Abstract
Background
Among the Mangifera species, mango (Mangifera indica) is an important commercial fruit crop. However, very few studies have been conducted on the Mangifera mitochondrial genome. This study reports and compares the newly sequenced mitochondrial genomes of three Mangifera species.
Results
Mangifera mitochondrial genomes showed partial similarities in the overall size, genomic structure, and gene content. Specifically, the genomes are circular and contain about 63–69 predicted functional genes, including five ribosomal RNA (rRNA) genes and 24–27 transfer RNA (tRNA) genes. The GC contents of the Mangifera mitochondrial genomes are similar, ranging from 44.42–44.66%. Leucine (Leu) and serine (Ser) are the most frequently used, while tryptophan (Trp) and cysteine (Cys) are the least used amino acids among the protein-coding genes in Mangifera mitochondrial genomes. We also identified 7–10 large chloroplast genomic fragments in the mitochondrial genome, ranging from 1407 to 6142 bp. Additionally, four intact mitochondrial tRNAs genes (tRNA-Cys, tRNA-Trp, tRNA-Pro, and tRNA-Met) and intergenic spacer regions were identified. Phylogenetic analysis based on the common protein-coding genes of most branches provided a high support value.
Conclusions
We sequenced and compared the mitochondrial genomes of three Mangifera species. The results showed that the gene content and the codon usage pattern of Mangifera mitochondrial genomes is similar across various species. Gene transfer from the chloroplast genome to the mitochondrial genome were identified. This study provides valuable information for evolutionary and molecular studies of Mangifera and a basis for further studies on genomic breeding of mango.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08383-1.
Keywords: Mangifera, Mitochondrial genome, Gene transfer, Phylogenetic analysis
Introduction
Mitochondria are semi-autonomous organelles found in virtually all eukaryotic cells, and their main function is to generate adenosine triphosphate (ATP) [1]. With the rapid development of genome assembly and sequencing techniques, various complete organellar genomes are being assembled. According to GenBank Organelle Genome Resources (https://www.ncbi.nlm.nih.gov/genome/browse/), the complete mitochondrial genomes currently assembled are less than one-tenth of the chloroplast genomes, indicating that mitochondrial genome assembly may be difficult and complex. Plant mitochondrial genomes are large and variable in size [2, 3], and their structural heterogeneity and gene sequences have meager base substitution rates [4, 5]. The sequences generally have large repeats that mediate recombinational isomerization within a species [4] and numerous non-tandem repeats of 50 bp and up that mediate recombination sometimes within a species, and likely the rearrangements seen between species [6, 7]. The evolution of the mitochondrial genome involves many structural rearrangements and gene transfer [8, 9]. An important feature of plant mitochondrial genome evolution is the transfer of genes between the mitochondria and the chloroplast genomes [10, 11]. Therefore, tracking the transfer of genes between organellar genomes is essential for understanding the evolution of plant mitochondrial genomes.
Among the genus Mangifera, mango (M. indica) is an important tropical fruit [12, 13], native to tropical and subtropical regions of Southeast Asia [14, 15]. Mango has a wide cultivation range [16], high nutritional value, esthetic appearance, and unique flavor [17]. Thus, it is well accepted by consumers, earning a reputation of “king of tropical fruits” [18]. However, very few studies have been conducted on the Mangifera mitochondrial genome. To date, the complete mitochondrial genome of only one Mangifera species has been deposited in GenBank [19].
In this study, the mitochondrial genomes of three Mangifera species were sequenced and compared with M. indica [19]. This study aimed to: (1) comparatively analyze the mitochondrial genome structures of four Mangifera species; (2) assess gene transfers between the chloroplast and mitochondrial genomes; (3) explore the evolutionary relationships among the Mangifera species based on the protein-coding genes of the mitochondrial genome.
Materials and methods
Plant material and sequencing
Fresh leaves of the three Mangifera species (M. persiciformis, M. longipes, and M. sylvatica) were collected from Xishuangbanna Tropical Flowers and Plants Garden. Total genomic DNA was extracted from all samples using cetyltrimethylammonium bromide (CTAB) method [20]. DNA samples were stored at − 80 °C until use.
