Abstract
Telomerase is unique among reverse transcriptases in containing a non-coding RNA (telomerase RNA, TER) that includes a short template used for processive synthesis of G-rich telomeric DNA repeats at the 3′ ends of most eukaryotic chromosomes1. Telomerase maintains genomic integrity, and its activity or dysregulation are critical determinants of human longevity, stem-cell renewal, and cancer progression2,3. Cryo electron microscopy (cryo-EM) structures at 4.8–10 Å resolution have established the general architecture, protein components and stoichiometries of Tetrahymena and human telomerase, but provided limited details of DNA– and RNA–protein interactions and insights into mechanism and recruitment4–6. Here we report 3.3, 3.8, and 4.4 Å resolution cryo-EM structures of active Tetrahymena telomerase with telomeric DNA at different steps of nucleotide addition. Details of interactions between telomerase reverse transcriptase (TERT), TER, and DNA reveal the structural basis of 5′ and 3′ template boundary determination, template–DNA duplex handling, and product chain separation during nucleotide addition. The structure and binding interface between TERT and telomerase protein p50, a homologue of human TPP17,8, define conserved interactions required for telomerase activation and recruitment to telomeres. Telomerase La-related protein p65 remodels multiple regions of TER, bridging the 5′- and 3′-ends and the conserved pseudoknot to facilitate assembly of the TERT–TER catalytic core.
Telomerase was first identified in the ciliated protozoan Tetrahymena9, which has continued to play a pioneering role in uncovering mechanistic details of chromosome end replication10,11. All telomerases contain a catalytic core of TERT and TER, which includes a conserved template/pseudoknot domain (t/PK) and stem-terminus element (STE)12. Additional TER-binding protein(s), i.e. p6513 in Tetrahymena and H/ACA scaRNP14 proteins in human, are required for TER biogenesis and core ribonucleoprotein (RNP) assembly. The 4.8 Å resolution cryo-EM structure of Tetrahymena telomerase revealed the first complete architecture of TERT5: its RNA binding (RBD), reverse transcriptase (RT) and C-terminal element (CTE) domains form a ring structure in common with Tribolium castaneum TERT-like protein15, while TERT-unique telomerase essential N-terminal (TEN) domain and TRAP motif (Fig. 1a) cross over the TERT ring from opposite sides, enclosing the t/PK and completing the catalytic cavity. Tetrahymena telomerase additionally comprises p50 and two Replication Protein A (RPA) related heterotrimeric complexes4,10,16, Teb1–Teb2–Teb3 (TEB) and p75–p45–p19 (Fig. 1a,b). p50, Teb1, and p75–p45–p19 are homologs of human telomere-associated proteins TPP17,8, POT117, and Ctc1–Stn1–Ten118 (CST), respectively, that transiently associate with telomerase for telomerase activation, single-stranded telomeric DNA (sstDNA) handling, and DNA polymerase α-primase recruitment for complementary strand synthesis19.
Fig. 1: 3.3 Å resolution structure of Tetrahymena telomerase with sstDNA.
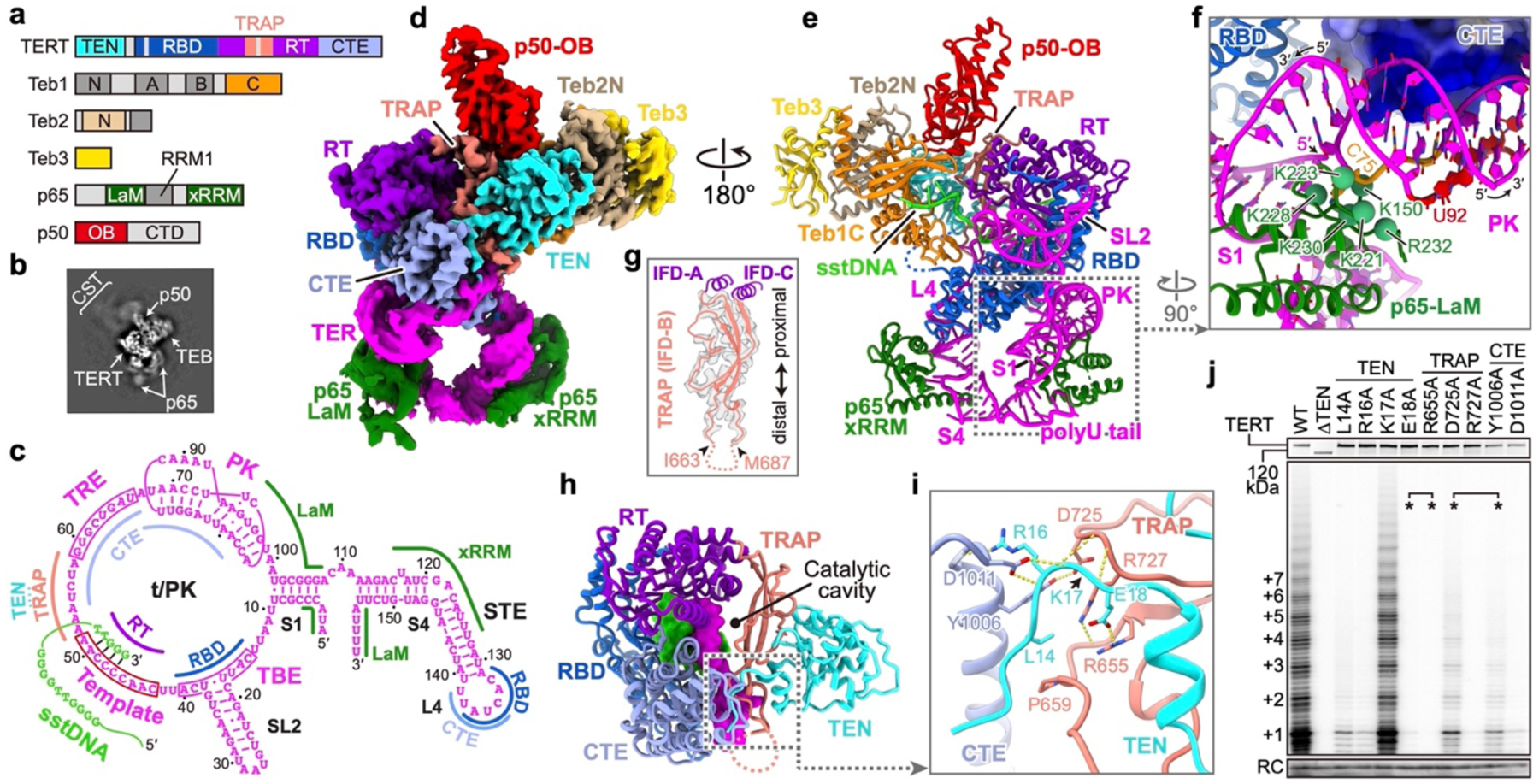
a, Domain organization of TERT, p65, p50, and TEB. Regions invisible in the cryo-EM map are colored in gray. b, Representative 2D class average image. c, Schematic of TER secondary structure. Interaction sites with TERT, sstDNA and p65 are indicated. d, Cryo-EM density. e, Molecular model of telomerase. f, TER PK and its interactions with TERT CTE and p65 LaM. Green spheres are positively charged p65 LaM residues on the interface. TERT CTE is shown as electrostatic surface. g, Superposition of ribbon diagram and cryo-EM density of TRAP. Dashed line indicates TRAP flexible linker. h, Structure of TERT, with DNA (green) and TER (magenta) within the catalytic cavity shown as surfaces. i, CTE–TRAP–TEN interface. j, Telomerase activity assays with alanine substitutions of residues shown in i. Asterisks indicate residues interacting with each other as pairs. The number of telomeric repeats synthesized are indicated at left. The top panel shows expression levels of 35S-Met labeled TERT mutants. RC, recovery control. Activity assays were successfully repeated 3 times. For gel source data for all Figures, see Supplementary Fig. 1.
Telomerase repetitively uses its integral template that is complementary to 1.5–1.8 telomere repeats (GGGTTG in Tetrahymena and GGTTAG in human) for multiple rounds of telomere repeat synthesis11 (Fig. 1c). This requires two types of template translocation: nucleotide translocation shifts the template by one register after each nucleotide addition, and repeat translocation resets the template back to its original position after synthesis of each telomeric repeat. During repeat translocation, the template must separate from the DNA, shift, and re-pair to the DNA with its alignment region. Structural details of almost all these steps are lacking. Here, the 3.3 Å resolution cryo-EM structure of telomerase with sstDNA at the second step of nucleotide addition, together with structures at the third, fourth, and fifth steps, reveal the details of protein, RNA, and DNA sidechain interactions that determine assembly, recruitment, and activity, and suggest a mechanism for processive telomeric repeat addition.
Overall structure
Endogenously expressed telomerase used for cryo-EM studies was purified from Tetrahymena with sstDNA (GTTGGG)5 added during the purification (Extended Data Fig. 1). By computationally excluding the dynamic p75–p45–p19 (Fig. 1b), we obtained a reconstruction of sstDNA-bound TERT–TER–p65 core RNP with p50 OB domain (p50-OB) and Teb1C–Teb2N–Teb3 at an average resolution of 3.3 Å (Fig. 1d,e, Extended Data Fig. 2, Extended Data Table 1, Supplementary Video 1).
