Abstract
The COVID-19 (coronavirus disease 2019) pandemic affected more than 186 million people with over 4 million deaths worldwide by June 2021. The magnitude of which has strained global healthcare systems. Chest Computed Tomography (CT) scans have a potential role in the diagnosis and prognostication of COVID-19. Designing a diagnostic system, which is cost-efficient and convenient to operate on resource-constrained devices like mobile phones would enhance the clinical usage of chest CT scans and provide swift, mobile, and accessible diagnostic capabilities. This work proposes developing a novel Android application that detects COVID-19 infection from chest CT scans using a highly efficient and accurate deep learning algorithm. It further creates an attention heatmap, augmented on the segmented lung parenchyma region in the chest CT scans which shows the regions of infection in the lungs through an algorithm developed as a part of this work, and verified through radiologists. We propose a novel selection approach combined with multi-threading for a faster generation of heatmaps on a Mobile Device, which reduces the processing time by about 93%. The neural network trained to detect COVID-19 in this work is tested with a F1 score and accuracy, both of 99.58% and sensitivity of 99.69%, which is better than most of the results in the domain of COVID diagnosis from CT scans. This work will be beneficial in high-volume practices and help doctors triage patients for the early diagnosis of COVID-19 quickly and efficiently.
Keywords: COVID-19, Artificial intelligence, Android application, Computed tomography, Lung, Deep learning
Graphical abstract
1. Introduction
After the outbreak in China in December 2019, the World Health Organization (WHO) identified Severe Acute Respiratory Syndrome CoronaVirus-2 (SARS-CoV-2) as a new type of coronavirus. COVID-19 is a disease caused by SARS-CoV-2, which primarily affects the respiratory system. The coronavirus 2019 breakout was declared a public health emergency at the international level by the World Health Organization on 30 January 2020. It was given pandemic status on 11 March 2020 [1]. Economies were ruined by the pandemic and it caused unrivaled challenges to healthcare and food systems across the globe. The pandemic has overwhelmed health care systems. As a result, diagnosing and treating other diseases have been postponed. After multiple waves of COVID-19, health care workers and society, in general, have become exhausted. The real-time reverse transcription-polymerase chain reaction (RT-PCR) test is the standard and used for detecting the presence of COVID-19 in an individual [2]. Due to the high false-negative rates, long turnaround times, and shortage of RT-PCR kits, chest CT scans were found to be an effective and fast alternative to diagnosing COVID-19 [3]. CT scans combine a series of X-ray images taken from different angles around the chest which are then post-processed by computer to create detailed cross-sectional images Chest CT scanning is valuable in the diagnosis of COVID-19 disease. In some cases, the RT-PCR gave negative results and is highly operator dependent, but CT scans confirmed the diagnosis of COVID-19 [4]. Overall due to the high specificity and fast diagnosis, chest CT findings can be a better option than RT-PCR [5]. On chest CT, COVID-19–associated pneumonia usually has a pattern of ground-glass opacification in a peripheral and lower lobe distribution in the lungs [[6], [7], [8]]. This common imaging pattern on CT was used as an aid to observe the COVID-19 in lungs through deep learning [9,10]. In this work, the neural network was trained to detect this finding with very high accuracy and specificity, and fewer parameters, as the model is to be ported on a mobile device. Most of the devices consisting of the Android Operating System (OS) consist of mobile phones and tablets. Given the dire situation caused by COVID-19, a portable, swift, and completely automated aid for diagnosing the disease on CT scans would be beneficial. The novel CovCT application proposed in this work can provide this service on a portable android device. Unlike time-consuming testing methods such as RT-PCR, the CovCT app would provide an optimal method of triaging patients with COVID-19. The CovCT android application is capable of detecting COVID-19 from chest CT scans and generating heatmaps to further illustrate COVID-19 affected regions in the lung parenchyma. The application is very lightweight due to fewer parameters in the neural network along with the best accuracy in the domain. Our results of the infection heatmap are also verified by expert cardiothoracic radiologists. A novel, swift and low-cost lung parenchyma segmentation algorithm is a part of the CovCT application. A novel approach for the faster generation of heatmaps on android devices is also developed in this work.
2. Related work
Artificial intelligence (AI) is one of the most popular approaches for automated disease detection. Many articles have been published in which AI is used to diagnose Covid-19 infection using chest computed tomography (CT) images. Some of the most important and recent advancements have been stated in Table 1 . To discover COVID-19 related characteristics from chest CT, some of them employed basic techniques such as hierarchical and spatial models [11]. Various deep learning approaches were employed in the direction of better accuracy for COVID-19 related findings from CT images [7,[12], [13], [14], [15], [16]]. Our group developed an AI-driven algorithm for detecting ground-glass opacities (GGOs) in COVID-19 patients' lung images [7]. In their analysis, they employed “MosMedData.” The authors segmented lungs and GGOs using point cloud and PointNet++ architectures. They are the first to deploy point cloud and PointNet++ architectures for medical image analysis with 98% evaluation accuracy. Gianchandani et al. proposed an ensemble method for COVID-19 diagnosis from chest X-rays through an ensemble of deep transfer learning models for better performance [17]. Hasan et al. proposed the Coronavirus Recognition Network (CVR-Net) in their work, which uses radiography images to detect COVID-19. The results from this showed an average accuracy of 78% [18].
Table 1.
The Study of various existing machine learning techniques for COVID-19 detection from chest CT scans and their corresponding results. All of these studies are aimed at COVID and Non-COVID classification of CT images.
| S. No. | Method | Results | Reference |
|---|---|---|---|
| 1 | Transfer learning on Inception Recurrent Residual Neural Network | Accuracy – 98.78% | [19] |
| 2 | Construction of AI model using Transfer learning on ShuffleNet V2 | The area under the curve (AUC), sensitivity of model and specificity of model were 0.968 9, 90.52% and 91.58% respectively. | [20] |
| 3 | Feature extraction done by DenseNet121 and bagging classifier trained on top of these | Accuracy – 99 ± 0.9% | [21] |
| 4 | Lesion-attention deep neural networks, using pretrained network weights including VGG16, ResNet18, and ResNet50 | with 0.94 of the AUC score | [22] |
| 5 | Comprehensive System using ResNet 50 | sensitivity, specificity, and the AUC score were 94%, 98%, and 0.994 0 respectively | [23] |
| 6 | Network based on regression of multi view point and 3-Dimensional U-Net | accuracy and sensitivity of 94% and 100% | [24] |
| 7 | Transfer learning on ResNet18 | AUC score – 0.996 5 | [25] |
Accuracies in models trained with transfer learning were also observed to be very good. Brunese et al. reported an average accuracy of 97% in their work. They used a pre-trained VGG-16 model and performed transfer learning on the model for automatic detection of COVID-19 using chest X-Ray images [26]. Jaiswal et al. performed transfer learning of the DenseNet201 Model for the classification of the COVID-19 infected patients. It extracted features by using its learned weights on the ImageNet dataset [27]. T. Anwar et al. used EfficientNet B4 to distinguish between COVID and normal CT-scan images with a 0.90 F1 score [28].
