]. Silencing ATF5 inhibited the induction of UPRmt-related genes during mitochondrial stress. Interestingly, overexpression of ATF5, but not ATF4, in worms lacking ATFS-1 restored the ability to induce the expression of HSP60 [3, 6, 94]. All the above studies indicated the overlapping network between the ISR and the UPRmt through the specific activation of CHOP, ATF4 and ATF5.
The imbalance of the ISR pathway is related to a variety of diseases ranging from neurodegenerative diseases to tumors, reflecting the importance of cell stress adaptation to maintain health [81–83]. The crosstalk between ISR and UPRmt promotes tumor progression. GCN2-eIF2α-ATF4 axis has been shown to enhance tumor cell proliferation by maintaining metabolic homeostasis [95]. Additionally, the GCN2-eIF2α-ATF4 pathway also induces the expression of xCT, which promotes the synthesis of glutathione (GSH), which is involved in ferroptosis and ultimately leads to cisplatin resistance in gastric cancer cells [96]. However, there are still some unresolved problems [97]. For example, the consequences of ISR activation in the mitochondrial stress response need to be further explored. Although the initial purpose of ISR and the mitochondrial stress response is to protect cells against stress and avoid death, which is more important for protective effects? An in-depth understanding of the specific roles and cross-pathways of CHOP, ATF4 and ATF5 in UPRmt and ISR may provide unexpected answers.
The UPRmt and cancer
The initiation and development of cancer is a multistep process that involves the acquisition of diverse functions, such as resistance to cell death, prevention of growth inhibition, and activation of proliferation signals [98]. Moreover, cancer cells are constantly under intracellular and extracellular pressure. Mutations in mtDNA, changes in metabolism, alterations in energy and oxygen requirements, and overload of mitochondrial unfolded and misfolded proteins lead to mitochondrial dysfunction. To manage various stresses, activation of the UPRmt in cancer cells maintains proteostasis and regulates metabolic reactions. Furthermore, mtDNA mutations mediate cancer metastasis by activating the UPRmt [99]. Notably, not all mtDNA mutations can be exploited by cancer cells, and only those specific genomic mtDNA landscapes that activate the UPRmt can be utilized by metastatic cancers [99–101].
Numerous studies have shown that activation of the UPRmt is indispensable for the development and progression of cancer [72, 82]. The UPRmt promoted by mitohormesis in cancer cells plays a critical role in stimulating the invasion and metastasis of cancer cells [102]. For example, high expression of UPRmt-related genes is significantly associated with poor overall survival and metastasis-free survival in breast cancer patients [102]. Evidence supporting the upregulation of UPRmt components in breast cancer suggests that UPRmt activation is involved in the progression of breast cancer [103]. In addition, single-nucleotide polymorphisms in genes encoding UPRmt components are associated with an increased risk of head and neck cancer [104]. Strikingly, under mitochondrial stress, UPRmt mediates the secretion of the mitokine GDF15, which in turn promotes the invasion of thyroid cancer cells [105].
In principle, the UPRmt facilitates cancer progression by inhibiting cancer cell death and promoting cancer growth. In the following section, we introduce the role of the signaling molecules and several proteins in the UPRmt transcriptional program in cancer development. A further understanding of the signaling molecules in the UPRmt and the role of UPRmt-related proteins in cancer development is expected to provide a new therapeutic strategy for cancer.
The roles of upstream signaling molecules in the UPRmt in cancer
Many signaling molecules in the UPRmt, including components of the CHOP/ATF4/ATF5, ERα and HSF1-SSBP1 axis, play an important role in tumorigenesis. These molecules promote tumorigenesis through various mechanisms in many tumors.
ATF5 and cancer
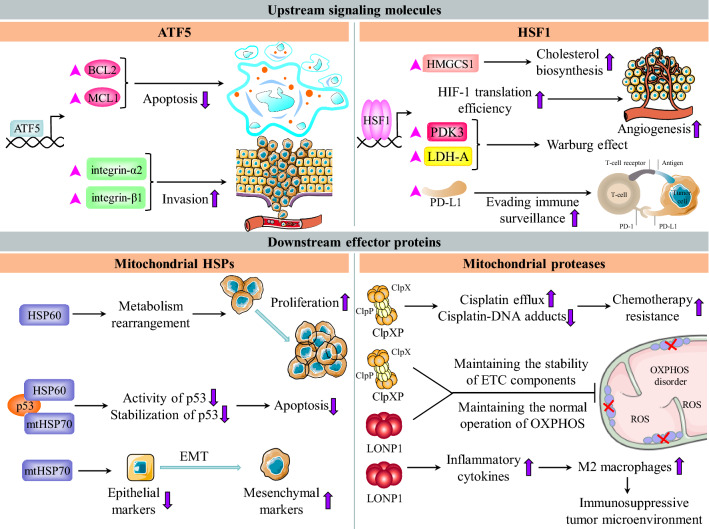
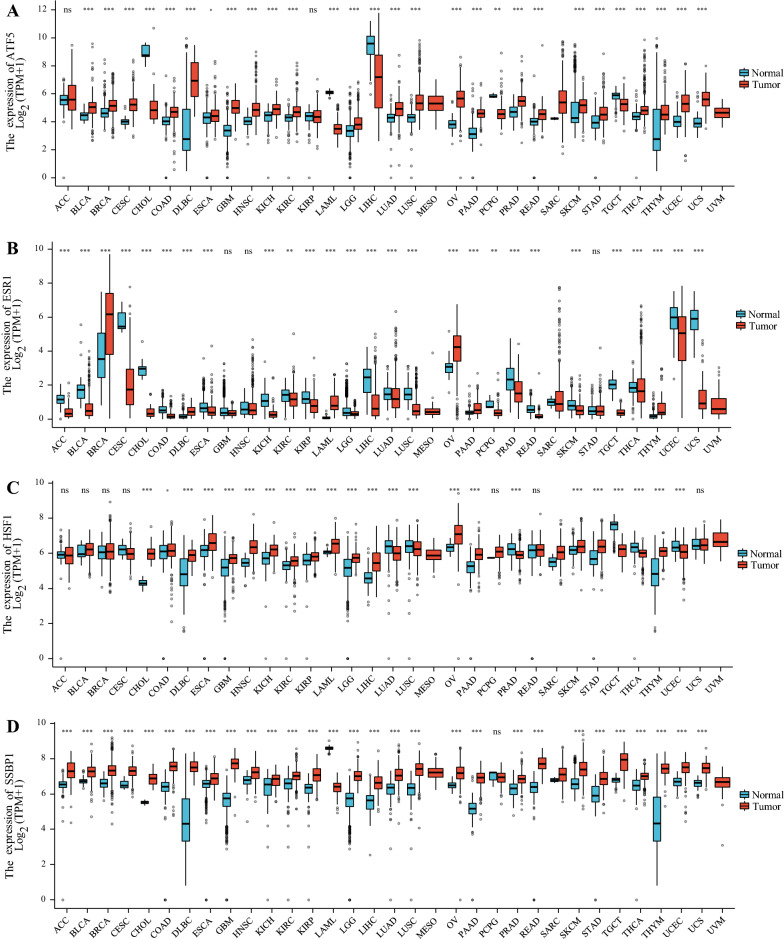
Previous studies have shown that ATF5 expression is significantly upregulated in a variety of cancers, such as epithelial ovarian cancer, glioblastoma, pancreatic cancer and chronic myeloid leukemia (Figs. 3, 4A) [106–109]. One mechanism underlying the upregulation of ATF5 expression in cancer is the alteration of epigenetic modification. For example, the methylation level in the promoter region of the ATF5 gene in glioma is significantly reduced compared with that in normal tissues. Moreover, this reduction in the methylation level is accompanied by a reduction in glioma differentiation [110]. In addition, upregulation of ATF5 transcription by the BCR-ABL/PI3K/AKT/FOXO4 signaling pathway and CREB3L2 is responsible for the high expression level of ATF5 in cancer [109, 111]. Mechanistically, ATF5 transcriptionally upregulates the expression of antiapoptotic proteins BCL2 and MCL1, hindering the apoptosis of cancer cells and reducing their chemosensitivity (Fig. 3) [106, 108, 111]. Activated ATF5 triggers the transcription of mTOR, a negative regulator of autophagy, inhibiting autophagy in cancer cells [109]. Additionally, ATF5-induced upregulation of integrin-α2 and integrin-β1 is conducive to cancer cell invasion (Fig. 3) [107]. In a previous study, a dominant-negative ATF5 mutant (DN-ATF5) lacking DNA binding ability, which blocked some signaling molecules in cells, was synthesized. The application of DN-ATF5 in cancer cell lines revealed its efficacy in reducing cancer cell viability [112]. Due to the promotive role of ATF5 in cancer, ATF5 expression is positively correlated with cancer progression in epithelial ovarian carcinomas and glioma [106, 111]. Targeting ATF5 is a potential strategy to kill cancer cells.
Fig. 3.
The roles of UPRmt components in cancer. The upstream signaling molecules of UPRmt play an important role in tumorigenesis. ATF5 transcribes BCL2 and MCL1 to inhibit cancer cell apoptosis. ATF5 upregulates integrin-α2 and integrin-β1 to promote invasion and migration. HSF1 suppresses the immune system by inducing PD-L1. HSF1 cooperates with PARP13 and PARP1 to repair the genome. The downstream effector proteins of UPRmt are conducive to the progression of cancer. HSP60 promotes the proliferation of cancer cells by regulating metabolic pathways such as glycolysis and the TCA cycle. mtHSP70 is involved in epithelial mesenchymal transition. HSP60 and mtHSP70 synergistically inhibit p53 to prevent it from exerting antitumor effects, thereby promoting the survival of cancer cells. ClpXP maintains the stability of mtDNA and genomic DNA, thus reducing the sensitivity to chemotherapy. LONP1 boosts the activation and M2 polarization of macrophages, thereby creating an immunosuppressive tumor microenvironment. ClpXP and LONP1 coordinately regulate mitochondrial bioenergetics in cancer
Fig. 4.
