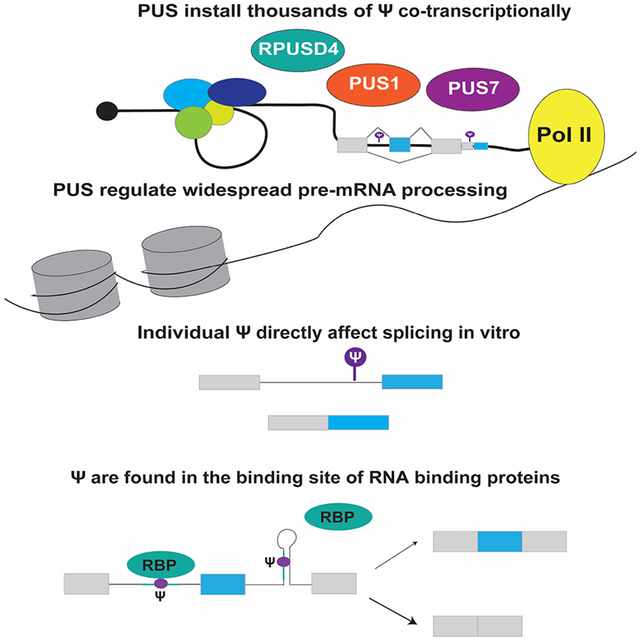
Summary
Pseudouridine is a modified nucleotide that is prevalent in human mRNAs and is dynamically regulated. Here we investigate when in their life cycle mRNAs become pseudouridylated to illuminate the potential regulatory functions of endogenous mRNA pseudouridylation. Using single-nucleotide resolution pseudouridine profiling on chromatin-associated RNA from human cells we identified pseudouridines in nascent pre-mRNA at locations associated with alternatively spliced regions, enriched near splice sites, and overlapping hundreds of binding sites for RNA binding proteins. In vitro splicing assays establish a direct effect of individual endogenous pre-mRNA pseudouridines on splicing efficiency. We validate hundreds of pre-mRNA sites as direct targets of distinct pseudouridine synthases, and show that PUS1, PUS7 and RPUSD4, three pre-mRNA modifying pseudouridine synthases with tissue-specific expression, control widespread changes in alternative pre-mRNA splicing and 3′ end processing. Our results establish the vast potential for co-transcriptional pre-mRNA pseudouridylation to regulate human gene expression via alternative pre-mRNA processing.
Graphical Abstract
Introduction
Pseudouridine is the most abundant modified nucleotide in RNA, and pseudouridylation of non-coding RNAs of the translation and splicing machineries is important for their functions. Recent transcriptome-wide methods for the detection of pseudouridine (Ψ) revealed that mRNAs in yeast and human cells contain pseudouridines that are dynamically regulated in response to cellular stress (Carlile et al., 2014; Lovejoy, Riordan and Brown, 2014; Schwartz et al., 2014; Li et al., 2015). However, while roles for other mRNA modifications have been uncovered, such as that of N6-methyladenosine (m6A) in regulating splicing, export, translation and decay, the endogenous functions of pseudouridine in mRNA are largely unknown (Gilbert, Bell and Schaening, 2016; Roundtree et al., 2017; Martinez and Gilbert, 2018).
The potential functions of RNA modifications are constrained by when the modification is installed during RNA biogenesis. Because previous studies profiled pseudouridines in mature poly(A)+ mRNAs (Carlile et al., 2014; Lovejoy, Riordan and Brown, 2014; Schwartz et al., 2014; Li et al., 2015), it was unknown when mRNA becomes pseudouridylated and which steps of the mRNA life cycle are affected by pseudouridines. In yeast, most mRNA pseudouridines have been genetically assigned to two conserved pseudouridine synthases (PUS), Pus1 and Pus7 (Carlile et al., 2014; Schwartz et al., 2014), which are nuclear localized during normal growth (Huh et al., 2003). Human PUS enzymes that have been reported to pseudouridylate mRNA targets (Li et al., 2015; Safra et al., 2017; Carlile, T.M., Martinez et al., 2019) also localize to the nucleus, or have nuclear isoforms (Fernandez-Vizarra et al., 2007; Ji et al., 2015; Safra et al., 2017). Further, PUS7 has been shown to be chromatin-associated and co-purifies with active Pol II promoters and enhancers (Ji et al., 2015). Pseudouridines are present in nuclear resident ncRNAs (Carlile et al., 2014; Schwartz et al., 2014). Thus, human PUS enzymes are present and active in the nucleus where they could target pre-mRNA. Because artificial RNA pseudouridylation has been shown to affect RNA-RNA(Newby and Greenbaum, 2001; Hudson, Bloomingdale and Znosko, 2013; Kierzek et al., 2014) and RNA-protein interactions (Chen et al., 2010; Delorimier et al., 2017; Vaidyanathan et al., 2017) known or likely to be relevant to splicing, we hypothesized that human PUS enzymes pseudouridylate nascent pre-mRNA where they could function in nuclear pre-mRNA processing.
Here, we used single-nucleotide resolution pseudouridine profiling, Pseudo-seq (Carlile et al., 2014; Carlile, Rojas-Duran and Gilbert, 2015), on chromatin-associated nascent RNA to discover thousands of candidate pseudouridines in pre-mRNA from the human hepatocellular carcinoma cell line HepG2. These pseudouridines are significantly enriched in the introns flanking sites of alternative splicing, suggesting regulatory potential. Consistently, we identified widespread differences in alternative splicing in response to genetic manipulation of pre-mRNA pseudouridylating enzymes PUS1, PUS7 and RPUSD4. Furthermore, we showed that site-specific installation of a single endogenous pseudouridine is sufficient to directly influence splicing in vitro. We also observed significant co-transcriptional deposition of pseudouridines in 3′ UTRs of pre-mRNAs and demonstrate prevalent PUS-dependent alternative cleavage and polyadenylation. Finally, pseudouridines overlap the experimentally validated binding sites of dozens of RNA binding proteins (RBPs) interrogated by eCLIP, providing a mechanistic link to altered pre-mRNA processing.
Results
Pre-mRNA is pseudouridylated co-transcriptionally in human cells
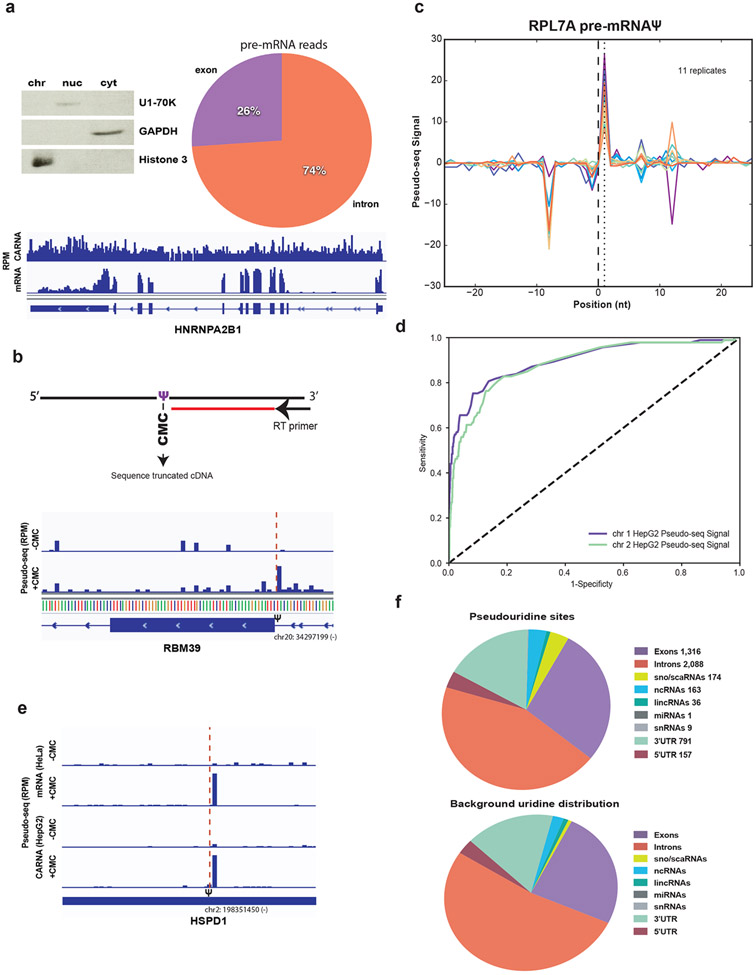
To determine if pseudouridine is added to pre-mRNA co-transcriptionally, we isolated chromatin-associated RNA from the hepatocellular carcinoma cell line HepG2 by a biochemical cellular fractionation that enriches for intron-containing unspliced pre-mRNA (Figure 1a) (Khodor et al., 2011; Bhatt et al., 2012). The majority (74%) of pre-mRNA sequencing reads mapped to introns, and read coverage of introns and exons was relatively uniform demonstrating efficient capture of intronic reads from unspliced pre-mRNA (Figure 1a). This result contrasts with the primarily exonic reads observed in poly(A)+ mRNA (Figure 1a). We then performed Pseudo-seq (Carlile et al., 2014) to identify the locations of pseudouridine in chromatin-associated RNA. In Pseudo-seq, pseudouridines are selectively modified with the chemical N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMC). The bulky covalent CMC-pseudouridine adduct blocks reverse transcriptase and allows for sequencing-based detection of pseudouridines from truncated cDNAs (Figure 1b). By design, Pseudo-seq detects only modification sites where a substantial fraction of the RNA is modified: because pseudouridine-containing RNAs are not pre-enriched during the Pseudo-seq protocol, high stoichiometry pseudouridylation is required to generate a block to reverse transcription that is sufficiently penetrant to produce detectable peaks of Pseudo-seq signal (Carlile, Rojas-Duran and Gilbert, 2015), which is the difference in normalized reads from the CMC condition compared to the mock-treated control (Figure 1c). Using this approach, Pseudo-seq signal identified known pseudouridine sites in ribosomal RNA (rRNA) with high sensitivity, specificity and reproducibility from the chromatin-associated RNA samples (Figure 1d, Supplemental Figure 1a, Table ST1A). We generated data on 11 biological replicates of chromatin-associated RNA to add a stringent reproducibility filter to our site calling and identify high-confidence sites of pre-mRNA modification (Figure 1c).
Figure 1. Pre-mRNA is pseudouridylated co-transcriptionally in human cells.
a) Left panel. Western blot of HepG2 cellular fractions, equal cell volumes were loaded and probed with antibodies against GAPDH (cytoplasm), U1-70K (nucleoplasm) and Histone3 (chromatin). Right panel. Distribution of pre-mRNA reads mapping to introns versus exons in the chromatin-associated RNA fraction. Bottom panel. Genome browser view of reads per million mapping across the highly expressed gene hnRNPA2B1 in a chromatin-associated RNA library compared to a poly(A)+ mRNA library from HepG2 cells. b) Detection of pseudouridine by Pseudo-seq with a representative genome browser view of Pseudo-seq reads mapping to RBM39, red dotted line indicates the location of the pseudouridine (chr20:34297199) identified by a CMC-dependent reverse transcriptase stop one nucleotide 3′ to the site. Reads per million (RPM). c) Pseudo-seq signal, equal to the difference in normalized reads between the +CMC and mock libraries. Traces for 11 biological replicates of chromatin-associated RPL7A pre-mRNA pseudouridine (chr9:136217792) are shown, d) ROC curve of true positive versus false positive rates of known pseudouridine locations in mature human rRNA for two representative chromatin-associated RNA replicates. The Pseudo-seq signal is displayed for both replicates, e) Representative genome browser view of Pseudo-seq reads mapping to HSPD1 from HepG2 chromatin-associated RNA and HeLa mRNA red dotted line indicates the location of this cell type conserved pseudouridine identified by a CMC-dependent reverse transcriptase stop one nucleotide 3′ to the site. f) Top panel. Summary of pseudouridines sites identified in chromatin associated RNAs. Bottom panel. Distribution of background uridines meeting the minimum read cutoff for site calling.
We identified expected sites in nascent rRNA corresponding to known positions in the mature rRNA sequences with an observed false positive rate of 0.01 and false discovery rate of 0.152 (Supplemental Figure 1a, Table ST1A). These results are consistent with the prevailing hypothesis that rRNA pseudouridines are added co-transcriptionally as has been shown for 2′-O-methylation in yeast (Koš and Tollervey, 2010; Birkedal et al., 2015). Intriguingly, pseudouridine profiling of chromatin-associated RNA allowed us to detect the precursor regions in pre-rRNA and identify putative sites of pseudouridylation in these regions (Table ST1A). These sites have complementarity to human snoRNAs that are predicted by snoGPS (Schattner, Brooks and Lowe, 2005) to direct their modification (Table ST1A). Hundreds more novel pseudouridines were found in nuclear resident non-coding RNAs that interact with chromatin, including the small nucleolar RNAs (snoRNAs), small cajal body-specific RNAs (scaRNAs) and long non-coding RNAs lncRNAs (Figure 1f, Table ST1B). These previously unknown sites were called based on observing highly reproducible enrichment of CMC-dependent read 3′ ends (Methods) using criteria that have previously generated site lists with high confirmation rates (>61%) in independent studies (Carlile et al., 2014; Carlile, T.M., Martinez et al., 2019; Khoddami et al., 2019) including using completely orthologous chemistry for pseudouridine detection (Khoddami and Cairns, 2013; Khoddami et al., 2019) (Supplemental Figure 1c-f, Table ST1D). Cell type specific pseudouridylation and distinct capture biases from technical approaches (e.g. Supplemental Figure 1d) influence which sites can be detected.
By these criteria, analysis of Pseudo-seq signal in pre-mRNA conservatively identified thousands of candidate pseudouridines, with the majority of pseudouridines found in introns (Figure 1b,c,f, Supplementary Table 3). This number likely represents a small fraction of the total pseudouridines in pre-mRNA since only ~1% of uridines in the nascent transcriptome had sufficient sequencing depth to meet our pseudouridine calling criteria (Methods). The distribution of pre-mRNA pseudouridines among gene features including introns, exons, 5′-UTRs and 3′-UTRs was similar to the distribution of uridines passing the minimum read cutoff for pseudouridine detection (Figure 1f). This apparently widespread modification of pre-mRNA introns with pseudouridine contrasts with the observed distribution of N6-methyladenosine (m6A), another abundant mRNA modification added to pre-mRNA, which is found primarily in exons (90% of m6As) despite a similar distribution of intronic to exonic reads as observed in our study (Ke et al., 2017). These results reveal that pseudouridylation is a co-transcriptional process that results in prevalent installation of pseudouridines in pre-mRNA, endowing this modification with the potential to affect nuclear pre-mRNA processing steps such as splicing and 3′ end processing.
Comparing pseudouridine sites in nascent pre-mRNA from HepG2 cells to sites previously identified in poly(A)+ mRNA from HeLa revealed both similarities and differences. Of 18 HeLa mRNA sites that were expressed at sufficient levels to assess pseudouridylation in HepG2 pre-mRNA, 5 were called in chromatin-associated RNA suggesting that they are added early and conserved across cell types (Figure 1e). Other sites that were expressed at levels with sufficient coverage in both cell lines were present exclusively in either HeLa or HepG2 cells (Supplemental Figure 1g), which is consistent with cell type-specific mRNA pseudouridylation and/or with cytoplasmic mRNA modification.
