Abstract
The terminal steps of lens cell differentiation require elimination of all organelles to create a central Organelle Free Zone (OFZ) that is required for lens function of focusing images on the retina. Previous studies show that the spatiotemporal elimination of these organelles during development is autophagy-dependent. We now show that the inhibition of PI3K signaling in lens organ culture results in the premature induction of autophagy within 24 hours, including a significant increase in LAMP1+ lysosomes, and the removal of lens organelles from the center of the lens. Specific inhibition of just the PI3K/Akt signaling axis was directly linked to the elimination of mitochondria and ER, while pan-PI3K inhibitors that block all PI3K downstream signaling removed all organelles, including nuclei. Therefore, blocking the PI3K/Akt pathway was alone insufficient to remove nuclei. RNAseq analysis revealed increased mRNA levels of the endogenous inhibitor of PI3K activation, PIK3IP1, in differentiating lens fiber cells preceding the induction of OFZ formation. Co-immunoprecipitation confirmed that PIK3IP1 associates with multiple PI3K p110 isoforms just prior to formation of the OFZ, providing a likely endogenous mechanism for blocking all PI3K signaling and activating the autophagy pathway required to form the OFZ during lens development.
Keywords: PI3K, Akt, autophagy, lens, Organelle Free Zone (OFZ), development
INTRODUCTION
The regulation of cell differentiation in the lens is a complex process that is finely tuned to provide this tissue with its distinct functions. The chick embryo is a classical organism for investigating the mechanisms that regulate such developmental processes. Its lens, whose mature function is to focus images on the retina, has provided a unique reductionist model for understanding the signaling pathways that promote cell differentiation events, from the signals that instruct a cell to begin its differentiation program to the signals required for its cells to acquire their terminally differentiated phenotype [1–9]. The lens plays an especially important role in such studies due to it being an avascular tissue, not innervated, and dominated by only two related cell types, the lens epithelial cells and their differentiated counterparts, the lens fiber cells [10, 11]. A particularly valuable aspect of this model is that at most stages of lens development the full spectrum of the lens cell differentiation program is present concurrently [2]. This property has made it possible to isolate distinct regions of differentiation from a single stage of development for molecular analyses and complement these studies with immunolocalization analyses, providing a detailed view of the gradient of events that occur as a cell transits from an undifferentiated to a terminally differentiated state.
The precise organization of lens cells during development is crucial to its function. Along its anterior surface is a monolayer of undifferentiated lens epithelial cells. These cells initiate their differentiation, expressing lens-specific proteins after they withdraw from the cell cycle and migrate posteriorly along the lens equator, maintaining their close association with the basement membrane capsule that surrounds the lens. These cells begin their morphogenetic transition into differentiated lens fiber cells in the transition zone, located where the lens equator meets the posterior aspect of the lens. These differentiating lens fiber cells become highly elongated with their basal surfaces attached to the posterior lens capsule and their anterior tips reaching up to and associating with the anterior surfaces of the overlying lens epithelial cells. New lens fiber cells are added continually during development, with each new fiber cell enveloping those that had formed before them, as the oldest fiber cells are compressed in the center of the lens. The lens fiber cells form the bulk of lens tissue. In their final stages of differentiation, beginning in the centermost region of the lens, these cells lose their mitochondria, endoplamic reticulum (ER), and Golgi followed by elimination of their nuclei, leading to the formation of the lens Organelle Free Zone (OFZ) [8, 12–16]. This feature is believed crucial to establishing lens clarity and the ability of the lens to focus images unimpaired on the retina.
There is evidence that many regulatory factors are required for the spatiotemporal removal of organelles from the developing lens to create its unique organelle free zone [17–20], with studies from our labs and others demonstrating the direct involvement of autophagic mechanisms [8, 21]. Further insight into the molecular pathway of autophagy-dependent organelle removal is provided by our studies showing the requirement for the mitophagy protein BNIP3L in the elimination of mitochondria during OFZ formation [12], and the role of this autophagy-promoting molecule in the removal of the ER and Golgi to form the OFZ [12]. Clinical relevance of autophagy-dependent organelle removal in the developing lens is provided by studies of human mutations in FYCO1, a bridging molecule that connects autophagosomes to microtubule molecular motors [22], which revealed that a failed autophagy process leads to the loss of lens transparency and congenital cataracts [21, 23, 24].
In previous studies, we identified a role for the MAP kinase JNK in regulation of the signaling pathways responsible for the induction of the autophagy mechanism that removes lens organelles to form the OFZ [8]. Those investigations revealed a previously unknown function for JNK in the phosphorylation of the mTORC1 complex proteins mTOR and RAPTOR and the activation of their downstream effector p70S6K [8]. The mTORC1/p70S6K pathway is a classical signaling axis for the regulation of autophagy [25, 26]. Inactivation of JNK blocked phosphorylation of mTOR, RAPTOR, and p70S6K, inducing OFZ formation by targeting lens organelles to autophagosomes [8]. Interestingly, while blocking JNK activity leads to loss of mitochondria, ER, and Golgi within 24hr, nuclear loss is not observed until a day later, suggesting that the role of JNK in regulating elimination of nuclei is indirect. Our studies with the BNP3L/NIX knockout mice provided further evidence that induction of autophagy alone is insufficient to remove lens nuclei as the elimination of mitochondria, ER, and Golgi are suppressed without impairing the timing of nuclear loss [12]. Despite current insights into the mechanisms of OFZ formation, including a link between hypoxia, HIF1α and BNIP3L-dependent non-nuclear organelle degradation [12, 27], and a role for the phospholipaseA/acyltransferase (PLAAT) family of phospholipases [28, 29] in organelle loss [30], there is still much to learn about the signals that regulate the process of organelle removal to form the OFZ.
Induction of the formation of the cell’s autophagic machinery has been linked to inhibition of the PI3K/Akt/mTORC1/p70S6K signaling axis [26, 31]. Inactivation of this pathway leads to degradation of protein aggregates and organelles to maintain cell homeostasis [31], and under conditions of nutrient starvation this pathway is inactivated to induce organelle “self-eating” [32–34]. The Class I PI3Ks are a family of receptor-activated signal transduction molecules with both catalytic (p110α, β, γ, and δ, the latter immune cell-specific) and regulatory (p85, p101) subunits [35]. The p110α, β, and δ catalytic subunits are activated by growth factor receptors like IGF-1 [36], while p110 γ is activated exclusively by G-protein coupled receptors [35]. Class I PI3Ks have many downstream effectors, the most predominant among them being Akt and Rac. They are linked to many essential cellular mechanisms including the regulation of cell growth, proliferation, differentiation, motility, survival, and intracellular trafficking, in addition to the suppression of autophagy [35]. Studies conducted with a lens conditional knockout of p110α has linked this PI3K catalytic subunit to the regulation of lens growth [37]. In previous studies we showed that exposure of primary, differentiating lens cell cultures to a pan-PI3K inhibitor prevented the organization of cortical actin structures, lens fiber cell morphogenesis, and the formation of differentiated lentoid organoids, correlated with suppression of a PI3K-dependent Rac signal [38]. Interestingly, inhibition of PI3K signaling after lentoids had formed induced premature appearance of TUNEL positive cells in the center of these differentiated structures [38], which normally occurs just prior to the elimination of nuclei during OFZ formation [14, 39]. These observations led us to investigation of the role of PI3K signaling in the mechanisms that regulate the removal of organelles, including nuclei, from lens fiber cells to form the OFZ.
MATERIALS AND METHODS
Animals
Animal studies performed were approved by Thomas Jefferson University’s Institutional Animal Care and Use Committee (IACUC). The investigations comply with all relevant guidelines. The animal studies also are in compliance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Embryonated chicken eggs were obtained from Poultry Futures (Lititz, PA), or SPAFAS, Charles River, (Wilmington, MA).
Lens Microdissection
Chick embryo lenses were removed from the chick embryo eyes and microdissected to isolateeither the epithelial (E), cortical fiber (FP), and nuclear fiber (FC) cell regions or the epithelial (E) and fiber (F) cell zones at E11, 13, and 15, as modeled in Figure 1A, as described previously [8].
Figure 1. Development and differentiation of the chick embryo lens.
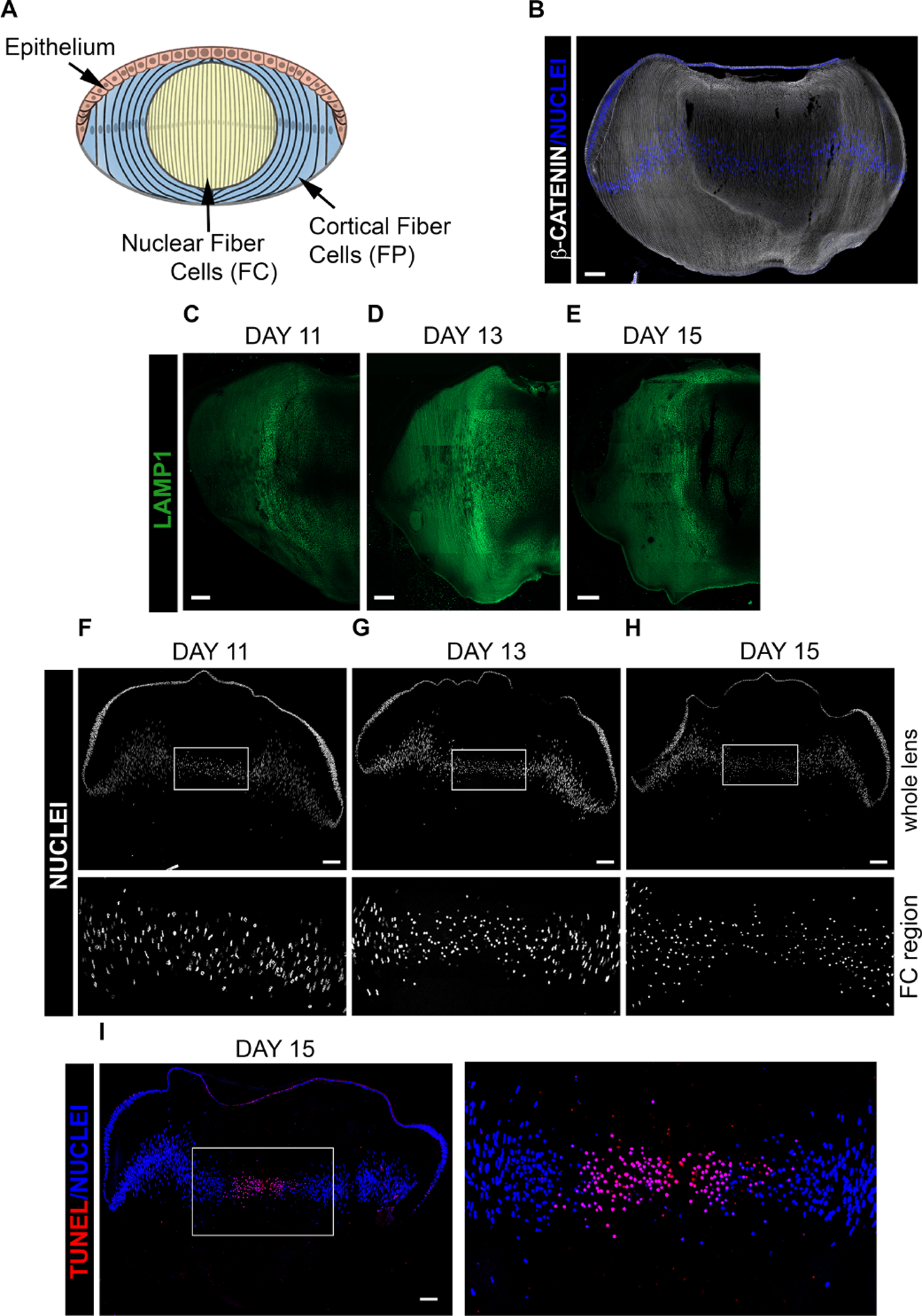
(A) Model depicting differentiation-specific zones of the developing lens. (B-I) Cryosections of chick embryo lenses at (C,F) D11, (B, D,G) D13 and (E,H,I) D15 were imaged by confocal microscopy following labeling for (B) β-catenin, revealing lens cytoarchitecture, and DAPI, (C-E) the lysosomal membrane protein LAMP1 to follow induction of lysosomes during developmental and their presence related to elimination of lens organelles during OFZ formation, (F-H) DAPI to follow the developmental stage specific elimination of nuclei, and (I) both TUNEL and DAPI to correlate DNA cleavage with chromatin removal during OFZ formation. Scale bar, 100μm. Results are representative of 3 independent studies.
Preparation of Ex Vivo Whole Lens Organ Cultures
Ex vivo lens organ cultures were prepared as described previously [8]. Briefly, E12 lenses were placed in culture in Complete Medium (Medium 199 [Life Technologies, 11150–059] with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin) at 37°C. After 1 h, PI3K signaling pathway inhibitors, or their vehicle control DMSO, was added to the media and the lenses cultured for 24 hrs. The inhibitors included the pan-PI3K inhibitors LY294002 (100 μM) and CH5132799 (25 μM or 100 μM), and the Akt allosteric inhibitor MK-2206 (10 μM). The concentration of inhibitors were determined based on their effective inhibition of PI3K/Akt signaling in both lens cortical and central fiber cells under the conditions of lens organ culture. Note that the concentration of the inhibitors added to the media will be higher than that which reaches the cells of the lens after diffusing across the thick matrix capsule that surrounds the lens [40].
Antibodies
Antibodies to calreticulin (sc-166837) TOM20 (for immunostaining, sc-17764), PIK3IP1 (sc-365778), and PI3K p110β (sc-602) were from Santa Cruz Biotechnology, Inc. Antibodies to TOM20 (for immunoblotting, 42406), pAkt ser473 (4060), pAkt thr308 (2965), Akt (9272), mTOR (2983), p-mTOR (2974), p70S6K (2708), p-p70S6K (97596), PI3K p110α (4249) were from Cell Signaling Technology. LC3B (L7543) was from Sigma Aldrich. Antibodies to LAMP1 (ab24170) and PI3K p110γ (ab154598) were from Abcam. Antibody to CP49 was a gift from Dr. Paul Fitzgerald (University of California, Davis). Antibody to PIK3IP1 (16826-1-AP) was from Proteintech. Secondary antibodies were from Jackson ImmunoResearch Laboratories. F-actin was from Invitrogen and nuclei were with labeled with DAPI from Biolegend.
Immunoblotting
WES – Simple Western system by Protein Simple:
Microdissected lens differentiation-state specific fractions were extracted in a TritonX100/Octylglucoside (OGT) buffer (44.4 mM n-octyl β-D-glucopyranoside, 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 5 mM EDTA, and 10 mM imidazole, containing 1 mM sodium vanadate, 0.2 mM H2O2 with protease and phosphatase inhibitors). The BCA assay (Thermo) was used to determine the protein concentrations of the extracts. Samples were prepared for WES immunoblot by mixing 2ugs of protein with a 5x Fluorescent Master Mix and Sample Buffer (ProteinSimple). The samples are denatured at 100°C for 5 minutes. Primary antibodies are diluted in Antibody Diluent II (ProteinSimple) and the secondary antibodies are used undiluted. The biotinylated ladder (5μl), protein lysates (3μl), primary antibodies (10μl), secondary antibodies (10μl), a luminol-peroxidase mix (15μl), and wash buffer (500μl) are loaded into designated wells of a WES plate. The plate comes preloaded with a separation matrix and a stacking matrix. The Simple Western, by ProteinSimple, uses capillary electrophoresis to detect proteins based on their size. A set of capillaries and the loaded WES plate are placed into the WES, in which immunodetection of proteins of interest occurs with a separation time of 25 minutes at 375 volts, primary and secondary antibodies were each incubated for 30 minutes. Data was analyzed using Compass for Simple Western software. Signals in each capillary are detected with chemiluminescence.
The data is shown in “lane” view created from an electropherogram that displays the intensity of the signals per second. The bands shown in the lane view are based on the signal intensity that the software calculates by quantifying the area under the curve of the acquired graph.