Many approaches have customized deep learning architectures for better detection. Ozturk et al. presented DarkCovidNet, which automatically detected COVID-19 using chest X-ray images. The classification accuracies obtained from this model were 98.08% for binary cases [29]. In another work, Mukherjee et al. developed a custom architecture for CNN which had nine layers for detecting COVID-19 cases. For the training of the model, they used X-Rays, and CT scans. The network achieved an overall accuracy of 96.28%, which was better than most of the CNN-based models [30]. In the Al-Karawi et al., Gabor filters extracted different texture features from CT images. Then these features were utilized for training support vector machines, which were further employed for classifying the COVID-19 cases. This approach got an average accuracy of 95.37% and a sensitivity of 95.99% [31]. N. Palaru et al. proposed Anam-Net, which is a CNN architecture based on depth embeddings. It detected an irregularity in COVID-19 chest CT images. The Anam-Net architecture was lightweight and can be used for inference generation in mobile or resource constraint platforms [32]. H. Alshazly et al. studied various deep network architectures such as SqueezeNet, Inception, ResNet, Xception, ShuffleNet, and DenseNet and proposed a transfer learning strategy to achieve the best performance. As a result, ResNet101 achieved an average accuracy of 99.4%, which is better than others, and had an average sensitivity of 99.1% [33]. N. Basantwani et al. used transfer learning on an Inception-V3 model and ported it to an android application with an accuracy of 94% [34].
A study by Hammad et al. suggests advancement in the classification accuracy of the neural network by introducing a feature extraction stage followed by a genetic algorithm which results in an increment of 0.95% of classification accuracy, compared to state of art models and approaches [35]. Another study gave an approach for myocardial infection detection using proposed CNN with a special focal loss function; results indicate that their approach has increased accuracy by 9% in detecting myocardial signals [36]. These both studies have proposed a method for improving the accuracy of detecting medical conditions from signals, while their focused intermediary steps can be tested for improving the accuracy of the deep learning model in COVID-19 detection from CT images.
Most of the models were observed to either have less accuracy or a very large number of parameters for the model to be ported to a mobile device. Some models were customized for mobile devices, but our approach is entirely different from them and outperforms them for a perfect blend of accuracy, parameters, and specificity. Fig. 1 shows an overall workflow of the proposed deep learning approach.
Fig. 1.
The figure shows an overall workflow of the proposed deep learning approach. Three different processing stages of a CT scan have been shown with help of dashed lines. The blue dashed line is used for denoting the steps of the lung parenchyma segmentation. The red dashed line represents the algorithm for the generation and augmentation of the infection heatmap. The green layer in the neural network is the last convolutional layer of the deep learning algorithm proposed in the work and the blue layer is the softmax layer of the deep neural network. The augmented heatmap is visible with the area of infection highlighted with a different color.
3. Overview of approach
This work is motivated to design a portable and accurate COVID-19 diagnosis system working on an Android application with a deep learning algorithm to analyze the chest CT scans for the presence of COVID-19 and further mark the COVID-19 presence in the segmented lungs' region through an attention map. The utmost focus lies in designing the deep neural network with high accuracy and fewer parameters that can be deployed to the Android Operating System (OS) using minimal memory, unlike many other works that render the trained models in hundreds of Megabytes (Mb). Chest CT images were normalized and converted to Portable Network Graphics (PNG) format for simple viewing and file storage on an Android smartphone. Then the image was fed to the lung parenchyma segmentation algorithm developed in this work using computer vision. The algorithm proposed for lung parenchyma segmentation is robust and involves very light processing on a mobile device. This segmentation algorithm first forms the contour around the detected regions of lung parenchyma and then segments the image for the lung region as shown in the blue dashed box region of Fig. 1. The images are then passed to the EfficientNetB0 deep learning model. If the prediction result is COVID-19 positive, it is allowed to move to the next stage, the heatmap generation. The heatmap is further augmented over segmented lungs, as shown in the red dashed box region of Fig. 1. This stage requires the output of the last convolutional layer (indicated by the green layer in Fig. 1) along with the model predictions to generate the heatmap. After testing a few algorithms to form heatmaps, the Score-CAM algorithm is used to generate the heat map of the COVID-19 class from the trained neural network due to its accurate and soft map formations. The algorithm was ported to Android using the Java programming language. The ScoreCAM algorithm takes a lot of time (9–10 min) to form the heat map, so a selective approach on activation maps and Multi-Threading environment in Android, as explained in this work, is applied before passing the neural network activations. This approach reduces the time taken to develop the heatmap to 50–60 s, which is further dependent on the Android device's performance. Finally, the heatmap is augmented over the chest CT scan and masked to show the segmented lungs region for better inference. The heatmap can be adjusted for its hue values and gradient values using image processing.
4. Material and methods
4.1. Dataset
In our study, we used the COVID-CT dataset mentioned in reference [37]. This dataset contains 63849 CT scan images from 377 patients. 15589 CT scan images belong to 95 patients affected with COVID-19 and 48260 CT scan images belonging to 282 non-COVID patients. The CT scans were gathered from Negin medical center, Sari, Iran. The original files in the dataset were in Tagged Image File Format (TIFF) format containing 16-bit grayscale data and did not include the patients' private information. Android devices or regular monitors do not visualize the 16-bit grayscale TIFF images. There is a separate algorithm for visualizing these TIFF files, given by the dataset authors. So, to make it accessible and simply visualize images, TIFF files were converted to 8-bit PNG images through a normalization process. Converting TIFF image files to PNG images gave a better view and analysis of these images on the Android platform. The tonal values of the TIFF image pixels range from 0 to over 5000, So if each image is scaled based on the maximum tonal value, it can cause data loss and reduce the performance of the network. For tackling this issue, we trained the neural network to detect the COVID-19 related findings not from the TIFF image files but the PNG images. This approach gave us more satisfactory results as any image uploaded to the android platform in PNG form was easy to visualize and process. Five-fold cross-validation was used to find the best hyper-parameters of the neural network and optimizer. The authors of the dataset provided training and testing data in five folds for this purpose. In each fold 20% of the data was used for testing. The model was trained on each fold of the data and tested on the corresponding test data fold. After searching the hyper-parameters for the best accuracy of the neural network, the whole dataset was rearranged by combining the five folds of data that were earlier distributed into train, test, and validation data. The distribution statics of train, validation and test data are shown in Table 2 .
Table 2.
The distribution statics of train, validation and test data.
| Dataset | COVID-19 Images | Normal Images |
|---|---|---|
| Train | 9128 | 9618 |
| Validation | 2282 | 39 262 |
| Test | 2282 | 2250 |
4.2. Normalization of TIFF images to PNG
The TIFF files contain pixel values ranging from 0 to 5000, which are rendered as a black image on android mobile phones. This makes the identification and processing complex. To solve this, a normalization process is applied to each TIFF image file in the dataset, and all images are converted into normalized PNG form, which is easily visible and processed on android OS. The process of normalization used here is the Min-Max Normalization applied with the help of OpenCV Library. Min-max normalization is applied, being the most common way to normalize the data. For every location in the TIFF image, the tonal value of the pixel is normalized according to the formula shown in (1). For this work, the maximum and minimum of the second function (g) are 65535 and 0, respectively. The wide range is chosen to enhance the contrast of the images before converting them to PNG. If the maximum of the second function is too small, all the images will appear black due to less contrast and tonal values. Hence, through normalization, the minimum value in the TIFF image gets transformed into 0, the maximum value gets changed to 65535, and every other value gets changed into a number between 0 and 65535, according to the formula in equation (1)
| (1) |
where f is the input function and g is the output normalized function. Here, v is the original value of pixel and v′ is the normalized value [38]. Now, the normalization of the TIFF image produces an output with values from 0 to 65535, so it is again divided by 255 to convert it into values between 0 and 255, as shown in Fig. 2 .
Fig. 2.
