Abstract
Enzymes are biomacromolecular protein catalysts that are widely used in a plethora of industrial-scale applications due to their high selectivity, efficiency and ability to work under mild conditions. Many industrial processes require the immobilization of enzymes to enhance their performance and stability. Encapsulation of enzymes in protein cages provides an excellent immobilization platform to create nanoreactors with enhanced enzymatic stability and desired catalytic activities. Here we show that the catalytic activity of nanoreactors, derived from the bacteriophage P22 viral capsids, can be finely-tuned by controlling the packaging stoichiometry and packing density of encapsulated enzymes. The packaging stoichiometry of the enzyme alcohol dehydrogenase (AdhD) was controlled by co-encapsulating it with wild-type scaffold protein (wtSP) at different stoichiometric ratios using an in vitro assembly approach and the packing density was controlled by selectively removing wtSP from the assembled nanoreactors. An inverse relationship was observed between the catalytic activity (kcat ) of AdhD enzyme and the concentration of co-encapsulated wtSP. Selective removal of the wtSP resulted in the similar activity of AdhD in all nanoreactors despite the difference in the volume occupied by enzymes inside nanoreactors, indicating that the AdhD enzymes do not experience self-crowding even under high molarity of confinement (Mconf ) conditions. The approach demonstrated here not only allowed us to tailor the activity of encapsulated AdhD catalysts but also the overall functional output of nanoreactors (enzyme-VLP complex). The approach also allowed us to differentiate the effects of crowding and confinement on the functional properties of enzymes encapsulated in an enclosed system, which could pave the way for designing more efficient nanoreactors.
Graphical Abstract
Introduction
In the past few decades biology has greatly influenced the process by which functional materials are designed and constructed.1 The complex organization of natural biological systems, at almost all length scales, has inspired the rapid growth of biomimetic approaches to tailor the synthesis of materials having defined structure-function relationships.2, 3 Cells frequently use self-assembled molecular complexes and supramolecular compartments to organize and regulate complex biological functions. Sequestration of enzymatic reactions into the confined environment of compartments, self-assembled from lipid molecules or protein subunits such as membrane organelles or bacterial micro-compartments, exerts spatial and temporal control over the pathways.4 This is believed to optimize the efficiency by enhancing local concentration of reacting species involved in the pathway, by regulating transport of molecules into and out of the compartment, and by isolating toxic or reactive intermediates from the rest of the cell.4–6 The precise control over biochemical reactions, selectivity, and efficient catalysis that these compartments afford has led to a growing interest in mimicking such systems for materials applications by encapsulating enzymes in well-defined self-assembled macromolecular compartments such as virus-like particles (VLPs). Encapsulation of enzymes in such compartments confers enzyme stability, ease of purification and reusability, and offers the potential to create useful designer nanoreactors with very high local concentration of enzymes, which would otherwise be impossible to attain without significant aggregation.7–37 The local concentration of enzymes can be increased further by condensing enzyme filled protein compartments into 3D superlattices in order to realize accelerated catalytic conversion.38 The high local concentration of enzymes in the nanoreactors can lead to modulation of the enzymatic activity.14–16, 26 The alteration in activity has been ascribed to slower diffusion of enzyme-VLP capsule, or to the combined effects arising from the crowded and confined microenvironment.14–16, 26 While crowding and confinement have often been used in conjunction to rationalize the outcome of encapsulated systems, statistical-thermodynamics models suggest that confinement and crowding can have different effects on the structural and functional properties of enzymes.39–42 Biological systems utilize confinement as a basic strategy to increase the local concentration of any encapsulated molecule, which can lead to enhanced reaction rates and increased intermolecular interactions between the encapsulated species.43 Crowding, on the other hand, is an intracellular phenomenon where the concentration of macromolecules, such as proteins, carbohydrates and nucleic acids can reach up to 300 mg/ml.44,45 At such high concentrations macromolecules experience excluded volume effects due to the inaccessibility of space occupied by other macromolecules of comparable size. These effects are such that, even in the absence of direct interactions between macromolecules, they can profoundly alter the stability, structural dynamics and kinetics of enzyme catalyzed reactions.46–48
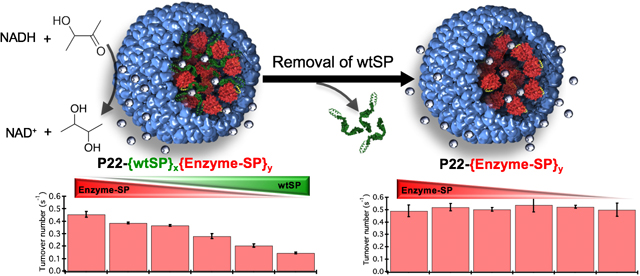
