Abstract
Since the vast majority of species solely rely on innate immunity for host defense, it stands to reason that a critical evolutionary trait like immunological memory evolved in this primitive branch of our immune system. There is ample evidence that vaccines such as bacillus Calmette-Guérin (BCG) induce protective innate immune memory responses (trained immunity) against heterologous pathogens. Here we show that while BCG vaccination significantly reduces morbidity and mortality against influenza A virus (IAV), it fails to provide protection against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). In contrast to IAV, SARS-CoV-2 infection leads to unique pulmonary vasculature damage facilitating viral dissemination to other organs, including the bone marrow (BM), a central site for BCG-mediated trained immunity. Finally, monocytes from BCG-vaccinated individuals mount an efficient cytokine response to IAV infection, while this response is minimal following SARS-CoV-2. Collectively, our data suggest that the protective capacity of BCG vaccination is contingent on viral pathogenesis and tissue tropism.
Keywords: SARS-CoV-2, influenza virus, BCG vaccination, animal models, trained immunity, hematopoietic stem cells, lung pathology, monocytes
Graphical abstract

Kaufmann et al. show that BCG vaccination protects mice and hamsters against influenza, but not SARS-CoV-2. The distinct tropism of SARS-CoV-2 for pulmonary endothelial cells may lead to lung hemorrhage and systemic viral dissemination. Thus, the protection of BCG against viral pathogens is mainly determined by pathogenesis of infections.
Introduction
While the concept of the innate memory response (termed trained immunity) is “young,” the evidence of innate memory in host defense against diverse infectious diseases is “old” (Divangahi et al., 2021). A growing body of literature indicates that live attenuated vaccines (LAVs) such as Bacillus Calmette-Guérin (BCG), measles-containing vaccines, smallpox, and oral poliovirus vaccines (OPVs) induce cross-protection against other infectious diseases (Namakula et al., 2020). While pathogen-associated molecular patterns (PAMPs) in LAVs activate an array of innate immune responses via pattern recognition receptors (e.g., Toll-like receptors [TLRs] or NODs), the basis for using adjuvants in non-live vaccines (e.g., inactivated vaccines against influenza) is to enhance the activity of innate immunity.
Many studies have shown that BCG vaccination provides heterologous protection against unrelated pathogens. For example, BCG vaccination in newborn children is associated with protection against not only tuberculosis but also respiratory tract infections, neonatal sepsis, and all-cause mortality (Aaby et al., 2011; Rieckmann et al., 2017). Additionally, both BCG and polio vaccinations at birth lead to 32% lower mortality than BCG vaccination alone, indicating that OPV has an additional beneficial effect on mortality (Lund et al., 2015). Pre-clinical data in murine models of infection further support these observations as it has been shown that BCG vaccination is associated with protection against fungal infections (e.g., Candida albicans) (Van 'T Wout et al., 1992), parasites (e.g., Schistosoma mansoni) (Tribouley et al., 1978), as well as DNA and RNA viruses (e.g., herpes and influenza virus) (Hippmann et al., 1992; Spencer et al., 1977; Mukherjee et al., 2017). It has also been shown that BCG vaccination in healthy adults is associated with protection against human experimental models of yellow fever (Arts et al., 2018) and malaria (Walk et al., 2019) infections, as well as all-cause respiratory infections in the elderly (Giamarellos-Bourboulis et al., 2020). Thus, epidemiological studies as well as pre-clinical and clinical data all support the cross-protective effects of BCG vaccination (Namakula et al., 2020). However, despite several large, ongoing clinical trials of BCG vaccination against the current coronavirus disease 2019 (COVID-19) pandemic, the protective efficacy of BCG against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) remains unknown.
Results
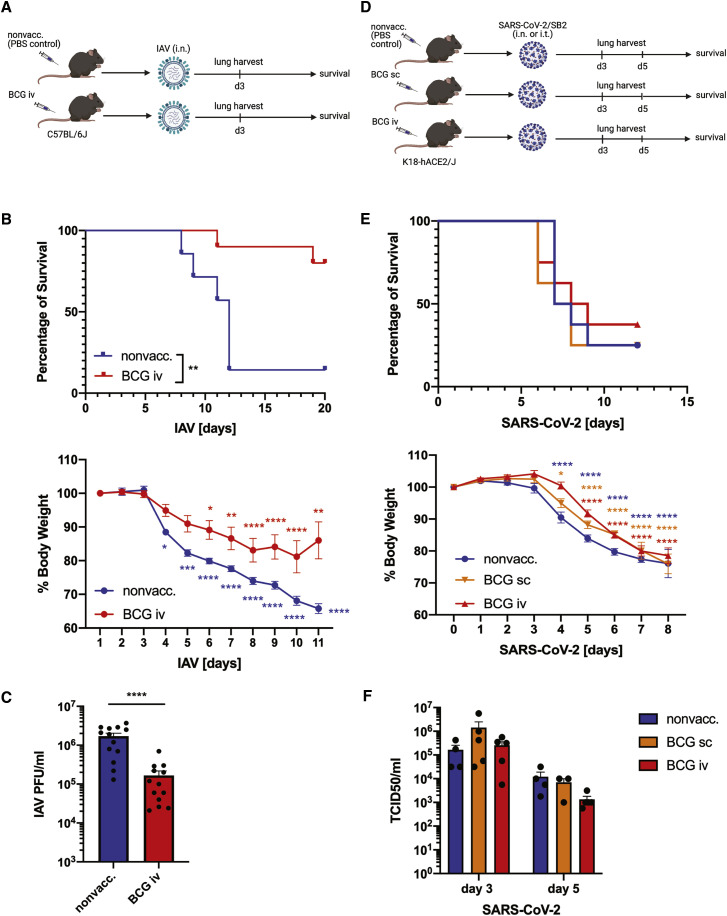
Previous studies have shown that intraperitoneal and intranasal (i.n.) BCG vaccinations provide protection in murine models of IAV infection (Mukherjee et al., 2017; Spencer et al., 1977). As we have recently shown that systemic (intravenous [i.v.]) BCG vaccination induces trained immunity (Kaufmann et al., 2018), we vaccinated wild-type C57BL/6J mice with BCG i.v.. Four weeks after BCG-i.v. vaccination, mice were challenged with a lethal or sublethal dose of IAV i.n. (Figure 1A). Remarkably, BCG-vaccinated mice showed a significant reduction in morbidity and mortality after IAV infection (Figure 1B). Likewise, the lung viral load was significantly lower at day 3 post IAV infection in BCG-i.v.-vaccinated mice compared with nonvaccinated controls (Figure 1C).
Figure 1.
In mice, BCG vaccination induces unspecific protection against IAV infection, but not against SARS-CoV-2
(A) Experimental model of BCG vaccination and intranasal (i.n.) IAV infection in C57BL/6J mice.
(B) Mortality (top) and morbidity (bottom) of 1-month BCG-i.v.-vaccinated or nonvaccinated control C57BL/6J mice after i.n. infection with a lethal dose of 90 plaque-forming units (PFU) influenza A/Puerto Rico/8/34 (IAV-PR8), n = 7–10/group.
(C) Lung viral load in BCG-i.v.-vaccinated and control nonvaccinated C57BL/6J mice at day 3 post sublethal IAV-PR8 infection (50 PFU), n = 13/group.
(D) Experimental model of BCG vaccination and i.n. or intratracheal (i.t.) SARS-CoV-2/SB2 infection in B6.Cg-Tg(K18-ACE2)2Prlmn/J mice.
(E) Mortality (top) and morbidity (bottom) (n = 8/group) of BCG-vaccinated and control K18-hACE2/J mice after i.n. infection with 1 × 104 TCID50/mL SARS-CoV-2.
(F) Lung viral load at day 3 and day 5 post sublethal i.t. SARS-CoV-2/SB2 infection (4,000 TCID50/mL), n = 3–5/group.
Stars in morbidity curves (B and E) indicate significant weight loss compared with day 0. Data are displayed as mean ± SEM. ∗p < 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (survival analyses and two-way ANOVA). See also Figure S1 and Table S1.
To test whether BCG vaccination likewise induces protection against SARS-CoV-2 infection in mice, we vaccinated transgenic mice expressing the human ACE2 receptor (B6.Cg-Tg(K18-ACE2)2Prlmn/J) with BCG subcutaneously (s.c.)—which is the equivalent of intradermal BCG vaccination in humans—or i.v. At 4 weeks post vaccination, we i.n. infected the vaccinated and nonvaccinated control K18-hACE2 mice with a lethal dose of SARS-CoV-2/SB2 (lineage B.4) (Figure 1D). However, we did not observe differences in morbidity and mortality between vaccinated and control groups (Figure 1E). In K18-hACE2 mice, a low level of hACE2 expression is detectable in the brain, and i.n. exposure to SARS-CoV-2 leads to rapid and severe neurological disorders that lead to death (Kumari et al., 2021). To overcome this limitation, we next infected the hACE2 mice intratracheally (i.t.) with SARS-CoV-2/SB2 to bypass the access of the virus to the brain via the olfactory system. However, BCG vaccination had no significant impact on either morbidity and survival (Figure S1A) or pulmonary SARS-CoV-2 replication at either day 3 or 5 post i.t. infection with lethal or sublethal doses, respectively (Figure 1F). Similarly, no differences in the systemic myeloid cell numbers were observed (Figure S1B, Table S1).