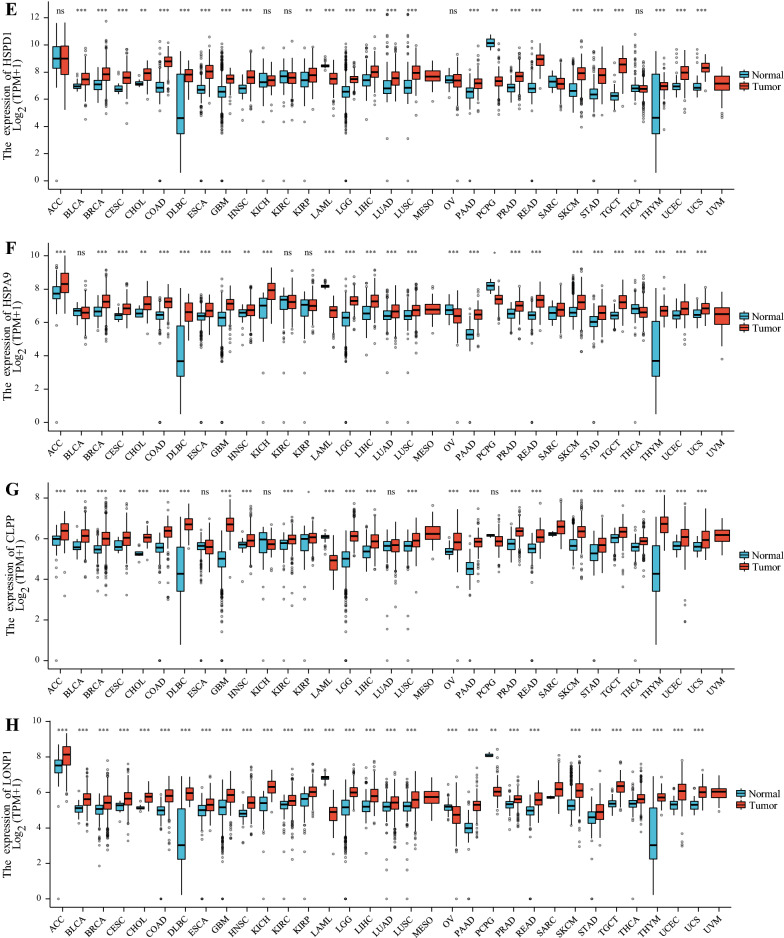
Pan-cancer analysis of the expression status of ATF5 (A), ERα (B), HSF1 (C), SSBP1 (D), HSP60 (E), mtHSP70 (F), ClpP (G) and LONP1 (H) in different cancers compared with adjacent normal tissues according to the TCGA and GTEX databases. The ggplot2 (3.3.3) package in R software (3.6.3) was used, and TPM represents transcription per million. *P < 0.05, **P < 0.01, ***P < 0.001
ERα and cancer
The contributions of ERα, an important component of the UPRmt, to tumors range far beyond its function in the UPRmt. As a member of the ER family, ERα has been indicated to play a critical role in a variety of tumors (Fig. 4B) [113]. ERα is expressed in 70% of breast tumors but in less than 10% of normal breast epithelium [114, 115]. One molecular mechanism underlying the increased ERα expression is that USP7 binds and deubiquitinates ERα to increase its stability [114]. Thus, mechanistically, ERα may be involved in tumorigenesis by regulating cell metabolism- and proliferation-related genes [116]. In prostate cancer, ERα facilitates the upregulation of genes related to epithelial-mesenchymal transition (EMT), thereby promoting the invasion and migration of cancer cells [117]. Due to the effect of ERα on tumor growth and invasion, ERα has been used as an effective target for endocrine cancer therapy [114, 115].
SSBP1, HSF1 and cancer
Previous studies have shown that the expression level of SSBP1 is upregulated in colorectal cancer (Fig. 4D) [118, 119]. Knockdown of SSBP1 leads to a decrease in mitochondrial content, suggesting that SSBP1 may promote cancer progression by influencing mitochondrial biogenesis [118]. In colorectal cancer, IL-6-STAT3-FOXP1 axis-mediated transcriptional activation of SSBP1 is beneficial for cancer cell proliferation and tumor growth [119]. Mechanistically, activated SSBP1 promotes mitochondrial biogenesis, enhances ROS production and activates the Akt/mTOR signaling pathway, resulting in telomerase activation and telomere elongation [119]. Therefore, upregulation of SSBP1 in tumor tissue predicts poor prognosis in patients with colorectal cancer [119]. Moreover, in non-small-cell lung cancer, SSBP1 enhances the resistance of cancer cells to ionizing radiation by inhibiting apoptosis [120]. CircZFR, a circular RNA, interacts with SSBP1 to promote the assembly of the CDK2/cyclin E1 complex. Subsequently, the activated CDK2/cyclin E1 complex phosphorylates Rb, thus releasing E2F1 from Rb-mediated inhibition. Consequently, E2F1 transcribes target genes to promote the G1/S transition and proliferation of cervical cancer cells [121]. However, in highly metastatic triple-negative breast cancer, SSBP1 expression is downregulated, and a low expression level of SSBP1 is associated with poor patient prognosis [122]. Low expression of SSBP1 leads to a decrease in the mtDNA copy number, thus enhancing calcineurin-dependent retrograde signaling and inducing c-Rel-mediated transcription of TGF-β. In turn, TGF-β drives EMT and metastasis of breast cancer cells [122]. The evidence indicating that SSBP1 can play either a procancer or anticancer role in different types of cancer suggests that further exploration of the function of SSBP1 in cancer development could deepen our understanding of cancer.
In addition to maintaining mitochondrial proteostasis, HSF1 can also participate in cancer initiation, development and progression by modulating the tumor microenvironment, inhibiting apoptosis, repairing the genome, promoting cell proliferation and migration and reprogramming metabolism (Figs. 3, 4C) [66, 123]. For example, PIM2-mediated activation of HSF1 induces transcriptional upregulation of PD-L1 to suppress the immune system, enabling cancer cells to evade immune surveillance (Fig. 3) [124]. In addition, HSF1 is involved in angiogenesis, which is accompanied by HuR-mediated enhancement of HIF-1 translation efficiency (Fig. 3) [125]. Consistent with its oncogenic roles, HSF1 is highly expressed in a variety of cancers (Fig. 4C). A high level of HSF1 expression predicts disease progression and a shortened survival time in patients with different types of cancer [66, 126]. Because of the dependence of cancer cells on HSF1, HSF1 can be used as an effective prognostic biomarker and is an attractive therapeutic target. Numerous screening studies have been performed to identify small-molecule inhibitors of HSF1 as next-generation anticancer chemotherapeutics. However, to date, no selective small molecule HSF1 inhibitors have been validated for clinical use. A recent study identified Direct Targeted HSF1 InhiBitor (DTHIB), a new HSF1 inhibitor that directly binds to the DNA binding domain of HSF1 and selectively promotes its degradation in the nucleus of cancer cells. More importantly, DTHIB significantly suppressed the HSF1 cancer gene signature and greatly inhibited tumor growth in mice [127]. In addition, our previous study revealed the mechanism by which cyclosporin A suppresses cancer progression by inhibiting HSF1 activity [128]. Cyclosporin A-mediated activation of ERK1/2, GSK3β and CK2 leads to phosphorylation of HSF1 at Ser303 and Ser307, which interferes with the formation of the HSF1-SSBP1 complex and reduces transcriptional activity of HSF1, resulting in the downregulation of HSP expression and inducing the death of cancer cells [128]. As HSF1 plays a pleiotropic role in cancer, its dysregulated expression in cancer and its relationship with the prognosis of cancer patients imply that HSF1 could be utilized as a biomarker for patient prognosis and a promising molecular target for cancer treatment and chemoprevention.
The roles of downstream effector proteins in the UPRmt in cancer
The transcriptional program induced by UPRmt influences multiple aspects of tumorigenesis. Cancer cells need these transcripts to maintain proteostasis and mitochondrial function. Among the proteins encoded by UPRmt-regulated genes, mitochondrial chaperones and proteases perform an indispensable function. In this chapter, we focus on the role of UPRmt-induced molecular chaperones and proteases in tumors.
Mitochondrial HSPs and cancer
HSP60 (HSPD1) and mtHSP70 (HSPA9) are mitochondria-localized cytoprotective proteins and are the main molecular chaperones induced by UPRmt. Accumulating studies have shown that the occurrence and development of cancer require HSP60 and mtHSP70, which can assist the refolding of unfolded and misfolded proteins and promote the depolymerization of aggregated proteins [129]. Moreover, increasing evidence indicates that the levels of HSP60 and mtHSP70 are significantly increased in a variety of tumors (Fig. 4E, F) [129–136]. In cancer cells, one mechanism underlying HSP60 overexpression is the mediation of its transcriptional regulation by the proto-oncogenes c-MYC and HSF1 [65, 66, 132]. In ovarian cancer, NF-κB p65 binds to the promoter of mtHSP70 to transcriptionally upregulate its expression [137].
HSP60 orchestrates various cell survival programs in cancer. For example, HSP60 binds and stabilizes survivin, protecting it from degradation and thus enabling it to play an inhibitory role in apoptosis [138, 139]. The molecular chaperone complex containing HSP60 interacts with cyclophilin D, a component of the mitochondrial permeability transition pore, inhibiting cyclophilin D-dependent cancer cell death [140]. Additionally, the interaction between HSP60 and p53 reduces the stability and activity of p53 to antagonize caspase-mediated apoptosis (Fig. 3) [138]. In the cytoplasm, HSP60 interacts with IKK to boost activation-related serine phosphorylation of IKK, thus activating the prosurvival IKK/NF-κB pathway [141]. A previous study demonstrated that HSP60 modulated protein translation to facilitate the growth of ovarian cancer and glioblastoma, which requires the AMPK/mTOR pathway [142, 143]. In addition, a recent study revealed the relationship between the HSP60 expression level and tumor lymph node metastasis, and high levels of HSP60 have been linked to resistance to hormone therapy in prostate cancer [133]. Furthermore, HSP60 regulates a variety of metabolic processes, including glycolysis and the TCA cycle (Fig. 3). For example, HSP60 promotes multiple myeloma development via metabolic reprogramming [144]. HSP60 has also been shown to play a role in the extracellular environment of cancer cells. Secretion of HSP60 requires exosomes and lipid rafts [145]. In fibrosarcoma, HSP60 is transported extracellularly via the ER-Golgi secretory pathway and is therefore involved in modulating the tumor microenvironment [146].