Pseudouridines are enriched near alternative splice regions and splice sites
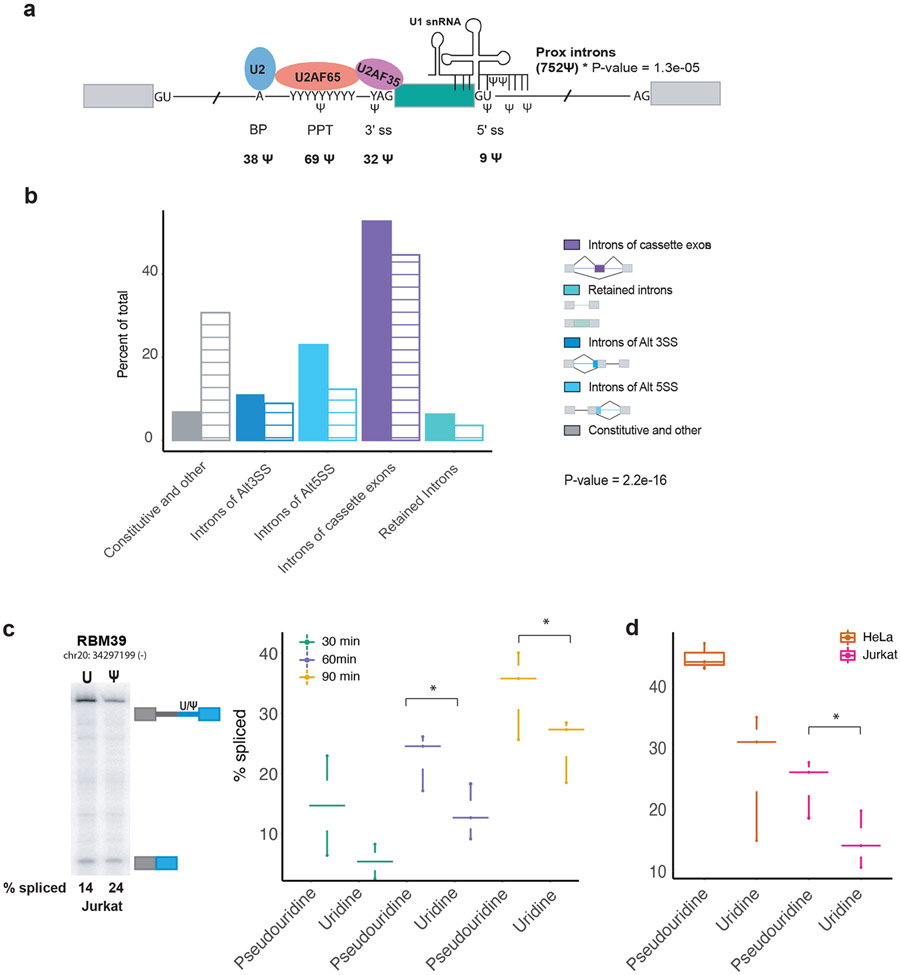
To explore the possibility of a role for pseudouridines in pre-mRNA processing we interrogated the location of pseudouridines relative to splicing features. We compared the fraction of called pseudouridines within splice sites (6 nt from exon ends) and in proximal introns (within 500nt of splice sites) to the fraction of uridines meeting our coverage cutoffs for detection in these regions (Figure 2a). A hypergeometric test (Fisher’s exact test) found that intronic pseudouridines are enriched compared to uridines within 6nt of exon ends (p-value < 0.55e−06). We also found that 752 pseudouridines are enriched in proximal introns within 500nt of splice sites (p-value = 1.3e−05) where splicing regulatory elements such as intronic splicing enhancers and silencers are often found (Figure 2a, Table ST1C). We identified pseudouridines at 3′ (32) and 5′ (9) splice sites, polypyrimidine tracts (PPT) (72) and branch site regions (37) (Figure 2a). Pseudouridines occur at critical residues for splicing including the conserved pyrimidine before the 3′ splice site (YAG), in the region of the 5′ splice site that base pairs with U1 snRNA, and in the branch site region that base pairs with U2 snRNA.
Figure 2. Pseudouridines are enriched around splicing regulatory features and directly affect splicing.
a) Schematic of a spliced exon including the core signals for splice site recognition: branch point region (BP), polypyrimidine tract (PPT) and 5′ and 3′ splice site (ss). Number of pseudouridines identified in each splice site region is summarized below the schematic. Pseudouridines are enriched in proximal introns (within 500nt) of splice sites, p-value = 1.3e−05 from Fisher’s exact test and within splice sites (within 6 nt from intron ends), p-value = 0.55e-06 from Fisher’s exact test. b) Distribution of pseudouridines (filled bars) versus uridines with adequate read coverage in the introns (line bars) of annotated alternatively spliced regions. P-value = 2.2e−16 denotes a significant change in the distribution of pseudouridine relative to the uridine distribution as determined by chi-squared test for the overall change in proportions across regions. Alternative splice sites (Alt SS). c) Representative RT-PCR gel of in vitro splicing of RBM39 two-exon reporter (Supplemental Figure 2a,c) that was either unmodified or site specifically modified with pseudouridine (−/+ Ψ) in splicing competent wildtype Jurkat nuclear extract. d) ) Quantification of % spliced of in vitro splicing of the RBM39 reporter in a splicing timecourse (30, 60 and 90 minutes) in Jurkat nuclear extract. Data is displayed as a stripchart with box plot, where the dots represent the value for each sample for a given condition ((30 (n=2), 60 (n=3) and 90 minutes (n=3)). P-values were calculated by a paired t-test and difference considered significant if p-value < 0.05. An asterisk denotes significance. e) Quantification of in vitro splicing of the RBM39 reporter that was either unmodified or site specifically modified with pseudouridine (−/+ Ψ) in Jurkat and HeLa nuclear extract. Quantification of % spliced from n=3 is displayed as a stripchart with box plot, where the dots represent the value for each sample for a given cell type. P-values were calculated by a paired t-test and difference considered significant if p-value < 0.05. An asterisk denotes significance.
We further explored the potential for co-transcriptional pre-mRNA pseudouridylation to regulate splicing by determining the distribution of intronic pseudouridines with respect to alternatively spliced regions. We compared the distribution of called pseudouridines to uridines meeting our coverage cutoffs for detection in introns flanking alternatively spliced region categories (Figure 2b). Pseudouridines are notably enriched around alternative splice sites including in the introns flanking cassette exons, introns of alternative 5′ and 3′ splice sites and in retained introns (Figure 2b). In contrast, pseudouridines are depleted from the introns of constitutive and other exons (Figure 2b). The observed difference in the overall distribution of pseudouridines compared to uridine in alternatively spliced was found to be statistically significant by a chi-squared test (p-value = 2.2e−16). Together, the distribution of pseudouridines in alternatively spliced regions and near splice sites is consistent with a role for pseudouridine in splicing regulation by pre-mRNA modifying pseudouridine synthases.
Site-specific pseudouridylation directly affects splicing in vitro
We site-specifically pseudouridylated pre-mRNA sequences (Figure 2c-d, Supplemental Figure 2) to determine whether individual pseudouridines identified in cells are sufficient to directly affect pre-mRNA splicing in vitro. We generated chimeric two-exon pre-mRNA splicing reporters containing intronic PUS7 target sites in RBM39 and MDM2 by in vitro transcription, site-specifically pseudouridylated in vitro with recombinant PUS7, and isolated the modified pre-mRNA by purification (Supplemental Figure 2 a-c). Modified and unmodified control pre-mRNAs were incubated with wildtype nuclear extract under splicing conditions. Remarkably, a single endogenous intronic pseudouridine installed upstream of the 3′ splice site in the RBM39 pre-mRNA was sufficient to directly enhance splicing as quantified by RT-PCR and compared to splicing of the unmodified control (Figure 2c-d). This effect of pseudouridine on splicing was observed in nuclear extracts from two different cell lines (Figure 2d, Supplemental Figure 2d) and across time points (Figure 2c). Similarly, modifying an MDM2 splicing reporter with a pseudouridine at an endogenously modified 3′ splice site UAG enhanced splicing in vitro (Supplemental Figure 2e-f). These results demonstrate a direct biochemical effect of individual endogenous pre-mRNA pseudouridines on splicing.
Pseudouridines are enriched in RNA binding protein binding sites
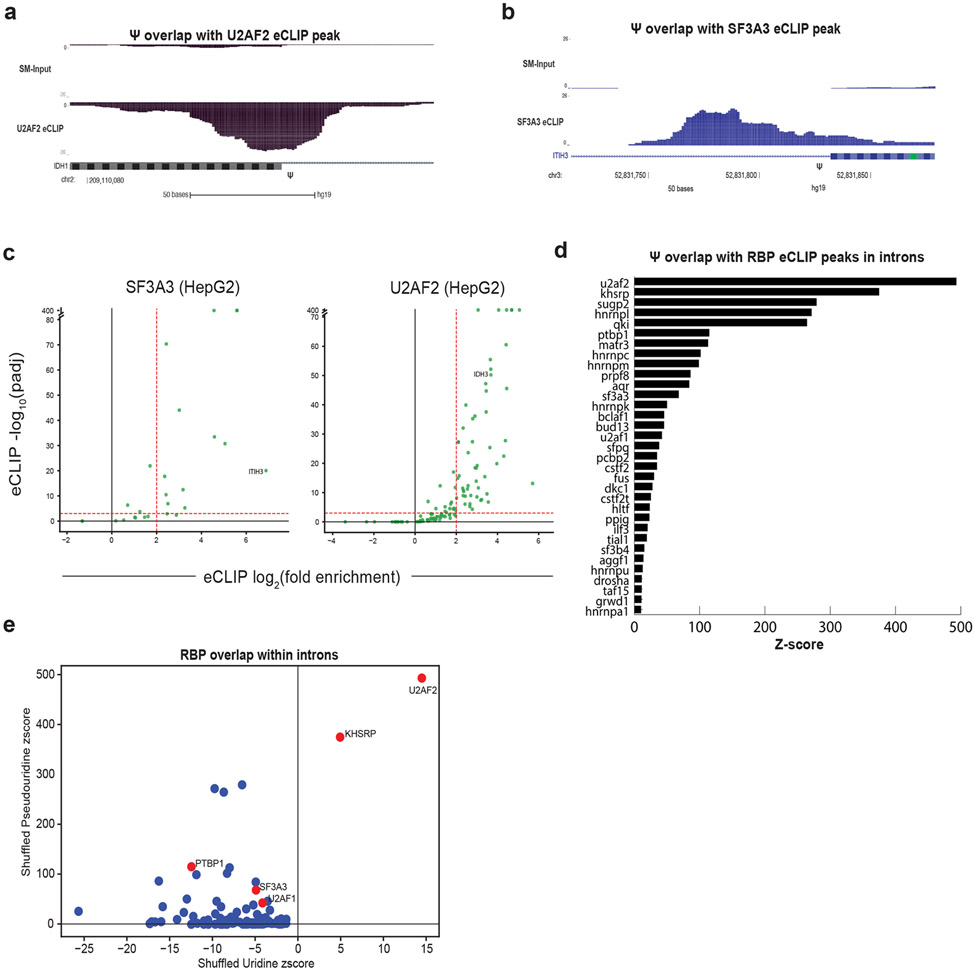
How might a single intronic pseudouridine alter splicing outcome? Given that diverse RNA-binding proteins (RBPs) show altered affinity for their target RNAs following artificial incorporation of pseudouridine (Chen et al., 2010; Delorimier et al., 2017; Vaidyanathan et al., 2017), we investigated the overlap of pre-mRNA pseudouridines with the binding sites of regulatory RBPs. We computationally compared pre-mRNA pseudouridines with RBP binding sites identified by enhanced UV crosslinking followed by immunoprecipitation and sequencing (eCLIP-seq) for 103 RBPs from HepG2 cells (Van Nostrand et al., 2016; Nussbacher and Yeo, 2018) (Figure 3a-d). Binding sites for each examined RBP overlap tens to hundreds of pseudouridines as demonstrated by eCLIP peaks with significant enrichment over the size matched input (> 4-fold enrichment and adjusted p-value < 0.001) (Figure 3a-c). Strikingly, we found 5,359 significant eCLIP clusters (RBP binding sites) that overlap pseudouridines and 40% (1,922/4789) of unique pseudouridines overlapped validated RBP binding sites across the transcriptome including 386 Ψ -RBP overlaps located in introns (Table ST1E). Of these 386 Ψ - RBP overlaps, 249 are in introns flanking annotated alternatively spliced exons.
Figure 3. Pseudouridines are enriched in RNA binding protein binding sites.
a) Genome browser views of U2AF2 and b) SF3A3 eCLIP peaks and size-matched input controls (SMI) on IDH1 and ITIH3 respectively. The location of pseudouridine relative to the eCLIP peak is denoted by (Ψ). c) Volcano plots of pseudouridines overlapping RBP eCLIP peaks displaying the fold enrichment (IP over size matched input) versus the SMI-normalized adjusted p-value (Van Nostrand et al. 2016 Nat Methods). The overlap between eCLIP peaks and pseudouridines is shown for two RBPs U2AF2 and SF3A3. Proximal introns refer to intronic sequences <500 nt from splice sites and distal introns refer to intronic sequences >500 nt from splice sites. d) Z-scores were generated by comparing the fraction of eCLIP peaks overlapping pseudouridines to the calculated overlap after shuffling pseudouridines within intronic regions 1000 times. e) The z score of pseudouridines shuffled 1000 times within intronic regions plotted against the z score of uridines shuffled 1000 times within intronic regions.
We determined the statistical significance of pseudouridine co-localization within the binding sites for individual RBPs by comparing the fraction of eCLIP peaks that overlap pseudouridines to the expected overlap for sites that were randomly located (shuffled) within intronic regions (Figure 3d, Table ST1F). Z-scores were generated for each RBP by performing a thousand shuffles (Table ST1F), revealing 33 RBPs with a Z-score above 10 (p<10−5) (Figure 3d). As another control, to account for the possibility of preferential detection of binding to abundant uridines, we performed the same experiment with all unmodified uridines that met the criteria for pseudouridine detection. The Z-scores for pseudouridines and uridines overlapping RBP binding sites for each RBP are provided in Table ST1F and plotted for comparison (Figure 3e). All RBPs were under-represented for intersection with unmodified uridines, except for U2AF2, which is known to bind a uridine-rich motif (Figure 3e). Thus, pseudouridines frequently occur within the binding sites for regulatory RBPs, and these features overlap more often than expected by chance.
Many of the highest scoring RBPs with binding sites that overlap intronic pseudouridines have documented roles in splicing regulation, including core splicing factors such as U2AF2, U2AF1, SF3A3 and PRPF8 (Figure 3a-d, Table ST1E). Other high-scoring RBPs include polyuridine and polypyrimidine binding splicing factors such as hnRNP C, PTBP1 and TIA1 (Figure 3d, Table ST1E). We observe an enrichment for polyuridine and polypyrimidine in the sequences flanking pseudouridines consistent with the enrichment of pseudouridine in the binding sites of RBPs (e.g. U2AF2) known to bind to these sequences (Supplemental Figure 3b, Table ST1F). These and other RBPs are known to be associated with nuclear RNAs and have diverse roles in RNA metabolism (Gratenstein et al., 2005; Pan et al., 2008; Fu and Ares, 2014; Zong et al., 2014; Attig et al., 2018). We also find pseudouridines enriched within pre-mRNAs encoding RBPs and splicing factors suggesting another way by which pseudouridines might affect RBP activity (Supplemental Figure 3c, Table ST1G). Altogether, these results establish widespread co-occurrence of newly identified pre-mRNA pseudouridines at sites likely to affect splicing including the binding sites of regulatory RBPs, in proximal introns of alternatively spliced regions, and at splice sites.
