Abstract
Background:
Long-term alcohol drinking is associated with numerous health complications including susceptibility to infection, cancer, and organ damage. However, due to the complex nature of human drinking behavior, it has been challenging to identify reliable biomarkers of alcohol use that could be used to determine drinking behavior prior to signs of overt organ damage. Recently, extracellular vesicle-bound microRNA (EV-miRNA) have been discovered to be consistent biomarkers of conditions including cancer and liver disease.
Methods:
In this study, we profiled the plasma EV-miRNA content by miRNA-Seq from 80 non-human primates after 12 months of voluntary ethanol drinking.
Results:
We identified a list of up- and downregulated EV-miRNA candidate biomarkers of both heavy drinking as well as those positively correlated with ethanol dose. We further overexpressed these candidate miRNA in control primary peripheral immune cells to assess potential functional mechanisms of these EV-miRNA. We report that overexpression of miR-155, miR-154, miR-34c, miR-450a, and miR-204 led to increased inflammatory TNFα or IL-6 production in PBMC after stimulation.
Conclusion:
This exploratory study identified several EV-miRNA that could serve as biomarkers of long-term alcohol drinking as well as provided a mechanism for alcohol-induced peripheral inflammation.
Keywords: Alcohol, extracellular vesicles, miRNA, non-human primates
INTRODUCTION
Alcohol consumption is widespread in the United States with 60% of the population aged 18 and older reporting alcohol use in the last month(NIAAA). Chronic alcohol consumption has been associated with numerous adverse health outcomes, including severe organ damage (Szabo and Bala, 2010) and cancers (Grewal and Viswanathen, 2012, Fedirko et al., 2011). Heavy alcohol consumption is also correlated with poor vaccine response(Messaoudi et al., 2013) and increased susceptibility to many bacterial (Saitz et al., 1997, Kline et al., 1995, Hudolin, 1975) and viral pathogens(Baum et al., 2010, Bhattacharya and Shuhart, 2003) indicating dysregulation of the immune system. However, the molecular mechanisms underlying immune dysfunction with chronic drinking are difficult to study because human alcohol consumption is unpredictable and challenging to monitor. Currently, blood ethanol content (BEC), aspartate aminotransferase (AST), alanine amino transferase (ALT), gamma-glutamyltransferase (GGT), carbohydrate-deficient transferrin (CDT)(Conigrave et al., 2002), phosphatidylethanol (PEth) (Viel et al., 2012), and mean corpuscular volume (MCV), are used as clinical indicators of alcohol use(Conigrave et al., 2002, Mundle et al., 2000). However, these biomarkers are limited in their sensitivity and specificity to alcohol consumption. For example, elevated AST, CDT and MCV levels were observed in only 56%, 69%, and 47%, respectively, of hospitalized patients who disclosed participation in heavy alcohol consumption(Anttila et al., 2004). This creates a need for new measurable, consistent biomarkers of alcohol consumption. Recent studies have highlighted the utility of extracellular vesicle bound micro RNA (EV-miRNA) as biomarkers of different disease states (Jia et al., 2014). miRNA are stable, regulatory small non-coding RNA, typically 22 nucleotides long, and are responsible for the targeting and silencing of messenger RNA (mRNA) by transcriptional blockade for degradation(Bartel, 2004). These extracellular miRNA can exist independently in biofluids or packaged in extracellular vesicles where they are stable over time(Ferracin et al., 2015, Valadi et al., 2007b). Extracellular vesicles such as exosomes and microvesicles are found in all biofluids including serum and plasma and facilitate cell-to-cell communication by horizontal transfer of vesicle content, including proteins, mRNA, and many types of non-coding RNA(Huang et al., 2013).
Alcohol exposure may lead to increased production of exosomes as well as altered expression of EV-miRNA content (Momen-Heravi et al., 2015). For example, miR-122, a liver-specific miRNA, is upregulated in exosomes isolated from human sera immediately following the consumption of 2 ml vodka 40% v/v ethanol/kg body weight and in mouse models of binge (5 g/kg 50% v/v ethanol diluted in water via oral gavage) and chronic drinking (a diet consisting of 36% of calories coming from ethanol for 5 weeks) (Momen-Heravi et al., 2015). Also, increased abundance of miR-122 results in monocyte hypersensitivity to lipopolysaccharides (LPS) (Momen-Heravi et al., 2015). Treatment of THP-1 cells in vitro with alcohol increases miR-27a abundance in EVs, which in turn induces M2 polarization in naïve THP-1 cells (Saha et al., 2016). Additionally, studies have reported miR-155, miR-122, and miR-34a as potential biomarkers of alcohol-induced liver disease (Momen-Heravi et al., 2015, Torres et al., 2018, Zhang et al., 2010). Moreover, the expression of circulating or tissue-residing miRNAs has been linked to alcohol-induced organ injury in the pancreas, liver, gut, and brain, the mechanisms of which are thought to be linked to induction of inflammatory cytokine production (Natarajan et al., 2015). The identification of EV-miRNA that could be used as reliable biomarkers of chronic ethanol consumption in the absence of overt organ damage is challenging when using strictly clinical samples, rodent models, or treated cell lines. Clinical studies are limited by inaccurate self-reporting of alcohol intake and medical histories, concurrent drug/tobacco use, and environmental factors. Data from rodent model studies is confounded by the stress caused by either forced feeding with surgically placed gavage tubes, access to high concentration ethanol only diets, and only short periods of ethanol exposure.
In the presented study, we utilized a rhesus macaque model of voluntary ethanol self-administration to identify potential extracellular vesicle bound miRNA (EV-miRNA) biomarkers of alcohol drinking. This model allows for elucidation of the impact of chronic alcohol consumption in an outbred species while avoiding confounding factors such as smoking or drug use. Using this model, we have reported large transcriptomic changes in peripheral blood mononuclear cells (PBMC) and tissue resident macrophages with alcohol consumption (Sureshchandra et al., 2016, Sureshchandra et al., 2019, Lewis et al., 2021). These studies revealed altered expression of genes associated with host defense, inflammation, and wound repair (Barr et al., 2016, Asquith et al., 2014). This transcriptional dysregulation could be partially explained by altered miRNA expression that we have observed in PBMC from the same animals (Barr et al., 2016, Asquith et al., 2014, Sureshchandra et al., 2019), however, global alterations in circulating and extracellular vesicle bound miRNA have not been explored. Therefore, we examined the impact of alcohol drinking on EV-miRNA expression using samples obtained from 80 rhesus macaques after 12 months of open access to 4% ethanol solution or isocaloric drink. Our analysis revealed that chronic ethanol consumption altered EV-miRNA cargo. We identified several EV-miRNA that were up- or down-regulated with alcohol drinking and that regulated genes with roles in myeloid cell activation, angiogenesis, and regulation of gene expression. Eight EV-miRNA were selected for additional functional studies. Over-expression of candidates miR-155, miR-154, miR-34c, miR-450a, and miR-204, which are upregulated in EV with chronic alcohol drinking, resulted in a heightened inflammatory response in PBMC after stimulation. In contrast, over-expression of mi-625, which was downregulated with chronic ethanol consumption decreased inflammatory cytokine production by PBMC from CHD animals. These findings both provide candidates that can be further validated as biomarkers of chronic alcohol drinking as well as provide a potential mechanism for increased inflammation associated with alcohol use.
