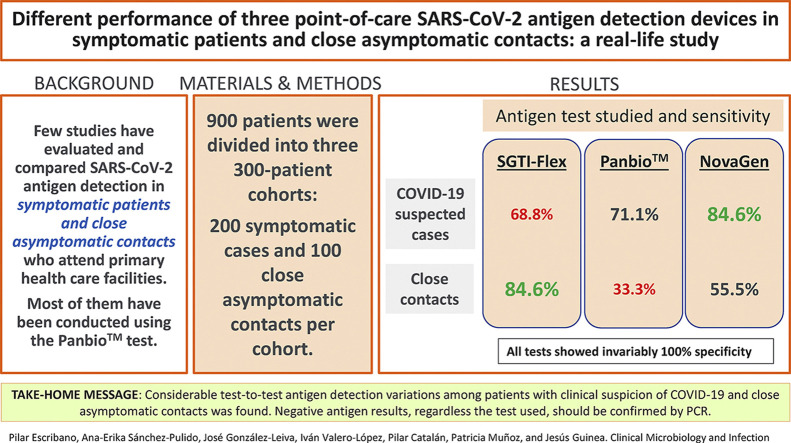
Abstract
Objectives
PCR on nasopharyngeal exudates, the cornerstone of the detection of SARS-CoV-2, is time-consuming and commonly unavailable at primary health care centres. Detection of viral nucleocapsid antigens using lateral flow point-of-care tests is helpful for the early triage of patients who attend health care facilities.
Methods
This was a prospective study carried out in clinically suspected cases and close asymptomatic contacts who attended a primary care centre (Madrid, Spain) for SARS-CoV-2 detection. Patients were divided into three 300-patient cohorts (n = 200 symptomatic cases and n = 100 close asymptomatic contacts per cohort). Three antigen detection tests (SGTI-Flex COVID-19 Ag, Panbio COVID-19 Ag Rapid Test Device, and GSD NovaGen SARS-CoV-2 Ag Rapid Test) were used and compared. Paired nasopharyngeal exudates were obtained, one swab for PCR and the other for antigen detection. Each antigen detection test was evaluated on one cohort.
Results
All tests showed invariably 100% specificity. Sensitivity was 68.9% (95% CI: 55.7–80; SGTI-Flex), 71.1% (95% CI: 55.6–83.6; Panbio), and 84.6% (95% CI: 72–93.1; NovaGen) in clinically suspected patients and 84.6% (95% CI: 54.5–98.1), 33.3% (95% CI: 11.8–61.6), and 55.6% (95% CI: 30.7–78.4) in close asymptomatic contacts, respectively. Sensitivity was systematically higher in samples yielding positive PCR results with Ct ≤ 20.
Discussion
We found considerable test-to-test antigen detection variations among patients with clinical suspicion of COVID-19 and close asymptomatic contacts. Negative antigen results, regardless of the test used, should be confirmed by PCR.
Keywords: Antigen detection, COVID-19, Close asymptomatic contact, Point of care, SARS-CoV-2
Graphical abstract
Introduction
The clinical presentation of COVID-19 ranges from asymptomatic or nonspecific mild illness to severe pneumonia with acute respiratory distress syndrome, progression to severe and fatal respiratory failure, and death [1]. COVID-19 spreads easily, and quick laboratory testing is key in helping halt the disease by recommending that infected patients self-isolate.
Detection of SARS-CoV-2 viral RNA in nasopharyngeal exudate samples is the cornerstone of COVID-19 diagnosis [2]. However, PCR is a time-consuming procedure commonly unavailable at primary health care centres; it alters laboratory routine by increasing the workload [3]. Detection of viral nucleocapsid antigens using immunoassays based on lateral flow point-of-care tests is helpful for the early triage of patients who attend health care facilities. The WHO supports antigen testing for the detection of SARS-CoV-2 in areas where PCR is not available, when PCR implies long turnaround times in reporting results, in the context of outbreak investigations or areas where prevalence is high, or for screening of asymptomatic positive contacts [4].
Few studies have evaluated and compared antigen detection in symptomatic patients and close asymptomatic contacts who attend hospital facilities and mostly have been conducted using the Panbio test [[5], [6], [7], [8], [9]]; furthermore, studies that have evaluated different tests in these two groups are insufficient [10,11].
It is thus unknown if the performance of antigen detection differs in patients with mild disease who attend primary care centres. Here, we report a real-life study in which we assessed the performance of three antigen detection tests in both patients with clinical suspicion of mild forms of COVID-19 and close asymptomatic contacts who attended a primary health care centre for SARS-CoV-2 detection.