Approaches such as physical entrapment of enzyme molecules during cage assembly, or directed encapsulation involving the covalent (e.g. coat protein fusion, SpyTag/SpyCatcher, or Sortase A enzyme mediated) and non-covalent interactions (e.g. protein-protein, nucleic acid-protein) have been used to achieve the packaging of a variable number of enzymes in VLPs both in vivo and in vitro.14, 15, 26, 49–56 In vivo encapsulation is a simple and efficient approach as the entire packaging process takes place in a host bacterial cell. However, it relies on the transcriptional-translational machinery of the host cell for encapsulation and can result in little to no control over the amount of cargo encapsulation with the added possible complication of co-encapsulation of (unwanted) host macromolecules. In contrast, in vitro assembly, requiring separate purification of VLP and enzyme, offers better control over cargo encapsulation. Despite recent advances in in vitro encapsulation, controlling the composition (i.e. protein-protein interactions) and packing density (crowding) of guest molecules in the interior volume remains difficult to achieve. Here we have engineered nanoreactors utilizing VLPs derived from the P22 bacteriophage and have controlled the composition and packing density of enzymatic cargo within the interior of the VLP affording fine-tuning of the activity of the resulting nanoreactors. The P22 VLP assembles from 420 subunits of coat protein (CP) and 100–300 subunits of scaffolding protein (SP) to form a T = 7 icosahedral capsid, 58 nm in diameter.57 The SP templates the assembly of the CP into the P22 capsid through a positively charged helix-turn-helix domain at its C-terminus and gets encapsulated during the assembly process.57, 58 Treatment of the P22 with 0.5 M guanidine hydrochloride (GuHCl) disrupts the non-covalent interactions between the CP and the SP helix-turn-helix domain, and the SP freely diffuses out to generate an empty shell.59 The SP can be extensively truncated to a minimal assembly domain and be modified at the N- or C-terminus without affecting its templating ability for capsid assembly.49, 60, 61 This truncated scaffold, fused to a protein cargo (cargo-SP), has been used to direct the P22 assembly and encapsulation of a number of cargo proteins through heterologous expression of both the components (CP and cargo-SP) in a host bacterial cell in an in vivo approach.49 We have previously shown encapsulation of many different gene products, including enzymes, with very high efficiency within the P22 VLPs. This in vivo directed encapsulation approach does not allow control over the stoichiometry and packing density of the encapsulated cargoes and suffers from some limitations such as encapsulation of misfolded proteins, or partial inactivation of proteins due to inherently high confinement molarity (Mconf ~ 6–7 mM) of P22 SP-directed encapsulation system.16, 49, 62, 63 However, using an in vitro assembly approach it is possible to overcome some of these limitations and control packaging stoichiometry and the packing density of protein cargos inside P22.64–66 Premixing of wild type (wt) SP and cargo-SP in a range of defined stoichiometric ratios and then adding them to CP subunits in a fixed stoichiometric ratio results in capsid assembly having an average composition determined by the input stoichiometry of SP and cargo-SP.63 The subsequent selective removal of wtSP from the series of co-assembled nanoreactors, upon mild treatment with a chaotrope, enables control over packing density.64 The pore size (~2.5 nm)67 of the P22 capsid shell allows the escape of wtSP but not the bulky folded cargo-SP.16, 49, 63
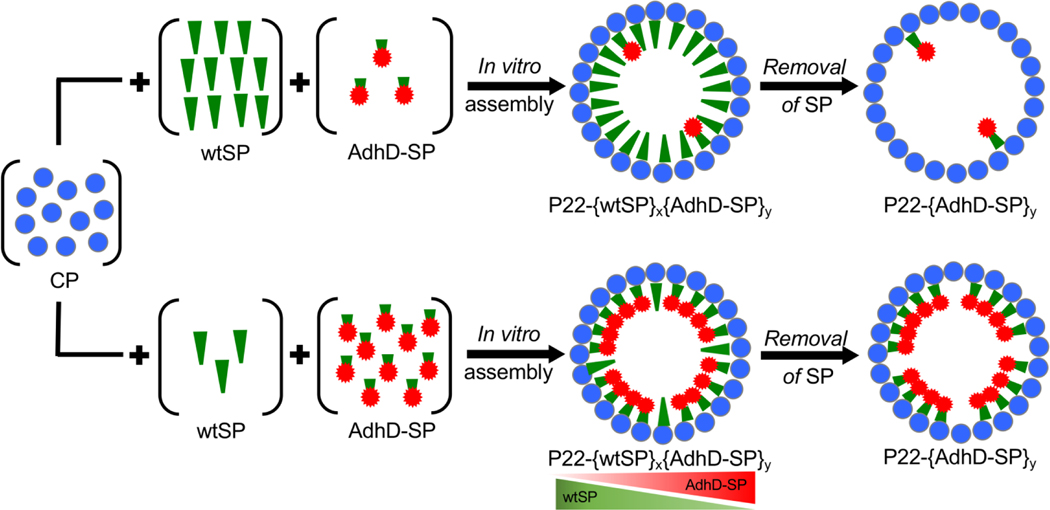
This methodology was used to design and construct P22 nanoreactors with controlled composition using the enzyme alcohol dehydrogenase (AdhD) as a model system (Figure 1). The AdhD is a monomeric thermostable, metal-ion independent enzyme from the aldo-keto reductase (AKR) family and presents a useful class of enzymes with considerable interest in the production of biofuels, agrochemicals and pharmaceutical products.68, 69
Figure 1.
Schematic showing the methodology used to control the composition of P22 nanoreactors. Nanoreactors were co-assembled in vitro with AdhD-SP and wtSP to control the loading of cargo macromolecules. A constant amount of CP was added to variable ratios of wtSP and AdhD-SP and allowed to self-assemble in vitro. Subsequently, the wtSP was selectively removed from these co-assembled nanoreactors to alter the packing density of enzymes inside P22.
Our results demonstrate that the compositional control, achieved through the encapsulation of variable amounts of AdhD and wtSP, influences the functional output at two levels: the molecular level (i.e. individual AdhD catalysts) and nanoreactor level (VLP nanoparticles encapsulating these catalysts). The wtSP not only acts as scaffold to co-template the assembly but exerts macromolecular crowding in a concentration dependent manner, leading to the tuning of enzymic output. Further, similar enzymic output of wtSP depleted nanoreactors despite differential packing density of AdhD suggested that the AdhD enzymes do not experience self-crowding even at high Mconf and that the resultant activity is influenced largely by the confinement inside P22 as the activities were lower than the free (unencapsulated) enzyme. Although the local environment of co-assembled and wtSP depleted nanoreactors impacted the functional output of AdhD differently, the overall activity of nanoreactors (with or without SP) increased with the increase in copy number of AdhD inside these nanoreactors. Thus, the ability to control composition has allowed us to create a range of nanoreactors that are modelled on a cell-like system for mimicking the crowding and confinement experienced by catalytic enzymes. To our knowledge this is the first experimental investigation where the underlying effects of crowding and confinement have been used in the design, construction, and modulation of the behavior of functional catalytic nanomaterials. These findings broaden the scope for experimental and theoretical work aimed at developing methodologies to realize better control over the catalytic activity of biomimetic nanomaterials.
Results and Discussion
Controlling the loading of AdhD in P22 nanoreactors using in vitro assembly.
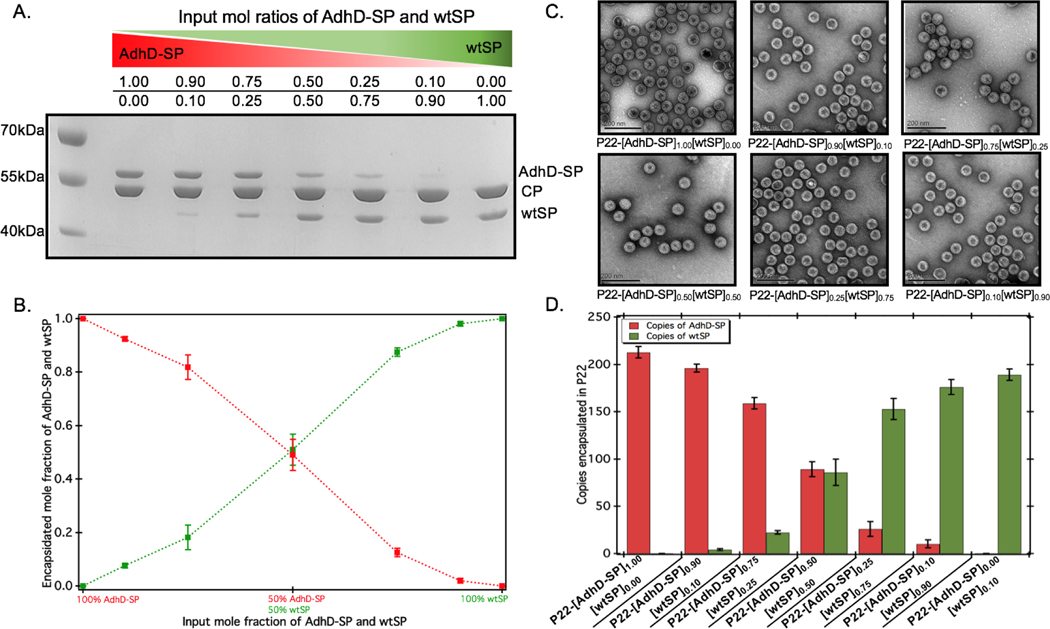