Knockdown of mtHSP70 induces the death of melanoma cells via a mechanism related to the MEK/ERK signaling pathway and the mitochondrial permeability regulators cyclophilin D and ANT [147]. In addition, previous studies have shown that depletion of mtHSP70 stimulates the death of KRAS mutant pancreatic ductal adenocarcinoma and colon cancer cells via a mechanism associated with an increase in mitochondrial membrane permeability [148]. Under hypoxic conditions, mtHSP70 interacts with HIF-1 and colocalizes with HIF-1 to the outer mitochondrial membrane. Subsequently, VDAC1 is truncated and activated, endowing cancer cells with resistance to apoptosis induced by chemotherapy [149]. In various types of thyroid cancer, upregulation of mtHSP70 promotes the proliferation of cancer cells, while inhibition of mtHSP70 induces cell cycle arrest [150, 151]. In tumor cells with mutations in KRAS or BRAF, mtHSP70 facilitates the interaction between PP1α and MEK1/2, which modulates MEK/ERK signaling activity, thereby promoting tumor cell proliferation. In addition, mtHSP70, as a negative regulator of the Raf/MEK/ERK signaling pathway, suppresses the anticancer function of Raf/MEK/ERK signaling [152, 153]. In oral squamous cell carcinoma, mtHSP70 is secreted by cancer cells in an autocrine manner; subsequently, extracellular mtHSP70 binds to PDPN, which plays a role in cell adhesion, participating in regulating the growth and invasiveness of cancer cells [154]. Overexpression of mtHSP70 modulates the activity of the PI3K/AKT signaling pathway to accelerate tumor EMT, which is accompanied by a decrease in epithelial markers and an increase in mesenchymal markers [134]. High expression of mtHSP70 also promotes the stemness of cancer cells [155]. In hepatocellular carcinoma (HCC), overexpression of mtHSP70 is closely associated with venous infiltration and disease progression [135]. Furthermore, mtHSP70 is involved in cancer metastasis by activating hnRNP-K and inactivating p53 (Fig. 3) [156].
Taken together, these observations indicate that the mitochondrial chaperones HSP60 and mtHSP70 rely on their protein-folding ability to maintain the stability of master signaling pathways in cancer cells, thus facilitating invasion and migration. HSP60 and mtHSP70 contribute to the survival of cancer cells by inhibiting proapoptotic proteins and activating antiapoptotic proteins (Fig. 3). Thus, HSP60 and mtHSP70 are widely exploited by various cancers (Fig. 4E, F). Accumulating studies have demonstrated that high expression of HSP60 and mtHSP70 is significantly associated with poor patient outcomes in various types of cancer [157–163]. Therefore, the levels of HSP60 and mtHSP70 can be used as prognostic indicators in cancer patients. Moreover, HSP60 and mtHSP70 are employed as potential therapeutic targets due to their diagnostic role [164, 165]. For example, shRNA-mediated silencing of HSP60 inhibits the growth of HCC [166]. An anti-HSP60 antibody exhibited cytotoxicity in ovarian cancer cells. Notably, combined downregulation of HSP60 and treatment with chemotherapy exhibits a significant synergistic tumoricidal effect [167]. Previous studies have shown the efficacy of SHetA2 and PRIMA-1MET in the treatment of ovarian cancer. SHetA2, a small-molecule drug, interferes with the mtHSP70-p53 complex, thereby relieving the inhibitory effect of mtHSP70 on p53. PRIMA-1MET binds and modifies mutant p53 to restore the proper protein conformation of p53 and reactivate the wild-type function of mutant p53. Therefore, p53 can exhibit tumor-suppressive effects [165]. In summary, further development of drugs targeting HSP60 and mtHSP70 may provide novel insights into the treatment of various cancers.
Mitochondrial proteases and cancer
The UPRmt-induced mitochondrial matrix proteases ClpP and LONP1 maintain mitochondrial homeostasis by removing harmful proteins. As major mitochondrial proteases, ClpP and LONP1 play important roles in tumors [168, 169]. Multiple studies have demonstrated that ClpP and LONP1 levels are markedly increased in numerous cancers (Fig. 4G, H) [130, 168, 170–174].
ClpP interacts with multiple respiratory chain proteins and metabolic enzymes in mitochondria that are essential for metabolic regulation in cancer cells [172]. For example, the ClpPX complex containing ClpP binds and stabilizes the SDHB subunit of respiratory chain complex II, maintaining the normal functioning of OXPHOS and promoting the production of ATP. Inhibition of ClpPX leads to an imbalance in the mitochondrial ETC and oxidative stress, which ultimately reduces the proliferation and motility of cancer cells (Fig. 3) [171]. In addition, ClpP regulates the proliferation and invasion of breast cancer cells via a mechanism associated with the Src/PI3K/AKT cascade [173]. A previous study revealed that robust ClpP activity endows cancer cells with resistance to cisplatin [175]. Mechanistically, activation of ClpP increases the levels of ATP7A and ATP7B, which are involved in the elimination of cisplatin, and ClpP-mediated cisplatin efflux blocks the production of cisplatin-mtDNA/genomic DNA adducts, thereby inhibiting cancer cell death (Fig. 3) [175].
LONP1 has been found to be involved in tumor metabolic reprogramming, which is related to remodeling the components of the ETC and antagonizing cellular senescence [176]. AKT-mediated phosphorylation of LONP1 increases its protease activity. Subsequently, LONP1 ensures the stabilization of ETC complex II and complex V and, thus, protects cancer cells from damage caused by ROS accumulation [177, 178]. In fact, ClpP and LONP1 coordinately regulate mitochondrial bioenergetics in cancer, which is reflected in the observation that ClpP and LONP1 have many common substrates. The substrates regulated by ClpP and LONP1 participate in processes such as OXPHOS, the TCA cycle, and amino acid and lipid metabolism [179]. LONP1 reduces the sensitivity of colon cancer cells to apoptosis and stimulates EMT in pancreatic cancer cells (Fig. 3) [170, 174]. Furthermore, LONP1 binds and stabilizes the HSP60-mtHSP70 complex, thereby facilitating HSP60-mediated p53 inhibition and promoting cancer cell survival [180]. The application of proteasome inhibitors inhibits the progression of multiple myeloma, whereas upregulation of LONP1 decreases the efficacy of proteasome inhibitors. Mechanistically, LONP1 functions outside mitochondria to partially compensate for the lack of proteasome activity, reducing the level of damaged intracellular proteins [181]. In addition, elevated LONP1 expression increases the level of ROS to promote the production of inflammatory cytokines, including TGF-β and IL-6, thus boosting the activation of M2 macrophages and establishing an immunosuppressive tumor microenvironment (Fig. 3) [182].
Cancer cells exploit the functions of ClpP and LONP1 in mitochondrial homeostasis and energy metabolism to accelerate their own invasion and metastasis. Several studies have proven that high levels of ClpP and LONP1 in colorectal cancer, prostate cancer, breast cancer and melanoma are notably correlated with poor prognosis in cancer patients (Fig. 4G, H) [173, 176, 179]. Accordingly, targeting ClpP and LONP1 is anticipated to reveal a new therapeutic perspective for cancer due to the oncogenic functions of these proteins [183, 184]. Indeed, inhibition of LONP1 mediated by triterpenoids leads to alterations in normal mitochondrial morphology and dysregulation of mitochondrial function, which ultimately triggers the death of cancer cells [185]. Although its activation plays a crucial role in maintaining cancer cell proteostasis, the oncogenic effects of ClpP may be dose dependent. Dysregulation of ClpP also disrupts proteostasis [186]. Previous studies have shown that ONC201 and its TR compound analogs, which belong to the imipridone class of small molecules, can specifically bind to ClpP, consequently inhibiting the proliferation of cancer cells [187]. Mechanistically, imipridones noncovalently bind ClpP and cause structural changes in ClpP, which induce its hyperactivation. Subsequently, hyperactivated ClpP accelerates the degradation of ETC substrates to interfere with mitochondrial structure and function, thus killing cancer cells [186]. Additionally, dysfunction of ClpP can be caused by acyldepsipeptide analogs, leading to nonspecific hydrolysis of model substrates of ClpP, thereby triggering caspase-dependent apoptotic cell death [188].
The roles of the UPRmt in physiological processes and other diseases
Accumulating studies have indicated that the UPRmt is related to many physiological and pathological processes as well as human diseases. The UPRmt plays an important role in aging, the immune response, cancer, heart disease and neurodegenerative diseases [46, 189–194]. In C. elegans, the transcriptional program induced by UPRmt involves numerous genes, which are enriched in mitochondrial chaperones, OXPHOS complex assembly factors and components, and glycolytic genes [4]. UPRmt also boosts the expansion of the mitochondrial network, which is active during normal development, thus satisfying the physiological requirements of individual cells in C. elegans [195]. CBP-1 in nematodes and CBP/p300 in mammals are involved in the transcriptional activation of UPRmt genes, thereby promoting extension of the lifespan and enhancement of immune responses [42]. The roles of the UPRmt in various human diseases have been gradually revealed. For instance, in decompensated cirrhosis, aging liver cells show the characteristics of mitochondrial dysfunction and an impaired UPRmt. ClpP, an effector in the UPRmt, promotes the elimination of ROS and, thus, delays the senescence of liver cells [196]. The weak regenerative ability of hematopoietic stem cells (HSCs) is associated with inactivation of the UPRmt effector protein SIRT7, and SIRT7 expression is significantly downregulated in senescent HSCs, demonstrating that UPRmt-mediated dysregulation of cell metabolism is one reason for the senescence of HSCs [63]. In contrast, during the transition of HSCs from quiescence to proliferation, UPRmt is activated to promote metabolic adaptation [197]. Evidence indicating that the enhancement of UPRmt activity inhibits the death of cardiomyocytes induced by chronic pressure overload demonstrates the cardioprotective function of the UPRmt [22]. Activation of the PGC-1α/ATF5 axis is beneficial for the alleviation of pathological cardiac hypertrophy [198]. Activation of UPRmt signaling promotes neurogenesis in the brains of mice with amyotrophic lateral sclerosis (ALS) [23]. Additionally, the UPRmt is involved in the differentiation of myoblasts [55, 56]. In summary, the UPRmt plays a critical role in a variety of physiological processes, most of which are accompanied by slight perturbations in mitochondrial homeostasis, thus leading to UPRmt activation. In turn, activated UPRmt maintains mitochondrial function. Specifically, moderate mitochondrial stress contributes to activation of the UPRmt and stabilization of mitochondrial function [41]. However, the UPRmt also acts as a promoter of the maintenance and propagation of deleterious mtDNA. The OXPHOS defect caused by the mutant mtDNA induces the UPRmt. Conversely, to promote mitochondrial recovery, UPRmt activation leads to intracellular accumulation of harmful mtDNA, which ultimately results in cellular dysfunction. This harmful mtDNA hijacks the UPRmt to facilitate its own dissemination [199, 200].
Conclusions
As an indispensable cytoprotective mechanism, activation of the UPRmt promotes the recovery of mitochondria from damage, maintains proteostasis, remodels the ETC, and eliminates accumulated ROS in response to various intracellular and extracellular stresses. Mitocytosis, a migrasome-mediated mitochondrial quality control mechanism, maintains mitochondrial membrane potential and mitochondrial respiration by removing damaged mitochondria from migrating cells [201]. It is worth exploring whether there is an association between UPRmt and mitocytosis. In a sense, the UPRmt in C. elegans is very similar to the UPRmt in mammals. In both, the efficiency of mitochondrial protein transport determines the activation state. In addition, both of these UPRmts involve intricate cell signaling pathways and dynamic epigenetic regulation. However, the UPRmt in mammals is more complex than the UPRmt in C. elegans because it involves more factors and regulatory branches. Although the UPRmt in C. elegans is well understood, the UPRmt in mammals needs more exploration. What are the other potential branches of the mammalian UPRmt? What functions do these branches have? Is there any crosstalk between the different branches of the UPRmt, resulting in an extensive signaling network? What is the precise nature of the UPRmt-induced transcriptional program, and what roles do these transcripts play? Is there a signaling node connecting the UPRmt, UPRER and HSR that regulates mitochondrial, ER and cytoplasmic homeostasis? The answers to these questions require further research.