Methods
Study patients and definitions
This was a prospective study carried out with patients who attended the Pavones primary care centre (Madrid, Spain) between 3 February and 14 April 2021, for either a COVID-19 diagnosis or screening of SARS-CoV-2 in close asymptomatic contacts. Patients with suspected COVID-19 were those with mild symptoms compatible with the disease. Close asymptomatic contacts were individuals for whom household or social contact with one (or more) individual(s) with positive SARS-CoV-2 PCR detection occurred in the absence of protection measures such as social distancing or wearing masks. A proven case of COVID-19 was a patient with mild compatible symptoms and a positive PCR detection of SARS-CoV-2. An asymptomatic carrier was a patient without any symptoms compatible with COVID-19 and with a positive SARS-CoV-2 PCR detection.
Consecutive patients who attended the health facility were interviewed by a study investigator and invited to participate if the onset of symptoms/close contact and sampling was within the prior 8 days. Patients who agreed to participate (n = 900) were interviewed by the aforementioned investigator and classified as clinically suspected COVID-19 cases (n = 600) or close asymptomatic contacts (n = 300).
Antigen and SARS-CoV-2 viral RNA detection in nasopharyngeal samples
Patients were divided into three 300-patient cohorts (200 symptomatic cases and 100 close asymptomatic contacts per cohort). The total number of patients was chosen based on the following criteria: sufficient patients in each cohort to avoid inconclusive results due to small sample size, and capacity of the research team to carry out the study in a scenario of high health care pressure. Each of the three antigen detection tests was evaluated in one cohort. Paired nasopharyngeal exudates were collected from the participating patients, one swab to detect the presence of viral SARS-CoV-2 RNA (TaqPath COVID-19-RT-PCR kit; Applied Biosystems, Pleasanton, CA, USA) and another for antigen detection. The nurses in charge of collecting samples performed antigen detection onsite, and results were interpreted following the manufacturer's instructions. PCR viral detection was carried out in the clinical microbiology department of Gregorio Marañón hospital; the latter samples were kept at 4°C upon sample reception (no later than 12 hours since sample collection).
The three evaluated point-of-care antigen detection tests were the SGTI-Flex COVID-19 Ag (Sugentech, Inc, Daejeon, Republic of Korea), the Panbio COVID-19 Ag Rapid Test Device (Abbott, Abbott Rapid Diagnostic, Jena GmbH, Jena, Germany), and the GSD NovaGen SARS-CoV-2 Ag Rapid Test (Eurofins-INGENASA, Daejeon). Sample processing for antigen detection was carried out.
Clinical information and data analysis
The following demographic and clinical data were collected: age, sex, presence of symptoms compatible with COVID-19 (fever >38°C, headache, cough, asthenia, myalgia, sore throat, runny nose, dyspnoea, anosmia, dysgeusia, abdominal pain, or diarrhoea), and number of days from the onset of symptoms (for symptomatic cases) or from close contact with a SARS-CoV-2 PCR positive case (for close asymptomatic contacts). The amplification cycle threshold (Ct) was obtained for samples with positive PCR results. Clinical data were collected in a pre-established protocol filled out during patient interview, and the staff of the health care centre carried out sample collection. The study did not interfere with routine clinical practice of the health centre, and PCR results were reported following the ordinary workflow. As part of the study, patients were informed about their antigen detection results and invited to follow the following recommendations: A positive result should be interpreted as a positive SARS-CoV-2 detection, and the patient was invited to comply with self-isolation without waiting for the PCR result; a negative detection should be interpreted with caution until confirmatory PCR result is reported.
Clinical categorical variables were described and compared using the χ2 or Fisher exact tests; continuous variables were compared using the nonparametric Mann-Whitney U test (for comparisons between two groups) and the Kruskal-Wallis test (for comparisons among three groups) (IBM SPSS Statistics for Windows, version 21.0; Armonk, NY, USA). The prevalence of individuals with SARS-CoV-2 (proportion of patients with positive PCR results), sensitivity, specificity, positive predictive value, and negative predictive value (95% CI) were calculated separately for each of the three antigen detection tests using PCR as the reference standard. Viral load analyzed is based on median Ct values, and differences were compared using the nonparametric Mann-Whitney U test; sensitivity was reassessed based on Ct results (Ct ≤ 20 or Ct > 20). Scatter plots were used to represent Ct values (Graph Pad Prism 5.02 statistical software; GraphPad, La Jolla, CA, USA).