MATERIALS AND METHODS
Animal studies and sample collection:
These studies used samples from a macaque model of voluntary ethanol self-administration established through schedule-induced polydipsia (Baker et al., 2014, Grant et al., 2008, Jimenez et al., 2015). Briefly, in this model, rhesus macaques are introduced to a 4% w/v ethanol solution during a 90-day induction period followed by concurrent access to the 4% w/v solution and water for 22 hours/day for one year. During this time, the macaques adopt a stable drinking phenotype defined by the amount of ethanol consumed per day and the pattern of ethanol consumption (g/kg/day) (Baker et al., 2014). Blood samples were taken from the saphenous vein every 5–7 days at 7 hours after the onset of the 22 hours/day access to ethanol and assayed by headspace gas chromatography for blood ethanol concentrations (BECs).
For these studies, plasma from a total of 80 rhesus macaques (22 controls, 33 moderate, and 25 heaving ethanol drinking animals); 17 females and 71 males across 8 cohorts collected after 12 months of open access (22 hours/day) to 4% ethanol in water solution was utilized. Drinking classifications were defined as chronic moderate drinking (CMD) or chronic heavy drinking (CHD) based on 12 months of ethanol self-administration (tissue and drinking data obtained from the Monkey Alcohol Tissue Research Resource: www.matrr.com). These cohorts of animals (Cohorts 4, 5, 6a, 6b, 7a, 7b, 10a, and 14 on matrr.com) were described in two previous studies of peripheral immune system response to alcohol (Sureshchandra et al., 2016, Sureshchandra et al., 2019). Blood plasma was isolated by centrifugation over histopaque (Sigma, St Louis, MO) as per manufacturer’s protocol and stored at −80C until they could be analyzed as a batch. The average daily ethanol intake for each animal is outlined in Supp. Table 1.
Plasma EV Isolation and Nanoparticle Tracking Assay
Plasma samples were thawed and centrifuged at 3,000 x g for 15 minutes. The SmartSEC Single Kit for EV Isolation (System Biosciences, Palo Alto, CA) was utilized for EV purification following manufacturer’s instructions. A Nanosight NS300 (Malvern Panalytical, Malvern, United Kingdom) was used for nanoparticle tracking analysis of isolated EVs. Size distribution and concentration of EVs were measured in triplicate based on 60s video recordings with constant syringe flow.
Plasma EV Isolation and miRNA library preparation
Plasma was filtered using Viviclear Mini Centrifugal Filters (Sartorius, Göttingen, Germany), a 0.8μm porous membrane. Extracellular vesicle (EV)-bound total RNA was isolated from the vesicles using the exoRNeasy Serum/Plasma Midi Kit (Qiagen, Valencia, CA) by membrane affinity column purification per the manufacturer’s protocol. EV-miRNA were selected by specific ligation of adaptors to the 3’ hydroxyl and 5’ phosphate groups, unique to mature miRNA, using the QIAseq miRNA Library Kit (Qiagen, Valencia, CA). Briefly, with a 5 μl total RNA input, cDNA synthesis was performed using universal reverse transcription, library amplification, and library cleanup.
Sequencing, identification of DEmiRNA, DEmiRNA targets, and enrichment
Each library was indexed using a unique barcode for multiplexing and sequenced on the NextSeq2000 or Hiseq4000 platform (Illumina, San Diego, CA) to yield single-end 100 bp sequences. Quality reports for the raw miRNA reads were generated using FASTQC. The reads were trimmed 10–50 bp. Followed by alignment to the Macaca mulatta genome from Ensembl using splice aware short read aligner suite Bowtie 2 (Langmead and Salzberg, 2012). The transcript counts per gene were then summarized using the summarizeOverlaps function. Samples were required to have a minimum of 1,500,000 reads post alignment and 400 detected miRNA to ensure good coverage of the genome. Differentially expressed miRNA (DEmiRNAs) were identified using the edgeR package (Robinson et al., 2010) with batch correction and were defined as those with a fold change ≥ 1.5, FDR corrected p-value ≤ 0.05. DEmiRNA from macaques were converted to their human homologs. The miRTarBase database (Huang et al., 2020) was scanned with the list of DEmiRNAs to identify validated gene targets. Functional enrichment of targeted genes was done using Metascape (Zhou et al., 2019) to identify clusters of genes mapping to specific biological or molecular pathways. ggplot2 and miRNet (Chang et al., 2020) were used for enrichment and target visualization.
PBMC stimulation with EV
22Total PBMC from a control macaque were thawed in RPMI supplemented with 10% FBS, 1% L-Glutamine, and 1% Penicillin/Streptomycin. EV from control and CHD macaques were isolated from plasma as described above using the SmartSEC Single Kit for EV Isolation (System Biosciences, Palo Alto, CA). 1×106 PBMC were plated per condition and ~1×107 isolated EV were added to the PBMC and incubated for 6 hours at 37°C in an incubator with 5% CO2. Cells were then stimulated for 16 h with or without PMA and Ionomycin at 37°C in an incubator with 5% CO2. Supernatants were collected stored at −80 °C short-term.
Electroporation with miRNA mimics and stimulation of PBMC
Total PBMC from control male and female macaques were thawed and washed four times in Optimem (ThermoFisher, Waltham, MA) to remove traces of serum. 5×106 cells were resuspended in 200ul Optimem containing 2ug miRNA mimic (Dharmacon, Lafayette, CO). The Gene pulser II System (Bio-Rad Laboratories, Berkeley, CA) was used to electroporate the sample in a 0.4cm gap cuvette using 500V for 0.05 seconds. Cells were moved to a 48 well plate containing 500uL RPMI supplemented with 10% FBS, 1% L-Glutamine, and 1% Penicillin/Streptomycin and allowed to recover for 48 hours in an incubator with 5% CO2 at 37C. Cells were then stimulated for 16 h with or without PMA and Ionomycin at 37°C in an incubator with 5% CO2. Supernatants were collected and cells were resuspended in Qiazol (Qiagen, Valencia CA) for RNA extraction. Both cells and supernatants were stored at −80 °C until they could be processed as a batch. A non-targeting negative control (miRIDIAN microRNA Mimic Negative Control #1; Dharmacon, Lafayette, CO) was included to show no effect of mimic electroporation alone.
TNF and IL-6 ELISA
Supernatants were collected following 16 h of incubation. Samples were analyzed in duplicates using Monkey IL6 ELISA and Monkey TNFα Total ELISA kits (ThermoFisher, Waltham, MA) per the manufacturer’s instructions. Unstimulated condition measured values were subtracted from stimulated condition values to plot a corrected value for each test group and control.
qPCR for electroporation efficiency
Cells were lysed in QIAzol Lysis reagent (Qiagen, Valencia, CA) and total RNA was extracted using the miRNeasy mini kit (Qiagen, Valencia, CA) per manufacturer’s instructions. We used TaqMan miRNA Assays (ThermoFisher, Waltham, MA) for detection of miRNA. Briefly, reverse transcription (30 min, 16 °C; 30 min, 42 °C; 5 min 85 °C) was done using 10ng RNA, TaqMan primers and High Capacity cDNA Reverse Transcription kit (ThermoFisher, Waltham, MA). qPCR was done on the StepOnePlus Real-Time PCR System (ThermoFisher, Waltham, MA) using TaqMan Universal PCR Master Mix.
