Abstract
The classic mode of STING activation is through binding the cyclic dinucleotide 2'3'-cyclic GMP–AMP (cGAMP), produced by the DNA sensor cyclic GMP–AMP synthase (cGAS), which is important for the innate immune response to microbial infection and autoimmune disease. Modes of STING activation that are independent of cGAS are much less well understood. Here, through a spatiotemporally resolved proximity labelling screen followed by quantitative proteomics, we identify the lysosomal membrane protein Niemann–Pick type C1 (NPC1) as a cofactor in the trafficking of STING. NPC1 interacts with STING and recruits it to the lysosome for degradation in both human and mouse cells. Notably, we find that knockout of Npc1 ‘primes’ STING signalling by physically linking or ‘tethering’ STING to SREBP2 trafficking. Loss of NPC1 protein also ‘boosts’ STING signalling by blocking lysosomal degradation. Both priming and boosting of STING signalling are required for severe neurological disease in the Npc1−/− mouse. Genetic deletion of Sting1 (the gene that encodes STING) or Irf3, but not that of Cgas, significantly reduced the activation of microglia and relieved the loss of Purkinje neurons in the cerebellum of Npc1−/− mice, leading to improved motor function. Our study identifies a cGAS- and cGAMP-independent mode of STING activation that affects neuropathology and provides a therapeutic target for the treatment of Niemann–Pick disease type C.
The cGAS–STING pathway has been implicated in multiple systemic autoimmune and inflammatory diseases, several of which affect the central nervous system1–5. In almost all cases, the immunopathology in the central nervous system is initiated by the abnormal release of self-DNA derived from the nucleus or the mitochondrion, which activates cGAS to produce cGAMP1. cGAMP then directly activates STING signalling. Activation of STING in a cGAS- and cGAMP-independent manner is also possible and has been described previously in studies of STING gain-of-function mutants6–9. Both ligand-dependent and ligand-independent activation of STING require the translocation of STING from the endoplasmic reticulum (ER) to the ER–Golgi intermediate compartment (ERGIC) and the Golgi. Activation-induced trafficking recruits TANK-binding kinase 1 (TBK1), which activates interferon regulatory factor 3 (IRF3)–interferon (IFN) signalling as well as IFN-independent activities10–12. STING is also recruited to the lysosome for degradation, both at homeostasis and after activation and trafficking, which dampens downstream IFN signalling13,14. Despite the importance of cellular organelles in modulating STING trafficking and immune signalling, we know very little about membrane cofactor proteins on each organelle and how they regulate STING signalling.
A screen of cofactors in STING trafficking
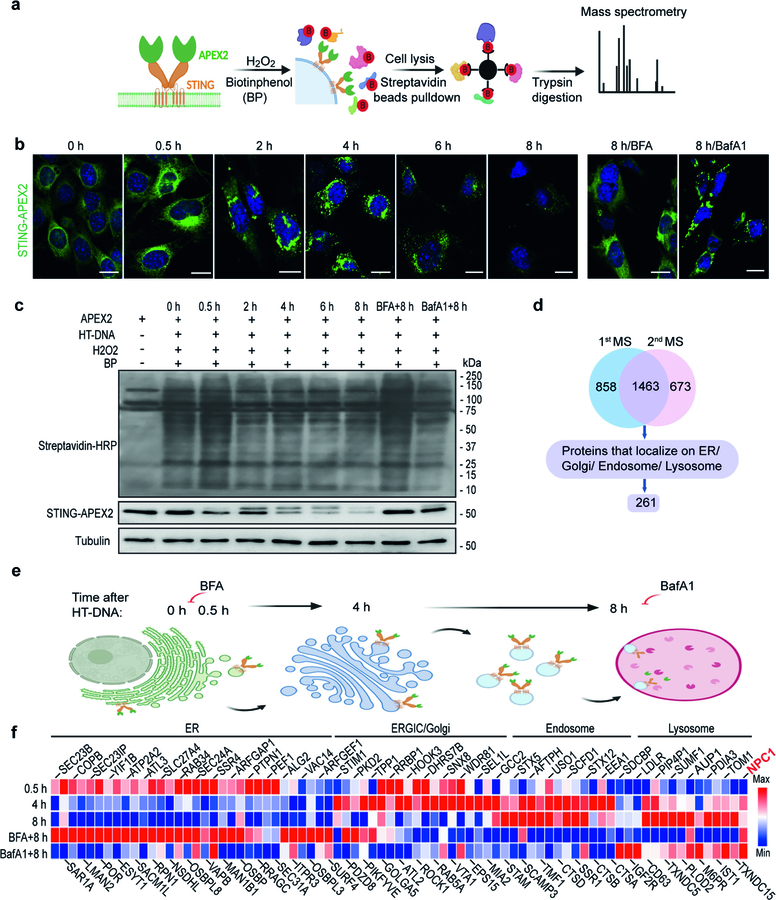
We performed spatiotemporally resolved proximity labelling using STING fused with engineered ascorbic acid peroxidase 2 (STING–APEX2) to define STING interactomes on various organelles along its trafficking route. APEX2 biotinylates near-neighbour proteins when the cells are treated with biotin-phenol and hydrogen peroxide15 (Extended Data Fig. 1a). This method allowed us to take ‘snapshots’ of the STING interactome during uninterrupted trafficking in live cells. We used streptavidin beads to pull down proteins that were biotinylated due to close proximity with STING–APEX2 at each time point, repeated the experiment three times and analysed two best sets of samples by tandem mass tag mass spectrometry (TMT-MS; Extended Data Fig. 1b–d). We then selected 261 candidate proteins on the basis of their known localization to intracellular organelles (https://compartments.jensenlab.org), normalized their abundance values to the unstimulated condition (0-h time point) and generated a heat map (Extended Data Fig. 1e, f).
We made several observations. First, the composition of the STING complex changes markedly as STING moves from one organelle to the next. Second, as expected, both the 0.5-h hits and the 8-h plus brefeldin A (BFA, which blocks the exit of STING from the ER) hits were enriched with ER proteins, and both the 8-h hits and the 8-h plus bafilomycin A1 (BafA1, which blocks the degradation of STING by lysosomes) hits were enriched with endosome and lysosome proteins, which validated our approach. Third, a mixture of ER, ERGIC–Golgi and endosome proteins was found at 4 h, which is likely to be due to the heterogeneity of STING trafficking at this intermediate time point. Finally, most of the known STING interactors localize to the ER, and our screen rediscovered many of them—such as SEC24A, SAR1A and STIM116,17—demonstrating the functionality and power of this screen. Very little is known about STING interactors on other organelles. We were particularly interested in a lysosomal candidate protein NPC1 and went on to investigate its role in STING signalling.
Knockout of Npc1 activates STING signalling
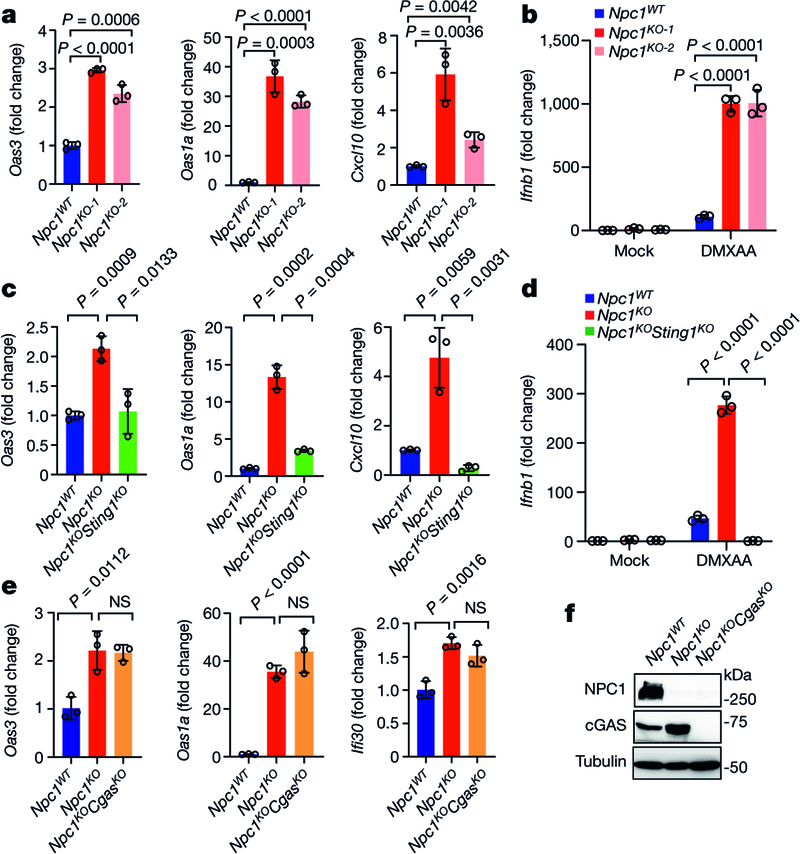
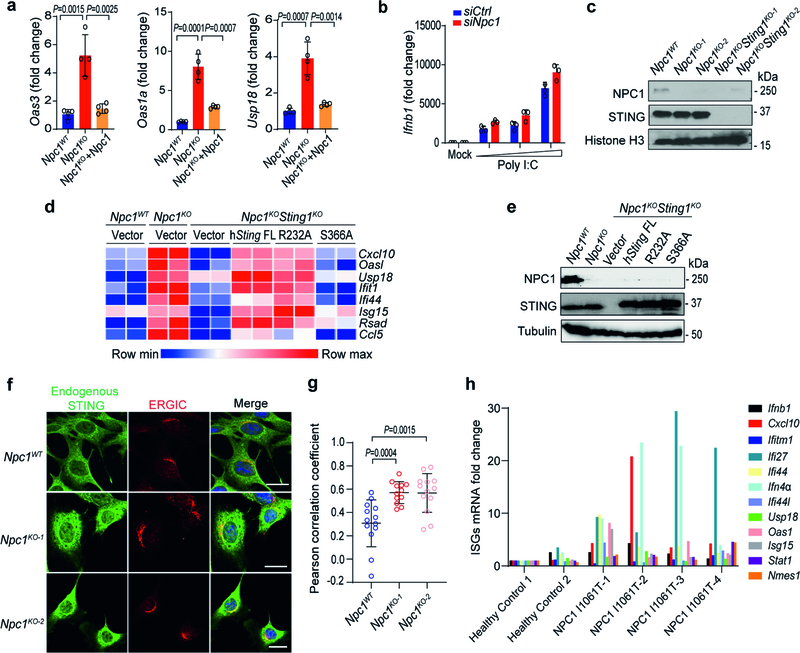
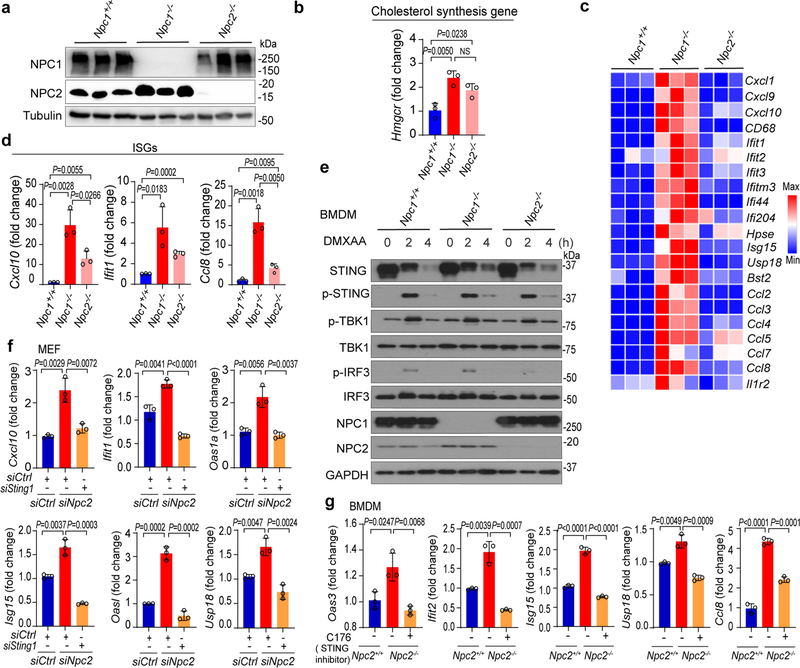
Two independent Npc1-knockout (Npc1KO) clonal mouse cell lines generated by CRISPR–Cas9 showed significantly increased expression of several IFN-stimulated genes (ISGs) at the resting state compared to non-targeted wild-type (Npc1WT) mouse embryonic fibroblasts (MEFs) (Fig. 1a). Reconstitution with wild-type NPC1 in Npc1KO MEFs reduced the expression of ISGs back to that of Npc1WT MEFs (Extended Data Fig. 2a). Stimulation with the STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA), but not with the RIG-I and MDA5 agonist poly(I:C), induced significantly higher expression of Ifnb1 mRNA in Npc1KO MEFs compared to Npc1WT MEFs (Fig. 1b, Extended Data Fig. 2b). Further knockout of Sting1 (Npc1KOSting1KO) significantly reduced tonic ISG expression in Npc1KO MEFs, as well as reducing DMXAA-induced Ifnb1 mRNA expression (Fig. 1c, d, Extended Data Fig. 2c). By contrast, additional knockout of Cgas (Npc1KOCgasKO) did not reduce tonic ISG expression compared to Npc1KO MEFs (Fig. 1e, f).