The conversion process of TIFF image to PNG image using Min-Max normalization and further intensity leveling by division with 255 to make tonal values between 0 and 255.
When saving the image, the output's decimal data values are rounded to the nearest integer. This gives us the image intensity values that can be displayed on standard monitors and Android devices. As a result, the data can be saved as PNG images. This is done with entire dataset images, and TIFF files are converted to PNG files before being fed into the pipelines that will be used to train the model. This process is useful not only for training, but it also makes it easier to display the files in PNG format on Android OS-dependent devices.
4.3. Lung parenchyma segmentation
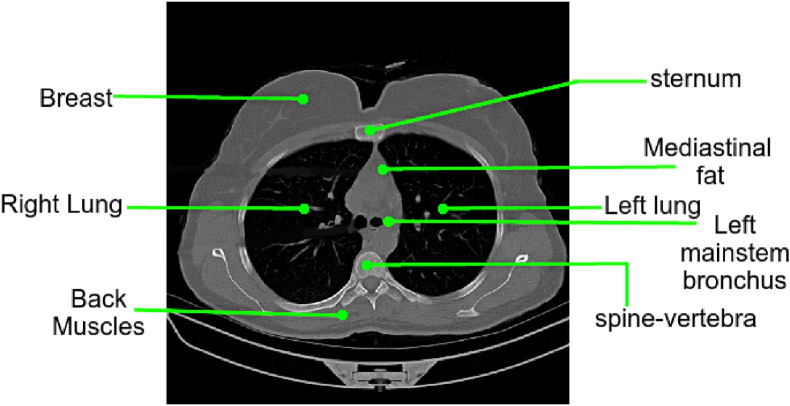
Lung parenchyma refers to the portion of the lungs that is involved in the gas transfer and includes alveoli, alveolar ducts, bronchioles, and other essential tissues. The esophagus, trachea, heart, lungs, diaphragm, thymus gland, aorta, spine, nerves, veins, and arteries are all imaged by the CT scan. Furthermore, the method proposed in this paper generates a heatmap only for detecting COVID-19 findings in the lungs. As a result, the region concerned with COVID-19 diagnosis is the lungs, and the rest organs must be segmented from the CT scan for a clear view of any COVID-19-related findings. An annotated image of the chest CT scan is shown in Fig. 3 . This algorithm is proposed to resolve this complex orientation into a simpler view of the lungs.
Fig. 3.
A CT image slice from the dataset shows the complex orientation of a regular CT image which makes the diagnosis difficult owing to stressed visibility of lungs parenchyma.
For a proper diagnosis, this segmentation algorithm takes out the region of the lungs parenchyma for a better view and analysis of the chest CT Image in an android device and magnifies it to image dimensions by a series of image processing operations. This segmented mask was used to augment the generated heatmap to show COVID-19 affected regions in the lung parenchyma. This algorithm takes a constant amount of time to run on an android device as compared to other approaches which use complex algorithms [[39], [40], [41], [42], [43]]. The chest CT scan uploaded by user on the application is read in form of bitmap with alpha, red, green, and blue channels. This is converted into an OpenCV n-dimensional array mat using the Utils package for Android. The mat image is still in ARGB form, is converted to grayscale. This conversion is necessary for further operations to take place. After converting the ARGB image to grayscale, the global thresholding algorithm, Otsu [44], is applied to get a binary image as shown in Fig. 4 . A binary image has either white or black pixels, which determine the foreground and background, respectively.
Fig. 4.
Image labeled (a) is the original CT image and image (b) is the result after Otsu thresholding is applied to the input image.
As the image is normalized during its conversion to PNG, the area representing lung parenchyma, diaphragm, and small other areas are highlighted in the images. Due to which the histogram of the image shows two clearly expressed peaks. The value which minimizes the weighted variance of these two clusters of the histogram is taken as the threshold value.
The thresholded binary image is subjected to some morphological image processing operations in order to remove impurities from the foreground objects. Morphological opening with a 3 × 3 pixel kernel is performed on the binary output of the thresholding algorithm, which first dilates the image to remove the holes and impurities inside the foreground mask and then erodes it to keep the size of the foreground constant. Fig. 5 (b) shows the outcome. The resulting foreground mask is filled with holes but does not cover the lung parenchyma boundary. To allow the foreground mask to cover the entire region, it is dilated twice more, as shown in Fig. 5(c). The resulting image is a binary mask with foreground pixels representing the lung parenchyma, diaphragm, and possibly 1–2 small findings. Contour detection is now used to segment the lung parenchyma from the binary mask. The contour finding function receives a binary mask as an input. A binary mask is provided as an input to the contour finding function. The function finds the complete contours of the foreground regions in the binary image along with the image's border and makes a tree-like hierarchy. The output of the function is the list of all detected regions in the foreground.
Fig. 5.
Image (a) is the otsu binarisation result and image (b) is the result when morphological opening operation is performed on (a) image, to remove impurities inside it. Image (c) is the result after dilating the (b) image, which have less number of holes inside lung parenchyma region.
This list is iterated to find lung contours using a selection construct that checks each contour for the area and sorts out contours with areas less than 512 × 512 pixels and greater than 100 × 100 pixels. This iteration process chooses the lung parenchyma contour from a list, as shown in Fig. 6 . The contour is then used to generate a binary mask that will be used to segment the lung parenchyma region from the CT image using the bitwise and operation. Finally, the segmented lung parenchyma from the CT scan is enlarged and displayed using a rectangle approximation from the detected contour. This enlarged view clearly shows the lung parenchyma, making it easier for radiologists to diagnose COVID-19.
Fig. 6.
Image labeled (a) contains the detected contours drawn on the original CT. Image (b) shows the contour filled and is used for final mask generation of lung parenchyma.
The asymptotic time complexity of the algorithm is O(1), which is big-O notation giving upper bound on the running time. Therefore, the algorithm takes constant time to run, which makes it best in case of time complexity. The algorithm in steps is provided in the supplementary material for this work. Implementation of this algorithm can be found at the code URL in the Data and Code Availability section.
4.4. Deep learning model architecture
This section explains the different approaches used in this work to find the optimal architecture for the deep learning model. The hyperparameters in the neural network are wisely calculated by hit and trial along with some bilinear interpolation. The size and interim top layers of the model are also decided by training and testing the model on five folds of data and cross-validating it. Finally, all the determined and calculated parameters are used to train the model with all the data present in five folds. Results from various works are analyzed for the selection of the best deep learning architecture that must be used as a base model for transfer learning tasks. Resnet50V2 and EfficientNetB0 deep neural architectures are observed to be best for the classification task of CT scans in to COVID and No-COVID images [45,46]. After changing the input layer in both the models to accept the grayscale 512 × 512 pixels CT scan image, both the models are trained and evaluated on the five folds of data without any hyper-parameter optimization. This process is done to select one out of these two architectures for final training and optimization. Observing the results in Table 3 , the EfficientNetB0 outperforms the Resnet50V2 model in four out of five folds of the data. Further, the number of parameters in EfficientNet B0 is 5.3 Million (Approx.), which is significantly less than Resnet50 V2 with 23 Million params (Approx.). Being less in number of parameters and more accurate, EfficientnetB0 is very suitable for being ported to mobile devices.
Table 3.