As seen with a different sstDNA at 4.8 Å resolution5, the TERT–TER catalytic core adopts an interlocked architecture (Fig. 1e, Extended Data Fig. 3, Supplementary Video 2). In the catalytic cavity the sstDNA and template form a 4 bp A-form duplex with the last base pair shifted out of the active site (Fig. 1c,h). Almost all of the t/PK circle has direct interactions with TERT, including the PK that is anchored on CTE by a cluster of positively charged residues (Fig. 1f). The rest of TER, including stem 1 (S1) and stem-loop 4, form a U-shape bound by p65 below the TERT ring (Fig. 1e). Loop 4 (L4) is inserted between and specifically interacts with RBD and CTE to close the TERT ring (Extended Data Fig. 3d–f).
TEN–TRAP interacts with the TERT ring at only two regions: the N- and C-termini of TRAP are covalently connected to IFD-A and IFD-C12 of RT, respectively, while the visible distal end of TRAP is anchored on CTE together with the N-terminal tail of TEN (Fig. 1g–i, Extended Data Fig. 4a–c). Beyond the distal anchor, TRAP has a 23-residue flexible loop (Fig. 1g), whose truncation has almost no effect on telomerase activity (Extended Data Fig. 4d), consistent with its absence in human TERT (Extended Data Fig. 5a). Behind the anchor, TER runs between and interacts with TRAP and CTE (Fig. 1h). We investigated the importance of the TEN–TRAP interface and its anchor point on CTE by assaying the effect of individual amino acid substitutions on telomerase activity. Except for K17A, which has only backbone interactions, all substitutions at the CTE–TRAP–TEN three-way anchor dramatically decrease overall activity and repeat addition processivity (RAP) (Fig. 1j, Extended Data Fig. 4f). Substitutions of residues at the extensive (~1900 Å2) TEN–TRAP interface generally decrease activity, although less dramatically (Extended Data Fig. 4e). These results demonstrate that TEN–TRAP and its anchor on CTE are essential for telomerase activities, including RAP.
p65 binds multiple regions of TER
p65 is a LARP7 protein20 whose C-terminal xRRM bends TER stem 4 (S4) during assembly of the TERT–TER–p65 core RNP21,22, but how the rest of p65 interacts with TER remains uncharacterized. Here, despite lower resolution in this region, p65 La motif (LaM) could be modeled with the help of its sequence and structural similarities to human La protein23. p65 RRM1, that completes the La module (LaM–RRM1) (Fig. 1a), is almost invisible in the cryo-EM map (Fig. 1d), indicating its flexibility. As expected, p65 LaM binds TER 3′-UUU-OH using a cleft lined with conserved aromatic residues20 (Extended Data Fig. 3g). Unexpectedly, p65 LaM also wedges into the junction between the PK and S1 (Fig. 1e,f, Extended Data Fig. 3h). The PK loop nucleotide C75 is flipped out to interact with the positively charged C-terminus of p65 LaM (Fig. 1f, Extended Data Fig. 3h). TER 5′-end binds to an adjacent region of p65 LaM near the PK (Extended Data Fig. 3h). Thus, p65 not only bends S4 to position L4 for interaction with TERT21, but its LaM also interacts simultaneously with TER 5′-end, 3′-end, and PK, consistent with a role as a chaperone for TER folding and RNP assembly.
p50 (TPP1) binds TERT on TEN–TRAP
p50 is functionally equivalent to the human telomerase activating role of TPP18. Taking advantage of the improved resolution, we rebuilt the model of p50-OB (aa. 1–184), and found it is highly similar to TPP1-OB7 (Fig. 2a, Extended Data Fig. 6a,b). p50 binds TERT at adjacent regions of TEN–TRAP (Fig. 2b). Two structurally conserved p50-OB loops Lα2-β4 and Lβ5-β6 bind TEN on a basic surface, while TRAP interacts with p50-OB β-barrel and α0 (Fig. 2c, Extended Data Fig. 6c). Previous studies had defined TEL patch and NOB on human TPP1-OB as critical for telomerase recruitment and RAP stimulation18,24–27. Docking TPP1-OB onto our cryo-EM structure reveals that its NOB and TEL patch residues are all located on the same interface with TERT as p50 (Fig. 2d, Extended Data Fig. 6d). S111, whose phosphorylation is important for the cell-cycle regulated TPP1 association with human telomerase28, is also located on the interface (Fig. 2d). Notably, TPP1-OB NOB is extended in the crystal structure (Fig. 2a), but based on sequence similarity (Extended Data Fig. 6d) likely also forms a α0 when bound to TERT. This structure provides the first atomic details of the TERT–p50 interface, and by structural homology that for human TERT–TPP1.
Fig. 2: Structure of p50-OB and interactions with TERT.
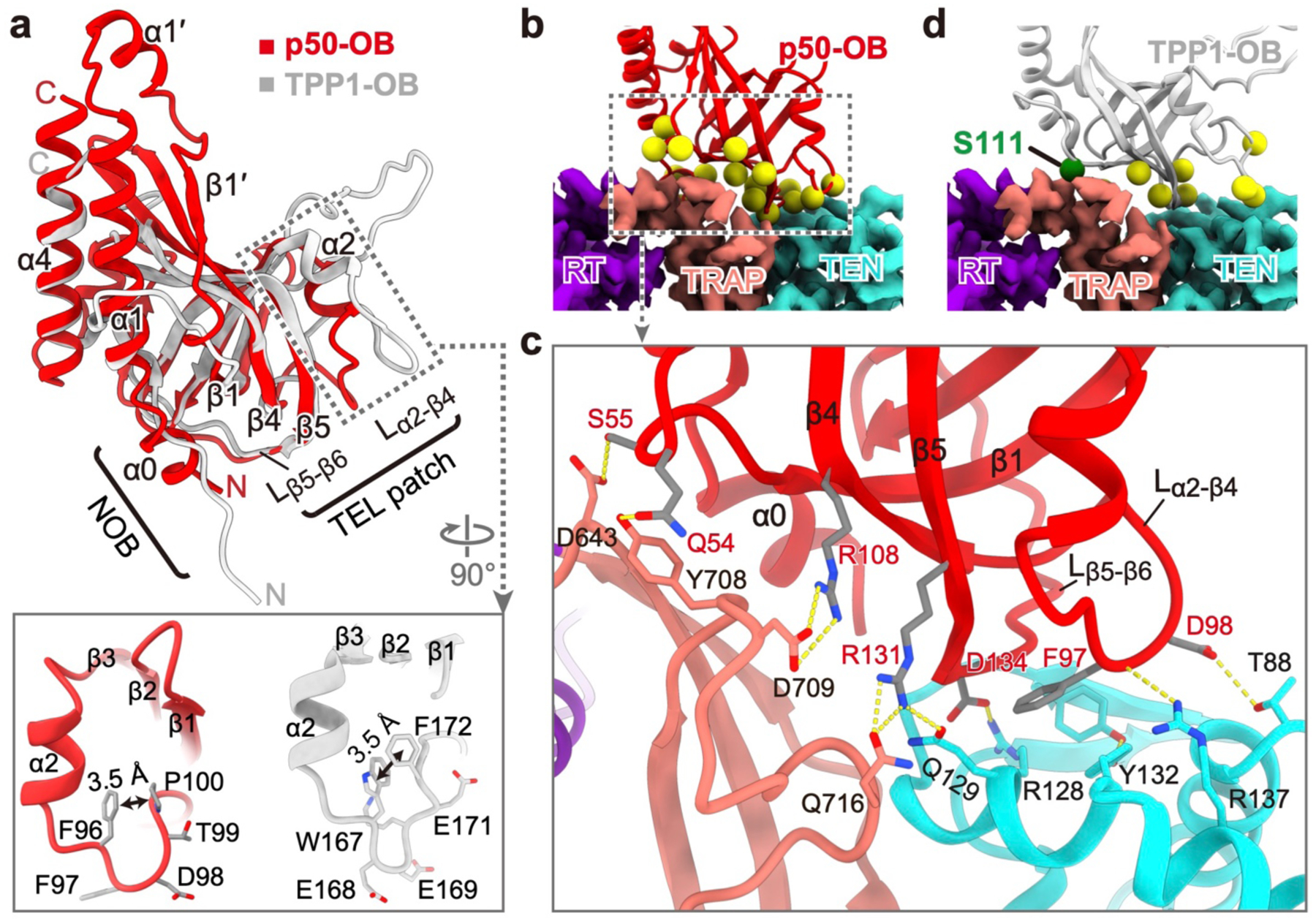
a, Comparison of p50-OB (red) and human TPP1-OB (gray, PDB ID: 2I46) structures. Zoomed-in views show Lα2-β4 in p50-OB (left) and TPP1-OB (right). b, Interface between p50-OB (ribbon, with yellow spheres for residues at interface) and TERT (cryo-EM density). c, Sidechain interactions between p50-OB and TRAP–TEN, as in b. d, TPP1-OB docked onto Tetrahymena TERT. Yellow spheres are TEL patch residues. Green sphere is S111.
