Abstract
This review focuses on the most reliable and up-to-date methods for diagnosing trypanosomoses, a group of diseases of wild and domestic mammals, caused by trypanosomes, parasitic zooflagellate protozoans mainly transmitted by insects. In Africa, the Americas and Asia, these diseases, which in some cases affect humans, result in significant illness in animals and cause major economic losses in livestock. A number of pathogens are described in this review, including several Salivarian trypanosomes, such as Trypanosoma brucei sspp. (among which are the agents of sleeping sickness, the human African trypanosomiasis [HAT]), Trypanosoma congolense and Trypanosoma vivax (causing “Nagana” or animal African trypanosomosis [AAT]), Trypanosoma evansi (“Surra”) and Trypanosoma equiperdum (“Dourine”), and Trypanosoma cruzi, a Stercorarian trypanosome, etiological agent of the American trypanosomiasis (Chagas disease). Diagnostic methods for detecting zoonotic trypanosomes causing Chagas disease and HAT in animals, as well as a diagnostic method for detecting animal trypanosomes in humans (the so-called “atypical human infections by animal trypanosomes” [a-HT]), including T. evansi and Trypanosoma lewisi (a rat parasite), are also reviewed. Our goal is to present an integrated view of the various diagnostic methods and techniques, including those for: (i) parasite detection; (ii) DNA detection; and (iii) antibody detection. The discussion covers various other factors that need to be considered, such as the sensitivity and specificity of the various diagnostic methods, critical cross-reactions that may be expected among Trypanosomatidae, additional complementary information, such as clinical observations and epizootiological context, scale of study and logistic and cost constraints. The suitability of examining multiple specimens and samples using several techniques is discussed, as well as risks to technicians, in the context of specific geographical regions and settings. This overview also addresses the challenge of diagnosing mixed infections with different Trypanosoma species and/or kinetoplastid parasites. Improving and strengthening procedures for diagnosing animal trypanosomoses throughout the world will result in a better control of infections and will significantly impact on “One Health,” by advancing and preserving animal, human and environmental health.
Graphical Abstract
Keywords: Antibody-detection, Cross-reactions, DNA detection, Microscope examination, Trypanosome, Undetected infection
Background
The family Trypanosomatidae (phylum Protozoa, class Kinetoplastida) comprises 14 monoxenous genera infecting insects (e.g. Leptomonas, Herpetomonas) and five dixenous genera having invertebrates as vectors (one genus infecting plants [Phytomonas] and the other four infecting animals and humans). Among the four genera infecting animals and humans, two are anecdotic, i.e. Endotrypanum found in sloths and Porcisia found in porcupine; the other two genera, namely Trypanosoma and Leishmania, contain pathogenic parasites of medical and veterinary importance. Both Trypanosoma and Leishmania are widely distributed over the world, and they affect humans, animals or both, as anthroponotic or zoonotic agents [1]. In humans, leishmaniasis has a high medical impact, with approximately 70,000 deaths recorded annually and an estimated 350 million individuals at risk [2]. In animals, leishmanioses seriously affect dogs, while in cattle and horses, the impact is limited [3, 4]. Trypanosomoses are significant diseases of domestic and wild mammals, affecting millions of livestock in Africa, the Americas and Asia [5], as well as of humans in Africa and Latin America [6–10].
Among the 125 Trypanosoma species found in mammals, 10% are considered to be pathogenic to humans and/or other mammals [11]. These pathogenic trypanosomes mainly inhabit the host’s blood and lymph, but do occur sometimes in the cerebrospinal fluid (CSF) and other host tissues, and some of them have intracellular stages. Trypanosomes are mainly transmitted by insects, but alternative means of transmission include mammals as vectors (e.g. vampire bats for Trypanosoma evansi and marsupials for Trypanosoma cruzi) and transcutaneous and transmembrane routes, such as peroral, venereal, intraplacental, iatrogenic, routes, among others, which allow occasional horizontal and vertical transmission.
The major pathogenic trypanosomes, approximately 10 species, subspecies or types, originate from Africa, where they are mainly cyclically transmitted by the bite of tsetse flies (as Salivarian trypanosomes) [11]; two subspecies are zoonotic, while the others are “animal parasites,” infecting wild and domestic mammals, including livestock, despite scarce occurrences in humans [12]. One other important pathogenic trypanosome of mammals occurs in the Americas, i.e. T. cruzi (subgenus Schizotrypanum) where it is responsible for a neglected tropical disease (NTD) named “Chagas disease” that extends into South and Central America. As a Stercorarian trypanosome, T. cruzi is biologically transmitted through the feces of triatomine bugs, and host contamination occurs by transmembrane or transcutaneous penetration. Trypanosoma cruzi is not only a human pathogen but also a zoonotic parasite, affecting a huge range of domestic and wild mammals, including livestock, as reported in a recent review [13]. In this review, we focus on the diagnosis of trypanosomes in animals; however, in a “One Health concept” [14], this focus includes human pathogens, providing that animals carry and/or are affected by these pathogens, and/or play a role in the epidemiology of the human diseases (reservoir or screen). We also cover the so-called “atypical human infections by animal trypanosomes” [12].
Definitions and geographical distributions
African trypanosomoses
Animal trypanosomoses of African origin (ATAO) are known under several disease names that are associated with one or several Trypanosoma species involved. “Nagana” is a disease complex caused by one or several Salivarian trypanosomes belonging to subgenera Nannomonas (Trypanosoma congolense, T. simiae and T. godfreyi) Duttonella (Trypanosoma vivax and T. uniforme; the occurrence of the latter needs to be confirmed) and Trypanozoon (Trypanosoma brucei brucei, T. brucei gambiense and T. brucei rhodesiense) “Surra” is a disease caused by T. evansi, and “Dourine” is caused by Trypanosoma equiperdum (sometimes referred as T. brucei evansi and T. brucei equiperdum, due to their phylogenetic relations) [11, 15–19].
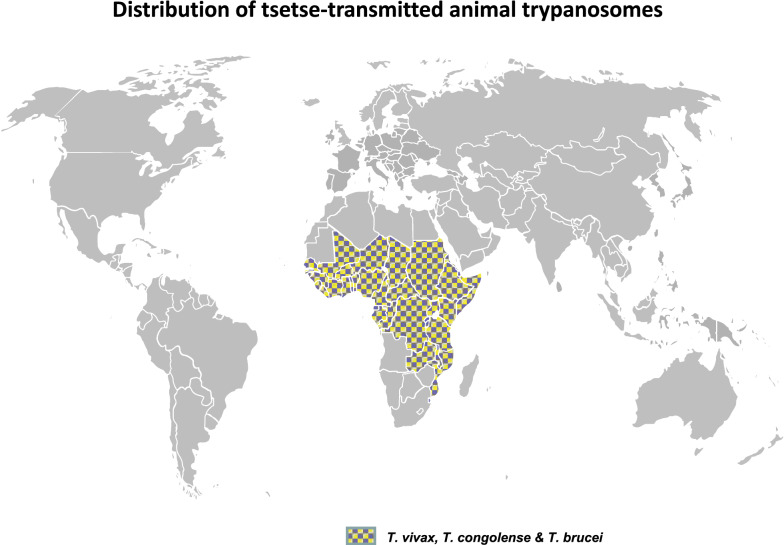
Apart from the last two species that will be discussed below, Trypanosoma spp. responsible for Nagana are mainly cyclically transmitted by flies of the genus Glossina (tsetse flies). Their geographical distribution is determined by the geographical distribution of the tsetse fly, which is restricted to specific areas of Africa (Fig. 1). Tsetse flies have been reported to occur in an area estimated to be 10 million km2 in size, in 37 countries, in humid and sub-humid sub-Saharan part of Africa (from latitude 10° N to 20°–30° S and in some limited areas of the Arabian Peninsula [5, 20, 21]. Nagana affects both wild and domestic mammals, but the impact of infection is the highest in cattle, with about 3 million livestock dying annually despite the administration of approximately 35 million doses of trypanocidal drugs. The economic losses in cattle production is estimated to be about US$ 1.0–1.2 billion [22]. Although the most pathogenic agent of Nagana for livestock is T. congolense type Savannah [23], T. vivax is the most prevalent. To a lesser extent and with a somehow enigmatic impact, T. brucei brucei also contributes to the disease complex. Moreover, the human pathogens T. b. gambiense and T. b. rhodesiense have been found in livestock [24–29], but their pathogenic effects and impact are not yet thoroughly clarified. However, experimental infections of cattle with T. b. rhodesiense were found to lead to a fatal central nervous syndrome in half of the cases tested [30]. In addition to cattle, other species, such as goats, sheep, pigs and dogs, may be affected. Horses and camels are very susceptible to Nagana; indeed, in the past, tsetse flies used to constitute a natural barrier preventing the introduction of camels and horses into the southern Sahel regions of Africa [31].
Fig. 1.
Geographical distribution of the “Nagana” disease complex (Trypanosoma congolense, T. vivax and T. brucei) [32–36]
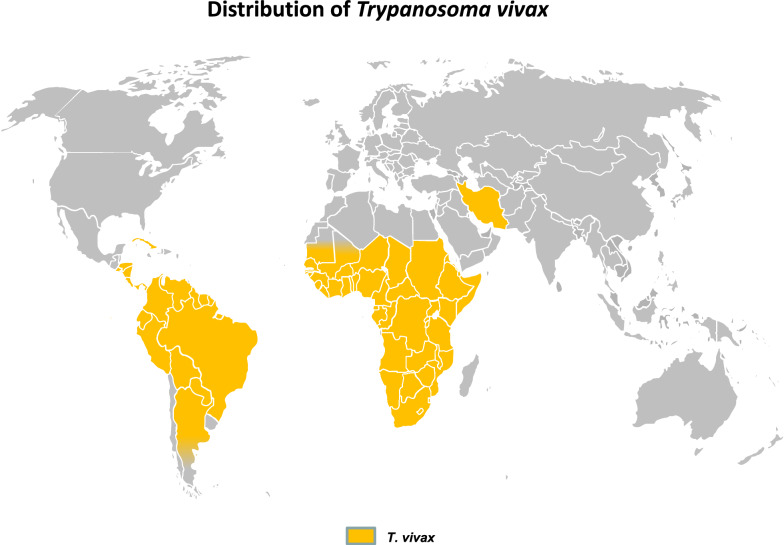
Some of these Trypanosoma spp. may also be mechanically transmitted by biting flies, such as tabanids and Stomoxyine flies [37, 38], especially T. vivax, as first suspected [39] and later confirmed in semi-liberty conditions [40], and also T. evansi, which is discussed in subsequent sections. Mechanical transmission has allowed T. vivax to spread in some areas of Africa that are free of or were cleared of tsetse (e.g. in Ethiopia [41]). Similarly, during the eighteenth century, T. vivax invaded South and Central America [6], and more recently it was reported in Iran [42]. Geographical distribution of T. vivax is presented in Fig. 2.
Fig. 2.
Geographical distribution of Trypanosoma vivax [6, 32–34, 36, 42]
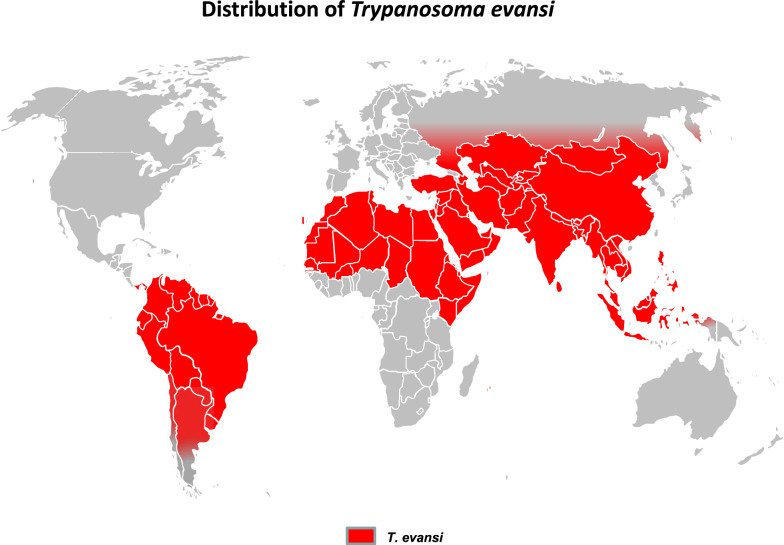
The other members of the ATAO are the two Trypanozoon derived from the T. brucei lineage, but these are not transmitted cyclically. Trypanosoma evansi, the causative agent of Surra, is mechanically transmitted by tabanids and Stomoxyine flies and found in tropical areas, but also ranges up to Mongolia. Surra significantly affects camels and horses in Africa and Latin America, but also cattle and buffaloes in Asia [43]. Surra is the most widely distributed animal trypanosomosis, ranging from South to Central America, the upper half of Africa, Middle East and Asia (Fig. 3). Trypanosoma equiperdum is the causative agent of Dourine, a venereal disease transmitted worldwide among equids [44]. In the last decade it was described in Italy [45], Mongolia [46], Ethiopia [47] and Iran [48], but its geographical distribution is mostly unknown. These two species may be found in the same hosts and in the same areas as the agents of Nagana, which interfers with species-specific diagnosis. Together with T. vivax, these trypanosomes are responsible for the “non-tsetse-transmitted animal trypanosomoses” (NTTAT) [49].
Fig. 3.
Geographical distribution of Trypanosoma evansi (“Surra”) [6, 33, 43, 50]
Clinical signs of ATAO may include intermittent fever, anemia, edema, abortion, decreased fertility, emaciation and death; genital and neurological symptoms are also possible. However, none of these symptoms are pathognomonic. Clinically, ATAO can be confused with other parasitic diseases (i.e. babesiosis, anaplasmosis, among others), rabies, plant intoxications and T. cruzi infection in Latin America. Therefore, case identification must rely on diagnostic techniques that: (i) confirm the presence of trypanosomes, with by microscopic visualization or by obtaining evidence of trypanosome DNA, or (ii) demonstrate a host—parasite contact through antibody detection techniques [51]. Differential diagnosis is based on observations and evolution of clinical signs, epidemiological context and, most critically, by laboratory test results.
Human African trypanosomiasis (HAT), or sleeping sickness, is caused by two T. brucei subspecies transmitted by tsetse flies, with a substantial socio-economic impact in humans, although it is still classified as a NTD [9, 52]. If not treated, sleeping sickness is usually a fatal disease. Trypanosoma b. gambiense is responsible for a primarily anthroponotic disease form that accounts for 98% of HAT cases [53, 54]. It results in a chronic disease that can last several years, and occurs in West and Central Africa. Some animals have been reported as potential reservoirs [26, 29], but their exact role in the epidemiology of the human disease is not clear [28, 55]. Based on molecular characterization, a variant of T. b. gambiense has been described recently, leading to the consideration of variants Tbg1 and Tbg2, and possibly more, within T. b. gambiense [56]. In East Africa, where the zoonotic T. b. rhodesiense is responsible for an acute disease form in humans, representing 2% of HAT cases [53, 54], the parasite is also found in a wide range of wild and domestic animals, including cattle, goats and pigs, which act as reservoirs [27, 57–59]. About 70 million people worldwide are at risk of sleeping sickness [53]. In addition to the socio-economic impact on exposed populations, the presence of these two subspecies creates a risk of infection for farmers, veterinarians and slaughter-house workers as well as laboratory technicians, when handling meat, carcasses and blood. Consequently, in these areas of Africa, animal samples should be handled with appropriate biosafety and containment procedures. The existence of these two forms of HAT also creates a need for subspecies-specific diagnosis methods in order to evaluate the human risk and to investigate, and possibly control, the animal reservoirs. Under the expanding “One Health” concept, identifying these zoonotic agents in livestock (and wild fauna) may prove useful to control HAT [60, 61].