PUS1, RPUSD4 and PUS7 are predominant pre-mRNA modifying enzymes
Human cells, including HepG2, express up to 13 pseudouridine synthases (PUSs). To identify which enzymes target pre-mRNA and potentially regulate splicing, we took an in vitro approach. This strategy overcomes a critical limitation of cell-based endogenous pseudouridine assignment, which requires very deep sequencing of PUS-depleted cells to avoid false negatives and accurately interpret absence of reads as evidence for modification by that PUS in wild type cells. The very large size of the human pre-mRNA transcriptome makes sequencing cellular RNA to sufficient depth unfeasible for our purpose. A recently developed high-throughput in vitro pseudouridylation assay (Carlile, T.M., Martinez et al., 2019),(Martinez, Nicole M., Gilbert, 2021) using purified PUS overcomes this limitation to identify which PUS(s) directly pseudouridylate sites of interest, including in lowly expressed RNAs. We verified excellent agreement between genetic and in vitro assignment approaches by cross-validating 85% of yeast PUS1-dependent pseudouridine sites in mRNA (Carlile et al., 2014; Carlile, T.M., Martinez et al., 2019) (Supplemental Figure 4a). Although, human PUS7 and TRUB1 recognize their targets in the context of a sequence motif, the features required for targeting by the other 10 human PUS have not been characterized making it difficult to predict the enzymes that direct modification of identified sites in pre-mRNA.
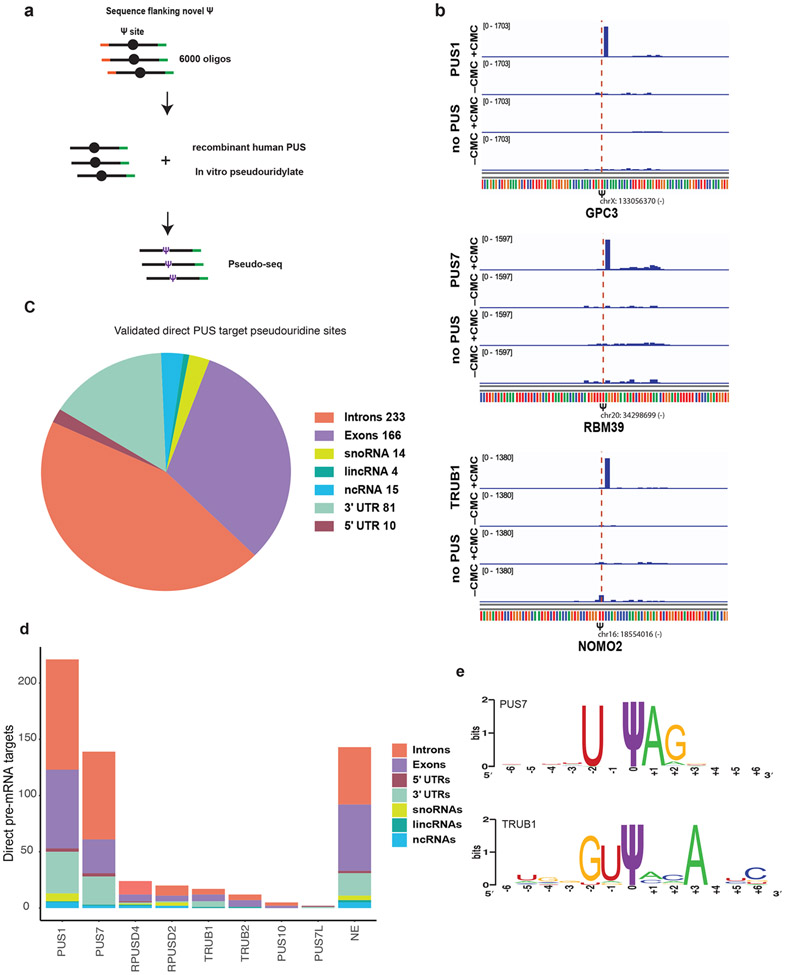
To identify human PUSs that directly pseudouridylate pre-mRNA sequences, we synthesized a pool of RNA containing each identified pseudouridine site flanked by 130 nucleotides of endogenous sequence (Figure 4a). The resulting RNA pool was incubated in separate reactions with individual recombinant human pseudouridine synthases PUS1, PUS7, PUS7L, PUS10, RPUSD2, RPUSD4, TRUB1, TRUB2 or HepG2 nuclear extract and pseudouridylation of the RNA from the pool was detected by Pseudo-seq. Pseudouridine sites were classified as direct targets of individual PUS using a statistically principled analysis pipeline(Martinez, Schaening-Burgos and Gilbert, 2021). This approach considers RT stops at the position of interest relative to surrounding positions and the significance of a peak is determined relative to the positions surrounding it by means of a Z-score calculation (Methods). PUS-dependent pseudouridine sites are then identified as those that have a high Z-score in the CMC-treated library, and a low Z-score in a no PUS CMC-treated control library (Supplemental Figure 4b). We identified hundreds of endogenous pre-mRNA pseudouridine sites as direct targets of one of the 8 tested pseudouridine synthases (Figure 4b-d, Table ST1H, Supplemental Figure 4b-e). Importantly, the in vitro validated sites had a similar distribution across RNA features and classes as the total list of candidate pseudouridines identified in chromatin associated RNA from cells (Figure 4c, Figure 1f).
Figure 4. Multiple PUS pseudouridylate pre-mRNA sequences.
a) Schematic of in vitro pseudouridylation assay with RNA made from a pool of 6000 oligos containing all the sites identified in HepG2 chromatin-associated RNA. In vitro pseudouridylation was carried by incubating pool RNA with recombinant human pseudouridine synthases (PUS) and pseudouridines were identified by Pseudo-seq. b) Genome browser view of Pseudo-seq reads at pseudouridine sites following RNA incubation with a recombinant PUS or no PUS control. Plots for three intronic pre-mRNA pseudouridines: a PUS1 target GPC3, PUS7 target in RBM39 and a TRUB1 target in NOMO2. c) Combined distribution of pseudouridines validated as direct targets of all tested PUS by in vitro Pseduo-seq assay. d) Summary of pseudouridines assigned as direct targets of each PUS protein from in vitro Pseudo-seq assay. e) Weblogo summarizing frequency of motifs identified among targets of PUS7 and TRUB1.
We identified PUS1, PUS7 and RPUSD4 as pre-mRNA pseudouridylating enzymes (Figure 4d). Orthologous yeast proteins Pus1 and Pus7 pseudouridylate the majority of known yeast mRNA sites (Carlile et al., 2014; Schwartz et al., 2014), suggesting broad conservation of mRNA targeting by these PUS. RPUSD4 is a member of a PUS family that has expanded to include 4 paralogs in higher eukaryotes (chordates) and was not previously known to have mRNA targets. Importantly, PUS1, PUS7 and RPUSD4 are present in the nucleus in human cells(Fernandez-Vizarra et al., 2007; Stadler et al., 2013; Ji et al., 2015) where they have access to pre-mRNA. There is almost no redundancy among the in vitro targets of each PUS (Supplemental Figure 4b-d), suggesting distinct targeting mechanisms and potentially specific regulatory programs. The fact that PUS1, RPUSD4 and PUS7 pseudouridylate the majority of identified pre-mRNA sites is consistent with their relatively high expression in HepG2 cells (Supplemental Figure 4c-e). Some sites were pseudouridylated in nuclear extract but not by purified PUS implying that at least one additional human PUS also pseudouridylates pre-mRNA and/or that some sites require co-factors that are supplied in the nuclear extract (Figure 4d, Table Table ST1H). Although in vitro pseudouridylation allows confident assignment of successfully modified RNAs to specific PUS, there are many potential mechanisms producing false negative results (see Discussion). Thus, some unassigned sites could still be cellular targets of one of the tested PUS. These results identify site-specific RNA targets for previously uncharacterized PUS (e.g. RPUSD2, PUS7L) and substantially expand the known targets for others (e.g. PUS1, PUS7) to include pre-mRNAs.
Most PUS proteins pseudouridylate their targets in diverse sequence contexts and do not display strong sequence preferences (Supplemental Figure 4f). This may reflect a predominantly structural mode of mRNA target recognition as shown for yeast Pus1 (Carlile, T.M., Martinez et al., 2019). By contrast, the targets of PUS7 are highly enriched for a UNΨAR motif, which is similar to, but a more permissive version of the motif recognized by yeast Pus7 (Figure 4e). This motif also overlaps with the YAG of the 3′ splice site for multiple PUS7 targets (Table ST1H). The PUS7 motif is enriched among all chromatin-associated pseudouridines identified in HepG2 cells (Supplemental Figure 4g) including some sites that are not modified by the recombinant protein in vitro (Discussion). We identified comparatively few targets of the mRNA targeting PUS, TRUB1 (Figure 4d), which occur primarily in the preferred TRUB1 sequence context GUΨCNANNC (Safra et al., 2017) (Figure 4e). It is possible that TRUB1 targets were under captured due to sequence bias in the ligation efficiency of circLigase (used for library preparation). Together, our in vivo pseudouridine profiling and in vitro PUS target validation identify pre-mRNA sites as the largest class of PUS targets and reveal the potential for multiple pseudouridine synthases to influence pre-mRNA processing by pseudouridylating diverse nascent pre-mRNA sequences.
Pseudouridine synthases PUS1, RPUSD4 and PUS7 regulate alternative splicing
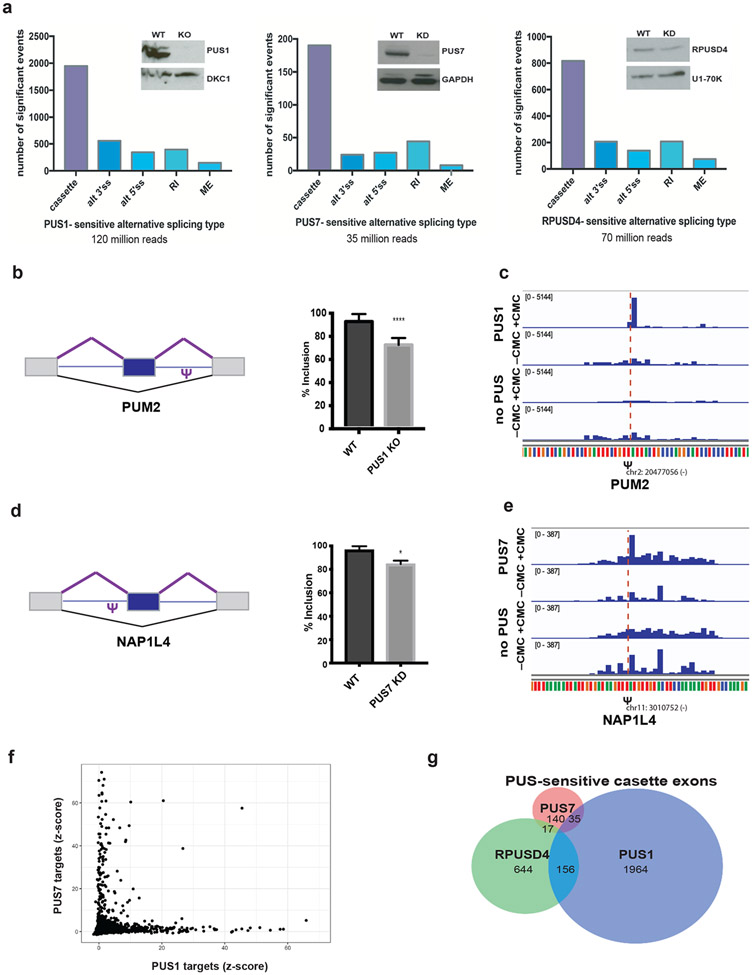
The prevalence of pseudouridines in alternatively spliced pre-mRNAs (Figure 2a) and within splicing factor binding sites (Figure 3a-d), together with our evidence that pseudouridine directly affects splicing in vitro (Figure 2c-d) and evidence from the literature that diverse RNA-RNA (Newby and Greenbaum, 2001; Hudson, Bloomingdale and Znosko, 2013; Kierzek et al., 2014) and RNA-protein (Chen et al., 2010; Delorimier et al., 2017; Vaidyanathan et al., 2017) interactions are sensitive to pseudouridine, suggested that altering PUS activity could cause changes in splicing. PUS1, PUS7 and RPUSD4 emerged as likely candidates to influence splicing from our in vitro pre-mRNA pseudouridylation assay (Figure 4d). We first examined whether PUS1-dependent pseudouridylation influenced splicing by making PUS1 knockout HepG2 cells using CRISPR/Cas9 (Figure 5a). We obtained highly reproducible RNA-seq data from poly(A)+ mRNA from PUS1 knockout and wildtype cells (R2>0.96), and quantified differences in alternative splicing and mRNA abundance (Methods). Strikingly, PUS1 knockout leads to thousands of changes in alternative splicing, as defined by splicing events that were statistically significant (FDR < 0.05) and exhibited greater than 10% difference in inclusion levels compared to wildtype cells (Figure 5a-b, Table ST1I). Of these differential splicing changes 195 exhibited a greater than 50% difference in inclusion levels between conditions. These changes in splicing affected 1617 genes and included cassette exons, alternative 5′ and 3′ splice sites, retained introns and mutually exclusive exons. We observed both differential inclusion and skipping of cassette exons upon PUS1 knockout (Figure 5b, Supplemental Figure 5e-f, Table ST1I), showing that the effect of PUS1 on splicing is pre-mRNA dependent. In contrast to these broad effects on alternative splicing, mature mRNA abundance changed very little, with approximately one hundred mRNAs significantly altered in cells lacking PUS1 (Supplemental Figure 6a). Manual inspection of the affected mRNAs did not reveal any splicing factors or RBPs that would be expected to indirectly affect splicing (Table ST1J).
