Abstract
Arterial medial calcification is a common disease in patients with type 2 diabetes, end-stage renal disease and hypertension, resulting in high incidence and mortality of cardiovascular event. H19 has been demonstrated to be involved in cardiovascular diseases like aortic valve diseases. However, role of H19 in arterial medial calcification remains largely unknown. We identified that H19 was upregulated in ß-glycerophosphate (β-GP) induced vascular smooth muscle cells (VSMCs), a cellular calcification model in vitro. Overexpression of H19 potentiated while knockdown of H19 inhibited osteogenic differentiation of VSMCs, as demonstrated by changes of osteogenic genes Runx2 and ALP as well as ALP activity. Notably, H19 interacted with miR-140-5p directly, as demonstrated by luciferase report system and RIP analysis. Mechanistically, miR-140-5p attenuated osteoblastic differentiation of VSMCs by targeting Satb2 and overexpression of miR-140-5p blocked H19 induced elevation of Satb2 as well as the promotion of osteoblastic differentiation of VSMCs. Interestingly, over-expression of Satb2 induced phosphorylation of ERK1/2 and p38MAPK. In conclusion, H19 promotes VSMC calcification by acting as competing endogenous RNA of miR-140-5p and at least partially by activating Satb2-induced ERK1/2 and p38MAPK signaling.
Keywords: H19, miR-140-5p, Satb2, smooth muscle cell differentiation, arterial calcification
Introduction
Arterial medial calcification (AMC) is a common cardiovascular disease, especially in patients with type 2 diabetes mellitus (T2DM), end-stage renal disease (ESRD), obesity-related disease state and ageing (Sarnak et al., 2019; Bourron et al., 2020). Nowadays, AMC has growing incidence because of the elevated aging population worldwide (McClelland et al., 2006). The epidemiologic data have demonstrated that the occurrence of AMC is an important risk factor for incidence and mortality of cardiovascular event (Budoff et al., 2019). In the past decades, AMC used to be thought as a negative and degenerative process resulting from deposition of hydroxyapatite in the arterial walls because of aging. However, with the advent of recent advance, AMC has now been illustrated to be a biological regulated active process that mainly involve the osteogenic transition of vascular smooth cell phenotype, resembling the process of osteoblast differentiation of pre-osteoblast cells (Xu et al., 2020). Notably, there exists significant ectopic expression of osteogenic phenotype genes such as alkaline phosphatase (ALP), runt related transcription factor 2 (Runx2), osteocalcin (OC), and bone morphogenetic protein 2 (BMP2) in calcified arterial walls (Rashdan et al., 2020; Xu et al., 2020). However, the underlying mechanism of AMC remains largely unknown and treatment strategies are limited.
H19, a well-known imprinted gene derived long non-coding RNA (lncRNA), was initially observed in liver and embryonic tissues as a key regulator of growth and development (Zhou et al., 2019). However, with the incoming brand-new various evidence, H19 has now been demonstrated to be widely involved in various physiological and pathological processes, such as stem cell development (Zhou et al., 2019), skeletal muscle differentiation and regeneration (Dey et al., 2014), cholestatic liver fibrosis (Song et al., 2017), aging and cancer progression (Lee et al., 2015). Recent advances have shown that increased circulating H19 level is closely related to higher risk of coronary artery diseases (Zhang et al., 2017; Bitarafan et al., 2019). A more recent study shown that H19 was increased in calcific aortic valve disease and promoted mineralization of valve interstitial cells (Hadji et al., 2016). However, the role of H19 in AMC is still largely unknown.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (20–22 nucleotides) and has been demonstrated to affect gene expression by binding to 3′ untranslated region (3’ UTR) of target genes. A growing body of evidence have also demonstrated that lncRNAs may exert their biological functions by acting as competitive endogenous RNA (ceRNA) to sponge specific miRNAs (Thomson and Dinger, 2016). For example, H19 antagonized the expression of miRNA let-7 to increase muscle differentiation (Kallen et al., 2013). Similar effects were reported in miR-106a (Imig et al., 2015) and miR-194-5p (Li et al., 2018), et al. Intriguingly, miR-675, embedded in H19 exon 1, is the production of H19 transcript and has been demonstrated in mediating functions of H19 in placental growth, BMSCs differentiation and bladder cancer proliferation (Liu et al., 2016a).
Satb2, short for special AT-rich sequence-binding protein 2, is a novel identified DNA binding transcription factor that plays an important role in tissue development, such as bone regeneration, and cancer progression (Dobreva et al., 2006; Ni et al., 2021). Indeed, some studies have demonstrated that Satb2 shares similar pathway with Runx2 and works together to induce bone formation (Dobreva et al., 2006; Hu et al., 2015). More recently, Hao et al. found that down-regulation of satb2 decreased VSMC calcification, suggesting satb2 might be pro-calcification factor in VSMC calcification (Hao et al., 2016).
In the present study, we characterize the role H19 in AMC. The results showed that H19 was elevated in calcified vascular smooth muscle cells (VSMCs). Knockdown of H19 increased while overexpression of H19 decreased the osteogenic differentiation of VSMCs. Mechanistically, H19 increased the expression of Satb2 through sponging miR-140-5p. These data suggest that H19 promotes VSMC calcification by acting as competing endogenous RNA of miR-140-5p to increase the expression of Satb2.
Materials and Methods
Reagents
The fetal bovine serum (FBS), DMEM and penicillin/streptomycin mixture were purchased from Gibco-BRL Co., Ltd (Grand Island, NY, United States). Antibodies for Satb2 (sc-81376, 1:1,000), phospho-p38 MAPK (sc-166182, 1:1,000), p38 MAPK (sc-81621, 1:1,000), phospho-ERK1/2 (sc-81492, 1:1,000), and ERK1/2 (sc-135900, 1:1,000) were purchased from Santa Cruz Biotechnology Inc (Waltham, MA, United States). Runx2 (ab23981, 1:1,000) and ALP (ab229126, 1:1,000) antibodies were purchased from Abcam (Cambridge, United Kingdom). Anti-alpha smooth muscle Actin (α-SMA) antibody (bs-10196R, 1:1,000) was bought from Beijing Biosynthesis Biotechnology CO., LTD. (Beijing, China). Horseradish peroxidase (HRP)-conjugated Goat Anti-Mouse IgG, HRP-conjugated Goat Anti-Rabbit IgG as well as anti-GAPDH monoclonal antibody were purchased from Proteintech (Rosemont, United States). Satb2 specific siRNA oligonucleotides and the control siRNA oligonucleotides were purchased from Shanghai GenePharma Co., Ltd (Shanghai, China). MiR-140-5p mimics, miR-140-5p inhibitor, the control mimics, the control inhibitor, H19 specific smart silence and the control smart silence were purchased from Ribobio (Guangzhou, China). H19-expressing lentivirus were constructed based on Ubi-MCS-SV40-EGFP system (sequence based on NR_002196, gene ID: 283120) by Shanghai Genechem Co., Ltd (Shanghai, China). Satb2-expressing lentivirus (sequence based on NM_001172509) were bought from Shanghai Genechem Co., Ltd (Shanghai, China). The ALP assay kits were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Alizarin Red S and ß-glycerophosphate (β-GP) were purchased from Sigma Chemical Co., Ltd (St.Louis, MO, United States).