Teb1C binds three sstDNA nucleotides
TEB large subunit Teb1 handles the sstDNA, similar to human POT117, and Teb2 and Teb3 are shared with Tetrahymena RPA29. Among the 4 OB-folds of Teb1 (Fig. 1a), Teb1A and Teb1B bind sstDNA with high affinity, but details of the lower affinity binding mode to Teb1C were unknown30. In the structure, four nucleotides (T27G28G29G30) pair with the template and seven (T20-T26) protrude out of the catalytic cavity passing near the template 5′-end (Fig. 3a–c, Extended Data Fig. 6e,f), but not pairing with it as previously suggested5. The continuous sstDNA density observed from the duplex to Teb1C argues against a G-quadruplex in the catalytic cavity or an anchor point on TEN31–33. At Teb1C, G17G18G19 are held in its C-shape oligonucleotide-binding cleft (Fig. 1e, Extended Data Fig. 6e–g) by stacking interactions and three H-bonds to G19 Watson-Crick edge (Extended Data Fig. 6h). Alanine substitutions of F603 that stacks on G17 and K660 that H-bonds G19 show significant defects in sstDNA binding and telomerase activity stimulation30. The 16 remaining sstDNA nucleotides (G1-G16), expected to interact with Teb1AB, are invisible in the cryo-EM map.
Fig. 3: Interactions between TERT and template–DNA duplex.
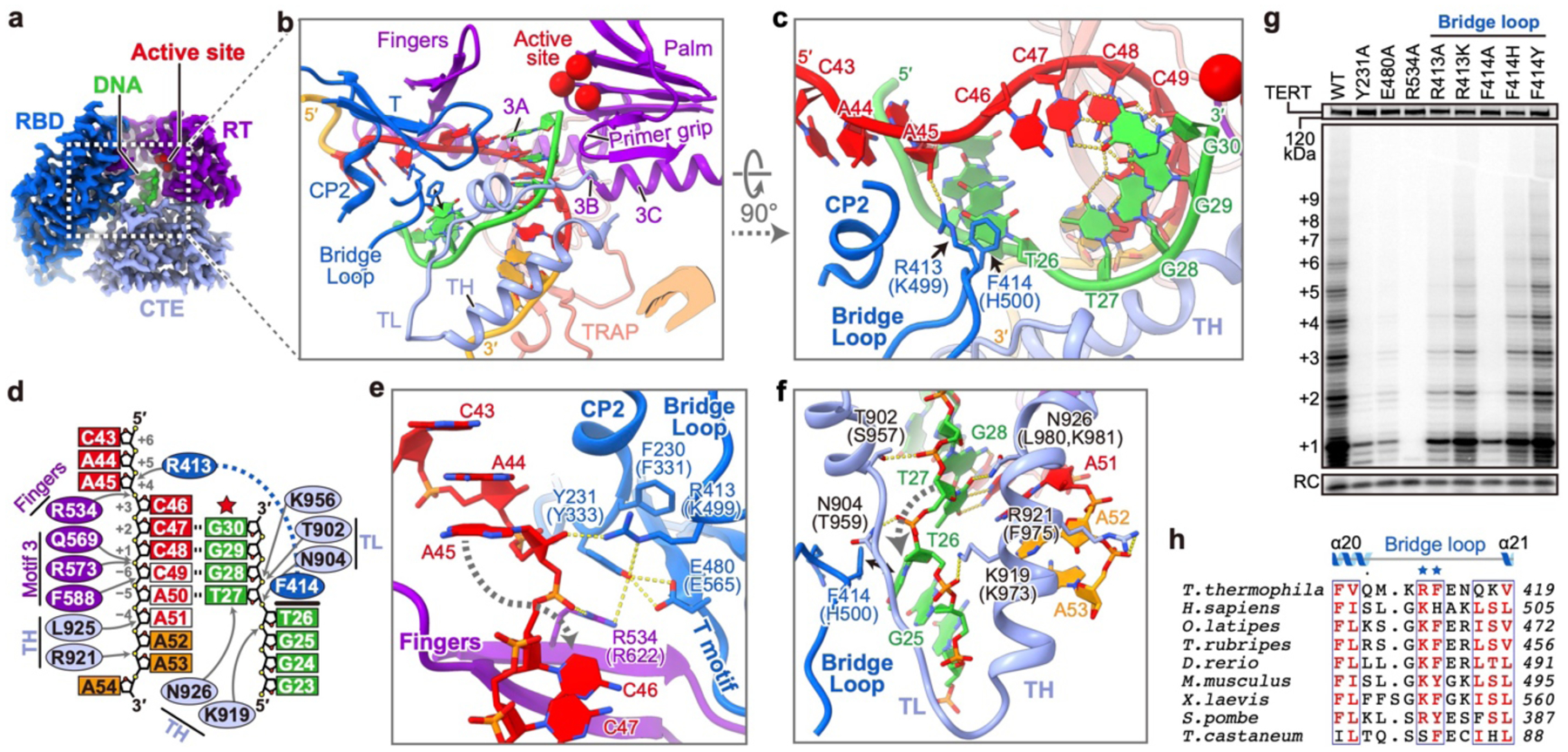
a, Cryo-EM densities of TERT and template–DNA. Red spheres are active site residues (D618,D815,D816). b, c, Ribbon depictions of template–DNA and TERT motifs involved in duplex handling. Template and adjacent nucleotides are red and orange, respectively. Bridge loop R413 and F414 are shown as sticks. The hand (inset in b) shows orientation relative to other polymerases. d, Schematic showing specific interactions between TERT and template–DNA. Template alignment nucleotides are red in white background. Arrows indicate sites of polar interactions. Bold line indicates F414 and T26 stacking interaction. e, f, Sidechain interactions surrounding the template (e) and DNA (f) nucleotide flipping regions. The corresponding human TERT residues are in parentheses. g, Telomerase activity assays with TERT substitutions on CP2, T motif, fingers and bridge loop. n=3 independent experiments. h, Sequence alignment of the bridge loop, with conserved residues in red.
Stabilization of the template–DNA duplex
In the 3.3 Å resolution structure, the template +3 nucleotide (C46) is at the active site ready for an incoming dGTP and the last base pair of the duplex is at position +2 (C47) (hereafter called telomerase T3D2, where T is template and D is duplex) (Fig. 3a–d). Starting from the active site, eight TER nucleotides stack continuously with their backbone adopting an A-form conformation (Fig. 3d, Extended Data Fig. 3k). Template nucleotides downstream of the active site (C43A-44A45) are rotated ~180° away from the duplex and stack together out of the catalytic cavity (Fig. 3d,e). Below the duplex, the unpaired DNA nucleotides are rotated outward ~120° in a helical stack (Fig. 3d,f). The duplex and adjacent nucleotides are handled by TERT with structural elements in common with most polymerases (palm, fingers, primer grip, thumb helix (TH), thumb loop (TL))34 and motifs specific to a subset of reverse transcriptases (motif 335, T36) or TERT (CP2/TFLY37–39, TRAP5) (Fig. 3b–d, Extended Data Fig. 5). Specifically, motif 3 interacts with the template (Extended Data Fig. 3j), TL interacts with the DNA backbone, and TH contacts DNA and TER nucleotides 3′ of the template (Fig. 3f).
Spatially located between CP2 and TL, a newly identified motif (here named bridge loop, aa. 411–418) in the RBD is inserted into the major groove of the duplex with its conserved tip residues (R413, F414) (Fig. 3h) bridging both ends of the duplex (Fig. 3b–d). On the DNA side, F414 breaks the duplex by stacking with T26, and the backbone turn is further stabilized by TH K919 and TL N904 (Fig. 3f). Alanine substitution of F414 dramatically decreases activity and abolishes RAP, while substitutions F414Y and F414H have relatively small effects (Fig. 3g, Extended Data Fig. 4g), consistent with a requirement for aromatic stacking interaction. On the RNA side, R413 contributes to a network of interactions among conserved CP2 (Y231), T motif (E480), and fingers (R534) residues that position the template nucleotide downstream of the active site (A45) and flip in the template nucleotide opposite the active site (C46) during nucleotide translocation (Fig. 3e). Individual alanine substitution of these residues all decrease activity to <20% and decrease RAP to <60% for R413A and E480A and undetectable for R534A and Y231A (Fig 3g, Extended Data Fig. 4g). The locations of these bridge loop and other residues at either end of the duplex, their sequence conservation, and effects of substitutions on telomerase activity suggest they play essential roles in template guidance and duplex stabilization, as discussed further below.
Template–DNA duplex length and handling
We next obtained telomerase structures with the template at position +5 (telomerase T5D5) at 3.8 Å and +4 (telomerase T4D4) at 4.4 Å resolution (Extended Data Fig. 7) and refined the duplex model in the previously published 4.8 Å resolution structure (telomerase T3D3)5. By using native sstDNA (telomerase T3D2) and locked nucleic acid40 (LNA)-containing sstDNA (telomerase T3D3, T4D4, T5D5), we could trap the duplex in the post- and pre-nucleotide translocation states, respectively (Extended Data Fig. 8). There is little conformational change in TERT between these two states, as also observed for Tribolium TERT-like protein41.
