Abstract
Background
The T2-fluid-attenuated inversion recovery (FLAIR) mismatch sign, has been considered a highly specific imaging biomarker of IDH-mutant, 1p/19q noncodeleted low-grade glioma. This systematic review and meta-analysis aimed to evaluate the diagnostic performance of T2-FLAIR mismatch sign for prediction of a patient with IDH-mutant, 1p/19q noncodeleted low-grade glioma, and identify the causes responsible for the heterogeneity across the included studies.
Methods
A systematic literature search in the Ovid-MEDLINE and EMBASE databases was performed for studies reporting the relevant topic before November 17, 2020. The pooled sensitivity and specificity values with their 95% confidence intervals were calculated using bivariate random-effects modeling. Meta-regression analyses were also performed to determine factors influencing heterogeneity.
Results
For all the 10 included cohorts from 8 studies, the pooled sensitivity was 40% (95% confidence interval [CI] 28–53%), and the pooled specificity was 100% (95% CI 95–100%). In the hierarchic summary receiver operating characteristic curve, the difference between the 95% confidence and prediction regions was relatively large, indicating heterogeneity among the studies. Higgins I2 statistics demonstrated considerable heterogeneity in sensitivity (I2 = 83.5%) and considerable heterogeneity in specificity (I2 = 95.83%). Among the potential covariates, it seemed that none of factors was significantly associated with study heterogeneity in the joint model. However, the specificity was increased in studies with all the factors based on the differences in the composition of the detailed tumors.
Conclusions
The T2-FLAIR mismatch sign is near-perfect specific marker of IDH mutation and 1p/19q noncodeletion.
Keywords: astrocytic, biomarker, glioma, molecular genetics
Key Points.
T2-FLAIR mismatch sign is a specific marker for 1p/19q noncodeleted low-grade glioma in adult patients.
Considerable heterogeneity exists in the diagnostic performance across studies.
Improvement is possible at the risk of bias (patient selection, flow, and timing).
Importance of the Study.
Our study confirms that the T2-FLAIR mismatch sign is a near-perfect specific marker of IDH mutation and 1p/19q noncodeletion. Despite considerable heterogeneity was proven for the diagnostic performance of the imaging marker, no statistically demonstrable cause was detected by meta-regression. However, a wide variability was noted in the composition of the detailed tumors across the studies, and considerable room for improvement exists at the risk of bias in the domains of patient selection, and flow and timing.
In conventional magnetic resonance imaging (MRI), several radiographic features, such as contrast enhancement, spatial heterogeneity, size, location, and necrosis of tumor were considered factors correlating to the histologic type and prognosis of gliomas.1–3 Advanced imaging techniques, such as diffusion-weighted imaging and perfusion imaging, are widely adopted in the field of neuro-oncology to estimate the cellularity of tumor and vascularity associated with neoangiogenesis more precisely than conventional imaging.4,5 However, these research trends are rapidly changing their target to excavate a new reliable predictive imaging biomarker for the molecular status of glioma since the molecular phenotypic and genotypic information, such as isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion status, is integrated into the WHO 2016 classification.6–9 R-2-hydroxyglutarate magnetic resonance spectroscopy is a representative example of recently spotlighted imaging biomarker for predicting IDH mutation status by detecting metabolite R-2-hydroxyglutarate, which is accumulated in the IDH-mutant gliomas with favorable diagnostic accuracy.10,11 Moreover, in the WHO 2021 classification, various molecular data were further included in the classification of glioma, including MYB, MAPK, TERTp, EGFR, H3 K27, or H3 G34.12 Simultaneously considering the emerging importance of molecular information and the preoperative setting of diagnostic imaging timing, the need for a reliable imaging biomarker for evaluating the molecular status of glioma has been emphasized for patient counseling, and treatment planning and management during the disease course.13
In particular, the T2-fluid-attenuated inversion recovery (FLAIR) mismatch sign, has been considered a highly specific imaging biomarker of IDH-mutant, 1p/19q noncodeleted (IDHmut-Noncodel, equivalent astrocytoma) low-grade glioma (LGG) since 2017.14–21 T2-FLAIR mismatch sign is defined as a presence of a complete/near-complete hyperintense signal of the tumor on T2 weighted image (T2WI), in combination with a relative hypointense signal except for a hyperintense peripheral rim on FLAIR.21 Considering the simplicity and essentiality of the two routine conventional MRI sequences (T2WI and FLAIR), which can be freely available as a cost-effective imaging biomarker worldwide even without advanced imaging techniques.13 Owing to the recent rapid publication in the topic on T2-FLAIR mismatch sign, systematic review and meta-analysis are warranted to discuss the cause of heterogeneity. Therefore, this systematic review and meta-analysis aimed to evaluate and determine the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG, and identify the causes responsible for the heterogeneity across the included studies.
Materials and Methods
This systematic review and meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.22
Literature Search
A search of MEDLINE and EMBASE databases was performed to find original literature reports aimed at identifying the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG. The following search terms were used: (T2-FLAIR mismatch) OR (T2-FLAIR suppression) AND (glioma) OR (LGG). For this search, no start date was set, and the included literature was updated until November 17, 2020. Additionally, the bibliographies of the retrieved studies were searched to identify any other appropriate studies.
Inclusion Criteria
Studies were included if they satisfied the following criteria: (1) involved pathologically diagnosed patients with LGG encompassing IDHmut-Noncodel LGG, (2) had molecular (IDH and 1p/19q) information of included tumor, (3) pretreatment MRI was the index test, and (4) contained sufficient information for the reconstruction of 2 × 2 tables to estimate the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG.
Exclusion Criteria
Exclusion criteria for the enrollment of studies were as follows: (1) case reports or case series including less than 5 patients, (2) conference abstracts, editorials, letters, consensus statements, guidelines, or review articles, (3) studies not focusing on the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG, (4) studies with, or with suspicion of, overlapping populations, and (5) insufficient data for the reconstruction of 2 × 2 tables.
