Abstract
Cancer cell‐derived extracellular vesicles (EVs) are promising biomarkers for cancer diagnosis and prognosis. However, the lack of rapid and sensitive isolation techniques to obtain EVs from clinical samples at a sufficiently high yield limits their practicability. Chimeric nanocomposites of lactoferrin conjugated 2,2‐bis(methylol)propionic acid dendrimer‐modified magnetic nanoparticles (LF‐bis‐MPA‐MNPs) are fabricated and used for simple and sensitive EV isolation from various biological samples via a combination of electrostatic interaction, physically absorption, and biorecognition between the surfaces of the EVs and the LF‐bis‐MPA‐MNPs. The speed, efficiency, recovery rate, and purity of EV isolation by the LF‐bis‐MPA‐MNPs are superior to those obtained by using established methods. The relative expressions of exosomal microRNAs (miRNAs) from isolated EVs in cancerous cell‐derived exosomes are verified as significantly higher than those from noncancerous ones. Finally, the chimeric nanocomposites are used to assess urinary exosomal miRNAs from urine specimens from 20 prostate cancer (PCa), 10 benign prostatic hyperplasia (BPH), patients and 10 healthy controls. Significant up‐regulation of miR‐21 and miR‐346 and down‐regulation of miR‐23a and miR‐122‐5p occurs in both groups compared to healthy controls. LF‐bis‐MPA‐MNPs provide a rapid, simple, and high yield method for human excreta analysis in clinical applications.
Keywords: clinical applications, extracellular vesicles, magnetic nanoparticle, prostate cancer, urinary exosomes
Abbreviations
- APTES
3‐aminopropyl triethoxysilane
- 3‐MPS
3‐mercaptopropyl trimethoxysilane
- Ab
antibodyCCM cell culture medium
- DTS
dimethyl pimelimidate/thin film sample processing
- DMEM
Dulbecco's modified Eagle's medium
- EV
extracellular vesicle
- FBS
fetal bovine serum
- FTIR
Fourier transform infrared
- GMBS
N‐(γ‐maleimidobutyryloxy)succinimide ester
- LF‐bis‐MPA‐MNPs
Lactoferrin conjugated bis‐MPA dendrimers modified magnetic nanoparticles
- LF
lactoferrin
- MNPs
magnetic nanoparticles
- NTA
nanoparticle tracking analysis
- PBS
phosphate‐buffered saline
- RT‐qPCR
quantitative reverse transcription polymerase chain reaction
- SEM
scanning electron microscopy
- TEI
total exosome isolation
- UC
ultracentrifugation
- UV‐Vis
ultraviolet–visible spectrophotometry
- WB
Western blot
1. INTRODUCTION
Extracellular vesicles (EVs), such as exosomes (30–150 nm in diameter, EVs originating from multivesicular endosomes) (Zhang et al., 2019), microvesicles (100–1000 nm in diameter, EVs originating from plasma membrane budding) (Muralidharan‐Chari et al., 2010), and apoptotic bodies (1–4 μm in diameter, EVs originating from apoptosis) (Caruso & Poon, 2018), are released by most types of cells. They are found in abundance in human excreta including blood, saliva, sputum, and urine (Doyle & Wang, 2019). EVs contain DNA, messenger RNA (mRNA), microRNA (miRNA), and cell‐type‐specific proteins(Zhang et al., 2019), which can be used as biomarkers for many types of diseases (including cancer) because their expression levels are significantly different from those in normal cells (Hoshino et al., 2020). Among the EVs, exosomes play a particularly important role as noninvasive biomarkers for early disease detection (Marcuello et al., 2019), with more exosomes being released from cancerous cells than noncancerous ones (Whiteside, 2016). They can be used as noninvasive or minimally invasive biomarkers. Among the body fluids, urine can be collected in large volumes and frequently. Moreover, it is less complex and more stable than serum or plasma and is a promising biological fluid for detecting biomarkers for diseases such as prostate cancer (PCa) (Erdbrügger et al., 2021).
miRNAs are small noncoding RNA molecules of ∼18–25 nucleotides in length that downregulate mRNA expression either by impairing mRNA transcription or by promoting its degradation (Negrini et al., 2009). Exosomal miRNAs such as miR‐21, miR‐23a, and miR‐1246 for colorectal cancer (CRC) (Ogata‐Kawata et al., 2014) and miR‐21, miR‐155, miR‐222, and miR‐29 for hepatic cancer have been reported as potential biomarkers for clinical diagnosis (Braconi et al., 2011). Furthermore, studies have shown that protein markers are up or downregulated differently in the serum exosomes of CRC (Chen et al., 2017), hepatic cancer (Wang et al., 2019), and PCa patients (Zhou & Zhu, 2019).
Despite the potential of using EVs as tumor markers, one of the limitations is the lack of standardization in EV isolation and analysis via quick and accurate techniques. Current isolation methods, including ultracentrifugation (UC) (Gupta et al., 2018), size‐based filtration (Heinemann et al., 2014), size‐exclusion chromatography (Stranska et al., 2018), polymer precipitation (Brown & Yin, 2017), immune affinity‐based techniques (Stranska et al., 2018), microfluidic‐based isolation techniques (Iliescu et al., 2019), are limited by being time‐consuming, require purification to remove impurities before analysis, and/or require heavy and expensive devices (Brennan et al., 2020; Gurunathan et al., 2019). Moreover, contaminating proteins and RNAs often need to be removed and standards used to improve the accuracy of an exosome‐based diagnosis. Thus, more efficient isolation/enrichment methods with large sample sizes should be developed and applied in clinical exosome‐based detection.
Nanoparticles (NPs) have been used in many applications (DeFrates et al., 2018). The small size, high surface‐area‐to‐volume ratio (Ali et al., 2016), magnetism, and stability after surface coating (Zhu et al., 2018) make iron oxide (Fe3O4) NPs an easy and convenient platform for isolation. Furthermore, many types of ligand (such as peptides (Zhang et al., 2009), antibodies (Xu et al., 2011), folic acid (Landmark et al., 2008), etc.) can be conjugated with the surfaces of the magnetic NPs (MNPs). Various novel Fe3O4 NPs with surface coatings (Wu et al., 2020) and immobilized ligands (Xu et al., 2014) have been fabricated for use in the point‐of‐care field (Xianyu et al., 2018). Various methods have been developed to synthesize MNPs such as coprecipitation (Yazdani & Seddigh, 2016), microemulsion (Lakshmanan et al., 2014), thermal decomposition (Unni et al., 2017), solvothermal (Kim et al., 2018), sonochemical (Wang et al., 2015), microwave‐assisted (Kostyukhin et al., 2020), chemical vapor deposition (Farhanian et al., 2018), combustion (Kooti & Sedeh, 2013), carbon arc (Brunsman et al., 1994), and laser pyrolysis (Alexandrescu et al., 2010). Among these methods, coprecipitation is the most widely used and most proper method for MNP synthesis with which the sizes of the particles can be controlled to around 9 nm (Soytaş et al., 2019). The basic protocol is the precipitation reaction of Fe(II) and Fe(III) salts with a molar ratio of 2:1 under alkaline conditions to produce black Fe3O4 NPs (Ali et al., 2016). These are precipitated out by adding sodium hydroxide (NaOH) or ammonium hydroxide (NH4OH) to maintain the pH between 9 and 14 at an elevated temperature (Mascolo et al., 2013). The simple chemical reaction is as follows (Setyawan & Widiyastuti, 2019):
One of the disadvantages of Fe3O4 is oxidization in air due to its high chemical activity (Eftekhari et al., 2019), which can lead to degradation or a loss of magnetism. Polymers such as dextran (Unterweger et al., 2018), polyethylene glycol (PEG) (Illés et al., 2018), polyvinylpyrrolidone (PVP) (Lee et al., 2008), polyethyleneimine (PEI) (Steitz et al., 2007), chitosan (Kievit et al., 2009), and dendrimers (Sun et al., 2016) are chemical compounds used for coating MNPs. The highly branched well defined 3D architecture of dendrimers (Singh et al., 2016) with multiple functional groups such as amine, carboxyl, thiol, hydroxyl, etc., (Lyu et al., 2019) enable them to become stabilized and functionalized on MNPs. Dendrimers are used in many applications, such as theranostics (Ray et al., 2018), biosensors (Erdem et al., 2018), optics (Wu et al., 2018), adhesives (Maturavongsadit et al., 2016), and coating (Nonahal et al., 2018). The surfaces of proteins display many cationic groups, such as amine, imidazole, and guanidinium. Dendrimers and 2,2‐bis(methylol)propionic acid (bis‐MPA) dendrimers (Carlmark et al., 2013) for the coating of NPs are modified onto the surfaces of the MNPs by using MNPs as core molecules for divergent growth of dendrimers with amine termini (Fu et al., 2013; Li et al., 2011). More functional hydroxyl groups on the surface can be converted to thiol functional groups and bind to the amine group of lactoferrin (LF) by cross‐linking (Dao et al., 2018).
LF, a member of the transferrin family (Kell et al., 2020), is a cationic iron‐binding glycoprotein with a molecular weight of 77–80 kDa (Baker & Baker, 2012). The biological functions of LF are antimicrobial, antibacterial, antiviral, antiparasitic, and antitumor activities (Jahani et al., 2015). It can bind to Fe, Cu, Zn, and Mn ions, along with lipopolysaccharides (LPSs), lipoteichoic acid, heparan sulfate, proteoglycan, oligodeoxynucleotides, DNA, and RNA (Sohrabi et al., 2014). Using LF as a nanocarrier has many advantages (Elzoghby et al., 2020), and owing to its physical properties, it can be used for EV isolation. Although the main function of LF is its ability to bind to free iron (Kell et al., 2020), its application to biological target isolation is noteworthy. The tertiary structure of LF consists of two homologous lobes – an N‐ and a C‐terminal lobes, connected by an α‐helical bridge, with numerous disulfide bonds making it stable (Sabra & Agwa, 2020). The N‐terminus of LF is cationic and hydrophobic. The α‐helical N terminal with an arginine side chain forms an anion‐binding pocket (Sabra & Agwa, 2020). It shows a net positive charge at high pH values of 8.0–8.5 (Sohrabi et al., 2014), so it can maintain a positive charge over a wide pH range. Similar to the plasma membranes of cells, EVs carry a net negative surface charge, which is mostly from phosphatidylserine (PS, a negatively charged lipid) (Beit‐Yannai et al., 2018). Recent studies have shown that PS is exposed on the plasma membrane of cancer cells and cancerous EV but not on noncancer cell membranes (Riedl et al., 2011). In addition to PS, LF has also been shown to interact with glycosaminoglycans (GAGs, a type of proteoglycan – mainly heparan sulfate proteoglycans (HSPs) and chondroitin sulfate proteoglycans) (El Yazidi‐Belkoura et al., 2001). The studies showed that the N‐terminal of human lactoferrin (hLF) with a unique cluster (G1RRRR5) played a key role in the interaction between hLF and GAGs (Cutone et al., 2020). HSPs are located on the surface of producer and recipient cells, as well as on EVs (Cerezo‐Magaña et al., 2020). One of the reasons for the binding of LF with the exosome biological target is the expression of glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), which acts as an LF receptor (Malhotra et al., 2016). The N domain of LF physically attaches to GAPDH on the exosome surface, while incubation of the LF N protein (recombinant protein designed to express the N‐terminal region of LF) with exosomes enhances its binding to the exosome surface (Dar et al., 2021). Altogether, electrostatic, hydrophobic, and biorecognition interactions are possible key elements for the binding of LF to exosomes.
Herein, we report the fabrication of chimeric nanocomposites of LF‐conjugated bis‐MPA dendrimer‐coated MNPs (LF‐bis‐MPA‐MNPs) for EV isolation based on a combination of electrostatic interaction, physical absorption, and biorecognition. Electrostatic interaction occurs between the positively charged LF and negatively charged surface of the EV, while Van der Waals forces (interaction between LF and other molecules such as lipoproteins on the plasma membrane) provide the driving force for physical adsorption. In addition, the LF receptors on the EV surface can bind with LF via an immunoaffinity mechanism. Therefore, based on the electrostatic interaction, physical absorption, and targeted binding of LF to the EV, we speculate that LF has great potential for EV isolation. The bis‐MPA dendrimer‐coated MNPs have a stable surface and do not undergo aggregation. The high isoelectric point (pI) of LF (∼8.5–10.0) (Brock et al., 1997) allows it to interact with EV over a wide pH range. Cell culture media (CCM) or body fluid has a pH < pI of LF (PBS pH 7.4, CCM pH 7.2–7.4, urine pH 4–8, blood 7.35–7.45, etc.), so positively charged LF strongly binds to the negatively charged surface of EV. After the isolation, EV was eluted in the elution buffer (pH ∼10.6), which has a higher pH than the pI of LF, leading to the dissociation of LF–EV complexes for EV release and collection. Our method was compared with UC (Skottvoll et al., 2018), the commercially available total exosome isolation (TEI) kit‐based precipitation method (Skottvoll et al., 2018), dimethyl pimelimidate thin‐film sample processing (DTS) chips (Jin, Koo, et al., 2018; Jin, Lee, et al., 2018) and cationic polymer‐coated MNPs (poly‐L‐lysine (PLL)‐coated MNPs (PLL‐MNPs) and polyethylenimine (PEI)‐coated MNPs (PEI‐MNPs)) (Farrell et al., 2007) to isolate EVs. The size, morphology, yield, and biomarker cargoes of the isolated EVs were characterized. LF‐bis‐MPA‐MNPs displayed excellent performance and efficiency for isolating exosomes from different biofluids, such as CCM and human urine. LF‐bis‐MPA‐MNPs provide a rapid, simple, and high yield method for urinary EV isolation in the laboratory and clinics.
