Abstract
Mu-opioid peptide receptor (MOPR) stimulation alters respiration, analgesia, and reward behavior, and can induce substance abuse and overdose. Despite its evident importance, the endogenous mechanisms for MOPR regulation of consummatory behavior have remained unknown. Here we report that endogenous MOPR regulation of reward consumption in mice acts through a specific dorsal raphe to nucleus accumbens projection. MOPR-mediated inhibition of raphe terminals is necessary and sufficient to determine consummatory response while select enkephalin-containing NAc ensembles are engaged prior to reward consumption, suggesting that local enkephalin release is the source of endogenous MOPR ligand. Selective modulation of NAc enkephalin neurons and CRISPR-Cas9-mediated disruption of enkephalin substantiate this finding. These results isolate a fundamental endogenous opioid circuit for state-dependent consumptive behavior and suggest alternative mechanisms for opiate modulation of reward.
For centuries opioids and their derivatives have been used as potent analgesics5. However, opioids—particularly those affecting the MOPR system—have both therapeutic and undesirable effects1,2,3. The effects of MOPR agonism on reward-related behaviours are driven through mesocorticolimbic circuits—including nucleus accumbens medial shell (mNAcSh)4,6,7,8, which is composed primarily of two projection populations expressing either preprodynorphin (encoded by the Pdyn gene) and dopamine-1 receptor (D1R) or preproenkephalin (encoded by the Penk gene) and dopamine-2 receptor (D2R). Preprodynorphin and preproenkephalin cleave into potent, efficacious endogenous MOPR agonists9,10. Exogenous agonism of MOPRs in mNAcSh is known to modulate a variety of behaviours11,12,13,14,15,16, including reward. However, the precise identity of the endogenous MOPR circuit and its cellular mechanisms has remained essentially unknown owing to the limited ability to isolate neuropeptidergic circuitry. Here we have identified these endogenous opioid mechanisms using a series of complementary, high-resolution approaches (Extended Data Table 1).
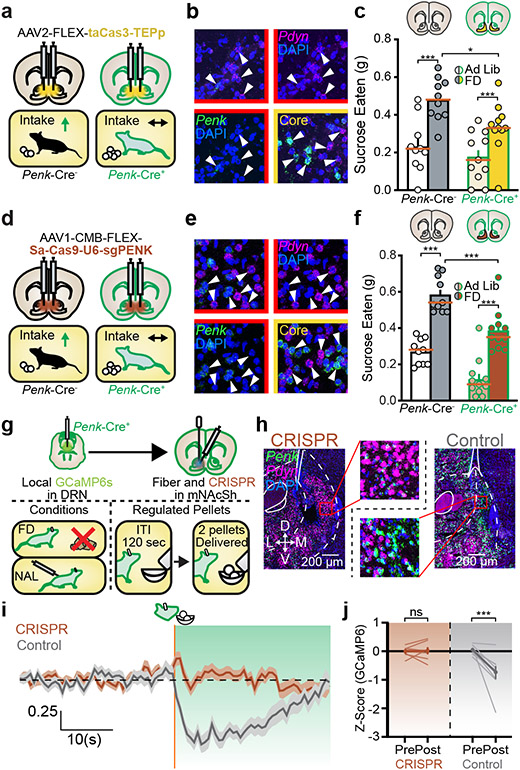
First, we determined whether MOPRs in mNAcSh are important for state-dependent reward consumption. We used a voluntary sucrose consumption paradigm in which mice were tested ad libitum or after 24 hours of food deprivation (FD, potentiated state) (Fig. 1a). Intracerebral microinjections of the MOPR selective antagonist CTAP17 into mNAcSh, but not nearby regions, reduced FD sucrose consumption (Fig. 1b, Extended Data Fig. 1a-c).
Fig. 1. Endogenous MOPR activation in mNAcSh is necessary for potentiating state-dependent consummatory behavior.
a. Schematic of voluntary sucrose consumption assay. b. Microinjections of CTAP (blue) into mNAcSh reduced FD intake compared to vehicle (gray, n = 11). c. Oprm1 KO mice (blue) show full suppression of FD intake (n = 11 WT, 12 KO). d and e. Oprm1fl/fl x Penk-Cre+ mice (green) show suppressed FD intake compared to Cre− mice (gray, n = 13 Cre−, 11 Cre+). f. Retro MOPR deletion reduced FD intake compared to wildtype (gray, n = 13 wildtype, 12 local NAc, 11 retro-NAc). All error bars represent ± SEM and n = biologically independent mice or cells (e). Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (b, c, e, f, Extended Data Table 4).
To determine the extent to which MOPRs enhance consumption, we tested Oprm1 constitutive knockout mice in the same sucrose consumption paradigm (Fig. 1c). Oprm1 KO mice had normal ad libitum sucrose intake, but did not increase consumption after FD, indicating that MOPRs in mNAcSh account for a significant portion of all MOPR-mediated potentiation of sucrose consumption.
Fluorescent in situ hybridization (FISH) experiments found that MOPRs were expressed on ~30-35% of all neurons within mNAcSh, and on about ~50% of Pdyn or Penk subpopulations (Fig. 1d-e, Extended Data Fig. 1d). To test the function of MOPRs on these subpopulations, we crossed MOPR conditional knockout mice (Oprm1fl/fl) with Pdyn-Cre or Penk-Cre mouse lines to selectively delete MOPRs from each cell-type (Fig. 1f, Extended Data Fig. 1e-j). Loss of MOPRs on Penk-Cre+, but not Pdyn-cre+, cells reduced FD sucrose consumption (Fig. 1g, Extended Data Fig. 1h). Oprm1fl/fl x Penk-Cre mice showed selective deficits in reward behaviors, displaying normal avoidance of the open arms in an elevated zero maze after restraint stress. In contrast, Oprm1 KO mice or wildtype mice given systemic naloxone showed disrupted avoidance behaviors. These results suggest that MOPR-dependent appetitive and avoidance behaviors are mediated by separable systems (Extended Data Fig. 2).
To test whether FD potentiation of sucrose consumption was mediated by accumbens or non-accumbens Penk+ cells, we conditionally deleted MOPRs in mNAcSh of Oprm1fl/fl mice (Extended Data Fig. 1m-o). mNAcSh MOPR deletion did not reduce FD intake, suggesting that MOPRs may act presynaptically. Consistent with this hypothesis, we found that retrograde MOPR deletion reduced FD sucrose consumption18 (Fig. 1f). These data indicate that presynaptic MOPRs in mNAcSh are recruited in a state-dependent manner to potentiate consumption, and are located in Penk+ neurons.
Fluorescent retrograde viral tracing (Fig. 2a, Extended Data Fig. 3a) of mNAcSh Penk afferents labeled several brain regions, including a lateralized population of dorsal raphe nucleus (LDRN). Selective inhibition experiments in other labeled inputs failed to modulate FD intake (Extended Data Fig. 4a-d). We therefore focused efforts on understanding LDRN MOPRs. Using FISH, we found that DRN cells expressed Penk (28%), Oprm1 (31%), and Tph2 (19%). About half of DRNPenk neurons coexpressed Oprm1 (14%), but only a small subset additionally expressed Tph2 (4%) (Fig. 2b-c). Combinatorial viral tracing and FISH experiments confirmed high coexpression of Penk and Oprm1 in neurons that project to mNAcSh (60% overlap) (Extended Data Fig. 3d-j). Considering the high degree of overlap, we will refer to this specific projection as LDRNMOPR-mNAcSh. We then determined that LDRNMOPR-mNAcSh projections also make functional monosynaptic connections with mNAcSh using optogenetics and ex vivo patch-clamp electrophysiology (Extended Data Fig. 4e-i). Taken together, these results indicate that a MOPR-expressing LDRN-mNAcSh circuit is distinct from reported canonical opioid/serotonin systems19-22.
Fig. 2. LDRNPenk-mNAcSh projections make monosynaptic connections and display opioid-dependent spatiotemporal signaling.
a. Schematic (left) of a fluorescently tagged retrograde virus injection into mNAcSh of a Penk-Cre+ mouse (middle, scale bar = 200 μm) and labeled neurons in dorsal raphe nucleus (right). b. Fluorescent in situ hybridization of Oprm1, Tph2 and Penk in dorsal raphe nucleus (scale bar = 200 μm). c. Quantification of b. h. Schematic of regulated lickometer paradigm. i. Average Z-scored trace aligned to onset of licking of LDRNPenk-mNAcSh terminals in WD/saline (green) and WD/naloxone conditions (blue). j. Heatmap of individual trials across all mice in WD/saline (green) or WD/naloxone conditions. Orange lines indicate onset of licking. k. Quantification of the average Z-score prior to lick onset versus after lick onset in WD/saline (green) and WD/naloxone (blue) conditions. Error bars or shaded region surrounding photometry trace represent ± SEM and n = biologically independent mice or cells (c). Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (f, k, Extended Data Table 4).
We next determined whether the in vivo activity of the LDRNMOPR-mNAcSh projection was: 1) related to time-locked reward consumption, and 2) endogenous opioid dependent. First, we expressed the calcium indicator GCaMP6s into LDRN of Penk-Cre+ mice23 and implanted an optical fiber into mNAcSh to measure changes in terminal fluorescent GCaMP6s activity. We then tested the mice in the voluntary sucrose paradigm after systemic saline or naloxone injections (Extended Data Fig. 5a, b). When GCaMP activity of LDRNMOPR-mNAcSh terminals was aligned to the onset of multi-pellet bouts, we observed a sustained inhibition of GCaMP activity (~20 seconds) coinciding with the total duration of the eating bout, which was naloxone sensitive (Extended Data Fig. 5c-h).
Due to variability of consummatory behavior within and between freely feeding mice, we next determined whether an alternative consummatory task with greater temporal resolution could produce similar MOPR-mediated LDRNMOPR-mNAcSh inhibition (Fig. 2d). Mice received non-contingent, intermittent access to a sucrose solution via lickometer to induce distinct lick bouts (similar to multi-pellet bouts). We observed immediate and sustained inhibition of LDRNMOPR-mNAcSh terminals when mice initiated licking following water deprivation (WD), which was blocked by naloxone (Fig. 2e, f). Quantification of the average of the trace (Z-score) prior to versus after the onset of a lick bout showed a reduction in GCaMP activity; this reduction was absent with naloxone pretreatment (Fig. 2g). Similar experiments using sucrose pellets showed similar patterns of time-locked GCaMP activity (Extended Data Fig. 5i-m). These results indicate that temporally precise recruitment of an endogenous opioid circuit acts to potentiate multiple forms of reward consumption in response to state changes in specific behaviors.
To test whether MOPRs on the LDRN-mNAcSh projection causally mediates the potentiation of reward consumption, we selectively restored MOPRs within LDRNPenk neurons in Oprm1 KO x Penk-Cre+ mice via targeted viral injection of a cre-dependent MOPR (Fig. 3a-c, Extended Data Fig. 6a). Rescue of MOPR restored FD potentiation of sucrose intake (Fig. 3d). Similarly, MOPR rescue partially restored morphine conditioned place preference (Fig. 3e). However, MOPR rescue was unable to restore morphine analgesia (Extended Data Fig. 6a-b). This indicates that MOPRs on LDRNPenk neurons are necessary for modulating endogenous or exogenous opioid reward, but do not mediate opioid analgesia.