The literature search and selection were independently performed by two radiologists, and consensus was obtained (S.J.C. and Y.A.D. with 8 and 3 years of experience in neuroradiology, respectively).
Data Extraction
The following data were extracted using standardized forms according to the PRISMA guideline:22 We extracted the following data from included studies: (1) characteristics of the included studies: authors, year of publication, institution (or source of database), country of origin, study period, study design (prospective vs. retrospective), number of included patients, mean age of included patients, male to female ratio, number of included tumor, whether all patients were adult or not, detailed number of included tumor according to the WHO grade (II or III), IDH mutation (or not), and 1p/19q codeletion (or noncodeletion), reference standard, and inclusion and exclusion criteria, (2) characteristics of MRI: vendor (and model), field strength, slice thickness, included sequences into MRI protocol, (3) analytic method for MRI interpretation: definition of T2-FLAIR mismatch sign, subjective vs. objective, multiple reader, the department of reader, and experience of reader, and (4) diagnostic performance of T2-FLAIR mismatch sign data for prediction of a patient with IDHmut-Noncodel LGG.
Quality Assessment
The methodological quality of the enrolled studies was evaluated using tailored questionnaires and Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria.23 Two reviewers (S.J.C. and Y.A.D) independently performed data extraction and quality assessment. Any disagreement between the two authors was resolved following expert-consultation of the third author (B.S.C, 20 years of experience in neuroradiology).
Data Synthesis and Analyses
The primary endpoint of the current systematic review and meta-analysis was to determine the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG. The secondary endpoint was to identify the causes responsible for the heterogeneity across the included studies by systematic evaluation and performing meta-regression analyses.
The pooled sensitivity and specificity values and their 95% confidence intervals (CIs) were obtained using bivariate random-effects modeling.24–28 The pooled results were graphically presented using hierarchic summary receiver operating characteristic (HSROC) curves with 95% confidence and prediction regions. Publication bias was analyzed using Deeks’ funnel plot, with Deeks’ asymmetry test being used to calculate the P-value, and determine statistical significance.29
Heterogeneity across the selected studies was evaluated using the Cochran Q test, which tests the presence of heterogeneity with P < 0.05.30 Heterogeneity was classified according to the Higgins I2 statistic as follows: 0–40%, might not be important; 30–60%, moderate heterogeneity; 50–90%, substantial heterogeneity; and 75–100%, considerable heterogeneity.25 The presence of a threshold effect (a positive correlation between sensitivity and false-positive rate) was sequentially evaluated: initially by visual assessment of coupled forest plots of sensitivity and specificity; and secondarily by Spearman correlation, with a correlation coefficient between the sensitivity and false-positive rate of greater than 0.6 being considered to indicate a threshold effect.31
To determine the causes of heterogeneity across the studies, meta-regression analyses were performed on the following covariates: (1) proportion of IDH-wild type among parent group (≥ 40%; ≥ 20%, and < 40%; and ≥ 10%, and < 20%), (2) proportion of IDHmut-Noncodel LGG among parent group (≥ 50%), (3) proportion of WHO grade II tumor among parent group (≥ 50%), (4) the definition of T2-FLAIR mismatch sign (group of “near-complete signal nulling (except rim) on FLAIR comparative to T2WI” vs. others), and (5) the number of readers.
The MIDAS and METANDI modules in STATA 16.0 (StataCorp, College Station, TX, USA) were used for all the statistical analyses, which were performed by one of the authors (S.J.C., with 4 years of experience in performing systematic reviews and meta-analyses).
Results
Literature Search
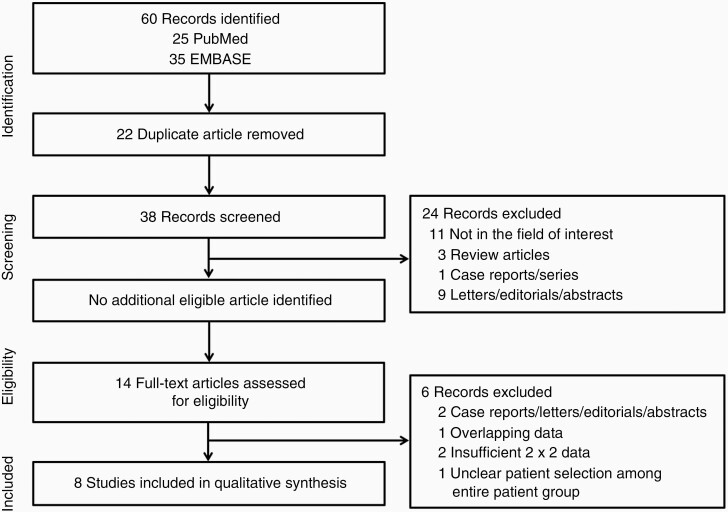
A systematic literature search (Figure 1) initially identified 60 studies. After eliminating 22 duplicates, the screening of the remaining 38 titles and abstracts yielded 14 potentially eligible studies. No additional study was identified after searching the bibliographies of these studies. After full-text reviews of the 14 provisionally eligible studies, one study was excluded since unclear data selection for T2/FLAIR mismatch evaluation among entire patient group,32 two because they contained insufficient data for the reconstruction of 2 × 2 tables,33,34 two because they were case reports, letters, editorials and abstracts,35,36 and one owing to suspected population overlap.37 Finally, 8 studies were included in the present systematic review and meta-analysis.14–21
Figure 1.
Flow diagram of the study selection process.
