Dear Editor,
In China, asthma affects 45.7 million adults with a prevalence of 4.2%, 1 while that of severe asthma ranges from 3.4% to 8.3% among asthmatic patients. 2 , 3 , 4 There is scarce information on the characteristics and biomarker expression, and the different phenotypes of severe asthma. The C‐BIOPRED consortium recruited patients with severe asthma from 33 university hospitals in 15 provinces in China. 5 These patients were receiving treatment with medium to high doses of inhaled corticosteroids and long‐acting β2‐agonists and experienced uncontrolled asthma, defined by the experience of two or more asthma exacerbations, requiring daily oral corticosteroids (OCS), and/or other asthma medication. The participants were all adults either with severe non‐smoking asthma (NSA; n = 342), current and former smokers with severe asthma with a smoking history of >5 pack‐years (SSA; n = 110), patients with mild to moderate asthma with controlled or partially controlled symptoms (MMA; n = 93) and healthy non‐smoking controls (HC; n = 100) (Figure S2; Table 1). The NSA and SSA groups had more symptomatic asthma as measured by the Asthma Control Questionnaire (ACQ) and Asthma Quality of Life Questionnaire (AQLQ) (Table S2) and they also reported more exacerbations in the previous year than MMA (Table 1). Measurement of forced expiratory volume in the first second (FEV1, litters or % predicted) and the ratio of FEV1 to forced vital capacity (FVC) (litters or % predicted), both measures of airflow obstruction were lower in severe asthma patients than in the MMA and HC groups (p < .001) (Table 1). The quality of life may be affected by airflow obstruction and exacerbations are supported by the significant correlations between AQLQ scores and FEV1 (L) (r = 0.09; p < .05) and exacerbations in the previous year (r = −0.11; p = 0064) (Figure S3).
TABLE 1.
Demographic characteristics of C‐BIOPRED
| Characteristic | Severe nonsmoking asthma (n = 342) | Smokers and ex‐smokers with severe asthma (n = 110) | Mild/moderate nonsmoking asthma (n = 93) | Healthy nonsmoking controls (n = 100) | p‐Value |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 101 (29.5) | 106 (96.4) | 43 (46.2) | 36 (36.0) | <.001 |
| Female | 241 (70.5) | 4 (3.6) | 50 (53.8) | 64 (64.0) | |
| Age (years) | 53.39 ± 11.45 | 57.31 ± 9.46 | 48.65 ± 12.07 | 33.18 ± 14.20 | <.001 |
| Age of diagnosis (years) | 40.08 ± 16.42 | 44.46 ± 17.36 | 35.76 ± 16.07 | NA | <.001 |
| BMI (kg/m2) | 24.31 ± 3.65 | 25.05 ± 3.25 | 24.21 ± 3.08 | 22.38 ± 2.81 | <.001 |
| BMI Normal (< 28) | 293 (85.7) | 90 (81.8) | 84 (90.3) | 98 (98.0) | <.001 |
| BMI Obesity (> = 28) | 49 (14.3) | 20 (18.2) | 9 (9.7) | 2 (2.0) | |
| Smoking (Pack‐years) | NA | 28.71 (22.76) | NA | NA | NA |
| Current Smoker | NA | 44 (40.0) | NA | NA | NA |
| Ex‐smoker | NA | 66 (60.0) | NA | NA | NA |
| pre‐BD FVC, L | 2.57 ± 0.76 | 3.11 ± 0.80 | 3.35 ± 0.94 | 3.81 ± 0.97 | <.001 |
| pre‐BD FVC % pred, L | 83.43 ± 17.38 | 80.76 ± 16.54 | 97.21 ± 17.54 | 101.50 ± 12.35 | <.001 |
| pre‐BD FEV1, L | 1.55 ± 0.58 | 1.79 ± 0.71 | 2.21 ± 0.81 | 3.20 ± 0.79 | <.001 |
| pre‐BD FEV1% pred | 64.79 ± 21.16 | 60.87 ± 21.22 | 81.69 ± 23.13 | 106.01 ± 11.44 | <.001 |
| pre‐BD FEV1/FVC, % | 60.17 ± 12.29 | 56.72 ± 12.43 | 65.38 ± 11.65 | 84.19 ± 5.83 | <.001 |
| Post‐BD, n | 329 | 108 | 80 | ||
| Post‐BD FEV1, L | 1.82 ± 0.60 | 2.07 ± 0.72 | 2.45 ± 0.78 | NA | <.001 |