Standard Western Blot:
Western blots were performed as described previously [8]. Briefly, microdissected lens fractions were extracted in OGT and 15μg loaded onto an SDS-PAGE 4–12% precast tris/glycine gels (Invitrogen). Proteins were electrophoretically transferred onto Immobilon-P membranes (Millipore), blocked in 5% milk for 1 hr at room temperature, and probed for primary antibodies at 4°C overnight. Secondary antibodies conjugated to horseradish peroxidase (Bio-Rad) were incubated for 1 hr at room temperature. HRP is incubated with the ECL plus reagent (Thermo Fisher Scientific) and the bands detected using the FluorChem E & M Imager (ProteinSimple).
Immunofluorescence and Histology
Lenses are isolated and fixed for 2 or 24 hrs at 4°C in 4% paraformaldehyde, washed in PBS, and then cryoprotected (30% sucrose) prior to freezing in Polyfreeze Tissue Freezing Media Red (Polyscience #25115). 20μm thick serial cryosections were cut using a Microm HM 550 Cryostat, and only the central sections used for these studies. Immunolabeling was performed as described previously [41]. Briefly, the sections were permeabilized with 0.25% Triton X-100 in PBS buffer (Corning) for 30 min and the incubated in block buffer (5% goat serum, 1% BSA in PBS) for 1 hr prior to incubation in primary antibody for either 3 hrs at 37 C or overnight at 4 C, followed by incubation with secondary antibody for 2 hrs at 37 C (Jackson ImmunoResearch Laboratories). F-actin was localized with Alexa 647-conjugated phalloidin (Invitrogen-Molecular Probes). Nuclei were stained with DAPI (Biolegend). For histological analyses, lenses were fixed in 4% paraformaldehyde, cryoprotected (30% sucrose) for 24 hrs and frozen with tissue freezing media prior to cryosectioning. Sections were stained with hematoxylin and eosin.
Images were acquired with a Nikon Eclipse Ti microscope with a Nikon Photometrics Cool Snap HQ camera.
TUNEL Assay
DNA strand breaks were analyzed using a TUNEL assay kit (Sigma Aldrich). Sections were costained with DAPI and examined by confocal microscopy.
Confocal Image Analysis
Confocal imaging was performed using the Zeiss LSM800 confocal microscope with 4 laser lines: 405, 488, 561, and 640, on a Zeiss Axio Imager Z2 microscope with a motorized XY scanning stage. Images were acquired with the Zeiss Plan-Apochomat 40x/1.3 Oil objective. Zen software drives this confocal microscope. 40X tiles were acquired with an optical plane thickness of 1μm.
Quantification
Line scan intensity analyses were performed with the profile tool in ZEN blue software. For these studies, the line was acquired at a position half-way between the lens anterior and posterior surfaces from the outer edge of the lens equator through to the middle of the lens nuclear fiber zone. Staining intensities were determined along this line, subsequent to which the region was divided into 5 zones (A, B, C, D, E) to quantify and compare the fluorescence intensity across all regions of lens fiber cell differentiation.
Live Labeling of Active Mitochondria
The green fluorescent dye Rhodamine 123 (Thermo Fisher Scientific) was used to label active mitochondria across all regions of lens fiber cell differentiation. For these studies, whole E12 chick lenses were incubated with R123 for 1 hr at 37 C, mounted on an agarose stand, covered with buffer, and confocal z-stacks acquired using the Zeiss 40X/0.8 W water dipping objective on a Zeiss LSM800 confocal microscope.
Immunoprecipitation
For each immunoprecipitation, epithelial (E) and fiber (F) cells were isolated by microdissection from fifty lenses. The tissue was extracted in OG/T buffer, incubated with antibody to PIK3IP1 at 4°C overnight and the immunoprecipitate pulled down with TrueBlot immunoprecipitation beads (Rockland) at 4°C for 1 hr. The immunoprecipitated PK3IP1complexes were subjected to electrophoresis and blotted for each of the PI3K isoforms p110α, β, and γ, as well as for PIK3IP1 from which the ratio of PI3K 110 isoform/PIK3IP1 was calculated.
RNA sequencing
RNAseq data from E13 microdissected chicken lenses was data-mined from our previous study (GSE53976). RNA levels were analyzed using GraphPad PRISM 8 (La Jolla, CA, USA). The heatmaps were drawn using Morpheus software (Broad Institute).
Statistical Analysis
Statistical analysis was performed using t test on 3 or more independent experiments. Error bars represent SEM. Differences were considered significant when *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
RESULTS
Development-specific elimination of organelles including nuclei to form the lens Organelle Free Zone
In the developing lens there is a gradient of differentiation from undifferentiated lens epithelial cells to terminally differentiated lens fiber cells, including the elimination of non-nuclear organelles and nuclei (modeled, Fig. 1A). The unique organization of these cells in the developing chick lens is highlighted by labeling lens cryosections at E13 for β-catenin, which localizes to their cell-cell borders (Fig. 1B). Our previous studies showed that induction of autophagic vesicles in the developing lens was temporally coincident with OFZ formation and that these vesicles were actively involved in removing non-nuclear organelles [8]. While an essential element of this autophagic process is the fusion of organelle-containing autophagosomes with lysosomes to form the autophagolysosomes in which the organelles are digested, the differentiation-state specific induction of lysosomes has not been previously studied in the developing lens. To examine this question, cryosections of lenses at different stages of the chick embryo developmental window during which autophagy is induced, E11, E13 and E15, were immunolabeled for LAMP1, a glycoprotein associated with lysosomal membranes (Fig. 1C–E). Confocal image analysis showed that the expression of LAMP1 is greatly increased at E13, brightest in the region of the border of the nuclear and cortical lens fiber cells zones (Fig. 1D). The differentiation-state specifc induction of LAMP1+ lysosomes is consistent with that of autophagic vesicles [8]. Perhaps the best studied feature of OFZ formation is the elimination of lens nuclei from the central light path. The process involves nuclear condensation and DNA cleavage prior to nuclear loss [14, 42]. Developmentally, nuclear elimination follows that of mitochondria, ER and Golgi [43]. DAPI labeling of lens cryosections at E11, E13, and E15 shows the timing of both nuclear condensation and loss during chick embryo development (Fig. 1F–H). While most nuclei are eliminated by E15, chromatin fragments often remained in the center of E15 lenses (Fig. 1H). As DNA cleavage is a key element of the loss of nuclei to form the OFZ [17, 18], TUNEL assay was performed on lens cryosections at E15 and the sections co-labeled for DAPI. Confocal image analysis showed that nuclei condense prior to becoming TUNEL+ and the subsequent removal of nuclei from the center of the developing lens (Fig. 1I).
The spatiotemporal elimination of mitochondria and ER in the developing chick embryo lens was determined at E11, E13, and E15 with biochemical and immunolocalization approaches using antibodies to the outer mitochondrial import receptor [44, 45] TOM20 (Fig. 2), and the ER resident protein [46] calreticulin (Fig. 3). This developmental period spans the timing of OFZ formation, with the spatiotemporal loss of organelles beginning in the center of the lens and moving outward in the direction of the nascent fiber cells located in the lens cortex [8, 43]. For biochemical analyses, lens fiber cells were separated from the lens epithelium and a microdissection approach used to separate the two differentiation state-specific fiber cell zones; the nascent, cortical fiber cells located at the lens periphery (FP), and the central, nuclear fiber cells (FC) (modeled in Fig. 1A). Differentiation-state specific fractions were analyzed by immunoblot analysis using the WES Simple Western system by ProteinSimple. The FP and FC samples were immunoblotted for TOM20 (Fig. 2A, mitochondria), calreticulin (Fig. 3A, ER), and the lens fiber cell-specific intermediate filament protein CP49 (Fig. 2A). The results showed that expression of both TOM20 and calreticulin are diminished in the central FC region of the lens as early as E11, while expression of CP49 remained stable. At later stages of lens development, the levels of these organelle markers decreased in both the cortical (FP) and central (FC) fiber cell zones with TOM20 and calreticulin barely detectable in the FC by E15. The maintenance of expression of CP49 shows that the elimination of organelles occurs without impacting the expression of differentiation-state specific proteins of lens fiber cells.
Figure 2. Elimination of mitochondria to form the OFZ during development of the embryonic chick lens.
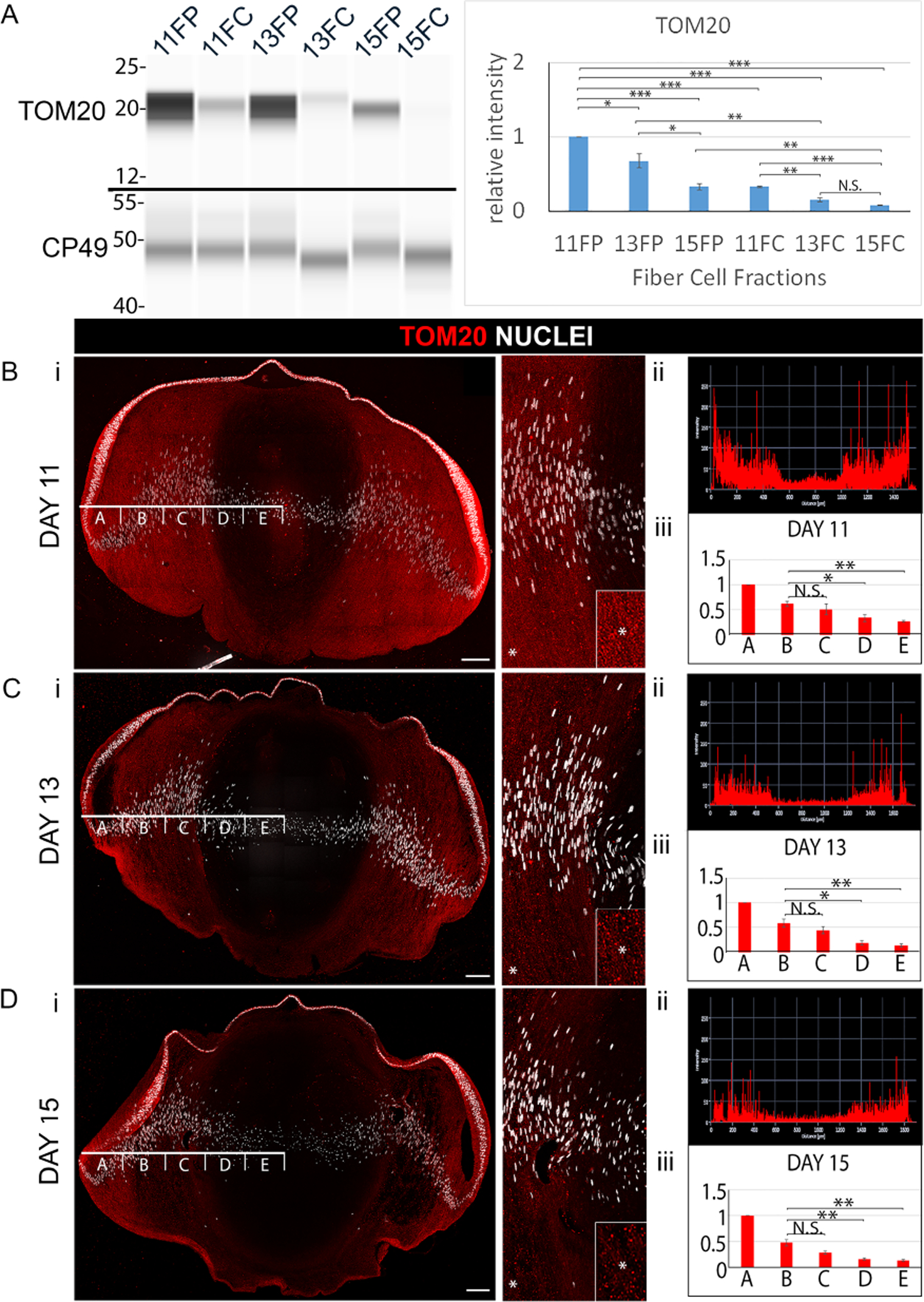
(A) Cortical fiber (FP) and central fiber (FC) regions of the chick embryo lens obtained by microdissection at E11, E13, and E15 were immunoblotted for the mitochondrial protein TOM20 and the lens fiber cell differentiation-specific protein CP49. The quantification of immunoblots for TOM20 from a minimum of 3 independent studies is shown in the panel to the right. Lens cryosections were immunolabeled for TOM20 and co-labeled with DAPI at (Bi) E11, (Ci) E13, and (Di) E15. Each confocal image is shown on the left for the whole lens section and on the right as a zoomed in view in the region of the border of the cortical and central lens fiber cells. Inserts are of the regions denoted by an asterisk, shown at higher magnification. (Bii, Cii, Dii) Line scan analyses across the entire width of the lenses immunolabeled for TOM20 in Bi, Ci, Di, respectively. (Biii, Ciii, Diii) Bar graphs quantifying the the fluorescence intensity from 3 independent studies over the left half of lenses as in Bi, Ci, Di, respectively, showing the staining intensities in the regions identified as A,B,C,D,E. These studies show the progressive removal of mitochondria from fiber cells with lens development. Scale bars, 100 μm. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test; N.S., not significant.
Figure 3. Elimination of endoplasmic reticulum (ER) to form the OFZ during development of the embryonic chick lens.
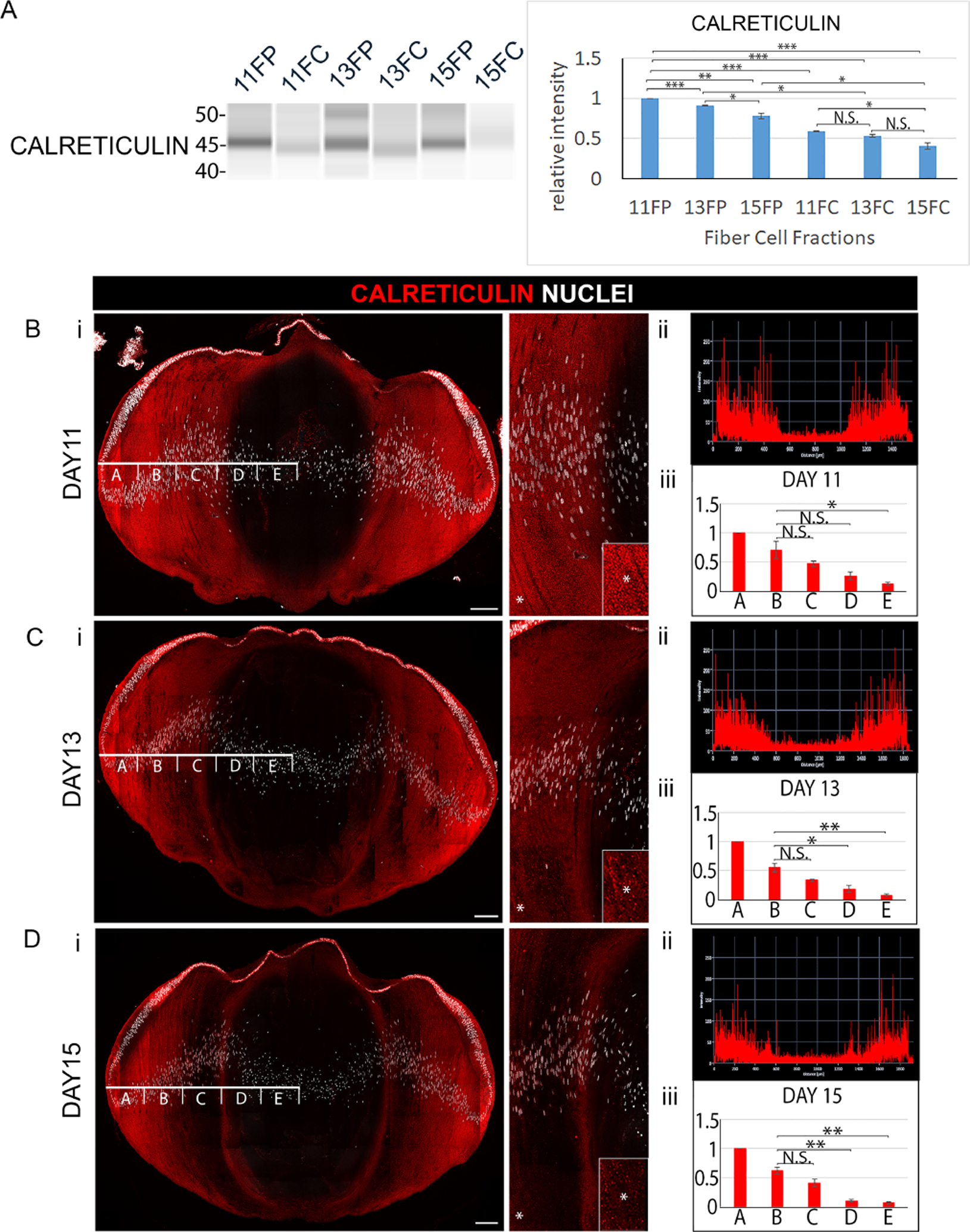
(A) Cortical fiber (FP) and central fiber (FC) regions of the chick embryo lens obtained by microdissection at E11, E13, and E15 were immunoblotted for the ER protein calreticulin. The quantification of immunoblots for calreticulin from a minimum of 3 independent studies is shown in the panel to the right. Lens cryosections were immunolabeled for calreticulin and co-labeled with DAPI at (Bi) E11, (Ci) E13, and (Di) E15. Each confocal image is shown on the left for the whole lens section and on the right as a zoomed in view in the region of the border of the cortical and central lens fiber cells. Inserts are of the regions denoted by an asterisk, shown at higher magnification. (Bii, Cii, Dii) Line scan analyses across the entire width of the lenses immunolabeled for calreticulin in Bi, Ci, Di, respectively. (Biii, Ciii, Diii) Bar graphs quantifying the fluorescence intensity from 3 independent studies over the left half of lenses as in Bi, Ci, Di, respectively, showing the staining intensities in the regions identified as A,B,C,D,E. These studies show the progressive removal of ER from fiber cells with lens development. Scale bars, 100 μm. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test; N.S., not significant.