To control the loading of AdhD in P22 nanoreactors, the AdhD was genetically fused to the N-terminus of truncated SP (amino acids 142–303). The fusion protein (AdhD-SP) was heterologously expressed in E. coli BL21(DE3) cells and purified by Ni-NTA column chromatography. The purified protein was characterized by sodium-dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass-spectrometry (Figure S1). The nanoreactors were self-assembled in vitro, where AdhD-SP and wtSP were mixed together at molar ratios of 0.0:1.0, 0.1:0.9, 0.25:0.75, 0.5:0.5, 0.75:0.25, 0.9:0.1, 1.0:0.0 and added to a molar equivalent of CP. The co-assembled nanoreactors (P22-[AdhD-SP]x[wtSP]y, where x and y represents the input molar ratios) were purified using ultracentrifugation, and analyzed by SDS-PAGE (Figure 2A). The gel electrophoresis confirmed the presence of both the components in co-assembled nanoreactors. Densitometric profiling of each lane in SDS-PAGE was used to calculate the average amount of AdhD-SP and wtSP encapsulated into the co-assembled nanoreactors. The densitometric profiles were analyzed with respect to known standards of each component and the area under each peak was determined for CP, wtSP and AdhD-SP (Figure S2). A calibration curve from the standards was used to calculate the mole ratios of wtSP, and AdhD-SP (relative to CP) encapsulated in the purified co-assembled nanoreactors. The output mole ratios encapsulated in the purified co-assembled nanoreactors, as a function of input stoichiometry used for assembly, is shown in Figure 2B. The data indicate that both the components, AdhD-SP and wtSP, were incorporated into the capsids in proportion to the input stoichiometric ratios. When imaged by transmission electron microscopy (TEM), the co-assembled nanoreactors were found to be homogeneous and monodispersed, showing an average size of 56.1±3.1 nm (≈100 particles analyzed) and intact spherical morphology, indicating that in vitro co-assembly did not alter the P22 VLP structure (Figure 2C). The particles were further analyzed by size exclusion chromatography (SEC) coupled with multi-angle light scattering (MALS)/quasi-elastic light scattering (QELS) to determine their molecular weight and sizes (Table 1 and Figure S3). P22 empty shell (ES) particles, having no SP present, were also analyzed to determine the molecular weight of the capsid alone, which was then subtracted from the observed molecular weight of co-assembled nanoreactors to calculate the total amount of cargo (AdhD-SP and wtSP) encapsulated. The number of copies of each component was calculated from the ratio of the two SP components, as determined by the densitometric analysis of the SDS-PAGE (Figure 2D and Table 1). The radius of gyration (Rrms) and hydrodynamic radius (Rh) of the co-assembled nanoreactors, determined from the angular dependence of the scattered light and their diffusion coefficients, respectively, were found to be similar to those previously reported for encapsulated P22 materials (Table 1).16, 17, 49 The value of Rrms/Rh reflects the distribution of densities from the center of the particle to radially outward; values around 0.78 indicate filled particles while values closer to 1.0 represent spherical shells with infinitely thin walls. 70, 71 The empty shell (ES) was found to have value of 1.0 and the co-assembled nanoreactors exhibited Rrms to Rh values ranging from 0.82–0.89, indicating cargo filled capsids. These findings show that in vitro assembly is a robust and generic approach for controlling the loading of cargo without affecting the integrity of P22 VLPs. The co-assembled nanoreactors were also analyzed by dynamic light scattering (DLS) to investigate the stability of capsids at 50°C as the kinetics measurements were performed at that temperature and were found to have the same size distribution as P22 measured at room temp (Table S1).63 After assembly and characterization, the kinetics of the co-assembled nanoreactors were measured at 50°C.
Figure 2.
Characterization of co-assembled nanoreactors (P22-[AdhD-SP]x[wtSP]y, where, x and y represents the input molar ratios). (A) SDS-PAGE analysis of isolated co-assembled nanoreactors shows gradations in the band intensities of AdhD-SP and wtSP, as the input molar ratios are changed. (B) Graph of the encapsulated AdhD-SP and wtSP over a range of input stoichiometric ratios of each component, generated from densitometric analysis of SDS-PAGE. (C) TEM images show the sizes and intact spherical morphology of co-assembled nanoreactors. (D) Number of copies of AdhD-SP and wtSP encapsulated in P22 calculated using SEC-MALS data.
Table 1.
Molar mass and size of co-assembled nanoreactors determined by SEC-MALS/QELS. The subscripts x and y represent molar ratios of AdhD-SP and wtSP used for assembly.
| Sample P22-[AdhD-SP]x[wtSP]y | Molar mass (MDa) | Copies of AdhD-SP | Copies of wtSP | Rrms (nm) | Rh (nm) | Rrms/ Rh |
|---|---|---|---|---|---|---|
|
| ||||||
| Empty Shell (ES) | 19.4 ± 0.4 | -------- | -------- | 25.9 ± 0.1 | 25.8 ± 0.2 | 1 ± 0.004 |
| P22-[AdhD-SP]1.00 [wtSP]0.0 | 31.10 ± 0.96 | 219 ± 6 | -------- | 20.9 ± 2.1 | 25.4 ± 1.1 | 0.82 ± 0.05 |
| P22-[AdhD-SP]0.90 [wtSP]0.10 | 30.22 ± 0.92 | 200 ± 4 | 3 ± 1 | 20.9 ± 2.3 | 25.4 ± 1.1 | 0.82 ± 0.05 |
| P22-[AdhD-SP]0.75 [wtSP]0.25 | 28.50 ± 0.74 | 154 ± 6 | 22 ± 2 | 21.6 ± 1.9 | 25.4 ± 0.8 | 0.85 ± 0.05 |
| P22-[AdhD-SP]0.50 [wtSP]0.50 | 26.94 ± 1.33 | 98 ± 8 | 74 ± 14 | 22.3 ± 1.7 | 25.4 ± 0.5 | 0.88 ± 0.05 |
| P22-[AdhD-SP]0.25 [wtSP]0.75 | 26.55 ± 1.03 | 43 ± 8 | 143 ± 11 | 22.5 ± 1.7 | 25.3 ± 0.5 | 0.89 ± 0.05 |
| P22-[AdhD-SP]0.10 [wtSP]0.90 | 26.34 ± 1.01 | 14 ± 4 | 183 ± 8 | 22.4 ± 1.7 | 25.5 ± 0.6 | 0.88 ± 0.05 |
| P22-[AdhD-SP]0.00 [wtSP]1.00 | 26.08 ± 0.83 | --------- | 194 ± 6 | 22.6 ± 1.8 | 25.6 ± 0.5 | 0.88 ± 0.05 |
The composition dictates the activity of co-assembled nanoreactors.