| Post‐BD FEV1% pred | 76.02 ± 21.16 | 70.47 ± 21.93 | 91.03 ± 21.79 | NA | <.001 |
| Post‐BD FEV1 (% increase) | 21.60 ± 15.45 | 17.88 ± 14.73 | 14.74±10.56 | NA | <.001 |
| Exacerbations in prior year | 1.50 ± 1.80 | 1.36 ± 1.60 | 0.33 ± 0.54 | NA | <.001 |
| Exacerbation in the previous year, n(%) | |||||
| Yes | 113 (33.0) | 36(32.7) | 64 (68.8) | NA | <.001 |
| No | 219 (64.0) | 72 (65.5) | 27 (29.0) | NA | |
| Healthcare resource utilization, n(%) | |||||
| Yes | 64 (18.7) | 14 (12.7) | 5 (5.4) | NA | <.001 |
| No | 278 (81.3) | 96 (87.3) | 88 (94.6) | 100 (100.0) | |
Abbreviations: BD, bronchodilator; BMI, body mass index; FEV1, forced expiratory volume in one second; FVC, forced vital capacity; Max, maximum; Min, Minimum, N, number of subjects in the cohort, n, Number of subjects included in the analysis; NA, Not applicable; SD, Standard deviation.
Note: Data are shown as mean ± standard deviation, unless as n (%). The subject level data used for FVC, pre‐BD FEV1, pre‐BD FEV1% pred, pre‐BD FEV1/FVC ratio, % analysis follows the algorithm below: If baseline data are available, then baseline data are used. Otherwise, if screening data are available, then screening data are used.
The proportion of patients who had nasal polyps and/or nasal polypectomy and gastro‐oesophageal reflux disease in severe asthma was higher than that in the MMA and HC (Table S3). Atopy defined by at least one positive specific IgE level and total serum IgE was higher in the three asthma groups than among HCs (p < .001) (Table 2). Regarding asthma treatments, the NSA and SSA groups, 9.94% of NSA and 12.73% of SSA received daily OCS, and those in the MMA group received none (Table S1). Therefore, severe asthma patients have more symptoms, more exacerbations, more airflow obstruction, and more nasal polyps even if they are on high‐dose asthma medication regimens, including oral corticosteroids.
TABLE 2.
Biomarkers in blood and exhaled air
| Severe nonsmoking asthma(n = 342) | Smokers and ex‐smokers with severe asthma(n = 110) | Mild/moderate nonsmoking asthma(n = 93) | Healthy nonsmoking controls (n = 100) | p‐Value | |
|---|---|---|---|---|---|
| Subjects with FeNO, n | 320 | 103 | 75 | 86 | |
| FeNO(ppb) | 31.00 (20.00, 54.00) | 27.00 (18.00, 59.50) | 28.00 (19.00, 47.00) | 15.00 (12.00, 19.75) | <.001 |
| Subjects with blood results, n | 341 | 107 | 93 | 100 | |
| Neutrophil count(10∧9/L) | 3.72 (2.91, 4.61) | 4.10 (3.30, 4.98) | 3.78 (2.91, 4.74) | 3.45 (2.86, 4.34) | 0.004 |
| Eosinophil count(10∧9/L) | 0.24 (0.11, 0.45) | 0.24 (0.13, 0.46) | 0.21 (0.13, 0.30) | 0.08 (0.05, 0.13) | <.001 |
| Neutrophil (%) | 57.40 (51.80, 64.04) | 61.00 (53.15, 66.65) | 59.40 (53.60, 65.50) | 58.70 (54.35, 62.75) | 0.066 |
| Eosinophil (%) | 3.60 (1.90, 7.50) | 3.50 (2.05, 6.45) | 3.50 (2.00, 5.00) | 1.20 (0.80, 2.22) | <.001 |
| Subjects with IgE and ECP results, n | 281 | 90 | 77 | 73 | |
| ECP*(μg/L) | 7.29 (4.00, 13.70) | 7.71 (4.36, 15.13) | 7.38 (3.73, 12.50) | 3.37 (2.35, 5.54) | <.001 |
| IgE Total (KU/L) | 161.00 (58.50, 395.00) | 232.00 (97.38, 534.50) | 171.00 (68.00, 393.00) | 32.40 (18.50, 94.30) | 0.001 |
| Atopy* | |||||
| above normal | 137 (48.8) | 49 (54.4) | 48 (62.3) | 19 (26.0) | <.001 |
| normal | 144 (51.2) | 41 (45.6) | 29 (37.7) | 54 (74.0) |
Data are presented as median values (interquartile range) for continuous variables and as n (%) for categorical variables.