The UPRmt has emerged as a protective response in various human diseases, including neurodegenerative diseases [202]. Alzheimer’s disease (AD) is a multifactorial brain disorder characterized by loss of memory and aggregation of two insoluble proteins, including tau neurofibrillary tangles and β-amyloid plaques [203]. Mitochondrial dysfunction, such as changes in mitochondrial enzyme activity, damaged mitochondrial ultrastructure, excessive ROS generation, altered mtDNA, reduced mitochondrial oxygen consumption and mitophagy impairment, is a common pathological hallmark in AD patients [204]. Several lines of evidence suggest that UPRmt is associated with the progression of familial and sporadic AD [192, 205]. Activation of UPRmt was observed in the brains of APP/PS1 transgenic mice and SHSY5Y cells treated with Aβ [206]. Recently, an important study identified a conserved mitochondrial stress response feature in AD patients and AD animal models [207]. More importantly, pharmacological or genetic activation of UPRmt attenuated cognitive impairment and decreased deposition of Aβ in an AD model [207]. Therefore, UPRmt is important for the maintenance of mitochondrial proteostasis and provides potential targets for AD therapy.
Accumulating evidence indicates that the UPRmt is activated in many types of tumors. The UPRmt promotes the development of cancer and boosts its progression through various mechanisms. Cancer cells utilize signaling molecules and transcriptional products in the UPRmt, such as ATF5, HSP60 and ClpP, to promote their proliferation, growth, invasion and metastasis. Therefore, targeting components of the UPRmt may be a potential, reliable, and effective method for the treatment of cancer [129, 168, 208]. For example, CP-d/n-ATF5-S1, a cell-penetrating peptide, has been exploited as an inhibitor of ATF5. CP-d/n-ATF5-S1 inhibits tumor growth by inducing apoptosis and has demonstrated excellent anticancer effects against glioblastoma, melanoma, prostate cancer and triple-negative breast cancer in a series of in vitro and in vivo experiments [208]. The relationship between UPRmt and cancer needs further interpretation. Can the UPRmt be activated only by mitochondrial dysregulation, or can cancer cells also promote constitutive activation of the UPRmt (via, for example, upstream regulator-mediated signaling activation and epigenetic changes in UPRmt-related components)? Which oncogenes and tumor suppressor genes interact with the components of the UPRmt in cancer? Although the role of the UPRmt in cancer biology has been clarified through many years of research, more hidden mysteries are still awaiting exploration.
Acknowledgements
Not applicable.
Abbreviations
- ACC
Adrenocortical carcinoma
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HCC
Hepatocellular carcinoma
- HNSC
Head and neck squamous cell carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute myeloid leukemia
- LGG
Brain lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin cutaneous melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- UVM
Uveal melanoma
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- ATF5
Activating transcription factor 5
- ATFS-1
Activating transcription factor associated with stress-1
- C. elegans
Caenorhabditis elegans
- ClpP
Caseinolytic mitochondrial matrix peptidase proteolytic subunit
- DN-ATF5
Dominant-negative ATF5 mutant
- DTHIB
Direct Targeted HSF1 InhiBitor
- eIF2α
α Subunit of eukaryotic translation initiation factor 2
- EMT
Epithelial-mesenchymal transition
- ER
Endoplasmic reticulum
- ERα
Estrogen receptor α
- ETC
Electron transport chain
- FUNDC1
FUN14 domain containing 1
- GCN2
General control nonderepressible 2
- H3K9me2
H3K9 dimethylation
- HRI
Heme-regulated inhibitor
- HSCs
Hematopoietic stem cells
- HSF1
Heat shock factor 1
- HSP60
Heat shock protein 60
- HSR
Heat shock response
- IGFBP7
Insulin-like growth factor-binding protein 7
- ISR
Integrated stress response
- LONP1
Lon peptidase 1
- MAM
Mitochondria associated membranes
- Met-tRNAi
Methionylated initiator transfer RNA
- mitophagy
Mitochondrial autophagy
- mtDNA
Mitochondrial DNA
- mtHSP70
Mitochondrial heat shock protein 70
- MTS
Mitochondrial targeting sequence
- NLS
Nuclear localization sequence
- OXPHOS
Oxidative phosphorylation
- PERK
PKR-like endoplasmic reticulum kinase
- PGAM5
Phosphoglycerate mutase 5
- PKR
Protein kinase R
- polyQ
Polyglutamine repeat protein
- ROS
Reactive oxygen species
- S. cerevisiae
Saccharomyces cerevisiae
- SSBP1
Single-strand DNA binding protein 1
- TCA
Tricarboxylic acid
- uORFs
Upstream open reading frames
- UPRER
Endoplasmic reticulum unfolded protein response
- UPRmt
Mitochondrial unfolded protein response
Authors’ contributions
GW, YF, PC and KT drafted the manuscript and prepared the figures. GW and KT discussed the revised the manuscript. All authors have read and approved the final manuscript.
Funding
This study was supported by the One Hundred Person Project of Hebei Province (E2016100019), China Postdoctoral Science Foundation (2017M621099) and Natural Science Foundation of Hebei Province (C2020205003).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have read the manuscript and approved for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ge Wang, Yumei Fan and Pengxiu Cao contributed equally
References
- 1.Ruan L, Wang Y, Zhang X, Tomaszewski A, McNamara JT, Li R. Mitochondria-associated proteostasis. Annu Rev Biophys. 2020;49:41–67. doi: 10.1146/annurev-biophys-121219-081604. [DOI] [PubMed] [Google Scholar]
- 2.Anderson AJ, Jackson TD, Stroud DA, Stojanovski D. Mitochondria-hubs for regulating cellular biochemistry: emerging concepts and networks. Open Biol. 2019;9(8):190126. doi: 10.1098/rsob.190126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol CB. 2016;26(15):2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58(1):123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quirós PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216(7):2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng P, Haynes CM. Mitochondrial dysfunction in cancer: potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol. 2017;47:43–49. doi: 10.1016/j.semcancer.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Münch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018;16(1):81. doi: 10.1186/s12915-018-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath E, Moschetta A, Haller D. Mitochondrial function—gatekeeper of intestinal epithelial cell homeostasis. Nat Rev Gastroenterol Hepatol. 2018;15(8):497–516. doi: 10.1038/s41575-018-0021-x. [DOI] [PubMed] [Google Scholar]
- 9.Morrow G, Kim HJ, Pellerito O, Bourrelle-Langlois M, Le Pécheur M, Groebe K, Tanguay RM. Changes in Drosophila mitochondrial proteins following chaperone-mediated lifespan extension confirm a role of Hsp22 in mitochondrial UPR and reveal a mitochondrial localization for cathepsin D. Mech Ageing Dev. 2016;155:36–47. doi: 10.1016/j.mad.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Tran HC, Van Aken O. Mitochondrial unfolded protein-related responses across kingdoms: similar problems, different regulators. Mitochondrion. 2020;53:166–177. doi: 10.1016/j.mito.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, Liu Y, Dillin A. Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell. 2016;166(6):1553–1563.e1510. doi: 10.1016/j.cell.2016.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO J. 2012;31(8):1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Wang D. Intestinal mitochondrial unfolded protein response induced by nanoplastic particles in Caenorhabditis elegans. Chemosphere. 2021;267:128917. doi: 10.1016/j.chemosphere.2020.128917. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Li D, Rui Q, Wang D. Toxicity induction of nanopolystyrene under microgravity stress condition in Caenorhabditis elegans. Sci Total Environ. 2020;703:135623. doi: 10.1016/j.scitotenv.2019.135623. [DOI] [PubMed] [Google Scholar]
- 15.Oks O, Lewin S, Goncalves IL, Sapir A. The UPR(mt) protects Caenorhabditis elegans from mitochondrial dysfunction by upregulating specific enzymes of the mevalonate pathway. Genetics. 2018;209(2):457–473. doi: 10.1534/genetics.118.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shamalnasab M, Gravel SP, St-Pierre J, Breton L, Jäger S, Aguilaniu H. A salicylic acid derivative extends the lifespan of Caenorhabditis elegans by activating autophagy and the mitochondrial unfolded protein response. Aging Cell. 2018;17(6):e12830. doi: 10.1111/acel.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolland SG, Schneid S, Schwarz M, Rackles E, Fischer C, Haeussler S, Regmi SG, Yeroslaviz A, Habermann B, Mokranjac D, et al. Compromised mitochondrial protein import acts as a signal for UPR(mt) Cell Rep. 2019;28(7):1659–1669.e1655. doi: 10.1016/j.celrep.2019.07.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Wu X, Chen P, Liu L, Xin N, Tian Y, Dillin A. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell. 2018;174(4):870–883.e817. doi: 10.1016/j.cell.2018.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler F, Müller-Rischart AK, Conradt B, Rolland SG. The loss of LRPPRC function induces the mitochondrial unfolded protein response. Aging. 2015;7(9):701–717. doi: 10.18632/aging.100812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runkel ED, Baumeister R, Schulze E. Mitochondrial stress: balancing friend and foe. Exp Gerontol. 2014;56:194–201. doi: 10.1016/j.exger.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Smyrnias I, Gray SP, Okonko DO, Sawyer G, Zoccarato A, Catibog N, López B, González A, Ravassa S, Díez J, et al. Cardioprotective effect of the mitochondrial unfolded protein response during chronic pressure overload. J Am Coll Cardiol. 2019;73(14):1795–1806. doi: 10.1016/j.jacc.2018.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q, Zhu L, Qiu W, Liu Y, Yang F, Chen W, Xu R. Nicotinamide riboside enhances mitochondrial proteostasis and adult neurogenesis through activation of mitochondrial unfolded protein response signaling in the brain of ALS SOD1(G93A) Mice. Int J Biol Sci. 2020;16(2):284–297. doi: 10.7150/ijbs.38487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta D, Delmotte P, Sieck GC. Inflammation-induced protein unfolding in airway smooth muscle triggers a homeostatic response in mitochondria. Int J Mol Sci. 2020;22(1):363. doi: 10.3390/ijms22010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Wang X, Qu JH, Liu B, Zhang P, Zhang T, Fan PC, Wang XM, Xiao GY, Su Y, et al. Caloric restriction induces microRNAs to improve mitochondrial proteostasis. iScience. 