American trypanosomosis
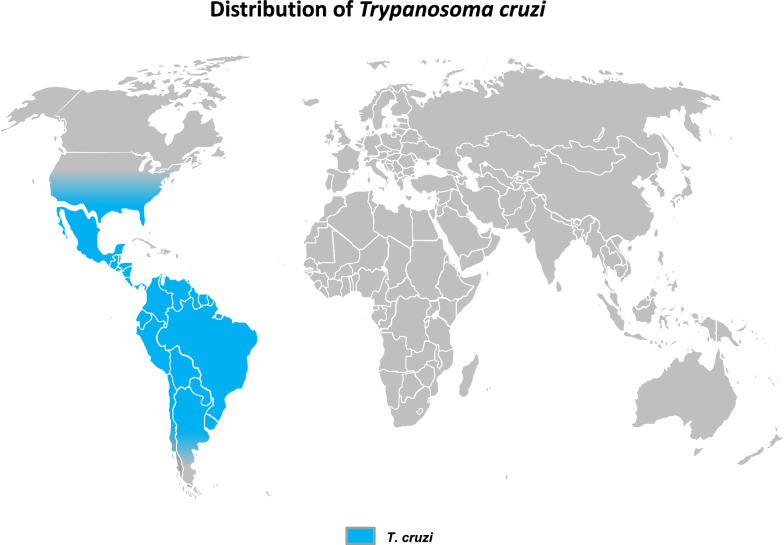
Trypanosoma cruzi, the agent of the American human trypanosomiasis, or Chagas disease, is mainly transmitted via feces of triatomine bugs, affecting 6–8 million people, mainly in Latin America, most of whom are chronic carriers [62], while 65–100 million people are at risk [63]. Other ways of transmission are of variable importance, such as vertical (mother to fetus and mother to child) and iatrogenic transmission, especially through blood transfusion and organ transplantation [64]. Detecting the potential presence of T. cruzi in blood collected from Latin American people who lived previously in endemic areas is a serious concern, notably in Europe and the USA [10]. Acute human cases were reported recently to be linked to peroral infection via fruit juices contaminated by the bug’s feces. This route of infection, possibly under-detected in the past, now appears to be a significant mode of transmission [65–68]. Although T. cruzi is considered to be a human pathogen, it has a large wild animal reservoir that includes marsupials, armadillos, raccoons, squirrels, wild pigs, rats, among others, as well as a domestic reservoir, including dogs, cats, pigs, sheep, goats, cattle and horses [69]. The presence of T. cruzi may be a source of interference in animals being investigated for trypanosomoses, as already observed for T. evansi/T. cruzi infections in horses in Argentina [70]. Additionally, the possible presence of a human pathogen in animal samples constitutes a potential risk for human health at the farm and laboratory levels. Trypanosoma cruzi is a neglected but true pathogen in animals, and even though the production cycle of livestock is mostly too short to allow clear clinical expression of the disease (especially for short-cycle species such as pigs [71]) it may undergo a complete evolution and cause deleterious clinical signs in other animals; for example, myocarditis has been observed in dogs and nervous invasion with ataxia in horses [72–74]. The geographical distribution of Chagas disease in humans extends from mid-Argentina and Chile to Mexico; however, the geographical distribution of T. cruzi extends more northwards to include most of the southern USA and a broad area extending from northern California to northern Pennsylvania [75] (Fig. 4). Consequently, in these areas, special care should be taken when handling animal samples; laboratory work should be performed with appropriate biosafety and containment procedures, especially when horse, cattle, pig, dog or wild fauna samples are being handled as these may be infected with T. cruzi [69].
Fig. 4.
Atypical human infections by animal trypanosomes
Very rare human cases caused by animal Trypanosoma species, including T. vivax, T. congolense, T. b. brucei, T. evansi and T. lewisi or T. lewisi-like, have been reported; these are referred to as as “atypical human infections by animal trypanosomes” (a-HT) [12, 77]. Among these, a growing number of human cases have been reported, particularly in Asia due to T. evansi, and T. lewisi (subgenus Herpetosoma). Trypanosoma lewisi is a cosmopolitan parasite of rats which has low pathogenicity in its original host [12]. Diagnostic methods for detecting T. lewisi in rats are thus needed for risk analysis and, conversely, diagnosis of this species may also be required in humans. Several human infections by T. congolense were recently exposed by PCR examination in a survey in Maro, southern Chad [78], suggesting that human infections with T. congolense might be more frequent than previously thought. In any case, these surprising results certainly need confirmation.
Distribution of pathogenic trypanosomes
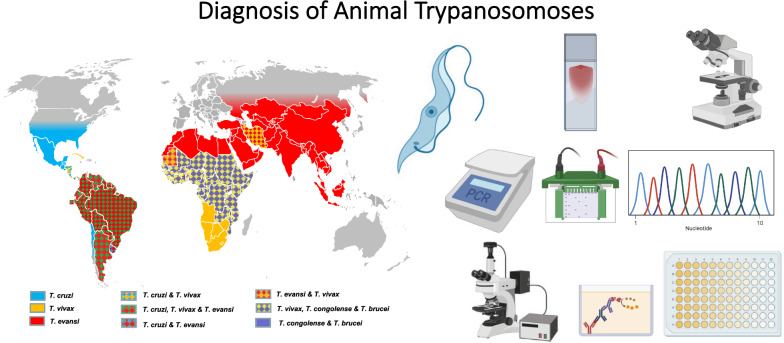
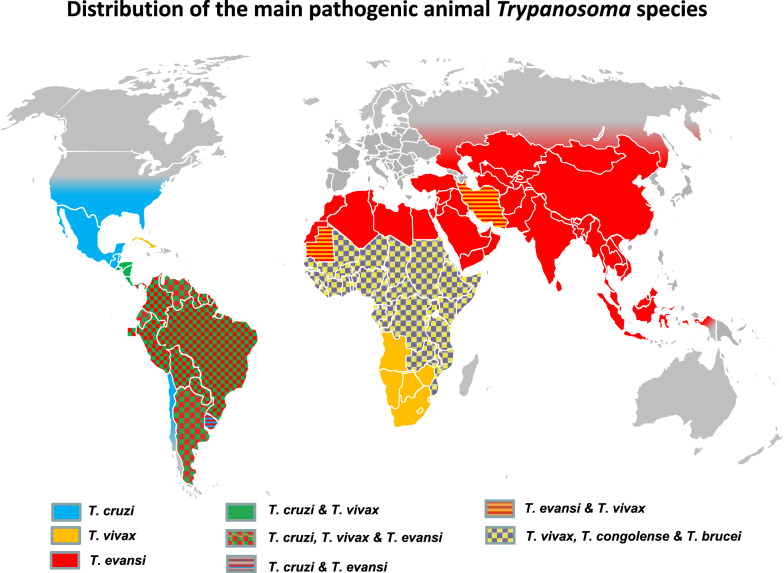
In addition to vector control, in the absence of protective vaccines or efficient prophylactic strategy, control of the diseases mentioned above strongly relies on detecting and treating positive cases. Diagnostic results not only support treatment decisions, but they are the basis for epidemiological studies, monitoring and evaluation of the efficiency of disease control measures. Furthermore, in the field of animal husbandry, diagnosis is also a tool used to implement health policy, including slaughter-hous policy, and to define the health status of animals prior to international movements. Finally, knowledge of those Trypanosoma species that are potentially present in the investigated area is necessary to consider interference in diagnosis and to prevent the risk of infection to humans handling these samples. To support such awareness, a tentative world distribution of pathogenic animal trypanosomes is represented in Fig. 5, mostly based on publications, including geographical reviews [6, 34–36, 43, 50].
Fig. 5.
Geographical distribution of pathogenic mammal trypanosomes [6, 33–36, 42, 43, 50, 69, 76]
Review and characterization of available diagnosis techniques
Clinical signs of animal trypanosomoses are not sufficiently specific to support a clinical diagnosis and, therefore, laboratory tests are required to confirm clinical suspicions. As a consequence, case confirmations and epidemiological studies can only be carried out using laboratory facilities.
The course of a Salivarian trypanosome infection can vary from acute to chronic, but also be asymptomatic, depending on the intrinsic pathogenicity of the parasite and susceptibility, immune competence and health history of the host. One of the characteristics of pathogenic Salivarian trypanosome infections is a highly fluctuating parasitemia, which reflects the affinity of the parasite to tissues, and the control of the parasite population by the host immune system, which the parasite cyclically escapes by developing a population with new variable surface glycoprotein (VSG), the generation of which is referred to as “variable antigen type” (VAT). Consequently, when a VAT is recognized by the host immune system, the trypanosome population exhibiting this VAT is destroyed by the immune system and the parasitemia decreases drastically to undetectable levels for some time; when a new VAT population multiplies, parasitemia once again increases. This cycle is the reason why parasite concentrations in the host blood are highly variable from one day to another and sometimes even nil, or undetectable. As a general rule, parasitemia is high in early infections, lower and less frequent in chronic infections and nil or erratic in the case of subclinical evolution of the disease. Therefore, diagnosis efficacy can be seriously affected when performed at times of low parasitemia levels.
Numerous diagnostic tests are available to detect trypanosomes or diagnose trypanosomoses [79]. Current diagnostic tests vary in their sensitivity and specificity, the ease with which they can be implemented and their cost [80]. The choice of one or several particular tests is guided by epidemiologically-adapted diagnostic requirements, availability of equipment and expertise and economic principles. These choices will be discussed in another article in this journal (“Proper use and perspective on diagnosis methods for animal trypanosomoses”). In the present review we describe the characteristics of the methods currently available for diagnosing animal trypanosomes. Apart from antigen detection, which despite numerous attempts remains unsuccessful, three types of diagnostic techniques for animal trypanosomes can be distinguished: (i) parasite detection; (ii) DNA detection; and (iii) antibody detection (Tables 1, 2). Technical information on these tests is available on the websites of the World Organization for Animal Health (OIE) [81] and of the Food and Agriculture Organization of the United Nations (FAO) [32]. A recently published “Compendium of standard diagnosis protocols for animal trypanosomoses of African origin” is available on the OIE website [13]. These documents can be found at the following links: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.14_NAGANA.pdf; https://www.fao.org/3/X0413E/X0413E00.htm; https://www.oie.int/app/uploads/2021/06/compendiumstandarddiagnosticprotocolsanimaltrypansomosesafricanorigin-en.pdf , respectively.
Table 1.
Summary of diagnostic techniques used for animal African trypanosomosis and non-tsetse transmitted animal trypanosomosis
| Target | Techniquea | Analytic sensitivity (parasite/ml) | Sensitivity (percentage estimation if available) | Specificity | Logistic needs: field/laboratory | Cost | Infection step | Scale | Livestock /fauna | Test objective and/or context |
|---|---|---|---|---|---|---|---|---|---|---|
| Parasite | Wet blood film | 104–105 [80] | No data | + | Field: microscope | Low | Acute infection | Individual | Livestock (fresh blood) | Epidemiology or experimental follow-up |
| HTC/BCM | 2.5 × 102–5 × 103 | + (14–24) [82] | + + | Field: microcentrifuge + microscope | Low | Acute sub-acute infection | Individual (group) | Livestock (fresh blood) | Individual diagnostic (epidemiology) | |
| mAECT | + | + + | Field: microcentrifuge + microscope | High | Acute sub-acute infection | Individual | Livestock (fresh blood) | For research: parasite isolation | ||
| Rodents inoculation | 10–103 | + + + | + + + | Laboratory with animal facilities | High | Acute sub-acute infection | Individual | Livestock (fresh blood) | For research: parasite isolation | |
| DNA | Pan-species primers (ITS1) | 50 [83] | + + (54–74) [84] | + + may require sequencing (99–100) [84] | Laboratory for molecular biology | High | Acute sub-acute infection | Individual and group | Livestock and wildlife | Epidemiology |
| Genus/species specific primers | 1–10 [85] | + + + (65–88) [84] | + + + + (99–100) [84] | Laboratory for molecular biology | High | Acute sub-acute infection | Individual and group | Livestock and wildlife | Epidemiology | |
| Antibodies | ELISA against total antigens | + + + (90.5) [82] | + + | Laboratory for serology | Low | Acute to chronic and past infection | Group (individual) | Livestock | Epidemiology (individual diagnostic) | |
| Protein specific ELISA | + + + (98.9) [86]; (85.9–98.1) [87] | + + + (98.9) [86]; (90.4–100) [87] | Laboratory for serology | Medium | Acute to chronic and past infection | Group (individual) | Livestock | Epidemiology (individual diagnostic) | ||
| Agglutination test (CATT) | + + (87) [88]; (44.5–95.2) [89]; (76–88) [90]; (32.4–52.9) [91] | + + (81) [88]; (79.5–99.5) [89]; (92–98) [90]; (77.8–84.6) [91] | Field | Low | Acute to chronic and past infection | Individual (group) | Livestock | Individual diagnostic (epidemiology) | ||
| IFAT | + + | + | Laboratory for serology | Medium | Acute to chronic and past infection | Individual (group) | Livestock | Individual diagnostic | ||
| CFT | + + (31.5–73.5) [89]; (90–100) [92] | + + (89.2–98.5) [89]; (89–99) [92] | Laboratory for serology | Medium | Acute to chronic and past infection | Group (individual) | Livestock | Individual diagnostic (epidemiology) | ||
| TL | No data | + + | Laboratory with animal facilities | Very high | Acute to chronic and past infection | Individual | Livestock | Individual diagnostic |
Main test characteristics are indicated. Qualitative information is given for sensitivity and specificity, ranging from low (−) to high (+ + +) performance. Quantitative data are given when available. It should be noted that these values are not directly comparable since tests were performed on different samples sets, with different techniques and in different laboratories. Primary use is indicated first and additional use is given in parentheses
aSee Abbreviation List for the full description of each abbreviation
Table 2.
Summary of diagnostic uses for main Tryanosoma species responsible for animal African trypanosomosis and non-tsetse transmitted animal trypanosomosis
| Target | Techniquea | Subgenus Nannomonas | Subgenus Duttonella | Subgenus Trypanozoon | ||
|---|---|---|---|---|---|---|
| Trypanosoma congolense | Trypanosoma vivax | Trypanosoma brucei brucei | Trypanosoma evansi | Trypanosoma equiperdum | ||
| Parasite | Wet blood films | +b | + | + | + Trypanozoon | |
| HTC/BCT | + + + | + + + | + + + | + + + Trypanozoon | ||
| mAECT | − | − | − | − | − | |
| Inoculation of rodents | + | 0 | + | + + | + | |
| DNA | Pan-species primers (ITS1) | + + | + + | + + | + + Trypanozoon | + |
| Genus/species/type-specific primers | + + + | + + + | + + + | + + + Trypanozoon | + + | |
| Specific primers among Trypanozoon | 0 | + | 0 | |||
| Antibodies | ELISA on WCLSA | + + + | + + + | + + + | + + + Trypanozoon | + |
| Protein-specific ELISA | Under test | Under test | 0 | + | 0 | |
| Agglutination test (CATT) | 0 | 0 | + | + + + | 0 | |
| IFAT | − | − | − | − | + | |
| CFT | 0 | 0 | 0 | 0 | + + | |
| TL | 0 | 0 | 0 | + | 0 | |
aSee Abbreviation List for the full description of each abbreviation
bQualitative uses: + + + = well adapted and used; + + = adapted and used; + = can be used; 0 = not used in practice
Finally, the four OIE reference laboratories on animal trypanosomoses may provide biological materials and technical training to support diagnostics (Table 3).
Table 3.
World Organization for Animal Health reference laboratories for trypanosomes
| Topics | OIE reference laboratories |
|---|---|
| Trypanosomoses (tsetse transmitted)/animal trypanosomes of African origin | CIRAD-Bios, UMR InterTryp (CIRAD-IRD), 34,398 Montpellier, cedex 5, France |
| Surra (Trypanosoma evansi) | National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine, Obihiro, Japan |
| Institute of Tropical Medicine Antwerp, B-2000 Antwerpen, Belgium | |
| Dourine (Trypanosoma equiperdum) | ANSES, Laboratory for animal health, Normandy site, Unit Physiopathology & Epidemiology of Equine Diseases (PhEED), RD675, 14430 Goustranville, France |
In this review, we primarily consider the methods recommended by the OIE (fully demonstrated and validated by large field applications), presenting their characteristics, performance, advantages, disadvantages and limitations.
Parasite detection techniques
In the hosts
Several direct parasite detection techniques based on microscopic examination can be used; ranked from the lowest to highest sensitivity these include: (i) microscopic examination of fresh wet blood films (simplest technique); (ii) the Giemsa-stained thin blood smear (GSBS), which allows identification to the subgenus level based on parasite morphology (Fig. 6); (iii) the hematocrit concentration technique (HCT), which uses a capillary tube, or the Woo method [93]; and (iv) the buffy coat method (BCM, Murray method) [94]. Thelatter is derived from the HCT, but the capillary tube is cut after centrifugation to extrude the buffy coat onto a microscope slide for examination. Concentration techniques exhibit a higher sensitivity (Table 1); however, the BCM has lower repeatability and reproducibility rates than the HCT due to varying levels of technician skills linked to the delicate extrusion and dropping of the buffy coat onto the slide that can in some instances vary in quality.
Fig. 6.