2. MATERIALS AND METHODS
2.1. Materials and reagents
Ferric chloride hexahydrate/iron (III) chloride (FeCl3.6H2O), ferrous sulfate heptahydrate /iron (II) sulfate (FeSO4.7H2O), 28% ammonia solution (NH4OH), Igepal® CO‐520, tetraethyl orthosilicate (TEOS, 98%), (3‐aminopropyl) triethoxysilane (APTES), 2,2‐bis(hydroxymethyl)propionic acid) (bis‐MPA), p‐toluenesulfonic acid monohydrate (p‐TSA), 3‐mercaptopropyl trimethoxysilane (3‐MPS), dimethyl pimelimidate dihydrochloride (DMP), glutaraldehyde (GAD), lead citrate, sodium citrate, sulfuric acid (H2SO4), hydrochloric acid (HCl), sodium hydroxide solution (NaOH), sodium bicarbonate (NaHCO3), ammonium chloride (NH4Cl), ethanol, methanol, cyclohexane, toluene, PLL, PEI, hLF (L4040), Bradford reagent (B6916), and formvar/carbon‐supported copper grids were purchased from Sigma‐Aldrich, USA.
Radioimmunoprecipitation assay (RIPA) buffer, bovine serum albumin (BSA), skimmed milk, tris‐buffered saline (TBS), Tween 20, phosphate‐buffered saline (PBS 10×, pH 7.4), N‐(γ‐maleimidobutyryloxy)succinimide ester (GMBS), DMP, Dulbecco's modified Eagle medium (DMEM), Roswell Park Memorial Institute (RPMI), DMEM/Nutrient Mixture F‐12 (DMEM/F12), fetal bovine serum (FBS), exosome‐depleted FBS, and antibiotic‐antimycotic were obtained from Thermo Fisher Scientific.
A TEI kit (4478359), exosomal miRNA and protein isolation kit (4478545) were purchased from Invitrogen. High Capacity cDNA Reverse Transcription Kit (Applied Biosystem 4374967) was purchased from Thermo Fisher Scientific. Brilliant III Ultra‐Fast SYBR Green qPCR master mix (600882‐51) was purchased from Agilent Technologies Inc., USA. Primers were synthesized by Macrogen Inc., Korea (Table S1).
For Western blot, rabbit monoclonal anti‐CD9 (ab236630), mouse monoclonal anti‐CD63 (ab193349), mouse monoclonal anti‐CD81 (ab79559), mouse monoclonal anti‐GAPDH (ab59164), rabbit polyclonal anti‐GAG (ab100970), mouse monoclonal anti‐Hsp60 (Abcam, ab59457), rabbit monoclonal anti‐ARF6 (ADP ribosylation factor 6) (ab131261), and rabbit monoclonal anti‐GRP78 (ab108615) were used as primary antibodies and polyclonal goat antimouse and antirabbit IgG‐horseradish peroxidase (HRP) were used as secondary antibodies (ab6789 and ab205718) according to the manufacturer's guidelines (Abcam, UK). For immunolabeled TEM, gold 10 nm goat antimouse (IgG) secondary antibody (ab39619) was purchased from Abcam.
All of the chemicals were used directly without any further purification.
2.2. Synthesis of Fe3O4 NPs
For the synthesis of Fe3O4 NPs, approximately 14.02 g FeCl3.6H2O and 8.6 g FeSO4.7H2O (2:1 molar ratio) were dissolved in 600 ml of distilled water, followed by adding 34 ml NH4OH and stirring at 600 rpm at 85°C for 1 h (the pH was adjusted from 9 to 14), which produced a black precipitate of Fe3O4.NPs. The precipitate was collected by using a magnet and washed several times with distilled water until the pH of the solution was neutral. The collected particles were dried at 56°C overnight and ready for the next modification steps.
2.3. Synthesis of PLL‐coated and PEI‐coated MNPs
Approximately 100 mg Fe3O4 was added to 100 ml of water, sonicated for 10 min, and the water discarded. Then, a solution containing PLL or PEI in 100 ml of water (0.9 mg/ml) was mixed with Fe3O4, and the mixture was sonicated overnight. Next, the mixture was stirred at 900 rpm for 1 h and centrifuged. After the supernatant was discarded, the modified PLL‐MNPs were dispersed in 100 ml of water and separated into 10 tubes (10 mg of particles in 1 ml per reaction).
2.4. Synthesis of the LF‐bis‐MPA‐MNPs
For modification of the Fe3O4 NPs with bis‐MPA, homofunctional bis‐MPA dendrimer‐coated MNPs were synthesized via combined inorganic sol–gel reaction and polycondensation. Briefly, Fe3O4 NPs, cyclohexane, Igepal® CO‐520, and ammonia solution (28%) were stirred in a flask at 1000 rpm for 30 min. TEOS (98%) was added dropwise to the reaction mixture, which was then stirred overnight. Next, APTES (99%) was added dropwise to the mixture, which was then stirred overnight (Li et al., 2011). Fe3O4‐NH2 NPs were washed with methanol and dried in a vacuum desiccator for 1 h. The Fe3O4‐NH2 NPs, bis‐ MPA, and p‐TSA were mixed and agitated at 200°C for 1 h (Fu et al., 2013). The mixture was cooled to room temperature and ultrasonically dispersed in methanol for 30 min. The MNPs were centrifuged, washed, and redispersed in methanol.
LF was immobilized on the thiol‐modified surfaces of the MNPs by attaching it with the heterobifunctional crosslinker GMBS (Dao et al., 2021). Hydroxyl groups on the surface of the MNPs were converted to thiol groups by using a silanization solution of 4% (v/v) 3‐MPS overnight at room temperature, followed by washing with ethanol and incubation with 1 mM GMBS solution at room temperature for 1 h. LF was attached to the GMBS‐immobilized surface at room temperature for 1 h. The modified MNPs were immediately used or stored at 4°C.
2.5. Optimization of the conditions for EV isolation
2.5.1. Optimization of the amount of LF
One hundred milligrams of Fe3O4‐bis‐MPA was added to 4% 3‐MPS (100 ml in ethanol) in 1 h at 1000 rpm. Precipitates were washed with ethanol several times and added to 1 mM GMBS (in 100 ml ethanol at 1000 rpm). The particles were washed with water several times and dried at 65°C for 3 h. The dried particles were combined with various concentrations of LF (1.0, 2.5, and 10 mg in 100 ml of water) 1 h at 1000 rpm. Then, the particles were washed with water, resuspended in 10 ml of water and separated into aliquots of 5 mg NPs per reaction tube. One milliliter of CCM was mixed with LF‐bis‐MPA‐MNPs at three different amounts using a rotator for 30 min. After the reaction, a magnetic stand was used to separate the CCM and MNPs. Reaction tubes were washed with PBS, and EVs were eluted in 200 μl of elution buffer (10 mM NaHCO3 pH ∼10.6). The miRNAs from eluted EVs were extracted and analyzed by RT‐PCR.
2.5.2. Optimization of the amount of MNPs
Dried particles, prepared as described above (Section 2.5.1), were added to 2.5 mg LF (100 ml in water) in 1 h at 1000 rpm. Then, the particles were washed with water and resuspended in 10 ml of water. The LF‐bis‐MPA‐MNPs were diluted to 5, 10, 20, and 30 mg for each reaction. One milliliter of CCM was mixed with LF‐bis‐MPA‐MNPs using a rotator for 30 min. Subsequent steps were the same as described above (Section 2.5.1).
2.5.3. Optimization of the isolation time
Dried particles, prepared as described above (Section 2.5.1), were added to 2.5 mg LF (100 ml in water) in 1 h at 1000 rpm. Then, the particles were washed with water, resuspended in 10 ml of water, and separated into five tubes (10 mg LF‐bis‐MPA‐MNPs/tube). One milliliter of CCM was mixed with LF‐bis‐MPA‐MNPs using a rotator for 5, 10, 15, and 30 min. Subsequent steps were the same as described above (Section 2.5.1).
2.6. Characterization of LF‐bis‐MPA‐MNPS
The morphology and size distribution of the different types of NPs (synthesized Fe3O4 MNPs, bis‐MPA‐MNPs, and LF‐bis‐MPA‐MNPs) were analyzed by using scanning electron microscopy (SEM; Hitachi S‐4700, Japan) and the ImageJ software, respectively. Approximately 1‐ml solutions of MNPs and LF‐bis‐MPA‐MNPs were prepared. Approximately 20 μl of MNPs and LF‐bis‐MPA‐MNP suspensions were dropped onto a silicon substrate, and the surfaces were dried in a fume hood at room temperature overnight. After Pt coating, the dried suspensions were placed in the SEM analysis chamber for imaging. Finally, the size distribution was analyzed with ImageJ by randomly measuring the size of 200 particles.
The zeta potentials of pure LF, synthesized Fe3O4 MNPs, and LF‐bis‐MPA‐MNPs were measured using a Brookhaven NanoBrook 90 Plus zeta potential analyzer (Brookhaven Instruments Corp., Holtsville, NY, USA). Approximately 1‐ml solutions of MNPs and LF‐bis‐MPA‐MNPs were prepared in distilled water (pH 7.0). The solutions were diluted at the ratio of 1:400 with water before analysis by the zeta potential analyzer at 25°C. All measurements were performed in triplicate.
Functional groups of pure LF, synthesized MNPs, and LF‐bis‐MPA‐MNPs were identified by using Fourier‐transform infrared (FTIR; Bruker Vertex 70 FTIR spectrometer, Germany) and an ultraviolet‐visible (UV‐Vis) spectrophotometer (Varian Cary 100 UV‐Vis).
2.7. The specimens for EV isolation
Cells from the HCT116 (colon), AGS (stomach), and HepG2 (liver) cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and maintained in DMEM, while those from the SNU449 liver cancer cell line (ATCC) were maintained in RPMI. Cells from the CCD‐18Co (colon), HepaRG (liver), and AMLF12 (liver) noncancerous cell lines (ATCC) were maintained in DMEM and DMEM/F12. All media for cell lines contained 1% antibiotic‐antimycotic and 10% exosome‐depleted FBS or vesicle‐depleted FBS prepared by centrifuging FBS at 100,000 × g overnight. Cells were cultured in a humidified incubator at 37°C under 5% CO2. CCM were collected after 72 h or 80% confluence (∼10 ml) and immediately used for experiments or stored at ‐20°C for up to 4 weeks. For clinical analysis, urine samples were collected from 20 patients with PCa, 10 patients with benign prostatic hyperplasia (BPH), and 10 healthy controls from the Asan Medical Center (AMC), Seoul, South Korea (Table S2). The use of patient information in this study was approved by the AMC (IRB no. 2019‐1312).
2.8. EV isolation methods
UC, TEI, DTS chip, PLL‐MNPs, and PEI‐MNPs were used for a comparative study with the LF‐bis‐MPA‐MNPs. The workflows of exosome isolation from CCM samples using UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs are reported in Table S3.
For ultracentrifugation, ∼1–40 ml CCM was centrifuged at 300 × g at 4°C for 10 min; 2000 G for 20 min; and 10000 G for 30 min and then ultracentrifuged at 110,000 × g (SW28 rotors, Beckman Coulter, Brea, CA and PA rotors, Hitachi Ultracentrifugation, Japan) for 70 min. After discarding the supernatant, the pellets were washed with PBS and ultracentrifuged at 110,000 × g for 70 min. The pellets that contain EVs were resuspended in 200 μl of 1× PBS before RNA extraction and EV analysis. UC can be used for various body fluids (blood, urine, saliva, etc.) but requires a large amount of starting material (Patel et al., 2019).
For exosome isolation using the TEI kit, CCM volumes (1, 5, 10, and 40 ml) were centrifuged at 300 × g for 10 min and then 2000 G for 20 min before TEI reagent volumes (0.5, 2.5, 5.0, and 20 ml) were added, followed by vortexing to obtain a homogenous solution. The mixture was incubated at 4°C overnight followed by centrifugation at 10,000 × g for 1 h at 4°C. The supernatant was discarded, and the pellets were resuspended in 200 μl of 1X PBS. The collected EVs were immediately used for experiments or kept at ‐20°C for long‐term storage. The TEI kit can be used for various body fluids and with starting volumes as low as 200 μl (Patel et al., 2019).
The DTS chip developed by our research group for bacterial isolation and nucleic acid extraction was used for the microfluidic‐based DTS chip experiment (Shin et al., 2015). The chip surface was immobilized with hydroxyl groups by using oxygen plasma. DMP was used as a crosslinker between the chip surface and the EVs. Mixtures of DMP, LF, and CCM (1–40 ml) were stirred at 60 rpm for 10 min and flowed onto the DTS chip at a flow rate of 400 μl/min. After washing, the isolated EVs were eluted with elution buffer (200 μl of NaHCO3, pH 10.6). The DTS chip can be used for various body fluids and with small starting volumes.
