Summary
Background
Currently, evaluation of the IgG antibodies specific for the SARS-CoV-2 Spike protein following vaccination is used worldwide to estimate vaccine response. Limited data are available on vaccine-elicited IgM antibodies and their potential implication in immunity to SARS-CoV-2.
Methods
We performed a longitudinal study to quantify anti-S SARS-CoV-2 IgG and IgM (IgG-S and IgM-S) in health care worker (HCW) recipients of the BNT162b2 vaccine. Samples were collected before administration (T0), at the second dose (T1) and three weeks after T1 (T2). The cohort included 1584 immunologically naïve to SARS-CoV-2 (IN) and 289 with history of previous infection (PI).
Findings
IN showed three patterns of responses: (a) IgG positive/IgM negative (36.1%), (b) coordinated IgM-S/IgG-S responses appearing at T1 (37.4%) and (c) IgM appearing after IgG (26.3%). Coordinated IgM-S/IgG-S responses were associated with higher IgG titres. In IgM-S positive PI, 64.5% were IgM-S positive before vaccination, whereas 32% and 3.5% developed IgM-S after the first and second vaccine dose, respectively. IgM-S positive sera had higher pseudovirus neutralization titres compared to the IgM-S negative.
Interpretation
Coordinated expression of IgG-S and IgM-S after vaccination was associated with a significantly more efficient response in both antibody levels and virus-neutralizing activity. The unconventional IgG-S positive/IgM-S negative responses may suggest a recruitment of cross coronaviruses immunity by vaccination, warranting further investigation.
Funding
Italian Ministry of Health under “Fondi Ricerca Corrente”- L1P5 and “Progetto Ricerca Finalizzata COVID-2020-12371675”; FUR 2020 Department of Excellence 2018-2022, MIUR, Italy; The Brain Research Foundation Verona.
Keywords: SARS-CoV-2, COVID-19, BTN162b2 vaccine, IgG, IgM, Spike
Abbreviations: IgM, Immunoglobulins M; IgG, Immunoglobulins G
Research in context.
Evidence before this study
It is generally accepted that IgM antibodies provide an early-stage response during viral infections prior to the maturation of the class-switched, high affinity IgG response for long-term immunity and immunological memory. The humoral response following SARS-CoV-2 vaccination is still under intensive investigation, with the main confounder being previous exposures to SARS-CoV-2 and the resulting presence of pre-existing immunity towards the Spike protein used in the vaccine formulation. Thus, the definition of correlates of protective immunity to SARS-CoV-2 infection and vaccination are urgently needed for guiding vaccine management and informing public health decisions. Nonetheless, most research to date has focused on the development and maintenance of the RBD-specific IgG, with little attention to IgM.
Added value of this study
We investigated a population of 1873 health care worker (HCW) recipients of the BNT162b2 (Comirnaty) vaccine, with 1584 immunologically naïve to SARS-CoV-2 (IN) and 289 with history of previous infection (PI). We performed a longitudinal analysis of the humoral response (IgG and IgM antibodies specific for the SARS-CoV-2 spike protein, IgG-S and IgM-S) in samples collected before administration (T0), at the second dose (T1) and 3 weeks after the second dose (T2). Furthermore, we analysed the vaccine response in a small group of subjects vaccinated with Vaxzevria (Astra Zeneca) or Spikevax (Moderna). We observed three unconventional patterns of antibody response: absence of IgM, development of IgM following IgG appearance and simultaneous presence of IgM and IgG. Among the three, the latter was associated with a more efficient response in both anti-SARS-CoV-2 IgG-S levels and virus-neutralizing activity, following vaccination.
Implications of all the available evidence
Our study highlights the importance of IgM in assessing response after SARS- CoV-2 vaccination. We demonstrated that SARS-CoV-2 vaccination can induce a humoral response that appears to be unconventional. This is suggestive of a response that recalls IgG developed against other coronaviruses. Indeed, only individuals that developed SARS-CoV-2 specific IgM together with SARS-CoV-2 specific IgG showed the better response and probably higher levels of protection, following vaccination. These findings are innovative, timely and significantly improve current knowledge by suggesting a crucial role of IgM in the development of anti-SARS-CoV-2 humoral response, following vaccination.
Alt-text: Unlabelled box
Introduction
Correlates of protective immunity to SARS-CoV-2 infection are under intensive investigation in COVID-19 patients and vaccinees and are urgently needed for guiding vaccine management and informing public health decisions.1,2 It is generally accepted that IgM antibodies provide an early-stage response during viral infections prior to the maturation of the class-switched, high affinity IgG response for long-term immunity and immunological memory.3 During SARS-CoV-2 infection, antigen (Ag)-specific IgM antibodies can be detected as soon as four days after infection with a peak at around 20 days, while Ag-specific IgG increase around 7 days after infection with a peak at approximately 25 days.4,5 Rapid deployment of SARS-CoV-2 specific IgM was reported to be associated with milder disease course compared with severe cases that experienced a later raise in IgM,6 although the question remains controversial.7 Several studies reported that a proportion of patients never develop IgM, while others develop IgG prior to IgM.2,5,8, 9, 10, 11, 12 Overall, these data suggest both a potential role of Ag-specific IgM in preventing severe disease but also the possibility that SARS-CoV-2 infection may trigger unconventional humoral responses, possibly generated by pre-existing immunity to other human coronaviruses.13,14
The humoral response following SARS-CoV-2 vaccination is still under intensive investigation, as it is not yet clear the role played by pre-existing immunity in the response to vaccination. Previously infected (PI) individuals have been shown to develop a more efficient antibody response to COVID-19 vaccines than immunologically naïve individuals (IN).15 Notably, neutralizing activity 7 days following the first vaccine dose in PI vaccinees was not significantly different from that observed in IN vaccinees 7 days after the second vaccine dose.15 Furthermore, the kinetic of both anti SARS-CoV-2 receptor-binding domain (RBD) IgG and live-virus neutralization capacity was faster in PI than in IN vaccinees.15 With regard to IgM, one study reported that about 50% of IN vaccinees did not develop IgM after the first dose of BNT162b2 vaccine.16
Nonetheless, most research thus far has concentrated on the development and maintenance of the RBD-specific IgG, with little attention to IgM.