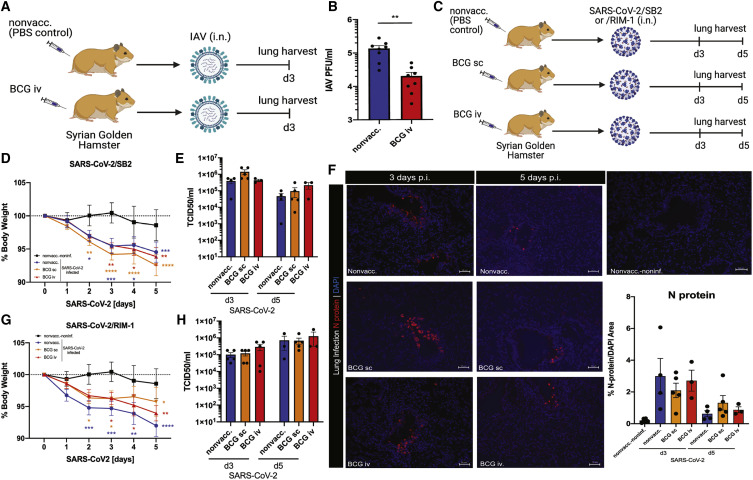
Given the limitations of K18-hACE2 transgenic mice expressing non-physiological levels of hACE2 receptor (McCray et al., 2007), we next examined the effects of BCG vaccination on IAV and SARS-CoV-2 infections in a Syrian golden hamster model (Chan et al., 2020). Syrian golden hamsters naturally express ACE2 receptor, are susceptible to SARS-CoV-2, and develop a moderate form of COVID-19-like disease (Imai et al., 2020). Likewise, they are moderately susceptible to IAV infection (Iwatsuki-Horimoto et al., 2018). At day 3 post IAV infection (i.n.) (Figure 2A), BCG-i.v.-vaccinated Syrian golden hamsters displayed significantly lower pulmonary viral load (Figure 2B) and less weight loss (Figure S2A) compared with nonvaccinated controls. We then infected BCG (s.c. and i.v.)-vaccinated Syrian golden hamsters with SARS-CoV-2/SB2 strain (i.n.) (Figure 2C). In contrast to IAV infection, there were no significant differences in weight loss between BCG-vaccinated and nonvaccinated hamsters after SARS-CoV-2 infection (Figure 2D). Additionally, BCG vaccination had no effect on pulmonary viral loads at day 3 and 5 post infection, as determined by TCID50 (Figure 2E), qPCR (Brandolini et al., 2021) (Figure S2B), and immunofluorescence (Figure 2F). The total lung leukocyte numbers were also similar between groups, with the exception of higher numbers in the BCG-i.v.-vaccinated animals after 3 days of SARS-CoV-2 infection (Figure S2C). This increase was also reflected in the cellular populations of bronchoalveolar lavage (BAL), lung, spleen, and blood (Figure S2D, Table S1).
Figure 2.
BCG vaccination protects Syrian golden hamsters against IAV, but not SARS-CoV-2 infection
(A) Experimental model of BCG vaccination and i.n. infection with IAV-H3N2 in Syrian golden hamsters.
(B) Lung viral load in BCG-i.v.-vaccinated and nonvaccinated control Syrian golden hamsters at day 3 post i.n. infection with 1 × 105 PFU IAV-H3N2, n = 8/group.
(C) Experimental model of BCG vaccination and i.n. infection with SARS-CoV-2/SB2 or/RIM-1 in Syrian golden hamsters.
(D–F) Morbidity (n = 7–10/group) (D), lung viral load by TCID50/mL measurement (n = 3–5/group) (E), and representative images and quantification of viral N protein staining in the lungs of golden hamsters infected with 1 × 105 PFU SARS-CoV-2/SB2 (n = 3–5/group) (F).
(G and H) Morbidity (n = 7–10/group) (G) and lung viral load determined by TCID50/mL assay (n = 3–5/group) (H) in BCG-vaccinated and control Syrian golden hamsters at 3 and 5 days post i.n. infection with 1 × 105 PFU SARS-CoV-2/RIM-1.
Stars in morbidity curves (D and G) indicate significant weight loss compared with day 0. Data are displayed as mean ± SEM. ∗p < 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (t test and two-way ANOVA). See also Figure S2, Table S2.
Recently, sequence analyses of SARS-CoV-2/SB2 strain revealed several mutations within the spike coding region (Banerjee et al., 2020). Thus, we next infected BCG-vaccinated (s.c. or i.v.) or nonvaccinated Syrian golden hamsters with another strain of SARS-CoV-2 that is more similar to the original SARS-CoV-2 strain isolated in Wuhan, China (SARS-CoV-2/RIM-1, formerly known as SARS-CoV-2/cp13.32, lineage B.1.147; Murall et al., 2021; Table S2). Like in SARS-CoV-2/SB2 infection, BCG vaccination had no impact on morbidity or lung viral loads in Syrian golden hamsters infected with SARS-CoV-2/RIM-1 (Figures 2G–2H and S2E). Likewise, serum interferon (IFN)-β and interleukin (IL)-6 levels showed no differences between SARS-CoV-2/SB2 and RIM-1 infections (Figures S2F–S2G).
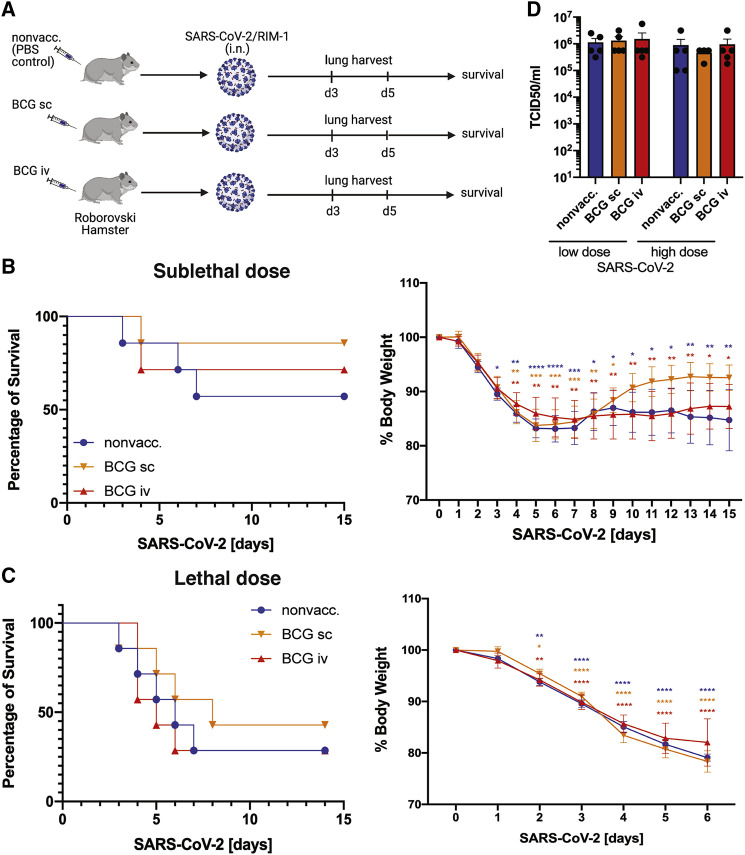
In contrast to the mild disease phenotype displayed by Syrian golden hamsters, Roborovski hamsters develop severe disease and can succumb to SARS-CoV-2 infection (Trimpert et al., 2020). However, similar to the Syrian golden hamsters, BCG vaccination (s.c. or i.v.) did not provide any protection against sublethal or lethal doses of SARS-CoV-2/RIM-1 infection in Roborovski hamsters when infected at 4 weeks post vaccination (Figures 3A–3D and S3A–S3C), or when infected with a sublethal dose of SARS-CoV-2/RIM-1 at 6 months post BCG vaccination (Figures S3D–S3E). Collectively, these data indicate that BCG vaccination fails to provide protection against mild or severe forms of infection with SARS-CoV-2.
Figure 3.
In Roborovski hamsters, BCG vaccination provides no protection against sublethal and lethal SARS-CoV-2 infection
(A) Experimental model of BCG vaccination and i.n. infection with SARS-CoV-2/RIM-1 in Roborovski hamsters.
(B and C) Mortality and morbidity of BCG-vaccinated and control Roborovski hamsters after i.n. infection with a sublethal (B) or lethal dose (C) of SARS-CoV-2/RIM-1, n = 7/group.
(D) Lung viral load in BCG-vaccinated and nonvaccinated Roborovski hamsters at day 3 post infection with a sublethal (low dose) or lethal dose (high dose) of SARS-CoV-2/RIM-1 as determined by TCID50/mL assay, n = 4–5/group.
