Abstract
Triple-negative breast cancer (TNBC) is an aggressive, metastatic/invasive sub-class of breast cancer (BCa). Cell surface protein-derived multi-epitope vaccine-mediated targeting of TNBC cells could be a better strategy against the disease. Literature-based identified potential cell surface markers for TNBC cells were subjected to expression pattern and survival analysis in BCa patient sample using TCGA database. The cytotoxic and helper T-lymphocytes antigenic epitopes in the test proteins were identified, selected and fused together with the appropriate linkers and an adjuvant, to construct the multi-epitope vaccine (MEV). The immune profile, physiochemical property (PP) and world population coverage of the MEV was studied. Immune simulation, cloning in a suitable vector, molecular docking (against Toll-like receptors, MHC (I/II) molecules), and molecular dynamics simulations of the MEV was performed. Cell surface markers were differentially expressed in TNBC samples and showed poor survival in TNBC patients. Satisfactory PP and WPC (up to 89 and 99%) was observed. MEV significant stable binding with the immune molecules and induced the immune cells in silico. The designed vaccine has capability to elicit immune response which could be utilized to target TNBC alone/combination with other therapy. The experimental studies are required to check the efficacy of the vaccine.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-022-03140-3.
Keywords: Triple-negative breast cancer, Multi-epitope vaccine, Immune simulation, Molecular docking, Molecular dynamics simulation
Introduction
Triple-negative breast cancer (TNBC) is characteristically devoid of estrogen/progesterone and HER2 receptors. TNBC accounts about 20% of the diagnosed breast cancer patients and occurs mostly in younger women. It is an aggressive phenotype of breast cancer and generally shows chemotherapy resistance, low overall and disease-free survival, and relapse in patients. TNBC is hard to manage clinically as after diagnosis it showed higher tendency for the recurrence at about 3 years (Kwon et al. 2017; Cîmpean et al. 2017). All these factors results in poor therapy outcome in TNBC. Moreover, the oncogenic potential of a single molecular alteration has not yet been proven clinically to be associated with the pathophysiology of TNBC subtypes. Due to its molecular heterogeneity, the established therapeutic target for the disease is not available till date. Recent advances in bioinformatics data analysis as well as molecular techniques allowed us to identify the therapeutic targets of clinical relevance. Cell surface markers targeted therapeutics has been found successful against breast cancer. The studies are in progress to develop the candidate therapeutic to target the cell surface markers-mediated toxicity in TNBC (Cubas et al. 2011; Tursynbay et al. 2016; Turdo et al. 2016; Hassan et al. 2016; M-Rabet et al. 2017; Quintero et al. 2017; Ling et al. 2017; Wan et al. 2021). Tumor antigens having potential to induce cytotoxic T lymphocytes play crucial role in the development of anti-cancer vaccine. Tumor-related antigenic epitopes are known to stimulate T cells which allow us to target/eliminate tumor cells by anti-tumor vaccine and are currently in clinical practice. Thus, it is the urgent need of time to develop the potential candidate vaccine which targets the differentially expressed surface proteins on TNBC cells in comparison to normal and/or other tissue—a designed multi-epitope-based peptide vaccine in a series of epitopes having adjuvant and linker sequences. These epitopes have the ability to elicit CTL, Th, and B cells and thereby produce potential cellular and humoral immune responses. The vaccine is processed in the cytoplasm of antigen presenting cells (APCs) and released free immunogenic epitopes, recognized by MHC class I and MHC class II molecules present in APCs. MHC class I receptor activates the cytotoxic T cells, similarly, MHC class II receptor stimulates the T helper cells. Activated T cells produce cytokines including tumor necrosis factors which stimulates their division into respective clones. These cells also differentiate into memory cells and kill the cancer cells possessing the antigen signature. The released cytokines activate the B cells which differentiated into plasma and memory B cells and perform the antitumor activity by producing antibodies and complement mediated cytotoxicity (Zhang 2018). Recently, Zhang (2018) reviewed the multi-epitope vaccine design and their characteristic advantage over classical/single epitope vaccine. Now a days computer-based vaccine design is a popular strategy to find and develop candidate vaccines against cancer. Publically available data on proteomics and genomics of cancer patients allow us to retrieve valuable information for vaccine design. Using online tools/databases one can predict T/B-cell epitopes and their immunological profile. Furthermore, the tools generally used in computer-based drug discovery such as molecular docking and molecular dynamics simulation allow us to study the vaccine–immune molecule interaction, stability, and energetics. Moreover, the availability of in silico immune simulation and vaccine cloning platforms allows us to predict these complicated biological events using advanced computation. The overall integration of imuno-informatics, bioinformatics and computer-based drug discovery tools greatly accelerated the development of anti-cancer candidate vaccines (Kardani et al. 2020)
NECTIN4 or PVLR4 is a tumor-associated antigen (TAA) and act as cell adhesion molecule found in different cancer including breast. Recently, it has been reported that about 60% TNBC sample showed the over-expressed NECTIN4 protein and was absent in normal breast tissue. Also, worse prognosis has been found associated with the NECTIN4 expression in TNBC patients (M-Rabet et al. 2017). PIM1 (proviral integration site for Moloney murine leukemia virus-1) is a Ser/Thr kinase and proto-oncogene which regulates various hallmarks of the cancer such as cellular proliferation, cell cycle, and apoptosis. It has been shown that PIM1 increased copy number and expression levels are significantly associated with the TNBC in comparison to non-TNBC. Targeting PIM1 with the monoclonal antibody showed decreased cell growth, induced apoptosis, and decreased drug resistance in cancer pre-clinical models (Tursynbay et al. 2016). Mesothelin (MTN) is a membrane protein significantly highly expressed in TNBC and recently found to be associated with the decreased disease-free survival and increased metastasis. The protein express only on mesothelial cells, but its increased expression has been identified in TNBC and significantly correlated with the poor survival and aggressiveness of the disease (Hassan et al. 2016). Guanylate-binding protein-1 (GBP1) has been found to be up-expressed in breast cancer and mediates its effect by promoting cellular proliferation, increased metastasis and invasion, and glycolysis in cancer cells (Wan et al. 2021). Recently, Quintero et al. reported GBP1 as a therapeutic target in the TNBC sample in comparison to non-TNBC cell line. Matrix metalloproteinase-14 (MMP-14) a cell surface protein required to form invadopodia which degrades the ECM during invasion process and increase the metastasis (Quintero et al. 2017). Monoclonal antibody-mediated immune-targeting of MMP-14 decreases hypoxia, suppresses immune system, and inhibits metastasis in experimental TNBC models (Ling et al. 2017). CUB domain containing protein 1 (CDCP1) is a membrane-bound protein whose up-expression results in loss of anchorage and thereby increased metastasis/invasion in TNBC (Turdo et al. 2016). TROP-2 a cell surface glycoprotein is up-express in various solid cancers and in comparison to low levels in the normal tissues. In Phase I and II clinical trials the monoclonal antibody-mediated targeting of TROP-2 showed positive therapeutic efficacy in TNBC patients (Cubas et al. 2011). The present study was aimed to identify the antigenic epitopes in TNBC cell surface markers to design the multi-epitope vaccine against TNBC using integrated bioinformatics approach.
Materials and methods
Literature search for target proteins
To design the multi-epitope vaccine against TNBC, we searched the target by considering the two principal components of vaccine against particular type of cancer: (1) selected protein should highly differentially over-expressed in cancer patient samples in comparison to matched control samples; (2) should expressed on the membrane of the TNBC cell surface. Keeping these things in our mind, we have searched the literature using various search engines namely, Pubmed, Google Scholar, Web of Sciences and Scopus database, etc.
Differential expression analysis using TCGA database
Protein encoding genes of the identified proteins were subjected to their differential expression profiling in TNBC samples in comparison to normal samples of breast invasive carcinoma data set of TCGA database. For this, UALCAN interactive web tool (Chandrashekhar et al. 2017). was utilized that provides a comprehensive interactive web services for analyzing the cancer OMICS data present in various databases such as TCGA, MET500 and CPTAC.
Survival analysis of the test proteins
Further the test proteins were subjected to their overall survival analysis in TNBC patients. The survival effects of the proteins in TNBC patients of BRCA data set of TCGA database was analyzed using Kaplan–Meier Plotter Database (KMPD) survival tool that is capable to assess the association of proteins with the OS (overall survival) of Breast cancer (BCa) patients and it subtypes (Nagy et al. 2021). OS analysis of the test genes was performed by submitting the gene symbol of each protein into Kaplan–Meier Plotter Database (KMPD) survival tool. TNBC samples of TCGA database were selected for the study. The tool generates the overall survival association graph of high and low expression levels of the query gene in a given set of clinical sample with respect to time (in months). The graph also indicates the hazard ratio (HR) and statistical significance (p) values for the calculation.
Sequence retrieval
To extract the multi-epitopes for vaccine construction from the test protein, first of all the amino acids sequence of the identified proteins were retrieved from the UniProtKB protein database (https://www.uniprot.org/) in FASTA format.
Immunoinformatics analysis
MHC class I and II epitopes prediction
Antigenic epitopes prediction is a critical step in the vaccine designing, because the CTL and HTL both the cells play major role in the induction of immune response against any foreign antigen. Both T-cell populations are stimulated by the binding of antigen to their respective MHC molecules. Therefore, it is important to identify the antigenic fragment that can bind with the MHC molecule and induce the immune response. For this we used the IEDB server which contains vigorous tools to predict the MHC class I and II binding epitopes. IEDB server hosts a different immunoinformatics tools and provide various methods for epitope prediction like average relative binding (ARB), artificial neural network (ANN), SMM with a peptide: MHC binding energy covariance matrix (SMMPMBEC), and stabilized matrix method (SMM). We used an IEDB recommended method NetMHCpan EL 4.1 to predict the epitopes for both the MHC molecules using a criteria IC50 less than or equal to 50 nM (http://tools.iedb.org/tepitool/).