Characteristics of the Included Studies
The detailed patient characteristics are presented in Table 1. Among them, 2 studies were simultaneously included The Cancer Imaging Archive (TCIA) database and their own dataset separately for the purpose of comparison.14,21 The two TCIA databases of the two studies were considered as separate cohorts in the analysis. Finally, we assessed 10 cohorts from 8 studies in the present systematic review and meta-analysis. All the studies (except one) were of retrospective design.14–18,20,21 A study tried to amass the two independent patient groups (each retrospective and prospective manners).19 The total included study population was 1342 with individual cohorts ranging from 59 to 408 patients. The patients had mean ages of 43–57 years. The patients were all adults in five cohorts, ranging from 19 to 82 years; however, two cohorts only report mean age,19,20 and three cohorts of two studies did not report age ranges.14,17
Table 1.
Characteristics of the Included Studies
| Source | Affiliation | Duration of patient recruitment | Study design | Patients | No. of | included tumor | Reference standard | Criteria | Criteria | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mean age (range) | M: F | All adult | WHO II: III | IDH mutant | IDH mutant | IDH wild | Inclusion | Exclusion | |||||
| 1p/19q noncodel | 1p/19q codel | |||||||||||||
| Aliotta E et al 2020 | University of Virginia, USA | N/A | Retro. | 134 | N/A | N/A | N/A | N/A | 49 | 54 | 31 | Histopathology, molecular information | LGG | Lack of image sequence and genetic information |
| Aliotta E et al 2020 | TCIA | N/A | Retro. | 93 | N/A | N/A | N/A | N/A | 49 | 26 | 18 | N/A | LGG | Lack of image sequence |
| Broen MPG et al 2018 | University Medical Center Rotterdam and Maastricht University Medical Center, Netherlands | N/A | Retro. | 154 | 43 (20–82) |
86: 68 |
Yes | 133:9 | 75 | 67 | 12 | Histopathology, molecular information | Supratentorial, LGG | Lack of image sequence and genetic information |
| Corell A et al 2020 | Sahlgrenska University Hospital, Sweden | 2010–2018 | Retro. + Pros. | 135 | 46.1 (N/A) | 76: 59 |
N/A | N/A | 50 | 85 | 0 | Histopathology, molecular information | Supratentorial, LGG (Retro. group), + suspicious LGG (Pros. group) * | Lack of image sequence and genetic information, IDH wild type |
| Deguchi S et al 2020 | Shizuoka Cancer Center, Japan | June 2009–November 2018 | Retro. | 64 | 47.4 (20–80) |
38: 26 |
Yes | 38:26 | 22 | 20 | 22 | Histopathology, molecular information | LGG | Lack of image sequence and genetic information |
| Foltyn M et al 2020 | The Heidelberg University Hospital, Germany | February 2009–March 2018 | Retro. | 408 | 57 (N/A) |
228:180 | N/A | 61:52 | 66 | 44 | 285 | Histopathology, molecular information | Entire glioma cohort† | Lack of image sequence and genetic information, multiple |
| Lasocki A et al 2018 | The Royal Melbourne Hospital, Australia | August 2010–August 2016 | Retro. | 59 | N/A | N/A | N/A | 43:16 | 26 | 21 | 12 | Histopathology, molecular information | LGG | Lack of image sequence and genetic information |
| Lee MK et al 2020 | Asan Medical Center, Korea | May 2015–May 2017 | Retro. | 110 | 47.4 (19–82) |
56: 54 |
Yes | 45:65 | 19 | 46 | 45 | Histopathology, molecular information | LGG | Lack of image sequence and genetic information |
| Patel SH et al 2017 | NYU Langone Medical Center, USA | 2011–2014 | Retro. | 60 | 45 (20–75) |
44: 38 |
Yes | 35:47 | 22 | 31 | 7 | Histopathology, molecular information | Supratentorial, LGG | Lack of image sequence and genetic information, pediatrics |
| Patel SH et al 2017 | TCIA | N/A | Retro. | 125 | 45.5 (21–82) |
62: 63 |
Yes | 58:67 | 68 | 34 | 23 | N/A | Supratentorial, LGG | Lack of image sequence and genetic information |
Codel, codeletion; IDH, isocitrate dehydrogenase; LGG, low-grade glioma (WHO grade II and III based on histopathology); N/A, not available; M: F, Male: Female; No., number; noncodel, noncodeletion; Retro., retrospective; TCIA, The cancer imaging archive.
*Consists of independent two (retrospective and prospective) patients group, the prospective cohort encompassed 4 glioblastoma cases
†The cohort encompassed 287 glioblastomas
The composition of the detailed tumors was variable across the studies based on the proportion of IDH-wild type, proportion of IDHmut-Noncodel LGG, and proportion of WHO grade. Two cohorts of the two studies consisted of ≥ 40% of IDH-wild type,18,20 three cohorts of three studies with ≥ 20% and < 40% of IDH-wild type,14,16,17 and three cohorts of two studies (including two TCIA databases) with ≥ 10% and < 20% of IDH-wild type.14,21
Six cohorts of six studies consist of ≥ 50% of IDHmut-Noncodel LGG,14–17,20,21 and four cohorts of four studies with < 50% of IDHmut-Noncodel LGG.14,18,19,21 Six cohorts of five studies consist of ≥ 50% of WHO grade II,15–18,21 and four cohorts of three studies with < 50% of WHO grade II.14,19,20
The reference standard was histopathology following surgical excision in all studies. The enroll criteria was mostly similar across the studies that included nearly LGGs with the presence of adequate image sequences and genetic information; however, two studies partly included glioblastoma since a study partially contained prospective patients group that comprised suspicious for LGG,19 and a study included the entire glioma cohort.19,20
Analysis and MRI Characteristics of the Included Studies
Considering MRI characteristics, six cohorts from five studies did not specify the information about the vendor.14–17,21 MR images were obtained at 3 T in 2 of the 8 enrolled studies,18,20 at either 1.5 T or 3 T in 4 studies,15–17,19 and this information was not available in 2 studies.14,21 All other MRI characteristics, including the vendor, and reference sequences other than T2WI and FLAIR, are described in detail in Table 2.