Stars in morbidity curves (B and C) indicate significant weight loss compared with day 0. Data are displayed as mean ± SEM. ∗p < 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (two-way ANOVA). See also Figure S3 and Table S1.
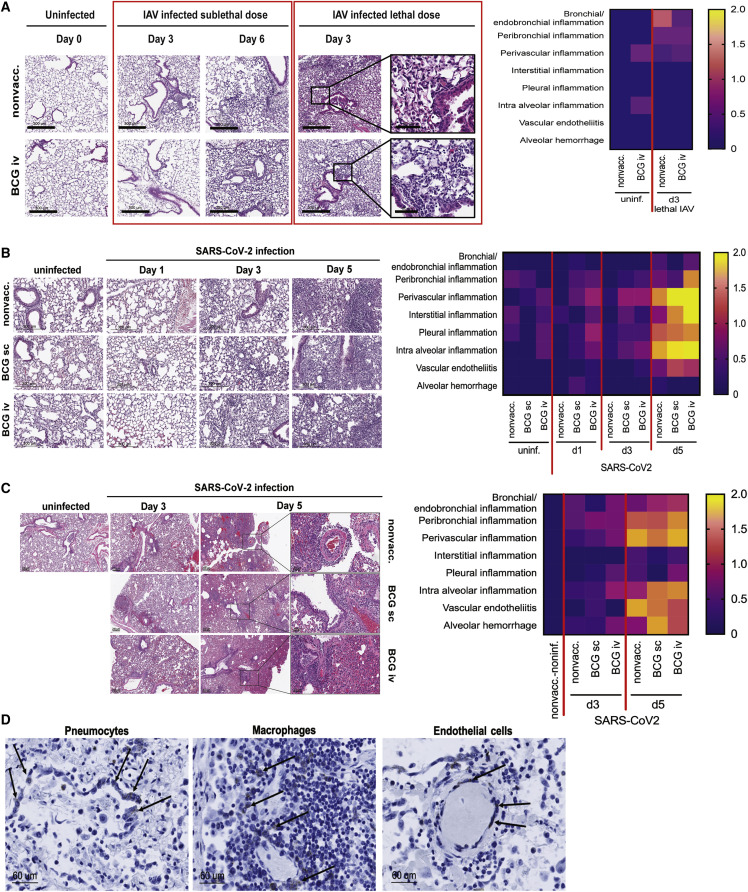
To investigate why BCG vaccination offers protection against IAV but not SARS-CoV-2 infections, we conducted in-depth comparative lung histological analyses. Our data indicate that after sublethal or lethal IAV infection, BCG-vaccinated mice revealed less lung damage in comparison with nonvaccinated mice (Figure 4A, Table S1). In contrast, BCG vaccination in K18-hACE2 mice did not improve lung pathology at days 1, 3, or 5 following SARS-CoV-2 infection (Figure 4B and Table S1). Likewise, after SARS-CoV-2 infection in BCG-vaccinated and nonvaccinated Syrian golden hamsters that exhibit only a clinically mild disease phenotype, severe lung lesions were observed, with extensive damage to the pulmonary vasculature after 5 days of infection (Figure 4C and Table S1). Pulmonary hemorrhage also occurs in 6-month BCG-vaccinated and nonvaccinated Roborovski hamsters at day 3 post sublethal SARS-CoV-2 infection (Figure S4A). In particular, we found that SARS-CoV-2 infection presented with (1) focal vasculitis and profound perivascular inflammatory infiltrates as well as severe endotheliitis; (2) pulmonary hemorrhage; and (3) a large quantity of lymphocytes infiltrating the lungs. Vascular damage is relatively unique to SARS-CoV-2 infection as it is not frequently described in other pulmonary infections. In line with these findings, SARS-CoV-2 RNA was detected in pneumocytes, pulmonary macrophages, and endothelial cells of a patient who died of COVID-19-induced acute respiratory distress syndrome (ARDS) (Figure 4D). The presence of SARS-CoV-2 within the endothelial cells is consistent with another study (Varga et al., 2020) suggesting that endothelial injury contributes to both the systemic effects of SARS-CoV-2 infection as well as perivascular inflammation (Gustafson et al., 2020). Systematic reviews of the lung histopathological reports also identified that microthrombi are a dominant feature of COVID-19 patients compared with patients with H1N1 influenza (Bonaventura et al., 2021; Hariri et al., 2021).
Figure 4.
SARS-CoV-2 induces significantly worse lung histological damage than IAV
(A) One-month BCG-vaccinated and nonvaccinated (PBS) C57BL/6J mice were i.n. infected with a sublethal or lethal dose of IAV-PR8. Representative images of lung histopathology on days 0, 3, and 6 post IAV infection (left) and scoring (right), n = 4–5/group, refer to Table S1.
(B) One-month BCG-vaccinated and nonvaccinated (PBS) K18-hACE2 mice were i.t. infected with 4,000 TCID SARS-CoV-2/SB2. Representative images of lung histopathology on days 1, 3, and 5 post SARS-CoV-2 infection (left) and scoring (right), n = 3–4/group, refer to Table S1.
(C) BCG-vaccinated and control Syrian golden hamsters were i.n. infected with 1 × 105 PFU SARS-CoV-2 and exhibited a clinically mild disease phenotype. Representative images of lung histopathologies on days 0, 3, and 5 post SARS-CoV-2 infection (left) and scoring (right), n = 3–6/group, refer to Table S1.
(D) RNAscope analysis of SARS-CoV-2 viral particles in human lung cells. Heatmap data are displayed as mean. See also Figure S4 and Table S1.
While the presence of other pulmonary viruses, such as IAV, in extrapulmonary tissues is rare, SARS-CoV-2 and extensive inflammation have been detected in kidneys, liver, heart, and brain, implicating the dissemination of SARS-CoV-2 from pulmonary endothelial damage. SARS-CoV-2 viremia (Di Cristanziano et al., 2020; Puelles et al., 2020) and the presence of viral RNA in the bone marrow (BM) of COVID-19 patients (Deinhardt-Emmer et al., 2021) has also been reported. Thus, we next hypothesized that viral dissemination secondary to pulmonary vascular damage impairs BCG-mediated trained immunity against SARS-CoV-2. We and others have recently shown that, in both mice and humans, BM is the site of BCG-induced trained immunity (Kaufmann et al., 2018; Cirovic et al., 2020). Interestingly, we have also demonstrated that access of a pathogen into the BM can prevent trained immunity (Khan et al., 2020). Thus, we next determined whether the extensive pulmonary vascular damage facilitates the dissemination of SARS-CoV-2 to the BM. Indeed, while influenza viral RNA in the BM of IAV-infected mice was barely detectable (Figure 5A), significant levels of SARS-CoV-2 RNA were detected in the BM of infected mice (Figures 5B–5C and S5A) as well as Roborovski (Figure 5D) and Syrian golden hamsters (Figure S5B). However, no differences in dissemination to the BM was observed between BCG-vaccinated and nonvaccinated animals (Figure S5B). Detection of viral replication-associated genes upE and N2 (Banerjee et al., 2021; Zeng et al., 2020; Al-Qaaneh et al., 2021) was potentially due to amplification of the virus systemically or directly in the BM. This indication is supported by a recent publication describing that SARS-CoV-2 infects and persists in the BM erythroid progenitors due to their expression of ACE2 and TMPRSS2 receptors (Huerga Encabo et al., 2021).
Figure 5.
BCG-induced trained immunity provides protection against IAV, but not SARS-CoV-2
(A) IAV viral load in the BM of C57BL/6J mice (day 3 and 6 post infection with 90 PFU IAV-PR8, n = 3/group).
(B and C) SARS-CoV-2 viral load in the BM of K18-hACE2/J mice (day 3 and 5 post i.t. infection with 4,000 TCID SARS-CoV-2/SB2, n = 3–5/group).
(D) SARS-CoV-2 viral load in the BM of Roborovski hamsters (day 3 post infection with 1 × 105 PFU SARS-CoV-2/RIM-1, n = 3–4/group).
(E) Experimental model of BCG vaccination in humans and infection of blood monocyte-derived macrophages (MDMs) with IAV and SARS-CoV-2/RIM-1 for assessment of cytokine responses.
(F) Expression of IL-1β, TNF-α, CCL2, type I IFNs, and IL-8 in MDMs from human donors before and after BCG vaccination, at 24 h post in vitro IAV-H3N2 or SARS-CoV-2/RIM-1 infection, n = 5/group.
Data are displayed as mean ± SEM. ∗p < 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (t test and one-way ANOVA). See also Figure S5.
Considering that BCG reprograms both mouse and human hematopoietic stem cells (HSCs) in the BM to generate trained monocytes/macrophages (Kaufmann et al., 2018; Cirovic et al., 2020), we next investigated whether monocytes from BCG-vaccinated individuals react differently to IAV and SARS-CoV-2 infection. We infected blood monocytes from healthy volunteers before and 90 days after BCG vaccination with live IAV or SARS-CoV-2 (Figure 5E). At 24 h post infection, IL-1β, tumor necrosis factor (TNF)-α, and CCL2 expression was higher in response to IAV infection post BCG vaccination, while SARS-CoV-2 induced only minimal levels of cytokines either pre or post BCG vaccination (Figure 5F). Thus, BCG-trained monocytes mounted an efficient cytokine response to IAV infection, while this response was minimal during SARS-CoV-2 infection.