Our group has previously shown that IN vaccinees fail to develop IgM against the SARS-CoV-2 spike glycoprotein (IgM-S) before IgG against the SARS-CoV-2 spike glycoprotein (IgG-S)14; more specifically, following the first vaccine dose, we observed the simultaneous development of IgM-S and IgG-S in 54% of the vaccinees, and an unconventional IgG-S response without detectable IgM-S in the remaining 46%. We observed a similar trend in PI vaccinees.
In this study, we analysed a cohort of Health Care Workers (HCW) including 1584 IN and 289 PI vaccinees to study the IgM-S response following BNT162b2 vaccination and assess its association with the development and maintenance of IgG responses. We leveraged the availability of two groups of PI vaccinees who had been infected in the first and the second pandemic wave in Italy to assess the antibody profile at different times after infection. In available subgroups of IN vaccinees, we evaluated humoral response following other types of vaccines, including Vaxzevria (AstraZeneca) and Spikevax (Moderna).
Methods
Population
The sera of 1989 HCW with and without pre-existing infection for SARS-CoV-2 (as per former nasal swab positivity) who had received their first vaccine dose (BNT162b2 mRNA, Pfizer-BioNTech) in January 2021 were analysed. Samples were collected before vaccine administration (T0), at the second dose (T1) and three weeks after T1 (T2) and tested for IgG against the Spike glycoprotein (IgG-S), IgG against the Nucleocapsid protein (IgG-N) and IgM against the Spike glycoprotein (IgM-S). All individuals who had received two doses of BNT162b2 vaccine and had complete serological data were included in the study. Among the 1957 individuals having complete information, 84 were negative at the swab test but had positive serology (IgM-S or IgG-S or IgG-N) at T0; they were considered as false negatives in accordance with a recent study17 and were not included in the present study. Antibody response analyses were conducted on 1584 IN subjects and 289 PI subjects.
Ethics
Samples were collected and stored in the University of Verona biobank (Ethics Committee approval prot. N. 1538) and in Tropica Biobank of the IRCCS Sacro Cuore Don Calabria Hospital (Ethics Committee approval prot. N. 17985). All participants signed informed consent.
Serology and neutralization
IgM-S and IgG-N were measured using the SARS-CoV-2 IgG-N assay and the SARS-CoV-2 IgM-S assay (Abbott, Ireland); IgG-S(RBD) were tested using the SARS-CoV-2 IgG II Quant assay (Abbott, Ireland) as previously described.14,17
Briefly, SARS-CoV-2 IgG-N, IgM and SARS-CoV-2 IgG II Quant (IgG-S) assays (Abbott, Ireland) were performed according to the manufacturer's procedure, using the ARCHITET i System (Abbott). The resulting chemiluminescent reaction was measured as a relative light unit (RLU) by the system optics. The RLU of the sample (S) was automatically compared with the RLU of a specific calibrator (C), resulting in a IgG assay index (S/C). As per manufacturer's instructions, the interpretation of the results were as follow: for IgG-N, index (S/C)<1.4 = negative, index (S/C)≥1.4 = positive. For IgM-S, index (S/C)<1 = negative, index (S/C)≥1 = positive. For IgM-S assay the reported positive predicted value (PPV) is 92.07% (IC 95%: 87.07, 95.24) and the reported negative predicted value (NPV) is 99.82% (IC 95%: 99.47, 99.94).
For IgG-S the Ab quantification was automatically performed by the system using a calibration curve, a fitting system and interpolation with 4 parameters (4PLC, Y weighted). The results in Arbitrary Unit (AU)/mL, is converted in the WHO international binding antibody unit (BAU)/mL according to the following equation: 1BAU = 0.142*AU, with BAU/mL<7.1 = negative and BAU/mL≥7.1 = positive. For IgG II Quant the manufacturer reports a PPV of 92.11% (IC 95%: 85.87, 95.73) and a NPV of 99.97% (IC 95%: 99.76, 100.00). Samples with values >5680 BAU/mL (upper limit of quantification) were diluted 1:2 and measured again. Concentrations were reported considering the dilution factor. Samples were run in single replicate.
Neutralizing activity of sera was tested using lentiviral particles pseudotyped with SARS-CoV-2 spike, as previously described.14,18
Statistical analysis
Kruskal-Wallis rank test and Fisher's exact test were used when needed in the descriptive analysis. Pseudovirus neutralization assay expressed as infectious dose (ID50) and IgG-S levels were ln-transformed [ln(ID50) and ln(IgG-S)] to resemble normal distributions. Two-level linear regression models (measurement: level 1 unit; subject: level 2 unit) were used to predict the mean of ln(ID50) and ln(IgG-S) levels according to time of examination (T0, T1, T2) and IgM-S group (for Figs. 1, 4 and 5) or serology group (for Figure 6), separately for IN and PI subjects. The models had a random intercept term at level 2 and time of examination, IgM-S/serology group, their interaction term, age at T0, sex and pandemic wave (1st or 2nd, for PI only) as fixed effect covariates. A first-order autoregressive error was included at level 1 in order to take the correlation of the within-subject observations over time into account. All statistical analyses were performed by using STATA software (release 17; StataCorp, College Station, TX).
Figure 1.
Neutralization assays in naïve and previously infected vaccinees.
Pseudovirus neutralization assay expressed as infectious dose (ID50) in naïve (panel a) and previously infected (panel b) vaccinees according to time of examination (T0, T1 and T2) and IgM-S development after two doses of the BNT162b2 vaccine (IgM-SPOS, red dots and lines; IgM-SNEG subjects, blue dots and lines). Predicted means of ln(ID50) levels (with the 95% confidence interval) according to time of examination and IgM-S group in naïve (panel c) and previously infected (panel d) vaccinees were obtained by a two-level linear regression model. Statistically significant p-values of the difference in the predicted means between consecutive times of examination in the same IgM-S group and between IgM-S groups at the same time of examination are reported in panels a and b.
Figure 4.
IgG-S response in naïve vaccinees.