Table 2.
MRI Characteristics and Criteria of the Included Studies
| Source | MRI information | MRI interpretation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Vendor (model) | Tesla | Sequences | Definition of T2-FLAIR mismatch sign | Decision | Multi-reader | Reader | Reader experience | Interreader agreement (kappa) | |
| Aliotta E et al 2020 | Siemens, Germany (N/A) | N/A | T1WI; CETIWI; T2WI; FLAIR; DWI | By radiologist’s decision * | Subj. | No | Neuroradiologist | 7 | N/A |
| Aliotta E et al 2020 | Variable (N/A) | N/A | T1WI; CETIWI; T2WI; FLAIR; DWI | By radiologist’s decision * | Subj. | No | Neuroradiologist | 7 | N/A |
| Broen MPG et al 2018 | N/A (N/A) | 1.5; 3 | T1WI; CETIWI; T2WI; FLAIR; DWI | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | Subj. | Yes | Neuroradiologist, Neurosurgeon | 3, 10 | 0.75 |
| Corell A et al 2020 | GE, USA; Philips, Netherland; Siemens, Germany (N/A) | 1.5: 3 | N/A | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | Subj. | Yes | Neuroradiologist, Neurosurgeon | N/A | 0.74–0.77 |
| Deguchi S et al 2020 | N/A (N/A) | 1.5; 3 | T2WI; FLAIR | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | Subj. | Yes | Neurosurgeon | 13, 26 | 0.73 |
| Foltyn M et al 2020 | Siemens, Germany (Magnetom TIM Trio/Prisma Fit, Verio, Skyra) |
3 | T1WI; CETIWI; T2WI; FLAIR; DWI | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | Subj. | Yes | Radiology resident | 3, 5 | 0.75 |
| Lasocki A et al 2018 | N/A (N/A) | 1.5; 3 | N/A | Over 50% area of signal nulling on FLAIR comparative to T2WI | Obj. | Yes | Neuroradiologist | N/A | 0.88 |
| Lee MK et al 2020 | Philips, Netherland (Achieva) | 3 | T1WI; CETIWI; T2WI; FLAIR; DWI; DSC | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | Subj. | Yes | Neuroradiologist | 2, 5 | N/A |
| Patel SH et al 2017 | N/A (N/A) | N/A | T2WI; FLAIR | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | N/A | Yes | Neuroradiologist | N/A | 0.75 |
| Patel SH et al 2017 | Variable (N/A) | N/A | T2WI; FLAIR | Near complete signal nulling (except rim) on FLAIR comparative to T2WI | N/A | Yes | Neuroradiologist | 3, 17 | 0.73 |
MRI, magnetic resonance imaging; CE, contrast-enhanced; DSC, dynamic susceptibility contrast; FLAIR, fluid attenuated inversion recovery; T1WI, T1-weighted image’; T2WI, T2-weighted image; N/A, not available; Subj., subjective; Obj., objective
*The articles did not specifically present the definition of T2/FLAIR mismatch sign
All but two studies used the same definition of T2-FLAIR mismatch sign proposed by Patel et al.: the subjective evaluation of the presence of a complete/near-complete hyperintense signal of the tumor on T2WI, in combination with a relative hypointense signal except for a hyperintense peripheral rim on FLAIR.21 A study aimed to assess the presence of T2-FLAIR mismatch sign objectively, when over 50% area of the tumor showed signal nulling on FLAIR comparative to T2WI, and the other study did not specifically presented the definition of T2-FLAIR mismatch sign.14 The result of consensus reading was extracted by multiple readers, including neuroradiologist, neurosurgeon, and radiology resident from 7 studies,15–21 and in two cohorts of one study, data extraction was performed by a single reader.14 Among the 7 studies that interpreted the T2-FLAIR mismatch sign by multiple readers,15–21 6 studies yielded interreader agreement.14–17,19,21
Diagnostic Performance of the MRI
