Abstract
Purpose:
Chemical shift-encoded magnetic resonance imaging enables accurate quantification of liver fat content though estimation of proton density fat-fraction (PDFF). Computed tomography (CT) is capable of quantifying fat, based on decreased attenuation with increased fat concentration. Current quantitative fat phantoms do not accurately mimic the CT number of human liver. The purpose of this work was to develop and validate an optimized phantom that simultaneously mimics the MRI and CT signals of fatty liver.
Methods:
An agar-based phantom containing 12 vials doped with iodinated contrast, and with a granular range of fat fractions was designed and constructed within a novel CT and MR compatible spherical housing design. A four-site, three-vendor validation study was performed. MRI (1.5T and 3T) and CT images were obtained using each vendor’s PDFF and CT reconstruction, respectively. An ROI centered in each vial was placed to measure MRI-PDFF (%) and CT number (HU). Mixed-effects model, linear regression, and Bland-Altman analysis were used for statistical analysis.
Results:
MRI-PDFF agreed closely with nominal PDFF values across both field strengths and all MRI vendors. A linear relationship (slope=−0.54±0.01%/HU, intercept=37.15±0.03%) with an R2 of 0.999 was observed between MRI-PDFF and CT number, replicating established in vivo signal behavior. Excellent test-retest repeatability across vendors (MRI: mean = −0.04%, 95% limits of agreement = [−0.24%, 0.16%]; CT: mean = 0.16 HU, 95% limits of agreement = [−0.15HU, 0.47HU]) and good reproducibility using GE scanners (MRI: mean = −0.21%, 95% limits of agreement = [−1.47%, 1.06%]; CT: mean = −0.18HU, 95% limits of agreement = [−1.96HU, 1.6HU]) were demonstrated.
Conclusions:
The proposed fat phantom successfully mimicked quantitative liver signal for both MRI and CT. The proposed fat phantom in this study may facilitate broader application and harmonization of liver fat quantification techniques using MRI and CT across institutions, vendors and imaging platforms.
Keywords: Phantom, Magnetic resonance imaging, Computed tomography, Liver, Fat
Introduction:
Abnormal accumulation of intracellular triglycerides is the hallmark feature of non-alcoholic fatty liver disease (NAFLD)1–3, which is emerging as the leading cause of liver disease in the Western world. NAFLD is widely expected to become the leading indication for liver transplantation in the near future4. Liver fat is also recognized as a major independent contributor to cardiovascular mortality5, cancer, and type 2 diabetes6. The prevalence of NAFLD is high in the general population (30%) and NAFLD can progress to cirrhosis7, with an associated increased risk of liver failure and liver cancer. For these reasons, non-invasive, rapid and accurate quantification of liver fat is needed for early detection, and quantitative staging and treatment monitoring of NAFLD.
Chemical shift-encoded magnetic resonance imaging (CSE-MRI) methods are well-established as reliable, accurate and reproducible methods for confounder-corrected measurement of proton density fat-fraction (PDFF)8–10, a well-validated quantitative MR imaging biomarker8. CSE-MRI methods are widely used for research and clinical purposes. However, the availability of MRI remains limited compared to X-ray computed tomography (CT). CT is widely available and used more frequently than MRI for abdominal imaging11.
CT is sensitive to the presence of liver fat. Increasing liver fat concentration is quantitatively related to lower X-ray attenuation, and abnormal levels of fat are reflected in a reduction of CT number (Hounsfield units or HU)12–15. For these reasons, there is emerging interest in the use of CT for the detection and staging of NAFLD, as well as harmonization of unenhanced CT-based measurements of liver fat with CSE-MRI based measurement of PDFF14,16,17.
Quantitative phantoms can enable quality assurance (QA) across imaging modalities, locations, sites, and vendors. In large multi-center drug discovery studies that rely on quantitative imaging as primary endpoints, phantom-based QA plays an essential role 14,16–18. MRI is widely used to quantify PDFF as a biomarker of liver fat content. However, the relationship between CT attenuation and MRI-PDFF is not known across different vendors and protocols. To avoid the potential for clinical misdiagnosis, quantitative phantoms may be helpful to set steatosis grading thresholds for different vendors and CT acquisition protocols. Considering the emerging rise of both quantitative CT and MRI techniques, there is an unmet need to develop quantitative phantoms that accurately mimic both the CT and MRI signal properties of fatty liver.
Current quantitative MRI fat phantoms have been developed and used for validation of MRI-based fat quantification18. Promising results show accurate and reproducible fat quantification across sites, vendors, field strengths and protocols using quantitative MRI fat phantoms18. However, MRI fat phantoms may not accurately represent the X-ray attenuation properties of fatty liver when evaluated with unenhanced CT. Given that MRI fat phantoms were not designed for use with CT and that the underlying mechanisms for quantifying fat are different, a quantitative CT fat phantom is essential for the development of CT-based liver fat quantification techniques. A single phantom that simultaneously mimics the MRI and CT signals of fatty liver would also facilitate the harmonization of MRI-PDFF and CT number measurements. Though a simple design phantom showed similar PDFF vs CT number relationship observed in human data from a previous study14, a more comprehensive study including design of quantitative MRI and CT compatible phantom and multi-center, multi-vendor validation is still needed.
Therefore, the overall purpose of this work was to design and develop a MRI and CT compatible phantom that simultaneously mimics the signal behavior of fatty liver using both modalities. This includes an optimized design for the spherical housing that uses MRI and CT compatible materials and a geometry that mitigates magnetic field inhomogeneities, as well as a finer range of fat fraction levels to enable accurate and robust validation. Further, multi-site, multi-vendor evaluation with both MRI and CT was performed to evaluate the accuracy and reproducibility of the phantom and to demonstrate its potential utility. A multi-site, multi-vendor study with both MRI and CT was performed to evaluate the accuracy and reproducibility of the phantom and to demonstrate its potential utility.
Materials and Methods:
Phantom construction:
The proposed phantom aims to enable high quality images free from potential artifacts, while replicating in vivo fat signals, for both MRI and CT, simultaneously.
As shown in Figure 1, the proposed phantom contains 12 vials (Volume: 25mL; Diameter: 20mm) that were built by mixing peanut oil (to mimic liver triglycerides in MRI) with an agar-based emulsion, as previously described by Hines et al19. The base contents of the emulsion included 2% (weight/volume) agar gel (i.e., agar mixed with de-ionized (DI) water), 43mM (millimolar) sodium dodecyl sulfate as surfactant, and 3mM sodium benzoate as preservative (Sigma-Aldrich). The oil-emulsion volume ratio was adjusted to obtain 12 different nominal PDFF levels (0%, 2.5%, 5%, 7.5%, 10%, 15%, 20%, 25%, 30%, 40%, 50%, and 100% (pure oil). Further, compared to past work we increased the number of vials with PDFF values less than 10% to increase the granularity of fat measurements at low fat concentrations. Fat concentrations below 10% PDFF are the most clinically relevant with clinically relevant thresholds of approximately 3.0–6.5%20–22. Further, past work has also shown that the greatest variability with CT attenuation occurs at low liver fat content14. In order to slightly increase the X-ray attenuation coefficients of the agar mixtures16 so that they mimic the attenuation coefficient of human liver, a small amount of iodinated contrast agent (7.3μL iohexol (Omnipaque 300, GE Healthcare) per 1mL of oil-emulsion mixture) was added to each vial. The concentration of the iodinated contrast agent was iteratively adjusted until the vial with 0% PDFF generated the same CT number as that of a non-steatotic liver (65.9HU at 120kVp)14.
Figure 1.