Cell Culture
Human VSMCs were acquired and identified by a previously established method (Xu et al., 2019). Human VSMCs from excess donor normal arterial tissues after kidney transplantations were grown, which was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University, China. Briefly, the tissue was fragmented (1,2 mm3), the aortas were minced and digested in digestion solution (0.125 mg/ml elastase, 0.25 mg/ml soybean trypsin inhibitor, 10 mg/ml collagenase I, 2.0 mg/ml crystallized bovine albumin, and 15 mM HEPES) at 37°C for 45 min. The cellular digests were filtered through a sterile 100 mM nylon mesh, centrifuged at 1,000 rpm for 10 min, and then washed twice in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin before cultured in the same medium. All cells were incubated at 37°C with a humidified atmosphere containing 5% CO2. Cells passaged between 3 and 6 times were chosen for the experiments. VSMCs were demonstrated by typical peak-valley phenotype and expression of SMC marker a-SMA. Free of mycoplasma infection in VSMCs was confirmed by mycoplasma testing kit (Shanghai Seo Biotechnology Co., Ltd). Medium containing 10 mM ß-GP was used to induce osteoblastic differentiation of VSMCs as reported previously (Xu et al., 2019).
Quantitative RT-PCR
Cells were incubated with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and total RNA was extracted as guided by the manufacturer’s instructions (Wu et al., 2020). For detection of mRNA and LncRNA expression, cDNA was synthesized with the guidance of the PrimeScript RT reagent Kit (Takara, Japan) and then Real-time PCR detection was performed by using the TB Green™ Premix Ex Taq™ II kit (Takara, Japan) with GAPDH as normalized control. For miRNA detection, miRNA was re-transcribed and analysed by All-in-One™ miRNA quantitative RT-PCR Detection System (GeneCopoeia, Rockville, United States) with guidance by the manufacturer’s protocol and using U6 as the normalized control. H19 primers (forward: 5- GCGTCCGGCCTTCCTGAAC-3, reverse: 5-GAGCTGGGTAGCACCATTTCTT-3) were as designed and synthesized by Shanghai Genechem Co., Ltd (Shanghai, China). U6 (HmiRQP9001), miR-140-5p (HmiRQP0181), miR-675-5p (HmiRQP0782), miR-22-3p (HmiRQP0332), miR-17-3p (HmiRQP0229), miR-29b-3p (HmiRQP0373), miR-106b-5p (HmiRQP0029), miR-214-5p (HmiRQP0321), GAPDH (HQP006940), Satb2 (HQP096946), ALP (HQP006440), and Runx2 (HQP016478) primers were purchased from GeneCopoeia.
Western Blot
Proteins analysis by western blot was performed in the same way as described previously (Liang et al., 2012; Peng et al., 2017; Lin et al., 2020). Briefly, total protein were lysed by using of RIPA lysate (Beyotime Biotechnology, Shanghai, China) supplemented with cocktail protease inhibitor (Sellect, Shanghai, China) and the extraction was measured by BCA Protein Assay kit (Beyotime Biotechnology, Shanghai, China). Total protein were then separated by 12% SDS-PAGE and transferred to 0.2 µm Polyvinylidene fluoride Membranes (PVDF, Millipore, Billerica, MA). The membranes were blocked by 5% milk for 1 h at room temperature, followed by incubation with specific primary antibody overnight. And then the blots were rewarmed to room temperature for 30 min, washed by phosphate-buffered saline (PBS) for 3 times with 10 min interval and followed by incubation with HRP-labeled secondary antibody for 1 h at room temperature. Finally, the reaction was detected by chemiluminescence assay with Luminata™ Crescendo Western HRP Substrate (Millipore, Billerica, MA) and was analyzed by ABI system.
Analysis of the ALP Activity
The cell layers were subjected to a lysis solution containing 20 mM Tris–HCl, pH 8.0, and 150 mM NaCl, 1% Triton X-100, 0.02% NaN3 and 1 mM PMSF. And then the lysates were sonicated for 4 times with 5 s interval. The ALP activity was measured by detecting p-nitrophenol release using the ALP kit (Liu et al., 2016b). The ALP activity was normalized by the total protein level of the cell lysate.
Detection of Alizarin Red S. Staining Level
Alizarin Red S. staining was conducted in a similar way as described previously (Liao et al., 2013). Briefly, VSMCs were cultured in 24-well plates for indicated time. Then, the content of mineralized matrix was examined by Alizarin Red S staining. Briefly, cells were fixed in 70% ethanol for 1 h at room temperature and then stained with 40 mM Alizarin Red S for 10 min. Next, cells were washed with PBS for 3 times to reduce nonspecific staining. The stained matrix was digitally photographed.
Luciferase Reporter Gene Assay System
The fragment of the wild type or mutant Satb2 3′ UTR was ligated into a pmirGLO -dual-luciferase miRNA target expression vector (Promega, United States). The constructed vectors were then identified by sequencing, cloned, and purified. VSMCs were treated with co-transfection of the luciferase reporter system that carried wild-type Satb2 3′ UTR sequence, mutant Satb2 3′ UTR sequence, and miR-140-5p mimics or scramble oligonucleotides. For detection of interaction between H19 and miR-140-5p, plasmid with wild-type H19 (with binding site to miR-140-5p) or mutant H19 sequence (with mutant binding site to miR-140-5p) were co-transfected with miR-140-5p mimics or scramble control. Subsequently, luciferase activities were determined by the luciferase assay system (Promega) at 48 h after transfection in accordance with the manufacturer’s protocol. The ratio of firefly to Renilla was used to normalize the firefly luciferase values. The specific primers for synthesis of wild type or mutant 3′UTR Satb2 mRNA, or wild type or mutant H19 were purchased from Ribobio.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
RIP experiments were performed using the Magna RIP™ Kit (Millipore, Billerica, MA). Briefly, VSMCs were treated with magnetic beads that were conjugated with anti-Ago2 antibody (Cambridge, United Kingdom) or negative control IgG (Cambridge, United Kingdom). Thereafter, the beads with conjugated molecules were treated by protein A/G beads and H19 and miR-140-5p levels were determined by qRT-PCR analysis.