2019;17:155–166. doi: 10.1016/j.isci.2019.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng J, Wang P, Chen X, Cheng H, Liu J, Fushimi K, Zhu L, Wu JY. FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc Natl Acad Sci USA. 2018;115(41):E9678–e9686. doi: 10.1073/pnas.1806655115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Deng J, Dong J, Liu J, Bigio EH, Mesulam M, Wang T, Sun L, Wang L, Lee AY, et al. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response. PLoS Genet. 2019;15(5):e1007947. doi: 10.1371/journal.pgen.1007947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jazwinski SM, Kriete A. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front Physiol. 2012;3:139. doi: 10.3389/fphys.2012.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 30.Torelli NQ, Ferreira-Júnior JR, Kowaltowski AJ, da Cunha FM. RTG1- and RTG2-dependent retrograde signaling controls mitochondrial activity and stress resistance in Saccharomyces cerevisiae. Free Radical Biol Med. 2015;81:30–37. doi: 10.1016/j.freeradbiomed.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 32.da Cunha FM, Torelli NQ, Kowaltowski AJ. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxid Med Cell Longevit. 2015;2015:482582. doi: 10.1155/2015/482582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science (New York, NY) 2012;337(6094):587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13(4):467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Gao K, Li Y, Hu S, Liu Y: SUMO peptidase ULP-4 regulates mitochondrial UPR-mediated innate immunity and lifespan extension. eLife 2019;8. [DOI] [PMC free article] [PubMed]
- 36.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matilainen O, Quirós PM, Auwerx J. Mitochondria and epigenetics—crosstalk in homeostasis and stress. Trends Cell Biol. 2017;27(6):453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt) Cell. 2016;165(5):1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao LW, Peng Q, Dong M, Gao K, Li Y, Li Y, Li CY, Liu Y. Histone deacetylase HDA-1 modulates mitochondrial stress response and longevity. Nat Commun. 2020;11(1):4639. doi: 10.1038/s41467-020-18501-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell. 2016;165(5):1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li TY, Sleiman MB, Li H, Gao AW, Mottis A, Bachmann AM, El Alam G, Li X, Goeminne LJE, Schoonjans K, et al. The transcriptional coactivator CBP/p300 is an evolutionarily conserved node that promotes longevity in response to mitochondrial stress. Nat Aging. 2021;1(2):165–178. doi: 10.1038/s43587-020-00025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatar M, Sedivy JM. Mitochondria: masters of epigenetics. Cell. 2016;165(5):1052–1054. doi: 10.1016/j.cell.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao LW, Niu R, Liu Y. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 2016;26(11):1182–1196. doi: 10.1038/cr.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bar-Ziv R, Bolas T, Dillin A. Systemic effects of mitochondrial stress. EMBO Rep. 2020;21(6):e50094. doi: 10.15252/embr.202050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naresh NU, Haynes CM. Signaling and regulation of the mitochondrial unfolded protein response. Cold Spring Harbor Perspect Biol. 2019;11(6):a033944. doi: 10.1101/cshperspect.a033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Sieburth D. FSHR-1/GPCR regulates the mitochondrial unfolded protein response in Caenorhabditis elegans. Genetics. 2020;214(2):409–418. doi: 10.1534/genetics.119.302947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Sieburth D. Sphingosine kinase activates the mitochondrial unfolded protein response and is targeted to mitochondria by stress. Cell Rep. 2018;24(11):2932–2945.e2934. doi: 10.1016/j.celrep.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan J, Rollins JA, Zang X, Wu D, Zou L, Wang Z, Ye C, Wu Z, Kapahi P, Rogers AN, et al. Translational regulation of non-autonomous mitochondrial stress response promotes longevity. Cell Rep. 2019;28(4):1050–1062.e1056. doi: 10.1016/j.celrep.2019.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155(3):699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernkopf DB, Jalal K, Brückner M, Knaup KX, Gentzel M, Schambony A, Behrens J. Pgam5 released from damaged mitochondria induces mitochondrial biogenesis via Wnt signaling. J Cell Biol. 2018;217(4):1383–1394. doi: 10.1083/jcb.201708191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng M, Lin N, Dong D, Ma J, Su J, Sun L. PGAM5: a crucial role in mitochondrial dynamics and programmed cell death. Eur J Cell Biol. 2021;100(1):151144. doi: 10.1016/j.ejcb.2020.151144. [DOI] [PubMed] [Google Scholar]
- 52.Yu B, Ma J, Li J, Wang D, Wang Z, Wang S. Mitochondrial phosphatase PGAM5 modulates cellular senescence by regulating mitochondrial dynamics. Nat Commun. 2020;11(1):2549. doi: 10.1038/s41467-020-16312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borch Jensen M, Qi Y, Riley R, Rabkina L, Jasper H: PGAM5 promotes lasting FoxO activation after developmental mitochondrial stress and extends lifespan in Drosophila. eLife. 2017;6. [DOI] [PMC free article] [PubMed]
- 54.Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T. ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells. Biochim Biophys Acta. 2015;1853(10 Pt A):2580–2591. doi: 10.1016/j.bbamcr.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 55.Deepa SS, Bhaskaran S, Ranjit R, Qaisar R, Nair BC, Liu Y, Walsh ME, Fok WC, Van Remmen H. Down-regulation of the mitochondrial matrix peptidase ClpP in muscle cells causes mitochondrial dysfunction and decreases cell proliferation. Free Radical Biol Med. 2016;91:281–292. doi: 10.1016/j.freeradbiomed.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE. 2007;2(9):e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34(4):699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radical Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 59.Fu B, Zhao J, Peng W, Wu H, Zhang Y. Resveratrol rescues cadmium-induced mitochondrial injury by enhancing transcriptional regulation of PGC-1α and SOD2 via the Sirt3/FoxO3a pathway in TCMK-1 cells. Biochem Biophys Res Commun. 2017;486(1):198–204. doi: 10.1016/j.bbrc.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 60.He C, Hart PC, Germain D, Bonini MG. SOD2 and the mitochondrial UPR: partners regulating cellular phenotypic transitions. Trends Biochem Sci. 2016;41(7):568–577. doi: 10.1016/j.tibs.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124(Pt 9):1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (New York, NY) 2015;347(6228):1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Münch C, Harper JW. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature. 2016;534(7609):710–713. doi: 10.1038/nature18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun. 2015;6:6580. doi: 10.1038/ncomms7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katiyar A, Fujimoto M, Tan K, Kurashima A, Srivastava P, Okada M, Takii R, Nakai A. HSF1 is required for induction of mitochondrial chaperones during the mitochondrial unfolded protein response. FEBS Open Bio. 2020;10(6):1135–1148. doi: 10.1002/2211-5463.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang G, Cao P, Fan Y, Tan K. Emerging roles of HSF1 in cancer: cellular and molecular episodes. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188390. doi: 10.1016/j.bbcan.2020.188390. [DOI] [PubMed] [Google Scholar]
- 67.Jia H, Mo W, Hong M, Jiang S, Zhang YY, He D, Yu D, Shi Y, Cao J, Xu X, et al. Interferon-α inducible protein 6 (IFI6) confers protection against ionizing radiation in skin cells. J Dermatol Sci. 2020;100(2):139–147. doi: 10.1016/j.jdermsci.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Williams R, Laskovs M, Williams RI, Mahadevan A, Labbadia J. A mitochondrial stress-specific form of HSF1 protects against age-related proteostasis collapse. Dev Cell. 2020;54(6):758–772.e755. doi: 10.1016/j.devcel.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 69.Rossi A, Pizzo P, Filadi R. Calcium, mitochondria and cell metabolism: a functional triangle in bioenergetics. Biochim Biophys Acta. 2019;1866(7):1068–1078. doi: 10.1016/j.bbamcr.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Szabadkai G, Duchen MR. Mitochondria: the hub of cellular Ca2+ signaling. Physiology (Bethesda) 2008;23:84–94. doi: 10.1152/physiol.00046.2007. [DOI] [PubMed] [Google Scholar]
- 71.Godoy JA, Rios JA, Picón-Pagès P, Herrera-Fernández V, Swaby B, Crepin G, Vicente R, Fernández-Fernández JM, Muñoz FJ. Mitostasis, calcium and free radicals in health, Aging and Neurodegeneration. Biomolecules. 2021;11(7):1012. doi: 10.3390/biom11071012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Malley J, Kumar R, Inigo J, Yadava N, Chandra D. Mitochondrial stress response and cancer. Trends Cancer. 2020;6(8):688–701. doi: 10.1016/j.trecan.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hori O, Ichinoda F, Tamatani T, Yamaguchi A, Sato N, Ozawa K, Kitao Y, Miyazaki M, Harding HP, Ron D, et al. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Biol. 2002;157(7):1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zurita Rendón O, Shoubridge EA. LONP1 is required for maturation of a subset of mitochondrial proteins, and its loss elicits an integrated stress response. Mol Cell Biol. 2018;38(20):e00412–e00417. doi: 10.1128/MCB.00412-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quiles JM, Gustafsson ÅB. Mitochondrial quality control and cellular proteostasis: two sides of the same coin. Front Physiol. 2020;11:515. doi: 10.3389/fphys.2020.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pellegrino MW, Haynes CM. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC Biol. 2015;13:22. doi: 10.1186/s12915-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 79.Killackey SA, Philpott DJ, Girardin SE. Mitophagy pathways in health and disease. J Cell Biol. 2020;219(11):e202004029. doi: 10.1083/jcb.202004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Jasper H, Toan S, Muid D, Chang X, Zhou H. Mitophagy coordinates the mitochondrial unfolded protein response to attenuate inflammation-mediated myocardial injury. Redox Biol. 2021;45:102049. doi: 10.1016/j.redox.2021.102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson NS, Haynes CM. Folding the mitochondrial UPR into the integrated stress response. Trends Cell Biol. 2020;30(6):428–439. doi: 10.1016/j.tcb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang SF, Chen S, Tseng LM, Lee HC. Role of the mitochondrial stress response in human cancer progression. Exp Biol Med (Maywood) 2020;245(10):861–878. doi: 10.1177/1535370220920558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science (New York, NY). 2020;368(6489). [DOI] [PMC free article] [PubMed]