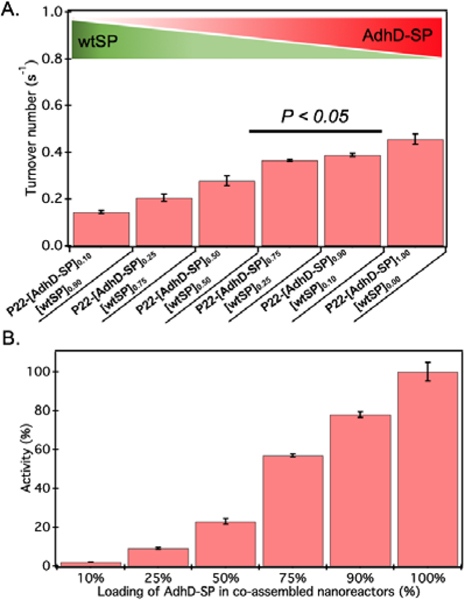
The activity of AdhD was measured by monitoring the rate of disappearance of the cofactor, NADH, upon reduction of acetoin at 50°C under slightly acidic pH (6.2) conditions to minimize any interference of the oxidation (reverse) reaction that takes place at alkaline pH (8.8).68 The activity of nanoreactors encapsulating variable amounts of AdhD-SP and wtSP (Figure 2B) was measured under conditions where the AdhD concentration was constant for all samples. The dependence of the reaction rate on acetoin followed Michaelis-Menten kinetics. The AdhD kinetic data from all the nanoreactors were fit to a Michaelis-Menten equation and the kinetic parameters kcat and KM were calculated from the equation. The data indicated that the increase in concentration of co-encapsulated wtSP significantly reduces the kcat of AdhD in these nanoreactors (Figure 3A), but the overall activity (kcat x number of copies of AdhD) of nanoreactor particles was increased as the packaging amount of AdhD was increased inside the nanoreactors (Figure 3b). The concentration of encapsulated wtSP ranges from 0.1 – 5.5 mM (4 – 193 mg/ml) inside these nanoreactors. At such high concentrations, macromolecules can affect the conformation and kinetics of enzymes either through macromolecular crowding (excluded volume effects) and/or through soft (weak non-covalent) interactions.44, 72–76 Soft interactions between the SP and AdhD are less likely given their similar pIs (AdhD-SP1–266 : 6.01 and wtSP1–266 : 5.13), and thus overall charge at the working pH conditions. Similar activity exhibited by both free (AdhD) and SP-fused enzyme (AdhD-SP) as a function of their increasing concentrations further suggests that the wtSP do not interact chemically with the enzyme and therefore do not influence kinetic parameters (Figure S4). Some researchers have reported that macromolecular crowding increases enzymatic rate because of the higher activity coefficients of reacting molecules, potentially overcoming the slowed diffusion in a crowded medium.77–79 A decrease in enzymatic rate is normally associated with the changes in conformational dynamics of the active site of enzyme induced by crowded surroundings.80–83 Coarse-grained simulations of HIV-1 protease revealed that the presence of crowder significantly reduced the fraction of protease with open-state conformation through the continuous collisions, leading to slowing down of enzymatic activity.83 The observed reduction in reaction rates of AdhD in these nanoreactors with the increase in wtSP is suggestive of a process inhibiting conformational dynamics, leading to either perturbation of the docking site or product release. Studies also show that a crowded medium generally favors molecular compaction, such as the adoption of globular forms as opposed to expanded conformations that occupy larger volumes, and molecular association to reduce the excluded volume and increase the entropy of the system.40, 72, 76, 84 Thus, if AdhD adopts an expanded conformation during its catalytic cycle, as reported in the literature for some members of the AKR family that undergo a conformational change upon cofactor binding and release,85 then we would expect to see a reduction in the activities as a consequence of the macromolecular crowding induced by wtSP, which would stabilize compact states of the enzyme in comparison to expanded conformations. The stabilization of compact states would thus lead to a reduction in activity of AdhD in nanoreactors as the concentration of co-encapsulated wtSP was increased. The crowders can affect the kinetics by slowing down the diffusion, however the catalytic efficiency of AdhD enzyme (kcat/KM ) is on the order of 102 M−1 s−1 (Figure S8), which is far lower than the enzymatic reactions that has kcat /KM on the order of 108 M−1 s−1 or greater and are susceptible to diffusional limitations.86 At such a low catalytic efficiency, the kinetics is unlikely to be affected by substrate diffusion, especially at a higher temperatures (50°C) where the kinetic measurements were done and a previous study showed that the P22 capsid does not present a barrier to the diffusion of small molecules used in this study and therefore does not influence the rate of reaction.16
Figure 3.
(A) The turnover number (kcat) of AdhD in co-assembled nanoreactors. Error bars represent standard deviations of triplicate measurements. The P-value (<0.05) indicates the statistically significant difference between the activities of those two data sets. (B) Graph showing the increase in overall activity of nanoreactor particle as the input packaging stoichiometry of AdhD-SP was increased in co-assembled nanoreactors.
We tested the behaviour of the free (unencapsulated) AdhD enzyme under macromolecular crowding conditions generated using poly-ethylene glycol (PEG) (Figure S5). PEG 8k was chosen because it is an elongated crowding agent with no significant secondary structure and thus mimics the wtSP, which is partially disordered (37 ± 3% α-helix, 30 ± 6% random coil, 33 ± 6 % β-turns)87, 88 at the N-terminus region and elongated structurally with estimated dimensions 24.7 nm in length and 2.2 nm in diameter.89 The free AdhD activity was measured with PEG present at three different concentrations (5%, 15%, and 25% w/v) and a decrease in kcat was observed with increasing PEG concentration. This is likely due to the adoption of compact conformation or the formation of self-associated forms of the AdhD under the influence of excluded volume effects. This correlates well with our data from the co-assembled nanoreactors and suggests that wtSP, inside the P22 nanoreactor, can act analogously to PEG as a molecular crowding agent to AdhD resulting in a reduction of activity of the enzyme inside co-assembled nanoreactors.
Removal of wtSP resulted in the restoration of activities of AdhD in the nanoreactors.
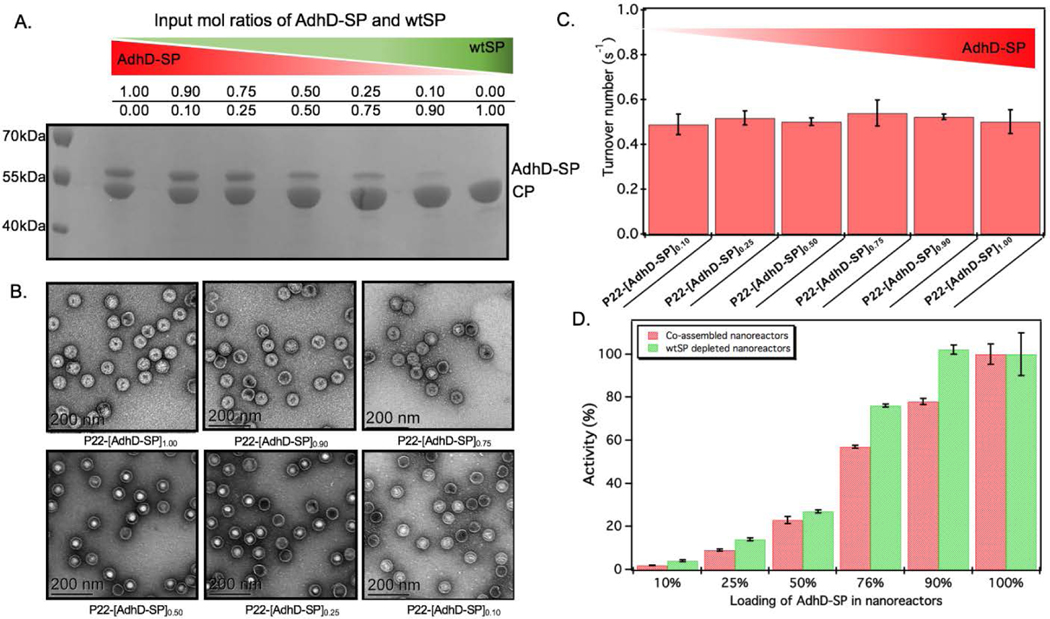
The approach utilized here for the removal of wtSP takes advantage of the fact that wtSP, but not bulky cargo-fused SP, can escape across the capsid shell upon treatment with 0.5 M GuHCl.49, 63 Thus, the wtSP was selectively removed from co-assembled nanoreactors containing AdhD-SP and wtSP (P22-[AdhD-SP]x[wtSP]y) encapsulated at different stoichiometric ratios and the resulting materials were analyzed by SDS-PAGE to determine the degree of wtSP removal. As shown in Figure 4A, we were able to remove all of the wtSP from all of the co-assembled nanoreactors without losing AdhD. TEM of these samples showed no change in morphology after the treatment (Figure 4B). The capsids were further analyzed by SEC-MALS/QELS to determine the size and number of enzymes in each capsid (Table 2). Molecular weight determined by SEC-MALS was used to calculate the number of copies of enzyme after subtracting the molecular weight of the P22 empty shell. Both, the radius of gyration and the hydrodynamic radius of the GuHCl treated capsids were comparable to non-treated capsids, indicating no change in size after the GuHCl treatment (Table 2 and Figure S6). Our results are supported by the literature that shows only very weak density differences in the areas surrounding hexons in three-dimensional reconstruction (3DR) images obtained from the cryo-EM of P22 capsids and the capsids that were treated with GuHCl to remove wtSP.90 Further analysis by DLS confirmed the integrity of particles at 50°C and the particle sized matched well with the size of co-assembled nanoreactors (Table S1). The enzymes, in GuHCl treated nanoreactors, followed Michaelis-Menten kinetic behavior and the data were fit to obtain kcat and KM (Figure 4C and Figure S7). Removal of wtSP from all the co-assembled nanoreactors resulted in an increase in kcat to a level similar to P22 encapsulating only the AdhD enzymes (Figure 4C). A similar trend was observed with KM in co-assembled and wtSP depleted capsids (Figure S7). We found that the increase in concentration of wtSP resulted in a concentration-dependent decrease in substrate affinity (increased KM), but when the wtSP was selectively removed the KM approached similar values for all nanoreactors. This suggests that the crowding induced by wtSP may have affected the conformational dynamics of the active site of the enzymes to carry out catalysis, thereby effectively increasing the KM. Interestingly, the kcat of AdhD in all the wtSP depleted nanoreactors approached a similar value, despite differences in their packing density, but remained lower than the kcat of the free enzyme (Figure S8). These results strongly suggest that the activity of enzymes in wtSP depleted nanoreactors is influenced by the enzymatic confinement in P22 and that the AdhD molecules do not undergo self-crowding even at local concentrations as high as 6.3 mM (~334 mg/ml, Table S2) present inside the nanoreactors assembled with AdhD only. Despite the similar kcat values of the encapsulated AdhD, the overall activity of SP depleted nanoreactors increased as the loading of AdhD-SP was increased, thus recapitulating the trend observed with co-assembled nanoreactors, but the activities were slightly enhanced at each loading in comparison to co-assembled nanoreactors as shown in the Figure 4D.