Statistical Analysis
SPSS 23.0 (SPSS, Chicago, IL, United States) was used for all statistical comparisons. Data are presented as means ± standard deviation (SD) unless specially stated. Student’s t test was performed when comparing two groups. One-way or two-way ANOVA followed by the Tukey’s HSD post hoc analysis was performed for multiple comparisons. Differences at p value under 0.05 were considered statistically significant.
Results
H19 Promotes Osteoblastic Differentiation and Mineralization of VSMCs
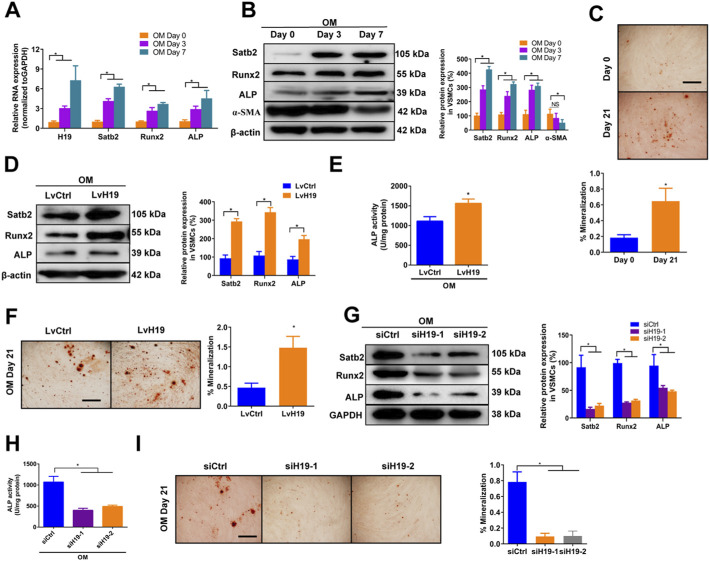
To assess the expression of H19 during the osteoblastic differentiation of VSMCs, we firstly used osteogenic medium (OM) -induced VSMCs that contained 10 mM ß-GP as in vitro calcification model and then observed the change of H19 and calcification markers such as Runx2 and ALP as well as Satb2. In the control medium (CM) treated VSMCs, no obvious change of H19 expression in VSMCs among Day 0, 3 and 7 (Supplementary Figure S1A). Similar results were found in both mRNA and proteins level of osteogenic marker such as Runx2 and ALP as well as Satb2 (Supplementary Figures S1A,B). In addition, contractile marker a-SMA did not change over time (Supplementary Figure S1B). Moreover, no difference in Alizarin Red S staining level in CM-treated VSMCs for Day 0 and Day 21 (Supplementary Figure S1C). However, Runx2 and ALP as well as Satb2 were increased but a-SMA was reduced in VSMCs after the treatment of ß-GP (Figures 1A,B), accompanied with increased Alizarin Red S Staining level (Figure 1C). These data suggest the successful establishment of in vitro calcification model. Moreover, the expression of H19 were increased significantly in OM-induced VSMCs in a time-dependent manner (Figure 1A), suggesting that H19 may be involved in osteoblastic differentiation of VSMCs.
FIGURE 1.
| H19 promotes osteogenic differentiation and mineralization of VSMCs treated with osteogenic medium. (A,B) Expression of H19, calcification marker genes Runx2 and ALP, and Satb2 during the osteogenic differentiation of VSMCs. VSMCs were treated with osteogenic medium (OM) that contains 10 mM ß-glycerophosphate (β-GP) for 7 days to induce osteogenic differentiation. (A) Expression of H19, Runx2, ALP, and Satb2 were determined by qRT-PCR. (B) Expression of Runx2, ALP, a-SMA and Satb2 were determined by Western blotting. The data were presented as densitometric ratios normalized to ß-actin [(B), right panel]. (C) Alizarin Red S staining were measured in VSMCs incubated with ß-GP for 21 days. Representative microscopic views are shown. Data were presented as ratio of positive staining area (lower panel). (D–F) VSMCs were treated with OM along with either lentivirus containing full length of H19 cDNA (LvH19) or control cDNA (LvCtrl). The protein expression of Runx2, ALP, and Satb2 were determined by Western blotting in VSMCs after treatment for 2 days (D). Data were presented as densitometric ratios normalized to GAPDH [(D), right panel]. The ALP activity was determined by an Elisa kit in VSMCs after treatment 2 days (E). Alizarin Red S staining were measured in VSMCs after treatment for 21 days. Representative microscopic views are shown (F). Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(F), right panel]. (G–I) VSMCs were incubated with OM medium along with either H19 siRNA (siH19) or control siRNA (siCtrl). The protein expression of Runx2, ALP, and Satb2 were determined by Western blotting after treatment for 2 days (G). Data were presented as densitometric ratios normalized to GAPDH [(G), right panel]. The ALP activity was determined by an Elisa kit in VSMCs after treatment for 2 days (H). Alizarin Red S staining were measured in after treatment for 21 days. Representative microscopic views are shown (I). Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(I), right panel]. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
Next, we determined whether H19 regulated VSMCs calcification by using loss- and gain-function of H19 (Supplementary Figures S2A,B). Overexpression of H19 by lentivirus containing H19 sequence (LvH19) resulted in increased protein expression of Runx2, ALP and Satb2, and ALP activity as well as Alizarin Red S staining level in both CM-treated VSMCs and OM-treated VSMCs when compared with control group (Figures 1D–F and Supplementary Figures S3A–C). In contrast, siRNA-mediated knockdown of H19 (siH19) decreased the expression of Runx2, ALP and Satb2 as well as ALP activity in OM-treated VSMCs when compared with control group (Figures 1G,H). Moreover, Alizarin Red S staining level of VSMCs were also decreased after knockdown of H19 (Figure 1I). In CM-treated VSMCs, reduced expression of Satb2 and Runx2 were also observed but no difference was found in ALP expression as well as ALP activity (Supplementary Figures S3D,E). Notably, due to the low base line lever, reduced Alizarin red S staining was observed but with no significance after down-regulation of H19 (Supplementary Figure S3F). Taken together, these data indicate that H19 promotes osteoblastic differentiation and mineralization of VSMCs.