Figure 5. Pseudouridine synthases regulate alternative splicing.
a) Left - Western blot of the CRISPR knockout PUS1 HepG2 cell line probed for PUS1 and a loading control. RNA was isolated from PUS1 knockout (KO) and wild type (WT) cells and mRNA-seq libraries were prepared from poly(A)+ mRNA. The number of significant alternative splicing changes in PUS1 KO versus WT (n=2 biological replicates) is displayed by type of alternative splicing: cassette exons (cassette), alternative 3′ splice sites (alt 3′ss), alternative 5′ splice sites (alt 5′ ss), retained introns (RI) and mutually exclusive exons (ME). Significant alternative splicing events were determined from rMATS as those events that changed by greater than 10% difference in percent inclusion and a false discovery rate (FDR) of less than or equal to 0.05. Middle - Western blot of representative RPUSD4 knockdown (~60%) at 96h following shRNA induction. RPUSD4-sensitive alternative splicing changes determined from RNA-seq analysis (n=2 biological replicates) as above. Right - Western blot of representative PUS7 knockdown (~90%) at 96h following shRNA induction. PUS7-sensitive alternative splicing changes determined from RNA-seq analysis (n=3 biological replicates) as above. b) Left - schematic of a cassette exon in PUM2 and location of pseudouridine. Right - quantification of exon inclusion in WT and PUS1 KO based on junction spanning reads from RNA-seq. Asterisk denotes statistical significance based on p-value < 0.05 as calculated by rMATS. c) Genome browser view of Pseudo-seq reads of the intronic PUM2 pseudouridine site (Figure 5b) following pseudouridylation with recombinant PUS1 or in the absence of PUS. d) Left - schematic of a cassette exon in NAP1L4 and location of pseudouridine. Right - quantification of exon inclusion in WT and PUS7 KD based on junction spanning reads from RNA-seq. Asterisk denotes statistical significance based on p-value < 0.05 as calculated by rMATS. e) Genome browser view of Pseudo-seq reads of the intronic NAP1L4 pseudouridine site (Figure 5d) following pseudouridylation with recombinant PUS7 or in the absence of PUS. f) Scatter plot showing pairwise comparisons of Z-score values at candidate pseudouridine sites incubated with recombinant PUS1 versus PUS7. g) Venn diagram of overlap among cassette exons regulated by PUS1, PUS7 and RPUSD4.
We considered the possibility that loss of site-specific pseudouridylation of U2 snRNA might underlie splicing changes in PUS1 KO cells. In budding yeast, a single pseudouridine, Ψ44, in the U2 snRNA is installed by Pus1 (Massenet et al., 1999). However, the corresponding position in the human U2 snRNA, Ψ43, was not affected in PUS1 KO cells based on primer extension with U2-specific primers (Supplemental Figure 6a). This result is consistent with a previous report showing PUS1-independent pseudouridylation of the homologous U2 snRNA site in mouse cells lacking PUS1 (Deryusheva and Gall, 2017) and with computational and biochemical evidence that human snRNAs are modified by the snoRNA/scaRNA-dependent pseudouridine synthase, DKC1 (Karijolich and Yu, 2010; Wu et al., 2011). Ψ43 is predicted to be modified by DKC1 in complex with scaRNA8 (Deryusheva and Gall, 2017). No other detected U2 snRNA pseudouridines were affected in PUS1 KO HepG2 cells (Supplemental Figure 6a-b) or mouse cells (Deryusheva and Gall, 2017). Thus, a mechanism other than loss of U2 snRNA pseudouridylation is responsible for widespread PUS1-sensitive alternative splicing.
Consistent with the possibility of direct effects of PUS1-dependent pre-mRNA pseudouridylation on splicing, in vitro validated PUS1 targets in cassette exons or flanking introns of PUM2, ARHGAP5, C14orf159, SNHG12, ANKRD10 and TANK showed differential splicing in PUS1 KO cells (Figure 5b-c, Supplemental Figure 5c). Other unassigned pseudouridines that were identified in cells overlap PUS1-sensitive cassette exons or their flanking introns (Supplemental Figure 5e). Some of these sites could be PUS1 targets that were not recapitulated in vitro due to limitations of the in vitro pseudouridylation assay (Discussion). Alternatively, their splicing may be affected indirectly by the absence of PUS1. Unfortunately, we were unable to interrogate pseudouridine status in cells for the vast majority of PUS1-sensitive cassette exons and flanking introns (Supplementary Figure 5d,f, Table ST1I). This blind spot arises because orders of magnitude fewer reads are sufficient to quantify alternative splicing in mature poly(A)+ mRNA compared to the coverage required for pre-mRNA pseudouridine discovery.
We used inducible shRNA expression to deplete the essential (Shalem et al., 2014; Wang et al., 2014) pre-mRNA pseudouridylating enzyme RPUSD4 and also PUS7 to determine their impact on alternative splicing by RNA-seq. Replicate experiments were reproducible (R2>0.98 and R2>0.94). Partial depletion of RPUSD4 (60 %) and PUS7 (90%) produced widespread effects on alternative splicing including 817 RPUSD4-sensitive cassette exons in 696 genes and 190 PUS7-sensitive cassette exons in 175 genes (Figure 5a,d, Table ST1K and ST1L). The RPUSD4 and PUS7 samples were sequenced at lower depth than the PUS1 KO cells possibly contributing to fewer identified splicing changes. NAP1L4 is an example of a PUS7-sensitive alternatively spliced exon where PUS7 directly pseudouridylates a position near the exon, in the upstream intron (Figure 5d-e). As with PUS1, we lacked data for the pseudouridine status of most RPUSD4- and PUS7-sensitive exons in cells due to coverage limitations (Supplemental Figure 5d,g). PUS7 depletion resulted in 374 differentially expressed genes (Supplemental Figure 6b, Table ST1M), consistent with previous studies showing altered PUS7-dependent mRNA levels in yeast (Schwartz et al., 2014). In contrast, RPUSD4 depletion resulted in almost no significant changes in mRNA levels (Supplemental Figure 6c, Table ST1N). As expected for shRNA mediated depletion, PUS7 and RPUSD4 mRNAs were significantly downregulated (Supplemental Figure 6b-c).
Each PUS pseudouridylates distinct pre-mRNA targets with little overlap (Figure 5f, Supplemental Figure 4c-e). Consistently, depletion of each of the predominant pre-mRNA targeting PUS produced distinct changes in the pattern of alternative splicing (Figure 5g). Notably, the pervasiveness and magnitude of PUS-dependent splicing changes are comparable to the effects of depleting canonical splicing regulators (Nostrand et al., 2018). These results support a widespread and non-redundant role for multiple PUSs in alternative splicing regulation. Given the enrichment of pseudouridines around alternatively spliced regions and our demonstration that endogenous pseudouridines can have direct biochemical effects on splicing in vitro (Figure 2b-d and Supplemental Figure 2), we expect a subset of the splicing changes in PUS knockout/knockdown cells to be a consequence of direct pre-mRNA pseudouridylation. However, we cannot rule out that some of the PUS-sensitive events are an indirect consequence of reduced pseudouridylation of other targets. Taken together, these findings support the broad potential for pre-mRNA pseudouridylation to impart widespread PUS-dependent alternative splicing.
Pseudouridine synthases PUS1, RPUSD4 and PUS7 regulate 3′ end processing
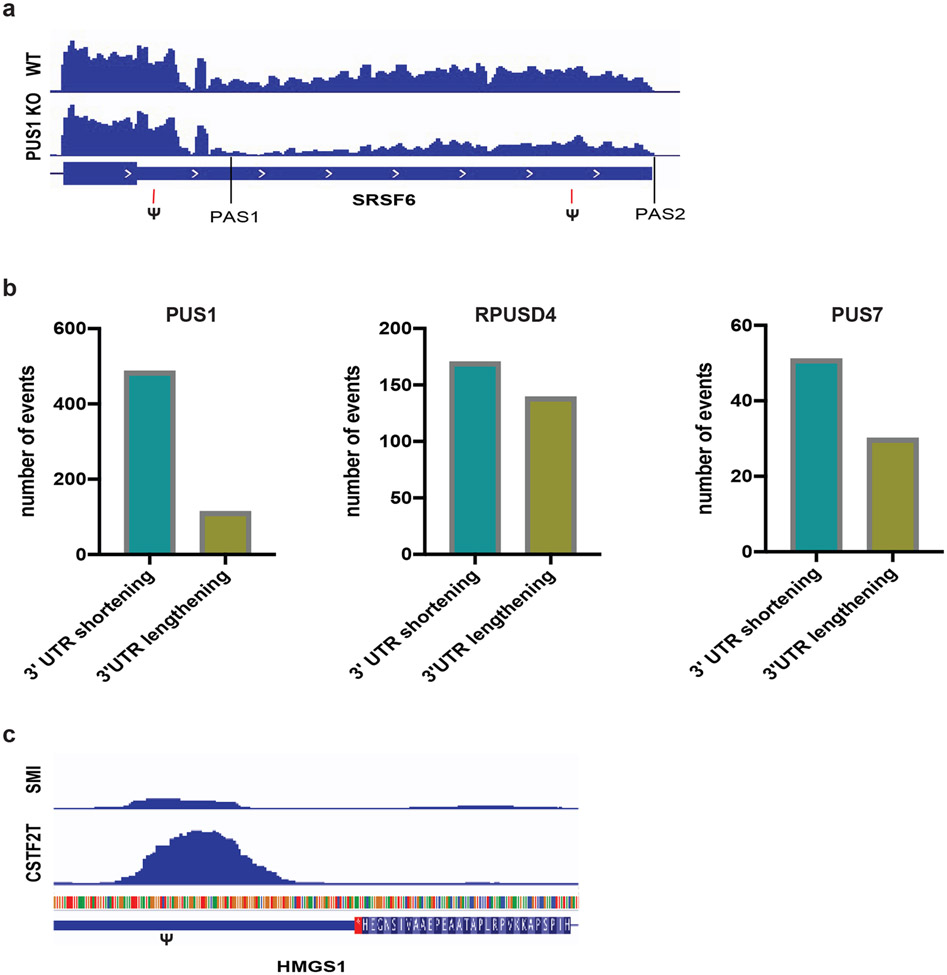
The finding that 3′ UTR pseudouridines are added to nascent pre-mRNA (Figure 1f) led us to hypothesize that pseudouridines might also influence another regulated step in pre-mRNA processing, that of cleavage and polyadenylation. We therefore analyzed the RNA-seq data from PUS-depleted cells to find evidence of alternative cleavage and polyadenylation events. SRSF6 is an example of a shift toward usage of a proximal polyA site and consequent expression of a shorter 3′ UTR isoform in the absence of PUS1 (Figure 6a). This pre-mRNA is directly modified by PUS1 at two locations near to and upstream of the proximal and the distal poly(A) sites (Table ST1H). Overall, this analysis suggests hundreds of instances of alternative cleavage and polyadenylation that exhibited reproducible and greater than 10% differences in polyA site usage (PAU) in PUS-depleted compared to wildtype cells (Figure 6b, Table ST1O-Q). Applying 3′ end sequencing methods to PUS-depleted cells will facilitate quantification the full extent of PUS-dependent APA and allow precise annotation of the 3′ ends of affected transcripts in the future. However, many studies have used conventional RNA-seq data to infer changes in APA (Xia et al., 2014; Kim, You and Nam, 2015; Grassi et al., 2016; Ha, Blencowe and Morris, 2018; Goering et al., 2021) and in cases were quantification of APA from conventional RNA-seq has been compared to 3′ end seq strong correlations have been observed (Ha, Blencowe and Morris, 2018; Goering et al., 2021). Pre-mRNA pseudouridines were enriched in the binding sites of canonical cleavage and polyadenylation factors, CSTF2T and CSTF2, and in binding sites of several RBPs known to regulate 3′ end processing (Figure 6c, Figure 3d, Table ST1E) suggesting a mechanism that could mediate altered 3′ end processing in response to PUS activity. These results extend the known roles of nuclear PUS to include controlling alternative 3′ end processing in human cells. The installation of pseudouridine co-transcriptionally positions it to influence multiple steps of pre-mRNA processing to establish distinct gene expression programs.
Figure 6. Pseudouridine synthases regulate 3′ end processing.
a) Genome browser view of a PUS1-dependent alternative cleavage and polyadenylation (APA) in the 3′ UTR of SRSF6. Upon PUS1 KO there is a shift toward usage of a proximal polyA site (PAS1) and away from the diatal polyA site (PAS2) resulting in expression of a shorter 3′ UTR isoform. The location of a two pseudouridine in this 3′ UTR upstream of each PAS is indicated. b) The number of significant alternative cleavage and polyadenylation events in PUS1 KO (n=2 biological replicates), RPUSD4 KD (n=2 biological replicates) or PUS7 KD (n=3 biological replicates) compared to WT is displayed by type of APA: 3′ UTR shortening and 3′ UTR lengthening. Significant APA events were determined from QAPA as those events that changed by greater than 10% difference in polyA site usage and were reproducible across replicates. c) Genome browser views of CSTF2T eCLIP peak and size-matched input controls (SMI) in the 3′ UTR of HMGS1. The location of a pseudouridine relative to the eCLIP peak is denoted by (Ψ).
Discussion
Mature human mRNAs were previously shown to be pseudouridylated, but the timing of pseudouridylation and the role of this modification in mRNA metabolism and gene regulation remained unaddressed. Here, by investigating chromatin-associated RNA we uncovered widespread modification of nascent pre-mRNA with pseudouridine, positioning pseudouridine to influence virtually all steps of mRNA processing. Consistent with this broad potential, we observed widespread changes in alternative splicing and 3′ end processing in response to changes in the expression of specific pre-mRNA modifying pseudouridine synthases. In further support of the function of pre-mRNA pseudouridines, we show that site-specific pre-mRNA pseudouridylation is sufficient to alter splicing outcomes in vitro. Notably, we find that pre-mRNA pseudouridines are significantly enriched within the experimentally determined binding sites of multiple splicing and processing factors and other regulatory RNA-binding proteins (RBPs). This finding, in light of previous reports that the presence of pseudouridine changes the affinity of diverse RBPs for their target RNAs, suggests a likely mechanism for pre-mRNA pseudouridylation to affect nuclear RNA processing. We note that pre-mRNA pseudouridylation is likely to be substantially more extensive than the sites identified here because stringent coverage criteria for detection and the large size of the nascent transcriptome restricted pseudouridine discovery to ~1% of highly expressed genes.
We validated hundreds of candidate pseudouridines identified in cells as direct targets of one of 8 purified human PUS proteins tested in vitro. These results establish PUS1, PUS7 and RPUSD4 as pre-mRNA modifying enzymes and add pre-mRNA as a new class of RNA targets for each of the tested PUS enzymes. Our data suggest that additional human PUS, among the 5 not tested here, modify pre-mRNA sites. Alternatively, some of the PUS tested may modify sites that were not recapitulated in our minimal in vitro system. Failure to be pseudouridylated in vitro could reflect the need for additional RNA sequences, cellular co-factors, low activity of recombinant protein, protein modifications not present in recombinant protein, or other features present in the endogenous context such as co-transcriptional PUS recruitment. While the PUS that modify the remaining candidate pseudouridine sites remain to be identified, unassigned sites resemble the in vitro validated sites in Pseudo-seq signal intensity and reproducibility in cells (Figure 1c,e, Table ST1C) suggesting that we have validated only a subset of true pseudouridine sites. Future work will likely reveal which of the other human PUS pseudouridylate pre-mRNA sequences and whether co-factors or co-transcriptional PUS recruitment are required for installation of some pseudouridines.
About half of the newly identified candidate pseudouridines are located in introns, where pseudouridine was not previously known to occur. These pseudouridines are well positioned to affect alternative splicing due to their enrichment in proximal introns, alternatively spliced regions and within splicing factor binding sites. Consistent with this regulatory potential, we find that installation of a single intronic pseudouridine is sufficient to affect splicing outcome in vitro. An advantage of these in vitro experiments is avoidance of all potentially confounding indirect effects of genetic manipulation of PUS activity in cells, providing strong support for a direct mechanistic effect of individual endogenous pre-mRNA pseudouridines on splicing.