Fig. 1. NPC1 deficiency chronically activates STING-mediated type I IFN signalling independently of cGAS.
a, c, Quantitative PCR with reverse transcription (qRT–PCR) analysis of the mRNA expression of resting-state ISGs (Oas3, Oas1a and Cxcl10) in Npc1WT and two independent Npc1KO MEF cell lines (a) or in Npc1WT, Npc1KO and Npc1KOSting1KO MEFs (c) (n = 3). b, d, qRT–PCR analysis of Ifnb1 mRNA expression in Npc1WT and two independent Npc1KO MEF cell lines (b) or in Npc1WT, Npc1KO and Npc1KOSting1KO MEFs (d) after stimulation with the STING agonist DMXAA (50 μg ml-1) for 2 h (n = 3). e, qRT–PCR analysis of the baseline mRNA expression of ISGs (Oas3, Oas1a, Ifi30) in Npc1WT, Npc1KO and Npc1KOCgasKO MEFs (n = 3). f, Immunoblot analysis of the indicated proteins in Npc1WT, Npc1KO and Npc1KOCgasKO MEFs. a–e, Data are mean ± s.d. a, c, e, Unpaired two-tailed Student’s t-test. b, d, Two-way ANOVA. NS, not significant. Data are representative of at least three independent experiments.
We next examined the role of cGAMP. We reconstituted Npc1KOSting1KO MEFs with wild-type STING, mutant STING(R232A) (which disrupts cGAMP binding) or mutant STING(S366A) (which disrupts IFN signalling). Both wild-type STING and STING(R232A), but not STING(S366A), increased the expression of ISGs in Npc1KOSting1KO MEFs, suggesting that cGAMP binding is not required, but recruitment of TBK1–IRF3 is required, for STING-mediated immune activation in Npc1KO MEFs (Extended Data Fig. 2d, e). Confocal microscopy revealed that STING co-localized with the ERGIC to a substantial extent in Npc1KO cells, consistent with the activation of STING during homeostasis (Extended Data Fig. 2f, g). Furthermore, we observed higher expression of several ISGs at the resting state in four unrelated lines of NPC1I1061T fibroblasts from patients with Niemann–Pick disease type C compared to two lines of fibroblasts from healthy control individuals (Extended Data Fig. 2h). Together, these data suggest that loss of Npc1 activates STING trafficking and signalling independent of cGAS and cGAMP.
SREBP2 primes STING signalling
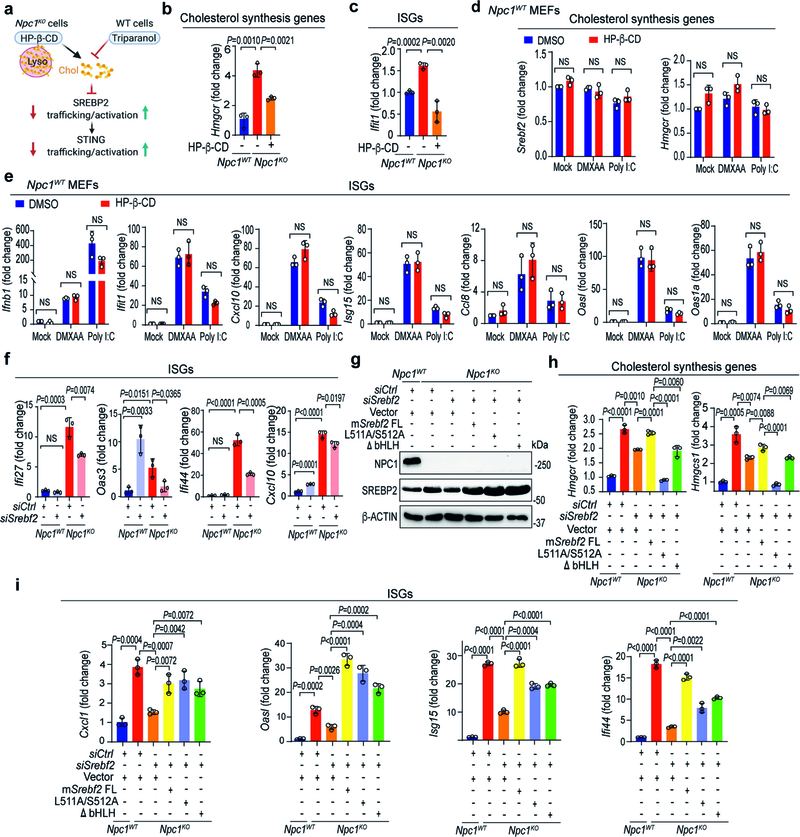
NPC1 deficiency causes the accumulation of cholesterol and other lipids in the lysosome, resulting in low levels of cholesterol on the ER and activating the translocation of SREBP2–SCAP from the ER to the Golgi. SREBP2 is cleaved on the Golgi, and the transcriptionally active fragment of SREBP2 translocates to the nucleus to activate cholesterol-synthesis genes such as Srebf2, Hmgcr and Hmgcs118. STING interacts with the SREBP2–SCAP complex19. Thus, we hypothesized that SREBP2 primes STING signalling by ‘tethering’ the trafficking of STING from the ER to the Golgi in Npc1KO cells.
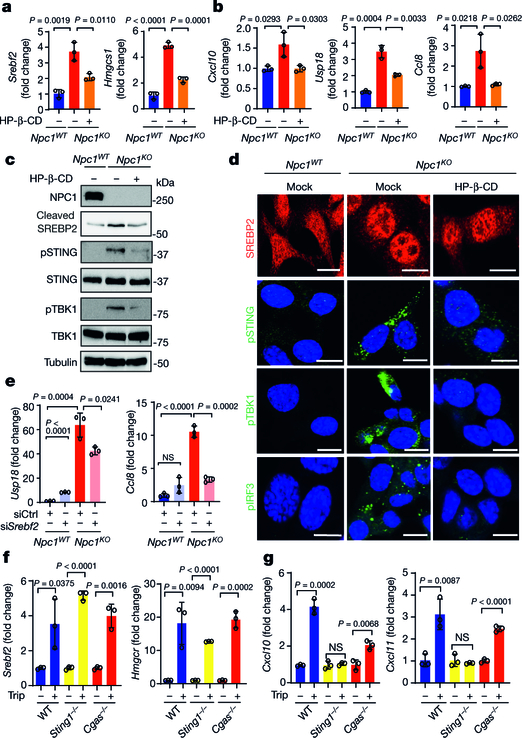
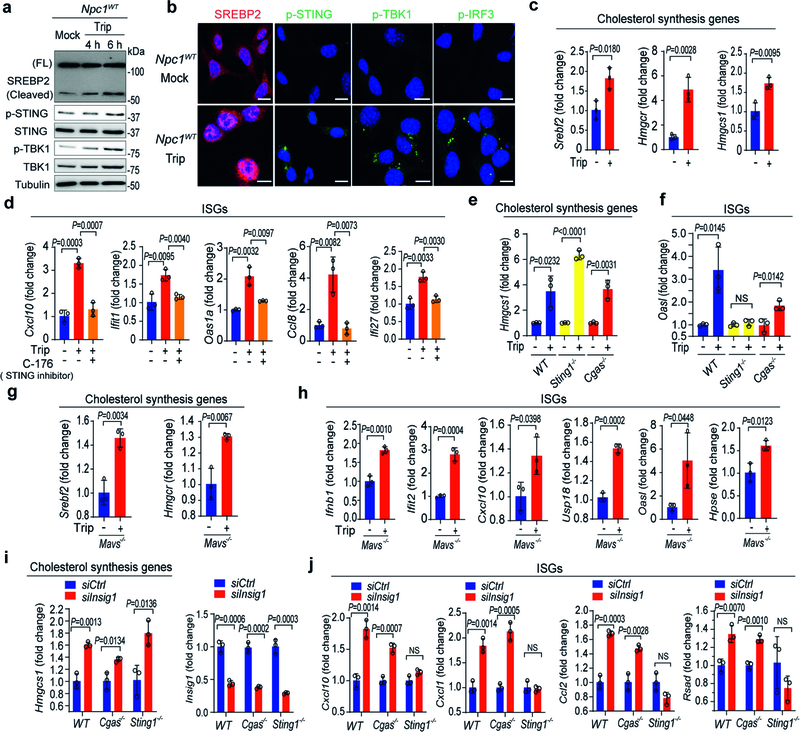
We performed several experiments to test this hypothesis (Extended Data Fig. 3a). First, we compared the activation status of SREBP2 and STING in Npc1WT cells, Npc1KO cells and Npc1KO cells treated with 2-hydroxypropyl-β-cyclodextrin (HP-β-CD). HP-β-CD promotes the exit of lysosomal cholesterol to the ER in NPC1-deficient cells, thus reducing SREBP2 trafficking and activation20 (Extended Data Fig. 3a). The mRNA expression of cholesterol-synthesis genes (Srebf2,Hmgcr and Hmgcs1) and ISGs (Cxcl10, Usp18, Ccl8 and Ifit1) was significantly higher in Npc1KO compared to Npc1WTMEFs, indicating that both SREBP2 and STING are activated (Fig. 2a, b, Extended Data Fig. 3b, c). Western blot and confocal microscopy also showed substantially higher levels of cleaved SREBP2 and nuclear translocation, as well as higher levels of phosphorylated (p)STING, pTBK1 and pIRF3 in Npc1KO compared to Npc1WTMEFs (Fig. 2c, d). Treatment with HP-β-CD reduced the activation of both SREBP2 and STING in Npc1KO cells, but not in wild-type cells (Fig. 2a–d, Extended Data Fig. 3b–e).