Comparison of these structures reveals an identical conformation of RNA nucleotides within the catalytic cavity (Fig. 3d, Extended Data Fig. 8). At each step, as the template–DNA duplex moves, the downstream template nucleotide flips into the active site, while the previously stacked 3′ nucleotide flips out, maintaining the total 8 nucleotide stack. Concurrently, the sstDNA interacts with the template to form a 4 or 5 base pair duplex depending on whether it is in the post- or pre-nucleotide translocation state, respectively. For each register of the duplex, interactions with the bridge loop, TH, and TL are maintained (Extended Data Fig. 8). These structures provide the first direct evidence for template–DNA duplex length and handling at different steps of telomere repeat synthesis, and suggest that only 4 stable (post-nucleotide translocation) base pairs, stabilized by the telomerase unique bridge loop, are present at each step.
Template boundaries and movement
The 5′ and 3′ template boundaries are determined by TERT interactions with the template boundary element (TBE)42 and template recognition element (TRE)43, which are connected to the template by TBEL linkers5 and TREL, respectively (Fig. 4a). Comparison of the other structures with T3D2 reveals that TBE and TRE remain fixed on TERT as the template moves from the +3 to +5 position (Fig. 4a–e, Extended Data Fig. 9a–c). On the template 5′-side, the TBE is anchored on RBD by CP2/TFLY, CP and T motifs37, plus additional stacking and hydrogen-bonding interactions for A40-C39 at the TBE 3′-end (Fig. 4d, Extended Data Fig. 3c,9b). On the template 3′-side, the TRE (G59-U66) wraps along and has numerous stacking interactions with a stripe of 5 aromatic residues on CTE (Fig. 4e, Extended Data Fig. 9c). The TRE backbone adopts a compressed zigzag conformation with a U60•U63 base pair in the middle. Unlike the fixed TBE and TRE, TBEL (U41U42) and TREL (A52-A58) exhibit conformational changes consistent with concerted stretching and looping44 as the template moves through the active site (Fig. 4b,c). In telomerase T3D2, the TBEL and +4,+5,+6 template nucleotides are looped out, with U41 and U42 pointing outward in opposite directions (Fig. 4d). However, in telomerase T5D5, where only the +6 template nucleotide is out of the catalytic cavity, the backbone of TBEL is more stretched (Fig. 4c, Extended Data Fig. 9b). Basic residues (R550,K551,K553) located on an extended finger-shaped linker between motif 2 and 3A (aa. 550–560, here named motif 3N) provide a surface for TBEL to slide over (Fig. 4d, Extended Data Fig. 3a). Individual alanine substitutions of these residues decrease telomerase activity to <40% and deletion of motif 3N abolishes activity (Fig. 4i), confirming the importance of motif 3N interaction with TBEL.
Fig.4. Structural details for template boundary determination.

a, Telomerase T3D2 catalytic cavity with TER (gray), DNA (green ribbon), and TERT (colored surface). b, c, Comparison of TER and template–DNA in telomerase T3D2 (b) and T5D5 (c). Red star marks TERT active site. d-g, Detailed TERT–TER interactions in regions indicated with dashed boxes in a. Hydrogen-bonds (dashed yellow lines) and stacking interactions (black lines) are indicated. TRAP–TH channel is shown as electrostatic surface in f. h, Schematic of interactions between TERT and TREL-TRE. i, Telomerase activity assays with TERT substitutions on the interface with TBEL and TREL. Telomerase activity and RAP were determined relative to WT. The data represent the mean ± s.d. from 3 independent experiments.
On the template 3′-end, TER passes through a positively charged hourglass-shaped channel formed by TH and TRAP (Fig. 4f). The upper part of the channel is lined by basic residues (K657,R658,R921) that interact with the backbone of the last three TER nucleotides in the catalytic cavity. In telomerase T3D2 these are template A51 and TREL A52A53 (Fig. 4f–h). Below the neck, A54 makes a sharp turn from the helical stack, and TREL A54-A58 form a loop, with A 54,C56,A58 stacked in and U55,U57 flipped out. In telomerase T5D5, all three template alignment nucleotides (C49A59A51) are in the TRAP–TH channel (Extended Data Fig. 9d–f). Of the 7 TREL nucleotides extruded through the neck only A52 and A53 are visible in the cryo-EM map; A54-A58 can apparently no longer be accommodated in the channel and loop out. Individual alanine substitution of basic residues along the TRAP–TH channel (Fig. 4h) all decrease telomerase activity to <25%, while Y694A has a less severe effect (Fig. 4i). Additionally, K657A, R658A and R921A located above the neck decrease RAP to undetectable or <50% (Fig. 4i). These results indicate that the interactions between the positively charged surface of the channel and TREL are essential for regulating telomeric repeat synthesis, and specifically interactions above the neck are involved in RAP. Together with the constriction at the neck they may create an energy barrier that prevents backwards movement of TREL nucleotides that have passed through the neck.
Mechanism of telomere repeat synthesis
Based on the four sstDNA-bound telomerase structures, especially the highest resolution structures T3D2 and T5D5, a model for telomere repeat synthesis emerges (Fig. 5). The sstDNA initially base pairs with the template alignment region and this short helix is stabilized in the catalytic cavity by the TEN–TRAP above it and by interactions with TH, TL, and bridge loop (Fig. 5, step 0). As the template moves through the activity site during nucleotide addition, the template–TBEL loop decreases in size while TREL nucleotides pushed through the TRAP–TH neck begin to loop out (Fig. 5, step 2 to 5). Although tension might accumulate as the TREL loop grows44, the TRAP–TH neck acts like a ratchet, preventing the nucleotides that have passed through from moving back.
Fig. 5: A model for telomere repeat synthesis.

At step 0/10, six TREL nucleotides are in the TRAP–TH channel, with 3 stacked above the neck and 3 fully stretched between the neck and TRE. Five template nucleotides beyond the +1 position and 2 TBEL nucleotides are looped out. The template moves from +1 to +6 position during the synthesis of one telomere repeat (steps 0–6), with concomitant stretching of TBEL and looping of TREL. At step 6, the TBEL is fully stretched and all 7 TREL plus one template nucleotide are looped out below the neck of the TRAP–TH channel. Following addition of the last dGTP a series of events leads to template translocation: duplex distortion (7), duplex melting and TRAP opening (8), template re-positioning (9), and DNA re-pairing and TRAP closing (10). TBE and TRE anchor sites, locations of TH, TL, F414, and template position at active site are illustrated. The pawl-and- ratchet cartoons illustrate the movement of TRAP (salmon) and TREL nucleotides (orange) through the TRAP–TH neck. Step 2 corresponds to the 3.3 Å resolution structure T3D2.
Following addition of the last telomeric repeat nucleotide (Fig. 5, step 6), the template has to translocate to its starting position. We propose that repeat translocation could share a fundamental mechanism with nucleotide translocation. Nucleotide translocation intermediate structures captured in a viral RNA dependent-RNA polymerase suggest an asymmetric movement of the duplex, where the product chain shifts first, distorting and weakening the base pairs, then the template chain follows45. A similar mechanism could apply to telomerase. However, after addition of the last nucleotide, only the DNA strand can move; the template cannot follow since TBEL is fully stretched (Fig. 5, step 7). This nucleotide translocation intermediate would destabilize the duplex, contributing to strand separation prior to template translocation. TERT CTE movement has been proposed to pull the DNA off the template46,47; however, our structures suggest that CTE rotation would be highly restricted. Instead, TEN–TRAP, which exhibits conformational dynamics (Extended Data Fig. 2d), could rotate around its two points of contact with the TERT ring, perhaps facilitated by the buildup of TREL nucleotides in the lower part of the TRAP–TH channel, thereby disrupting the narrow channel and releasing the tension (Fig 5, step 8). Then, the template would slip back along motif 3 (Extended Data Fig. 3j) and reset the +1 position at the active site (Fig. 5, step 9), where TREL on template 3′ side is fully stretched (Extended Data Fig. 9g). Since TH and TL interact with the sstDNA throughout synthesis (Extended Data Fig. 8), they could hold the DNA backbone during template translocation, consistent with mutagenesis results for human telomerase47. Stacking of bridge loop F414 on the flipped DNA bases could prevent the DNA chain from moving, thereby positioning it to re-pair with the template alignment region. After the template–DNA duplex reforms, the TRAP–TH channel closes (Fig. 5, step 10/0). This unified mechanism may also explain why LNA-containing primers, which stabilize the nucleotide pre-translocation state, prevent repeat translocation.
We note that since only a 4–5 bp duplex forms throughout the steps of telomere repeat synthesis (Extended Data Fig. 8), template–DNA strand separation should not require much energy; rather the duplex needs to be stabilized in the catalytic cavity. We propose that TEN–TRAP and the RBD bridge loop have evolved to retain the short template–duplex throughout nucleotide addition as well as play an essential role in template translocation. Modeling suggests a similar set of TERT–TER interactions for human telomerase48. Comparison of p50-OB and TPP1-OB structures and interactions also provides evidence for a conserved TERT interface for telomerase activation and recruitment to telomeres.