Antigenicity prediction of epitopes and vaccine construct
Selected epitopes and the vaccine construct should also be antigenic in nature. Therefore, prior to construct the vaccine each selected epitope was submitted to vaxiJen v2.0 online server to predict the antigenic score (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). VaxiJen server has highest accuracy at the threshold of 0.5 but we sorted only those epitopes who showed a threshold value of more than or equal to 1.0. Further, we also predicted the antigenicity value of the final vaccine construct using the same server.
Multi-epitope vaccine construction
Selected CTL and HTL epitopes those were passed from above prediction steps were used in the final vaccine construction. Final vaccine construct was prepared by connecting the CTL and HTL epitopes together with the help of alanine–alanine–tyrosine (AAY) and glycine–proline rich, GPGPG linkers. Additionally, we used the GM-CSF as adjuvant and helper sequence to enhance the vaccine immunogenicity for long lasting in TNBC patients. GM-CSF adjuvant was attached with the N-terminal end of vaccine using EAAAK linker. Constructed vaccine sequence was also flanked by the padre sequence.
B-cell epitopes prediction
ElliPro online tool of IEDB server (http://tools.iedb.org/ellipro) was used to obtain linear (continuous) and discontinuous epitopes at default parameters setting. To predict the B-cell epitopes, PDB file of final vaccine construct was uploaded in the ElliPro online tool. ElliPro predicts the amino acids sequences of protein that are expected to be recognized as epitopes in the context of B-cell response. Ellipro used a method to predict the epitopes on the basis of solvent-accessibility, protein structural protrusion and flexibility.
Allergenicity prediction
Amino acids sequence of final multiepitopes vaccine was submitted to online web server AlgPred 2.0 to predict the allergenicity of the vaccine (Sharma et al. 2020). Allergenicity prediction of vaccine was performed on the basis of similarity of the query sequence to known allergen protein and known validated antibody IgE Ab epitopes. AlgPred 2.0 web server is designed to predict the combined allergenic score of a protein by using different approaches namely, Basic Local Alignment Search Tool (BLAST), IgE epitopes and motif-based approaches.
Characterizations of immune profile of the multi-epitope vaccine construct
To induce the whole immune response efficiently against cancer cells, it is very important to determine the immune profile of the constructed vaccine using immune simulation approach. Therefore, we determined the immune profile of final multi-epitope vaccine by submitting the vaccine construct sequence to C-ImmSim 10.1 web server (Rapin et al. 2010). C-ImmSim functioning is based on the hypothesis of various immune system specific elements like, antigen processing and presentation to CTL and HTL, cell to cell cooperation, B-cell and T-cell maturation, response and memory cell formation, clonal selection by antigen affinity, clonal deletion theory, hypermutation of the antibodies, the T-cell replicative senescence and anergy in B and T lymphocytes, etc.
Population coverage
After all the immunoinformatics analysis successfully completed, we determined the population coverage score of the designed multi-epitope vaccine construct using the IEDB population coverage analysis in order to check the that our construct effectively covers the entire world population or not by using HLA class I/II alleles of the different population in the world.
Bioinformatics analyses of multi-epitopes vaccine construct
Two-dimensional and three-dimensional structures prediction of multi-epitope vaccine construct
The two-dimensional (2D) and three-dimensional (3D) structures of the multi-epitope vaccine construct were generated using PSIPRED 4.0 tool of PSIPRED workbench web server (http://bioinf.cs.ucl.ac.uk/psipred/) and RosettaCM online computational protein design software, respectively (Song et al. 2013). PSIPRED server uses the PSIBLAST to generate the sequence profile and normalize it and then predict the secondary structure using neural networking. RosettaCM predicts the tertiary structure of a given sequence by assembling the topologies of aligned segments in Cartesian space and build the unaligned regions de novo in torsion space. It retained the most representative low-energy model by doing the energy optimization of predicted structure by all-atom refinement. RosettaCM generates the model with more favorable side-chain and backbone conformations.
Refinements of tertiary structure of the MEV construct
The 3D model obtained by RosettaCM online computational protein design software was also subjected to the refinement procedure. To refine the obtained 3D structure, GalaxyRefine (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) online server was utilized. GalaxyRefine applies a refinement method based on the establishment and repacking of the side chains. As a result, the refinement achieves overall relaxed structure by using dynamic simulations.
Tertiary structure model validation
PRO-CHECK, ERRAT, and ProSA-web tool were used to assess the validation of tertiary structure of multi-epitope vaccine construct. PROCHECK provides a Ramachandran plot of residues based on stereochemical qualities of a protein model and predict the backbone conformation on the basis of Psi/Phi angles in the Ramachandran plot. ERRAT verifies the tertiary structure of a given protein using the statistics of the non-bonded atom–atom interactions in compression to a suitable high-resolution crystallography structures present in the database.
Analysis of physiochemical properties of final multi-epitope vaccine construct
Further multi-epitope vaccine construct was subjected to determine its physicochemical properties like amino acids sequence length, molecular weight, aliphatic index, instability index, half-life, hydropathicity and solubility using ProtParam web tool (Wilkins et al. 1999). The ProtParam works on the basis of pK value of each amino acid in a given protein. It predicts the stability, half-life and hydropathicity of a given protein using instability index (if instability is < 40 considered as stable), ‘N-end rule’ (amino acids at N-terminal utilized for protein degradation determination) and aliphatic index (volume occupied by aliphatic side chains, along with hydrophobic amino acids like alanine, valine, leucine, and isoleucine), respectively.
Molecular docking of multi-epitope vaccine construct with Tlrs and MHC class I and II molecules
The binding affinity of the designed multi-epitope vaccine construct with specific immune receptors MHC class I (HLA-A/B/C) and MHC class II (DRB1, DQA1 and DQB1) and TLRs (TLR2 and TLR4) was checked using molecular docking approach. For this purpose, an online HDOCK server was utilized.
Molecular dynamics (MD) simulation
The molecular dynamics (MD) simulation of the vaccine construct was performed using GROMACS 2020.4 package (Lemkul 2019; Lindahl et al. 2020). GROMOS 54A7 force field and single point charge (SPC) water model was used to perform MD simulation. System was solvated inside a cubic box with a buffer distance of 1 Å. Following solvation, appropriate numbers of counter ions were added in order to neutralize the system. Energy minimization was produced using 5000 steps of steepest descent method. Furthermore, system was equilibrated using, 100 ps for NVT (at 300 K) and 100 ps for NPT (at 1 bar). This energy minimized and equilibrated system was used to compute trajectories of 10,000 ps. For temperature and pressure coupling, V-Rescale and Parrinello–Rahman method was applied, respectively (Parrinello and Rahman 1981). For bond length conservation LINCS algorithm was used (Bussi et al. 2007; Hess et al. 1997). Analysis of computed trajectories was performed using various GROMACS analysis tools (Gupta et al. 2020; Kushwaha et al. 2021; Singh et al. 2021).
Reverse translation, codon optimization and computer-based cloning of the MEV
Designed multi-epitope vaccine was under taken to codon optimization to express in large quantity in suitable expression vector because incorrect mRNA structure and more GC content reduce the translation rate of protein biosynthesis. Therefore, codon optimization of vaccine construct is essential to get the large expression. For codon optimization as well as reverse translation, Codon Adaptation Tool (JCat) server 31 (https://www.jcat.de/) was used. JCat server 31 predicts the mRNA secondary structure and calculates the codon optimization index (CIA) score and GC content and reverse translated multi-epitope vaccine amino acids sequence into complementary DNA sequence. CIA score and percentage of GC content was used as indicative of protein translation of vaccine construct. The codon optimized MEV sequence was introduced into the pET-30a (+) vector using SnapGene tool and expressed in E. coli strain K12.