Ethical considerations
This study was approved by the Ethics Committee of Hospital Gregorio Marañón (CEIm; study no. MICRO.HGUGM.2020-033). The research was performed in accordance with relevant guidelines/regulations. Informed consent was obtained from all participating patients.
Results
Comparisons of three SARS-CoV-2 antigen detection diagnostic tests in patients with clinical suspicion of infection
Of the 600 patients with symptoms compatible with COVID-19 included in the study, 63% were female with median age of 44 (IQR, 30–58) years. Fever (>38°C), headache, sore throat, arthromyalgia, and cough were the most frequent symptoms (Table 1 ). Patients with positive test results (PCR, antigen detection, or both) had fever significantly more frequently and more cough, anosmia, ageusia, and arthromyalgia than patients with negative determinations (p < 0.05); in contrast, they had less dyspnoea, sore throat, and thoracic pain (p < 0.05) (Table 1). Variables in patients in the three groups of tests were comparable, with the exception of cough, dyspnoea, headache, and number of days from symptom onset to sampling, which showed intergroup variations (Table 2 ). The median number of days for the collection of nasopharyngeal exudates after the onset of symptoms was 2 (IQR, 1–4), with statistically significant differences among tests: SGTI-Flex test (2, IQR 1–4) compared with the Panbio (3, IQR 1–4) and the NovaGen tests (2, IQR 1–3) (p < 0.05) (Table 2). Samples for antigen detection were collected within the recommended period from the onset of symptoms or close contact.
Table 1.
Clinical characteristics of the 600 study participants suspected of having active COVID-19
| COVID-19 suspected cases (%) | PCR result (%) |
Antigen detection result (%) |
Positive PCR result (%) |
||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive antigen | Negative antigen | ||
| Fever (>38°C) | 33.2 | 46.2a | 28.5a | 50a | 29a | 41a | 23.3a |
| Cough | 44.3 | 51.3a | 41.9a | 53.4a | 42.1a | 43.8a | 30a |
| Dyspnoea | 10.5 | 3.8a | 12.9a | 4.2a | 12a | 3.5 | 1.7 |
| Sore throat | 33.7 | 25.9a | 36.4a | 27.1 | 35.3 | 22.2 | 15 |
| Anosmia | 7.3 | 19a | 3.2a | 18.6a | 4.6a | 15.3 | 13.3 |
| Ageusia | 6.3 | 16.5a | 2.7a | 16.9a | 3.7a | 13.9 | 10 |
| Arthromyalgia | 32.5 | 40.5a | 29.6a | 40.7a | 30.5a | 33.3 | 26.7 |
| Diarrhoea | 18.7 | 17.7 | 19 | 16.1 | 19.3 | 13.2 | 15 |
| Thoracic pain | 9.3 | 4.4a | 11.1a | 2.5a | 11a | 2.1 | 6.7 |
| Headache | 56.7 | 55.1 | 57.2 | 57.6 | 56.4 | 47.2a | 31.7a |
Frequencies of symptoms are shown, with comparisons of frequencies between patients with detectable and undetectable SARS-CoV-2 RNA, patients with detectable and undetectable antigen, and patients with positive PCR and detectable and undetectable antigen results.
aStatistically significant difference (p < 0.05).
Table 2.
Comparison of patients suspected of having COVID-19 assorted by the studied test
| SGTI-Flex | Panbio | NovaGen | |
|---|---|---|---|
| Cough | 38% | 41% | 54% |
| Dyspnoea | 7.5% | 9% | 15% |
| Headache | 51.5% | 49.5% | 69% |
| Time since symptom onset (d), median (IQR) | 2 (1–4) | 3 (1–4) | 2 (1–3) |
Only variables with statistically significant differences are shown.
SARS-CoV-2 positive detection prevalence was 26.3%, ranging from 22.5% (Panbio) to 30.5% (SGTI-Flex) (p > 0.05) among the cohorts. The sensitivity of the SGTI-Flex, Panbio, and NovaGen tests was 68.9% (95% CI: 55.7–80), 71.1% (95% CI: 55.6–83.6), and 84.6% (95% CI: 72–93.1), respectively. All tests invariably showed 100% specificity and positive predictive values. Sensitivity was systematically higher in samples yielding positive PCR results with Ct ≤ 20 (Table 3 ).
Table 3.