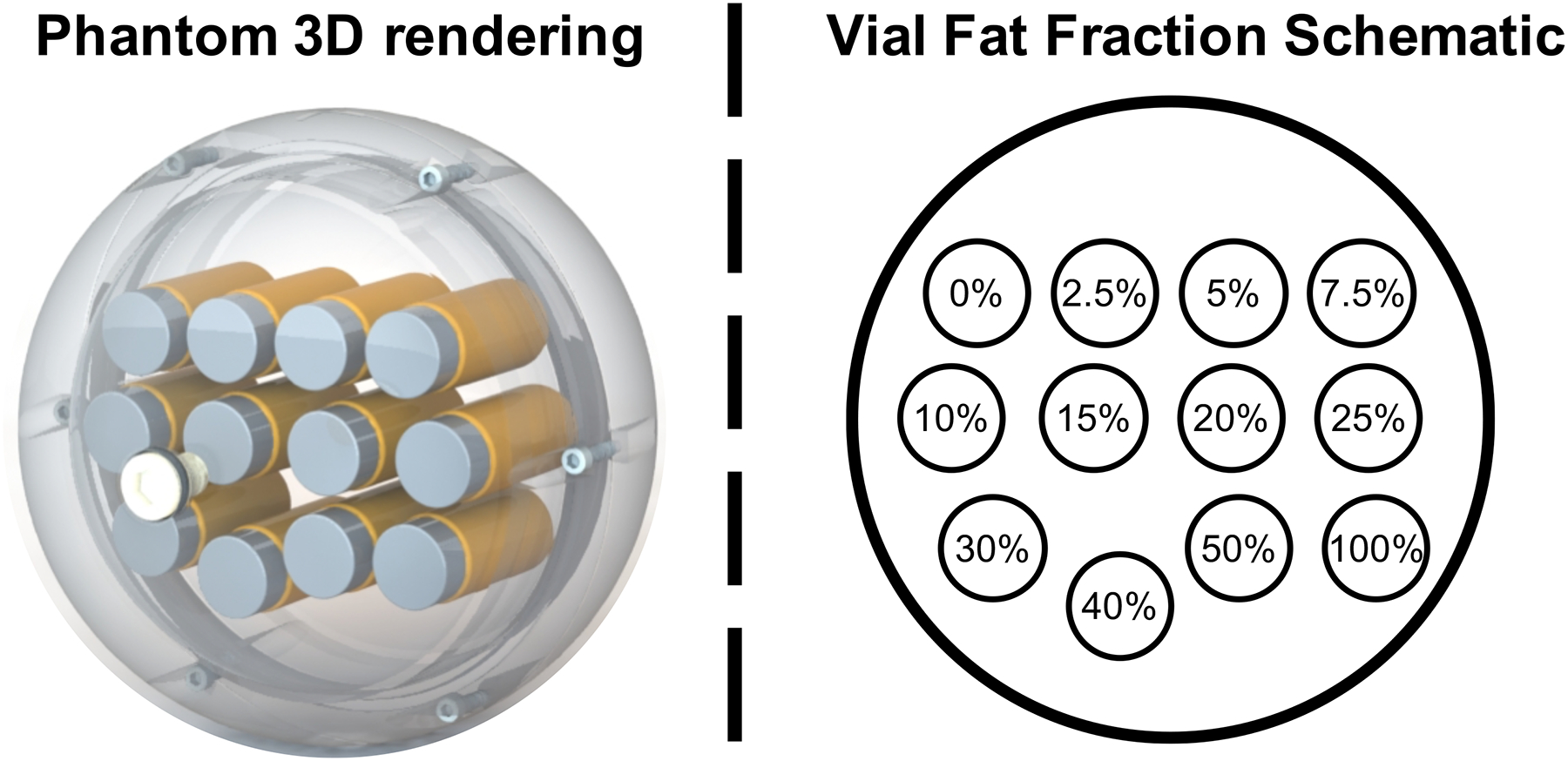
3D rendering phantom image (left) and schematic of the fat-fraction vial layout in the axial plane (right).
The vials were enclosed in a custom designed acrylic spherical housing (Calimetrix) filled with deionized water to mimic the X-ray attenuation environment of a typical adult abdomen and also to optimize magnetic field homogeneity. Low-density nylon screws were used in the sphere housing to avoid CT streak artifacts.
Data acquisition:
A multi-site, multi-vendor (round-robin) validation study was performed using the proposed phantom to study the reproducibility, including test-retest repeatability of fat quantification for both MRI and CT. A total of four sites participated in this study (Site I, Site II, Site III, and Site IV), all located within the United States, using two modalities (MRI and CT), three vendors (GE Healthcare, Siemens Healthineers, and Philips Healthcare) and two magnetic field strengths (1.5T and 3T). Details regarding the sites and vendors are summarized in Table 1 (MRI) and Table 2 (CT). The phantom was placed in a custom designed shipping case (Calimetrix) and shipped overnight between sites.
Table 1:
MRI systems and MRI acquisition parameters across the four sites.
| Site | Scanner # | Vendor | Field Strength (T) | Model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | 1 | GE | 1.5 | Optima MR450w | ||||||
| 2 | GE | 3 | Discovery MR750 | |||||||
| II | 1 | Siemens | 3 | MAGNETOM Skyra | ||||||
| III | 1 | Siemens | 1.5 | MAGNETOM Aera | ||||||
| 2 | Siemens | 3 | MAGNETOM Skyra | |||||||
| IV | 1 | Philips | 1.5 | Ingenia 1.5T | ||||||
| 2 | Philips | 3 | Ingenia 3.0T | |||||||
| MRI Acquisition Parameters | ||||||||||
| Sequence | 3D SGRE | Flip angle | 5° (1.5T) | 3° (3T) | ||||||
| Echo number | 6 | Slice thickness | 3 mm | |||||||
| FOV | 20×20 cm2 | Matrix | 130×130×10 | |||||||
| Echo train (GE) | 1 (1.5T) | 2 (3T) | ETL (GE) | 6 (1.5T) | 3 (3T) | |||||
| Echo train (Siemens) | 1 (1.5T) | 2 (3T) | ETL (Siemens) | 6 (1.5T) | 3 (3T) | |||||
| Echo train (Philips) | 1 (1.5T and 3T) | ETL (Philips) | 6 (1.5T and 3T) | |||||||
Note: ETL, echo train length.
Table 2:
CT systems and CT acquisition parameters across the four sites.
| Site | Scanner # | Vendor | Model | |||||
|---|---|---|---|---|---|---|---|---|
| I | 1 | GE | Optima CT580 | |||||
| 2 | GE | Optima CT660 | ||||||
| 3 | GE | Revolution CT | ||||||
| 4 | GE | Discovery CT750 HD | ||||||
| 5 | GE | Discovery CT750 HD | ||||||
| 6 | Siemens | SOMATOM Definition Edge | ||||||
| II | 1 | Siemens | SOMATOM Definition Flash | |||||
| III | 1 | Siemens | SOMATOM Definition Flash | |||||
| 2 | Siemens | SOMATOM Force | ||||||
| IV | 1 | Siemens | SOMATOM Definition Flash | |||||
| 2 | Siemens | SOMATOM Force | ||||||
| 3 | Philips | Brilliance iCT256 | ||||||
| 4 | Philips | Ingenuity CT | ||||||
| CT Acquisition Parameters | ||||||||
| Protocol | 1 | 2 | 3 | 4 | 5 | |||
| Axial | Axial | Helical | Helical | AAPM | ||||
| kV | 120 | |||||||
| mAs | 500 | 250 | 250 | 125 | AEC | |||
| Slice thickness (mm) | 1.25 | 5 | ||||||
| Recon FOV (cm 2 ) | 20×20 | |||||||
| CTDIvol (mGy) | ~46 | ~23 | ~40 | ~20 | ~18 | |||
| Recon Kernel | Standard of soft tissue-equivalent | |||||||
The proposed phantom was shipped sequentially from site to site (Site I, Site II, Site III, and Site IV) for data acquisition over a duration of four months. At Site I, the same data acquisition was repeated after the phantom was returned, for a time lag of four months between the first and last acquisitions.
At each site, all MRI and CT acquisitions were performed by experienced MRI and CT operators, respectively. The phantom was allowed to equilibrate indoors for at least 12 hours and stored in the scanner room for at least 1 hour before imaging, in order to stabilize the phantom temperature. All acquisitions (both MRI and CT) were repeated after removing the phantom from the scanner, to validate repeatability. Note that all MRI and CT scanners from the four sites followed required protocols to perform quality control (QC) tests. For all four sites, the CT QC program performed QC testing that follows American College of Radiology (ACR) CT Accreditation requirements and recommendations. A certified medical physicist performed annual QC testing including radiation dose and dose display accuracy. Certified CT technologists performed daily CT QC testing including scanning a water phantom and verifying the CT number accuracy and the image uniformity. For all four sites, certified MRI technologists performed annual and weekly QC testing following the procedures outlined by the ACR including scanning an ACR phantom to ensure adequate imaging performance.