Fig. 3. MOPR activation on LDRNMOPR-mNAcSh is sufficient to potentiate reward consumption.
a. Schematic of Oprm1 KO x Penk-Cre rescue of MOPRs in LDRN. b. In situ micrograph (left; Oprm1, Penk, Cre) and associated fluorescent micrograph (right; GFP-tagged viral vector) of DRN in an Oprm1 KO x Penk-Cre+ mouse (scale bar = 200 μm). c. Quantification of b. d. Oprm1 KO x Penk-Cre+ mice (red) show restored FD intake compared to Cre− (blue) mice (n = 8 Cre−, 11 Cre+). e. Morphine conditioned place preference (CPP) assay. (top) Schematic of CPP procedure. (bottom left) Heat map of time spent in each chamber during pretest and posttest. Warmer colors indicate more time spent in that area. (bottom right) Wildtype and Oprm1 KO x Penk-Cre+ (red) mice spent more time in the morphine paired side compared to Cre− (blue) mice (n = 6 WT, 8 Cre−, 8 Cre+). f. Schematic of opto-MOR experiments. g. Expression of EYFP-tagged opto-MOR in LDRNPenk (left, scale bar = 200 μm) and fiber placement (red dashed line) in mNAcSh (right, scale bar = 200 μm). Zoomed in image shows labeled nuclei in DRN. h. Saline/No Laser treated mice licked more compared to the naloxone/No Laser treated test day. Opto-MOR stimulation restored licking after naloxone injections (n = 8). All error bars represent ± SEM and n = biologically independent mice. Medians are in orange. Post hoc p-values are derived from One-way (d) or Two-way ANOVA with Sidak multiple comparisons (c, g, Extended Data Table 4).
Local LDRN MOPR rescue does not necessarily exclude the potential effects of MOPR on cell bodies or other potential downstream targets. Thus, we determined whether time-locked activation of MOPRs on LDRNMOPR-mNAcSh terminals could potentiate sucrose consumption. First, we expressed the light activated chimeric receptor opto-MOR on LDRNPenk neurons and implanted fibers into mNAcSh. Next, mice were trained on the intermittent sucrose solution paradigm24 (Fig. 3f, g). We then tested whether selective stimulation of presynaptic MOPR signaling via opto-MOR on LDRNPenk-mNAcSh terminals could circumvent naloxone mediated opioid antagonism and restore lick behavior. Naloxone alone reduced licks compared to saline (Fig. 3h). Photo-activation of opto-MOR after naloxone was sufficient to restore licking back to WD levels. Further analysis revealed that behaviorally initiated and time-locked opto-MOR stimulation restored sustained licking behavior that was blunted by naloxone (Extended Data Fig. 6c). An opposing experiment using ChR2 photostimulation of LDRNPenk-mNAcSh terminals resulted in reduced sucrose consumption (Extended Data Fig. 6d-h). These findings indicate that time locked, MOPR-mediated inhibition of the LDRNPenk-mNAcSh circuit is both necessary and sufficient for potentiating reward consumption.
We next sought to determine the source of the endogenous ligand for the LDRNMOPR-mNAcSh circuit. The high density of enkephalin within mNAcSh suggests that the presynaptic MOPRs could be activated via retrograde transmission. To determine whether mNAcShPenk neurons are recruited during consummatory behavior, we expressed GCaMP6s in mNAcShPenk neurons and performed endoscopic single-cell calcium imaging during voluntary sucrose intake (Fig. 4a-b, Extended Data Fig. 7a-b). We reliably tracked single-cell calcium activity in 281 of the same neurons across both test days (Fig. 4c-e). mNAcShPenk neurons were not uniformly responsive to sucrose consumption, but instead showed one of several distinct response patterns (Fig. 4f). K-means clustering isolated four unique subpopulations of neurons (clusters), which included: 1) Onset activated (2%), 2) Pre-onset activated (7%), 3) Onset inhibited (15%), and 4) Non-responsive (76%) (Fig. 4g-j). These clusters were separable from additional clusters that were most responsive to sucrose sniffing, rearing, or grooming (Extended Data Fig. 7d-j). These results suggest that local mNAcShPenk neuronal populations are selectively recruited to enhance consumptive behavior and may provide the endogenous opioid for the LDRNMOPR-mNAcShPenk circuit.
Fig. 4. mNAcSh enkephalinergic ensembles are modulated by physiological state and reward consumption.
a. Schematic of in vivo 1-photon imaging experiments. b. Expression of GCaMP6s and GRIN lens placement (red dashed line) in mNAcSh of a Penk-Cre+ mouse (scale bar = 200 μm). c. Example cell map (left) for cells that were matched (right) across ad libitum and FD conditions. d and e. Heat plot of all 281 matched cells on the ad libitum or FD test day. Change in Z-scored fluorescence shown in white (more) or purple (less). Cells aligned to onset of pellet consumption. f. TSNE plot of neural clusters determined using kmeans clustering analysis. g. Heat plot of the three behaviorally modulated clusters. Clusters separated by black dashed lines and color coded to match. Change in Z-scored fluorescence displayed by white (more) or purple (less). Cells aligned to onset of multi-pellet bouts. h-j. Average trace of Onset activated (h, cluster 1), Pre-onset activated (i, cluster 2) and Onset inhibited neurons (j, cluster 3).
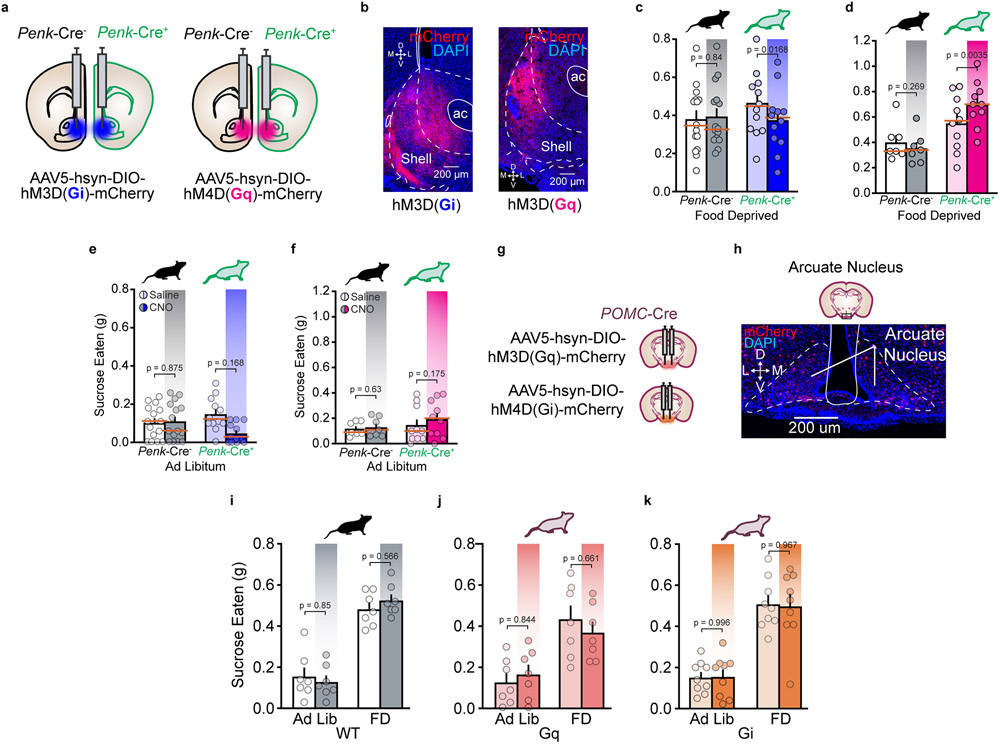
To test the necessity of local mNAcShPenk in potentiating reward consumption, we induced cell-type specific ablations of mNAcShPenk neurons via caspase viral injections (Fig. 5a-c) and then tested in the voluntary sucrose consumption paradigm. Caspase ablation of mNAcShPenk neurons significantly reduced FD sucrose consumption, but had no impact on ad libitum intake (Fig. 5d). We next sought to determine whether modulation, rather than ablation, of local mNAcShPenk neurons could bidirectionally alter voluntary sucrose consumption using the inhibitory DREADD HM4D(Gi) or excitatory DREADD HM3D(Gq) during the sucrose consumption tests (Extended Data Fig. 8a-f). Consistent with caspase results, Gi-DREADD activation reduced, and Gq-DREADD activation increased, FD sucrose consumption relative to saline test days. DREADD modulation of arcuate nucleus POMC neurons failed to alter FD sucrose consumption (Extended Data Fig. 8g-k). These results indicate that mNAcSh enkephalin is the likely endogenous opioid source for potentiating reward consumption.
Fig. 5. Endogenous mNAcSh enkephalin is necessary for potentiating reward consumption.
a. Schematic of food intake paradigm after caspase-me ablation. b. FISH of Pdyn and Penk in mNAcSh after caspase injections. c. Caspase injections in mNAcSh of Penk-Cre+ (yellow) mice reduced FD intake compared to Cre− (gray) mice (n = 10 Cre−, 11 Cre+). d. Schematic of food intake paradigm after CRISPR disruption. e. FISH of Pdyn and Penk in mNAcSh after CRISPR viral injections. f. CRISPR virus injections in Penk-Cre+ (brown) mice reduced FD intake compared to Cre− (gray) mice (n = 10 Cre−, 11 Cre+). g. Schematic of regulated food intake after GCAMP6s virus injections into LDRN and CRISPR/Control virus injections into mNAcSh of Penk-Cre+ mice. h. FISH (scale bar = 200 μm) of Pdyn and Penk in mNAcSh after CRISPR or Control injections. i. Average Z-scored trace aligned to onset of consumption in Control (gray) or CRISPR (brown) treated mice not (CRISPR n = 3, Control n = 3, repeated 3 times). j. Quantification of the average Z-score prior to pellet onset versus after onset in CRISPR (brown) or Control (gray) mice. All error bars or shaded region surrounding photometry traces represent ± SEM and n = biologically independent mice (d, h), and mice tested multiple times (l). Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (d, h, l, Extended Data Table 4).