H19 Interacts With miR-140-5p
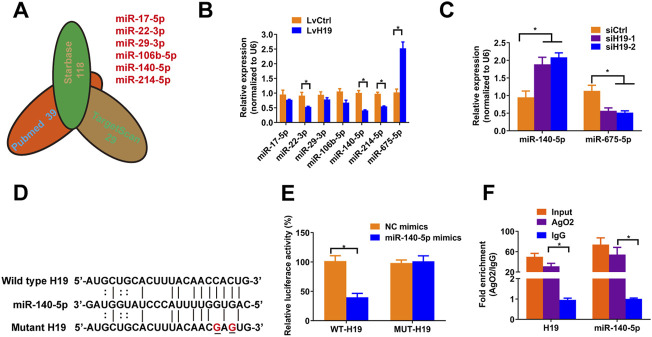
To verify the mechanism by which H19 regulates VSMCs calcification, we screened Pubmed databases for 39 miRNAs that were reported to be associated with osteoblastic differentiation of BMSCs and VSMCs. Then we assessed the potential interaction between these miRNAs and H19 by using starbase databases. Thereafter, we used TargetScan (Version 7.2) to confirm miRNAs that have conserved binding sequence with Runx2, BMP2 and Satb2 (Figure 2A). We selected miR-17a-5p, miR-22-3p, miR-29-3p, miR-106b-5p, miR-140-5p, and miR-214-5p for further analysis. Then, we assessed the expression of these miRNAs after overexpression of H19. The results shown that overexpression of H19 increased the expression of miR-675-5p, which has been demonstrated to be H19-derived miRNAs, and decreased the expression of miR-22-3p, miR-140-5p and miR-214-5p in VSMCs, with the most significant reduction in miR-140-5p (Figure 2B). In contrast, knockdown of H19 increased the expression of miR-140-5p in VSMCs (Figure 2C). Importantly, luciferase activity reporter assay system revealed that transfection of miR-140-5p mimics significantly decreased luciferase activity in wild type H19 but not mutant H19 that was devoid of conserved miR-140-5p binding site (Figures 2D,E). Interestingly, RIP analysis also shown enrichment of H19 and miR-140-5p in Ago2 pull-down precipitate (Figure 2F), suggesting miR-140-5p might mediate degradation of H19. These results suggest that H19 functions as a “sponge” of miR-140-5p in VSMCs.
FIGURE 2.
H19 interacts with miR-140-5p. (A) Predication of potential targets of H19 by using of bioinformatic analysis methods starbase and Targetscans in combination with Pubmed database. The number indicated potential miRNAs that may inhibit osteogenic differentiation of VSMC or interact with H19. (B) VSMCs were treated with lentivirus containing full length of H19 cDNA (LvH19) or control cDNA (LvCtrl). qRT-PCR was performed to evaluate expression of miR-17-5p, miR-22-3p, miR-29-3p, miR-106b-5p, miR-140-5p, miR-214-5p, and miR-675-5p in VSMCs transfected with LvH19 or LvCtrl. (C) The expression of H19 in VSMCs after infected with H19 siRNA (siH19) or control siRNA (siCtrl). Expression of miR-140-5p and miR-675-5p were evaluated by qRT-PCR in VSMCs infected with siH19 or siCtrl. (D) Schematic view of miR-140-5p putative target sites in WT and MUT H19 3′-UTR. (E) VSMCs were transfected with luciferase report vector containing WT or MUT H19 3′-UTR following by transfection of miR-140-5p mimics or control mimics. Luciferase activity was determined 48 h post transfection and luciferase activity was normalised to Renilla luciferase activity. (F) RIP analysis was performed to evaluate enrichment of H19 and miR-140-5p in Ago2 immunoprecipitates. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
MiR-140-5p Restrains VSMCs Calcification by Targeting Satb2
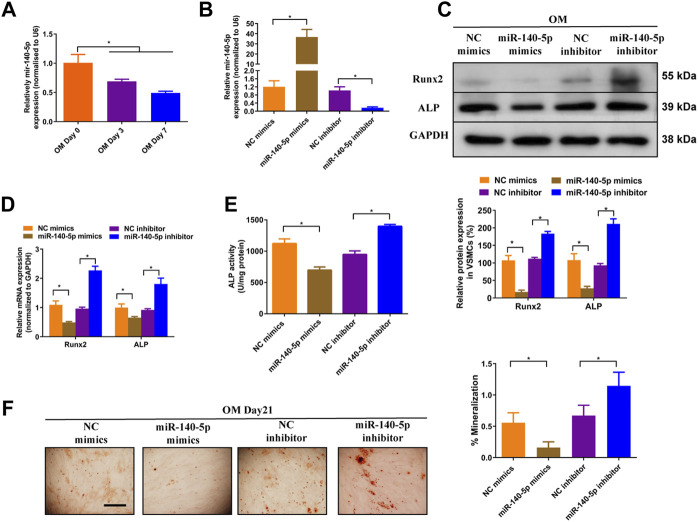
Previous studies have shown that miR-140-5p inhibited osteoblast differentiation of human BMSCs and bone development in zebrafish embryonic by targeting BMP2 (Hwang et al., 2014; Gan et al., 2016). However, role of miR-140-5p in VSMCs calcification is largely unknown. To assess the effects of miR-140-5p on osteoblastic differentiation of VSMCs, we firstly determined the expression of miR-140-5p in 10 mM ß-GP induced VSMCs. The results shown that miR-140-5p was decreased during the process of VSMCs calcification in a time-dependent manner (Figure 3A). Moreover, overexpression of miR-140-5p reduced the expression of Runx2 and ALP (Figures 3B–D) as well as ALP activity (Figure 3E). In contrast, down-regulation of miR-140-5p increased mRNA and protein expression of Runx2 and ALP (Figures 3B–D) as well as ALP activity (Figure 3E). Similar change was also found in VSMCs by using Alizarin Red S staining (Figure 3F). Taken together, these data validates the inhibition effects of miR-140-5p on VSMCs calcification.
FIGURE 3.