Notably, the expression levels of the PUS proteins vary across tissues and cell types (Supplemental Figure 7a-c) highlighting the regulatory potential for pre-mRNA pseudouridylation by these factors to control human gene expression. In support of this idea we present evidence for cell type specific regulation of pre-mRNA pseudouridylation in HepG2 compared to HeLa (Supplementary Figure 1g) and show that genetic manipulation of pre-mRNA pseudouridylating enzymes PUS1, PUS7 and RPUSD4 in cells leads to thousands of changes in alternative splicing as well as alternative cleavage and polyadenylation that altered 3′ UTR length for hundreds of mRNAs. The mechanisms guiding cell type specific pre-mRNA modification and their functional outcome on cell type-specific gene regulation are open areas for investigation.
Mechanistically, pseudouridines in pre-mRNA have the potential to influence splicing by three main mechanisms: altering pre-mRNA-snRNA interactions, modulating pre-mRNA-protein interactions, or by influencing pre-mRNA secondary structure (Martinez and Gilbert, 2018). We identified pseudouridines in regions poised to function by any of these modes. Single pseudouridines stabilize synthetic RNA duplexes by 1-2 kcal/mol as compared to uridine base pairs (Hudson, Bloomingdale and Znosko, 2013; Kierzek et al., 2014). This stabilizing effect is predicted to enhance splice site recognition by promoting binding of spliceosomal snRNAs to the pseudouridylated 5′ splice site or branch site. Pseudouridines have also been shown to alter the affinities of various RNA binding proteins (e.g. PUM2, MBNL1, U2AF2) for RNA in vitro (Chen et al., 2010; Delorimier et al., 2017; Vaidyanathan et al., 2017). We identify 3′ splice site recognition factor U2AF2 as one of the factors that has significant overlap between its binding sites and pseudouridine locations in pre-mRNA. U2AF1, the other component of the U2AF heterodimer also overlaps pseudouridines locations in pre-mRNA including at the 3′ splice site. The sensitivity of most RBPs to pseudouridylation of their binding sites remains to be determined, but many are likely to be affected given that diverse RBPs show 2 to 100-fold differences in affinity for artificially pseudouridylated compared to unmodified RNA (Chen et al., 2010; Delorimier et al., 2017; Vaidyanathan et al., 2017). Finally, pseudouridylation could indirectly alter splice site accessibility and/or splicing factor binding by changing pre-mRNA secondary structure, as has recently demonstrated for certain intronic m6A modification sites (Liu et al., 2015).
Altogether, our results implicate pseudouridine and the pre-mRNA modifying pseudouridine synthases as novel regulators of pre-mRNA processing. This may have clinical relevance for the multiple pseudouridine synthases implicated in mitochondrial myopathy (Bykhovskaya et al., 2004; Fernandez-Vizarra et al., 2007), digestive disorders (Barrett et al., 2008; Dubois et al., 2010; McGovern et al., 2010; Festen et al., 2011; Repnik and Potočnik, 2016), intellectual disability (Shaheen et al., 2016; de Brouwer et al., 2018), resistance to viral infection (Marceau et al., 2016; Zhao et al., 2016), X-linked dyskeratosis congenita(Heiss et al., 1998; Knight et al., 1999), and cancer (Mannoor, Liao and Jiang, 2012; Williams and Farzaneh, 2012; Thorenoor and Slaby, 2014; McMahon, Contreras and Ruggero, 2015).
Limitations of the study
Our study reveals that pre-mRNAs are extensively pseudouridylated co-transcriptionally, which positions pseudouridine to influence any step of mRNA processing. We recognize that this represents a subset of true sites as this approach is limited to highly expressed genes that were detected with sufficient coverage by pseudouridine profiling of chromatin-associated RNA. Therefore, lack of evidence of pseudouridylation cannot be interpreted as lack of pseudouridylation. This limitation also prevented us from interrogating the pseudouridine status of many PUS-dependent pre-mRNA processing changes. Alternative methods for pseudouridine detection, such as SCARLET (Liu and Pan, 2016) could be useful for additional validation of candidate pseudouridines in pre-mRNA by an orthogonal method and for estimating stoichiometry at sites of interest.
We find a new role for PUS in regulating widespread pre-mRNA processing and present evidence that pseudouridines can have a direct biochemical effect on splicing. However, we cannot rule out that some of the identified PUS-sensitive alternative splicing events are an indirect consequence of chronic PUS depletion. Additionally, our data suggests that PUS regulate 3′ end processing. Future characterization and quantification of exact mRNA 3′ ends by 3′ end sequencing will be valuable for further study of PUS-dependent APA regulation because the variability in the capture of mRNA 3′ ends in standard RNA-seq limits its application.
Finally, we show that multiple pseudouridine synthases directly modify pre-mRNA sequences in high throughput in vitro pseudouridylation assays. This work demonstrates a new class of targets for many PUS that are associated with diseases and whose expression is regulated in different cell types. We caution that a negative result in this assay should not be interpreted strongly since failure to be pseudouridylated in this approach could result from the need for additional RNA sequences, the absence of cellular co-factors, low activity of recombinant protein, protein modifications that are not present in recombinant protein, or other features present in the endogenous context such as co-transcriptional PUS recruitment.
Star Method
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Wendy V. Gilbert (wendy.gilbert@yale.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All sequencing and annotations data have been deposited at GEO and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cell Culture
Human hepatocellular carcinoma cells HepG2 from ATCC HB8065 (lot 59635738) were grown in DMEM (HyClone SH30022.FS) supplemented with 10% fetal bovine serum (FBS HyClone SH30071.03). Cells were grown at 37C with 5% CO2 and maintained at subconfluency.
Method details
Total RNA Isolation
HepG2 cells were harvested by pelleting and resuspending fresh or frozen (−80C) pellets in 1mL of QIAzol (Qiagen). Total RNA was harvested according to the manufacturer’s procedure.
Western Blotting
Whole cell lysates were made by pelleting HepG2 cells and re-suspending fresh or frozen (−80C) pellets in RIPA buffer (50mM Tris pH 8, 150 mM NaCl, sodium deoxycholate 0.5%, sodium dodecyl sulfate 0.1%, NP-40 1%), lysed on ice for 10 min with vortexing. Spun down at 4C and maximum speed (13,200 rpm) for 15 min and collected supernatant as lysate. Approximately, 20ug of whole cell lysates, as determined by Bradford assay, were loaded in 10% SDS-PAGE for western blot of PUS1 knockout and wildtype HepG2 cells. HepG2 fractions were isolated as described below and treated with Benzonuclease to release proteins from nucleic acid. Equal cell volumes of cellular fractions (3%) were loaded in 12% SDS-PAGE gels for Western blots of cell compartments to determine fraction purity. Gels were transferred to nitrocellulose membranes. Membranes were blocked in 5% milk and incubated with primary antibodies overnight at 4C in 5% milk low-salt TBST (50 mM Tris pH 7.5 150 mM NaCl 0.1% Tween-20). Antibodies used for Western blot were as follows: anti-PUS1 at 1:1000 (Bethyl Labs A301-651A), anti-U1-70K at 1:2000 (Millipore 05-1588), anti-GAPDH at 1:10,000 (Sigma-Aldrich G9545), anti-H3 at 1:20,000 and anti-DKC1 at 1:1000 (GeneTex GTX109000). Secondary antibody incubation was for 1 hour at room temperature using HRP conjugated antibodies: anti-mouse IgG at 1:3000(Invitrogen 62–6520) or goat anti-rabbit IgG at 1:3000 (Promega W4011). Washes were with high-salt TBST (50 mM Tris pH 7.5 400 mM NaCl 0.1% Tween-20).
CRISPR knockout generation
PUS1 CRISPR knockout HepG2 cells were generated using a two-guide strategy to take out the first exon containing the translation start codon. Oligos for the upstream PUS1upF 5′-CACCGCGCAGGGTCCACCGTCCGA -3′ and PUS1upR 5′-AAACTCGGACGGTGGACCCTGCGC -3′, and for the downstream guide PUS1dnF 5′- CACCGATAACAGCGGTTAGCGGCA -3′ and PUS1dnR 5′- AAACTGCCGCTAACCGCTGTTATC -3′ were phosphorylated and annealed and then cloned into px458 (Addgene) digested with BbsI. HepG2 cells were transfected with both plasmids for the upstream guide and downstream guide (1.25 μg of each) with Lipofectamine 2000 according to the manufacturer’s protocol in 6-well plates. After 24h the cells were visually inspected for GFP fluorescence and prepared for sorting by trypsinizing and re-suspended in FACS buffer (PBS without Mg2+ and Ca2+ and supplemented with 2% FBS). Single GFP positive cells were sorted on an Aria I sorter and plated into each well of a 96 well plate. PUS1 knockout clones were expanded and knockout verified by PCR of genomic DNA and Western Blot with anti-PUS1.
PUS protein depletion
shRNAs targeting PUS7 and RPUSD4 were cloned into the lentiviral vector pLKO-Tet-On (Addgene) digested with AgeI-HF and EcoRI-HF to remove the stuffer sequence. Two oligos containing the complementary shRNA targeting sequence with the corresponding overhangs were annealed and ligated into the vector by standard cloning. Oligos sequences for RPUSD4 and PUS7 were:
shRPUSD4_2F
5′-CCGGGCTTCGAGTTCACTTGTCCTTCTCGAGAAGGACAAGTGAACTCGAAGCTTTTT G-3′, shRPUSD4_2R, 5′- AATTCAAAAAGCTTCGAGTTCACTTGTCCTTCTCGAGAAGGACAAGTGAACTCGAAG C-3′
shPUS7_4F
5′- CCGGTCTTAGTTCAGACTCATATATCTCGAGATATATGAGTCTGAACTAAGATTTTTG -3′
shPUS7_4R
5′- AATTCAAAAATCTTAGTTCAGACTCATATATCTCGAGATATATGAGTCTGAACTAAG A-3′.
Lentiviral particles were prepared by transfecting a 10cm dish of HEK293T cells with 5μg pLKO-Tet-On, 4.5μg pCMV-dR8.2, 500ng pCMV-VSV-G and transfection reagent X-tremeGENE 9 according to the manufacturer’s protopcol. Viral supernatant was harvested 48h after transfection. HepG2 cells were transduced in 6-well plate with 1mL of viral supernatant in 6 well plates with 1mL of cell suspension. Cells stably integrated with the lentiviral vector were selected 48h post-transduction with 3μg/mL of puromycin until cells in the untransduced cells did not survive. shRNA expression was induced in HepG2 cells with Doxocycline to a final concentration of 500 ng/mL. Cells were maintained in Doxoxycline containing media for 96h and whole cell lysates were prepared for Western Blot analysis of depletion and RNA was isolated for RNA-seq library construction.
Nuclear extract preparation
HepG2 cells were pelleted by spinning down at 1000 rpm for 5 min, washed with PBS. Cells were transferred to 1.5 mL tube and centrifuged at 3000 rpm for 1 minute. The pellet was resuspended in cytoplasmic extract buffer (10 mM HEPES pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 0.15% NP-40) ~100 μL per 2x106 cells and incubated on ice for 5 minutes and spun down for 3 min at 6500 rpm for 3 minutes. Supernatant was collected as cytoplasmic fraction. The nuclear pellet was re-suspended in equal volume of nuclear extract buffer (20 mM HEPES pH 7.6, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 20% glycerol). Three cycles of 15 minutes at −80C followed by thawing at 37C with vortexing for 1 minute in between cycles. Spun down at max speed for 15 minutes at 4C. Collected supernatant as nuclear extract.
Cellular Fractionation
Biochemical fractionation was performed essentially as described in (Bhatt et al., 2012; Pandya-Jones et al., 2013) for 11 biological replicates of HepG2 cells. All fractions were prepared from fresh cell pellets. Two 10 cm dishes with ~10x106 each of HepG2 cells were trypsinized and spun down at 500xg and washed with cold PBS (1mM EDTA). Cell pellets were re-suspended in 400 μL of cytoplasmic NP-40 lysis buffer (10 mM Tris-HCl pH 7.5, 0.15% NP-40, 150 mM NaCl) by flicking tube and incubated on ice for 5 minutes. Lysate was layered over 1 mL of sucrose cushion (24% RNAse-free sucrose in cytoplasmic lysis buffer) and spun down 10 minutes at 15,000xg at 4°C. Supernatant was collected as cytoplasmic fraction. Nuclear pellet was washed 2x with PBS without displacing pellet (1mM EDTA) and re-suspended in 200 μL glycerol buffer (20 mM Tris-HCl pH 7.9, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, 50% glycerol) by flicking tube followed by addition of 200 μL of nuclei lysis buffer (10 mM HEPES pH 7.6, 1 mM DTT, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M UREA, 1% NP-40). Samples were then vortexed on high 2x 2 seconds, incubated on ice for 2 minutes, centrifuged for 2 min at 4°C 15,000xg and the supernatant collected as nucleoplasmic fraction. The remaining chromatin pellet was washed 2x with PBS without displacing pellet (1mM EDTA) and re-suspended in 100uL of PBS (1mM EDTA). DNase I (2uL NEB) was added to resuspended chromatin-pellet and incubated at 37°C for 5-10 minutes to dissolve pellet. Collected 10uL for western blot of fractions and added 1mL of QIAzol (Qiagen) to remaining chromatin and incubated at 50°C for 10 min to solubilize chromatin. Followed manufacturer’s instruction to finish isolating chromatin-associated RNA with QIAzol. Samples were DNase I treated (NEB) treated after QIAzol isolation according to manufacturer’s instructions.
rRNA depletion
Ribosomal RNA (rRNA) was depleted using one Ribozero (Illumina MRZH11124) reaction per 20ug of chromatin-associated RNA following the manufacturer’s protocol.
Recombinant PUS plasmids and purification
Human TRUB1, PUS7, PUS7L, TRUB2, RPUSD2 and PUS10 were cloned from human cDNA obtained from Human ORFeome (cDNA from Mammalian Gene Collection) with Gibson assembly into the BamH1 site of pET15b expression. Recombinant hPus1 was purified as described (Czudnochowski et al., 2013). Expression was induced in BL21 (DE3) Gold cells [Agilent] with 0.1 mM IPTG at OD600 0.6-0.8. Cells were grown overnight at 16°C, then harvested by centrifugation and resuspended in lysis buffer (50 mM HEPES-KOH pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 1x Protease Inhibitor Cocktail (Roche)) and lysed by sonication. Human PUS1 was induced and the bacterial lysate was centrifuged at 12,000 rpm for 30 min, bound on a HisTrap column (GE Healthcare) and eluted off the column with 250 mM imidazole. The protein was then dialyzed overnight at 4°C into storage buffer (50 mM HEPES-KOH pH 7.0, 100 mM NaCl, 1 mM β-mercaptoethanol) and further purified by gel filtration over a Superdex-200 column (GE Healthcare). The protein product was concentrated with a centrifugal filter unit (MD Millipore) and concentration determined by Bradford staining against a BSA standard. Human TRUB1, PUS7, PUS7L, TRUB2, RPUSD2 and PUS10 were purified as follows. Rosetta 2 BL21 (DE3) pLysS cells were transformed with the expression vector and an individual colony was grown at 37°C in LB to OD600 0.8. Induction of expression was overnight at 18°C with 1mM IPTG. Protein was affinity purified using the HisTrap HP 5mL column (GE) on an FPLC. Bound protein was washed with wash buffer (50mM potassium phosphate buffer pH 8, 0.5M NaCl, 30mM Imidazole) and then eluted with elution buffer (50mM potassium phosphate buffer pH 8, 0.5M NaCl, 300mM Imidazole). Protein was concentrated with Amicon Ultra Centrifugal Filter Units and stored in storage buffer containing 20mM HEPES (pH 7.5), 200mM NaCl, 10% glycerol, and 1mM DTT. Concentration was determined by Bradford with BSA standards.
