Abstract
Shifting range limits are predicted for many species as the climate warms. However, the rapid pace of climate change will challenge the natural dispersal capacity of long-lived, sessile organisms such as forest trees. Adaptive responses of populations will, therefore, depend on levels of genetic variation and plasticity for climate-responsive traits, which likely vary across the range due to expansion history and current patterns of selection. Here, we study levels of genetic and plastic variation for phenology and growth traits in populations of red spruce (Picea rubens), from the range core to the highly fragmented trailing edge. We measured more than 5000 offspring sampled from three genetically distinct regions (core, margin and edge) grown in three common gardens replicated along a latitudinal gradient. Genetic variation in phenology and growth showed low to moderate heritability and differentiation among regions, suggesting some potential to respond to selection. Phenology traits were highly plastic, but this plasticity was generally neutral or maladaptive in the effect on growth, revealing a potential liability under warmer climates. These results suggest future climate adaptation will depend on the regional availability of genetic variation in red spruce and provide a resource for the design and management of assisted gene flow.
This article is part of the theme issue ‘Species’ ranges in the face of changing environments (Part II)’.
Keywords: quantitative genetics, local adaptation, phenotype, provenance, conservation
1. Background
Geographical ranges vary greatly among species with regard to size and degree of fragmentation [1]. The distribution of a species reflects its biogeographic history of range expansion and migration as well as its pattern of adaptation to different environments within the range. These processes play a major role in shaping the current fit between a species' functional traits and its climatic niche, as well as the ability of species to respond to changing climates [2].
For widespread, mobile species with little range fragmentation, the response to climate warming can involve migration poleward in latitude [3] or upwards in altitude [4]. However, the current rapid pace of climate change is a major problem for geographically restricted or fragmented species, especially those that are sessile and have long generation times, such as forest trees in which genetic adaptation may be slow [5]. In the past, trees exhibited remarkably high migration rates according to pollen records, but current climate change predictions indicate that trees require implausibly high migration rates to match the climate shifts of the future [6]. In the face of migration lag, mature trees may be exposed to long periods of unfavourable climatic conditions including effects of phenological mismatches, heat and drought stress and/or failure to meet chilling requirements [7,8]. Thus, tree responses to rapid climate change will depend more on standing genetic variation within current populations, as well as the ability of existing genotypes to plastically respond to climate stress, both of which may depend on patterns of past migration and population fragmentation in different parts of the range [9].
When a species cannot migrate rapidly, then the current pattern of local adaptation—that is, how phenotypes have genetically diverged across the range to match their local climate conditions—may increasingly become mismatched from the optimal phenotypic value as the climate shifts. In these cases, the amount of standing genetic variation in climate-adaptive traits will be critical to responding to shifting selection pressures. Evolutionary adaptation along climatic gradients requires the presence of genetic variation in functional traits that underlie the response of plant fitness to the abiotic environment [10]. Functional traits such as phenology (i.e. the timing of growth and dormancy) are known to be highly responsive to climate and often exhibit clinal differentiation along climatic gradients [11,12]. Adaptive genetic variation among individuals resulting from natural selection on traits such as phenology is expected to vary across landscapes, but the spatial distribution of this variation is also shaped by historical processes such as migration and range fragmentation [13].
There are multiple schools of thought regarding how adaptive genetic variance might be partitioned across the range. According to the centre-periphery hypothesis, marginal populations that typically occur in unfavourable environments and at lower densities at the periphery of the range are predicted to have lower adaptive genetic variation than populations in the core of the range, and therefore are more prone to extinction [14]. However, the validity of this hypothesis at broader geographical scales has been questioned, as phylogeographic surveys indicate that range-wide patterns of genetic variation are strongly shaped by historical processes during past climate-driven range shifts and not just current range configuration [15]. For example, post-glacial range expansions often produced founding events during colonization that reduced genetic variation of populations expanding into new regions [16]. Additional effects of history can be evident in ‘rear edge’ populations that occur near areas of former glacial refugia in many species. These rear edge populations may be small and fragmented, but also can support high regional diversity as a consequence of their greater age and persistence [15]. Where the genetic variance for climate-adaptive traits is located within the range has important implications for where evolutionary responses to climate change may be most constrained.
In addition to knowing where within the range adaptive genetic variation for individual traits is distributed, predicting selection responses to climate change may be impacted by genetic correlations between suites of traits. When the direction of genetic correlation between traits is antagonistic to the direction of selection [17], this imposes an additional constraint that slows down a population's ability to track spatio-temporal environmental heterogeneity [18]. Genetic correlations between traits arise at the genotype level due to features of genetic architecture such as pleiotropy and physical linkage, although in rare cases correlations may also reflect assortative mating and correlational selection within a population. At the among-population level, genetic correlations can similarly reflect pleiotropy and linkage disequilibrium (LD) caused by linkage and drift in allele frequencies, but there is an additional component of correlational selection for combinations of trait values that may be selected together (i.e. ‘selection covariance’ [19]). Despite their potential to impose constraint on the evolutionary response of populations to future changes in selection, few studies have investigated how genetic correlations between suites of traits may impact climate change responses [20].
While spatial patterns of adaptive genetic variation and covariation in traits will influence the potential for evolutionary change, in the short-term, more immediate responses to climate change may be mediated by phenotypic plasticity. Phenotypic plasticity is the ability of a genotype to produce distinct phenotypes in response to different environments. Plasticity is hypothesized to be important for a species to respond to climate change, as plastic changes can happen within a generation unlike evolutionary processes that occur across generations [5,21,22]. Thus, phenotypic plasticity may be crucial for short-term response to climate change, and may provide a population enough time for an adaptive response to a novel environment [23]. Evolution of plasticity is predicated on the amount of genetic variation in the plastic responses of different genotypes within a population, known as genotype-by-environment (G × E) interaction, which can vary greatly by species and trait [24,25]. As such, the magnitude of GxE determines how much the norm of reaction can evolve in response to selection favouring genotypes whose change in trait expression maximizes fitness under the new environment. Thus, GxE provides a reservoir of different plastic responses to environmental variation, some of which may be adaptive, and thus has wide-ranging implications for how populations may respond to environmental change [26].
However, the mere occurrence of plasticity or GxE does not guarantee that plastic responses will help maintain homeostasis for fitness under changing climates. The plastic responses of genotypes within a population to a novel environment can be adaptive or maladaptive depending on the direction of change in the environment and its effect on the phenotype. Adaptive plasticity is the change in expression of a phenotype by a genotype that allows it to tolerate a range of environmental conditions without a loss in fitness that would otherwise arise if the phenotype remained static [27]. However, there are theoretical studies that indicate numerous constraints and limits on being plastic both at ecological and genetic levels and as such can result in the expression of maladaptive phenotypes [28]. This can occur when unreliable environmental cues trigger a non-optimal phenotypic response, which might reduce fitness in that environment. There are also genetic costs for maintaining the machinery for plastic responses, which require energy and material expenses [29]. In sum, evidence of plasticity in itself does not confer it to be adaptive, as plasticity can also be non-adaptive or maladaptive, resulting in reduced fitness [30].
While many studies have begun investigating species for their ability to respond to climate change through adaptive evolution or phenotypic plasticity, there remain few examples that address these issues in species with highly fragmented ranges, where the spatio-temporal history of isolation, drift and selection may result in spatially complex patterns of adaptive potential. As a result, there is currently a major gap in our knowledge of how different regions within a fragmented species range may respond similarly or differently to climate change, and consequently, where resource managers should begin focusing conservation efforts. In this study, we aim to help fill this gap with a case study of adaptive genetic variation in red spruce (Picea rubens Sarg.)—a prime example of a climate-sensitive tree species with a highly fragmented range. The range of suitable climatic environments for red spruce is predicted to shift dramatically northwards in latitude by the end of the twenty-first century, with severe range contraction and loss of suitable climate predicted for the southern fragmented part of its range [31]. While climate-based suitability models can help inform how species occurrence (expressed as a probability) may be affected by climate change, they generally do not predict the response of functional traits like phenology or growth. Here, we use red spruce to address how climate-adaptive variation is distributed across a species range. We focus on variation in the timing of seasonal phenology (spring bud break and late-summer bud set) and their relation to height growth (a proxy for fitness in forest trees), as these traits often show clinal divergence along climatic gradients in trees, exhibit heritable genetic variation and are known to be phenotypically plastic to environmental cues [32]. Specifically, we address the following questions:
Q1: How do genetic, plastic and GxE variation for phenology and growth traits vary in different regions of a species range that differ in the degree of fragmentation, demographic history and local climate conditions?