Sensitivity, specificity, and negative predictive values of antigen detection on nasopharyngeal exudates for the detection of SARS-CoV-2 in suspected cases or close asymptomatic contacts
| Patients tested | Antigen detection kit | Prevalence (% patients with positive PCR) | PCR Ct analysis | SARS-CoV-2 detection performance |
|
|---|---|---|---|---|---|
| Sensitivity (95% CI) | NPV (95% CI) | ||||
| COVID-19 suspected cases | SGTI-Flex | 30.5 | Overall | 68.8 (55.7–80) | 88 (81.8–92.6) |
| PCR Ct ≤ 20 (67.2%) | 87.8 (73.8–96) | ND | |||
| PCR Ct > 20 | 30 (11.9–54.3) | ND | |||
| Panbio | 22.5 | Overall | 71.1 (55.6–83.6) | 92.2 (87.1–95.8) | |
| PCR Ct ≤ 20 (46.7%) | 100 (100–100) | ND | |||
| PCR Ct > 20 | 45.8 (25.5–67.2) | ND | |||
| NovaGen | 26 | Overall | 84.6 (72–93.1) | 95 (90.1–97.8) | |
| PCR Ct ≤ 20 (61.5%) | 93.7 (79.2 – 99.2) | ND | |||
| PCR Ct > 20 | 70 (45.7–88.1) | ND | |||
| Close asymptomatic contacts | SGTI-Flex | 13 | Overall | 84.6 (54.5–98.1) | 97.7 (92.1–99.7) |
| PCR Ct ≤ 20 (61.5%) | 100 (100–100) | ND | |||
| PCR Ct > 20 | 60 (14.6–94.7) | ND | |||
| Panbio | 15 | Overall | 33.3 (11.8–61.6) | 89.5 (81.4–94.8) | |
| PCR Ct ≤ 20 (20%) | 100 (100–100) | ND | |||
| PCR Ct > 20 | 16.7 (2.1–48.4) | ND | |||
| NovaGen | 18 | Overall | 55.5 (30.7–78.4) | 91.1 (83.2–96) | |
| PCR Ct ≤ 20 (33.3%) | 83.3 (35.9–99.6) | ND | |||
| PCR Ct > 20 | 41.7 (15.2–72.2) | ND | |||
Ct, cycle threshold; ND, not done; NPV, negative predictive value.
Comparisons of three SARS-CoV-2 antigen detection tests in close asymptomatic contacts
Of the 300 close asymptomatic contacts, 53.7% were female with median age of 47 (IQR, 29–60) years. Nasopharyngeal exudates were obtained within a median of 5 days (IQR, 3–7) from close contact with a proven case. Statistically significant differences among the tests were seen: SGTI-Flex test (4.5, IQR 2.25–6) compared with the Panbio (5, IQR 4–7) and the NovaGen tests (5, IQR 3–7) (p < 0.05).
SARS-CoV-2 positive detection prevalence (asymptomatic carriers) was 15.3%, ranging from 13% (SGTI-Flex test) to 18% (NovaGen test) (p > 0.05) among the cohorts. The sensitivity of SGTI-Flex, Panbio, and NovaGen was 84.6% (95% CI: 54.5–98.1), 33.3% (95% CI: 11.8–61.6), and 55.6% (95% CI: 30.7–78.4), respectively. All tests invariably showed 100% specificity and positive predictive values. Sensitivity was systematically higher in samples yielding positive PCR results with Ct ≤ 20 (Table 3).