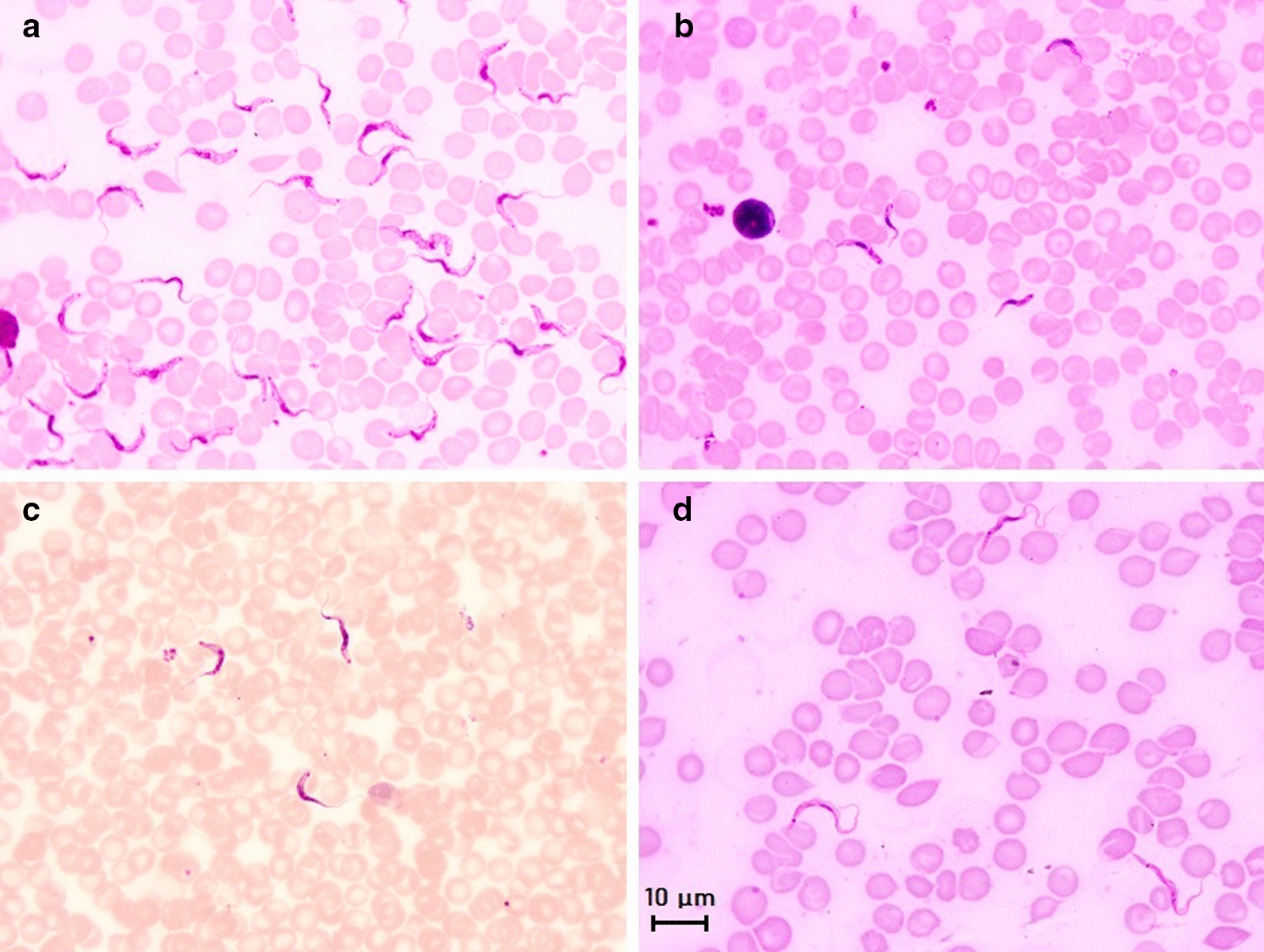
Main morphological features of four subgenera of mammal trypanosomes on Giemsa-stained thin blood smears. a Microscopic image of Trypanosoma brucei brucei in mice blood; morphology of the subgenus Trypanozoon: large-sized trypomastigote (17–30 µm), slender form, free flagellum, small sub-terminal kinetoplast, sharp posterior extremity, central nucleus and large undulating membrane; b Microscopic image of Trypanosoma congolense-type savanna in mice blood; morphology of the subgenus Nannomonas: small-sized trypomastigote (8–22 µm), no free flagellum, terminal sub-lateral kinetoplast, round posterior extremity, central nucleus and no undulating membrane. c Microscopic image of Trypanosoma vivax in cattle blood; morphology of the subgenus Duttonella: large-sized trypomastigote (20–27 µm), slender form, free flagellum, large terminal kinetoplast, round posterior extremity, central nucleus and large undulating membrane. d Microscopic image of Trypanosoma lewisi in rat blood; morphology of the subgenus Herpetosoma: very large-sized trypomastigote (21–36 µm), slender form, free flagellum, very large sub-terminal kinetoplast, very long sharp posterior extremity, anterior nucleus and large undulating membrane. Scale bar: 10 µm
Parasite detection techniques require little equipment and are generally inexpensive, fast and easy to carry out although blood smear observation is time-consuming. Therefore, they are the techniques of choice to ascertain a Trypanosoma infection. The GSBS is subgenus specific, i.e. it is able to distinguish the subgenera Nannomonas, Duttonella, Trypanozoon, Megatrypanum (such as Trypanosoma theileri, a non-pathogenic trypanosome found in bovines and cyclically transmitted by tabanids as a Stercorarian parasite [11]), Schizotrypanum and Herpetosoma (Fig. 6). In addition, GSBS allows the study of parasite morphology and can faciliatedifferential diagnosis of other haemoparasitoses (e.g. Babesia, Theileria, Anaplasma). Both the HCT and BCM give immediate results, but HCT is better suited to screen large numbers of animals since it is the faster and more reproducible of the two tests. HCT and BCM also provide the packed cell volume (PCV), which estimates the level of anemia, one of the most critical indicators of trypanosomosis in cattle, among other hemoparasitoses, hemonchosis, etc. As trypanosomosis is a herd problem, the PCV profile can be used as a marker to orientate investigations. The PCV can also be used to decide whether a sample should be submitted for PCR examination, thereby limiting the total number of samples needed to be tested and increasing the probability of detecting infected animals. Nevertheless, the main drawbacks of these techniques are their very low analytic sensitivity, which depends on the concentration of the parasite in the sample and the volume of the sample examined (as low as 70 µl in capillary tubes and around 3–5 µl in direct blood examination and GSBS). Consequently, the sensitivity of parasitological techniques may vary from “very high” in early infection, when the animals are unable to control the parasitemia (91% for T. evansi in cattle [95, 96]), to “low” in chronic infections, when parasitemia is lower and transient (30–60% in sheep infected with T. vivax), and “nil” in healthy carrier situations when the animals can maintain the parasite at undetectable level in the blood [6] or in extravascular foci [97], including the skin [98]. BCM has been reported to have a very low individual sensitivity of around 14–24% [82]. Parasitological techniques are thus likely to miss chronic infections and asymptomatic carriers. At a population level, the sensitivity of parasite detection techniques is “high” during epizootic outbreaks, but “low” or “very low” in stable enzootic areas where most of the animals are in chronic or subclinical stages of the disease evolution. As an example of the latter: in French Guiana, despite regular sampling of about one-third of the cattle farms per year, T. vivax may not be observed during a 3- to 5-year period, before relapse occurs and the parasite being detected again on several farms [6].
There are other techniques based on parasite detection, and while these may be more sensitive, they are also more expensive and/or more time-consuming. In addition, they require specific skills and equipment that are not generally available. The use of anion exchange chromatography was thoroughly investigated and described in a study published in the 1970s [99]. It was recently reviewed and described again for the diagnosis of sleeping sickness [100]. The mini anion exchange centrifugation technique (mAECT), currently the most sensitive method to diagnose sleeping sickness [101, 102], can be applied to animal samples. However, the technique is cumbersome and is not suitable for the examination of a large number of samples. In vivo isolation of trypanosomes through intra-peritoneal injection of blood from a suspect animal to rodents, usually mice or rats, preferably immunosuppressed using cyclophosphamide [103], is expensive and time-consuming, diagnosis is not immediate and the method raises serious animal welfare concerns. Nevertheless, blood inoculation into rodents is more sensitive than HCT and some PCR procedures (e.g. tests targeting the internal transcribed spacer [ITS]); thus, blood inoculation is beneficial in revealing sub-patent infections and for parasite isolation [104]. However, the success rate of in vivo cultures depends on the Trypanosoma species involved: it is “highly sensitive” for the detection of Trypanozoon infections (especially T. evansi), of “medium sensitivity” for T. congolense strains, and generally “nil” or “scarcely effective” for T. vivax. On the other hand, the method is currently an essential tool for parasite isolation (parasite purification and enrichment before cryo-conservation) and the massive production of trypanosomes to prepare trypanosome antigens used in serological diagnosis or for subsequent molecular characterization. Procedures for in vitro cultivation of Trypanosoma spp. have also been described, but these require sophisticated equipment and protocols, the test results are not immediate (time delay) and they are certainly not suitable for large-scale studies. Competition among field stocks may also affect the results of in vitro cultures; for example, in the case of mixed infections, T. theileri easily overgrows T. b. brucei [105].
Overall, a negative result from parasitological examination does not unequivocally mean the absence of infection, as a negative result is not conclusive in terms of carrier/non-carrier since a Trypanosoma parasite may still be present and remain undetected. Such long periods of undetectable infection (1 year) despite a daily blood test (HTC) were observed in French Guiana, in a sheep experimentally infected with T. vivax and kept under a mosquito net [6].
Conversely, a positive parasitological result may verify the infection of an animal by a specific Trypanosoma parasite depending on the level of specificity allowed by the technique and the epizootiological context. For example, microscopic observation of a Trypanozoon on a GSBS from a cattle blood sample in Southeast Asia indicates T. evansi, while the same result in Africa can only indicate a Trypanozoon due to the potential presence of up to five species or subspecies of Trypanozoon on the African continent.
Consequently, detecting a single taxon cannot exclude the presence of a mixed infection, according to the geographical origin of the sample. Thus, in areas of possible mixed infections (Africa, Latin America), detection of an infection in an animal by a Trypanosoma will always leave open the possibility of mixed infection by one (or several) other Trypanosoma species or type, which may remain undetected, for several reasons (parasitemia below the detection threshold, extravascular refuge of the parasite, etc.). In conclusion, the status of an animal whose parasitological test results are positive should be considered as “animal infected by at least (the trypanosome subgenus, species or type detected), and possibly others”. Potential concomitant infection(s) will always lead to dubious situations in the field.
In the vectors
Microscopic observation of pathogenic trypanosomes may also be applied to cyclical vectors (tsetse flies and triatomine bugs); however, the morphology of trypanosomes in their cyclical vector is not characteristic, and the location of the parasite in the vector’s organs, such as gut, salivary glands or proboscis, may be confusing; consequently, such diagnoses have a very low species specificity. When molecular techniques were also applied, many parasitological diagnoses were shown to be wrong, probably due to a lack of sensitivity in the vector’s organ detection, but also due to the potential presence of many unidentified Trypanosoma spp. that may be found in the same vector’s organs, thus interfering with the diagnosis [106]. In addition, the dissection procedure is subject to a high risk of contamination, mainly between organs of the same vector, which could result in false positive organs by PCR due to the high sensitivity of the latter. Consequently, it is recommended that microscopic observation be used first to detect the trypanosomes in the vector’s organ, followed by molecular tools on these organs to identify the species or subspecies, similar to procedures used in hosts (see section In the hosts).
DNA detection techniques
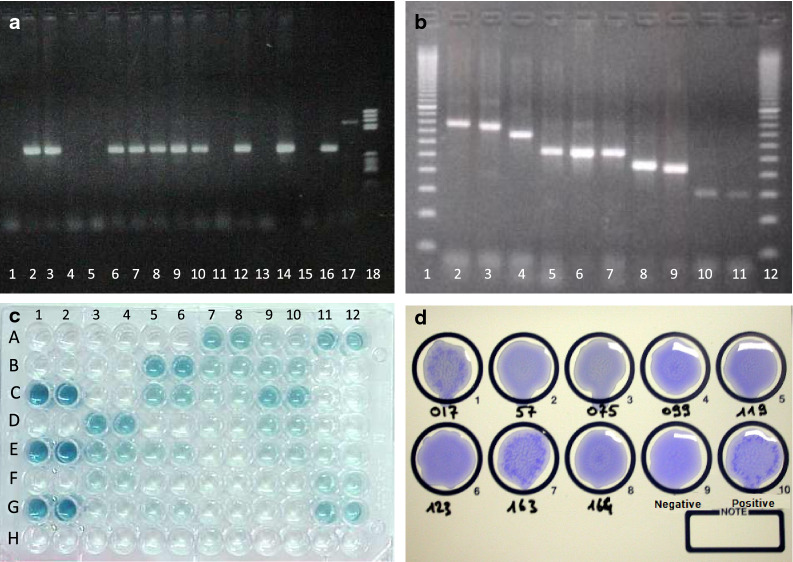
DNA detection techniques used for trypanosomes can be applied to both the hosts and vectors. Several primer pairs have been designed for PCR amplification of trypanosome DNA. Based on highly repetitive satellite DNA (10,000–20,000 tandem repeats per genome), the gold standard primer sets available for the different trypanosome subgenera, species and types are, based to the OIE “Compendium of standard diagnosis protocols for animal trypanosomoses of African origin” [51]: TBR1 and TBR2 (subgenus Trypanozoon); TCS1 and TCS2 (T. congolense savannah type); TCF1 and TCF2 (T. congolense forest type); TCK1 and TCK2 (T. congolense Kenya Coast type [or Kilifi]); TSM1 and TSM2 (T. simiae); DGG1 and DGG2 (T. godfreyi) [107]; and TVW1 and TVW2 (T. vivax) [108, 109]. These monospecific PCRs are positive when the specific weight product expected is visible on the gel (Fig. 7a).
Fig. 7.
Molecular and serological tests for the detection of trypanosomes and trypanosomoses: a Ethidium bromide-stained electrophoresis gel of a monospecific PCR; the result can be considered to be positive when a visible PCR product exhibits the specific weight expected (here: lanes 2, 3, 6–10, 12, 14, 16); otherwise, when the product is non-specific (lane 17) or non-visible (lanes 1, 4, 5, 11, 13, 15), the PCR is negative. Lane 18 is the DNA ladder. b Ethidium bromide-stained electrophoresis gel of a multi-specific PCR based on the amplification of the internal transcribed spacer 1 (ITS1); species-specific results are deduced from the weight of the visible PCR products obtained (here: lanes 1, 12 are the DNA ladder; lanes 2–4 are T. congolense; lanes 5–7 are Trypanozoon; lane 8 is T. theileri; lane 9 is T. simiae; lanes 10, 11 are T. vivax. c Trypanosoma vivax antibody detection ELISA plate; first 2 rows are blanks (A, B), positive controls (C, E, G) and negative controls (D, F, H); all samples are tested in duplicate and appear to be positive, doubtful or negative, according to their mean optical density. d Picture of the card of a CATT/T. evansi exhibiting parasite agglutinations in the positive control and samples 163 and 017; other samples are considered to be negative
For T. vivax, several other primer sets have been published since the development of TVW primers [110–113], but none have proved to exhibit better sensitivity than the gold standard. In addition, it has been claimed by a few authors that some strains of T. vivax, notably from East Africa, may not be detected using TVW primers [110]; however, such parasites have never been isolated, and the results were never confirmed [114]. Consequently, to date, TVW primers remain the gold standard method for detecting and identifying T. vivax, but a well-designed comparison of the different published primers pairs on samples from different regions deserves to be performed.
Similarly, several primers have been developed to detect T. evansi [95, 115, 116], including nested and TaqMan primers [117, 118], but their sensitivity was found to be lower than that of TBR primers [85]. As per their specificity regarding other subspecies of the subgenus Trypanozoon, it was never fully documented. More interestingly, some primers were developed to distinguish Type A from Type B of T. evansi [83, 119]; these primers are helpful for obtaining accurate epidemiological information. More specific methods are also available to identify T. b. gambiense and T. b. rhodesiense [120–123], which could provide new information on the role of domestic and wild fauna in the maintenance of some sleeping sickness foci [26, 27, 29, 59, 124]. However, these single-gene targets are of low sensitivity [125]. Due to the diversity of taxon-specific primers in tsetse flies or mammalian hosts within the tsetse belt in Africa, a complete identification of Trypanosoma species may require three to six or even more PCR tests to be carried out per sample, which considerably increases the cost of the diagnosis.
In the Americas, and outside the tsetse belt in Africa, primers TBR and TVW are recommended for detecting T. evansi and T. vivax, respectively, for the diagnosis of trypanosomes in livestock. However, in horses, it is not possible tomake a definitive distinction between T. equiperdum and T. evansi using standard diagnostic tools. Clinical observations, analysis of the presence of vectors and information on the mode of transmission and overall epidemiological context are necessary to differentiate these two species. In fact, these parasites are so close that even molecular biology techniques barely differentiate them. At the present time, these two species tend to be considered as subspecies of T. brucei; T. brucei evansi and T. brucei equiperdum have polyphyletic origins, as shown by genomic studies [18, 19, 126]. In the Americas, TCZ1 and TCZ2 primers for detecting T. cruzi should also be used in addition to primers TBR and TVW [127]. Finally, a differential diagnosis for Trypanosoma rangeli may also be necessary in wild mammals [128].