Q2: Do genetic correlations exist between phenological and growth traits, and how may these impact evolutionary potential to respond to warming climates?
Q3: Is phenotypic plasticity in phenology traits adaptive or maladaptive under warming climates?
2. Methods
(a) . Study system
Red spruce is a long-lived temperate, coniferous tree species, often achieving ages of over 350 years [33]. Beginning in the 1980s, red spruce experienced widespread growth decline as a result of anthropogenic pollution and climate warming [34]. Red spruce has a highly fragmented distribution in the Central and Southern Appalachians, where populations at the ‘rear edge’ of the range are often isolated from each other and limited to elevations above 1066 m in West Virginia and above 1371 m in North Carolina and Tennessee [35]. Further to the north, red spruce forms more contiguous stands and is a common member of the cool-temperate forests of New England and maritime Canada. The species shows strong signals of decline in effective population size (Ne) and genetic subdivision across its range, based on analysis of whole exome sequencing [36]. The genetic subdivision closely follows geography and is divided into three distinct ancestry groups (henceforth referred to as regions) viz. core, margin and edge (figure 1), demonstrating a lack of connectivity between the current range core of the species in the northeast and the fragmented margin and trailing edge regions in the southeast. These three geo-genetic regions diverged from each other during the Holocene as red spruce expanded from a southern refugium near the present-day edge to track the receding glacier northward towards the present-day core [31,36]. Thus, the regions differ in terms of their geography, their degree of population fragmentation and in their expansion history, but it is unknown how they differ for quantitative genetic variation and plasticity in climate-adaptive phenological traits. For populations in the edge region, the lack of connectivity with the core may prevent natural migration northwards, and migration upwards in altitude may be limited by maximum elevations available in this region. Meanwhile, populations in the core may have greater connectivity, but the predicted shifts in climate within the lifetimes of single individuals draws into question whether natural migration will keep pace with climate change velocity.
Figure 1.
Map of the geographical locations of 65 Picea rubens populations (circles) sampled for this study. Colours indicate the assignment to geo-genetic regions based on genomic analyses. The three common garden sites at Vermont, Maryland and North Carolina are indicated as grey squares along with the elevation (m.a.s.l.) at which they are located. The brown shaded area on the map indicates the known range extent of red spruce [37].
(b) . Study site and experimental design
Seeds were collected primarily during late summer and autumn of 2017 from naturally occurring stands across red spruce's range, as far south as North Carolina and Tennessee and north to New England and New Brunswick, Canada. Care was taken to avoid planted or reforested areas. Seeds were collected from open-pollinated maternal trees. Since red spruce is wind-pollinated and predominantly outcrossing, the identity of the sire is unknown and we thus consider offspring as maternal half-siblings (hereafter referred to as families) in this study. The number of cones sampled varied by tree depending on accessibility and reproductive output, averaging 34 cones per tree. We sampled 340 families, typically occurring as sets of three to five families from each of 65 populations across its range in North America (figure 1). Here, populations refer to the local area where a group of nearby families were sampled. We included seed sources from elevations as a low as 776 m in West Virginia, 1036 m in Tennessee and 1251 m in North Carolina; locations outside those reported by the USDA [35]. The seeds from all trees were cleaned, germinated in petri dishes with wetted sand in a controlled germination cabinet, and grown in a soil-less potting media (ProMix MX) in 164 ml ‘conetainers’ (SC10R; Steuwe and Sons) in 98-cell racks at the University of Vermont greenhouse from April 2018 to early May 2019 [38]. The positions of the seedlings were randomized in each rack, and rack position was randomized once per week within the greenhouse. Seedlings received a liquid fertilizer application once per week using a mixture of Jack's LX All Purpose (21-5-20) and Jacks Dark Weather plus Mag (15-0-14) at a ratio that delivered 150 ppm of N. At least 15 seedlings were grown for each maternal family, plus extra seedlings used for replacements, border rows and fillers (see below).
In spring 2019, seedlings were planted into three common garden sites located near Burlington, Vermont (VT: N 44.4759°, W −73.2121°), Frostburg, Maryland (MD: N 39.642483°, W −78.939213°) and Asheville, North Carolina (NC: N 35.504163°, W −82.5995°) (figure 1). The VT garden is located at the northern part of the range, the MD garden towards the latitudinal mid-point of the range and the NC garden at the southern portion of the range, with elevations of each garden being 59 m, 588 m and 665 m, respectively. This latitudinal gradient in garden sites was thus chosen to provide an expected gradient of increasing temperature and growing season length from north to south. Five seedlings from each of the 340 families were planted at each garden in a randomized block design, with one seedling/family in each of five raised beds (blocks). Timing of plantings was staggered by garden site climate based on historic average date of last frost (NC: 17 April 2019; MD: 6 May 2019; VT: 8 May 2019). To maintain density and avoid edge effects on experimental seedlings, a border row was planted around the perimeter within each bed. Beds at each garden were filled with the same soil-less growth media (General Purpose Mix 703F from ProSource Plus LLC) supplemented with a fertilizer bead application (Osmocote Plus 15-9-12; applied at a rate of 1.02 kg per m2). Beds were located in open areas, and overhead light intensity was reduced by 50% through use of a shade cloth cover. Shade cloths were taken down before the onset of winter snow in late autumn 2019 after all plants were dormant, and erected again in late spring 2020.
(c) . Phenotypic trait measurements
Field Book v. 4.2.1 [39] software on an android tablet was used to record and maintain the database of phenotypic and phenological traits in the field. The initial height of the seedlings was measured to the nearest 0.5 cm immediately after planting in 2019, and again at the end of each growing season in 2019 and 2020. These data were used to calculate seasonal height increment growth as the difference between final and initial height for each growing season. Observation of bud set (recorded as Julian day of year (DOY)) began in mid-July each year as elongation of the main stem ceased and was assessed 2–3 times per week until all buds had set. In 2019, we scored bud set as stage 4 of Dhont et al. [40], characterized by the emergence of fully formed apical bud that is brown in colour and clearly visible with whorls of needles beginning to open outwards. In 2020, we revised this approach to recording bud set stages 1–4 (on the Dhont et al. scale) for each seedling during every census and using nonlinear regression to fit the estimated timing when the plant reached stage 4 (see below). This was implemented to avoid errors from attempting to score a single discrete date for bud set. In spring 2020 (late March–early June), bud break was recorded as the DOY when the needles emerged from the bud scales and was converted to physiological units of growing degree days (GDD) frequently reported in phenology studies [41]. The equation for estimating GDD was as follows:
where TMAX and TMIN are the daily maximum and minimum temperature and TBASE was set to 0°C. The daily temperature at each garden site was obtained from the gridMET dataset [42]. Cumulative growing degree days (cGDD) were derived by summing the GDD for each day from 1 January to the DOY of bud break.
(d) . Question 1: variability in climate and climate-adaptive traits across the range
In order to address research question 1, we quantified climate transfer distance, trait variability and heritability of the traits as described below.
(i) . Climate relationships of sources and garden sites
To describe how historic climatic conditions varied among families sourced from different regions, we used principal component analysis (PCA) in R [43] using the prcomp function on multiple climatic variables available from the climate NA database [44] and annual potential evapotranspiration from ENVIREM [45]. A total of 11 climate variables were selected to generate the principal component (PC) scores: chilling degree days (DD_0), heating degree days (DD18), mean annual solar radiation (MAR) in MJ m−2 d−1, precipitation as snow (PAS) in mm, May to September precipitation (MSP) in mm, per cent mean annual relative humidity (RH), extreme maximum temperature (EXT) over 30 years in °C, Hargreaves climatic moisture deficit (CMD) in mm, continentality (TD) in °C, day of the year on which frost-free period ends (eFFP) and annual potential evapotranspiration (PET) in mm. Our variable selection process aimed to select ecologically relevant variables that show variation across the climatic space occupied by red spruce and delineate this space well while minimizing intercollinearity (see [46] for more details). Variables were centred and scaled prior to analysis.