The spatiotemporal pattern of elimination of mitochondria and ER was determined by confocal microscopy image analysis of cryosections from E11, E13, and E15 lenses immunolabeled for TOM20 (Fig. 2Bi–Di) or calreticulin (Fig. 3Bi–Di). Each section was co-labeled with DAPI to provide context of nuclear condensation/elimination relative to the loss of mitochondria and ER. High-magnification, high-resolution confocal images of TOM20 and calreticulin immunolabeling of lens sections shows the specificity of these antibodies for their targets (Supplemental Figure 1B,C). For all studies, immunolabeling was performed on the centermost section of serial sections cut across the entire lens. This approach ensured comparison of the same regions of the lens at different stages of development for different organelle proteins, and the precise determination of the spatiotemporal removal of organelles from the lens. Using the Zeiss Zen software profile tool, fluorescence intensity was quantified across the fiber cells at a location midway between the anterior and posterior of the lens starting from a position adjacent to the equatorial epithelium through to the center of the developing lens (Figs. 2Bii–Dii; 3Bii–Dii). The line scan was divided into 5 equal zones across the differentiating fiber cell region of the lens (designated as A-E in Figs. 2Bii–Dii and 3Bii–Dii) for a minimum of three independent studies, and total fluorescence intensity was quantified across each of these zones (Figs. 2Biii–Diii, and 3Biii–Diii). The results showed that significant loss of mitochondria and ER occurred in the FC between E11 and E13, followed by the continued loss of these organelles in the center of the lens that expanded into the cortical fiber cell region through E15, by which time there is significant elimination of nuclei from the center of the lens, although often it is not complete (Fig. 2Di, 3Dii). While organelle loss has occurred in the FP as well as the FC by E15, at this developmental time nuclear loss is limited to the FC.
PI3K/Akt signaling is suppressed and autophagy pathways induced in the differentiating fiber cells in the region of OFZ formation
Our previous studies of lens fiber cell differentiation show that the spatiotemporal removal of non-nuclear organelles from the center of the lens to create the OFZ is an autophagy-dependent process [8, 21]. Classically, suppression of the PI3K/Akt/p70S6K signaling axis has been considered a principal pathway for autophagy induction [47]. It is this pathway that is linked to autophagy, or “self-eating”, that degrades organelles to maintain homeostasis in response to stresses such as serum starvation [48, 49]. However, the potential involvement of the PI3K/Akt signaling axis in regulating the autophagy-dependent removal of lens organelles had not been investigated. PI3K is typically assayed by examining the phosphorylation of its downstream effector Akt at either its thr308 or ser473 sites [50–52]. Phosphorylation of both is required for full activation of Akt. We investigated whether there could be a link between the inhibition of PI3K/Akt activation and the induction of OFZ formation in the central lens fiber cells. For these studies, we performed WES immunoblot analysis for the phospho-serine473 Akt site in cortical (FP) and central (FC) fiber cell fractions isolated by microdissection from lenses at E11, E13, and E15 days of development (Fig. 4A). This analysis showed a significant decrease in Akt phosphorylation in central fiber cells (FC) compared to the fiber cells in the cortical zone (FP) as well as a decrease in the total level of Akt at all stages examined. The decrease in the level of phosphorylated Akt in the central region of the developing lens is consistent with a link between this pathway, the induction of autophagy, and the formation of the OFZ.
Figure 4. Induction of autophagy during lens OFZ formation.
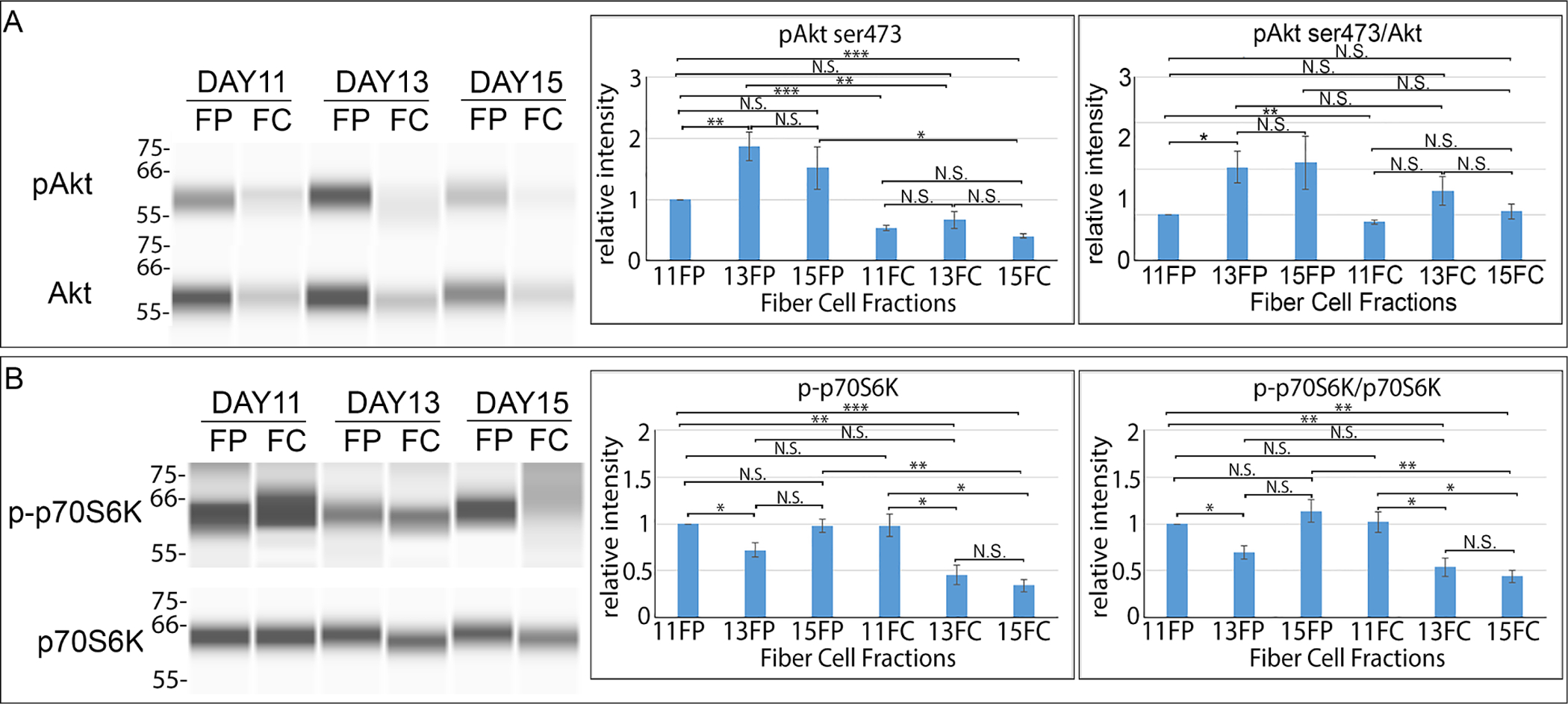
Chick embryo lenses were microdissected at E11, E13, and E15 to isolate cortical fiber (FP) and central fiber (FC) zones, and were subjected to immunoblot analysis (A,B) to determine the activation state of molecules downstream of the PI3K-regulated autophagy induction pathway including (A) phosphoSer473-Akt (pAkt) and total Akt and (B) phospho-p70S6K (p-p70S6K) and total p70S6K. Quantification of the results for a minimum of 3 independent studies are shown for pAkt ser473, pAkt ser473/Akt, p-p70S6K, and p-p70S6K/p70S6K. Results are consistent with the induction of autophagy signaling during the period of lens OFZ formation. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test; N.S., not significant.
The developmental timing of the induction of autophagy signaling was determined relative to the the formation of the OFZ. For these studies, cortical (FP) and central (FC) fiber cells isolated from chick embryo lenses at E11, E13, and E15 were immunoblotted for phospho-p70S6K, and p70S6K (Fig. 4B). While at E11 there was little difference in the activation state of p70S6K between fiber cells in the center of the lens and the cortical fiber cells that span through to the lens equator, by E13 there is a significant suppression of p70S6K phosphorylation in the central fiber cells (Fig. 4B), a developmental time just prior to establishment of an organelle free region in the center of the lens. The inactivation p70S6K and the increase in lysosome formation as mitochondria and ER are removed from the developing lens highlights the importance of autophagy to the formation of the OFZ. The sustained Akt signaling observed in the peripheral fiber cell region of the E13 lens as its downstream effector p70S6K activity is suppressed and lysosomes are induced is consistent with the many different biological pathways that PI3K regulates in the cell and indicates that some of these pathways must be retained at this stage of fiber cell differentiation.
PI3K/Akt inhibition promotes premature induction of autophagy-signaling in the developing lens
We examined whether there was a direct link between the suppression of PI3K/Akt signaling and the induction of autophagy in lens fiber cells during lens development. Chick embryo lenses were placed in organ culture at E12, prior to the establishment of the OFZ, in the presence of PI3K pathway inhibitors and examined after 24 hours. The inhibitors for this study included the pan-PI3K inhibitors LY294002 (Fig. 5A,B; 100 μM) and CH5132799 (Fig. 5A; 25 μM), and the allosteric Akt inhibitor MK-2206 (10 μM) (Fig. 5B), with DMSO as vehicle control. The efficacy of the inhibitors was determined by immunoblot of isolated fiber cell fractions for phosphorylation of Akt at its ser473 (Fig. 5A,B) and thr308 (Fig. 5A,B) sites. Total expression of Akt was also determined and the data quantified and presented graphically as the ratio of pAkt/Akt (Fig. 5A,B). The results showed that blocking activation of all PI3K downstream signaling effectors with LY24002 or CH5132799, or directly inhibiting activation the PI3K downstream effector Akt with the MK-2206 inhibitor, significantly reduced Akt phosphorylation. Both inhibition of all PI3K downstream signaling or just the PI3K/Akt signaling axis blocked activation of p70S6K in lens fiber cells (Fig. 5C), a key downstream effector in the PI3K/Akt/mTORC1/p70S6K pathway, and a result consistent with induction of autophagy.
Figure 5. Suppression of the PI3K/Akt signaling axis causes a premature induction of autophagy in the developing lens.
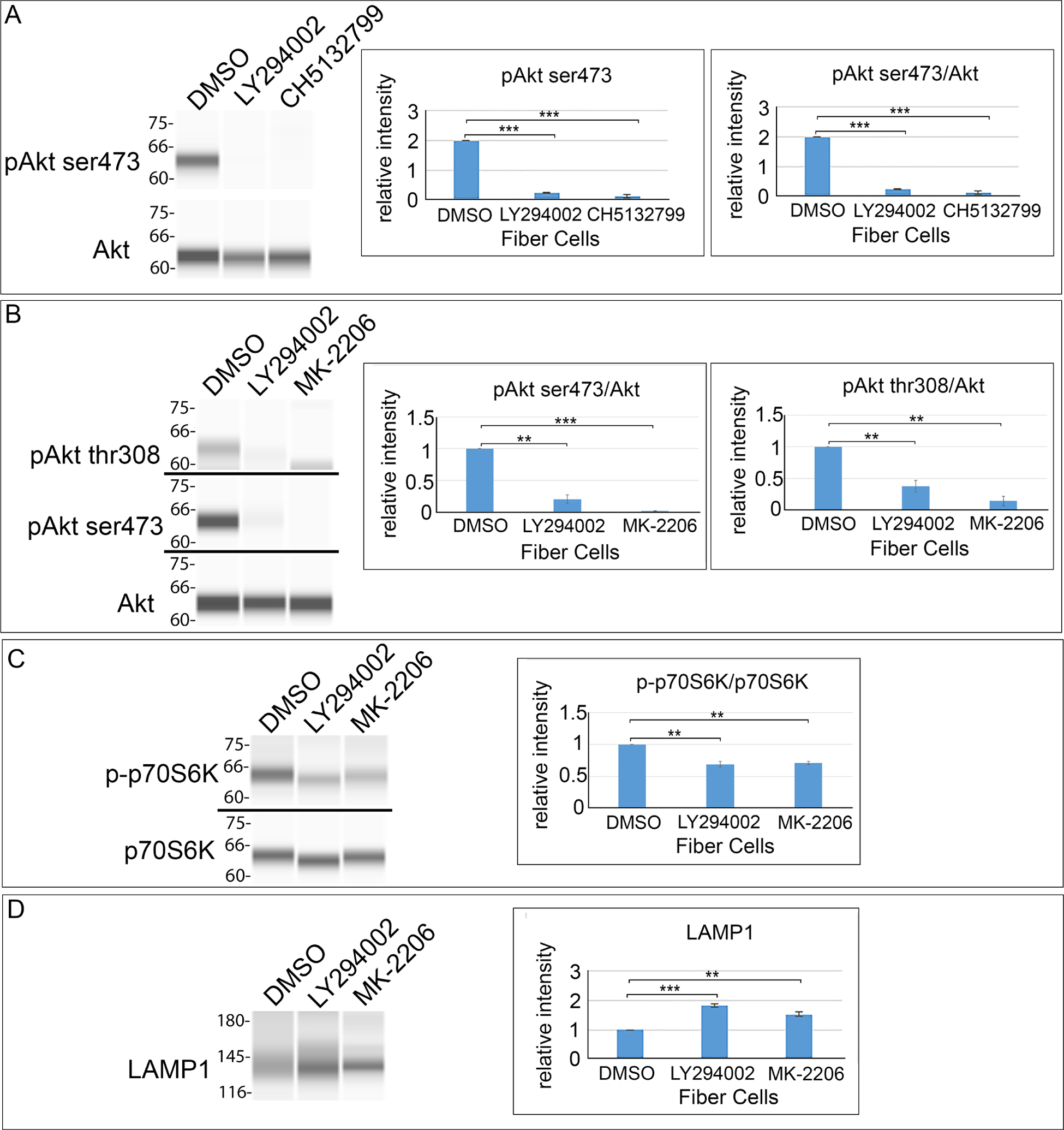
(A-D) Lens fiber cells were isolated from E12 lenses that were treated for 24 hrs in organ culture with PI3K pathway inhibitors or their vehicle DMSO and analyzed by immunoblot with quantification performed on a minimum of three independent studies. (A) Fiber cell fractions exposed to the pan-PI3K inhibitors LY294002 and CH5132799 were immunoblotted for phosphoAkt ser473 (pAkt ser473) and total Akt. (B-D) Fiber cell fractions exposed to the pan-PI3K inhibitor LY294002 and the Akt-specific inhibitor MK-2206 were immunoblotted for (B) phosphoAkt ser473 (pAkt ser473), phosphoAkt thr308 (pAkt thr308), and total Akt; (C) phospho-p70S6K (p-p70S6K) and total p70S6K; and (D) the lysosomal protein LAMP1. The results show that blocking the PI3K/Akt signaling axis prematurely induces autophagy in lens fiber cells. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test.