Statistical Analysis
All statistical analyses were conducted in Prism 7(GraphPad). Data sets were first tested for normality. Nine group comparisons were tested for statistical significance using One-way ANOVA (〈=0.05) followed by Holm Sidak’s multiple comparisons tests. Two group comparisons were tested for statistical significance using either paired t-test or unpaired t-test with a Welch’s correction. Error bars for all graphs are defined as ± SEM. Multivariate linear regression analysis compared significant shifts in curve over horizontal line, with R-squared reported. Statistical significance of functional enrichment was defined using hypergeometric tests. P-values less than or equal to 0.05 were considered statistically significant. Values between 0.05 and 0.1 are reported as trending patterns.
RESULTS
Extracellular vesicle associated miRNA (EV-miRNA) expression changes with heavy alcohol drinking
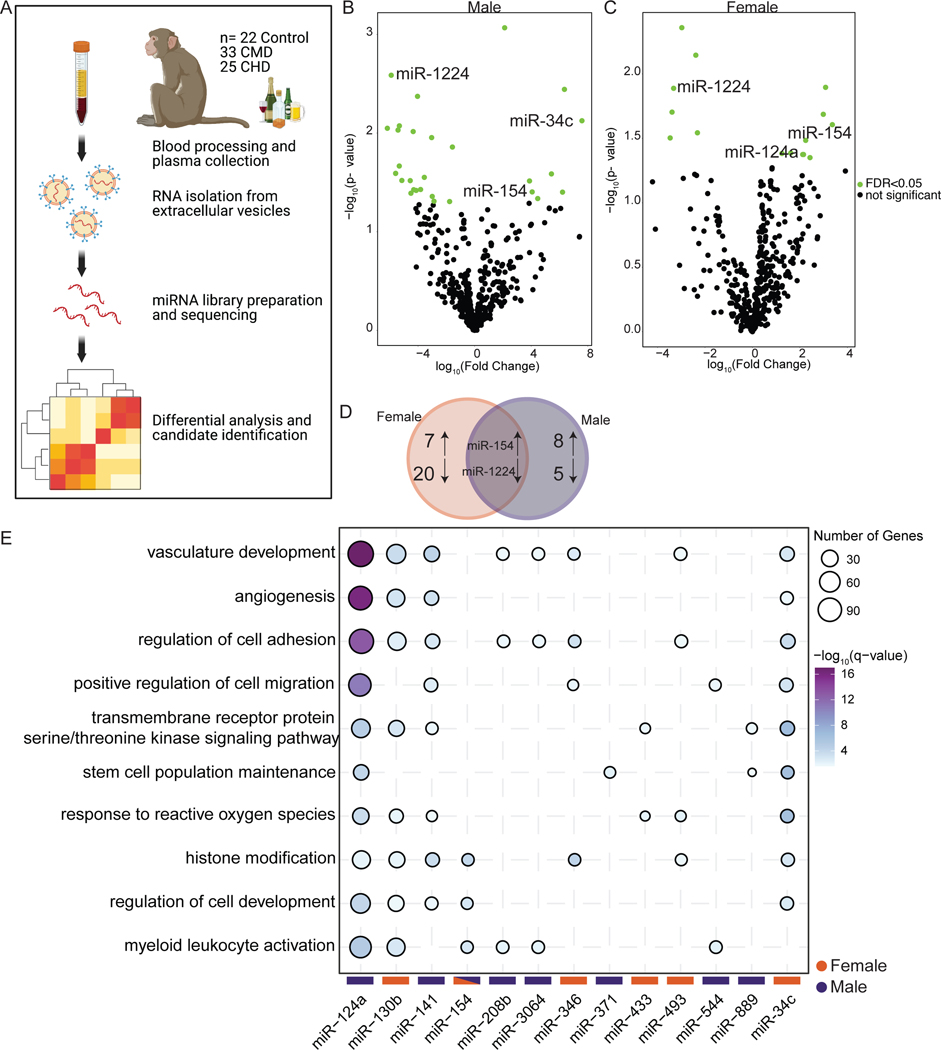
To identify candidate biomarkers of alcohol drinking, we profiled miRNA from circulating extracellular vesicles (EV-miRNA) isolated from macaque plasma after 12 months of voluntary alcohol drinking (Figure 1A). There were no differences in concentration of EVs between control and alcohol-exposed plasma (Supp. Figure 1A,B). Total RNA was extracted from the EVs and miRNA libraries were constructed for 22 control, 33 Chronic Moderate Drinking (CMD), and 25 Chronic Heavy Drinking (CHD) animals (Supp. Table 1). Initial analysis of the sequencing data revealed significant differences between male and female CHD animals, therefore separate analyses were carried out for each. We first sought to identify in EV-miRNA that were differentially expressed with CHD (>3g EtOH/kg bodyweight/day) by comparing miRNA-Seq data from CHD and control animals. This analysis revealed a number of differentially expressed miRNA (DEmiRNA) in both males and females (Figure 1B,C). Only one upregulated (miR-154) and one downregulated (miR-1224) DEmiRNA were found to be in common between males and females by this analysis (Figure 1D). We next identified validated gene targets of the human homologs of these DEmiRNA using miRTarBase and performed functional enrichment on those genes using Metascape (Zhou et al., 2019) (Figure 1E, Supp. Figure 2 and Supp. Table 2). The numbers of validated gene targets varied greatly between EV-miRNA (Supp. Figure 2A,B). The targets of several upregulated EV-miRNA enriched to gene ontology (GO) terms associated with blood vessel development such as “angiogenesis” and “vasculature development” (e.g., miR-124a, 130b, 141 and 34c) as well as “response to reactive oxygen species” (e.g. miR-124a, 433, 493 and 34c). Other genes regulated by EV-miRNA upregulated with chronic ethanol consumption played a role in “myeloid leukocyte activation” (e.g. miR-124a, 130b, 154, 208b, 544) (Figure 1E). We also performed functional enrichment on the targets of the downregulated EV-miRNA to assess processes that may be upregulated with CHD. The targets of these downregulated EV-miRNA mapped to GO terms associated with signaling (e.g., “regulation of protein kinase activity”), cell cycle (e.g., “mitotic DNA damage checkpoint”), and tissue/epithelial homeostasis (e.g., “tissue homeostasis” and “response to hypoxia”) (Supp. Figure 2C). These data show that CHD alters expression of several EV-miRNA that can potentially serve as biomarkers of chronic ethanol consumption.
Figure 1: Extracellular vesicle associated miRNA (EV-miRNA) expression changes with heavy alcohol drinking.