(ii) . Genetics, environment and genotype-by-environment interaction
To investigate the effect of region, planting environment and GxE on phenology (DOY for bud set and cGDD and DOY for bud break) and growth, we ran a series of linear mixed effects models (LMMs) for each trait estimating the fixed effects of source region (core, margin and edge) and garden (VT, MD and NC), as well as the random effects of population nested within region, bed nested within garden and family nested within population and region using the R package lme4 [47]. We modelled random intercepts for the population and bed effects, while for family we modelled random intercepts and slopes among the gardens (e.g. 1 + garden|family) representing the G × E interaction effect (model I, electronic supplementary material, table S1). Simplified LMMs were performed without the garden|family random effect and likelihood ratio tests (LRTs) used to determine statistical significance of G × E within populations (model I.a, electronic supplementary material, table S1). We tested for the significance of quantitative genetic variation within populations (G + G × E) by fitting a simplified model that dropped the family and garden|family random effects (model I.b, electronic supplementary material, table S1) and compared this to the full model using LRTs to determine statistical significance. A second simplified model was performed dropping just the garden|family random effect and compared to the full model using LRTs to determine significance of G × E.
(iii) . Heritability of the traits
Broad-sense heritability estimates for the traits were calculated through a Bayesian approach using the R package MCMCglmm [48]. Broad-sense heritability was estimated as the proportion of genetic variance over the total phenotypic variance:
where VG is the broad-sense genetic variance estimated from the half-sib families () and populations (), and VP is the total phenotypic variance. Because the expression of phenotypic variance, and hence heritability, is environment-specific, we estimated H2 separately for each garden. The Bayesian model for each trait at each garden was run for 1 × 107 iterations after a burn-in of 1 × 104 and thinning interval of 1000 iterations with weakly informative priors (variance, ‘V’ = 1 and degree of belief parameter, ‘nu’ = 0.002 for the residual and random variance) for all the traits, except bud set in 2020 where parameters expanded priors (prior means, ‘alpha.mu’ = 0 and prior covariance matrix, ‘alpha.V’ = 100 for random variance) were used [49] (model II, electronic supplementary material, table S1).
(iv) . Trait plasticity
To estimate phenotypic plasticity for each trait, we ran LMMs (model IV, electronic supplementary material, table S1) that included the fixed effect of garden climate (gPC1, based on each garden's eigenvector score along PC1 of the climate PCA for the year of measurement) and random effects of bed (1|bed) and family with random intercepts and slopes (gPC1|family). We used the ‘coef()’ function to extract the random slopes for each family with gPC1, which provides a measure of plasticity to garden climate [30]. To test if the magnitude of plasticity reflected differences due to source climate variability, we modelled the absolute value of plasticity as a function of source climate (sPC1), with a random effect of population (model V, electronic supplementary material, table S1).
(e) . Question 2: genetic correlations between phenology and growth
We estimated genetic correlations between pairs of traits using Bayesian MCMCglmm models. Genetic correlations at the family level were estimated by first extracting family-level BLUPs (best linear unbiased predictors) for each trait from initial LMMs run with a fixed effect of garden and random effects of family and bed (model III.a, electronic supplementary material, table S1). These BLUPs were then scaled by dividing by the standard deviation and used as input to a multi-response MCMCglmm model with random effect of population (model III.b, electronic supplementary material, table S1). The correlation among traits estimated from the unit covariance was taken as the family level genetic correlation, while the correlation among traits from the estimated population covariance was taken as the genetic correlation at the population level. While analysing variation among BLUPs may underestimate the variability within a given trait [50], here we use BLUPs to investigate correlations between traits, as implemented in similar studies looking at genetic correlations [51].
(f) . Question 3: assessing evidence for adaptive phenotypic plasticity
To understand if plasticity in trait expression was adaptive, neutral or maladaptive, we correlated plasticity for each phenology trait to the corresponding family BLUPs for height growth for the season, which we used as a proxy for early-life fitness (model VI, electronic supplementary material, table S1). Height was selected as a fitness proxy because there was limited variability in survivorship in this experiment, and height is arguably a primary early-life fitness component for red spruce seedlings [38]. We predicted either a positive or negative relationship with height growth for a season to indicate if plasticity in the trait was adaptive or maladaptive, respectively.
3. Results
(a) . Seed source and common garden climates
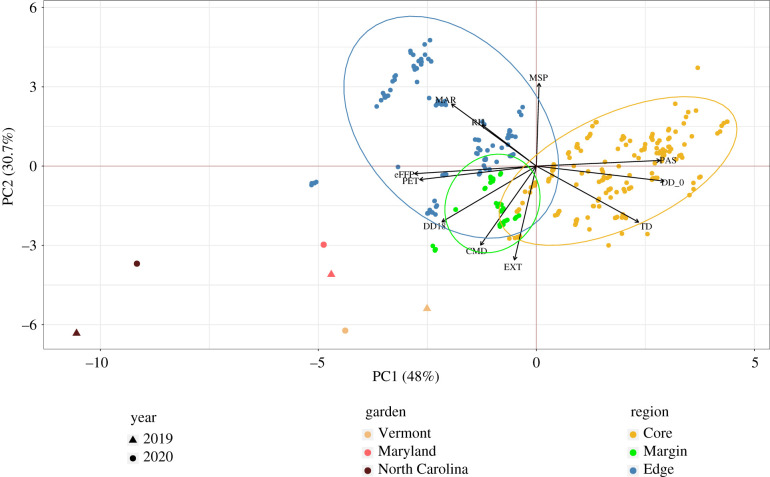
PC1 of the climate PCA explained 48.5% of the climate variance among families (figure 2). Higher PC1 scores corresponded to colder climates with higher seasonality. Chilling degree days (DD_0), PAS and TD increased with increasing PC1 score, while day of the year marking the eFFP, annual PET, MAR and heating degree days (DD18) increased with decreasing PC1 score.
Figure 2.
Principal component analysis (PCA) based on 11 selected variables characterizing the climate of origin (1961–1990 normals) for 340 Picea rubens seed families used in this study. The common garden sites are also plotted based on the selected climate variables for 2019 (triangles) and 2020 (circles). Selected climate variables are chilling degree-days (DD_0), heating degree days (DD18), mean annual solar radiation (MAR) in MJ m−2 d−1, precipitation as snow (PAS) in mm, May to September precipitation (MSP) in mm, per cent mean annual relative humidity (RH), extreme maximum temperature (EXT) over 30 years in °C, Hargreaves climatic moisture deficit (CMD) in mm, continentality (TD) in °C, day of the year on which frost-free period ends (eFFP) and annual potential evapotranspiration (PET) in mm.
PC2 explained an additional 31.8% of the variation in climate among families, with higher scores corresponding to cooler, wetter climates. MSP loaded positively on PC2, while EXT over 30 years and Hargreaves CMD loaded negatively on PC2. Within this overall PCA climate space, the families from the core region occupied the cooler axis with higher seasonality, while those from the edge region covered the warmer climate space with lower seasonality, and the margin was intermediate along PC1, but characterized by the warmest environments with highest moisture deficit along PC2.
For the climate years of this experiment (2019–2020), the three gardens occupied areas of climate space that were generally warmer, drier and less seasonal than the multi-decadal average source climates of the families (figure 2). The VT and MD gardens were generally more similar to the source climates of the families, while the NC garden was notably warmer and drier (lower PC1 and PC2 scores).