Figure 4.
Characterization and activity of AdhD in wtSP depleted nanoreactors. (A) SDS-PAGE analysis shows selective removal of wtSP from all the co-assembled nanoreactors. (B) TEM images show that morphology is maintained after chaotrope treatment. (C) Graph showing the similar activities of AdhD in nanoreactors after removing wtSP from co-assembled nanoreactors. (D) Graph comparing the activity of co-assembled and wtSP depleted nanoreactors. The nanoreactors containing AdhD only (100 percent loading level) showed similar activities before and after GuHCl treatment, indicating that GuHCl does not affect the enzymatic activity and the capsid porosity.
Table 2.
Molar mass and size of wtSP depleted nanoreactors determined by SEC-MALS/QELS.
| Sample P22-[AdhD-SP]x | Copies of AdhD-SP | Rrms (nm) | Rh (nm) |
|---|---|---|---|
|
| |||
| Empty Shell (ES) | -------- | 25.9 ± 0.1 | 25.8 ± 0.2 |
| P22-[AdhD-SP]1.00 | 175 ± 13 | 20.5 ± 2.3 | 25.4 ± 0.9 |
| P22-[AdhD-SP]0.90 | 152 ± 6 | 20.5 ± 2.7 | 25.0 ± 0.4 |
| P22-[AdhD-SP]0.75 | 108 ± 6 | 21.5 ± 2.4 | 24.9 ± 0.8 |
| P22-[AdhD-SP]0.50 | 42 ± 13 | 22.3 ± 2.8 | 25.2 ± 0.8 |
| P22-[AdhD-SP]0.25 | 21 ± 7 | 23.5 ± 2.1 | 24.9 ± 0.7 |
| P22-[AdhD-SP]0.10 | 7 ±1 | 22.2 ± 1.0 | 25.2 ± 0.3 |
Concentration dependent activity of the free (unencapsulated) AdhD enzyme.
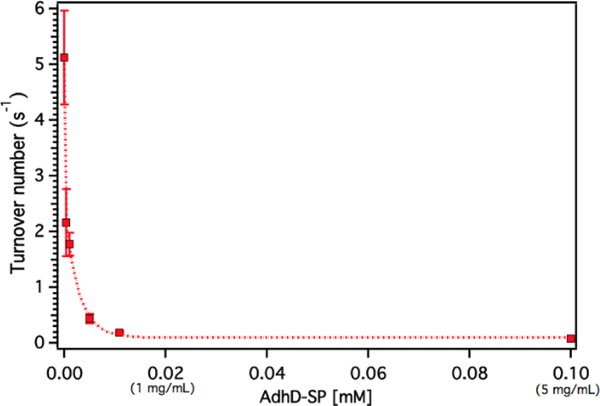
The confinement molarity (Mconf ), or the local concentration of the encapsulated AdhD enzyme inside these wtSP depleted nanoreactors, ranges from 0.2 – 5.8 mM (10 – 285 mg/ml). High local concentration in an enclosed system can lead to enhanced molecular associations.91–93 Therefore, to probe the effects of high local concentration on the AdhD enzyme (by analogy to Mconf ), we measured the activity of the free enzyme (AdhD-SP and AdhD), as a function of bulk concentration. The concentration of substrate and cofactor used were at least 30-fold over the highest concentration of enzyme used in the experiment. The kcat for the reaction exhibited a strong concentration dependence and decreased markedly with increasing enzyme concentration (Figure 5). Free AdhD, without the SP fusion, also showed concentration dependent activity with a similar trend (Figure S4). This suggests that some self-association interactions between individual AdhD monomers could be responsible for the diminished activity. The association may have led to the formation of clusters in which the buried enzymes may not be involved in catalysis due to restricted substrate accessibility or the dynamic movement of the active site, whereas the surface exposed enzymes and some freely diffusing enzymes may still be contributing towards the activity.
Figure 5.
Graph showing the concentration dependence of free AdhD enzyme activity.
Dynamic light scattering (DLS) measurements revealed the formation of higher order clusters of the AdhD enzymes with increasing concentration, supporting the self-association hypothesis (Figure S9). The fit of the activity versus concentration data indicates that the kinetic behavior of the free AdhD plateaus at high concentrations. This behavior of the free enzyme at high concentration potentially allows us to interpret the role of confinement in the AdhD encapsulated in the P22 nanoreactors. Under the high local concentrations of enzymes inside the P22 there could be significant self-association resulting in confinement effects that contribute to the lowered activity of the encapsulated AdhD when compared to the free enzyme at low concentrations. Just as observed in the free AdhD, where the activity plateaus at higher concentrations, the encapsulated AdhD shows the same activity at all loadings (after wtSP removal) where the Mconf for all nanoreactors is at or above the highest concentrations measured for the free AdhD where the decrease in activity has already plateaued.
These experiments show that crowding and confinement can influence the outcome of an encapsulation system independently. The experiments are, however, ensemble averages, and previously Dashti et al.52 showed a wide distribution in the amount of co-encapsulated cargoes in single particle measurements of mixed in vitro assemblies despite a clear trend observed in ensemble measurements. Therefore, complementing ensemble-averaged measurements with single-particle analysis will enable appropriate interpretation of the data and the insight into the structural dynamics of enzymes with better sensitivity and resolution under crowded and confined conditions.
CONCLUSION
Previously we have shown that a wide range of cargo molecules can be encapsulated in P22 using genetically programmed in vivo assembly approach. This approach, however, suffers from some limitations such as the lack of ability to control packaging amount of a cargo (or multiple cargos), encapsulation of misfolded proteins, or partial inactivation of proteins upon encapsulation in P22. To overcome some of these challenges, an in vitro assembly approach was developed for cargo encapsulation in P22. Using this approach, we have designed and constructed P22 nanoreactors, whose catalytic properties could be fine-tuned by controlling the composition of encapsulated cargos. A variety of nanoreactors were assembled in the presence of AdhD-SP and wtSP mixed at different stoichiometric ratios. The composition of assembled nanoreactors followed the input stoichiometry, and contained AdhD-SP and wtSP encapsulated in well-regulated and variable amounts, which was then translated into the control over functional output of AdhD catalyst molecules and the nanoreactors (i.e. enzyme-VLP complex). The selective removal of wtSP from the co-assembled nanoreactors allowed us to gain further control over the catalytic performance of the nanoreactors and independently investigate the effects of microenvironment on the functional properties of encapsulated AdhD enzyme. The insights gained from this research could prove useful in designing new materials with cell-like complexity using bio-inspired and biomimetic approaches.