A) Experimental Design for the study. B,C) Volcano plots showing significant (FDR≤0.05) differentially expression miRNA with heavy drinking in female (B) and male (C) macaques. D) Venn diagram comparing DEmiRNA between male and female macaques with heavy drinking. E) Bubble plot depicting GO Biological processes associated with the validated target genes of the indicated upregulated EV-miRNA from males and females with heavy drinking. The size of the bubble represents the number of target genes, and the color represents the -log10(q-value) significance of enrichment.
Dose-dependent changes in extracellular vesicle associated miRNA expression
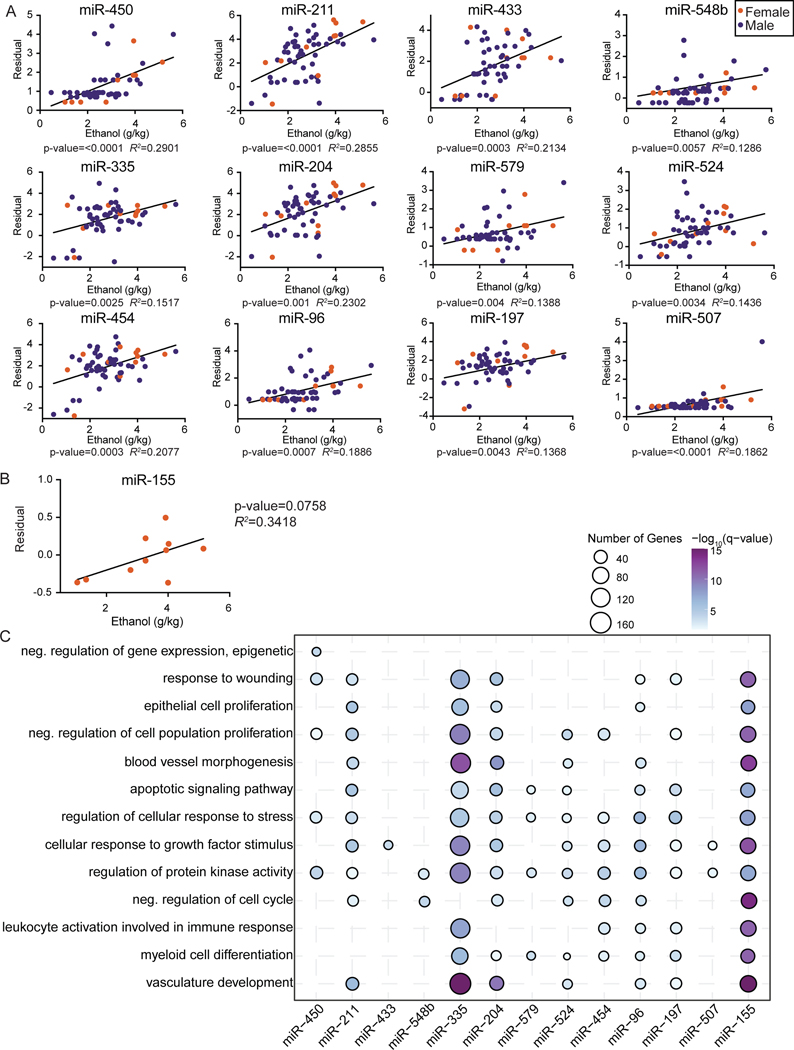
We next identified EV-bound miRNA whose expression was significantly correlated with dose of ethanol (g/kg/day). Similar trends in male and female animals were noted; therefore, data from both sexes were pooled for this analysis (Figure 2A). We identified 12 miRNAs whose expression was positively correlated with ethanol dose (Figure 2A). The expression of miR-155, had a modest positive correlation with ethanol consumption in female animals (p=0.07) (Figure 2B). We used miRTarBase to identify validated gene targets of each of these miRNAs (Supp. Figure 3A) and carried functional enrichment to understand the biological processes impacted by these DEmiRNA (Figure 2C). The targets of several EV-miRNA enriched to gene ontology (GO) terms associated with blood vessel development such as “Blood vessel morphogenesis” and “vasculature development” (e.g., miR-335, 204, 155) (Figure 2C). A few EV-miRNA targeted genes regulate tissue repair as indicated by enrichment to GO terms “response to wounding”, “epithelial cell proliferation” and “cellular response to growth factor stimulus” (e.g., miR-211, 335, 204, 96) (Figure 2B). Finally, some DE-miRNA targeted genes mapping to “myeloid cell differentiation” (e.g., miR-335, 579, 454, 197, 155). This analysis indicates that chronic ethanol consumption potentially disrupts pathways associated with angiogenesis, tissue repair and inflammation in a dose-dependent manner by altering expression of EV-miRNA.
Figure 2: Dose-dependent changes in extracellular vesicle associated miRNA expression.
A) Scatter plots of partial residual expression values for selected miRNA and ethanol dose (g EtOH/ kg body weight/ day). B) Scatter plots of partial residual expression values for miR-155 and ethanol dose (g EtOH/ kg body weight/ day) in female animals. C) Bubble plot depicting GO Biological processes associated with the validated target genes of the indicated positively correlated EV-miRNA with ethanol dose. The size of the bubble represents the number of target genes, and the color represents the -log10(q-value) significance of enrichment.