Discussion
The central mechanisms involved in innate memory responses depend on epigenetic reprogramming that modifies the chromatin accessibility and thereby the readable gene information. In the context of LAVs, there are three known factors that can affect the epigenetic programming of an immune cell: (1) direct infection; (2) PAMPs from the microorganisms; and (3) endogenous cytokines released during the induction of the host response. The impact of these key factors occurs centrally, at the level of HSCs in the BM, and peripherally at the tissue-specific level. Recently, it has been demonstrated in pre-clinical (Kaufmann et al., 2018) and clinical (Cirovic et al., 2020) studies that BCG reprograms BM-HSCs towards myelopoiesis to generate trained immunity, while the access of virulent Mycobacterium tuberculosis to the BM prevents trained immunity (Khan et al., 2020). These studies provide a logical explanation of why short-lived innate immune cells can acquire memory and how trained immunity induced by LAVs (e.g., BCG) can provide cross-protection against other infectious diseases. Here our data suggest that, compared with IAV infection, the unique pulmonary vascular damage induced during SARS-CoV-2 infection may allow for viral dissemination to the BM, effectively preventing the ability of BCG to generate trained immunity. Furthermore, our results from monocytes of BCG-vaccinated humans suggest that, in contrast to IAV infection, potential virulence factors from SARS-CoV-2 can substantially suppress the inflammatory programming of trained immune cells. Thus, identifying these virulence factors can lead us closer to uncovering how SARS-CoV-2 hijacks our immune responses.
Our results contrast with a recent study demonstrating that BCG-i.v. vaccination increases survival of K18-hACE2 transgenic mice following SARS-CoV-2 challenge (Hilligan et al., 2022). The approach used by Hilligan et al., i.n. SARS-CoV-2 infection, has been observed by us and reported by others (Kumari et al., 2021) to result in pulmonary and neurological pathology, whereas we did not observe neurological symptoms following i.t. challenge. However, in our K18-hACE2 transgenic mouse model, BCG-i.v. vaccination did not provide significant protection after either i.n. or i.t. infection with SARS-CoV-2. Our results from the BCG vaccination in murine models of COVID-19 are furthermore supported by two physiological hamster models of SARS-CoV-2 infection, demonstrating that BCG vaccination has no significant impact on protection against mild or severe forms of COVID-19. One possible explanation for this discrepancy is the strains of BCG (Tice versus Pasteur) used between our two groups that may have different protective capacities.
These preclinical studies are currently being validated in humans, with several large, ongoing clinical trials of both BCG vaccination given for the first time and following revaccination. Using three animal models of SARS-CoV-2 infection, our results strongly argue for the vaccination of all individuals enrolled in BCG clinical trials with currently available, SARS-CoV-2-specific effective vaccines. Our observations are also supported by published data on the efficacy of BCG vaccination against COVID-19 in human cohorts (Pépin et al., 2021; Hensel et al., 2020). Importantly, it has been shown that BCG vaccination prior to influenza vaccination enhances the antigen (HA)-specific antibody responses against the 2009 pandemic influenza virus (Leentjens et al., 2015). Thus, the potential of BCG vaccination in augmenting the immunogenicity or prolonging the protective effects of a subsequent vaccination can be a novel approach for improving vaccination strategies. Considering dysregulated immunity is the major cause of morbidity and mortality during SARS-CoV-2 and variants of SARS-CoV-2 infection, understanding the pathogenesis of COVID-19 is essential for developing novel host-targeted therapies.
Limitations of the study: in this study, we show that the protection of BCG to pulmonary viral pathogens is mainly determined by pathogenesis of infections. In contrast to IAV infection, the distinct tropism of SARS-CoV-2 to infect pulmonary endothelial cells can be a major cause of lung hemorrhage and dissemination of the virus to other organs including BM. Given our recent studies showing that a pulmonary pathogen (M. tuberculosis) can access the BM to reprogram hematopoietic stem and progenitor cells (HSPCs) and prevent trained immunity (Khan et al., 2020), we hypothesize that the presence of SARS-CoV-2 in the BM can potentially erase the BCG-epigenetic imprinting of HSPCs. However, we have not performed either transcriptomic or epigenomic experiments to support this hypothesis. Thus, further epigenomic/transcriptomic investigation is required to precisely test this plausible hypothesis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Immunofluorescence primary antibodies against N protein | Genetex | GTX135361; RRID:AB_2887484 |
| Immunofluorescence primary antibodies against CD45 | abcam | ab10558; RRID:AB_442810 |
| Immunofluorescence secondary antibody | Jackson ImmunoResearch | 715-606-151; RRID:AB_2340866 |
| Immunofluorescence secondary antibody | Jackson ImmunoResearch | 711-166-152; RRID:AB_2313568 |
| DApI | Sigma | Cat# D9542 |
| Mouse anti-CD11b-Pacific blue (clone M1/70) | ThermoFisher | RRID:AB_10372795 |
| Mouse anti-CD11c-PE-Cy7 (clone N418) | BD Bioscience | RRID:AB_2870343 |
| Mouse anti-Siglec-F-PE-CF594 (clone E50–2440) | BD Biosciences | RRID:AB_2687994 |
| Mouse anti-F4/80-APC (clone BM8) | ThermoFisher | RRID:AB_469452 |
| Mouse anti-Ly6C-FITC (clone HK1.4) | BD Bioscience | RRID:AB_394628 |
| Mouse anti-Ly6G-PerCP-efluor710 (clone 1A8) | ThermoFisher | RRID:AB_2573893 |
| Universal Innovex FcReceptor Blocker | Fisher | Cat# 50-486-803 |
| Viability dye eF506 | ThermoFisher | Cat# 65-0866-14 |
| Hamster anti-Pan-leukocytes | Washington University Monoclonal Antibody Center | HAT13A |
| Hamster anti-T cells | Washington University Monoclonal Antibody Center | HAT19A |
| Hamster anti-B cells | Washington University Monoclonal Antibody Center | HASA7A |
| Hamster anti-CD18 | Washington University Monoclonal Antibody Center | BAQ30A |
| Zenon Kit AF647 | Invitrogen ThermoFisher Scientific | Z25208; RRID:AB_2736961 |
| Zenon Kit AF488 | Invitrogen ThermoFisher Scientific | Z25102; RRID:AB_2736942 |
| Zenon Kit PE | Invitrogen ThermoFisher Scientific | Z25055; RRID:AB_2736983 |
| Zenon Kit AF700 | Invitrogen ThermoFisher Scientific | Z25011; RRID:AB_2736964 |
| anti-CD11b-PE-Cy7 | Novusbio | Cat# NB110-89474PECY7 |
| anti-CD8b-BV650 (clone 341) | BD | Cat# 742918; RRID: AB_2741146 |
| anti-CD4-BUV395 (clone GK1.5) | BD | Cat# 563790; RRID: AB_2738426 |
| anti-MHCII-BV421 (clone 14-4-4S) | BD | Cat# 744830; RRID: AB_2742516 |
| Bacterial and virus strains | ||
| Mycobacterium bovis BCG-TICE (TMC 1028) | ATCC | ATCC Number: 35743 |
| SARS-CoV-2/SB2 | Dr. Samira Mubareka, University of Toronto, ON, Canada | N/A |
| SARS-CoV-2/RIM-1 | This study | GenBank accession number MW599736 |
| Influenza A/Puerto Rico/8/34 (H1N1, PR8) | Dr. Jonathan A. McCullers (St. Jude Children Research Hospital) | N/A |