About 5–10 μg of total DNA from each sample was used to construct a shotgun library with an average insertion size of 350 bp. Illumina Novaseq6000 (Illumina, USA) was used to sequence the DNA samples in the paired-end sequencing mode. The generated mitochondrial genome raw reads were approximately 0.7–1.2 Gb per sample. Meanwhile, the mitochondrial genome depth of coverage was more than 900 × .
Mitochondrial genome assembly, annotation, and sequence analysis
The raw sequencing data were filtered using Trimmomatic v0.38 [21], and the SPAdes v3.5.0 was used to de novo assembled with different K-mer parameters [22]. For the regions with complex structures or low sequencing coverage, polymerase chain reaction (PCR) with Sanger sequencing was used for validation to ensure that the circular genome sequence was complete and accurate.
Mitochondrial genome annotation was performed using BLASTN and BLASTX alignment tools of the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) using Dicotyledoneae species mitochondrial genomes as the reference sequences. The tRNAs were predicted using the tRNAscan-SE 2.0 [23] software, and their secondary structure maps were generated using ARWEN [24]. A circular diagram of the mitochondrial genomes of Mangifera was drawn using OGDRAW v1.3.1 [25].
MEGA v7.0.26 was used to calculate the amino acid composition of the protein-coding genes and their relative synonymous codon usage (RSCU) values [26]. The GC and AT skews were calculated according to the formula: GC skew = (G − C)/(G + C), AT skew = (A − T)/(A + T). Meanwhile, simple sequence repeats (SSR) were detected using MISA software (http://pgrc.ipk-gatersleben.de/misa/misa.html) [27]. Analysis of non-tandem repeats in mitochondrial genomes larger than 50 bp was performed using REPuter [28].
Identification of chloroplast gene insertion in the mitochondrial genome
We mapped the mitochondrial genome to the plastid genome using the BLASTN tool with the default settings. Circos v0.69 [29] software was then used to map the mitochondrial and chloroplast genomes and gene transfer segments of Mangifera plants.
Phylogenetic analysis
Phylogenetic analyses were performed for the 15 Dicotyledoneae species using Carica papaya as outgroups. Protein-coding genes common to the mitochondrial genomes of all species were extracted to construct phylogenetic trees. MUSCLE v.3.8.31 [30] was used to align the mitochondrial gene sequences of the Dicotyledoneae species. Phylogenetic analysis was performed using the maximum likelihood (ML) method via RAxML v8.1.5 with 1000 bootstrap replicates [31].
Results
Basic characteristics of the Mangifera mitochondrial genomes
Raw sequence data were obtained from M. longipes (MZ751075), M. persiciformis (MZ751076), and M. sylvatica (MZ751077). The three newly sequenced Mangifera mitochondrial genomes have been deposited in the GenBank database.
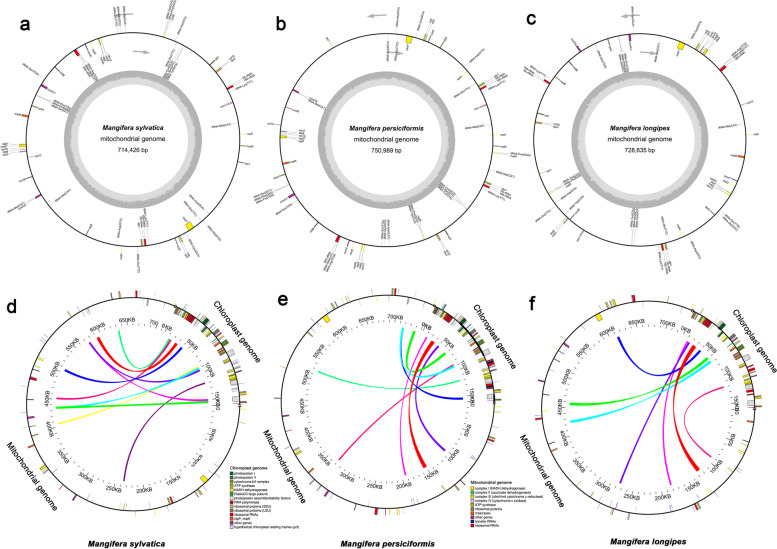
The mitochondrial genome sizes of M. longipes, M. persiciformis, and M. sylvatica were 728,635 bp, 750,898 bp, and 714,426 bp, respectively (Fig. 1a-c; Table 1). Furthermore, the GC contents of the various Mangifera mitochondrial genomes were similar, ranging from 44.42–44.66%. The four Mangifera mitochondrial genomes contained 63–69 predicted functional genes, including five ribosomal RNA (rRNA) genes and 24–27 transfer RNA (tRNA) genes (Table 1). The number and type of genes were partially similar among the four Mangifera mitochondrial genomes. The three newly sequenced mitochondrial genomes of Mangifera species were submitted to GenBank with the accession numbers; MZ751075, MZ751076, and MZ751077.