Fig. 2. SREBP2 trafficking primes STING trafficking and activation in Npc1-deficient cells.
a, b, qRT–PCR analysis of SREBP2 and STING activation. SREBP2 activation induces cholesterol-synthesis genes (a; Srebf2 and Hmgcs1) and STING activation induces ISGs (b; Cxcl10, Usp18 and Ccl8). Npc1WT and Npc1KO MEFs were mock-treated or treated with HP-β-CD (0.3 mM) overnight (n = 3). c, Immunoblot analysis of SREBP2 and STING activation. SREBP2 cleavage indicates activation. STING activation induces pSTING and pTBK1. d, Fluorescent microscopy analysis of SREBP2 and STING activation. Cleaved SREBP2 translocates to the nucleus (red). The STING activation markers pSTING, pTBK1 and pIRF3 are shown in green. The nucleus dye DAPI is shown in blue. Scale bars, 10 μm. e, STING activation in Npc1KO cells requires SREBP2. qRT–PCR analysis of ISG expression in Npc1WT and Npc1KO MEFs transfected with control siRNA (siCtrl) or siSrebf2 for 48 h (n = 3). f, g, qRT–PCR analysis of cholesterol-synthesis genes (f) and ISGs (g) in wild-type, Sting1−/− and Cgas−/−BMDMs that were mock-treated or treated with triparanol (Trip, 14 μM) for 12 h (n = 3). a, b, e–g, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
To directly assess SREBP2 trafficking versus transcriptional activities, we knocked down Srebf2 (which encodes SREBP2) and reconstituted cells with wild-type SREBP2 or transcriptionally inactive mutants (SREBP2(L511A/S512A), which cannot be cleaved at the Golgi21; or SREBP2(ΔbHLH), in which the leucine basic helix-loop-helix zipper transcriptional domain is deleted22). Knockdown of Srebf2 in Npc1KO MEFs significantly reduced the expression of ISGs (Fig. 2e, Extended Data Fig. 3f), which was restored by reconstitution with either wild-type SREBP2 or transcription-inactive mutants (Extended Data Fig. 3g–i). These data suggest that SREBP2 primes STING trafficking by tethering, which activates STING signalling in Npc1KO cells.
To further corroborate our findings, we sought other pharmacological and non-pharmacological means to induce SREBP2 trafficking, and then measured their effect on STING signalling. Triparanol inhibits the conversion of desmosterol into cholesterol. Treating wild-type MEFs or bone-marrow-derived macrophages (BMDMs) with triparanol increased SREBP2 cleavage, nuclear translocation, the levels of pSTING, pTBK1 and pIRF3, and the expression of cholesterol-synthesis genes, confirming SREBP2 trafficking and STING activation (Extended Data Fig. 4a–c). Treating wild-type BMDMs with triparanol also increased the expression of several ISGs, and this effect was attenuated by treatment with C-176, a small-molecule inhibitor of STING (Extended Data Fig. 4d). We also treated wild-type, Sting1−/− and Cgas−/− BMDMs, as well as wild-type and Mavs−/− MEFs, with triparanol. Triparanol induced the expression of both cholesterol-synthesis genes and ISGs, and only Sting1−/− cells lost the ability to induce ISGs (Fig. 2f, g, Extended Data Fig. 4e–h). These data show that neither cytosolic DNA sensing by cGAS nor RNA sensing by RIG-I and MDA5 is required for the induction of ISGs by triparanol. Instead, triparanol activates SREBP2 trafficking, which then primes STING signalling.
In a similar fashion, small interfering RNA (siRNA)-mediated knockdown of INSIG1, an ER retention factor for SREBP2–SCAP, also induced the expression of Hmgcs1 mRNA in wild-type, Cgas−/− and Sting1−/− MEFs (Extended Data Fig. 4i). The expression of ISGs induced by knockdown of INSIG1 was abrogated in Sting1−/− but not in Cgas−/− MEFs (Extended Data Fig. 4j). Together, these data suggest that pharmacological or non-pharmacological induction of SREBP2 trafficking primes STING signalling independently of cytosolic nucleic acid sensing.
NPC1 is the lysosomal adaptor for STING
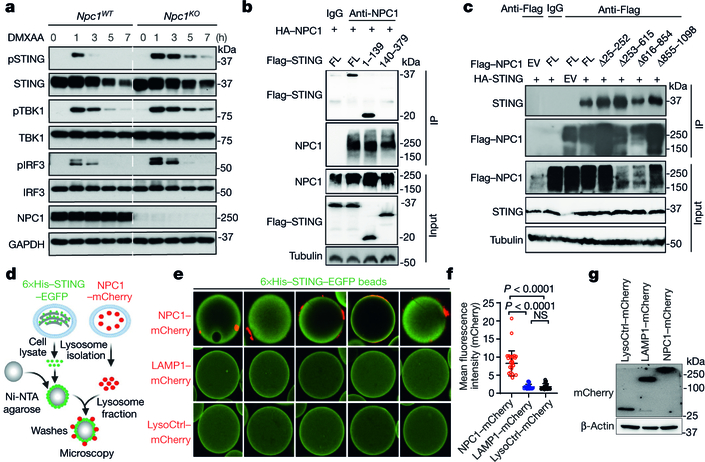
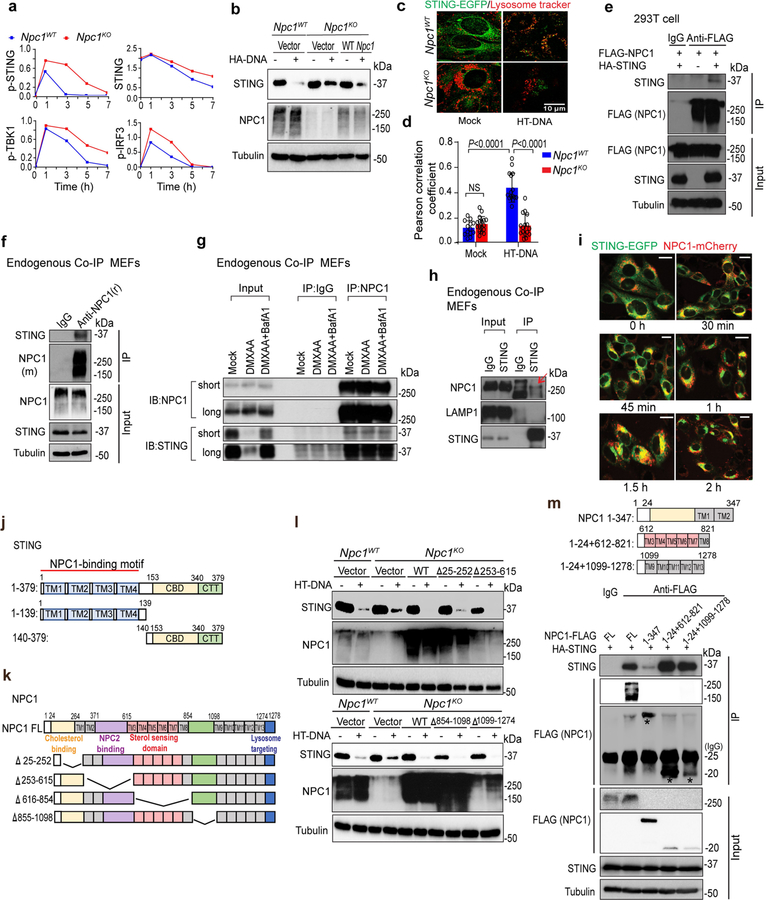
Because we identified NPC1 in the STING–APEX2 proximity labelling screen, we next investigated whether NPC1 interacts with STING and whether NPC1 is required for the degradation of STING by the lysosome, which if blocked should boost STING signalling. After stimulation with the STING agonist DMXAA, we observed stronger and more prolonged activation of pSTING, pTBK1 and pIRF3 in Npc1KO compared to Npc1WT MEFs. STING protein was rapidly degraded in Npc1WT MEFs after stimulation with DMXAA but became more stable in Npc1KO MEFs (Fig. 3a, Extended Data Fig. 5a). Ectopic expression of wild-type NPC1 in Npc1KO cells rescued STING degradation (Extended Data Fig. 5b). Live-cell microscopy with stably expressed STING–EGFP and LysoTracker Red revealed extensive co-localization of STING–EGFP with lysosomes in Npc1WT but not in Npc1KO MEFs after stimulation (Extended Data Fig. 5c, d). These data suggest that NPC1 is required for recruiting STING vesicles to the lysosome for degradation.