To directly determine whether the local mNAcShPenk neuropeptide is recruited to potentiate reward consumption, we designed and packaged a single-viral Staphylococcus aureus Cas9 (SaCas9)-guided CRISPR mRNA construct25,26 to disrupt enkephalin neuropeptide (Extended Data Fig. 9c-e). Fluorescence-activated cell sorting (FACS) and PCR analyses indicated that this virus disrupts enkephalin mRNA production by 90% via indel mutations (Extended Data Fig. 9f-i). Viral injections into mNAcSh reduced Penk expression by 30-50% (Fig. 5e) and behaviourally reduced ad libitum and FD sucrose consumption (Fig. 5f). Penk-Cre+ mice with unilateral CRISPR infection did not reduce consumption (Extended Data Fig. 9j). To test whether MOPRs on LDRNPenk terminals function as Penk-sensitive autoreceptors, we expressed the CRISPR virus directly in LDRNPenk neurons. We did not observe deficits in FD sucrose consumption, suggesting that an autoreceptor mechanism is not the primary role of MOPRs in this circuit (Extended Data Fig. 9i-k). We also examined a potential role for local Pdyn or its derivitives27 by locally deleting them in mNAcSh of Pdynfl/fl mice (Extended Data Fig. 9m). This approach did not reduce FD sucrose consumption. Finally, we examined whether there is a direct interaction between postsynaptic mNAcShPenk and presynaptic LDRNMOPR by locally deleting enkephalin in mNAcSh using CRISPR and simultaneously recording GCaMP6s signal in LDRN-mNAcSh terminals during sucrose consumption (Fig. 5gj, Extended Data Fig. 9n-q, Extended Data Table 2). Control mice with intact enkephalin production showed similar consummatory induced inhibition of calcium traces as observed in Fig. 2, whereas mice expressing the CRISPR virus targeting enkephalin production did not (Fig. 5 i, j). Naloxone treatment blunted control inhibitory GCaMP6s traces, but not those in CRISPR-expressing mice. Together, these results indicate that enkephalin production in mNAcSh is necessary for potentiating reward consumption via retrograde action on LDRNMOPR.
In this study, we have isolated an endogenous mechanism underlying MOPR modulation of state-dependent consummatory behaviours in mNAcSh (further discussion is presented in Supplementary Discussion, Extended Data Fig. 10). While the studies described above were designed to investigate neuropeptidergic systems, it is important to consider how GABA (γ-aminobutyric acid) and glutamate additionally contribute to reward potentiation in this circuit. Recent work has shown that local DRNVgat neurons are sensitive to food deprivation21. If this is related to downstream projections to mNAcSh, then it could suggest that MOPRs act on glutamatergic inputs to bias neurotransmission toward inhibition. This hypothesized mechanism is consistent with pharmacological reports28, and is likely to be embedded within other mNAcSh-GABA circuits29. Similarly, MOPRs in mNAcSh are also thought to interact with other neuropeptides, especially those of lateral hypothalamic origin (for example, melanin-concentrating hormone (MCH) or orexin (also known as hypocretin))30,31,32,33 which modulate reward-related behaviours4,34. Similarly, it seems plausible that coincidental stimulation of MOPRs and dopamine signalling could manifest as potentiated motivation, or even pathological compulsive-like behaviors35,36. Investigations into these complementary circuits will be an important continuation of this work.
We also demonstrated that MOPRs are necessary for potentiating avoidance (Extended Data Fig. 2). However, this effect is not driven by the LDRN-mNAcSh circuit described here, suggesting that an alternative “MOPR avoidance” circuit may exist. This multi-regional selectivity would be consistent with current trends in neuropeptide research37-43. Beyond differences in circuit dynamics, an additional consideration for how neuropeptides generate dramatically dissimilar effects is cell-type variations in receptor signaling. While likely a key factor for understanding opioids27,44,45, the ability to causally isolate specific receptor features has been stymied by technological limitations10. Using new technologies27,46-51 for understanding specific peptide/receptor signaling in discrete circuits will be crucial for parsing how widely expressed neuropeptides can mediate complimentary and opposing behaviors.
The results presented in this manuscript reveal new avenues for research on opioid regulation of reward and substance abuse. Future work will necessitate expanding circuit, regional, and peptidergic investigations to include traditional and nontraditional roles of opioids, providing insights for the development of effective therapeutics for neuropsychiatric disorders.
Methods
Animals
Adult (18–35 g) male and female wildtype, preprodynorphin-IRES-Cre (Pdyn-Cre), preproenkephalin-IRES-Cre (Penk-Cre), Oprm1 knockout (Oprm1 KO), Oprm1 conditional knockout (Oprm1fl/fl), Oprm1fl/fl x Penk-Cre, Oprm1fl/fl x Pdyn-Cre, Oprm1 KO x Penk-Cre, proopiomelanocortin-cre (POMC-Cre) and preprodynorphin conditional knockout (Pdynfl/fl) mice were group housed, given access to food pellets and water ad libitum, and maintained on a 12 hr:12 hr light:dark cycle (lights on at 7:00 AM). All mice were kept in a sound-attenuated, isolated holding facility one week prior to surgery, post-surgery, and throughout the duration of the behavioral assays to minimize stress. For cell-type conditional deletion, ablation, chemogenetic, and optogenetic experiments, we used Cre− cage and littermate controls. For Oprm1 KO, Oprm1fl/fl and associated crosses, experimental mice were compared to age-matched wild-type or Cre− littermate controls. Unless otherwise noted, animals had ad libitum access to food and water. Any variation from these approaches was due to behavioral attrition from off-target injections/implants or headcap failures. The mice were bred at Washington University in Saint Louis or the University of Washington. Where needed, Oprm1fl/fl mice were crossed to Ai14-tdTomato mice on C57BL/6 background, bred, and backcrossed for three generations. All animals were drug and test naive, individually assigned to specific experiments as described, and not involved with other experimental procedures. Statistical comparisons did not detect any significant differences between male and female mice, and were therefore combined to complete final group sizes. All animals were monitored for health status daily and before experimentation for the entirety of the study. All procedures were approved by the Animal Care and Use Committee of Washington University, Animal Care and Use Committee of the University of Washington, and conformed to US National Institutes of Health guidelines.
Tissue processing
Unless otherwise stated, animals were transcardially perfused with 0.1 M phosphate-buffered saline (PBS) and then 40 mL 4% paraformaldehyde (PFA). Brains were dissected and post-fixed in 4% PFA overnight and then transferred to 30% sucrose solution for cryoprotection. Brains were sectioned at 30 mM on a microtome and stored in a 0.01M phosphate buffer at 4°C prior to immunohistochemistry and tracing experiments. For behavioral cohorts, viral expression and optical fiber placements were confirmed before inclusion in the presented datasets.
RNAscope Fluorescent In Situ Hybridization
Following rapid decapitation of WT, Oprm1 KO, Oprm1 KO x Penk-Cre, Oprm1fl/fl, Oprm1fl/fl x Penk-Cre, Oprm1fl/fl x Pdyn-Cre, or Penk-Cre mice brains were rapidly frozen in 100mL −50°C isopentane and stored at −80°C. Coronal sections corresponding to the site of interest or injection plane used in the behavioral experiments were cut at 20uM at −20°C and thaw-mounted onto SuperFrost Plus slides (Fisher). Slides were stored at −80°C until further processing. Fluorescent in situ hybridization was performed according to the RNAscope 2.0 Fluorescent Multiple Kit User Manual for Fresh Frozen Tissue (Advanced Cell Diagnostics, Inc.) as described by Wang et al. (2012). Briefly, sections were fixed in 4% PFA, dehydrated, and treated with pretreatment 4 protease solution. Sections were then incubated for target probes for mouse mu opioid receptor (Oprm1, accession number NM_001039652.1, probe region 1135 - 2162), proenkephalin (Penk, accession number NM_001002927.2, probe region 106 – 1332), prodynorphin (Pdyn, accession number NM_018863.3, probe region 33 - 700), tryptophan hydroxylase 2 (tph2, NM_173391.3, probe region 1640 - 2622), vesicular glutamate transporter type 2 (slc17a6, NM_080853.3, probe region 86 - 2998), vesicular GABA transporter (slc32a1, NM_009508.2, probe region 894 - 2037), or Cre (KC845567.1, probe region 1058 – 2032) All target probes except mu consisted of 20 ZZ oligonucleotides and were obtained from Advanced Cell Diagnostics. Following probe hybridization, sections underwent a series of probe signal amplification steps followed by incubation of fluorescently labeled robes designed to target the specific channel associated with the probes. Slides were counterstained with DAPI, and coverslips were mounted with Vectashield Hard Set mounting medium (Vector Laboratories). Images were obtained on an Olympus Fluoview 3000 confocal microscope and analyzed with HALO software. To analyze the images, each image was opened in the HALO software and boundaries were set for the area to be analyzed. DAPI positive cells were then registered and used as markers for individual cells. The maximum area around each cell for probes to be detected was then set, approximately 3 microns. An observer blind to the brain tissue origin and probes used then set thresholds for each channel which determines the minimum intensity of fluorescence for a probe to be counted. These thresholds were validated by manual spot check throughout the image to ensure cells and probes were being appropriately counted. A positive cell consisted of an area within the radius of a DAPI nuclear staining that measured at least 3 positive pixels for receptor probes, or 10 total positive pixels for neurotransmitter probes. The HALO software reported the total counts of cells and levels of overlap, which are reported in the data. Two - three separate slices from the NAc or DRN were used for each animal and that total is presented in the data.
Stereotaxic Surgery
After mice were acclimated to the holding facility for at least seven days, the mice were anaesthetized in an induction chamber (1%-4% isoflurane) and placed into a stereotaxic frame (Kopf Instruments, model 1900) where they were mainlined at 1%-2% isoflurane. For mice receiving viral injections, a blunt needle (86200, Hamilton Company) syringe was used to deliver the vector at a rate of 100 μL/min. The type of virus, injection volume, and stereotaxic coordinates for each experiment are listed in Table S1 interest (NAc medial shell: AP +1.4, ML ± 0.6, DV −4.3; Dorsal raphe nucleus: AP −4.5, ML ± 0.0, DV −2.6). For mice receiving intracranial implants (i.e., cannulas, fiber photometry or optogenetic optic fibers, or GRIN lens), a small hole was drilled above the site of interest (NAc medial shell: AP +1.4, ML ± 0.6, DV −4.3; Dorsal raphe nucleus: AP −4.5, ML ± 0.0, DV −2.6) and the implant was slowly lowered to the coordinates. Cannulas were secured to the skull using one bone screw and super glue (Lang Dental). All other implants were secured using MetaBond (C & B Metabond).
Food Intake Test (FI)
Behaviorally tested mice were habituated to a 26x26cm chamber for three days for 1hr. Mice were given free access to ~0.8g of sucrose pellets during this habituation period. After the final habituation day, mice were either left ad libitum, or food deprived (animals still had access to water). Twenty-four hours later, animals were placed into the food intake chamber for 1hr and had free access to sucrose pellets. Total weight of the sucrose pellets was recorded before and after the test session. After testing, food deprived animals were returned to an ad libitum diet. 48 hours later, animals received the other testing condition (order counter-balanced across cages). Each test session was recorded and behavior scored offline using Ethovision. Wild-type/control mice that did not consume more than 0.01g during the food deprived condition were removed from analysis. Animals that did not lose at least 5% bodyweight after food deprivation were also removed from analysis (animals lost ~11% bodyweight on average). While most mice were tested twice (in either ad libitum or food deprived states), there were a few instances in which mice were tested in each state multiple times due to intersecting manipulations (e.g., multiple drug conditions per state). These unique instances are listed in Extended Data Table 3. We specifically note one unique experiment wherein two groups of 3 mice were tested 3 times for a total of 6 six test days in the mNAcSh CRISPR/LDRN photometry experiment (Fig. i-l). This was done to increase the total number of comparable trials; since the data collected each test day was indistinguishable across test days, we pooled the data together.