Candidate EV-miRNA target genes align with PBMC gene expression changes in CHD macaques
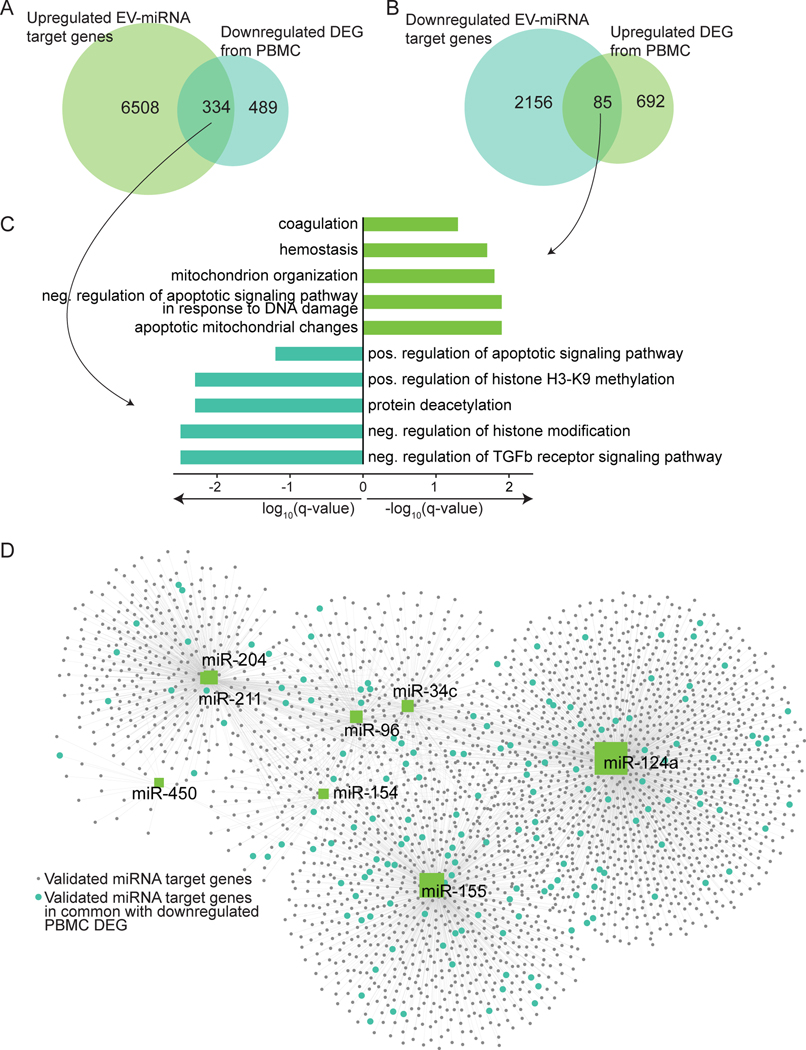
The analyses described above identified a total of 28 upregulated and 26 downregulated unique potential candidate EV-miRNA to predict alcohol consumption (Supp. Table 2). As chronic alcohol drinking has been associated with an inflammatory phenotype in circulating immune cells (Lewis et al., 2021, Sureshchandra et al., 2019, Szabo and Saha, 2015), we next asked whether these circulating EV-miRNA could impact the gene expression within PBMC. We compared the list of down- and up-regulated differentially expressed genes (DEG) reported in our previous studies of PBMC (Sureshchandra et al., 2019) to that of the validated target genes of the up- and down-regulated EV-miRNA, respectively (Figure 3A,B). This analysis showed that 334 of the validated targets of upregulated EV-miRNA were previously identified as downregulated DEG in PBMC from alcohol consuming animals (Sureshchandra et al., 2019) (Figure 3A). Functional enrichment of these common DEG enriched to GO terms associated with regulation of TGFβ receptor signaling and chromatin modification (Figure 3C). The 85 validated targets of downregulated EV-miRNA that were shared with DEG previously reported (Sureshchandra et al., 2019) to be upregulated with chronic heavy drinking enriched to apoptotic/DNA damage and coagulation related processes (Figure 3B,C). A majority of the common PBMC DEG are validated targets of miRs-204, 211, 450, 154, 96, 34c, 155, and 124a (Figure 3D).
Figure 3: Candidate EV-miRNA target genes comparison to PBMC gene expression changes in CHD macaques.
A) Venn diagram comparison of target genes from upregulated EV-miRNA with downregulated DEG from PBMC (Sureshchandra et al., 2019). B) Venn diagram comparison of target genes from downregulated EV-miRNA with upregulated DEG from PBMC. C) GO Biological process enrichment of genes in common from (A) and (B) where the X-axis is log10(q-value) or -log10(q-value), respectively. D) miRNet network visualization generated using miRNet of candidate upregulated miRNA (green squares), their validated gene targets from miRTarBase (grey circles), and the validated targets that are common with the downregulated PBMC DEG (blue circles). The size of the miRNA square represents how many validated targets that miRNA has.
To investigate the potential source of these DE EV-miRNA, we compared the up- and downregulated EV-miRNA to differentially expressed cellular miRNA from male and female PBMC with alcohol drinking (Sureshchandra et al., 2019) (Supp. Figure 3B). We identified only 2 upregulated and 1 downregulated miRNA in common, suggesting that the increased or decreased EV-miRNA in circulation do not correlate with miRNA expression changes within PBMC themselves (Supp. Figure 3B).
Upregulated EV-miRNA associated with heightened inflammatory response in PBMC
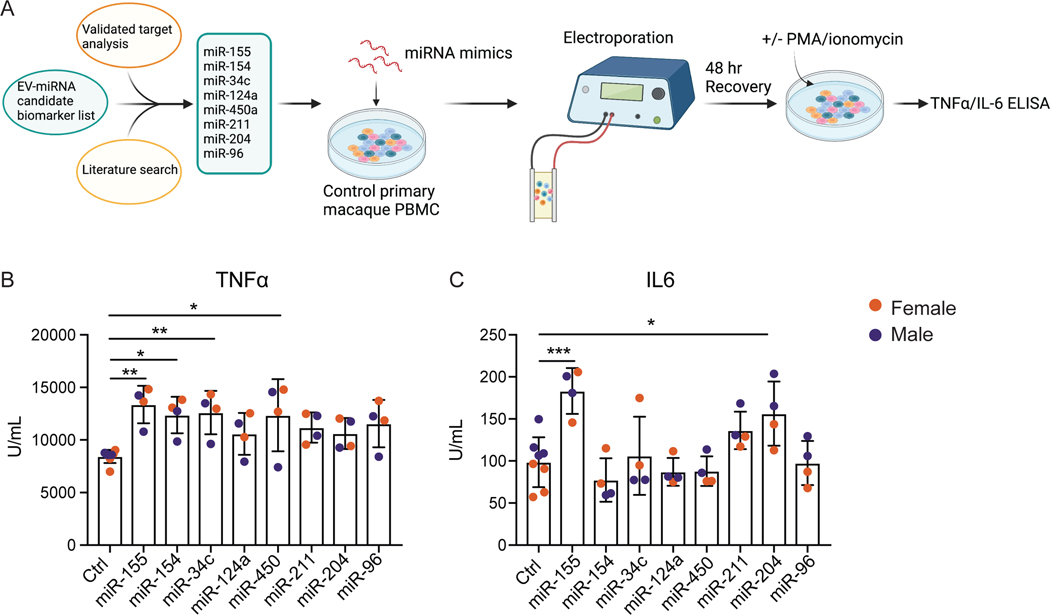
We set out to determine whether circulating EVs from CHD animals could lead to heightened inflammation in the periphery as has been observed in patients with alcohol use disorders and rodent models of alcoholic liver disease (Hong et al., 2002, Karatayli et al., 2019, Miguez et al., 2012). We isolated EVs from control and CHD plasma and added them to PBMC from ethanol-naïve animals followed by stimulation with PMA/ionomycin. The addition of EVs from CHD plasma led to a greater increase in IL-6 levels following PMA stimulation (Supp. Figure 3C). To identify EV-miRNA that may be contributing to this inflammatory phenotype, we selected 8 upregulated EV-miRNA from the candidate list based on their validated target genes and previously studied biological implications (Figure 4A, Figure 3D, and Supp. Table 3). We electroporated PBMC obtained from ethanol-naïve male and female macaques (n=2/sex) with mimics of the 8 selected EV-miRNA (Supp. Figure 3D). We then stimulated the cells with PMA/ionomycin and assessed production of canonical pro-inflammatory cytokines TNFα and IL-6. Electroporation of negative control miRNA alone did not affect the response to stimulation (Supp. Figure 3E). PBMC transfected with miR-155, generated a heightened IL-6 and TNFα response, whereas PBMC transfected with miR-154, miR-34c, or miR450 mimics produced increased amounts of TNFα and those transfected with miR-204 produced increased levels of IL-6 only in response to PMA/Ionomycin stimulation (Figure 4B,C). Finally, to determine whether we could rescue the heightened inflammatory response in CHD PBMC with re-introduction of the downregulated EV-miRNA, we electroporated miR-625 into CHD PBMC followed by PMA/ionomycin stimulation. This miRNA was selected because its validated target genes play a role in signaling and regulation of gene expression. Over-expression of miR-625 slightly reduced IL-6 production in CHD PBMC (Supp. Figure 3F). These results indicate that circulating EV miRNA play an important in modulating immune function with chronic drinking.