(b) . Quantitative genetic trait variation and genotype × environment interaction
(i) . Bud set
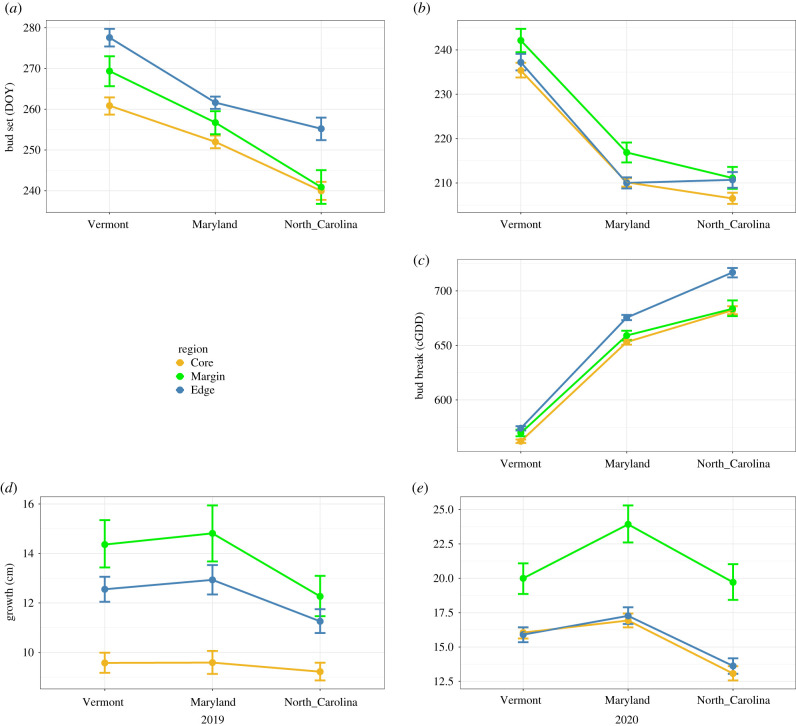
Bud set timing was strongly impacted by both region and garden (figure 3a,b). In 2019, there was a strong environmental effect of garden on bud set timing (p < 0.001), with the earliest bud set at NC (on average 21 days earlier than the other gardens) and latest at VT. Genetic differences among regions (p < 0.001) revealed the core tended to set bud earlier (approx. 240–260 DOY), while plants from the edge tended to set later into the season (approx. 255–280 DOY), irrespective of garden site. During 2020, bud set timing was again strongly influenced by region (p < 0.0001) and garden (p < 0.0001), similar to what was observed during the first growing season (figure 3b). The strong effect of the environment was most pronounced at the MD and NC gardens, where buds set 25 and 29 days earlier (respectively) compared to the VT garden. However, in 2020 genetic divergence among regions was primarily driven by the margin, which tended to have a significantly later bud set timing than the core and edge. In both 2019 and 2020, there were significant garden × region interactions, but these generally reflected small shifts in effect size and not large rank-order changes in means (figure 3).
Figure 3.
Reaction norms for bud set (day of year, DOY) (a,b), bud break (cumulative growing degree days, cGDD) (c) and height growth (
cm) (d,e). Reaction norms for bud set across three common gardens represented by their region of origin (core, margin, edge) assignments based on results of genetic clustering analyses [36]. Colour is coded based on the region ancestry, with yellow, green and blue for core, margin and edge, respectively.
There was abundant within-population genetic variation for bud set in both 2019 and 2020 (electronic supplementary material, figure S2). For 2019, comparing model I (with G and GxE effects) to model I.b (without G or GxE effects) returned a highly significant difference: LRT χ2 = 128.08, d.f. = 6, p < 0.0001. Breaking this down further revealed the presence of broad-sense genetic variation (model I.a versus I.b: LRT χ2 = 77.107, d.f. = 1, p < 0.0001) as well as significant GxE (model I versus I.a: LRT χ2 = 50.972, d.f. = 5, p < 0.0001). The effect of GxE on bud set was evident in the norm-of-reaction plots, which clearly show variability in reaction norms of families with respect to garden sites (electronic supplementary material, figure S2). In 2020, comparing model I to model I.b returned a highly significant difference: LRT χ2 = 80.538, d.f. = 6, p < 0.0001, with the presence of broad-sense genetic variation (model I.a versus I.b: LRT χ2 = 45.36, d.f. = 1, p < 0.0001) as well as significant GxE for bud set (model I versus I.a: LRT χ2 = 35.178, d.f. = 5, p < 0.0001).
(ii) . Bud break
cGDD to bud break in 2020 showed strong effects of both region and garden (figure 3c). Plants in NC and MD broke bud after significantly greater accumulated heat sums (120.18 and 90.76 cGDD, respectively) compared to the more northern VT garden (figure 3c). While bud break occurred with fewer cGDD in the VT garden, this corresponded to a later DOY compared to the two more southern gardens (electronic supplementary material, figure S1). Genetic differences among regions (p < 0.0001) revealed higher cGDD requirements for the edge and margin regions, breaking bud an average of 12.03 and 7.47 cGDD later than plants from the core (figure 3c). Bud break also showed significant interaction effects between gardens and regions (p = 0.001), with edge plants requiring proportionally greater heat sums to break buds at the NC and MD gardens compared to core and margin plants in these gardens. There was abundant within-population genetic variation for cGDD requirement (model I versus model I.b: LRT χ2 = 312.79, d.f. = 6, p < 0.0001), including both significant broad-sense genetic variation (model I.a versus I.b: LRT χ2 = 159.29, d.f. = 1, p < 0.0001) as well as GxE (model I versus I.a: LRT χ2 = 153.5, d.f. = 5, p < 0.0001).
(iii) . Height growth
There was notable differentiation in height growth among regions (p < 0.001), especially between the core compared to the margin and edge in 2019 (figure 3d), with higher growth for the margin and edge evidenced by an increase of 4.74 cm and 3.02 cm, respectively, compared to the core (figure 3d). This trend changed slightly in 2020, with margin exhibiting significantly greater growth (p < 0.001) compared to core and edge, with an increase of 3.76 cm (figure 3e). The height growths for core and edge were in the range of approximately 15–20 cm, while the growth for margin ranged from approximately 15 cm to well beyond 35 cm during 2020; however, this growth increase was primarily attributed to a single population (XBM) extending the growth range for the margin.
The difference in height growth among gardens was non-significant in 2019, indicating a reduced influence of the environment on the final height that plants achieved by season's end. However, there was an overall trend towards decreased growth at the NC garden that most affected plants from the margin, resulting in a significant garden × region interaction (p = 0.001). During 2020, growth was lower at the NC garden, with a significant garden × region interaction (figure 3e).
LRTs for height growth in 2019 revealed abundant within-population genetic variation (model I versus I.b: LRT χ2 = 129.89, d.f. = 6, p < 0.0001), primarily attributable to broad-sense genetic variance among families (model I.a versus I.b: LRT χ2 = 113.39, d.f. = 1, p < 0.0001) as well as smaller but still significant GxE effects (model I versus I.a: χ2 = 16.499, d.f. = 5, p = 0.01). The large genetic variance among families and GxE variance was evident in the norm of reaction plots (supplementary figure S2). Height growth in 2020 also revealed abundant within-population genetic variation (model I versus I.b: LRT χ2 = 63.317, d.f. = 6, p < 0.0001) consisting of variance among families (model I.a versus I.b: LRT χ2 = 57.164, d.f. = 1, p < 0.0001), but with non-significant GxE effects (model I versus I.a: χ2 = 6.1533, d.f. = 5, p = 0.2916) (supplementary figure S2).
(iv) . Heritabilities and genetic correlations between traits
Broad-sense heritability (H2) for phenology and height growth varied across sites and years (table 1 and electronic supplementary material, table S2). Bud break in 2020 had the highest H2 estimate out of all the traits measured, with heritability highest at MD (0.408), followed by VT (0.294) and NC (0.283). H2 for bud set was highest in 2019 at VT (0.252), followed by NC (0.199) and MD (0.12), while in 2020 bud set H2 was markedly lower at VT (0.075), but comparable with 2019 values at MD (0.176) and NC (0.174). H2 for growth was similar across sites in 2019 (VT: 0.195, MD: 0.186, NC: 0.187) and was lower across sites in 2020, decreasing by 45.41%, 33.25% and 39.11% in VT, MD and NC, respectively.
Table 1.
Broad-sense genetic correlations at family (above diagonal) and population level (below diagonal) between functional traits as estimated with Bayesian linear mixed effects models (MCMCglmm, electronic supplementary material, table S1 model III.b). The heritability ranges for each trait are shown along the shaded diagonal. Family- and population-level genetic correlations were estimated among traits from the unit covariance and population covariance of the model, respectively. The lower and upper limits of the 95% confidence interval are given in the square brackets. Bold entries are significant at p < 0.05. Correlations with ‘n.a.’ were not estimated.