IgM-S (panel a) and IgG-S (panel b) measures in naïve vaccinees according to time of examination (T0, T1 and T2) and time of IgM-S positivity (IgM-SNEG, n = 572, blue dots; IgM-SPOST1, n = 593, red dots; IgM-SPOST2, n = 418, purple dots). Being all naïve subjects, no individuals had detectable IgM-S at T0. Predicted means of ln(IgG-S) measures (with the 95% confidence interval) according to time of examination and time of IgM-S positivity (panel c) were obtained by a two-level linear regression model. Statistically significant p-values of the difference in the predicted means between consecutive times of examination at the same time of IgM-S positivity and between different times of IgM-S positivity at the same time of examination are reported in panel b. The horizontal lines indicate the cut-off value to discriminate positive and negative samples for each assay.
Figure 5.
IgG-S response in previously infected vaccinees.
IgM-S (panel a) and IgG-S (panel b) measures in previously infected vaccinees according to time of examination (T0, T1 and T2) and time of IgM-S positivity (IgM-SNEG, n = 117, blue dots; IgM-SPOST0, n = 111, orange dots; IgM-SPOST1, n = 55, red dots; IgM-SPOST2, n = 6, purple dots). Predicted means of ln(IgG-S) measures (with the 95% confidence interval) according to time of examination and time of IgM-S positivity (panel c) were obtained by a two-level linear regression model. Statistically significant p-values of the difference in the predicted means between consecutive times of examination at the same time of IgM-S positivity and between different times of IgM-S positivity at the same time of examination are reported in panel b. The horizontal lines indicate the cut-off value to discriminate positive and negative samples for each assay.
Figure 6.
IgG-S response in previously infected vaccinees producing IgM-S.
IgG-S measures (panel a) in previously infected vaccinees who produced IgM-S at T1 or at T2 following BNT162b2 vaccination according to time of examination (T0, T1 and T2) and negative or positive serology at T0 (SerologyNEG, n = 32, green dots; SerologyPOS, n = 29 magenta dots). Predicted means of ln(IgG-S) measures (with the 95% confidence interval) according to time of examination in (i) previously infected subjects who did not elicit IgM-S or had IgM-S at T0 (PI*, red line), (ii) subjects who did not have detectable IgM-S at T0 but produced them at T1 or at T2 following vaccination, and who had detectable IgG-S and/or IgG-N at T0 (SerologyPOS, magenta line), (iii) subjects as the previous ones, but with negative serology at T0 (SerologyNEG, green line), and (iv) naïve vaccinees (blue line) (panel B) were obtained by a two-level linear regression model. For SerologyNEG and SerologyPOS subjects, statistically significant p-values of the difference in the predicted means between consecutive times of examination in the same subject group and between different subject groups at the same time of examination are reported in panel b. For all four group of subjects, statistically significant p-values of the difference in the predicted means between different groups of subjects at the same time of examination are reported in panel b table. The horizontal lines indicate the cut-off value to discriminate positive and negative samples for each assay.
Role of funding source
This work was supported by the Italian Ministry of Health under “Fondi Ricerca Corrente”- L1P5 and “Progetto Ricerca Finalizzata COVID-2020-12371675” to IRCCS Sacro Cuore Don Calabria Hospital, by FUR 2020 Department of Excellence 2018-2022, MIUR, Italy and by The Brain Research Foundation Verona. The funding source had no role in the development of this study.
Results
Development of IgM-S is associated with higher neutralizing activity in naïve vaccinees
We initially tested the neutralizing activity against SARS-CoV-2 of sera from IN (n = 48) and PI (n = 50) vaccinees, in a cohort described in our previous study14 collected at the time of first vaccine dose (T0), at the second dose (T1) and 3 weeks after the second dose (T2). Among IN vaccinees, IgM-S were detected in 35/48 (72.9%) after the two vaccine doses (IgM-SPOS) while the remaining 13/48 (27.1%) had undetectable IgM-S (IgM-SNEG) (Figure 1a and c). IgM-SPOS IN vaccinees had higher neutralizing activity than IgM-SNEG IN vaccinees (blue dots) at T2 (p = 0.008).
Among PI vaccinees, 22/50 (44.0%) had undetectable IgM-S while the remaining 28/50 (56.0%) resulted positive at any of the timepoints. No significant differences in neutralization activity were observed when comparing the two groups of PI vaccines at each timepoint (Figure 1b and d). This first set of data on a limited number of vaccinees confirmed our previous observation of the absence of detectable IgM-S in a significant fraction of IN vaccinees and expanded on the association of IgM-S responses with higher serum neutralizing activity.
IgM-S development following BNT162b2 vaccine
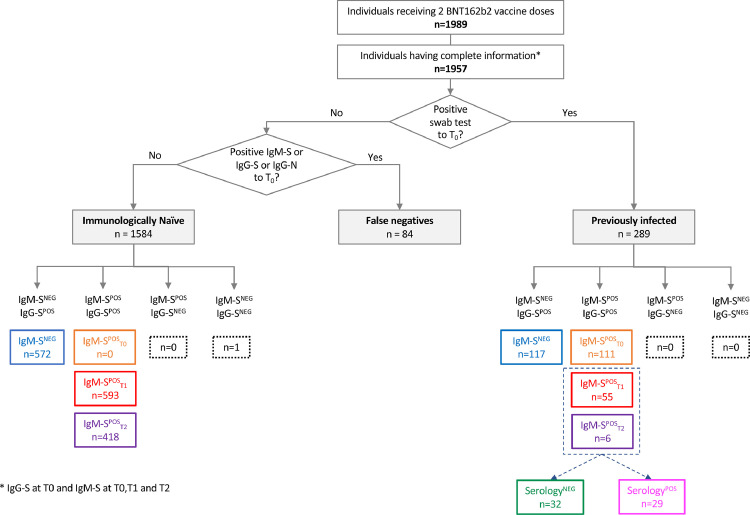
We further tested these initial observations on a larger cohort of 1989 HCW who had been vaccinated with two doses of BNT162b2 vaccine (Figure 2 depicts a flowchart of the patients' groups that were analysed in this study). The study included longitudinal samples collected at the day the first dose of vaccine was administered (T0), at the second dose (3 weeks after the first one, T1) and 3 weeks after the second dose (T2). Among those 1989 subjects, complete information (IgG-S, IgM-S and IgG-N at T0, T1 and T2) was available for 1957 vaccinees. Vaccinees with negative swab and no infection history but positive serology at T0 (n = 84) were considered as false negatives and were not included in this analysis. Of the evaluable 1873 patients, 289 were previously infected (PI), with a history of SARS-CoV-2 infection documented by a positive swab test; 1584 were immunologically naïve (IN) with no documented history of infection, negative swab test and negative serology (IgM-S, IgG-S and IgG-N) at T0. For all these patients we had access to serum samples that were used to quantify IgG-S(RBD) as proxy of neutralization activity.19 We divided the two initial groups (PI and IN) into four sub-groups, according to the time of IgM-S positivity: (a) IgM-S never detected (IgM-SNEG); (b) IgM-S detected before the first vaccine dose (IgM-SPOST0); (c) IgM-S detected after the first vaccine dose (IgM-SPOST1); (d) IgM-S detected after the second vaccine dose (IgM-SPOST2). We further explored whether the development of IgM-S before, after or at the same time of IgG-S could reflect a gain in the load of IgG-S thus providing a putative proxy of protection from future infections in IN or PI.