The training and testing results of ResNet50 V2 and EfficientNet B0 models on five folds data.
| Model | Fold | Training Accuracy | Testing Accuracy |
|---|---|---|---|
| ResNet50 V2 | Fold 1 | 96.72 | 95.88 |
| EfficientNet B0 | Fold 1 | 97.13 | 96.21 |
| ResNet50 V2 | Fold 2 | 96.32 | 95.91 |
| EfficientNet B0 | Fold 2 | 97.05 | 96.00 |
| ResNet50 V2 | Fold 3 | 97.19 | 96.20 |
| EfficientNet B0 | Fold 3 | 96.73 | 96.07 |
| ResNet50 V2 | Fold 4 | 97.71 | 96.23 |
| EfficientNet B0 | Fold 4 | 97.98 | 97.19 |
| ResNet50 V2 | Fold 5 | 96.85 | 95.93 |
| EfficientNet B0 | Fold 5 | 97.82 | 97.01 |
Hence, The deep learning model is built on the top of EfficientNetB0 architecture.
The EfficientnetB0 model was developed for three channels RGB image, so we make an input layer with our defined shape, i.e., 512 × 512 pixels, and add the base as efficientnetB0 layers without including the top layers. The top layer is excluded in order to configure model output for two classes. After adding the Input layer and EfficientNetB0, a global average pooling layer is added on the top of the base of the classification model. The average of each of these feature maps is taken, and the vector which comes as result is fed directly into the dense layer with a dropout layer in between, as shown in Fig. 7 . Now, to select proper dropout from 0 to 1, the EfficientNet model is again passed through a test in which the same model is trained with different dropouts on the data, and the dropout of 0.3 is found to be optimum. For preventing the overfitting of the model, a dropout is placed and is experimentally verified to give better results in this case.
Fig. 7.
Finalized Architecture of the neural network, to be trained for detection of COVID-19 from PNG images of CT scans. The complete architecture of EfficientNetB0 is explained in supplementary section.
The dense layer contains the softmax activation function, which further renders the final output of the classification model to two probabilities, the first value gives the probability of COVID-19 presence, and second value gives the probability of no COVID-19.
The algorithm for development and training of the deep neural network is given in the supplementary material of this work. The implementation of the algorithm can be found at the code given in the Data and Code Availability section.
4.5. Model training and testing
Data from the dataset is organized into three data generators (i.e., train, test, and validation) which feed the data to the training algorithm of the model. The data from these generators are sent to the model in the form of batches. Each batch contains a fixed number of input data samples. The batch size used for train and validation generators is 10 each, and for test generator is 20. The model is created using Keras according to the specified architecture in section 4.4, and weights are initialized for the base layers as the Imagenet weights. Imagenet weights are weights of the EfficientNetB0 model after training it on the Imagenet dataset [47]. These weights are available from Keras itself. The optimizer used for updating the neural network weights to minimize the cost function used in this work is Nesterov-accelerated Adaptive moment Estimation or Nadam with a learning rate of 0.0001 [48]. Optimizers help us in knowing, how to change weights of the model and learning rate to reduce the occurring loss in training process. Two callbacks are added before training the model for better accuracy these are, Early Stopping and Reduce Learning Rate on Plateau. Early stopping callback is monitored with the validation loss. If validation loss is not decreasing in 5 continuous epochs, the training is stopped by the callback, and epochs with the least validation loss are saved. This step is taken to prevent the model from overfitting the data and further analyze what can be done to better the model. Reduce Learning rate on the plateau is a callback that also monitors the validation loss and if it does not decrease in a fixed number of epochs which is 3 in this work, reduces the model learning rate by a constant factor F, which is 0.2 in the work.
5. Android implementation
This section explains the android implementation of the approach. Any requirement other than an Android device is not necessary according to the implementation that has been used. The whole approach has been ported to the android application using the Android Studio. The algorithm for the implementation of the android features is available in the supplementary section and the related implementation with the codes can be found at the URL given in Data and Code Availability section. Further detailed explanations are listed in the given section.
5.1. Re-scaling and grayscale conversion of CT image
The input image is read in the form of an ARGB (Alpha, Red, Green, Blue) Bitmap by default, shown in Fig. 8 [49]. The uploaded image is checked for its dimensions, which, if not square in shape, is rejected and again prompted for input. This bitmap is processed with the Image processing module of the OpenCV library to convert it into a grayscale image with a single channel. This grayscale image is then resized into a 512 × 512 pixels, which is used for further operations (see Fig. 9).
Fig. 8.
The pre-processing algorithm for input image through user before moving to inference generation.
Fig. 9.
Image (a) represents the Logo and Splash screen of CovCT Application. Image (b) Is the Home screen and (c) is the upload activity for accepting inputs from user.
The application activity prompts the user to submit the input as a CT image. After uploading is completed, a check is run on the input data to make it in a format acceptable by the further processes, including the segmentation algorithm and the neural network.
5.2. Lung parenchyma segmentation
For this stage, the processed input image is passed through the algorithm explained in section 4.3. The image processing module of OpenCV is again put to use. The grayscale single-channel image in the form of a mat object is run through the application. This algorithm is processed on a worker thread without the main looper in order to prevent it from interrupting or stopping the User Interface (UI) Thread. Message from the worker thread is used to indicate that the algorithm has processed the mat, and upon reception of the message, another routine call is made to the function which handles the inference generation from the original CT image as shown in Fig. 10 . The process involves using the Imgproc module and core module of OpenCV Software Development Kit (SDK) for implementation into the android application.
Fig. 10.
The process of lung parenchyma segmentation takes place on a separate worker thread. The handler object is use to send and receive message from runnable highlighted in yellow and when the task is complete, the inference generation stage is called.
5.3. Deployment of neural network and inference
The trained model is deployed to the application for offline inference generation instead of relying on the web servers and API programming. As the inference is On-device, it ensures no concern for data privacy, and along with it, fast inference timings are recorded as no data is to be sent or received over the network. For getting the inference from the Neural Network API (NNAPI) of Android OS, the model file is to be bundled into the application. The trained model file is 45 Mb which must be reduced in size to be bundled into the Android Application Package (APK) as shown in Fig. 11 . For this purpose, the model is converted into a Tensorflow lite flat buffer file (.tflite). With the use of Tensorflow lite, not only is the size-reduced, but the model is optimized for speed and latency on the edge devices, as shown in Table 4 . The number of threads for the generation of inference are tested on multiple devices for optimal performance in terms of speed and processor efficiency. Starting from 2 threads, the inference timings were reduced till 8 number of threads, but the reduction in timing between 6 and 8 number of threads was insignificant for real-world so, the optimum number of threads for inference generation is chosen to be 6.
Fig. 11.
Graphical representation of deployment and inference generation steps in android application. The tflite model file is teh flatbuffer verison of trained model file and inference from this are generated using Java API of Tensorflow Lite which interacts with the Neural Network API of Android and processes the inputs. Input image after conversion to float array is passed to interpreter which process it.
Table 4.