For exosome isolation by PLL‐MNPs and PEI‐MNPs, 1–40 ml CCM was mixed with PLL‐MNPs or PEI‐MNPs (∼10 mg per reaction) by rotating for 30 min. The CCM was separated with PLL‐MNPs or PEI‐MNPs using a magnetic stand, and the solution was discarded. The particles were washed several times with PBS. Approximately 200 μl of elution buffer (10 mM NaHCO3 pH ∼10.6) or RIPA buffer was added to collect the pure EVs or EV proteins for EV characterization.
For exosome isolation by using the LF‐bis‐MPA‐MNPs, CCM (1–40 ml) was collected and either used immediately or stored at ‐20°C for up to 4 weeks. The CCM was thoroughly mixed with LF‐bis‐MPA‐MNPs (approximately 10 mg per reaction) with a rotator for 30 min, after which the reaction tube was briefly centrifuged. The CCM and MNPs were separated by using a magnet stand, while PBS was used for washing steps and then discarded, after which 200 μl of elution buffer (10 mM NaHCO3 pH ∼10.6) or 50–200 μl of RIPA buffer was added and incubated for 3 min. The eluted exosomes were collected in a fresh tube and immediately used or stored at ‐20°C. LF‐bis‐MPA‐MNPs, as well as PLL‐MNPs or PEI‐MNPs, can be used for various body fluids. In this study, the minimum sample volume was 1 ml, but smaller starting volumes are feasible due to the benefit of magnetic separation (Oksvold et al., 2015).
2.9. EV characterization
The morphology of the isolated EVs was characterized by using SEM/transmission electron microscopy (TEM), the concentration was analyzed by using NP tracking analysis (NTA), the surface charges were determined by using a zeta potential analyzer (Malvern Instruments, UK), the protein constituents were identified by using western blotting, and the miRNA content was analyzed by using reverse transcription‐quantitative real‐time polymerase chain reaction (RT‐qPCR). The purity of the isolated EVs was determined as the ratio of the nanovesicle count (via NTA) to the protein concentration (via a Bradford assay) (Webber & Clayton, 2013).
2.9.1. SEM and TEM
EVs isolated by LF‐bis‐MPA‐MNPs were fixed in 4% paraformaldehyde solution. Subsequently, the samples were diluted in distilled water, pipetted onto silicon chips, and dried under a ventilation hood, after which a coating of Pt was applied by sputtering. Images were captured using a Hitachi S‐4700 scanning electron microscope (Japan).
The immunogold staining antibodies of specific proteins were used to identify exosomal proteins and verify the appearance of exosomal surface proteins likely to interact with LF. In this study, CD63 (exosome marker) (Andreu & Yáñez‐Mó, 2014), and GAPDH (LF receptor) (Rawat et al., 2012) markers were confirmed by TEM. Briefly, isolated exosomes were dropped onto the formvar/carbon‐supported copper grids and incubated at 37°C for 30 min. Then, the grids were blocked with 5% BSA for 20 min and washed. For immunogold labeling of CD63 and GAPDH, the grids were incubated with anti‐CD63 monoclonal antibody and anti‐GAPDH monoclonal antibody at 4°C overnight (1:25 dilution ratio). Next, the grids were washed and incubated with antimouse IgG conjugated to 10‐nm gold particles (1:100 dilution ratio) and streptavidin conjugated to 10‐nm gold particles) for 2 h. The grids were washed and fixed in 2.5% GAD for 5 min before contrasting with 3% lead citrate solution for 5 min. Images were acquired by TEM (JEM‐F200 transmission electron microscope).
2.9.2. NTA and zeta potential
For NTA analysis, isolated exosomes were diluted at the ratio of 1:100, 1:200, or 1:400 in PBS. The size distribution and concentration of isolated exosomes were carried out via NTA using a Nanosight NS300 (Malvern Instruments, UK); the camera level was set at 9 and the detection threshold at 11. Filtered PBS without NPs was used as the background. Three video recordings with duration per 60 s were taken.
Surface charges were assessed by zeta potential measurements, acquired using a Zetasizer Lab (Malvern Instruments, UK). Before the measurements, exosomes isolated by UC and TEI (the pellets after centrifugation) were added to 1 ml of distilled water, while exosomes isolated by DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs in 200 μl of elution buffer were completed to 1 ml by adding 800 μl of distilled water. All EV samples from different isolation methods were diluted in distilled water (pH 7.0).
2.9.3. Western blotting
Western blot analysis of specific proteins was used to confirm the purity of the isolated EVs: CD63, CD9, and CD81 (belongs to the tetraspanin family and is an exosome‐specific protein) (Andreu & Yáñez‐Mó, 2014), GAPDH (LF receptor on exosome surface) (Rawat et al., 2012), GAGs, and proteoglycans on cancerous cell surfaces has shown the interaction with LF (El Yazidi‐Belkoura et al., 2001), Hsp60 (a chaperonin promising for CRC diagnosis) (Campanella et al., 2015), ARF‐6 (ADP‐ribosylation factor 6, which is microvesicle protein marker) (Akers et al., 2013), and GRP78 (a member of the heat shock protein family of molecular chaperons associated with the endoplasmic reticulum and cellular stress and found in apoptotic bodies) (Gennebäck et al., 2013).
The western blot protocol is described as follows. Cells and isolated EVs by different methods were lysed by RIPA buffer on ice in 5 min. The protein concentration was determined by serially diluting BSA as the standard for the Bradford assay. The same amount of protein (10 μg) was loaded onto a 10% sodium dodecyl sulfate (SDS)‐polyacrylamide gel electrophoresis (PAGE) gel. Afterward, the protein was transferred to a polyvinylidene difluoride (PVDF) membrane via a Bio‐Rad Mini Transblot electrophoretic transfer cell by following the manufacturer's guidelines. The blot was blocked in 5% skimmed milk in TBS containing 0.1% (v/v) Tween 20 (TBS‐T). Rabbit monoclonal anti‐CD9, mouse monoclonal anti‐CD63, mouse monoclonal anti‐CD81, mouse monoclonal anti‐GAPDH, rabbit polyclonal anti‐GAG, mouse monoclonal anti‐Hsp60, rabbit monoclonal anti‐ARF6, and rabbit monoclonal anti‐GRP78 were used as primary antibodies according to the manufacturer's guidelines. Polyclonal goat antimouse and antirabbit IgG‐horseradish peroxidase (HRP) secondary antibodies were added for 1 h. After washing with TBS‐T, spots were detected by using a mixture of peroxidase and chemiluminescent substrates (1:1). Chemo‐luminescence signals were detected with a molecular imager (Bio‐Rad).
2.9.4. Exosomal miRNA extraction
The non‐EV‐associated miRNA was tested after EV isolation by LF‐bis‐MPA‐MNPs. We analyzed the miRNA‐21 amount from the non‐EV‐associated miRNA by directly taking eluted EV (200 μl) into RT‐qPCR without RNA extraction. For exosomal miRNA isolation, miRNAs were collected from isolated exosomes by using a total exosome RNA and protein isolation kit. Briefly, 200 μl of sample in PBS or elution buffer was thoroughly mixed with 200 μl of denaturing solution (preheated at 37°C) and incubated on ice for 5 min. After incubation, 400 μl of acid‐phenol:chloroform was added to the mixture and vortexed for 60 s. The mixture was centrifuged for 5 min at 13,000 × g at room temperate to separate it into aqueous and organic phases. Afterward, the aqueous phase was moved to a fresh tube. A total amount of 100% ethanol (1/3 volume) was added to the aqueous phase and vortexed for 15 s. Approximately 700 μl of volume was placed onto a filter cartridge in a collection tube each time and then spun at 13,000 × g for 30 s to move the sample through the filter cartridge. A total amount of 100% ethanol (1/3 volume) was added to the flow‐through containing small RNA and vortexed for 15 s. Approximately 700 μl of volume was placed onto the filter cartridge in a collection tube and then spun at 13,000 × g for 30 s. Samples were washed once with 700 μl wash solution 1 and twice with 500 μl wash solutions 2 and 3. The filter cartridge was dried by spinning for 1 min at 13,000 G and then placed in a fresh collection tube, after which 50 μl of preheated (95°C) nuclease‐free water was applied to the filter. Samples were centrifuged for 30 s at 13,000 × g to recover the RNA, which was repeated once. The eluate containing small RNA (100 μl) was collected and stored at −20°C.
2.9.5. RT‐qPCR
For RT‐qPCR, an RT Master Mix was prepared for each RNA sample by using the High Capacity cDNA Reverse Transcription Kit and protocol (Applied Biosystems) using unique stem‐loop RT primers for various miRNA targets (mi21, mi31, miR‐23a, miR‐135b, hsa‐let7c, and U6). Approximately 12 μl of RT Master Mix was added to PCR tubes, to which 3 μl of each RNA sample was added. The tubes were spun down to remove air bubbles and incubated on ice for 5 min, followed by placing in a thermocycler and reacted as follows: 4°C for 5 min, 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. cDNA products were used immediately for qPCR performed according to the following standard protocol: 5 μl of cDNA was amplified in a 20 μl mixture volume of 10 μl Brilliant III Ultra‐Fast SYBR Green qPCR master mix (Agilent Technologies Inc., USA), 25 pmol of each primer, and deionized water. Plates were spun down at a fast speed to remove air bubbles and the PCR was run by using a preincubation cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s, and a cooling step of 40°C for 30 s. The cycle threshold (Ct) value was obtained via the Bio‐Rad CFX Maestro software. The average and standard deviation of the Ct values (performed in triplicated) were calculated. All of the primer sequences are present in Table S1. U6 sRNA was used as the internal reference to normalize the expression levels of the target genes (Brow & Guthrie, 1988; Duan et al., 2018). The ratio of the Ct values of the miRNAs and U6 were calculated as inversely proportional to the relative quantity of the miRNAs. The relative expression (RQ) of miRNA was analyzed by using the ΔCt and 2‐ΔΔCT methods (Han et al., 2015).
ΔCt is calculated as
| (1) |
which shows the difference between the Ct values of the miRNAs and U6.
ΔΔCt was calculated as
| (2) |
RQ = 2–ΔΔCT represents the value of the ratio of miRNA expression in cancer cell/clinical samples to that in noncancerous cells/clinical samples.
2.9.6. Cytotoxicity test
To assess the quality of the EVs for functional applications, ∼200 μl of isolated EVs from HCT116 CCM and HepG2 using the LF‐bis‐MPA‐MNPs were added to CCD‐18Co and HepaRG cell cultures in a 96‐well plate. CCD‐18Co and HepaRG cells were cultured in a 96‐well plate without EV addition as the control group. The cell densities for both the experimental and control groups were 104 cells/100 μl culture media. Cell proliferation and expression of miRNA were measured after 3 days of culturing.
Approximately 200 μl of isolated EVs from HCT116 CCM (via the LF‐bis‐MPA‐MNPs) were added to the CCD‐18Co cell culture in a 96‐well plate and incubated at 37°C for 72 h under 5% CO2. The densities of the EVs were equivalent to 10–100 μg/ml of protein components found in the EVs. In addition, CCD‐18Co cells were cultured in a 96‐well plate without EV addition as the control group. For both groups, the cell density was 104 cells in 100 μl of CCM. Cell proliferation was measured after 3 days of culturing by adding WST‐1 cell proliferation reagent (10 μl per well) and incubating at 37°C for 4 h under 5% CO2. After incubation, the absorbance of the formazan product was read at 460 nm, with a reference wavelength of 600 nm.
2.10. Statistical analysis
Differences between groups and method comparison were analyzed using two‐tailed Student's t‐tests. p‐Value ≤ 0.05 was considered to be significant. Error bars in the graphs represent the standard deviation of the mean for triplicate experiments. Analyses and graphs were performed by using GraphPad Prism 8 (GraphPad Software, La Jolla, California, USA).
3. RESULTS AND DISCUSSION
3.1. Characterization of the LF‐bis‐MPA‐MNPs
Scheme 1 shows a schematic illustration of the application of MNPs for EV isolation and detection, along with the synthesis and modification of LF‐bis‐MPA‐MNPs. Figure 1a shows SEM images that confirmed uniform size distribution and shape of the pure synthesized Fe3O4 MNPs and LF‐bis‐MPA‐MNPs. The average sizes of the pure MNPs and LF‐bis‐MPA‐MNPs were around 54.1 ± 9.1 and 66.5 ± 17 nm, respectively (Figure 1b), which were uniformly distributed. Figure 1c exhibits the zeta potentials of commercial Fe3O4 MNPs, synthesized Fe3O4 MNPs, bis‐MPA‐MNPs, and LF‐bis‐MPA‐MNPs. The commercial Fe3O4 MNPs, synthesized Fe3O4 MNPs, and bis‐MPA‐MNPs without an LF coating on the surface had negative charge values of –54.5 ± 1.27, –32.03 ± 1.31, and –16.2 ± 1.45 mV, respectively. The zeta potential of the bis‐MPA‐MNPs dispersed in deionized water was negative, which indicates the existence of carboxyl groups on their surfaces (Carlmark et al., 2013), whereas that of the LF‐bis‐MPA‐MNPs was positive (20.97 ± 0.83 mV) due to the LF conjugation.