- 84.Tian X, Zhang S, Zhou L, Seyhan AA, Hernandez Borrero L, Zhang Y, El-Deiry WS. Targeting the integrated stress response in cancer therapy. Front Pharmacol. 2021;12:747837. doi: 10.3389/fphar.2021.747837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eckl EM, Ziegemann O, Krumwiede L, Fessler E, Jae LT. Sensing, signaling and surviving mitochondrial stress. Cell Mol Life Sci CMLS. 2021;78(16):5925–5951. doi: 10.1007/s00018-021-03887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, Kim VN. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol Cell. 2018;71(6):1051–1063.e1056. doi: 10.1016/j.molcel.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 87.Tremblay BP, Haynes CM. Mitochondrial distress call moves to the cytosol to trigger a response to stress. Nature. 2020;579(7799):348–349. doi: 10.1038/d41586-020-00552-0. [DOI] [PubMed] [Google Scholar]
- 88.Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579(7799):427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579(7799):433–437. doi: 10.1038/s41586-020-2076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21(17):4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE. 2007;2(9):e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vasudevan D, Neuman SD, Yang A, Lough L, Brown B, Bashirullah A, Cardozo T, Ryoo HD. Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat Commun. 2020;11(1):4677. doi: 10.1038/s41467-020-18453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bohlen J, Harbrecht L, Blanco S, Clemm von Hohenberg K, Fenzl K, Kramer G, Bukau B, Teleman AA. DENR promotes translation reinitiation via ribosome recycling to drive expression of oncogenes including ATF4. Nat Commun. 2020;11(1):4676. doi: 10.1038/s41467-020-18452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang YT, Lim Y, McCall MN, Huang KT, Haynes CM, Nehrke K, Brookes PS. Cardioprotection by the mitochondrial unfolded protein response requires ATF5. Am J Physiol Heart Circ Physiol. 2019;317(2):H472–h478. doi: 10.1152/ajpheart.00244.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29(12):2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang SF, Wung CH, Chen MS, Chen CF, Yin PH, Yeh TS, Chang YL, Chou YC, Hung HH, Lee HC. Activated integrated stress response induced by salubrinal promotes cisplatin resistance in human gastric cancer cells via enhanced xCT expression and glutathione biosynthesis. Int J Mol Sci. 2018;19(11):3389. doi: 10.3390/ijms19113389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keerthiga R, Pei DS, Fu A. Mitochondrial dysfunction, UPR(mt) signaling, and targeted therapy in metastasis tumor. Cell Biosci. 2021;11(1):186. doi: 10.1186/s13578-021-00696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 99.Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MAK, Eliceiri KW, LaFramboise T, Grandhi S, Santos J, et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPR(mt) to promote metastasis. Oncogene. 2017;36(31):4393–4404. doi: 10.1038/onc.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kenny TC, Germain D. mtDNA, metastasis, and the mitochondrial unfolded protein response (UPR(mt)) Front Cell Dev Biol. 2017;5:37. doi: 10.3389/fcell.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kenny TC, Gomez ML, Germain D. Mitohormesis, UPR(mt), and the complexity of mitochondrial DNA landscapes in cancer. Can Res. 2019;79(24):6057–6066. doi: 10.1158/0008-5472.CAN-19-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kenny TC, Craig AJ, Villanueva A, Germain D. Mitohormesis primes tumor invasion and metastasis. Cell Rep. 2019;27(8):2292–2303.e2296. doi: 10.1016/j.celrep.2019.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen FM, Huang LJ, Ou-Yang F, Kan JY, Kao LC, Hou MF. Activation of mitochondrial unfolded protein response is associated with Her2-overexpression breast cancer. Breast Cancer Res Treat. 2020;183(1):61–70. doi: 10.1007/s10549-020-05729-9. [DOI] [PubMed] [Google Scholar]
- 104.Ahmed MW, Mahjabeen I, Gul S, Khursheed A, Mehmood A, Kayani MA. Relationship of single nucleotide polymorphisms and haplotype interaction of mitochondrial unfolded protein response pathway genes with head and neck cancer. Future Oncol (London, England) 2019;15(33):3819–3829. doi: 10.2217/fon-2019-0365. [DOI] [PubMed] [Google Scholar]
- 105.Kang YE, Kim JM, Lim MA, Lee SE, Yi S, Kim JT, Oh C, Liu L, Jin Y, Jung SN, et al. Growth differentiation factor 15 is a cancer cell-induced mitokine that primes thyroid cancer cells for invasiveness. Thyroid Off J Am Thyroid Assoc. 2021;31:772. doi: 10.1089/thy.2020.0034. [DOI] [PubMed] [Google Scholar]
- 106.Chen A, Qian D, Wang B, Hu M, Lu J, Qi Y, Liu DX. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int J Gynecol Pathol Off J Int Soc Gynecol Pathol. 2012;31(6):532–537. doi: 10.1097/PGP.0b013e31824df26b. [DOI] [PubMed] [Google Scholar]
- 107.Nukuda A, Endoh H, Yasuda M, Mizutani T, Kawabata K, Haga H. Role of ATF5 in the invasive potential of diverse human cancer cell lines. Biochem Biophys Res Commun. 2016;474(3):509–514. doi: 10.1016/j.bbrc.2016.04.131. [DOI] [PubMed] [Google Scholar]
- 108.Hu M, Wang B, Qian D, Li L, Zhang L, Song X, Liu DX. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 2012;32(10):4385–4394. [PubMed] [Google Scholar]
- 109.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118(10):2840–2848. doi: 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hua XM, Wang J, Qian DM, Song JY, Chen H, Zhu XL, Zhou R, Zhao YD, Zhou XZ, Li L, et al. DNA methylation level of promoter region of activating transcription factor 5 in glioma. J Zhejiang Univ Sci B. 2015;16(9):757–762. doi: 10.1631/jzus.B1500067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16(6):671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun X, Jefferson P, Zhou Q, Angelastro JM, Greene LA. Dominant-Negative ATF5 compromises cancer cell survival by targeting CEBPB and CEBPD. Mol Cancer Res MCR. 2020;18(2):216–228. doi: 10.1158/1541-7786.MCR-19-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jia M, Dahlman-Wright K, Gustafsson J. Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29(4):557–568. doi: 10.1016/j.beem.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 114.Xia X, Liao Y, Huang C, Liu Y, He J, Shao Z, Jiang L, Dou QP, Liu J, Huang H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019;465:118–128. doi: 10.1016/j.canlet.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 115.Huang B, Omoto Y, Iwase H, Yamashita H, Toyama T, Coombes RC, Filipovic A, Warner M, Gustafsson J. Differential expression of estrogen receptor α, β1, and β2 in lobular and ductal breast cancer. Proc Natl Acad Sci USA. 2014;111(5):1933–1938. doi: 10.1073/pnas.1323719111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kowalska K, Habrowska-Górczyńska DE, Urbanek KA, Domińska K, Piastowska-Ciesielska AW. Estrogen receptor α is crucial in zearalenone-induced invasion and migration of prostate cancer cells. Toxins. 2018;10(3):98. doi: 10.3390/toxins10030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Y, Pan C, Yu L, Ruan H, Chang L, Yang J, Zheng Z, Zheng F, Liu T. SSBP1 upregulation in colorectal cancer regulates mitochondrial mass. Cancer Manage Res. 2019;11:10093–10106. doi: 10.2147/CMAR.S211292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang G, Wang Q, Huang Q, Chen Y, Sun X, He L, Zhan L, Guo X, Yin C, Fang Y, et al. Upregulation of mtSSB by interleukin-6 promotes cell growth through mitochondrial biogenesis-mediated telomerase activation in colorectal cancer. Int J Cancer. 2019;144(10):2516–2528. doi: 10.1002/ijc.31978. [DOI] [PubMed] [Google Scholar]
- 120.Wang Y, Hu L, Zhang X, Zhao H, Xu H, Wei Y, Jiang H, Xie C, Zhou Y, Zhou F. Downregulation of mitochondrial single stranded DNA binding protein (SSBP1) induces mitochondrial dysfunction and increases the radiosensitivity in non-small cell lung cancer cells. J Cancer. 2017;8(8):1400–1409. doi: 10.7150/jca.18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou M, Yang Z, Wang D, Chen P, Zhang Y. The circular RNA circZFR phosphorylates Rb promoting cervical cancer progression by regulating the SSBP1/CDK2/cyclin E1 complex. J Exp Clin Cancer Res CR. 2021;40(1):48. doi: 10.1186/s13046-021-01849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiang HL, Sun HF, Gao SP, Li LD, Huang S, Hu X, Liu S, Wu J, Shao ZM, Jin W. SSBP1 suppresses TGFβ-driven epithelial-to-mesenchymal transition and metastasis in triple-negative breast cancer by regulating mitochondrial retrograde signaling. Can Res. 2016;76(4):952–964. doi: 10.1158/0008-5472.CAN-15-1630. [DOI] [PubMed] [Google Scholar]
- 123.Zhang B, Fan Y, Cao P, Tan K. Multifaceted roles of HSF1 in cell death: a state-of-the-art review. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188591. doi: 10.1016/j.bbcan.2021.188591. [DOI] [PubMed] [Google Scholar]
- 124.Yang T, Ren C, Lu C, Qiao P, Han X, Wang L, Wang D, Lv S, Sun Y, Yu Z. Phosphorylation of HSF1 by PIM2 induces PD-L1 expression and promotes tumor growth in breast cancer. Can Res. 2019;79(20):5233–5244. doi: 10.1158/0008-5472.CAN-19-0063. [DOI] [PubMed] [Google Scholar]