Results and discussion
Identification of candidate TNBC antigenic protein, expression pattern and survival analysis
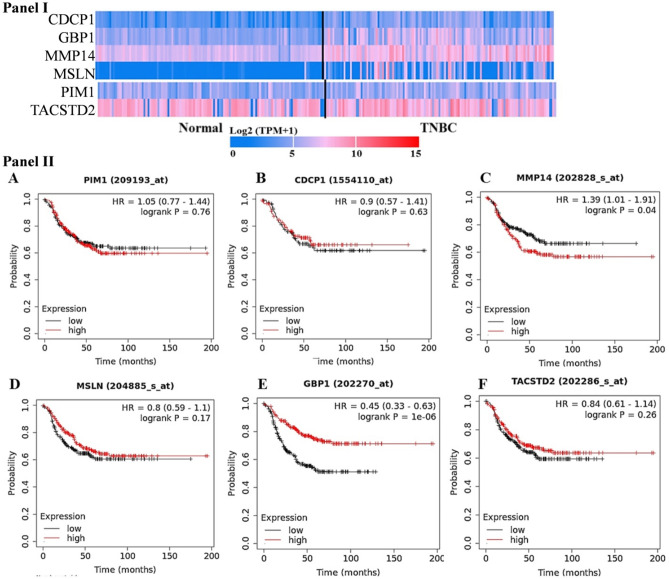
In the present study, we utilized the integrated immunoinformatics principle to design a candidate vaccine against the triple-negative breast cancer. For this, an exhaustive literature was searched to identify the potential therapeutic cell surface proteins expressed on the cell membrane of TNBC cells. The immune-based targeting of the cancer cell requires that the candidate antigenic protein should be present at the cancer cell surface and their expression should be higher in cancer cells with little or low expression in the normal cells (Jiang et al. 2019). We keep these facts in our mind during identification of the candidate proteins for the development of TNBC vaccine. The pre-clinical and clinical research on TNBC cell surface proteins was searched to get the potential markers for the vaccine design. We identified CDCP1, GBP1, MMP14, MSLN, NECTIN4, PIM1 (serine/threonine protein kinase PIM1) and TROP2 (tumor-associated calcium signal transducer 2) proteins as candidate markers for the present study. The gene expression profile of the target proteins were studied by using TCGA database which showed the significant over expression in TNBC samples in comparison to normal tissue (Fig. 1, panel I). Some of the proteins (GBP1, MMP14 and MSLN) showed significant higher expression in the TNBC sample in comparison to normal tissue. We did not find the expression pattern of NECTIN4 in ULCAN database, but the published literature showed its higher expression in TNBC patients (M-Rabet et al. 2017). It has been reported that the cancer related deaths are mostly associated with the relapse of the disease (Riggio et al. 2021). On the basis of this fact we further explored the association of the selected proteins with the survival in TNBC patients. For this, the survival analysis of the TNBC patient was performed using the clinical data present in the TCGA database and the results are shown in (Fig. 1, panel II). The survival analysis of the PIM1 and MMP14 proteins showed the hazard ratio (HR) > 1. Moreover, CDCP1, MSLN, and TROP2 proteins showed HR value more than 0.8. The results of expression pattern (normal versus TNBC samples) and the survival analysis of the test proteins showed their potentiality for candidate protein for the development of vaccine against TNBC.
Fig. 1.
Expression profile of target gene in normal and triple-negative breast cancer patient. Data were curated from ULCAN and TCGA database. The official symbol of the selected protein was submitted in the search options of the ULCAN tool utilizing TCGA dataset. The heatmap for the query was generated by the tool. NECTIN4 expression pattern was not found in the database (Panel I). Panel II shows overall survival (OS) analysis of the target gene in normal and triple-negative breast cancer patient. Data were curated from TCGA database (Panel II). NECTIN4 was not found in the database. OS of the test proteins in TNBC patients was performed by submitting the gene symbol of each protein into Kaplan–Meier Plotter Database (KMPD) survival tool of TCGA database. In the figure, the red and black line represents (high and low expression pattern, respectively) overall survival probability across the 200 months’ time in triple-negative breast cancer patients. HR and log rank P indicate the hazard ratio and association significance, respectively
Identification of potential CTL epitopes
The antigenic peptide sequence binds to the MHC molecules and appears on the cell surface of the cancer/infected cells. The CTLs recognize the peptide–protein complex and kills the cells. Thus, the CTL epitopes rendered as a potential candidate in the scope of in silico cancer vaccine design. The CTL epitopes for the CDCP1, GBP1, MMP14, MSLN, NECTIN4, PIM1 and TROP2 vaccine candidate proteins were identified by IEDB server using appropriate algorithms (Rankpep, Bimas, NetMHC 4.0, Syfpeithi, and MHCPred) and the results are shown in Table 1. The amino acids sequence of identified proteins namely CDCP1 (UniProt ID: Q9H5V8), GBP1 (UniProt ID: P32455), MMP14 (UniProt ID: P50281), MSLN (UniProt ID: Q13421), NECTIN4 (UniProt ID: Q96NY8), PIM1 (UniProt ID: P11309) and TACSTD2 (UniProt ID: P09758) were retrieved in FASTA format as per methodology provided. The position of the identified epitopes in the protein and the type of MHC class I alleles to which the epitopes showed potential binding are shown in Table 1. The IC50 values less than 50 nM (for binding against MHC class I alleles) and their respective percentile ranks were considered to filter the antigenic epitopes of the different test proteins. Further, we tabulated only those epitopes which showed antigenicity score more than 1 (Table 1). The selected epitopes for the PIM1, CDCP1, MMP14, MSLN, and NECT4 showed IC50 values in the range of 6–38, 13–45, 22–41, 4–46, and 17–40 nM, respectively. The TROP2 protein showed only one epitope with more than 1 antigenicity score. The GBP1 derived epitope did not showed the antigenicity score even more than the normal threshold value (i.e., 0.4), thus we excluded it for the CTL epitope list. The final CTL epitopes for the vaccine design were selected by using the criteria (1) lowest IC50 values against MHC class I allele binding, (2) antigenicity score > 1, and (3) the epitope should bind more than one MHC class I allele. The proteins for which lesser CTL epitopes and/or only one allele binding potential recorded, having low IC50 values were also considered for the vaccine construct designing. We selected the DLFDFITER, KINSLAHLR, and MLLSKINSL (from PIM1); KVYLRTPNW, and LPRESNITV (from CDCP1); RFREVPYAY, and PPRCLLLPL (from MMP14); SVSTMDALR, FYPGYLCSL, and TQMDRVNAI (from MSLN); AVTSEFHLV, and GTTSSRSFK (from NECT4); and YLDEIPPKF antigenic epitopes from TROP2 proteins for the vaccine construct. The finally selected CTL epitopes showed binding with the HLA-A (68:01; 33:01; 33:03; 31:01; 02:01; 02:06; 32:01; and 11:01), HLA-B (08:01; 57:01; 58:01; and 07:02), and HLA-C (14:02; 07:02 and 05:01) alleles.
Table 1.
CD8+ cytotoxic T lymphocytes (CTLs) epitope selection for PIM1, CDCP1, MMP14, MSLN, TROP2, and NECT4
| Proteins | Epitopes | Positions | MHC class I alleles | IC50 values | Percentile rank | Antigenicity score | |
|---|---|---|---|---|---|---|---|
| PIM1 | DLFDFITER | 128–136 | HLA-A*68:01 | 8.76 | 0.07 | 1.1942 | |
| HLA-A*33:01 | 12.09 | 0.02 | |||||
| HLA-A*33:03 | 23.33 | 0.07 | |||||
| KINSLAHLR | 5–13 | HLA-A*31:01 | 6.96 | 0.03 | 1.851 | ||
| KLIDFGSGA | 183–191 | HLA-A*02:06 | 24.44 | 0.23 | 1.3963 | ||
| HLA-A*02:01 | 37.53 | 0.34 | |||||
| LILERPEPV | 118–126 | HLA-A*02:06 | 21.66 | 0.2 | 1.1386 | ||
| HLA-A*02:01 | 37.83 | 0.34 | |||||
| MLLSKINSL | 1–9 | HLA-B*08:01 | 6.83 | 0.02 | 1.3426 | ||
| HLA-A*02:01 | 10.89 | 0.1 | |||||
| HLA-A*02:06 | 27.02 | 0.25 | |||||
| CDCP1 | KTISCTDHR | 413–421 | HLA-A*31:01 | 30.27 | 0.22 | 1.0479 | |
| RIKMQEGVK | 185–193 | HLA-A*30:01 | 35.32 | 0.17 | 1.0898 | ||
| KVYLRTPNW | 556–564 | HLA-A*32:01 | 13.87 | 0.02 | 1.1621 | ||
| HLA-B*57:01 | 24.08 | 0.06 | |||||
| HLA-B*58:01 | 31.67 | 0.15 | |||||
| LLAKPCYIV | 51–59 | HLA-A*02:01 | 45.46 | 0.4 | 1.4682 | ||
| LPRESNITV | 34–42 | HLA-B*07:02 | 18.58 | 0.06 | 1.3577 | ||
| MMP14 | RFREVPYAY | 158–166 | HLA-C*14:02 | 22.21 | 0.1 | 1.0207 | |
| HLA-A*30:02 | 37.07 | 0.05 | |||||
| HLA-A*29:02 | 37.69 | 0.11 | |||||
| PPRCLLLPL | 7–15 | HLA-B*07:02 | 34.05 | 0.1 | 1.277 | ||
| NYTPKVGEY | 130–138 | HLA-C*14:02 | 41.84 | 0.17 | 1.6215 | ||
| MSLN | DLATFMKLR | 527–535 | HLA-A*68:01 | 25.71 | 0.28 | 1.0888 | |
| HLA-A*33:01 | 26.12 | 0.04 | |||||
| HLA-A*33:03 | 44.4 | 0.14 | |||||
| SVSTMDALR | 241–249 | HLA-A*68:01 | 6.14 | 0.04 | 1.1239 | ||
| HLA-A*33:03 | 46.95 | 0.15 | |||||
| FYPGYLCSL | 444–452 | HLA-C*14:02 | 4.28 | 0.02 | 1.2151 | ||
| HLA-C*07:02 | 28.76 | 0.01 | |||||
| STMDALRGL | 243–251 | HLA-A*68:02 | 28.66 | 0.18 | 1.3541 | ||
| HLA-A*02:06 | 28.78 | 0.26 | |||||
| TQMDRVNAI | 334–342 | HLA-A*02:06 | 11.31 | 0.11 | 1.7285 | ||
| NECT4 | QRITHILHV | 234–242 | HLA-C*06:02 | 31.33 | 0.02 | 1.0053 | |
| GSVAEMSSY | 305–313 | HLA-C*03:02 | 40.14 | 0.17 | 1.0168 | ||
| AVTSEFHLV | 203–211 | HLA-A*02:06 | 26.51 | 0.25 | 1.0379 | ||
| GTTSSRSFK | 189–197 | HLA-A*11:01 | 17.98 | 0.08 | 1.2842 | ||
| HLA-A*30:01 | 29.54 | 0.13 | |||||
| SSRDSQVTV | 322–330 | HLA-A*30:01 | 34.49 | 0.16 | 1.555 | ||
| EVKGTTSSR | 186–194 | HLA-A*68:01 | 21.45 | 0.22 | 1.9232 | ||
| HLA-A*33:03 | 40.26 | 0.13 | |||||
| TROP2 | YLDEIPPKF | 260 | 268 | HLA-C*05:01 | 14.66 | 0.02 | 1.0799 |
Identification of potential HTL epitopes
The 15 mer HTL epitopes for the vaccine candidate proteins were identified on IEDB server using appropriate algorithms (Rankpep, Bimas, NetMHC 4.0, Syfpeithi, and MHCPred) and the results are shown in Table 2. The position of the identified HTL epitopes in the test protein and the type of MHC class II alleles (HLA super types) to which the epitopes showed potential binding are shown in Table 2. Less than 50 nM IC50 values for binding against MHC class II alleles and percentile ranks were considered to filter the antigenic HTL epitopes. The epitopes which showed antigenicity score more than 1 were only considered for the tabulation (Table 1). The selected epitopes for the PIM1, GBP1, MSLN, and NECT4 showed IC50 values in the range of 12–45, 17–11, 25–34, and 23–50 nM, respectively. The TROP2 protein showed only one HTL epitope with more than 1 antigenicity score. The CDCP1 and MMP14 derived HTL epitope did not showed the antigenicity score even more than the normal threshold value (i.e., 0.4), thus we excluded these proteins for the HTL epitope list. The final HTL epitopes for the vaccine design were selected by using the criteria (1) lowest IC50 values against MHC class II allele binding, (2) antigenicity score > 1, and (3) the epitope should bind more than one MHC class II allele. The proteins for which lesser HTL epitopes and/or only one allele binding potential recorded, having low IC50 values were also considered for the vaccine construct designing. LARSFFWQVLEAVRH, ARSFFWQVLEAVRHC, RSFFWQVLEAVRHCH, PDSFVLILERPEPVQ (from PIM1); EVERVKAESAQASAK, SADFVSFFPDFVWTL (from GBP1); LDTLTAFYPGYLCSL, TEQLRCLAHRLSEPP (from MSLN), PCFYRGDSGEQVGQ, YEEELTLTRENSIRR (from NECT4); and LVITNRRKSGKYKKV antigenic HTL epitopes from TROP2 proteins for the vaccine construct. The finally selected HTL epitopes showed binding with the HLA-DPA1*02:01/DPB1*01:01; HLA-DPA1*03:01/DPB1*04:02; HLA-DPA1*01:03/DPB1*02:01; HLA-DRB1 (04:05, 13:02, 09:01, 11:01, and 07:01); HLA-DQA1*05:01/DQB1*03:01; and HLA-DRB3*01:01 HLA super types.