SCHEME 1.
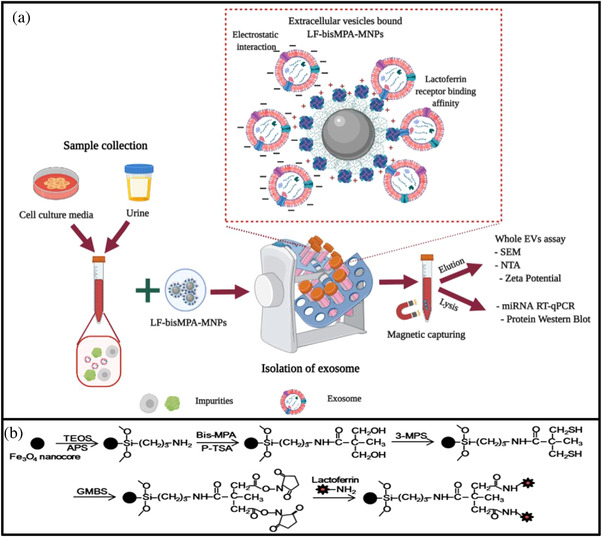
A schematic illustration and the mechanism of EV isolation by LF‐bis‐MPA‐MNPs. (a) Schematic diagram of the LF‐bis‐MPA‐MNPs for EV isolation and detection in CCM and human urine (created with BioRender.com). (b) Schematic diagram of synthesis and modification of the LF‐bis‐MPA‐MNPs
FIGURE 1.
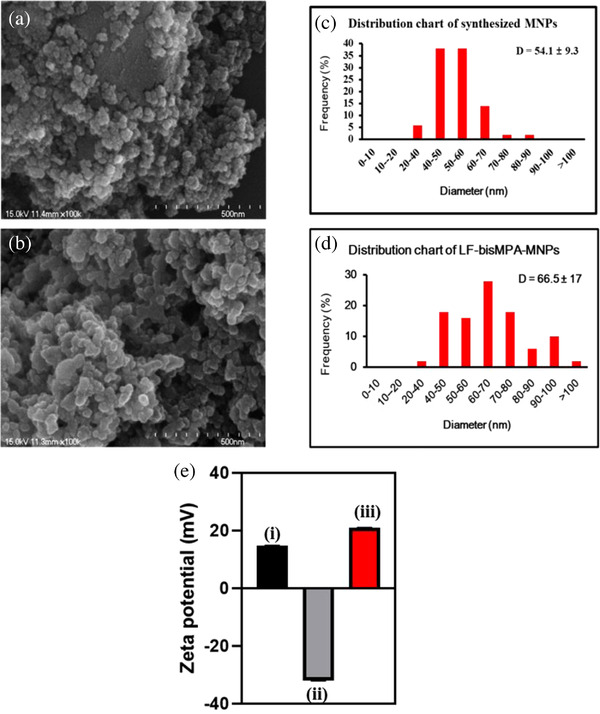
Characterization of the LF‐bis‐MPA‐MNPs. SEM images showed the uniform size distribution of Fe3O4 NPs (a) and LF‐bis‐MPA‐MNPs (b). Size distribution of Fe3O4 NPs and LF‐bis‐MPA‐MNPs were analyzed by ImageJ by measuring the sizes of 200 particles (c and d). Zeta potential analysis (E) showed the positive charge of LF (i), negative charge of Fe3O4 NPs (ii), and positive charge of LF‐bis‐MPA‐MNPs (iii)
FTIR spectroscopy was used to identify the chemical bonding and molecular structure of the materials. Figure 2a shows FTIR spectra of the as‐synthesized Fe3O4 NPs, LF, and LF‐bis‐MPA‐MNPs. Characteristic absorption peaks of Fe‐O were observed at 552.92 and 549.62 cm–1 in the FTIR spectra for the as‐synthesized Fe3O4 NPs and LF. Moreover, the FTIR spectrum of LF showed absorption bands associated with histidine, lysine, phenylalanine, tyrosine, aspartic acid, tryptophan, serine, and threonine (Duca et al., 2018). FTIR spectra of the as‐synthesized Fe3O4 NPs, LF, and LF‐bis‐MPA‐MNPs are shown in Figure S1A–C. The stretch vibration of histidine at 1638.82 cm–1 overlapped with the absorption band of amide I. The strong absorption band near 1450.49 cm–1 indicates aspartic acid and the band at 1399.65 cm–1 indicates the carboxyl group of aspartic acid. The presence of tyrosine was shown by the peak at 1243.37 cm–1, while the absorption peak at 1050.88 cm–1 indicated N≡C or C═C and C—H deformation vibrations in the tryptophan side‐chains that are linked to LF. The peaks indicating pure LF were observed at 1636.03, 1449.38, and 1404.96 cm–1 after LF had been conjugated on MNPs. The FTIR spectrum for LF shows the ν N‐H stretching frequency (3282.85 cm–1), while the characteristic absorption band at 3255.25 cm–1 was attributed to the stretching frequency of NH2 in the LF‐bis‐MPA‐MNPs. These results indicate that LF was successfully attached to the bis‐MPA‐MNPs by cross‐linking.
FIGURE 2.
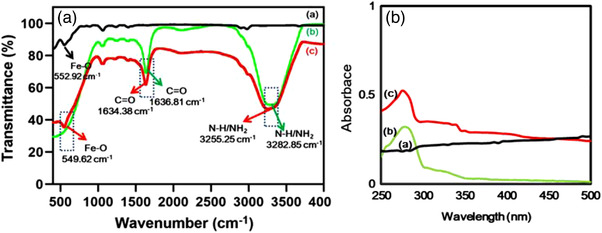
Chemical bonding and molecular structure analysis via FTIR and UV‐Vis. (a) FTIR spectra of the as‐synthesized MNPs (curve a), pure LF (curve b), and LF‐bis‐MPA‐MNPs (curve c). The major differences between as‐synthesized MNPs (curve a) and LF‐bis‐MPA‐MNPs (curve c) are shown by the peaks at 3255.25 and 1634.38 cm–1, which indicate the stretching frequencies of NH2 and amide group, respectively. (b) UV‐Vis spectra of the as‐synthesized MNPs (curve a), pure LF (curve b), and LF‐bis‐MPA‐MNPs (curve c)
The characterization of the synthesized Fe3O4 MNPs by UV‐Vis spectroscopy is shown in Figure 2b. In our synthesis protocol, we used FeSO4 (Fe2+) and FeCl3 (Fe3+) to form the MNPs (so‐called Fe3O4 NPs). A UV‐Vis spectrum of core Fe3O4 NPs showed no characteristic absorbance peaks (Figure 2b, curve a). It was similar to some other published papers that found no peaks in the UV‐Vis spectrum of Fe3O4 NPs (Vollath, 2013). Meanwhile, LF showed a peak in the UV region at the wavelength of 280 nm (Figure 2b, curve b), in agreement with other reported studies (Sun et al., 2018). The LF‐bis‐MPA‐MNPs showed a peak at the wavelength of 275 nm (Figure 2b, curve c), which indicated that LF was successfully coated on Fe3O4 NPs. However, after the core Fe3O4 NPs were modified and coated with bis‐MPA and LF, the absorbance was shifted to shorter wavelengths with increasing particle size (Vollath, 2013). Thus, we observed the shift to a shorter wavelength (blue‐shift).
3.2. Characterization of the isolated EVs
EVs from HCT116 CCM were isolated by using UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs (Table S3). Overall, EV isolation by LF‐bis‐MPA‐MNPs took less time (∼30 min for the binding reaction) than UC (∼130 min for multiple steps of centrifugation), TEI (overnight reaction), and the DTS chip (∼40 min) and the same reaction time with PLL‐MNPs and PEI‐MNPs (30 min).
The concentration of LF for coating the MNPs and the concentration of the LF‐bis‐MPA‐MNPs for EV isolation were optimized (Figure S2). Briefly, we optimized the LF concentration, the amount of LF‐bis‐MPA‐MNPs, and the incubation time based on the RT‐qPCR results of exosomal miR‐21 extracted from exosomes isolated from various conditions during the synthesis of LF‐bis‐MPA‐MNPs. To optimize the LF concentration, various concentrations of LF (1.0, 2.5, and 10 mg in 100 ml of distilled water) were tested during the synthesis of LF‐bis‐MPA‐MNPs. We randomly used 5 mg of LF‐bis‐MPA‐MNPs for exosome isolation from 1 ml of CCM (see Section 2). At this point, the LF concentration was chosen as 2.5 mg in 100 ml of distilled water (Figure S2‐A). Compared to UC, the LF‐bis‐MPA‐MNPs synthesized from 2.5 and 10 mg LF in 100 ml of distilled water showed similar Ct values. However, there was no significant difference between 2.5 and 10 mg LF. Thus, the LF concentration was chosen as 2.5 mg in 100 ml of distilled water. To optimize the amount of LF‐bis‐MPA‐MNPs, we tested various amounts of synthesized LF‐MNPs (5, 10, 20, and 30 mg of LF‐bis‐MPA‐MNPs for exosome isolation from 1 ml of CCM). The amount of LF was chosen as 2.5 mg per 100 ml of distilled water (Figure S2‐A), and the amount of LF‐bis‐MPA‐MNPs was chosen as 10 mg (Figure S2‐B). The Ct value of exosomal miR‐21 extracted from exosomes isolated by LF‐bis‐MPA‐MNPs (10 mg) was lower than that of exosome miR‐21 extracted from exosomes isolated by UC. Thus, we chose 10 mg LF‐bis‐MPA‐MNPs as the optimal amount. To optimize the isolation time, exosomes were isolated by mixing LF‐bis‐MPA‐MNPs and 1 ml of CCM for 5, 10, 15, and 30 min. The Ct value of exosomal miR‐21 extracted from exosomes isolated by LF‐bis‐MPA‐MNPs during 30 min of isolation was lower than the Ct value of exosome miR‐21 extracted from exosomes isolated by LF‐bis‐MPA‐MNPs during 5, 10, and 15 min of isolation and the UC control. Thus, 30 min was the optimal isolation time. Finally, the optimized amount of LF was 2.5 mg per 100 ml of distilled water, the optimized amount of LF‐bis‐MPA‐MNPs was 10 mg per reaction, and the isolation time was set at 30 min (Figure S2A–C).
SEM images in Figure 3a show the binding of EVs on the surface of the LF‐bis‐MPA‐MNPs as well as round‐shaped EVs in solution with a diameter range from 30–170 nm (Figure 3b). The average diameters of the EVs isolated by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs were 163.1 ± 27.7, 165.9 ± 32.5, 167.6 ± 56.6, 180.5 ± 0.71, 177.3 ± 2.08, and 164.5 ± 21.9, respectively (Figure 3c and Figure S3 and Table S4). The NTA analysis showed that the average concentration (particles/mL) of EVs isolated by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs was (5.19 ± 0.36) × 1010, (5.95 ± 0.7) × 1010, (0.61 ± 0.13) × 1010, (2.98 ± 0.17) × 1010, (2.6 ± 0.26) × 1010, and (7.3 ± 2.37) × 1010, respectively ( Table S4). The average concentration of EVs isolated by LF‐bis‐MPA‐MNPs was 1.42 ± 0.41, 1.37 ± 0.64, 12.36 ± 4.11, 2.47 ± 0.7, and 2.8 ± 0.84 fold higher than that of the EVs isolated by UC, TEI, DTS chip, PLL‐MNPs, and PEI‐MNPs, respectively. However, exosome aggregation might affect the average size and concentration during NTA measurement.
FIGURE 3.
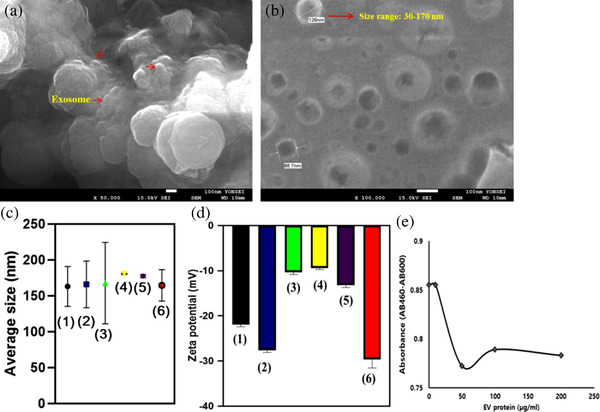
Whole EVs assay. (a and b) SEM image showed the binding of EVs to the surface of LF‐bis‐MPA‐MNPs, and the round‐shaped vesicles from the eluate were observed with a diameter range from 30 to 170 nm. (c and d) Size profile and zeta potential of EVs isolated from CCM by different methods: (1) UC, (2) TEI, (3) DTS chip, (4) PLL‐MNPs, (5) PEI‐MNPs, and (6) LF‐bis‐MPA‐MNPs. (e) Determination of cell proliferation: WST‐1 was used to determine the number of viable CCD‐18Co cells after treatment with isolated EVs. The EVs densities were equivalent to 10 to 100 μg/ml of the protein component found in EVs
The distribution of charge density around particles causes electrostatic potential differences (Weiner, 2013). The zeta potential of the isolated EVs was measured to evaluate their stability and integrity; those isolated by using UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs had zeta potentials of –21.80 ± 0.51, –27.61 ± 0.46, –10.35 ± 0.52, –9.4 ± 0.32, –13.23 ± 0.49, and –29.64 ± 1.88 mV, respectively (Figure 3d). The isolated EVs from different methods had negative zeta potentials. The zeta potentials of the isolated EVs were in the range of –9.4 to –29.64 mV due to their plasma membrane structure (Beit‐Yannai et al., 2018), thereby suggesting their good stability in solution.