For MRI, multi-echo 3D spoiled gradient echo (SGRE) CSE-MRI data were collected at both 1.5T and 3T at each site and for each vendor using product CSE-MRI methods (IDEAL IQ, GE Healthcare; LiverLab, Siemens Healthineers; mDixonQuant, Philips Healthcare). For each acquisition, MRI-PDFF maps were acquired using the vendor acquisition and reconstruction software. Detailed acquisition parameters are shown in Table 1. Note that the acquisition parameters were replicated as closely as possible across different sites and vendors. The 3T CSE-MRI acquisition was used to provide the MRI-PDFF reference values for each site in order to compare MRI-PDFF and CT attenuation in later analyses.
For CT, five different abdominal unenhanced CT protocols were performed at 120kVp with different scan modes (i.e., Axial and Helical) covering different dose levels (Axial High Dose: 500mAs; Axial Low Dose: 250mAs; Helical High Dose: 250mAs; Helical Low Dose: 125mAs; American Association of Physicists in Medicine (AAPM) abdomen standard protocol23). For each acquisition, CT images were reconstructed using vendor specific soft tissue reconstruction kernel, which was consistent with clinical protocol for non-contrast abdominal CT imaging. Detailed acquisition parameters are shown in Table 2. Note that the acquisition parameters were replicated as closely as possible across different sites and vendors.
Data and Statistical analysis:
A 1.5cm2 circular region of interest (ROI) was placed in the center of the central slice through each vial for both MRI-PDFF and CT images to obtain mean estimates of PDFF (%) and CT number (HU). Note that all data used in the following statistical analyses described below (with the exception of the reproducibility study) were from the average of “test” set and “retest” set of acquisition measurements for both MRI and CT. For MRI data analysis, linear regression and Bland-Altman analysis (Mean with 95% Limits of Agreement (LoA) defined as ±1.96×standard deviation) were performed to study the relationship between MRI-PDFF and nominal PDFF for both 1.5T and 3T across different vendors and platforms.
To evaluate the relationship between CT number and potential contributing factors (i.e., fat-fraction, CT vendor, and CT protocol), linear mixed-effects statistical modeling was applied to determine the sources of variation and correlation for the observed CT measurements24. In this model, the CT number measurement depends on the true fat-fraction of each vial, represented as PDFF, with fixed effects from vendor and protocol of each acquisition (i.e., effect on slope and intercept of the CT number and PDFD calibration relationship). As part of the linear mixed-effects statistical modeling, GE (vendor) and the Helical High Dose (protocol) were chosen as the references. Based on the results from linear mixed-effects model analysis (see Results), the Helical High Dose protocol was chosen as the reference CT acquisition for the following further analyses.
In order to illustrate the need to add iodine contrast agent in phantom to mimic in vivo CT attenuation characteristics, a previous developed quantitative MRI fat phantom18 with no iodine added was also scanned with CT using the same acquisition protocol used for the proposed phantom. We note that the performance regarding MRI-PDFF vs CT number was discussed briefly by Pickhardt et al16. Linear regression analysis (i.e. slope with 95% confidence interval, intercept with 95% confidence interval, and coefficient of determination (R2)) was performed between MRI-PDFF and CT number to facilitate direct comparisons between our proposed phantom data with measurements using the prior MRI fat phantom mentioned above and in vivo datasets14. A one-sample t-test was applied to test the difference between 0% nominal PDFF vial measured CT number (corrected to true 0% PDFF with linear regression results) and the intercept (65.9HU) for the in vivo PDFF vs CT attenuation relationship for a PDFF value of 0%, in order to demonstrate whether the proposed phantom is able to mimic the CT attenuation behavior of a non-steatotic liver. CT number difference was calculated as the difference between the measured CT number and the averaged value across all CT measurements for each PDFF level.
Bland-Altman analysis (Mean with 95% LoA) was also performed to evaluate test-retest repeatability at all sites for both MRI and CT. The stability of the phantom was also evaluated by comparing the first and last CT and MRI measurements made at Site I, also using Bland-Altman analysis (Mean with 95% LoA).
All data measurements were made using OsiriX (Pixmeo), and all statistical analyses were implemented using Python (numpy 1.18.1, panda 0.3.1, and matplotlib 3.2.0).
Results:
Figure 1 depicts a 3D rendering of the phantom model on the left and a diagram (axial plane) of the vial fat-fraction arrangement on the right. In Figure 2, representative CT images (first row) and MRI-PDFF maps (1.5T and 3T, second and third row, respectively) obtained from four sites (Site I, Site II, Site III, and Site IV), with the three different vendors are shown. The CT images shown were from the Helical High Dose CT protocol. High quality MRI and CT images from the proposed phantom were consistently observed across all sites, vendors, platforms and magnetic field strength.
Figure 2.
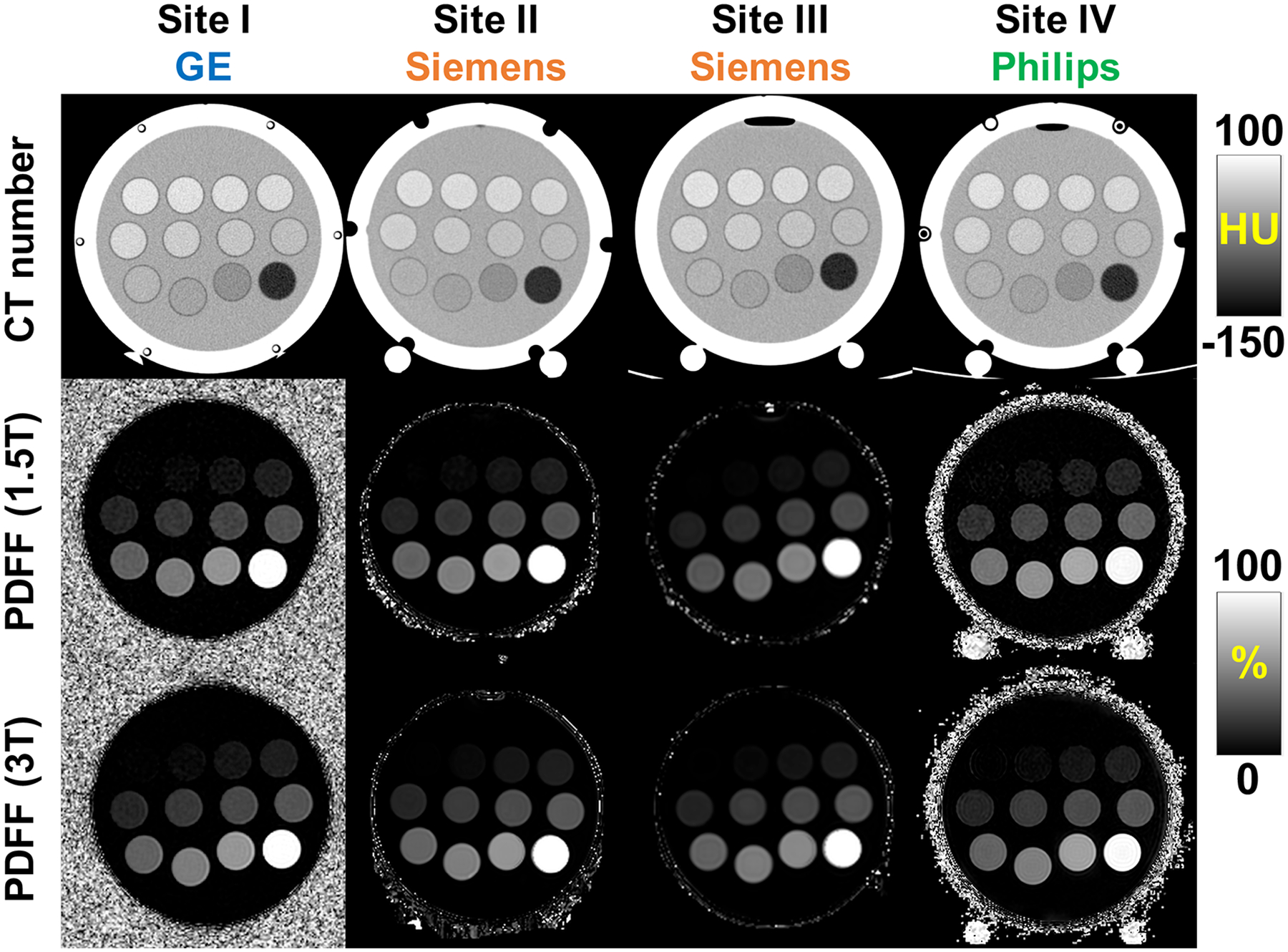
High quality MRI and CT images were obtained for the proposed MR and CT compatible phantom across different sites and vendors. Example CT images (first row) and MRI-PDFF maps (1.5T and 3T, second row and third row, respectively) obtained from four sites (Site I, Site II, Site III, and Site IV), with three different vendors (GE, Siemens, and Philips). The CT images shown were acquired using the Helical High Dose CT protocol (120 kV, 250 mAs). Note that the two white circles on the bottom of the sphere are an external phantom support used at some sites. PDFF, proton density fat-fraction.