| Influenza A virus isolate A/H3N2 A/Hong-Kong/1/68 | Dr. Salman Qureshi, McGill University, Montreal | N/A |
| Biological samples | ||
| LPS from E.coli O26:B6 | Sigma | Cat# L2654 |
| Chemicals, peptides, and recombinant proteins | ||
| Middlebrook 7H9 broth | Fisher Scientific | Cat# 271310 |
| Glycerol | Wisent | Cat# 800-040-LL |
| Tween80 | Fisher | Cat# 338-500; CAS Number 9005-65-6 |
| BSA for ADC | Wisent | Cat# 800-195-EG |
| Dextrose for ADC | Fisher Scientific | Cat# D16-10; CAS Number 50-99-7 |
| NaCl for ADC | Fisher Scientific | Cat# S6713 |
| PBS | Gibco | Cat# 10-010-049 |
| DMEM | Gibco | Cat# 11965-092 |
| FBS | Gibco | Cat# 26140 |
| Pen/Strep | Gibco | Cat# 15240062 |
| 10% Formalin | Sigma | HT501128-4L |
| Crystal violet (0.1% w/v in 10% EtOH) | Sigma | Cat# C0775; CAS Number 548-62-9 |
| QIAzol Lysis Reagent | Qiagen | Cat# 79306 |
| Chloroform | Sigma | Cat# 528730; CAS Number 67-66-3 |
| 1X EvaGreen qPCR mastermix | ABM | Cat# ABMMastermix-S |
| SYBR Select MasterMix | Applied Biosystems | Cat# 4472908 |
| 10mM Tris Base | Sigma | Cat# T1503-250G |
| 1mM EDTA | Sigma | Cat# E7889-100ML |
| Tween 20 | Sigma | Cat# P7949-500ML |
| DMSO | Sigma | Cat# D2438 |
| TritonX | Sigma | Cat# T9284-500ML |
| donkey serum | Sigma | Cat# D9663-10ML |
| Bovine serum albumin | Cedarlane | Cat# AK8917-0500 |
| DAKO | Cedarlane | Cat# S302380-2 |
| DNAse | Stemcell | Cat# 7469 |
| RPMI | Gibco | Cat# 11875119 |
| HEPES | Wisent | Cat# 330-050-EL |
| Nonessential amino acids | Wisent | Cat# 321-010-EL |
| human Serum | Sigma | Cat# H4522-100ML |
| TPCK | Fisher | Cat# 20233 |
| Collagenase IV | Sigma | Cat# C5138 |
| Paraformaldehyde 16% | Thermo Fisher | Cat# 28908 |
| Critical commercial assays | ||
| Viral DNA/RNA kit | N/A | |
| QuantiTect Reverse Transcription Kit | ABM | Cat# 205311 |
| Genezol TriRNA Pure Kit | Geneaid | Cat# GZX050 |
| LunaScript RT SuperMix Kit | NEB | Cat# E3010L |
| 2.5 HD Brown Detection Kit | Advanced Cell Diagnostics | Cat# 322300 |
| Hamster IFN-β ELISA | MyBioSource | Cat# MBS014227 |
| Hamster IL-6 ELISA | MyBioSource | Cat# MBS7606648 |
| Deposited data | ||
| SARS-CoV-2/RIM-1 (formerly known as SARS-CoV-2/cp13.32, Lineage B.1.147) | GenBank | GenBank accession number MW599736 |
| Experimental models: Cell lines | ||
| VeroE6 cells | ATCC | ATCC Number CRL-1586 |
| MDCK cells | ATCC | ATCC Number CRL-2936 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | JAX | Cat# 000664 |
| B6.Cg-Tg(K18-ACE2)2Prlmn/J mice | JAX | Cat# 034860 |
| Syrian Golden Hamsters (Mesocricetus auratus) | Envigo | Cat# 089 |
| Roborovski Hamsters (Phodopus roborovskii) | Breeder for Pet Shop Trade | N/A |
| Human PBMC | 300BCG cohort between April 2017 and June 2018 at Radboud umc, Nijmegen, The Netherlands | N/A |
| Oligonucleotides | ||
| mouse hprt Fwd: 5′-aggacctctcgaagtgttgg-3′ | IdT | N/A |
| mouse hprt Rev: 5′-aacttgcgctcatcttaggc-3′ | IdT | N/A |
| mouse gapdh Fwd: 5′-ctgaggaccaggttgtctcc-3′ | IdT | N/A |
| mouse gapdh Rev: 5′-ctccttggaggccatgtagg-3′ | IdT | N/A |
| hamster gapdh Fwd: 5′-aaggccaacaccatcttcc-3′ | IdT | N/A |
| hamster gapdh Rev: 5′-gaaggtgtggagatgatgacc-3′ | IdT | N/A |
| viral SARS-CoV-2-RBD Fwd: 5′-CAATGGTTTAACAGGCACAGG-3′ | IdT | N/A |
| viral SARS-CoV-2-RBD Rev: 5′-CTCAAGTGTCTGTGGATCACG-3′ | IdT | N/A |
| viral SARS-CoV-2-UpE Fwd: 5′-attgttgatgagcctgaag-3′ | IdT | N/A |
| viral SARS-CoV-2-UpE Rev: 5′-ttcgtactcatcagcttg-3′ | IdT | N/A |
| IAV-PR8 NS1-Fwd: 5′-AGAAAGTGGVAGGCCCTCTTTGTA-3′ | IdT | N/A |
| IAV-PR8 NS1-Rev: 5′-GGGCACGGTGAGCGTGAACA-3′ | IdT | N/A |
| viral SARS-CoV-2-N2 primers were purchased from IdT as commercial 2019-nCoV_N2 Combined Primer/Probe Mix (Cat #RV202002 and #RV202016) as per In vitro Diagnostic Guideline by CDC (Catalog # 2019-nCoVEUA-01, Date 3/30/2020, CDC-006-00019, Revision 03) | IdT | N/A |
| human Il1b-Fwd: TACATCAGCACCTCTCAAGCA | IdT | N/A |
| human Il1b-Rev: CCACATTCAGCACAGGACTCT | IdT | N/A |
| human Il8-Fwd: CTGCGCCAACACAGAAATTAT | IdT | N/A |
| human Il8-Rev: CATCTGGCAACCCTACAACAG | IdT | N/A |
| human Ifnb-Fwd: ATTGTCAGTGTCAGAAGCTCC | IdT | N/A |
| human Ifnb-Rev: CTCAGGGATGTCAAAGTTCCTC | IdT | N/A |
| human Ccl2-Fwd: TCCTCGCAACTTTGTGGTAG | IdT | N/A |
| human Ccl2-Rev: TTCAGTTCCAGGTCATACACG | IdT | N/A |
| human Tnfa-Fwd: AGTGAAGTGCTGGCAACCAC | IdT | N/A |
| human Tnfa-Rev: GAGGAAGGCCTAAGGTCCAC | IdT | N/A |
| human Ifna-Fwd: TGGCTGTGAAGAAATACTTCCG | IdT | N/A |
| human Ifna-Rev: TGTTTTCATGTTGGACCAGATG | IdT | N/A |
| human Gapdh-Fwd: GGCTGTTGTCATACTTCTCATGG | IdT | N/A |
| human Gapdh-Rev: GGAGCGAGATCCCTCCAAAAT | IdT | N/A |
| Software and algorithms | ||
| GraphPad Prism v9.0.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v10 | BD | https://www.flowjo.com/solutions/flowjo/downloads |
| Aperio ImageScope | Leica | https://www.leicabiosystems.com/digital-pathology/manage/aperio-imagescope/ |
| ZEISS ZEN 2.3 Pro | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| ImageJ | NIH | https://imagej.net/software/fiji/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Maziar Divangahi (maziar.divangahi@mcgill.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. Additional Supplemental Items (Table S1) are available from Mendeley Data at https://doi.org/10.17632/v9nmjfz94b.1. Whole Genome Sequencing analysis of SARS-CoV-2/RIM-1 has been posted under the GenBank accession number MW599736. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Animals
Six-to 8-week old female C57BL/6J and B6.Cg-Tg(K18-ACE2)2Prlmn/J mice were purchased from Jackson Laboratories or bred at the RI-MUHC, Montreal, QC, Canada. Three-to four-week-old male Syrian Golden Hamsters (Mesocricetus auratus) were purchased from Envigo. Three-to four-week old female and male Roborovski Hamsters (Phodopus roborovskii) were purchased from a breeder for the pet shop trade. All animals were clinically examined and tested for pathogens upon arrival.
All animal studies were conducted in accordance with the guidelines of and approved by the Animal Research Ethics Board of McGill University (project ID: 5860). Animals were housed under SPF conditions with ad libitum access to food and water, and were randomly assigned to experimental groups. Experiments were performed using age- and sex-matched animals.
Human subjects
Healthy male and female adult volunteers of Western European ancestry (18-19 years old) were included in the 300BCG cohort between April 2017 and June 2018 at Radboud umc, Nijmegen, The Netherlands. Written informed consent was obtained. Blood was collected before and 3 months after BCG administration (0.1ml BCG-Bulgaria, InterVax, intradermal). PBMC were isolated using Ficoll Gradient Centrifugation and frozen in FBS +20% DMSO.
Microbe strains
BCG-Tice
BCG-TICE was grown in 7H9 broth (BD) supplemented with 0.2% glycerol (Wisent), 0.05% Tween80 (Fisher), and 10% albumin-dextrose-catalase (ADC) under constant shaking at 37°C. For vaccination, bacteria in log growing phase (OD 0.4–0.9) were centrifuged (4000 RPM, 10 min) and resuspended in sterile PBS. Single cell suspensions were obtained by passing the bacteria 10-15 times through a 22G needle (Terumo). Animals were vaccinated with 1 × 106 CFU BCG-TICE in 100μl sterile PBS subcutaneously or intravenously. Infection experiments were carried out at 4-6 weeks post vaccination or as indicated.