Figure 2.
Study population.
Classification and distribution of the different types of IgM-S and IgG-S responses in naïve and previously infected subjects who received the BNT162b2 vaccine. NEG: negative; POS: positive.
IgM-S serotyping identifies three patterns of responses in naïve vaccinees
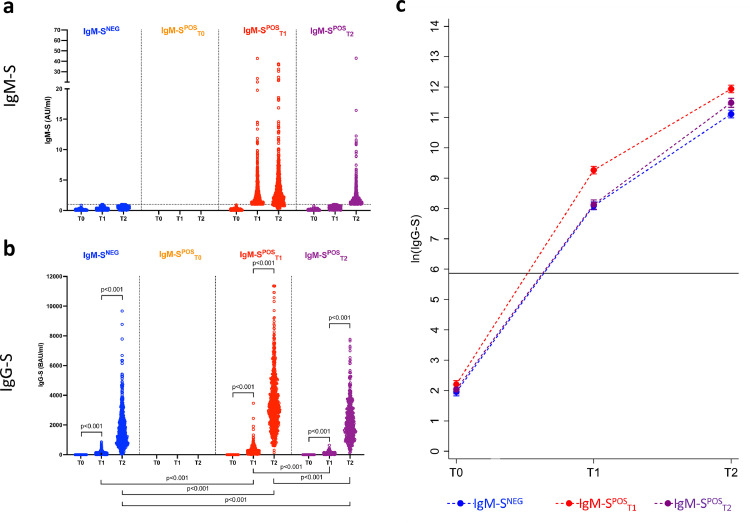
Of the 1584 IN vaccinees, 1011 (63.8%) developed both IgM-S and IgG-S (IgM-SPOS), 572 (36.1%) developed IgG-S but not IgM-S (IgM-SNEG), none had IgM-S but not IgG-S and only one (0.1%) was negative for both isotypes (Figure 2). Among the 1011 IgM-SPOS vaccinees, 593 (58.7%) developed both IgG-S and IgM-S at T1 (IgM-SPOST1), 418 (41.3%) developed IgG-S at T1 and IgM-S at T2 (IgM-SPOST2). Among the 572 IgM-SNEG vaccinees (excluding the single subject who did not elicit IgG-S), 550 (96.2%) developed IgG-S at T1 and the rest (n = 22, 3.8%) at T2 (Figs. 3 and 4a). All vaccinees who were IgM-S positive at T1 were also IgG-S positive at the same time point (Figure 3). Only eight vaccinees with undetectable IgM-S/IgG-S at T1 (Figure 3, row IgM-SPOST2, column T1) became positive for both at T2. Therefore, the patterns of IgM-S/IgG-S responses can be interpreted as follows: (a) IgM-S negative (IgM-SNEG, 572/1584, 36.1%, blue dots in Figs. 3 and 4), (b) IgG-S/IgM-S coordinated (IgM-SPOST1, 593/1584, 37.4%, red dots in Figs. 3 and 4); and (c) IgM-S delayed responses (IgM-SPOST2, 418/1584, 26.4%, purple dots in Figs. 3 and 4). We defined as coordinated (pattern b) the IgG-S and IgM-S responses that appeared in the same time window regardless of whether IgM-S appeared before or at the same time of IgG-S, a pattern that can be considered a canonical primary antibody response. Conversely, patterns (a) and (c) can be considered as non-canonical.
Figure 3.
Development of IgM-S and IgG-S following vaccination.
Scatterplots of IgM-S (y axis) and IgG-S (x axis) measures in naïve vaccines according to time of examination (T0, T1 and T2) and time of IgM-S positivity (IgM-SNEG, blue dots; IgM-SPOST1, red dots; IgM-SPOST2, purple dots).
In Figure 4b and c, the IgM-SPOST1 (red dots) group had statistically significantly higher IgG-S levels than groups IgM-SNEG (blue dots) and IgM-SPOST2 (purple dots) after both the first (p < 0.001) and the second (p < 0.001) vaccines dose. Thus, of the three groups of vaccinees identified in our analysis, the IgM-S/IgG-S coordinated group (IgM-SPOST1) displayed a more efficient response to the vaccine, at least as measured by the levels of IgG-S antibodies elicited by the first and second vaccine dose. Of note, IN vaccinees who displayed the delayed IgM-S pattern (IgM-SPOST2) were older (median 47 years) and had a higher frequency of males (43%) than the other two groups (Table 1). In all subgroups, we also observed a statistically significant lower IgG-S antibody response with increasing age (difference in ln IgG-S for one-year increase of age = -0.015, p < 0.001) and a higher IgG-S response in females (difference in ln IgG-S between females and males = 0.1, p = 0.007).
Table 1.
