Abstract
Using the automated Riboprinter system, we have initiated the construction of an electronic Riboprint database composed of 72 ECOR reference strains and 15 archetypal virulent strains in order to provide a new simple molecular characterization method. More than 90% of the ECOR strains clustered in their original phylogenetic group. All but one of the archetypal virulent strains had a profile identical to that of one of the ECOR strains and could be easily affiliated with a phylogenetic group. This method appears to be an accurate and practical tool especially for investigating the genetic relationship between clinical extraintestinal pathogenic strains and B2 subgroup ECOR strains or archetypal pathotype strains.
Escherichia coli is both the most common commensal bacterium and the most frequent community-acquired pathogen in humans. E. coli belongs to the normal fecal flora but can cause various intestinal (gastroenteritis and colitis) and extraintestinal (urinary tract infection, septicemia, and neonatal meningitis) infections. The genetic structure of E. coli is considered clonal, and phylogenetic analyses have shown that strains of this species fall into four main phylogenetic groups (A, B1, B2, and D) (11, 24). Recent attempts to establish a link between phylogeny and virulence suggest that extraintestinal pathogenic E. coli strains are mostly derived from phylogenetic group B2 and, to a lesser extent, group D (3, 4, 18). In contrast, most human commensal strains originate from phylogenetic groups A and B1. Contrary to extraintestinal pathogenic strains, each pathotype of intestinal pathogenic strains shows phylogenetic diversity: enteropathogenic E. coli, enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAggEC), diffuse adherent E. coli (DAEC), and enteroinvasive E. coli are distributed among all the phylogenetic groups (8, 20, 22). Studies based on molecular characterization and genetic relatedness of pathogenic and commensal strains have improved our understanding of the pathogenicity and acquisition of virulence traits in E. coli.
The reference techniques for phylogenetic grouping are multilocus enzyme electrophoresis (MLEE) (11, 23) and ribotyping (3, 9, 16, 19), but both techniques are complex and time-consuming. Moreover, neither method is standardized. Thus, to determine the phylogenetic group of a given strain, a collection of typed reference strains, such as the ECOR collection, must be tested in parallel (3, 9, 12, 20). The ECOR collection is a set of 72 reference E. coli strains isolated between 1973 and 1983 from a variety of animal hosts and geographic locations. Those strains are well characterized and represent the entire range of genotypic variation in the species as a whole (17).
Being automated and standardized, the Riboprinter microbial characterization system is suited to rapid and high-throughput typing of bacterial strains. The Riboprinter can yield 32 ribotypes a day directly from fresh colonies. Given their worldwide distribution and interconnection, Riboprinters permit immediate comparisons of ribotypes through connection of their databases. The purpose of the present study was to initiate the construction of an electronic Riboprint database of archetypal virulent strains and the ECOR collection, in order to provide the scientific community with a rapid tool for investigating genetic relationships between clinical and reference E. coli strains.
Bacterial isolates.
The 72 strains of the ECOR collection (17) were kindly provided by R. Selander (Department of Biology, University of Rochester, Rochester, N.Y.). Of these, 68 belong to the four main phylogenetic groups (A, B1, B2, and D) and 4 are unclassified (group E) (11, 24). Information concerning these strains can be obtained at the T. Whittam laboratory web site (http://foodsafe.msu.edu/whittam/ecor/index.html). We also used several archetypal strains representing different E. coli pathotypes, including neonatal meningitis strains RS218 (kindly provided by K. Kim, Johns Hopkins University School of Medicine, Baltimore, Md.), C5 and RS176 (obtained from R. Bortolussi, Dhalousie University, Halifax, Nova Scotia, Canada), uropathogenic strains J96 and E536 (provided by J. Hacker, Institut für Molekulare Infektionsbiologie, Würzburg, Germany), uropathogenic strain CFT073 (obtained from H. Mobley, University of Maryland, Baltimore), enteropathogenic strain E2348/69, EHEC strain EDL933, ETEC strains EDL1493, E2539-C1, and TX-1, EAggEC strains O42 and JM221, and DAEC A30 and C1845. All the diarrheagenic strains were kindly provided by C. Le Bouguenec (Unité de Pathogénie Bactérienne des Muqueuses, Institut Pasteur, Paris, France). E. coli laboratory K-12 strain MG1655, which belongs to phylogenetic group A, was also studied (11).