Analysis of samples with false negative antigen detection results
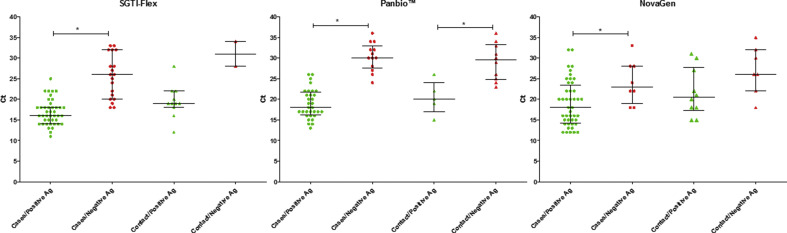
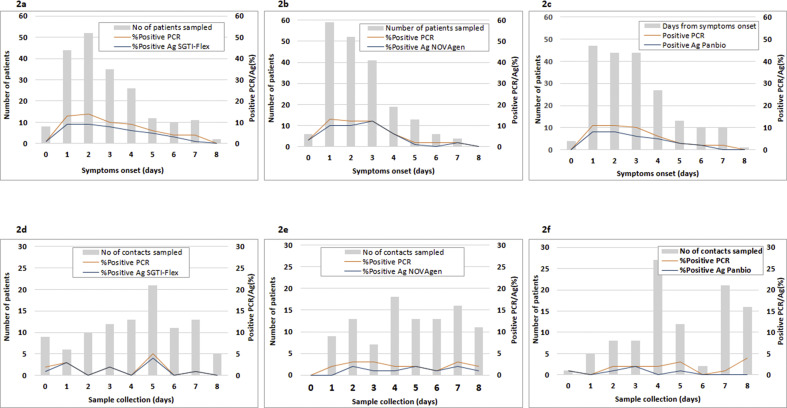
We observed that the lower the viral load of SARS-CoV-2 in nasopharyngeal exudate, the higher the probability of obtaining a false antigen result. Comparisons of median Ct values in PCR-positive samples and grouped by positive antigen detection versus negative antigen detection were assessed. Differences reached statistical significance (p < 0.05) in the following comparisons: clinically suspected cases (SGTI-Flex test, 16 vs 26; Panbio test, 18 vs 30; and NovaGen test, 18 vs 23) and close asymptomatic contacts (Panbio test, 20 vs 29.5) (Fig. 1 ). Moreover, the number of samples with positive antigen or positive PCR results trended higher in the first 3 to 4 days from the onset of clinical symptoms to sample collection, regardless of the test used. There was also a tendency of false negative antigen determinations to cluster in the first days (Fig. 2 ). In contrast, no such pattern was found for the close asymptomatic contacts group (Fig. 2).
Fig. 1.
Scatter plots representing cycle threshold (Ct) values (median and interquartile range) obtained in SARS-CoV-2 PCR positive samples from patients clinically suspected of having COVID-19 and close asymptomatic contacts using SGTI-Flex (1), Panbio (b), and NovaGen (c). Samples are grouped by antigen results; plots indicate Ct values of samples from patients with positive (green) and negative antigen results (red). ∗Statistically significant difference (p < 0.05).
Fig. 2.
Comparisons of the number of samples with positive PCR results and positive antigen detection from clinically suspected patients using SGTI-Flex (a), Panbio (b), and NovaGen (c) or close asymptomatic contacts using SGTI-Flex (d), Panbio (e), or NovaGen (f).
No statistically significant differences were found in median Ct values of positive PCR samples for symptomatic patients (SGTI-Flex test, 18 (IQR 15.5–22); Panbio test, 21 (IQR, 17–26.5); NovaGen test, 20 (IQR, 15–24); p > 0.05) or close asymptomatic contacts (SGTI-Flex, 19 (IQR, 18.5–25); Panbio, 26 (IQR, 22–31); NovaGen, 24 (IQR, 18–30); p > 0.05).
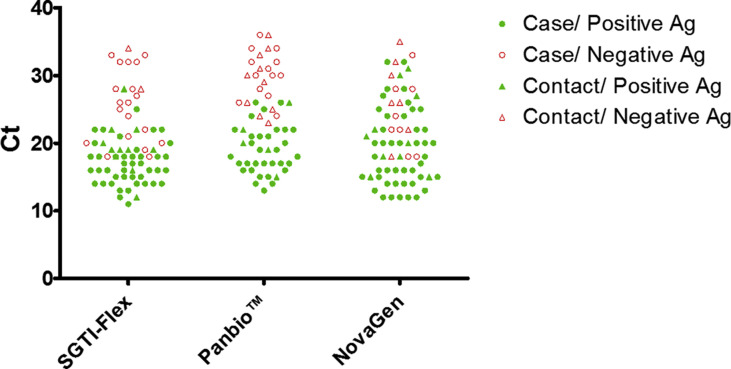
False negatives grouped by test were as follows: SGTI-Flex test (n = 21 (66.6% female); n = 19 cases, 1–7 days of symptoms, Ct values 18–33; n = 2 close asymptomatic contacts, 0–5 days from contact, Ct values 28–34); Panbio test (n = 23 (30.4% female); n = 13 cases, 1–7 days of symptoms, Ct values 24–36; n = 10 close asymptomatic contacts, 2–8 days from contact, Ct values 23–36); and NovaGen test (n = 16 (81% female); n = 8 cases, 1–6 days of symptoms, Ct values 18–33; n = 8 close asymptomatic contacts, 1–8 days from contact, Ct values 15–35) (Fig. 3 ).
Fig. 3.
Representation of patients with positive PCR results and their corresponding Ct, grouped into the cohorts of patients tested with SGTI-Flex, Panbio, and NovaGen. Patients clinically suspected of having COVID-19 and close asymptomatic contacts are indicated, as well as the antigen result for each patient.