The inference timings on four different specification android devices for trained tensorflow model converted to flatbuffer.
| Device | Inference Timing (4 Threads) | Inference Timing (6 Threads) | Inference Timing (8 Threads) |
|---|---|---|---|
| Nokia 5.1 Plus | 291 ms | 273 ms | 276 ms |
| Xiaomi Note4 | 303 ms | 270 ms | 270 ms |
| Samsung A 30 | 276 ms | 253 ms | 259 ms |
| Samsung Galaxy S2 | 312 ms | 281 ms | 282 ms |
5.4. Heatmap generation using selective approach and multi-threading
For the generation of the heatmap and saving time and extra computation, gradient-free Class activation Maps (CAM) based visualization method Score-CAM is used [50]. Score-CAM is ported into android using Java in this work. For faster computations, N-Dimensional Arrays for Java are used. There are 320 activation maps coming as the output of the convolutional layer to be visualized. Each of these activation maps has the size 512 × 512 pixels. Processing all of these 320 activation maps by the Scorecam algorithm on the android OS takes 600–700 s (Based on mobile performance), as shown in Table 5 . This processing time is too much for the application to be in proper use. To reduce the processing time, a combination of selective approach and multi-threading is used, which significantly reduces the heatmap generation timings, as seen in Table 5. In the selective approach, the number of activation maps was decreased from 320 to 80 by selecting every fourth map starting from the first one, as shown in Fig. 12 . This rendered the activation map quickly, compromising a very little clarity of the heat map. Choosing every fourth activation map was proven to give the best and clear results for forming the final heatmap with a reduction of about 74% in heatmap generation timing. Initially the complexity of the heatmap generation process is θ(n), in which n is the required total number of operations. After the selection approach the complexity becomes θ(n/4), which is evident from the results in Table 5. Still, the processing time is not suitable for proper usage of the application, as visible from Table 5. So multi-threading comes in the role.
Table 5.
The effect of multi-threading and selection approach on heatmap generation timings as tested on Xiaomi Note4 android device.
| Heatmap Generation Time (Seconds) | Multi-threading | Selection Approach |
|---|---|---|
| 950–1020 | No | No |
| 230–250 | No | Yes |
| 370–390 | Yes | No |
| 60–80 | Yes | Yes |
Fig. 12.
The representation of Selection and multi-threading approach for generation of Heatmap using ScoreCAM algorithm in Android Device. The background threads keep running without interrupting the Main (UI) thread. The combination of both approaches, reduce the processing time of heatmap significantly and gives an efficient use of CovCT application.
Eight worker threads are initialized for processing approximately 9 activation maps, which further reduces the time, as shown in Fig. 12. These threads are run parallel on the android operating system and maximize the use of resources and computation power. The threads are separate from the Main looper, which renders a clean working of the UI thread and keeps the worker thread in the background, reducing about 59% of processing time. Hence, the total average case complexity of the algorithm is θ(n) which gets reduced to θ(n/4) using the selection approach and further using combination of multi-threading it is reduced to θ(n/10). Table 5 shows the experimental results after both the approaches are used together, resulting in a significant reduction of about 92% in heatmap generation process timing.
5.5. Augmentation of heatmap and gradient changing
This stage involves the augmentation of the generated heatmap over the grayscale CT image segmented with the help of a binary mask. The generated heatmap is blended with the grayscale CT image. This blending operation takes place according to equation (2)
| (2) |
where, f(x) and g(x) are the source images which are to be blended and h(x) is the final blended image. The c factor is the blending factor in the equation. After the blending is completed, the resultant image of blending operation is masked with help of the binary mask generated in lung segmentation process. This highlights the infected region only on lung parenchyma and is easy to observe by radiologists. For further analysis and highlighting the infected regions using different colors, an option for changing the hue of the resultant image is provided.
A track bar is added for changing the colors in the resultant heatmap and helps to observe the infection in various colors easily. The augmented image is converted to the Hue, Saturation, and Value (HSV) color space. The value from the track bar is added to the hue channel by scalar addition for color change. An option to augment the heatmap on the original CT image is also provided, which replaces the masked image with a full CT image as visible in the image (b) of Fig. 13 .
Fig. 13.
Figure indicating the last steps of the process of COVID-19 detection from CT scans. (a) image is the processing stage of heatmap generation. (b) is the heatmap augmented over full CT image using linear blending. (c) is the segmented and augmented image using mask.
6. Results and discussion
6.1. Results
Some of the results of our trained model are displayed in Fig. 14. These results are generated directly from the python scripts for a swift generation. The results generated from the CovCT android application are also the same (in both position and numerically) but may differ in the color schemes of the detected region in the heatmap. Being of importance in medical diagnosis, our results are also verified by two radiologists, which are discussed in the end of this section. The training of the model is performed on Google Collaboratory (URL: https://colab.research.google.com/?utm_source=scs-index) for an uninterrupted computing and GPU instance. The model is loaded with pre-trained weights on the Imagenet dataset, and transfer learning is done on the training dataset of CT scans.
Fig. 14.
A sample of data given to radiologists for reviewing the results of our trained model. There are two original CT scans (a1) and (a2), the corresponding heatmaps for detected COVID-19 infection in lung parenchyma is seen in images (b1) and (b2) respectively. Further (c1) and (c2) are their augmented heatmap over original CT scans.
The training accuracy, validation accuracy, training loss, and validation loss are monitored at each epoch while training the model on CT scan data. The training accuracy is a fraction of correct classifications from a total number of classifications. It can also be said as the accuracy which a model would receive if applied on training data. While, the validation accuracy is the accuracy which model would receive if applied to data that is not used for training, therefore, validation data. The training accuracy in the first epoch itself went to 79.61%, which increased with each epoch. Fig. 15 shows the graph plotted for the training and validation accuracy of the model in blue and orange color respectively. It is observed that the training accuracy increased with each epoch and went to 99.62% in the fifteenth epoch. Validation accuracy of the model kept on increasing from 73.34% in the first epoch to 99.58% in the fifteenth epoch, as visible from the graph in Fig. 15. The training loss and validation loss are plotted using blue and orange lines in Fig. 16 , respectively. It is observed that the training loss kept on decreasing till the 15th epoch to 2.11%. The validation loss is decreasing from 28.09% to 2.51%, the minimum at the 15th epoch, after which it does not decrease further. The observance and the early stopping callbacks saved the best-trained model from the 15th epoch, which has the best validation accuracy of 99.58% and the least validation loss of 2.51%.
Fig. 15.
A graphical representation of training and validation accuracy of the deep learning model after training. The x-axis represents the training epochs and y-axis, the accuracy on a scale of 0–1.
Fig. 16.
A graphical representation of training and validation loss of the deep learning model. The x-axis represents the epochs and the x-axis represents the loss on a scale of 0–1.
The trained neural network is evaluated on a test partition of the dataset with 4532 images. 2282 images are from COVID-19 class and 2250 images from normal patients. Taking the COVID-19 CT image as a positive result with class 0 and no-COVID as a negative result with class 1, the model evaluation resulted in 2275 true positives (TP), 12 false positives (FP), 2238 true negatives (TN), and only 7 false negatives (FN) as seen through the confusion matrix in Fig. 17 . Hence, 4513 images are correctly classified and 19 images are wrong classified.
Fig. 17.
Confusion Matrix plotted on the result of testing the trained model with the test partition of dataset containing 4532 images.
To detect the percentage of actual COVID-19 affected people detected correctly by the model, the sensitivity of the trained model is calculated. Sensitivity tells us what proportion of the positive class were correctly classified by our trained model. The sensitivity is also known as True Positive Rate (TPR) or Recall and it is calculated according to formula (3). The sensitivity of the model trained in this work comes out to be 99.69%.
| (3) |
As another metric of performance, the specificity of the model is calculated which tells us about the proportion of the No-COVID samples which are correctly classified by our model. In other words, specificity also known as True Negative Rate (TNR) calculates the proportion of the negative class which correctly gets classified. It is calculated according to formula (4) and comes out to be 99.46%.
| (4) |
The precision of the model is calculated in this work which tells the ability of a classification model to identify only the relevant samples out of all samples classified. This metric is calculated according to formula (5) and comes out to be 99.47%.
| (5) |
To summarize, the precision of the model is calculated to be 99.47% and a sensitivity of 99.69%. The testing accuracy and specificity of the model are 99.58 and 99.46, respectively. The whole summary of testing the model on all five folds of testing unseen data is presented in Table 6 . This concludes that the model is very accurate and efficient while classifying the CT scans into COVID and No-COVID classes.