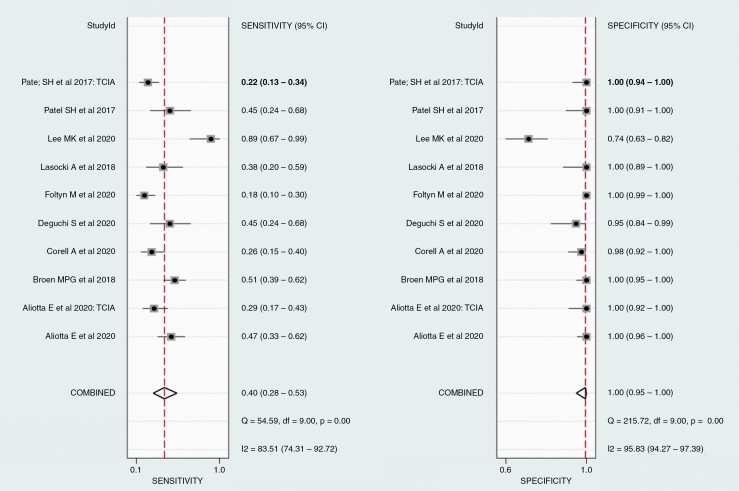
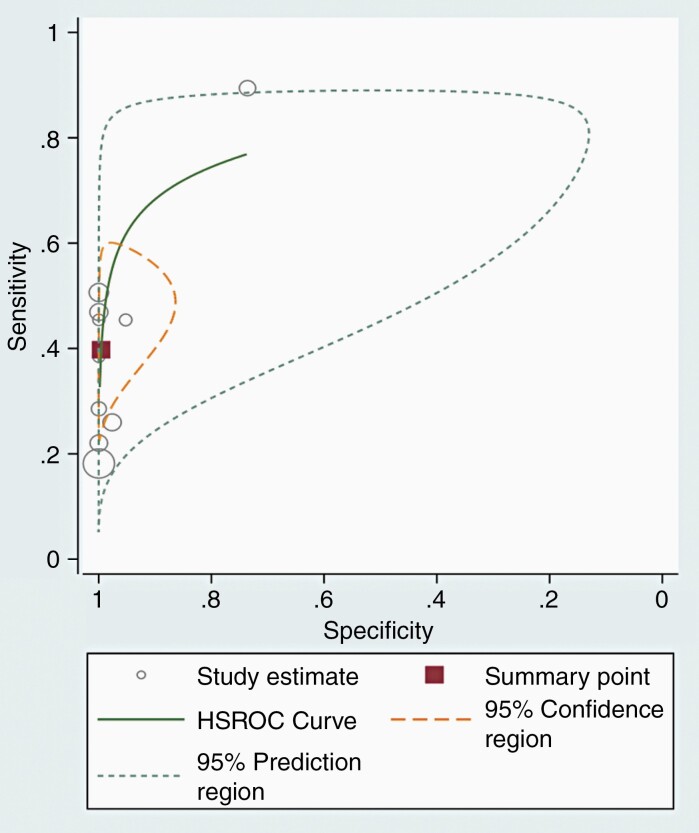
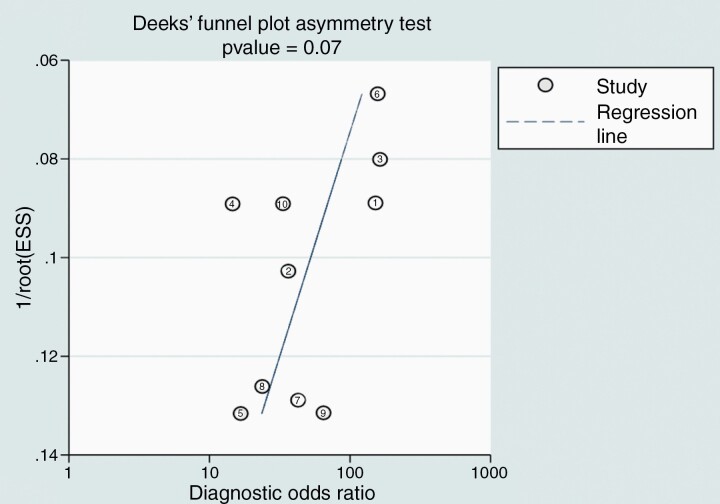
For all the 10 included cohorts, the pooled sensitivity was 40% (95% CI 28–53%), and the pooled specificity was 100% (95% CI 95–100%; Figure 2). A study presented outlier results in terms of sensitivity and specificity.18 In the HSROC curve, the difference between the 95% confidence and prediction regions was relatively large, indicating heterogeneity among the studies (Figure 3), while the area under the HSROC curve was 0.81 (95% CI 0.77–0.84). According to the Q test, heterogeneity was present (P < .01). Higgins I2 statistics demonstrated considerable heterogeneity in sensitivity (I2 = 83.5%) and in specificity (I2 = 95.8%), and there was no threshold effect (Spearman correlation coefficient of 0.53, i.e., < 0.6). When the outlier was removed, heterogeneity in sensitivity (I2 = 72.9%) and specificity (I2 = 74.5%) was decreased. According to Deeks’ funnel plot, the likelihood of publication bias was low, with a p-value 0.07 for the slope coefficient (Figure 4).
Figure 2.
Pooled sensitivity and specificity for the predictive accuracy of T2/ fluid-attenuated inversion recovery (FLAIR) mismatch sign. Horizontal lines indicate 95% confidence intervals of the individual studies.
Figure 3.
Hierarchical summary receiver operating characteristic (HSROC) curve for the predictive accuracy of T2/FLAIR mismatch sign.
Figure 4.
Deeks’ funnel plot to evaluate publication bias.
Meta-Regression
We also performed a meta-regression analysis to determine the cause of heterogeneity (Table 3). Among the potential covariates, none of these factors was significantly associated with study heterogeneity in the joint model. However, factors including proportion of IDH-wild type of ≥ 10% and < 20% (vs. ≥ 20% and < 40%, or ≥ 40%), proportion of IDHmut-Noncodel LGG of ≥ 50% (vs. < 50%), proportion of WHO grade II of ≥ 50% (vs. < 50%), definition of T2-FLAIR mismatch and multiple reader number (vs. single reader) show the statistical significance on the specificity on studies, even though the results were almost near identical (99% or 100%).
Table 3.
Meta-Regression of T2-FLAIR Mismatch Sign for Prediction of the IDH-Mutant, 1p/19q Noncodeleted Low-Grade Glioma
| Covariate | Subgroup | Meta-analytic summary estimate | P-value by Joint model | |||
|---|---|---|---|---|---|---|
| Sensitivity [95% CI] | P-value | Specificity [95% CI] | P-value | |||
| Proportion of IDH-wild type | ≥ 40% | 0.51 [0.20–0.83] | .43 | 0.99 [0.94–1.00] | .04 | .71 |
| ≥ 20% and < 40% | 0.47 [0.28–0.65] | .24 | 0.99 [0.97–1.00] | .03 | .57 | |
| ≥10% and <20% | 0.40 [0.25–0.55] | .77 | 1.00 [0.99–1.00] | .00 | .79 | |
| Proportion of IDH-mutant, 1p/19q noncodeletion | ≥50% | 0.33 [0.19–0.46] | .43 | 1.00 [0.99–1.00] | .00 | .26 |
| Proportion of WHO grade II | ≥50% | 0.48 [0.32–0.64] | .08 | 0.99 [0.97–1.00] | .02 | .27 |
|
Definition
of T2-FLAIR mismatch sign |
Near complete signal nulling (except rim) on FLAIR comparative to T2WI | 0.41 [0.25–0.56] | .63 | 0.99 [0.97–1.00] | .00 | .26 |
| Reader number | Multiple | 0.40 [0.26–0.55] | .70 | 0.99 [0.98–1.00] | .00 | .36 |
CI, confidence interval; IDH, isocitrate dehydrogenase; FLAIR, fluid attenuated inversion recovery
Quality Assessment
The bias risk and its applicability were assessed according to the QUADAS-2 criteria (Supplementary Figure S1).