Discussion
Our study shows that the three evaluated SARS-CoV-2 antigen detection tests do not perform equally in patients with clinical suspicion of infection or close asymptomatic contacts. Two of the three tests showed higher sensitivity in symptomatic patients, whereas the third performed better in close asymptomatic contacts. Differences are not attributable to dissimilarities in prevalence among cohorts or viral load quantified by Ct values.
A recent systematic review conducted by the Cochrane Library reports that the sensitivity of antigen detection for COVID-19 diagnosis is higher in symptomatic patients (72.0%, 95% CI 63.7%–79.0%) than in asymptomatic patients (58.1%, 95% CI 40.2%–74.1%). Sensitivity was higher over the first week after symptom onset and/or in patients with positive PCR determinations with higher viral loads [5,12,13]. Low sensitivity may be due to the high heterogeneity found among studies, populations studied, prevalence of COVID-19 in the population studied, or timing of sample collection [12], among other factors.
Here, we confirm the high specificity of antigen detection—it did not detect any case undetected by PCR—for the detection of SARS-CoV-2 [6,[13], [14], [15]]. Antigen detection alleviates the workload in the microbiology laboratory in settings with a high prevalence of the disease, given that with positive results there is no need for PCR confirmation. However, negative results in antigen determinations do require PCR confirmation, given that sensitivity values are not 100%. Data on the performance of antigen detection in close asymptomatic contacts are scarce, despite being of great importance in epidemiology. Studies conducted in primary care facilities also report higher sensitivity values (72%–92%) in patients with a clinical suspicion of COVID-19 [6,[16], [17], [18], [19]] in comparison to close asymptomatic contacts (54%–76%) [6,17]. However, the aforementioned studies evaluated one test each, Panbio, widely used for SARS-CoV-2 screening.
Another very important point is the time since the contact occurred or symptoms appeared. We preselected patients in whom symptom onset or close contact with a SARS-CoV-2 positive case had occurred within the prior 8 days and evaluated three antigen detection tests in three cohorts of patients. One of the most important observations drawn from our study is that not all antigen detection tests perform equally well in patients with clinically suspected COVID-19 and in close asymptomatic contacts. NovaGen shows the highest sensitivity values in clinically suspected cases, whereas the SGTI-Flex test shows the highest sensitivity in close asymptomatic contacts. Being aware of the differences in test-to-test performance is key when screening for SARS-CoV-2, particularly in close asymptomatic contacts and symptomatic patients, for whom antigen-negative determinations must be confirmed by PCR.
We do not have a clear explanation for the test-to-test variations observed. Patients tested with NovaGen, which performed better in clinically suspected cases, had fever, headache, and dyspnoea more frequently than patients in the other two cohorts. Better performance of the three tests was seen when positive PCR samples were grouped by Ct values (Ct ≤ 20), although these differences among samples within each cohort of patients do not explain performance dissimilarities. An alternative explanation for the discrepancies between PCR and antigen detection may be poor quality of the sample collected for antigen detection, given that both swabs were not dipped into viral transport medium. However, had this been the case, the three tests would have been equally affected. The SARS-CoV-2 nucleocapside protein is a large 419–amino acid protein with three conserved domains: an N-terminal RNA-binding domain, a C-terminal dimerization domain, and an intrinsically disordered central Ser/Arg (SR)-rich linker [20,21]. Differences in sensitivity results among tests may be due to detection of antigens sourced from different parts of the nucleocapsid protein. Expression of antigens may be affected by infection stage or even by some viral variants. Previous studies have showed that the higher sensitivity of antigen detection in samples with low Ct may be a good system for detecting individuals who are contagious [7,22]. In light of our results, we cannot hold with certainty that negative antigen detection can be automatically translated as either a negative PCR result or a positive low viral load PCR, as some of the false antigen detections occurred in samples with positive PCR and low Ct values.
Our study is limited by the fact that we did not study the performance of the three tests in the same pool of samples. However, we opted for such an approach to preserve the sensitivity of antigen detection, as testing the three tests simultaneously would have required dilution of samples upon discharge into a broth viral transport medium. Furthermore, we do not know whether asymptomatic carriers could have developed symptoms in the days after sample collection.
We conclude there are considerable test-to-test antigen detection variations among patients with clinical suspicion of COVID-19 and close asymptomatic contacts. Such differences should be taken into account when choosing tests for SARS-CoV-2 screening in patients attending primary care. Negative antigen results, even when the test with the highest sensitivity in detecting infected individuals is used, do not preclude PCR confirmation.