Figure 4: Upregulated EV-miRNA associated with heightened inflammatory response in PBMC.
A) Experimental design for miRNA candidate selection and electroporation created using Biorender.com. B,C) PBMC from control macaques were transfected with miRNA mimics and further stimulated with PMA/ionomycin. The production of TNFα (D) and IL-6 (E) was measured from supernatants by ELISA. P-value significance is denoted by *=<0.05, **=<0.01, ***=<0.001.
DISCUSSION
Long term alcohol drinking is a complicating factor for numerous health conditions including infections, cancer, and organ damage (NIAAA, Szabo and Saha, 2015). However, a lack of unbiased methods for measuring alcohol drinking over time makes assessing the impact of chronic ethanol consumption challenging. The first goal of this study was to utilize a rhesus macaque model of voluntary ethanol self-administration to discover candidate extracellular vesicle bound miRNA (EV-miRNA) biomarkers of chronic alcohol drinking. We explored potential candidates of heavy drinking as well as candidates that correlated with average dose of ethanol. In this model, chronic heavy drinking is defined by The Monkey Alcohol Tissue Research Resource (MATRR) as >3 grams of ethanol per kilogram body weight per day. As one standard drink is 14 grams of ethanol, this equates to more than 12 drinks per day in an average 60kg human. Due to known differences in alcohol metabolism and long-term effects of alcohol between males and females, we first examined the EV-miRNA profiles of macaques that engaged in 12 months of heavy drinking separated by sex. This allowed us to identify sex-dependent and independent EV-miRNA. A total of 16 EV-miRNA were upregulated while 26 were downregulated with chronic heavy drinking with only 2 EV-miRNA shared between the two sexes (miR-154 was upregulated and miR-1224 was downregulated). Validated targets of the upregulated EV-miRNA regulated expression of genes involved in pathways associated with blood vessel development as well as cell migration and adhesion. As these EV-miRNA are in circulation, their functional implications could be widespread throughout the body.
The macaque model allows for a variety of drinking patterns and behaviors and therefore we profiled the levels of plasma EV-miRNA as a function of ethanol dose, which led to the identification of an additional 13 miRNA. One of these miRNA, miR-433, was also identified in the CHD-control comparison. Several studies reported a role for miR-433 in cancer cell and hematopoietic stem cell proliferation, migration, and differentiation (Lin et al., 2013, Yang et al., 2013). miR-433 has not been studied in the context of chronic alcohol use, but our data suggest it could be a promising candidate biomarker for alcohol use, particularly in women. Despite the limited overlap, miRNA that correlated with ethanol consumption, like those associated with heavy drinking, also targeted genes involved in blood vessel development and wounding processes. Accordingly, both moderate and heavy alcohol drinking have been linked to hypertension, cardiovascular disease and heart failure in men and women (Djousse and Gaziano, 2008, Malinski et al., 2004, Sesso et al., 2008) and EV-miRNA have been implicated in cardiovascular maintenance and development of disease (Pfeifer et al., 2015). Whether these alcohol-induced EV-miRNA are directly targeting these processes requires further investigation.
As alcohol and its metabolites circulate through the body and act on both peripheral and tissue-resident cell populations, the cellular origins of these EV-miRNA are difficult to parse. To account for peripheral immune cells as a potential source, we compared the differential EV-miRNA to differential miRNA identified in PBMC with alcohol drinking (Sureshchandra et al., 2019). No significant overlap was seen between EV-miRNA and PBMC miRNA, suggesting that the source of the EV-miRNA may be non-immune tissues. However, some miRNA are made in cells and targeted directly for secretion, thus it is still possible these EV-miRNA are produced by PBMC. Previous studies have identified the pancreas, liver, intestine, brain, and heart to have altered miRNA expression with ethanol exposure (Natarajan et al., 2015). Further, studies have identified liver-derived EV- bound miRNA-122 and let-7f as indicators of alcohol-induced liver injury (Odegaard et al., 2020). While it is known that miRNA packaging in EVs is selective and can have targeted effects on recipient cells, many of the functional mechanisms and downstream impacts of these processes remain elusive (Groot and Lee, 2020, Valadi et al., 2007a).
We and others have shown chronic alcohol drinking to alter the function of peripheral immune cells, poising them towards a hyper-inflammatory response (Lewis et al., 2021, Sureshchandra et al., 2019, Szabo and Saha, 2015). We sought to determine whether the EV-miRNA differentially expressed with ethanol consumption could modulate response of circulating immune cells to stimulus. We first compared the validated gene targets of the EV-miRNA with the DEG identified from PBMC obtained from some of the same animals (Sureshchandra et al., 2019). We identified a substantial number of downregulated genes in PBMC that were validated targets of upregulated EV-miRNA. We narrowed our focus to 8 potential candidate EV-miRNA that were either upregulated with heavy drinking or positively correlated with daily ethanol consumption dose and targeted significant numbers of genes that were downregulated with chronic drinking in PBMC and played a role in inflammation/tissue repair (miR-155, miR-154, miR-34c, miR-124, miR-450, miR-211, miR-204, and miR-96). Overexpression of these miRNA led to enhanced inflammatory response following PMA stimulation. The mechanisms by which these miRNAs modulate production of IL-6 and TNFα could be occurring through either a direct inhibition of NFκB regulators or via an indirect/secondary target. For instance, a number of genes targeted by these miRNAs are involved in TNFα production and signaling pathways (ex. STAT3, C5AR2, THBS1). More specifically, miR-155 has previously been shown to increase inflammatory responses through direct targeting of NFKB pathway regulators (BCL6) and pro-inflammatory macrophage activation (Alivernini et al., 2017, Bala et al., 2017). The miR-34 family members have been linked to inflammation in the skin and liver and have been associated with non-alcoholic fatty liver disease (Li et al., 2021, Torres et al., 2018, Wu et al., 2020). While we electroporated the mimics and further stimulated total PBMC, previous studies on the disproportionate effects of alcohol on innate immune cells leads us to hypothesize that this heightened expression of TNFα and IL-6 is due to changes in monocytes with the EV-miRNA. Increased expression of these miRNA in EV could indicate early signs of alcohol-induced organ damage prior to increased levels of AST or ALT and could lead to aberrant hyper-activation of monocytes in the periphery (Momen-Heravi et al., 2015, Natarajan et al., 2015).