In the patient selection domain, a TCIA cohort showed a high risk of bias owing to inappropriate exclusion.14 In the index test domain, three cohorts of two studies were considered to have an unclear risk of bias due to the unclear information of blinding to the index test and nulled information of pre-specified threshold.14,19 In the flow and timing domain, a cohort was considered to have a high risk of bias because a group of patients (IDH-wild type) were not included in the analysis.19 The bias risks in the reference standard domain were regarded as low in all the studies. In terms of applicability, three cohorts of two studies were considered to have indeterminate concern regarding the applicability in the index test14,19; however, other studies were considered to have low concern regarding applicability in the patient selection, index test, and reference standard domains.
Discussion
This systematic review and meta-analysis evaluated the diagnostic performance of T2-FLAIR mismatch sign in predicting a patient with IDHmut-Noncodel LGG. Our analysis demonstrated a near-perfect pooled specificity of 100%, and a relatively low power of pooled sensitivity of 40%. The considerable heterogeneities were proven among the enrolled studies by the results of the Q test and the Higgins I2 statistics for both sensitivity and specificity; however, the degree of heterogeneities were markedly decreased in the analysis following removal of data of the outlier study. Even though there was no proven significant factor associated with study heterogeneity in the joint model of the meta-regression analysis among the potential covariates, the composition of the detailed tumors showed wide variation across the studies. We found that at least some studies were considered to have a high risk of bias in the domains of patient selection, and flow and timing. These results shed light on the current state of the research regarding this topic, as well as the need for quality improvement.
From the molecular phenotypic and genotypic information, such as IDH and 1p/19q status, which is integrated into the revised WHO 2016 classification of the glioma, considerable interest has been aroused for the imaging biomarkers that can predict the molecular status of glioma.38 In particular, The T2-FLAIR mismatch sign, has been proved as a near-perfect specific imaging marker of IDHmut-Noncodel LGG.14–21 The sign has an advantage that it can predict not only merely predict the molecular type of tumor but also the behavior of the tumor, and the prognosis considering that the status of both IDH mutation and 1p/19q codeletion is known to be associated with slower tumor growth and better prognosis in LGGs.39,40 Even though the pathophysiology of the T2-FLAIR mismatch sign and the reason for this finding frequently associated with IDHmut-Noncodel LGG are not yet clear, several suggestions are present. Abundant microcystic change, histopathologically, of the tumor was known to be significantly well correlated with the T2-FLAIR mismatched region of the tumor.16,21,33 Since microcystic change is a histological hallmark of protoplasmic astrocytoma (which was a subtype of diffuse low-grade glioma before revised WHO 2016 classification, and now removed from the WHO classification as an entity), and owing to the signal intensity of the fluid within the microcysts may nulled on FLAIR, it is reasonable theory.16 In addition, Patel et al. assumed that this phenomenon could be associated with increased levels of proteins in the mammalian target of rapamycin pathway.21 However, this hypothesis needs further study.
Our analysis showed a near-perfect pooled specificity of 100%, and a relatively low power of pooled sensitivity of 40%. The reason for this low sensitivity remains unclear. We suggest, and hope further molecular and genetic clarification may solve the reason for these radiologically heterogeneous phenotypes within the IDHmut-Noncodel LGG. In a few reports, the combination of apparent diffusion coefficient (ADC) or cerebral blood volume (CBV) to the T2-FLAIR mismatch sign have improved the predictable power of the sign.14,18 Aliotta et al. reported that ADC value more than 1.5 × 10−3mm2/s exhibited a strong concordance with the T2-FLAIR mismatch sign and the combination of both parameters improved the sensitivity in IDHmut-Noncodel LGG. Lee et al. emphasized the combination of the ADC or CBV histogram parameters on T2-FLAIR mismatch sign as a reinforcement stratagem to improve the sensitivity in clinical practice. Further trials like those kinds of reinforcement and complementary combination of imaging biomarker may enhance the sensitivity for the T2-FLAIR mismatch sign.