As shown in Figure 3, high correlation was observed for the linear fit between measured and nominal PDFF values across different vendor scanners for both 1.5T (slope=0.99±0.09×10−2, intercept=0.82±0.04, R2=0.999) and 3T (slope=0.99±0.08×10−2, intercept=1.04±0.03, R2=0.999) (top row). Bland-Altman analysis shows no significant bias between measured PDFF and nominal PDFF at 1.5T (mean=0.26% and 95% LoA = [−1.78%,2.29%]) and at 3T (mean=0.65% and 95% LoA = [−1.36%,2.66%]) across all vendors (bottom row).
Figure 3.
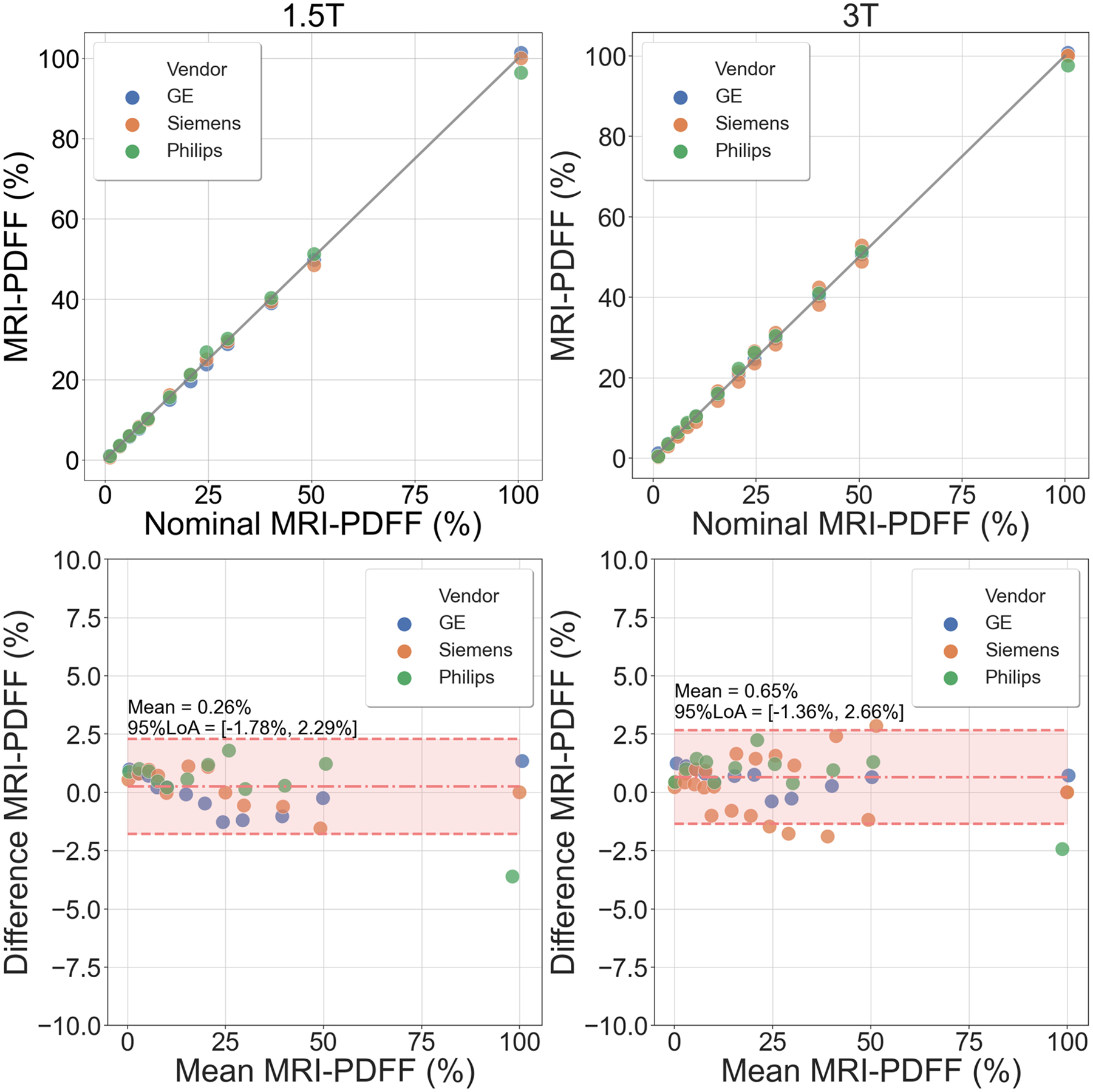
High correlation and agreement were observed between the measured MRI-PDFF and nominal PDFF across vendor imaging systems for both 1.5T and 3T (top row). Bland-Altman analysis results demonstrate no significant bias between measured MRI-PDFF and nominal PDFF for both 1.5T (mean = 0.26% and 95% LoA = [−1.78%, 2.29%]) and 3T (mean = 0.65% and 95% LoA = [−1.36%, 2.66%]) across vendors (bottom row). PDFF, proton density fat-fraction; LoA, limits of agreement.
Linear mixed-effects modeling results are shown in Table 3, examining the relationship between CT number (as the dependent variable) and MRI-PDFF (as the independent variable). CT number has strong correlation and a linear relationship with PDFF, with slope of −1.84±0.01HU/%, intercept of 67.8±2.9HU, and R2=0.997. As shown in Table 3, scanner vendor has a significant effect (p<0.005) on CT measurements, i.e., the vendor selection has a significant effect on slope and intercept for CT number and MRI-PDFF calibration, although CT attenuation is insensitive to CT protocol selection (p>0.1 with narrow 95% confidence interval), i.e., the protocol selection has no significant effect on slope and intercept for CT number and MRI-PDFF calibration. Since the protocol selection has no significant effect on CT number, the following analysis focuses on the data acquired with the Helical High Dose protocol.
Table 3:
Linear mixed-effects model results using GE and Helical High Dose protocol as the reference for vendor and protocol, respectively. In this model, CT measurement depends on the true fat-fraction of each vial with fixed effects from vendor and protocol. CT number has high correlation with MRI-PDFF in a relationship with slope of −1.84±0.01 HU/%, intercept of 67.8±2.9 HU, and R2=0.997. CT vendor selection has a significant contribution (p<0.005) to the CT number versus MRI-PDFF calibration relationship (i.e., slope and intercept), however, CT protocol selection has no significant contribution (p>0.1) to the CT number versus MRI-PDFF calibration relationship (i.e., slope and intercept). PDFF, proton density fat-fraction.
| Coef. | Std.Err. | p | 95% CI | |||
|---|---|---|---|---|---|---|
| Slope (HU/%) | Reference (GE) | −1.841 | 0.007 | <0.001 | [−1.855, −1.827] | |
| Vendor | Siemens | 0.055 | 0.006 | <0.001 | [0.043, 0.067] | |
| Philips | 0.024 | 0.008 | <0.005 | [0.008, 0.040] | ||
| Protocol | Axial High Dose | −0.003 | 0.009 | 0.774 | [−0.020, 0.015] | |
| Axial Low Dose | −0.001 | 0.009 | 0.968 | [−0.018, 0.017] | ||
| Helical Low Dose | 0.004 | 0.009 | 0.685 | [−0.014, 0.021] | ||
| AAPM | 0.013 | 0.009 | 0.154 | [−0.005, 0.031] | ||
| Intercept (HU) | Reference (Helical High Dose) | 67.8 | 2.9 | <0.001 | [62.1, 73.4] | |
| Vendor | Siemens | −5.6 | 0.2 | <0.001 | [−6.0, −5.1] | |
| Philips | −3.0 | 0.3 | <0.001 | [−3.6, −2.4] | ||
| Protocol | Axial High Dose | −0.1 | 0.3 | 0.798 | [−0.7, 0.6] | |
| Axial Low Dose | −0.4 | 0.3 | 0.188 | [−1.1, 0.2] | ||
| Helical Low Dose | −0.4 | 0.3 | 0.231 | [−1.1, 0.3] | ||
| AAPM | −0.4 | 0.3 | 0.288 | [−1.0, 0.3] | ||
Note: Coef., coefficient; Std.Err., standard error; 95% CI, 95% confidence interval; AAPM, American Association of Physicists in Medicine.