PI3K/Akt inhibition promotes induction of autophagy in the developing lens
A key indicator of the induction of autophagy is the lipidation of LC3BI to form LC3BII, a molecule involved in the formation of autophagic vesicles and the delivery of these vesicles to lysosomes [53, 54]. As LC3B lipidation is an early and sustained event in autophagy induction, E12 lenses were exposed to either pan-PI3K or Akt-specific inhibitors in organ culture for both 6 (Fig. 6A) and 24 (Fig. 6B) hours. Following microdissection of the fiber cell region of the lens into cortical fiber (FP) and central fiber (FC) cell fractions, the samples were immunoblotted for LC3BI/II to examine the link between PI3K/Akt inhibition and the activation of autophagy (Fig. 6). The results showed that formation of LC3BII was induced in both FP and FC regions of lens differentiation in response to inhibition of PI3K/Akt signaling.
Figure 6. Inhibition of PI3K/Akt in lens fiber cells induces autophagy.
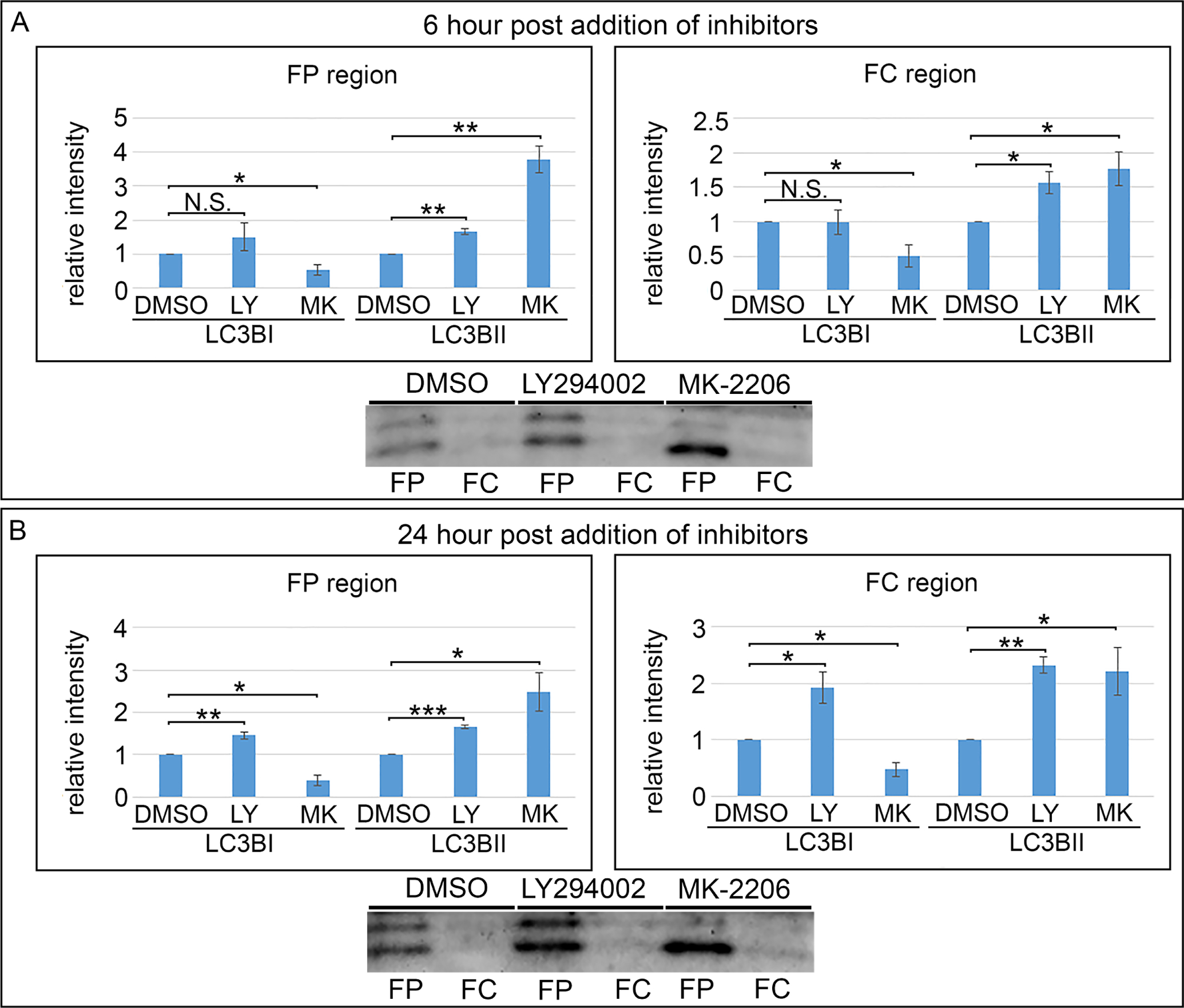
E12 lenses treated for (A) 6 hrs or (B) 24 hrs in organ culture with the pan-PI3K inhibitor, LY294002, or the Akt-specific inhibitor, MK-2206, or their vehicle DMSO were microdissected to isolate the cortical fiber (FP) and central fiber (FC) zones and immunoblotted for LC3BI and the autophagy marker LC3BII. Quantification of the relative intensity of LC3BI and LC3BII normalized to DMSO is displayed in bar graphs. LC3BII increases in the FP and FC regions at both 6 and 24 hours post treatment. The results show that blocking PI3K/Akt signaling induces autophagy in lens fiber cells. Results are representative of 3 independent studies. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test; N.S., not significant.
To examine the impact of blocking PI3K/Akt signaling on the presence of lysosomes, an organelle that fuses with autophagosomes to digest their contents, both immunoblot (Fig. 5D) and immunolocalization (Fig. 7A) analyses were conducted for the lysosomal membrane protein LAMP1. The immunoblot results showed a significant induction of LAMP1 expression in response to both PI3K and Akt inhibitors. The immunolocalization studies confirms this increase in LAMP1 expression and revealed a specificity to the regions where lysosomes are induced, with the regions of greatest increase in LAMP1+ lysosomes consistent with their role in the autophagic removal of lens organelles. High-magnification, high-resolution confocal images of LAMP1 immunolabeling of lens sections shows the specificity of this antibody for lysosomes (Supplemental Fig. 1A). These studies show that blocking the PI3K/Akt signaling axis results in premature induction of autophagy in the developing lens. H&E histological staining shows that the activation of these autophaghic pathways occurs with little impact on lens morphology (Fig. 7B). In LY294002-treated lenses a separation between lens equatorial epithelial cells and their adjacent nascent lens fiber cells was observed at the epithelial/fiber cell interface.
Figure 7. Induction of autophagy following inhibition of all PI3K downstream pathways or just the PI3K/Akt signaling axis has no effect on lens morphogenesis.
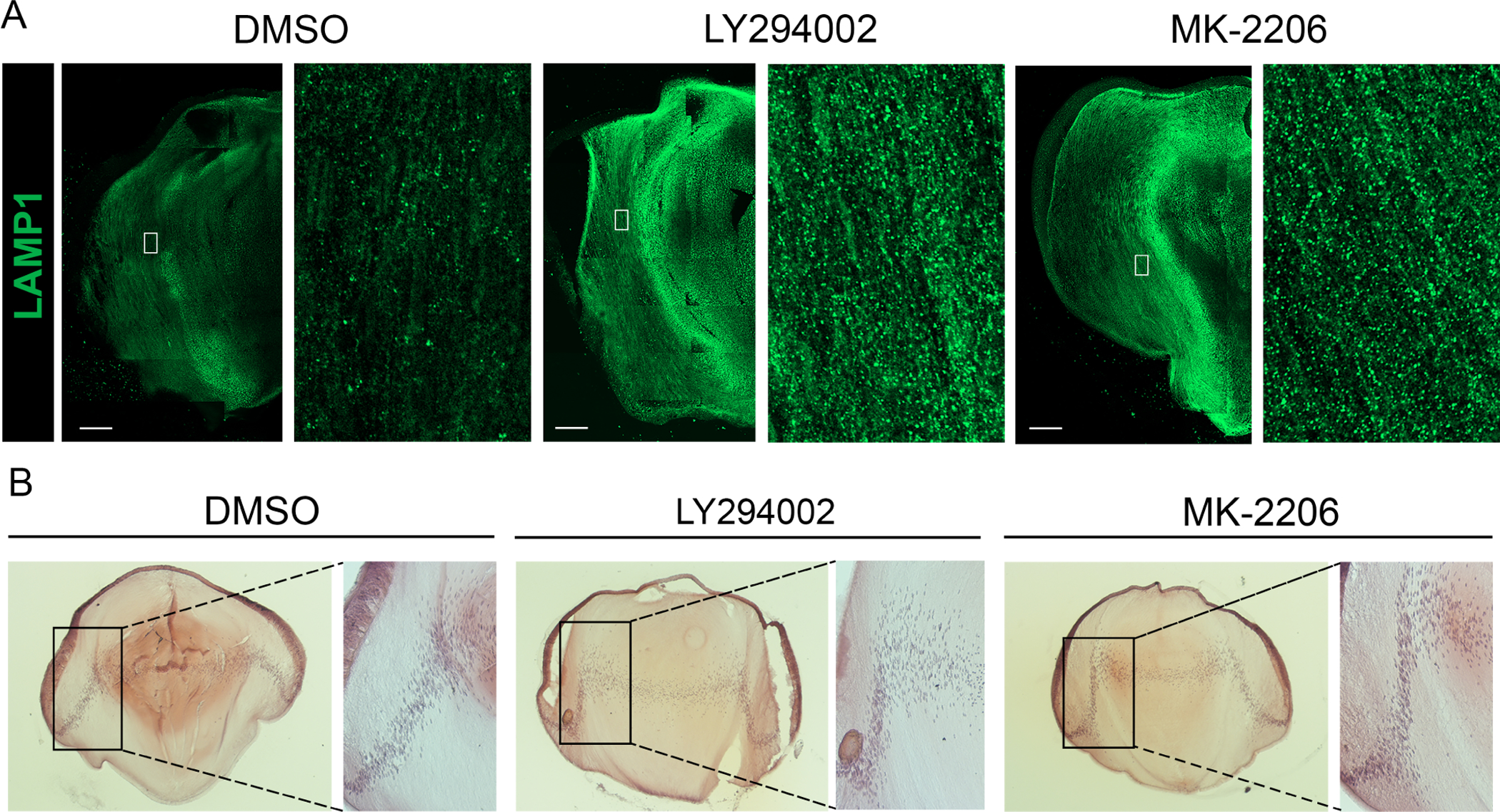
Cryosections of E12 lenses were exposed in organ culture to the pan-PI3K inhibitor LY294002, the Akt-specific inhibitior MK-2206, or their vehicle DMSO for 24 hrs. (A) Lens sections immunolabeled for the lysosomal protein LAMP1 and imaged by confocal microscopy. Boxed in regions are shown on the right at higher magnification. Scale bar, 100 μm. (B) Lens sections stained with H&E and imaged by phase microscopy. Boxed regions are shown to the right at higher magnification. Results are representative of 3 independent studies.
Inhibition of PI3K signaling does not impact mitochondria membrane potential
We have previously shown that mitochondria become depolarized during the transition of lens equatorial epithelial cells to their differentiated fiber cell phenotype and that they are eliminated subsequently during the fiber cell maturation process [55]. Since loss of mitochondrial membrane potential precedes mitochondrial removal, we investigated whether membrane potential was lost when PI3K signaling was suppressed. For this study, E12 chick lenses were cultured for 24 hours with either DMSO or LY294002 as above and live labeled with Rhodamine 123, a green fluorescent dye sequestered by active mitochondria. Whole lenses were imaged live by confocal microscopy using an immersion lens with the embryonic lenses positioned with the equator positioned vertically and the lens anterior epithelium located on the right. Images were acquired at different depths beginning at the top-facing edge of the lens equator through to the center of the lens at 700μm in a field that spans from the lens anterior epithelium to midway between the anterior and posterior aspects of the lens. Images are shown for both control and PI3K inhibitor treated lenses at depths of 100μm, 400μm, and 700μm (Fig. 8), which spans fiber cells from nascent, cortical fiber cells to the mature fiber cells in the center of the lens. Quantification of the fluorescence intensity of Rhodamine123 labeling at each depth was determined by line scan analysis and plotted graphically. High-magnification, high-resolution confocal images of Rhodamine123 labeling show its specificity for mitochondria (Supplemental Fig. 1D). The results show that inhibition of PI3K signaling had no significant effects on mitochondrial polarity (Fig. 8).
Figure 8. Inhibition of PI3K signaling does not impact mitochondrial membrane potential.
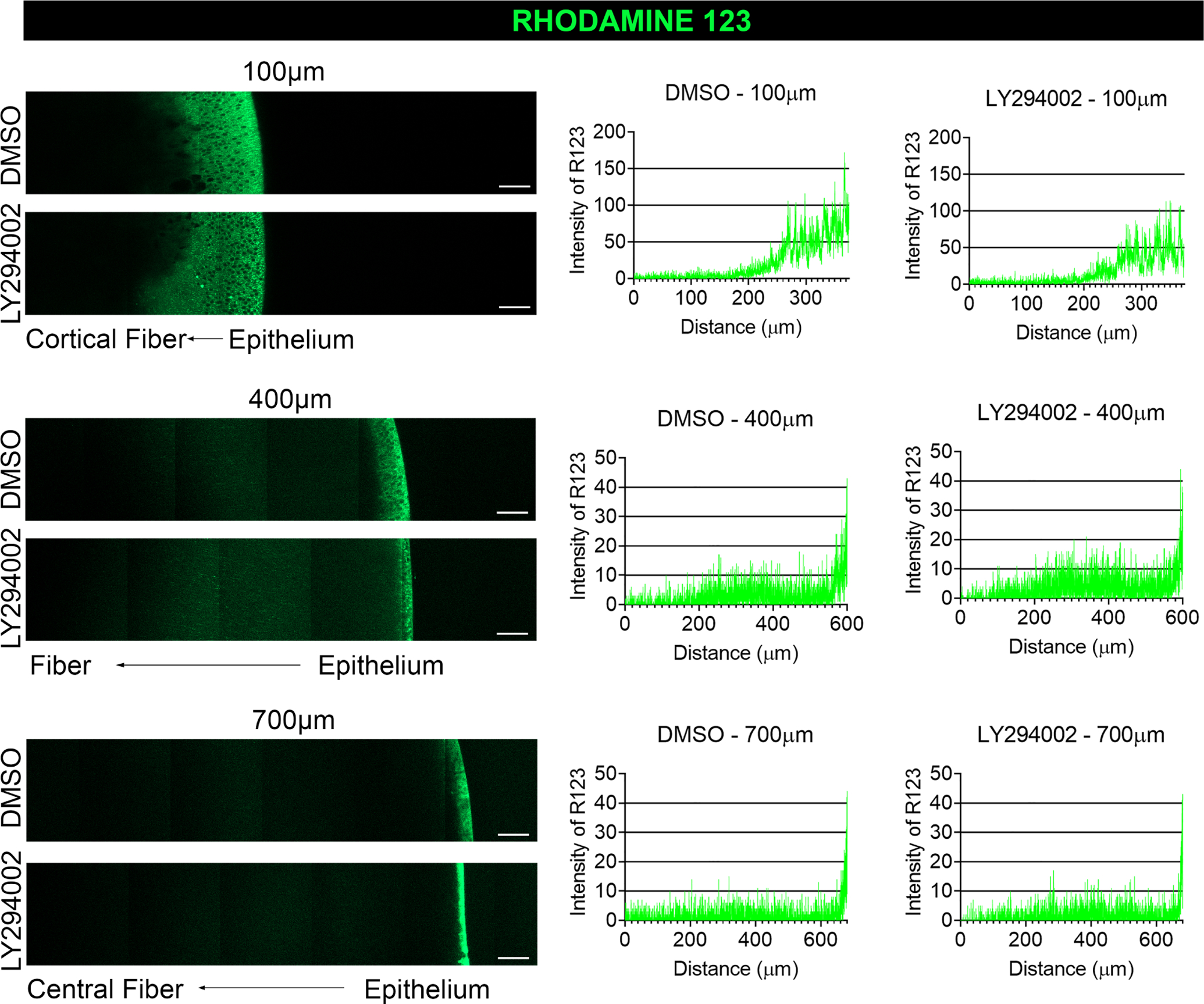
E12 lenses were treated for 24 hrs in organ culture with the pan-PI3K inhibitor, LY294002, or its vehicle DMSO and live labeled with Rhodamine 123 which is sequestered by active mitochondria and fluoresces green. The lens equatorial axis is positioned vertically with the anterior epithelium facing to the right and the lenses imaged by confocal microscopy at depths of 100 μm, 400 μm, and 700 μm. Fluorescence intensities were determined by line scan analyses and presented in graphical form to the right of the confocal images. The inhibition of PI3K signaling had no impact on mitochondrial membrane potential in the developing lens. Results are representative of 3 independent studies. Scale bar, 50 μm.