miR-140-5p antagonises osteogenic differentiation of VSMCs. (A) qRT-PCR was performed to evaluate the expression of miR-140-5p in VSMCs cultured in OM medium. (B) qRT-PCR was performed to evaluate the expression of miR-140-5p in VSMCs transfected with specific miR-140-5p mimics or inhibitor. (C) Western blotting was performed to determine the protein expression of Runx2 and ALP in VSMCs transfected with specific miR-140-5p mimics or inhibitor. The data were presented as densitometric ratios normalized to GAPDH [(C), lower panel]. (D) qRT-PCR was performed to determine the protein expression of Runx2 and ALP in VSMCs transfected with specific miR-140-5p mimics or inhibitor. (E) The ALP activity was evaluated by specific Elisa kits in VSMCs transfected with specific miR-140-5p mimics or inhibitor. (F) Alizarin Red S staining were measured in ß-GP-treated VSMCs transfected with specific miR-140-5p mimics or inhibitor for 21 days. Representative microscopic views are shown. Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(F), right panel]. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
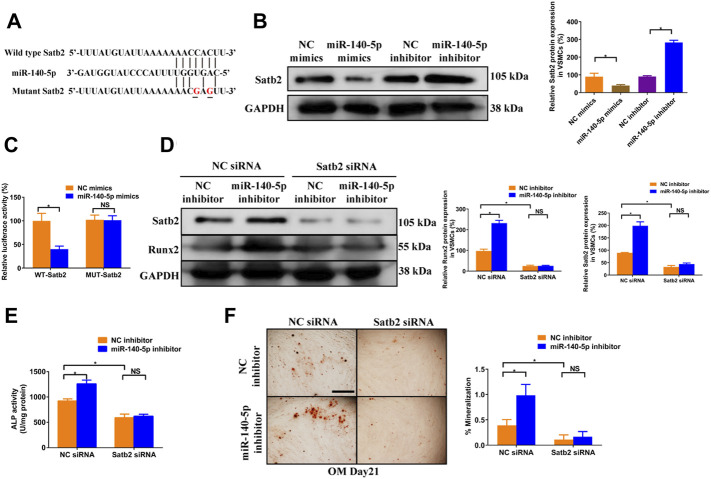
To gain insight into the mechanism by which miR-140-5p restrained VSMCs calcification, online bioinformatics tool TargetScan (version 7.2, http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org/mirdb/index.html) as well as miRmap (https://mirmap.ezlab.org/app/) were adopted to predict potential miR-140-5p targeting genes. Of these, Satb2 has been shown to possess properties as the key regulator for the process of bone information as well as VSMC calcification (Hao et al., 2016; Liu et al., 2020). Figure 4A illustrated the potential binding sites of miR-140-5p with Satb2, suggesting that Satb2 may be targeting gene of miR-140-5p. Furthermore, western blot showed that Satb2 protein level was down-regulated by miR-140-5p mimics and up-regulated by miR-140-5p inhibitor (Figure 4B). Luciferase report assay also demonstrated that overexpression of miR-140-5p reduced wild type Satb2 promotor activity but not that of mutant Satb2 (Figures 4A,C). These data imply that Satb2 may be the target of miR-140-5p in VSMCs. To make clear whether Satb2 mediates the inhibition effect of miR-140-5p on VSMCs calcification, we also used Satb2 specific siRNA to block its expression. Down-regulation of Satb2 reduced the expression of Runx2 and decreased ALP activity as well as Alizarin Red S staining (Figures 4D–F), suggesting that Satb2 plays a important role in VSMCs calcification. Of note, inhibition of miR-140-5p increased the expression of Runx2 and enhanced ALP activity as well as Alizarin Red S staining, but these effects were abolished by suppression of Satb2 (Figures 4D–F). Taken together, these results demonstrate that miR-140-5p inhibits VSMCs calcification by targeting Satb2.
FIGURE 4.
miR-140-5p antagonises osteogenic differentiation of VSMCs by targeting Satb2. (A) Schematic view of miR-140-5p putative binding sites in WT and MUT Satb2 3′-UTR. (B) Western blotting was performed to determine the protein expression of Satb2 in VSMCs transfected with specific miR-140-5p mimics or inhibitor. The data were presented as densitometric ratios normalized to GAPDH [(B), lower panel]. (C) VSMCs were transfected with luciferase report vector containing WT or MUT Satb2 3′-UTR following by transfection of miR-140-5p mimics or control mimics. Luciferase activity was determined 48 h post transfection and luciferase activity was normalised to Renilla luciferase activity. (D) Western blotting was performed to determine the protein expression of Satb2 and Runx2 in VSMCs transfected with specific miR-140-5p inhibitor with or without Satb2 siRNA. The data were presented as densitometric ratios normalized to GAPDH [(D), lower panel]. (E) The ALP activity was determined by an Elisa kit in VSMCs infected with inhibitor of miR-140-5p in the presence of Satb2 siRNA or not. (F) Alizarin Red S staining were measured in ß-GP-treated VSMCs infected with inhibitor of miR-140-5p in the presence of Satb2 siRNA or not for 21 days. Representative microscopic views are shown. Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(F), right panel]. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
To further validate the role of Satb2 on VSMC calcification, we determined the change of calcification markers after knockdown or over-expression of Satb2. Satb2 siRNA mediated down-regulation of Satb2 significantly reduced the expression of Runx2 and ALP activity as well as Alizarin Red S staining when compared with control group in OM-incubated VSMCs (Figures 5A–C). In addition, in CM-treated VSMCs, over-expression of Satb2 induced expression of Runx2 (Figure 5D) and increased ALP activity (Figure 5E). Interestingly, over-expression of Satb2 induced phosphorylation of ERK1/2 and p38MAPK (Figure 5D), two important kinases in cellular differentiation. Taken together, these data indicate that Satb2 is responsible for the process of VSMC calcification.
FIGURE 5.
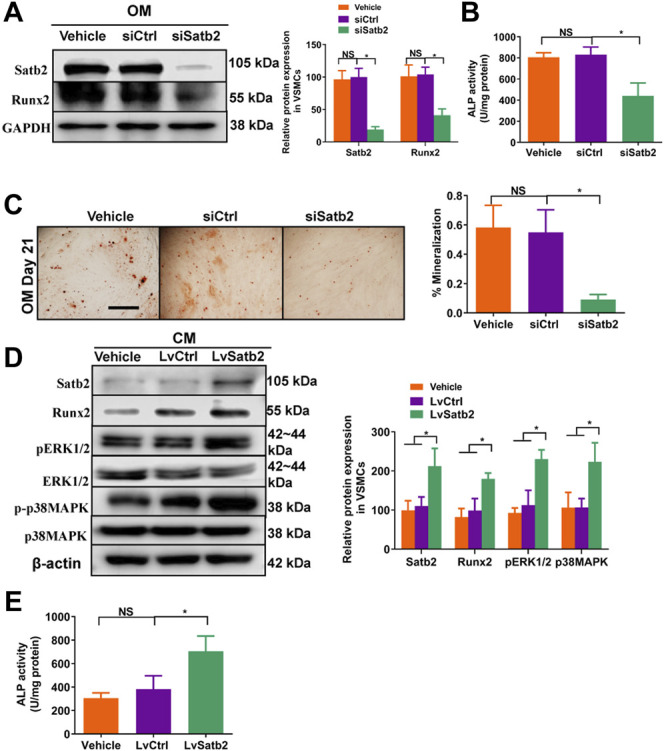
Knockdown of Satb2 inhibits osteogenic differentiation of VSMCs. OM-treated VSMCs were transfected with control siRNA (siCtrl), Satb2 siRNA (siSatb2), or vehicle control for 48 h. (A) Western blotting was performed to evaluate Satb2 and Runx2 expression. The data were presented as densitometric ratios normalized to GAPDH [(A), right panel]. (B) The ALP activity was determined by an Elisa kit. (C) Alizarin Red S staining were measured in ß-GP-treated VSMCs infected with siCtrl or siSatb2 for 21 days. Representative microscopic views are shown. Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(C), right panel]. CM-treated VSMCs were infected with control lentivirus (LvCtrl), Satb2-expressing lentivirus (LvSatb2), or vehicle control for 48 h. (D) Western blotting was performed to evaluate Satb2, Runx2, pERK1/2, ERK1/2, p38MAPK, and p38MAPK expression. The data were presented as densitometric ratios normalized to ß-actin or phosphorylation/total protein [(D), right panel]. (E) The ALP activity was determined by an Elisa kit. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
H19 Enhances Osteoblastic Differentiation of VSMCs by Sponging miR-140-5p
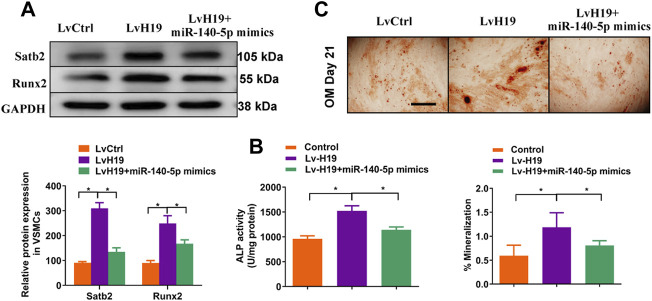
To further validate the role of miR-140-5p in H19-induced calcification of VSMC, we upregulated the expression of miR-140-5p in VSMCs in the presence of H19. The results showed that overexpression of miR-140-5p partially abolished H19 induced expression of Runx2 (Figure 6A) and ALP activity (Figure 6B) as well as Alizarin Red S staining (Figure 6C), suggesting miR-140-5p is involved in H19 induced VSMC calcification. Interestingly, H19-induced expression of Satb2 was also partially blocked by overexpression of miR-140-5p (Figures 6A–C). These data suggest that H19 induces VSMC calcification via miR-140-5p/Satb2 axis.