Amplifications of the ITS1 of ribosomal DNA have been developed to allow the identification of all African Trypanosoma spp. in single or mixed infections using one single test [129–134], based on the specific weight of the PCR products obtained (Fig. 7b). These tests are helpful for screening; however, the sizing the PCR product(s) on gels can sometime be unreliable. Consequently, sequencing is most often required to confirm species identification, a procedure that is not suitable for routine diagnosis. Alternatively, ITS1 amplification can be used for sample screening and followed by monospecific PCR for species identification, when required. Finally, based on the highly conserved regions from which the primers have been designed, amplification of the ITS1 with TRYP1 primers allows the detection of all African pathogenic trypanosomes, as well as of T. lewisi [96, 134, 135] and even T. cruzi and Leishmania spp. (S. Ravel, personal communication) [136]. Loop-mediated isothermal amplification (LAMP) was also developed for trypanosome diagnosis [137]; however, the limited use of this technique does not fully validate veterinary usage.
PCR techniques require well-equipped laboratories and well-trained technicians. The quality of the results depends on the quality and quantity of DNA preparations and the choice of adequate primers, and the cost remains higher than parasitological techniques. However, depending on the context and the question to be answered, PCR techniques offer significant improvement in terms of sensitivity and specificity. The PCR is a highly sensitive method and typically provides a two- to threefold higher prevalence than parasitological methods when applied to field samples [6, 29, 138, 139]. In the best cases, analytical sensitivity of the PCR test for trypanosomes reaches as low as one to two parasites, or even less, per reaction [85, 139]. The sensitivity depends on the DNA targeted by the primers (repeated sequence or gene vs single gene) and the sample preparation method [140].
The specificity of PCR methods is theoretically very high, ranging from the subgenus to subspecies or type levels depending on the primer set used. PCR can also be used in vectors or in wildlife [141]. The gold standard PCRs are extremely sensitive. False-positive results may occur due to sample contamination with trypanosome DNA from real positive samples. False-negative results may occur when the parasitemia is very low (< 1–10 trypanosome/ml of blood), which is frequent in chronic infections and healthy carriers, when too much DNA was used in the PCR reaction or because of remaining inhibiting factors due to extraction of poor quality DNA. False-negative results may also be obtained when the specificity of the primers is too high, so that not all isolates of a particular Trypanosoma species are recognized. For example, in East Africa, the use of primers for the detection of the RoTat 1.2 VSG gene of T. evansi has undoubtedly left undetected the T. evansi Type B strains that are devoid of this gene [33, 142, 143].
LAMP techniques, sometimes promoted as “field methods,” are also very promising [144]; however, they remain in the hands of those laboratories that implemented their initial development and have never reached the stage of practical and widespread use, so they have never been really validated [143, 145, 146].
Real-time PCR (RT-PCR) methods have been developed for T. evansi [147], T. brucei [148] and T. congolense [149]; they were applied to hosts and vectors with high sensitivity and specificity, but their use has to date been very limited, probably due to associations with high technical skills and equipment and cost. These techniques have not yet been validated for routine diagnostic tests. Other attempts have been made for T. b. gambiense DNA detection, but the results of quantitative RT-PCRs were disappointing, with tests exhibiting a low sensitivity [150].
Spliced Leader trypanosome RNA (SL-RNA) detection was recently developed for the diagnosis of living T. b. gambiense in humans, but this technique has not been evaluated in animals so far [151].
As stated earliers in this review, the detection of pathogenic trypanosomes may be performed in the vectors using the same molecular techniques as used in hosts. However, sample preparation must be adapted to the insect organs of interest in order to get eliminate any PCR inhibitors present in insect samples, such as salivary glands, proboscis and midguts [109, 152] or fecal drop [153].
Finally, sample collection has been simplified by using blood or buffy coats spotted on filter papers [154]; such methods are greatly recommended nowadays, especially for the international shipment of samples. Many samples can be processed simultaneously, making them potentially suitable for large-scale surveys. However, currently, the cost and complex technology of PCR analyses are still limiting factors for generalized routine use of the test in remote enzootic/endemic areas.
Although DNA detection methods exhibit a higher sensitivity and specificity than parasitological methods, they are affected by the same limitations. Indeed, (i) a negative test cannot ascertain the absence of infection (serological tests are better adapted to do so); and (ii) a positive test ascertains the presence of a specific DNA taxon, but in geographical areas where several pathogenic Trypanosoma species co-exist, an animal positive to one taxon may be carrier of one or several others. Users of molecular detection methods must be fully aware of these limitations, which are too often overlooked.
Antigen detection methods
In addition to detecting either the parasites themselves, or their DNA, antigen detection can also be implemented to evidence active infection. ELISAs for antigen detection based on monoclonal antibodies, which were developed in the 1990s, initially showed promise [155]. However, in field evaluations they were found to present a severe lack of sensitivity and specificity and, hence, were abandoned [6, 156, 157]. Nevertheless, antigen detection methods using monoclonal antibodies would be beneficial and likely to be suitable for the development of rapid tests; if based on cautiously pre-identified trypanosome antigens circulating in the bloodstream (constitutive or secreted), this avenue of research should be encouraged. Such a rapid test could be useful for deciding on treatment, probably alongside HCT and Card agglutination tests, providing they can be developed in other species than T. evansi, to complete the panel of ATAO diagnostic tools. For the time being, serological methods for trypanosome are focused on antibody detection.
Antibody detection methods based on native antigens
Antibody detection methods can provide evidence of a contact between the host and the parasite with very high sensitivity. However, due to the persistence of the antibodies in the blood serum after parasite elimination, such techniques cannot ascertain active infection. They are thus valuable tools in epidemiological surveys and for the detection of suspect animals. They exhibit a high specificity for other genera, such as Anaplasma, Babesia and Theileria, among others, but among the Trypanosomatidae, the specificity is rather low due to patent cross-reactions, as detailed in a subsequent section.
The OIE validated four main antibody detection techniques for routine use: the indirect fluorescent antibody test (IFAT), the whole-cell lysate soluble antigens (WCLSA) antibody-detection ELISA, the complement fixation test (CFT; used for Dourine) and the Card Agglutination Test for Trypanosomes (CATT/T. evansi used for Surra). Among these OIE-recommended tests, only the latter test is commercially available (Table 1), making the availability of trypanosome serological diagnostics quite limited. IFAT, CFT and classical ELISAs for the diagnosis of trypanosomosis detect immunoglobulin G (IgG), the levels of which are fairly stable during infection course.
Indirect fluorescent antibody tests
The IFATs for trypanosomes exhibit good sensitivity but limited specificity. The main drawbacks of the IFAT is the need for sophisticated microscopy, the subjectivity of the interpretation, which makes the comparison of results quite tricky, and the tiredness of technician’s eyes it generates [158]. Consequently, the performance of the IFAT remains subjective and as such it is not adapted to large-scale studies. The technique is generally used for individual diagnosis, as an alternative to ELISA, and mostly for Dourine.
Enzyme-linked immunosorbent assay
The original antibody ELISA for trypanosomoses [159] has been further developed for large-scale surveys in bovines, buffalos, camels, horses and pigs [160, 161]. The standard antigens for ELISA (nowadays called “classical ELISA”) are derived from native trypanosome bloodstream forms produced in laboratory rats and purified by diethylaminoethanol (DEAE) anion-exchange chromatography [99]. They include an extensive range of native antigens that confer the tests a high sensitivity [6, 160, 162, 163]. The classical ELISAs recommended by the OIE have been standardized for camels, cattle, buffalo, elephants, among others [104, 164–166]. The result of an ELISA is considered to be positive when the optical density of the sample is higher than the positive threshold previously defined (Fig. 7c).
Using the ELISA, positive seroconversion occurs generally and, on average, around 10–20 days after infection. After a fully curative treatment, negative seroconversion has been observed within 3–4 months in young and adult animals [167] and after 6 months in older individuals [6, 168, 169], although some authors claim it might take up to 13 months [170]. Based on these data, adequate sampling and proper knowledge of trypanocide use facilitate the correct interpretation of the test results; for example, a serological test implemented 6 months after treatment would confirm treatment efficacy.
Antibody-detection ELISAs have high sensitivity and are better suited than parasitological and PCR techniques to establish the prevalence of infected animals. In addition, their genus specificity is high, meaning that infection with other hemoparasites, such as Theileria theileri, Theileria mutans, Babesia divergens or Anaplasma marginale, do not cause cross-reactions in serological tests against pathogenic trypanosomes [159]; even Trypanosoma theileri, the non-pathogenic Megatrypanum found in Bovinae, does not cross-react [171, 172]. However, species-specificity among the pathogenic trypanosomes is generally low due to strong cross-reactions between the main parasites T. vivax, Trypanozoon and T. congolense sensu lato (s.l.) [173].
Immunodiagnostics by ELISA requires expertise and relatively expensive and sophisticated equipment, both of which are not always readily available. The technique also involves the production of native parasites to prepare soluble antigens from whole-cell lysate of the trypanosomes, which are not commercially available. In practice, trypanosome antigen production is limited to specialized laboratories. At the individual level in the field, in terms of test implementation there is a substantial delay between the actual sampling and the availability of the results. Moreover, a cut-off value must be defined and the ELISA adapted, evaluated and validated for each host species. All of these factors may present an obstacle to accurate interpretation of the results, given the difficulty of acquiring reference samples for the various species of interest. Nevertheless, the antibody ELISA lends itself to a high degree of automation and standardization that is suitable for sero-epidemiological studies. After collection, serum samples can be stored at − 20 °C or blotted onto filter papers (then stored as dried serum or blood spots) to make the samples more suitable for international shipment. Taken together, the antibody ELISA is an instrumental tool for large-scale surveys to determine the distribution of ATAO (including NTTAT), as well as for post-treatment or post-control campaign follow-ups. Recent work on lyophilized reagents and serum samples has demonstrated that lyophilization is a convenient way to store and ship reagents for ELISAs [174], which should help considerably in further wide-spread implementation of the ELISA technique for trypanosomes in Africa.
Based on in vivo-produced parasites, the classical ELISA is difficult to standardize for high throughput, and this aspect needs improvement. Quality and standardization of the native antigens for ELISAs could be significantly enhanced through the production of the parasite in vitro. WCLSA prepared this way will guarantee a high sensitivity due to the rich panel of native antigens they exhibit and the lower degradation occurring during preparation. This method will allow high standardization and reproducibility of the antigens produced and also solve the ethical problem of using living animals for parasite production.
Agglutination tests
Agglutination tests have been developed for trypanosomosis detection; however, most have been abandoned due to poor standardization, with the exception of the CATT for T. evansi (CATT/T. evansi), which is commercially available from the Institute of Tropical Medicine, Antwerp, Belgium [175, 176]. The antigen of this CATT consists of fixed and stained T. evansi Rode Trypanozoon antigen type (RoTat) 1.2 parasites produced in rats. The test mainly detects IgMs, which are early-circulating antibodies. IgMs are pentavalent immunoglobulins presenting very high antigen binding affinity; one advantage of these IgMs is that they are prone to form lattices with the antigen. The CATT is carried out on white plastic cards that are rotated for 5 min at 70 revolutions per minute (rpm); the agglutination of stained parasites can be observed by reading with the naked eye (Fig. 7d). A disadvantage of IgMs is that being phagocytized as immune-complexes, their concentration in the serum fluctuates over time, thereby being responsible for false negative test results [177]. As the IgMs have a short half-life, they are a good indicator of a recent infection, or at least of a recent circulation of trypanosomes in the blood. CATT/T. evansi is rather inexpensive, fast, simple and can be implemented in the field on any host species. Its sensitivity is generally high in equids, buffalo, camels, sheep, goat and dogs, with a medium specificity. However, once again, although genus specificity is high, species specificity is limited and cross-reactions with other Trypanozoon and with T. vivax have been reported [41]. Since both specificity and sensitivity of CATT/T. evansi are low in cattle and pigs [95, 96], the test is not adapted to large-scale studies of these animals.
A specific CATT/T. brucei gambiense is available for diagnostic purposes in humans [178]; however, it has not been largely used in animals [26]. Its level of specificity in animals has not been determined, but it can be presumed to be low because of strong cross-reactions already observed among Trypanozoon and between T. brucei and other Salivarian trypanosomes [173, 179].
Complement fixation test
The CFT allows the detection of T. equiperdum (Doflein, 1901) antibodies in both asymptomatic equids and in individuals with clinical signs based on the use of crude antigens derived from the T. equiperdum OVI strain ITMAS 241199C that is adapted to rodents [180]. This test has been used to confirm cases of Dourine [181], a disease that is considered by the OIE to be non-treatable [182]. An inter-laboratory ring trial to evaluate CFT for Dourine diagnosis that involved 25 reference laboratories for Dourine confirmed the reliability of this method and the importance of standardizing critical reagents, including the crude antigens and the use of a standard T. equiperdum serum across multiple laboratories [92, 183].
Trypanolysis test
The trypanolysis test (TL) assesses the presence of specific antibodies through exposure to live T. evansi RoTat 1.2 previously grown in mice [184]. The test has been shown to exhibit high specificity, although comparative studies are scarce. As for tsetse-transmitted trypanosomes, other strains, such as LiTat 1.3, 1.5 and 1.6, used for detection of T. b. gambiense in humans, can also be used for detection of T. b. brucei [29], which tends to show a limited specificity. The TL is rarely used for diagnosis in animals due to it being highly time consuming and costly, with substantial technical constraints and the ethical issue of growing parasites in live animals. Experimental studies are needed to accurately determine the performance of trypanolysis in cattle infected with different Trypanozoon [29].
Antibody detection methods based on purified or recombinant antigens
Several attempts have been made to improve the potential for the standardization of ELISAs through the use of purified or recombinant antigens. Some of these target the VSG, a strategy that could be limited to clonal parasites presenting a highly predominant VAT, such as RoTat 1.2 [184]. The potential limitations of the tests based on VSGs have been recently discussed, with the authors concluding that they cannot be the best tools [33]. Indeed, for T. evansi, the ELISA using RoTat 1.2 VSG might be too specific to be able to detect all variants of the taxon, since T. evansi type B does not express this gene [185], while it is absent in some other isolates [142]. Surprisingly, a new isolate of T. equiperdum was also recently classified as a type B [126]. The reliance on RoTat 1.2 VSG as the basis for diagnosis thus undoubtedly means that non-RoTat 1.2 T. evansi will not be detected [119, 143]. Regarding other Trypanosoma spp., Auty et al. [33] concluded that “the overall within-species diversity in both T. congolense and T. vivax [repertoire] probably means that a ‘catch-all’ diagnostic test based on VSGs is unlikely to be successful”.
Attempts at developing diagnostic tests for T. evansi using invariant antigens were recently made. Although the authors claimed the tests were very sensitive and specific, comprehensive evaluation is still needed, and it is therefore too early to draw conclusions on their value [186].
Other attempts at recombinant antigen-based techniques have been made for the development of rapid tests; promising as far as standardization is concerned, they generally present a lower sensitivity, but exhibit a higher specificity when a highly species-specific antigen is selected [86, 87, 187–189]. However, recombinant antigen techniques are generally based on a single molecule harboring a very limited epitope diversity; as such, they can hardly compete with native antigens in terms of sensitivity. Still, such methods could be helpful to develop highly species-specific tests to be used as a second diagnosis step. For example, in the case of T. cruzi, such tests could allow the diagnostics after screening to be refined with a more sensitive tool based on cross-reactions with native antigens of T. evansi [190]. On the other hand, a recent study has shown that the sensitivity of recombinant antigen-based ELISA could be markedly enhanced by combining several recombinant antigens, which opens the door to further improvements [191].
Other tests have been developed or are under development to target more specific antigens, such as the RDT (rapid diagnosis test) for T. vivax and T. congolense [87]. Recently, an antigen capture-ELISA test for T. vivax and T. congolense using TvGM6 and TcCB1 proteins as antigens, respectively, has been introduced commercially as an RDT (CEVA; Ceva Santé Animale, Libourne, France) [82, 87]. It is the only RDT to reach this stage for the diagnosis of animal African trypanosomosis (AAT), but is still not widely available. In addition, it lacks sensitivity in very early and late infections, and it has the same drawbacks as other antibody-detecting tests in terms of specificity as it will also detect recent other than past infections, which significantly reduces its relevance for treatment decision-making. As the RDT format is primarily relevant for diagnosis at the individual level in the field (pen-side test), it is to be feared that only tests based on antigen detection will be of real commercial interest.