Table 2.
CD4+ helper T cell (HTLs) epitope selection PIM1, CDCP1, GBP1, MSLN, TROP2, and NECT4
| Proteins | Epitopes | Positions | MHC class II allele | IC50 | PR | AS |
|---|---|---|---|---|---|---|
| PIM1 | SGIRVSDNLPVAIKH | 54–68 | HLA-DRB1*13:02 | 12 | 0.95 | 1.025 |
| LARSFFWQVLEAVRH | 143–157 | HLA-DPA1*02:01/DPB1*01:01 | 19 | 0.29 | 1.038 | |
| HLA-DPA1*03:01/DPB1*04:02 | 23 | 0.7 | ||||
| HLA-DPA1*01:03/DPB1*02:01 | 26 | 0.47 | ||||
| ARSFFWQVLEAVRHC | 144–158 | HLA-DPA1*02:01/DPB1*01:01 | 20 | 0.31 | 1.197 | |
| HLA-DPA1*03:01/DPB1*04:02 | 23 | 0.9 | ||||
| HLA-DPA1*01:03/DPB1*02:01 | 31 | 0.57 | ||||
| RSFFWQVLEAVRHCH | 145–159 | HLA-DPA1*02:01/DPB1*01:01 | 22 | 0.65 | 1.294 | |
| HLA-DPA1*03:01/DPB1*04:02 | 24 | 1.5 | ||||
| HLA-DPA1*01:03/DPB1*02:01 | 32 | 0.99 | ||||
| LSKINSLAHLRAAPC | 3–17 | HLA-DRB1*01:01 | 30 | 6 | 1.274 | |
| PDSFVLILERPEPVQ | 113–127 | HLA-DRB1*04:05 | 30 | 4.4 | 1.319 | |
| HLA-DRB1*13:02 | 45 | 3.3 | ||||
| FERPDSFVLILERPE | 110–124 | HLA-DRB1*04:05 | 31 | 15 | 1.238 | |
| ERPDSFVLILERPEP | 111–125 | HLA-DRB1*04:05 | 32 | 7 | 1.255 | |
| GBP1 | EVERVKAESAQASAK | 490–504 | HLA-DQA1*05:01/DQB1*03:01 | 11 | 44 | 1.1151 |
| DLQTKMRRRKACTIS | 578–592 | HLA-DRB1*11:01 | 8.9 | 32 | 1.0282 | |
| NEIQDLQTKMRRRKA | 574–588 | HLA-DRB1*11:01 | 7.8 | 32 | 1.1932 | |
| QLYYVTELTHRIRSK | 141–155 | HLA-DRB1*01:01 | 9.7 | 50 | 1.2099 | |
| LYYVTELTHRIRSKS | 142–156 | HLA-DRB1*01:01 | 9.7 | 50 | 1.3288 | |
| SADFVSFFPDFVWTL | 168–182 | HLA-DRB1*09:01 | 7 | 50 | 1.3754 | |
| DQLYYVTELTHRIRS | 140–154 | HLA-DRB1*01:01 | 9.7 | 50 | 1.4230 | |
| MSLN | LDTLTAFYPGYLCSL | 438–452 | HLA-DRB1*09:01 | 33 | 0.23 | 1.008 |
| DTLTAFYPGYLCSLS | 439–453 | HLA-DRB1*09:01 | 34 | 0.24 | 1.021 | |
| TEQLRCLAHRLSEPP | 98–112 | HLA-DRB1*11:01 | 25 | 4.7 | 1.094 | |
| NECT4 | EEELTLTRENSIRRL | 87–401 | HLA-DRB1*07:01 | 45 | 21 | 1.165 |
| EELTLTRENSIRRLH | 388–402 | HLA-DRB1*07:01 | 45 | 23 | 1.288 | |
| KYEEELTLTRENSIR | 385–399 | HLA-DRB1*07:01 | 50 | 20 | 1.114 | |
| LPCFYRGDSGEQVGQ | 50–64 | HLA-DRB3*01:01 | 45 | 1.6 | 1.013 | |
| YEEELTLTRENSIRR | 27–41 | HLA-DRB1*07:01 | 23 | 49 | 1.096 | |
| TROP2 | LVITNRRKSGKYKKV | 295–309 | HLA-DRB1*11:01 | 16 | 1.5 | 1.008 |
PR percentile rank, AS antigenicity score
Construction of the subunit vaccine
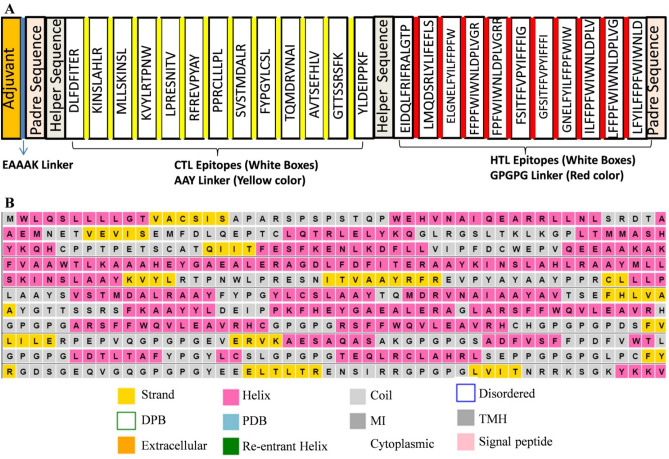
The selected antigenic CTL and HTL epitopes were considered for the construction of the TNBC vaccine. A total of 13 CTL, and 11 HTL epitopes were selected and linked together with the aid of linker respective sequences. The “AAY” linker is known to provide binding for the transporter associated antigenic peptides (TAP), and also increase the epitopic presentation while the “GPGPG” linker enhance the HTL mediated responses and conserve conformation mediated immunogenicity. Moreover, AAY and GPGPG linkers allow the dissociation and identification of individual epitopes during antigen processing and presentation. Thus, the AAY and GPGPG linkers were used to fuse the CTL and HTL antigenic epitopes of the different test proteins, respectively. The GM-CSF (granulocyte–macrophage colony-stimulating factor) was attached at the N-terminal of the vaccine construct, utilized as an adjuvant to increase the immunogenicity of the vaccine. The adjuvant sequence was followed by the EAAK linker in the construct. The pan-HLA DR binding epitopes (PADRE) sequence is a 13 aa synthetic peptide acts as universal T helper epitope. The PADRE sequence enhances the potential of vaccines by activating the CD4 T lymphocytes (CD4 cells). The helper sequence (HEYGAEALERAG) was incorporated in the vaccine construct for the better separation and presentation of the epitopes (Fig. 2A). The final vaccine construct was composed of 563 amino acids in length. The sequence homology of the MEV construct to the human protein sequence showed no significant alignments.
Fig. 2.