Materials and Methods
Materials.
Reagents for buffer preparation were purchased from Fisher Scientific, unless otherwise specified. DNA primers were purchased from Eurofins MWG Operon. E. coli BL21(λDE3) were purchased from Lucigen. Protease and phosphatase inhibitor mini tablets were purchased from Roche. For gene assembly, NEBuilder® HiFi DNA assembly master mix was purchased from New England Biolabs. Gene block were purchased from IDT.
Cloning of AdhD into pBad vector.
The AdhD gene from Pyrococcus furiosus was ordered from the IDT after codon optimization for BL21(DE3). The AdhD gene was cloned into a pBad vector containing gene for truncated scaffold protein (SPt) using Gibson assembly and the primers 5’ CGCAGCAATGCCGTAGCAGAACAGGGC 3’and 5’ GCCGCTGTGATGGTGATGATGGGC 3’ for linearization of SPt vector. The Gibson assembly was carried out using HiFi DNA assembly master mix. After verifying the DNA sequence from Eurofins Genomics, the assembled vector was transformed into BL21(DE3) cells for protein expression.
Expression and purification of free AdhD-SP.
Transformed BL21(DE3) cells were grown in LB medium at 37 °C in the presence of ampicillin to maintain selection for the vector containing cells. The expression of AdhD-SP was induced by addition of L-arabinose to a final concentration of 13 mM at OD600 = 0.6. The cultures were grown for 4 h post induction. The cells were harvested at 4500 xg in a benchtop centrifuge and re-suspended in PBS (50 mM phosphate, 100 mM sodium chloride, pH 7.8) with lysozyme, DNase, RNase and protease mini tablets for 30 min. The cell suspension was lysed by sonication and centrifuged at 12,000 xg for 45 min at 4 °C. The supernatant was removed and filtered through 0.45-micron filter. For purification, the filtered supernatant was loaded on to Ni-NTA column at 1 mL min−1, followed by washing of the column with wash buffer (50 mM phosphate, 100 mM sodium chloride, pH 7.8) at 1 ml min−1. The protein was eluted with a linear gradient of imidazole (20–500 mM imidazole) at 2 ml min−1. The protein was dialyzed into an assembly buffer (50 mM Tris-HCl, 25 mM NaCl, 2 mM EDTA, 3 mM β-mercaptoethanol, 1% glycerol, and pH 7.4) for in vitro assembly.
Preparation of coat protein (CP) subunits for in vitro assembly.
CP subunits were obtained by dissociating the ES in 3 M GuHCl (final concentration) for 30 min at room temperature. To obtain ES, the P22 procapsids were incubated with 0.5 M GuHCl for 1.5 h at 4 °C so as to extract SP. Capsids were pelleted in ultracentrifuge at 45 000 rpm for 50 min followed by resuspension and incubation in 0.5 M GuHCl for 1 h. This extraction process was repeated until the supernatant was free from SP. The ES were then purified over an S-500 Sephadex size exclusion column. Assembly buffer was used in the background to make GuHCl solution for dissociation of ES and removal of SP.
In vitro assembly.
CP subunits (~2 mg ml−1), as prepared above in 3 M GuHCl, were mixed with purified AdhD-SP (or pre-mix of AdhD-SP and wtSP) subunits in a 1 : 1 molar ratio. The volume of the AdhD-SP was adjusted such that the final concentration of GuHCl was 1.5 M in all assembly reaction mixtures. The mixture was dialyzed into assembly buffer for 24 h at room temperature with a buffer exchange in between.
Removal of wtSP from co-assembled nanoreactors.
To remove wtSP, the co-assembled nanoreactors were dialyzed into the assembly buffer containing 0.5M GuHCl using 100kDa MWCO dialysis membrane. The dialysis buffer was exchanged three times with a dialysis time of 4h each time. The final dialysis was done with the PBS buffer with no GuHCl added.
SDS-PAGE.
Protein samples were mixed with 4× loading buffer containing 100 mM DTT, heated in a boiling water bath for 10 min, removed, and spun down on a benchtop centrifuge. Samples were separated on a 15% acrylamide gel at a constant current of 35 mA for approximately 1 h. Gels were stained with an Instant Blue protein stain, rinsed with water, and imaged. Images were recorded on a UVP MultDoc-IT Digital Imaging System. A 10–180 kDa PageRuler prestained ladder was used for reference.
Transmission electron microscopy.
Samples (4 μL, 0.3 mg mL−1 protein) were applied to glow-discharged carbon coated grids and incubated for 45 s. Excess liquid was wicked away with filter paper. The sample loaded grids were then washed on a droplet of water. Grids were stained with 4 μL 2% uranyl acetate for 10 s and excess stain was wicked away with filter paper. Images were taken on a JEOL 1010 transmission electron microscope at accelerating voltage of 80 kV.
Size exclusion chromatography with multiangle light scattering and refractive index detection.
Samples were separated over a WTC-0200S (Wyatt Technologies) size exclusion column utilizing an Agilent 1200 HPLC system at a flow rate 0.7 ml min−1 of MALS buffer (50 mM phosphate, 100 mM NaCl and 200 ppm NaN3, pH 7.2). Sample volume of 25 μL per injection was loaded onto the column, and the column was run for 35 min. Eluted peaks were detected using a UV-Vis detector (Agilent), a Wyatt HELEOS multi-angle laser light scattering (MALS) detector, and an Optilab rEX differential refractometer (Wyatt Technology Corporation). The number-average particle molecular weight, Mn, was measured across FWHM of each peak with Astra 6.0.3.16 (Wyatt Technology Corporation) using a previously calculated dn/dc value of 0.185 mL g−1.
Kinetics assay of free and P22 encapsulated enzyme.