In vitro pseudouridylation
A pool of 6000 DNA oligos (Twist Biosciences) was designed to contain all the sites identified as pseudouridines in chromatin-associated RNA from HepG2 cells. The DNA oligos in the pool contained 65 nucleotides upstream and 64 nucleotides downstream of the pseudouridine. In addition, the sequences contained the T7 promoter and a 3′ adapter sequence to serve as handles for PCR amplification. The pool was amplified by PCR with primers oBZ131 (GCTAATACGACTCACTATAGGG) and oTC_pool2_rev (GTCCTTGGTGCCCGAGTG) using Phusion Polymerase (NEB). PCR reactions were supplemented with 3% DMSO and gel purified prior to in vitro transcription. RNA was in vitro transcribed with the MEGAshortscript T7 transcription kit (Thermo Fisher) and gel purified. RNA was re-suspended in water, denatured at 75C for 2 min, cooled on ice for 5 min and folded at 37C for 20 min following addition of 5X pseudouridylation buffer (500 mM Tris pH 8.0, 500 mM Ammonium Acetate, 25 mM MgCl2, 0.5 mM EDTA) to 1X for the final reaction volume. In vitro pseudouridylation reactions were carried out by incubating 15-30 pmol of folded RNA with 600nM recombinant human pseudouridine synthases: PUS1, PUS7, PUS7L PUS10, RPUSD2, RPUSD4, TRUB1, TRUB2, HepG2 nuclear extract or no PUS for 45 minutes at 30C in 500uL final reaction volumes extract in (1X pseudouridylation buffer, 2mM DTT). RNA was purified immediately by phenol chloroform extractions and isopropanol precipitated.
Pseudo-seq
Pseudo-seq libraries were prepared as previously described in detail (Carlile, Rojas-Duran and Gilbert, 2015). Briefly, rRNA-depleted chromatin-associated RNA was fragmented in 10mM ZnOAc for 2 minutes at 60C. Fragmentation was quenched by addition of EDTA to 20mM. RNA was then either treated with CMC (0.4M final) or mock treated (−CMC) in BEU buffer for 45 min at 40C and CMC was reversed from Us and Gs by incubation in Sodium carbonate buffer for 2 hours at 50C. RNA fragments 120-140 nucleotides in length were size selected from denaturing polyacrylamide gels for library preparation. The ends of the fragmented RNA were repaired by treatment with T4 Polynucleotide Kinase (NEB) and Calf intestinal alkaline phosphatase (NEB) in 1X PNK buffer (NEB). A 3′ adenylated adapter (AppTGGAATTCTCGGGTGCCAAGG/3ddC/) was ligated to the RNA fragments with T4 RNA ligase in 1X T4 RNA ligase buffer (NEB), followed by reverse transcription with AMV RT (Promega) and RT primer:
/5Phos/GATCGTCGGACTGTAGAACTCTGAACCTGTCGGTGGTCGCCGTATCATT/iSp18/CACTCA/iSp18/GCCTTGGCACCCGAGAATTCCA. Truncated cDNAs (120-170nt) were size selected from denaturing polyacrylamide gels and gel purified cDNAs were circularized with CircLigase ssDNA ligase (Epicentre). Circularized cDNAs were PCR amplified with forward primer (AATGATACGGCGACCACCGA) and BC reverse primer (CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCCTTGGCACCCGA GAATTCCA where Xs represent the unique barcode sequence). In vitro Pseudo-seq libraries were prepared as described above (Martinez, Nicole M., Gilbert, 2021) except with full length in vitro transcribed RNA recovered from in vitro pseudouridylation assays. After CMC modification and reversal, full length RNA was gel purified in denaturing polyacrylamide gels. Reverse transcription was performed using the 3′ adapter sequence appended to the pool oligos as a handle with RT primer ONM_RT-L2
(/5Phos/NNNNNNNNNGATCGTCGGACTGTAGAA CTCTGAACGTGTAGATC/iSp18/CACTCA/iSp18/CCTTGGCACCCGAGAATTCCAGTCCT TGGTGCCCGAGTG), truncated cDNAs from 140-170 nts were size selected and circularized for PCR amplification with primers RP1 (AATGATACGGCGACCACCGAGATCTACACGTT CAGAGTTCTACAGTCCGA) and BC reverse primer. Libraries were sequenced on an Illumina HiSeq in single-end mode to 20-30 million reads per sample for in vivo Pseudo-seq and 15-20 million reads for in vitro Pseudo-seq libraries.
RNA-seq
Total RNA was isolated for two replicates of wildtype and PUS1 knockout HepG2 cell as described above and stranded poly(A)+ selected mRNA-seq libraries were performed by the MIT BioMicroCenter using the TruSeq Stranded mRNA Library Prep Kit from Illumina according to the manufacturers’ protocol. Libraries were sequenced on an Illumina Next-seq with paired end 40-bp reads to a depth of ~150 million paired reads per replicate. mRNA purity was comparable across libraries with 1.4% (WT1), 1.7% (WT2), 1.3% (KO1) and 1.3% (KO2) rRNA contamination. PUS7 (n=3) and RPUSD4 (n=2) knockdown stranded poly(A)+ selected mRNA-seq libraries were prepared by Genewiz and sequenced on a HiSeq with paired end 150-bp reads.
Sequencing Analysis
All sequencing data has been deposited in GEO, accession GSE123613. mRNA-seq reads were mapped to the human genome using STAR (Dobin et al., 2013) aligner. Reads were mapped to genome assembly GRCh37 with ENSEMBL GRCh37.75 annotations. STAR alignment was carried out using the following parameters: -- outFilterType BySJout -- outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 -- alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --alignEndsType EndToEnd. Pseudo-seq sequencing data was analyzed using in house Bash and Python scripts. Cutadapt (Martin, 2011) was used to trim the 3′ adapter sequence from the reads. In vitro Pseudo-seq sequencing reads were also PCR duplicate collapsed using fastx_collapser (http://hannonlab.cshl.edu/fastx_toolkit/). Processed reads were mapped to a bowtie index of ENSEMBL GRCh37.75, 45S rRNA and snRNAs for in vivo Pseudo-seq libraries and the pool of sequences for in vitro Pseudo-seq libraries using tophat2 (Kim et al., 2013). The mapped reads were processed with in house Python scripts. Pseudo-seq signal or peak height was calculated as follows for all possible used annotated in GRCh37.75 excluding repetitive elements. For each U position centered on a 51-nucleotide window, the fraction of reads whose 5′ends map to the position was divided by the reads mapping to the window was calculated. Pseudo-seq signal is the difference in the fraction of reads between the +CMC and −CMC multiplied by the window size. For each Ψ the Pseudo-seq signal corresponds to that of the expected RT stop 1 nt 3′ of the Ψ site. Sites were called as pseudouridines if an RT stop with a peak height >1, met a read cutoff (reads/nt in the window) of 0.1 and was present in 7 out of 11 replicate libraries of chromatin-associated RNA.
PUS protein assignment
Peak heights for in vitro pseudouridylated sites were determined using the pipeline described previously in detail (Martinez, Schaening-Burgos and Gilbert, 2021). Briefly, peak heights were calculated based on a Z-score of read 5′ ends accumulating at the modified site relative to the background of other positions in a selected window in CMC-treated libraries as shown below.
Where:
readsi = 5′ends at position
μ = avg 5’ ends at each position in window
σ = standard deviation of 5’ ends counts in window
Size selection of cDNA can lead to coverage biases. A window from positions 60 to 95 was selected as the background for the Z-score calculation based on the distribution of reads covering the RNA oligos. Z-scores are calculated for each putative psi position in the RNA pool and sites are assigned as modified by a particular PUS by selecting cutoffs such that the modified sites have Z-scores greater than the background of all other sites within each library and compared to a library prepared without addition of a pseudouridine synthase (no PUS). We plotted the distribution of Z-scores for each library as an inverse CDF. We chose cutoffs corresponding to the inflection point where the modified sites diverged from background sites within the same library and from the CMC-treated no PUS) library (Supplemental Figure 4b). We also required that the Z-score be higher than in a mock treated (−CMC) library to ensure the peak is CMC-dependent. For the libraries in (Supplemental Figure 4b) the cutoffs were set as follows: PUS7_plusCMC > 5, noPUS_plusCMC < 4 and Zdiff (PUSplusCMC − minusCMC) >= 2. This approach allows us to assign sites as targets of more than one PUS in the case that more than one enzyme modifies the same site and allows us to compare data generated from different experiments.
Alternative splicing analysis
Analysis of differential alternative splicing between wildtype and PUS1 KO HepG2 mRNA-seq was carried out by rMATS (version 3.) using Ensembl GRCh37.72 annotations. rMATS reported differences in alternative splicing of types skipped exons (SE), alternative 3′ splice sites (A3SS), alternative 5′ splice sites (A5SS), retained introns (RI) and mutually exclusive exons. Percent inclusion differences were determined from the rMATS junction only output files. Events with an absolute difference in percent inclusion between PUS1 KO and WT cells of greater than or equal to 10% and with and false discovery rate (FDR) of equal to or less than 0.05 are considered significant and reported.
Differential gene expression analysis
HTseq (version 0.6.1) was used to generate read counts tables that were submitted to DESeq2 (Love, Huber and Anders, 2014)(version 1.14.1) to determine differences in mRNA abundance between PUS1 KO, RPUSD4 KD or PUS7 KD and WT HepG2 cells from mRNA-seq data. After DESeq analysis genes with 0 counts in at least one sample were filtered out. Differences in mRNA levels with Padj < 0.05 were considered significant.
Enrichment Analysis
Bedtools was used for overlapping pseudouridines and uridines with genomic features. As a background set for calculating the enrichment of pseudouridines in different regions we used all the uridines that met our read cutoff of 0.1 in our replicate cutoff of 7 out of 11, as was used for pseudouridine calling. Detected pseudouridines were classified into features according to gencode v19 annotations according to the following priority snoRNAs > lincRNAs > snRNA > miRNAs > antisense ncRNAs > 5′ UTR > 3′ UTR > exons > introns. Retained introns were filtered as introns. Enrichments of pseudouridines in the introns of alternatively spliced regions was carried out by overlapping intronic pseudouridines compared to uridines with MISO version 2 annotations (https://miso.readthedocs.io/en/fastmiso/annotation.html) which were generated from Ensembl genes, known Genes (UCSC) and RefSeq genes annotations. We compared the distribution of pseudouridines to uridines that we detected in introns flanking alternatively spliced region categories obtained as described above (Figure 2b). A a chi-squared test for the overall difference in proportions across categories for pseudouridines compared to uridines was applied to evaluate the significance of the difference in the distribution.(p-value = 2.2e−16). We compared the observed pseudouridine distribution to the uridine distribution for uridines that were captured in our sequencing assay to correct for uridine content and coverage bias. The distribution for pseudouridines relative to splice sites was compared to the distribution of detected uridines in proximal introns (within 500nt of splice sites) and splice sites (within 6 nt from exon ends). A hypergeometric test was applied (Fisher’s exact test) to determine the significant in the observed enrichment of pseudouridines in these regions. P-values < 0.05 were considered significant. Pseudouridines were considered as present in branch site regions if they overlapped annotated branch site regions (Mercer et al., 2015; Taggart et al., 2017; Pineda and Bradley, 2018). Pseudouridines were classified as present in putative PPT tracts if they were within 50 nucleotides of the 3′ splice site AG and by manual inspection.
Motif Enrichment
131 nucleotides of sequence surrounding pseudouridine was used as the input to MEME (Bailey et al., 2015) version 5.0.2 in discriminative mode to identify motifs enriched in pseudouridine compared to the 131 nucleotides of sequence flanking the background set of uridines that met our read cutoff for pseudouridine calling in the Pseudo-seq libraries. Significantly enriched motifs are presented.
GO analysis
Gene ontology analysis was performed for pseudouridine containing genes using PANTHER version 11 (Mi, Muruganujan and Thomas, 2013). All Ensembl gene IDs for pseudouridine containing genes were used as the test set and the Ensembl gene IDs for all genes that contained uridines matching our read cutoff for pseudouridine identification (Supplementary Table 4). Reported enriched GO terms correspond to those that were significant after Bonferroni correction for multiple hypothesis testing.
Analysis of RBP eCLIP overlap with pseudouridines
Size matched input-normalized bed files for eCLIP biological replicates were combined into a single bedtool of shared peaks using bedtools intersect, where a shared peak was defined as at least one intersecting nucleotide. eCLIP peaks at pseudouridines in HepG2 cells were then identified by using bedtools intersect to determine eCLIP peaks where the peak overlapped with a pseudouridine. Volcano plots of these pseudouridine intersecting eCLIP peaks were then generated using the eCLIP log2(fold change) and Padj (Van Nostrand et al., 2016) values. We selected significance cutoffs of log2(fold change) of 2 and −log10(Padj) of 3. For the volcano plots, if an RBP had multiple pseudouridines within an eCLIP peak, we plotted the best cluster as defined by first the lowest Padj and highest fold change values. Introns were determined by the genic location using gencode hg19 annotations. To identify pseudouridine-interacting RBPs we ranked RBPs for preferential binding to pseudouridine sites. We calculated the fraction of significant eCLIP peaks that intersected pseudouridine sites over the total number of significant eCLIP peaks, then compared this ratio to one calculated using pseudouridine sites shuffled within introns. We performed this calculation with 1000 iterations of shuffling the pseudouridine location in introns to calculate a standard distribution of this ratio, and then calculated the z-score of the observed ratio. To filter for RBPs that bind uridine-rich motifs, we performed the same calculation with all high-confidence uridines that did not have a pseudouridine and repeated the 1000 iterations of shuffling and measuring the intersection with the RBPs.
In vitro splicing from nuclear extracts
The splicing reporter backbone pcAT7-Glo1 was digested with NdeI and XbaI and Gblocks including the entire endogenous exon and 100nt of intronic sequence (including the pseudouridylated position) and 15nt of vector overlapping sequence. The Gblock sequences are:
RBM39
TAGAAACTGGGCATATGATTTGAATGAACCCTGCTATTGTAGTCCTCTTTTATTAATG CTTTCCTGACATTTACCCTGTTAGTTGAGGCTCTTCATTGTTCCTGCACTGAGCTGTA GAATTCTCTTTTGTTATAGATTTGCAAACAAGACTTTCCCAGCAGACTGAAGTCTAG AGGGCCCGTTT
MDM2
TAGAAACTGGGCATATGGCTTTAGTTTTAACTGTTGTTTATGTTCTTTATATATGATG TATTTTCCACAGATGTTTCATGATTTCCAGTTTTCATCGTGTCttttttttCCTTGTAGGCAA ATGTGCAATACCAACATGTCTGTACCTACTGATGGTGCTGTAACCACCTCACAGATT CCAGCTTCGGAACAAGAGACCCTGTCTAGAGGGCCCGTTT.