FIGURE 6.
H19 promotes osteogenic differentiation of VSMCs by sponging miR-140-5p. VSMCs were transfected with LvH19, miR-140-5p, or corresponding negative control. (A) Western blotting was performed to evaluate Satb2 and Runx2 expression. The data were presented as densitometric ratios normalized to GAPDH [(A), lower panel]. (B) The ALP activity was determined by an Elisa kit. (C) Alizarin Red S staining were measured in ß-GP-treated VSMCs infected with LvCtrl or LvH19 in the presence of miR-140-5p mimics or not for 21 days. Representative microscopic views are shown. Scale bar represents 200 µm. Data were presented as ratio of positive staining area [(C), lower panel]. Each experiment was repeated for three times. The data represent the mean ± SD of triplicates. *p < 0.05.
Discussion
In the present study, we provide evidence that H19 plays an important role in the process of VSMC calcification. Over-expression of H19 increased while knockdown of H19 decreased the osteogenic phenotype transition of VSMC. Notably, H19 directly interacted with miR-140-5p and inhibition of miR-140-5p partially blocked H19-induced VSMC calcification. Taken together, these data show that H19 induces VSMC calcification by regulating miR-140-5p/Satb2 loop (Figure 7).
FIGURE 7.
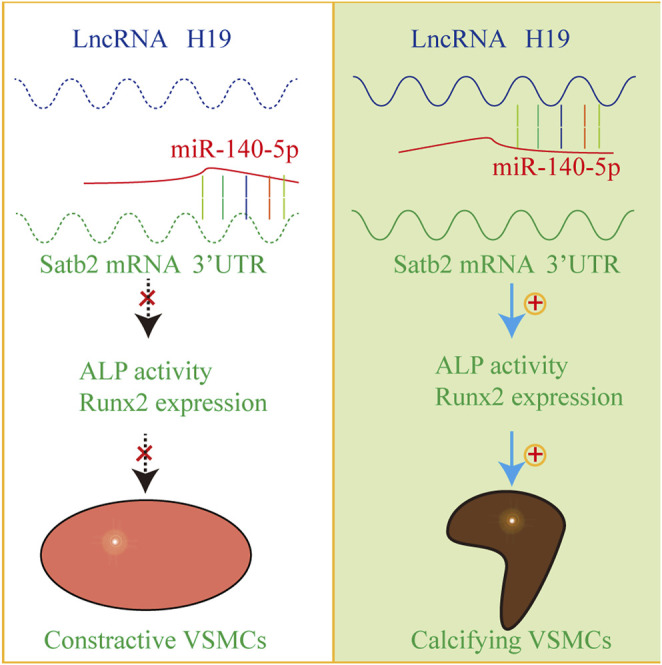
The mechanism diagram about H19/miR-140-5p/Satb2 axis in the regulation of osteogenic differentiation of VSMCs. Normally, miR-140-5p antagonizes the expression of Satb2 to inhibit osteogenic differentiation of VSMCs in absence of H19. However, overexpression of H19 can sponge miR-140-5p and release Satb2. As a result, VSMCs undergo phenotype transition into calcifying VSMCs and induce vascular calcification.
We firstly demonstrated that H19 induced osteogenic differentiation of human VSMCs. We reported that H19 was increased in calcifying VSMCs in a similar trend with calcification marker genes Runx2, which suggested that H19 may be associated with the process of VSMC calcification. Moreover, we found that over-expression of H19 promoted while downregulation of H19 alleviated VSMC calcification, suggesting that H19 play an essential role in VSMC calcification. Our results are in consistent with previous studies that knockdown of H19 reduced calcification of VSMCs (Liu et al., 2020). In their study, knockdown of H19 reduced phosphorylation of p38 and ERK1/2. However, how H19 regulates p38 MAPK and ERK is unclear. It seems that activation of p38 and ERK1/2 might be the common signaling for VSMC calcification but not specific for H19. Moreover, H19, a well-known long non-coding, acts mostly by long-term transcriptional regulation instead of rapid and transient signaling pathway. Mostly, activation of signaling pathway might by caused by classical ligand-receptor method. In our present work, however, we have demonstrated clear clues for H19-miR-140-5p-Satb2 loops in the regulation of VSMC calcification, which provides novel understanding that H19 regulate VSMC calcification by acting as the sponging RNA of miR-140-5p. Notably, we found that over-expression of Satb2 induced phosphorylation of p38 MAPK and ERK1/2, which suggest that Satb2 might be responsible for H19-induced activation of p38 MAPK and ERK1/2. However, it is unclear how Satb2 affects p38 MAPK and ERK signaling pathway.
By acting as a ceRNA to sponging miRNAs, H19 has been demonstrated to communicate with various miRNAs, including let-7, miR-17-5p, and miR-106a, to affect the expression of protein-coding mRNAs indirectly and thus regulate multiple physiological and pathological processes (Zhang et al., 2019). In consistent with these findings, we characterized that miR-140-5p interacted with H19 directly and inhibition of miR-140-5p abolished H19-induced VSMC calcification, as demonstrated by decreased Runx2 expression and ALP activity. Similar findings for the interaction between H19 and miR-140-5p were found in osteoarthritis and bone formation (Bi et al., 2020; Yang et al., 2020). Importantly, our work provides insight into the understanding of vascular calcification.