The activity assays were carried out on an Agilent’s 8453 UV-Visible spectrophotometer fitted with an 8 position multicell transport and a VWR temperature controller. All assays were conducted at 50 °C in triplicates and an assay volume of 110 μL was used for measurement. Kinetic assays were performed in 100mM sodium phosphate, pH 6.2 buffer. Reaction mixtures containing acetoin substrate (ranging from 0.3mM – 100mM), NADH (A340 ~1.8) and buffer were preheated in a quartz cuvette for 2 minutes prior to the addition of free or encapsulated enzyme. NADH thermal degradation was monitored for 80–100 seconds then enzyme was added to the solution. Reactions were mixed thoroughly by pipetting and the enzyme activity was measured by monitoring the rate of disappearance of NADH at A340 nm. Concentrations were calculated using extinction coefficients of NADH, P22 coat protein, AdhD-SP, and wtSP as 6220 M−1cm−1, 44920 mM−1cm−1, 61310 M-1cm-1, 17420 M−1cm−1, respectively (ExPASy’s ProtParam tool). Total concentration of the encapsulated enzyme was calculated using densitometry and molar mass data from SEC-MALS. From the densitometry, ratio of AdhD-SP to wtSP was determined, which was then applied to SEC-MALS data to calculate number of copies of AdhD-SP and wtSP encapsulated inside the capsids. From the number of copies and A280, the concentration of encapsulated enzyme was calculated using the set of equations given in the supplementary information. Kinetics plots were fit to Michaelis-Menten kinetics or substrate inhibition model using Igor pro 6.37.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Science Foundation (BMAT DMR-1507282) and the National Institutes of Health (R01 AI104905). We thank the Indiana University Electron Microscopy Center, Physical Biochemistry Instrumentation facility and Biological mass-spectrometry lab for access to instrumentation used in this work.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Electronic Supplementary Information (ESI) available. DOI: 10.1039/C9NR08348K
Notes and references
- 1.Meyers MA, Chen PY, Lin AYM and Seki Y, Prog Mater Sci, 2008, 53, 1–206. [Google Scholar]
- 2.Naik RR and Singamaneni S, Chem Rev, 2017, 117, 12581–12583. [DOI] [PubMed] [Google Scholar]
- 3.Lepora NF, Verschure P. and Prescott TJ, Bioinspir Biomim, 2013, 8, 013001. [DOI] [PubMed] [Google Scholar]
- 4.Giessen TW and Silver PA, Journal of Molecular Biology, 2016, 428, 916–927. [DOI] [PubMed] [Google Scholar]
- 5.Chen AH and Silver PA, Trends Cell Biol, 2012, 22, 662–670. [DOI] [PubMed] [Google Scholar]
- 6.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F. and Sutter M, Nat Rev Microbiol, 2018, 16, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azuma Y, Edwardson TGW and Hilvert D, Chemical Society Reviews, 2018, 47, 3543–3557. [DOI] [PubMed] [Google Scholar]
- 8.Giessen TW, Curr Opin Chem Biol, 2016, 34, 1–10. [DOI] [PubMed] [Google Scholar]
- 9.Kim EY and Tullman-Ercek D, Curr Opin Biotechnol, 2013, 24, 627–632. [DOI] [PubMed] [Google Scholar]
- 10.Vriezema DM, Aragones MC, Elemans JAAW, Cornelissen JJLM, Rowan AE and Nolte RJM, Chemical Reviews, 2005, 105, 1445–1489. [DOI] [PubMed] [Google Scholar]
- 11.Jakobson CM, Chen Y, Slininger MF, Valdivia E, Kim EY and Tullman-Ercek D, J Mol Biol, 2016, 428, 2989–2996. [DOI] [PubMed] [Google Scholar]
- 12.Wagner HJ, Capitain CC, Richter K, Nessling M. and Mampel J, Eng Life Sci, 2017, 17, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tetter S. and Hilvert D, Angewandte Chemie, 2017, 56, 14933–14936. [DOI] [PubMed] [Google Scholar]
- 14.Glasgow JE, Capehart SL, Francis MB and Tullman-Ercek D, ACS nano, 2012, 6, 8658–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiedler JD, Brown SD, Lau JL and Finn MG, Angewandte Chemie-International Edition, 2010, 49, 9648–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson DP, Prevelige PE and Douglas T, ACS nano, 2012, 6, 5000–5009. [DOI] [PubMed] [Google Scholar]
- 17.Patterson DP, Schwarz B, Waters RS, Gedeon T. and Douglas T, ACS Chem Biol, 2014, 9, 359–365. [DOI] [PubMed] [Google Scholar]
- 18.Worsdorfer B, Pianowski Z. and Hilvert D, Journal of the American Chemical Society, 2012, 134, 909–911. [DOI] [PubMed] [Google Scholar]
- 19.Colletier JP, Chaize B, Winterhalter M. and Fournier D, BMC Biotechnol, 2002, 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z, Fu J, Dhakal S, Johnson-Buck A, Liu M, Zhang T, Woodbury NW, Liu Y, Walter NG and Yan H, Nat Commun, 2016, 7, 10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dulieu C, Moll M, Boudrant J. and Poncelet D, Biotechnol Progr, 2000, 16, 958–965. [DOI] [PubMed] [Google Scholar]
- 22.Bornscheuer UT, Angewandte Chemie-International Edition, 2003, 42, 3336–3337. [DOI] [PubMed] [Google Scholar]
- 23.Datta S, Christena LR and Rajaram YR, 3 Biotech, 2013, 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Sanchez L, Cadena-Nava RD, Palomares LA, Ruiz-Garcia J, Koay MS, Cornelissen JJ and Vazquez-Duhalt R, Enzyme Microb Technol, 2014, 60, 24–31. [DOI] [PubMed] [Google Scholar]
- 25.Comellas-Aragones M, Engelkamp H, Claessen VI, Sommerdijk NAJM, Rowan AE, Christianen PCM, Maan JC, Verduin BJM, Cornelissen JJLM and Nolte RJM, Nature nanotechnology, 2007, 2, 635–639. [DOI] [PubMed] [Google Scholar]
- 26.Minten IJ, Claessen VI, Blank K, Rowan AE, Nolte RJM and Cornelissen J, Chemical Science, 2011, 2, 358–362. [Google Scholar]
- 27.Schoonen L, Nolte RJ and van Hest JC, Nanoscale, 2016, 8, 14467–14472. [DOI] [PubMed] [Google Scholar]
- 28.Maity B, Fujita K. and Ueno T, Current Opinion in Chemical Biology, 2015, 25, 88–97. [DOI] [PubMed] [Google Scholar]
- 29.Wilkerson JW, Yang SO, Funk PJ, Stanley SK and Bundy BC, N Biotechnol, 2018, 44, 59–63. [DOI] [PubMed] [Google Scholar]
- 30.Frey R, Mantri S, Rocca M. and Hilvert D, J Am Chem Soc, 2016, 138, 10072–10075. [DOI] [PubMed] [Google Scholar]
- 31.Brasch M, Putri RM, de Ruiter MV, Luque D, Koay MS, Caston JR and Cornelissen JJ, J Am Chem Soc, 2017, 139, 1512–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson DP, McCoy K, Fijen C. and Douglas T, J Mater Chem B, 2014, 2, 5948–5951. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Zheng G. and Zhu S, Microb Cell Fact, 2018, 17, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren H, Zhu S. and Zheng G, Int J Mol Sci, 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL and Rome LH, Proc Natl Acad Sci U S A, 2005, 102, 4348–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selivanovitch E. and Douglas T, Curr Opin Virol, 2019, 36, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi H, Choi B, Kim GJ, Kim HU, Kim H, Jung HS and Kang S, Small, 2018, 14, e1801488. [DOI] [PubMed] [Google Scholar]
- 38.Uchida M, LaFrance B, Broomell CC, Prevelige PE Jr. and Douglas T, Small, 2015, 11, 1562–1570. [DOI] [PubMed] [Google Scholar]