Abbreviations: ECP, eosinophil cationic protein; FeNO, fractional level of nitric oxide in exhaled breath; HX2, house dust mix; MX2, mold mix; TX4, tree pollen mix; WX5, weed pollen mix; FX5, food allergens.
Specific IgE (HX2, FX5, MX2, TX4, and WX5 Phadiatop) <0.35 Ku/L is considered non‐atopic.
Note: The Kruskal‐Wallis test was here used for continuous data and Fisher's exact test for discrete data.
Note: Atopy (above normal) means the subjects have at least one allergen above normal (> .35).
To understand the role of inflammatory factors in severe asthma, we measured blood eosinophil and neutrophil counts (BEC and BNC) and fractional level of nitric oxide in exhaled breath (FeNO), and the level of granulocytes in sputum (SEC and SNC) collected after inhalation of hypertonic saline solutions in a smaller group (260 out of 545 asthma participants). Levels of BEC, BNC, and serum eosinophil cationic protein (ECP), a product released from activated eosinophils, were higher in the NSA, SSA, and MMA than in the HC (Table 2). FeNO levels were also higher in the three asthma groups (Table 2). BEC (%) was correlated with FeNO (r = 0.37; p < .001) (Figure S4), as were sputum eosinophils (%) (r = 0.38; p < .0001) (Figure 1). Median SEC of NSA and SSA were 11.6% and 8.1%, respectively, which were higher than those of MMA (5.1%) and HC (0.9%) (p < .0001) (Table S4; Figure S5). There was no difference in SNC among the four groups. Using BEC ≥ 300/μl as a marker of Type‐2 inflammation, 38.4% of severe asthma can be categorized as severe eosinophilic asthma, while this would be 76.8% using a cut‐off for SEC (%) ≥ 2.5%. There was a correlation between SEC (%) and the number of exacerbations in the previous year (r = 0.24; p = 0.005), AQLQ score (r = −0.18; p = 0.012), and ACQ5 score (r = 0.17; p = .015) (Figure 1). SNC (%) was correlated with that of BNC (%) (r = 0.26; p 0.0002). In NSA and SSA groups, SNC (%) was negatively correlated with FEV1 (% predicted) (r = −0.28; p < .001) and FEV1/FVC ratio (r = −0.27; p < .001), but there was no correlation of SEC (%) with these parameters (Figure 1).
FIGURE 1.
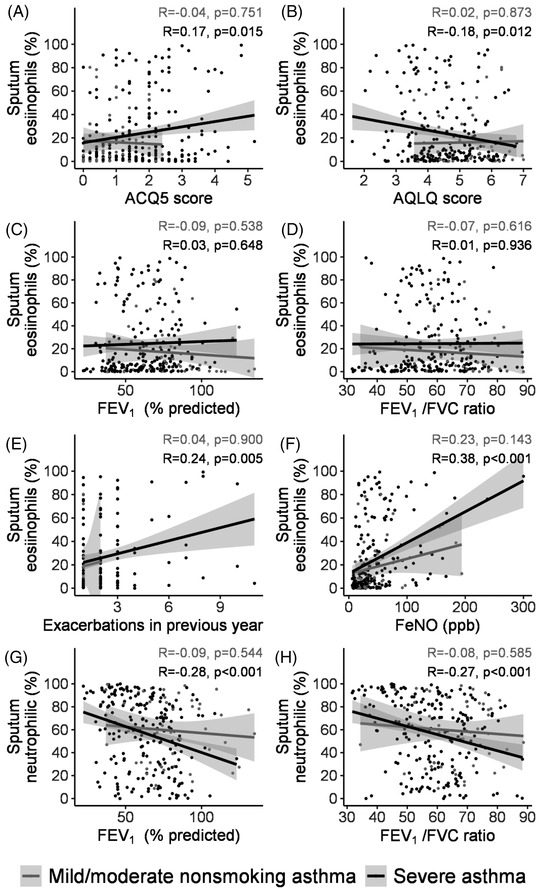
Correlations between sputum eosinophils and neutrophil (%), and ACQ‐5 score, AQLQ score, FEV1 (% predicted), FEV1/FVC ratio, exacerbations in the previous year, and FeNO for severe asthma and mild‐moderate asthma. The correlation coefficients (R) and p‐values are indicated
At the one‐year review in the NSA and SSA, there were no changes in airflow obstruction or the biomarkers of inflammation (BEC, BNC, and FeNO) indicating biostability of the phenotype. However, improvement in symptoms and asthma control was observed in NSA (Table 3).