SARS-CoV-2
SARS-CoV-2/SB2 (lineage B.4) was kindly provided by Dr. Samira Mubareka, University of Toronto, ON, Canada. SARS-CoV-2/RIM-1 was isolated from a patient at MUHC, Montreal, QC (formerly known as SARS-CoV-2/cp13.32, Lineage B.1.147 (Murall et al., 2021), Table S2). Whole Genome Sequencing was performed and posted under the GenBank accession number MW599736. SARS-CoV-2 were propagated in VeroE6 cells. Mice were intranasally or intratracheally infected as previously described (Kaufmann et al., 2018) with 4000 TCID50 SARS-CoV-2/SB2 (sublethal infection) or 1 × 104 TCID50 SARS-CoV-2/SB2 (lethal infection). Syrian Golden Hamster were intranasally infected with 1 × 105 PFU SARS-CoV-2/SB2 or SARS-CoV-2/RIM-1. Roborovski Hamsters were infected with SARS-CoV-2/RIM-1 at a sublethal dose of 1.4 × 104 PFU and a lethal dose of 1 × 105 PFU.
Influenza A virus
Influenza A/Puerto Rico/8/34 (H1N1, PR8) virus was kindly provided by Dr. Jonathan A. McCullers (St. Jude Children Research Hospital). Influenza A virus isolate A/H3N2 A/Hong-Kong/1/68 was kindly provided by Dr. Salman Qureshi, McGill University, Montreal. IAV was propagated in and isolated from MDCK cells and titrated using standard plaque assay in MDCK cells (Gaush and Smith, 1968). Mice were challenged intranasally (in 25μL PBS) with IAV at a sublethal dose of 50 PFU or a lethal dose of 90 PFU of PR8. Golden Hamsters were intranasally infected with 1 × 105 PFU IAV-H3N2.
Method details
TCID50 analysis
Viral titers in cell cultures and homogenized organs were quantified in triplicates using TCID50/ml assay. To this end, VeroE6 were infected for 1h at 37°C in DMEM (Gibco) containing 10-fold serial dilutions of the virus (0.1mL final volume). DMEM + virus was removed and 0.5mL DMEM+2% FBS (Gibco)+1% Pen/Strep (Wisent) was added. Plates were incubated for 3 days and then fixed with 10% Formalin and stained with Crystal violet (0.1% w/v in 10% EtOH). TCID50/mL was calculated using the method by Reed and Muench.
Isolation of nucleic acids and reverse transcription (RT)
TRIzol-chloroform method (Genezol TriRNA Pure Kit) was used for viral and total RNA isolation from VeroE6 cell culture supernatants or mouse and hamster bone marrow and lung tissues, respectively. cDNA samples were prepared with QuantiTect Reverse Transcription Kit (ABM), according to the manufacturer’s instructions. In order to assess the TCID50/ml values in each sample, standards using VeroE6 cells infected with a known amount of viral SARS-CoV-2 were prepared and serially diluted (Brandolini et al., 2021), and equivalent amounts of RNA from each sample were reverse-transcribed.
From human MDMs, RNA was extracted using Trizol (Qiagen) and Genezol TriRNA Pure Kit (Geneaid) according to manufacturer’s instructions. Five hundred ng of RNA was reverse transcribed using the LunaScript RT SuperMix Kit (NEB), as directed by the manufacturer.
qPCR analysis
Viral RNA was quantified using qPCR (CFX96 Touch Real-Time PCR Detection System, Bio-Rad). cDNA was amplified using 1X EvaGreen qPCR mastermix (ABM) and the cycling conditions were as follows: 10 min at 95°C for enzyme activation, followed by 40 cycles of 15s at 95°C and 1 min at 60°C. The TCID50/ml in each sample was calculated using a standard curve from a serial dilution of the viral SARS-CoV-2 cDNA (Brandolini et al., 2021). In addition, relative expression of the gene of interest was measured from individual cDNA samples and normalized to the reference housekeeping genes Hprt or Gapdh, and expressed as simple power calculations (2ˆ-dCT). The TCID50/ml values were normalized either by the total RNA amount or tissue weight.
Primer Sequences: Mouse hprt, mouse gapdh, hamster gapdh, and viral SARS-CoV-2 primers (RBD, N2, UpE) were specifically designed for the target genes and synthesize by IdT: mouse hprt Fwd: 5′-aggacctctcgaagtgttgg-3′, and mouse hprt Rev: 5′-aacttgcgctcatcttaggc-3′; mouse gapdh Fwd: 5′-ctgaggaccaggttgtctcc-3′, and mouse gapdh Rev: 5′-ctccttggaggccatgtagg-3′; hamster gapdh Fwd: 5′-aaggccaacaccatcttcc-3′, and hamster gapdh Rev: 5′-gaaggtgtggagatgatgacc-3′; viral SARS-CoV-2-RBD Fwd: 5′-CAATGGTTTAACAGGCACAGG-3′, viral SARS-CoV-2-RBD Rev: 5′-CTCAAGTGTCTGTGGATCACG-3′; viral SARS-CoV-2-UpE Fwd: 5′-attgttgatgagcctgaag-3′, and viral SARS-CoV-2-UpE Rev: 5′-ttcgtactcatcagcttg-3′; IAV-PR8 NS1-Fwd: 5′-AGAAAGTGGVAGGCCCTCTTTGTA-3′, and IAV-PR8 NS1-Rev: 5′-GGGCACGGTGAGCGTGAACA-3′. The viral SARS-CoV-2-N2 primers were purchased from IdT as commercial 2019-nCoV_N2 Combined Primer/Probe Mix (Cat #RV202002 and #RV202016) as per In vitro Diagnostic Guideline by CDC (Catalog # 2019-nCoVEUA-01, Date 3/30/2020, CDC-006-00019, Revision 03).
Human MDM cDNA was generated by qPCR using SYBR Select MasterMix (Applied Biosystems) and the following primers: GAPDH, Il1b, Il8, Ccl2, Tnfa, Ifna, Ifnb. Cq values obtained on CFX96 PCR System (Biorad) were analyzed using the formula 2-ΔCq formula normalizing target gene expression to GAPDH. Primer sequences: Il1b-F: TACATCAGCACCTCTCAAGCA. Il1b-R: CCACATTCAGCACAGGACTCT. Il8-F: CTGCGCCAACACAGAAATTAT. Il8-R: CATCTGGCAACCCTACAACAG. Ifnb-F: ATTGTCAGTGTCAGAAGCTCC. Ifnb-R: CTCAGGGATGTCAAAGTTCCTC. Ccl2-F: TCCTCGCAACTTTGTGGTAG. Ccl2-R: TTCAGTTCCAGGTCATACACG. Tnfa-F: AGTGAAGTGCTGGCAACCAC. Tnfa-R: GAGGAAGGCCTAAGGTCCAC. Ifna-F: TGGCTGTGAAGAAATACTTCCG. Ifna-R: TGTTTTCATGTTGGACCAGATG. Gapdh-F: GGCTGTTGTCATACTTCTCATGG. Gapdh-R: GGAGCGAGATCCCTCCAAAAT.
Histopathological analyses
Lung samples were incubated in 10% Formalin for 48h before embedding in paraffin and sectioning. H&E staining for pathohistological analysis was performed by the Histopathology Core of RI-MUHC, Montreal, QC, Canada. Slides were imaged using Aperio ImageScope and scored by a pulmonary pathologist who was blinded to the experimental groups. The scoring system for both IAV and SARS-CoV-2 was based on previous publications with a maximal score of 3 (Coulombe et al., 2014; Downey et al., 2017; Tzelepis et al., 2018).
Immunofluorescence
For Immunofluorescence, formalin-fixed, paraffin-embedded lungs were sectioned at 4μm and deparaffinized and rehydrated in decreasing xylene and increasing alcohol gradients, respectively. Slides were immersed in Tris-EDTA-Tween (10mM Tris Base, 1mM EDTA, 0.05% Tween 20, pH 9) and antigen retrieval was achieved using a pressure cooker (20min, 98°C, 50kPa). Slides were cooled down at room temperature for 20 min, immersed in tap water for 10 min and then transferred in PBS. Sections were pre-treated with 10% DMSO +0.5% TritonX in PBS for 5 min. Following three washes with PBS, sections were blocked with 10% donkey serum +1% Bovine serum albumin (BSA) in PBS for 30 min at room temperature (RT). Primary antibodies against N protein (GTX135361, Genetex) or CD45 (ab10558, abcam) were incubated overnight at 4°C (1:100, diluted in 5% donkey serum +1% BSA). Following washes, secondary antibodies (1:400, 715-606-151, 711-166-152, Jackson ImmunoResearch) along with DAPI (1:1000) were incubated for 2 h at RT. Following washes, slides were mounted with DAKO (Agilent Technologies) and coverslipped. Samples were imaged using an Axio Imager 2 (Zeiss) microscope at 20x magnification, with identical imaging settings for each marker or interest (i.e., N protein, CD45). For N protein, in every lung section, images from non-overlapping fields of view depicting all areas with detectable signal were collected. For samples that had no detectable N protein, images from non-overlapping fields of view covering 80-90% of the lung surface were collected. For CD45, images from non-overlapping fields of view covering 80-90% of lung surface were acquired. Acquired images were analysed using ImageJ (NIH). Similar thresholding was applied across all images, depending on the marker of interest. For DAPI, the option of auto-threshold was selected. The % area for each marker was calculated as the percent of marker area/DAPI area.