Finally, RDTs for the detection of the human parasites T. b. gambiense were evaluated in animals, while the sensitivity was medium to high, specificity was not satisfying, with the authors concluding that “the SD BIOLINE HAT® is not suitable for screening of T. b. gambiense in domestic livestock” [192, 193].
The OIE-recommended ELISA based on WCLSA remains the best tool for antibody detection with optimal sensitivity. The improvements required on this technique can now be fulfilled, thanks to the use of lyophilized reagents and dry samples, and to the in vitro production of most of the ATAO. It can then be expected, in the near future, that WCLSA will be prepared from well-standardized in vitro-produced parasites.
Conclusions on serological diagnosis
Serological methods for trypanosomoses appear useful, thanks to their high sensitivity and specificity regarding the pathogenic trypanosomes (T. theileri-infected animals do not show seropositivity). However, cross-reactions are very strong between pathogenic trypanosomes [173], or even Leishmania, in humans and animals [69, 194, 195]. Consequently, routine antibody detection methods for trypanosomoses are not species specific. Thus, the seropositivity of an animal to one of the antibody detection methods used (T. vivax, T. evansi, T. brucei, T. congolense, Leishmania, T. cruzi, etc.) must be interpreted as a positivity to one or several of the Trypanosoma and/or Leishmania species present in the geographical area (see Fig. 5). For example, in the Americas, a dog testing seropositive to Leishmania may be—or may have been—infected “either or/and” by Leishmania, T. evansi and T. cruzi. In Africa, when seropositive to T. vivax ELISA, bovines can be considered as “are currently, or have been infected”, “either or/and” by T. vivax, Leishmania, T. congolense s.l., T. brucei spp. and/or T. evansi. Users of serological detection methods should be fully aware of this limitation, which is too often overlooked.
Tools for the detection of animal trypanosomes in humans
Typical human trypanosomes are T. b. gambiense and T. b. rhodesiense in Africa, and T. cruzi in the Americas. Their specific diagnosis methods, quite similar to those used in animals, is reviewed elsewhere (see “Proper use and perspective on diagnosis methods for animal trypanosomoses” [51]). Briefly, they include: (i) clinical suspicions; (ii) parasitological techniques (direct blood or lymph microscopic examination, HCT and mAECT) [196, 197]; and (iii) serological techniques, such as IFAT, ELISA, CATT/T. b. gambiense, RDTs and TL specific for sleeping sickness [198–202]. With the exception of the latter, which exhibits high species specificity, these serological tests have low species specificity. However, in a given epidemiological setting, where only one pathogenic trypanosome species or subspecies is expected in humans, a genus-specific diagnosis is sufficient for population screening. Molecular techniques are also useful in conjunction with mAECT to detect T. brucei spp. [203]. Therefore, the use of these tools must be tailored to the final purpose of the investigation, which may be an epidemiological study, blood bank screening or individual diagnosis for treatment decision-making or follow-up.
Although humans have innate immunity against most animal trypanosomes [204], a few cases of human infections with animal trypanosomes have been reported. A small part was due to T. vivax and T. congolense, but most of the confirmed cases were due to T. evansi (agent of the Surra in animals) and T. lewisi (parasite of rats transmitted by fleas), which were detected occasionally in Africa, but mostly in Asia [12]. In particular, two T. evansi cases of febrile episodes were detected by microscope examination of blood and confirmed by molecular assays in India in 2004 [205, 206] and in Vietnam in 2015 [207]. For the diagnosis of T. evansi in humans, CATT/T. evansi, ELISA T. evansi, parasitological techniques and PCR commonly used for animals were applied (with only slight modifications for ELISA) [207].
For T. lewisi, a dozen cases have been described with variable issues: self-cure in some cases, successful treatment using drugs effective for HAT (pentamidine, suramine, melarsoprol) or death [208]. However, recent observations demonstrated that none of these trypanocides was efficient against a T. lewisi isolated in Thailand [209]. Moreover, resistance testing to normal human serum reveals that T. lewisi is a potentially underestimated human pathogen [210]. The increase of anthropization and the presence of invasive rodents dwelling with humans in poor living conditions reinforces the hypothesis of in-door contamination of humans by T. lewisi [211].
Pertaining to the diagnosis of T. lewisi in humans, most of the known cases have so far been identified by direct microscope observation (Fig. 6) and by molecular techniques; specific primers hybridizing inside the ITS1 were published to detect T. lewisi DNA [96]. In Thailand, a 45-day-old infant infected with a T. lewisi-like was indeed diagnosed based on the ITS1 sequence [212]. Attempts to develop ELISA T. lewisi were recently made with promising results [213]; however, human reference serum samples are still lacking for the standardization of such tests.
Conclusions
The diagnosis of trypanosomoses can proceed based on evidence of: (i) the parasite itself, with limited sensitivity and specificity; (ii) its DNA, with higher sensitivity and potentially high specificity (although limited within the Trypanozoon); (iii) immunoglobulins directed against more or less specific parasite antigens. The characteristics of these tests must be carefully considered for effective application according to the different epidemiological situations. In addition, a comprehensive analysis of the proper use of these diagnostic techniques will be necessary to adapt recommendations and to support the prospects of developing complementary diagnostic methods, including rapid tests that could be applied individually for decision making in the field.
Acknowledgements
We are very grateful to Veerle Lejon (Directeur de Recherche, Institut de Recherche pour le Développement [IRD] for her perusal revision of the manuscript.
Abbreviations
- AAT
Animal African trypanosomosis
- a-HT
Atypical human infections by animal trypanosomes
- ATAO
Animal trypanosomoses of African origin
- BCM
Buffy coat method
- CATT
Card agglutination test
- CFT
Complement fixation test
- DEAE
Diethylaminoethanol
- ELISA
Enzyme-linked immunosorbent assay
- FAO
Food and Agriculture Organization of the United Nations
- GSBS
Giemsa-stained thin blood smear
- HAT
Human African trypanosomiasis
- HTC
Hematocrit concentrations technique
- IFAT
Indirect fluorescent antibody test
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- ITS1
Internal transcribed spacer 1
- LAMP
Loop-mediated isothermal amplification
- mAECT
Mini anion exchange centrifugation technique
- NTD
Neglected tropical disease
- NTTAT
Non-tsetse transmitted animal trypanosomosis
- PCV
Packed cell volume
- OIE
World Organization for Animal Health
- RoTat
Rode Trypanozoon antigen type
- rpm
Revolutions per minute
- RT-PCR
Real-time PCR
- SL-RNA
Spliced-leader trypanosome RNA
- TL
Trypanolysis test
- VAT
Variable antigen type
- VSG
Variable surface glycoprotein
- WCLSA
Whole-cell lysate-soluble antigens
Authors' contributions
Conceptualization: MD. Writing and preparing the original draft of the manuscript: MD. Writing, reviewing and editing the manuscript: MD, MG, AS, ST, GB, AB, GG, PT, SH, SR, DS, VJ, SJ, PJ, PS, DB. All authors read and approved the final manuscript.
Funding
No funding.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marc Desquesnes, Email: marc.desquesnes@cirad.fr.
Marisa Gonzatti, Email: mgonzat@usb.ve.
Alireza Sazmand, Email: alireza.sazmand@basu.ac.ir.
Sophie Thévenon, Email: sophie.thevenon@cirad.fr.
Géraldine Bossard, Email: geraldine.bossard@cirad.fr.
Alain Boulangé, Email: abk32a@gmail.com.
Geoffrey Gimonneau, Email: geoffrey.gimonneau@cirad.fr.
Philippe Truc, Email: philippe.truc@ird.fr.
Stéphane Herder, Email: stephane.herder@ird.fr.
Sophie Ravel, Email: Sophie.ravel@ird.fr.
Denis Sereno, Email: denis.sereno@ird.fr.
Vincent Jamonneau, Email: vincent.jamonneau@ird.fr.
Sathaporn Jittapalapong, Email: sjittapalapong@gmail.com.
Philippe Jacquiet, Email: philippe.jacquiet@envt.fr.
Philippe Solano, Email: philippe.solano@ird.fr.
David Berthier, Email: david.berthier@cirad.fr.
References
- 1.Kaufer A, Ellis J, Stark D, Barratt J. The evolution of trypanosomatid taxonomy. Parasit Vectors. 2017;10:287. doi: 10.1186/s13071-017-2204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Res. 2017;6:750. doi: 10.12688/f1000research.11120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller N, Welle M, Lobsiger L, Stoffel MH, Boghenbor KK, Hilbe M, et al. Occurrence of Leishmania sp. in cutaneous lesions of horses in Central Europe. Vet Parasitol. 2009;166:346–351. doi: 10.1016/j.vetpar.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Lobsiger L, Muller N, Schweizer T, Frey CF, Wiederkehr D, Zumkehr B, et al. An autochthonous case of cutaneous bovine leishmaniasis in Switzerland. Vet Parasitol. 2010;169:408–414. doi: 10.1016/j.vetpar.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Murray M, Gray AR. The current situation on animal trypanosomiasis in Africa. Prev Vet Med. 1984;2:23–30. doi: 10.1016/0167-5877(84)90045-X. [DOI] [Google Scholar]
- 6.Desquesnes M. Livestock trypanosomoses and their vectors in Latin America. Paris: World Organization for Animal Health (OIE); 2004.
- 7.Kennedy PG. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness) Lancet Neurol. 2013;12:186–194. doi: 10.1016/S1474-4422(12)70296-X. [DOI] [PubMed] [Google Scholar]
- 8.Gonzatti MI, González-Baradat B, Aso PM, Reyna-Bello A. Trypanosoma (Duttonella) vivax and typanosomosis in Latin America: Secadera/Huequera/Cacho Hueco. In: Magez S, Radwanska M, editors. Trypanosomes and trypanosomiasis. Vienna: Springer; 2014. pp. 261–285. [Google Scholar]
- 9.Buscher P, Cecchi G, Jamonneau V, Priotto G. Human African trypanosomiasis. Lancet. 2017;390:2397–2409. doi: 10.1016/S0140-6736(17)31510-6. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Molina JA, Molina I. Chagas disease. Lancet. 2018;391:82–94. doi: 10.1016/S0140-6736(17)31612-4. [DOI] [PubMed] [Google Scholar]
- 11.Hoare CA. The trypanosomes of mammals. A Zoological Monograph. Oxford: Blackwell; 1972. [Google Scholar]
- 12.Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, Joshi P, et al. Atypical human infections by animal trypanosomes. PLoS Negl Trop Dis. 2013;7:e2256. doi: 10.1371/journal.pntd.0002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desquesnes M. Compendium of diagnostic protocols of the OIE reference laboratory for animal trypanosomoses of African origin. Montpellier: Organization for Animal Health (OIE); 2017.
- 14.Molia S, Saillard J, Dellagi K, Cliquet F, Bart J-M, Rotureau B, et al. Practices in research, surveillance and control of neglected tropical diseases by One Health approaches: a survey targeting scientists from French-speaking countries. PLoS Negl Trop Dis. 2021;15:e0009246. doi: 10.1371/journal.pntd.0009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zweygarth E, Rehbein G, Ahmed J. In vitro culture of infective blood forms of Trypanosoma brucei evansi (Steel 1885) Berl Munch Tierarztl Wochenschr. 1982;95:407–408. [PubMed] [Google Scholar]
- 16.Wells EA. Animal trypanosomiasis in South America. Prev Vet Med. 1984;2:31–41. doi: 10.1016/0167-5877(84)90046-1. [DOI] [Google Scholar]
- 17.Sazmand A, Desquesnes M, Otranto D. Trypanosoma evansi. Trends Parasitol. 2022;S1471–4922:00001. doi: 10.1016/j.pt.2021.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Wei Y, Wen Y-Z, Desquesnes M, Lun Z-R. Molecular epidemiology of Trypanosoma evansi and T. equiperdum and atypical human infection by animal trypanosomes. In: Hide G, editor. The molecular epidemiology of trypanosomes and Leishmania. New York: Landes Bioscience and Springer Science; 2011. pp. 1–14. [Google Scholar]
- 19.Carnes J, Anupama A, Balmer O, Jackson A, Lewis M, Brown R, et al. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis. 2015;9:e3404. doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsen P, Amoudi M, Leclerq M. First record of Glossina fuscipes fuscipes Newstead, 1910 and Glossina morsitans submorsitans Newstead, 1910 in southwestern Saudi Arabia. Ann Soc Belg Med Trop. 1990;70:281–287. [PubMed] [Google Scholar]
- 21.Itty P. Economics of trypanosomiasis control: research implications. In: Kategile JA, Mubi S, editors. Future of livestock industries in east and southern Africa. Addis Ababa: International Livestock Centre for Africa; 1992. p. 227. [Google Scholar]
- 22.Food and Agricultural Organization of the United Nations (FAO). Programme against African trypanosomosis (PAAT). https://www.fao.org/paat/the-programme/the-disease/en/. Accessed 10 Feb 2022.
- 23.Bengaly Z, Sidibe I, Ganaba R, Desquesnes M, Boly H, Sawadogo L. Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Vet Parasitol. 2002;108:1–19. doi: 10.1016/S0304-4017(02)00164-4. [DOI] [PubMed] [Google Scholar]
- 24.Truc P, Mathieu-Daudé F, Tibayrenc M. Multilocus isozyme identification of Trypanosoma brucei stocks isolated in Central Africa: evidence for an animal reservoir of sleeping sickness in Congo. Acta Trop. 1991;49:127–135. doi: 10.1016/0001-706X(91)90060-W. [DOI] [PubMed] [Google Scholar]
- 25.Pepin J, Meda HA. The epidemiology and control of human African trypanosomiasis. Adv Parasitol. 2001;49:71–132. doi: 10.1016/S0065-308X(01)49038-5. [DOI] [PubMed] [Google Scholar]
- 26.Njiokou F, Nimpaye H, Simo G, Njitchouang G, Asonganyi T, Cuny G, et al. Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite. 2010;17:61–66. doi: 10.1051/parasite/2010171061. [DOI] [PubMed] [Google Scholar]
- 27.Hamill L, Kaare M, Welburn S, Picozzi K. Domestic pigs as potential reservoirs of human and animal trypanosomiasis in northern Tanzania. Parasit Vectors. 2013 doi: 10.1186/1756-3305-6-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buscher P, Bart JM, Boelaert M, Bucheton B, Cecchi G, Chitnis N, et al. Do cryptic reservoirs threaten gambiense-sleeping sickness elimination? Trends Parasitol. 2018;34:197–207. doi: 10.1016/j.pt.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N'Djetchi MK, Ilboudo H, Koffi M, Kabore J, Kabore JW, Kaba D, et al. The study of trypanosome species circulating in domestic animals in two human African trypanosomiasis foci of Cote d'Ivoire identifies pigs and cattle as potential reservoirs of Trypanosoma brucei gambiense. PLoS Negl Trop Dis. 2017;11:e0005993. doi: 10.1371/journal.pntd.0005993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellde BT, Reardon MJ, Kovatch RM, Chumo DA, Williams JS, Boyce WL, et al. Experimental infection of cattle with Trypanosoma brucei rhodesiense. Ann Trop Med Parasitol. 1989;83:133–150. doi: 10.1080/00034983.1989.11812418. [DOI] [PubMed] [Google Scholar]
- 31.Desquesnes M, Gutierrez CA. Animal trypanosomosis: an important constraint for livestock in tropical and sub-tropical regions. In: Javed T, editor. Livestock: rearing, farming practices and diseases. Hauppauge: Nova Science Publishers; 2012. pp. 127–144. [Google Scholar]
- 32.Uilenberg G. A field guide for the diagnosis, treatment and prevention of African animal trypanosomosis. Rome: FAO; 1998. https://www.fao.org/3/x0413e/x0413e00.htm. Accessed 10 Feb 2022.