TABLE 3.
Data for baseline and longitudinal visits
| Severe nonsmoking asthma (n = 205) | Smokers and ex‐smokers with severe asthma (n = 58) | SNA | SSA | |||
|---|---|---|---|---|---|---|
| Baseline | Longitudinal | Baseline | Longitudinal | p‐Value | pValue | |
| Longitudinal visit subjects with pulmonary test, n | 205 | 204 | 58 | 58 | ||
| pre‐BD FVC, L (Mean (SD)) | 2.61 (0.75) | 2.64 (0.74) | 3.04 (0.70) | 3.15 (0.76) | 0.640 | 0.501 |
| pre‐BD FEV1, L (Mean (SD)) | 1.56 (0.56) | 1.61 (0.54) | 1.71 (0.60) | 1.81 (0.72) | 0.208 | 0.545 |
| pre‐BD FEV1% predicted (Mean (SD)) | 65.62 (21.54) | 68.85 (21.09) | 58.38 (18.89) | 62.21 (22.39) | 0.093 | 0.436 |
| pre‐BD FEV1/FVC, %(Mean(SD)) | 59.50 (12.34) | 60.92 (12.20) | 55.74 (12.30) | 56.23 (13.13) | 0.222 | 0.804 |
| Longitudinal visit subjects with bronchodilator reversibility test, n | 190 | 190 | 56 | 56 | ||
| FEV1 reversibility (L), median(IQR) | 0.28 (0.22, 0.38) | 0.18 (0.09, 0.27) | 0.27 (0.18, 0.42) | 0.23 (0.16, 0.37) | <.001 | 0.322 |
| FEV1 reversibility (%), median (IQR) | 20.74 (13.66, 31.11) | 12.12(5.91, 20.75) | 15.98 (8.78, 31.10) | 14.90 (8.31, 22.28) | <.001 | 0.480 |
| Healthcare resource utilization,n(%) | 205 | 205 | 58 | 58 | ||
| Yes | 43 (21.0) | 64 (31.2) | 7 (12.1) | 16 (27.6) | 0.024 | 0.061 |
| No | 162 (79.0) | 141 (68.8) | 51 (87.9) | 42 (72.4) | ||
| Subjects with questionnaire, n | 205 | 205 | 57 | 57 | ||
| ACQ | ||||||
| ACQ5 | 1.65 (1.03) | 1 | 1.81 (0.98) | 1.56 (1.07) | 0.012 | 0.158 |
| ACQ7 | 1.85 (0.86) | 1.65 (0.82) | 2.08 (0.85) | 1.84 (0.94) | 0.017 | 0.121 |
| AQLQ | ||||||
| AQLQ | 4.54 (1.02) | 4.83 (1.07) | 4.71 (1.06) | 4.91 (1.02) | 0.005 | 0.296 |
| Symptoms | 4.76 (1.07) | 5.06 (1.06) | 4.78 (1.06) | 5.09 (1.01) | 0.004 | 0.129 |
| Activity limitation | 4.46 (1.02) | 4.76 (1.09) | 4.67 (1.21) | 4.87 (1.16) | 0.004 | 0.392 |
| Emotional | 4.44 (1.37) | 4.75 (1.35) | 4.69 (1.33) | 4.79 (1.48) | 0.018 | 0.716 |
| Environmental | 4.19 (1.46) | 4.40 (1.50) | 4.60 (1.46) | 4.62 (1.50) | 0.106 | 0.925 |