Inhibition of PI3K signaling induces premature elimination of mitochondria and ER from differentiating lens fiber cells
The studies above show that inhibition of the PI3K/Akt signaling axis in differentiating lens fiber cells induces autophagy (Figs. 5–7). To examine whether suppression of PI3K signaling is responsible for the elimination of mitochondria and ER to form the lens OFZ, E12 chick embryo lenses were exposed for 24 hrs to the pan-PI3K inhibitors LY294002 or CH5132799, with vehicle DMSO as control (Figs. 9,10). The impact of inhibiting PI3K signaling on mitochondria and ER in lens fiber cells was evaluated with antibodies to the mitochondrial outer membrane protein TOM20 (Fig. 9) and the ER resident protein careticulin (Fig. 10), respectively, using both biochemical (Figs. 9AB; 10A,B) and immunolocalization (Figs. 9C,D and 10C,D) approaches. WES immunoblot results were quantified using Compass for Simple Western software (Figs. 9B, 10B). Confocal image analysis was performed on immunolabeled lens cryosections representing the centermost of serial sections cut across the entire lens (Fig. 9C,D and 10C,D). Quantification of the confocal imaging based on fluorescence intensity was determined for both the FP region (Figs. 9Ci,Di and 10Ci,Di; in the region designated as A in Figs. 9C,D and 10C,D) and the FC region (Figs. 9Cii,Dii and 10Cii,Dii; in the regions designated as B in Figs. 9C,D and 10C,D) using Zeiss’s Zen software line scan tool.
Figure 9. Inhibition of PI3K signaling induces premature loss of mitochondria.
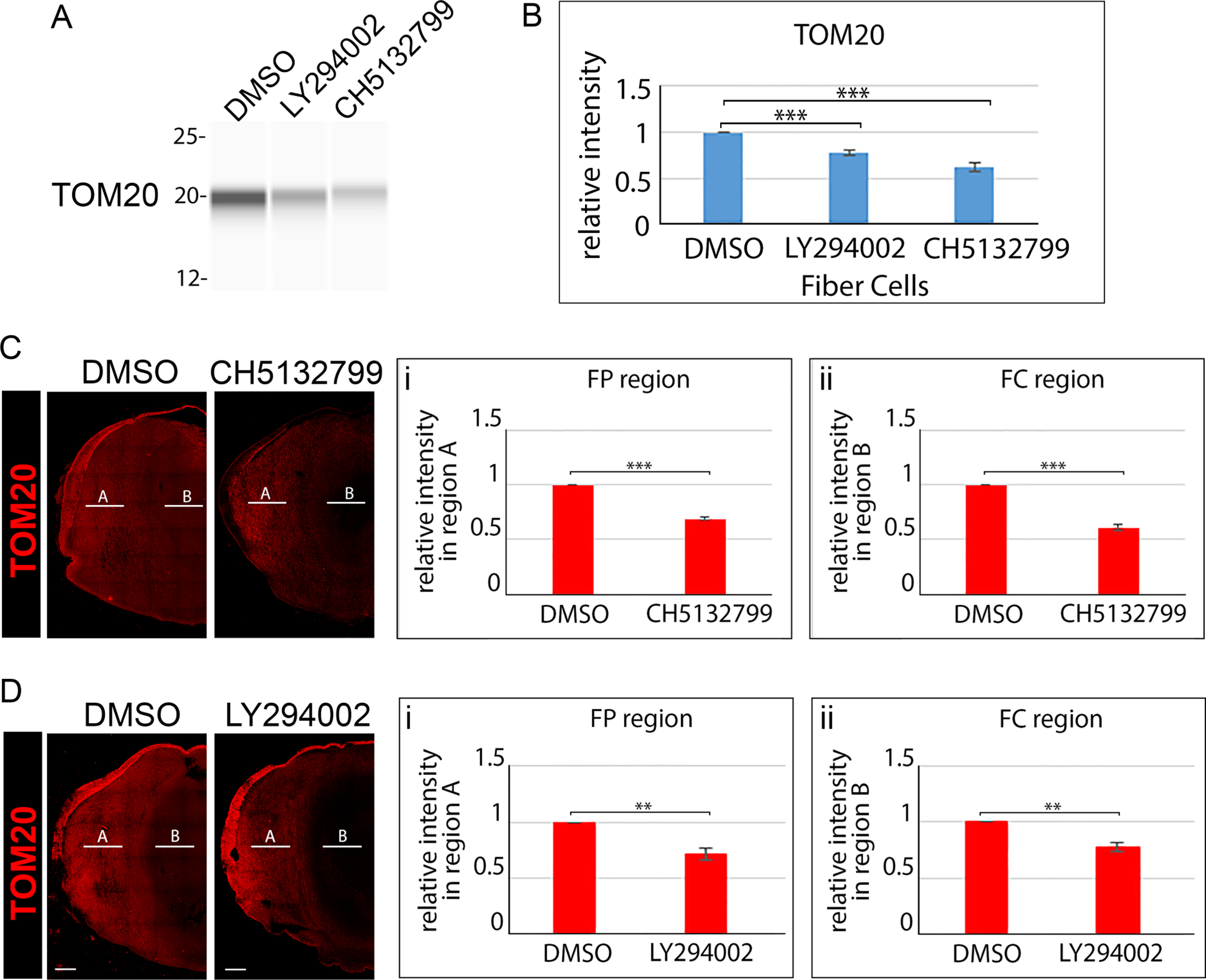
E12 lenses were treated for 24 hrs in organ culture with the pan-PI3K inhibitor LY294002, the pan-PI3K inhibitor CH5132799, or their vehicle DMSO. (A) Lens fiber cell fractions isolated from control and inhibitor treated lenses were immunoblotted for TOM20 and (B) the immunoblot results quantified from 3 independent studies. (C) Confocal imaging of cryosections of E12 lenses following exposure for 24 hrs to the the pan-PI3K inhibitor CH5132799 or its vehicle DMSO immunolabeled for TOM20. (D) Confocal imaging of cryosections of E12 lenses following exposure for 24 hrs to the the pan-PI3K inhibitor LY294002 or its vehicle DMSO immunolabeled for TOM20. Line scan analyses were performed across the entire width of the lenses and the fluorescence intensity quantified from 3 independent experiments in the cortical fiber region FP (Ci,Di) denoted as A, and the central fiber region FC (Cii, Dii) denoted by B. The data with two independent PI3K inhibitors shows that blocking PI3K signaling induces premature loss of the mitochondria to form the OFZ. Scale bar, 100 μm. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test.
Figure 10. Inhibition of PI3K signaling induces premature loss of ER.
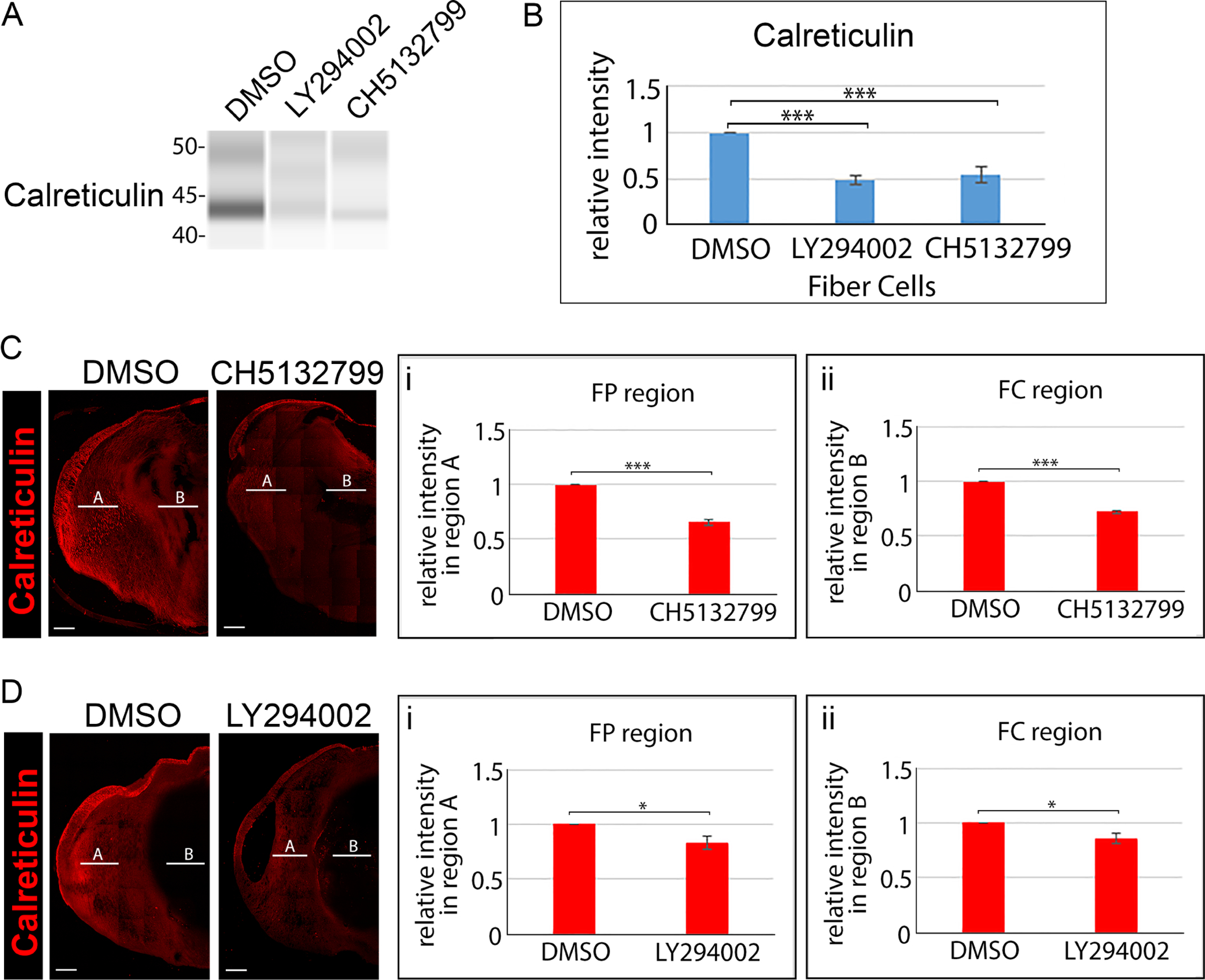
E12 lenses were treated for 24 hrs in organ culture with the pan-PI3K inhibitor LY294002, the pan-PI3K inhibitor CH5132799, or their vehicle DMSO. (A) Lens fiber cell fractions isolated from control and inhibitor treated lenses were immunoblotted for calreticulin and (B) the immunoblot results quantified from 3 independent studies. (C) Confocal imaging of cryosections of E12 lenses following exposure for 24 hrs to the the pan-PI3K inhibitor CH5132799 or its vehicle DMSO immunolabeled for calreticulin. (D) Confocal imaging of cryosections of E12 lenses following exposure for 24 hrs to the the pan-PI3K inhibitor LY294002 or its vehicle DMSO immunolabeled for calreticulin. Line scan analyses were performed across the entire width of the lenses and the fluorescence intensity quantified from 3 independent experiments in the cortical fiber region FP (Ci,Di) denoted as A, and the central fiber region FC (Cii, Dii) denoted by B. The data with two independent PI3K inhibitors shows that blocking PI3K signaling induces premature loss of ER to form the OFZ. Scale bar, 100 μm. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test.
The immunoblot analysis showed premature loss of both mitochondria (Fig. 9A,B) and ER (Fig. 10A,B) from lens fiber cells was induced by both pan-PI3K inhibitors within just 24 hours. Confocal image analysis confirmed that the suppression of PI3K signaling induced elimination of both mitochondria (Fig. 9C,D) and ER (Fig. 10C,D), and showed that blocking this signaling pathway extended loss of these organelles from the central fiber cell zone where OFZ formation begins into the cortical fiber region. The findings that PI3K inhibitors promoted premature elimination of mitochondria and ER from lens fiber cells links the classical PI3K autophagy-regulating pathway to the induction of organelle loss in the developing lens to form the OFZ.
Suppression of the PI3K/Akt signaling axis leads to premature elimination of mitochondria and ER in the developing lens
Since Akt is considered a crucial signaling intermediate of PI3K whose suppression leads to activation of autophagy [47], we examined whether suppression of the PI3K/Akt signaling axis alone was sufficient to induce premature elimination of mitochondria and ER to form the OFZ during lens development. For these studies, E12 chick embryo lenses were exposed for 24 hrs to the Akt-specific inhibitor MK-2206, with vehicle DMSO as control. The impact of inhibiting Akt signaling on mitochondria and ER in lens fiber cells was evaluated by immunoblot (Fig. 11Aa,Ba) and immunolocalization (Figs. 11Ac,d; 11Bc,d) analyses, with results quantified as described above for both the immunoblot (Figs. 11Ab,Bb) and confocal (Figs. 11Ae,f; 11Be,f) studies. The results of blocking only the PI3K/Akt signaling axis paralleled those obtained with the pan-PI3K inhibitors that inhibit other PI3K downstream effectors as well as Akt, showing that elimination of mitochondria and ER for formation of the OFZ during lens development is linked to the specific inhibition of the PI3K/Akt signaling axis.
Figure 11. Inhibition of the PI3K/Akt signaling axis is sufficient to induce premature loss of mitochondria and ER.