Automated ribotyping.
All E. coli isolates and ECOR strains were characterized by automated ribotyping with the Riboprinter (Qualicon Inc., Wilmington, Del.). Ribotyping was performed under the conditions recommended by the manufacturer (6, 25), with the following modifications. The validated EcoRI restriction enzyme was replaced by HindIII (New England BioLabs, Beverly; Mass.) at 100 U/μl in standardized reagents in 0.5-ml tubes (Sarstedt, Orsay, France). The other steps were unmodified and automated, and up to 32 isolates could be analyzed per day.
For each strain analyzed, one fresh colony was picked and resuspended in sample buffer and added to the processing module for a heat treatment step at 80°C for 10 min in order to inhibit endogenous DNA-degrading enzymes. The temperature was then reduced, and two lytic enzymes (lysostaphin and N-acetylmuramidase) were added to the sample. The sample carrier was then loaded onto the Riboprinter system with the other commercial reagents. Restriction enzyme digestion, gel electrophoresis, and blotting steps were completely automated. Briefly, bacterial DNA was digested with the chosen restriction enzyme and loaded onto an agarose gel; restriction fragments were separated by electrophoresis and simultaneously transferred to a nylon membrane. After a denaturation step, the blotted nucleic acids were hybridized with a sulfonated DNA probe harboring the genes for the small and large rRNA subunits of E. coli. The hybridized probe was detected with alkaline phosphatase-labeled antibodies directed against sulfonated DNA. Bound labeled antibodies were then detected by capturing light emission from a chemiluminescent substrate with a charge-coupled device camera. The output consisted of a densitometric scan depicting the distribution of the restriction fragments and their molecular weights and was saved in the Riboprinter computer.
Ribotype analysis.
For each batch of eight samples, ribotypes were normalized to the position of the molecular weight standards by using the Qualicon software. Computerized ribotypes were exported for analysis in .txt files, converted to .int files with GelConvert 1.01 software (Qualicon), and imported into Gel Compar software, version 4.1 (Applied Maths, Ghent, Belgium). Clustering analysis was performed with the unweighted pair group method using arithmetic averages (UPGMA) based on the Dice coefficient for the band matching (10), with a position tolerance setting of 0.8% and an optimization setting of 0.25% (default values are 1% for position tolerance and 0.5% for optimization). Bands for analysis with the Dice coefficient were assigned manually, according to densitometric curves and the accompanying hard-copy photograph. As previously reported, HindIII generates a few bands of very low intensity, especially under 1 kb (15). One third of the ECOR reference strains, which were ribotyped twice, showed that these faint bands were nonreproducible (data not shown). Thus, these bands were not taken into account in the band-based cluster analysis. On the other hand, major bands were always reproducible and were perfectly in accordance with manual ribotypes patterns previously obtained (3) (data not shown).
Access to the Riboprinter database for downloading ECOR ribotypes will be made available upon request (edouard.bingen@rdb.ap-hop-paris.fr or marc.lange@pasteur-lille.fr). Furthermore, all data (including the dendrogram) are available at the Molecular Typing Center website (http://www.pasteur-lille.fr /english/techno/ctm/ecor.html).