- 39.Long S KJ, Asuri P Biochem Anal Biochem, 2018, 7 355. [Google Scholar]
- 40.Zhou HX, Rivas G. and Minton AP, Annu Rev Biophys, 2008, 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou HX, J Mol Recognit, 2004, 17, 368–375. [DOI] [PubMed] [Google Scholar]
- 42.Ping G, Yuan JM, Sun Z. and Wei Y, J Mol Recognit, 2004, 17, 433–440. [DOI] [PubMed] [Google Scholar]
- 43.Oehler S. and Muller-Hill B, Journal of Molecular Biology, 2010, 395, 242–253. [DOI] [PubMed] [Google Scholar]
- 44.Ellis RJ, Trends in biochemical sciences, 2001, 26, 597–604. [DOI] [PubMed] [Google Scholar]
- 45.Ellis RJ and Minton AP, Nature, 2003, 425, 27–28. [DOI] [PubMed] [Google Scholar]
- 46.Ralston GB, J Chem Educ, 1990, 67, 857–860. [Google Scholar]
- 47.Kuznetsova IM, Turoverov KK and Uversky VN, Int J Mol Sci, 2014, 15, 23090–23140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittal S, Chowhan RK and Singh LR, Biochimica et biophysica acta, 2015, 1850, 1822–1831. [DOI] [PubMed] [Google Scholar]
- 49.O’Neil A, Reichhardt C, Johnson B, Prevelige PE and Douglas T, Angewandte Chemie-International Edition, 2011, 50, 7425–7428. [DOI] [PubMed] [Google Scholar]
- 50.Comellas-Aragones M, Engelkamp H, Claessen VI, Sommerdijk NA, Rowan AE, Christianen PC, Maan JC, Verduin BJ, Cornelissen JJ and Nolte RJ, Nature nanotechnology, 2007, 2, 635–639. [DOI] [PubMed] [Google Scholar]
- 51.Minten IJ, Hendriks LJA, Nolte RJM and Cornelissen J, Journal of the American Chemical Society, 2009, 131, 17771–17773. [DOI] [PubMed] [Google Scholar]
- 52.Dashti NH, Abidin RS and Sainsbury F, ACS nano, 2018, 12, 4615–4623. [DOI] [PubMed] [Google Scholar]
- 53.Azuma Y. and Hilvert D, Methods Mol Biol, 2018, 1798, 39–55. [DOI] [PubMed] [Google Scholar]
- 54.Azuma Y, Zschoche R, Tinzl M. and Hilvert D, Angewandte Chemie, 2016, 55, 1531–1534. [DOI] [PubMed] [Google Scholar]
- 55.Schoonen L, Nolte RJM and van Hest JCM, Nanoscale, 2016, 8, 14467–14472. [DOI] [PubMed] [Google Scholar]
- 56.Schoonen L, Pille J, Borrmann A, Nolte RJM and van Hest JCM, Bioconjugate Chem, 2015, 26, 2429–2434. [DOI] [PubMed] [Google Scholar]
- 57.Botstein D, Waddell CH and King J, J Mol Biol, 1973, 80, 669–695. [DOI] [PubMed] [Google Scholar]
- 58.Padilla-Meier GP, Gilcrease EB, Weigele PR, Cortines JR, Siegel M, Leavitt JC, Teschke CM and Casjens SR, The Journal of biological chemistry, 2012, 287, 33766–33780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greene B. and King J, Virology, 1994, 205, 188–197. [DOI] [PubMed] [Google Scholar]
- 60.Parker MH, Casjens S. and Prevelige PE Jr., J Mol Biol, 1998, 281, 69–79. [DOI] [PubMed] [Google Scholar]
- 61.Qazi S, Miettinen HM, Wilkinson RA, McCoy K, Douglas T. and Wiedenheft B, Molecular pharmaceutics, 2016, 13, 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Neil A, Prevelige PE, Basu G. and Douglas T, Biomacromolecules, 2012, 13, 3902–3907. [DOI] [PubMed] [Google Scholar]
- 63.Sharma J, Uchida M, Miettinen HM and Douglas T, Nanoscale, 2017, 9, 10420–10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anand G, Sharma S, Dutta AK, Kumar SK and Belfort G, Langmuir, 2010, 26, 10803–10811. [DOI] [PubMed] [Google Scholar]
- 65.Fuller MT and King J, Biophysical Journal, 1980, 32, 381–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prevelige PE Jr., Thomas D. and King J, J Mol Biol, 1988, 202, 743–757. [DOI] [PubMed] [Google Scholar]
- 67.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King J. and Chiu W, Proc Natl Acad Sci U S A, 2011, 108, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machielsen R, Uria AR, Kengen SWM and van der Oost J, Applied and environmental microbiology, 2006, 72, 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solanki K, Abdallah W. and Banta S, Biotechnol J, 2016, 11, 1483–1497. [DOI] [PubMed] [Google Scholar]
- 70.Hotz J. and Meier W, Langmuir, 1998, 14, 1031–1036. [Google Scholar]
- 71.Egelhaaf SU and Schurtenberger P, J Phys Chem-Us, 1994, 98, 8560–8573. [Google Scholar]
- 72.Minton AP and Wilf J, Biochemistry-Us, 1981, 20, 4821–4826. [DOI] [PubMed] [Google Scholar]
- 73.Miklos AC, Sarkar M, Wang YQ and Pieak GJ, Journal of the American Chemical Society, 2011, 133, 7116–7120. [DOI] [PubMed] [Google Scholar]
- 74.Sarkar M, Smith AE and Pielak GJ, Proceedings of the National Academy of Sciences of the United States of America, 2013, 110, 19342–19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang M. and Guo ZH, Journal of the American Chemical Society, 2007, 129, 730–731. [DOI] [PubMed] [Google Scholar]
- 76.Norris MG and Malys N, Biochem Biophys Res Commun, 2011, 405, 388–392. [DOI] [PubMed] [Google Scholar]
- 77.Wenner JR and Bloomfield VA, Biophys J, 1999, 77, 3234–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran-Zorzano MT, Viale A, Munoz F, Alonso-Casajus N, Eydallin G, Zugasti B, Baroja-Fernandez E. and Pozueta-Romero J, Febs Lett, 2007, 581, 1035–1040. [DOI] [PubMed] [Google Scholar]
- 79.Sasaki Y, Miyoshi D. and Sugimoto N, Nucleic Acids Res, 2007, 35, 4086–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pastor I, Vilaseca E, Madurga S, Garces JL, Cascante M. and Mas F, J Phys Chem B, 2011, 115, 1115–1121. [DOI] [PubMed] [Google Scholar]
- 81.Olsen SN, Thermochim Acta, 2006, 448, 12–18. [Google Scholar]
- 82.Olsen SN, Ramlov H. and Westh P, Comp Biochem Physiol A Mol Integr Physiol, 2007, 148, 339–345. [DOI] [PubMed] [Google Scholar]
- 83.Minh DD, Chang CE, Trylska J, Tozzini V. and McCammon JA, J Am Chem Soc, 2006, 128, 6006–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hou S, Ziebacz N, Kalwarczyk T, Kaminski TS, Wieczorek SA and Holyst R, Soft Matter, 2011, 7, 3092–3099. [Google Scholar]
- 85.Sanli G, Dudley JI and Blaber M, Cell Biochem Biophys, 2003, 38, 79–101. [DOI] [PubMed] [Google Scholar]
- 86.Blanch HW, Clark DS Biochemical engineering; New York: Marcel Dekker, 1997. [Google Scholar]
- 87.Tuma R, Prevelige PE Jr. and Thomas GJ Jr., Biochemistry-Us, 1996, 35, 4619–4627. [DOI] [PubMed] [Google Scholar]
- 88.Teschke CM, King J. and Prevelige PE, Biochemistry-Us, 1993, 32, 10658–10665. [DOI] [PubMed] [Google Scholar]
- 89.Greene B. and King J, Journal of Biological Chemistry, 1999, 274, 16135–16140. [DOI] [PubMed] [Google Scholar]
- 90.Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W. and Prevelige PE Jr., Biophys J, 1999, 76, 3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jordan PC, Patterson DP, Saboda KN, Edwards EJ, Miettinen HM, Basu G, Thielges MC and Douglas T, Nat Chem, 2016, 8, 179–185. [DOI] [PubMed] [Google Scholar]
- 92.Martin GL, Minteer SD and Cooney M, Chemical Communications, 2011, 47, 2083–2085. [DOI] [PubMed] [Google Scholar]
- 93.Wu F. and Minteer SD, Biomacromolecules, 2013, 14, 2739–2749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.