Transparency declaration
The authors declare that they have no conflicts of interest.
Author contributions
Study design: PE, JG; development and methodology: PE, JG; collection of the data: ASP, JGL, IVL, PC; data analysis and interpretation: JG, PE; writing all sections of the manuscript: PE, JG; manuscript revision: PE, ASP, JGL, PC, IVL, PM, JG.
Acknowledgements
The authors are grateful to Dainora Jaloveckas for manuscript editing assistance. Genomica (SGTI-Flex COVID-19 Ag) and Eurofins-INGENASA (GSD NovaGen SARS-CoV-2 Ag Rapid Test) kindly provided the antigen detection kits. This study was supported by grants from Instituto de Investigación Sanitaria Gregorio Marañón (II-PI-1-2020). The funders and companies supplying the antigen tests had no role in the study design, data collection, analysis, decision to publish, or preparation/content of the manuscript. PE (CPII20/00015) is a recipient of a Miguel Servet contract supported by the FIS. JG is a steady researcher contracted by Fundación para Investigación Sanitaria del Hospital Gregorio Marañón.
We would like to thank the following members of the COVID-19 study group Hospital Gregorio Marañón (Adán (Iván), Adán (Javier), Alcalá (Luis), Alonso (Roberto), Álvarez-Uría (Ana), Bouza (Emilio), Catalán (Pilar), Escribano (Pilar), Estévez (Agustín), Fernández-Del Rey (Rocío), Galar (Alicia), García de Viedma (Darío), González (Pedro), Gómez-Núñez (Ana), Guinea (Jesús), Herranz (Marta); Kestler (Marta), Machado (Marina), Marín (Mercedes), Muñoz (Patricia), Olmedo (María), Ortiz (Javier), Palomo (María), Pérez-Granda (María Jesús), Pérez (Laura), Rincón (Cristina), Rodríguez (Sara), Rodríguez (Belén), Ruiz-Serrano (María Jesús), Valerio (Maricela), Vidán (Maite) (and COVID-19 study group Pavones care centre (María Alonso Blanco (Ana), Coca Escribano (Elena), Blázquez Andrés (Susana), Sastre Páez (María Sonsoles), Vega López (Patricia), Díez Vaquero (Lorena), Sandoval Montejo (María Carmen), Pinto Zurita (Jannet), Jiménez Otero (Estefanía), Cañón Cañón (Carolina), Castro Romaní (Marisol), Villamañán Lobo (David), Roca Sánchez (Susana), López Rodríguez (M. Estrella), Jiménez Morillo (Raquel), Plaza Rivera (Sara), Zaragoza Vargas (Natalia), Sánchez Sánchez (José Alejandro), Sánchez Carrasco (Cristina), Corral Martínez (Pedro), Valero López (Iván), Ramos del Río (Lourdes), Huertas Uhagón (María), Martín Francisco (María de la Peña), Cañas Zuluaga (Ángela Bibiana), Heras Martín (Laura), García García (Ana Isabel), Sevilla Aguerre (Ainhoa), López Alonso (Ana Belén), Sánchez Sánchez (Luis Alberto), Cánovas Martínez (Miriam), Roy Zafra (Rut), and García Ramos (Aitor)).