Fig. 1.
Sequence diagram of Mangifera mitochondrial genomes (a-c), and schematic representation of gene transfers between chloroplast and mitochondrial genomes in Mangifera species (d-f). The chloroplast and mitochondrial genomes used for alignment were from the same species [32]. Genes within a circle are transcribed clockwise, while those outside the circle are transcribed counterclockwise. Colored lines within the circle show the points of chloroplast genome insertion in the mitochondrial genome. (The assembly may best fit on a circular map, but there is no evidence for a circular genomic molecule so far)
Table 1.
Genomic features of Mangifera mitochondrial genome
Gene transfer between the mitochondrial and chloroplast genomes
Mangifera mitochondrial genomes are approximately 4.5–5.5 times larger than the chloroplast genomes. We identified 7–10 large chloroplast genomic fragments in the mitochondrial genomes, including genes and intergenomic regions (Fig. 1d-f; Table S1). These fragments ranged from 1407 to 6142 bp, and the sequences had more than 96% similarity in the mitochondrial and chloroplast genomes. The chloroplast genome segments transferred into the mitochondrial genome observed in the three Mangifera species included five intact chloroplast genes (petN, psaA, atpI, trnI-CAU, and trnC-GCA) and many partial genes and intergenic spacers. Four mitochondrial tRNAs genes (tRNA-Cys, tRNA-Trp, tRNA-Pro, and tRNA-Met) and intergenic spacer regions were also identified (Table S1).
Codon usage and AT-skew analysis of the protein-coding genes
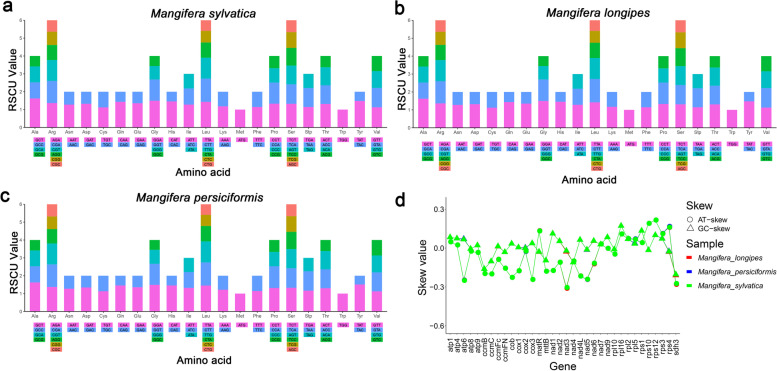
Relative synonymous codon usage (RSCU) analysis of the Mangifera mitochondrial genomes are shown in Fig. 2a-c, indicating that all codons are present in the protein-coding genes. The 34–36 protein-coding genes of the three newly sequenced Mangifera mitochondrial genomes contain 9401–10,102 codons (Table S2, S3, S4). The results showed that A or T nucleotides were used in high frequency in the third codon position compared to other nucleotides. The most frequent codons used were TTT (Phe), ATT (Ile), and GAA (Glu) and were used ≥278 times in the protein-coding genes of the three newly sequenced Mangifera mitochondrial genomes. In contrast, codons with a third codon G or C were used rarely (≤ 73), such as TGC (Cys), CAC (Arg) and TAC (Tyr). This may be a kind of AT mutation bias that has an obvious influence on codon. Notably, the codon usage pattern was highly consistent across the Mangifera mitochondrial genomes. The protein-coding genes of the majority strands showed positive AT and negative GC skews, while those of the minority strands showed positive GC and negative AT skews. The AT skews of the protein-coding genes was highly consistent across the Mangifera mitochondrial genomes (Fig. 2d; Table S5).