A previous trial of meta-analysis was conducted by Goyal et al,41; however, the analysis only focused on the pool of the data of the predictive power of T2-FLAIR mismatch sign, and did not discuss the causes of heterogeneity across each original articles. The sample size, also, was too small to acquire confidence of the pooled data since the number of included original articles were 4, and more original articles with the same purpose were published recently.14,16,18-20 Therefore, there is a need to update the research and add the newly available information to a systematic review and re-analysis encompassing a larger sample size, and the addition of discussion in terms of cause of heterogeneity. In our meta-analysis, considerable heterogeneities were proven among enrolled studies by the results of the Q test and the Higgins I2 statistics for both sensitivity and specificity. Although there is no proven covariate in the meta-regression joint model, the degrees of heterogeneities were markedly decreased in the analysis following the removal of data of the outlier study (from I2 = 83.5% to 72.9% in sensitivity, from I2 = 95.8% to 74.5% in specificity). This result indirectly fears that the heterogeneity of diagnostic performance could arise from an outlier. A possible cause of outlier is a detailed composition of the tumors. Actually, tumor composition, including the proportion of IDH-wild/mutant type, 1p/19q noncodeletion or WHO grade, was variable across the studies. As T2-FLAIR mismatch sign was not detected in glioblastoma,20 inclusion of IDH-mutant glioblastoma (equivalent to WHO grade 4 astrocytoma) may affect the diagnostic performance. Instead, Patel et al. reported “fluid attenuation in noncontrast-enhancing tumor”, which is a novel neuroimaging metric and modified version of T2-FLAIR mismatch sign, that predicts IDH status in glioblastoma.42 This kind of specific imaging biomarker could have great potential as countermeasure to heterogeneous tumors such as glioblastomas. Other possible cause is the definition of T2-FLAIR mismatch sign. The interreader disagreement was nulled in the study by Lee et al.18 If the researcher regards the small area of T2-FLAIR mismatch sign in case without near-complete signal nulling (except rim), it may cause both lowering the specificity and enhancing sensitivity (threshold effect). Furthermore, we found that at least some studies were considered to have a high risk of bias in the domains of patient selection, and flow and timing, which were mainly contributed by inappropriate management of composition of the detailed tumors. Therefore, enrollment of well-designed cohorts would be helpful for more realistic assessment of diagnostic performance. Our study demonstrated that specificity, as well as sensitivity, showed heterogeneity statistically. This phenomenon is probably because the specificities of the included cohorts were compactly concentrated in a too narrow zone (near 100%); thus, even a small difference would have resulted in a statistically significant heterogeneity. Lastly, variety of MRI machines, and inclusion of various field strength of magnets could be factors for heterogeneity.
This study had several limitations. First, we assumed that MRI sequence acquisition parameters could be confounders in interpreting the presence of T2-FLAIR mismatch sign; however, most studies did not present the detailed parameters. The degree of fluid suppression on FLAIR may vary depending on the parameters,43 and it is likely that these parameters varied in the studies. An optimization study for the parameters of T2 and FLAIR in predicting the IDHmut-Noncodel LGG is timely needed. Second, the definition of T2-FLAIR mismatch sign was not identical across the studies, despite being similar. In addition, all cohorts of the included studies were based on retrospective design consisting of patients with pathologically proven gliomas. However, considering the real-world clinical setting where the radiologists face on the MRI images before the pathologic confirmation further evaluation based on prospective design is necessary to improve confidence in the real clinical utility. Third, the predictive accuracy of T2-FLAIR mismatch sign in entire population is still warranted to be further evaluated considering the worthy report of false-positive cases in children or young adults.36 Fourth, included studies were based on the WHO 2016 classification of gliomas. Further studies investigating the relation between T2-FLAIR mismatch sign or other novel imaging biomarker and CDKN2A/B homozygous deletion which as newly included in the diagnosis of IDH-mutant astrocytoma on the WHO 2021 classification of gliomas seem necessary. Finally, as we announced before, the effort reducing variability of tumor composition across the study is required to minimize the heterogeneity.
In conclusion, The T2-FLAIR mismatch sign is a near-perfect specific marker of IDH mutation and 1p/19q noncodeletion. Further improvements are warranted in terms of a well-designed cohort.
Supplementary Material
Acknowledgments
We would like to thank Editage (www.editage.co.kr) for English language editing.
Funding
This study was financially supported by Seoul National University Bundang Hospital Research Fund (Grant No14-2020-026)
Conflict of interest statement. The authors thank to two experts for statistical analysis: S.Y.A. who in Division of Statistics in Medical Research Collaborating Center at Seoul National University Bundang Hospital; K.H.H. who in Department of Radiology, Severance Hospital, Research Institute of Radiological Science, Center for Clinical Imaging Data Science, Yonsei University College of Medicine
Authorship statement. Implementation: Y.A.D. and B.S.C.; interpretation of the data: Y.A.D., S.J.C., B.S.C., S.H.B., Y.J.B., L.S., C.J., and J.H.K.; writing the draft manuscript: Y.A.D. and S.J.C., approval of the final version: Y.A.D., S.J.C., B.S.C., S.H.B., Y.J.B., L.S., C.J., and J.H.K.; experimental design: S.J.C. and B.S.C.; analysis: S.J.C., S.H.B., Y.J.B., L.S., C.J., and J.H.K.; experimental design: B.S.C.; and writing the manuscript at the revision stage: B.S.C., S.H.B., Y.J.B., L.S., C.J., and J.H.K.