SARS-CoV-2 RNA in situ hybridization (ISH)
RNA ISH was performed using RNA scope® technology (Advanced Cell Diagnostics [ACD], Newark, CA, USA). The assay was performed using the 2.5 HD Brown Detection Kit in a HybEZ™ oven (ACD) according to the manufacturer’s instructions. Briefly, four micron sections of formalin-fixed paraffin-embedded tissue mounted on charged slides were deparaffinized in xylene and dehydrated in an ethanol series. Sections were subjected to antigen retrieval in Target Retrieval Solution, dehydration and Protease Plus treatment. Sections were then incubated with two set of probes (V-nCoV-N [nucleocapsid] and V-nCoV2019-S [spike], ACD) for 2h at 40°C followed by six steps of amplification using a specific set of ready-to-use solutions (AMP1-6). The signal was detected using DAB solution and sections were counterstained with Mayer’s Hematoxylin, blued with ammonium hydroxide in water before mounting and visualisation on a brightfield microscope.
Flowcytometric analysis
Single cell suspensions of mouse blood, and Syrian Golden hamster blood, BAL, lung, and spleen were RBC-lysed and stained with the following antibodies:
Mouse: anti-CD11b-Pacific blue (clone M1/70, eBioscience), anti-CD11c-PE-Cy7 (clone N418, BD Bioscience), anti-Siglec-F-PE-CF594 (clone E50–2440, BD Biosciences), anti-F4/80-APC (clone BM8, eBioscience), anti-Ly6C-FITC (clone HK1.4, BD Bioscience), anti-Ly6G-PerCP-efluor710 (clone 1A8, eBioscience).
Hamster: Hamster antibodies were obtained from Washington University Monoclonal Antibody Center and freshly conjugated using Zenon Kits (Invitrogen ThermoFisher Scientific). Commercial antibodies were tested for hamster cross-specificity. Universal Innovex FcReceptor Blocker (Fisher) was used to prevent unspecific binding. Viability dye eF506 (eBioscience), anti-Pan-leukocytes-AF647 (HAT13A + Z25208), anti-CD11b-PE-Cy7 (Novusbio, NB110-89474), anti-T cells-AF488 (HAT19A + Z25102), anti-B cells-PE (HASA7A + Z25055), anti-CD8b-BV650 (BD, clone 341), anti-CD4-BUV395 (BD, clone GK1.5), anti-MHCII-BV421 (BD, clone 14-4-4S), anti-CD18-AF700 (BAQ30A + Z25011).
Samples were fixed with 1% PFA and acquired on BD Fortessa-X20 within 3 days. Data were analyzed using FlowJo v10.
Human PBMC stimulation and cytokine measurement
PBMC from young healthy volunteers (18-19 years of age) from the 300BCG cohort were thawn with DNAse (Stemcell). 1.1 × 106 cells were seeded per well in 96-well flat bottom plates in serum-free RPMI (Gibco) + 1% HEPES +1% Pen/Strep +1% NEAA (all Wisent). Cells adhered for 2h at 37°C and were subsequently washed with PBS. Cells were incubated in RPMI +1% HEPES +1% Pen/Strep +1% NEAA +10% human Serum (Stemcell) for 5 days, with 50% media exchange every 2nd day. Assuming that 6% of initially seeded cells are adherent monocytes/macrophages on day 5, cells were infected with IAV/H3N2 A/Hong-Kong/1/68 or SARS-CoV-2/RIM-1 at MOI 1 PFU for 6h in DMEM +0.1% BSA. Media only and LPS (100ng/ml) stimulations served as controls. After 6h, the stimulation media was replaced with RPMI +1% Hepes +1% Pen/Strep +1% NEAA +0.1% BSA, 2μg/ml TPCK was added in IAV infection wells. At 24h post infection, cells were harvested with 200μl Trizol (Qiagen) for RNA extraction.
Hamster ELISA
Hamster IFN-β and IL-6 ELISA kits were purchased from MyBioSource and used according to the manufacturer’s protocol.
Quantification and statistical analysis
Statistics were calculated using GraphPad Prism v9.0.0 and are displayed as mean +/- SEM. N numbers indicate biological replicates per group. Student’s t-test was used for pairwise comparisons, ONE- and Two-way ANOVA for multiple group comparisons, as appropriate. p values <0.05 ∗, ≤0.01 ∗∗, ≤0.001 ∗∗∗, ≤0.0001 ∗∗∗∗.
Acknowledgments
The authors are grateful for the permission to use RNAscope images from a COVID-19 patient in this report. The authors acknowledge the support by the Histopathology Platforms at RI-MUHC, UHN Toronto, and IUCPQ Quebec City, as well as the support by the Animal Research Division at RI-MUHC. The authors thank Anisha Hundal for her assistance with ImageJ analysis for CD45 quantification. This work was supported by the Canadian Institutes of Health Research (CIHR) Project Grant (MM1-174910) to M.D. and I.L.K. as well as McGill Interdisciplinary Initiative in Infection and Immunity (MI4) Emergency COVID-19 Research Funding awards to M.D. M.D. holds a Fonds de Recherche du Québec - Santé (FRQS) Award and the Strauss Chair in Respiratory Diseases. I.L.K. holds a Canada Research Chair in Barrier Immunity. C.S.R. holds a Peter Munk Chair in Aortic Disease Research. P.J. holds a FRQS Award. E.K. and N.K. were funded by postdoctoral Fellowships from FRQS and CIHR. A.U. is supported by a postdoctoral fellowship from the University of Toronto's Medicine by Design initiative. All models and the graphical abstract have been generated using BioRender.
Author contributions
E.K., N.K., K.T., A.U., E.P., G.F., A.L., P.M., F.M., A.A., D.K.A., M.S., P.J., and M.D. performed experiments and analyzed data. S.J.C.F.M., P.D., K.M., A.B., and S.M. contributed cells, virus, reagents, and expertise. E.K., N.K., D.C.V., I.L.K., C.S.R., M.A.B., M.G.N., P.J., and M.D. designed experiments and discussed results and strategy. E.K., N.K., P.J., and M.D. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: February 21 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110502.
Supplemental information
and S2
References
- Aaby P., Roth A., Ravn H., Napirna B.M., Rodrigues A., Lisse I.M., Stensballe L., Diness B.R., Lausch K.R., Lund N., et al. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? J. Infect Dis. 2011;204:245–252. doi: 10.1093/infdis/jir240. [DOI] [PubMed] [Google Scholar]
- Al-Qaaneh A.M., Alshammari T., Aldahhan R., Aldossary H., Alkhalifah Z.A., Borgio J.F. Genome composition and genetic characterization of SARS-CoV-2. Saudi J. Biol. Sci. 2021;28:1978–1989. doi: 10.1016/j.sjbs.2020.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B., et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100. doi: 10.1016/j.chom.2017.12.010. e5. [DOI] [PubMed] [Google Scholar]
- Banerjee A., El-Sayes N., Budylowski P., Jacob R.A., Richard D., Maan H., Aguiar J.A., Demian W.L., Baid K., D'agostino M.R., et al. Experimental and natural evidence of SARS-CoV-2-infection-induced activation of type I interferon responses. iScience. 2021;24:102477. doi: 10.1016/j.isci.2021.102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Nasir J.A., Budylowski P., Yip L., Aftanas P., Christie N., Ghalami A., Baid K., Raphenya A.R., Hirota J.A., et al. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerg. Infect Dis. 2020;26:2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura A., Vecchié A., Dagna L., Martinod K., Dixon D.L., Van Tassell B.W., Dentali F., Montecucco F., Massberg S., Levi M., Abbate A. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 2021;21:319–329. doi: 10.1038/s41577-021-00536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandolini M., Taddei F., Marino M.M., Grumiro L., Scalcione A., Turba M.E., Gentilini F., Fantini M., Zannoli S., Dirani G., Sambri V. Correlating qRT-PCR, dPCR and viral titration for the identification and quantification of SARS-CoV-2: a new approach for infection management. Viruses. 2021;13:1022. doi: 10.3390/v13061022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirovic B., De Bree L.C.J., Groh L., Blok B.A., Chan J., Van Der Velden W., Bremmers M.E.J., Van Crevel R., Händler K., Picelli S., et al. BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe. 2020;28:322–334. doi: 10.1016/j.chom.2020.05.014. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F., Jaworska J., Verway M., Tzelepis F., Massoud A., Gillard J., Wong G., Kobinger G., Xing Z., Couture C., et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity. 2014;40:554–568. doi: 10.1016/j.immuni.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Deinhardt-Emmer S., Wittschieber D., Sanft J., Kleemann S., Elschner S., Haupt K.F., Vau V., Häring C., Rödel J., Henke A., et al. Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. Elife. 2021;10:e60361. doi: 10.7554/eLife.60361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristanziano V., Meyer-Schwickerath C., Eberhardt K.A., Rybniker J., Heger E., Knops E., Hallek M., Klein F., Holtick U., Jung N. Detection of SARS-CoV-2 viremia before onset of COVID-19 symptoms in an allo-transplanted patient with acute leukemia. Bone Marrow Transpl. 2020;56:716–719. doi: 10.1038/s41409-020-01059-y. [DOI] [PubMed] [Google Scholar]
