Abstract
Among various diarrheagenic Escherichia coli strains from clinical sources, we found that the urease gene was specifically associated with enterohemorrhagic E. coli (EHEC) strains irrespective of their serogroups. The results suggest that the urease gene can be a useful genetic marker for the detection of EHEC strains and for the diagnosis of infections caused by EHEC strains in the clinical situation.
In 1996, a large outbreak of food-borne infection due to enterohemorrhagic Escherichia coli (EHEC) O157:H7 occurred in Sakai City, Osaka Prefecture, Japan. The outbreak involved more than 6,000 people and resulted in three deaths (11). Recently, Hayashi et al. (7) reported the complete genome sequence of an EHEC O157:H7 strain isolated from the Sakai outbreak (referred to as strain O157 Sakai). The study demonstrated that a 1,460-kb DNA sequence is specifically present in O157 Sakai but is not found in the E. coli K-12 genome (2). Most of the sequence consisted of prophage genomes or regions with prophage-like features (designated Sakai prophage-like elements [SpLEs]) (7). Among the SpLEs, the largest one, SpLE1 (ca. 86 kb long), possesses several genes potentially related to the virulence of the organism. Here we describe the results of our investigation on the distribution of the genes identified on SpLE1 among various diarrheagenic E. coli strains.
Strain O157 Sakai (strain RIMD0509952) (7) was used as the standard strain in the present study. The strains examined in the present study consisted 55 diarrheagenic E. coli strains including 22 strains of EHEC, 12 strains of enterotoxigenic E. coli (ETEC), 8 strains of enteropathogenic E. coli (EPEC), 3 strains of enteroaggregative E. coli (EAggEC), and 10 strains of enteroinvasive E. coli (EIEC) and 4 other enteropathogenic bacteria including 2 strains of Shigella spp. (Shigella flexneri and S. dysenteriae) and 2 strains of Salmonella spp. (Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium). All strains were from our laboratory collection, and all strains except one EHEC strain were clinical isolates, the one exception was derived from a calf. E. coli JM109 (13) was also included in the examination as a negative control strain.
All DNA probes used in the study were prepared by PCR amplification. The sequences of the oligonucleotides used as PCR primers are described in Table 1. PCR was performed in a reaction mixture with a total volume of 50 μl. The reaction mixture contained the following components: 0.5 μg of genomic DNA which was extracted from O157 Sakai by a standard method (12), 5 μl of 10× PCR buffer, 4 μl of a deoxynucleoside triphosphate mixture (containing dATP, dCTP, dTTP, and dGTP at concentrations of 10 mM each), 20 pmol of each primer, and 2.5 U of Taq DNA polymerase (Wako Pure Chemical Industry Ltd., Osaka, Japan), with the volume completed to 50 μl with distilled water. PCR conditions were as follows: after 3 min of denaturation at 94°C, a cycle of denaturation at 94°C for 1 min, annealing at the optimum temperature for 1 min (Table 1), and extension at 72°C for 1 min was repeated 30 times. The PCR product was separated on a 2% agarose gel and was extracted from the agarose gel with an QIAEXII gel extraction kit (QIAGEN, Hilden, Germany). The DNA probes were labeled with the PCR DIG Probe Synthesis kit, and the hybridized DNAs were detected with alkaline phosphatase-labeled anti-digoxigenin monoclonal antibody (Roche, Indianapolis, Ind.). Hybridization was carried out at 42°C under high-stringency conditions (50% concentration of formamide in hybridization solution) and with washing at 55°C.
TABLE 1.