Tetrahymena telomerase biogenesis protein p65 binds the t/PK enclosing stem, PK, and STE, that are elements common to almost all TERs. Pof8, a fission yeast LARP7, is a component of telomerase, and has recently been proposed to bind the PK49. In humans, Larp7 deficiency affects telomere length50. Together, this suggests that LARP7s may play a common role in TER assembly with TERT. In summary, the results presented here provide insights into telomerase assembly, mechanism of telomeric repeat synthesis, telomerase recruitment, and telomeric DNA handling.
METHODS
Telomerase sample preparation.
Tetrahymena thermophila CU522 TERT-FZZ strain with a replacement of the endogenous TERT gene with a C-terminally TAP-tagged (3xFLAG-TEV-ZZ) TERT51 (provided by Dr. Kathleen Collins) was used for telomerase purification. Tetrahymena telomerase was purified following the previously described protocol16, with minor modifications. Sixteen liters of Tetrahymena TERT-FZZ cells were grown in PPYS media and harvested by centrifugation. The cell pellet was washed with 20 mM HEPES·NaOH pH 8.0, and then lysed in lysis buffer (20 mM HEPES·NaOH pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mM TCEP, 10% glycerol, 0.2% IGEPAL CA-630, 0.1% Triton X-100) at 4 °C for 30 min. The cell lysate was clarified by ultracentrifugation at 230,000 × g for 1 hour. After ultracentrifugation, the supernatant of the lysate was incubated with Rabbit-IgG agarose slurry (Sigma) overnight at 4 °C. Then, the resin was washed with wash buffer (20 mM HEPES·NaOH pH 8.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP, 10% glycerol, 0.1% IGEPAL CA-630) and eluted with TEV protease. During the TEV elution step, single-stranded telomeric DNA (sstDNA) was added to a final concentration of 5 μM to saturate telomerase, and excess sstDNA was washed away in the following steps of purification. The elution fraction from IgG resin was incubated with 30 μL of pre-washed anti-Flag M2 affinity gel (Sigma) for 1 hour at 4 °C. After that, the anti-Flag resin was washed extensively with wash buffer and eluted using a small volume (30–50 μL) of elution buffer (20 mM HEPES·NaOH pH 8.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP, 0.1% IGEPAL CA-630) supplemented with 1 mg/mL 3× FLAG peptide. Telomerase samples bound with different sstDNAs were prepared separately with DNA primers (GTTGGG)5 (telomerase T3D2), (GTTGGG)2GTTGGLGLGLT (telomerase T4D4), and (GTTGGG)2GTTGGGLGLTLT (telomerase T5D5), where TL/GL represents locked nucleic acid (LNA) oligonucleotide (Exiqon).
Cryo-EM specimen preparation and data collection.
For cryo-EM sample, 3 μL of the purified telomerase was applied to glow-discharged lacey carbon grids with a supporting ultrathin carbon film (Ted Pella). The grids were then blotted with filter paper and flash-frozen in liquid ethane using an FEI Vitrobot Mark IV. Cryo-EM grids were screened in an FEI Tecnai TF20 transmission electron microscope while optimizing freezing conditions.
Vitrified cryo-EM grids were loaded into an FEI Titan Krios electron microscope at 300 kV for automated image acquisition with SerialEM52. Movies of dose-fractionated frames were acquired with a Gatan K2 Summit direct electron detector operated in super-resolution mode, yielding a pixel size of 0.68 Å on the sample level). A Gatan Imaging Filter (GIF) was installed between the electron microscope and the K2 camera with the slit width setting to 20 eV. The electron microscope was carefully aligned prior to each imaging session and the parallel beam was optimized using coma-free alignment in SerialEM52. The dose rate on the detector was set to ~ 6 electrons/Å2/s and the total exposure time of each movie was 8 s, which fractionated into 40 frames of images with 0.2 s exposure time for each frame. In total, 13,097 movies for telomerase T3D2, 5,020 movies for telomerase T4D4, and 12,922 movies for telomerase T5D5 were collected in separate imaging sessions.
Cryo-EM data processing of telomerase T3D2.
Dose-fractionated frames except for the first of each movie were 2× binned (pixel size of 1.36 Å) and aligned for correction of beam-induced drift using MotionCor253. Two averaged images, one with dose weighting and the other without, were generated after drift correction. The averaged micrographs without dose weighting were used for the determination of CTF parameters and particle picking, and the averaged micrographs with dose weighting were used for particle extraction and further data processing. The defocus and astigmatism values of each micrograph were determined by CTFFIND454. All micrographs after motion correction and their power spectra were visually inspected, and the micrographs with too much thick carbon area, ice contamination or defocus value outside the range from −0.8 to −4.0 μm were discarded. Finally, a total of 10,849 micrographs were kept. Particles in those averaged images were automatically picked with Gautomatch (mrc-lmb.cam.ac.uk/kzhang/) using 17 projections from previously reported cryo-EM map of Tetrahymena telomerase (EMD-7821)5. To avoid losing good particles on the low contrast micrographs collected with carbon coated cryo-EM grids, we used a low threshold for particle picking (over 350 particles on each micrograph) and then utilized our previously developed data processing protocol5 to select good particles (as detailed below). Finally, a total of 3,816,856 particles were picked, extracted in dimensions of 256 × 256 square pixels and 2× binned to 128 × 128 square pixels (pixel size of 2.72 Å) to speed up following data processing with RELION 3.0 (Ref55).
The cryo-EM data processing procedure is outlined in Extended Data Fig. 2a. Two datasets, one for each data collection session, were initially processed separately in two batches. For each dataset, extracted particles were classified into 6 subsets using 3D classification. The previously reported DNA-bound telomerase density map (EMD-7821) was low-pass filtered to 60 Å before using as the initial model. The particles in each 3D class were then classified into 100 classes using reference-free 2D classification. Particles in 2D classes with fuzzy or uninterpretable features were discarded, and most of them were pure noise from background carbon film instead of real telomerase particles. By doing the above 3D and 2D classification steps, we could keep more particles that were in the rare views. After the initial particle screening step, a total of 1,015,378 particles (26.6% of all particles) were combined, re-centered and re-extracted from dose-weighted micrographs in dimensions of 256 × 256 square pixels (pixel size of 1.36 Å). From those re-extracted particles, 945,254 particles (24.8% of all particles) were further selected using an additional round of 2D classification. Refinement of those particles with a spherical mask generated a 3D reconstruction with clear secondary structure features for the majority of telomerase (including TERT–TER–p65 core, p50-OB and TEB subcomplex) and weak density for the flexible p75–p45–p19 subcomplex, which could only be partially observed at a lower threshold of the density map (CL=3σ). To improve the overall resolution, a soft mask (mask1) was used to exclude the flexible p75–p45–p19 subcomplex during the following data processing. Then, we performed another round of 3D classification with local angular search (RELION options: --sigma_ang 12), using the orientation parameters determined by the above 3D refinement as inputs. The particles were classified into 6 classes, and those in the two good classes with clear features for the catalytic core were selected.
Refinement of the combined 466,385 particles (12.2% of all particles) from the two good classes generated an overall 3.3 Å resolution reconstruction with variable local resolutions within the map. An additional round of 3D classification with a higher regularization parameter T (RELION options: --tau2_fudge 10 --sigma_ang 12) was performed to separate the movements of the TEN–TRAP region. Refinement of particles in the three major classes generated three reconstructions (P1, P2 and P3) with their TEN–TRAP at slightly different positions relative to the TERT ring (Extended Data Fig. 2d). 193,117 particles in the P2 class were refined again, followed by multiple rounds of CTF refinement to correct the CTF parameters, anisotropic magnification and higher-order aberrations. Beam-induced particle motion was corrected in RELION3.0 using Bayesian polishing module. The resulted “shiny” particles were refined with mask1, resulting in a final 3.3 Å resolution reconstruction with improved overall densities. To improve local resolution for model building, focused 3D classifications without alignment (RELION options: --skip_align --tau2_fudge 10) were performed using soft masks separately around the PK–LaM and Teb3, followed by 3D refinements with mask1. The resulting two reconstructions and the 3.3 Å resolution reconstruction were used for model building, as detailed below.
Resolutions of the cryo-EM maps were estimated on the basis of the “gold-standard” Fourier Shell Correlation (FSC) = 0.143 criterion56. The cryo-EM maps were corrected for the modulation transfer function (MTF) of the detector, sharpened with a negative B-factor and low-pass filtered to the stated resolution using the relion_postprocess program in RELION55. Local resolution evaluations were determined by ResMap57 with two independently refined half-maps. Data collection and processing statistics are given in Extended Data Table 1.
Cryo-EM data processing of telomerase T4D4 and T5D5.