Fig. 2.
Relative synonymous codon usage (RSCU) (a-c) and AT-skew analysis of the protein-coding genes (d) in the three Mangifera mitochondrial genomes
Repeat sequence analysis of the Mangifera mitochondrial genomes
A total of 82–85 SSRs were identified in the Mangifera mitochondrial genomes. Mononucleotide repeats of A/T were more prevalent than the other repeat types, dinucleotide repeats (AT/TA) were the second most numerous, while tri- and hexa-nucleotide repeats were less numerous and occurred only in intergenic or intronic regions (Table S6, S7, S8). Besides SSRs, 56–76 non-tandem repeats with lengths ≥50 bp (total length: 26,115–32,557 bp; 3.5–4.5% of the genome) were also identified in the Mangifera mitochondrial genomes (Table S9, S10, S11).
Phylogenetic relationship among the Mangifera species
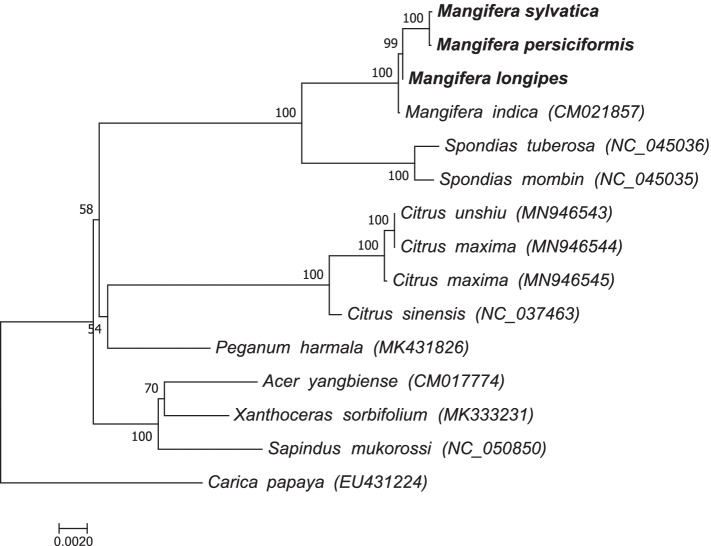
The protein-coding genes common to the mitochondrial genomes of the Dicotyledoneae species were used to infer the phylogenetic location of Mangifera species. The ML trees were constructed based on the 19 shared protein-coding genes (PCGs) (RPS3, COX1, COX2, COX3, NAD9, CCMFN, CCMFC, CCMC, CCMB, ATP9, ATP1, ATP6, ATP4, RPL16, MTTB, NAD2, NAD4L, NAD6, and NAD7). The ML analyses revealed that most branches had very high support. Within the Mangifera genus, M. sylvatica was evolutionarily closer to M. persiciformis (Fig. 3).
Fig. 3.
ML phylogenetic tree of four Mangifera species with 11 species in Dicotyledoneae based on the common protein-coding genes. Numbers related to the branches are ML bootstrap values
Discussion
Plant mitochondrial genomes have undergone rapid and tremendous structural changes since the initial endosymbiotic event [33, 34]. Because of this evolutionary pattern, the genomic composition of mitochondria has become complex, making plant mitochondrial genome studies comparatively challenging [35, 36]. Here, we sequenced and assembled mitochondrial genomes of three Mangifera species. Because of the high recombination frequency, plant mitochondrial genomes have a dynamic structure with multiple configurations such as major loops, sub loops and linear molecules in mitochondria [7]. In the present study, after the gap-filling step, the three Mangifera mitochondrial genomes was assembled into a single, circular molecule. A comparative analysis with the M. indica mitochondrial genome revealed that the GC content of the four mitochondrial genomes is similar among the Mangifera species [19]. Moreover, the rRNA (represented by three genes; 5S subunits, small 18S, and large 26S) and tRNA varies in number (from 24 to 27) and origin (mitochondrial or chloroplast) in the Mangifera mitochondrial genomes. The mitochondrial genomes of the common ancestor of angiosperms consist of 41 protein-coding genes [37]. About 34–38 genes of the 41 protein-coding genes were detected in the Mangifera mitochondrial genomes, implying that the deleted relevant genes might have been transferred to the nuclear genome, a common phenomenon during angiosperm evolution [38]. Thus, the three newly sequenced Mangifera mitochondrial genomes provide new insights into the function and structure of the mitochondrial genomes in Mangifera species.