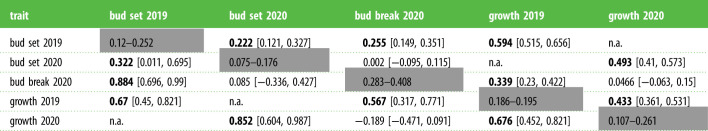 |
Growth and bud set exhibited significantly positive genetic correlations at the population and family levels across both 2019 and 2020 (table 1), indicating that timing of bud set was a strong developmental determinant of height growth. Growth variation among families and populations also showed high repeatability, evidenced by strong positive genetic correlations across years (table 1). Bud set timing was also genetically correlated across years, though not as strong as seen in growth (table 1). Interestingly, there was a highly significant genetic correlation between 2019 bud set and 2020 bud break (population rg = 0.884, family rg = 0.255), indicating that genetic variability for height growth late into the season was associated with delayed resumption of growth the following year. By contrast, there was no significant correlation between bud break and bud set timing within the same year (2020: population rg = 0.085, family rg = 0.002). This cross-year trend was again observed when considering previous year height growth with subsequent year bud break timing (population rg = 0.567, family rg = 0.339), but again this correlation was absent within the same year (2020: population rg = −0.189, family rg = 0.0466).
(v) . Trait plasticity
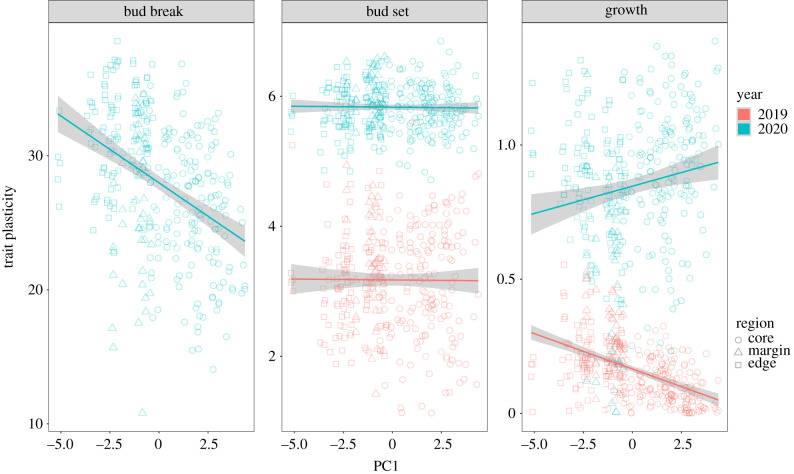
Trait plasticity showed significantly negative association with source climate for bud break (2020) and height growth (2019) (figure 4; model V electronic supplementary material, table S3). Plants from the edge that occupied warmer but less seasonal environments (lower PC1 scores) exhibited higher plasticity for bud break and height growth (2019) compared to plants originating from cooler more seasonal environments found in the margin and core. Plasticity for bud set and height growth in 2020 did not vary significantly with source climate.
Figure 4.
Phenotypic plasticity of Picea rubens phenological (bud set (2019, 2020) and bud break (2020)) and growth traits in relation to the seed families' climate of origin (PC1, figure 2). Plasticity was calculated from the random effects slopes of family values on common garden climates (gPC1|family). Points represent families; lines show the model predictions of the linear mixed effects model (model V, electronic supplementary material, table S1) and shaded areas represent the standard deviation. Points in red are for measurement in 2019 and cyan for measurements in 2020. The regions core, margin and edge are represented by the shapes circle, triangle and square, respectively.
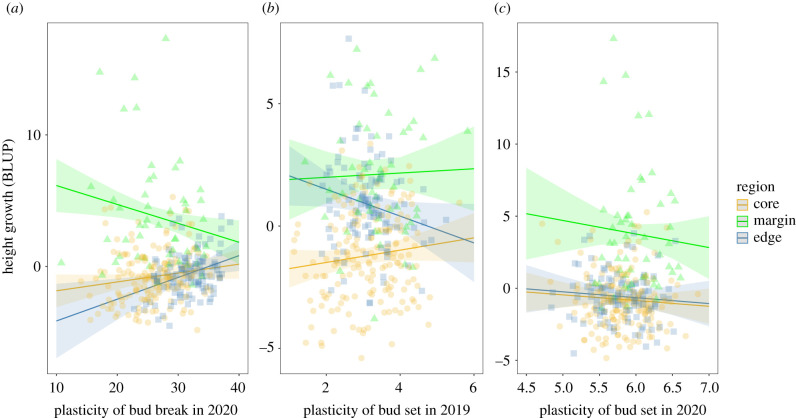
Using height growth as a fitness proxy, we found that levels of plasticity in phenological traits were generally neutral or maladaptive (table 2 and figure 5). For bud break, the plasticity-fitness showed a significant region × plasticity interaction (Model VI, table 2). The plasticity in bud break was significantly maladaptive at the population (p < 0.001) and family (p < 0.01) levels for the margin region, while bud break plasticity was not significantly associated with height growth (i.e. was neutral) for the core and edge regions (figure 5a and electronic supplementary material, figure S3A). In 2019, plasticity of bud set at the family level was significantly maladaptive for the edge (p < 0.05), while core and margin plants showed neutral plasticity (figure 5b). Moreover, plasticity was neutral for all regions for bud set in 2020 (figure 5c), and bud set plasticity for all three regions was neutral at the population level for 2019 and 2020 (electronic supplementary material, figure S3B,C).
Table 2.
F-test results of the linear models analysing the effects of phenotypic plasticity, region and their interaction on height growth (as a proxy for early-life fitness). The F-values are indicated with their significance levels. See electronic supplementary material, table S1 (model VI) for model details.
| trait | plasticity | region | plasticity × region |
|---|---|---|---|
| family level | |||
| bud set (2019) | F1,328 = 2.2125 | F2,328 = 74.4775*** | F2,328 = 3.0416* |
| bud set (2020) | F1,328 = 1.3927 | F2,328 = 77.1650*** | F2,328 = 0.1142 |
| bud break (2020) | F1,328 = 0.0010 | F2,328 = 81.0307*** | F2,328 = 7.0109** |
| population level | |||
| bud set (2019) | F1,59 = 7.2142** | F2,59 = 36.6626*** | F2,59 = 0.2738 |
| bud set (2020) | F1,59 = 2.9188 | F2,59 = 26.7479*** | F2,59 = 0.8523 |
| bud break (2020) | F1,59 = 4.3731* | F2,59 = 36.0075*** | F2,59 = 11.5023*** |
*p < 0.05, **p < 0.01, ***p < 0.001.
Figure 5.
Plasticity for bud break in 2020 (a) and bud set in 2019 (b) and 2020 (c) plotted against height growth BLUP (best linear unbiased prediction) to estimate the adaptive or non-adaptive nature of plasticity present in the trait at the family level. Points represent families; lines show the model predictions of the linear model (model VI, electronic supplementary material, table S1) and shaded areas represent the standard deviation. Corresponding statistical results are shown in table 2.
4. Discussion
(a) . Environmental and genetic variability in phenology and growth
Species with highly fragmented distributions may face limitations on their ability to migrate with climate change, creating a pressing need to assess the potential for adaptive genetic or plastic responses of populations in situ. Phenological traits are an important component of this adaptive response, and the timing of phenology must be well-matched with the local climatic environment to avoid maladaptation at present, and importantly, in the future. The climatic environments inhabited by red spruce vary dramatically in different parts of its highly fragmented range and are coincident with the distribution of three geographically and genetically differentiated regional ancestry groups (figure 1). Thus, the degree to which phenology traits like bud break and bud set are environmentally flexible versus under divergent genetic control has important implications for how red spruce forests in each region are likely to respond to climate change.