In summary, we comprehensively profiled the blood plasma of macaques to find potential EV-miRNA biomarkers of long-term alcohol drinking and provide a mechanism of alcohol-associated inflammation in peripheral immune cells. Our exploratory examination led to the discovery of potential candidate biomarkers of both heavy drinking and those that increase with ethanol dose in macaques. To validate these candidates as true biomarkers, a confirmatory study would need to be performed in a new cohort of animals and then further in humans. While miRNA are generally well-conserved across species, the differences between the EV-miRNA in macaques and humans has yet to be investigated.
Supplementary Material
Acknowledgements
We are grateful to the members of the Grant laboratory for expert animal care and sample procurement. We thank Dr. Jennifer Atwood for assistance with sorting in the flow cytometry core at the Institute for Immunology, UCI. We thank Dr. Melanie Oakes from UCI Genomics and High-Throughput Facility for assistance with 10X library preparation and sequencing. We thank Dmitry Fishman for assistance with the NTA Assay and Bridget Ratitong for running the assay.
Funding
This study was supported by NIH 1R21AA025839–01A1 (Messaoudi), 5U01AA013510–20 (Grant), and 2R24AA019431–11 (Grant). S.A.L is supported by NIH 1F31A028704–01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests
No competing interests reported.
Data availability
The datasets supporting the conclusions of this article are available on NCBI’s Sequence Read Archive (SRA# PRJNA769716).
REFERENCES
- ALIVERNINI S, GREMESE E, MCSHARRY C, TOLUSSO B, FERRACCIOLI G, MCINNES IB & KUROWSKA-STOLARSKA M. 2017. MicroRNA-155-at the Critical Interface of Innate and Adaptive Immunity in Arthritis. Front Immunol, 8, 1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANTTILA P, JÄRVI K, LATVALA J. & NIEMELÄ O. 2004. Method-dependent characteristics of carbohydrate-deficient transferrin measurements in the follow-up of alcoholics. Alcohol Alcohol, 39, 59–63. [DOI] [PubMed] [Google Scholar]
- ASQUITH M, PASALA S, ENGELMANN F, HABERTHUR K, MEYER C, PARK B, GRANT KA & MESSAOUDI I. 2014. Chronic ethanol consumption modulates growth factor release, mucosal cytokine production, and microRNA expression in nonhuman primates. Alcohol Clin Exp Res, 38, 980–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAKER EJ, FARRO J, GONZALES S, HELMS C. & GRANT KA 2014. Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcohol Clin Exp Res, 38, 2835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALA S, CSAK T, KODYS K, CATALANO D, AMBADE A, FURI I, LOWE P, CHO Y, IRACHETA-VELLVE A. & SZABO G. 2017. Alcohol-induced miR-155 and HDAC11 inhibit negative regulators of the TLR4 pathway and lead to increased LPS responsiveness of Kupffer cells in alcoholic liver disease. J Leukoc Biol, 102, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARR T, GIRKE T, SURESHCHANDRA S, NGUYEN C, GRANT K. & MESSAOUDI I. 2016. Alcohol Consumption Modulates Host Defense in Rhesus Macaques by Altering Gene Expression in Circulating Leukocytes. J Immunol, 196, 182–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTEL DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–97. [DOI] [PubMed] [Google Scholar]
- BAUM MK, RAFIE C, LAI S, SALES S, PAGE JB & CAMPA A. 2010. Alcohol use accelerates HIV disease progression. AIDS Res Hum Retroviruses, 26, 511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHATTACHARYA R. & SHUHART MC 2003. Hepatitis C and alcohol: interactions, outcomes, and implications. J Clin Gastroenterol, 36, 242–52. [DOI] [PubMed] [Google Scholar]
- CHANG L, ZHOU G, SOUFAN O. & XIA J. 2020. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res, 48, W244–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONIGRAVE KM, DEGENHARDT LJ, WHITFIELD JB, SAUNDERS JB, HELANDER A, TABAKOFF B. & GROUP WIS 2002. CDT, GGT, and AST as markers of alcohol use: the WHO/ISBRA collaborative project. Alcohol Clin Exp Res, 26, 332–9. [PubMed] [Google Scholar]
- DJOUSSE L. & GAZIANO JM 2008. Alcohol consumption and heart failure: a systematic review. Curr Atheroscler Rep, 10, 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEDIRKO V, TRAMACERE I, BAGNARDI V, ROTA M, SCOTTI L, ISLAMI F, NEGRI E, STRAIF K, ROMIEU I, LA VECCHIA C, BOFFETTA P. & JENAB M. 2011. Alcohol drinking and colorectal cancer risk: an overall and dose-response meta-analysis of published studies. Ann Oncol, 22, 1958–72. [DOI] [PubMed] [Google Scholar]
- FERRACIN M, LUPINI L, SALAMON I, SACCENTI E, ZANZI MV, ROCCHI A, DA ROS L, ZAGATTI B, MUSA G, BASSI C, MANGOLINI A, CAVALLESCO G, FRASSOLDATI A, VOLPATO S, CARCOFORO P, HOLLINGSWORTH AB & NEGRINI M. 2015. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget, 6, 14545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANT KA, LENG X, GREEN HL, SZELIGA KT, ROGERS LS & GONZALES SW 2008. Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol Clin Exp Res, 32, 1824–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREWAL P. & VISWANATHEN VA 2012. Liver cancer and alcohol. Clin Liver Dis, 16, 839–50. [DOI] [PubMed] [Google Scholar]
- GROOT M. & LEE H. 2020. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG F, KIM WH, TIAN Z, JARUGA B, ISHAC E, SHEN X. & GAO B. 2002. Elevated interleukin-6 during ethanol consumption acts as a potential endogenous protective cytokine against ethanol-induced apoptosis in the liver: involvement of induction of Bcl-2 and Bcl-x(L) proteins. Oncogene, 21, 32–43. [DOI] [PubMed] [Google Scholar]
- HUANG HY, LIN YC, LI J, HUANG KY, SHRESTHA S, HONG HC, TANG Y, CHEN YG, JIN CN, YU Y, XU JT, LI YM, CAI XX, ZHOU ZY, CHEN XH, PEI YY, HU L, SU JJ, CUI SD, WANG F, XIE YY, DING SY, LUO MF, CHOU CH, CHANG NW, CHEN KW, CHENG YH, WAN XH, HSU WL, LEE TY, WEI FX & HUANG HD 2020. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res, 48, D148–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG X, YUAN T, TSCHANNEN M, SUN Z, JACOB H, DU M, LIANG M, DITTMAR RL, LIU Y, KOHLI M, THIBODEAU SN, BOARDMAN L. & WANG L. 2013. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics, 14, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUDOLIN V. 1975. Tuberculosis and alcoholism. Ann N Y Acad Sci, 252, 353–64. [DOI] [PubMed] [Google Scholar]
- JIA S, ZOCCO D, SAMUELS ML, CHOU MF, CHAMMAS R, SKOG J, ZAROVNI N, MOMEN-HERAVI F. & KUO WP 2014. Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev Mol Diagn, 14, 307–21. [DOI] [PubMed] [Google Scholar]