Fig. 3. NPC1 is a lysosomal adaptor that mediates STING degradation.
a, Immunoblot analysis of the STING signalling cascade. Npc1WT and Npc1KO MEFs were stimulated with the STING agonist DMXAA (30 μg ml−1) for 0, 1, 3, 5 or 7 h. The total and phosphorylated proteins immunoblotted are identified on the left. b, c, STING and NPC1 interaction domain mapping. HEK293T cells were transfected with the indicated plasmids (top), and 24 h later, the indicated antibodies were used for the pull-down. b, Full-length (FL) HA–NPC1 co-immunoprecipitated with Flag–STING truncated variants. c, Full-length HA–STING co-immunoprecipitated with Flag–NPC1 truncated variants. EV, empty vector; IP, immunoprecipitation. d–g, Cell-free lysosome recruitment assay. d, Diagram of the assay. e, Representative images of 6×His–STING–EGFP beads incubated with LysoCtrl–mCherry-, LAMP1–mCherry- and NPC1-mCherry-labelled lysosomes. f, Quantification of the recruitment of lysosomes to 6×His–STING–EGFP beads. For quantification, NPC1–mCherry, n = 33 beads; LAMP1–mCherry, n = 20 beads; LysoCtrl–mCherry, n = 20 beads. Data are mean ± s.d. Unpaired two-tailed Student’s t-test. g, Immunoblot analysis of the indicated proteins in different MEFs. Data are representative of at least two independent experiments.
We next confirmed the interaction between NPC1 and STING using co-immunoprecipitation of ectopically or endogenously expressed proteins (Extended Data Fig. 5e–h). We also reconstituted Npc1KO−Sting1KO MEFs with NPC1–mCherry and STING–EGFP and observed co-localization by live-cell microscopy (Extended Data Fig. 5i). Truncation experiments suggested that the STING transmembrane (TM) domain is required for interaction with NPC1 (Fig. 3b, Extended Data Fig. 5j). None of the NPC1 truncations disrupted its interaction with HA–STING or STING degradation, suggesting that multiple motifs in NPC1 have redundant roles for STING degradation (Fig. 3c, Extended Data Fig. 5k, l). NPC1 has 13 TM domains grouped into three TM bundles that are adjacent to one another in the three-dimensional space23. When each TM bundle was expressed individually, TM3–TM8 and TM9–TM13 exhibited robust interactions with HA–STING (Extended Data Fig. 5m). These data collectively show that NPC1 interacts with STING through TM bundles.
We next used a cell-free assay to mimic the recruitment of STING vesicles to the lysosome (Fig. 3d). We isolated 6×His–STING–EGFP-labelled ER microsomes and attached them to Ni-NTA agarose beads to immobilize STING vesicles. We also isolated fluorescently labelled lysosomes by fractionation of Npc1KOSting1KO MEFs expressing NPC1–mCherry, LAMP1–mCherry or lysosome-localized control peptide (LysoCtrl-mCherry)24. We then incubated STING–EGFP-coated beads with either NPC1–mCherry-, LAMP1–mCherry- or LysoCtrl–mCherry-labelled lysosomes. Only NPC1–mCherry-labelled lysosomes were recruited to STING–EGFP-coated beads (Fig. 3d–g). Together, our data suggest that NPC1 is the lysosomal adaptor for STING and that Npc1KO blocks the degradation of STING, thus boosting STING signalling.
STING drives neuropathology in Npc1−/− mice
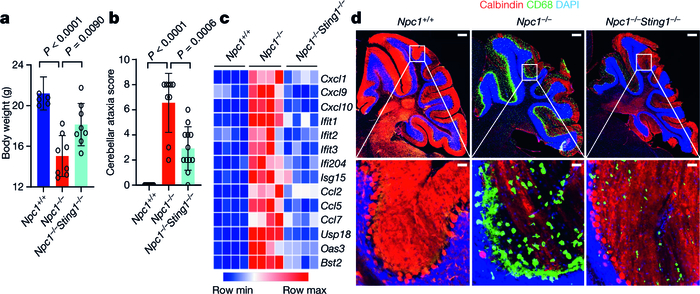
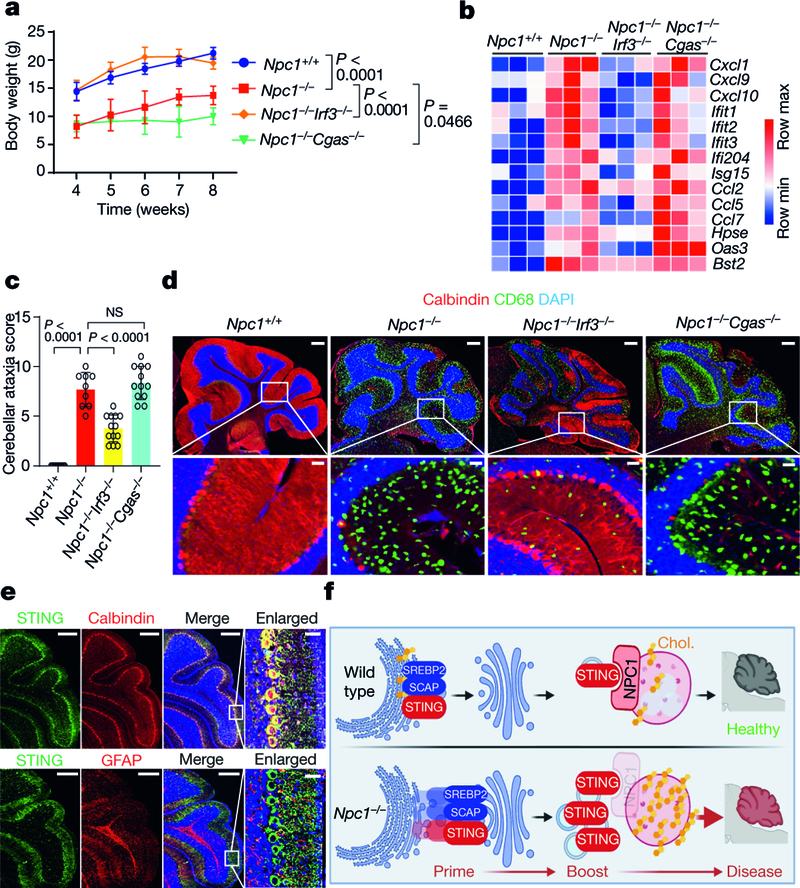
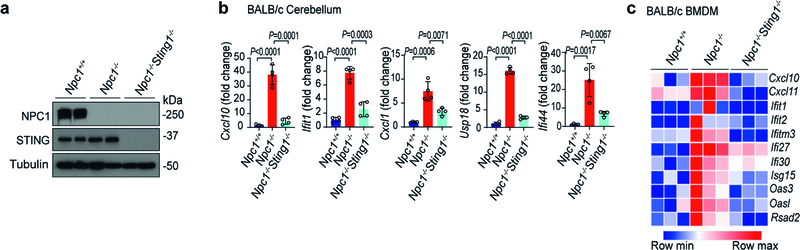
Npc1−/− mice exhibit many features of human Niemann–Pick disease type C, including impaired motor function due to progressive loss of Purkinje neurons, and reduced survival25,26. The proximity of Sting1 to the Npc1 gene on mouse chromosome 18 (around 24 Mb apart) precludes interbreeding. We thus deleted Sting1 on the BALB/c Npc1 allele using CRISPR–Cas9 (Extended Data Fig. 6a). Npc1−/− mice exhibited reduced body weight compared to wild-type mice, and body weight was significantly improved in Npc1−/−Sting1−/− mice (Fig. 4a). We next performed cerebellar ataxia scoring, which measures mouse motor function and coordination using a composite score of four tests: hindlimb clasping, ledge test, gait and kyphosis27. Npc1−/−mice showed severe ataxia at eight weeks old, and motor function was notably improved in Npc1−/−Sting1−/−mice, with a significantly lower ataxia score (Fig. 4b). An increased IFN gene signature has been observed in Npc1−/− mouse brain but the mechanism was unknown28. We also observed higher expression of ISGs and inflammatory genes in the cerebellum and BMDMs of Npc1−/− compared to Npc1+/+ mice (wild type). The activation of ISGs and inflammatory genes was eliminated in Npc1−/−Sting1−/− mice (Fig. 4c, Extended Data Fig. 6b, c).
Fig. 4. STING drives neuroinflammation and neuropathology in Npc1−/−mice.
a, Body weight of mice of the indicated genotypes (8 weeks old). All mice are on the BALB/c background (Npc+/+ mice, n = 5; Npc−/− mice, n = 8; Npc1−/−Sting1−/− mice, n = 8). b, Cerebellar ataxia scores for BALB/c mice of the indicated genotypes (8 weeks old). Mice were scored 0–12, which reflects a combination of four measurements (each 0–3): hindlimb clasping, ledge test, gait and kyphosis (Npc1+/+ mice, n = 10; Npc1−/− mice, n = 9; Npc1−/−Sting1−/− mice, n = 12). c, Heat map showing ISG expression in the cerebellum of mice of the indicated genotypes (8 weeks old). All mice are on the BALB/c background. The mRNA expression of each ISG was measured by qRT–PCR (n = 4). Min, minimum; max, maximum. d, Fluorescent IHC staining of the cerebellum of mice of the indicated genotypes (8 weeks old). All mice are on the BALB/c background. Calbindin (Purkinje neuron marker) is shown in red, CD68 (microglia activation marker) is shown in green and DAPI (nucleus marker) is shown in blue. Representative images are shown (n = 3). The bottom images are enlarged views. Scale bars, 200 μm (top); 30 μm (bottom).a, b, Data are mean ± s.d. Unpaired two-tailed Student’s t-test.
We next performed fluorescent immunohistochemistry (IHC) analysis of the mouse cerebellum. The cerebellum of Npc1+/+ mice is healthy with robust calbindin staining (a Purkinje cell marker) and minimal CD68 staining (a marker for microglia activation) (Fig. 4d). The cerebellum of Npc1−/− mice showed an opposite pattern, with many fewer Purkinje cells and numerous activated microglia (Fig. 4d). Cerebellum pathology was significantly improved in Npc1−/−Sting1−/−mice compared to Npc1−/− mice, with increased numbers of Purkinje cells and decreased microgliosis (Fig. 4d). Together, these data suggest that STING is required for the neuroinflammation, neuropathology and motor defects that are observed in Npc1−/− mice.
The Npc1−/− phenotype is cGAS-independent
We next examined the role of cGAS (upstream of STING) and IRF3 (downstream of STING) in Npc1−/− disease pathology in mice. Both Cgas−/− and Irf3−/− mice are healthy on the C57BL/6J background. C57BL/6J Npc1−/−mice develop more severe disease than BALB/c Npc1−/− mice although the cerebellum neuropathology is similar29.