Plasmids were in vitro transcribed and subsequently linearized by restriction digest with Xba1. Splicing reporter RNA (30pmol) were mock treated for unmodified or in vitro pseudouridylated with recombinant human PUS7 as described in the in vitro pseudouridylation section. Purified RNA substrates (15 pmol) were incubated with 32% Jurkat or HeLa nuclear extract in a total volume of 13 uL under splicing conditions 11.5 mM Tris-HCl, pH7.5, 3.0 mM MgCl2, 1 mM ATP, 20 mM CP, 0.5 mM DTT, 58 mM KCl, 3% PVA, 0.1 mM EDTA, and 11.5% glycerol. Reactions were incubated for 30-60 min at 30°C or indicated time points. RNA was recovered from the reactions by proteinase K treatment, phenol- chloroform extraction and ethanol precipitation. The RNAs from the splicing reactions were analyzed by RT-PCR performed and analyzed as previously described in using reporter and gene-specific primers and a low cycle number (Lynch and Weiss, 2000). Primer sequences for reporter specific primers GE1-F – 5′-GCAAGGTGAACGTGGATGAAGTTGG – 3′ and for gene-specific primers MDM2_E-R 5′-CAGGGTCTCTTGTTCCGAAGCTGG -3′ or RBM39 E-R 5′-CAGTCTGCTGGGAAAGTCTTGTTTGC -3′.
Site lists and reproducibility analyses
We obtained lists of pseudouridine sites called in two publications: the initial list from our group, published in supplemental table 8 of Carlile et al. 2014, and those reported in Table ST1G of Khoddami et al. 2019. For Extended Data Figure 1B, we compared the lists reported by either publication and categorized them into sets, based on whether they had been called by either or both of these methods. For Extended Data Figure 1C, we additionally determined the sites that had been called in any of the following publications: (a) Schwartz 2014, which called sites transcriptome wide in HEK293 cells, rather than HeLa; (b) Carlile 2019, which used an in vitro approach to validate sites (c) Li 2015. We then used the UpSetR package (version 1.4.0) to visualize the reproducibility of the Carlile 2014 sites across these datasets. ROC curves for HepG2 CARNA were generated based on a list of true positive sites corresponding know pseudouridine positions in the mature human 28S, 18S and 5.8S rRNA (Lestrade and Weber, 2006; Taoka et al., 2018) as previously described (Carlile et al., 2014). Pseudo-seq signal was calculated 1 nt 3′ to each known pseudouridine. A range of 500 equally spaced cutoff scores were chosen spanning the range of observed peak values. At each cutoff score, the true positive and false positive rates were calculated, and plotted. False positive rate was calculated for chosen cutoffs used for site calling in HepG2 chromatin-associated RNA (see Supplemental Figure 1a and Table S1).
Sequence context analyses
For each set of sites in Extended Data Figure 1B, we generated a fasta file that contained the 11-nt sequence surrounding the pseudouridine site. We then used Weblogo 3 to visualize the frequency of each nucleotide relative to the pseudouridine site.
RBS-Seq signal analysis
We downloaded the raw reads from GSE90963, using reads obtained from RiboMinus-treated total RNA (GSM2418439 and GSM2418440) and polyA-selected RNA (GSM2418443 and GSM2418444). Adapters were trimmed using bbduk. Reads were mapped to hg19 using novoalign, using parameters -t 60 -h 120 -b 4 -H 20 -r A 2 -s 2. For bisulfite-treated samples, the -b 4 parameter was also included. Python scripts using the pysam package were used to determine coverage, the number of deletions, and the number of read 5′ ends at each position, which were then stored as wig files for visualization.
Alternative cleavage and polyadenylation analysis
Analysis of alternative cleavage and polyadenylation was performed with QAPA (Ha, Blencowe and Morris, 2018), which takes in pseudoalignments. For this purpose we mapped the Illumina RNA-seq reads from WT, PUS1 KO, PUS7 KD and RPUSD4 KD using ENSEMBL GRCh37.75 genome fasta files and Gencode hg19 3′ UTR annotations. Fasta files of 3′ UTR annotations was generated with:
qapa fasta -f genome_fasta_file gencode_3UTR_annotations.bed output_sequences_qapa.fasta A salmon index was generated with command: salmon index -t output_sequences_qapa.fa -I utr_library. The paired sequencing reads for each sample were then mapped using command: salmon quant.sf -I utr library −1 ISR −1 Readl.fastq.gz −2 Read2.fastq.gz --validateMappings -o sample_transcript quant. QAPA was then run on samples to quantify polyA site usage (PAU) at annotated 3′UTRs using the following command: qapa quant --db ensembl.identifiers.txt sample_transcript_quant*/quant.sf > PAU_results.txt. We then calculated mean PUA per condition and filtered for >5 TPM in at least 2 samples. An event was classified as an isoform switch if the average difference in PUA between conditions was greater than 10% and the difference between replicates was less than 10%.
Primer extension
Total RNA from WT and PUS1 KO cells was isolated as described above and 20 μg of RNA for each sample was treated with CMC (final concentration 0.1 M) treated and reversed as described. Reverse primers to the U2 snRNA (U2snRNA_ext_sh and U2 snRNA_ext_lo were radiolabeled with γ-ATP (Perkin-Elmer) by treatment with T4 PNK (NEB). Primer extension was carried out with AMV RT (Promega). Briefly, primers were annealed in 1× AMV RT buffer by heating to 90 °C then cooling to 42 °C over 30 minutes in thermocycler. Reverse transcription was carried out at 42 °C for 1 h. Reactions were quenched with 2× stop solution (0.5× TBE, 90% formamide, 0.05% w/v bromophenol blue, 0.05% w/v xylene cyanol). Reactions were then run on 10% TBE–urea PAGE sequencing gel.
Quantification and statistical analysis
Statistical analysis was performed in R unless otherwise stated in the methods. Student’s T Tests were performed using the t.test() function in R. The hypergeometric tests were performed using the fisher.test(), chisq.test function in R. All other statistics data is specified in the methods and figure legends.
Supplementary Material
Supplementary Table 1, Pseudo-seq, eCLIP, in vitro PUS sites, gene expression, exon skipping, and APA data (related to Figures 1, 3-6)
(A) HepG2 45S rRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(B) HepG2 ncRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(C) HepG2 pre-mRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(D) HeLa mRNA pseudouridine validation analysis (related to Figure 1) (E) RBP eCLIP peaks with significant enrichment over the size matched input (> 4-fold enrichment and adjusted p-value < 0.001) and overlapping pseudouridine locations (related to Figure 3).
(F) Z scores comparing the fraction of RBP eCLIP peaks overlapping pseudouridines to the calculated overlap after shuffling pseudouridines within intronic regions 1000 times and the fraction of RBP eCLIP peaks overlapping uridines to the calculated overlap after shuffling uridines within intronic regions 1000 times (related to Figure 3).
(G) Genes containing pseudouridine versus a background set of genes that met uridine read cutoffs which were used for gene ontology analysis (related to Figures 1 and 3).
(H) In vitro pseudouridylated sites by recombinant human PUSs. Z-score is listed for each source of PUS. PUS assignment was determined by Z-score analysis (related to Figure 4).
(L) Cassette exons or skipped exons (SE) that are alternatively spliced in PUS1 KO compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(J) Differentially expressed genes in PUS1 KO compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(K) Cassette exons or skipped exons (SE) that are alternatively spliced in RPUSD4 KD compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(L) Cassette exons or skipped exons (SE) that are alternatively spliced in PUS7 KD compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(M) Differentially expressed genes in PUS7 KD compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(N) Differentially expressed genes in RPUSD4 KD compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(O) Genes showing alternative polyadenylation (APA) in PUS1 KO compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
(P) Genes showing alternative polyadenylation (APA) in RPUSD4 KD compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
(Q) Genes showing alternative polyadenylation (APA) in PUS7 KD compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-PUS1 | Bethyl Labs | A301-651A |
| Mouse monoclonal anti-U1-70K | Millipore | 05-1588 |
| Rabbit polyclonal anti-GAPDH | Sigma-Aldrich | G9545 |
| Rabbit polyclonal Anti-H3 | Abcam | ab1791 |
| Rabbit polyclonalanti-DKC1 | GeneTex | GTX109000 |
| Goat anti-mouse IgG, HRP | Invitrogen | 62-6520 |
| Goat anti-rabbit IgG, HRP | Promega | W4011 |
| Bacterial and virus strains | ||
| One Shot™ Stbl3™ Chemically Competent E. coli | Invitrogen | C737303 |
| NEB® Stable Competent E. coli | NEB | C3040H |
| BL21 (DE3) Gold cells | Agilent | 230130 |
| NEB® 5-alpha Competent E. coli | NEB | C2988J |
| Rosetta 2(DE3)pLysS Competent Cells | Novagen | 71403-M |
| Chemicals, peptides, and recombinant proteins | ||
| Polybrene | Millipore Sigma | TR-1003-G |
| TRIzol Reagent | Invitrogen | 15596026 |
| QIAzol Reagent | Qiagen | 79306 |
| X-tremeGene 9 Transfection Reagent | Millipore Sigma | 6365787001 |
| Lipofectamine 2000 | Invitrogen | 11668027 |
| Benzonase Nuclease | Millipore Sigma | E1014-25KU |
| T4 PNK | NEB | M0201L |
| T4 DNA Ligase | NEB | M0202L |
| AMV RT | Promega | M5108 |
| MMLV RT | NEB | M0253S |
| Taq Polymerase | NEB | M0273S |
| Phusion Polymerase | NEB | M0530L |
| Doxycycline hyclate | Sigma-Aldrich | D9891-1G |
| Puromycin dihydrochloride | Sigma-Aldrich | P7255-100MG |
| N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide metho-p- toluenesulfonate (CMC) | Sigma-Aldrich | C106402 |
| BbsI | NEB | R0539S |
| NdeI | NEB | R0111S |
| XbaI | NEB | R0145S |
| hTRUB1 | This paper | N/A |
| hPUS7 | This paper | N/A |
| hPUS7L | This paper | N/A |
| TRUB2 | This paper | N/A |
| RPUSD2 | This paper | N/A |
| hPUS10 | This paper | N/A |
| hPUS1 | This paper | N/A |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | 4693159001 |
| Hyclone Fetal Bovine Serum | GE Healthcare | SH30071.03 |
| HyClone Dulbecco's Modified Eagle Medium (DMEM) | GE Healthcare | SH3022.FS |
| Critical commercial assays | ||
| T7 MegaShortScript kit | Thermo Fisher Scientific | AM1345 |
| rRNA depletion | Illumina | MRZH11124 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE123613 |
| Experimental models: Cell lines | ||
| HepG2 | ATCC | HB-8065 |
| HEK293T | ATCC | CRL-1573 |
| Oligonucleotides | ||
| PUS1upF 5′- CACCGCGCAGGGTCCACCGTCCGA -3′ | This paper (IDT) | N/A |
| PUS1upR 5′- AAACTCGGACGGTGGACCCTGCGC -3′ | This paper (IDT) | N/A |
| PUS1dnF 5′- CACCGATAACAGCGGTTAGCGGCA -3′ | This paper (IDT) | N/A |
| PUS1dnR 5′- AAACTGCCGCTAACCGCTGTTATC -3′ | This paper (IDT) | N/A |
| shPUS7_4F 5′- CCGGTCTTAGTTCAGACTCATATATCTCGAGATATATGAGTCTGAACTAAGATTTTTG -3′ | This paper (IDT) | N/A |
| shPUS7_4R 5′- AATTCAAAAATCTTAGTTCAGACTCATATATCTCGAGATATATGAGTCTGAACTAAGA-3′ | This paper (IDT) | N/A |
| shRPUSD4_2F 5′- CCGGGCTTCGAGTTCACTTGTCCTTCTCGAGAAGGACAAGTGAACTCGAAGCTTTTTG -3′ | This paper (IDT) | N/A |
| shRPUSD4_2R 5′- AATTCAAAAAGCTTCGAGTTCACTTGTCCTTCTCGAGAAGGACAAGTGAACTCGAAGC -3′ | This paper (IDT) | N/A |
| DNA pool of 6000 oligos | Twist Biosciences | GSE123613 |
| Pool PCR F primer (oBZ131) 5′-GCTAATACGACTCACTATAGGG -3′ | IDT | N/A |
| Pool PCR R primer (oTC_pool2_rev) 5′- GCTAATACGACTCACTATAGGG -3′ | IDT | N/A |
| Pool RT primer (ONM_RT-L2) 5′- /5Phos/NNNNNNNNNGATCGTCGGACTGTA GAACTCTGAACGTGTAGATC/iSp18/CACTC A/iSp18/CCTTGGCACCCGAGAATTCCAGTCCTTGGTGCCCGAGTG -3′ |
IDT | N/A |
| RP1 primer—Library PCR F 5′- AATGATACGGCGACCACCGAGATCTACAC GTTCAGAGTTCTACAGTCCGA -3′ |
IDT | N/A |
| Barcoded reverse PCR primers (XXXXXX indicates unique barcodes)—Library PCR R 5′- CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA -3′ | IDT | N/A |
| In cell Pseudo-seq RT primer 5′- /5Phos/GATCGTCGGACTGTAGAACTCTGAACCTGTCGGTGGTCGCCGTATCATT/iSp18/CACTCA/iSp18/GCCTTGGCACCCGAGAATTCCA -3′ | IDT | N/A |
| In cell Pseudo-seq–Library PCR F 5′-AATGATACGGCGACCACCGA -3′ | IDT | N/A |
| RBM39 splicing reporter GBlock TAGAAACTGGGCATATGATTTGAATGAACCCTGCTATTGTAGTCCTCTTTTATTAATG CTTTCCTGACATTTACCCTGTTAGTTGAGGCTCTTCATTGTTCCTGCACTGAGCTGTA GAATTCTCTTTTGTTATAGATTTGCAAACAAGACTTTCCCAGCAGACTGAAGTCTAG AGGGCCCGTTT |
This paper (IDT) | N/A |
| RBM39 splicing reporter GBlock TAGAAACTGGGCATATGGCTTTAGTTTTAACTGTTGTTTATGTTCTTTATATATGATG TATTTTCCACAGATGTTTCATGATTTCCAGTTTTCATCGTGTCttttttttCCTTGTAGGCAA ATGTGCAATACCAACATGTCTGTACCTACTGATGGTGCTGTAACCACCTCACAGATT CCAGCTTCGGAACAAGAGACCCTGTCTAGAGGGCCCGTTT. |
This paper (IDT) | N/A |
| GE1-F 5′- GCAAGGTGAACGTGGATGAAGTTGG – 3′ |
IDT | N/A |
| MDM2_E-R 5′- CAGGGTCTCTTGTTCCGAAGCTGG -3′ |
IDT | N/A |
| RBM39_E-R 5′- CAGTCTGCTGGGAAAGTCTTGTTTGC -3′ |
IDT | N/A |
| U2snRNA_ext_sh 5′- CCTCGGATAGAGGACGTATCAG -3′ |
IDT | N/A |
| U2snRNA_ext_lo 5′- TACCAGGTCGATGCGTGG -3′ |
IDT | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | 48138 |
| Tet-pLKO-puro | Addgene | 21915 |
| pCMV-dR8.2 | Addgene | 8455 |
| pCMV-VSV-G | Addgene | 8454 |
| pcAT7-Glo1 | Gift from Kristen Lynch | N/A |
| Software and algorithms | ||
| Bowtie 2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Tophat2 | Kim, 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
| STAR | Dobin, 2013 | https://github.com/alexdobin/STAR |
| cutadapt | Martin, 2011 | https://cutadapt.read/thedocs.io/en/stable/ |
| rMATS | Shen, 2014 | http://rnaseq-mats.sourceforge.net |
| MEME Suite | Bailey, 2015 | https://meme-suite.org/meme/ |
| HTseq | Anders, 2015 | https://htseq.readthedocs.io/en/master/ |
| DEseq2 | Love, 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| QAPA | Ha, 2018 | https://github.com/morrislab/qapa |
| Bedtools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
Acknowledgements
We thank Brenton R. Graveley for HepG2 cells, Tristan A. Bell for purified hPUS1, and Matthew G. Thompson and Kristen W. Lynch for splicing competent nuclear extracts and splicing reporter backbone plasmids. We thank Karla M. Neugebauer and members of the Gilbert lab for helpful discussions and critical reading of the manuscript. This work was supported by NIH GM101316 and American Cancer Society RSG-13-396-01-RMC to W.V.G; NIH HG004659 and HG009889, to G.W.Y.; and Jane Coffin Childs Post-doctoral Fellowship 161624T and NIH K99 GM135537 to N.M.M.