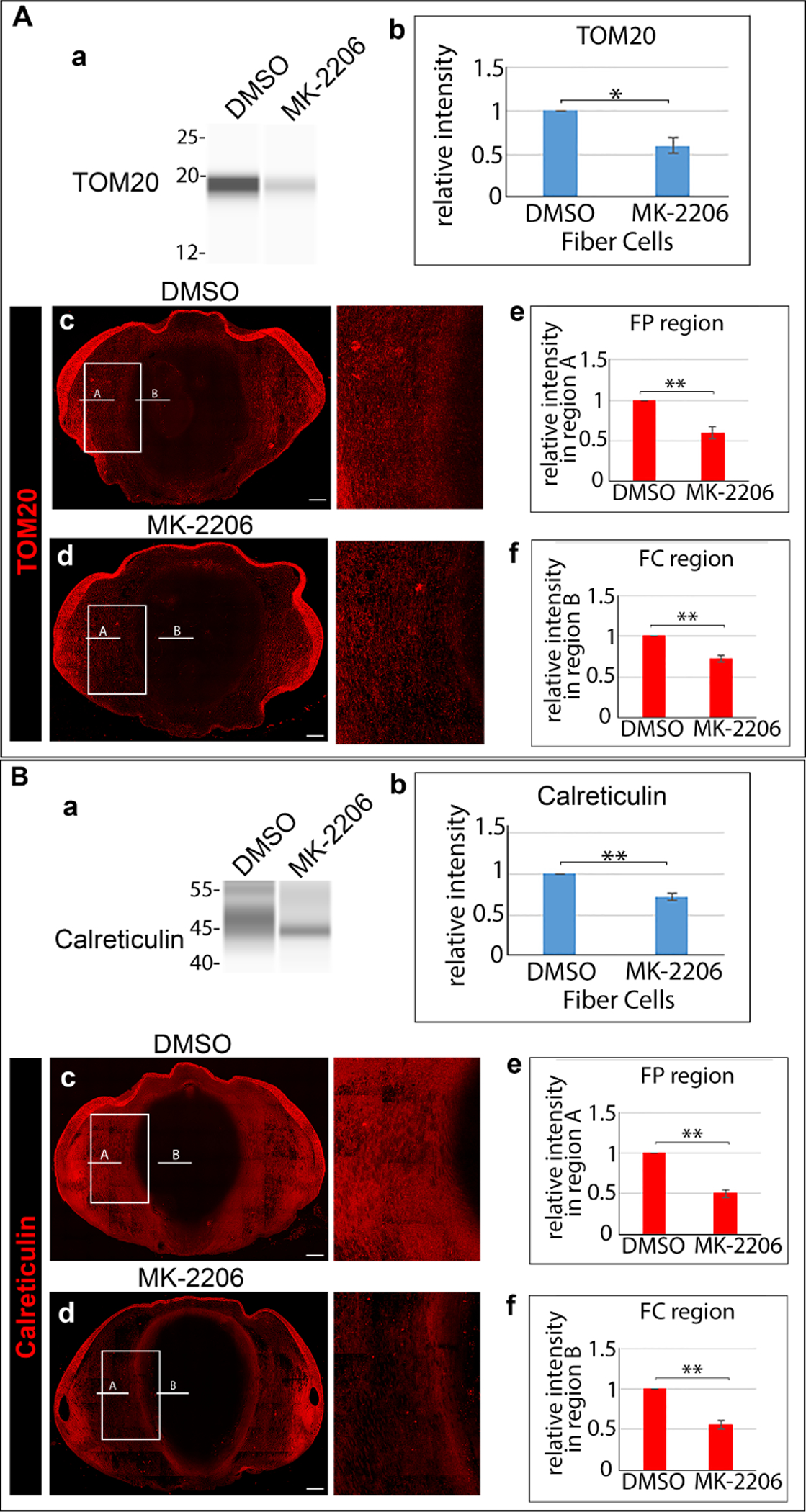
E12 lenses were treated for 24 hrs in organ culture with the Akt-specific inhibitor MK-2206 or its vehicle DMSO. Control and inhibitor treated lenses were microdissected to isolate lens fiber cells, immunoblotted for (Aa) TOM20 or (Ba) calreticulin, and (Ab, Bb) the results quantified from 3 independent studies. Cryosections of E12 lenses exposed for 24 hrs in organ culture to MK-2206 or its vehicle DMSO were immunolabeled for (Ac, Ad) TOM20 or (Bc, Bd) calreticulin and imaged by confocal microscopy. Images are shown on the left for the whole lens section and on the right as a zoomed in view in the region of the border of the cortical and central lens fiber cells. Line scan analyses were performed across the entire width of the lenses and the fluorescence intensity quantified from 3 independent experiments in the cortical fiber region FP (Ae, Be) denoted as A, and the central fiber region FC (Af, Bf) denoted by B. The data shows that inhibition of the PI3K/Akt signaling axis induces premature loss of the mitochondria and ER to form the OFZ. Scale bar, 100 μm. Error bars represent S.E., *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ .001, t test.
Impact of PI3K signaling on the elimination of nuclei to form the OFZ
The removal of nuclei during the last stages of lens fiber cell differentiation to form the OFZ must require a higher level of regulation than elimination of other lens organelles to insure that their loss does not impact the survival of the terminally differentiated lens fiber cells, a cell type that is retained throughout life. The impact on nuclear elimination of the pan-PI3K inhibitors LY294002 and CH5132799 were compared to the Akt specific inhibitor MK-2206 and DMSO vehicle control. These studies were performed in organ culture as above, with the CH5132799 inhibitor used at 100μM, a concentration that effectively removes mitochondria without impacting lens tissue morphology (Supplemental Figure S2). Confocal image analysis was performed following TUNEL assay and DAPI labeling of lens cryosections that represent the centermost of serial sections cut across the entire lens (Fig. 12). Inhibition of the Akt signaling was alone insufficient to cause premature removal of lens fiber cell nuclei from the center of the lens (Fig. 12D). In lenses exposed to the MK-2206 inhibitor nuclear condensation appeared similar to controls; however, while no TUNEL labeling was detected in control cultures (Fig. 12A), blocking Akt signaling did induce DNA cleavage in a low number of fiber cell nuclei in the central fiber zone. With just a 24 hour exposure, both the LY294002 (Fig. 12B) and CH5132799 (Fig. 12C) pan-PI3K inhibitors were highly effective in inducing premature DNA cleavage of the condensed nuclei of fiber cells located specifically in the center of the lens, with the LY294002 inhibitor also promoting nuclear elimination within this time frame. There was no impact of inhibiting PI3K/Akt signaling on the nuclei of the cortical fiber cells of the lens. These results suggest that nuclear removal from the center of the lens involves the inhibition of multiple downstream signaling effectors of PI3K in addition to Akt and that the signals involved in regulating nuclear condensation, DNA cleavage, and nuclear/chromatin removal may involve spatiotemporal suppression of distinct PI3K signaling pathways.
Figure 12. Inhibition of PI3K signaling induces premature spatiotermporal DNA cleavage and nuclear elimination.
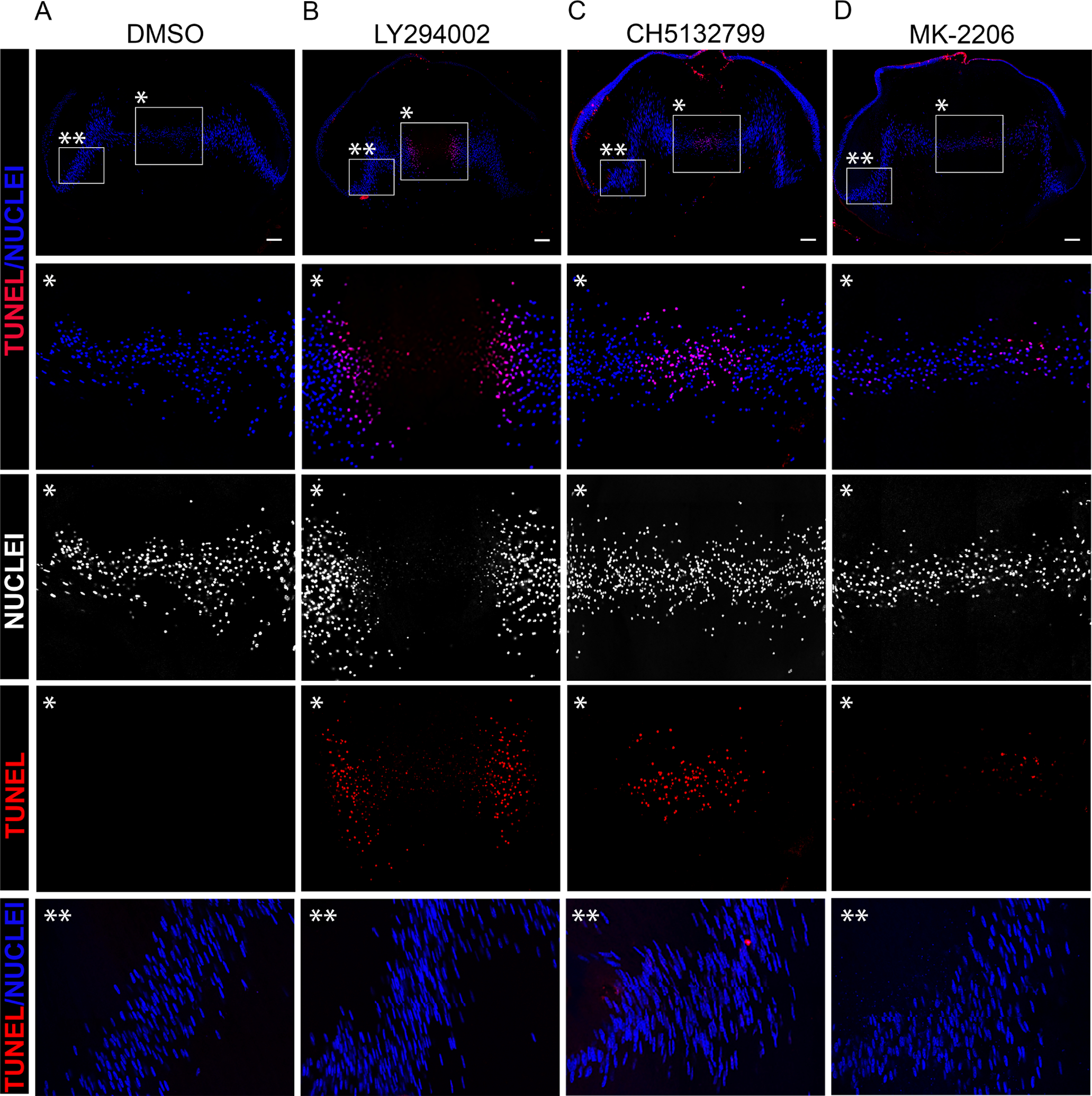
TUNEL assay was performed on cryosections of E12 lenses exposed for 24 hrs in organ culture to (A) DMSO, the pan-PI3K inhibitors (B) LY294002 or (C) CH5132799, or (D) the Akt-specific inhibitor MK-2206, and the sections co-labeled with the nuclear marker DAPI. Images were acquired by confocal microscopy. Boxed in regions with a single asterisk indicate the region of the central fiber zone (FC) shown at higher magnification below. Boxed in regions with a double asterisk indicate the region of the cortical fiber region (FP) shown at higher magnification below. The data shows significant premature induction of DNA cleavage and nuclear loss with the pan-PI3K inhibitors. Scale bar, 100 μm. Results are representative of 3 independent studies.
The PI3K inhibitor PIK3IP1 is a candidate for the endogenous regulation of PI3K signaling to induce OFZ formation during lens development
We previously produced an RNAseq database to analyze differentiation-state specific message expression in four distinct differentiation-state specific lens fractions of E13 chick embryo lenses isolated by microdissection of E13 lenses [3], with the GEO ascension number GSE53976. These regions of the developing lens included 1) the undifferentiated lens anterior epithelium (EC), 2) the equatorial epithelium, the region of lens differentiation initiation located along the lens equator (EQ), 3) the nascent, differentiating fiber cells located in the lens cortex (FP), and 4) the maturing fiber cells in the center of the lens (FC). We now performed data mining of this data base to determine the relative expression of the molecular components of PI3K (catalytic and regulatory). The results are displayed both as a line plot (Fig. 13A) and a Morpheus heat map (Fig. 13B). The detected expression levels used to generate the line chart and heat maps are presented as Supplemental Information in Table S1. This analysis revealed that there are many changes in the expression of both catalytic and regulatory subunits of PI3K during lens differentiation, the most striking being an exponential increase in PIK3IP1 in fiber cells about to undergo the process of eliminating their organelles. PIK3IP1 is an endogenous inhibitor of PI3K signaling that functions by competitively inhibiting the PI3K p85 regulatory subunit from binding to the p110 catalytic subunit, thereby preventing PI3K activation [56]. We then examined the potential link between PIK3IP1 expression in lens fiber cells and its spatiotemporal association with PI3K 110 catalytic subunits. For these studies, we immunoblotted fiber cell fractions isolated from lenses at E12-E15 of development to determine the levels of expression of the PI3K catalytic subunits p110α, p110β, and p110γ. There was no significant change in the ubiquitously expressed PI3K subunits p110α and p110β during this developmental period (Figs. 13C, D), and decreased expression of the p110γ isoform (Fig. 13E). Our studies also showed that PIK3IP1 protein was highly expressed in lens fiber cells between E12-E15 (Fig. 13F), consistent with a potential regulatory role for this molecule in inhibiting PI3K activity to induce autophagy-dependent OFZ formation.
Figure 13. Endogenous PI3K inhibitor PIK3IP1 association with PI3K catalytic subunits is consistent with induction of OFZ formation.
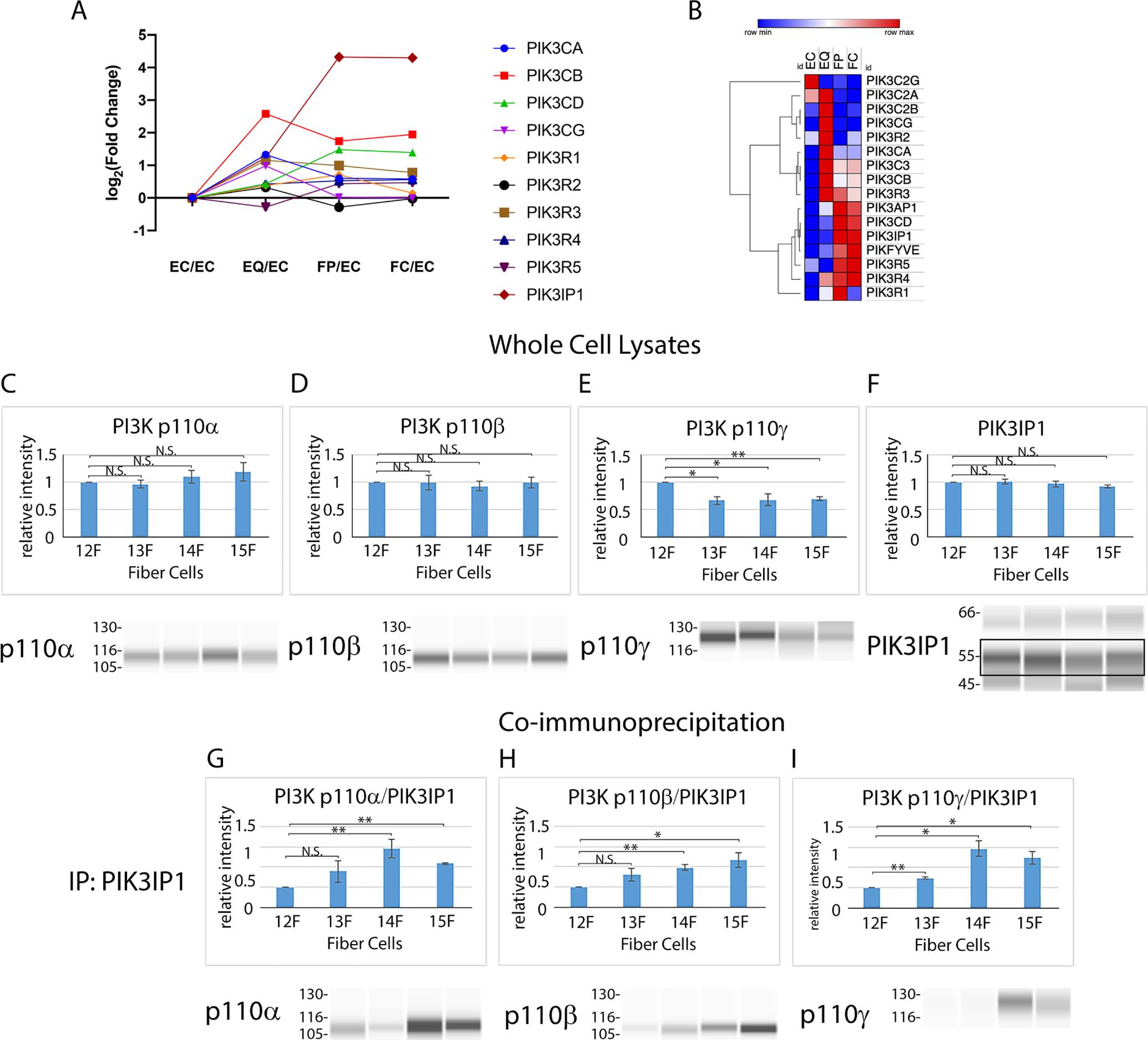
Detected mRNA levels of PI3K subunits by (A) RNAseq in microdissected fractions of the E13 lens and (B) a clustered heatmap of that data scaled by row. Immunoblot analysis of chick embryo lens fiber cells isolated by microdissection at developmental stages E12, E13, E14, and E15 for the PI3K p110 isoforms (C) p110α, (D) p110β, and (E) p110γ and for the endogenous PI3K inhibitor (F) PIK3IP1, quantified and represented as bar graphs above the immunoblots. (G-H) Co-immunoprecipitation analysis in which chick embryo lens fiber cells isolated by microdissection at developmental stages E12, E13, E14, and E15 were immunoprecipitated for PI3KIP1 and immunoblotted for p110α, p110β, p110γ and PIK3IP1, and quantified as the ratio of p110/PIK3IP1 which is presented as bar graphs that represent a minimum of 3 independent studies. EC-anterior lens epithelium, EQ-lens equatorial epithelium, FP-cortical fiber cells, FC-central fiber cells, F-fiber cells. Error bars represent S.E., *P ≤ 0.05 and **P ≤ 0.01, t test; N.S., not significant.