In practice, when MRI-PDFF is considered to be the reference, CT number becomes the independent variable (x-axis) used to the predict MRI-PDFF as the dependent variable (y-axis). This analysis is summarized in Figure 4, which demonstrates high correlation (slope=−0.54±0.01%/HU, intercept=37.15±0.10%, R2=0.999) between the reference MRI-PDFF and CT number using the proposed phantom from a specific acquisition (MRI: Site I, GE, Discovery MR750, 3T; CT: Site I, GE, Optima CT580, Helical High Dose protocol). The phantom measurements closely replicated the relationship between PDFF and CT attenuation data previously observed in vivo (slope=−0.58±0.01%/HU, intercept=38.23±0.60%, R2=0.828)14. In contrast, MRI and CT measurements collected using a conventional MRI fat phantom which was developed for a previous study18. Linear regression analysis demonstrates slope=−0.78±0.01%/HU, intercept=17.93±0.44%, and R2=0.998, which are also plotted in Figure 4, demonstrating explicitly that the previous MRI fat phantom does not mimic the in vivo relationship between PDFF and CT number. Note that both the phantom data and the in vivo data shown in Figure 4 were collected using the same MRI and CT scanners at the same site in order to mitigate potential bias introduced by differences in site and vendor. The one-sample t-test result (p=0.45) demonstrated no statistically significant difference between the CT number of the zero-fat vial in the phantom and the intercept (65.9HU) for the in vivo PDFF vs CT attenuation relationship for a PDFF value of 0%.
Figure 4:
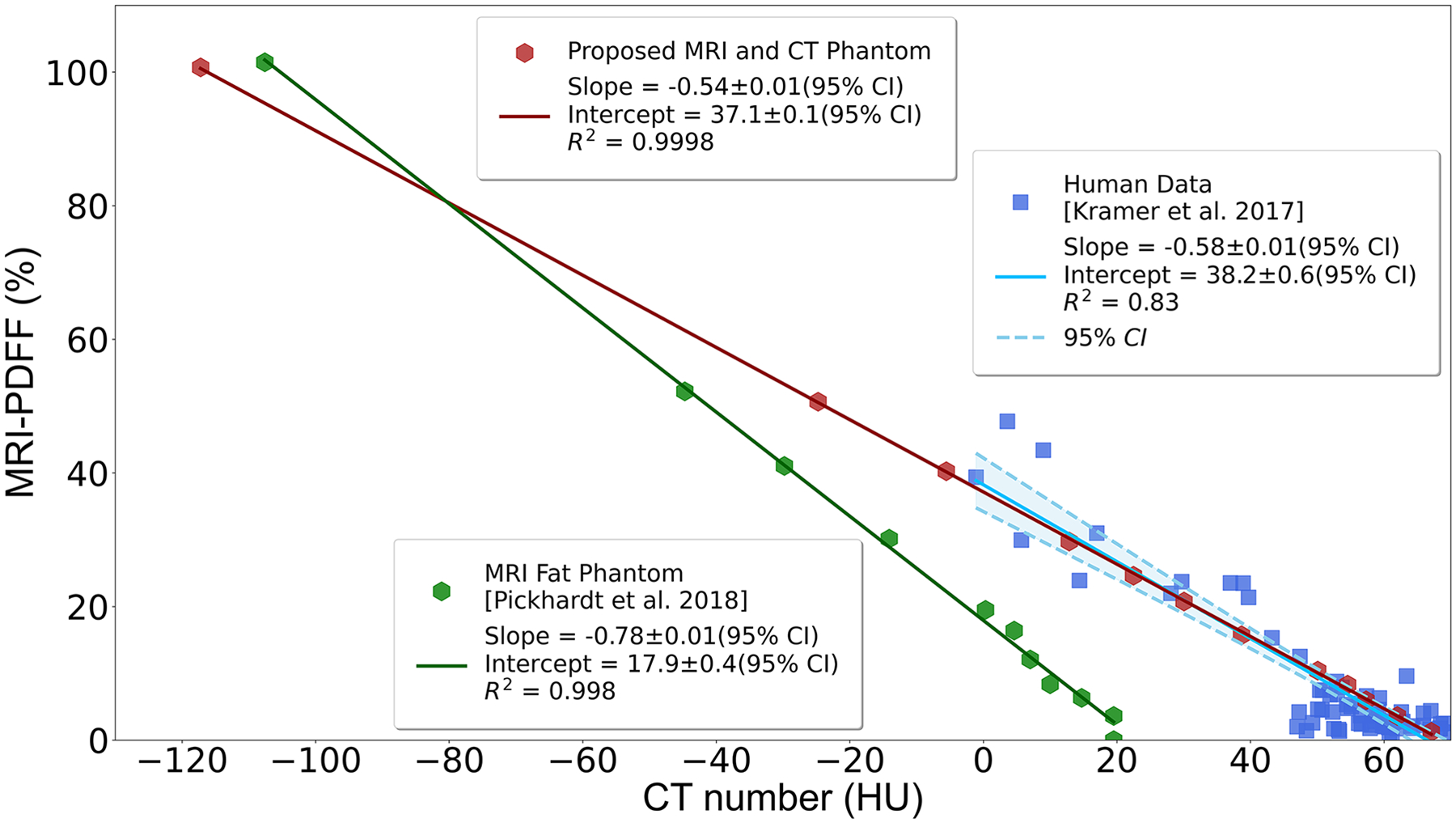
High correlation between measured MRI-PDFF and CT attenuation number for the proposed phantom from two specific acquisitions (MRI: Site I, GE Discovery MR750, 3T; CT: Site I, GE Discovery CT750 HD, Helical High Dose protocol). The proposed phantom closely mimics the signal behavior observed in vivo during a previous human study [14]. A conventional MRI fat phantom developed for a previous study [18] does not mimic in vivo relationship between PDFF and CT number. Note that all phantom data and in vivo data were collected using the same vendor’s machines (both MRI and CT) at the same site. PDFF, proton density fat-fraction; CI, confidence interval.
As shown in Figure 5, CT number measurement differences (i.e., the difference between CT number measurements from each acquisition and the averaged CT number measurement from all acquisitions) are within a range of −5 to 5HU across different PDFF levels measured by MRI. Note the standard deviation of the CT number difference shown here depends on the number of scanners available for each vendor.
Figure 5.