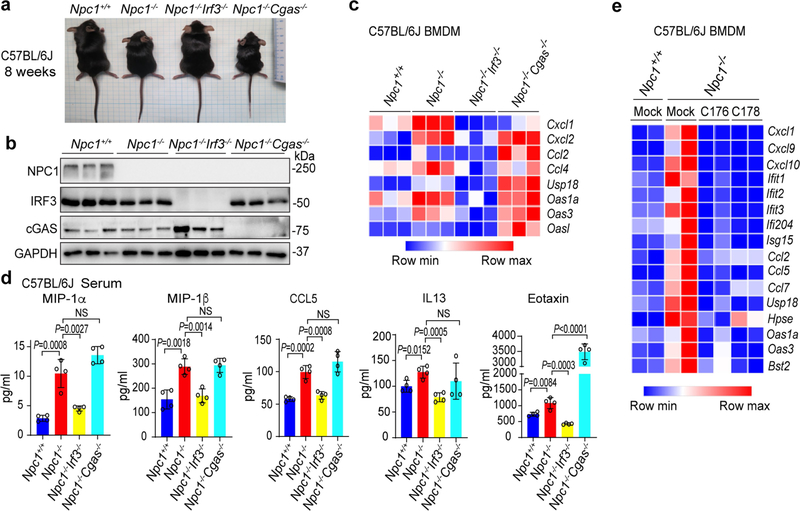
We compared Npc1+/+, Npc1−/−, Npc1−/−Irf3−/− and Npc1−/−Cgas−/−mice (all on the C57BL/6J background), and found that loss of Irf3, but not that of Cgas, ameliorated many Npc1−/− disease phenotypes in mice. First, body size and weight was rescued in Npc1−/−Irf3−/− but not in Npc1−/−Cgas−/− mice (Fig. 5a, Extended Data Fig. 7a, b). Npc1−/− cerebellum and BMDMs showed broadly increased expression of innate immune genes, especially the ISGs, most of which were significantly reduced in Npc1−/−Irf3−/− but not in Npc1−/−Cgas−/− mice (Fig. 5b, Extended Data Fig. 7c). The increase in the production of inflammatory cytokines and chemokines that was observed in Npc1−/− mice was also reduced in Npc1−/−Irf3−/− but not in Npc1−/−Cgas−/− mice (Extended Data Fig. 7d).
Fig. 5. IRF3, but not cGAS, is required for disease pathogenesis in Npc1−/−mice.
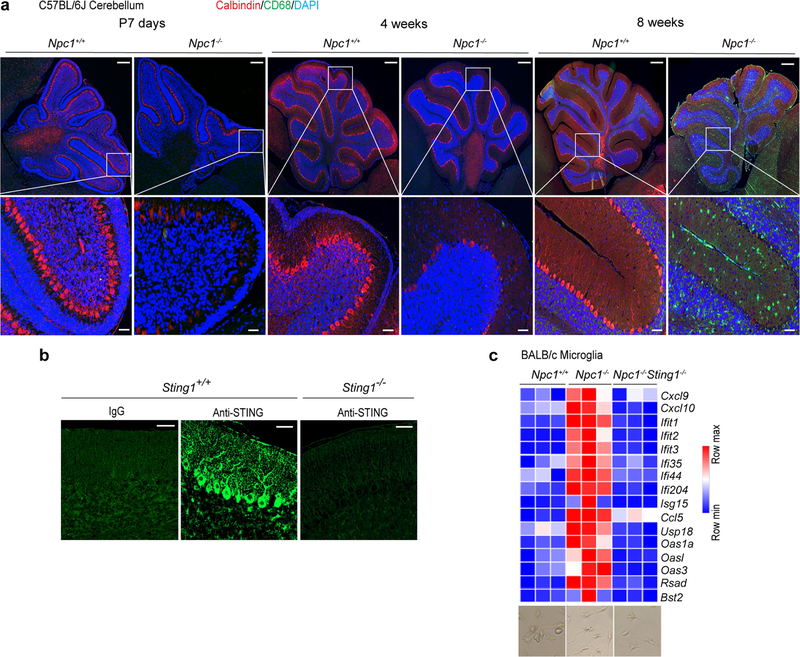
a, Body weight of mice of the indicated genotypes. All mice are on the C57BL/6J background (Npc1+/+ mice, n = 5; Npc1−/− mice, n = 10; Npc1−/−Irf3−/− mice, n = 10; Npc1−/−Cgas−/− mice, n = 10). b, Heat map showing ISG expression in the cerebellum of mice of the indicated genotypes (n = 3; 8 weeks old). The mRNA expression of each ISG was measured by qRT–PCR. c, Cerebellar ataxia scores of mice of the indicated genotypes (8 weeks old). All mice are on the C57BL/6J background. Mice were scored as in Fig. 4b (Npc1+/+ mice, n = 9; Npc1−/− mice, n = 9; Npc1−/−Irf3−/− mice, n = 13; Npc1−/−Cgas−/− mice, n = 12). d, Fluorescent IHC staining of the cerebellum of mice (C57BL/6J) of the indicated genotypes. Calbindin is shown in red, CD68 is shown in green and DAPI is shown in blue. Representative images are shown (n = 3). The bottom images are enlarged views. Scale bars, 200 μm (top); 30 μm (bottom). e, Fluorescent IHC co-staining of neural markers with STING in the cerebellum of wild-type mice (C57BL/6J; 4 weeks old). Top, STING is shown in green and calbindin (Purkinje cell marker) is shown in red. Bottom, STING is shown in green and GFAP (astrocyte marker) is shown in red. DAPI is shown in blue. Representative images are shown (n = 3). Scale bars, 200 μm (main panels); 30 μm (enlarged views; far right). f, Graphical abstract of the study. Chol., cholesterol. a, c, Data are mean ± s.d. a, Two-way ANOVA. c, Unpaired two-tailed Student’s t-test.
Npc1−/−Irf3−/− mice showed significantly improved motor function with reduced cerebellar ataxia, whereas Npc1−/−Cgas−/− mice were similar to Npc1−/− mice (Fig. 5c). Npc1−/−Irf3−/− mouse cerebellum showed an increased number of Purkinje neurons and decreased microgliosis compared to Npc1−/− mice (Fig. 5d). Treating Npc1−/− BMDMs with a STING inhibitor suppressed the expression of ISGs (Extended Data Fig. 7e). The pathology of the Npc1−/−Cgas−/− mouse cerebellum was indistinguishable from that of the Npc1−/− mouse cerebellum, further demonstrating that cGAS is not required for NPC1 pathogenesis.
Both Purkinje cell death and the activation of microglia contribute to Npcl−/− neuropathology in mice26,30. We observed the loss of Purkinje cells as early as postnatal day (P)7, with no signs of microglia activation (Extended Data Fig. 8a). The cerebellum of four-week-old Npc1−/− mice showed substantial loss of Purkinje cells, with very low levels of microglia activation, whereas that of eight-week-old Npcl−/− mice lacked nearly all Purkinje cells, with strong activation of microglia (Extended Data Fig. 8a). We next examined the expression of STING protein in the cerebellum, and found that STING is almost exclusively expressed in Purkinje cells and not expressed in astrocytes (Fig. 5e, Extended Data Fig. 8b). This is consistent with Purkinje cells as known initiators of Npcl−/− cerebellar neuropathology26. We also isolated microglia from mouse cerebellum shortly after birth (P10-P12) and found that Npcl−/− microglia express significantly higher levels of ISGs and inflammatory genes compared to Npc1+/+ and Npc1−/−Stlng1−/− microglia (Extended Data Fig. 8c). Together, these data implicate a cell-intrinsic function of STING in both Purkinje cells and microglia in driving Npcl−/− neuropathology. The relative contribution of STING activity in these two cell types requires further study.
A STING prime-boost model
We next wanted to determine whether both STING priming and boosting are required for driving severe neurological disease in vivo. We took advantage of the fact that Npc1 and Npc2 deficiency cause identical biochemical defects in cholesterol and lipid distribution that should lead to similar priming of STING signalling; however, Npc2 deficiency will lack a boost due to the presence of Npc1. Notably, Npc2−/− mice develop less severe disease than Npc1−/− mice31,32. We confirmed that Npc2−/− cells and mouse tissues show similar SREBP2 activation to Npc1−/−, and that Npc2−/− cells express intact NPC1 protein (Extended Data Fig. 9a, b).
Both Npc1−/− and Npc2−/− mouse cerebellum show increased expression of immune genes compared to the wild type. However, levels of immune gene expression in the Npc2−/− cerebellum were much lower than those in the Npc1−/− cerebellum (Extended Data Fig. 9c, d). The degradation of STING protein is delayed only in Npc1−/− BMDMs and not in Npc2−/− BMDMs (Extended Data Fig. 9e). The moderate increase in ISG expression in Npc2-knockdown MEFs was eliminated by the additional knockdown of Sting1 (Extended Data Fig. 9f). Treatment with a STING inhibitor also decreased the expression of ISGs in Npc2−/− BMDMs (Extended Data Fig. 9g). These data suggest that both Npc1−/− and Npc2−/−could prime STING signalling, but because NPC1 is required for recruiting STING to the lysosome,Npc1−/− offers an additional boost (by blocking STING degradation), and that both priming and boosting of STING signalling are required for driving the severe neurological disease in Npc−/−-mice (Fig. 5f).