- 125.Gabai VL, Meng L, Kim G, Mills TA, Benjamin IJ, Sherman MY. Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR. Mol Cell Biol. 2012;32(5):929–940. doi: 10.1128/MCB.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen F, Fan Y, Cao P, Liu B, Hou J, Zhang B, Tan K. Pan-cancer analysis of the prognostic and immunological role of HSF1: a potential target for survival and immunotherapy. Oxid Med Cell Longev. 2021;2021:5551036. doi: 10.1155/2021/5551036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dong B, Jaeger AM, Hughes PF, Loiselle DR, Hauck JS, Fu Y, Haystead TA, Huang J, Thiele DJ. Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Sci Transl Med. 2020;12(574):eabb5647. doi: 10.1126/scitranslmed.abb5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shao J, Han B, Cao P, Zhang B, Liu M, Li D, Zhou N, Hao Q, Duan X, Chang Y, et al. HSF1 phosphorylation by cyclosporin A confers hyperthermia sensitivity through suppression of HSP expression. Biochim Biophys Acta. 2019;1862(8):846–857. doi: 10.1016/j.bbagrm.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 129.Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5(7):714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 130.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cappello F, Bellafiore M, Palma A, Marciano V, Martorana G, Belfiore P, Martorana A, Farina F, Zummo G, Bucchieri F. Expression of 60-kD heat shock protein increases during carcinogenesis in the uterine exocervix. Pathobiol J Immunopathol Mol Cell Biol. 2002;70(2):83–88. doi: 10.1159/000067304. [DOI] [PubMed] [Google Scholar]
- 132.Tsai YP, Teng SC, Wu KJ. Direct regulation of HSP60 expression by c-MYC induces transformation. FEBS Lett. 2008;582(29):4083–4088. doi: 10.1016/j.febslet.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 133.Castilla C, Congregado B, Conde JM, Medina R, Torrubia FJ, Japón MA, Sáez C. Immunohistochemical expression of Hsp60 correlates with tumor progression and hormone resistance in prostate cancer. Urology. 2010;76(4):1017.e1011–1016. doi: 10.1016/j.urology.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 134.Na Y, Kaul SC, Ryu J, Lee JS, Ahn HM, Kaul Z, Kalra RS, Li L, Widodo N, Yun CO, et al. Stress chaperone mortalin contributes to epithelial–mesenchymal transition and cancer metastasis. Can Res. 2016;76(9):2754–2765. doi: 10.1158/0008-5472.CAN-15-2704. [DOI] [PubMed] [Google Scholar]
- 135.Yi X, Luk JM, Lee NP, Peng J, Leng X, Guan XY, Lau GK, Beretta L, Fan ST. Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol Cell Proteomics MCP. 2008;7(2):315–325. doi: 10.1074/mcp.M700116-MCP200. [DOI] [PubMed] [Google Scholar]
- 136.Takano S, Wadhwa R, Yoshii Y, Nose T, Kaul SC, Mitsui Y. Elevated levels of mortalin expression in human brain tumors. Exp Cell Res. 1997;237(1):38–45. doi: 10.1006/excr.1997.3754. [DOI] [PubMed] [Google Scholar]
- 137.Li S, Lv M, Qiu S, Meng J, Liu W, Zuo J, Yang L. NF-κB p65 promotes ovarian cancer cell proliferation and migration via regulating mortalin. J Cell Mol Med. 2019;23(6):4338–4348. doi: 10.1111/jcmm.14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283(8):5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 139.Kim W, Ryu J, Kim JE. CCAR2/DBC1 and Hsp60 positively regulate expression of survivin in neuroblastoma cells. Int J Mol Sci. 2019;20(1):131. doi: 10.3390/ijms20010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ghosh JC, Siegelin MD, Dohi T, Altieri DC. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010;70(22):8988–8993. doi: 10.1158/0008-5472.CAN-10-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chun JN, Choi B, Lee KW, Lee DJ, Kang DH, Lee JY, Song IS, Kim HI, Lee SH, Kim HS, et al. Cytosolic Hsp60 is involved in the NF-kappaB-dependent survival of cancer cells via IKK regulation. PLoS ONE. 2010;5(3):e9422. doi: 10.1371/journal.pone.0009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo J, Li X, Zhang W, Chen Y, Zhu S, Chen L, Xu R, Lv Y, Wu D, Guo M, et al. HSP60-regulated mitochondrial proteostasis and protein translation promote tumor growth of ovarian Cancer. Sci Rep. 2019;9(1):12628. doi: 10.1038/s41598-019-48992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tang H, Li J, Liu X, Wang G, Luo M, Deng H. Down-regulation of HSP60 suppresses the proliferation of glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci Rep. 2016;6:28388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu X, Guo J, Chen Y, Liu X, Yang G, Wu Y, Tian Y, Liu N, Yang L, Wei S, et al. The 60-kDa heat shock protein regulates energy rearrangement and protein synthesis to promote proliferation of multiple myeloma cells. Br J Haematol. 2020;190(5):741–752. doi: 10.1111/bjh.16569. [DOI] [PubMed] [Google Scholar]
- 145.Merendino AM, Bucchieri F, Campanella C, Marcianò V, Ribbene A, David S, Zummo G, Burgio G, Corona DF, Conway de Macario E, et al. Hsp60 is actively secreted by human tumor cells. PLoS ONE. 2010;5(2):e9247. doi: 10.1371/journal.pone.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hayoun D, Kapp T, Edri-Brami M, Ventura T, Cohen M, Avidan A, Lichtenstein RG. HSP60 is transported through the secretory pathway of 3-MCA-induced fibrosarcoma tumour cells and undergoes N-glycosylation. FEBS J. 2012;279(12):2083–2095. doi: 10.1111/j.1742-4658.2012.08594.x. [DOI] [PubMed] [Google Scholar]
- 147.Wu PK, Hong SK, Park JI. Mortalin depletion induces MEK/ERK-dependent and ANT/CypD-mediated death in vemurafenib-resistant B-Raf(V600E) melanoma cells. Cancer Lett. 2021;502:25–33. doi: 10.1016/j.canlet.2020.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu PK, Hong SK, Starenki D, Oshima K, Shao H, Gestwicki JE, Tsai S, Park JI. Mortalin/HSPA9 targeting selectively induces KRAS tumor cell death by perturbing mitochondrial membrane permeability. Oncogene. 2020;39(21):4257–4270. doi: 10.1038/s41388-020-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mylonis I, Kourti M, Samiotaki M, Panayotou G, Simos G. Mortalin-mediated and ERK-controlled targeting of HIF-1α to mitochondria confers resistance to apoptosis under hypoxia. J Cell Sci. 2017;130(2):466–479. doi: 10.1242/jcs.195339. [DOI] [PubMed] [Google Scholar]
- 150.Starenki D, Sosonkina N, Hong SK, Lloyd RV, Park JI. Mortalin (GRP75/HSPA9) promotes survival and proliferation of thyroid carcinoma cells. Int J Mol Sci. 2019;20(9):2069. doi: 10.3390/ijms20092069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Starenki D, Hong SK, Lloyd RV, Park JI. Mortalin (GRP75/HSPA9) upregulation promotes survival and proliferation of medullary thyroid carcinoma cells. Oncogene. 2015;34(35):4624–4634. doi: 10.1038/onc.2014.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu PK, Hong SK, Park JI. Steady-state levels of phosphorylated mitogen-activated protein kinase kinase 1/2 determined by Mortalin/HSPA9 and protein phosphatase 1 alpha in KRAS and BRAF tumor cells. Mol Cell Biol. 2017;37(18):e00061-17. doi: 10.1128/MCB.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu PK, Hong SK, Veeranki S, Karkhanis M, Starenki D, Plaza JA, Park JI. A mortalin/HSPA9-mediated switch in tumor-suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol Cell Biol. 2013;33(20):4051–4067. doi: 10.1128/MCB.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsuneki M, Maruyama S, Yamazaki M, Xu B, Essa A, Abé T, Babkair H, Cheng J, Yamamoto T, Saku T. Extracellular heat shock protein A9 is a novel interaction partner of podoplanin in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2013;434(1):124–130. doi: 10.1016/j.bbrc.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 155.Yun CO, Bhargava P, Na Y, Lee JS, Ryu J, Kaul SC, Wadhwa R. Relevance of mortalin to cancer cell stemness and cancer therapy. Sci Rep. 2017;7:42016. doi: 10.1038/srep42016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ryu J, Kaul Z, Yoon AR, Liu Y, Yaguchi T, Na Y, Ahn HM, Gao R, Choi IK, Yun CO, et al. Identification and functional characterization of nuclear mortalin in human carcinogenesis. J Biol Chem. 2014;289(36):24832–24844. doi: 10.1074/jbc.M114.565929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheng W, Zhang B, Zikeliyar M, Wang J, Jian H, Wu K, Zhang Y, Ding J. Elevated Mortalin correlates with poor outcome in hepatocellular carcinoma. Ann Diagn Pathol. 2019;42:59–63. doi: 10.1016/j.anndiagpath.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 158.Xu M, Jin T, Chen L, Zhang X, Zhu G, Wang Q, Lin Z. Mortalin is a distinct bio-marker and prognostic factor in serous ovarian carcinoma. Gene. 2019;696:63–71. doi: 10.1016/j.gene.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 159.Cui X, Li Z, Piao J, Li J, Li L, Lin Z, Jin A. Mortalin expression in pancreatic cancer and its clinical and prognostic significance. Hum Pathol. 2017;64:171–178. doi: 10.1016/j.humpath.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 160.Jin H, Ji M, Chen L, Liu Q, Che S, Xu M, Lin Z. The clinicopathological significance of Mortalin overexpression in invasive ductal carcinoma of breast. J Exp Clin Cancer Res CR. 2016;35:42. doi: 10.1186/s13046-016-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dundas SR, Lawrie LC, Rooney PH, Murray GI. Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J Pathol. 2005;205(1):74–81. doi: 10.1002/path.1672. [DOI] [PubMed] [Google Scholar]
- 162.Hjerpe E, Egyhazi S, Carlson J, Stolt MF, Schedvins K, Johansson H, Shoshan M, Avall-Lundqvist E. HSP60 predicts survival in advanced serous ovarian cancer. Int J Gynecol Cancer Off J Int Gynecol Cancer Soc. 2013;23(3):448–455. doi: 10.1097/IGC.0b013e318284308b. [DOI] [PubMed] [Google Scholar]
- 163.Li XS, Xu Q, Fu XY, Luo WS. Heat shock protein 60 overexpression is associated with the progression and prognosis in gastric cancer. PLoS ONE. 2014;9(9):e107507. doi: 10.1371/journal.pone.0107507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meng Q, Li BX, Xiao X. Toward developing chemical modulators of Hsp60 as potential therapeutics. Front Mol Biosci. 2018;5:35. doi: 10.3389/fmolb.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ramraj SK, Elayapillai SP, Pelikan RC, Zhao YD, Isingizwe ZR, Kennedy AL, Lightfoot SA, Benbrook DM. Novel ovarian cancer maintenance therapy targeted at mortalin and mutant p53. Int J Cancer. 2020;147(4):1086–1097. doi: 10.1002/ijc.32830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang YH, Lin KH, Yu JS, Wu TJ, Lee WC, Chao CC, Pan TL, Yeh CT. Targeting HSP60 by subcutaneous injections of jetPEI/HSP60-shRNA destabilizes cytoplasmic survivin and inhibits hepatocellular carcinoma growth. Mol Carcinog. 2018;57(9):1087–1101. doi: 10.1002/mc.22827. [DOI] [PubMed] [Google Scholar]
- 167.Harper AK, Fletcher NM, Fan R, Morris RT, Saed GM. Heat Shock Protein 60 (HSP60) serves as a potential target for the sensitization of chemoresistant ovarian cancer cells. Reprod Sci (Thousand Oaks, Calif) 2020;27(4):1030–1036. doi: 10.1007/s43032-019-00089-2. [DOI] [PubMed] [Google Scholar]
- 168.Goard CA, Schimmer AD. Mitochondrial matrix proteases as novel therapeutic targets in malignancy. Oncogene. 2014;33(21):2690–2699. doi: 10.1038/onc.2013.228. [DOI] [PubMed] [Google Scholar]