Table 6.
The summary of testing the model on five folds of unseen testing data with total 4532 images. The average row shows the average of result of all five folds of data.
| Fold | Performance Metrics (%) |
||||
|---|---|---|---|---|---|
| Accuracy | Specificity | Sensitivity | Precision | F1 | |
| 1 | 99.12 | 98.88 | 99.35 | 98.92 | 99.13 |
| 2 | 99.67 | 99.33 | 100.0 | 99.35 | 99.67 |
| 3 | 99.55 | 99.33 | 99.77 | 99.33 | 99.55 |
| 4 | 99.88 | 99.77 | 100.0 | 99.78 | 99.89 |
| 5 | 99.66 | 100.0 | 99.33 | 100.0 | 99.66 |
| Avg. | 99.58 | 99.46 | 99.69 | 99.47 | 99.58 |
In this work the confidence intervals are also calculated which provides the upper and lower bounds between which our model accuracy can vary. The selection of a particular confidence level for an interval determines the probability that the interval will contain the true accuracy of the model. The confidence intervals for our trained classifier are shown in Table 7 which are calculated according to formula (6).
| (6) |
Table 7.
Confidence intervals on various number of observations for accuracy of trained model in this work.
| Confidence | Number of Observations |
||
|---|---|---|---|
| 4532 | 1000 | 100 | |
| 90% | 99.43–99.73 | 99.25–99.91 | 95.58–100 |
| 95% | 99.39–99.76 | 99.18–99.98 | 98.32–100 |
| 99% | 99.33–99.82 | 99.05–100 | 97.91–100 |
Three confidence intervals are calculated at 90%, 95%, and 99% for which the number of standard deviations from the Gaussian distribution is 1.64, 1.96, 2.58 along with different sizes of samples from the validation dataset. The various size of sample is whole validation dataset, 1000 and 100 observations. The confidence interval at each cell in Table 7 covers the true classification accuracy of the trained model on validation dataset, which is unseen data to the model.
A Receiver Operating Characteristic (ROC) curve is plotted for the trained model. ROC is a graphic plot that demonstrates the diagnostic ability of a binary classifier system. In this graph the TPR is plotted against the FPR at various values of classifier thresholds. As our task is binary classification, ROC is plotted, and the respective Area Under Curve (AUC) is shown on the graph itself in Fig. 18 . The AUC is the percentage of area under the ROC curve on a scale of 0–1. The more the AUC, the more the ability of the model to distinguish between positive and negative classes. Here, in our case the AUC is 0.999843, which shows that our model is highly able to distinguish COVID-19 from normal cases in CT scans and is an excellent classifier. Along with this, the model is working efficiently with the PNG images, which do not have features as clear as in TIFF images. The model is ported to an android application and is performing with brilliance on devices with no bugs. The heatmap generation timing range from 60 to 80 s, which is based on the device performance.
Fig. 18.
Top figure represents the ROC curve of the trained model generated with the test data as input. The bottom image is the enlarged view of the top image.
Two cardiothoracic trained radiologists reviewed the results of our trained model. They found the model to be very sensitive and accurate in detecting ground glass opacities (GGOs). The primary false positives were due to detecting ground glass opacities from non-COVID-19 causes such as pulmonary edema and partially collapsed airspaces. Given the high sensitivity, this could be beneficial in high volume practices and/or resource-poor settings where this application can mitigate delays in diagnosing and triaging.
6.2. Discussion
This work proposed the development of a novel AI-driven android application that can not only detect COVID-19 from chest CT scans with very high accuracy and sensitivity but also identify the regions affected by the COVID-19 infection in the lungs. This proposed application is the only work that uses color gradients and visualization algorithms to mark the regions of COVID-19 infection in the lung parenchyma. The classification accuracy of the trained AI model is found to be 99.58% along with a combination of very few parameters. This combination makes the model file and application installation package very lightweight and proven to give the best results as compared to the other works in COVID-19 detection from CT scans. The visualization results for COVID-19 infection, of the application, are also verified through two cardiothoracic radiologists who find the application to be a very useful and accurate tool for the doctors to be used in mass settings. At the start of the application, the lung parenchyma is also segmented through a robust, swift, and novel image processing algorithm which helps in better envisioning CT scan and understanding of infection heatmap which is augmented over this segmented parenchyma. As the CT scans are of high resolution, the processing time for the heatmap generation from the neural network layers is very long and impractical. To reduce the processing time for generating heatmaps, a novel approach combining selection criteria and multi-threading is proposed which is successful in reducing the processing time up to 93% and making the process extremely fast.
The android application is practical due to its high accuracy, specificity, portability, and easy user interface. There is no need for any exceptional environment created by the user to utilize the application's capabilities. While the algorithm may lead to very few false positive or false negative results, it should serve as a useful tool for healthcare workers to identify patients with COVID-19. While some other studies have classified COVID-19 from chest CT scans with good accuracy and specificity, in many works results were not verified by a physician or radiologist.
Sedik A. et al. propose CNN and ConvLSTM models for detection of COVID-19 from Chest CT and X-Ray images. They use a total of 3000 image datasets in which they have combined both CT and X-Ray images, which is very less when compared to the dataset used in this work. Also, there is no verification of their results from any doctor or radiologist. Their proposed modalities are not deployed to any application for use. The metrics reported by them are tested on part of augmented data which is not generalized and their model seem to perform poorly in some settings owing to its overfitting, as marked by its authors [51]. Another study [52] in the same domain, tries to improve the accuracy and performance of CNN and convLSTM by proposing a data augmentation framework. They use generative networks and transformations to augment the dataset up to 100% and the highest accuracy and F1 score of data augmented deep learning models they achieve is 99% along with the highest specificity of 98.7%. Our proposed work outperforms their approach as can be seen from the results. Also, this work does not rely on generative networks for data augmentation as medical data is crucial, and using artificially synthesized data to structure the deep neural network for COVID-19 detection may prove detrimental in practical settings. Our algorithm has an accuracy of 99.58% and specificity of 99.46% and outlines regions of the COVID-19 presence in the lung parenchyma using heatmaps for easier visualization and verified by radiologists. Also in the domain of COVID-19 detection from CT scans using transfer learning methods, our work proves to be more accurate, efficient and of more utility than other works [19,[25], [26], [27], [28], [29]]. Our application is also convenient due to its portable nature. It can reach every part of the globe, helping many doctors and radiologists in the diagnosis of COVID-19.
As future research, percent lung involvement can be quantified and help grade patients on severity of disease. Along with this segmentation, better accuracy and mass data adaptation are the areas where more research work can be contributed. An interesting follow-up project would be to see which of these patients developed progressive lung damage and fibrosis on follow up CT scans. By reviewing this data using AI, we can identify patients at higher risk and alter treatment plans accordingly.
7. Conclusion
The CovCT Application is developed with a very accurate neural network at its base and various techniques applied for its smooth and fast working in this work. The neural network is trained and tested on a vast and balanced dataset of CT images and found to be more accurate and having less number of parameters than other works in the domain. Another feature of the application is the lung parenchyma segmentation which is deployed to it using the proposed novel and low-cost algorithm running in O(1), constant time. The lung parenchyma segmentation removes the outer parts of the lungs and works robustly as tested on the dataset. Following the classification on CT image, heatmap is also generated to depict the areas in which COVID-19 infection may be present in the lungs. This heatmap is augmented over the segmented part of the lungs which gives a clear view of the infection heatmap. A novel approach for the reduction in heatmap generation timing is developed and tested which is successful in reducing running time complexity from θ(n) to θ(n/10). The results of the heatmap are also verified by expert cardiothoracic radiologists.