Our previous work and other reports have shown that miRNAs, such as miR-34b and miR-204, play a key role during the osteogenic differentiation of VSMCs (Lin et al., 2018; Lin et al., 2019). miR-140-5p has been reported to suppress osteoblast differentiation of mesenchymal stem cell by targeting osteogenesis marker gene BMP2 (Hwang et al., 2014). However, role of mir-140-5p in VSMC calcification remains unclear. In the present study, we found that inhibition of miR-140-5p increased while over-expression of miR-140-5p reduced the expression of Runx2 and ALP activity as well as Alizarin Red S staining. Notably, Satb2 was demonstrated to be the target of miR-140-5p in the regulation of VSMC calcification. Little has been clarified in the role of Satb2 in the regulation of vascular calcification. Recently, Hao et al. found that down-regulation of satb2 decreased VSMC calcification (Hao et al., 2016). But how Satb2 affect vascular calcification remains unclear. In the present work, we found that over-expression of Satb2 induced phosphorylation of both p38MAPK and ERK1/2, which indicates that Satb2 might regulate p38 and ERK1/2 signaling.
There exist some limitations for the present study. For example, all the study are performed in vitro, further study are needed to examine in vivo effects of H19 or miR-140-5p. Well-designed animal model might be helpful to demonstrate potential pro-calcification role of H19, which will provide evidence for potential drug targets to block arterial calcification.
In conclusion, our data indicate that H19 enhances osteoblastic differentiation of vascular smooth muscle cell by targeting miR-140-5p/Satb2 axis. Our study provides insight into understanding of vascular calcification and potential therapeutic targets for cardiovascular diseases.
Acknowledgments
We thank Professor Xu-Biao Xie for his help and support.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Xiang-Ya Hospital, Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work is funded by the National Natural Science Foundation of China (grant numbers 81870623, 81770881, 82070910, 82100494, and 82100944).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.774363/full#supplementary-material
References
- Bi H. U., Wang D., Liu X., Wang G., Wu X. (2020). Long Non-coding RNA H19 Promotes Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Regulating microRNA-140-5p/SATB2 axis. J. Biosci. 45, 56. 10.1007/s12038-020-0024-y [DOI] [PubMed] [Google Scholar]
- Bitarafan S., Yari M., Broumand M. A., Ghaderian S. M. H., Rahimi M., Mirfakhraie R., et al. (2019). Association of Increased Levels of lncRNA H19 in PBMCs with Risk of Coronary Artery Disease. Cell J 20, 564–568. 10.22074/cellj.2019.5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourron O., Phan F., Diallo M. H., Hajage D., Aubert C.-E., Carlier A., et al. (2020). Circulating Receptor Activator of Nuclear Factor kB Ligand and Triglycerides Are Associated with Progression of Lower Limb Arterial Calcification in Type 2 Diabetes: a Prospective, Observational Cohort Study. Cardiovasc. Diabetol. 19, 140. 10.1186/s12933-020-01122-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budoff M., Backlund J.-Y. C., Bluemke D. A., Polak J., Bebu I., Schade D., et al. (2019). The Association of Coronary Artery Calcification with Subsequent Incidence of Cardiovascular Disease in Type 1 Diabetes: The DCCT/EDIC Trials. JACC: Cardiovasc. Imaging 12, 1341–1349. 10.1016/j.jcmg.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B. K., Pfeifer K., Dutta A. (2014). The H19 Long Noncoding RNA Gives Rise to microRNAs miR-675-3p and miR-675-5p to Promote Skeletal Muscle Differentiation and Regeneration. Genes Dev. 28, 491–501. 10.1101/gad.234419.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., et al. (2006). SATB2 Is a Multifunctional Determinant of Craniofacial Patterning and Osteoblast Differentiation. Cell 125, 971–986. 10.1016/j.cell.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Gan S., Huang Z., Liu N., Su R., Xie G., Zhong B., et al. (2016). MicroRNA-140-5p Impairs Zebrafish Embryonic Bone Development via Targeting BMP-2. FEBS Lett. 590, 1438–1446. 10.1002/1873-3468.12190 [DOI] [PubMed] [Google Scholar]
- Hadji F., Boulanger M.-C., Guay S.-P., Gaudreault N., Amellah S., Mkannez G., et al. (2016). Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation 134, 1848–1862. 10.1161/circulationaha.116.023116 [DOI] [PubMed] [Google Scholar]
- Hao J., Zhang L., Cong G., Ren L., Hao L. (2016). MicroRNA-34b/c Inhibits Aldosterone-Induced Vascular Smooth Muscle Cell Calcification via a SATB2/Runx2 Pathway. Cell Tissue Res 366, 733–746. 10.1007/s00441-016-2469-8 [DOI] [PubMed] [Google Scholar]
- Hu N., Feng C., Jiang Y., Miao Q., Liu H. (2015). Regulative Effect of Mir-205 on Osteogenic Differentiation of Bone Mesenchymal Stem Cells (BMSCs): Possible Role of SATB2/Runx2 and ERK/MAPK Pathway. Ijms 16, 10491–10506. 10.3390/ijms160510491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Park S.-K., Lee H. Y., Kim S. W., Lee J. S., Choi E. K., et al. (2014). miR-140-5p Suppresses BMP2-Mediated Osteogenesis in Undifferentiated Human Mesenchymal Stem Cells. FEBS Lett. 588, 2957–2963. 10.1016/j.febslet.2014.05.048 [DOI] [PubMed] [Google Scholar]
- Imig J., Brunschweiger A., Brümmer A., Guennewig B., Mittal N., Kishore S., et al. (2015). miR-CLIP Capture of a miRNA Targetome Uncovers a lincRNA H19-miR-106a Interaction. Nat. Chem. Biol. 11, 107–114. 10.1038/nchembio.1713 [DOI] [PubMed] [Google Scholar]
- Kallen A. N., Zhou X.-B., Xu J., Qiao C., Ma J., Yan L., et al. (2013). The Imprinted H19 lncRNA Antagonizes Let-7 microRNAs. Mol. Cell 52, 101–112. 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-F., Su J., Kim H. S., Chang B., Papatsenko D., Zhao R., et al. (2015). Modeling Familial Cancer with Induced Pluripotent Stem Cells. Cell 161, 240–254. 10.1016/j.cell.2015.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-f., Li Y.-c., Wang Y., Sun L.-b. (2018). The Effect of LncRNA H19/miR-194-5p Axis on the Epithelial-Mesenchymal Transition of Colorectal Adenocarcinoma. Cell Physiol Biochem 50, 196–213. 10.1159/000493968 [DOI] [PubMed] [Google Scholar]