| ESS | 6.87 (4.2) | 6.66 (4.4) | 7.46 (4.42) | 7.58 (4.73) | 0.393 | 0.889 |
| MARS | 21.44 (2.92) | 21.45 (3.33) | 21.77 (2.8) | 21.84 (2.69) | 0.599 | 0.961 |
| Subjects with FeNO, n | 192 | 194 | 54 | 54 | ||
| FeNO(ppb) | 29.50 [20.00, 52.00] | 28.00 [18.25, 48.00] | 26.50 [18.00, 63.50] | 29.00 [16.25,48.50] | 0.758 | 0.775 |
| Subjects with blood results, n | 205 | 197 | 58 | 57 | ||
| Neutrophil count(10∧9/L) | 3.60 [3.00, 4.50] | 3.54 [2.85, 4.59] | 3.90 [3.27,4.95] | 3.89 [2.90,5.21] | 0.580 | 0.775 |
| Eosinophil count(10∧9/L) | 0.23 [0.11, 0.40] | 0.23 [0.11, 0.44] | 0.26 [0.13,0.40] | 0.20 [0.12,0.42] | 0.501 | 0.745 |
| Neutrophil (%) | 57.70 [52.20, 64.20] | 58.90 [52.30, 64.70] | 60.85 [52.85, 66.68] | 59.10 [50.40, 67.00] | 0.703 | 0.576 |
| Eosinophil (%) | 3.60 [1.90, 6.50] | 4.20 [1.90, 7.00] | 3.60 [2.18, 6.37] | 3.30 [1.80, 6.00] | 0.320 | 0.724 |
| Subjects with IgE and ECP results, n | 169 | 162 | 47 | 50 | ||
| ECP (μg/L) | 7.77 [4.03, 16.20] | 7.05 [4.10, 13.73] | 6.98 [4.55, 14.95] | 6.99 [4.30, 14.95] | 0.389 | 0.745 |
| IgE Total (Ku/L) | 165.00 [57.80, 407.00] | 168.50 [62.98, 378.75] | 320.00 [128.50, 909.00] | 321.00 [94.57, 1189.00] | 0.760 | 0.940 |
| Atopy* | ||||||
| above normal | 88 (52.07) | 89 (54.94) | 29 (61.70) | 24 (48.00) | 0.659 | 0.222 |
| normal | 81 (47.93) | 73 (45.06) | 18 (38.30) | 26 (52.00) | ||
Data are presented as mean (standard deviation) or median (interquartile range) for continuous variables, n (%) for categorical variables.
Note: Using all severe patients without missing value.
Abbreviations: FEV1, forced expiratory volume in one second; FVC, forced vital capacity; BD. bronchodilator.
Note: P‐values are based on the comparison between baseline and longitudinal values. The Kruskal‐Wallis test was used for continuous data and the Fisher exact test for discrete data.
Atopy (above normal) means the subjects have at least one above normal (>0.35) records for HX2, FX5, MX2, TX4, WX5, and Phadiatop.