This study has several implications for both STING and NPC1 biology. First, this is an example of cGAS-independent activation of STING signalling in a physiologically relevant neurodegenerative disease. Second, the degradation of STING by the lysosome is at least in part adaptor-mediated and not completely stochastic. We showed previously that blocking lysosomal acidification through treatment with BafA1 enhances the STING-mediated anti-tumour response13. The STING–NPC1 interface uncovered here could offer a unique therapeutic opportunity for selectively blocking the degradation of STING to boost anti-viral and anti-tumour immunity. Third, STING can be primed by SREBP2-tethered trafficking during cholesterol synthesis. Trafficking-mediated activation of STING could also have different properties compared to canonical ligand-mediated activation. Fourth, we observed the expression of STING protein in Purkinje neurons, and these neurons were lost in the cerebellum of Npc1−/− mice before any evidence of microglia activation. This raises the intriguing possibility that the activation of STING in neuronal cells may directly cause cell death, which merits further investigation. Finally, the difference in STING activation between Npc1−/− and Npc2−/− mice may explain why Npc2−/− mice develop less severe disease and why NPC2 mutations are less prevalent in humans. Despite the fact that Niemann–Pick disease type C was discovered over a century ago, no drug for its treatment has yet been approved by the US Food and Drug Administration. STING antagonists have been developed for treating autoimmune and autoinflammatory diseases33. Therefore, our study provides a rationale for targeting the STING pathway as an avenue of therapy for Niemann–Pick disease type C.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03762-2.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Mice and cells
BALB/c and C57BL/6J background Npc1+/− mice were purchased from the Jackson laboratory (003092, 030097). Irf3−/− mice (C57BL/6J) were provided by K. Fitzgerald with permission from T. Taniguchi. Cgas−/− and Mavs−/− mice (C57BL/6J) were provided by Z. Chen. Npc2+/−(BALB/c) mice were provided by J. Dietschy. Npc1−/−Sting1−/− (BALB/c) mice were generated by CRISPR–Cas9 technology at the University of Texas Southwestern Transgenic core on the BALB/c Npc1+/− allele. All mice were housed in pathogen-free barrier facilities under 12-h light–dark cycles. Temperatures were set to 21 ± 2 °C with humidity of 40–60%. Animal work was approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. Primary MEFs and BMDMs were isolated and cultured as described previously12. HEK293T cells were obtained from ATCC. Fibroblasts from patients with Niemann–Pick disease type C (NPC1I1061T) were obtained from the Coriell Institute for Medical Research. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) fetal bovine serum (FBS) (heat-inactivated), 10 mM HEPES, 2 mM l-glutamine and 1 mM sodium pyruvate with the addition of 100 U ml−1 penicillin and 100 mg ml−1 streptomycin, at 37 °C with 5% CO22. Mycoplasma tests were conducted monthly and confirmed to be negative.
Reagents and antibodies
Biotin-phenol (Sigma-Aldrich, SML2135), streptavidin magnetic beads (Pierce, 88817), hydrogen peroxide (H2 O2), 30% (w/w) (Sigma-Aldrich, H1009–100ML), sodium ascorbate (VWR International, 95035–692), Trolox (Sigma-Aldrich, 238813–5G), phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, 10837091001), sodium azide (VWR International, AA14314–22), DMXAA (Invivogen, tlrl-dmx), herring testis DNA (Sigma-Aldrich, D6898), lipofectamine 2000 (Thermo Fisher Scientific, 11668019), lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, 13778150), RIPA lysis buffer (Millipore, 20–188), protease inhibitor cocktail (Sigma-Aldrich, 4693116001), GelCode Blue Stain (Thermo Fisher Scientific, 24590), C-176 STING antagonist (BioVision, B2341), C-178 STING antagonist, (BioVision, B2396),bafilomycin A1 (Sigma-Aldrich, B1793), brefeldin A (Sigma-Aldrich, B6542), HP-β-CD (Sigma-Aldrich, C0926–5G), triparanol (Cayman, 20918), Lysosome Purification Kit from Tissue and Cultured Cells (BioVision, K235–50), LysoTracker Red DND-99 (Thermo Fisher Scientific, L7528), Ni-NTA agarose (Qiagen, 30210), streptavidin–HRP (1:1,000, Life Technologies, S-911), anti-NPC1 (1:1,000, Abcam, rabbit ab134113), anti-NPC1 (1:100, for endogenous co-immunoprecipitation, Thermo Fisher Scientific, rabbit PA1–16817), anti-NPC1 (1:1,000, LSbio, mouse B3437), anti-cGAS (mouse specific) (1:1,000, D3O8O, CST31659), anti-STING (1:1,000, D2P2F, CST13647), STING rabbit mAb (rodent preferred) (1:1,000, D1V5L, CST50494), anti-STING (1:200, for endogenous STING staining, Proteintech 19851–1-AP), anti-pSTING (Ser365) (1:1,000 for western blot (WB), 1:100 for immunofluorescence (IF), D8F4W, CST 72971 s), anti-TBK1 (1:1,000, D1B4, CST 3504), anti-pTBK1 (1:1,000 for WB, 1:100 for IF, D52C2, CST 5483), anti-IRF3 (1:1,000, D83B9, CST 4302), anti-pIRF3 (Ser396) (1:1,000 for WB, 1:100 for IF, 4D4G, CST 4947), anti-α-tubulin (1:5,000, Sigma T5168), anti-GAPDH (1:5,000, Thermo Fisher Scientific, PA1–987), anti-NPC2 (1:1,000, Abcam 186829), goat anti–rabbit IgG–HRP conjugate (1:5,000, BioRad Laboratories 1706515), goat anti–mouse IgG–HRP conjugate (1:5,000, BioRad Laboratories 1706516), monoclonal anti-calbindin-D-28K antibody produced in mouse (1:200, Sigma-Aldrich C9848), anti-CD68 (1:200, Abcam, ab125212), ERGIC-53 antibody (B-9) (1:100, Santa Cruz, sc-271517), anti-SREBP2 (1:1,000 for WB, 1:100 for IF, Abcam, ab30682), anti-SREBP2 (1:1,000, Novus, NB100–74543), anti-LAMP1 (1:1,000, D2D11, CST 9091), anti-GFAP (1:200, Abcam, ab53554), donkey anti-mouse IgG(H+L) Alexa Fluor-568 (1:500, Thermo Fisher Scientific, A10037), donkey anti-mouse IgG(H+L) Alexa Fluor-647 (1:500, Thermo Fisher Scientific, A31571), donkey anti-rabbit IgG(H+L) Alexa Fluor-488 (1:500, Thermo Fisher Scientific, A21206), donkey anti-goat IgG(H+L) Alexa Fluor-568 (1:500, Thermo Fisher Scientific, A11057), Neural Tissue Dissociation Kit (P) (Miltenyi Biotec, 130–092-628), Myelin Removal Beads II, human, mouse, rat (Miltenyi Biotec, 130–096-733), CD11b MicroBeads, human and mouse (Miltenyi Biotec, 130–049-601), Fungizone (Sigma-Aldrich, A2411), recombinant mouse macrophage colony-stimulating factor (M-CSF, R&D Systems, 416-ML), recombinant mouse granulocyte macrophage colony-stimulating factor (GM-CSF, R&D Systems, 415-ML) and recombinant mouse TGF-β1 (BioLegend, 763104).
APEX2 proximity labelling
APEX2 proximity labelling was performed as previously described15. In brief, Sting1−/− MEFs stably expressing STING–APEX2 cultured in 10-cm dishes were stimulated with or without HT-DNA (1 μg ml−1) for the indicated time. Before collection, cells were incubated with 6 ml biotin-phenol (500 μM) in complete medium for 30 min at 37 °C, then treated with H2 O2 (1 mM) at room temperature for 1 min. Cells were then washed three times with fresh quencher solution (10 mM sodium ascorbate, 10 mM sodium azide and 5 mM Trolox solutionin phosphate-buffered saline (PBS)).After washes,cells were lysed in fresh RIPA lysis buffer supplemented with 1× protease inhibitor cocktail (1 mM PMSF, 5 mM Trolox, 10 mM sodium azide and 10 mM sodium ascorbate, 600 μl for each 10-cm dish) by gentle pipetting on ice for 5 min. Whole-cell lysates were centrifuged at 15,000g for 10 min at 4 °C to clear cellular debris. The Pierce 660-nm assay was used to quantify the amount of protein in each cleared whole-cell lysate.
To pull down biotinylated proteins, streptavidin magnetic beads (50 μl for each sample) were washed two times with 1 ml of RIPA lysis buffer each. Cleared whole-cell lysates were incubated with streptavidin magnetic beads at 4 °C overnight. Then, beads were washed twice with RIPA lysis buffer, once with KCl, once with 0.1 M Na2CO3, once with 2 M urea in 10 mM Tris-HCl (pH 8.0) and twice with RIPA lysis buffer at 4 °C. Biotinylated proteins were eluted by boiling the beads in 50 μl of 3× protein loading buffer (6 times: 10.5 ml dH2O, 10.8 ml glycerol, 10.5 ml 1 M Tris-HCl (pH 6.8), 2.79 g DTT, 3.0 of sodium dodecylsulfate and 3.6 mg bromophenol blue) supplemented with 2 mM biotin and 20 mM DTT for 10 min. Eluates were collected by pelleting the beads using a magnetic rack. For mass spectrometry, proteins were run into NuPAGE Novex Bis-Tris 4–12% (w/v) gels for around 1 cm. Gels were stained with GelCode Blue Stain and destained with ddH2O for 1 h. Gel slices containing proteins were cut and submitted to the University of Texas Southwestern Proteomics Core for the TMT-MS.
Target guide sequence for CRISPR–Cas9, siRNA, RNA and cytokine analysis
LentiCRISPRv2 vector was used to generate the knockout cell lines. Target guide sequences used are: Npc1 1: GGAGAGTGTGGAATTGC GAC; Npc1 2: ACTAAGTCATAGCCGTCCTT; Mb21d1 (Cgas) 1: GCGGACG GCTTCTTAGCGCG; Mb21d1 (Cgas) 2: AAAGGGGGGCTCGATCGCGG; Sting1 1:GTCCAAGTTCGTGCGAGGCT; Sting1 2: GCATGCTTAGGGACT TATAG.
Predesigned siRNA oligomers were purchased from Sigma-Aldrich and dissolved in water to 20 μM. Reverse transfection was conducted by using lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) with siRNA in Opti-MEM media (Thermo Fisher Scientific). siRNAs used in this study are: siNpc1 1: SASI_Mm01_00075301, siNpc1 2: SASI_Mm01_00075303, siNpc1 3: SASI_Mm01_00075304. siCgas: SASI_Mm01_00129826. siSrebf2: SASI_Mm01_00020952. siInsig1: SASI_ Mm01_00056250. Pooled siRNAs for secondary screen were purchased from Sigma-Aldrich.
Total RNA was extracted with TRI reagent according to the manufacturer’s protocol (Sigma-Aldrich). cDNA was generated using iScript Reverse Transcription Supermix kit (Bio-Rad, 1708840). qRT–PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, 1725120) and ABI-7500 Fast Real-Time PCR system (Applied Biosystems).
Serum from C57BL/6J background wild-type, Npc1−/−, Npc1−/−Irf3−/− and Npc1−/−Cgas−/− mice at 4 weeks old was collected for cytokine array analysis using the Bio-Plex Pro Mouse Cytokine Group I Panel 23-Plex Assay kit (Bio-Rad). The Bio-Plex Pro Assay was performed at the University of Texas Southwestern Medical Center Genomics and Microarray Core according to the manufacturer’s protocol.