This work has proposed three novel contributions which are, one of its kind CovCT application with an exceptionally accurate and most lightweight deep neural network, low cost and swift lung parenchyma segmentation algorithm, and the combination of selection approach with multi-threading to reduce the timing of heatmap generation of mobile phone device.
To analyze the performance of the CovCT application eight different typical cases of lung parenchyma visibility in CT scans from different patients are taken and processed, results are in Fig. 19 . In (a1) and (a2) Image of Fig. 19, both the lung regions are clearly visible in the CT scan, which is processed efficiently and perfectly by the segmentation algorithm as seen in (b1) and (b2), and further (c1) and (c2) are the results of augmentation of heatmap on segmented lungs in which pink shades are representing the COVID-19 affected region. In (a3) image, there is a blur region present in the right lung; still, the segmentation algorithm performs well and segments the lung parenchyma carefully, as seen from resulting image (b3). In images (a4) and (a5), the right lung is seen with a portion of liver in the CT scan image. Our segmentation algorithm clearly segments the lung from surrounding structures, as seen in results (b4) and (b5) respectively, further the deep learned model also treats that structure as an organ, not opacification, which can be seen in image result (c4) and (c5). In image (a6) one lung is partially visible for the high opacification in lung parenchyma due to parenchyma structures. The prediction of the deep learning model carefully judges the region of COVID-19 infection as seen in the resulting image (c6). The structures are not taken as COVID-19 related findings. In image (a7), both the lungs are partially visible. Still, they are processed equally well by the segmentation algorithm, as visible from the result (b7). Further, the deep learning model can also detect the affected region in this small portion, as seen in image (c7). The test case (a8) is of minimal lung parenchyma visibility and a complex case where the majority of the image contains non-lung structures. The segmentation algorithm does not get confused at any point and segments the parenchyma without mixing it with muscles and fat, as seen in the result (b8). Further, the neural network also judges the opacification and does not get confused due to other structures, visible through the result (c8). This analysis of the CT scans shows that both, the segmentation algorithm and the neural network perform accurately and efficiently on CT images, including those CT images which do not contain majority lung parenchyma. Hence, the application is not only a theoretical research work but a pragmatic application of verified research which can prove beneficial in mass screening of COVID-19 patients whenever needed.
Fig. 19.
Analysis of Application's segmentation algorithm and deep neural network of eight typical different lung visibility cases from different patients of COVID-19 where in the first case (a1) and second case (a2), lung parenchyma is fully visible, third case (a3) have a blur in upper part of right lung. Fourth case (a4) shows the lung parenchyma with one lung inflated. Fifth case (a5) is the CT scan with right lung having an organ view in right lung. (a6) image shows right lung inflated, seventh case (a7) shows both lungs partially visible in the image due to inflation. In eight case (a8) again both lungs are inflated and parenchyma is slightly visible. The (b) images are corresponding extracted lungs for each case and (c) images are output with highlighted regions showing COVID-19 related findings using neural network.
Data and Code Availability
For interested scholars and researchers who wish to use this work for education, research, and improvement purposes, we have maintained an open-source repository containing the codes of the CovCT android application and the deep learning scripts used to train and test the model. The repository contains the implementation of all the novel algorithms proposed in the work. The repositories mentioned in this section contain the python implementation of algorithms and also the Java implementation of all algorithms in application development. The repository contains code files, trained models, deep learning results, and an android application package (APK). A demonstration video is also provided in the repository, along with the android test results. A readme file is embedded into the repository along with the history of whole version control using git. The repository is located at application code and results https://github.com/monjoybme/CovCT_application. Along with it a repository for available datasets and resource code files for processing the data is available at pre-processing code repositoryhttps://github.com/monjoybme/CovCT. For researchers and scholars who wish to use and get insights from the dataset we used in this work, the authors of the dataset have maintained a well-explained repository having all data and its reports. It can be reached out to data repository link https://github.com/mr7495/COVID-CTset.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
Aryan Verma would like to thank Google Summer of Code (GSoC) 2021 program for funding the work and the organization, Department of Biomedical Informatics, Emory School of Medicine for selecting him as a GSoC student and providing constant support, guidance and mentoring for the work. The details of the organization and work done under GSoC can be explored at https://summerofcode.withgoogle.com/archive/2021/projects/6468381577838592/
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105298.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., Schluger N.W., Volpi A., Yim J.-J., Martin I.B., et al. The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. Radiology. 2020;296:172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hope M.D., Raptis C.A., Henry T.S. 2020. Chest Computed Tomography for Detection of Coronavirus Disease 2019 (Covid-19): Don't Rush the Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and rt-pcr testing for coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the coronavirus disease (covid-19): rrt-pcr or ct? Eur. J. Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanne J.P. 2020. Chest Ct Findings in 2019 Novel Coronavirus (2019-ncov) Infections from Wuhan, china: Key Points for the Radiologist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha M., Amin S.B., Sharma A., Kumar T.S., Kalia R.K. 2021. AI-driven Quantification of Ground Glass Opacities in Lungs of Covid-19 Patients Using 3d Computed Tomography Imaging, medRxiv: the Preprint Server for Health Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z., et al. vol. 295. 2020. Ct Imaging Features of 2019 Novel Coronavirus (2019-ncov) Radiology; pp. 202–207. 2020 apr. 110.1148/radiol. 2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest ct assessment of covid-19: a deep learning approach. Radiology: Cardiothorac. Imaging. 2020;2 doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Y., Liu Y., Li S., Xu J. 2020. Miniseg: an Extremely Minimum Network for Efficient Covid-19 Segmentation. arXiv preprint arXiv:2004. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6:1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D., Menotti D. Covid-19 detection in ct images with deep learning: a voting-based scheme and cross-datasets analysis. Inform. Med. Unlocked. 2020;20:100427. doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebdani A.M., Mostafavi A. 2021 5th International Conference on Pattern Recognition and Image Analysis (IPRIA) IEEE; 2021. Medical image processing and deep learning to diagnose covid-19 with ct images; pp. 1–6. [DOI] [Google Scholar]
- 16.Elaziz M.A., Hosny K.M., Salah A., Darwish M.M., Lu S., Sahlol A.T. New machine learning method for image-based diagnosis of covid-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gianchandani N., Jaiswal A., Singh D., Kumar V., Kaur M. Rapid covid-19 diagnosis using ensemble deep transfer learning models from chest radiographic images. J. Ambient Intell. Hum. Comput. 2020:1–13. doi: 10.1007/s12652-020-02669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan M., Alam M., Elahi M., Toufick E., Roy S., Wahid S.R., et al. 2020. Cvr-net: A Deep Convolutional Neural Network for Coronavirus Recognition from Chest Radiography Images. arXiv preprint arXiv:2007.11993. [Google Scholar]