Primers used in the present study
| Primer designation | Targeta | Similarityb | Probe no.c | Sequence | Product size (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|---|
| 1299-1 | ECs1299 | Integrase | 1 | 5′-TAAGGCAGTGGTTATCGACG-3′ (20-mer) | 801 | 63 |
| 1299-2 | 5′-ATGCCTGCATCATCGGTACA-3′ (20-mer) | |||||
| 1312-1 | ECs1312 | TraT | 2 | 5′-GGAATTCTTGAGATTTTCTACAATCG-3′ (26-mer) | 759 | 50 |
| 1312-2 | 5′-GTCGACTCAGAGTATATTGGCGATT-3′ (25-mer) | |||||
| 1324-1 | ECs1324 | UreC | 3 | 5′-TCTAACGCCACAACCTGTAC-3′ (20-mer) | 397 | 60 |
| 1324-2 | 5′-GAGGAAGGCAGAATATTGGG-3′ (20-mer) | |||||
| 1360-1 | ECs1360 | Iha | 4 | 5′-ATGATAACCGGGATGGGCAA-3′ (20-mer) | 964 | 58 |
| 1360-2 | 5′-ATGATGCCACCTCTTCGGTG-3′ (20-mer) | |||||
| 1391-1 | ECs1391 | BfpM | 5 | 5′-GAACAGGGAAATTCAGCAGC-3′ (20-mer) | 475 | 60 |
| 1391-2 | 5′-ATCGACGATTGCTGGAAAGG-3′ (20-mer) | |||||
| 1396-1 | ECs1396 | Fluffing protein (AIDA-1) | 6 | 5′-ACTGGTTACCAGTACTGCTG-3′ (20-mer) | 883 | 60 |
| 1396-2 | 5′-ACCAGTCTTCATCGCTGTCA-3′ (20-mer) | |||||
| 1409-1 | ECs1409 | L0010 | 7 | 5′-ATATCACAATCTCCCGTCCG-3′ (20-mer) | 790 | 63 |
| 1409-2 | 5′-AGTCTGTCAACCAGTTCTGG-3′ (20-mer) |
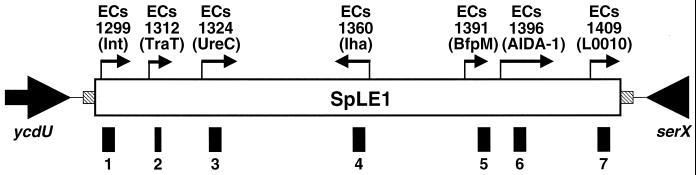
SpLE1 encodes 111 open reading frames (ORFs) (>150 bp) ; several ORFs likely encode proteins potentially related to bacterial pathogenesis such as TraT, Iha, AIDA-1, and urease (1, 6, 8–10). We selected seven ORFs found on SpLE1 and examined their distributions among various types of diarrheagenic E. coli strains, Shigella spp., and Salmonella spp. by colony hybridization (Table 2 and Fig. 1).
TABLE 2.
Distributions of ORFs located on SpLE1 in diarrheagenic E. coli strains and enteropathogens
| Type | Serogroup | No. of strains | No. (%) of strains positive with the following ORF probe:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| EHEC | O157 | 15 | 15 (100) | 15 (100) | 15 (100) | 15 (100) | 15 (100) | 13 (86.7) | 15 (100) |
| EHEC | O26 | 4 | 4 (100) | 4 (100) | 4 (100) | 3 (75.0) | 4 (100) | 3 (75.0) | 4 (100) |
| EHEC | O111 | 3 | 3 (100) | 2 (66.7) | 3 (100) | 2 (66.7) | 3 (100) | 0 | 3 (100) |
| ETEC | 12 | 0 | 0 | 0 | 0 | 0 | 2 (16.7) | 1 (8.3) | |
| EPEC | 8 | 0 | 1 (12.5) | 0 | 2 (25.0) | 7 (87.5) | 6 (75.0) | 0 | |
| EIEC | 10 | 0 | 8 (80.0) | 0 | 7 (70.0) | 9 (90.0) | 1 (10.0) | 4 (40.0) | |
| EAggEC | 3 | 1 (33.3) | 1 (33.3) | 0 | 3 (100) | 0 | 1 (33.3) | 3 (100) | |
| Shigellaa | 2 | 0 | 2 (100) | 0 | 0 | 2 (100) | 1 (50.0) | 1 (50.0) | |
| Salmonellab | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| E. coli JM109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
S. flexneri and S. dysenteriae.
S. enterica serovar Enteritidis and S. enterica serovar Typhimurium.
FIG. 1.
Locations of the DNA probes on SpLE1. Arrows and shaded boxes indicate the direction of transcription and the direct repeated sequences, respectively. The black boxes below SpLE1 represent the locations of the DNA probes.