Isolated exosomes have been used for many biological assays, so the cytotoxicity of the isolated EV was also assessed (Patel et al., 2019). We analyzed if cancerous exosomes can affect noncancerous cells. A cytotoxicity assay was conducted to determine the proliferation effect of EVs from cancerous cells on normal human cells. Isolated EVs from the CCM of HCT116 cells were added to CCD‐18Co cells that had been precultured in a 96‐well plate. Cell proliferation was measured after 3 days of culturing (Figure 3e). Following 72 h of incubation, WST‐1 reagent was added to the cell culture medium to determine the number of viable CCD‐18Co cells after treatment with isolated cancerous EVs. The viable cells were analyzed by the cleavage of tetrazolium salts (WST‐1) by cellular enzymes to produce formazan. An increase in formazan is equivalent to an increase in cell number. After WST‐1 incubation, the absorbance of the formazan product was read at 460 nm and the reference wavelength at 600 nm via a microplate reader (PerkinElmer 2030 Multilabel Reader). The value of AB460–AB600 decreased after normal cells were treated with isolated EVs from a cancerous cell line (Table S5). The result indicated that exosomes isolated by LF‐bis‐MPA‐MNPs would contain cytotoxic factors that affect the viability of noncancerous cells (Patel et al., 2019). Next, EVs were characterized through immunolabeled electron microscopy against CD63 and GAPDH for the different methods (UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs). Immunogold labeling of the exosome samples isolated by each isolation method showed round‐shaped particles with the morphology of exosomes and size range of 30–200 nm (Figure 4 and Figure S4). The results verified that exosomes were successfully isolated by the isolation methods. The exosomes were characterized for size range, morphology, and exosome‐specific protein (CD63). Furthermore, GAPDH was analyzed to verify the binding of LF with LF receptors on the exosomal membrane.
FIGURE 4.
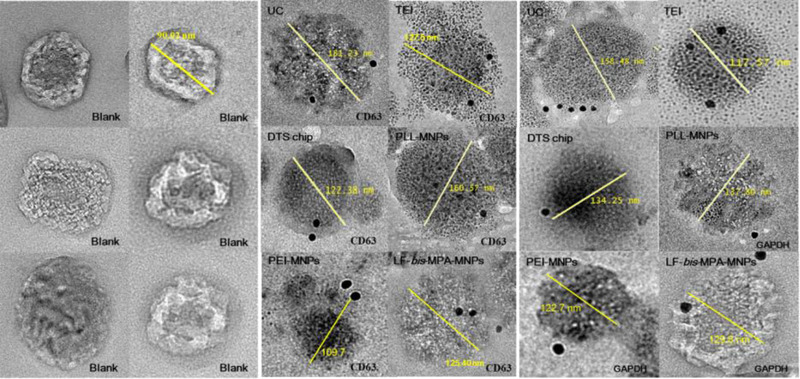
Validation of exosome isolation from colon cancer CCM by immunogold electron microscopy. TEM micrographs of exosomes from six different isolation methods: UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs. Data for six different isolation methods were revealed by CD63‐labeling and GAPDH‐labeling. Blank (no labeling) corresponded to exosomes isolated by LF‐bis‐MPA‐MNPs. Exosome size ranged from 30 to 200 nm
3.3. Proteins from isolated EVs
Exosomes have specific proteins (such as CD9, CD63, and CD81) on their surface that are recognized by specific antibodies (Hikita et al., 2018). We examined the EV purity by immunoblotting for marker proteins specific to EVs and colorectal and hepatic cancer cells. CD63, CD9, and CD81 are specific exosome markers, ARF6 is a microvesicle‐related protein, and Grp78 is an endoplasmic reticulum‐localized protein (indicates apoptotic bodies). Although CD63, CD9, and CD81 were found in the EV protein isolated by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs, the expression of those proteins differed among the isolation methods (Figure 5a). CD63, CD9, and CD81 were detected at high intensities from EVs isolated by UC and TEI. However, the intensities of those proteins were not different from EVs isolated by PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs and very low from EVs isolated by the DTS chip. Although ARF6 was found in exosomal protein isolated by UC (low intensity), TEI, DTS chip, PLL‐MNPs, and PEI‐MNPs, it was not found in EVs isolated by LF‐bis‐MPA‐MNPs. It indicated very few amounts of microvesicles bound to LF‐bis‐MPA‐MNPs. GRP78 was not found in exosomal proteins isolated by six different isolation methods. It indicated that apoptotic bodies were not contained in the EV samples.
FIGURE 5.
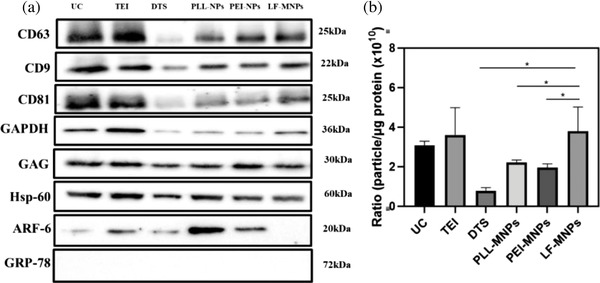
Analysis of exosomal proteins from isolated exosomes by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs (LF‐MNPs). (a) Expression of exosomal protein markers (CD63, CD9, and CD81), proteins indicative of the interaction between exosomes and LF (GAPDH, GAG), cancer‐associated protein marker (Hsp60), microvesicle protein marker (ARF6), and apoptotic bodies protein marker (Grp78) in EVs isolated by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs (LF‐MNPs). (b) Purity of EV determined by the ratio of EV particle concentration (NTA) to protein concentration measured by the Bradford assay. Each bar represents the mean ± standard deviation (n = 3, * p‐value ≤ 0.05)
The exosome is an important vesicle for diagnostic and therapy (Hon et al., 2017). However, the functionality of EVs was strongly affected by their purity (Webber & Clayton, 2013). The International Society of Extracellular Vesicles (ISEV) has issued guidelines for classifying EVs as exosomes (Théry et al., 2018). First, their guidelines include the calculation of the ratio between the number of particles and the protein or lipid content. Second, the amounts of some membrane proteins, cytosolic/periplasmic protein, and proteins that are not contained in exosomes are determined (Théry et al., 2018). Over 3 × 1010 particles/μg of proteins and under 2 × 109 signify high and poor purity of the isolated EVs, respectively (Webber & Clayton, 2013). The ratios of particle/protein from isolated EVs by UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs were 3.07 ± 0.22 × 1010, 3.6 ± 1.38 × 1010, 0.78 ± 0.16 × 1010, 2.21 ± 0.13 × 1010, 1.95 ± 0.19 × 1010, and 3.79 ± 1.23 × 1010, respectively (Figure 5b and Table S4). The results demonstrated that the EVs isolated by UC, TEI, and LF‐bis‐MPA‐MNPs were of high purity, the EVs isolated by the DTS chip were impure, and the EVs isolated by PLL‐MNPs and PEI‐MNPs were of low purity.
3.4. MiRNAs in the isolated EVs
Exosomal miRNAs were extracted from CCM‐derived exosomes of HCT116 cells. LF can bind to RNA, so the free miRNA available to bind to LF‐bis‐MPA‐MNPs was tested. The miRNA‐21 amounts from isolated EV‐associated miRNA and non‐EV‐associated miRNA were compared by RT‐qPCR. The results showed that very little free miRNA was bound to LF‐bis‐MPA‐MNPs (Figure S5), indicating that the miRNA was mostly from EVs.
The RT‐qPCR results allowed to compare the amount of exosomal miRNAs among different isolation methods. Exosomal miRNAs extracted from EVs isolated by LF‐bis‐MPA‐MNPs were compared with those isolated by UC, TEI, DTS chip, PLL‐MNPs, and PEI‐MNPs. MiR‐21 and miR‐31 represent potential markers for the diagnosis and prognosis of colon cancer (Schee et al., 2012; You et al., 2019) and U6 is one of the most common miRNA housekeeping genes (Duan et al., 2018). In this study, Ct values after cDNA synthesis and RT‐qPCR toward targets miR‐21, miR‐31, and U6 were used to compare the isolation methods. At this point, there were no significant differences in the Ct values of miR‐21, miR‐31, and U6 (Figure 6a–c) from exosomes isolated by LF‐bis‐MPA‐MNPs compared to exosomes isolated by UC and TEI. However, the Ct values of exosomal miRNAs extracted from exosomes isolated by the DTS chip, PLL‐MNPs, and PEI‐MNPs were higher than those obtained for exosomal miRNAs extracted from exosomes isolated by UC, TEI, and LF‐bis‐MPA‐MNPs (Figure 6a–c). We assumed the amount of exosomal miRNAs extracted from exosomes isolated by LF‐bis‐MPA‐MNPs was comparable to the amount extracted from exosomes isolated by UC and TEI and higher than the amount extracted from exosomes isolated by the DTS chip, PLL‐MNPs, and PEI‐MNPs. It means the amount of exosomes isolated by LF‐bis‐MPA‐MNPs can be comparable to that isolated by UC and TEI and higher than that isolated by the DTS chip, PLL‐MNPs, and PEI‐MNPs. These results indicated that LF‐bis‐MPA‐MNPs represent a suitable method for EV isolation comparable to UC and TEI and with better efficiency than the DTS chip, PLL‐MNPs, and PEI‐MNPs.
FIGURE 6.
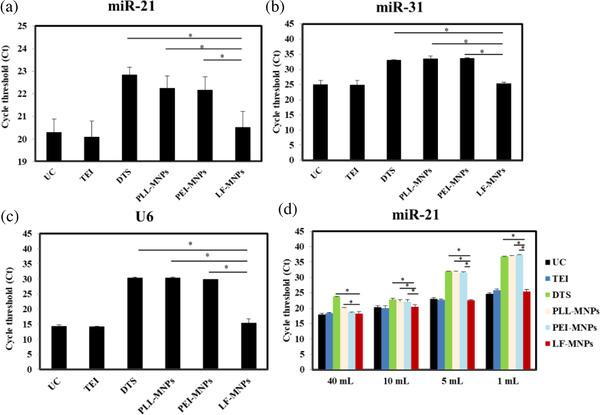
miRNA characterization. (a) Exosomal miR‐21, miR‐31, and U6 released from the HCT116 cell line were examined by RT‐qPCR in EVs isolated from 10 ml of CCM from cancer cell line HCT116 via UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs (LF‐MNPs). (b) Exosomal miR‐21 was examined by RT‐qPCR in EVs isolated from different volumes of CCM (1–40 ml) from cancer cell line HCT116 via UC and LF‐bis‐MPA‐MNPs. Data are presented as the mean ± standard deviation (n = 3, *p‐value ≤ 0.05)
Exosomal miR‐21 was extracted from four different volumes of CCM (1, 5, 10, and 40 ml) using UC, TEI, DTS chip, PLL‐MNPs, PEI‐MNPs, and LF‐bis‐MPA‐MNPs. Figure 6d shows that compared to the DTS chip and PLL‐MNPs, more miR‐21 was obtained from exosomes isolated by LF‐bis‐MPA‐MNPs in a sample volume range of 1–40 ml. Compared to PEI‐MNPs, more miR‐21 was obtained from exosomes isolated by LF‐bis‐MPA‐MNPs in 1, 5, and 10 ml of sample, but the 40 ml sample was not significant. It indicated that LF‐bis‐MPA‐MNPs had higher efficiency in EV isolation than the DTS chip, PLL‐MNPs, and PEI‐MNPs. However, there are no significant differences among LF‐bis‐MPA‐MNPs, UC, and TEI in the amount of miRNA extracted from isolated EV in a sample volume range of 1–40 ml. These results indicated that LF‐bis‐MPA‐MNPs represent a suitable method for EV isolation with rapidity, simplicity, and high efficiency.
Taken together, although the NTA analysis of LF‐bis‐MPA‐MNPs showed that the isolated exosomes were larger than the range of 30–150 nm in diameter, more specific exosomal proteins and miRNAs were discovered by using western blotting and RT‐qPCR. Hence, LF‐bis‐MPA‐MNPs is a rapid and simple EV isolation method with high efficiency and purity comparable with other established methods without the need for cumbersome and expensive devices.
3.5. Verify application of LF‐bis‐MPA‐MNPs in relative expression of cancer‐related EV miRNA
We investigated the relative expression of miRNAs in the exosomes from the CCM of CRC and liver cancer cells. We analyzed the relative amounts of miRNAs that can be used as cancer markers in the isolated EVs by LF‐bis‐MPA‐MNPs as follows:
| (3) |
Next, the relative expression of miR‐21 was calculated via the 2–ΔΔCt method (Han et al., 2015). The results were used to verify that miRNAs from exosomes isolated by LF‐bis‐MPA‐MNPs could be used for comparing the expression of cancerous exosomal miRNAs to noncancerous exosomal miRNAs.