Gene transfer in cells occurs between different organelles, including the chloroplast, mitochondria, and nucleus [39, 40]. Gene transfer from chloroplasts to mitochondrial genomes is common during long-term plant evolution [39, 41], both intracellular gene transfer and horizontal gene transfer are involved in the process of gene transfer between the chloroplast and mitochondrial genomes [42]. Compared to our previous study [32], the three newly mitochondrial genomes provide new findings in respect to gene transfer from the Mangifera chloroplast genome to the mitogenome. First, we found that the chloroplast genome segments transferred into the mitochondrial genome observed in the Mangifera species were relatively conserved (Fig. 1d-f; Table S1), these chloroplast genome segments transferred to the mitogenome were also found in species of other genera [39, 43]. In addition, we also found that Mangifera has a broadly divergent segment location resulting from chloroplast gene insertion into the mitochondrial genome. Intracellular gene transfers may account for the high degree of rearrangements among the mitochondrial genomes [44]. Because the chloroplast genome segments transferred into the mitochondria highly aligned with the original chloroplast genome sequences, such gene transfers might have caused disassembly of the mitochondrial genomes.
The codon usage bias was measured by calculating the relative synonymous codon usage (RSCU). The results indicate a strong A or T bias in the third position of the codon in the protein-coding genes of the Mangifera mitochondrial genomes, which is commonly observed in plant mitochondrial genomes [1, 44]. Codon usage pattern in the Mangifera mitochondrial genomes were highly consistent.
Plant mitochondrial genomes are rich in repeat sequences [45], the vast majority of differences in the size of plant mitochondrial genomes can be explained by differences in the size of the repeat sequences. SSRs and non-tandem repeats from three newly sequenced Mangifera mitochondrial genomes were investigated in this study. SSRs are important molecular markers for species identification, evolutionary analysis and studying genetic diversity [46]. Almost all angiosperm mitochondrial genomes have large (> 1 kb) non-tandem repeats and they are recombinationally active [6]. The longest non-tandem repeat in the mitochondrial genomes of Mangifera are 8348–13,655 bp and may be one of the reasons for isomerization. Previous studies have uncovered the genetic relationships among the Mangifera species through morphological, nuclear, internal ribosomal transcribed spacer (ITS), amplified fragment length polymorphism (AFLP), and chloroplast gene analyses [32, 47–50]. Plant mitochondria have evolved rapidly, resulting in heterogeneity, large-scale genomic reorganization, and gene mosaicism in the mitochondrial genomes of various species [34, 51]. Size and structural variations of plant mitochondrial genomes are evident, but functional genes remain conserved [36, 52]. Here, sequence-based phylogenetic tree was constructed using the protein-coding genes to explore the evolutionary relationship between Mangifera and Dicotyledoneae species. There were several inconsistencies between the chloroplast and mitochondrial phylogenetic trees regarding the phylogenetic topology [32]. Differential inheritance of organelles in the same cytoplasm can disrupt the linkage disequilibrium between mitochondrion and chloroplast [53, 54], and if this occurs, phylogenetic reconstruction of the two organelle genomes could conflict. In addition, limited sampling, incomplete lineage classification, and differences in the evolutionary rates could account for the conflicting phylogenetic reconstruction of the two organelle genomes [55]. Therefore, larger-scale sampling is required better to understand the evolution of the mitochondrial genome of Mangifera. Sequencing and assembling the three new complete mitochondrial genomes of Mangifera is the first step towards understanding the mitochondrial genome variation in this genus.