For telomerase T4D4, 1,749,767 particles were picked from 4,885 micrographs. 111,603 particles were selected by similar procedures as described above for telomerase T3D2. To separate DNA-bound and DNA-free particles, an additional round of focused 3D classification without alignment (RELION options: --skip_align --tau2_fudge 10) was performed using a spherical mask surrounding the template–DNA duplex region. Finally, a 4.4 Å resolution reconstruction with clear duplex density was generated with selected 28,297 DNA-bound particles (Extended Data Fig. 7c). For telomerase T5D5, 3,951,672 particles were picked from 11,612 micrographs. 120,360 DNA-bound particles were selected, which yielded a 3.8 Å resolution reconstruction (Extended Data Fig. 7d). By using a smaller spherical mask surrounding the template–DNA duplex region during focused 3D classification (RELION options: --skip_align --tau2_fudge 20), we could further separate these DNA-bound particles into two subsets, 51,617 particles with a shorter duplex and 68,743 particles with a longer duplex (Extended Data Fig. 7d). Overall and local resolutions of the T4D4 and T5D5 reconstructions were evaluated as described above for telomerase T3D2.
Model building and refinement.
The atomic model of telomerase T3D2 was built and refined manually in COOT58. Initially, the previously reported model (PDB: 6D6V) generated based on a 4.8 Å resolution cryo-EM map5 was fitted into the current 3.3 Å resolution cryo-EM map with UCSF Chimera59 as the starting point for the model building. With the aid of high-resolution features, we manually adjusted side chain conformation of TERT, when necessary, moved the main chain to ensure a close fit between the density and the model. Sequence assignment of TRAP and CTE were mainly guided by visible densities of amino acid residues with bulky side chains, such as Phe, Tyr and Trp. However, the flexible linker between TEN and RBD (aa. 180–215) was invisible and could not be modeled, as well as two linkers within RBD (aa. 252–280) and TRAP (aa. 664–686). Modeling of the TEB subcomplex, including Teb1C (aa. 511–697), Teb2N (aa. 29–175) and Teb3 (aa. 5–121), was achieved in similar way. The main chain of p50-OB (aa. 1–184) was manually re-traced and de novo built with the help of secondary structure prediction generated by PSIPRED60. The C-terminal region of p50 (aa. 185–249) is masked out at least in part along with p75–p45–p19 subcomplex during cryo-EM data processing. Homology model of p65 LaM (aa. 114–238) was generated using PHYRE261 and manually adjusted according to the cryo-EM map, including the removal of an invisible internal linker (aa. 170–201). TER was initially modeled piece by piece, and then connected manually in COOT. For the regions contacting TERT, including the t/PK domain and the distal part of SL4, well-defined nucleotide densities facilitated the de novo model building process. For the rest of TER, including SL1 and the proximal part of SL4, previous model (PDB: 6D6V) was adjusted for their base conformation and, when necessary, for their backbones to fit into the density map. The apical loop of SL2 was modeled with the help of its NMR structure (PDB: 2M22)62.
The cryo-EM density of telomeric DNA was traced from its 3′ end all the way to Teb1C (Extended Data Fig. 6e, f). Eight nucleotides were modeled unambiguously inside the catalytic cavity, including G30-T27 which pair with the template and T26-G23 which stack with TERT F414. Nucleotides densities binding to the C-shape cleft of Teb1C could be assigned to three guanosines instead of thymidine according to their size and shape (Extended Data Fig. 6g, h). Eventually, we modeled them as G19-G17 since the weak density connecting them to G23 fitted better with a flexible linker of three instead of four nucleotides (Extended Data Fig. 6e). Nucleotides from G16 to G1 were invisible in the cryo-EM map.
The model obtained above was used as the starting point for the modeling of other lower resolution reconstructions. The model was first rigid-body fitted into these maps using Chimera and then manually checked in COOT. The template–DNA duplex and adjacent TBEL/TREL regions were manually adjusted to fit the densities. Based on the knowledge gained from the 3.3 Å resolution T3D2 structure, the template–DNA duplex in the previously published model of telomerase with (GTTGGG)2GTLTLGLGLGG (telomerase T3D3, PDB: 6D6V) was reevaluated, and new modeling showed a 5 bp template–DNA duplex (Extended Data Fig. 8).
All models were refined using Phenix63 in real space with secondary structure, Ramachandran, and rotamer restraints. The structures were validated using the Molprobity scores and statistics of the Ramachandran plots. Refinement statistics of the models were summarized in Extended Data Table 1. Model/map FSC validation was shown in Extended Data Fig. 2f. Visualization of the atomic models, including figures and movies, were prepared using UCSF Chimera59 and ChimeraX64.
In vitro transcription and purification of TER.
TER was in vitro transcribed with a linearized DNA template containing a hammerhead ribozyme sequence at the 3′ end to allow self-cleavage of TER at a precise position after the transcription65. Briefly, homemade T7 RNA polymerase was added to a reaction containing 40 mM MgCl2, 4–6 mM of each NTP, and 1 μM DNA template in 40 mM Tris·HCl pH 8.0, 1 mM spermidine, 2.5 mM DTT and 0.01% Triton X-100. The reaction was incubated overnight at 37 °C for transcription of the DNA template and cleavage of the hammerhead ribozyme construct. TER was subsequently purified by electrophoresis on a 10% denaturing polyacrylamide gel followed by electroelution of the excised RNA band66.
In vitro reconstitution of telomerase and direct activity assays.
The expression and assembly of telomerase was performed in vitro in rabbit reticulocyte lysates (RRL) as previously described4,67, with minor changes. TERT and p65 were co-expressed using TNT® quick-coupled transcription/translation systems (Promega) in the presence of TER. The expression of TERT was monitored by 35S-methionine incorporation. p50N30 (aa. 1–251) was expressed in another RRL synthesis reaction. Teb1C (aa. 505–701) was expressed in E. coli and purified by size-exclusion chromatography (SEC) before use. The expressed TERT–TER–p65 core, p50N30 and Teb1C were incubated together in RRL at 30 °C for 30 min before activity assays. The final concentrations of TER and Teb1C in the RRL mixtures were 370 nM and 450 nM, respectively. The activity assay was carried out in a 20 μL solution containing 200 μM dTTP, 3 μM dGTP, ~ 5–10 uCi dGTP-[α32P] (Perkin-Elmer), 2 mM Mg2+, 10 μM (GTTGGG)5 primer and 10 μL RRL mixture. Reactions were performed at 30 °C for 15 min and stopped with quench buffer (10 mM Tris·HCl pH 8.0 and 10 mM EDTA). The products were phenol/chloroform extracted and ethanol precipitated together with a 15 nt 32P-end-labelled DNA oligonucleotide as a recovery control (RC) and resolved on a 10% denaturing polyacrylamide gel. The gel was dried and exposed to a phosphor imaging screen and scanned on a PharosFX system (Bio-Rad).
Band intensities were quantified using QuantityOne software (Bio-Rad). Relative activities of TERT mutations were determined by normalizing the integrated density of each lane relative to the RC then comparing to that of wild-type TERT (WT, as 100%). Relative RAP of TERT mutations were determined by the “fraction left behind” (FLB) method32,68. For each lane, the counts for each RAP band, which represented the extension of one telomere repeat, were divided by the number of radioactive guanosines incorporated and normalized against the RC. Then, the FLB of each RAP band was calculated by taking the sum of counts for each RAP band and all RAP bands below it divided by the total counts of RAP bands in this lane. Then, the ln(1-FLB) of each RAP band was plotted against repeat number added and the scattered points were fitted with a linear regression to get the slope value. Finally, the relative RAP for each mutation was determined by −ln(2)/slope and compared to that of WT (as 100%). Activity assays for each TERT mutation were repeated 3–4 times.
Extended Data
Extended Data Fig. 1: Biochemical and biophysical evaluation of endogenously purified Tetrahymena telomerase with single-stranded telomeric DNA (sstDNA).

a, Silver-stained SDS-PAGE gel of the tandem affinity purified telomerase. Serial diluted BSA samples were loaded together to assist concentration estimation of the telomerase sample. Gel image is representative of independent biological replicates (n=3). b, Direct telomeric DNA extension assays of the purified telomerase bound with different sstDNA primers. A standard telomere addition pattern is observed when using (GTTGGG)5 or (GTTGGG)3 primer (P1 and P2). However, the translocation of product is inhibited when using (GTTGGG)2GTTGGLGLGLT primer (P3), resulting in a single dark band (red asterisk). GL denotes LNA nucleotide instead of DNA nucleotide. Note that the LNA containing product (red asterisk) migrates slightly slower through the gel than non-modified DNA. RC, recovery control. Gel image is representative of independent biological replicates (n=3). c, Motion corrected cryo-EM micrograph. d, Representative 2D class averages of telomerase particles. All results from sample purification (a), activity assays (b), and cryo-EM experiments (c) were successfully reproduced at least three times. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2: Cryo-EM data processing workflow of telomerase with sstDNA (GTTGGG)5 (telomerase T3D2) and the evaluation of the reconstruction.