Editor: M. Cevik
Contributor Information
COVID-19 study group Hospital Gregorio Marañón:
Iván Adán, Javier Adán, Luis Alcalá, Roberto Alonso, Ana Álvarez-Uría, Emilio Bouza, Pilar Catalán, Pilar Escribano, Agustín Estévez, Rocío Fernández-Del Rey, Alicia Galar, Darío García de Viedma, Pedro González, Ana Gómez-Núñez, Jesús Guinea, Marta Herranz, Marta Kestler, Marina Machado, Mercedes Marín, Patricia Muñoz, María Olmedo, Javier Ortiz, María Palomo, María Jesús Pérez-Granda, Laura Pérez, Cristina Rincón, Sara Rodríguez, Belén Rodríguez, María Jesús Ruiz-Serrano, Maricela Valerio, and Maite Vidán
COVID-19 study group Pavones care centre:
Ana María Alonso Blanco, Elena Coca Escribano, Susana Blázquez Andrés, María Sonsoles Sastre Páez, Patricia Vega López, Lorena Díez Vaquero, María Carmen Sandoval Montejo, Jannet Pinto Zurita, Estefanía Jiménez Otero, Carolina Cañón Cañón, Marisol Castro Romaní, David Villamañán Lobo, Susana Roca Sánchez, M. Estrella López Rodríguez, Raquel Jiménez Morillo, Sara Plaza Rivera, Natalia Zaragoza Vargas, José Alejandro Sánchez Sánchez, Cristina Sánchez Carrasco, Pedro Corral Martínez, Iván Valero López, Río Lourdes Ramos del, María Huertas Uhagón, Peña Martín Francisco María de la, Ángela Bibiana Cañas Zuluaga, Laura Heras Martín, Ana Isabel García García, Ainhoa Sevilla Aguerre, Ana Belén López Alonso, Luis Alberto Sánchez Sánchez, Miriam Cánovas Martínez, Rut Roy Zafra, and Aitor García Ramos
References
- 1.Andrew M., Searle S.D., McElhaney J.E., McNeil S.A., Clarke B., Rockwood K., et al. COVID-19, frailty and long-term care: implications for policy and practice. J Infect Dev Ctries. 2020;14:428–432. doi: 10.3855/jidc.13003. [DOI] [PubMed] [Google Scholar]
- 2.Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:e00512–e00520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg A., Ghoshal U., Patel S.S., Singh D.V., Arya A.K., Vasanth S., et al. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. 2021;93:2281–2286. doi: 10.1002/jmv.26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO . 2020. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays interim guidance.https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays [Google Scholar]
- 5.Muhi S., Tayler N., Hoang T., Ballard S.A., Graham M., Rojek A., et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: a validation and implementation study. Lancet Reg Health West Pac. 2021;9:100115. doi: 10.1016/j.lanwpc.2021.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linares M., Perez-Tanoira R., Carrero A., Romanyk J., Perez-Garcia F., Gomez-Herruz P., et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alemany A., Baro B., Ouchi D., Rodo P., Ubals M., Corbacho-Monne M., et al. Analytical and clinical performance of the Panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect. 2021;82:186–230. doi: 10.1016/j.jinf.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruger C. Call for a pan-European COVID-19 response must be comprehensive. Lancet. 2021;397:1540–1541. doi: 10.1016/S0140-6736(21)00471-2. [DOI] [PubMed] [Google Scholar]
- 9.Masia M., Fernandez-Gonzalez M., Sanchez M., Carvajal M., Garcia J.A., Gonzalo-Jimenez N., et al. Nasopharyngeal panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infect Dis. 2021;8:ofab059. doi: 10.1093/ofid/ofab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruana G., Croxatto A., Kampouri E., Kritikos A., Opota O., Foerster M., et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE Study. Microorganisms. 2021;9:798. doi: 10.3390/microorganisms9040798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favresse J., Gillot C., Oliveira M., Cadrobbi J., Elsen M., Eucher C., et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Med. 2021;10:265. doi: 10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinnes J., Deeks J.J., Berhane S., Taylor M., Adriano A., Davenport C., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3:CD013705. doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Garcia F., Romanyk J., Moya Gutierrez H., Labrador Ballestero A., Perez Ranz I., Gonzalez Arroyo J., et al. Comparative evaluation of Panbio and SD Biosensor antigen rapid diagnostic tests for COVID-19 diagnosis. J Med Virol. 2021;93:5650–5654. doi: 10.1002/jmv.27089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jääskeläinen A.J., Kekäläinen E., Kallio-Kokko H., Mannonen L., Kortela E., Vapalahti O., et al. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill. 2020;25:2000603. doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres I., Poujois S., Albert E., Colomina J., Navarro D. Evaluation of a rapid antigen test (Panbio COVID-19 Ag rapid test device) for SARS-CoV-2 detection in close asymptomatic contacts of COVID-19 patients. Clin Microbiol Infect. 2021;27:e1–e4. doi: 10.1016/j.cmi.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., et al. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study. Sci Rep. 2021;11:10519. doi: 10.1038/s41598-021-90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., et al. The evaluation of a newly developed antigen test (QuickNavi-COVID19 Ag) for SARS-CoV-2: a prospective observational study in Japan. J Infect Chemother. 2021;27:890–894. doi: 10.1016/j.jiac.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernandez-Fuentes M.A., et al. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2021;27:e7–e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulilete O., Lorente P., Leiva A., Carandell E., Oliver A., Rojo E., et al. Panbio rapid antigen test for SARS-CoV-2 has acceptable accuracy in symptomatic patients in primary health care. J Infect. 2021;82:391–398. doi: 10.1016/j.jinf.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Z., Cao Y., Liu W., Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13:1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]