- 169.Nouri K, Feng Y, Schimmer AD. Mitochondrial ClpP serine protease-biological function and emerging target for cancer therapy. Cell Death Dis. 2020;11(10):841. doi: 10.1038/s41419-020-03062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gibellini L, Pinti M, Boraldi F, Giorgio V, Bernardi P, Bartolomeo R, Nasi M, De Biasi S, Missiroli S, Carnevale G, et al. Silencing of mitochondrial Lon protease deeply impairs mitochondrial proteome and function in colon cancer cells. FASEB J Off Publ Fed Am Soc Exp Biol. 2014;28(12):5122–5135. doi: 10.1096/fj.14-255869. [DOI] [PubMed] [Google Scholar]
- 171.Seo JH, Rivadeneira DB, Caino MC, Chae YC, Speicher DW, Tang HY, Vaira V, Bosari S, Palleschi A, Rampini P, et al. The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol. 2016;14(7):e1002507. doi: 10.1371/journal.pbio.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N, et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27(6):864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Luo J, Zeng B, Tao C, Lu M, Ren G. ClpP regulates breast cancer cell proliferation, invasion and apoptosis by modulating the Src/PI3K/Akt signaling pathway. PeerJ. 2020;8:e8754. doi: 10.7717/peerj.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu C, Wang H, Li H, Chen X, Wu X, Lu B, Zhang W, Zhou Y, Xiao GG, Gao G. Inhibition of LONP1 suppresses pancreatic cancer progression via c-Jun N-terminal kinase pathway-meditated epithelial-mesenchymal transition. Pancreas. 2019;48(5):629–635. doi: 10.1097/MPA.0000000000001321. [DOI] [PubMed] [Google Scholar]
- 175.Zhang Y, Maurizi MR. Mitochondrial ClpP activity is required for cisplatin resistance in human cells. Biochem Biophys Acta. 2016;1862(2):252–264. doi: 10.1016/j.bbadis.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Quirós PM, Español Y, Acín-Pérez R, Rodríguez F, Bárcena C, Watanabe K, Calvo E, Loureiro M, Fernández-García MS, Fueyo A, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8(2):542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 177.Ghosh JC, Seo JH, Agarwal E, Wang Y, Kossenkov AV, Tang HY, Speicher DW, Altieri DC. Akt phosphorylation of mitochondrial Lonp1 protease enables oxidative metabolism and advanced tumor traits. Oncogene. 2019;38(43):6926–6939. doi: 10.1038/s41388-019-0939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li J, Agarwal E, Bertolini I, Seo JH, Caino MC, Ghosh JC, Kossenkov AV, Liu Q, Tang HY, Goldman AR, et al. The mitophagy effector FUNDC1 controls mitochondrial reprogramming and cellular plasticity in cancer cells. Sci Signal. 2020;13(642):eaaz8240. doi: 10.1126/scisignal.aaz8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee YG, Kim HW, Nam Y, Shin KJ, Lee YJ, Park DH, Rhee HW, Seo JK, Chae YC. LONP1 and ClpP cooperatively regulate mitochondrial proteostasis for cancer cell survival. Oncogenesis. 2021;10(2):18. doi: 10.1038/s41389-021-00306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kao TY, Chiu YC, Fang WC, Cheng CW, Kuo CY, Juan HF, Wu SH, Lee AY. Mitochondrial Lon regulates apoptosis through the association with Hsp60-mtHsp70 complex. Cell Death Dis. 2015;6(2):e1642. doi: 10.1038/cddis.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Maneix L, Sweeney MA, Lee S, Iakova P, Moree SE, Sahin E, Lulla P, Yellapragada SV, Tsai FTF, Catic A. The Mitochondrial protease LonP1 promotes proteasome inhibitor resistance in multiple myeloma. Cancers. 2021;13(4):843. doi: 10.3390/cancers13040843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kuo CL, Chou HY, Chiu YC, Cheng AN, Fan CC, Chang YN, Chen CH, Jiang SS, Chen NJ, Lee AY. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020;474:138–150. doi: 10.1016/j.canlet.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 183.Wong KS, Houry WA. Chemical modulation of human mitochondrial ClpP: potential application in cancer therapeutics. ACS Chem Biol. 2019;14(11):2349–2360. doi: 10.1021/acschembio.9b00347. [DOI] [PubMed] [Google Scholar]
- 184.Wong KS, Houry WA. Recent advances in targeting human mitochondrial AAA+ poteases to develop novel cancer therapeutics. Adv Exp Med Biol. 2019;1158:119–142. doi: 10.1007/978-981-13-8367-0_8. [DOI] [PubMed] [Google Scholar]
- 185.Gibellini L, Pinti M, Bartolomeo R, De Biasi S, Cormio A, Musicco C, Carnevale G, Pecorini S, Nasi M, De Pol A, et al. Inhibition of Lon protease by triterpenoids alters mitochondria and is associated to cell death in human cancer cells. Oncotarget. 2015;6(28):25466–25483. doi: 10.18632/oncotarget.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ishizawa J, Zarabi SF, Davis RE, Halgas O, Nii T, Jitkova Y, Zhao R, St-Germain J, Heese LE, Egan G, et al. Mitochondrial ClpP-mediated proteolysis induces selective cancer cell lethality. Cancer Cell. 2019;35(5):721–737.e729. doi: 10.1016/j.ccell.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Graves PR, Aponte-Collazo LJ, Fennell EMJ, Graves AC, Hale AE, Dicheva N, Herring LE, Gilbert TSK, East MP, McDonald IM, et al. Mitochondrial protease ClpP is a target for the anticancer compounds ONC201 and related analogues. ACS Chem Biol. 2019;14(5):1020–1029. doi: 10.1021/acschembio.9b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wong KS, Mabanglo MF, Seraphim TV, Mollica A, Mao YQ, Rizzolo K, Leung E, Moutaoufik MT, Hoell L, Phanse S, et al. Acyldepsipeptide analogs dysregulate human mitochondrial ClpP protease activity and cause apoptotic cell death. Cell Chem Biol. 2018;25(8):1017–1030.e1019. doi: 10.1016/j.chembiol.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 189.Zhu L, Zhou Q, He L, Chen L. Mitochondrial unfolded protein response: an emerging pathway in human diseases. Free Radical Biol Med. 2021;163:125–134. doi: 10.1016/j.freeradbiomed.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 190.Zhu L, Luo X, Fu N, Chen L. Mitochondrial unfolded protein response: A novel pathway in metabolism and immunity. Pharmacol Res. 2021;168:105603. doi: 10.1016/j.phrs.2021.105603. [DOI] [PubMed] [Google Scholar]
- 191.Pérez MJ, Ivanyuk D, Panagiotakopoulou V, Di Napoli G, Kalb S, Brunetti D, Al-Shaana R, Kaeser SA, Fraschka SA, Jucker M, et al. Loss of function of the mitochondrial peptidase PITRM1 induces proteotoxic stress and Alzheimer's disease-like pathology in human cerebral organoids. Mol Psychiatry. 2020;267:24193. doi: 10.1038/s41380-020-0807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol. 2018;19(2):109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 193.Smyrnias I. The mitochondrial unfolded protein response and its diverse roles in cellular stress. Int J Biochem Cell Biol. 2021;133:105934. doi: 10.1016/j.biocel.2021.105934. [DOI] [PubMed] [Google Scholar]
- 194.Muñoz-Carvajal F, Sanhueza M. The mitochondrial unfolded protein response: a hinge between healthy and pathological aging. Front Aging Neurosci. 2020;12:581849. doi: 10.3389/fnagi.2020.581849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shpilka T, Du Y, Yang Q, Melber A, Uma Naresh N, Lavelle J, Kim S, Liu P, Weidberg H, Li R, et al. UPR(mt) scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nat Commun. 2021;12(1):479. doi: 10.1038/s41467-020-20784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sen B, Rastogi A, Nath R, Shasthry SM, Pamecha V, Pandey S, Gupta KJ, Sarin SK, Trehanpati N, Ramakrishna G. Senescent hepatocytes in decompensated liver show reduced UPR(MT) and its key player, CLPP, attenuates senescence in vitro. Cell Mol Gastroenterol Hepatol. 2019;8(1):73–94. doi: 10.1016/j.jcmgh.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mohrin M, Widjaja A, Liu Y, Luo H, Chen D. The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell. 2018;17(3):e12756. doi: 10.1111/acel.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang B, Tan Y, Zhang Z, Feng P, Ding W, Wang Q, Liang H, Duan W, Wang X, Yu S, et al. Novel PGC-1α/ATF5 axis partly activates UPR(mt) and mediates cardioprotective role of tetrahydrocurcumin in pathological cardiac hypertrophy. Oxid Med Cell Longev. 2020;2020:9187065. doi: 10.1155/2020/9187065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, Patel MR. Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab. 2016;24(1):91–103. doi: 10.1016/j.cmet.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lin YF, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature. 2016;533(7603):416–419. doi: 10.1038/nature17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jiao H, Jiang D, Hu X, Du W, Ji L, Yang Y, Li X, Sho T, Wang X, Li Y, et al. Mitocytosis, a migrasome-mediated mitochondrial quality-control process. Cell. 2021;184(11):2896–2910.e2813. doi: 10.1016/j.cell.2021.04.027. [DOI] [PubMed] [Google Scholar]
- 202.Hu D, Liu Z, Qi X. Mitochondrial quality control strategies: potential therapeutic targets for neurodegenerative diseases? Front Neurosci. 2021;15:746873. doi: 10.3389/fnins.2021.746873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Knopman DS, Amieva H, Petersen RC, Chételat G, Holtzman DM, Hyman BT, Nixon RA, Jones DT. Alzheimer disease. Nat Rev Dis Primers. 2021;7(1):33. doi: 10.1038/s41572-021-00269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 205.Beck JS, Mufson EJ, Counts SE. Evidence for Mitochondrial UPR gene activation in familial and sporadic Alzheimer's disease. Curr Alzheimer Res. 2016;13(6):610–614. doi: 10.2174/1567205013666151221145445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shen Y, Ding M, Xie Z, Liu X, Yang H, Jin S, Xu S, Zhu Z, Wang Y, Wang D, et al. Activation of mitochondrial unfolded protein response in SHSY5Y expressing APP cells and APP/PS1 mice. Front Cell Neurosci. 2019;13:568. doi: 10.3389/fncel.2019.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D'Amico D, Moullan N, Potenza F, Schmid AW, Rietsch S, et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature. 2017;552(7684):187–193. doi: 10.1038/nature25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Karpel-Massler G, Horst BA, Shu C, Chau L, Tsujiuchi T, Bruce JN, Canoll P, Greene LA, Angelastro JM, Siegelin MD. A Synthetic cell-penetrating dominant-negative ATF5 peptide exerts anticancer activity against a broad spectrum of treatment-resistant cancers. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22(18):4698–4711. doi: 10.1158/1078-0432.CCR-15-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.