We found that bud phenology was highly responsive to environmental differences across our garden sites. Across both years, trees set buds much later in the year (ca 20–30 days) in the northern VT garden than the southern NC garden, with the MD site generally intermediate between these two. A similar difference between sites was observed for the timing of spring bud break in terms of DOY, which occurred 20–30 days earlier at NC compared to VT (electronic supplementary material, figure S1). Thus, growing season length, when measured as the number of days between bud break and bud set, was quite similar between southern and northern sites despite the differences in absolute timing. However, the amount of cGDD required to break buds was much higher at NC than VT (figure 3c), revealing that trees were physiologically slower to break dormancy at NC in spring while also resuming dormancy earlier in autumn. Thus, despite the climatically longer frost-free period in NC compared to VT, the effective growing season (physiologically) was actually more constrained, effectively leading to slightly lower average height growth under the warmest conditions of NC (figure 3). While timing of bud break is known to be strongly temperature-driven, timing of bud set is most likely dependent on an interaction between temperature and photoperiod [52,53]. This might be the reason for the convergence between core and edge in bud set timing during the second growing season at VT and MD, while later bud set was only revealed at the NC site. The NC site was climatically much further away on the warmer side of the climate PCA compared to the other garden sites as well as the average climate of the seed sources (figure 2). Thus, our results for phenology and growth at the NC site, which exists outside the historic climatic range of red spruce, may serve as a good proxy for evaluating responses under a warming climate. The reduced growth and shorter effective growing season at the NC site are indicative of how red spruce seedlings may respond to warmer climates, with varied outcomes depending on region of origin (figure 3).
In addition to the strong environmental effects, there were also clear genetic differences in means, as well as GxE variation, for bud phenology. The significant effect of region indicates the evolution of divergent phenology and growth patterns between the core, margin and edge regions since their divergence following range expansion out of the southern Appalachian refugium, about 8800 years ago [31,36]. Plants from the core set bud up to 20 days earlier under identical conditions compared to plants from the more southern margin and edge regions, consistent with earlier bud set having evolved in response to shorter growing seasons. Interestingly, the pattern of divergence between core and edge was exactly inverse to the environmental influence on bud set: plants with northern (core) ancestry set bud earlier than more southern ancestry sources, while the environmental effect of growing in a northern (VT) environment was to delay bud set, relative to more southern growth environments. This trend was not evident for bud break, which showed an alignment between cGDD at bud break across extremes of ancestry and garden site; that is, less cGDD at bud break for core and in VT compared to edge and in NC. These results are consistent with the core region having adapted its phenology to accommodate the reduced growing season length in the north (figure 3). This was likely an essential adaptation as red spruce expanded away from lower latitudes during post-glacial expansion.
Adaptation to future climate change will require genetic variation in climate-adaptive traits [12] and also lack of strong genetic constraints or trade-offs between traits [17]. There was abundant within-population genetic variation during both seasons, with significant heritability of all traits in each garden site and year. This indicates that red spruce in general has potential for short-term adaptative responses to climate change. There were also abundant genetic correlations among phenology traits and between phenology and growth. Genetic correlations can either facilitate or constrain climate adaptation, since the high correlation between traits means that selection on one trait will produce indirect selection on the correlated trait [17]. Among the strongest and most consistent genetic correlations were those between bud set and height growth for each season, which were significantly positive at both family and population levels (table 1). This indicates that, within the environments of this experiment, genotypes that delayed bud set consistently achieved greater seasonal growth relative to earlier-setting genotypes, pointing to benefits from growing late into the season when conditions permit. Bud set in 2019 also exhibited a strong cross-season genetic correlation with cGDD at bud break the following year (table 1). Plants that set buds later tended to require higher cGDD for bud break in the following year.
The population-level genetic correlation was much higher than the family-level genetic correlations for the traits studied. Genetic correlations can arise due to multiple processes at among-population and among-family levels that may constrain or facilitate selection responses to climate change [17], including pleiotropy, LD (physical and statistical LD) and coordinated selection on multiple traits. The traits we're investigating are likely to be highly polygenic; thus we consider the influence of LD to be minimal, and thus infer correlations are most likely due to pleiotropy (family level) or as a consequence of coordinated selection (‘selection covariance’ sensu [19]). The strong population-level correlation we observed would seem to suggest selection has favoured a coordinated strategy in edge populations of delayed dormancy onset to take advantage of late season growing conditions, while requiring higher cGDD before breaking dormancy the following spring, presumably to avoid pre-mature bud break during unpredictable winter or early spring warm periods. Additionally, the weaker but still significant family-level correlation points to a pleiotropic effect, indicating that genotypes that developmentally delay bud set in the current year will be forced to delay the start to the growing season the following year. This constraint could mean that the timing of spring and autumn transitions may not be able to respond independently to selection, even if future climate conditions might favour their decoupling. By contrast to the effects of environment on bud phenology, height growth was more uniform across garden sites, differing primarily among the regions.
During 2019, plants from the core had the lowest amount of height growth compared to edge and margin plants. This stark difference in growth between core and edge, the two extremes of the species distribution, faded during the second growing season when both regions had comparable height growth. Margin plants on the other hand exhibited consistently high growth during both growing seasons across all garden sites. This robust growth response by plants from the margin could reflect hybrid vigour as a result of introgression with black spruce (Picea mariana), which is known to occur in the margin ([36,54], SR Keller and T Capblancq 2021, unpublished data). This suggests that height growth in our experiment was under greater genetic control and less affected by environmental differences among the test sites, although there was a trend towards lower mean growth in NC for both years. Overall, this pattern is indicative of determinate growth in spruce, in which buds are set after reaching the determined height growth for the season [52].
(b) . The adaptive value of plasticity varies among traits and reflects source climate and ancestry
In addition to the identification of significant genetic (co)variation for phenology and growth, a key finding of this study is GxE for phenology traits and the varying degrees of plasticity expressed by families from different regions. The greatest levels of plasticity occurred in cGDD for bud break (figure 4), and we observed significant associations between source climate (PC1) and the magnitude of plasticity for bud break (2020) and growth (2019) (figure 4, electronic supplementary material, table S3). These results suggest that bud break is highly plastic and that genotypes from warmer environments maintain higher plasticity levels than genotypes from colder environments, but they are not sufficient to say whether or not that plasticity is adaptive since adaptive plasticity must be associated with maintenance of fitness across variable environments [55], and not all phenotypic plasticity ends up being adaptive [28]. When using height growth as a fitness proxy, we found the plasticity for bud set in 2019 was significantly maladaptive for edge plants, but remained neutral for core and margin plants. However, in 2020 bud set plasticity was generally neutral for all three regions. For bud break, plasticity of margin plants was significantly maladaptive while plasticity was neutral or (non-significantly) positive for core and edge plants.
Overall, these results indicate the presence of abundant plasticity in phenological traits; however, plasticity seemed to provide little adaptive benefit during the experiment. While edge plants exhibited higher growth in 2019 than the core, this gap largely closed the following year in VT and MD. The reduced growth of edge plants in 2020 (relative to core plants) may have reflected the higher degree of maladaptive plasticity in edge plants for bud set in 2019. We recognize that we have only captured a snapshot of the range of environmental conditions and life-history stages of this long-lived species, and plasticity for phenology may show different effects on fitness under different conditions (e.g. late-spring frosts) or at later life-history stages. Nevertheless, our results points to a largely non-adaptive role for plasticity during response to environmental change in red spruce, in which trait responses may be highly plastic but do not necessarily buffer fitness against environmental variation.
(c) . Management implications
Assisted gene flow has been proposed as a management practice to counter the effect of rapid climate warming and maintain local adaptation [56]. Successful implementation of assisted gene flow requires the development of transfer guidelines that can evaluate the effect on plant fitness of transferring genotypes from one climate to another, e.g. planting seed sources from warmer climates northwards or upwards in altitude to combat climate change [57]. Such a practice, if properly calibrated and implemented, could help facilitate local adaptation and reduce maladaptation under future novel environments.
Our study shows that the core is currently adapted to shorter growing seasons and as a result sets bud earlier and achieves reduced growth relative to edge genotypes under identical environments. Somewhat paradoxically, the plastic effect of transferring a given genotype to a warmer climate is also to hasten bud set. Therefore, plasticity without adaptation (or assisted gene flow) is unlikely to lead to bud set phenology that is well-matched to future warmer climates, and this appears true for all parts of the range. Under the warmest garden in our experiment (NC), edge genotypes were shown to have later bud set relative to the core in both years of growth, which allowed them to attain higher height growth in both seasons, but particularly in 2019. This is also borne out by the significant genetic correlation between bud set timing and height growth observed across years. For spring phenology, a warming climate may be expected to increase the cGDD until bud break (as seen in NC), and edge genotypes tended to have the largest cGDD requirements across gardens. Whether this reflects an adaptation to warm and/or variable springs to avoid premature release from dormancy is unknown, but we did not observe any genetic correlation between bud break and height growth during our experiment. So at least for the early-life stages monitored in this experiment, higher cGDD to bud break seems to not have major consequence for seedling performance, and may actually be a pleiotropic effect of later bud set in the previous season. Taken together, the case could then be made that assisted gene flow from edge to core could have benefits of introducing genotypes with delayed bud set and therefore greater height growth under expected warming climates, with little cost in terms of bud break phenology.