Schematic diagram of the final vaccine construct and secondary structure. A The vaccine consists of the adjuvant, the Padre, the helper, the CTL and the HTL epitope sequences. B The predicted secondary structure of the final vaccine construct. DPB disordered protein binding, PDB putative domain boundary, MI membrane interaction, TMH transmembrane helix
Prediction, refinement, and quality assessment of the MEV 3D structure
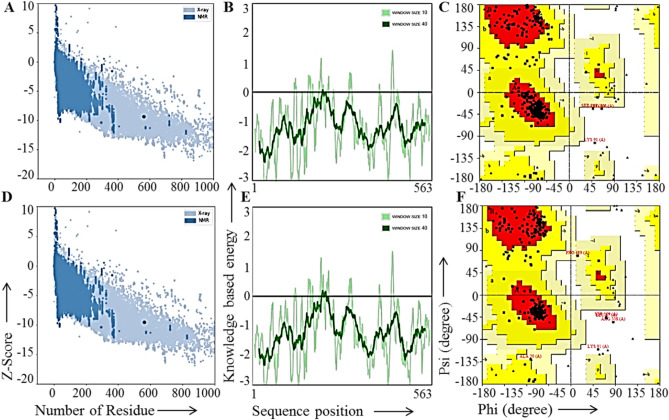
First of all the secondary structure of the MEV was obtained using PSIPRED 4.0 tool. The results of the secondary structure prediction for the vaccine construct possessed 18% alpha-helix, 21% transmembrane helices, 44% β-sheets, and 27% disordered structure (Fig. 2B). The 3-D structure of the final vaccine construct sequence was predicted by using trRosetta sever. GalaxyRefine server was used for the refinement of the obtained initial tertiary structure of the vaccine and used for further analysis. The five models generated by the GalaxyRefine server showed the MolProbity and root mean square deviation (RMSD) in the range of 1.950–1.984 and 0.292–0.325 (data not shown). We considered the first model with 1.961 MolProbity score, 96.1% Ramachandran favored region and 0.292 RMSD value for further analysis. It has been reported that the protein models having more favored region, lesser outliners/rotamer region should be considered as an ideal model. The initial vaccine construct model produced from trRosetta server and the refine model obtained from the GalaxyRefine were studied with the aid of the ERRAT workspace. The initial and the refined vaccine construct model showed 90.8% and 93.6% amino acid residues in the Ramachandran favored region, respectively (Fig. 3C, F). Further, the ProSA-web tool was utilized to compare the quality and significant errors in the final vaccine construct refined model. The quality and possible errors in the final vaccine 3D model were determined by ProSA-web tool. The model with more negative Z-score shows the overall good quality of the test protein. In the present study, the initial and the refined vaccine construct models showed − 9.37 and − 9.56 Z-score (Fig. 3A, D) which indicates the overall good quality of the refined structure. Also, the knowledge-based energy was found satisfactory in both the models in the ProSA-web analysis given in Fig. 3B, E.
Fig. 3.
Comparison of non-refined and refined 3D structure of the final vaccine construct. A Z-score plot for the initial vaccine construct using ProSA-web tool. B Knowledge-based energy plot for the initial vaccine construct using ProSA-web tool. C Ramachandran plot of the initial vaccine construct using Procheck tool. D Z-score plot for the final refined vaccine construct using ProSA-web tool. E Knowledge-based energy plot for the final refined vaccine construct using ProSA-web tool. F Ramachandran plot of the final refined vaccine construct using Procheck tool
Identification of linear and dis-continuous B-cell epitopes
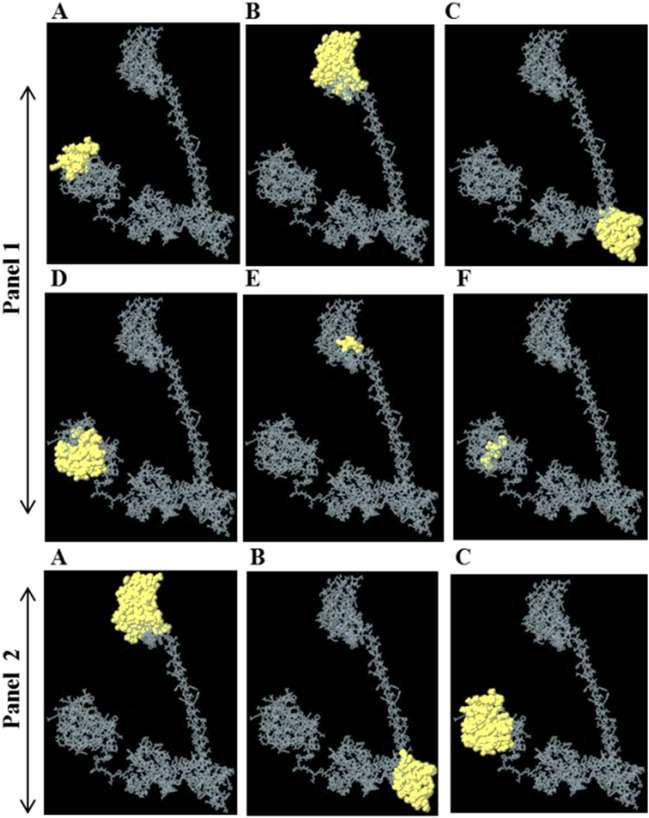
In comparison to T cell, the B cells recognize antigens in a different way. They simply recognize the antigens (solvent exposed) with the help of membrane-bound B-cell receptors (immunoglobulins) which results in to their activation and release of soluble immunoglobulins. In the present study, we identified the B-cell linear (occur locally in the peptide) and dis-continuous (occur at different parts of the peptide and comes together due to its folding in 3D space) epitope in the final vaccine construct. Both the linear and dis-continuous B-cell epitopes are involved in the antigen–antibody reaction. A total of six linear epitopes were found in the construct with score range of 0.514–0.802 (Fig. 4A–F, panel I). The number of residues, start and end points are summarized in the (Supplementary Table 1). Furthermore, three dis-continuous B-cell epitopes were found with score ≈ 0.7 (Supplementary Table 2, Fig. 4A–C, panel II). The number of residues in each predicted epitope is provided in the (Supplementary Table 2).
Fig. 4.
B-cell linear and discontinuous epitopes (Panel 1) A–F corresponds to linear epitope 1–6. (Panel 2) A–C corresponds to discontinuous epitope 1–3 predicted in final refine vaccine construct through Ellipro server
Antigenicity, allergenicity, physiochemical parameters, and population coverage of the final vaccine construct
According to the allergenicity evaluation, the multi-epitope peptide vaccine was not allergen. The vaccine showed a value of 0.684 for antigenicity prediction. The molecular weight (Mw) and theoretical isoelectric point value (pI) of the multi-epitope peptide vaccine were 61,860.72 Da and 7.90, respectively. The solubility tendency value of the peptide vaccine upon overexpression in E. coli was 0.799. The total number of the obtained positively (Arg + Lys) and negatively (Asp + Glu) charged amino acid residues was 56 and 54. The computed instability index for a protein should be < 40. In the present study the computed instability index was non-significantly higher (41.18) than the normal value which indicates the vaccine construct as a stable protein. The computed half-life of the multi-epitope peptide vaccine in the E. coli (> 10 h), yeast (> 20 h), and mammalian reticulocytes (30 h) were found satisfactory. The aliphatic index and GRAVY were found 78.10 and − 0.173, respectively. The multi-epitope peptide vaccine had low GRAVY which represents its comparatively hydrophilic property. The predicted aliphatic index indicates the thermal stability of the multi-epitope peptide vaccine. The population coverage of the designed vaccine is an important parameter for a new candidate vaccine. The population coverage of the epitopes (CTL and HTL) present in the final vaccine construct was predicted on IEDB server and the results are tabulated in (Supplementary table 3). The CTL epitopes of the vaccine showed 4–89% coverage among different population of the world. The East Asia, Europe, North America, Northeast Asia, South Asia, Southeast Asia, and the West Indies population showed more than 80% coverage. Further, the HTL epitope showed the population coverage range of 4–99.99%. The Central Africa, Central America, East Africa, Europe, North America, South America, South Asia, and West Africa population showed more than 99% coverage. The broad population coverage of the designed vaccine showed satisfactory results and substantiates its potential vaccine candidature for TNBC patients.