Since the endogenous inhibitory activity of PIK3IP1 on PI3K activity requires its association with the p110 catalytic subunit of PI3K, we investigated whether there was a developmental-state specific association of PIK3IP1 with the PI3K 110 catalytic subunits in lens fiber cells which would effectively block PI3K activity in vivo. For these studies, co-immunoprecipitation analysis was performed on lens fiber cell fractions isolated from lenses at developmental stages E12-E15 in which PIK3IP1 was immunoprecipitated and the immunoprecipitates subjected to immunoblot for the PI3K p110α, p110β, and p110γ isoforms to determine the level of association of PIK3IP1 with these different 110 isoforms. Immunoprecipitates were also immunoblotted for PIK3IP1. The results were quantified and are presented as the ratio of p110 subunit to PIK3IP1 (Figs. 13G, H, I). These studies revealed that there is a significant increase in the linkage of the inhibitory PIK3IP1 protein to these three PI3K p110 isoforms at E14, consistent with a role for PIK3IP1 in inhibiting PI3K signaling to induce autophagy and create the OFZ.
CONCLUSIONS
A hallmark of the maturation of lens fiber cells, the differentiated cell type that comprises the mass of lens tissue, is the developmental-state specific, spatiotemporal removal of organelles from the fiber cells located in the center of the lens. This unique phenomenon includes the removal of all organelles, with elimination of fiber cell mitochondria, ER, and Golgi followed by fiber cell nuclei. The establishment of this Organelle Free Zone (OFZ) during development is essential to the lens function of focusing clear images on the retina. Previous studies from our lab showed that the mechanism for removing lens mitochondria, ER, and Golgi involves an autophagic process dependent on suppression of a JNK signaling pathway [8]. Blocking JNK activation induced premature OFZ formation through the induction of classical autophagy signaling pathways including dephosphorylation/inactivation of mTORC1 complex proteins and their downstream target p70S6 kinase [8]. Further evidence for the requirement for autophagy to form the OFZ was provided by our studies showing a role for the mitophagy protein BNIP3L in the elimination of mitochondria, ER and Golgi during OFZ formation [12], and that HIF1a regulates BNIP3L’s function in non-nuclear organelle elimination [27]. The studies presented here expand our understanding of the lens autophagy activation signaling pathway involved in induction of differentiation-state specific organelle removal, and suggest an essential role for suppression of the classical autophagy regulator PI3K in the autophagy-dependent removal of mitochondria and ER to form the OFZ.
PI3K has many downstream signaling effectors that impact many distinct cell functions, with its regulation of Akt key to the PI3K-regulated autophagy signaling pathway in which PI3K inactivation induces autophagy, or “self-eating” of organelles in response to stresses such as nutrient starvation [32–34]. Our new studies suggest that the spatiotemporal suppression of PI3K signaing is likely the master regulator of the pathways that eliminate lens organelles during development to create the OFZ, with the specific inhibition of the PI3K/Akt signaling axis, the pathway that targets lens mitochondria and ER for degradation in autophagosomes. While PI3K and JNK have been reported to participate in coordinated signaling pathways [57–59], how they act together for the removal of lens organelles is still unknown and an interesting area for future studies.
The Class I family of PI3Ks has many different roles and downstream effectors, the best studied of which is Akt, whose inactivation is a well-known intermediate step in the link between PI3K and autophagy induction [26, 31]. Our studies now show that inhibition of the PI3K/Akt signaling axis specifically induces premature removal of mitochondria and ER in the developing chick lens (modeled, Fig. 13). The loss of these organelles is associated with induction of an autophagy signaling pathway that induces lipidation of LC3BI to form LC3BII, a crucial autophagy mediator required for autophagosome biogenesis and the delivery of autophagosomes to lysosomes. We now show the induction of LAMP1+ lysosomes during lens development with an spatiotemporal pattern of expression consistent with our previous findings of the presence of organelle-containing autophagosomes during formation of the OFZ [8]. Furthermore, inhibition of the PI3K/Akt signaling axis leads to a premature increase in LAMP1-positive lysosomes in lens fiber cells and the elimination of mitochondira and ER from these cells.
Under normal, nutrient-rich conditions, autophagy processes are important for removal/recycling of damaged organelles, including mitochondria after they lose their membrane potential and become depolarized [60–64]. Our previous studies demonstrate that mitochondria in lens fiber cells lose membrane potential before they are removed to form the OFZ [55]. We now show that the loss of PI3K signaling does not induce loss of mitochondrial membrane potential, and that the link between PI3K and mitochondrial removal during OFZ formation is associated with its regulation of autophagy induction. We found that inhibition of all PI3K signaling in the developing lens with pan-PI3K inhibitors induced removal of mitochondria and ER, as well as nuclear condensation followed by DNA cleavage and nuclear loss in the center of the lens within a day of exposure to the inhibitors. In contrast, specifically inhibiting only the PI3K/Akt signaling axis induced the premature removal of mitochondria and ER from the center of the lens and nuclear condensation within 24 hrs, with only low levels of TUNEL+ nuclei and no evidence of nuclear removal (modeled, Fig. 13). These results show that while autophagy induced by blocking the PI3K/Akt signaling axis is sufficient to remove non-nuclear lens organelles, the enhanced DNA cleavage and elimination of nuclei observed downstream of blocking PI3K signaling involves effectors of PI3K that are distinct from and/or in addition to Akt. Since the induction of autophagy by blocking the PI3K/Akt signaling axis under conditions of cell stress or to remove damaged organelles does not result in nuclear removal, it was not surprising to find that the autophagy process alone is not sufficient for the removal of lens nuclei to form the OFZ.
Nuclear elimination to form the OFZ is a multistep, highly regulated process. The PI3K-regulated autophagy pathway is most likely to function at the stages in this process that require the removal of fragmented nuclear material. Another major downstream target of PI3K, the RhoGTPase Rac [65], can regulate changes in nuclear morphology [66], and therefore is likely another key mediator of PI3K-mediated induction of nuclear loss in the developing lens. Nuclear elimination from lens fiber cells has been viewed as the hallmark of OFZ formation, with more studies focused on the process of nuclear loss than that of other organelles. The best studied aspect of nuclear removal is the cleavage and fragmentation of its chromatin, particularly in terms of the nuclear localization and activation of DNaseIIβ in differentiating lens fiber cells [17–20] and its regulation by the proteasome-dependent degradation of p27 and activation of CDK1 [17]. Interestingly, when DNaseIIβ is knocked out, the process of removing the nuclear membrane appears to be unimpaired [20]. Without the action of DNaseIIβ in these knockout mice chromatin is retained in the central lens fiber cells, with progressive morphological changes in chromatin morphology including the presence of DNA in globular and fragmented forms [18, 20], suggesting other chromatin remodeling pathways are also involved. Future studies will examie the upstream signaling regulators of nuclear loss in order to fully understand the process of lens OFZ formation during development.
Our data revealed an important role for suppression of PI3K signaling in the spatiotemporal induction of OFZ formation in studies of ex vivo lens organ cultures exposed to PI3K pathway inhibitiors. However, it was also important to identify the potential endogenous regulator(s) that could rapidly turn off all PI3K signaling during the maturation of fiber cells in the center of the lens and lead to the massive spatiotemporal removal of organelles in this region in vivo. Our discovery that PIK3IP1, a negative regulator of PI3K, is induced in lens fiber cells just prior to OFZ formation and associates with multiple Class I PI3K catalytic subunits in these lens fiber cells coincident with the induction of loss of organelles, suggests this regulatory molecule as the endogenous inducer of the autophagic process required to create the OFZ. In support of such a pathway, studies in cardiac tissue show that the association of PIK3IP1 with the PI3K p110α catalytic subunit induces autophagy to reduce hypertrophic growth [67]. It is likely that both Akt-dependent and -independent pathways involved in non-nuclear organelle and nuclear loss to form the OFZ are regulated by different p110 isoforms, which in turn are impacted upstream by receptor tyrosine kinases (RTKs) and G-protein coupled receptors (GPCRs) in their environment [35, 36]. Our findings reveal that PI3K signaling pathways are precisely regulated in order to execute the finely tuned processes required to remove different lens fiber cell organelles while maintaining cell viability and function. The results of our studies with the lens model of OFZ formation are expected to have significant implications for the regulation of similar processes in other tissues and show that the lens provides a unique reductionist model in which to understand shared mechanisms for both the maintenance and turnover of organelles. In addition, these discoveries open new avenues of study for determining how nuclei can be removed while maintaining cellular integrity.
Supplementary Material
Figure S1. Organelle-specificity of labeling approaches. Cryosections of embryonic chick lenses were stained for (A) LAMP1, (B) calreticulin, or (C) TOM20. (D) Chick embryo lenses live labeled with Rhodamine 123 (R123). Boxed in regions are shown at higher magnification in the panels to the right. Images were acquired by high-resolution confocal microscopy.
Figure S2. The pan-PI3K inhibitor CH5132799 induces elimination of mitochondria without impacting lens morphogenesis. E12 lenses were treated for 24 hrs in organ culture with the pan-PI3K inhibitor CH5132799 at concentrations of 25μM and 100 μM or its vehicle DMSO. Cryosections were immunolabeled for TOM20 (top row) or labeled with fluorescent phalloidin and DAPI (bottom row) and imaged by confocal microscopy. The data shows that this inhibitor of PI3K signaling did not affect structure or organization of lens fiber cells at concentrations that induce elimination of mitochondria to form the OFZ. Scale bar, 100 μm. Results are representative of 3 independent studies.
Table S1. FPKMs of mRNAs encoding PI3K proteins from differentiation-state specific chicken lens regions. FPKM, Fragments per kilobase of exon per million reads mapped.
Figure 14. Model comparing the impact of pan-PI3K inhibitors to specific inhibition of the PI3K/Akt signaling axis on inducing the premature loss of mitochondira, ER and nuclei to form the lens OFZ.
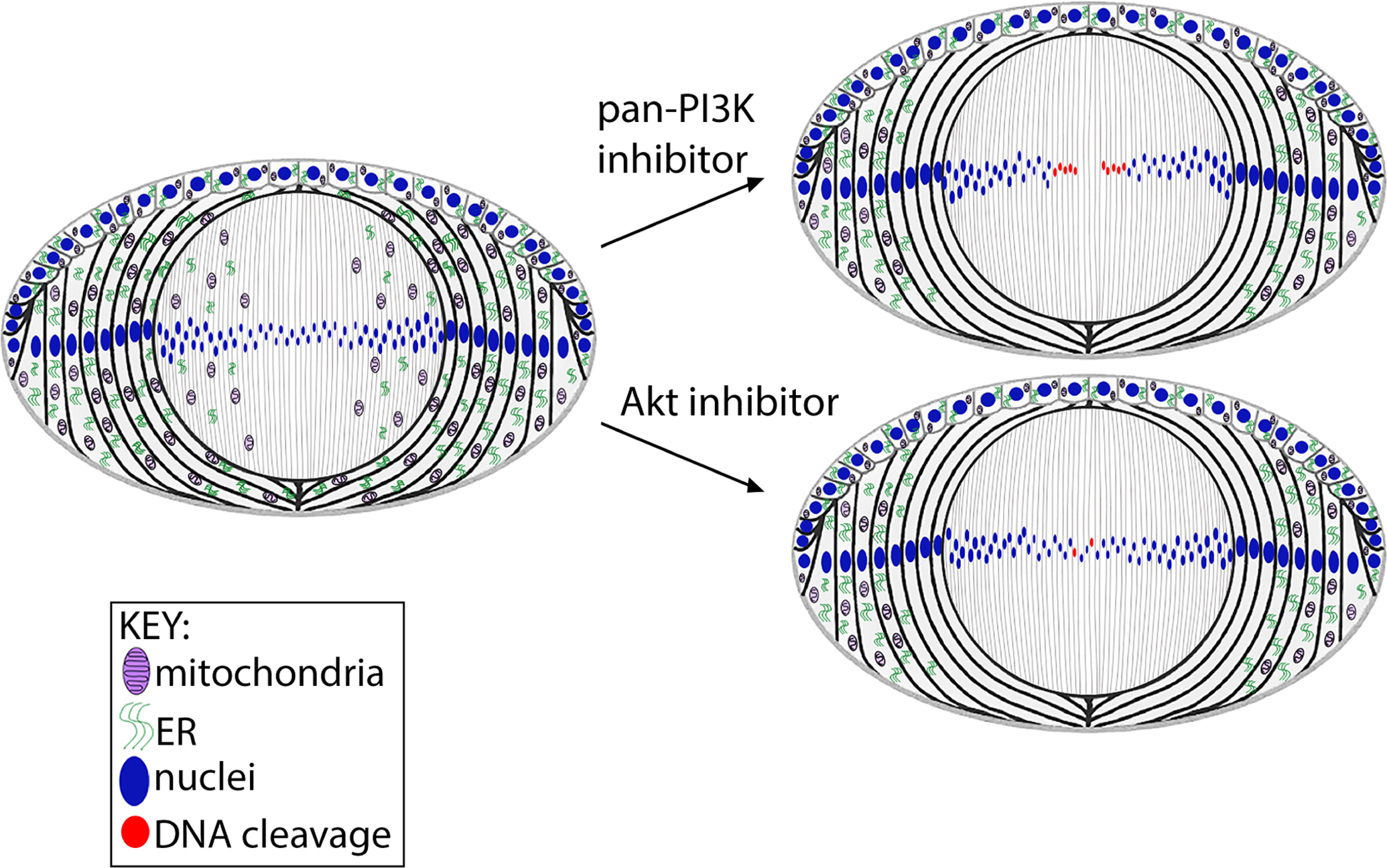
The model shows that suppression of all PI3K downstream signaling effectors with pan-PI3K inhibitors induces premature spatiotemporal elimination of lens organelles, including nuclei, and the specificity of inhibiting the PI3K/Akt signaling axis that activates autophagy on the elimination of mitochondria and ER. As inhibition of the PI3K/Akt signaling axis alone is insufficient to eliminate nuclei, these results show that inhibition of multiple downstream effectors of PI3K are necessary to induce nuclear loss.
Highlights.
PI3K inhibition is linked to spatiotemporal autophagy induction in embryonic lenses
PI3K inhibition induces premature formation of the lens Organelle Free Zone (OFZ)
Suppression of the PI3K/Akt axis induces mitochondria and ER loss in OFZ formation
Blocking Akt is alone insufficient to induce nuclear elimination in OFZ formation
ACKNOWLEDGEMENTS
We thank Paul FitzGerald for his generous gift of CP49 antibody.
FUNDING
This work was supported by NEI/NIH grant RO1 EY02478.