- 33.Auty H, Torr SJ, Michoel T, Jayaraman S, Morrison LJ. Cattle trypanosomosis: the diversity of trypanosomes and implications for disease epidemiology and control. Rev Sci Tech. 2015;34:587–598. doi: 10.20506/rst.34.2.2382. [DOI] [PubMed] [Google Scholar]
- 34.Giordani F, Morrison LJ, Rowan TG, De Koning HP, Barrett MP. The animal trypanosomiases and their chemotherapy: a review. Parasitology. 2016;143:1862–1889. doi: 10.1017/S0031182016001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan-Kadle AA, Ibrahim AM, Nyingilili HS, Yusuf AA, Vieira RF. Parasitological and molecular detection of Trypanosoma spp in cattle, goats and sheep in Somalia. Parasitology. 2020;147:1786–1791. doi: 10.1017/S003118202000178X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fetene E, Leta S, Regassa F, Büscher P. Global distribution, host range and prevalence of Trypanosoma vivax: a systematic review and meta-analysis. Parasit Vectors. 2021;14:80. doi: 10.1186/s13071-021-04584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldacchino F, Muenworn V, Desquesnes M, Desoli F, Charoenviriyaphap T, Duvallet G. Transmission of pathogens by Stomoxys flies (Diptera, Muscidae): a review. Parasite. 2013;20:26. doi: 10.1051/parasite/2013026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldacchino F, Desquesnes M, Mihok S, Foil LD, Duvallet G, Jittapalapong S. Tabanids: neglected subjects of research, but important vectors of disease agents! Infect Genet Evol. 2014;28:596–615. doi: 10.1016/j.meegid.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Bouet G, Roubaud E. Expériences de transmission des trypanosomiases animales d'Afrique occidentale Française par les stomoxes. Bull Soc Path Exot. 1912;5:544–550. [Google Scholar]
- 40.Desquesnes M, Dia ML. Trypanosoma vivax: mechanical transmission in cattle by one of the most common African tabanids Atylotus agrestis. Exp Parasitol. 2003;103:35–43. doi: 10.1016/S0014-4894(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 41.Birhanu H, Fikru R, Said M, Kidane W, Gebrehiwot T, Hagos A, et al. Epidemiology of Trypanosoma evansi and Trypanosoma vivax in domestic animals from selected districts of Tigray and Afar regions, northern Ethiopia. Parasit Vectors. 2015;8:212. doi: 10.1186/s13071-015-0818-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asghari MM, Rassouli M. First identification of Trypanosoma vivax among camels (Camelus dromedarius) in Yazd, central Iran, jointly with Trypanosoma evansi. Parasitol Int. 2022;86:102450. doi: 10.1016/j.parint.2021.102450. [DOI] [PubMed] [Google Scholar]
- 43.Desquesnes M, Holzmuller P, Lai DH, Dargantes A, Lun ZR, Jittaplapong S. Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. BioMed Res Int. 2013;2013:194176. doi: 10.1155/2013/194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claes F, Agbo EC, Radwanska M, Te Pas MF, Baltz T, De Waal DT, et al. How does Trypanosoma equiperdum fit into the Trypanozoon group? A cluster analysis by RAPD and multiplex-endonuclease genotyping approach. Parasitology. 2003;126:425–431. doi: 10.1017/S0031182003002968. [DOI] [PubMed] [Google Scholar]
- 45.Calistri P, Narcisi V, Atzeni M, De Massis F, Tittarelli M, Mercante MT, et al. Dourine reemergence in Italy. J Equine Vet Sci. 2013;33:83–89. doi: 10.1016/j.jevs.2012.05.057. [DOI] [Google Scholar]
- 46.Suganuma K, Narantsatsral S, Battur B, Yamasaki S, Otgonsuren D, Musinguzi SP, et al. Isolation, cultivation and molecular characterization of a new Trypanosoma equiperdum strain in Mongolia. Parasit Vectors. 2016;9:481. doi: 10.1186/s13071-016-1755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gizaw Y, Ashenafi H, Demssie T, Bekana M, Govaere J, Abei G. Pathological observations in horses naturally infected with Trypanosoma equiperdum in Western Arsi Zone, Ethiopia. J Adv Vet Res. 2021;11:9–16. [Google Scholar]
- 48.Sazmand A, Bahari A, Papi S, Otranto D. Parasitic diseases of equids in Iran (1931–2020): a literature review. Parasit Vectors. 2020;13:586. doi: 10.1186/s13071-020-04472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touratier L. The Office International des epizooties ad hoc group on non tsetse animal trypanosomoses: its origin, scope and perspectives. Mem Inst Oswaldo Cruz. 1999;94:191–194. doi: 10.1590/S0074-02761999000200012. [DOI] [PubMed] [Google Scholar]
- 50.Aregawi WG, Agga GE, Abdi RD, Büscher P. Systematic review and meta-analysis on the global distribution, host range, and prevalence of Trypanosoma evansi. Parasit Vectors. 2019;12:67. doi: 10.1186/s13071-019-3311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desquesnes M. Nagana: infections with salivarian trypanosomoses (excluding Trypanosoma evansi and T. equiperdum). In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2021. Paris: OIE. https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/. Accessed 10 Feb 2022.
- 52.Bukachi SA, Wandibba S, Nyamongo IK. The socio-economic burden of human African trypanosomiasis and the coping strategies of households in the South Western Kenya foci. PLoS Negl Trop Dis. 2017;11:e0006002. doi: 10.1371/journal.pntd.0006002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franco JR, Cecchi G, Priotto G, Paone M, Diarra A, Grout L, et al. Monitoring the elimination of human African trypanosomiasis at continental and country level: Update to 2018. PLoS Negl Trop Dis. 2020;14:e0008261. doi: 10.1371/journal.pntd.0008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao JM, Qian ZY, Hide G, Lai DH, Lun ZR, Wu ZD. Human African trypanosomiasis: the current situation in endemic regions and the risks for non-endemic regions from imported cases. Parasitology. 2020;147:922–931. doi: 10.1017/S0031182020000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehlitz D, Molyneux D. The elimination of Trypanosoma brucei gambiense? Challenges of reservoir hosts and transmission cycles: Expect the unexpected. Parasit Epidemiol. 2019;6:e00113. doi: 10.1016/j.parepi.2019.e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jamonneau V, Truc P, Grébaut P, Herder S, Ravel S, Solano P, et al. Trypanosoma brucei gambiense Group 2: the unusual suspect. Trends Parasitol. 2019;35:983–995. doi: 10.1016/j.pt.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Welburn SC, Picozzi K, Fèvre EM, Coleman PG, Odiit M, Carrington M, et al. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001;358:2017–2019. doi: 10.1016/S0140-6736(01)07096-9. [DOI] [PubMed] [Google Scholar]
- 58.Ng'ayo MO, Njiru ZK, Kenya EU, Muluvi GM, Osir EO, Masiga DK. Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness? Kinetoplastid Biol Dis. 2005;4:5. doi: 10.1186/1475-9292-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franco JR, Simarro PP, Diarra A, Jannin JG. Epidemiology of human African trypanosomiasis. Clin Epidemiol. 2014;6:257–275. doi: 10.2147/CLEP.S39728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmes P. On the road to elimination of Rhodesiense human African trypanosomiasis: first WHO meeting of stakeholders. PLoS Negl Trop Dis. 2015;9:e0003571. doi: 10.1371/journal.pntd.0003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simo G, Rayaisse JB. Challenges facing the elimination of sleeping sickness in west and central Africa: sustainable control of animal trypanosomiasis as an indispensable approach to achieve the goal. Parasit Vectors. 2015;8:640. doi: 10.1186/s13071-015-1254-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hemmige V, Tanowitz H, Sethi A. Trypanosoma cruzi infection: a review with emphasis on cutaneous manifestations. Int J Parasitol. 2012;51:501–508. doi: 10.1111/j.1365-4632.2011.05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Álvarez-Hernández D-A, Franyuti-Kellya G-A, Díaz-López-Silvac R, González-Chávezd A-M, González-Hermosillo-Cornejo D, Vázquez-López R. Chagas disease: Current perspectives on a forgotten disease. Rev Med Hosp Gen Mex. 2016;81:154–164. doi: 10.1016/j.hgmx.2016.09.010. [DOI] [Google Scholar]
- 64.Coura JR. The main sceneries of Chagas disease transmission. The vectors, blood and oral transmissions—a comprehensive review. Mem Inst Oswaldo Cruz. 2015;110:277–282. doi: 10.1590/0074-0276140362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardoso AV, Lescano SA, Amato Neto V, Gakiya E, Santos SV. Survival of Trypanosoma cruzi in sugar cane used to prepare juice. Rev Inst Med Trop Sao Paulo. 2006;48:287–289. doi: 10.1590/S0036-46652006000500009. [DOI] [PubMed] [Google Scholar]
- 66.Steindel M, Kramer Pacheco L, Scholl D, Soares M, de Moraes MH, Eger I, et al. Characterization of Trypanosoma cruzi isolated from humans, vectors, and animal reservoirs following an outbreak of acute human Chagas disease in Santa Catarina State. Brazil Diagn Microbiol Infect Dis. 2008;60:25–32. doi: 10.1016/j.diagmicrobio.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 67.AlarcondeNoya B, Diaz-Bello Z, Colmenares C, Ruiz-Guevara R, Mauriello L, Zavala-Jaspe R, et al. Large urban outbreak of orally acquired acute Chagas disease at a school in Caracas. Venezuela. J Infect Dis. 2010;201:1308–1315. doi: 10.1086/651608. [DOI] [PubMed] [Google Scholar]
- 68.de Noya AB, Colmenares C, Dıaz-Bello Z, Ruiz-Guevara R, Medina K, Munoz-Calderon A, et al. Orally-transmitted Chagas disease: Epidemiological, clinical, serological and molecular outcomes of a school microepidemic in Chichiriviche de la Costa, Venezuela. Parasite Epidemiol Control. 2016;1:188–198. doi: 10.1016/j.parepi.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desquesnes M. Veterinary aspects. In: Telleria J, Tibayrenc M, editors. American Trypanosomiasis Chagas disease: one hundred years of research. Amsterdam: Elsevier; 2017. pp. 283–298. [Google Scholar]
- 70.Monzon CM, Colman OLR. Estudio seroepidemiologique de la tripanosomiasis equina (O. Mal de Caderas) mediante la prueba de immunofluorescencia indirecta en la Provincia de Formosa (Argentina) Anos 1983 à 1987. Arq Bras Med Vet Zootec. 1988;1988:279–285. [Google Scholar]
- 71.Yauri V, Castro-Sesquen YE, Verastegui M, Angulo N, Recuenco F, Cabello I, et al. Domestic pig (Sus scrofa) as an animal model for experimental Trypanosoma cruzi infection. Am J Trop Med Hyg. 2016;94:1020–1027. doi: 10.4269/ajtmh.15-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raccurt CP. Trypanosoma cruzi en Guyane Française: revue des données accumulées depuis 1940. Méd Trop. 1996;56:79–87. [PubMed] [Google Scholar]
- 73.Caldas IS, da Matta Guedes PM, dos Santos FM, de Figueire do Diniz L, Martins TA, Nascimento AF, et al. Myocardial scars correlate with eletrocardiographic changes in chronic Trypanosoma cruzi infection for dogs treated with benznidazole. Trop Med Int Health. 2013;18:75–84. doi: 10.1111/tmi.12002. [DOI] [PubMed] [Google Scholar]
- 74.Bryan LK, Hamer SA, Shaw S, Curtis-Robles R, Auckland LD, Hodo CL, et al. Chagas disease in a Texan horse with neurologic deficits. Vet Parasitol. 2016;216:13–17. doi: 10.1016/j.vetpar.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Browne AJ, Guerra CA, Alves RV, da Costa VM, Wilson AL, Pigott DM, et al. The contemporary distribution of Trypanosoma cruzi infection in humans, alternative hosts and vectors. Sci Data. 2017;4:170050. doi: 10.1038/sdata.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bern C, Kjos S, Yabsley MJ, Montgomery SP. Trypanosoma cruzi and Chagas' disease in the United States. Clin Microb Rev. 2011;24:655–681. doi: 10.1128/CMR.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sazmand A, Joachim A, Otranto D. Zoonotic parasites of dromedary camels: so important, so ignored. Parasit Vectors. 2019;12:610. doi: 10.1186/s13071-019-3863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibrahim MAM, Weber JS, Ngomtcho SCH, Signaboubo D, Berger P, Hassane HM, et al. Diversity of trypanosomes in humans and cattle in the HAT foci Mandoul and Maro, southern Chad—a matter of concern for zoonotic potential? PLoS Negl Trop Dis. 2021;15:e0009323. doi: 10.1371/journal.pntd.0009323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toure SM. Diagnostic des trypanosomiases animales. Rev Elev Med Vet Pays Trop. 1977;30:1–10. doi: 10.19182/remvt.8099. [DOI] [PubMed] [Google Scholar]
- 80.Paris J, Murray M, McOdimba F. A comparative evaluation of the parasitological techniques currently available for the diagnosis of African trypanosomiasis in cattle. Acta Trop. 1982;39:307–316. [PubMed] [Google Scholar]
- 81.World Organization for Animal Health (OIE). Manual of diagnostic tests and vaccines for terrestrial animals 2021. Paris: OIE; 2021. https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/. Accessed 10 Feb 2022.