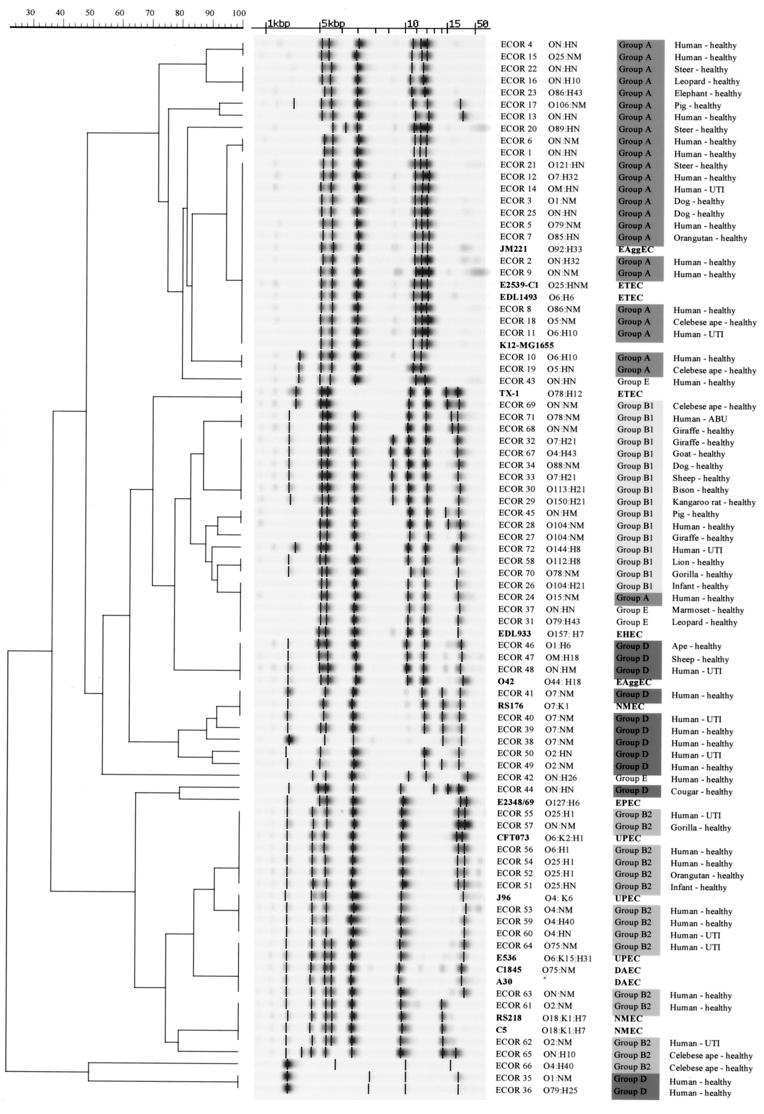
Ribotyping of the 72 ECOR strains using the HindIII enzyme yielded 32 ribotypes after exclusion of the faint bands generated by this enzyme (see Materials and Methods). The number of strains presenting a given ribotype ranged from 1 to 12. All strains with the same pattern belonged to the same phylogenetic group. Only one exception was noted: the patterns for ECOR 24, 26, 31, and 37 belonged to groups A, B1, E, and E, respectively. A phylogenetic tree of the 72 ECOR strains, 15 pathogenic reference strains, and K-12 strain MG1655 was obtained by UPGMA (Fig. 1). Three major clusters containing 69 ECOR strains were clearly distinguished on the tree. Strains belonging to groups A and B2 were clearly separated from each other and from a cluster containing the B1 and D strains. Three strains (ECOR 35, 36, and 66, belonging to groups D, D, and B2, respectively) displayed an atypical ribotype and were separated from the three major clusters. Group B1 and D strains clustered very close together but remained distinguishable from each other. In subcluster B1, one pattern was yielded by strains belonging to group A and the group of unclassified strains (Fig. 1). Thus, strains yielding this pattern could not be unambiguously grouped. Apart from this exception, each of the other 31 patterns always contained strains belonging to a given phylogenetic group. Thus, a clinical strain yielding one of these 31 patterns can be unambiguously categorized in one of the four phylogenetic groups.
FIG. 1.