We compared the expression of miR‐21, miR‐135b, miR‐23a, and hsa‐let7c from HCT116 (colon), HepG2 (liver), and SNU449 (liver) cancerous cells lines and noncancerous cell lines CCD‐18Co (control, colon cell) and AML/F12 (control, liver cell). Some studies showed that miR‐21, miR‐23a and miR‐135b are overexpressed in human colorectal and liver cancer (Huang et al., 2018; Necela et al., 2011; You et al., 2019) and let‐7c is downregulated in CRC and liver cancer (Chen et al., 2014; Shimizu et al., 2010). For CRC, the relative expressions of exosomal miR‐21 and miR‐135b from the HCT116 cancerous cells were higher than in the CCD‐18Co normal cells (fold changes ∼2.52 and ∼1.83, respectively) (Figure 7a–d), while the relative expression of exosomal hsa‐let7c was lower (fold change ∼8.3) (Figure 7b). However, the relative expression of miR‐23a between the cancerous and normal cells was not significantly different (Figure 7c). For liver cancer, Figure 7a–d showed that the exosomal miR‐21, miR‐23a, and miR‐135b levels from Hep‐G2 and SNU‐449 cancerous cells were significantly higher than those in the AML/F12 normal cells (fold changes of ∼11.74, ∼5.59, and ∼26.97 in miR‐21, miR‐23a, and miR‐135b from HepG2, respectively, and fold changes of ∼1.67, ∼8.92, and ∼56.36 in miR‐21, miR‐23a, and miR‐135b from SNU‐449, respectively). Moreover, the exosomal hsa‐let7c from HepG2 and SNU‐339 was significantly lower than in AML/F12 (Figure 7c). Exosome isolation by LF‐bis‐MPA‐MNPs was proved to be suitable for miRNA profiles.
FIGURE 7.
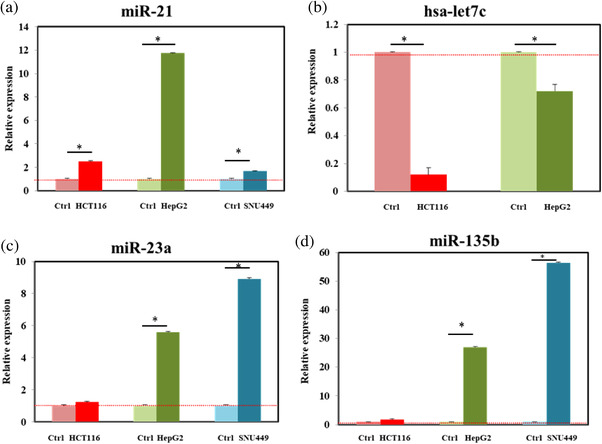
Relative expression of miRNA from isolated CCM EVs by using LF‐bis‐MPA‐MNPs. EVs were isolated from the CCM of colon cancer cell line HCT116 and liver cancer cell lines HepG2 and SNU 449 and noncancerous colon cell line CCD‐18Co (normal control) and noncancerous liver cell AMLF12 (normal control) by using LF‐bis‐MPA‐MNPs. The relative expressions of miR‐21 and miR‐23a were calculated by using the 2(‐ΔΔCt) method. Data were presented as the mean ± standard deviation (n = 3, * p‐value ≤ 0.05)
3.6. PCa diagnosis using isolated urinary EVs
Urine‐based exosomal miRNAs have been studied as an alternative diagnostic tool in prostate disease diagnosis (Erdbrügger et al., 2021). The limitations of previous studies involved the low concentration of miRNAs and the lack of a standard for validation (Paiva et al., 2020). Therefore, we aimed to distinguish urinary EV‐derived miRNAs as markers associated PCa and BPH after EV isolation by using LF‐bis‐MPA‐MNPs. Urine specimens from 20 patients with PCa, 10 patients with BPH, and 10 healthy controls were analyzed. We profiled the expression levels of four EV‐derived miRNAs as a panel of prostate disease diagnosis (miR‐21, miR‐346 miR‐23a, and miR‐122‐5p (Foj et al., 2017; Jackson et al., 2014; Wang et al., 2019; Zhu et al., 2020)) via their relative expression by using RT‐qPCR (Figure 8). The four EV‐derived miRNAs were found to be differentially expressed in both BPH and PCa compared to urine from healthy controls. Based on the results, miRNA‐21 and miRNA‐346 were significantly upregulated (Figure 8a and b) while miRNA‐23a and miRNA‐122‐5p were downregulated (Figure 8c and d) in urinary EVs from BPH and PCa patients compared to healthy controls. These results were in agreement with those from a previous study showing that miRNA‐21 is an oncogenic miRNA in the urinary EVs of PCa patients compared to healthy controls (Bautista‐Sánchez et al., 2020). To further investigate distinguishing the miRNA expression levels of PCa and BPH, we compared the relative expression levels of four miRNAs in the two groups of patients. However, no significant difference was found in the levels of four miRNAs between BPH and PCa groups. There are some factors could affect to the results compare to previous publishes. One of them is the choice of reference genes, which is critical for miRNA expression. U6 has been used as a reference gene in urinary miRNA in this study. Moreover, size of samples and sample collection methods could affect to the results. Therefore, a panel of miRNAs as a potential diagnostic tool is still needed to investigate for a standard validation in the further studies.
FIGURE 8.
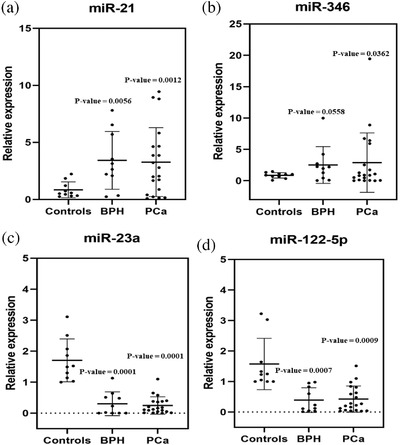
Relative expression of miR‐21, miR‐346, miR‐23a, and miR‐122‐5p from EVs from BPH, PCa, and normal urine samples isolated by using LF‐bis‐MPA‐MNPs. (a) The relative expressions of miR‐21 from BPH and PCa urine samples were upregulated compared to the control group (p ≤ 0.05). (b) The relative expressions of miR‐346 from BPH and PCa urine samples were upregulated compared to the control group (p ≤ 0.05). (c) The relative expression of miR‐23a from BPH and PCa urine samples was downregulated compared to the control group (p ≤ 0.01). (d) The relative expression of miR‐122‐5p from BPH and PCa urine samples was downregulated compared to the control group (p ≤ 0.01) (n = 10 for BPH and n = 20 for PCa and n = 10 for healthy controls). The data were compared by using two‐tailed t‐tests
4. CONCLUSIONS
Some issues remain for EV isolation for clinical applications: (1) how to create a novel method for exosome isolation that is sufficiently rapid and sensitive for clinical applications and (2) how to distinguish exosomes in EV pools including microvesicles. To solve issue (1), we report a rapid and simple EV isolation method using LF‐bis‐MPA‐MNPs as chimeric nanocomposites employing electrostatic interaction, physical absorption, and receptor and ligand biorecognition that overcomes the drawbacks of existing methods, such as complex steps, time‐consuming, requiring expensive instruments, and low accuracy. The simplified approach with LF‐bis‐MPA‐MNPs can be used to isolate EVs from multiple samples in CCM and clinical specimens within 30 min. The LF‐bis‐MPA‐MNPs were synthesized via a one‐pot approach (Scheme 1). Due to strong binding between the LF and EVs, LF‐bis‐MPA‐MNPs comprise a fast and efficient method for EV isolation. The LF‐bis‐MPA‐MNPs exhibited high EV capture efficiency and purity while maintaining their biological functions, as was verified by SEM, zeta potential analysis, NTA, cytotoxicity testing, western blots, and RT‐qPCR. The LF‐bis‐MPA‐MNPs present a centrifuge‐free, antibody‐free method that can be manually or automatically carried out over a short period of time (a comparison of EV isolation techniques is shown in Table S6).
Despite the advantages of LF‐bis‐MPA‐MNPs for EV isolation, our study has some limitations. First, the specificity of the LF‐bis‐MPA‐MNPs for exosome isolation cannot perfectly distinguish them from other types of EVs. Thus, we are focusing on EV isolation including all types of EVs using the LF‐bis‐MPA‐MNPs. Nevertheless, the isolated EV excluded apoptotic bodies according to the western blot results (Grp78). Second, we validated the clinical ability of the LF‐bis‐MPA‐MNPs with 40 urine samples of BPH and PCa patients and healthy controls. The expression levels of the urinary exosomal miRNAs were significantly up‐ and downregulated in the patient samples compared to the healthy samples. The panel of the miRNAs could be useful for diagnosis of the PCa and the BPH compared to healthy control. However, it could not distinguish the expression levels of miRNAs between the BPH and PCa samples. We expected to be able to detect differences between the BPH and PCa samples for the early detection of PCa. However, we failed in this endeavor, and so the diagnostic and prognostic performance of urinary exosomal miRNAs for distinguishing between PCa and BPH requires further clinical study to obtain a wider miRNA profile using the LF‐bis‐MPA‐MNPs method. Taken together, the chimeric nanocomposites offer a promising method for EV isolation from human specimens. The developed method is rapid, simple, and highly efficient. We envision that there is great potential for EV‐related applications, including stem cell treatment, transplantation, immune‐based treatment, and theranostics.
CONFLICTS OF INTEREST
There are no conflicts to declare.
AUTHOR CONTRIBUTIONS
Yong Shin supervised the whole project. Thuy Nguyen Thi Dao conceived the research and designed the experiments. Thuy Nguyen Thi Dao and Myoung Gyu Kim worked on experiments. Thuy Nguyen Thi Dao performed the analysis and made interpretations of data. Yoon Ok Jang, Huifang Liu, Bonhan Koo, Hyo Joo Lee, Myoung Gyu Kim, Yunlim Kim, Yun‐Yong Park, and Hyun Soo Kim provided chemicals and supported data analysis. Yong Shin, Thuy Nguyen Thi Dao made comments, suggested appropriate modifications. Choung‐Soo Kim, Thuy Nguyen Thi Dao and Yong Shin wrote and edited the manuscript. All authors read and approved the final manuscript.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGEMENTS
This study was supported by the Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation of Korea (NRF) (2020R1A2C2007148 and 2016R1A5A1010148), and also supported by the Yonsei University Research Fund of 2021 (2021‐22‐0051).
Dao, T. N. T. , Kim, M. G. , Koo, B. , Liu, H. , Jang, Y. O. , Lee, H. J. , Kim, Y. , Park, Y.‐Y. , Kim, H. S. , Kim, C.‐S. , & Shin, Y. (2022). Chimeric nanocomposites for the rapid and simple isolation of urinary extracellular vesicles. Journal of Extracellular Vesicles, 11, e12195. 10.1002/jev2.12195
Contributor Information
Choung‐Soo Kim, Email: cskim@amc.seoul.kr.
Yong Shin, Email: shinyongno1@yonsei.ac.kr.