Regulated Food Intake Test
Mice were habituated to a Med-Associates operant conditioning box for three days, during which non-contingent deliveries of two sucrose pellets occurred on a VI of 120sec (±30sec) for 30 minutes. On the final habituation day, mice received an i.p. injection of saline to habituate them to the injection process. Mice were then placed back in the home cage for 15 minutes. At the end of 15 minutes, mice were placed into the test apparatus and behavioral testing proceeded. The same procedure was conducted on each test day, for injections of either saline or naloxone (2mg/kg i.p.). Test days were recorded scored offline by an observer blind to the conditions. Drug order and physiological state were counterbalanced across test days and cages for all experiments. Mice that were used in the free intake photometry experiment were also used for the regulated intake experiments.
Regulated Lickometer Test
Mice were habituated to a Med-Associates operant conditioning box for three days, during which non-contingent presentation of a lickometer, filled with 10% sucrose solution, occurred on a VI of 120sec (±30sec) for 30 minutes. On the final habituation day, mice received an i.p. injection of saline to habituate them to the injection process. Mice were then placed back in the home cage for 15 minutes. At the end of 15 minutes, mice were placed into the test apparatus and behavioral testing proceeded. The same procedure was conducted on each test day, for injections of either saline or naloxone (2mg/kg i.p.). For opto-MOR and ChR2 experiments, mice received 20Hz stimulation (473 nm, 10 ms pulse width, ~1-3mW light power) upon the first lick of the lickometer and was continued until the lickometer was retracted. Drug order, laser stimulation, and physiological state were all counterbalanced across test days and cages for all experiments.
Restraint Stress
For Elevated Zero Maze testing (described below), mice were habituated to the testing room for 1 day prior to testing for 1 hour. On the test day, mice were brought to the test room and allowed 30 additional minutes to habituate. After the second habituation, mice were either placed into a 50mL conical tube for 30 minutes (restrained) or left in the home cage (unrestrained). After the restraint period, mice were released from the tube back into the home cage for 30 minutes. Finally mice were tested on the elevated zero maze or food intake experiments, after which they were returned to the home cage.
Elevated Zero Maze (EZM)
The EZM (Harvard Apparatus, Holliston, MA) was made of grey plastic, 200 cm in circumference, comprised of four 50 cm sections (two opened and two closed). The maze was elevated 50 cm above the floor and had a path width of 4 cm with a 0.5 cm lip on each open section. Room lighting was maintained at 4 lux. Mice were positioned head first into a closed arm, and allowed to roam freely for 7 min. For the naloxone experiment, mice were injected prior to the 30 minute restraint. Mean open arm time was the primary measure of anxiety-like behavior.
Drug Microinjections and DREADD Experiments
For drug microinjection experiments, animals were habituated for food intake (described below) one week after cannula implantation. DREADD tested mice were allowed to recover for 4 weeks after surgery before habituation. The day before testing, microinjection animals received an infusion of vehicle (ACSF) to habituate them to the microinjection process. Mice were scruffed and a double 32G microinjector was inserted through the guide cannulas. Animals were then placed into a box in which they could freely walk around during the infusion of vehicle. Mice received a total injection of 200uL per/side over the course of 1 minute. Microinjections were then left in the cannulas for an additional minute to ensure complete diffusion of vehicle. Mice were then placed back in the home cage for 15 minutes. At the end of 15 minutes, mice were placed into the test apparatus and behavioral testing proceeded. The same procedure was conducted on each test day, for injections of either vehicle or the competitive and selective mu opioid receptor antagonist D-Phe-Cys-Tyr-D-Trp-Arg-Thr-Pen-Thr-NH2 (CTAP (1ug/1uL)). On the final day of habituation, DREADD mice received an i.p. injection of saline to habituate them to the injection process. Mice were then placed back in the home cage for 15 minutes. At the end of 15 minutes, mice were placed into the test apparatus and behavioral testing proceeded. The same procedure was conducted on each test day, for injections of either saline or clozapine-N-oxide (CNO, 3mg/kg i.p.) Drug order and physiological state was counterbalanced across test days and cages all experiments.
Patch-Clamp Electrophysiology
Mice (n = 5; 4-6 months; 2 males) were anesthetized with pentobarbital (50 mg/kg) before transcardial perfusion with ice-cold sucrose cutting solution containing the following (in mM): 75 sucrose, 87 NaCl, 1.25 NaH2P04, 7 MgCl2, 0.5 CaCl2, 25 NaHCO3, 306-308 mOsm. Brains were then rapidly removed, and coronal sections 300 μm thick were taken using a vibratome (Leica, VT 1200). Sections were then incubated in aCSF (32°C) containing the following (in mM): 126 NaCl, 2.5 KCl, 1.2 NaH2P04, 1.2 MgCl2, 2.4 CaCl2, 26 NaHCO3, 15 glucose, 305 mOsm.. After an hour of recovery, slices were constantly perfused with aCSF (32°C) and visualized using differential interference contrast through a 40x water-immersion objective mounted on an upright microscope (Olympus BX51WI). Whole-cell patch-clamp recordings were obtained using borosilicate pipettes (4–5.5 MΩ) back-filled with internal solution containing the following (in mM): 117 Cs-Methanesulfonate, 20 HEPES, 0.4 EGTA, 2.8 NaCl, 5 TEA, 5 ATP, and 0.5 GTP (pH 7.35, 285 mOsm). To assess connectivity between DRNPenk and NACsh, voltage clamp recordings were performed from cells located near eYFP-expressing axons within the NAC. 5 ms blue light pulses were delivered through the objective while holding each cell at −70 mV and +10 mV to assess glutamatergic and GABAergic input, respectively. During voltage clamp recordings, TTX (1 μM) and 4-AP (1 mM) (Sigma) were applied to the bath, and then the AMPA/kainate receptor antagonist, DNQX (10 μM), or the GABAA antagonist, gabazine (10 μM), were applied to test for glutamate and GABA mediated currents, respectively. Data acquisition occurred at 10 kHz sampling rate through a MultiClamp 700B amplifier connected to a Digidata 1440A digitizer (Molecular Devices). Data were processed using Clampfit v11.0.3.03 (Molecular Devices) and analyzed using GraphPad Prism v8.3.0. All tests were two-sided and corrected for multiple comparisons or unequal variance where appropriate.
In Vivo Fiber Photometry
Fiber photometry recordings were made throughout the entirety of 60-minute Food Intake, 30-minute Regulated Food Intake, and 30-minute Lickometer Tests. Prior to recording, an optic fiber was attached to the implanted fiber using a ferrule sleeve (Doric, ZR_2.5). Two LEDs were used to excite GCaMP6s. A 531-Hz sinusoidal LED light (Thorlabs, LED light: M470F3; LED driver: DC4104) was bandpass filtered (470 ± 20 nm, Doric, FMC4) to excite GCaMP6s and evoke Ca2+-dependent emission. A 211-Hz sinusoidal LED light (Thorlabs, LED light: M405FP1; LED driver: DC4104) was bandpass filtered (405 ± 10 nm, Doric, FMC4) to excite GCaMP6s and evoke Ca2+-independent isosbestic control emission. Prior to recording, a 120 s period of GCaMP6s excitation with 405 nm and 470 nm light was used to remove the majority of baseline drift. Laser intensity for the 470 nm and 405 nm wavelength bands were measured at the tip of the optic fiber and adjusted to ~50 μW before each day of recording. GCaMP6s fluorescence traveled through the same optic fiber before being bandpass filtered (525 ± 25 nm, Doric, FMC4), transduced by a femtowatt silicon photoreceiver (Newport, 2151) and recorded by a real-time processor (TDT, RZ5P). The envelopes of the 531-Hz and 211-Hz signals were extracted in real-time by the TDT program Synapse at a sampling rate of 1017.25 Hz.
Photometry Analysis
Custom MATLAB scripts were developed for analyzing fiber photometry data in context of mouse behavior and can be accessed via GitHub (https://github.com/BruchasLab). The isosbestic 405 nm excitation control signal was subtracted from the 470 nm excitation signal to remove movement artifacts from intracellular Ca2+-dependent GCaMP6s fluorescence (see Figure S3B). Baseline drift was evident in the signal due to slow photobleaching artifacts, particularly during the first several minutes of each hour-long recording session. A double exponential curve was fit to the raw trace and subtracted to correct for baseline drift. After baseline correction, the photometry trace was z-scored relative to the mean and standard deviation of the test session. The post-processed fiber photometry signal was analyzed in the context of animal behavior during food intake and lickometer tests.
Tail-immersion Test
Tail-immersion tests52 were performed by submerging 2-4cm of the mouse tail into hot water (54°C) and recording the total time between tail immersion and withdrawal. The cut-off was 10sec after immersion in order to prevent permanent damage to the tail tissue. After the initial immersion response (time 0), mice were injected with morphine (5mg/kg, s.c.) and tested every 10 minutes for 1 hour, then again at 90 minutes.
Morphine Conditioned Place Preference (CPP)
Mice were trained in an unbiased, balanced three compartment conditioning apparatus as previously described (McCall et al., 2015). Briefly, mice were pre-tested by placing individual animals in the small central compartment and allowing them to explore the entire apparatus for 30 min. Time spent in each compartment was recorded with a video camera (ZR90; Canon, Tokyo, Japan) and analyzed using Ethovision 11 (Noldus). The drug paired side was randomly assigned to the mice. For the three conditioning days, mice received a subcutaneous injection of saline in the morning, and six hours later received an injection of morphine (5mg/kg, s.c.). CPP was assessed on day 5 by allowing the mice to roam freely in all three compartments and recording the time spent in each. The difference in preference for context A and B from the posttest to the pretest was used as the ultimate preference score.
In vivo Ca2+ Imaging and Behavior
Mice were habituated to the food intake chamber for 3 days. During these habituation sessions, they were also mounted with an Inscopix miniature microscope (nVista) that was attached to a commutator to prevent the cord from tangling. Prior to the first test day, scope focus, power, and gain were optimized individually for each mouse. These settings were then kept constant for each test day. During test days, mice were allowed to freely consume sucrose pellets for 30 minutes, during which GCaMP6s fluorescence was recorded. When the test day was over, mice were returned to the home cage.
In vivo Ca2+ Imaging Data Processing
Inscopix data acquisition software (IDAS; Inscopix) was used to acquire TIFF images of fluorescence dynamics at 20 frames per second. Calcium frame acquisition was triggered via Ethovision XT (v10) and began at the onset of the session. Calcium acquisition was automatically terminated after 30 minutes.
Inscopix data processing software (IDPS; Inscopix) was used to preprocess Ca2+ data from each imaging session as previously described 53,54. Briefly, for each day, calcium imaging data from ad libitum and food deprived test days were down-sampled temporally (2x temporal bin) and spatially (4x spatial bin) and rigid motion correction was applied. After preprocessing, putative single neuron activity was segmented using Constrained Non-negative Matrix Factorization for Endoscopic data (CNMFe) using custom MATLAB (MathWorks, Natick, MA, USA) scripts (https://github.com/BruchasLab). Individual putative striatal neurons were tracked between ad libitum and food deprived sessions using CellReg (cite PMID: 29069591). For each mouse the spatial correlation registration threshold performed best, and thresholds were determined by the algorithm. Final registration utilized the probabilistic model with a P_same (probability of cells being the same) > 0.6 for all mice.
In Vivo Ca2+ Imaging Data Analysis
Raw fluorescence traces for individual putative neurons were Z-normalized using the mean and standard deviation of the entire imaging session. Individual neurons were grouped together based on similar activity during approach and consumption of sucrose rewards using a standard k-means clustering approach. The optimal number of clusters was determined by silhouette analysis. Discrete timestamps for the onset of sucrose consumption, sniffing and rearing were manually scored from simultaneously recorded behavioral videos. The mean activity of neuronal clusters was then plotted relative to the onset of behavioral events. Normalization and analysis of individual neuron calcium dynamics was performed in MatLab using custom scripts (https://github.com/BruchasLab).
Generation and validation of AAV1-FLEX-SaCas9-U6-sgPenk
The sgRNA targeted to the Penk locus (sgPenk) was designed as previously described (Hunker et al., 2020). The following oligos (Sigma) were used to clone into pAAV-FLEX-SaCas9-U6-sgRNA (Addgene Cat#: 124844). Penk forward: CACCGTTTGCACCTGGCTGCTGGCGC; Penk reverse: AAACGCGCCAGCAGCCAGGTGCAAAC.
Targeted deep sequencing of Penk locus
Nuclei isolation, FACS, and targeted deep sequencing were performed as described previously (Hunker et al., 2020). Tissue punches of the ventral striatum from 4 mice co-injected with AAV1-FLEX-SaCas9-U6-sgPenk and AAV1-FLEX-EGFP-KASH were pooled into a single group and homogenized in 2mL of homogenization buffer containing (in mM): 320 Sucrose (sterile filtered), 5 CaCl (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 EDTA pH 8 (sterile filtered), 0.1% NP40, 0.1 Protease Inhibitor Cocktail (PIC, Sigma Cat#: P8340), 1 β-mercaptoethanol. Homogenization was performed using 2mL glass dounces (Sigma Cat#: D8938-1SET); 25 times with pestle A, then 25 times with pestle B. The volume of the homogenate was transferred to a 15mL conical tube and brought up to 5mL using homogenization buffer, mixed by inversion, and incubated on ice for 5 minutes. 5mL of 50% Optiprep density gradient medium (Sigma Cat#: D1556-250ML) containing (in mM): 5 CaCl (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 PIC, 1 β-mercaptoethanol was added to the homogenate and mixed by inversion. The mixture was gently loaded on 10mL of 29% iso-osmolar Optiprep solution in a 1x3 ½ in Beckman centrifuge tube (SW32 Ti rotor) and spun at 7500 RPM for 30min at 4°C. The floating cell debris was removed using a KimWipe and the supernatant was gently poured out. The nuclei pellet was vigorously resuspended in sterile 1xPBS. 500 GFP-positive and 500 GFP-negative nuclei were sorted directly into 3uL of REPLI-g Advanced Storage buffer (Qiagen Cat#: 150365) in an PCR tube strip (Genessee Cat #: 24-706) using a BD AriaFACS III. Whole genome amplification (WGA) was performed directly following FACS using the REPLI-g Advanced DNA Single Cell kit (Qiagen Cat#: 150365) according to manufacturer’s instructions.
For generation of the specific amplicons, 1ul of WGA DNA was diluted 1:50 and amplified (PCR 1) with Phusion High Fidelity Polymerase (Thermo Fisher Cat#: F530L) using the following thermocycler protocol: initial denaturation (30sec, 95°C); denaturation (10sec, 95°C); annealing (20sec, 66°C); extension (10sec, 72°C); cycle repeated x34; final extension (5min, 72°C). PCR forward: GCTCAGGAAAGACTGTCC, PCR reverse: TGACCACTAGAAGTCTGC.
1uL of PCR 1 was amplified again (PCR 2) with the same set of primers using the same thermocycler protocol. The 310bp amplicon from PCR 2 was gel extracted using the MinElute gel extraction kit (Qiagen Cat#: 28606) and sent to Genewiz for Amplicon-EZ targeted deep sequencing and Sanger sequencing. Reads received from Amplicon-EZ were trimmed in Excel up to the sgRNA and PAM sequence to avoid false mutational reads due to PCR error. The number of unique reads containing specific insertions, deletions, and base changes within the targeted region were then summed in Excel.
Statistical analyses
All data collected were averaged and expressed as mean ± SEM. Statistical significance was taken as *p < 0.05, **p < 0.01, and ***p < 0.001, as determined by Pearson’s correlation, Student’s t test, one-way ANOVA or a two-way repeated-measures ANOVA followed by Sidak post hoc tests as appropriate. For electrophysiology data, we used Student’s t test. For photometry experiments, we used Pearson’s correlation and Student’s t tests, as appropriate. For microinjection, genetic deletion, photometry, 1-photon, ablation, chemogenetic, and optogenetic behavioral experiments, we used one-way or two-way repeated-measures ANOVA followed by a Tukey or Sidak post hoc tests. All n values for each experimental group are described in the appropriate figure legend. For behavioral experiments, group size ranged from n = 3 to n = 15. For in situ hybridization quantification experiments, slices were collected from 2-3 mice, with data averaged from 2-3 slices per mouse. For electrophysiology experiments, the number of cells recorded were as follows: n = 107 total recorded cells, 8 cells that showed oEPSPs, and 5 cells that showed oIPSPs. Statistical analyses were performed in GraphPad Prism 8.0 (Graphpad, La Jolla, CA) and MATLAB 9.6 (The MathWorks, Natick, MA).
Data Availability
The behavioral dataset supporting the current study are available as Source Data or from the author upon request.
Code Availability
Custom MATLAB analysis and code was created to appropriately organize, process, and combine photometry and single-photon recording data with associated behavioral data. Analysis code for photometry and single-photon imaging from Figures 2 and 4 will be made available on Github (https://github.com/BruchasLab).
Extended Data
Extended Data Figure 1. Endogenous MOPR activation in mNAcSh is necessary for potentiating state-dependent consummatory behavior.
a. Placement map of each microinjector tip (blue = intake suppression compared to vehicle-deprived, green = intake enhancement compared to vehicle-deprived). b. Schematic of vehicle (ACSF, gray, top) or drug (CTAP, blue, bottom) microinjections into areas surrounding nucleus accumbens (NAc) medial shell. c. CTAP (blue) had no effect on ad libitum or hunger enhanced intake compared to vehicle (gray) control days when injections were outside NAc medial shell (n = 8). d. In situ hybridization of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200μm). e and f. Quantification of MOPR expression in mNAcSh. g. Schematic of Oprm1fl/fl x Pdyn-Cre mouse line cross. h and i. In situ hybridization (h) and quantification (i) of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200 μm) in Oprm1fl/fl x Pdyn-Cre mouse line. j. Loss of MOPRs on Pdyn-Cre+ neurons did not disrupt normal ad libitum or food deprived enhanced intake compared to Pdyn-Cre− littermate control mice (n = 9 Cre−, 10 Cre+). k and l. In situ hybridization (k) and quantification (l) of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200 μm) in Oprm1fl/fl x Penk-Cre mouse line. m. Schematic and image of rAAV5-CMV-Cre-GFP injections into NAc medial shell of Oprm1fl/fl mice. n. Schematic of combined viral spread map of local MOPR deletion. o. Schematic (top) and image (bottom) of AAV2retro-CMV-myc-NLS-Cre or AAV2retro-GFP-Cre injections into NAc (left); retrogradely labeled cells in dorsal raphe nucleus (right). All error bars represent ± SEM and n = biologically independent mice or cells (f, i, l). Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, j).
Extended Data Figure 2. Endogenous MOPR activation in mNAcSh is necessary for potentiating state-dependent avoidance behavior.
a. Schematic of elevated zero maze (EZM) test. Mice were tested after habituation to the test room (Unrestrained) or after 30 minutes of restraint stress (Restraint). b. Example heat plots of time spend in the open arms of the EZM in wildtype (WT, left) or Oprm1 KO (KO, right) after no restraint (top) or 30 minutes of restraint (bottom). c. Unrestrained WT mice spent ~30% of the EZM test in the open arms. Mice exposed to restraint stress significantly reduced their exploration to 10%. Pretreatment with naloxone prevented Restraint induced avoidance. Oprm1 KO mice did not display open arm avoidance after Restraint. Penk-Cre x cKO mice displayed normal avoidance of the open arms after Restraint (n = 9 WT Unrestrained, 9 WT Restrained, 8 Oprm1 KO Unrestrained, 7 Oprm1 KO Restrained, 8 Oprm1fl/fl x Penk-Cre− Restrained, 9 Oprm1fl/fl x Penk-Cre+ Restrained). d. Schematic of food intake assay after food deprivation or Restraint. e. Food deprived mice showed normal increase in intake relative to their ad libitum test day. Mice exposed to Restraint did not increase food intake relative to their Unrestrained test day (n = 11 deprived, 10 Restrained). All error bars represent ± SEM and n = biologically independent mice. Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, h).
Extended Data Figure 3. LDRNPenk-NAc projections express MOPRs.
a. Retrograde fluorescently tagged cells in amygdala of Penk-Cre+ mouse after injections into nucleus accumbens medial shell. b. (left) In situ hybridization of Oprm1, Slc32a1 and Slc17a6 in DRN (scale bar = 200 μm). Zoomed in and channel separated images (right) of the red square in the left panel. c. Quantification of in situ from panel b. d and e. Schematic and image of a local injection of AAV2retro-GFP-Cre into nucleus accumbens shell (scale bar = 200 μm). Zoomed in images of e are designated as red (i) and yellow (ii) boxes. f. (left) In situ hybridization of Oprm1, Penk and Cre in dorsal raphe nucleus (scale bar = 200 μm). Zoomed in and channel separated images (right) of the red square in the left panel. g. Quantification of in situ from panel f. h. Schematic of CTb experiment. i. Fluorescently tagged CTb was injected into mNAcSh (scale bar = 200 μm). j. CTb tagged cells were observed in dorsal raphe nucleus, including the lateral sites in which enkephalin neurons were labeled in Fig. 2a (scale bar = 200 μm). Red square shows zoomed in image (right) with labeled cells (depicted by white arrows). All error bars represent ± SEM and n = biologically independent cells.
Extended Data Figure 4. Non-DRN sites do not mediate food deprived potentiation of sucrose consumption.
a. Local MOPR deletion in paraventricular thalamus of Oprm1fl/fl mice did not reduce food deprived enhanced intake (n = 8, paired t-test t(7) = 9.634, p < 0.001). b. Schematic of local caspase ablations in either ventral pallidum or basomedial amygdala. c. Local and cell-type specific ablation of enkephalin neurons in VP or BMA did not reduce food deprived enhanced intake relative to Cre− control mice (n = 8 Cre−, 9 VP, 7 BMA). d. Caspase injection site confirmation in ventral pallidum (scale bar = 200 μm). AAV2-FLEX-taCas3-TEPp and AAV5-hsyn-EYFP were coinfused for cell-type specific deletion, and non-specific labeling. e and f. Schematic and image of a local injection of AAV5-Ef1a-DIO-EYFP into DRNPenk (scale bar = 200 μm). g. Image of dorsal raphe projection fibers from e (scale bar = 200 μm). h. oEPSC amplitude was reduced by the application of DNQX (n = 8). Blue shaded region indicates duration of optical stimulation. i. oIPSC amplitude was reduced by the application of gabazine (n = 5). All error bars represent ± SEM and n = biologically independent mice or cells (h, i). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c) or two tailed paired t-test (h, i).
Extended Data Figure 5. LDRNMOPR-mNAcSh projection activity is negatively modulated by sucrose consumption in an opioid receptor dependent manner.
a. Schematic of photometry and voluntary sucrose consumption paradigm. b. Expression of GCaMP6s in DRNPenk (left, scale bar = 200 μm) and fiber placement in mNAcSh (scale bar = 200 μm). c. FD increases food intake (green) relative to ad libitum intake (white), and is reduced by systemic naloxone (blue, n = 8). d. Eating microstructure across ad libitum/saline (white), food deprived/saline (green) and food deprived/naloxone (blue) test days. After food deprivation, the majority of sucrose pellets consumed during multi-pellet bouts (left), and more pellets are eaten per multi-pellet bout (right). This shift in eating behavior is blunted by naloxone. e. Example of raw 405nm and 470nm channels from photometry experiments. f. Example of raw df/f trace highlighting specific single pellet (black) or multi-pellet (green) intake events during food deprived test day. Colored boxes on top expand color matched portions of the trace below. Orange lines indicate onset of pellet consumption. g. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh terminals in FD/saline condition (green) and FD/naloxone condition (blue).
h. Average Z-scored trace (dark line) and error in on the ad libitum test day. When GCaMP activity is aligned to the onset of each pellet eaten in the ad libitum condition, no significant deviation in activity is observed (black, top). When aligned to only multi-pellet bout onset, there is no deviation from baseline activity (brown, bottom). i. Schematic of regulated food intake paradigm. Two pellets were non-contingently delivered every 90-150 seconds for 30 minutes. Mice were tested in either FD/saline or FD/naloxone conditions. j. Total number of sucrose pellets eaten in the regulated intake paradigm. Mice ate significantly more pellets on the food deprivation/saline test day (green), which was reduced to baseline levels after systemic naloxone (blue) (n = 10) k. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh. Food deprived/saline trace (green) shows rapid and sustained inhibition. Food deprived/naloxone trace (blue) shows blunted response. l. Heatmap of individual trials across all tested mice in food deprived/saline (green) or food deprived/naloxone test days. Orange lines indicate onset of pellet consumption. m. Quantification of the average Z-score twenty seconds prior to the onset of multi-pellet pellet bouts versus twenty seconds after the onset. FD/saline traces (green) show significant reductions in GCaMP6s activity whereas FD/naloxone traces (blue) do not (n = 10). All error bars represent ± SEM and n = biologically independent mice. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (f, i).
Extended Data Figure 6. MOPR activation on LDRNMOPR-mNAcSh is sufficient to enhance consummatory behavior.
a. Schematic of the Tail Immersion Test. Mice were tested at time 0, were injected with morphine (5mg/kg, s.c.), then tested every 10 min for up to 60 min, then again at 90 min. b. WT, Oprm1 KO, and Oprm1 KO x Penk-Cre rescue mice all show similar baseline responses at time 0. After morphine administration, WT mice significantly increase their latency to flick their tail, whereas Oprm1 KO and Oprm1 KO x Penk-Cre show no analgesic response to morphine mice (n = 5 WT, 2 Oprm1 KO, 6 Oprm1 KO x Penk-Cre rescue; WT/T0 vs Oprm1 KO/T0 p = 0.116, WT/T0 vs Oprm1 KO x Penk-Cre rescue/T0 p = 0.063, WT/T10 vs Oprm1 KO/T10 p < 0.001, WT/T10 vs Oprm1 KO x Penk-Cre rescue/T10 p < 0.001, WT/T90 vs Oprm1 KO/T90 p < 0.001, WT/T90 vs Oprm1 KO x Penk-Cre rescue/T90 p < 0.001). Statistical differences between WT and Oprm1 KO designated as (***) and differences between WT and Oprm1 KO x Penk-Cre rescue designated as (+++). c. Raster plots of individual licking events for one mouse, separated by trial. Only trials in which mice licked were included. d. Schematic of ChR2 experiments. e. Expression of EYFP-tagged ChR2 in LDRNPenk cell bodies (left, scale bar = 200 μm) and fiber placement in mNAcSh (right, scale bar = 200 μm). f. Penk-Cre− and Penk-Cre+ mice licked similar amounts for a sucrose solution in the ad libitum/No Laser condition. ChR2 photo-stimulation did not reduce licking (n = 7 Cre−, 6 Cre+). g. Penk-Cre− and Penk-Cre+ mice licked similar amounts for a sucrose solution in the WD/No Laser condition. By contrast, ChR2 photo-stimulation significantly reduced Cre+ licking, but not Cre− licking. h. Raster plots of individual licking events for one Penk-Cre+ mouse in the WD condition, separated by trial. Only trials in which mice licked were included. ChR2 photo-stimulation disrupted lick bout behavior compared to No Laser test days. All error bars represent ± SEM and n = biologically independent mice or cells (a). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, f, g).
Extended Data Figure 7. mNAcSh enkephalinergic ensembles are modulated by physiological state and potentiate consummatory behavior.
a. Sucrose consumed during ad libitum and food deprived test days (n = 5). Miniscope headmount did not disrupt normal intake behaviors. b. Examples of individual cell traces aligned to initiation of a multi-pellet bout. c. Average trace of all tracked cells aligned to bout consumption on food deprived test day. d. TSNE plot of clusters for multi-pellet bouts during the ad libitum state. e. Average trace of Onset activated neurons (cluster 1). f. Average trace of Pre-onset activated neurons (cluster 2). g. Total proportion and overlap of enkephalin neuron subpopulations modulated by multi-pellet bouts (pink), food sniffs (orange), rearing (blue), and grooming (brown). h. TSNE (left) and mean Z-scored traces (right) of food sniffing behavior sorted by kmeans clustering. i. TSNE (left) and mean Z-scored traces (right) of rearing behavior sorted by kmeans clustering. j. TSNE (left) and mean Z-scored traces (right) of grooming behavior sorted by kmeans clustering. All error bars represent ± SEM (a) or SEM is represented by the shaded region surrounding the trace (c, e, f, h, i, j).
Extended Data Figure 8. Modulation of mNAcSh or POMC-containing neurons during sucrose consumption.
a. Schematic of hM3D(Gi) (left) and hM3D(Gq) (right) DREADD experiments. b. Fluorescent micrograph of mCherry-tagged enkephalin cells in mNAcSh for hM3D(Gi) (left) and hM3D(Gq) (right) experiments (scale bar = 200μm). c. CNO injections suppressed hunger enhanced intake in Cre+ mice, but had no effect in Cre− mice (n = 15 Cre−, 12 Cre+). d. CNO injections increased intake above the already elevated food deprived intake in Penk-Cre+ mice, but had no effect in Penk-Cre− mice (n = 7 Cre−, 10 Cre+). e. Systemic CNO administration (3mg/kg, i.p.) suppressed the already low ad libitum intake in Penk-Cre+ mice, but did not reduce intake in Penk-Cre− mice. f. Systemic CNO administration (3mg/kg, i.p.) had no effect on ad libitum intake in Penk-Cre− or Penk-Cre+ mice). g. Gq or Gi DREADD injections into arcuate nucleus of POMC-Cre mice. h. Micropictograph of mCherry-tagged, DREADD-expressing cells in arcuate nucleus (scale bar = 200 μm). i-k. Neither Gi nor Gq stimulation had an effect on ad libitum or food deprived intake in Cre− or Cre+ mice (n = 7 Cre−, 7 Gq, 9 Gi). All error bars represent ± SEM and n = biologically independent mice. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, d, e, f, i, j, k).
Extended Data Figure 9. Endogenous mu-opioid peptide within mNAcSh is necessary for potentiating consummatory behavior.
a. FISH (scale bar = 200 μm) of mNAcSh caspase injections. b. FISH quantification of (a). c. FISH (scale bar = 200 μm) of mNAcSh CRISPR injections. d. FISH quantification of (c). e. Schematic of CRISPR virus development and validation. f. Sequencing of GFP+ nuclei: (Top) sgPenk sequence with PAM underlined and SaCas9 cut site indicated by black arrow. (Middle) Sanger sequencing results displaying multiple peaks beginning at the SaCas9 predicted cut site. (Bottom) Top ten mutations at cut site with the percent of occurrence on the left. Insertions: underlined. Deletions: marked with “−“. Affected sites after SaCas9 insertion: shaded brown. g. Percent of wild type (black), deletions (brown), insertions (pink), and base changes (white) as percent of total reads for GFP+ and GFP− nuclei. h. Frequency distribution of insertions (pink) and deletions (brown) for Penk from GFP+ nuclei. i. Sanger sequencing results displaying no unusual peaks beginning at the SaCas9 predicted cut site. j. Unilateral hits or bilateral misses of NAc medial shell with the CRISPR mediated deletion of enkephalin did not reduce food deprived enhanced intake (n = 4). k. Fluorescent micrograph of CRISPR virus expression in dorsal raphe nucleus. l. Deletion of enkephalin from dorsal raphe nucleus did not reduce food deprived enhanced intake (n = 8) m. Deletion of dynorphin in NAc medial shell did not reduce food deprived enhanced intake (n = 7). n. Heatmap of individual trials across all tested mice in CRISPR/saline (orange) or Control/saline (gray) test days. Orange lines indicate onset of pellet consumption. o. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh after systemic naloxone injections. Control-treated mice (blue) show blunted inhibition. CRISPR-expressing trace (brown) shows negligible and phasic inhibition. p. Heatmap of individual trials across all tested mice in CRISPR/naloxone (orange) or Control/naloxone (blue) test days. q. Quantification of the average Z-score twenty seconds prior to the onset of multi-pellet pellet bouts versus twenty seconds after the onset. Neither Control traces (blue) nor CRISPR traces (brown) show significant deviations in GCaMP activity. Some mice did not lick on naloxone treated days and were therefore not included in this analysis (CRIPSR n = 3, Control n = 3). All error bars represent ± SEM and n = biologically independent mice or cells (b, d). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (q) or two-tailed paired t-test (j, l, m).
Extended Data Figure 10. A LDRNMOPR-mNAcShPenk circuit mediates potentiation of consummatory behaviors.
Schematic of voluntary sucrose consumption task (upper left) and LDRN-mNAcSh projection (upper right). Effects of LDRNMOPR-mNAcShPenk manipulations on behavior (lower left) and schematic of hypothesized physiology (lower right).
Extended Data Table 1.
Key Resources
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken-anti-GFP 1:500 | Abcam | Ab13970 |
| Anti-Chicken 488 1:500 | Abcam | Ab150169 |
| Bacterial and Virus Strains | ||
| rAAV5-CMV-Cre-GFP | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV2retro-CMV-myc-NLS-Cre | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV2retro-GFP-Cre | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV2retro-EF1α-DIO-EYFP | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV5-EF1α-ChR2-EYFP | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV5-DJ-EF1α-DIO-GCAMP6s | Stanford University Gene Vector and Viral Core | N/A |
| AAV5-EF1α-DIO-rMOR-GFP | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV5-EF1α-DIO-oMOR-EYFP | The Hope Center Viral Core – Washington University at St. Louis | N/A |
| AAV5-hsyn-DIO-HM4D(Gq)-mCherry | Addgene | N/A |
| AAV5-hsyn-DIO-HM3D(Gi)-mCherry | addgene | N/A |
| AAV2-FLEX-taCas3-TEVp | UNC Vector Core | N/A |
| AAV1-CMB-FLEX-Sa-Cas9U6-sgPenk | This Manuscript | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CTAP | Tocris | CAT#1560; CAS: 103429-32-9 |
| VECTASHIELD Hardset Antifade Mounting Medium | Vector Laboratories | CAT#H-1400 |
| VECTASHIELD Hardset Antifade Mounting Medium with DAPI | Vector Laboratories | CAT#H-1800 |
| Clozapine-N-Oxide | Enzo Sciences | CAT#BML-NS105-0025; CAS: 34233-69-7 |
| Naloxone hydrocholoride | Tocris | CAT#0599; CAS: 357-08-4 |
| Morphine | Tocris | CAT#5158 ; CAS: 52-26-6 |
| Critical Commercial Assays | ||
| RNAscope Fluorescent Multiplex Kit 2.0 | Advanced Cell Diagnostics | CAT#320850 |
| Mm-OPRM1 | Advanced Cell Diagnostics | CAT#315841 |
| Mm-Pdyn | Advanced Cell Diagnostics | CAT#318771 |
| Mm-Penk | Advanced Cell Diagnostics | CAT#318761 |
| Mm-TPH2 | Advanced Cell Diagnostics | CAT#318691 |
| Mm-Slc17a6 | Advanced Cell Diagnostics | CAT#319171 |
| Mm-Slc32a1 | Advanced Cell Diagnostics | CAT#319191 |
| Mm-Cre | Advanced Cell Diagnostics | CAT#312281 |
| Experimental Models: Organisms/Strains | ||
| Oprm1 KO | Jackson Laboratories | Stock No: 007559 |
| Oprm1fl/fl | (99) | Stock No: 030074 |
| Pdyn-IRES-Cre | Gift from Dr. Richard Palmiter | Stock No: 027958 |
| Penk-IRES-Cre | (101) | Stock No: 025112 |
| Pdyn-Cre x Oprm1fl/fl | This paper | N/A |
| Penk-Cre x Oprm1fl/fl | This paper | N/A |
| Oprm1 KO x Penk-Cre | This paper | N/A |
| POMC-Cre | Jackson Laboratories | Stock No: 005965 |
| Pdyn-Crefl/fl | Bred in house | Gift from Charley Chavkin |
| Software and Algorithms | ||
| FIJI/ImageJ | NIH | https://fiji.sc/ |
| MATLAB 2020b | Mathworks | https://www.mathworks.com/products.html |
| Med-PC V Software Suite | Med-Associates Inc. | https://www.med-associates.com/med-pc-v/ |
| Ethovision 10 | Noldus | https://www.noldus.com/ethovision-xt |
| Synapse 95-44132P | Tucker-Davis Technologies | https://www.tdt.com/files/manuals/SynapseManual.pdf |
| PRISM 8 | Graphpad | https://www.graphpad.com/ |
| Photometry analysis code | Parker et al. | https://github.com/BruchasLab |
| Clampfit v11.0.3.03 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| Olympus Fluoview 3000 v2.4.1.198 | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Illustrator CS6 | Adobe | https://www.adobe.com/products/illustrator.html |
| HALO v3.2.1851.229 | Indica Labs | https://indicalab.com/halo/ |
| nVista v3.0 | Inscopix | https://www.inscopix.com/nVista |
Extended Data Table 2.
Overview of Mouse Lines and Experiments
| Mouse Line | Behavior | Approach | Virus |
|---|---|---|---|
| Oprm1 KO | Sucrose Consumption (Figure 1) | N/A | N/A |
| Sucrose Consumption (Figure 3) | Viral Injection | AAV5-EF1α-DIO-rMOR-GFP | |
| Morphine CPP (Figure 3) | Viral Injection | AAV5-EF1α-DIO-rMOR-GFP | |
| Oprm1 KO x Penk-Cre | Sucrose Consumption (Figure 3) | Viral Injection | AAV5-EF1α-DIO-rMOR-GFP |
| Oprm1fl/fl | Sucrose Consumption (Figure 1) | Viral Injection Viral Injection Viral Injection |
rAAV5-CMV-Cre-GFP AAV2retro-GFP-Cre AAV2retro-CMV-myc-NLS-Cre |
| Pdyn-Cre x Oprm1fl/fl | Sucrose Consumption (Figure 1) | Genetic Cross | N/A |
| Penk-Cre x Oprm1fl/fl | Sucrose Consumption (Figure 1) | Genetic Cross | N/A |
| Penk-IRES-Cre | Anatomical Tracing (Figure 2) | Viral Injection | AAV2retro-EF1α-DIO-EYFP |
| Sucrose Solution Consumption (Figure 2) | Viral Injection, Fiber Photometry | AAV5-DJ-EF1α-DIO-GCAMP6s | |
| Sucrose Solution Consumption (Figure 3) | Viral Injection, Optogenetics | AAV5-EF1α-DIO-oMOR-EYFP | |
| Sucrose Consumption (Figure 4) | Viral Injection, 1-Photon Imaging | AAV5-DJ-EF1α-DIO-GCAMP6s | |
| Sucrose Consumption (Figure 5) | Viral Injection, Caspase | AAV2-FLEX-taCas3-TEVp | |
| Sucrose Consumption (Figure 5) | Viral Injection, CRISPR | AAV1-CMB-FLEX-Sa-Cas9U6-sgPenk | |
| Sucrose Pellet Consumption (Figure 5) | Viral Injection, Fiber Photometry | AAV5-DJ-EF1α-DIO-GCAMP6s AAV1-CMB-FLEX-Sa-Cas9U6-sgPenk |
Extended Data Table 3.
Total Number of Test Days per Behavior Test
| Figure | Behavior | Manipulation | # of Test Days per Mouse |
|---|---|---|---|
| 1b | Sucrose Consumption | Drug Microinjection | 4 |
| 1c | Sucrose Consumption | Genetic Deletion | 2 |
| 1e | Sucrose Consumption | Genetic Deletion | 2 |
| 1f | Sucrose Consumption | Genetic Deletion | 2 |
| 2d-g | Regulated Sucrose Solution | Systemic Drug Injection | 3 |
| 3d | Sucrose Consumption | Genetic Rescue | 2 |
| 3e | Place Preference | Systemic Drug Injection | 1 |
| 3h | Regulated Sucrose Solution | Optogenetic Stimulation/Systemic Drug Injection | 4 |
| 4 | Sucrose Consumption | 1—Photon imaging | 2 |
| 5c | Sucrose Consumption | Caspase Deletion | 2 |
| 5f | Sucrose Consumption | CRISPR Deletion | 2 |
| 5g-j | Regulated Sucrose Consumption | CRISPR Deletion/Systemic Drug Injection | 6 |
Extended Data Table 4.
Main Figure Exact Statistics
| Figure | Panel | Comparison | p-value |
|---|---|---|---|
| 1 | b | Ad Lib/Vehicle vs. FD/Vehicle FD/Vehicle vs. FD/CTAP Ad Lib/CTAP vs. FD/CTAP |
0.000014 0.000926 0.006 |
| c | Ad Lib/WT vs. FD/WT FD/WT vs. FD/Oprm1 KO Ad Lib/Oprm1 KO vs. FD/Oprm1 KO |
0.000000000037 0.000015 0.144 |
|
| e | Ad Lib/Cre− vs. FD/Cre− FD/Cre− vs. FD/Cre+ Ad Lib/Cre+ vs. FD/Cre+ |
0.00000018 0.00149 0.00056 |
|
| f | Ad Lib/WT vs. FD/WT Ad Lib/Local vs. FD/Local Ad Lib/Retro vs. FD/Retro FD/WT vs. FD/Retro FD/Local vs. FD/Retro |
0.00000006 0.00000000021 0.000306 0.000803 0.001337 |
|
| 2 | g | Pre/Saline vs. Post/Saline Pre/Naloxone vs. Post/Naloxone |
0.023 0.877 |
| 3 | d | Ad Lib/Cre− vs. FD/Cre− Ad Lib/Cre+ vs. FD/Cre+ FD/Cre− vs. FD/Cre+ |
0.114 0.000012 0.000829 |
| e | WT vs. Cre− WT vs. Cre+ Cre− vs. Cre+ |
0.002 0.231 0.046 |
|
| h | Saline/Off vs. Naloxone/Off Saline/Off vs. Saline/On Saline/Off vs. Naloxone/On |
0.041 0.99 0.962 |
|
| 5 | c | Ad Lib/Cre− vs. FD/Cre− FD/Cre− vs. FD/Cre+ Ad Lib/Cre+ vs. FD/Cre+ |
0.000001 0.029 0.00021 |
| f | Ad Lib/Cre− vs. FD/Cre− FD/Cre− vs. FD/Cre+ Ad Lib/Cre+ vs. FD/Cre+ |
0.000000000016 0.000148 0.00000000011 |
|
| j | Pre/CRISPR vs. Post/CRISPR Pre/Control vs. Post/Control |
0.987 0.000241 |
Supplementary Material
Acknowledgements
We thank Lamley Lawson, Dylan Blumenthal, Michelle Chung, Taylor Hobbs, and Carina Pizzano for animal colony maintenance. We thank the Bruchas lab, Stuber lab and multiple trainee’s and faculty within the NAPE Center for helpful discussions. We thank Dr. Brigitte Kieffer for the Oprm1fl/fl mice, and Dr. Greg Scherrer for the Penk-Cre mice.
Funding:
D.C.C was funded by NIH grants NS007205, DA043999, DA049862, DA051489. C.E.P. was funded by NIH grant DA051124. M.A.R. was funded by NIH grant DK121883 and a NARSAD Young Investigator Award. J.A.M. was funded by NIH grants DA041781, DA042499, DA045463. G.D.S. DA032750, DA038168 and DA048736. M.R.B. was funded by NIH grants R37DA033396, R61DA051489, and the Mallinckrodt Endowed Professorship. M.R.B. and LZ were funded by NIH grants P30DA048736.