However, it should be also be noted that although the edge region on average shows phenology that appears adapted to longer growing seasons, the range of family mean values shows broad overlap between core and edge within a given garden site, including the site with the warmest conditions (NC). This finding, along with the significant heritability for phenology and growth, suggests that standing levels of genetic variation within the core is already high, and edge genotypes may provide little additional benefits, at least for these specific traits.
Lastly, plants from the margin appear to behave quite differently from the other two regions. Plants from the margin express very high growth, likely attributable to hybrid vigour through introgression with its congener, black spruce. Despite the fact that bud break plasticity under warming was highly maladaptive for margin plants, their hybrid vigour outperformed core and edge plants across all environments. However, margin plants did suffer a decline in growth at the range edge sites (VT and NC) compared to the more central MD site. One could argue that past introgression of black spruce genes into margin populations represents another form of (unassisted) gene flow that has produced clear benefits in terms of growth across the entire range of climates tested in our experiment. Thus, natural hybrid genotypes such as those that appear in the margin and have been vetted by selection, may offer an additional tool to consider in the effort to select genotypes for restoration and conservation.
Acknowledgements
We thank the many people who have contributed to the collection and processing of the red spruce seed collections used here, including Barbara Crane, Robert Jetton, John Major, John Malcom, Brittany Verrico, Victor Vankus, Camcore, the PA Department of Conservation and Natural Resources. We especially appreciate the efforts of G. Scott Bryan, Thomas Christensen, Douglas Gross, Robert Eaton, Michael Kudish and Christopher Maier, to locate candidate trees and collect tissue samples. We also appreciate the help of those who helped with establishing, managing and monitoring the common gardens, including Thibaut Capblancq, Katie Bardsley, Noah Kaufman, Natalie Haydt, Robin Paulman, Erica Duda and John Piasecki. Lastly, we thank two anonymous reviewers for comments on this work, and the organizers of the symposium on ‘Evolution of Species Ranges' for their work in assembling this special issue and its contributions.
Data accessibility
Data and code for analysis are available at https://anoobvinu07.github.io/NSF_RedSpruce_CG/ with doi:10.5281/zenodo.5701667. Data are available at: https://github.com/anoobvinu07/NSF_RedSpruce_CG/tree/main/data. The data are provided in the electronic supplementary material [58].
Authors' contributions
A.P.: data curation, formal analysis, investigation, methodology, visualization, writing—original draft, writing—review and editing; S.D.Y.: investigation, project administration, resources; S.L.: investigation, writing—review and editing; J.L.A.: investigation; K.J.: investigation, project administration, supervision; J.R.B.: conceptualization, resources, writing—review and editing; D.M.N.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing; M.C.F.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing; S.R.K.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by National Science Foundation (NSF) grant nos. 1656099 to S.R.K. and 1655344 to M.C.F. and D.M.N.
References
- 1.Gaston KJ. 2003. The structure and dynamics of geographic ranges. New York, NY: Oxford University Press on Demand. [Google Scholar]
- 2.Parmesan C. 2006. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637-669. ( 10.1146/annurev.ecolsys.37.091305.110100) [DOI] [Google Scholar]
- 3.Jump AS, Mátyás C, Peñuelas J. 2009. The altitude-for-latitude disparity in the range retractions of woody species. Trends Ecol. Evol. 24, 694-701. ( 10.1016/j.tree.2009.06.007) [DOI] [PubMed] [Google Scholar]
- 4.Morueta-Holme N, Engemann K, Sandoval-Acuña P, Jonas JD, Segnitz RM, Svenning J-C. 2015. Strong upslope shifts in Chimborazo's vegetation over two centuries since Humboldt. Proc. Natl Acad. Sci. USA 112, 12 741-12 745. ( 10.1073/pnas.1509938112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savolainen O, Bokma F, García-Gil R, Komulainen P, Repo T. 2004. Genetic variation in cessation of growth and frost hardiness and consequences for adaptation of Pinus sylvestris to climatic changes. For. Ecol. Manag. 197, 79-89. ( 10.1016/j.foreco.2004.05.006) [DOI] [Google Scholar]
- 6.Davis MB, Shaw RG. 2001. Range shifts and adaptive responses to quaternary climate change. Science 292, 673-679. ( 10.1126/science.292.5517.673) [DOI] [PubMed] [Google Scholar]
- 7.Kimmins J, Lavender D. 1987. Implications of climate change for the distribution of biogeoclimatic zones in British Columbia and for the growth of temperate forest species. In Woody plant growth in a changing chemical and physical environment (ed. DP Lavender), pp. 209–309. Vancouver, BC: University of British Columbia Press.
- 8.McCreary D, Lavender D, Hermann R. 1990. Predicted global warming and Douglas-fir chilling requirements. Ann. Sci. For. 47, 325-330. ( 10.1051/forest:19900404) [DOI] [Google Scholar]
- 9.Valladares F, et al. 2014. The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecol. Lett. 17, 1351-1364. ( 10.1111/ele.12348) [DOI] [PubMed] [Google Scholar]
- 10.Bussotti F, Pollastrini M, Holland V, Brueggemann W. 2015. Functional traits and adaptive capacity of European forests to climate change. Environ. Exp. Bot. 111, 91-113. ( 10.1016/j.envexpbot.2014.11.006) [DOI] [Google Scholar]
- 11.Parmesan C, Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37-42. ( 10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 12.Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis-McLane S. 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95-111. ( 10.1111/j.1752-4571.2007.00013.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowry DB. 2010. Landscape evolutionary genomics. Biol. Lett. 6, 502-504. ( 10.1098/rsbl.2009.0969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vucetich JA, Waite TA. 2003. Spatial patterns of demography and genetic processes across the species' range: null hypotheses for landscape conservation genetics. Conserv. Genet. 4, 639-645. ( 10.1023/A:1025671831349) [DOI] [Google Scholar]
- 15.Hampe A, Petit RJ. 2005. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8, 461-467. ( 10.1111/j.1461-0248.2005.00739.x) [DOI] [PubMed] [Google Scholar]
- 16.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907-913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 17.Etterson JR, Shaw RG. 2001. Constraint to adaptive evolution in response to global warming. Science 294, 151-154. ( 10.1126/science.1063656) [DOI] [PubMed] [Google Scholar]
- 18.Nürnberger B. 2001. Ecological genetics. In Encyclopedia of biodiversity (ed. Levin SA), pp. 245-258. New York, NY: Elsevier. See https://www.sciencedirect.com/science/article/pii/B012226865200081X. [Cited 2 Nov. 2021.] [Google Scholar]
- 19.Armbruster WS, Schwaegerle KE. 1996. Causes of covariation of phenotypic traits among populations. J. Evol. Biol. 9, 261-276. ( 10.1046/j.1420-9101.1996.9030261.x) [DOI] [Google Scholar]
- 20.Shaw RG, Etterson JR. 2012. Rapid climate change and the rate of adaptation: insight from experimental quantitative genetics. New Phytol. 195, 752-765. ( 10.1111/j.1469-8137.2012.04230.x) [DOI] [PubMed] [Google Scholar]
- 21.Chevin L, Collins S, Lefèvre F. 2013. Phenotypic plasticity and evolutionary demographic responses to climate change: taking theory out to the field. Funct. Ecol. 27, 967-979. ( 10.1111/j.1365-2435.2012.02043.x) [DOI] [Google Scholar]
- 22.Franks SJ, Weber JJ, Aitken SN. 2014. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 7, 123-139. ( 10.1111/eva.12112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevin L-M, Lande R. 2010. When do adaptive plasticity and genetic evolution prevent extinction of a density-regulated population? Evolution 64, 1143-1150. ( 10.1111/j.1558-5646.2009.00875.x) [DOI] [PubMed] [Google Scholar]
- 24.Falconer DS. 1952. The problem of environment and selection. Am. Nat. 86, 293-298. ( 10.1086/281736) [DOI] [Google Scholar]
- 25.Murren CJ, et al. 2014. Evolutionary change in continuous reaction norms. Am. Nat. 183, 453-467. ( 10.1086/675302) [DOI] [PubMed] [Google Scholar]
- 26.Saltz JB, Bell AM, Flint J, Gomulkiewicz R, Hughes KA, Keagy J. 2018. Why does the magnitude of genotype-by-environment interaction vary? Ecol. Evol. 8, 6342-6353. ( 10.1002/ece3.4128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pigliucci M. 2001. Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: John Hopkins University Press. [Google Scholar]
- 28.Kleunen MV, Fischer M. 2005. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol. 166, 49-60. ( 10.1111/j.1469-8137.2004.01296.x) [DOI] [PubMed] [Google Scholar]
- 29.DeWitt TJ, Sih A, Wilson DS. 1998. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13, 77-81. ( 10.1016/S0169-5347(97)01274-3) [DOI] [PubMed] [Google Scholar]
- 30.Arnold PA, Nicotra AB, Kruuk LEB. 2019. Sparse evidence for selection on phenotypic plasticity in response to temperature. Phil. Trans. R. Soc. B 374, 20180185. ( 10.1098/rstb.2018.0185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capblancq T, Lachmuth S, Fitzpatrick MC, Keller SR. 2022. From the last glacial maximum to the end of the 21st century: the journey of a montane tree species. Manuscript in preparation. [DOI] [PMC free article] [PubMed]
- 32.Savolainen O, Pyhäjärvi T, Knürr T. 2007. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 38, 595-619. ( 10.1146/annurev.ecolsys.38.091206.095646) [DOI] [Google Scholar]
- 33.Blum BM. 1990. Picea rubens Sarg. red spruce. Silv. N. Am. 1, 250-259. [Google Scholar]
- 34.Reams GA, Deusen PCV. 1993. Synchronie large-scale disturbances and red spruce growth decline. Can. J. For. Res. 23, 1361-1374. ( 10.1139/x93-173) [DOI] [Google Scholar]
- 35.USDA, NRCS. 2021. The PLANTS Database. National Plant Data Team, Greensboro, NC, USA. See http://plants.usda.gov. [Cited Apr. 12 2021.]