- JIMENEZ VA, HELMS CM, CORNEA A, MESHUL CK & GRANT KA 2015. An ultrastructural analysis of the effects of ethanol self-administration on the hypothalamic paraventricular nucleus in rhesus macaques. Front Cell Neurosci, 9, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARATAYLI E, HALL RA, WEBER SN, DOOLEY S. & LAMMERT F. 2019. Effect of alcohol on the interleukin 6-mediated inflammatory response in a new mouse model of acute-on-chronic liver injury. Biochim Biophys Acta Mol Basis Dis, 1865, 298–307. [DOI] [PubMed] [Google Scholar]
- KLINE SE, HEDEMARK LL & DAVIES SF 1995. Outbreak of tuberculosis among regular patrons of a neighborhood bar. N Engl J Med, 333, 222–7. [DOI] [PubMed] [Google Scholar]
- LANGMEAD B. & SALZBERG SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS SA, SURESHCHANDRA S, DORATT B, JIMENEZ VA, STULL C, GRANT KA & MESSAOUDI I. 2021. Transcriptional, Epigenetic, and Functional Reprogramming of Monocytes From Non-Human Primates Following Chronic Alcohol Drinking. Front Immunol, 12, 724015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI B, LIU J, XIN X, ZHANG L, ZHOU J, XIA C, ZHU W. & YU H. 2021. MiR-34c promotes hepatic stellate cell activation and Liver Fibrogenesis by suppressing ACSL1 expression. Int J Med Sci, 18, 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN X, RICE KL, BUZZAI M, HEXNER E, COSTA FF, KILPIVAARA O, MULLALLY A, SOARES MB, EBERT BL, LEVINE R. & LICHT JD 2013. miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia, 27, 344–52. [DOI] [PubMed] [Google Scholar]
- MALINSKI MK, SESSO HD, LOPEZ-JIMENEZ F, BURING JE & GAZIANO JM 2004. Alcohol consumption and cardiovascular disease mortality in hypertensive men. Arch Intern Med, 164, 623–8. [DOI] [PubMed] [Google Scholar]
- MESSAOUDI I, ASQUITH M, ENGELMANN F, PARK B, BROWN M, RAU A, SHAW J. & GRANT KA 2013. Moderate alcohol consumption enhances vaccine-induced responses in rhesus macaques. Vaccine, 32, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIGUEZ MJ, ROSENBERG R, BURBANO-LEVY X, CARMONA T. & MALOW R. 2012. The effect of alcohol use on IL-6 responses across different racial/ethnic groups. Future Virol, 7, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOMEN-HERAVI F, BALA S, KODYS K. & SZABO G. 2015. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep, 5, 9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNDLE G, MUNKES J, ACKERMANN K. & MANN K. 2000. Sex differences of carbohydrate-deficient transferrin, gamma-glutamyltransferase, and mean corpuscular volume in alcohol-dependent patients. Alcohol Clin Exp Res, 24, 1400–5. [PubMed] [Google Scholar]
- NATARAJAN SK, PACHUNKA JM & MOTT JL 2015. Role of microRNAs in Alcohol-Induced Multi-Organ Injury. Biomolecules, 5, 3309–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAAA Alcohol facts and statistics. National Institute of Health.
- ODEGAARD KE, CHAND S, WHEELER S, TIWARI S, FLORES A, HERNANDEZ J, SAVINE M, GOWEN A, PENDYALA G. & YELAMANCHILI SV 2020. Role of Extracellular Vesicles in Substance Abuse and HIV-Related Neurological Pathologies. Int J Mol Sci, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PFEIFER P, WERNER N. & JANSEN F. 2015. Role and Function of MicroRNAs in Extracellular Vesicles in Cardiovascular Biology. Biomed Res Int, 2015, 161393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, MCCARTHY DJ & SMYTH GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAHA B, MOMEN-HERAVI F, KODYS K. & SZABO G. 2016. MicroRNA Cargo of Extracellular Vesicles from Alcohol-exposed Monocytes Signals Naive Monocytes to Differentiate into M2 Macrophages. J Biol Chem, 291, 149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITZ R, GHALI WA & MOSKOWITZ MA 1997. The impact of alcohol-related diagnoses on pneumonia outcomes. Arch Intern Med, 157, 1446–52. [PubMed] [Google Scholar]
- SESSO HD, COOK NR, BURING JE, MANSON JE & GAZIANO JM 2008. Alcohol consumption and the risk of hypertension in women and men. Hypertension, 51, 1080–7. [DOI] [PubMed] [Google Scholar]
- SURESHCHANDRA S, RAIS M, STULL C, GRANT K. & MESSAOUDI I. 2016. Transcriptome Profiling Reveals Disruption of Innate Immunity in Chronic Heavy Ethanol Consuming Female Rhesus Macaques. PLoS One, 11, e0159295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SURESHCHANDRA S, RAUS A, JANKEEL A, LIGH BJK, WALTER NAR, NEWMAN N, GRANT KA & MESSAOUDI I. 2019. Dose-dependent effects of chronic alcohol drinking on peripheral immune responses. Sci Rep, 9, 7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZABO G. & BALA S. 2010. Alcoholic liver disease and the gut-liver axis. World J Gastroenterol, 16, 1321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZABO G. & SAHA B. 2015. Alcohol’s Effect on Host Defense. Alcohol Res, 37, 159–70. [PMC free article] [PubMed] [Google Scholar]
- TORRES JL, NOVO-VELEIRO I, MANZANEDO L, ALVELA-SUÁREZ L, MACÍAS R, LASO FJ & MARCOS M. 2018. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J Gastroenterol, 24, 4104–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VALADI H, EKSTROM K, BOSSIOS A, SJOSTRAND M, LEE JJ & LOTVALL JO 2007a. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 9, 654–9. [DOI] [PubMed] [Google Scholar]
- VALADI H, EKSTRÖM K, BOSSIOS A, SJÖSTRAND M, LEE JJ & LÖTVALL JO 2007b. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 9, 654–9. [DOI] [PubMed] [Google Scholar]
- VIEL G, BOSCOLO-BERTO R, CECCHETTO G, FAIS P, NALESSO A. & FERRARA SD 2012. Phosphatidylethanol in blood as a marker of chronic alcohol use: a systematic review and meta-analysis. Int J Mol Sci, 13, 14788–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU J, LI X, LI D, REN X, LI, HERTER EK, QIAN M, TOMA MA, WINTLER AM, SEREZAL IG, ROLLMAN O, STAHLE M, WIKSTROM JD, YE X. & LANDEN NX 2020. MicroRNA-34 Family Enhances Wound Inflammation by Targeting LGR4. J Invest Dermatol, 140, 465–476 e11. [DOI] [PubMed] [Google Scholar]
- YANG Z, TSUCHIYA H, ZHANG Y, HARTNETT ME & WANG L. 2013. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem, 288, 28893–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, JIA Y, ZHENG R, GUO Y, WANG Y, GUO H, FEI M. & SUN S. 2010. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem, 56, 1830–8. [DOI] [PubMed] [Google Scholar]
- ZHOU Y, ZHOU B, PACHE L, CHANG M, KHODABAKHSHI AH, TANASEICHUK O, BENNER C. & CHANDA SK 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun, 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.