Comparison of the main characteristics among the four IgM-S subgroups of naïve subjects.
| IgM-SNEG (n = 572) | IgM-SPOST0 (n = 0) | IgM-SPOST1 (n = 593) | IgM-SPOST2 (n = 418) | p-value | |
|---|---|---|---|---|---|
| Age at T0, median | 45 | - | 42 | 47 | <0.001 (Kruskal-Wallis rank test) |
| Female, % (vs male) | 66.4 | - | 65.1 | 56.9 | <0.001 (Fisher's Exact test) |
Finally, 28 (1.8%) and 2 (0.1%) IN vaccinees became positive for IgG-N at T1 and T2, respectively (Supplementary Fig. 1). Because the nucleocapsid protein is not present in the BNT162b2 vaccine, these vaccinees most likely were infected during vaccination. The proportion of IgG-N positive vaccinees was not statistically different in the three IN subgroups (IgM-SNEG, IgM-SPOST1 and IgM-SPOST2; p = 0.200). We performed a sensitivity analysis by excluding IN vaccinees who became IgG-N positive at T1 or T2, and we observed the same results as in the main analysis (p < 0.001).
IgM-S and IgG-S responses in previously infected vaccinees
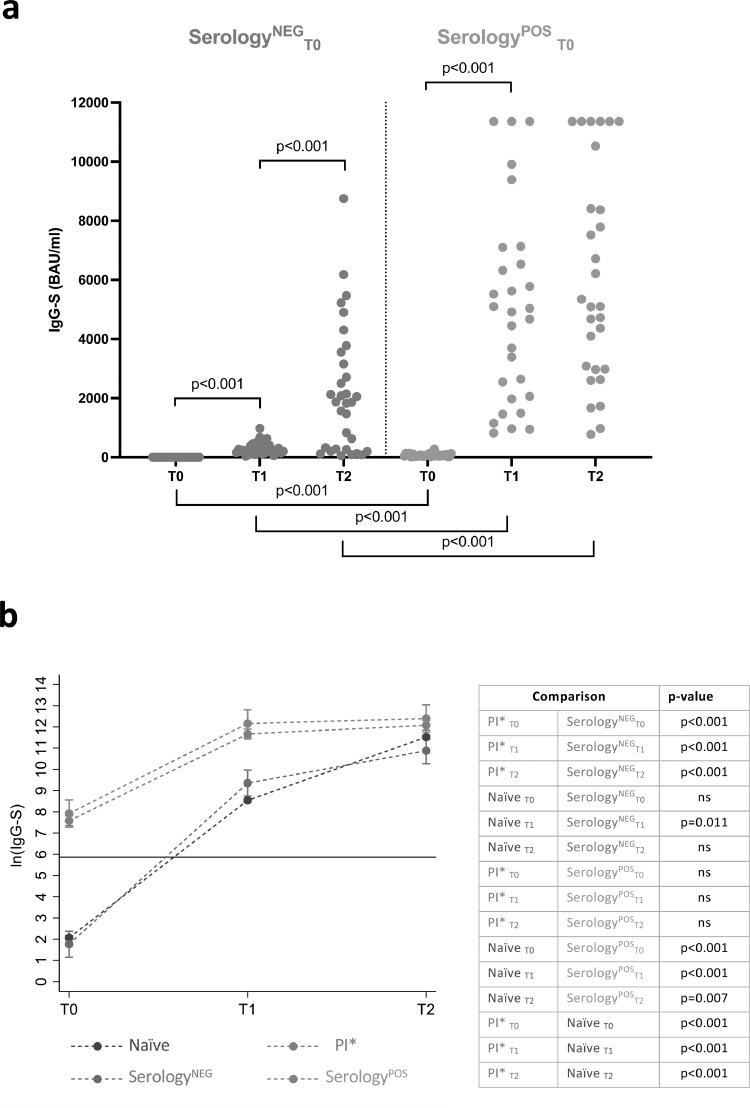
At T0, 117/289 (40.5%) PI vaccinees were IgM-S negative (IgM-SNEG) and 172/289 (59.5%) were IgM positive (IgM-SPOS) (Figure 2). Of these, 111 (64.5%) were positive at T0 (IgM-SPOST0), 55 (32.0%) at T1 (IgM-SPOST1), and 6 (3.5%) at T2 (IgM-SPOST2) (Figure 2). Among IgM-SPOST0 vaccinees, 24/111 (21.6%) and 87/111 (78.4%) had been infected during the first and second wave of the pandemic in Italy, respectively. The IgG-S levels significantly increased after both the first (p < 0.001) and second doses (p = 0.002) in all PI subgroups, except for IgM-SPOST0 individuals for whom the second dose did not significantly improve IgG-S levels as compared to the first vaccine dose (p-value=0.49) (Figure 5b). There were no significant differences of IgG-S levels between PI subgroups after the second vaccine dose (Figure 5b and c). The fact that IgM-SPOST2 vaccinees reached IgG-S levels similar to the other groups only after the second dose of vaccine (Figure 5c) suggests that in these subjects a single dose of vaccine induces suboptimal antibody levels.
Among the PI vaccinees who developed IgM-S after the first and second vaccine dose (55 IgM-SPOST1 and 6 IgM-SPOST2), 29 had undetectable IgM-S but were IgG-S and/or IgG-N positive at T0 and were classified as serology positive (SerologyPOS), whereas 32 were negative at T0 for IgM-S, IgG-S and IgG-N and were therefore classified as serology negative (SerologyNEG) (Figure 2). Comparison of the IgG-S levels elicited by the first and second dose of vaccine in these two groups revealed a faster and stronger IgG-S response in PI vaccinees classified as serology positive (p < 0.001) (Figure 6a). Next, we compared the two groups with the 1584 IN vaccinees and with the subgroup of PI vaccinees (PI*), from which the SerologyNEG and SerologyPOS PI vaccinees were excluded (Figure 2). SerologyNEG vaccinees were different from PI* vaccinees at all time points (p < 0.001) but similar to IN (except at T1, p = 0.011), while SerologyPOS were different from IN (p < 0.001 at T0 and T1, p = 0.007 at T2), and similar to PI* (Figure 6b). Thus, these data revealed the presence among PI vaccinees of subjects (SerologyNEG) who displayed a naïve serological profile and responded to vaccination with a coordinated IgM-S/IgG-S pattern similar to that of a primary response. Of note, these vaccinees were generally younger, had been mostly infected during the second wave and were mostly asymptomatic. In contrast, SerologyPOS PI vaccinees were generally older, had a slightly higher frequency of males, were mostly infected during the first wave, and reported symptomatic COVID-19 (Table 2).