Footnotes
Competing Interests: The authors declare no competing interests.
Data and materials availability: Plasmids generated in this study will be deposited to Addgene. Mouse lines generated in this study will be deposited to Jackson Laboratories. This study did not generate any new/unique reagents. Custom MATLAB analysis and code was created to appropriately organize, process, and combine photometry and single-photon recording data with associated behavioral data. Analysis code for photometry and single-photon imaging from Figures 2 and 4 will be made available on Github (https://github.com/BruchasLab). The behavioral dataset supporting the current study are available as Source Data or from the author upon request.
References
- 1.Mattison J The Treatment of the Morphine-Disease. Indian Med. Gaz 26, 65–68 (1891). [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC). CDC grand rounds: prescription drug overdoses - a U.S. epidemic. MMWR Morb. Mortal. Wkly. Rep 61, 10–13 (2012). [PubMed] [Google Scholar]
- 3.Volkow N, Benveniste H & McLellan AT Use and Misuse of Opioids in Chronic Pain. Annu. Rev. Med 69, 451–465 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wei AD & Ramirez J-M Presynaptic Mechanisms and KCNQ Potassium Channels Modulate Opioid Depression of Respiratory Drive. Front. Physiol 10, 1407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herkenham M & Pert CB In vitro autoradiography of opiate receptors in rat brain suggests loci of ‘opiatergic’ pathways. Proc. Natl. Acad. Sci. U. S. A 77, 5532–5536 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelley AE, Baldo BA & Pratt WE A proposed hypothalamic-thalamic-striatal axis for the integration of energy balance, arousal, and food reward. J. Comp. Neurol 493, 72–85 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Massaly N, Morón JA & Al-Hasani R A Trigger for Opioid Misuse: Chronic Pain and Stress Dysregulate the Mesolimbic Pathway and Kappa Opioid System. Front. Neurosci 10, 480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro DC & Bruchas MR A Motivational and Neuropeptidergic Hub: Anatomical and Functional Diversity within the Nucleus Accumbens Shell. Neuron 102, 529–552 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes J et al. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature 258, 577–580 (1975). [DOI] [PubMed] [Google Scholar]
- 10.Simantov R & Snyder H Isolation and structure identification of a morphine-like peptide ‘enkephalin’ in bovine brain. Life Sci. 18, 781–787 (1976). [DOI] [PubMed] [Google Scholar]
- 11.Bakshi VP & Kelley AE Feeding induced by opioid stimulation of the ventral striatum: role of opiate receptor subtypes. J. Pharmacol. Exp. Ther 265, 1253–1260 (1993). [PubMed] [Google Scholar]
- 12.Resendez SL et al. μ-Opioid receptors within subregions of the striatum mediate pair bond formation through parallel yet distinct reward mechanisms. J. Neurosci. Off. J. Soc. Neurosci 33, 9140–9149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro DC & Berridge KC Opioid hedonic hotspot in nucleus accumbens shell: mu, delta, and kappa maps for enhancement of sweetness ‘liking’ and ‘wanting’. J. Neurosci 34, 4239–4250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y et al. Targeted expression of μ-opioid receptors in a subset of striatal direct-pathway neurons restores opiate reward. Nat. Neurosci 17, 254–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodnar RJ, Glass MJ, Ragnauth A & Cooper ML General, mu and kappa opioid antagonists in the nucleus accumbens alter food intake under deprivation, glucoprivic and palatable conditions. Brain Res. 700, 205–212 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Kelley AE, Bless EP & Swanson CJ Investigation of the effects of opiate antagonists infused into the nucleus accumbens on feeding and sucrose drinking in rats. J. Pharmacol. Exp. Ther 278, 1499–1507 (1996). [PubMed] [Google Scholar]
- 17.Kramer TH et al. Novel peptidic mu opioid antagonists: pharmacologic characterization in vitro and in vivo. J. Pharmacol. Exp. Ther 249, 544–551 (1989). [PubMed] [Google Scholar]
- 18.Tervo DGR et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z-Q et al. Central serotonergic neurons are differentially required for opioid analgesia but not for morphine tolerance or morphine reward. Proc. Natl. Acad. Sci. U. S. A 104, 14519–14524 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Land BB et al. Activation of the kappa opioid receptor in the dorsal raphe nucleus mediates the aversive effects of stress and reinstates drug seeking. Proc. Natl. Acad. Sci. U. S. A 106, 19168–19173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nectow AR et al. Identification of a Brainstem Circuit Controlling Feeding. Cell 170, 429–442.e11 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Huang KW et al. Molecular and anatomical organization of the dorsal raphe nucleus. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunaydin LA et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siuda ER et al. Spatiotemporal control of opioid signaling and behavior. Neuron 86, 923–935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunker AC et al. Conditional Single Vector CRISPR/SaCas9 Viruses for Efficient Mutagenesis in the Adult Mouse Nervous System. Cell Rep. 30, 4303–4316.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Hasani R et al. In vivo detection of optically-evoked opioid peptide release. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maldonado-Irizarry CS, Swanson CJ & Kelley AE Glutamate receptors in the nucleus accumbens shell control feeding behavior via the lateral hypothalamus. J. Neurosci. Off. J. Soc. Neurosci 15, 6779–6788 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vachez YM et al. Ventral arkypallidal neurons inhibit accumbal firing to promote reward consumption. Nat. Neurosci 24, 379–390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castro DC, Terry RA & Berridge KC Orexin in Rostral Hotspot of Nucleus Accumbens Enhances Sucrose ‘Liking’ and Intake but Scopolamine in Caudal Shell Shifts ‘Liking’ Toward ‘Disgust’ and ‘Fear’. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol 41, 2101–2111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgescu D et al. The hypothalamic neuropeptide melanin-concentrating hormone acts in the nucleus accumbens to modulate feeding behavior and forced-swim performance. J. Neurosci. Off. J. Soc. Neurosci 25, 2933–2940 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim BK, Huang KW, Grueter BA, Rothwell PE & Malenka RC Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature 487, 183–189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connor EC et al. Accumbal D1R Neurons Projecting to Lateral Hypothalamus Authorize Feeding. Neuron 88, 553–564 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Berthoud H-R & Münzberg H The lateral hypothalamus as integrator of metabolic and environmental needs: from electrical self-stimulation to opto-genetics. Physiol. Behav 104, 29–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corre J et al. Dopamine neurons projecting to medial shell of the nucleus accumbens drive heroin reinforcement. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Chiara G & Imperato A Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc. Natl. Acad. Sci. U. S. A 85, 5274–5278 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith KS & Berridge KC The ventral pallidum and hedonic reward: neurochemical maps of sucrose ‘liking’ and food intake. J. Neurosci. Off. J. Soc. Neurosci 25, 8637–8649 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Land BB et al. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J. Neurosci. Off. J. Soc. Neurosci 28, 407–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zelikowsky M et al. The Neuropeptide Tac2 Controls a Distributed Brain State Induced by Chronic Social Isolation Stress. Cell 173, 1265–1279.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker KE et al. A Paranigral VTA Nociceptin Circuit that Constrains Motivation for Reward. Cell 178, 653–671.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Hasani R et al. Distinct Subpopulations of Nucleus Accumbens Dynorphin Neurons Drive Aversion and Reward. Neuron 87, 1063–1077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massaly N et al. Pain-Induced Negative Affect Is Mediated via Recruitment of The Nucleus Accumbens Kappa Opioid System. Neuron 0, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fetterly TL et al. Insulin Bidirectionally Alters NAc Glutamatergic Transmission: Interactions between Insulin Receptor Activation, Endogenous Opioids, and Glutamate Release. J. Neurosci. Off. J. Soc. Neurosci 41, 2360–2372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid CL et al. Bias Factor and Therapeutic Window Correlate to Predict Safer Opioid Analgesics. Cell 171, 1165–1175.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh J & Desiraju T Differential effects of opioid peptides administered intracerebrally in loci of self-stimulation reward of lateral hypothalamus and ventral tegmental area--substantia nigra. NIDA Res. Monogr 87, 180–191 (1988). [PubMed] [Google Scholar]
- 46.Lemos JC, Roth CA & Chavkin C Signaling events initiated by kappa opioid receptor activation: quantification and immunocolocalization using phospho-selective KOR, p38 MAPK, and K(IR) 3.1 antibodies. Methods Mol. Biol. Clifton NJ 717, 197–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banghart MR & Sabatini BL Photoactivatable neuropeptides for spatiotemporally precise delivery of opioids in neural tissue. Neuron 73, 249–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banala S et al. Photoactivatable drugs for nicotinic optopharmacology. Nat. Methods 15, 347–350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y et al. Battery-free, lightweight, injectable microsystem for in vivo wireless pharmacology and optogenetics. Proc. Natl. Acad. Sci. U. S. A (2019) doi: 10.1073/pnas.1909850116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patriarchi T et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun F et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matthes HW et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature 383, 819–823 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Resendez SL et al. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat. Protoc 11, 566–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou P et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental Discussion Citations
- 55.Stratford TR & Kelley AE GABA in the nucleus accumbens shell participates in the central regulation of feeding behavior. J. Neurosci. Off. J. Soc. Neurosci 17, 4434–4440 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baumgartner HM, Cole SL, Olney JJ & Berridge KC Desire or Dread from Nucleus Accumbens Inhibitions: Reversed by Same-Site Optogenetic Excitations. J. Neurosci. Off. J. Soc. Neurosci 40, 2737–2752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castro DC, Cole SL & Berridge KC Lateral hypothalamus, nucleus accumbens, and ventral pallidum roles in eating and hunger: interactions between homeostatic and reward circuitry. Front. Syst. Neurosci 9, 90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders BT, Yager LM & Robinson TE Cue-evoked cocaine ‘craving’: role of dopamine in the accumbens core. J. Neurosci. Off. J. Soc. Neurosci 33, 13989–14000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaughan CW, Ingram SL, Connor MA & Christie MJ How opioids inhibit GABA-mediated neurotransmission. Nature 390, 611–614 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Weibel R et al. Mu opioid receptors on primary afferent nav1.8 neurons contribute to opiate-induced analgesia: insight from conditional knockout mice. PloS One 8, e74706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corder G et al. Loss of μ opioid receptor signaling in nociceptors, but not microglia, abrogates morphine tolerance without disrupting analgesia. Nat. Med 23, 164–173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa E, Mocchetti I, Supattapone S & Snyder SH Opioid peptide biosynthesis: enzymatic selectivity and regulatory mechanisms. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 1, 16–21 (1987). [DOI] [PubMed] [Google Scholar]
- 63.DiFeliceantonio AG, Mabrouk OS, Kennedy RT & Berridge KC Enkephalin surges in dorsal neostriatum as a signal to eat. Curr. Biol. CB 22, 1918–1924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The behavioral dataset supporting the current study are available as Source Data or from the author upon request.