This C‐BIOPRED study of patients with severe asthma in China uniquely defines a population with the most severe disease characterized by poorer asthma control, frequent exacerbations, and chronic airflow obstruction despite taking maximal amounts of asthma medication in terms of oral corticosteroid therapy, with ≈10% needing oral corticosteroid therapy which is associated with systemic side‐effects. The use of biomarkers in severe asthma to define those with Type‐2 inflammation that is associated with eosinophilic inflammation indicates that SEC is a better biomarker than BEC or FeNO in that respect 6 because it could distinguish MMA from NSA and SSA. Thus, using SEC, a very high proportion of patients with severe asthma of up to 76.8% would have severe eosinophilic asthma, which is higher than in other severe asthma cohorts 7 , 8 , 9 of Caucasian populations. This has implications for future therapies because of the current availability of biologic therapies such as anti‐IgE, anti‐IL5, anti‐IL‐5Rα, and anti‐IL4Rα monoclonal antibodies for Type‐2 severe asthma. 10
These findings provide insight into the role of some of the factors driving various traits of severe asthma. Thus, the degree of airflow obstruction and the exacerbation rate may determine the level of deterioration while the eosinophilic inflammation may underlie the rate of exacerbations. Indeed, the amelioration in severe asthma provided by the anti‐IL5 or anti‐IL5Rα antibodies that suppress eosinophil inflammation led to a reduction in exacerbation rates. 10 However, the negative correlation between the sputum neutrophil count and FEV1 indicated a possible role of neutrophils in determining airflow obstruction. Since severe asthma has different molecular phenotypes other than Type‐2, 11 further exploration of molecular phenotypes in this Chinese cohort may facilitate precision medicine and allow further dissection of these multiple molecular pathways. 12
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This study was funded by Astra‐Zeneca, China.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
We are grateful to all the members of the C‐BIOPRED Study Group (listed in Supplementary Information) who took part in the design and in the recruitment of participants into the study. We thank Yicheng Fan, Yujing Liu, Assam Nkouibert Prysewley Assam, Zhaohui Zhou, Wei Jiang, and Jia Wang of Astra‐Zeneca for their help in data and statistical analyses and in the preparation of the figures and tables. We also thank all the participants across China who willingly took part in C‐BIOPRED study.
These authors contributed equally as co‐first authors.
A list of investigators in the C‐BIOPRED consortium is provided in the Supporting Information.
Contributor Information
Kian Fan Chung, Email: f.chung@imperial.ac.uk.
Nanshan Zhong, Email: nanshan@vip.163.com.
REFERENCES
- 1. Huang K, Yang T, Xu J, et al. Prevalence, risk factors, and management of asthma in China: a national cross‐sectional study. Lancet. 2019;394(10196):407‐418. [DOI] [PubMed] [Google Scholar]
- 2. Su N, Lin JT, Wang WY, et al. A cross‐section study of severe asthma in eight provinces of China. Zhonghua Nei Ke Za Zhi. 2016;55(12):917‐921. [DOI] [PubMed] [Google Scholar]
- 3. Wang WY, Lin JT, Zhou X, et al. A survey on clinical characteristics and risk factors of severe asthma in China. Zhonghua Yi Xue Za Zhi. 2020;100(14):1106‐1111. [DOI] [PubMed] [Google Scholar]
- 4. Wang G, Wang F, Gibson PG, et al. Severe and uncontrolled asthma in China: a cross‐sectional survey from the Australasian Severe Asthma Network. J Thorac Dis. 2017;9(5):1333‐1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung KF, Wenzel SE, Brozek JL, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343‐373. [DOI] [PubMed] [Google Scholar]
- 6. Pavlidis S, Takahashi K, Ng Kee Kwong F, et al. “T2‐high” in severe asthma related to blood eosinophil, exhaled nitric oxide and serum periostin. Eur Respir J. 2019;53(1). [DOI] [PubMed] [Google Scholar]
- 7. Moore WC, Bleecker ER, Curran‐Everett D, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw DE, Sousa AR, Fowler SJ, et al. Clinical and inflammatory characteristics of the European U‐BIOPRED adult severe asthma cohort. Eur Respir J. 2015;46(5):1308‐1321. [DOI] [PubMed] [Google Scholar]
- 9. Kupczyk M, Dahlen B, Sterk PJ, et al. Stability of phenotypes defined by physiological variables and biomarkers in adults with asthma. Allergy. 2014;69(9):1198‐1204. [DOI] [PubMed] [Google Scholar]
- 10. Chung KF. Targeting the interleukin pathway in the treatment of asthma. Lancet. 2015;386(9998):1086‐1096. [DOI] [PubMed] [Google Scholar]
- 11. Kuo CS, Pavlidis S, Loza M, et al. T‐helper cell type 2 (Th2) and non‐Th2 molecular phenotypes of asthma using sputum transcriptomics in U‐BIOPRED A transcriptome‐driven analysis of epithelial brushings and bronchial biopsies to define asthma phenotypes in U‐BIOPRED. Eur Respir J. 2017;49(2):443‐455. [Google Scholar]
- 12. Chung KF, Adcock IM. Precision medicine for the discovery of treatable mechanisms in severe asthma. Allergy. 2019;74(9):1649‐1659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