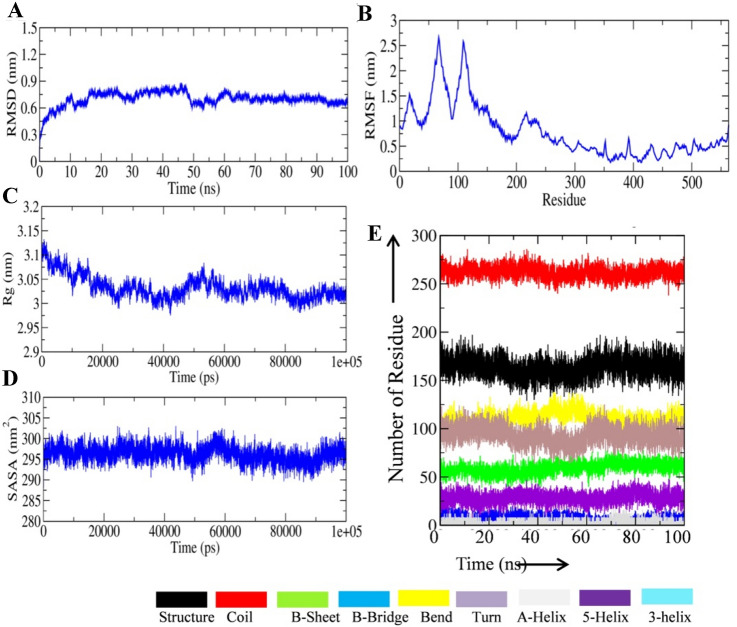
Molecular dynamics simulation of the MEV construct
The molecular dynamics (MD) simulation of the final designed MEV was performed to check the energetics and conformational stability. A total of 10 ns MD simulation was performed. The system was equilibrated for temperature, pressure, volume, density, and kinetic/potential/total energy (data not shown). Root-mean-square fluctuation (RMSF), root mean square deviation (RMSD), Solvent accessible surface area (SASA), and radius of gyration (Rg) parameters were studied by analyzing the 10 ns MD simulation trajectory obtained after the simulation run. Not much fluctuation in the RMSD was observed which indicates that the stability of the final vaccine construct (Fig. 5A, panel I). RMSF fluctuation in the vaccine construct was analyzed and the results are shown in (Fig. 5B, panel I). The result showed that the amino acid residues from 50 to 300 possess more fluctuation in comparison to other amino acids in the vaccine construct which indicates the flexible region in the vaccine. The Radius of gyration (Rg) of the vaccine construct was significantly reduced during the MD simulation period which indicates the compactness of the vaccine in the aqueous environment (Fig. 5C, panel I). The Solvent accessible surface area (SASA) parameter for the vaccine construct was stable during the entire period of the MD simulation which indicates the stable conformation of the final vaccine construct in the aqueous environment (Fig. 5D, panel I). Next we studied the hydrogen bond formation within the vaccine molecules and between vaccine and surrounding water molecules. The intra-molecular hydrogen bond pattern in the vaccine construct showed increase bond formation during the initial simulation period and vice-versa (Fig. 6A, panel II). It should be noted that the inter-molecular hydrogen bond formation showed opposite pattern (Fig. 6B, panel II). The rhythmic change in inter/intra-molecular hydrogen bond formation in the vaccine construct indicates the good interaction with the surrounding water molecule. Change in the secondary MEV structure was studied with the help of simulation trajectory and the results are shown in (Fig. 6C, panel II).
Fig. 5.
Molecular dynamics simulation trajectory plot of final vaccine construct. A The RMSD of the final vaccine construct during 100 ns MD simulation. B The RMSF of the final vaccine construct during 100 ns MD simulation. C The Rg values of the final vaccine construct during 100 ns MD simulation. D The SASA of the final vaccine construct during 100 ns MD simulation (Panel I). Hydrogen bond formation and secondary structure change in the final vaccine construct during the MD simulation period. A Hydrogen bond formation within the final vaccine structure. B Hydrogen bond formation between final vaccine structure and surrounding water molecules. C Change in secondary structure during the 100 ns MD simulation period of the final vaccine construct (Panel II)
Fig. 6.
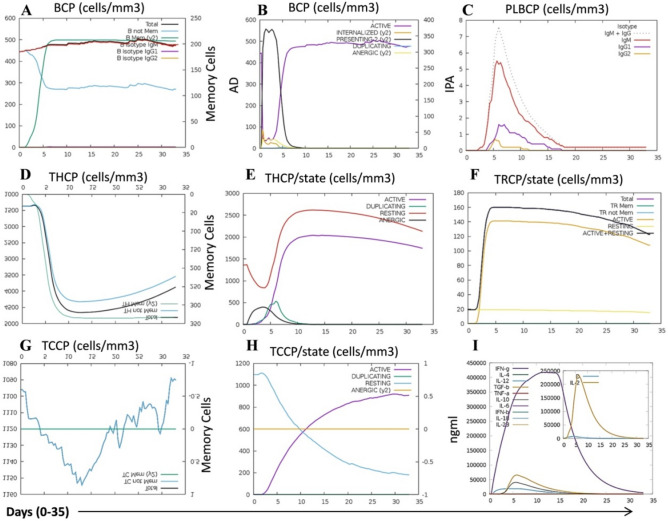
Immune simulation of the final construct vaccine. A B lymphocyte and its sub-population. B B lymphocytes population per entity-state (i.e., showing counts for active, presenting on class-II, internalized the Ag, duplicating and anergic. C Plasma B lymphocytes count sub-divided per isotype (IgM, IgG1 and IgG2). D CD4 T-helper lymphocytes count showing total and memory counts. E CD4 T-helper lymphocytes count sub-divided per entity-state (i.e., active, resting, anergic and duplicating). F CD4 T-regulatory lymphocytes count showing total/memory/per entity-state counts are plotted here. G CD8 T-cytotoxic lymphocytes count showing total and memory populations. H CD8 T-cytotoxic lymphocytes population count. I Cytokines and interleukin graph. BCP-B cell population, AD active duplicate, IPA internalized, presenting, anergic, PLBCP plasma B lymphocyte cell population, THCP-T helper cell population, TRCP T regulatory cell population, TCCP T cytotoxic cell population
The immune response profile of the vaccine in in silico immune simulation
The C-ImmSim online server was utilized to perform the immune stimulation of the refined vaccine model to obtain its immune profiles. The server utilizes the T helper/cytotoxic C/B lymphocytes, plasma B lymphocytes, macrophages, and dendritic cells are allowed to involve in 3D stochastic cellular automation. The interaction among these immune cells is governed by some specific rules which ensure the interaction should be in the line of phase specific recognition and response in immune system for a given antigen. The model simulates the production of new lymphoid/myeloid cells, avoidance of auto-reactivity, and antigen presenting to immune cells (Rapin et al. 2010). The dendritic cells and macrophages present antigen to MHC molecules and are involve in innate immunity. Additionally the dendritic cells are involved in the tissue surveillance process also. The immune simulation results showed that the vaccine increased the total, resting and antigen presenting dendritic cells and macrophage population significantly during the simulation time (35 days) (Supplementary Figs. 1–5). The T helper and T cytotoxic lymphocytes are cell-mediated responders and involve in adaptive immunity. The B lymphocytes is involved in antibody production, and produces adaptive and humoral immunity. The natural killer (NK) cell is involved in cell-mediated immunity and produced innate immunity (Folcik et al. 2007). The NK cells have potential to differentiate among cancer and normal cells. Previously, it has been reported that the vaccine epitope mediated immune response enhance the NK cell infiltration in the TNBC tumor and thereby killing of the cancer cells (Abdel-Latif and Youness, 2020). In the present immune simulation result the vaccine administration showed significant increase in the total count of NK cells (Supplementary Fig. 2). The immunoglobulin molecules are involved in the proliferation and activation of the lymphocytes. The vaccine-mediated immune simulation showed significant induction of IgM + IgG and IgM antibodies (Supplementary Fig. 1). The immune simulation result showed that the vaccine triggered the total B lymphocytes as well as the memory lymphocytes (Fig. 6A–C). Similarly the significant induction was observed in T helper, T cytotoxic, and T regulatory cells in the presence of vaccine during the simulation period of 35 days (Fig. 6D–H). IFN-γ is a pleiotropic cytokine with anti-tumor potential (granzyme B and perforin mediated apoptosis) and known to prevent the immune system over-activation (mechanism unknown). The T helper/T cytotoxic cells and NK cells are important player for the regulation of IFN-γ secretion in the innate and immunity response, respectively (Jorgovanovic et al. 2020). In the present study we found that the vaccine immune simulation revealed the significant increase in lymphocyte and NK cells population as well as the production of IFN-γ, and IL-2 (Fig. 6I). Thus, the overall immune simulation results for the designed vaccine indicate the immune stimulation mediated TNBC cancer cells targeting potential of the vaccine.
Molecular docking and molecular dynamics simulation of the vaccine and immune molecules
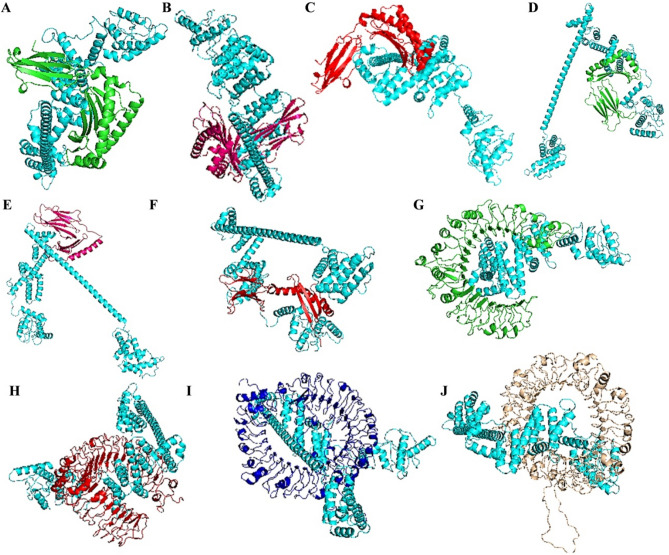
To study the binding efficacy of the final refined structure of the vaccine with the immune molecules we performed the molecular docking. There is some difference among the peptide binding potential of toll like receptors (TLR2/TLR4), MHC I (HLA/B/C) and II (HLA molecules) molecules which generally deals with the length of the peptide involve in the interaction with the MHC molecules. The MHC I and MHC II molecules bound on the surface of the antigen presenting cells (APC) presents the antigens to the CD8 and CD4 T-cell receptors (present on CD8 and 4 T cells), respectively. After recognizing the respective epitopes the CD8 and C4 T cells become cytotoxic T lymphocytes (CTL) and helper/regulatory (Th/Treg) T cells and perform their functions (Sanchez-Trincado et al. 2017). Forero et al. (2016) reported that the expressions of HLA-DQA1 and HLA-DRB1 MHC II molecules in TNBC patient samples are associated with the disease relapse-free survival in the patients. Based on these facts in the present study we performed the docking analysis to study the interaction between the vaccine and MHC I (HLA-A/B/C) and MHC II (HLA-DQA1/DQB1/DRB1) molecules. The results obtained from HDOCK server showed significant binding of the vaccine with the MHC I and II molecules (Supplementary Table 4). The docking pose of the vaccine with the MHC molecules are shown in (Fig. 7A–F). Molecular docking results showed that the docking score of vaccine construct docked with the immune molecules was in the range of − 296.25 to − 440.11 (Supplementary Table 4). Toll-like receptors or TLRs (expressed on antigen presenting cells) recognize specific pattern in the host molecules and mediate important role in the innate immunity during pathogen invasion and disease conditions including cancer. TLRs are known to express on cancer cells and play important role in tumor initiation, development and metastasis. It has been reported that the TNBC cells possess about tenfold less TLR2 expression in comparison to ER/PR+ cells. Recently, Shi et al. (2020) reported that, TLR 4, and TLR 7 was less expressed in the TNBC patient samples in comparison to luminal and HER2+ patient samples. Previously it was shown that the low expression of TLR9 is related with the aggressiveness and high risk of disease relapse in TNBC patients (Tuomela et al. 2012). The literature indicates that the low expression of TLR2/4/7 and 9 might help in the immune evade in TNBC and promote tumor development. Thus keeping these facts in our mind we performed the molecular docking analysis of the MEV with these immune molecules. The results obtained from HDOCK server showed significant binding with the TLR2, TLR4, TLR7 and TLR9 receptors (Supplementary Table 4). The docking pose of the vaccine with the TLR2, TLR4, TLR7 and TLR9 receptors are shown in (Fig. 7G–J). The interaction of vaccine with these TLRs might enhance the associated immune response and thus help to check the cancer cell evade.