Tissue histology and fluorescent immunohistochemistry
Mouse brains were collected and fixed in 4% paraformaldehyde solution in PBS (Thermo Fisher Scientific, J19943K2) for 7 days at 4 °C. Tissues were transferred to 10% sucrose in PBS for 1 day at 4 °C, followed by 20% sucrose in PBS for 1 day at 4 °C and finally 30% sucrose in PBS for 1 day at 4 °C. The brain samples were processed at the University of Texas Southwestern Whole Brain Microscopy Facility for embedding blocks, sections and immunofluorescence staining. For paraffin-embedded tissue, the brain samples were fixed in 4% paraformaldehyde solution in PBS then sent to the Tissue Management Core at the University of Texas Southwestern for paraffin embedding and sectioning. The monoclonal anti-calbindin-D-28K antibody produced in mouse (Sigma-Aldrich), anti-STING (Proteintech), anti-GFAP (Abcam) and anti-CD68 antibodies (Abcam) were used as primary antibodies. Donkey anti-mouse IgG(H+L) Alexa Fluor-568/647 (Thermo Fisher Scientific), donkey anti-rabbit IgG(H+L) Alexa Fluor-488 (Thermo Fisher Scientific) and donkey anti-goat IgG(H+L) Alexa Fluor-568 (Thermo Fisher Scientific) were used as secondary antibodies. Whole-brain images were captured using a Zeiss Axioscan Z1 slide scanner.
Fluorescent confocal microscopy
Cells were cultured on glass coverslips (for fixed-cell microscopy) or on glass-bottomed dishes (for live-cell microscopy) in complete medium. For fixed-cell microscopy, cells were fixed with 4% paraformaldehyde or 100% methanol (−20 °C) for 10 min, permeabilized with 0.2% Triton X-100 in PBS buffer and blocked with 3% BSA in PBS buffer. Coverslips were incubated with primary antibody for 1 h at room temperature followed by fluorescence-conjugated secondary antibody for 1 h at room temperature, then mounted on slides with VECTASHIELD mounting medium containing DAPI (Vector Laboratories). Slides were imaged with a Zeiss LSM 880 confocal microscope. Live-cell microscopy was imaged with an Andor spinning-disc confocal microscope with environmental CO2 and temperature controls. Image deconvolution was conducted using AutoQuant X software (Media Cybernetics), then analysed using Image J. Co-localization was analysed by the Image J plug-in EzColocalization.
Co-immunoprecipitation and immunoblot
Wild-type HEK293T cells were transfected with the indicated plasmids, collected 24 h later, washed once with PBS buffer, lysed in IP lysis buffer (20 mM Tris-HCl, pH 7.4, 0.5% Nonidet-P40, 150 mM NaCl and 1× protease inhibitor mixture) and centrifuged at 15,000g for 10 min at 4 °C. A small amount of supernatant was saved as input. The rest of the supernatant was incubated with primary antibody (or control IgG) and Dynabeads Protein G (Life Technology) overnight at 4 °C. Beads were washed twice with IP buffer, twice with high-salt IP buffer (500 mM NaCl) and twice with low-salt IP buffer (50 mM). IP complex was eluted in immunoblot sample buffer and boiled at 95 °C for 10 min. For immunoblot, equal amounts of protein were separated by SDS–PAGE, followed by transferring to nitrocellulose membrane. Membrane blots were blocked in 5% milk in TBST buffer for 1 h at room temperature, followed by incubation with primary antibodies in 3% milk TBST buffer at 4 °C overnight. After several washes, membrane blots were incubated with HRP-conjugated IgG secondary antibody for 1 h at room temperature. Then, membrane blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Cell-free lysosome recruitment assay
The cell-free lysosome recruitment assay was modified based on an assay described previously24. In brief, Npc1KOSting1KO MEFs stably expressing 6×His–STING–EGFP were lysed with lysis buffer (containing 20 mM Tris–HCl, pH 7.4, 0.5% NP-40, 150 mM NaCl and 1× protease inhibitor mixture) and centrifuged at 20,000g for 10 min at 4 °C. Supernatant containing 6×His–STING–EGFP vesicles (ER microsomes) were attached on Ni-NTA agarose beads by incubation for 4 h at 4 °C, followed by several PBS washes at 4 °C. Npc1KOSting1KO MEFs stably expressing NPC1–mCherry, LAMP1–mCherry or lysosome-localized control peptide (LysoCtrl–mCherry) were used to isolate intact functional lysosomes with the Lysosome Purification Kit (Biovision) according to the manufacturer’s protocol. Cells were washed with PBS and incubated with 500 μl lysosome isolation buffer (containing 1× protease inhibitor mixture) on ice for 2 min. A precooled glass Dounce homogenizer was used to stroke the samples at least 30 times on ice. The homogenate was added to 500 μl lysosome enrichment buffer and centrifuged at 500g for 10 min at 4 °C. Then, the five density gradient solutions were prepared with lysosome gradient and lysosome enrichment buffer in the kit. The highest density gradient solution was at the bottom and the lightest one on the top. The supernatant of each sample was mixed with lysosome gradient buffer at a ratio of 3:1, and the mixture was laid on the top, then centrifuged at 145,000g for 2 h at 4 °C. The lysosome fraction is visible on the top tenth of the gradient volume. Isolated lysosomes were resuspended in PBS and incubated with 6×His–STING–EGFP-coated Ni-beads for 1 h at 37 °C, and this was followed by several PBS washes. Images were acquired using a Zeiss LSM 880 confocal microscope.
Mouse behaviour analysis
Cerebellar ataxia score was determined according to a composite phenotype scoring system27. Eight-week-old mice were analysed unless noted otherwise. Evaluations include ledge test, hindlimb clasping, gait and kyphosis. Each measurement is recorded on a scale of 0–3, with a combined total of 0–12. In brief, in the ledge test, mice were placed on the cage’s ledge. Walking along the ledge without losing balance will score 0; occasionally losing footing but otherwise coordinated will score 1; unable to use hind legs or landing on head rather than paws during descent will score 2; unable to move on the ledge will score 3. In the hindlimb clasping test, mice were lifted by the tail and observed for 10 s. Hindlimbs consistently extended will score 0; one hindlimb retracted for more than 50% of the time will score 1; both hindlimbs retracted more than 50% of the time will score 2; both hindlimbs entirely retracted and touching the abdomen for more than 50% of the time will score 3. In the gait test, mice were placed on a flat surface to observe walking from behind. Mouse body weight supported by all limbs will score 0; limp or tremor while walking will score 1; severe tremor or limp and duck feet will score 2; difficulty moving forward with lowered abdomen will score 3. In the kyphosis test, mice were placed on a flat surface to observe walking from the side. Straightened spine while walking will score 0; mildly bent spine but straightens while walking will score 1; mildly bent spine that does not straighten while walking will score 2; severely bent spine that does not straighten while walking will score 3.
Microglia isolation and baseline ISG expression analysis
Ten-to-twelve day old mice (P10–P12) were anaesthetized and perfused with heparin in PBS (20 U ml−1). The isolated brains were dissociated using a Neural Tissue Dissociation Kit (P) (Miltenyi Biotec) on a GentleMACS Dissociator according to the manufacturer’s protocol. A single-cell suspension was obtained by passing through a 70-μm strainer followed by centrifugation at 300g for 10 min at 4 °C then resuspension with 0.5% BSA in PBS. Myelin was removed by the Myelin Removal Beads II Kit (Miltenyi Biotec) on the autoMACS separator with the ‘Deletes’ program. Next, cells were incubated with CD11b Micro-Beads (Miltenyi Biotec) and positively sorted through the autoMACS separator. CD11b-positive cells were plated (200,000 cells per well) in 12-well plates with microglia complete medium containing DMEM/F-12, 10% heat-inactivated FBS, 100 U ml−1 penicillin/100 mg ml−1 streptomycin, 0.25 μg ml−1 fungizone, 10 ng ml−1 rmM-CSF (R&D Systems), 10 ng ml−1 rmGM-CSF (R&D Systems) and 50 ng ml−1 TGF-β1 (BioLegend) for 6 days before collection with TRI reagent for RNA extraction and analysis of baseline ISG expression by qRT–PCR.
Statistical analysis
Statistical tests are noted in figure legends. All data are shown as means ± s.d., and all analyses were performed using Prism 8 software (Graphpad). Heat maps were generated using Morpheus (https://software.broadinstitute.org/morpheus/). Figures 4d, 5f and Extended Data Figs. 1a,e, 3a were created with BioRender.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Extended Data Fig. 1 |. Screen of STING trafficking cofactors using spatiotemporally resolved proximity labelling and quantitative proteomics.
a, Diagram for proximity labelling and proteomic discovery of trafficking cofactors using Sting1−/− MEFs stably expressing STING–APEX2. b, Representative microscopy images of STING–APEX2 trafficking at various time points after stimulation (HT-DNA, 1 μg ml−1). BFA blocks the exit of STING from the ER; BafA1 blocks the degradation of STING by lysosomes. Scale bars, 10 μm. c, Immunoblot analysis of cell lysates. STING–APEX2 MEFs were mock-treated or stimulated with HT-DNA (1 μg ml−1) for the indicated times with or without treatment with BFA or BafA1 (top). Then, proximity labelling was performed and biotinylated proteins were detected by streptavidin–HRP. Immunoblot is representative of at least three independent experiments. d, TMT-MS quantitative proteomics data filtering from two independent experiments and candidate discovery. e, Diagram of the STING trafficking route after stimulation, including time points (top) and organelles. f, Heat map showing selected STING trafficking cofactor candidates at each organelle (top). MS value was normalized to the 0-h time point. Data are representative of three independent experiments.