Conclusions
Here, we sequenced and compared the mitochondrial genomes of three Mangifera species. The results showed that the gene content and the codon usage pattern was highly consistent across the Mangifera mitochondrial genomes. We also identified 7–10 large fragments transferred from the chloroplast genome to the mitochondrial genome. The findings of this study provide valuable genetic resources for further studies on Mangifera species.
Supplementary Information
Acknowledgments
We are grateful to Zhangguang Ni for his assistance in sample collection.
Collection of plant material
We have permission to collect Mangifera species. The collection of plant material comply with relevant institutional, national, and international guidelines and legislation.
Authors’ contributions
Yingfeng Niu and Jin Liu conceived the study, wrote and revised the manuscript. Chengwen Gao performed the data analyses and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32160396), Youth Talent Growth Fund of YITC (No. QNCZ2020–3), Sci-tech Innovation System Construction for Tropical Crops Grant of Yunnan Province (No. RF2021), Technology Innovation Talents Project of Yunnan Province (No. 2018HB086), and Natural Science Foundation of Shandong Province (No. ZR2020QH321).
Availability of data and materials
The data supporting the findings of this study are freely available in GenBank on the NCBI website at https://www.ncbi.nlm.nih.gov, using the accession number MZ751075, MZ751076, and MZ751077. Raw sequencing data have been deposited at the NCBI Sequence Read Archive (SRA) under accession PRJNA778602.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chengwen Gao, Email: gaochengwen@qdu.edu.cn.
Jin Liu, Email: liujin06@126.com.
References
- 1.Bi CW, Lu N, Xu YQ, He CP, Lu ZH. Characterization and Analysis of the Mitochondrial Genome of Common Bean (Phaseolus vulgaris) by Comparative Genomic Approaches. Int J Mol Sci. 2020;21(11):3778. [DOI] [PMC free article] [PubMed]
- 2.Oldenburg DJ, Bendich AJ. DNA maintenance in plastids and mitochondria of plants. Front Plant Sci. 2015;6:883 [DOI] [PMC free article] [PubMed]
- 3.Bendich AJ. Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol Direct. 2010;5:42. [DOI] [PMC free article] [PubMed]
- 4.Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28(1–2):87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 5.Gualberto JM, Newton KJ. Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. In: Merchant SS, editor. Annual Review of Plant Biology, Vol 68. 2017. pp. 225–252. [DOI] [PubMed] [Google Scholar]
- 6.Wynn EL, Christensen AC. Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3-genes genomes. Genetics. 2019;9(2):549–559. doi: 10.1534/g3.118.200948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozik A, Rowan BA, Lavelle D, Berke L, Schranz ME, Michelmore RW, et al. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. Plos Genet. 2019;15(8):e1008373. [DOI] [PMC free article] [PubMed]
- 8.Allen JF. Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc Natl Acad Sci U S A. 2015;112(33):10231–10238. doi: 10.1073/pnas.1500012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu ZQ, Liao XZ, Zhang XN, Tembrock LR, Broz A. Genomic architectural variation of plant mitochondria-a review of multichromosomal structuring. J Syst Evol. 2022;60(1):160–8.