Fig. 7.
Docking pose of vaccine construct with target immune molecules. A HLA-A allele. B HLA-B allele. C HLA-C allele. D HLA-DQB1. E HLA-DQA1. F HLA-DRB1. G TLR2 receptor. H TLR4 receptor. I TLR7 receptor. J TLR9 receptor
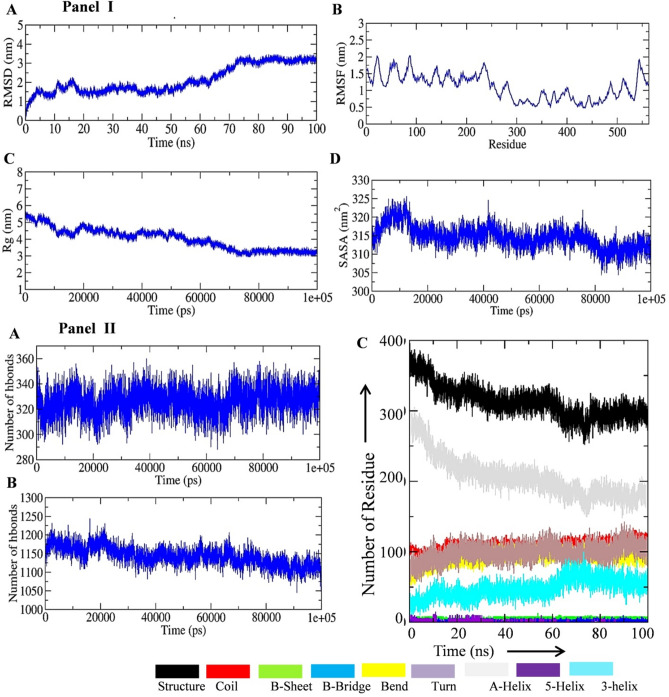
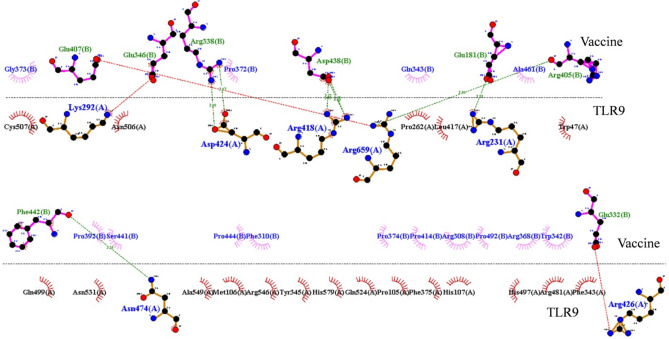
Docking results revealed that vaccine construct-TLR9 complex showed minimum Dock Score (− 440.11), thus we selected TLR-9 for further study. To get insight into the binding pattern and interaction of vaccine construct with immune targets, we performed MD simulation of MEV-TLR9 receptor for 100 ns of MD simulation. RMSD analysis of vaccine construct in complex with TLR9 receptor showed equilibrated pattern throughout 100 ns MD simulation and a significant reduction in structural deviation in comparison to the vaccine construct (Fig. 8A). RMSD analysis of vaccine construct in complex with TLR9 receptor showed that protein–protein interaction significantly reduced the structural deviation in vaccine construct. RMSF of the vaccine construct in complex with TLR9 receptor was also analyzed and the plot is shown in Fig. 8B. RMSF plot of vaccine construct showed that amino acid residues between 1 and 200 had high fluctuation in 100 ns MD simulation in comparison to the other parts of vaccine construct. Rg analysis of vaccine construct in complex with TLR9 receptor was conducted and plot is shown in Fig. 8C. Rg plot showed slight decrease in the Rg value up to the 40 ns mark and remained equilibrated throughout 100 ns MD simulation. Furthermore, the average Rg value of vaccine construct in complex with TLR9 receptor was lower than the average Rg value of vaccine construct alone which indicates comparatively more compact structure after binding with other proteins. SASA analysis of the vaccine construct in complex with TLR9 receptor was also conducted. Fairly stable SASA plot of vaccine construct in complex with TLR9 receptor was observed (Fig. 8D). Notably, SASA of vaccine-TLR9 receptor complex was significantly less than the SASA of vaccine construct which expected after the binding of another protein with vaccine construct. Secondary structure analysis was conducted to assess the effect of binding of immune molecule with the vaccine construct. A significant increase of amino acid residues in the coil region of vaccine construct was observed after the binding of TLR9. Furthermore, amino acid residues in the structured region of vaccine construct did not show any significant change (Fig. 8E). To assess the residual interaction between vaccine construct and TLR9 receptor we extracted the average PDB of vaccine construct and TLR9 complex by using GROMACS utility gmx_covar. Ligplot of the interacting amino acid residues between vaccine construct and TLR9 receptor is shown in Fig. 9.
Fig. 8.
MD simulation analysis of vaccine construct in complex with immune molecule. A RMSD plot of vaccine construct in complex with TLR9 receptor during 100 ns MD simulation. B RMSF plot of vaccine construct in complex with TLR9 receptor during 100 ns MD simulation. C Rg plot of vaccine construct in complex with TLR9 receptor during 100 ns MD simulation. D SASA plot of vaccine construct in complex with TLR9 receptor during 100 ns MD simulation. E Time-dependent evolution of secondary structure of vaccine construct in complex with TLR9 receptor during 100 ns of MD simulation
Fig. 9.
Picture depicting interacting residues between vaccine construct and TLR9 receptor. Interaction is generated using Ligplot online tool
In silico cloning and prediction of RNA secondary structure
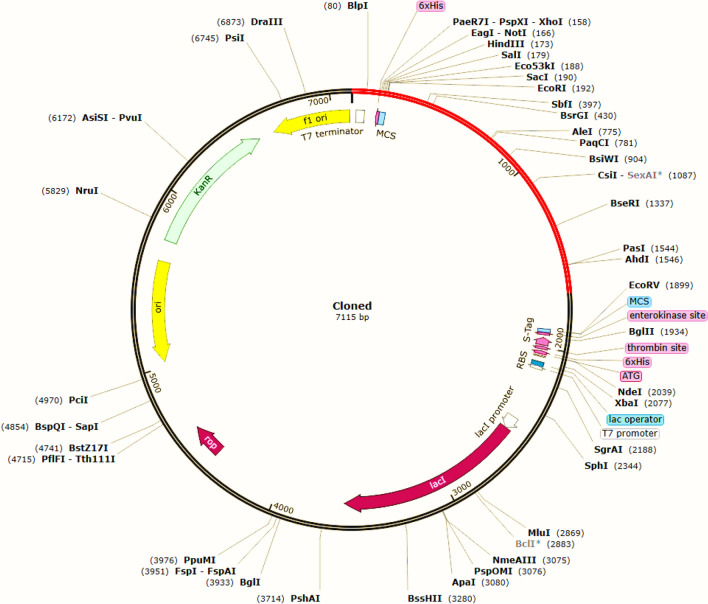
Java Codon Adaptation Tool was used to optimize the codon usage of the designed vaccine for maximum expression into the E. coli expression system. For this, the constructed linear vaccine t was reverse translated into specific cDNA sequence in Escherichia coli (E. coli) strain K12 (Fig. 10). This step was essential to ensure translation and elevates protein production. JCat tool generates 1689 nucleotides long cDNA sequence after codon optimization. A codon adaptation index (CAI) value more than 0.8 and the GC content value lie between 30 and 70% are taken as a prominent protein expression into the host environment. Our constructed vaccine possessed a CAI of 0.96483 and GC content in E. coli was recorded as 55.713%. Therefore, the results support efficient expression of the final constructed vaccine in the E. coli host. Further, recombinant plasmid was made by inserting the adapted codon sequences into the expression vector pET-30a (+) for in silico cloning using SnapGene software. The codon sequence was inserted between EcoR1 (192) and BamH1 (198), producing a clone with a total length of 7115 bp. Drabner and Guzman, (Drabner and Guzmán 2001) reviewed the importance of bacterial vaccine vectors used to deliver and express heterologous vaccine antigens in order to prevent cancer. Bacterial vaccines have the advantage to express multiple antigens. This type vaccine could easily produce and induces strong immune responses on oral or intranasal administration.