- 82.Pillay D, Izotte J, Fikru R, Buscher P, Mucache H, Neves L, et al. Trypanosoma vivax GM6 antigen: a candidate antigen for diagnosis of African animal trypanosomosis in cattle. PLoS ONE. 2013;8:e78565. doi: 10.1371/journal.pone.0078565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Claes F, Radwanska M, Urakawa T, Majiwa PA, Goddeeris B, Buscher P. Variable Surface Glycoprotein RoTat 1.2 PCR as a specific diagnostic tool for the detection of Trypanosoma evansi infections. Kinetoplastid Biol Dis. 2004;3:3. doi: 10.1186/1475-9292-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Clare Bronsvoort BM, Wissmann BV, Fevre EM, Handel IG, Picozzi K, Welburn SC. No gold standard estimation of the sensitivity and specificity of two molecular diagnostic protocols for Trypanosoma brucei spp in Western Kenya. PLoS ONE. 2010;5:e8628. doi: 10.1371/journal.pone.0008628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pruvot M, Kamyingkird K, Desquesnes M, Sarataphan N, Jittapalapong S. A comparison of six primer sets for detection of Trypanosoma evansi by polymerase chain reaction in rodents and Thai livestock. Vet Parasitol. 2010;171:185–193. doi: 10.1016/j.vetpar.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 86.Tran T, Claes F, Verloo D, De Greve H, Büscher P. Towards a new reference test for Surra in camels. Clin Vaccine Immunol. 2009;16:999–1002. doi: 10.1128/CVI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boulange A, Pillay D, Chevtzoff C, Biteau N, Come de Graca V, Rempeters L, et al. Development of a rapid antibody test for point-of-care diagnosis of animal African trypanosomosis. Vet Parasitol. 2017;233:32–38. doi: 10.1016/j.vetpar.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 88.Delafosse A, Doutoum AA. Comparaison de trois tests sérologiques pour le diagnostic de terrain du surra (trypanosomose à Trypanosoma evansi) chez le dromadaire au Tchad. Rev Elev Med Vet Pays Trop. 2000;53:249–256. doi: 10.19182/remvt.9720. [DOI] [Google Scholar]
- 89.Claes F, Ilgekbayeva G, Verloo D, Saidouldin T, Geerts S, Buscher P, et al. Comparison of serological tests for equine trypanosomosis in naturally infected horses from Kazakhstan. Vet Parasitol. 2005;131:221–225. doi: 10.1016/j.vetpar.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Reid S, Copeman D. The development and validation of an antibody-ELISA to detect Trypanosoma evansi infection in cattle in Australia and Papua New Guinea. Prev Vet Med. 2003;61:195–208. doi: 10.1016/j.prevetmed.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Fikru R, Andualem Y, Getachew T, Menten J, Hasker E, Merga B, et al. Trypanosome infection in dromedary camels in Eastern Ethiopia: Prevalence, relative performance of diagnostic tools and host related risk factors. Vet Parasitol. 2015;211:175–181. doi: 10.1016/j.vetpar.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 92.Cauchard J, Soldan A, Madeline A, Johnson P, Büscher P, Petry S. Inter-laboratory ring trials to evaluate serological methods for dourine diagnosis. Vet Parasitol. 2014;205:70–76. doi: 10.1016/j.vetpar.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 93.Woo PTK. The heamatocrit centrifuge technique for diagnosis of African trypanosomiasis. Acta Trop. 1970;27:384–386. [PubMed] [Google Scholar]
- 94.Murray M, Murray PK, McIntyre WIM. An improved parasitological technique for the diagnosis of African trypanosomiasis. Trans R Soc Trop Med Hyg. 1977;71:325–326. doi: 10.1016/0035-9203(77)90110-9. [DOI] [PubMed] [Google Scholar]
- 95.Holland WG, Thanh NG, Do TT, Sangmaneedet S, Goddeeris B, Vercruysse J. Evaluation of diagnostic tests for Trypanosoma evansi in experimentally infected pigs and subsequent use in field surveys in north Vietnam and Thailand. Trop Anim Hlth Prod. 2005;37:457–467. doi: 10.1007/s11250-005-1217-y. [DOI] [PubMed] [Google Scholar]
- 96.Desquesnes M, Kamyingkird K, Yangtara S, Milocco C, Ravel S, Wang M-H, et al. Specific primers for PCR amplification of the ITS1 (ribosomal DNA) of Trypanosoma lewisi. Infect Genet Evol. 2011;11:1361–1367. doi: 10.1016/j.meegid.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 97.Whitelaw D, Gardiner PR, Murray M. Extravascular foci of Trypanosoma vivax in goats: the central nervous system and aqueous humor of the eye as potential sources of relapse infections after chemotherapy. Parasitology. 1988;97:51–61. doi: 10.1017/S0031182000066737. [DOI] [PubMed] [Google Scholar]
- 98.Camara M, Soumah AM, Ilboudo H, Travaillé C, Clucas C, Cooper A, et al. Extravascular dermal trypanosomes in suspected and confirmed cases of gambiense human African trypanosomiasis. Clin Infect Dis. 2021;73:12–20. doi: 10.1093/cid/ciaa897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lanham SM, Godfrey DG. Isolation of salivarian trypanosomes from man and other mamals using DEAE-cellulose. Exp Parasitol. 1970;28:521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- 100.Lejon V, Buscher P, Nzoumbou-Boko R, Bossard G, Jamonneau V, Bucheton B, et al. The separation of trypanosomes from blood by anion exchange chromatography: from Sheila Lanham's discovery 50 years ago to a gold standard for sleeping sickness diagnosis. PLoS Negl Trop Dis. 2019;13:e0007051. doi: 10.1371/journal.pntd.0007051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lumsden W, Kimber C, Evans D, Doig S. Trypanosoma brucei: miniature anion-exchange centrifugation technique for detection of low parasitaemias: adaptation for field use. Trans R Soc Trop Med Hyg. 1979;73:312–317. doi: 10.1016/0035-9203(79)90092-0. [DOI] [PubMed] [Google Scholar]
- 102.Lumsden WH, Kimber CD, Dukes P, Haller L, Stanghellini A, Duvallet G. Field diagnosis of sleeping sickness in the Ivory Coast. I. Comparison of the miniature anion-exchange/centrifugation technique with other protozoological methods. Trans R Soc Trop Med Hyg. 1981;75:242–250. doi: 10.1016/0035-9203(81)90326-6. [DOI] [PubMed] [Google Scholar]
- 103.Smith CJ, Levine RF, Mansfield JM. Cloning of African trypanosomes in mice immunosuppressed by cyclophosphamide treatment. Am J Trop Med Hyg. 1982;31:1098–1102. doi: 10.4269/ajtmh.1982.31.1098. [DOI] [PubMed] [Google Scholar]
- 104.Desquesnes M, Bossard G, Thevenon S, Patrel D, Ravel S, Pavlovic D, et al. Development and application of an antibody-ELISA to follow up a Trypanosoma evansi outbreak in a dromedary camel herd in France. Vet Parasitol. 2009;162:214–220. doi: 10.1016/j.vetpar.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 105.Verloo D, Brandt J, Van Meirvenne N, Buscher P. Comparative in vitro isolation of Trypanosoma theileri from cattle in Belgium. Vet Parasitol. 2000;89:129–132. doi: 10.1016/S0304-4017(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 106.De La Rocque S, Lefrancois T, Reifenberg JM, Solano P, Kabore I, Bengaly Z, et al. PCR analysis and spatial repartition of trypanosomes infecting tsetse flies in Sideradougou area of Burkina Faso. Ann N Y Acad Sci. 1998;849:32–38. doi: 10.1111/j.1749-6632.1998.tb11030.x. [DOI] [PubMed] [Google Scholar]
- 107.Masiga DK, McNamara JJ, Gibson WC. A repetitive DNA sequence specific for Trypanosoma (Nannomonas) godfreyi. Vet Parasitol. 1996;62:27–33. doi: 10.1016/0304-4017(95)00847-0. [DOI] [PubMed] [Google Scholar]
- 108.Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE. Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology. 1989;99:57–66. doi: 10.1017/S0031182000061023. [DOI] [PubMed] [Google Scholar]
- 109.Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22:909–918. doi: 10.1016/0020-7519(92)90047-O. [DOI] [PubMed] [Google Scholar]
- 110.Masake RA, Majiwa PA, Moloo SK, Makau JM, Njuguna JT, Maina M, et al. Sensitive and specific detection of Trypanosoma vivax using the polymerase chain reaction. Exp Parasitol. 1997;85:193–205. doi: 10.1006/expr.1996.4124. [DOI] [PubMed] [Google Scholar]
- 111.Morlais I, Ravel S, Grebaut P, Dumas V, Cuny G. New molecular marker for Trypanosoma (Duttonella) vivax identification. Acta Trop. 2001;80:207–213. doi: 10.1016/S0001-706X(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 112.Ventura RM, Paiva F, Silva RA, Takeda GF, Buck GA, Teixeira MM. Trypanosoma vivax: characterization of the spliced-leader gene of a Brazilian stock and species-specific detection by PCR amplification of an intergenic spacer sequence. Exp Parasitol. 2001;99:37–48. doi: 10.1006/expr.2001.4641. [DOI] [PubMed] [Google Scholar]
- 113.Fikru R, Hagos A, Roge S, Reyna-Bello A, Gonzatti MI, Merga B, et al. A proline racemase based PCR for identification of Trypanosoma vivax in cattle blood. PLoS ONE. 2014;9:e84819. doi: 10.1371/journal.pone.0084819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Solano P, Desquesnes M, Sidibe I. Le diagnostic de Trypanosoma vivax : un problème non résolu dans l’épidémiologie des trypanosomoses. Rev Elev Med Vet Pays Trop. 1997;50:209–213. doi: 10.19182/remvt.9571. [DOI] [Google Scholar]
- 115.Diall O. Camel trypanosomiasis in Mali: contribution to the diagnosis and the epidemiology. Brussels: Vrije Universiteit Brussel; 1993. [Google Scholar]
- 116.Wuyts N, Chokesajjawatee N, Panyim S. A simplified and highly sensitive detection of Trypanosoma evansi by DNA amplification. Southeast Asian J Trop Med Public Health. 1994;25:266–271. [PubMed] [Google Scholar]
- 117.Aradaib IE, Majid AA. A simple and rapid method for detection of Trypanosoma evansi in the dromedary camel using a nested polymerase chain reaction. Kinetoplastid Biol Dis. 2006;5:2. doi: 10.1186/1475-9292-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taylor TK, Boyle DB, Bingham J. Development of a TaqMan PCR assay for the detection of Trypanosoma evansi, the agent of surra. Vet Parasitol. 2008;153:255–264. doi: 10.1016/j.vetpar.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 119.Njiru ZK, Constantine CC, Masiga DK, Reid SA, Thompson RC, Gibson WC. Characterization of Trypanosoma evansi type B. Infect Genet Evol. 2006;6:292–300. doi: 10.1016/j.meegid.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 120.Van Xong H, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, Pays A, et al. A VSG expression site–associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/S0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 121.Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, Magnus E, et al. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am J Trop Med Hyg. 2002;67:684–690. doi: 10.4269/ajtmh.2002.67.684. [DOI] [PubMed] [Google Scholar]
- 122.Njiru ZK, Ndung'u K, Matete G, Ndungu JM, Gibson WC. Detection of Trypanosoma brucei rhodesiense in animals from sleeping sickness foci in East Africa using the serum resistance associated (SRA) gene. Acta Trop. 2004;90:249–254. doi: 10.1016/j.actatropica.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 123.Picozzi K, Carrington M, Welburn S. A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008;118:41–46. doi: 10.1016/j.exppara.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 124.Solomon Ngutor K, Idris LA, Oluseyi OO. Silent human Trypanosoma brucei gambiense infections around the old Gboko sleeping sickness focus in Nigeria. J Parasitol Res. 2016;2016:2656121. doi: 10.1155/2016/2656121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cunningham LJ, Lingley JK, Tirados I, Esterhuizen J, Opiyo M, Mangwiro CT, et al. Evidence of the absence of human African trypanosomiasis in two northern districts of Uganda: Analyses of cattle, pigs and tsetse flies for the presence of Trypanosoma brucei gambiense. PLoS Negl Trop Dis. 2020;14:e0007737. doi: 10.1371/journal.pntd.0007737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oldrieve GR, Verney M, Jaron K, Hebert L, Matthews K. Monomorphic Trypanozoon: towards reconciling phylogeny and pathologies. Microb Genom. 2021 doi: 10.1099/mgen.0.000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moser DR, Kirchhof LV, Donelson JE. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol. 1989;27:1477–1482. doi: 10.1128/jcm.27.7.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dario MA, Pavan MG, Rodrigues MS, Lisboa CV, Kluyber D, Desbiez AL, et al. Trypanosoma rangeli genetic, mammalian hosts, and geographical diversity from five Brazilian biomes. Pathogens. 2021;10:736. doi: 10.3390/pathogens10060736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Desquesnes M, McLaughlin G, Zoungrana A, Dávila AM. Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. Int J Parasitol. 2001;31:610–614. doi: 10.1016/s0020-7519(01)00161-8. [DOI] [PubMed] [Google Scholar]
- 130.Desquesnes M, Davila AM. Applications of PCR-based tools for detection and identification of animal trypanosomes: a review and perspectives. Vet Parasitol. 2002;109:213–231. doi: 10.1016/S0304-4017(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 131.Delespaux V, Ayral F, Geysen D, Geerts S. PCR-RFLP using SSU-rDNA amplification: applicability for the diagnosis of mixed infections with different trypanosome species in cattle. Vet Parasitol. 2003;117:185–193. doi: 10.1016/j.vetpar.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 132.Geysen D, Delespaux V, Geerts S. PCR-RFLP using SSU-rDNA amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Vet Parasitol. 2003;110:171–180. doi: 10.1016/S0304-4017(02)00313-8. [DOI] [PubMed] [Google Scholar]
- 133.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–192. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 134.Ravel S, Mediannikov O, Bossard G, Desquesnes M, Cuny G, Davoust B. A study on African animal trypanosomosis in four areas of Senegal. Folia Parasitol. 2015;62:2015. doi: 10.14411/fp.2015.044. [DOI] [PubMed] [Google Scholar]
- 135.Desquesnes M, Ravel S, Cuny G. PCR identification of Trypanosoma lewisi, a common parasite of laboratory rats. Kinetoplastid Biol Dis. 2002;1:2. doi: 10.1186/1475-9292-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Medkour H, Varloud M, Davoust B, Mediannikov O. New molecular approach for the detection of kinetoplastida parasites of medical and veterinary interest. Microorganisms. 2020;8:356. doi: 10.3390/microorganisms8030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, et al. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Holland W, Claes F, My L, Thanh N, Tam P, Verloo D, et al. A comparative evaluation of parasitological tests and a PCR for Trypanosoma evansi diagnosis in experimentally infected water buffaloes. Vet Parasitol. 2001;97:23–33. doi: 10.1016/S0304-4017(01)00381-8. [DOI] [PubMed] [Google Scholar]
- 139.Fernandez D, Gonzalez-Baradat B, Eleizalde M, Gonzalez-Marcano E, Perrone T, Mendoza M. Trypanosoma evansi: A comparison of PCR and parasitological diagnostic tests in experimentally infected mice. Exp Parasitol. 2009;121:1–7. doi: 10.1016/j.exppara.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 140.Pruvot M, Kamyingkird K, Desquesnes M, Sarataphan N, Jittapalapong S. The effect of the DNA preparation method on the sensitivity of PCR for the detection of Trypanosoma evansi in rodents and implications for epidemiological surveillance efforts. Vet Parasitol. 2013;191:203–208. doi: 10.1016/j.vetpar.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 141.Masiga D, McNamara J, Laveissière C, Truc P, Gibson W. A high prevalence of mixed trypanosome infections in tsetse flies in Sinfra, Cote d'Ivoire, detected by DNA amplification. Parasitology. 1996;112:75–80. doi: 10.1017/S0031182000065094. [DOI] [PubMed] [Google Scholar]
- 142.Ngaira JM, Olembo NK, Njagi EN, Ngeranwa JJ. The detection of non-RoTat 12 Trypanosoma evansi. Exp Parasitol. 2005;110:30–38. doi: 10.1016/j.exppara.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 143.Njiru ZK, Ouma JO, Enyaru JC, Dargantes AP. Loop-mediated isothermal amplification (LAMP) test for detection of Trypanosoma evansi strain B. Exp Parasitol. 2010;125:196–201. doi: 10.1016/j.exppara.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 144.Thekisoe OM, Inoue N, Kuboki N, Tuntasuvan D, Bunnoy W, Borisutsuwan S, et al. Evaluation of loop-mediated isothermal amplification (LAMP), PCR and parasitological tests for detection of Trypanosoma evansi in experimentally infected pigs. Vet Parasitol. 2005;130:327–330. doi: 10.1016/j.vetpar.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 145.Thekisoe OM, Kuboki N, Nambota A, Fujisaki K, Sugimoto C, Igarashi I, et al. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 2007;102:182–189. doi: 10.1016/j.actatropica.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 146.Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, Kibona SN, et al. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol. 2008;38:589–599. doi: 10.1016/j.ijpara.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sharma P, Juyal P, Singla L, Chachra D, Pawar H. Comparative evaluation of real time PCR assay with conventional parasitological techniques for diagnosis of Trypanosoma evansi in cattle and buffaloes. Vet Parasitol. 2012;190:375–382. doi: 10.1016/j.vetpar.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 148.Becker S, Franco JR, Simarro PP, Stich A, Abel PM, Steverding D. Real-time PCR for detection of Trypanosoma brucei in human blood samples. Diagn Microbiol Infect Dis. 2004;50:193–199. doi: 10.1016/j.diagmicrobio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 149.Ahmed HA, MacLeod ET, Welburn SC, Picozzi K. Development of real time PCR to study experimental mixed infections of T. congolense Savannah and T. b. brucei in Glossina morsitans morsitans. PLoS ONE. 2015;10:e0117147. doi: 10.1371/journal.pone.0117147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Compaoré CFA, Ilboudo H, Kaboré J, Kaboré JW, Camara O, Bamba M, et al. Analytical sensitivity of loopamp and quantitative real-time PCR on dried blood spots and their potential role in monitoring human African trypanosomiasis elimination. Exp Parasitol. 2020;219:108014. doi: 10.1016/j.exppara.2020.108014. [DOI] [PubMed] [Google Scholar]