References
- 1. Pallud J, Capelle L, Taillandier L, et al. Prognostic significance of imaging contrast enhancement for WHO grade II gliomas. Neuro Oncol. 2009;11(2):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pignatti F, van den Bent M, Curran D, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20(8):2076–2084. [DOI] [PubMed] [Google Scholar]
- 3. Asari S, Makabe T, Katayama S, Itoh T, Tsuchida S, Ohmoto T. Assessment of the pathological grade of astrocytic gliomas using an MRI score. Neuroradiology. 1994;36(4):308–310. [DOI] [PubMed] [Google Scholar]
- 4. LaViolette PS, Mickevicius NJ, Cochran EJ, et al. Precise ex vivo histological validation of heightened cellularity and diffusion-restricted necrosis in regions of dark apparent diffusion coefficient in 7 cases of high-grade glioma. Neuro-oncology. 2014;16(12):1599–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 6. Delfanti RL, Piccioni DE, Handwerker J, et al. Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol. 2017;135(3):611601–611611. [Google Scholar]
- 7. Eichinger P, Alberts E, Delbridge C, et al. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep. 2017;7(1):13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bahrami N, Hartman SJ, Chang YH, et al. Molecular classification of patients with grade II/III glioma using quantitative MRI characteristics. J Neurooncol. 2018;139(3):633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fellah S, Caudal D, De Paula A, M, et al. Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? AJNR Am J Neuroradiol. 2013;34(7):1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de la Fuente M, I, Young RJ, Rubel J, et al. Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro Oncol. 2016;18(2):283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou M, Zhou Y, Liao H, et al. Diagnostic accuracy of 2-hydroxyglutarate magnetic resonance spectroscopy in newly diagnosed brain mass and suspected recurrent gliomas. Neuro Oncol. 2018;20(9):1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jain R, Johnson DR, Patel SH, et al. “Real world” use of a highly reliable imaging sign: “T2-FLAIR mismatch” for identification of IDH mutant astrocytomas. Neuro Oncol. 2020;22(7):936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aliotta E, Dutta SW, Feng X, et al. Automated apparent diffusion coefficient analysis for genotype prediction in lower grade glioma: association with the T2-FLAIR mismatch sign. J Neurooncol. 2020;149(2):325–335. [DOI] [PubMed] [Google Scholar]
- 15. Broen MPG, Smits M, Wijnenga MMJ, et al. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol. 2018;20(10):1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deguchi S, Oishi T, Mitsuya K, et al. Clinicopathological analysis of T2-FLAIR mismatch sign in lower-grade gliomas. Sci Rep. 2020;10(1):10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lasocki A, Gaillard F, Gorelik A, Gonzales M. MRI features can predict 1p/19q status in intracranial gliomas. AJNR Am J Neuroradiol. 2018;39(4):687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee MK, Park JE, Jo Y, et al. Advanced imaging parameters improve the prediction of diffuse lower-grade gliomas subtype, IDH mutant with no 1p19q codeletion: added value to the T2/FLAIR mismatch sign. Eur Radiol. 2020;30(2):844–854. [DOI] [PubMed] [Google Scholar]
- 19. Corell A, Ferreyra Vega S, Hoefling N, et al. The clinical significance of the T2-FLAIR mismatch sign in grade II and III gliomas: a population-based study. BMC Cancer. 2020;20(1):450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foltyn M, Nieto Taborda KN, Neuberger U, et al. T2/FLAIR-mismatch sign for noninvasive detection of IDH-mutant 1p/19q non-codeleted gliomas: validity and pathophysiology. Neurooncol Adv. 2020;2(1):vdaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel SH, Poisson LM, Brat DJ, et al. T2-FLAIR Mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res. 2017;23(20):6078–6085. [DOI] [PubMed] [Google Scholar]
- 22. Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med. 2009;151(4):W65–W94. [DOI] [PubMed] [Google Scholar]
- 23. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. [DOI] [PubMed] [Google Scholar]
- 24. Suh CH, Park SH. Successful publication of systematic review and meta-analysis of studies evaluating diagnostic test accuracy. Korean J Radiol. 2016;17(1):5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim KW, Lee J, Choi SH, Huh J, Park SH. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: a practical review for clinical researchers-part I. General guidance and tips. Korean J Radiol. 2015;16(6):1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J, Kim KW, Choi SH, Huh J, Park SH. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: a practical review for clinical researchers-part II. Statistical methods of meta-analysis. Korean J Radiol. 2015;16(6):1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reitsma JB, Glas AS, Rutjes AW, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–990. [DOI] [PubMed] [Google Scholar]
- 28. Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20(19):2865–2884. [DOI] [PubMed] [Google Scholar]
- 29. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. [DOI] [PubMed] [Google Scholar]
- 30. Hoaglin DC. Misunderstandings about Q and “Cochran’s Q test” in meta-analysis. Stat Med. 2016;35(4):485–495. [DOI] [PubMed] [Google Scholar]
- 31. Deville WL, Buntinx F, Bouter LM, et al. Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol. 2002;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juratli TA, Tummala SS, Riedl A, et al. Radiographic assessment of contrast enhancement and T2/FLAIR mismatch sign in lower grade gliomas: correlation with molecular groups. J Neurooncol. 2019;141(2):327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tay KL, Tsui A, Phal PM, Drummond KJ, Tress BM. MR imaging characteristics of protoplasmic astrocytomas. Neuroradiology. 2011;53(6):405–411. [DOI] [PubMed] [Google Scholar]
- 34. Throckmorton P, Graber JJ. T2-FLAIR mismatch in isocitrate dehydrogenase mutant astrocytomas. Variability and evolution. 2020;95(11):e1582–e1589. [DOI] [PubMed] [Google Scholar]
- 35. Trong PD, Jesser J, Deimling A, et al. Preoperative predictors of malignancy in non-enhancing glioma in the era of molecular classification. Neuro Oncol. 2018;20(vi182), Supplement 6. [Google Scholar]
- 36. Johnson DR, Kaufmann TJ, Patel SH, et al. There is an exception to every rule-T2-FLAIR mismatch sign in gliomas. Neuroradiology. 2019;61(2):225–227. [DOI] [PubMed] [Google Scholar]
- 37. Batchala PP, Muttikkal TJE, Donahue JH, et al. Neuroimaging-based classification algorithm for predicting 1p/19q-codeletion status in IDH-mutant lower grade gliomas. AJNR Am J Neuroradiol. 2019;40(3):426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 39. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. [DOI] [PubMed] [Google Scholar]
- 40. Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goyal A, Yolcu YU, Goyal A, et al. The T2-FLAIR-mismatch sign as an imaging biomarker for IDH and 1p/19q status in diffuse low-grade gliomas: a systematic review with a Bayesian approach to evaluation of diagnostic test performance. Neurosurg Focus. 2019;47(6):E13. [DOI] [PubMed] [Google Scholar]
- 42. Patel SH, Batchala PP, Muttikkal TJE, et al. Fluid attenuation in non-contrast-enhancing tumor (nCET): an MRI Marker for Isocitrate Dehydrogenase (IDH) mutation in Glioblastoma. J Neurooncol. 2021;152(3):523–531. [DOI] [PubMed] [Google Scholar]
- 43. Lu H, Nagae-Poetscher LM, Golay X, et al. Routine clinical brain MRI sequences for use at 3.0 Tesla. J Magn Reson Imaging. 2005;22(1):13–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.