- Liang Q. H., Jiang Y., Zhu X., Cui R. R., Liu G. Y., Liu Y., et al. (2012). Ghrelin Attenuates the Osteoblastic Differentiation of Vascular Smooth Muscle Cells through the ERK Pathway. Plos One 7, e33126. 10.1371/journal.pone.0033126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.-B., Zhang Z.-Y., Yuan K., Liu Y., Feng X., Cui R.-R., et al. (2013). MiR-133a Modulates Osteogenic Differentiation of Vascular Smooth Muscle Cells. Endocrinology 154, 3344–3352. 10.1210/en.2012-2236 [DOI] [PubMed] [Google Scholar]
- Lin X., Zhu T., Xu F., Zhong J. Y., Li F., Shan S. K., et al. (2020). Plasma Exosomes Derived from Patients with End-Stage Renal Disease and Renal Transplant Recipients Have Different Effects on Vascular Calcification. Front Cell Dev Biol 8, 618228. 10.3389/fcell.2020.618228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Li F., Xu F., Cui R.-R., Xiong D., Zhong J.-Y., et al. (2019). Aberration Methylation of miR-34b Was Involved in Regulating Vascular Calcification by Targeting Notch1. Aging 11, 3182–3197. 10.18632/aging.101973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Xu F., Cui R.-R., Xiong D., Zhong J.-Y., Zhu T., et al. (2018). Arterial Calcification Is Regulated via an miR-204/DNMT3a Regulatory Circuit Both In Vitro and in Female Mice. Endocrinology 159, 2905–2916. 10.1210/en.2018-00320 [DOI] [PubMed] [Google Scholar]
- Liu C., Chen Z., Fang J., Xu A., Zhang W., Wang Z. (2016a). H19-derived miR-675 Contributes to Bladder Cancer Cell Proliferation by Regulating P53 Activation. Tumor Biol. 37, 263–270. 10.1007/s13277-015-3779-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Yang X. C., Chen M. L., Zhuang Z. W., Jiang Y., Wang J., et al. (2020). LncRNA H19/Runx2 axis Promotes VSMCs Transition via MAPK Pathway. Am. J. Transl Res. 12, 1338–1347. [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu F., Pei H. X., Zhu X., Lin X., Song C. Y., et al. (2016b). Vaspin Regulates the Osteogenic Differentiation of MC3T3-E1 through the PI3K-Akt/miR-34c Loop. Sci. Rep. 6, 25578. 10.1038/srep25578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland R. L., Chung H., Detrano R., Post W., Kronmal R. A. (2006). Distribution of Coronary Artery Calcium by Race, Gender, and Age. Circulation 113, 30–37. 10.1161/circulationaha.105.580696 [DOI] [PubMed] [Google Scholar]
- Ni H., Chen Y., Xia W., Wang C., Hu C., Sun L., et al. (2021). SATB2 Defect Promotes Colitis and Colitis-Associated Colorectal Cancer by Impairing Cl-/HCO3- Exchange and Homeostasis of Gut Microbiota. J. Crohn's colitis 15, 2088–2102. 10.1093/ecco-jcc/jjab094 [DOI] [PubMed] [Google Scholar]
- Peng Y. Q., Xiong D., Lin X., Cui R. R., Xu F., Zhong J. Y., et al. (2017). Oestrogen Inhibits Arterial Calcification by Promoting Autophagy. Sci. Rep. 7, 3549. 10.1038/s41598-017-03801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashdan N. A., Sim A. M., Cui L., Phadwal K., Roberts F. L., Carter R., et al. (2020). Osteocalcin Regulates Arterial Calcification via Altered Wnt Signaling and Glucose Metabolism. J. Bone Miner Res. 35, 357–367. 10.1002/jbmr.3888 [DOI] [PubMed] [Google Scholar]
- Sarnak M. J., Amann K., Bangalore S., Cavalcante J. L., Charytan D. M., Craig J. C., et al. (2019). Chronic Kidney Disease and Coronary Artery Disease: JACC State-Of-The-Art Review. J. Am. Coll. Cardiol. 74, 1823–1838. 10.1016/j.jacc.2019.08.1017 [DOI] [PubMed] [Google Scholar]
- Song Y., Liu C., Liu X., Trottier J., Beaudoin M., Zhang L., et al. (2017). H19 Promotes Cholestatic Liver Fibrosis by Preventing ZEB1-Mediated Inhibition of Epithelial Cell Adhesion Molecule. Hepatology 66, 1183–1196. 10.1002/hep.29209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D. W., Dinger M. E. (2016). Endogenous microRNA Sponges: Evidence and Controversy. Nat. Rev. Genet. 17, 272–283. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- Wu F., Lin X., Shan S.-K., Li F., Xu F., Zhong J.-Y., et al. (2020). The Suppression of miR-199a-3p by Promoter Methylation Contributes to Papillary Thyroid Carcinoma Aggressiveness by Targeting RAP2a and DNMT3a. Front. Cell Dev. Biol. 8, 594528. 10.3389/fcell.2020.594528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhong J. Y., Lin X., Shan S. K., Guo B., Zheng M. H., et al. (2020). Melatonin Alleviates Vascular Calcification and Ageing through Exosomal miR-204/miR-211 Cluster in a Paracrine Manner. J. Pineal Res. 68, e12631. 10.1111/jpi.12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Li F.-X. -Z., Lin X., Zhong J.-Y., Wu F., Shan S.-K., et al. (2019). Adipose Tissue-Derived Omentin-1 Attenuates Arterial Calcification via AMPK/Akt Signaling Pathway. Aging 11, 8760–8776. 10.18632/aging.102251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Xu L., Wang S. (2020). Regulation of lncRNA-H19/miR-140-5p in Cartilage Matrix Degradation and Calcification in Osteoarthritis. Ann. Palliat. Med. 9, 1896–1904. 10.21037/apm-20-929 [DOI] [PubMed] [Google Scholar]
- Zhang L., Yang Z., Huang W., Wu J. (2019). H19 Potentiates Let-7 Family Expression through Reducing PTBP1 Binding to Their Precursors in Cholestasis. Cell Death Dis 10, 168. 10.1038/s41419-019-1423-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Gao W., Long Q.-Q., Zhang J., Li Y.-F., Liu D.-C., et al. (2017). Increased Plasma Levels of lncRNA H19 and LIPCAR Are Associated with Increased Risk of Coronary Artery Disease in a Chinese Population. Sci. Rep. 7, 7491. 10.1038/s41598-017-07611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Xu J., Zhang L., Liu S., Ma Y., Wen X., et al. (2019). Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell stem cell 24, 285 e5. 10.1016/j.stem.2018.11.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.