REFERENCES
- Akers, J. C. , Gonda, D. , Kim, R. , Carter, B. S. , & Chen, C. C. (2013). Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus‐like vesicles, and apoptotic bodies. Journal of Neuro‐ Oncology, 113, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrescu, R. , Bello, V. , Bouzas, V. , Costo, R. , Dumitrache, R. , García, M. A. , Giorgi, R. , Morales, M. P. , Morjan, I. , Serna, C. J. , & Veintemillas‐Verdaguer, S. (2010). Iron oxide materials produced by laser pyrolysis. AIP Conference Proceedings, 1275, 22 [Google Scholar]
- Ali, A. , Hira Zafar, M. Z. , ul Haq, I. , Phull, A. R. , Ali, J. S. , & Hussain, A. (2016). Synthesis, characterization, applications, and challenges of iron oxide nanoparticles. Nanotechnology Science and Applications, 9, 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu, Z. , & Yáñez‐Mó, M. J. (2014). Tetraspanins in extracellular vesicle formation and function. Frontiers in Immunology, 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, H. M. , & Baker, E. N. (2012). A structural perspective on lactoferrin function. Biochemistry and Cell Biology, 90(3), 320–328. [DOI] [PubMed] [Google Scholar]
- Bautista‐Sánchez, D. , Arriaga‐Canon, C. , & Pedroza‐Torres, A. (2020). The promising role of miR‐21 as a cancer biomarker and its importance in RNA‐based therapeutics. Molecular Therapy‐Nucleic Acids, 20, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beit‐Yannai, E. , Tabak, S. , & Stamer, W. D. (2018). Physical exosome: Exosome interactions. Journal of Cellular and MolecularMmedicine, 22(3), 2001–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi, C. , Henry, J. C. , & Kogure, T. (2011). The role of microRNAs in human liver cancers. Seminars in Oncology, 38 (6), 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, K. , Martin, K. , & FitzGerald, S. (2020). A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Scientific Reports, 10(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock, J. H. (1997). Lactoferrin structure‐function relationships. In Hutchens T. W. & Lönnerdal B. (Eds.), Lactoferrin: Interactions and biological functions (pp. 3–23). Humana Press. [Google Scholar]
- Brow, D. A. , & Guthrie, C. J. N. (1988). Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature, 334(6179), 213–218. [DOI] [PubMed] [Google Scholar]
- Brown, P. N. , & Yin, H. (2017). Polymer‐based purification of extracellular vesicles. Methods in Molecular Biology, 1660, 91–103. [DOI] [PubMed] [Google Scholar]
- Brunsman, E. , Sutton, R. , & Bortz, E. (1994). Magnetic properties of carbon‐coated, ferromagnetic nanoparticles produced by a carbon‐arc method. Journal of Applied Physics, 75(10), 5882–5884. [Google Scholar]
- Campanella, C. , Rappa, F. , Sciumè, C. , Marino Gammazza, A. , Barone, R. , Bucchieri, F. , Gammazza, A. M. , Barone, R. , Bucchieri, F. , David, S. , Curcurù, G. , Bavisotto, C. C. , Pitruzzella, A. , Geraci, G. , Modica, G. , Farina, F. , Zummo, G. , Fais, S. , de Macario, E. C. , … Cappello, F. (2015). Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer, 18, 3230–3229. [DOI] [PubMed] [Google Scholar]
- Carlmark, A. , Malmström, E. , & Malkoch, M. J. (2013). Dendritic architectures based on bis‐MPA: Functional polymeric scaffolds for application‐driven research. Chemical Society Reviews, 42(13), 5858–5879. [DOI] [PubMed] [Google Scholar]
- Caruso, S. , & Poon, I. K. (2018). Apoptotic cell‐derived extracellular vesicles: More than just debris. Frontiers in Immunology, 9, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo‐Magaña, M. , Bång‐Rudenstam, A. , & Belting, M. (2020). The pleiotropic role of proteoglycans in extracellular vesicle mediated communication in the tumor microenvironment. Seminars in Cancer Biology, 62, 99–107. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Hou, Y. , Wang, K. , Li, J. , Xia, Y. , Yang, X. , Lv, G. , Xing, X. , & Shen, F. (2014). Reexpression of Let‐7g microRNA inhibits the proliferation and migration via K‐Ras/HMGA2/Snail axis in hepatocellular carcinoma. BioMed Research International, 2014, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Xie, Y. , & Xu, L. (2017). Protein content and functional characteristics of serum‐purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Journal of Cancer, 140(4), 900–913. [DOI] [PubMed] [Google Scholar]
- Cutone, A. , Rosa, L. , & Ianiro, G. (2020). Lactoferrin's anti‐cancer properties: Safety, selectivity, and wide range of action. Biomolecules, 15, 10(3), 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao, T. N. T. , Reddy, A. S. , Zhao, F. , Liu, F. , Koo, B. , Moniruzzaman, M. , Kim, J. , & Shin, Y. (2021). Green synthesis‐based magnetic diatoms for biological applications. ACS Sustainable Chemistry & Engineering, 7 (5), 5440–5448. [Google Scholar]
- Dao, T. N. T. , Yoon, J. , Jin, C. E. , Koo, B. , Han, K. , Shin, Y. , & Lee, T. Y. (2018). Rapid and sensitive detection of Salmonella based on microfluidic enrichment with a label‐free nanobiosensing platform. Sensors and Actuators B, 262, 588–594. [Google Scholar]
- Dar, G. H. , Mendes, C. C. , & Kuan, W. L. (2021). GAPDH controls extracellular vesicle biogenesis and enhances the therapeutic potential of EV mediated siRNA delivery to the brain. Nature Communications, 12(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFrates, K. , Markiewicz, T. , & Gallo, P. (2018). Protein polymer‐based nanoparticles: Fabrication and medical applications. International Journal of Molecular Sciences, 19(6), 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, L. M. , & Wang, M. Z. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells, 8(7), 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Z. Y. , Cai, G. Y. , & Li, J. J. (2018). U6 can be used as a housekeeping gene for urinary sediment miRNA studies of IgA nephropathy. Scientific Reports, 8(1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca, G. , Anghel, L. , & Erhan, R. V. (2018). Structural aspects of lactoferrin and serum transferrin observed by FTIR spectroscopy. Chemistry Journal of Moldova 13(1), 111–116. [Google Scholar]
- Eftekhari, A. , Foroughifar, N. , & Hekmati, M. (2019). Fe3O4@ L‐arginine magnetic nanoparticles: A novel and magnetically retrievable catalyst for the synthesis of 1′ 3‐diaryl‐2N‐azaphenalene. Journal of the Chinese Chemical Society, 66(7), 761–768. [Google Scholar]
- El Yazidi‐Belkoura, I. , Legrand, D. , & Nuijens, J. (2001). The binding of lactoferrin to glycosaminoglycans on enterocyte‐like HT29‐18‐C1 cells is mediated through basic residues located in the N‐terminus. Biochimica et Biophysica Acta, 1568(3), 197–204. [DOI] [PubMed] [Google Scholar]
- Elzoghby, A. O. , Abdelmoneem, M. A. , & Hassanin, I. A. (2020). Lactoferrin, a multi‐functional glycoprotein: Active therapeutic, drug nanocarrier & targeting ligand. Biomaterials, 2020, 120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , & Bijnsdorp, I. V. (2021). Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles, 10(7), e12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdem, A. , Eksin, E. , & Kesici, E. (2018). Dendrimers integrated biosensors for healthcare applications. Nanotechnology and Biosensors, 2018, 307–317. [Google Scholar]
- Farhanian, D. , De Crescenzo, G. , & Tavares, J. R. (2018). Large‐scale encapsulation of magnetic iron oxide nanoparticles via syngas photo‐initiated chemical vapor deposition. Scientific Reports, 8(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell, L. L. , Pepin, J. , & Kucharski, C. (2007). A comparison of the effectiveness of cationic polymers poly‐l‐lysine (PLL) and polyethylenimine (PEI) for non‐viral delivery of plasmid DNA to bone marrow stromal cells (BMSC). European Journal of Pharmaceutics and Biopharmaceutics, 65(3), 388–397. [DOI] [PubMed] [Google Scholar]
- Foj, L. , Ferrer, F. , & Serra, M. (2017). Exosomal and non‐exosomal urinary miRNAs in prostate cancer detection and prognosis. The Prostate, 77(6), 573–583. [DOI] [PubMed] [Google Scholar]
- Fu, J. F. , Yu, W. Q. , & Dong, X. (2013). Mechanical and tribological properties of natural rubber reinforced with carbon blacks and Al2O3 nanoparticles. Materials and Designs, 49, 336–346. [Google Scholar]
- Gennebäck, N. , Hellman, U. , Malm, L. , Larsson, G. , Ronquist, G. , Walderstrom, A. , & Morner, S. (2013). Growth factor stimulation of cardiomyocytes induces changes in the transcriptional contents of secreted exosomes. Journal of Extracellular Vesicle, 2(1), :20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Rawat, S. , & Arora, V. (2018). An improvised one‐step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Research and Therapy, 9(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan, S. , Kang, M. H. , & Jeyaraj, M. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells, 8(4), 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Xu, G. X. , & Lu, H. (2015). Dysregulation of miRNA‐21 and their potential as biomarkers for the diagnosis of cervical cancer. International Journal of Clinical and Experimental Pathology, 8(6), 713. [PMC free article] [PubMed] [Google Scholar]
- Heinemann, M. L. , Ilmer, M. , Silva, L. P. , Hawke, D. H. , Recio, A. , Vorontsova, M. A. , Alt, E. , & Vykoukal, J. (2014). Benchtop isolation and characterization of functional exosomes by sequential filtration. Journal of Chromaltography A. 1371, 125–135. [DOI] [PubMed] [Google Scholar]
- Hikita, T. , Miyata, M. , Watanabe, R. , & Oneyama, C. (2018). Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Scientific Reports, 8(1), 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon, K. W. , Abu, N. , Ab Mutalib, N. S. , & Jamal, R. (2017). Exosomes as potential biomarkers and targeted therapy in colorectal cancer: A mini‐review. Frontiers in Pharmacololy, 8, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. , & Vatter, F. A. P. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell, 182 (4), 1044–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Liu, Y. , Yu, P. , Qu, J. , Guo, Y. , Li, W. , Wang, S. , & Zhang, J. (2018). MiR‐23a transcriptional activated by Runx2 increases metastatic potential of mouse hepatoma cell via directly targeting Mgat3. Scientific Reports, 8, 7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliescu, F. S. , Vrtačnik, D. , Neuzil, P. , & Iliescu, C. (2019). Microfluidic technology for clinical applications of exosomes. Micromachines, 10(6), 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illés, E. , Szekeres, M. , Tóth, I. Y. , Farkas, K. , Foldesi, I. , Szabo, A. , Ivan, B. , & Tombacz, E. (2018). PEGylation of superparamagnetic iron oxide nanoparticles with self‐organizing polyacrylate‐PEG brushes for contrast enhancement in MRI diagnosis. Nanomaterials, 8(10), 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, B. L. , Grabowska, A. , & Ratan, H. L. (2014). MicroRNA in prostate cancer: Functional importance and potential as circulating biomarkers. BMC cancer, 14(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahani, S. , Shakiba, A. , & Jahani, L. (2015). The Antimicrobial effect of lactoferrin on Gram‐negative and Gram‐positive bacteria. International Journal of Ingection, 2(3). [Google Scholar]
- Jin, C. E. , Koo, B. , Lee, T. Y. , Han, K. , Kim, S. B. , Park, I. J. , & Shin, Y. (2018a). Simple and low‐cost sampling of. cell‐free nucleic acids from blood plasma for rapid and sensitive detection of circulating tumor DNA. Advanced Science, 5, 1800614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C. E. , Lee, T. Y. , Koo, B. , Sung, H. , Kim, S. H. , & Shin, Y. (2018). Rapid virus diagnostic system using bio‐optical sensor and microfluidic sample processing. Sensors and Actuators B, 255, 2399–2406. [Google Scholar]
- Kell, D. B. , Heyden, E. L. , & Pretorius, E. (2020). The biology of lactoferrin, an iron‐binding protein that can help defend against viruses and bacteria. Frontiers in Immunology, 11, 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kievit, F. M. , Veiseh, O. , Bhattarai, N. , Fang, C. , Gunn, J. W. , Lee, D. H. , Ellenbogen, R. G. , Olson, J. M. , & Zhang, M. (2009). PEI–PEG–chitosan‐copolymer‐coated iron oxide nanoparticles for safe gene delivery: Synthesis, complexation, and transfection. Advanced Functional Materials, 19(14), 2244–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Tran, V. T. , Oh, S. , Kim, C. S. , Hong, J. C. , Kim, S. , Joo, Y. S. , Mun, S. , Kim, M. H. , Jung, J. W. , J, L. , Kang, Y. S. , Koo, J. W. , & Lee, J. B. (2018). Scalable Solvothermal synthesis of superparamagnetic Fe3O4 nanoclusters for bioseparation and Theragnostic probes. ACS Applied Materials & Interfaces, 10(49), 41935–41946. [DOI] [PubMed] [Google Scholar]
- Kooti, M. , & Sedeh, A. N. (2013). Synthesis and characterization of NiFe2O4 magnetic nanoparticles by combustion method. Journal of Materials Science & Technology, 29(1), 34–38. [Google Scholar]
- Kostyukhin, E. M. , Nissenbaum, V. D. , Abkhalimov, E. V. , Kustov, A. L. , Ershov, B. G. , & Kustov, L. M. (2020). Microwave‐assisted synthesis of water‐dispersible humate‐coated magnetite nanoparticles: Relation of coating process parameters to the properties of nanoparticles. Nanomaterials, 10(8), 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan, R. , Okoli, C. , Boutonnet, M. , Järås, S. , & Rajarao, G. K. (2014). Microemulsion prepared magnetic nanoparticles for phosphate removal: Time efficient studies. Journal of Enviroment Chemical Engineering, 2(1), 185–189. [Google Scholar]