- 10.Turmel M, Otis C, Lemieux C. Mitochondrion-to-chloroplast DNA transfers and Intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae) Genome Biol Evol. 2016;8(9):2789–2805. doi: 10.1093/gbe/evw190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan DB, Wu ZQ. History of plastid DNA insertions reveals weak deletion and AT mutation biases in angiosperm mitochondrial genomes. Genome Biol Evol. 2014;6(12):3210–3221. doi: 10.1093/gbe/evu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iquebal MA, Jaiswal S, Mahato AK, Jayaswal PK, Angadi UB, Kumar N, Sharma N, Singh AK, Srivastav M, Prakash J, et al. MiSNPDb: a web-based genomic resources of tropical ecology fruit mango (Mangifera indica L.) for phylogeography and varietal differentiation. Sci Rep. 2017;7:9. doi: 10.1038/s41598-017-14998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lora J, Hormaza JI. Pollen wall development in mango (Mangifera indica L., Anacardiaceae) Plant Reprod. 2018;31(4):385–397. doi: 10.1007/s00497-018-0342-5. [DOI] [PubMed] [Google Scholar]
- 14.Dutta SK, Srivastav M, Rymbai H, Chaudhary R, Singh AK, Dubey AK, Lal K. Pollen-pistil interaction studies in mango (Mangifera indica L.) cultivars. Sci Hortic. 2013;160:213–221. [Google Scholar]
- 15.Sherman A, Rubinstein M, Eshed R, Benita M, Ish-Shalom M, Sharabi-Schwager M, Rozen A, Saada D, Cohen Y, Ophir R. Mango (Mangifera indica L.) germplasm diversity based on single nucleotide polymorphisms derived from the transcriptome. BMC Plant Biol. 2015;15:11. doi: 10.1186/s12870-015-0663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajpai A, Muthukumar M, Ahmad I, Ravishankar KV, Parthasarthy VA, Sthapit B, Rao R, Verma SP, Rajan S. Molecular and morphological diversity in locally grown non-commercial (heirloom) mango varieties of North India. JEnvironBiol. 2016;37(2):221–228. [PubMed] [Google Scholar]
- 17.Surapaneni M, Vemireddy LR, Begum H, Reddy BP, Neetasri C, Nagaraju J, Anwar SY, Siddiq EA. Population structure and genetic analysis of different utility types of mango (Mangifera indica L.) germplasm of Andhra Pradesh state of India using microsatellite markers. Plant Syst Evol. 2013;299(7):1215–1229. [Google Scholar]
- 18.Khan AS, Ali S, Khan IA. Morphological and molecular characterization and evaluation of mango germplasm: an overview. Sci Hortic. 2015;194:353–366. [Google Scholar]
- 19.Wang P, Luo YF, Huang JF, Gao SH, Zhu GP, Dang ZG, et al. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020;21(1):60. [DOI] [PMC free article] [PubMed]
- 20.Li J, Wang S, Jing Y, Ling W, Zhou S. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013;48(1):72–8.
- 21.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe TM, Chan PP. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laslett D, Canback B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24(2):172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 25.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 32.Niu YF, Gao CW, Liu J. Comparative analysis of the complete plastid genomes of Mangifera species and gene transfer between plastid and mitochondrial genomes. PeerJ. 2021;9:e10774. doi: 10.7717/peerj.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sloan DB, Warren JM, Williams AM, Wu ZQ, Abdel-Ghany SE, Chicco AJ, Havird JC. Cytonuclear integration and co-evolution. Nat Rev Genet. 2018;19(10):635–648. doi: 10.1038/s41576-018-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 2015;112(33):10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloan DB. Using plants to elucidate the mechanisms of cytonuclear co-evolution. New Phytol. 2015;205(3):1040–1046. doi: 10.1111/nph.12835. [DOI] [PubMed] [Google Scholar]
- 36.Hong Z, Liao XZ, Ye YJ, Zhang NN, Yang ZJ, Zhu WD, et al. A complete mitochondrial genome for fragrant Chinese rosewood (Dalbergia odorifera, Fabaceae) with comparative analyses of genome structure and intergenomic sequence transfers. BMC Genomics. 2021;22(1):672. [DOI] [PMC free article] [PubMed]
- 37.Mower JP, Sloan D, Alverson AJ. Plant mitochondrial genome diversity: the genomics revolution: plant mitochondrial genome diversity: the genomics revolution. 2012. [Google Scholar]