ABBREVIATIONS
- OFZ
Organelle Free Zone
- R123
rhodamine 123
- PI3K
phosphoinositide 3-kinase
- PIK3IP1
phosphoinositide-3-kinase interacting protein 1
- mTOR
mammalian target of rapamycin
- pmTOR
phosphorylated mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- ER
endoplasmic reticulum
- P70S6K
ribosomal protein S6 kinase also known as p70S6K
- p-p70S6K
phosphorylated p70S6K
- RAPTOR
regulatory associated protein of mTORC1
- pRAPTOR
phosphorylated regulatory associated protein of mTORC1
- JNK
jun n-terminal kinase
- pJNK
phosphorylated jun n-terminal kinase
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick and labeling
- EC
anterior lens epithelium
- EQ
equatorial lens epithelium
- FP
cortical lens fiber cells
- FC
nuclear lens fiber cells
- DMSO
dimethyl sulfoxide
- LC3B
light chain 3B
- RTK
receptor tyrosine kinase
- GPCR
G-protein coupled receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- [1].Menko S, Philp N, Veneziale B, Walker J, Integrins and development: how might these receptors regulate differentiation of the lens, Ann N Y Acad Sci 842 (1998) 36–41. [DOI] [PubMed] [Google Scholar]
- [2].Walker J, Menko AS, Integrins in lens development and disease, Exp Eye Res 88 (2009) 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chauss D, Basu S, Rajakaruna S, Ma Z, Gau V, Anastas S, Brennan LA, Hejtmancik JF, Menko AS, Kantorow M, Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens, G3 (Bethesda) 4 (2014) 1515–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Walker JL, Menko AS, alpha6 Integrin is regulated with lens cell differentiation by linkage to the cytoskeleton and isoform switching, Dev Biol 210 (1999) 497–511. [DOI] [PubMed] [Google Scholar]
- [5].Walker JL, Zhang L, Menko AS, A signaling role for the uncleaved form of alpha 6 integrin in differentiating lens fiber cells, Dev Biol 251 (2002) 195–205. [DOI] [PubMed] [Google Scholar]
- [6].Walker JL, Zhang L, Menko AS, Transition between proliferation and differentiation for lens epithelial cells is regulated by Src family kinases, Dev Dyn 224 (2002) 361–372. [DOI] [PubMed] [Google Scholar]
- [7].Basu S, Rajakaruna S, Menko AS, Insulin-like growth factor receptor-1 and nuclear factor kappaB are crucial survival signals that regulate caspase-3-mediated lens epithelial cell differentiation initiation, The Journal of biological chemistry 287 (2012) 8384–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Basu S, Rajakaruna S, Reyes B, Van Bockstaele E, Menko AS, Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells, Autophagy 10 (2014) 1193–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Basu S, Rajakaruna S, De Arcangelis A, Zhang L, Georges-Labouesse E, Menko AS, α6 integrin transactivates insulin-like growth factor receptor-1 (IGF-1R) to regulate caspase-3-mediated lens epithelial cell differentiation initiation, J Biol Chem 289 (2014) 3842–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].COULOMBRE JL, COULOMBRE AJ, LENS DEVELOPMENT: FIBER ELONGATION AND LENS ORIENTATION, Science 142 (1963) 1489–1490. [DOI] [PubMed] [Google Scholar]
- [11].Mathias RT, Rae JL, Baldo GJ, Physiological properties of the normal lens, Physiol Rev 77 (1997) 21–50. [DOI] [PubMed] [Google Scholar]
- [12].Brennan LA, McGreal-Estrada R, Logan CM, Cvekl A, Menko AS, Kantorow M, BNIP3L/NIX is required for elimination of mitochondria, endoplasmic reticulum and Golgi apparatus during eye lens organelle-free zone formation, Exp Eye Res 174 (2018) 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wride MA, Lens fibre cell differentiation and organelle loss: many paths lead to clarity, Philosophical transactions of the Royal Society of London. Series B, Biological sciences 366 (2011) 1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bassnett S, Mataic D, Chromatin degradation in differentiating fiber cells of the eye lens, The Journal of cell biology 137 (1997) 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dahm R, Lens fibre cell differentiation - A link with apoptosis?, Ophthalmic Res 31 (1999) 163–183. [DOI] [PubMed] [Google Scholar]
- [16].Dahm R, Dying to see, Sci Am 291 (2004) 82–89. [DOI] [PubMed] [Google Scholar]
- [17].Rowan S, Chang ML, Reznikov N, Taylor A, Disassembly of the lens fiber cell nucleus to create a clear lens: The p27 descent, Exp Eye Res 156 (2017) 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].De Maria A, Bassnett S, DNase IIbeta distribution and activity in the mouse lens, Investigative ophthalmology & visual science 48 (2007) 5638–5646. [DOI] [PubMed] [Google Scholar]
- [19].Lyu L, Whitcomb EA, Jiang S, Chang ML, Gu Y, Duncan MK, Cvekl A, Wang WL, Limi S, Reneker LW, Shang F, Du L, Taylor A, Unfolded-protein response-associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation, leading to cataract, FASEB journal : official publication of the Federation of American Societies for Experimental Biology 30 (2016) 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nishimoto S, Kawane K, Watanabe-Fukunaga R, Fukuyama H, Ohsawa Y, Uchiyama Y, Hashida N, Ohguro N, Tano Y, Morimoto T, Fukuda Y, Nagata S, Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens, Nature 424 (2003) 1071–1074. [DOI] [PubMed] [Google Scholar]
- [21].Costello MJ, Brennan LA, Basu S, Chauss D, Mohamed A, Gilliland KO, Johnsen S, Menko S, Kantorow M, Autophagy and mitophagy participate in ocular lens organelle degradation, Exp Eye Res 116 (2013) 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T, FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport, J Cell Biol 188 (2010) 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Frost LS, Mitchell CH, Boesze-Battaglia K, Autophagy in the eye: implications for ocular cell health, Exp Eye Res 124 (2014) 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen J, Ma Z, Jiao X, Fariss R, Kantorow WL, Kantorow M, Pras E, Frydman M, Riazuddin S, Riazuddin SA, Hejtmancik JF, Mutations in FYCO1 cause autosomal-recessive congenital cataracts, American journal of human genetics 88 (2011) 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim YC, Guan KL, mTOR: a pharmacologic target for autophagy regulation, J Clin Invest 125 (2015) 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laplante M, Sabatini DM, mTOR signaling in growth control and disease, Cell 149 (2012) 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brennan L, Disatham J, Kantorow M, Hypoxia regulates the degradation of non-nuclear organelles during lens differentiation through activation of HIF1a, Exp Eye Res (2020) 108129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Uyama T, Tsuboi K, Ueda N, An involvement of phospholipase A/acyltransferase family proteins in peroxisome regulation and plasmalogen metabolism, FEBS Lett 591 (2017) 2745–2760. [DOI] [PubMed] [Google Scholar]
- [29].Mardian EB, Bradley RM, Duncan RE, The HRASLS (PLA/AT) subfamily of enzymes, J Biomed Sci 22 (2015) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Morishita H, Eguchi T, Tsukamoto S, Sakamaki Y, Takahashi S, Saito C, Koyama-Honda I, Mizushima N, Organelle degradation in the lens by PLAAT phospholipases, Nature 592 (2021) 634–638. [DOI] [PubMed] [Google Scholar]
- [31].Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J, The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration, Cell Signal 26 (2014) 2694–2701. [DOI] [PubMed] [Google Scholar]
- [32].Levine B, Klionsky DJ, Development by self-digestion: molecular mechanisms and biological functions of autophagy, Dev Cell 6 (2004) 463–477. [DOI] [PubMed] [Google Scholar]
- [33].Lum JJ, DeBerardinis RJ, Thompson CB, Autophagy in metazoans: cell survival in the land of plenty, Nat Rev Mol Cell Biol 6 (2005) 439–448. [DOI] [PubMed] [Google Scholar]
- [34].Mathew R, Karantza-Wadsworth V, White E, Role of autophagy in cancer, Nat Rev Cancer 7 (2007) 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vanhaesebroeck B, Stephens L, Hawkins P, PI3K signalling: the path to discovery and understanding, Nature reviews. Molecular cell biology 13 (2012) 195–203. [DOI] [PubMed] [Google Scholar]
- [36].Jia S, Roberts TM, Zhao JJ, Should individual PI3 kinase isoforms be targeted in cancer?, Curr Opin Cell Biol 21 (2009) 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sellitto C, Li L, Vaghefi E, Donaldson PJ, Lin RZ, White TW, The Phosphoinosotide 3-Kinase Catalytic Subunit p110alpha is Required for Normal Lens Growth, Investigative ophthalmology & visual science 57 (2016) 3145–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Weber GF, Menko AS, Phosphatidylinositol 3-kinase is necessary for lens fiber cell differentiation and survival, Invest Ophthalmol Vis Sci 47 (2006) 4490–4499. [DOI] [PubMed] [Google Scholar]
- [39].Ishizaki Y, Jacobson MD, Raff MC, A role for caspases in lens fiber differentiation, J Cell Biol 140 (1998) 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].DeDreu J, Walker JL, Menko AS, Dynamics of the lens basement membrane capsule and its interaction with connective tissue-like extracapsular matrix proteins, Matrix Biol 96 (2021) 18–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Menko AS, DeDreu J, Logan CM, Paulson H, Levin AV, Walker JL, Resident immune cells of the avascular lens: Mediators of the injury and fibrotic response of the lens, Faseb j 35 (2021) e21341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Modak SP, Perdue SW, Terminal lens cell differentiation. I. Histological and microspectrophotometric analysis of nuclear degeneration, Exp Cell Res 59 (1970) 43–56. [DOI] [PubMed] [Google Scholar]
- [43].Bassnett S, The fate of the Golgi apparatus and the endoplasmic reticulum during lens fiber cell differentiation, Invest Ophthalmol Vis Sci 36 (1995) 1793–1803. [PubMed] [Google Scholar]
- [44].Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D, Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20, Cell 100 (2000) 551–560. [DOI] [PubMed] [Google Scholar]
- [45].Iwahashi J, Yamazaki S, Komiya T, Nomura N, Nishikawa S, Endo T, Mihara K, Analysis of the functional domain of the rat liver mitochondrial import receptor Tom20, J Biol Chem 272 (1997) 18467–18472. [DOI] [PubMed] [Google Scholar]
- [46].Smith MJ, Koch GL, Multiple zones in the sequence of calreticulin (CRP55, calregulin, HACBP), a major calcium binding ER/SR protein, EMBO J 8 (1989) 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Carpentier S, N’Kuli F, Grieco G, Van Der Smissen P, Janssens V, Emonard H, Bilanges B, Vanhaesebroeck B, Gaide Chevronnay HP, Pierreux CE, Tyteca D, Courtoy PJ, Class III phosphoinositide 3-kinase/VPS34 and dynamin are critical for apical endocytic recycling, Traffic 14 (2013) 933–948. [DOI] [PubMed] [Google Scholar]
- [48].Zhao S, Li L, Wang S, Yu C, Xiao B, Lin L, Cong W, Cheng J, Yang W, Sun W, Cui S, H2O2 treatment or serum deprivation induces autophagy and apoptosis in naked mole-rat skin fibroblasts by inhibiting the PI3K/Akt signaling pathway, Oncotarget 7 (2016) 84839–84850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Saxton RA, Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease, Cell 168 (2017) 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Downward J, Mechanisms and consequences of activation of protein kinase B/Akt, Curr Opin Cell Biol 10 (1998) 262–267. [DOI] [PubMed] [Google Scholar]
- [51].Franke TF, Kaplan DR, Cantley LC, Toker A, Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate, Science 275 (1997) 665–668. [DOI] [PubMed] [Google Scholar]
- [52].Hemmings BA, PtdIns(3,4,5)P3 gets its message across, Science 277 (1997) 534. [DOI] [PubMed] [Google Scholar]
- [53].Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clavé C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WA, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fésüs L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, González-Estévez C, Gorski S, Gottlieb RA, Häussinger D, He YW, Heidenreich K, Hill JA, Høyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jäättelä M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovács AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, López-Otín C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Meléndez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Münz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nürnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Tallóczy Z, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcátegui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL, Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes, Autophagy 4 (2008) 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC, Regulation of mammalian autophagy in physiology and pathophysiology, Physiol Rev 90 (2010) 1383–1435. [DOI] [PubMed] [Google Scholar]
- [55].Weber GF, Menko AS, The canonical intrinsic mitochondrial death pathway has a non-apoptotic role in signaling lens cell differentiation, J Biol Chem 280 (2005) 22135–22145. [DOI] [PubMed] [Google Scholar]
- [56].Zhu Z, He X, Johnson C, Stoops J, Eaker AE, Stoffer DS, Bell A, Zarnegar R, DeFrances MC, PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein, Biochemical and biophysical research communications 358 (2007) 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kato T, Yamada A, Ikehata M, Yoshida Y, Sasa K, Morimura N, Sakashita A, Iijima T, Chikazu D, Ogata H, Kamijo R, FGF-2 suppresses expression of nephronectin via JNK and PI3K pathways, FEBS Open Bio 8 (2018) 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Goyal A, Poluzzi C, Willis CD, Smythies J, Shellard A, Neill T, Iozzo RV, Endorepellin affects angiogenesis by antagonizing diverse vascular endothelial growth factor receptor 2 (VEGFR2)-evoked signaling pathways: transcriptional repression of hypoxia-inducible factor 1α and VEGFA and concurrent inhibition of nuclear factor of activated T cell 1 (NFAT1) activation, The Journal of biological chemistry 287 (2012) 43543–43556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chekmarev J, Azad MG, Richardson DR, The Oncogenic Signaling Disruptor, NDRG1: Molecular and Cellular Mechanisms of Activity, Cells 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Youle RJ, Narendra DP, Mechanisms of mitophagy, Nat Rev Mol Cell Biol 12 (2011) 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B, The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy, Biochim Biophys Acta 1366 (1998) 177–196. [DOI] [PubMed] [Google Scholar]
- [62].Kim I, Rodriguez-Enriquez S, Lemasters JJ, Selective degradation of mitochondria by mitophagy, Arch Biochem Biophys 462 (2007) 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mizushima N, Autophagy: process and function, Genes Dev 21 (2007) 2861–2873. [DOI] [PubMed] [Google Scholar]
- [64].Yokota S, Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate, Eur J Cell Biol 61 (1993) 67–80. [PubMed] [Google Scholar]
- [65].Welch HC, Coadwell WJ, Stephens LR, Hawkins PT, Phosphoinositide 3-kinase-dependent activation of Rac, FEBS Lett 546 (2003) 93–97. [DOI] [PubMed] [Google Scholar]
- [66].Navarro-Lerida I, Pellinen T, Sanchez SA, Guadamillas MC, Wang Y, Mirtti T, Calvo E, Del Pozo MA, Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion, Dev Cell 32 (2015) 318–334. [DOI] [PubMed] [Google Scholar]
- [67].Song HK, Kim J, Lee JS, Nho KJ, Jeong HC, Kim J, Ahn Y, Park WJ, Kim DH, Pik3ip1 modulates cardiac hypertrophy by inhibiting PI3K pathway, PLoS One 10 (2015) e0122251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Organelle-specificity of labeling approaches. Cryosections of embryonic chick lenses were stained for (A) LAMP1, (B) calreticulin, or (C) TOM20. (D) Chick embryo lenses live labeled with Rhodamine 123 (R123). Boxed in regions are shown at higher magnification in the panels to the right. Images were acquired by high-resolution confocal microscopy.
Figure S2. The pan-PI3K inhibitor CH5132799 induces elimination of mitochondria without impacting lens morphogenesis. E12 lenses were treated for 24 hrs in organ culture with the pan-PI3K inhibitor CH5132799 at concentrations of 25μM and 100 μM or its vehicle DMSO. Cryosections were immunolabeled for TOM20 (top row) or labeled with fluorescent phalloidin and DAPI (bottom row) and imaged by confocal microscopy. The data shows that this inhibitor of PI3K signaling did not affect structure or organization of lens fiber cells at concentrations that induce elimination of mitochondria to form the OFZ. Scale bar, 100 μm. Results are representative of 3 independent studies.
Table S1. FPKMs of mRNAs encoding PI3K proteins from differentiation-state specific chicken lens regions. FPKM, Fragments per kilobase of exon per million reads mapped.