Comparative analysis of HindIII ribotypes obtained with the Riboprinter for the ECOR collection and a subset of E. coli pathogenic reference strains. Clustering was performed by UPGMA, and similarity analysis was based on the use of the Dice coefficient (see Materials and Methods). For each strain, name, serotype, phylogenetic group or pathovar, and source is indicated. O:H serotypes are as listed at the T. Whittam laboratory website. ON, HN, nontypeable with standard antisera; NM, nonmotile strain; EPEC, enteropathogenic E. coli; NMEC, neonatal meningitis E. coli; DAEC, diffuse adherent E. coli EAggEc, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EHEC, enterohemmorhagic E. coli; UPEC, uropathogenic E. coli; ABU, asymptomatic bacteriuria; UTI, symptomatic urinary tract infection.
To evaluate this grouping method, we tested 15 pathogenic E. coli strains. The automated ribotyping technique readily grouped these strains, all but one of which yielded patterns identical to one of those obtained with the ECOR collection. As expected, the six extraintestinal pathogenic strains were classified in groups B2 and D. Interestingly, the neonatal meningitis strains (RS218 and C5) and the three uropathogenic strains (J96, E536, and CFT073) belonged to four different subgroups within group B2. The intestinal pathogenic strains were distributed among all the phylogenetic groups, and strains belonging to one pathotype were distributed among different phylogenetic groups. For example, EAggEC strains JM221 and O42 belonged to groups A and D, respectively, while ETEC strains belonged to groups A (E2536-1 and EDL1493) and B1 (TX-1). However, EHEC strain EDL933 showed a ribotype associated with strains belonging to groups A and B1 and the group of unclassified strains and could not thus be affiliated with a given group. Nevertheless, this profile may be considered representative of the O157:H7 strains, since 10 other strains tested had an identical pattern (data not shown) (7).
Phylogenetic analysis is increasingly important in the analysis of bacterial virulence. Thus, for emerging pathogens for which virulence factors are partially known, determination of the genetic background could be of the utmost importance (5, 18, 21). New phylogenotyping methods have recently been developed, such as repetitive-element PCR fingerprinting (13), fluorescent amplified fragment length polymorphism (1), and multilocus sequence typing (14), but manual ribotyping and MLEE remain the gold standard in this field. However, these techniques are complex and time-consuming and are therefore unsuitable for studies of the relationship between virulence and genetic background. The availability of an automated, standardized ribotyping technique provides the opportunity to initiate a database of ECOR strains, making the Riboprinter a simple and rapid phylogenotyping tool.
We chose to ribotype the ECOR collection with HindIII, because of its discriminating power and the fact that the profiles it yields can easily be attributed to a given phylogenetic group (2, 3, 7). Most of the ECOR strains clustered in their original phylogenetic group (11), and the similarity level in the four major divisions (∼70%) was not different from those found by Herzer et al. (11). The group B1 and D strains were the most difficult to discriminate. However, strains belonging to these two groups never yielded identical patterns. Despite a lesser discriminating power of ribotyping compared to MLEE typing, only one ribotype pattern, containing ECOR 24, 26, 31, and 37, carried a risk of erroneous phylogenetic group attribution. Considering that more than 90% of the pathogenic strains tested here yielded profiles identical to that of one of the ECOR strains, the phylogenetic group of each strain could be determined without recourse to a statistical clustering method. Indeed, analyzing retrospectively the ribotypes obtained from our collection of 69 E. coli neonatal meningitis strains described previously (3), we observed that 88% of these strains yielded a profile identical to that of one of the ECOR strains (data not shown). A recently published method determines the phylogenetic group of a strain using a rapid and simple PCR-based technique (7) but appears to be more suited to screening studies. Indeed, the automated ribotyping method has the advantage of attributing a strain to a phylogenetic subgroup defined by one or a few ECOR strains. In this study we observed an excellent correlation between B2 subgroups described by Herzer et al. (11) and B2 ribotyping subgroups. Of particular interest, the five B2 archetypal extraintestinal pathogenic strains were distributed among the four major B2 subgroups (represented by more than one strain). Thus, this rapid method appears to be a more accurate tool for investigating the phylogenetic relationship of a strain, especially uropathogenic and neonatal meningitis strains, to the E. coli population as a whole. Moreover, the electronic ribopattern database that we have initiated provides rapid investigation of the genetic relationship between clinical extraintestinal and B2 subgroup ECOR or archetypal pathotype strains.