- 38.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 2002;99(15):9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen VB, Giang VNL, Waminal NE, Park HS, Kim NH, Jang W, Lee J, Yang TJ. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J Ginseng Res. 2020;44(1):135–144. doi: 10.1016/j.jgr.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123–U116. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 41.Gui ST, Wu ZH, Zhang HY, Zheng YZ, Zhu ZX, Liang DQ, Ding Y. The mitochondrial genome map of Nelumbo nucifera reveals ancient evolutionary features. Sci Rep. 2016;6:11. doi: 10.1038/srep30158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filip E, Skuza L. Horizontal Gene Transfer Involving Chloroplasts. Int J Mol Sci. 2021;22(9):4484. [DOI] [PMC free article] [PubMed]
- 43.Cui HA, Ding Z, Zhu QL, Wu Y, Qiu BY, Gao P. Comparative analysis of nuclear, chloroplast, and mitochondrial genomes of watermelon and melon provides evidence of gene transfer. Sci Rep. 2021;11(1):1595. [DOI] [PMC free article] [PubMed]
- 44.Shidhi PR, Biju VC, Anu S, Vipin CL, Deelip KR, Achuthsankar SN. Genome Characterization, Comparison and Phylogenetic Analysis of Complete Mitochondrial Genome of Evolvulus alsinoides Reveals Highly Rearranged Gene Order in Solanales. Life-Basel. 2021;11(8):769. [DOI] [PMC free article] [PubMed]
- 45.Morley SA, Nielsen BL. Plant mitochondrial DNA. Front Biosci Landmark. 2017;22:1023–1032. doi: 10.2741/4531. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Lor A, Luro F, Ollitrault P, Navarro L. Genetic diversity and population structure analysis of mandarin germplasm by nuclear, chloroplastic and mitochondrial markers. Tree Genet Genomes. 2015;11(6):123.
- 47.Nishiyama K, Choi YA, Honsho C, Eiadthong W, Yonemori K. Application of genomic in situ hybridization for phylogenetic study between Mangifera indica L. and eight wild species of Mangifera. Sci Hortic. 2006;110(1):114–117. [Google Scholar]
- 48.Sankaran M, Dinesh MR, Chaitra N, Ravishankar KV. Morphological, cytological, palynological and molecular characterization of certain Mangifera species. Curr Sci. 2018;115(7):1379–1386. [Google Scholar]
- 49.Eiadthong W, Yonemori K, Kanzaki S, Sugiura A, Utsunomiya N, Subhadrabandhu S. Amplified fragment length polymorphism analysis for studying genetic relationships among Mangifera species in Thailand. J Am Soc Hortic Sci. 2000;125(2):160–164. [Google Scholar]
- 50.Yonemori K, Honsho C, Kanzaki S, Eiadthong W, Sugiura A. Phylogenetic relationships of Mangifera species revealed by ITS sequences of nuclear ribosomal DNA and a possibility of their hybrid origin. Plant Syst Evol. 2002;231(1–4):59–75. [Google Scholar]
- 51.Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. Plos Biol. 2012;10(1):e1001241. [DOI] [PMC free article] [PubMed]
- 52.Wu ZX, Hu KN, Yan MJ, Song LP, Wen J, Ma CZ, et al. Mitochondrial genome and transcriptome analysis of five alloplasmic male-sterile lines in Brassica juncea. BMC Genomics. 2019;20:348. [DOI] [PMC free article] [PubMed]
- 53.Thyssen G, Svab Z, Maliga P. Exceptional inheritance of plastids via pollen in Nicotiana sylvestris with no detectable paternal mitochondrial DNA in the progeny. Plant J. 2012;72(1):84–88. doi: 10.1111/j.1365-313X.2012.05057.x. [DOI] [PubMed] [Google Scholar]
- 54.Tsujimura M, Mori N, Yamagishi H, Terachi T. A possible breakage of linkage disequilibrium between mitochondrial and chloroplast genomes during emmer and Dinkel wheat evolution. Genome. 2013;56(4):187–193. doi: 10.1139/gen-2012-0153. [DOI] [PubMed] [Google Scholar]
- 55.Trusty JL, Johnson KJ, Lockaby BG, Goertzen LR. Bi-parental cytoplasmic DNA inheritance in Wisteria (Fabaceae): evidence from a natural experiment. Plant Cell Physiol. 2007;48(4):662–665. doi: 10.1093/pcp/pcm036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are freely available in GenBank on the NCBI website at https://www.ncbi.nlm.nih.gov, using the accession number MZ751075, MZ751076, and MZ751077. Raw sequencing data have been deposited at the NCBI Sequence Read Archive (SRA) under accession PRJNA778602.