a, Data processing workflow (detailed in methods). Soft masks used in data processing are colored in orange. b, Euler angle distributions of telomerase particles used for the 3.3 Å resolution reconstruction. c, Local resolution evaluation of the 3.3 Å resolution cryo-EM map shown in surface views (left) and a slice view of the core region (right). d, Superposition of reconstructions P1, P2 and P3, that illustrate the rotation of TEN–TRAP. The three maps were lowpass-filtered to 6 Å and aligned on the TERT ring. p50 (red) and TEB bind to and move together with TEN–TRAP. e, Plot of the Fourier shell correlation (FSC) as a function of the spatial frequency with resolution of the final reconstruction indicated. f, FSC coefficients as a function of spatial frequency between model and cryo-EM density maps. Red curve: refined model versus half map 1 used for refinement; green curve: refined model versus half map 2 not used for refinement; black curve: refined model versus the combined final map. The generally similar appearances between the red and green curves suggests no substantial over-fitting. g, Representative cryo-EM densities (gray and mesh) encasing the related atomic models (color sticks and ribbons).
Extended Data Fig. 3: Detailed interactions between TERT and p65 with TER.

a, Close-up view of motif 3N (aa. 550–560). Motif 3A helix is bent toward motif 2, and motif 3N in between forms a finger shape architecture. b, Ribbon diagram of the TERT–TER “interlock” with TERT domains colored as indicated. c, Schematic of SL2, TBE and TBEL nucleotides and their interactions with TERT RBD domain. Arrows indicate sites of polar interactions. Bold line represents the stacking interaction between F242 and C39. d-f, Structure of TER L4 and its interactions with TERT RBD, CTE and p65 xRRM (green). g, Rainbow colored ribbon diagram of p65 LaM with secondary structural elements labeled. Positively charged and aromatic residues located on the interface between p65 LaM and TER are shown as spheres. h, Electrostatic surface representation of p65 LaM and its interactions with TER S1, PK and the 3′ polyU. p65 LaM in g and h are in the same orientation. i, Schematic of PK with regions that interact with TERT and p65 indicated. j, Interactions between motif 3 and the template. End of motif 3B and start of motif 3C are in minor groove of the duplex. k, The eight TER nucleotides that stack inside the catalytic cavity. Cryo-EM densities are shown as transparent meshes. Ideal A-form stacking of 8 nucleotides (white) is shown for comparison. Backbone of the last 3 TER nucleotides in the stacking deviate from ideal A-form conformation.
Extended Data Fig. 4: Interactions between TEN–TRAP and telomerase activity assays.

a, Ribbon representation of TERT with its domains colored as indicated. Unmodeled regions of TERT are shown as dashed lines, including the linker between TEN and RBD (aa. 180–215), flexible linkers within RBD (aa. 252–280), and TRAP (aa. 664–686). b, Hydrophobic interactions between the distal region of TRAP and the C-terminal helix of TEN domain, which is further stabilized by Q168 via two hydrogen bonds. c, The extended β sheet across TEN and TRAP. V791Y (V731 in Tetrahymena) mutation in hTERT that disrupts telomere length maintenance and cell immortalization is located at the interface69. d, e, In vitro reconstituted telomerase activity assays with TERT mutations on the TEN–TRAP interface. The top panels are SDS-PAGE gels showing the expression level of 35S-Met incorporated TERT mutants. RC, recovery control. Quantitation of activity and RAP for each mutant are shown in bar graphs below. f, g, Quantitation of activity and RAP for gel shown in Fig. 1j and 3g. The data represent the mean ± s.d. from 3 independent experiments.
Extended Data Fig. 5: Comparison between TERT from Tetrahymena (TtTERT), human (hTERT), and the TERT-like protein from Tribolium castaneum (TcTERT-like).

a, Sequence alignment of TtTERT and hTERT. Secondary structures and conserved motifs of TtTERT are shown on top, with unmodeled regions shown as dashed lines. The alignments of TEN, RBD, RT, CTE domains and TRAP motif were conducted separately with NIH COBALT70 and then merged together. The alignment of CP2/TFLY region was adjusted manually according to the previously reported alignment37. b, Structural comparison of the TERT-ring of TtTERT (color) and TcTERT-like protein (gray, PDB ID: 3KYL). TcTERT-like protein lacks TEN, TRAP, and TER, and was crystallized with an artificial template–DNA duplex. c, d, Ribbon diagrams of template–DNA duplexes and surrounding structural elements of TtTERT (c) and TcTERT-like protein (d). The palm, fingers, primer grip, thumb helix, thumb loop, motif 3 and T are structurally conserved between TtTERT and TcTERT-like protein. The “bridge loop” of TcTERT-like protein is in a similar position as the one in TtTERT, however the tip residues (S82 and F83) have no contact with the template–DNA duplex. CP2, which participates in template 5′ boundary definition and template nucleotides guidance in TtTERT, appears to be absent in TcTERT-like protein.
Extended Data Fig. 6: Details of p50-OB–TERT (a-d) and Teb1C–sstDNA (e-h) interactions.

a, Rainbow colored ribbon diagram of p50-OB with secondary structure elements labeled. b, Comparison of p50-OB (red) and human TPP1-OB (gray, PDB ID: 2I46) structures. c, TEN loop (aa. 121–126) passes through a hydrophobic cleft of p50-OB. This loop is disordered loop in the TEN domain crystal structure71.d, Structure based sequence alignment of p50-OB and human TPP1-OB. The secondary structure elements of p50-OB (red) and TPP1-OB (gray) are shown above and below the sequence alignment, respectively. Residues located at the interface between p50-OB and TERT are highlighted in yellow. The NOB and TEL patch residues on human TPP1-OB are indicated and colored in yellow as well. The phosphorylation site S111 of TPP1-OB is colored in green. Scaffold residues of Lα2-β4 shown in Fig. 2a (lower) are colored in blue. e, Path of sstDNA from active site to Teb1C. Low-pass filtered cryo-EM density of sstDNA (transparent surface) is superimposed with the unfiltered DNA density (green) to better show its flexible region from T20 to G22. Cryo-EM densities corresponding to TERT domains, TER and Teb1C are colored as in Fig. 1d. f, Sequence of the sstDNA used for the cryo-EM sample preparation with the template and Teb1C interacting regions indicated. Nucleotides from G1 to G16 are invisible in the cryo-EM map. g, Interactions between sstDNA nucleotides and Teb1C as indicated in e. Intermolecular hydrogen bonds and stacking interactions are shown as dashed yellow lines and black arrows, respectively. h, Specific interactions between Teb1C residues K660, E667 and sstDNA nucleotide G19 shown together with their cryo-EM densities. Hydrogen bonds and their lengths are indicated.
Extended Data Fig. 7: Cryo-EM reconstructions of telomerase with different sstDNA bound.

a, List of sstDNA primers used for cryo-EM sample preparation and their sequences. DNA/LNA nucleotides that pair with the template are underlined. b, Resolution of reconstructions determined by gold-standard FSC at the 0.143 criterion. c, d, Cryo-EM data processing workflow of telomerase T4D4 and T5D5, and evaluations of the final reconstructions. Initial particle screening processes are analogous to those described in the data processing workflow of telomerase T3D2 and omitted for brevity. Focused 3D classifications were performed to separate DNA-free and DNA-bound particles. Short duplexes were observed in both of T4D4 and T5D5 reconstructions. We note that for telomerase T5D5, there is a subset of particles with a longer duplex, which we attribute to the greater stability conferred on the duplex by LNA nucleotides at the thermodynamically most stable duplex (dGGGGT·rACCCC) formed in the previous step.
Extended Data Fig. 8: Template–DNA duplexes in telomerase structures at different steps of telomeric DNA synthesis.

Top, sequences of sstDNA primers. TL/GL denotes LNA nucleotide. DNA/LNA nucleotides pairing with the template are underlined. Middle, ribbon diagrams of the duplex, template adjacent nucleotides, bridge loop, thumb helix (TH) and thumb loop (TL) superimposed with cryo-EM densities (transparent surfaces). Bottom, schematics of the duplexes. The active site (red star), bridge loop residues (R413 and F414), and catalytic cavity (gray shade) in different structures are aligned to show the relative positions of the duplex. TER and DNA nucleotides are color coded as in Fig. 4.
Extended Data Fig. 9: Structural details of template boundary determination (TBE, TBEL, TREL, TRE) in telomerase T5D5.

a, Telomerase catalytic cavity in telomerase T5D5 with TER (gray) and DNA (green) shown as ribbon and TERT shown as surface (colored). TBE, TBEL, template, TREL and TRE nucleotides are highlighted as indicated. b-e, Detailed interactions between TERT and TER in regions as indicated in a. Intermolecular hydrogen bonds and stacking interactions are shown as dashed yellow lines and black arrows, respectively. The electrostatic surface of the TRAP–TH channel is shown in d. f, Schematic showing specific interactions between TERT and TREL-TRE as shown in c and e. Nucleotides from A54 to A58 are unmodeled and indicated as dashed orange lines. g, Predicted TRE and TREL conformation when the template is at the +1 position (template nucleotide C48 at the active site). TREL nucleotides C56U57A58 would be fully stretched (~5–6 Å phosphate-to-phosphate distance for each nucleotide) to span the distance from the “neck” of the TRAP–TH channel to the anchored TRE.