- Landmark, K. J. , DiMaggio, S. , Ward, J. , Kelly, C. , Vogt, S. , Hong, S. P. , Kotlyar, A. , Myc, A. , Thomas, T. P. , Penner‐Hahn, J. E. , Baker, J. R. Jr. , Holl, M. M. B. , & Orr, B. G. (2008). Synthesis, characterization, and in vitro testing of superparamagnetic iron oxide nanoparticles targeted using folic acid‐ conjugated dendrimers. ACS Nano, 2(4), 773–783. [DOI] [PubMed] [Google Scholar]
- Lee, H. Y. , Lee, S. H. , Xu, C. , Xie, J. , Lee, J. H. , Wu, B. , Koh, A. L. , Wang, X. , Sinclair, R. , & Wang, S. X. (2008). Synthesis and characterization of PVP‐coated large core iron oxide nanoparticles as an MRI contrast agent. Nanotechnology, 19(16), 165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Xu, L. Q. , Wang, L. , Wu, Y. P. , Li, J. , Neoh, K. G. , & Kang, E. T. (2011). Clickable poly (ester amine) dendrimer‐grafted Fe3O4 nanoparticles prepared via successive Michael addition and alkyne–azide click chemistry. Polymer Chemistry, 2(6), 1312–1321. [Google Scholar]
- Lyu, Z. , Ding, L. , Huang, A. T. , Kao, C. L. , & Peng, L. (2019). Poly (amidoamine) dendrimers: Covalent and supramolecular synthesis. Materials Today Chemistry, 13, 34–48. [Google Scholar]
- Malhotra, H. , Sheokand, N. , Kumar, S. , Chauhan, A. S. , Kumar, M. , Jakhar, P. , Boradia, V. M. , Raje, C. I. , & Raje, M. (2016). Exosomes: Tunable nano vehicles for macromolecular delivery of transferrin and lactoferrin to specific intracellular compartment. Journal of biomedical nanotechnology, 12(5), 1101–1114. [DOI] [PubMed] [Google Scholar]
- Marcuello, M. , Vymetalkova, V. , & Neves, R. P. (2019). Circulating biomarkers for early detection and clinical management of colorectal cancer. Molecular Aspects of Medicine, 69, 107–122. [DOI] [PubMed] [Google Scholar]
- Mascolo, M. C. , Pei, Y. , & Ring, T. A. (2013). Room temperature co‐precipitation synthesis of magnetite nanoparticles in a large pH window with different bases. Materials, 6(12), 5549–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturavongsadit, P. , Bi, X. , Gado, T. A. , Nie, Y. Z. , & Wang, Q. (2016). Adhesive peptides conjugated PAMAM dendrimer as a coating polymeric material enhancing cell responses. Chinese Chemical Letters, 27(9), 1473–1478. [Google Scholar]
- Muralidharan‐Chari, V. , Clancy, J. W. , Sedgwick, A. , & Schotey, C. D. (2010). Microvesicles: Mediators of extracellular communication during cancer progression. Journal of Cell Science, 123(10), 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necela, B. M. , Carr, J. M. , Asmann, Y. W. , & Thompson, E. A. (2011). Differential Expression of MicroRNAs in Tumors from Chronically Inflamed or Genetic (APCMin/+) Models of Colon Cancer. PLOS One, 6(4), 18501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini, M. , Nicoloso, M. S. , & Calin, G. A. (2009). MicroRNAs and cancer—New paradigms in molecular oncology. Current Opinion in Cell Biology, 21(3), 470–479. [DOI] [PubMed] [Google Scholar]
- Nonahal, M. , Rastin, H. , Saeb, M. R. , Sari, M. G. , Moghadam, M. H. , Zarrintaj, P. , & Ramezanzadeh, B. (2018). Epoxy/PAMAM dendrimer‐modified graphene oxide nanocomposite coatings: Nonisothermal cure kinetics study. Progress in Organic Coatings, 114, 233–243. [Google Scholar]
- Ogata‐Kawata, H. , Izumiya, M. , Kurioka, D. , Honma, Y. , Yamada, Y. , Furuta, K. , Gunji, T. , Ohta, H. , Okamoto, H. , Sonoda, H. , Watanabe, M. , Nakagama, H. , Yokota, J. , Kohno, T. , & Tsuchiya, N. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE, 9(4), e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksvold, M. P. , Neurauter, A. , & Pedersen, K. W. (2015). Magnetic bead‐based isolation of exosomes. Methods in Molecular Biology, 1218, 465–481. [DOI] [PubMed] [Google Scholar]
- Paiva, R. , Zauli, D. , Neto, B. , & Brum, I. S. (2020). Urinary microRNAs expression in prostate cancer diagnosis: A systematic review. Clinical and Translational Oncology, 22(11), 2061–2073. [DOI] [PubMed] [Google Scholar]
- Patel, G. K. , Khan, M. A. , Zubair, H. , Srivastava, S. K. , Khushman, M. , Singh, S. , & Singh, A. P. (2019). Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Scientific Reports, 9(1), 5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat, P. , Kumar, S. , Sheokand, N. , Raje, C. L. , & Raje, M. (2012). The multifunctional glycolytic protein glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) is a novel macrophage lactoferrin receptor. Biochemistry and Cell Biololy, 90(3), 329–338. [DOI] [PubMed] [Google Scholar]
- Ray, S. , Li, Z. , Hsu, C. H. , Hwang, L. P. , Lin, Y. C. , Chou, P. T. , & Lin, Y. Y. (2018). Dendrimer‐and copolymer‐based nanoparticles for magnetic resonance cancer theranostics. Theranostics, 8(22), 6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl, S. , Rinner, B. , Asslaber, M. , Schaider, H. , Walzer, S. , Novak, A. , Lohner, K. , & Zweytick, D. (2011). In search of a novel target – Phosphatidylserine exposed by non‐apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochimica et Biophysica Acta, 1808(11), 2638–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra, S. , & Agwa, M. M. (2020). Lactoferrin, a unique molecule with diverse therapeutical and nanotechnological applications. International Journal of Biological Macromolecules, 164, 1046–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schee, K. , Boye, K. , Abrahamsen, T. W. , & Fodstad, F. K. (2012). Clinical relevance of microRNA miR‐21, miR‐31, miR‐92a, miR‐101, miR‐106a and miR‐145 in colorectal cancer. BMC Cancer, 12(1), :505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyawan, H. , & Widiyastuti, W. (2019). Progress in the preparation of magnetite nanoparticles through the electrochemical method. KONA Powder and Particle Journal, 36, 145–155. [Google Scholar]
- Shimizu, S. , Takehara, T. , Hikita, H. , Kodama, T. , Miyagi, T. , Hosui, A. , Tatsumi, T. , Ishida, H. , Noda, T. , Nagano, H. , Doki, Y. , Mori, M. , & Hayashi, N. (2010). The let‐7 family of microRNAs inhibits Bcl‐xL expression and potentiates sorafenib‐induced apoptosis in human hepatocellular carcinoma. Journal of Hepatology, 52(5), 698–704. [DOI] [PubMed] [Google Scholar]
- Shin, Y. , Lim, S. Y. , Lee, T. Y. , & Park, M. K. (2015). Dimethyl adipimidate/T hin film S ample processing (DTS); A simple, low‐cost and versatile nucleic acid extraction assay for downstream analysis. Scientific reports, 5(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J. , Jain, K. , Mehra, N. K. , & jain, N. K. (2016). Dendrimers in anticancer drug delivery: Mechanism of interaction of drug and dendrimers. Articial Cells, Nanomedicine, and Biotechnology, 44(7), 1626–1634. [DOI] [PubMed] [Google Scholar]
- Skottvoll, F. S. , Berg, H. E. , Bjørseth, K. , Lund, K. , Lund, K. , Roos, N. , Bekhradnia, S. , Thiede, B. , Sandber, C. , Vik‐Mo, E. O. , Roberg‐Larsen, H. , Nyström, Bo , Lundanes, E. , & Wilson, S. R. (2018). Ultracentrifugation versus kit exosome isolation: NanoLC–MS and other tools reveal similar performance biomarkers, but also contaminations. Future Science, 5(1).359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi, S. M. , Niazi, A. , Chahardoli, M. , Hortamani, A. , & Setoodeh, P. (2014). In silico investigation of lactoferrin protein characterizations for the prediction of anti‐microbial properties. Molecular Biology Research Communications, 3(2), 85. [PMC free article] [PubMed] [Google Scholar]
- Soytaş, S. H. , Oğuz, O. , & Menceloğlu, Y. Z. (2019). Polymer nanocomposites with decorated metal oxides. Polymer Composites with Functionalized Nanoparticles, 1, 287–323. [Google Scholar]
- Steitz, B. , Hofmann, H. , Kamau, S. W. , Hassa, P. O. , Hottiger, M. O. , Rechenberg, B. , Amtenbrink, M. H. , & Fink, A. P. (2007). Characterization of PEI‐coated superparamagnetic iron oxide nanoparticles for transfection: Size distribution, colloidal properties and DNA interaction. Journal of Magnetism and Magnetic Materials, 311(1), 300–305. [Google Scholar]
- Stranska, R. , Gysbrechts, L. , Wouters, J. , Vermeersch, P. , Bloch, K. , Dierickx, D. , Andrei, G. , & Snoeck, R. (2018). Comparison of membrane affinity‐based method with size‐exclusion chromatography for isolation of exosome‐like vesicles from human plasma. Journal of Translational Medicine, 16(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Gao, X. , Bi, H. , Xie, Y. , & Tang, L. (2018). Assessment of binding interaction between bovine lactoferrin and tetracycline hydrochloride: Multi‐spectroscopic analyses and molecular modeling. Molecules, 23(8), 1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Mignani, S. , Shen, M. , & Shi, X. (2016). Dendrimer‐based magnetic iron oxide nanoparticles: Their synthesis and biomedical applications. Drug Discovery Today, 21(12), 1873–1885. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni, M. , Uhl, A. M. , Savliwala, S. , Savitzky, B. H. , Dhavalikar, R. , Garraud, N. , Arnold, D. P. , Kourkoutis, L. F. , Andrew, J. S. , & Rinaldi, C. (2017). Thermal decomposition synthesis of iron oxide nanoparticles with diminished magnetic dead layer by controlled addition of oxygen. ACS Nano, 11(2), 2284–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterweger, H. , Dézsi, L. , Matuszak, J. , Janko, C. , Poettler, M. , Jordan, J. , Bäuerle, T. , Szebeni, J. , Fey, T. , Boccaccini, A. R. , Alexiou, C. , & Cicha, I. (2018). Dextran‐coated superparamagnetic iron oxide nanoparticles for magnetic resonance imaging: Evaluation of size‐dependent imaging properties, storage stability and safety. International Journal of Nanomedicine, 13, 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollath, D. (2013). Nanoparticles‐nanocomposites–nanomaterials: An introduction for beginners. John Wiley & Sons. [Google Scholar]
- Wang, H. , Lu, Z. , & Zhao, X. (2019). Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. Journal of Hematology and Oncology, 12(1), 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Tan, H. Y. , Feng, Y. G. , Zhang, C. , Chen, F. , & Feng, Y. (2019). microRNA‐23a in human cancer: Its roles, mechanisms and therapeutic relevance. Cancers, 11(1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Nkurikiyimfura, I. , & Pan, Z. (2015). Sonochemical synthesis of magnetic nanoparticles. Chemical Engineering Communications, 202(5), 616–621. [Google Scholar]
- Webber, J. , & Clayton, A. (2013). How pure are your vesicles? Journal of extracellular vesicles, 2(1), 19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, B. B. (2013). Measuring the Size & Surface Charge of Exosomes, Microvesicles and Liposomes. Application Note, Brookhaven Instruments, a Nova Instruments company.
- Whiteside, T. L. (2016). Tumor‐derived exosomes and their role in cancer progression. Advances in clinical chemistry, 74, 103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Wu, B. , Wang, W. , Chiang, K. S. , Jen, A. K. Y. , & Luo, J. (2018). Ultra‐efficient and stable electro‐ optic dendrimers containing supramolecular homodimers of semifluorinated dipolar aromatics. Materials Chemistry Frontiers, 2(5), 901–909. [Google Scholar]
- Wu, Y. , Lu, Z. , Li, Y. , Yang, J. , & Zhang, X. (2020). Surface Modification of Iron Oxide‐Based Magnetic Nanoparticles for Cerebral Theranostics: Application and Prospection. Nanomaterials, 10(8), 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xianyu, Y. , Wang, Q. , & Chen, Y. (2018). Magnetic particles‐enabled biosensors for point‐of‐care testing. Trends in Analytical Chemistry, 106, 213–224. [Google Scholar]
- Xu, H. , Aguilar, Z. P. , Yang, L. , Kuang, M. , Duan, H. , Xiong, Y. , Wei, H. , & Wang, A. (2011). Antibody conjugated magnetic iron oxide nanoparticles for cancer cell separation in fresh whole blood. Biomaterials, 32(36), 9758–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Sun, J. , Wang, Y. , Sheng, J. , Wang, F. , & Sun, M. (2014). Application of iron magnetic nanoparticles in protein immobilization. Molecules, 19(8), 11465–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani, F. , & Seddigh, M. (2016). Magnetite nanoparticles synthesized by co‐precipitation method: The effects of various iron anions on specifications. Materials Chemistry and Physics, 184, 318–323. [Google Scholar]
- You, C. , Jin, L. , Xu, Q. , Shen, B. , Jiao, X. , & Huang, X. (2019). Expression of miR‐21 and miR‐138 in colon cancer and its effect on cell proliferation and prognosis. Oncology Letters, 17(2), 2271–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, Y. , Liu, H. , & Tang, W. H. (2019). Exosomes: Biogenesis, biologic function and clinical potential. Cell Bioscience, 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, M. , Park, J. H. , Singelyn, J. , Ma, H. , Sailor, M. J. , Ruoslahti, E. , Ozkan, M. , & Ozkan, C. (2009). A surface‐charge study on cellular‐uptake behavior of F3‐peptide‐conjugated iron oxide nanoparticles. Small, 5(17), 1990–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , & Zhu, X. (2019). MicroRNA‐21 and microRNA‐30c as diagnostic biomarkers for prostate cancer: A meta‐analysis. Cancer Management and Research, 11, 2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Ji, H. , Yu, P. , Niu, J. , Farooq, M. U. , Akram, M. W. , Udego, I. O. , Li, H. , & Niu, X. (2018). Surface modification of magnetic iron oxide nanoparticles. Nanomaterials, 8(10), 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Tang, G. , & Yan, J. (2020). MicroRNA‑122 regulates docetaxel resistance of prostate cancer cells by regulating PKM2. Experimental and Therapeutic Medicine, 20(6), 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION