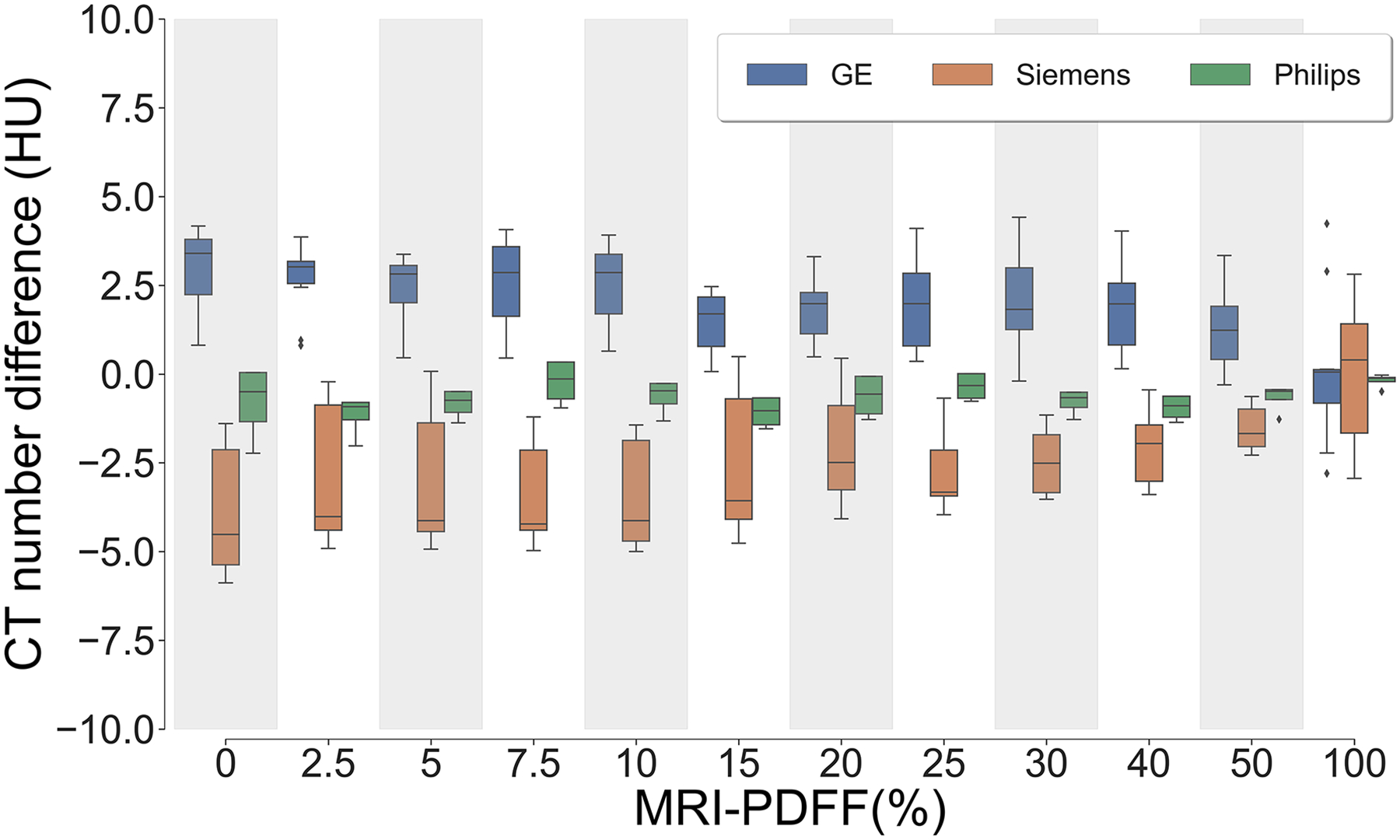
CT number difference (i.e., the difference between CT measurement from each acquisition and the averaged CT measurement from all acquisitions) were compared between vendors, across different PDFF levels. The CT number difference from all vendors fall within a range of −5 to 5 HU across all MRI-PDFF levels. Note that the standard deviation of CT number difference depends on the quantity of scanners for each vendor. PDFF, proton density fat-fraction.
Bland-Altman analysis results regarding test-retest acquisitions for both MRI and CT using the proposed phantom are shown in Figure 6 (top), demonstrating bias and variability between repeated acquisitions across all vendors and both modalities. MRI-PDFF differences (left) locate in a region with mean: −0.04% and 95% limits of agreement: [−0.24%,0.16%] across vendors. In addition, the CT number test-retest bias and variability (right) are within a region with mean: 0.16HU and 95% limits of agreement: [−0.15HU, 0.47HU] across vendors.
Figure 6.
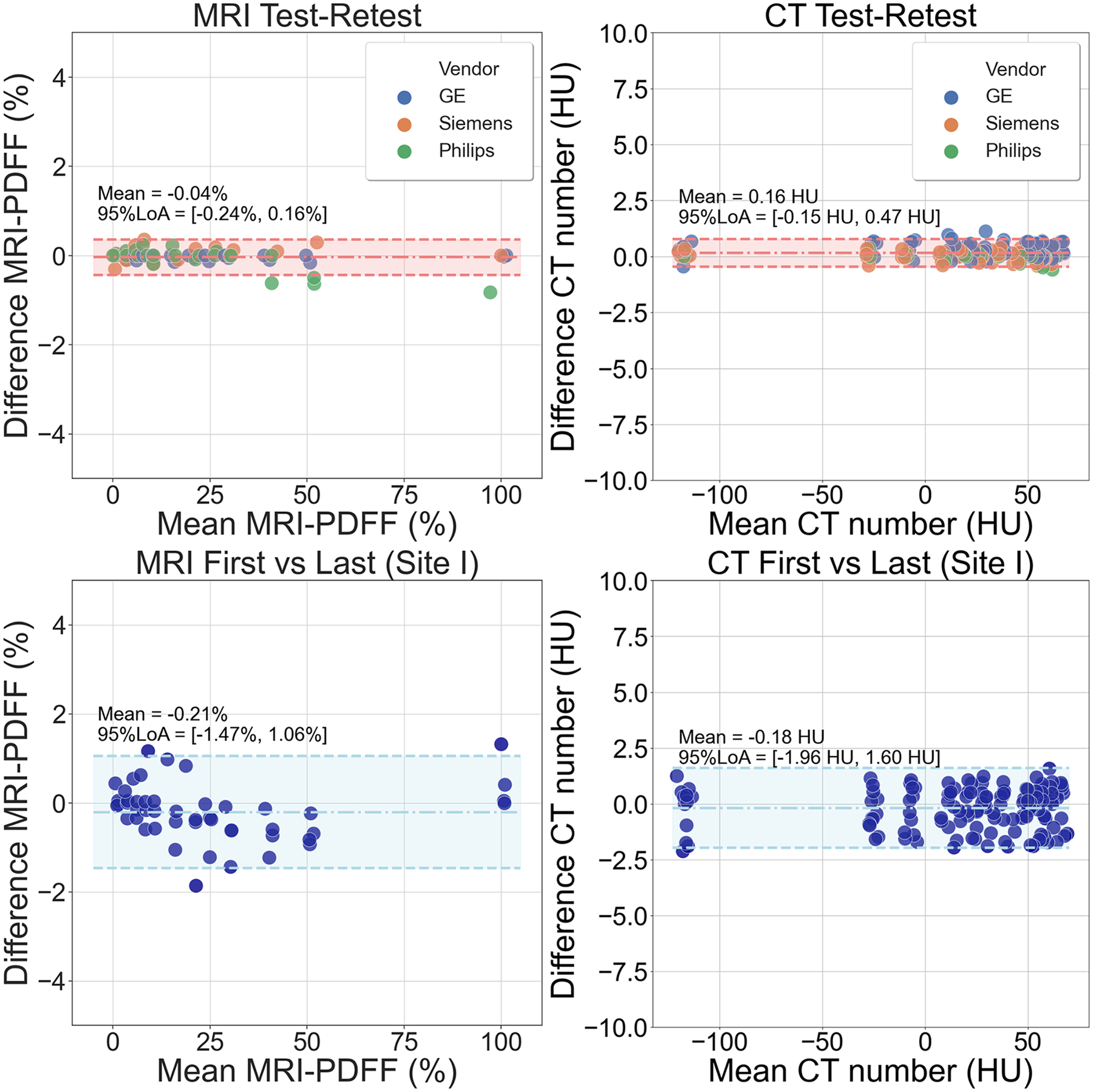
Bland-Altman analysis results regarding test and retest acquisitions for both MRI and CT using the proposed phantom shows bias and variability for both modalities (top row). On the top left, MRI-PDFF differences are within a region (mean: −0.04%; 95% limits of agreement: [−0.24%, 0.16%]) across different vendors. On the top right, CT number differences are within a region (mean: 0.16 HU; 95% limits of agreement: [−0.15 HU, 0.47 HU]) across different vendors. Bland-Altman analysis results regarding first and last acquisitions at Site I for both MRI and CT using the proposed phantom demonstrates that the phantom was stable over the 4-month study period, for both modalities (bottom row). On the bottom left, MRI-PDFF differences are within a region (mean: −0.21%; 95% limits of agreement: [−1.47%, 1.06%]). On the bottom right, CT number differences are within a region (mean: −0.18 HU; 95% limits of agreement: [−1.96 HU, 1.60 HU]). PDFF, proton density fat-fraction; LoA, limits of agreement.
Figure 6 (bottom) also depicts the Bland-Altman analysis comparing the first and last acquisitions for both MRI and CT using the proposed phantom at Site I, demonstrating the stability of these measurements over a 4-month period, and after shipping to three other sites. There was no significant change in MRI-PDFF (mean: −0.21%; 95% limits of agreement: [−1.47%,1.06%]) or CT number (mean: −0.18HU; 95% limits of agreement: [−1.96HU,1.60HU]) between the first and last measurements made at Site I.
Discussion:
In this work, we successfully developed, validated and demonstrated the potential utility of a fat phantom that simultaneously mimics liver fat with both MRI and CT. Validation was performed in a round-robin multi-site, multi-vendor study at four sites. Accurate estimation of fat biomarkers and high reproducibility across sites, vendors, and protocols were achieved for both MRI and CT, in the same phantom. Importantly, the phantom mimicked previously observed in vivo signal behavior14,16,17. Further, this study demonstrated the stability of the phantom over the duration of the study period. Based on these results, the proposed fat phantom may enable reproducible application of liver fat quantification techniques using MRI and CT across institutions, vendors and field strength. This phantom may also enable the calibration of CT systems to provide a one-to-one harmonization of CT number with MRI-PDFF.
A previously developed quantitative MRI fat phantom used in a previous multi-site multi-vendor MRI fat quantification study18 was examined with CT in order to illustrate the phantom CT attenuation characteristics. However, the prior MRI fat phantom did not mimic the CT attenuation properties of liver in MRI-PDFF and CT number correlation relationship. In the current study, the proposed phantom closely mimics the in vivo relationship between CT attenuation and MRI-PDFF observed in human liver over a wide range of liver fat content14 and agreed with a previous phantom study16. Importantly, our statistical analysis confirms that there was no significant difference between phantom attenuation behavior and the CT number in the liver for 0% PDFF.
There are several limitations of our study. First, all acquisitions at the different sites were performed at room temperature, not at body temperature. Bias in MRI-PDFF measurement may be introduced due to temperature variability between acquisitions, although this effect is small for the CSE-MRI methods used in this study25. The high accuracy and reproducibility observed in this study suggest that temperature or other confounders had minimal effect on MRI-PDFF quantification. Furthermore, the effect of temperature on CT-based fat quantification is unknown. A further limitation is that the proposed phantom did not mimic the effects of iron overload, which can be seen occasionally in human livers. Iron impacts MR signal substantially, although -correction strategies generally account for this effect26,27. Severe iron overload can lead to a very small increase in the CT attenuation28. Other factors, including glycogen 29 and iodine deposition from long-standing amiodarone therapy30 can increase the X-ray attenuation of liver.
In this study, the uncertainty of the measured PDFF values in the phantom was not fully evaluated. There are two potential sources of uncertainty in PDFF values that included (1) accuracy/precision of the scales used in phantom construction (2) MRI measurements. Accurate estimation of uncertainty could be achieved with multiple repetitions on both weighting chemicals during phantom construction, followed by analysis (MRI-based or otherwise) of the vial contents. However, the actual manufacturing uncertainty was likely small compared to imaging related uncertainty and is not the focus of this study. For the purpose of this study, which is to investigate the calibration relationship between PDFF, as measured by MRI, and CT number, the uncertainty of PDFF measurements, which serve as the ground truth, is not directly relevant. Further studies would be needed to determine the precise uncertainty of PDFF values in the phantom, although these are thought to be small. Also, the observed variability of CT measurements across different vendors may be relevant in clinical applications. In order to overcome this vendor dependence, vendor-specific calibrations of CT attenuation to fat content level may be needed.
A practical limitation was that the round-robin study did not include a comprehensive set of all possible MRI and CT vendors, platforms and acquisition protocols. This limited the ability to provide a comprehensive statistical analysis of all combinations of vendors, platforms, and protocols. By using mixed-effect model analysis, we were able to evaluate the vendor and protocol effect on fat quantification with CT. We note, however that the effect of vendors had a small, but measurable impact on fat quantification. Additional sites and vendors would be necessary for comprehensive evaluation of these effects and is beyond the scope and purpose of this study, which aimed to demonstrate the potential utility of the proposed phantom.
An additional limitation is that the housing may impact the apparent CT number measurements. Future studies may be needed in order to provide a general calibration relationship that includes the effects of the housing on CT number or to find other agents for mimicking attenuation characteristics of liver. The geometric design of the proposed phantom may also be a limitation. Anatomic CT phantoms often include bone, spine, or extra layers of exterior attenuating material to mimic the abdominal wall.
Finally, we note that this study did not investigate emerging dual energy CT (DECT) methods for quantifying tissue fat, as evaluation of DECT methods was beyond the scope of this study. The performance of DECT to quantify liver fat has shown conflicting results in the literatures. Specifically, Kramer et al14 and Artz et al31 demonstrated that DECT-based material decomposition showed poor performance for quantifying liver fat compared to attenuation measured using single energy CT. Other studies32,33, however, suggest that DECT may enable accurate and reproduceable liver fat quantification. These conflicting results suggest an uncertain role for the use of DECT methods to quantify liver fat. Practically, the use of DECT is limited by a small installed base of CT systems with dual energy capabilities, although evaluation of DECT for liver fat quantification should be considered for future studies. For example, virtual non-contrast enhanced images could be obtained with a simple DECT reconstruction. Another limitation of this study is that the relationship between PDFF and CT number at different X-ray energies (kVp) was not investigated. However, we note that the in vivo relationship between PDFF and CT number at different tube energies (kVp), is also unknown, although monochromatic reconstructions obtained in vivo using DECT suggest a weak dependence on X-ray energy in vivo14.
Conclusions:
In summary, we successfully developed and validated a novel MRI and CT compatible fat phantom. Using this proposed phantom, validation was performed at four sites with multiple vendors, models and field strength/protocols. Accurate MRI-PDFF and CT number measurements were observed across sites, vendor, and field strength (MRI). Further, the proposed phantom accurately mimicked the known in vivo relationship of CT liver attenuation and MRI-PDFF. The proposed phantom may provide a useful tool for site qualification in clinical trials, acceptance testing and periodic QA for both MRI and CT applications aimed at quantifying liver fat. Further, the proposed phantom may provide a useful means to harmonize MRI and CT data acquired as part of multi-center clinical trials.
Acknowledgements:
The authors acknowledge support from the Wisconsin State Economic Engagement and Development (SEED) Program, as well as the NIH (R01 DK088925, R01 DK100651, R41 EB025729, R44 EB025729, K24 DK102595, and R01 DK083380). Further, the authors wish to thank GE Healthcare, who provides research support to the University of Wisconsin and Duke University, Siemens Healthineers who provides research support to the University of Wisconsin-Madison, the Johns Hopkins University, Duke University, and the University of Texas-Southwestern, Philips Healthcare who provides research support to the University of Texas-Southwestern. Finally, Dr. Reeder is a Romnes Faculty Fellow, and has received an award provided by the University of Wisconsin-Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. The authors wish to thank David Rutkowski from Calimetrix for helpful discussions.
Conflicts of interest:
Jean Brittain is an employee and founder of Calimetrix, and Diego Hernando and Scott Reeder are founders of Calimetrix. Jean Brittain and Scott Reeder have ownership interests in Elucent Medical, Cellectar Biosciences, and Reveal Pharmaceuticals, Finally, GE Healthcare provides research support to the University of Wisconsin.
Data availability statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References:
- 1.Sanyal AJ, Banas C, Sargeant C, et al. Similarities and differences in outcomes of cirrhosis due to nonalcoholic steatohepatitis and hepatitis C. Hepatology. 2006;43(4):682–689. doi: 10.1002/hep.21103 [DOI] [PubMed] [Google Scholar]
- 2.Ekstedt M, Franzén LE, Mathiesen UL, et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44(4):865–873. doi: 10.1002/hep.21327 [DOI] [PubMed] [Google Scholar]
- 3.Rubinstein E, Lavine JE, Schwimmer JB. Hepatic, Cardiovascular, and Endocrine Outcomes of the Histological Subphenotypes of Nonalcoholic Fatty Liver Disease. Semin Liver Dis. 2008;28(4):380–385. doi: 10.1055/s-0028-1091982 [DOI] [PubMed] [Google Scholar]
- 4.Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62(1 Suppl):S47–64. doi: 10.1016/j.jhep.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 5.Targher G, Day CP, Bonora E. Risk of Cardiovascular Disease in Patients with Nonalcoholic Fatty Liver Disease. N Engl J Med. 2010;363(14):1341–1350. doi: 10.1056/NEJMra0912063 [DOI] [PubMed] [Google Scholar]
- 6.Hazlehurst JM, Woods C, Marjot T, Cobbold JF, Tomlinson JW. Non-alcoholic fatty liver disease and diabetes. Metabolism. 2016;65(8):1096–1108. doi: 10.1016/j.metabol.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinella ME. Nonalcoholic Fatty Liver Disease: A Systematic Review. JAMA. 2015;313(22):2263–2273. doi: 10.1001/jama.2015.5370 [DOI] [PubMed] [Google Scholar]
- 8.Reeder SB, Sirlin C. Quantification of Liver Fat with Magnetic Resonance Imaging. Magn Reson Imaging Clin N Am. 2010;18(3):337–357. doi: 10.1016/j.mric.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeder SB, Hu HH, Sirlin CB. Proton Density Fat-Fraction: A Standardized MR-Based Biomarker of Tissue Fat Concentration. J Magn Reson Imaging JMRI. 2012;36(5):1011–1014. doi: 10.1002/jmri.23741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoo T, Serai SD, Pirasteh A, et al. Linearity, Bias, and Precision of Hepatic Proton Density Fat Fraction Measurements by Using MR Imaging: A Meta-Analysis. Radiology. 2018;286(2):486–498. doi: 10.1148/radiol.2017170550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno CC, Hemingway J, Johnson AC, Hughes DR, Mittal PK, Duszak R. Changing Abdominal Imaging Utilization Patterns: Perspectives From Medicare Beneficiaries Over Two Decades. J Am Coll Radiol. 2016;13(8):894–903. doi: 10.1016/j.jacr.2016.02.031 [DOI] [PubMed] [Google Scholar]
- 12.Puchner SB, Lu MT, Mayrhofer T, et al. High-Risk Coronary Plaque at Coronary CT Angiography Is Associated with Nonalcoholic Fatty Liver Disease, Independent of Coronary Plaque and Stenosis Burden: Results from the ROMICAT II Trial. Radiology. 2015;274(3):693–701. doi: 10.1148/radiol.14140933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn L, Reeder SB, del Rio AM, Pickhardt PJ. Longitudinal Changes in Liver Fat Content in Asymptomatic Adults: Hepatic Attenuation on Unenhanced CT as an Imaging Biomarker for Steatosis. Am J Roentgenol. 2015;205(6):1167–1172. doi: 10.2214/AJR.15.14724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer H, Pickhardt PJ, Kliewer MA, et al. Accuracy of Liver Fat Quantification With Advanced CT, MRI, and Ultrasound Techniques: Prospective Comparison With MR Spectroscopy. AJR Am J Roentgenol. 2017;208(1):92–100. doi: 10.2214/AJR.16.16565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickhardt PJ, Hahn L, Muñoz del Rio A, Park SH, Reeder SB, Said A. Natural history of hepatic steatosis: observed outcomes for subsequent liver and cardiovascular complications. AJR Am J Roentgenol. 2014;202(4):752–758. doi: 10.2214/AJR.13.11367 [DOI] [PubMed] [Google Scholar]
- 16.Pickhardt PJ, Graffy PM, Reeder SB, Hernando D, Li K. Quantification of Liver Fat Content With Unenhanced MDCT: Phantom and Clinical Correlation With MRI Proton Density Fat Fraction. Am J Roentgenol. 2018;211(3):W151–W157. doi: 10.2214/AJR.17.19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Blake GM, Li K, et al. Liver Fat Content Measurement with Quantitative CT Validated against MRI Proton Density Fat Fraction: A Prospective Study of 400 Healthy Volunteers. Radiology. Published online November 5, 2019:190467. doi: 10.1148/radiol.2019190467 [DOI] [PubMed] [Google Scholar]
- 18.Hernando D, Sharma SD, Aliyari Ghasabeh M, et al. Multisite, multivendor validation of the accuracy and reproducibility of proton-density fat-fraction quantification at 1.5T and 3T using a fat-water phantom: Proton-Density Fat-Fraction Quantification at 1.5T and 3T. Magn Reson Med. 2017;77(4):1516–1524. doi: 10.1002/mrm.26228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hines CDG, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 Independent, T2* Corrected MRI with Accurate Spectral Modeling for Quantification of Fat: Validation in a Fat-Water-SPIO Phantom. J Magn Reson Imaging JMRI. 2009;30(5):1215–1222. doi: 10.1002/jmri.21957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczepaniak LS, Nurenberg P, Leonard D, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288(2):E462–468. doi: 10.1152/ajpendo.00064.2004 [DOI] [PubMed] [Google Scholar]
- 21.Tang A, Desai A, Hamilton G, et al. Accuracy of MR Imaging–estimated Proton Density Fat Fraction for Classification of Dichotomized Histologic Steatosis Grades in Nonalcoholic Fatty Liver Disease. Radiology. 2014;274(2):416–425. doi: 10.1148/radiol.14140754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehm JL, Wolfgram PM, Hernando D, Eickhoff JC, Allen DB, Reeder SB. Proton density fat-fraction is an accurate biomarker of hepatic steatosis in adolescent girls and young women. Eur Radiol. 2015;25(10):2921–2930. doi: 10.1007/s00330-015-3724-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AdultAbdomenPelvisCT.pdf. Accessed October 28, 2019. https://www.aapm.org/pubs/CTProtocols/documents/AdultAbdomenPelvisCT.pdf
- 24.Lindstrom MJ, Bates DM. Newton—Raphson and EM Algorithms for Linear Mixed-Effects Models for Repeated-Measures Data. J Am Stat Assoc. 1988;83(404):1014–1022. doi: 10.1080/01621459.1988.10478693 [DOI] [Google Scholar]
- 25.Hernando D, Kellman P, Haldar JP, Liang Z-P. Robust Water/Fat Separation in the Presence of Large Field Inhomogeneities Using a Graph Cut Algorithm. Magn Reson Med Off J Soc Magn Reson Med Soc Magn Reson Med. 2010;63(1):79–90. doi: 10.1002/mrm.22177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernando D, Kramer JH, Reeder SB. Multipeak Fat-Corrected Complex R2* Relaxometry: Theory, Optimization, and Clinical Validation. Magn Reson Med Off J Soc Magn Reson Med Soc Magn Reson Med. 2013;70(5):1319–1331. doi: 10.1002/mrm.24593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multi-Echo Water-Fat Separation and Simultaneous R2* Estimation with Multi-Frequency Fat Spectrum Modeling. Magn Reson Med Off J Soc Magn Reson Med Soc Magn Reson Med. 2008;60(5):1122–1134. doi: 10.1002/mrm.21737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence EM, Pooler BD, Pickhardt PJ. Opportunistic Screening for Hereditary Hemochromatosis With Unenhanced CT: Determination of an Optimal Liver Attenuation Threshold. AJR Am J Roentgenol. 2018;211(6):1206–1211. doi: 10.2214/AJR.18.19690 [DOI] [PubMed] [Google Scholar]
- 29.Leander P, Sjöberg S, Höglund P. CT and MR imaging of the liver. Clinical importance of nutritional status. Acta Radiol Stockh Swed 1987. 2000;41(2):151–155. doi: 10.1080/028418500127345172 [DOI] [PubMed] [Google Scholar]
- 30.Patrick D, White FE, Adams PC. Long-term amiodarone therapy: a cause of increased hepatic attenuation on CT. Br J Radiol. 1984;57(679):573–576. doi: 10.1259/0007-1285-57-679-573 [DOI] [PubMed] [Google Scholar]
- 31.Artz NS, Hines CDG, Brunner ST, et al. Quantification of hepatic steatosis with dual-energy computed tomography: comparison with tissue reference standards and quantitative magnetic resonance imaging in the ob/ob mouse. Invest Radiol. 2012;47(10):603–610. doi: 10.1097/RLI.0b013e318261fad0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyodo T, Yada N, Hori M, et al. Multimaterial Decomposition Algorithm for the Quantification of Liver Fat Content by Using Fast-Kilovolt-Peak Switching Dual-Energy CT: Clinical Evaluation. Radiology. 2017;283(1):108–118. doi: 10.1148/radiol.2017160130 [DOI] [PubMed] [Google Scholar]
- 33.Itaya S, Matsui T, Kamiyama T, Yoshino H. Evaluation of Fat Quantification in the Liver Using Dual Energy CT. Nihon Hoshasen Gijutsu Gakkai Zasshi. 2016;72(11):1084–1090. doi: 10.6009/jjrt.2016_JSRT_72.11.1084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