Fig. 10.
In silico cloning of the vaccine. Black color represents the vector pET-30a (+) and the insert of interest (vaccine sequence) are shown in red colour
Conclusion
The occurrences of global TNBCs cases are of concern due to its high mortality rate and early relapse of the disease. No proper therapeutic treatments are available for the patients yet. The discovery of immunotherapy could emerge as an effective treatment approach, where the application of in silico methods would be beneficial to design an effective vaccine against this havoc disease. In this study, immuno-informatic tools were used to construct TNBC multi-epitope peptide vaccine derived from over-expressed cell surface protein markers on TNBC cells. The designed multi-epitope vaccine triggers strong immune response that was observed in computer-based immunogenic and antigenic predictions. The final construct had 74.44% and 99.88% world population coverage for HLA I and II, respectively. Molecular docking studies showed potential interaction between the vaccine and immune molecules (MHCI, MHCII, and TLRs) and molecular dynamics simulation study revealed the conformational stability of the MEV. Immune simulation studies confirm the immune response activating efficacy of the vaccine. Lastly, the in silico expression of the constructed vaccine in bacterial host indicates the translational efficacy of the vaccine. The proposed vaccine requires future in vitro and in vivo studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
SK acknowledges Indian Council of Medical Research, India and Department of Science and Technology, India for providing financial support in the form Ad-hoc Project [No. 5/13/15/2020/NCD-III] and DST-SERB Grant [EEQ/2016/000350], respectively. SK also acknowledges DST-India for providing Departmental grant to the Department of Biochemistry, Central University of Punjab, Bathinda, India in the form of DST-FIST grant. KSP acknowledge Department of Biotechnology, India for providing DBT-Senior Research Fellowship. MS acknowledges Indian Council of Medical Research, India for providing ICMR-Senior Research fellowship [File No. 5/3/8/80/ITR-F/2020-ITR]. AKS acknowledges CSIR, India for providing CSIR-Senior Research Fellowship. SS and PC acknowledge Central Computing Facility, Indian Institute of Information Technology-Allahabad, India.
Author contribution
SK conceptualized, supervised, the study and wrote the original draft of the manuscript; KSP, MS, and AKS curated the data and analyzed them. PC and SS generated MD simulation data and AKS analyzed it. SG critically read the manuscript and provided his valuable suggestions.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Not applicable.
References
- Abdel-Latif M, Youness RA. Why natural killer cells in triple negative breast cancer? World J Clin Oncol. 2020;11:464–476. doi: 10.5306/wjco.v11.i7.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:1–8. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cîmpean AM, Ribatti D, Raica M. Triple negative breast cancer: the kiss of death. Oncotarget. 2017;8:46652–46662. doi: 10.18632/oncotarget.16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas R, Zhang S, Li M, Chen C, Yao Q. Chimeric Trop2 virus-like particles: a potential immunotherapeutic approach against pancreatic cancer. J Immunother. 2011;34:251–263. doi: 10.1097/CJI.0b013e318209ee72. [DOI] [PubMed] [Google Scholar]
- Drabner B, Guzmán CA. Elicitation of predictable immune responses by using live bacterial vectors. Biomol Eng. 2001;17:75–82. doi: 10.1016/s1389-0344(00)00072-1. [DOI] [PubMed] [Google Scholar]
- Folcik VA, An GC, Orosz CG. The basic immune simulator: an agent-based model to study the interactions between innate and adaptive immunity. Theor Biol Med Model. 2007;4:1–18. doi: 10.1186/1742-4682-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, Downs-Kelly E, Burwell TC, Vaklavas C, Buchsbaum DJ, Myers RM, LoBuglio AF, Varley KE. Expression of the MHC class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol Res. 2016;4:390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Senapati S, Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. 2020;2020:1–12. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan R, Thomas A, Alewine C, Le DT, Jaffee EM, Pastan I. Mesothelin immunotherapy for cancer: ready for prime time? J Clin Oncol. 2016;34:4171–4189. doi: 10.1200/JCO.2016.68.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- Jiang T, Shi T, Zhang H, Hu J, Song Y, Wei J, Ren S, Zhou C. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol. 2019;12:1–13. doi: 10.1186/s13045-019-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res. 2020;8:1–16. doi: 10.1186/s40364-020-00228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardani K, Bolhassani A, Namvar A. An overview of in silico vaccine design against different pathogens and cancer. Expert Rev Vaccines. 2020;19(8):699–726. doi: 10.1080/14760584.2020.1794832. [DOI] [PubMed] [Google Scholar]
- Kushwaha PP, Singh AK, Prajapati KS, Shuaib M, Gupta S, Kumar S. Phytochemicals present in Indian ginseng possess potential to inhibit SARS-CoV-2 virulence: a molecular docking and MD simulation study. Microb Pathog. 2021;157:1–11. doi: 10.1016/j.micpath.2021.104954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Eom KY, Koo TR, Kim BH, Kang E, Kim SW, Kim YJ, Park SY, Kim IA. A prognostic model for patients with triple-negative breast cancer: importance of the modified Nottingham prognostic index and age. J Breast Cancer. 2017;20:65–73. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemkul J. From proteins to perturbed Hamiltonians: a suite of tutorials for the GROMACS-2018 molecular simulation package. LiveCoMS. 2019;1:1–53. doi: 10.33011/livecoms.1.1.5068. [DOI] [Google Scholar]
- Lindahl A, Hess VDS, van der Spoel D (2020) GROMACS 2020.2 Source code 2020
- Ling B, Watt K, Banerjee S, Newsted D, Truesdell P, Adams J, Sidhu SS, Craig AWB. A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models. Oncotarget. 2017;8:58372–58385. doi: 10.18632/oncotarget.17702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrabet M, Cabaud O, Josselin E, Finetti P, Castellano R, Farina A, Agavnian-Couquiaud E, Saviane G, Collette Y, Viens P, Gonçalves A, Ginestier C, Charafe-Jauffret E, Birnbaum D, Olive D, Bertucci F, Lopez M. Nectin-4: a new prognostic biomarker for efficient therapeutic targeting of primary and metastatic triple-negative breast cancer. Ann Oncol. 2017;28:769–776. doi: 10.1093/annonc/mdw678. [DOI] [PubMed] [Google Scholar]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11:6047–6057. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1039/c9dt02916h. [DOI] [Google Scholar]
- Quintero M, Adamoski D, Reis LMD, Ascenção CFR, Oliveira KRS, Gonçalves KA, Dias MM, Carazzolle MF, Dias SMG. Guanylate-binding protein-1 is a potential new therapeutic target for triple-negative breast cancer. BMC Cancer. 2017;17:727–733. doi: 10.1186/s12885-017-3726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5:9862–9876. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggio AI, Varley KE, Welm AL. The lingering mysteries of metastatic recurrence in breast cancer. Br J Cancer. 2021;24:13–26. doi: 10.1038/s41416-020-01161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017:1–15. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Patiyal S, Dhall A, Pande A, Arora C, Raghava GPS. AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief Bioinform. 2020;17:294–306. doi: 10.1093/bib/bbaa294. [DOI] [PubMed] [Google Scholar]
- Shi S, Xu C, Fang X, Zhang Y, Li H, Wen W, Yang G. Expression profile of Toll-like receptors in human breast cancer. Mol Med Rep. 2020;21:786–794. doi: 10.3892/mmr.2019.10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Gupta S, Kumar S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1 pp domain inhibitor: an in silico study. Comp Biol Med. 2021;130:1–10. doi: 10.1016/j.compbiomed.2020.104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomela J, Sandholm J, Karihtala P, Ilvesaro J, Vuopala KS, Kauppila JH, Kauppila S, Chen D, Pressey C, Härkönen P, Harris KW, Graves D, Auvinen PK, Soini Y, Jukkola-Vuorinen A, Selander KS. Low TLR9 expression defines an aggressive subtype of triple-negative breast cancer. Breast Cancer Res Treat. 2012;135:481–493. doi: 10.1007/s10549-012-2181-7. [DOI] [PubMed] [Google Scholar]
- Turdo F, Bianchi F, Gasparini P, Sandri M, Sasso M, De Cecco L, Forte L, Casalini P, Aiello P, Sfondrini L, Agresti R, Carcangiu ML, Plantamura I, Sozzi G, Tagliabue E, Campiglio M. CDCP1 is a novel marker of the most aggressive human triple-negative breast cancers. Oncotarget. 2016;7:69649–69665. doi: 10.18632/oncotarget.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursynbay Y, Zhang J, Li Z, Tokay T, Zhumadilov Z, Wu D, Xie Y. Pim-1 kinase as cancer drug target: An update. Biomed Rep. 2016;4:140–146. doi: 10.3892/br.2015.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Qu J, Li L, Gao F. Guanylate-binding protein 1 correlates with advanced tumor features, and serves as a prognostic biomarker for worse survival in lung adenocarcinoma patients. J Clin Lab Anal. 2021;35:1–8. doi: 10.1002/jcla.23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol. 2018;15(2):182–184. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.