- 19.Alom M.Z., Rahman M., Nasrin M.S., Taha T.M., Asari V.K. 2020. Covid_mtnet: Covid-19 Detection with Multi-Task Deep Learning Approaches. arXiv preprint arXiv:2004. [Google Scholar]
- 20.Hu R., Ruan G., Xiang S., Huang M., Liang Q., Li J. medRxiv; 2020. Automated Diagnosis of Covid-19 Using Deep Learning and Data Augmentation on Chest Ct. [Google Scholar]
- 21.Kassania S.H., Kassanib P.H., Wesolowskic M.J., Schneidera K.A., Detersa R. Automatic detection of coronavirus disease (covid-19) in x-ray and ct images: a machine learning based approach. Biocybernet. Biomed. Eng. 2021;41:867–879. doi: 10.1016/j.bbe.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B., Gao X., He M., Liu L., Yin G. vol. 2020. 2020. A fast online covid-19 diagnostic system with chest ct scans. (Proceedings of KDD). [Google Scholar]
- 23.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid Ai Development Cycle for the Coronavirus (Covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning Ct Image Analysis. arXiv preprint arXiv:2003. [Google Scholar]
- 24.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., Zhang X., Yang L., Fang Y., Xing Z., et al. A deep learning approach to characterize 2019 coronavirus disease (covid-19) pneumonia in chest ct images. Eur. Radiol. 2020;30:6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning-based automated detection of covid-19 from lung ct scan slices. Appl. Intell. 2021;51:571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunese L., Martinelli F., Mercaldo F., Santone A. Machine learning for coronavirus covid-19 detection from chest x-rays. Procedia Comput. Sci. 2020;176:2212–2221. doi: 10.1016/j.procs.2020.09.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the covid-19 infected patients using densenet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 28.Anwar T., Zakir S. 2020 IEEE 23rd International Multitopic Conference (INMIC) 2020. Deep learning based diagnosis of covid-19 using chest ct-scan images; pp. 1–5. IEEE. [DOI] [Google Scholar]
- 29.Ozturk T., Talo M., et al. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee H., Ghosh S., Dhar A., Obaidullah S.M., Santosh K., Roy K. Deep neural network to detect covid-19: one architecture for both ct scans and chest x-rays. Appl. Intell. 2021;51:2777–2789. doi: 10.1007/s10489-020-01943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Karawi D., Al-Zaidi S., Polus N., Jassim S. MedRxiv; 2020. Machine Learning Analysis of Chest Ct Scan Images as a Complementary Digital Test of Coronavirus (Covid-19) Patients. [Google Scholar]
- 32.Paluru N., Dayal A., Jenssen H.B., Sakinis T., Cenkeramaddi L.R., Prakash J., Yalavarthy P.K. Anam-net: anamorphic depth embedding-based lightweight cnn for segmentation of anomalies in covid-19 chest ct images. IEEE Transact. Neural Networks Learn. Syst. 2021;32:932–946. doi: 10.1109/TNNLS.2021.3054746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alshazly H., Linse C., Barth E., Martinetz T. Explainable covid-19 detection using chest ct scans and deep learning. Sensors. 2021;21:455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basantwani N., Kumar A., Gangwar S., Olkha A., Mathur G., et al. Covid-19 detection android app based on chest x-rays & ct scans. INFOCOMP J. Comput. Sci. 2021;20 [Google Scholar]
- 35.Hammad M., Iliyasu A.M., Subasi A., Ho E.S.L., El-Latif A.A.A. A multitier deep learning model for arrhythmia detection. IEEE Trans. Instrum. Meas. 2021;70:1–9. doi: 10.1109/TIM.2020.3033072. [DOI] [Google Scholar]
- 36.Hammad M., Alkinani M.H., Gupta B.B., El-Latif A.A.A. Myocardial infarction detection based on deep neural network on imbalanced data. Multimed. Syst. 2021 doi: 10.1007/s00530-020-00728-8. [DOI] [Google Scholar]
- 37.Rahimzadeh M., Attar A., Sakhaei S.M. A fully automated deep learning-based network for detecting covid-19 from a new and large lung ct scan dataset. Biomed. Signal Process Control. 2021;68:102588. doi: 10.1016/j.bspc.2021.102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han J., Pei J., Kamber M. Elsevier; 2011. Data Mining: Concepts and Techniques. [Google Scholar]
- 39.Wei Y., Shen G., Li J.-j. A fully automatic method for lung parenchyma segmentation and repairing. J. Digit. Imag. 2013;26:483–495. doi: 10.1007/s10278-012-9528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armato S., III, MacMahon H. vol. 1256. Elsevier; 2003. Automated lung segmentation and computer-aided diagnosis for thoracic ct scans; pp. 977–982. (International Congress Series). [DOI] [Google Scholar]
- 41.Tan S. Segmentationoflunglesionson CT scans using watershed, active contours, and Markov randomfield. Med. Phys. 2013;40 doi: 10.1118/1.4793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Nunzio G., Tommasi E., Agrusti A., Cataldo R., De Mitri I., Favetta M., Maglio S., Massafra A., Quarta M., Torsello M., et al. Automatic lung segmentation in ct images with accurate handling of the hilar region. J. Digit. Imag. 2011;24:11–27. doi: 10.1007/s10278-009-9229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai J., Wei Q. Automatic lung fields segmentation in ct scans using morphological operation and anatomical information. Bio Med. Mater. Eng. 2014;24:335–340. doi: 10.3233/BME-130815. [DOI] [PubMed] [Google Scholar]
- 44.Otsu N. IEEE transactions on systems, man, and cybernetics. Vol. 25. 2006. A threshold selection method from gray-level histograms. 417–4. [Google Scholar]
- 45.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 46.Tan M., Le Q. International Conference on Machine Learning. PMLR; 2019. Efficientnet: rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 47.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [DOI] [Google Scholar]
- 48.Kingma D.P., Ba J. arXiv e-prints; 2014. Adam: A Method for Stochastic Optimization. arXiv:1412.6980. [Google Scholar]
- 49.Handling Bitmaps android developers. 2021. https://developer.android.com/topic/performance/graphics URL: 21-10-07.
- 50.Wang H., Wang Z., Du M., Yang F., Zhang Z., Ding S., Mardziel P., Hu X. arXiv e-prints; 2019. Score-CAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. arXiv:1910.01279. [Google Scholar]
- 51.Sedik A., Hammad M., Abd El-Samie F.E., Gupta B.B., Abd El-Latif A.A. Efficient deep learning approach for augmented detection of coronavirus disease. Neural Comput. Appl. 2021:1–18. doi: 10.1007/s00521-020-05410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedik A., Iliyasu A.M., El-Rahiem A., Abdel Samea M.E., Abdel-Raheem A., Hammad M., Peng J., El-Samie A., Fathi E., El-Latif A., et al. Deploying machine and deep learning models for efficient data-augmented detection of covid-19 infections. Viruses. 2020;12:769. doi: 10.3390/v12070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For interested scholars and researchers who wish to use this work for education, research, and improvement purposes, we have maintained an open-source repository containing the codes of the CovCT android application and the deep learning scripts used to train and test the model. The repository contains the implementation of all the novel algorithms proposed in the work. The repositories mentioned in this section contain the python implementation of algorithms and also the Java implementation of all algorithms in application development. The repository contains code files, trained models, deep learning results, and an android application package (APK). A demonstration video is also provided in the repository, along with the android test results. A readme file is embedded into the repository along with the history of whole version control using git. The repository is located at application code and results https://github.com/monjoybme/CovCT_application. Along with it a repository for available datasets and resource code files for processing the data is available at pre-processing code repositoryhttps://github.com/monjoybme/CovCT. For researchers and scholars who wish to use and get insights from the dataset we used in this work, the authors of the dataset have maintained a well-explained repository having all data and its reports. It can be reached out to data repository link https://github.com/mr7495/COVID-CTset.