Table 2.
Comparison of the main characteristics among the four IgM-S subgroups of previously infected subjects.
| IgM-SNEG (n = 117) | IgM-SPOST0 (n = 111) | IgM-SPOST1 (n = 55)1 | IgM-SPOST2 (n = 6) | p-value | |
|---|---|---|---|---|---|
| Age, median | 43.0 | 47.0 | 46.0 | 35 | 0.219 (Kruskal-Wallis rank test) |
| Female, % (vs male) | 70.9 | 62.2 | 56.4 | 33.3 | 0.086 (Fisher's exact test) |
| 2nd wave, % (vs 1st wave) | 44.4 | 78.4 | 65.5 | 66.7 | <0.001 (Fisher's exact test) |
| Symptoms, % (vs no symptoms) | 80.3 | 88.3 | 70.4 | 50 | 0.008 (Fisher's exact test) |
The majority of SerologyPOS subjects (18/29) had IgG-N at T0. The remaining where IgG-S positive. SerologyNEG subjects, on the contrary, did not present IgG-N at T0, which instead appeared at T1 in as many as 11/32 (34%) subjects (Supplementary Fig. 2). In IN subjects, however, we observed only 28/1584 subjects (1.8%) positive for IgG-N at T1 (Supplementary Fig. 1a).
Together these data defined three patterns of IgM-S responses in PI vaccinees (Figure 5): (a) negative IgM-S (IgM-SNEG) (b) persistent IgM-S (IgM-SPOST0) and (c) delayed IgM-S (IgM-S detected at T1, IgM-SPOST1 or at T2, IgM-SPOST2). Pattern (a) was consistent with that of a canonical anamnestic response after the natural decay of IgM-S that follows infection. Pattern (b) was observed in 21.6% of PI vaccinees who had been infected almost one year before vaccination and it was someway unexpected since IgM responses are usually short lived. There are however reports on the persistence of long-lived memory IgM B cells in other viral infections including influenza.20, 21, 22, 23 Pattern (c) revealed a proportion of PI vaccinees who may had experienced only a transient infection which was not sufficient to induce a fully matured class-switched response and responded to vaccination with a pattern typical of a primary response.
IgM response in naïve subjects vaccinated with Vaxzevria and Spikevax vaccines
We analysed a limited numbers of available naïve individuals vaccinated with the Vaxzevria (Astra Zeneca) and with the Spikevax (Moderna) vaccines. Among the 37 subjects vaccinated with Vaxzevria, all developed IgG-S following vaccination, but only 6 (16.2%) had detectable IgM-S (Table 3). Similarly, among the 15 subjects vaccinated with Spikevax, all elicited IgG-S and only 2 also had evidence of detectable IgM-S (13.3%), thus confirming, in albeit smaller numbers, a consistently non-canonical IgM response in other types of vaccinations as well.
Table 3.
IgM-S and IgG-S development following the two doses vaccination with BNT162b2, Vaxveria and Spikevax vaccines.
| IgM-SNEG | IgM-SPOS | Total | |
|---|---|---|---|
| BNT162b2 -Pfizer/BionTech | 573 (36.2%) | 1011 (63.8%) | 1584 |
| Vaxzevria-AstraZeneca | 31 (83.8%) | 6 (16.2%) | 37 |
| Spikevax-Moderna | 13 (86.7%) | 2 (13.3%) | 15 |
Discussion
The serological response to vaccination shows a relatively rapid decay as observed in natural infection/immunization.24,25 The extent of this decay is so pronounced that the vaccine efficacy itself has been questioned and a booster dose of BNT162b2 vaccine has been recently authorized by FDA. In this context, it is of paramount importance to gain further information on the patterns of antibody responses that are associated to protective immunity. Most studies have concentrated the attention on IgG responses, while a few have addressed the role of IgM in virus neutralization. One such study26 reported that in adults recovered from mild COVID-19, while IgG were maintained for long periods of time, the neutralization capacity decayed more rapidly and was most strongly associated with anti-S trimer IgM. Prevost et al.27 also reported that the virus neutralization capacity decreases significantly 6 weeks after the onset of symptoms, following a similar trend as anti-RBD IgM and found a stronger correlation with neutralization for IgM than IgG and IgA, suggesting that at least part of the neutralizing activity is mediated by IgM. There are limited data on the kinetic of appearance of IgM after vaccination and its association with virus neutralizing activity.16
Here we report that following BNT162b2 vaccination, higher neutralization activity correlates with the presence of both IgG-S and IgM-S in IN vaccinees, suggesting that IgM-S may contribute to protective immunity. On the other hand, we found that 36.1% of IN vaccinees responded to vaccination with IgG-S but not IgM-S. In addition, in vaccinees who responded with both isotypes, 41.3% developed IgM-S after IgG-S. Of note, of the three isotype patterns that we identified, only that with coordinated IgM-S/IgG-S responses, could be considered as a bona fide primary immune response pattern but it was represented in only 37.4% vaccine recipients while the others were either IgM-S negative (36.1%) or developed IgM-S after IgG-S (26.4%). More importantly, vaccinees exhibiting IgG-S without IgM-S or IgM-S after IgG-S had significantly lower IgG-S levels compared to those with coordinated IgM-S/IgG-S responses; this suggests that coordinated IgM-S/IgG-S responses are associated with increased immunity. Also, in the small group of HCW who received the adenovirus-based vaccine Vaxzevria (Astra Zeneca) or the RNA vaccine Spikevax (Moderna), as many as 80% did not develop IgM-S after vaccination. Thus, the unconventional isotype pattern follows SARS-CoV-2 spike vaccination regardless of the type of vaccine used.
Taken together our data suggest that vaccination elicits either a canonical primary response with coordinated IgM-S/IgG-S, associated with higher levels of IgG-S antibodies, or a non-canonical IgM-S negative response. We propose that these non-canonical responses may leverage on pre-existing immunity to cross-reactive human coronaviruses, or even both types of responses where the first to appear is the anamnestic cross-reactive response28,29 followed by the later appearance of IgM after recruitment of naïve B cells specific to SARS-CoV-2 epitopes.