- 151.Ngay Lukusa I, Van Reet N, Mumba Ngoyi D, Miaka EM, Masumu J, Patient Pyana P, et al. Trypanosome SL-RNA detection in blood and cerebrospinal fluid to demonstrate active gambiense human African trypanosomiasis infection. PLoS Negl Trop Dis. 2021;15:e0009739. doi: 10.1371/journal.pntd.0009739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lefrançois T, Solano P, Bauer B, Kaboré I, Touré SM, Cuny G, et al. Polymerase chain reaction characterization of trypanosomes in Glossina morsitans submorsitans and G. tachninoides collected on the game ranch of Nazinga, Burkina Faso. Acta Trop. 1999;72:65–77. doi: 10.1016/S0001-706X(98)00080-1. [DOI] [PubMed] [Google Scholar]
- 153.Pizarro JC, Lucero DE, Stevens L. PCR reveals significantly higher rates of Trypanosoma cruzi infection than microscopy in the Chagas vector, Triatoma infestans: high rates found in Chuquisaca, Bolivia. BMC Infect Dis. 2007;7:66. doi: 10.1186/1471-2334-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Katakura K, Lubinga C, Chitambo H, Tada Y. Detection of Trypanosoma congolense and T. brucei subspecies in cattle in Zambia by polymerase chain reaction from blood collected on filter paper. Parasitol Res. 1997;83:241–245. doi: 10.1007/s004360050240. [DOI] [PubMed] [Google Scholar]
- 155.Nantulya V, Lindqvist K. Antigen-detection enzyme immunoassays of diagnosis of Trypanosoma vivax, T. congolense and T. brucei infections in cattle. Trop Med Parasitol. 1989;40:267–272. [PubMed] [Google Scholar]
- 156.Desquesnes M. Evaluation of three antigen detection tests (monoclonal trapping ELISA) for African trypanosomes, with an isolate of Trypanosoma vivax from French Guyana. Ann N Y Acad Sci. 1996;791:172–184. doi: 10.1111/j.1749-6632.1996.tb53524.x. [DOI] [PubMed] [Google Scholar]
- 157.Eisler M, Lessard P, Masake R, Moloo S, Peregrine A. Sensitivity and specificity of antigen-capture ELISAs for diagnosis of Trypanosoma congolense and Trypanosoma vivax infections in cattle. Vet Parasitol. 1998;79:187–201. doi: 10.1016/S0304-4017(98)00173-3. [DOI] [PubMed] [Google Scholar]
- 158.Luckins AG, Melhitz D. Evaluation of an indirect fluorescent antibody test, enzyme-linked immunosorbent assay and quantification of immunoglobulins in the diagnosis of bovine trypanosomiasis. Trop Anim Health Prod. 1978;10:149–159. doi: 10.1007/BF02235328. [DOI] [PubMed] [Google Scholar]
- 159.Luckins AG. Detection of antibodies in trypanosome-infected cattle by means of a microplate enzyme-linked immunosorbent assay. Trop Anim Health Prod. 1977;9:53–62. doi: 10.1007/BF02297393. [DOI] [PubMed] [Google Scholar]
- 160.Desquesnes M. International and regional standardization of immunoenzyme tests: methods, concerns and limitations. Rev Sci Tech. 1997;16:809–823. doi: 10.20506/rst.16.3.1065. [DOI] [PubMed] [Google Scholar]
- 161.Hopkins JS, Chitambo H, Machila N, Luckins AG, Rae PF, van de Bossche P, et al. Adaptation and validation of antibody-ELISA using dried blood spots on filter paper for epidemiological surveys of tsetse-transmitted trypanosomosis in cattle. Prev Vet Med. 1998;37:91–99. doi: 10.1016/S0167-5877(98)00101-9. [DOI] [PubMed] [Google Scholar]
- 162.Wright PF, Nilsson E, Van Rooij EMA, Lelenta M, Jeggo MH. Standardisation and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev Sci Tech. 1993;12:435–450. doi: 10.20506/rst.12.2.691. [DOI] [PubMed] [Google Scholar]
- 163.Greiner M, Bhat TS, Patzelt RJ, Kakaire D, Schares G, Dietz E, et al. Impact of biological factors on the interpretation of bovine trypanosomisis serology. Prev Vet Med. 1997;30:61–73. doi: 10.1016/S0167-5877(96)01088-4. [DOI] [PubMed] [Google Scholar]
- 164.Camoin M, Kocher A, Chalermwong P, Yangtarra S, Thongtip N, Jittapalapong S, et al. Adaptation and evaluation of an ELISA for Trypanosoma evansi infection (Surra) in elephants and its application to a serological survey in Thailand. Parasitology. 2017;145:371–377. doi: 10.1017/S0031182017001585. [DOI] [PubMed] [Google Scholar]
- 165.Kocher A, Desquesnes M, Kamyingkird K, Yangtara S, Leboucher E, Rodtian P, et al. Evaluation of an indirect-ELISA test for Trypanosoma evansi infection (Surra) in buffaloes and its application to a serological survey in Thailand. BioMed Res Int. 2015;2015:361037. doi: 10.1155/2015/361037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Desquesnes M, Kamyingkird K, Pruvot M, Kengradomkij C, Bossard G, Sarataphan N, et al. Antibody-ELISA for Trypanosoma evansi: application in a serological survey of dairy cattle, Thailand, and validation of a locally produced antigen. Prev Vet Med. 2009;90:233–241. doi: 10.1016/j.prevetmed.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 167.Desquesnes M, Bengaly Z, Dia ML. Evaluation de la persistance des anticorps détectés par Elisa-indirect Trypanosoma vivax après traitement trypanocide chez des bovins naturellement infectés. Rev Elev Med Vet Pays Trop. 2003;56:141–144. doi: 10.19182/remvt.9855. [DOI] [Google Scholar]
- 168.Madruga CR, Araujo FR, Cavalcante-Goes G, Martins C, Pfeifer IB, Ribeiro LR, et al. The development of an enzyme-linked immunosorbent assay for Trypanosoma vivax antibodies and its use in epidemiological surveys. Mem Inst Oswaldo Cruz. 2006;101:801–807. doi: 10.1590/S0074-02762006000700016. [DOI] [PubMed] [Google Scholar]
- 169.Yadav SC, Kumar R, Manuja A, Goyal L, Gupta AK. Early detection of Trypanosoma evansi infection and monitoring of antibody levels by ELISA following treatment. J Parasit Dis. 2014;38:124–127. doi: 10.1007/s12639-012-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Van den Bossche P, Chigoma D, Shumba W. The decline of anti-trypanosomal antibody levels in cattle after treatment with trypanocidal drugs and in the absence of tsetse challenge. Acta Trop. 2000;77:263–270. doi: 10.1016/S0001-706X(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 171.Ferenc S, Siopinskif V, Courtney C. The development of an enzyme-linked immunosorbent assay for Trypanosoma vivax and its use in a seroepidemiological survey of the Eastern Caribbean Basin. Int J Parasitol. 1990;20:51–56. doi: 10.1016/0020-7519(90)90172-J. [DOI] [PubMed] [Google Scholar]
- 172.Desquesnes M, Gardiner PR. Epidémiologie de la trypanosomose bovine (Trypanosoma vivax) en Guyane française. Rev Elev Med Vet Pays Trop. 1993;46:463–470. doi: 10.19182/remvt.9447. [DOI] [PubMed] [Google Scholar]
- 173.Desquesnes M, Bengaly Z, Millogo L, Meme Y, Sakande H. The analysis of the cross-reactions occurring in antibody-ELISA for the detection of trypanosomes can improve identification of the parasite species involved. Ann Trop Med Parasitol. 2001;95:141–155. doi: 10.1080/00034983.2001.11813624. [DOI] [PubMed] [Google Scholar]
- 174.Bossard G, Millogo L, Thevenon S, Vitouley H, Bengaly Z, Desquesnes M. No more cold-chain failures, using dehydrated reagents in ELISA antibody-detection against animal trypanosomes of African origin. Vet Parasitol. 2021;299:109568. doi: 10.1016/j.vetpar.2021.109568. [DOI] [PubMed] [Google Scholar]
- 175.Bajyana-Songa E, Hamers-Casterman C, Hamers R, Pholpark M, Pholpark S, Leidl K, et al. The use of the card agglutination test (Testryp CATT) for the detection of T. evansi infection: a comparison with other trypanosomiasis diagnostic tests under field conditions in Thailand. Ann Soc Belg Med Trop. 1987;67:137–148. [PubMed] [Google Scholar]
- 176.Bajyana Songa E, Hamers R. A card agglutination test (CATT) for veterinary use based on an early VAT RoTat 1/2 of Trypanosoma evansi. Ann Soc Belg Med Trop. 1988;68:233–240. [PubMed] [Google Scholar]
- 177.Wernery U, Zachariah R, Mumford JA, Luckins T. Preliminary evaluation of diagnostic tests using horses experimentally infected with Trypanosoma evansi. Vet J. 2001;161:287–300. doi: 10.1053/tvjl.2000.0560. [DOI] [PubMed] [Google Scholar]
- 178.Truc P, Lejon V, Magnus E, Jamonneau V, Nangouma A, Verloo D, et al. Evaluation of the micro-CATT, CATT/Trypanosoma brucei gambiense, and LATEX/T b gambiense methods for serodiagnosis and surveillance of human African trypanosomiasis in West and Central Africa. Bull World Health Organ. 2002;80:882–886. [PMC free article] [PubMed] [Google Scholar]
- 179.Asonganyi T, Suh S, Tetuh M. Prevalence of domestic animal trypanosomiasis in the Fontem sleeping sickness focus. Cameroon Rev Elev Med Vet Trop. 1990;43:69–74. doi: 10.19182/remvt.8900. [DOI] [PubMed] [Google Scholar]
- 180.Hébert L, Guitton E, Madeline A, Géraud T, Carnicer D, Lakhdar L, et al. Validation of a new experimental model for assessing drug efficacy against infection with Trypanosoma equiperdum in horses. Vet Parasitol. 2018;263:27–33. doi: 10.1016/j.vetpar.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 181.Büscher P, Gonzatti MI, Hébert L, Inoue N, Pascucci I, Schnaufer A, et al. Equine trypanosomosis: enigmas and diagnostic challenges. Parasit Vectors. 2019;12:234. doi: 10.1186/s13071-019-3484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.World Organization for Animal Health (OIE). Dourine (infection with Trypanosoma equiperdum). Paris: OIE. 2021. https://www.oie.int/app/uploads/2021/03/dourine.pdf. Accessed 10 Feb 2022.
- 183.Büscher, P. Dourine in horses (Trypanosoma equiperdum infection). In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2021. OIE, Paris, France. https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/. Accessed 10 Feb 2022.
- 184.Verloo D, Holland W, My LN, Thanh NG, Tam PT, Goddeeris B, et al. Comparison of serological tests for Trypanosoma evansi natural infections in water buffaloes from north Vietnam. Vet Parasitol. 2000;92:87–96. doi: 10.1016/S0304-4017(00)00284-3. [DOI] [PubMed] [Google Scholar]
- 185.Birhanu H, Gebrehiwot T, Goddeeris BM, Buscher P, Van Reet N. New Trypanosoma evansi Type B isolates from Ethiopian dromedary camels. PLoS Negl Trop Dis. 2016;10:e0004556. doi: 10.1371/journal.pntd.0004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rudramurthy G, Sengupta P, Ligi M, Balamurugan V, Suresh K, Rahman H. Serodiagnosis of animal trypanosomosis using a recombinant invariant surface glycoprotein of Trypanosoma evansi. Indian J Exp Biol. 2017;55:209–216. [Google Scholar]
- 187.Lejon V, Claes F, Verloo D, Maina M, Urakawa T, Majiwa PA, et al. Recombinant RoTat 1.2 variable surface glycoprotein as antigen for diagnosis of Trypanosoma evansi in dromedary camels. Int J Parasitol. 2005;35:455–460. doi: 10.1016/j.ijpara.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 188.Bossard G, Boulange A, Holzmuller P, Thevenon S, Patrel D, Authie E. Serodiagnosis of bovine trypanosomosis based on HSP70/BiP inhibition ELISA. Vet Parasitol. 2010;173:39–47. doi: 10.1016/j.vetpar.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 189.Thuy NT, Goto Y, Lun ZR, Kawazu S, Inoue N. Tandem repeat protein as potential diagnostic antigen for Trypanosoma evansi infection. Parasitol Res. 2012;110:733–739. doi: 10.1007/s00436-011-2632-9. [DOI] [PubMed] [Google Scholar]
- 190.Desquesnes M, Bosseno MF, Breniere SF. Detection of Chagas infections using Trypanosoma evansi crude antigen demonstrates high cross-reactions with Trypanosoma cruzi. Infect Genet Evol. 2007;7:457–462. doi: 10.1016/j.meegid.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 191.Tounkara M, Boulangé A, Thonnus M, Bringaud FDR, Bélem AMG, Bengaly Z, et al. Novel protein candidates for serodiagnosis of African animal trypanosomosis: Evaluation of the diagnostic potential of lysophospholipase and glycerol kinase from Trypanosoma brucei. PLoS Negl Trop Dis. 2021;15:34. doi: 10.1371/journal.pntd.0009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Matovu E, Kitibwa A, Picado A, Biéler S, Bessell PR, Ndungu JM. Serological tests for gambiense human African trypanosomiasis detect antibodies in cattle. Parasit Vectors. 2017;10:546. doi: 10.1186/s13071-017-2487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vourchakbé J, Tiofack ZAA, Kante TS, Mpoame M, Simo G. Molecular identification of Trypanosoma brucei gambiense in naturally infected pigs, dogs and small ruminants confirms domestic animals as potential reservoirs for sleeping sickness in Chad. Parasite. 2020;27:63. doi: 10.1051/parasite/2020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vexenat AC, Santana JM, Teixeira AR. Cross-reactivity of antibodies in human infections by the kinetoplastid protozoa Trypanosoma cruzi, Leishmania chagasi and Leishmania (Viannia) braziliensis. Rev Inst Med Trop Sao Paulo. 1996;38:177–185. doi: 10.1590/S0036-46651996000300003. [DOI] [PubMed] [Google Scholar]
- 195.Zanette MF, Lima VMF, Laurenti MD, Rossi CN, Vides JP, Vieira RFC, et al. Serological cross-reactivity of Trypanosoma cruzi, Ehrlichia canis, Toxoplasma gondii, Neospora caninum and Babesia canis to Leishmania infantum chagasi tests in dogs. Rev Soc Bras Med Trop. 2014;47:105–107. doi: 10.1590/0037-8682-1723-2013. [DOI] [PubMed] [Google Scholar]
- 196.Lutumba P, Robays J, Miaka C, Kande V, Simarro PP, Shaw AP, et al. The efficiency of different detection strategies of human African trypanosomiasis by T. b. gambiense. Trop Med Int Health. 2005;10:347–356. doi: 10.1111/j.1365-3156.2005.01391.x. [DOI] [PubMed] [Google Scholar]
- 197.Mumba Ngoyi D, Ali Ekangu R, Mumvemba Kodi MF, Pyana PP, Balharbi F, Decq M, et al. Performance of parasitological and molecular techniques for the diagnosis and surveillance of gambiense sleeping sickness. PLoS Negl Trop Dis. 2014;8:e2954. doi: 10.1371/journal.pntd.0002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jamonneau V, Bucheton B, Kabore J, Ilboudo H, Camara O, Courtin F, et al. Revisiting the immune trypanolysis test to optimise epidemiological surveillance and control of sleeping sickness in West Africa. PLoS Negl Trop Dis. 2010;4:e917. doi: 10.1371/journal.pntd.0000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Enciso C, Montilla M, Santacruz MM, Nicholls RS, Rodriguez A, Mercado M, et al. Comparison of the indirect immunofluorescent (IFAT), ELISA test and the comercial Chagatek test for anti-Trypanosoma cruzi antibodies detection. Biomedica. 2004;24:104–108. doi: 10.7705/biomedica.v24i1.1254. [DOI] [PubMed] [Google Scholar]
- 200.Mitashi P, Hasker E, Lejon V, Kande V, Muyembe JJ, Lutumba P, et al. Human african trypanosomiasis diagnosis in first-line health services of endemic countries, a systematic review. PLoS Negl Trop Dis. 2012;6:e1919. doi: 10.1371/journal.pntd.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.World Health Organization (WHO). Control and surveillance of human African trypanosomiasis. Geneva: WHO Press; 2013. https://apps.who.int/iris/handle/10665/95732. Accessed 10 Feb 2022. [PubMed]
- 202.Lumbala C, Biéler S, Kayembe S, Makabuza J, Ongarello S, Ndungu JM. Prospective evaluation of a rapid diagnostic test for Trypanosoma brucei gambiense infection developed using recombinant antigens. PLoS Negl Trop Dis. 2018;12:e0006386. doi: 10.1371/journal.pntd.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Buscher P, Deborggraeve S. How can molecular diagnostics contribute to the elimination of human African trypanosomiasis? Expert Rev Mol Diagn. 2015;15:607–615. doi: 10.1586/14737159.2015.1027195. [DOI] [PubMed] [Google Scholar]
- 204.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 205.Joshi PP, Shegokar VR, Powar RM, Herder S, Katti R, Salkar HR, et al. Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. Am J Trop Med Hyg. 2005;73:491–495. doi: 10.4269/ajtmh.2005.73.491. [DOI] [PubMed] [Google Scholar]
- 206.Truc P, Gibson W, Herder S. Genetic characterization of Trypanosoma evansi isolated from a patient in India. Infect Genet Evol. 2007;7:305–307. doi: 10.1016/j.meegid.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 207.Van Vinh CN, Buu Chau L, Desquesnes M, Herder S, Phu H, Lan N, Campbell JI, et al. A clinical and epidemiological investigation of the first reported human infection with the zoonotic parasite Trypanosoma evansi in Southeast Asia. Clin Infect Dis. 2016;62:1002–1008. doi: 10.1093/cid/ciw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Doke P, Kar A. A fatal case of Trypanosoma lewisi in Maharashtra. India Ann Trop Med Public Health. 2011;4:91–95. doi: 10.4103/1755-6783.85759. [DOI] [Google Scholar]
- 209.Desquesnes M, Yangtara S, Kunphukhieo P, Jittapalapong S, Herder S. Zoonotic trypanosomes in South East Asia: Attempts to control Trypanosoma lewisi using human and animal trypanocidal drugs. Infect Genet Evol. 2016;44:514–521. doi: 10.1016/j.meegid.2016.07.041. [DOI] [PubMed] [Google Scholar]
- 210.Lun ZR, Wen YZ, Uzureau P, Lecordier L, Lai H, Lan YG, et al. Resistance to normal human serum reveals Trypanosoma lewisi as an underestimated human pathogen. Mol Biochem Parasitol. 2015;199:58–61. doi: 10.1016/j.molbiopara.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 211.Pumhom P, Morand S, Tran A, Jittapalapong S, Desquesnes M. Trypanosoma from rodents as potential source of infection in human-shaped landscapes of South-East Asia. Vet Parasitol. 2015;208:174–180. doi: 10.1016/j.vetpar.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 212.Sarataphan N, Vongpakorn M, Nuansrichay B, Autarkool N, Keowkarnkah T, Rodtian P, et al. Diagnosis of a Trypanosoma lewisi-like (Herpetosoma) infection in a sick infant from Thailand. J Med Microbiol. 2007;56:1118–1121. doi: 10.1099/jmm.0.47222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gao J-M, Truc P, Desquesnes M, Vincendeau P, Courtois P, Zhang X, et al. A preliminary serological study of Trypanosoma evansi and Trypanosoma lewisi in Chinese human population. Agr Nat Resour. 2018;52:612–616. doi: 10.1016/j.anres.2018.11.024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.