- 36.Capblancq T, Butnor JR, Deyoung S, Thibault E, Munson H, Nelson DM, Fitzpatrick MC, Keller SR. 2020. Whole-exome sequencing reveals a long-term decline in effective population size of red spruce (Picea rubens). Evol. Appl. 13, 2190-2205. ( 10.1111/eva.12985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little EL Jr. 1971. Atlas of United States trees. Conifers and important hardwoods. Vol. 1. Washington DC: US Department of Agriculture Forest Service Miscellaneous Publication 1146.
- 38.Capblancq T, Munson H, Butnor JR, Keller SR. 2021. Genomic drivers of early-life fitness in Picea rubens. Conserv. Genet. 22, 963–976. ( 10.1007/s10592-021-01378-7) [DOI]
- 39.Rife TW, Poland JA. 2014. Field book: an open-source application for field data collection on android. Crop Sci. 54, 1624-1627. ( 10.2135/cropsci2013.08.0579) [DOI] [Google Scholar]
- 40.Dhont C, Sylvestre P, Gros-Louis M-C, Isabel N. 2010. Guide-terrain pour l'identification des stades de débourrement et de formation du bourgeon apical chez l’épinette blanche / Field guide for identifying apical bud break and bud formation stages in white spruce. See http://cfs.nrcan.gc.ca/publications?id=32086. [Cited 26 Jul. 2021.]
- 41.McMaster GS, Wilhelm WW. 1997. Growing degree-days: one equation, two interpretations. Agric. For. Meteorol. 87, 291-300. ( 10.1016/S0168-1923(97)00027-0) [DOI] [Google Scholar]
- 42.Abatzoglou JT. 2013. Development of gridded surface meteorological data for ecological applications and modelling. Int. J. Climatol. 33, 121-131. ( 10.1002/joc.3413) [DOI] [Google Scholar]
- 43.R Core Team. 2021. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/. [Google Scholar]
- 44.Wang T, Hamann A, Spittlehouse D, Carroll C. 2016. Locally downscaled and spatially customizable climate data for historical and future periods for North America. PLoS ONE 11, e0156720. ( 10.1371/journal.pone.0156720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Title PO, Bemmels JB. 2018. ENVIREM: an expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 41, 291-307. ( 10.1111/ecog.02880) [DOI] [Google Scholar]
- 46.Capblancq T, Lachmuth S, Fitzpatrick MC, Keller SR. 2022. From common garden to adaptive genes: exploring local adaptation to climate in red spruce. Manuscript in preparation. [DOI] [PMC free article] [PubMed]
- 47.Bates D, Mächler M, Bolker B, Walker S. 2014. Fitting linear mixed-effects models using lme4. ArXiv14065823 Stat. See http://arxiv.org/abs/1406.5823. [Cited 11 Apr. 2021.]
- 48.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1-22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 49.Hadfield J. 2014. MCMCglmm course notes. See cran.nexr.com/web/packages/MCMCglmm/vignettes/CourseNotes.pdf.
- 50.Hadfield JD, Wilson AJ, Garant D, Sheldon BC, Kruuk LEB. 2010. The misuse of BLUP in ecology and evolution. Am. Nat. 175, 116-125. ( 10.1086/648604) [DOI] [PubMed] [Google Scholar]
- 51.Yan W, Wang B, Chan E, Mitchell-Olds T. 2021. Genetic architecture and adaptation of flowering time among environments. New Phytol. 230, 1214-1227. ( 10.1111/nph.17229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamilton JA, El Kayal W, Hart AT, Runcie DE, Arango-Velez A, Cooke JEK. 2016. The joint influence of photoperiod and temperature during growth cessation and development of dormancy in white spruce (Picea glauca). Tree Physiol. 36, 1432-1448. ( 10.1093/treephys/tpw061) [DOI] [PubMed] [Google Scholar]
- 53.Singh RK, Svystun T, AlDahmash B, Jönsson AM, Bhalerao RP. 2017. Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. New Phytol. 213, 511-524. ( 10.1111/nph.14346) [DOI] [PubMed] [Google Scholar]
- 54.de Lafontaine G, Prunier J, Gérardi S, Bousquet J. 2015. Tracking the progression of speciation: variable patterns of introgression across the genome provide insights on the species delimitation between progenitor–derivative spruces (Picea mariana × P. rubens). Mol. Ecol. 24, 5229-5247. ( 10.1111/mec.13377) [DOI] [PubMed] [Google Scholar]
- 55.Conover DO, Schultz ET. 1995. Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol. Evol. 10, 248-252. ( 10.1016/S0169-5347(00)89081-3) [DOI] [PubMed] [Google Scholar]
- 56.Aitken SN, Whitlock MC. 2013. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367-388. ( 10.1146/annurev-ecolsys-110512-135747) [DOI] [Google Scholar]
- 57.Rehfeldt GE, Jaquish BC, Sáenz-Romero C, Joyce DG, Leites LP, Bradley St Clair J, López-Upton J. 2014. Comparative genetic responses to climate in the varieties of Pinus ponderosa and Pseudotsuga menziesii: reforestation. For. Ecol. Manag. 324, 147-157. ( 10.1016/j.foreco.2014.02.040) [DOI] [Google Scholar]
- 58.Prakash A, DeYoung S, Lachmuth S, Adams JL, Johnsen K, Butnor JR, Nelson DM, Fitzpatrick MC, Keller SR. 2022. Genotypic variation and plasticity in climate-adaptive traits after range expansion and fragmentation of red spruce (Picea rubens Sarg.). Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Prakash A, DeYoung S, Lachmuth S, Adams JL, Johnsen K, Butnor JR, Nelson DM, Fitzpatrick MC, Keller SR. 2022. Genotypic variation and plasticity in climate-adaptive traits after range expansion and fragmentation of red spruce (Picea rubens Sarg.). Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data and code for analysis are available at https://anoobvinu07.github.io/NSF_RedSpruce_CG/ with doi:10.5281/zenodo.5701667. Data are available at: https://github.com/anoobvinu07/NSF_RedSpruce_CG/tree/main/data. The data are provided in the electronic supplementary material [58].