- Divangahi M., Aaby P., Khader S.A., Barreiro L.B., Bekkering S., Chavakis T., Van Crevel R., Curtis N., Dinardo A.R., Dominguez-Andres J., et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat. Immunol. 2021;22:2–6. doi: 10.1038/s41590-020-00845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey J., Pernet E., Coulombe F., Allard B., Meunier I., Jaworska J., Qureshi S., Vinh D.C., Martin J.G., Joubert P., Divangahi M. RIPK3 interacts with MAVS to regulate type I IFN-mediated immunity to Influenza A virus infection. Plos Pathog. 2017;13:e1006326. doi: 10.1371/journal.ppat.1006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaush C.R., Smith T.F. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl. Microbiol. 1968;16:588–594. doi: 10.1128/am.16.4.588-594.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Tsilika M., Moorlag S., Antonakos N., Kotsaki A., Domínguez-Andrés J., Kyriazopoulou E., Gkavogianni T., Adami M.E., Damoraki G., et al. Activate: randomized clinical trial of BCG vaccination against infection in the elderly. Cell. 2020;183:315–323. doi: 10.1016/j.cell.2020.08.051. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson D., Raju S., Wu R., Ching C., Veitch S., Rathnakumar K., Boudreau E., Howe K.L., Fish J.E. Overcoming barriers: the endothelium as a linchpin of coronavirus disease 2019 pathogenesis? Arterioscler Thromb. Vasc. Biol. 2020;40:1818–1829. doi: 10.1161/ATVBAHA.120.314558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri L.P., North C.M., Shih A.R., Israel R.A., Maley J.H., Villalba J.A., Vinarsky V., Rubin J., Okin D.A., Sclafani A., et al. Lung histopathology in coronavirus disease 2019 as compared with severe acute respiratory sydrome and H1N1 influenza: a systematic review. Chest. 2021;159:73–84. doi: 10.1016/j.chest.2020.09.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel J., Mcandrews K.M., Mcgrail D.J., Dowlatshahi D.P., Lebleu V.S., Kalluri R. Protection against SARS-CoV-2 by BCG vaccination is not supported by epidemiological analyses. Sci. Rep. 2020;10:18377. doi: 10.1038/s41598-020-75491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilligan K.L., Namasivayam S., Clancy C.S., O'mard D., Oland S.D., Robertson S.J., Baker P.J., Castro E., Garza N.L., Lafont B.A.P., et al. Intravenous administration of BCG protects mice against lethal SARS-CoV-2 challenge. J. Exp. Med. 2022;219:e20211862. doi: 10.1084/jem.20211862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippmann G., Wekkeli M., Rosenkranz A.R., Jarisch R., Götz M. [Nonspecific immune stimulation with BCG in Herpes simplex recidivans. Follow-up 5 to 10 years after BCG vaccination] Wien Klin Wochenschr. 1992;104:200–204. [PubMed] [Google Scholar]
- Huerga Encabo H., Grey W., Garcia-Albornoz M., Wood H., Ulferts R., Aramburu I.V., Kulasekararaj A.G., Mufti G., Papayannopoulos V., Beale R., Bonnet D. Human erythroid progenitors are directly infected by SARS-CoV-2: implications for emerging erythropoiesis in severe COVID-19 patients. Stem Cell Rep. 2021;16:428–436. doi: 10.1016/j.stemcr.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K., Nakajima N., Ichiko Y., Sakai-Tagawa Y., Noda T., Hasegawa H., Kawaoka Y. Syrian hamster as an animal model for the study of human influenza virus infection. J. Virol. 2018;92 doi: 10.1128/JVI.01693-17. e01693–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonca L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C., et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172:176–190. doi: 10.1016/j.cell.2017.12.031. e19. [DOI] [PubMed] [Google Scholar]
- Khan N., Downey J., Sanz J., Kaufmann E., Blankenhaus B., Pacis A., Pernet E., Ahmed E., Cardoso S., Nijnik A., et al. M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell. 2020;183:752–770 e22. doi: 10.1016/j.cell.2020.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P., Rothan H.A., Natekar J.P., Stone S., Pathak H., Strate P.G., Arora K., Brinton M.A., Kumar M. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses. 2021;13:132. doi: 10.3390/v13010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leentjens J., Kox M., Stokman R., Gerretsen J., Diavatopoulos D.A., van Crevel R., Rimmelzwaan G.F., Pickkers P., Netea M.G. BCG Vaccination Enhances the Immunogenicity of Subsequent Influenza Vaccination in Healthy Volunteers: A Randomized, Placebo-Controlled Pilot Study. J Infect Dis. 2015;212 doi: 10.1093/infdis/jiv332. 1930–1938. [DOI] [PubMed] [Google Scholar]
- Lund N., Andersen A., Hansen A.S., Jepsen F.S., Barbosa A., Biering-Sorensen S., Rodrigues A., Ravn H., Aaby P., Benn C.S. The effect of oral polio vaccine at birth on infant mortality: a randomized trial. Clin. Infect Dis. 2015;61:1504–1511. doi: 10.1093/cid/civ617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Subramaniam R., Chen H., Smith A., Keshava S., Shams H. Boosting efferocytosis in alveolar space using BCG vaccine to protect host against influenza pneumonia. PLoS One. 2017;12:e0180143. doi: 10.1371/journal.pone.0180143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murall C.L., Fournier E., Galvez J.H., N'guessan A., Reiling S.J., Quirion P.O., Naderi S., Roy A.M., Chen S.H., Stretenowich P., et al. A small number of early introductions seeded widespread transmission of SARS-CoV-2 in Québec, Canada. Genome Med. 2021;13:169. doi: 10.1186/s13073-021-00986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namakula R., De Bree L.C.J., Th A.T., Netea M.G., Cose S., Hanevik K. Monocytes from neonates and adults have a similar capacity to adapt their cytokine production after previous exposure to BCG and beta-glucan. PLoS One. 2020;15:e0229287. doi: 10.1371/journal.pone.0229287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pépin J., Labbé A.C., Carignan A., Parent M.E., Yu J., Grenier C., Beauchemin S., De Wals P., Valiquette L., Rousseau M.C. Does BCG provide long-term protection against SARS-CoV-2 infection? A case-control study in Quebec, Canada. Vaccine. 2021;39:7300–7307. doi: 10.1016/j.vaccine.2021.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann A., Villumsen M., Sorup S., Haugaard L.K., Ravn H., Roth A., Baker J.L., Benn C.S., Aaby P. Vaccinations against smallpox and tuberculosis are associated with better long-term survival: a Danish case-cohort study 1971-2010. Int. J. Epidemiol. 2017;46:695–705. doi: 10.1093/ije/dyw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J.C., Ganguly R., Waldman R.H. Nonspecific protection of mice against influenza virus infection by local or systemic immunization with Bacille Calmette-Guérin. J. Infect Dis. 1977;136:171–175. doi: 10.1093/infdis/136.2.171. [DOI] [PubMed] [Google Scholar]
- Tribouley J., Tribouley-Duret J., Appriou M. [Effect of Bacillus Callmette Guerin (BCG) on the receptivity of nude mice to Schistosoma mansoni] C R. Seances Soc. Biol. Fil. 1978;172:902–904. [PubMed] [Google Scholar]
- Trimpert J., Vladimirova D., Dietert K., Abdelgawad A., Kunec D., Dökel S., Voss A., Gruber A.D., Bertzbach L.D., Osterrieder N. The Roborovski dwarf hamster is A highly susceptible model for a rapid and fatal course of SARS-CoV-2 infection. Cell Rep. 2020;33:108488. doi: 10.1016/j.celrep.2020.108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis F., Blagih J., Khan N., Gillard J., Mendonca L., Roy D.G., Ma E.H., Joubert P., Jones R.G., Divangahi M. Mitochondrial cyclophilin D regulates T cell metabolic responses and disease tolerance to tuberculosis. Sci. Immunol. 2018;3:eaar4135. doi: 10.1126/sciimmunol.aar4135. [DOI] [PubMed] [Google Scholar]
- Van 'T Wout J.W., Poell R., Van Furth R. The role of BCG/PPD-activated macrophages in resistance against systemic candidiasis in mice. Scand. J. Immunol. 1992;36:713–719. doi: 10.1111/j.1365-3083.1992.tb03132.x. [DOI] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk J., De Bree L.C.J., Graumans W., Stoter R., Van Gemert G.J., Van De Vegte-Bolmer M., Teelen K., Hermsen C.C., Arts R.J.W., Behet M.C., et al. Outcomes of controlled human malaria infection after BCG vaccination. Nat. Commun. 2019;10:874. doi: 10.1038/s41467-019-08659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and S2
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. Additional Supplemental Items (Table S1) are available from Mendeley Data at https://doi.org/10.17632/v9nmjfz94b.1. Whole Genome Sequencing analysis of SARS-CoV-2/RIM-1 has been posted under the GenBank accession number MW599736. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.