Footnotes
Declaration of Interests
G.W.Y. is co-founder, member of the Board of Directors, on the scientific advisory board, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. GWY is a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The authors declare no other competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attig J et al. (2018) ‘Heteromeric RNP Assembly at LINEs Controls Lineage-Specific RNA Processing’, Cell. doi: 10.1016/j.cell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL et al. (2015) ‘The MEME Suite’, Nucleic Acids Research, 43(W1), pp. W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC et al. (2008) ‘Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease’, Nature Genetics, 40(8), pp. 955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DM et al. (2012) ‘Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions’, Cell, 150(2), pp. 279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal U et al. (2015) ‘Profiling of ribose methylations in RNA by high-throughput sequencing’, Angewandte Chemie - International Edition. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- de Brouwer APM et al. (2018) ‘Variants in PUS7 Cause Intellectual Disability with Speech Delay, Microcephaly, Short Stature, and Aggressive Behavior’, American Journal of Human Genetics, pp. 1045–1052. doi: 10.1016/j.ajhg.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykhovskaya Y et al. (2004) ‘Missense Mutation in Pseudouridine Synthase 1 (PUS1) Causes Mitochondrial Myopathy and Sideroblastic Anemia (MLASA)’, The American Journal of Human Genetics, 74(6), pp. 1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Martinez NM et al. (2019) ‘mRNA structure determines modification by pseudouridine synthase 1’, Nature Chemical Biology, 15(10), pp. 966–974. doi: 10.1038/s41589-019-0353-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM et al. (2014) ‘Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells’, Nature, 515(7525), pp. 143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF and Gilbert WV (2015) Pseudo-Seq, Methods in Enzymology. doi: 10.1016/bs.mie.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C et al. (2010) ‘A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo.’, Molecular and cellular biology, 30(17), pp. 4108–4119. doi: 10.1128/MCB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N et al. (2013) ‘In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities’, Journal of Molecular Biology. doi: 10.1016/j.jmb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorimier E et al. (2017) ‘Pseudouridine modification inhibits muscleblind-like 1 (MBNL1) binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility’, Journal of Biological Chemistry, 292(10), pp. 4350–4357. doi: 10.1074/jbc.M116.770768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S and Gall JG (2017) ‘Dual nature of pseudouridylation in U2 snRNA: Pus1p-dependent and Pus1p-independent activities in yeasts and higher eukaryotes’, RNA. doi: 10.1261/rna.061226.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A et al. (2013) ‘STAR: ultrafast universal RNA-seq aligner’, Bioinformatics, 29(1), pp. 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois PCA et al. (2010) ‘Multiple common variants for celiac disease influencing immune gene expression’, Nature Genetics, 42(4), pp. 295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra E et al. (2007) ‘Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA)’, Journal of Medical Genetics, 44(3), pp. 173–180. doi: 10.1136/jmg.2006.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festen EAM et al. (2011) ‘A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for crohn’s disease and celiac disease’, PLoS Genetics. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD and Ares M (2014) ‘Context-dependent control of alternative splicing by RNA-binding proteins’, Nature Reviews Genetics, pp. 689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA and Schaening C (2016) ‘Messenger RNA modifications: Form, distribution, and function’, Science, 352(6292), pp. 1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R et al. (2021) ‘LABRAT reveals association of alternative polyadenylation with transcript localization, RNA binding protein expression, transcription speed, and cancer survival’, BMC genomics. doi: 10.1186/s12864-021-07781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi E et al. (2016) ‘Roar: Detecting alternative polyadenylation with standard mRNA sequencing libraries’, BMC Bioinformatics. doi: 10.1186/s12859-016-1254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratenstein K et al. (2005) ‘The WD-repeat protein GRWD1: Potential roles in myeloid differentiation and ribosome biogenesis’, Genomics. doi: 10.1016/j.ygeno.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Ha KCH, Blencowe BJ and Morris Q (2018) ‘QAPA: A new method for the systematic analysis of alternative polyadenylation from RNA-seq data’, Genome Biology. doi: 10.1186/s13059-018-1414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS et al. (1998) ‘X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions’, Nature Genetics. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Hudson G. a, Bloomingdale RJ and Znosko BM (2013) ‘Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides.’, RNA (New York, N.Y.), 19(11), pp. 1474–82. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK et al. (2003) ‘Global analysis of protein localization in budding yeast’, Nature. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ji X et al. (2015) ‘Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions.’, Proceedings of the National Academy of Sciences of the United States of America, 112(12), pp. 3841–6. doi: 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J and Yu YT (2010) ‘Spliceosomal snRNA modifications and their function’, RNA Biology. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S et al. (2017) ‘m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover’, Genes and Development, 31(10), pp. 990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V et al. (2019) ‘Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution’, Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1817334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V and Cairns BR (2013) ‘Identification of direct targets and modified bases of RNA cytosine methyltransferases’, Nature Biotechnology, 31(5), pp. 458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL et al. (2011) ‘Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila’, Genes and Development, 25(23), pp. 2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E et al. (2014) ‘The contribution of pseudouridine to stabilities and structure of RNAs’, Nucleic Acids Research, 42(5), pp. 3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D et al. (2013) ‘TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions’, Genome Biology, 14(4), p. R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, You BH and Nam JW (2015) ‘Global estimation of the 3’ untranslated region landscape using RNA sequencing’, Methods. doi: 10.1016/j.ymeth.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Knight SW et al. (1999) ‘X-Linked Dyskeratosis Congenita Is Predominantly Caused by Missense Mutations in the DKC1 Gene’, The American Journal of Human Genetics. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koš M and Tollervey D (2010) ‘Yeast Pre-rRNA Processing and Modification Occur Cotranscriptionally’, Molecular Cell. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L and Weber MJ (2006) ‘snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs.’, Nucleic acids research. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X et al. (2015) ‘Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome’, Nature Chemical Biology, 11(8), pp. 592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- Liu N et al. (2015) ‘N6 -methyladenosine-dependent RNA structural switches regulate RNA-protein interactions’, Nature, 518(7540), pp. 560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N and Pan T (2016) ‘Probing n6-methyladenosine (M6a) RNA modification in total RNA with SCARLET’, in Methods in Molecular Biology. doi: 10.1007/978-1-4939-3067-8_17. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W and Anders S (2014) ‘Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2’, Genome Biology. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP and Brown PO (2014) ‘Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae’, PLoS ONE, 9(10). doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW and Weiss A (2000) ‘A Model System for Activation-Induced Alternative Splicing of CD45 Pre-mRNA in T Cells Implicates Protein Kinase C and Ras’, Molecular and Cellular Biology. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannoor K, Liao J and Jiang F (2012) ‘Small nucleolar RNAs in cancer.’, Biochimica et biophysica acta. NIH Public Access, 1826(1), pp. 121–8. doi: 10.1016/j.bbcan.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau CD et al. (2016) ‘Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens.’, Nature, 535(7610), pp. 159–63. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) ‘Cutadapt removes adapter sequences from high-throughput sequencing reads’, EMBnet.journal, 17(1), p. 10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martinez Nicole M., Gilbert WV (2021) ‘Investigating pseudouridylation mechanisms by high-throughput in vitro RNA pseudouridylation and sequencing’, Methods in Molecular Biology, 2298(RNA Modifications). [DOI] [PubMed] [Google Scholar]
- Martinez NM and Gilbert WV (2018) ‘Pre-mRNA modifications and their role in nuclear processing’, Quantitative Biology, 6(3), pp. 210–227. doi: 10.1007/s40484-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Schaening-Burgos C and Gilbert WV (2021) ‘Pseudouridine site assignment by high-throughput in vitro RNA pseudouridylation and sequencing’, in. Academic Press (Methods in Enzymology). doi: 10.1016/bs.mie.2021.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S et al. (1999) ‘Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA.’, Molecular and cellular biology, 19(3), pp. 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DPB et al. (2010) ‘Genome-wide association identifies multiple ulcerative colitis susceptibility loci’, Nature Genetics, 42(4), pp. 332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Contreras A and Ruggero D (2015) ‘Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease.’, Wiley Interdisciplinary Reviews: RNA. NIH Public Access, 6(2), pp. 173–89. doi: 10.1002/wrna.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR et al. (2015) ‘Genome-wide discovery of human splicing branchpoints’, Genome Research, 25(2), pp. 290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A and Thomas PD (2013) ‘PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees’, Nucleic Acids Research. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI and Greenbaum NL (2001) ‘A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture.’, RNA (New York, N.Y.), 7(6), pp. 833–845. doi: 10.1017/S1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL et al. (2016) ‘Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP)’, Nature Methods. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL et al. (2018) ‘A Large-Scale Binding and Functional Map of Human RNA Binding Proteins’, bioRxiv. doi: 10.1101/179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbacher JK and Yeo GW (2018) ‘Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels’, Molecular Cell. doi: 10.1016/j.molcel.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H et al. (2008) ‘Cyclophilin A is required for CXCR4-mediated nuclear export of heterogeneous nuclear ribonucleoprotein A2, activation and nuclear translocation of ERK1/2, and chemotactic cell migration’, Journal of Biological Chemistry. doi: 10.1074/jbc.M704934200. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A et al. (2013) ‘Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression.’, RNA (New York, N.Y.), 19(6), pp. 811–27. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda JMB and Bradley RK (2018) ‘Most human introns are recognized via multiple and tissue-specific branchpoints’, Genes and Development. doi: 10.1101/gad.312058.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnik K and Potočnik U (2016) ‘eQTL analysis links inflammatory bowel disease associated 1q21 locus to ECM1 gene’, Journal of Applied Genetics. doi: 10.1007/s13353-015-0334-1. [DOI] [PubMed] [Google Scholar]
- Roundtree IA et al. (2017) ‘Dynamic RNA Modifications in Gene Expression Regulation’, Cell, pp. 1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M et al. (2017) ‘TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code’, Genome Research, 27(3), pp. 393–406. doi: 10.1101/gr.207613.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN and Lowe TM (2005) ‘The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs’, Nucleic Acids Research. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S et al. (2014) ‘Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA’, Cell, 159(1), pp. 148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R et al. (2016) ‘A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition’, Human Genetics, 135(7), pp. 707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O et al. (2014) ‘Genome-scale CRISPR-Cas9 knockout screening in human cells’, Science. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler C et al. (2013) ‘Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells’, Nature Methods, 10(4), pp. 315–323. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]
- Taggart AJ et al. (2017) ‘Large-scale analysis of branchpoint usage across species and cell lines’, Genome Research. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M et al. (2018) ‘Landscape of the complete RNA chemical modifications in the human 80S ribosome’, Nucleic Acids Research. doi: 10.1093/nar/gky811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorenoor N and Slaby O (2014) ‘Small nucleolar RNAs functioning and potential roles in cancer’, Tumor Biology. doi: 10.1007/s13277-014-2818-8. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan PP et al. (2017) ‘Pseudouridine and N(6)-methyladenosine modifications weaken PUF protein/RNA interactions.’, RNA (New York, N.Y.), 23(5), pp. 611–618. doi: 10.1261/rna.060053.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T et al. (2014) ‘Genetic screens in human cells using the CRISPR-Cas9 system’, Science. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GT and Farzaneh F (2012) ‘Are snoRNAs and snoRNA host genes new players in cancer?’, Nature reviews. Cancer, 12(2), pp. 84–8. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- Wu G et al. (2011) ‘Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation’, Wiley Interdisciplinary Reviews: RNA, pp. 571–581. doi: 10.1002/wrna.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z et al. (2014) ‘Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’2-UTR landscape across seven tumour types’, Nature Communications. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y et al. (2016) ‘Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV- 1 transcription and escape from latency’, EMBO reports, 17(10), pp. 1441–1451. doi: 10.15252/embr.201642682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong FY et al. (2014) ‘The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing’, PLoS Genetics. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1, Pseudo-seq, eCLIP, in vitro PUS sites, gene expression, exon skipping, and APA data (related to Figures 1, 3-6)
(A) HepG2 45S rRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(B) HepG2 ncRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(C) HepG2 pre-mRNA pseudouridines identified in chromatin-associated RNA. Pseudo-seq signal is listed for each replicate (related to Figure 1).
(D) HeLa mRNA pseudouridine validation analysis (related to Figure 1) (E) RBP eCLIP peaks with significant enrichment over the size matched input (> 4-fold enrichment and adjusted p-value < 0.001) and overlapping pseudouridine locations (related to Figure 3).
(F) Z scores comparing the fraction of RBP eCLIP peaks overlapping pseudouridines to the calculated overlap after shuffling pseudouridines within intronic regions 1000 times and the fraction of RBP eCLIP peaks overlapping uridines to the calculated overlap after shuffling uridines within intronic regions 1000 times (related to Figure 3).
(G) Genes containing pseudouridine versus a background set of genes that met uridine read cutoffs which were used for gene ontology analysis (related to Figures 1 and 3).
(H) In vitro pseudouridylated sites by recombinant human PUSs. Z-score is listed for each source of PUS. PUS assignment was determined by Z-score analysis (related to Figure 4).
(L) Cassette exons or skipped exons (SE) that are alternatively spliced in PUS1 KO compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(J) Differentially expressed genes in PUS1 KO compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(K) Cassette exons or skipped exons (SE) that are alternatively spliced in RPUSD4 KD compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(L) Cassette exons or skipped exons (SE) that are alternatively spliced in PUS7 KD compared to WT HepG2 cells as determined by rMATS (related to Figure 5).
(M) Differentially expressed genes in PUS7 KD compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(N) Differentially expressed genes in RPUSD4 KD compared to WT HepG2 cells as determined by DESeq2 analysis of mRNA levels (related to Figure 5).
(O) Genes showing alternative polyadenylation (APA) in PUS1 KO compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
(P) Genes showing alternative polyadenylation (APA) in RPUSD4 KD compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
(Q) Genes showing alternative polyadenylation (APA) in PUS7 KD compared to WT HepG2 cells as determined by QAPA (related to Figure 6).
Data Availability Statement
All sequencing and annotations data have been deposited at GEO and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.