There is accumulating evidence that the immune response to SARS-CoV-2 is influenced by cross-coronavirus immunity, with some data pointing to the risk of immunopathogenic responses due to low affinity cross-reactive antibodies generated by an original antigenic sin30 and other data pointing to a potential protective role of cross-reactive antibodies. Chaudhury et al. recently reported12 that the IgM response is highly specific for SARS-CoV-2, while the IgG response is more cross-reactive. The same authors hypothesize that the IgM response is naïve-derived, while the IgG response is memory-derived, thus explaining the simultaneous appearance of IgM and IgG. Furthermore, Kaplonek et al. recently reported13 the near simultaneous evolution of IgG and IgM specific for the S2 subunit of SARS-CoV-2 spike at early time points in a cohort of COVID-19 survivors and proposed that it could be a reflection of expansion of pre-existing cross-coronavirus immunity to the conserved S2-domain. Furthermore, there is evidence that SARS-CoV-2 infection reactivates hCoVs-specific memory B cells29,31 concomitantly with the recruitment of SARS-CoV-2 naïve B cells and the appearance of virus-neutralizing antibodies specific for the RBD of the SARS-CoV-2 Spike protein. Our observation of IgM-S/IgG-S isotype patterns consistent with those of an anamnestic response following vaccination of naïve individuals is highly suggestive of the recruitment by the vaccine of cross-coronavirus immunity. Whether this would reflect in higher or lower vaccine efficacy remains speculative. However, the established safety of current SARS-CoV-2 vaccines with few signals of immunopathogenic events suggest that cross-coronavirus immunity may play, if any, a protective rather than a pathogenic role following vaccination.
The IgM-S response to vaccination of PI vaccinees also displayed some interesting features. Of the subgroup of PI vaccinees who were IgM-S positive at baseline, 21.6% had been infected during the first pandemic wave in Italy, almost one year before vaccination. While the persistence of IgM-S in these subjects was unexpected, there are reports that IgM antibodies may persist for long period of times after natural infection owing to the persistence of long-lived memory IgM positive B cells.21,22 Our data suggest that at least a subset of PI vaccinees developed these types of long-lived IgM responses. Of note, the presence of IgM-S before vaccination was associated to the most rapid kinetic of IgG-S responses when compared to those of vaccinees who were either IgM-S negative or had a delayed IgM-S response.
Unexpectedly, we observed a group of PI vaccinees who elicited IgM-S following vaccination. Among them, a subgroup classified as serology negative at baseline (SerologyNEG) showed an IgG-S response similar to that of IN vaccinees. These subjects may therefore have had a false-positive swab result. Of these, a consistent fraction (34%) displayed IgG-N after vaccination, suggestive of an infection event. However, in our cohort, only 1.8% of truly IN subjects showed evidence of infection (IgG-N positivity), providing a crude estimate of the occurrence of infection during the vaccination schedule. We speculate that the serology negative vaccinees who became IgG-N positive after vaccination may have experienced a recall response to cross reactive N epitopes similar to that reported in a study by Dobaño et al.,32 which suggests that anti-N antibodies may be produced following spike-based vaccines resulting from a cross-reactive response.
This observation deserves further investigation because it suggests that not all individuals with a previous history of SARS-CoV-2 infection develop an immunological memory sufficient to ensure a rapid class-switched response to a single dose of vaccine.
While the correlates of protection from SARS-CoV-2 infection have not yet been fully established, it is generally accepted that antibody-mediated neutralization of the virus is a key determinant.33 Assessing the presence of IgM before and after vaccination may therefore provide useful information on vaccine efficacy and, to some extent, guide decisions on the vaccine regimens in previously infected persons or in IgM non responder individuals. The combined examination of all three branches of adaptive immunity at the level of SARS-CoV-2-specific CD4+ and CD8+ T cell, as well as neutralizing antibody responses in COVID-19 patients, provided evidence that coordinated CD4+ T cell, CD8+ T cell, and antibody responses are protective, but uncoordinated responses may fail to control disease.34 Thus, while antibodies still represent the strongest correlate of immunity, it is plausible that coordinated T and B cell responses are needed to confer protection. In this context, studies assessing the expression of the different antibody isotypes may provide useful insights for the understanding of protective immunity in both natural infection and vaccination.
This study presents some limitations. Due to limited amount of serum samples collected, we could not determine which specific antibody subclasses correlates with neutralization. For the same reason, we did not address the fine specificity of IgG and IgM antibodies. Therefore, we cannot conclude on a potential priming effect of previous exposures to common human coronaviruses on the response to vaccination. Furthermore, we did not have access to cellular samples, and we could not determine the effect of the pre-existing cellular immunity on the development of the humoral response, following vaccination. The sensitivity of the assays detecting IgM-S and IgG-S could also be argued to be an issue, even though the assays that we used are fully validated and routinely used for clinical screening.35,36 It must be noted that the assays we used for IgG and IgM quantification is designed to measure Spike S1-specific immunoglobulins, and does not allow the detection of IgG and IgM against other epitopes. Finally, this study focuses on the humoral response within the first weeks following vaccination and a longer follow up is needed to confirm the current observations.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
Contributors
A.R., D.Z., A.B., L.L. conceived the paper design, analysed and discussed data and wrote the manuscript; C.P. and Z.B. designed the study, enrolled patients, collected and managed clinical data; L.C. and Sim. A. performed statistical analysis; M.T.V., L.D.C., G.S., Ni. Te., and AMSLV collected samples and clinical data for Vaxveria and Spikevax vaccines; Sil. A. and M.P. performed pseudovirus neutralization assays; Na. Ti., S.S.L. and T.F. participated in data collection and analysis. All authors read, critically revised, and approved the manuscript. D.Z., A.R., L.C., Sim. A. and C.P. have verified the underlying data.
Acknowledgments
We acknowledge the generous contribution of our health-care worker's colleagues, whose sera samples were essential to this study. We thank Dr. Stefania Tremolada for assistance in bleeding subjects vaccinated with Vaxzevria and Spikevax.
Data sharing statement
Raw data are available upon request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103888.