Extended Data Table 1:
Cryo-EM data collection, refinement and validation statistics.
| Telomerase T3D2 (EMDB-23437) (PDB 7LMA) | Telomerase T4D4 (EMDB-23438) | Telomerase T5D5 (EMDB-23439) (PDB 7LMB) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 105,000 | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e-/Å2) | 48 | 48 | 48 |
| Defocus range (μm) | −0.8 – −4.0 | −0.8 – −4.0 | −0.8 – −4.0 |
| Pixel size (Å) | 1.36 | 1.36 | 1.36 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 3,816,856 | 1,749,767 | 3,951,672 |
| Particle images after class2d (no.) | 945,254 | 373,757 | 764,707 |
| Final particle images (no.) | 193,117 | 28,297 | 120,360 |
| Map resolution (Å) | 3.3 | 4.4 | 3.8 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 3.0 – 6.0 | 4.4 – 8.0 | 3.5 – 7.0 |
| Refinement | |||
| Initial model used (PDB code) | 6D6V | 7LMA | |
| Model resolution (Å) | 4.1 | 4.4 | |
| FSC threshold | 0.5 | 0.5 | |
| Map sharpening B factor (Å2) | −82.0 | −130.8 | |
| Model composition | |||
| Non-hydrogen atoms | 18,672 | 18,498 | |
| Protein residues | 1,837 | 1,837 | |
| RNA/DNA Nucleotides | 170 | 162 | |
| Ligands | 1 | 1 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.005 | |
| Bond angles (°) | 0.858 | 0.915 | |
| Validation | |||
| MolProbity score | 1.66 | 2.02 | |
| Clashscore | 6.61 | 11.64 | |
| Poor rotamers (%) | 0.91 | 0.06 | |
| Ramachandran plot | |||
| Favored (%) | 95.69 | 93.31 | |
| Allowed (%) | 4.31 | 6.69 | |
| Disallowed (%) | 0.00 | 0.00 |
Supplementary Material
Supplementary Fig. 1: Source images for all data obtained by polyacrylamide gel electrophoresis.
Acknowledgements
This work was supported by NIH R35GM131901 and NSF MCB2016540 grants to J.F. and NIH grant GM071940 to ZHZ. We acknowledge use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111 and U24GM116792), NSF (DBI-1338135 and DMR-1548924). Some preliminary data was collected at the Stanford-SLAC Cryo-EM Center (S2C2) supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129541) and National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539,) and by grants from the Simons Foundation (SF349247) and NY State Assembly. We thank D. Weisman for help with illustration of Fig. 5.
Footnotes
Competing interests The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-021-03529-9
Data availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 23437 (telomerase T3D2), 23438 (telomerase T4D4) and 23439 (telomerase T5D5). The atomic models have been deposited in the Protein Data Bank (PDB) under accession codes 7LMA (telomerase T3D2) and 7LMB (telomerase T5D5). Atomic model and cryo-EM density map of telomerase T3D3 were retrieved from the PDB (accession code 6d6v) and EMDB (accession code EMD-7821). Other structures used in this study were retrieved from the PDB with accession codes 2I46 for TPP1-OB, 3KYL for Tribolium TERT-like protein and 2M22 for TER SL2. Uncropped version of all the gels are included as Supplementary Information Figure 1.
References
- 1.Blackburn EH & Collins K Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M & Blackburn EH The telomere syndromes. Nat Rev Genet 13, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shay JW Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov 6, 584–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J et al. Structure of Telomerase with Telomeric DNA. Cell 173, 1179–1190 e1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen THD et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Xin H et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445, 559–562 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Greider CW & Blackburn EH Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 (1985). [DOI] [PubMed] [Google Scholar]
- 10.Chan H, Wang Y & Feigon J Progress in Human and Tetrahymena Telomerase Structure Determination. Annu Rev Biophys 46, 199–225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu RA, Upton HE, Vogan JM & Collins K Telomerase Mechanism of Telomere Synthesis. Annu Rev Biochem 86, 439–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podlevsky JD, Bley CJ, Omana RV, Qi X & Chen JJ The telomerase database. Nucleic Acids Res 36, D339–343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prathapam R, Witkin KL, O’Connor CM & Collins K A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 12, 252–257 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell JR, Cheng J & Collins K A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3’ end. Mol Cell Biol 19, 567–576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillis AJ, Schuller AP & Skordalakes E Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455, 633–637 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Jiang J et al. The architecture of Tetrahymena telomerase holoenzyme. Nature 496, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei M, Podell ER & Cech TR Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 11, 1223–1229 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Lim CJ & Cech TR Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev Mol Cell Biol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lue NF, Chan J, Wright WE & Hurwitz J The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun 5, 5762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraia RJ, Mattijssen S, Cruz-Gallardo I & Conte MR The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh M et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 47, 16–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone MD et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446, 458–461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR & Curry S Structural analysis reveals conformational plasticity in the recognition of RNA 3’ ends by the human La protein. Structure 16, 852–862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar J et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong FL et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tesmer VM, Smith EM, Danciu O, Padmanaban S & Nandakumar J Combining conservation and species-specific differences to determine how human telomerase binds telomeres. Proceedings of the National Academy of Sciences of the United States of America (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grill S, Tesmer VM & Nandakumar J The N Terminus of the OB Domain of Telomere Protein TPP1 Is Critical for Telomerase Action. Cell Rep 22, 1132–1140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y et al. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proceedings of the National Academy of Sciences of the United States of America 110, 5457–5462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upton HE, Chan H, Feigon J & Collins K Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes. J Biol Chem 292, 217–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Z et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proceedings of the National Academy of Sciences of the United States of America 108, 20357–20361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shastry S, Steinberg-Neifach O, Lue N & Stone MD Direct observation of nucleic acid binding dynamics by the telomerase essential N-terminal domain. Nucleic Acids Res 46, 3088–3102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansson LI et al. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proceedings of the National Academy of Sciences of the United States of America (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patrick EM, Slivka JD, Payne B, Comstock MJ & Schmidt JC Observation of processive telomerase catalysis using high-resolution optical tweezers. Nat Chem Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Susac L & Feigon J Structural Biology of Telomerase. Cold Spring Harb Perspect Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie M, Podlevsky JD, Qi X, Bley CJ & Chen JJ A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res 38, 1982–1996 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura TM et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Jansson LI et al. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol 22, 883–888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama BM, Gomez A & Stone MD A conserved motif in Tetrahymena thermophila telomerase reverse transcriptase is proximal to the RNA template and is essential for boundary definition. J Biol Chem 288, 22141–22149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harkisheimer M, Mason M, Shuvaeva E & Skordalakes E A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure 21, 1870–1878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vester B & Wengel J LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43, 13233–13241 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Schaich MA et al. Mechanisms of nucleotide selection by telomerase. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai CK, Miller MC & Collins K Template boundary definition in Tetrahymena telomerase. Genes Dev 16, 415–420 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MC & Collins K Telomerase recognizes its template by using an adjacent RNA motif. Proceedings of the National Academy of Sciences of the United States of America 99, 6585–6590 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berman AJ, Akiyama BM, Stone MD & Cech TR The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol 18, 1371–1375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M et al. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat Commun 11, 2605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W & Lee YS A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat Struct Mol Biol 22, 844–847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu RA, Tam J & Collins K DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J 36, 1908–1927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Gallagher-Jones M, Sušac L, Song H & Feigon J A structurally conserved human and Tetrahymena telomerase catalytic core. Proceedings of the National Academy of Sciences, 202011684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X et al. Quality-Control Mechanism for Telomerase RNA Folding in the Cell. Cell Rep 33, 108568 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Holohan B et al. Impaired telomere maintenance in Alazami syndrome patients with LARP7 deficiency. BMC Genomics 17, 749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Min B & Collins K An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 36, 609–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Buchan DW, Minneci F, Nugent TC, Bryson K & Jones DT Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41, W349–357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richards RJ, Theimer CA, Finger LD & Feigon J Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res 34, 816–825 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cash DD & Feigon J Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res 45, 482–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrov A, Wu T, Puglisi EV & Puglisi JD RNA purification by preparative polyacrylamide gel electrophoresis. Methods Enzymol 530, 315–330 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Hong K et al. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol 33, 3962–3971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Latrick CM & Cech TR POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29, 924–933 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu TW, D’Souza Y & Autexier C The Insertion in Fingers Domain in Human Telomerase Can Mediate Enzyme Processivity and Telomerase Recruitment to Telomeres in a TPP1-Dependent Manner. Mol Cell Biol 36, 210–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papadopoulos JS & Agarwala R COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Jacobs SA, Podell ER & Cech TR Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13, 218–225 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: Source images for all data obtained by polyacrylamide gel electrophoresis.
Data Availability Statement
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 23437 (telomerase T3D2), 23438 (telomerase T4D4) and 23439 (telomerase T5D5). The atomic models have been deposited in the Protein Data Bank (PDB) under accession codes 7LMA (telomerase T3D2) and 7LMB (telomerase T5D5). Atomic model and cryo-EM density map of telomerase T3D3 were retrieved from the PDB (accession code 6d6v) and EMDB (accession code EMD-7821). Other structures used in this study were retrieved from the PDB with accession codes 2I46 for TPP1-OB, 3KYL for Tribolium TERT-like protein and 2M22 for TER SL2. Uncropped version of all the gels are included as Supplementary Information Figure 1.