A significant portion of the EHEC strains reacted with all seven DNA probes (13 of 15 serogroup O157 strains and 3 of 4 serogroup O26 strains), suggesting that SpLE1 or SpLE1-like elements may be widely distributed in EHEC strains, in particular, in serotype O157. In contrast, other types of diarrheagenic E. coli and Shigella spp. reacted with only some of the DNA probes, if any (Table 2). Among the four probes which reacted with all EHEC strains (probes 1, 3, 5, and 7), probe 3 reacted solely with EHEC strains and not with other E. coli strains, while the other probes reacted with some other types of pathogenic E. coli strains and Shigella spp. The sequence of probe 3 corresponds to the sequence of an internal part of the ureC gene, and thus, this finding suggests that the urease operon may be specifically and ubiquitously distributed in EHEC strains, at least in serogroups O157, O26, and O111.
Since it was demonstrated that all EHEC strains tested possess the ureC gene, irrespective of their serogroups, we examined the urease activities of these EHEC strains including O157 Sakai. Among 23 EHEC strains tested, only 1 strain (a serogroup O157 strain) showed urease activity when urea agar base (Becton Dickinson, Sparks, Md.) was used. These results suggest that, at least under the conditions used in the present study, urease production could not be detected in most of the EHEC strains tested, despite their possession of the ureC gene.
It is of particular importance that probe 3, whose sequence corresponds to the sequence of an internal part of ureC, reacted with all EHEC strains tested but none of the other types of diarrheagenic E. coli strains. This suggests that the urease operon is uniquely present in EHEC strains, irrespective of the serogroup. Although the production of urease was not detected in most EHEC strains examined in the present study, the urease gene will be a useful marker for differentiation of EHEC strains from other diarrheagenic E. coli strains.
E. coli infections are not limited to gastroenteritis. Therefore, it is noteworthy that uropathogenic E. coli (UPEC) was reported to contain the urease operon (3, 4). Although the full sequence of the UPEC ureC gene is not available, phylogenetic analysis of UreA, UreB, and UreG shows that the urease genes of EHEC strains exhibit the highest degree of similarity to those of Klebsiella aerogenes, while the urease genes of UPEC are most similar to those of Proteus mirabilis (data not shown). Furthermore, the gene organizations of the urease operons were different from each other in EHEC and UPEC strains (5). These data suggest that an appropriate primer set can differentiate the EHEC ureC gene from the UPEC ureC gene. We are now examining this possibility by analyzing a larger number of clinical E. coli isolates from patients with both gastrointestinal and urinary tract infections.
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science “Research for the Future Programs” (grants 97L00101 and 97L00704) and a grant for International Health Cooperation Research from the Ministry of Health, Labor and Welfare.
REFERENCES
- 1.Benz I, Schmidt M A. AIDA-1, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shoa Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Collins C M, Falkow S. Genetic analysis of an Escherichia coli urease locus: evidence of DNA rearrangement. J Bacteriol. 1988;170:1041–1045. doi: 10.1128/jb.170.3.1041-1045.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins C M, Falkow S. Genetic analysis of Escherichia coli urease genes: evidence for two distinct loci. J Bacteriol. 1990;172:7138–7144. doi: 10.1128/jb.172.12.7138-7144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Orazio S F, Collins C M. The plasimd-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J Bacteriol. 1993;175:3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finlay B B, Paranchych W. Nucleotide sequence of the surface exclusion genes traS and traT from IncF0 lac plasmid pED208. J Bacteriol. 1986;166:713–721. doi: 10.1128/jb.166.3.713-721.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Lee M H, Mulrooney S B, Renner M J, Markowitz Y, Hausinger R P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. Protein Sci. 1992;2:1042–1052. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulrooney S B, Hausinger R P. Sequence of Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarr P I, Bilge S S, Vary J C, Jr, Jelacic S, Habeeb R L, Ward T R, Baylor M R, Besser T E. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H, Wada A, Inagaki Y, Itoh K, Tamura K. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
- 12.Wilson K. Preparation of genomic DNA from bacteria. 1994. p. 2.4.1. .–2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1, 8th ed. Current Protocols, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 13.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]