Acknowledgments
This work was supported in part by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires (Appel d'offre 1998), “Recherche de déterminants génétiques de pathogénicité chez E. coli K1 responsable de méningite néonatale.”
We thank K. Kim, J. Hacker, R. Bortolussi, H. Mobley, and C. Le Bouguenec for providing some of the strains used in this study. We are grateful to Maryse De-Ré, Institut Pasteur de Lille, for her valuable technical assistance in parts of this study. We also thank Martine Alliot and Catherine Diesel-Klein (Qualicon Europe) for helpful discussions and Qualicon Europe for providing the Riboprinter reagents used in this work.
REFERENCES
- 1.Arnold C, Metherell L, Willshaw G, Maggs A, Stanley J. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J Clin Microbiol. 1999;37:1274–1279. doi: 10.1128/jcm.37.5.1274-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingen E, Denamur E, Brahimi N, Elion J. Genotyping may provide rapid identification of Escherichia coli K1 organisms that cause neonatal meningitis. Clin Infect Dis. 1996;22:152–156. doi: 10.1093/clinids/22.1.152. [DOI] [PubMed] [Google Scholar]
- 3.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 4.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisse S, Verduin C M, Milatovic D, Fluit A, Verhoef J, Laevens S, Vandamme P, Tummler B, Verbrugh H A, van Belkum A. Distinguishing species of the Burkholderia cepacia complex and Burkholderia gladioli by automated ribotyping. J Clin Microbiol. 2000;38:1876–1884. doi: 10.1128/jcm.38.5.1876-1884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce J L. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 1996;50:77–81. [Google Scholar]
- 7.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czeczulin J R, Whittam T S, Henderson I R, Navarro-Garcia F, Nataro J P. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–2699. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 10.Dice L R. Measures of the amount of ecological associations between species. J Ecol. 1945;26:297–302. [Google Scholar]
- 11.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilali F, Ruimy R, Saulnier P, Barnabe C, Lebouguenec C, Tibayrenc M, Andremont A. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect Immun. 2000;68:3983–3989. doi: 10.1128/iai.68.7.3983-3989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J R, O'Bryan T T. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin Diagn Lab Immunol. 2000;7:265–273. doi: 10.1128/cdli.7.2.265-273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 15.Machado J, Grimont F, Grimont P A. Computer identification of Escherichia coli rRNA gene restriction patterns. Res Microbiol. 1998;149:119–135. doi: 10.1016/s0923-2508(98)80027-2. [DOI] [PubMed] [Google Scholar]
- 16.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picard B, Garcia J S, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picard B, Journet-Mancy C, Picard-Pasquier N, Goullet P. Genetic structures of the B2 and B1 Escherichia coli strains responsible for extra-intestinal infections. J Gen Microbiol. 1993;139:3079–3088. doi: 10.1099/00221287-139-12-3079. [DOI] [PubMed] [Google Scholar]
- 20.Pupo G M, Karaolis D K, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogerie F, Marecat A, Gambade S, Dupond F, Beaubois M, Lange M. Characterization of shiga-toxin producing Escherichia coli and O157 serotype Escherichia coli isolated in France from healthy domestic cattle. Int J Food Microbiol. 2001;63:217–223. doi: 10.1016/s0168-1605(00)00422-0. [DOI] [PubMed] [Google Scholar]
- 22.Rolland K, Lambert-Zechovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of Escherichia coli. Microbiology. 1998;144:2667–2672. doi: 10.1099/00221287-144-9-2667. [DOI] [PubMed] [Google Scholar]
- 23.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 25.Sethi M R. Fully automated microbial characterization and identification for industrial microbiologists. Am Lab. 1997;5:31–35. [Google Scholar]