Appendix. Supplementary materials
References
- 1.Krammer F., Srivastava K., Alshammary H., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384(14):1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li K., Huang B., Wu M., et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun. 2020;11(1):6044. doi: 10.1038/s41467-020-19943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong S., Ruprecht R.M. Immunoglobulin M: an ancient antiviral weapon – rediscovered. Front Immunol. 2020;11(1943) doi: 10.3389/fimmu.2020.01943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y., Wang M., Zuo Z., et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.X., Liu B.Z., Deng H.J., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Wang J., Xu X. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Jighefee H.T., Yassine H.M., Nasrallah G.K. Evaluation of antibody response in symptomatic and asymptomatic COVID-19 patients and diagnostic assessment of new IgM/IgG ELISA kits. Pathogens. 2021;10(2) doi: 10.3390/pathogens10020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X., Sun J., Nie S., et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020;26(8):1193–1195. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
- 9.Pan Y., Li X., Yang G., et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81(1):e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu J., Wu C., Li X., et al. Profile of Immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71(16):2255–2258. doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z., Zhu F., Guo F., Yang B., Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J Med Virol. 2020;92(10):1735–1738. doi: 10.1002/jmv.25820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhury S., Hutter J., Bolton J.S., et al. Serological profiles of pan-coronavirus-specific responses in COVID-19 patients using a multiplexed electro-chemiluminescence-based testing platform. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0252628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplonek P., Wang C., Bartsch Y., et al. Early cross-coronavirus reactive signatures of humoral immunity against COVID-19. Sci Immunol. 2021;6(64):eabj2901. doi: 10.1126/sciimmunol.abj2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalle Carbonare L., Valenti M.T., Bisoffi Z., et al. Serology study after BTN162b2 vaccination in participants previously infected with SARS-CoV-2 in two different waves versus naïve. Commun Med. 2021;1(1):38. doi: 10.1038/s43856-021-00039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gobbi F., Buonfrate D., Moro L., et al. Antibody response to the BNT162b2 mRNA COVID-19 vaccine in subjects with prior SARS-CoV-2 infection. Viruses. 2021;13(3):422. doi: 10.3390/v13030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naaber P., Tserel L., Kangro K., et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg Health Eur. 2021;10 doi: 10.1016/j.lanepe.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonfrate D., Piubelli C., Gobbi F., et al. Antibody response induced by the BNT162b2 mRNA COVID-19 vaccine in a cohort of health-care workers, with or without prior SARS-CoV-2 infection: a prospective study. Clin Microbiol Infect. 2021;27(12):1845–1850. doi: 10.1016/j.cmi.2021.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizzato M., McCauley S.M., Neagu M.R., et al. Lv4 is a capsid-specific antiviral activity in human blood cells that restricts viruses of the SIVMAC/SIVSM/HIV-2 lineage prior to integration. PLoS Pathog. 2015;11(7) doi: 10.1371/journal.ppat.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salazar E., Kuchipudi S.V., Christensen P.A., et al. Convalescent plasma anti-SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J Clin Invest. 2020;130(12):6728–6738. doi: 10.1172/JCI141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Throsby M., van den Brink E., Jongeneelen M., et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohannon C., Powers R., Satyabhama L., et al. Long-lived antigen-induced IgM plasma cells demonstrate somatic mutations and contribute to long-term protection. Nat Commun. 2016;7(1):11826. doi: 10.1038/ncomms11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skountzou I., Satyabhama L., Stavropoulou A., et al. Influenza virus-specific neutralizing IgM antibodies persist for a lifetime. Clin Vaccine Immunol. 2014;21(11):1481–1489. doi: 10.1128/CVI.00374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auladell M., Jia X., Hensen L., et al. Recalling the future: immunological memory toward unpredictable influenza viruses. Front Immunol. 2019;10(1400):1400. doi: 10.3389/fimmu.2019.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicenti I., Basso M., Gatti F., et al. Faster decay of neutralizing antibodies in never infected than previously infected healthcare workers three months after the second BNT162b2 mRNA COVID-19 vaccine dose. Int J Infect Dis. 2021;112:40–44. doi: 10.1016/j.ijid.2021.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibarrondo F.J., Hofmann C., Fulcher J.A., et al. Primary, recall, and decay kinetics of SARS-CoV-2 vaccine antibody responses. ACS Nano. 2021 doi: 10.1021/acsnano.1c03972. [DOI] [PubMed] [Google Scholar]
- 26.Harrington W.E., Trakhimets O., Andrade D.V., et al. Rapid decline of neutralizing antibodies is associated with decay of IgM in adults recovered from mild COVID-19. Cell Rep Med. 2021;2(4) doi: 10.1016/j.xcrm.2021.100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevost J., Gasser R., Beaudoin-Bussieres G., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep Med. 2020;1(7) doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beretta A., Cranage M., Zipeto D. Is cross-reactive immunity triggering COVID-19 immunopathogenesis? Front Immunol. 2020;11(2695) doi: 10.3389/fimmu.2020.567710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen-Contant P., Embong A.K., Kanagaiah P., et al. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11(5) doi: 10.1128/mBio.01991-20. 2020.07.20.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatti A., Monsalve D.M., Pacheco Y., Chang C., Anaya J.M., Gershwin M.E. Original antigenic sin: a comprehensive review. J Autoimmun. 2017;83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Song G., He W.T., Callaghan S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Commun. 2021;12(1):2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobano C., Jimenez A., Rubio R., et al. Spike-based COVID-19 immunization increases antibodies to nucleocapsid antigen. Transl Res. 2022;240:26–32. doi: 10.1016/j.trsl.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 34.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins V., Fabros A., Wang X.Y., Bhandari M., Daghfal D.J., Kulasingam V. Anti-SARS-CoV-2 IgM improves clinical sensitivity early in disease course. Clin Biochem. 2021;90:1–7. doi: 10.1016/j.clinbiochem.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maine G.N., Lao K.M., Krishnan S.M., et al. Longitudinal characterization of the IgM and IgG humoral response in symptomatic COVID-19 patients using the abbott architect. J Clin Virol. 2020;133 doi: 10.1016/j.jcv.2020.104663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.