Extended Data Fig. 2 |. NPC1 deficiency primes STING trafficking and activation independently of cGAMP.
a, qRT–PCR analysis of the baseline expression of ISGs (Oas3, Oas1a and Usp18) in Npc1WT, Npc1KO and Npc1KO MEFs stably expressing wild-type Npc1 (n = 4). b, qRT–PCR analysis of Ifnb1 mRNA expression in wild-type and Npc1-knockdown MEFs after stimulation with an increasing amount of Poly I:C (0, 1, 2, 4 μg ml−1) for 3 h (n = 3). c, Immunoblot analysis of the indicated proteins (left) in Npc1WT, Npc1KO and Npc1KOSting1KO MEFs. d, Heat map showing the baseline expression of ISGs in Npc1WT, Npc1KO and Npc1KOSting1KO MEFs reconstituted with vector, STING wild type (FL), R232A or S366A mutants (n = 2). The mRNA expression of each ISG was measured by qRT–PCR. e, Immunoblot analysis of the indicated proteins (left) in different reconstituted MEFs (shown on top) as in d. f, Fluorescent microscopy analysis of endogenous STING localization at the resting state in Npc1WT and two independent clones of Npc1KO MEFs. Endogenous STING is shown in green, an ERGIC marker (ERGIC53) is shown in red and DAPI is shown in blue. Scale bars, 10 μm. g, Quantification of STING co-localization with the ERGIC in f (Npc1WT, n = 13; Npc1KO-1, n = 12; Npc1KO-2, n = 13). h, qRT–PCR analysis of the baseline expression of ISGs in fibroblasts from healthy control individuals (n = 2) and unrelated patients with Niemann–Pick disease type C (NPC1I1061T) (n = 4). a, b, g, Data are mean ± s.d. a, g, Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
Extended Data Fig. 3 |. SREBP2 trafficking primes STING signalling in Npc1KO cells independently of its transcriptional activity.
a, Diagram showing mechanisms of action for HP-β-CD and triparanol on cholesterol synthesis and SREBP2 activation. HP-β-CD promotes the egress of lysosomal cholesterol to the ER, thus limiting SREBP2 trafficking and activation in Npc1KO cells. Triparanol inhibits the biochemical conversion of desmosterol into cholesterol, thus promoting SREBP2 trafficking and activation in wild-type cells. b, c, qRT–PCR analysis of cholesterol-synthesis genes (b) and ISGs (c) in Npc1WT and Npc1KO MEFs mock-treated or treated with HP-β-CD (0.3 mM) overnight (n = 3). d, e, qRT–PCR analysis of mock-, DMXAA- (50 μg ml−1, 2 h) or poly(I:C)- (1 μg ml−1, 2 h) induced expression of cholesterol-synthesis genes (d) and ISGs (e) with mock- or HP-β-CD treatment (0.3 mM, 8 h) in Npc1WT MEFs (n = 3). f, qRT–PCR analysis of the baseline expression of ISGs in Npc1WT and Npc1KO MEFs transfected with siCtrl or siSrebf2 for 48 h (n = 3). g, Immunoblot analysis of the indicated proteins (left) in Npc1WT, Npc1KO, Npc1KOSrebf2KD and Npc1KOSrebf2KD MEFs reconstituted with SREBP2 wild type (FL) or transcription-inactive mutants (L511A/S512A, ΔbHLH). h, i, qRT–PCR analysis of the expression of cholesterol-synthesis genes (h) and ISGs (i) in Npc1WT, Npc1KO, Npc1KOSrebf2KD and Npc1KOSrebf2KD MEFs reconstituted with SREBP2 wild type (FL) or transcription-inactive mutants (L511A/S512A, ΔbHLH) (n = 3). b–f, h, i, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. NS, not significant. Data are representative of at least two independent experiments.
Extended Data Fig. 4 |. SREBP2 primes STING signalling independently of cGAS or MAVS.
a, Immunoblot analysis of the SREBP2 and STING activation in Npc1WT MEFs that were treated with mock or triparanol (14 μM) for the indicated times (shown on the top). SREBP2 cleavage indicates activation. STING activation induces pSTING and pTBK1. b, Fluorescent microscopy analysis of SREBP2 and STING activation. Cleaved SREBP2 translocates to the nucleus (red). The STING activation markers pSTING, pTBK1 and pIRF3 are shown in green. The nucleus dye DAPI is shown in blue. Scale bars, 10 μm. c, qRT–PCR analysis of cholesterol-synthesis genes in BMDMs that were either mock-treated or treated with triparanol (14 μM) for 12 h (n = 3). d, qRT–PCR analysis of the baseline expression of ISGs in BMDMs that were either mock-treated or treated with triparanol (14 μM) alone or in combination with the STING inhibitor C-176 (0.5 μM) for 12 h (n = 3). e, f, qRT–PCR analysis of cholesterol-synthesis genes (e) and ISGs (f) in wild-type, Sting1−/− and Cgas−/− BMDMs that were either mock-treated or treated with triparanol (14 μM) for 12 h (n = 3). g, h, qRT–PCR analysis of the expression of cholesterol-synthesis genes (g) and ISGs (h) in Mavs−/− MEFs that were either mock-treated or treated with triparanol (14 μM) for 12 h (n = 3). i, j, qRT–PCR analysis of the cholesterol-synthesis gene Hmgcs1, knockdown efficiency of Insig1 (i) and the expression of ISGs (j) in wild-type, Cgas−/− and Sting1−/− MEFs (n = 3). c–j, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
Extended Data Fig. 5 |. NPC1 interacts with STING and mediates the lysosomal degradation of STING.
a, Quantification of immunoblots in Fig. 3a. b, Immunoblot analysis of STING degradation in Npc1WT, Npc1KO or Npc1KO MEFs stably expressing wild-type NPC1. Cells were mock-treated or stimulated with HT-DNA (1 μg ml−1) for 8 h. c, d, Live-cell microscopy images of STING–EGFP and lysosomes. Npc1WT and Npc1KO MEFs stably expressing STING–EGFP were mock-treated (left) or stimulated with HT-DNA (right, 4 μg ml–1). STING–EGFP is shown in green and LysoTracker-Red is shown in red. Scale bar, 10 μm. Representative images in c and quantifications in d. Npc1WT 0 h (n = 13), Npc1KO 0 h (n = 17), Npc1WT 15 h (n = 15), and Npc1KO 15 h (n = 16) for quantification. Data are mean ± s.d. Unpaired two-tailed Student’s t-test. e, Immunoblot analysis of NPC1 and STING co-immunoprecipitation in HEK293T cells. HEK293T cells were transfected with the indicated plasmids (top), and 24 h later, anti-IgG (mouse) or anti-Flag was used for the pull-down. HA–STING co-immunoprecipitation was analysed by anti-STING immunoblot. Whole-cell lysates were blotted by anti-Flag (NPC1), anti-STING and anti-tubulin as input. f, Immunoblot analysis of endogenous STING and NPC1 co-immunoprecipitation in wild-type MEFs. Anti-IgG (rabbit) or anti-NPC1 (rabbit) was used for the pull-down. Both immunoprecipitation and lysate were blotted for endogenous STING (rabbit), NPC1 (mouse) and tubulin. g, Immunoblot analysis of endogenous STING and NPC1 co-immunoprecipitation in wild-type MEFs with mock-treated, DMXAA (30 μg ml−1, 8 h)-treated or DMXAA combined with BafA1-treated (to prevent STING degradation). Anti-IgG (rabbit) or Anti-NPC1 (rabbit) was used for the pull-down. Both immunoprecipitation and lysate input were blotted for endogenous STING (rabbit) and NPC1 (mouse). h, Immunoblot analysis of endogenous STING interaction with NPC1 or LAMP1 in wild-type MEFs. Anti-IgG (rabbit) or anti-STING (rabbit) was used for the pull-down. Both immunoprecipitation and lysate input were blotted for endogenous STING, NPC1 or LAMP1. Red arrow, co-immunoprecipitated NPC1 band. i, Live-cell fluorescent microscopy analysis of STING–EGFP and NPC1–mCherry localization. Npc1KOSting1KO MEFs stably expressing STING–EGFP (green) and NPC1–mCherry (red) were stimulated with HT-DNA (4 μg ml−1) and imaged at the indicated times. Scale bars, 10 μm. j, k, Diagrams showing STING (j) and NPC1 (k) full length and truncations. l, Immunoblot analysis of STING degradation in Npc1WT, Npc1KO or Npc1KO MEFs stably expressing indicated NPC1 truncations. Cells were mock-treated or stimulated with HT-DNA (1 μg ml−1) for 8 h. Cell lysates were analysed for the proteins indicated on the left. m, Immunoblot analysis of STING interaction with NPC1 transmembrane bundles in HEK293T cells. HEK293T cells were transfected with the indicated plasmids (top), and 24 h later, anti-IgG (mouse) or anti-Flag (mouse) was used for the pull-down. Both immunoprecipitation and whole-cell lysates were analysed by anti-Flag (NPC1), anti-STING and anti-tubulin. Data are representative of at least two independent experiments.
Extended Data Fig. 6 |. STING drives neuroinflammation in Npc1−/− mice.
a, Immunoblot analysis of the indicated proteins (left) in whole-brain lysates from BALB/c Npc1+/+, Npc1−/− and Npc1−/−Sting1−/− mice (n = 2). b, qRT–PCR analysis of the expression of ISGs in the cerebellum of BALB/c Npc1+/+, Npc1−/− and Npc1−/−Sting1−/− mice (n = 4). c, Heat map showing the expression of ISGs in BMDMs from BALB/c Npc1+/+, Npc1−/− and Npc1−/−Sting1−/− mice (n = 3). The mRNA expression level of each ISG was measured by qRT–PCR. b, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
Extended Data Fig. 7 |. STING and IRF3, but not cGAS, are required for Npc1−/− disease pathogenesis.
a, Representative image for the body size of eight-week-old mice. b, Immunoblot analysis of proteins (left) in whole-brain lysates of mice of the indicated genotypes (C57BL/6J; n = 3). c, Heat map showing the expression of ISGs in BMDMs from mice of the indicated genotypes (C57BL/6J; n = 3). The mRNA expression level of each ISG was measured by qRT–PCR. d, Serum cytokine levels in four-week-old mice of the indicated genotypes measured by multiplex ELISA (n = 4). e, Heat map showing the expression of ISGs in Npc1+/+ and Npc1−/− BMDMs that were mock-treated or treated with the STING inhibitor C-176 (0.5 μM) or C-178 (0.5 μM) overnight (n = 2). The mRNA expression of each ISG was measured by qRT–PCR. d, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
Extended Data Fig. 8 |. STING function in Purkinje cells and microglia.
a, Progression of Npc1−/− neuropathology. Fluorescent IHC staining of mouse cerebellum of Npc1+/+ and Npc1−/− mice at P7, four weeks old and eight weeks old. Calbindin is shown in red, CD68 is shown in green and DAPI is shown in blue. Representative images are shown (n = 3). Bottom panels are enlarged views. Scale bars, 200 μm (top); 30 μm (bottom). b, STING antibody validation for IHC. Fluorescent IHC staining with IgG (rabbit), anti-STING (rabbit) in wild-type C57BL/6J cerebellum or staining with anti-STING (rabbit) in Sting1−/− C57BL/6J cerebellum. Scale bars, 30 μm. Representative images are shown (n = 3). c, Heat map showing the baseline expression of ISGs in ex-vivo-cultured microglia isolated from P10–P12 mouse brains of the indicated genotypes (n = 3). The mRNA expression of each ISG was measured by qRT–PCR. Representative microscopy images of isolated microglia from indicated genotype of mice are shown at the bottom. Scale bars, 30 μm.
Extended Data Fig. 9 |. STING signalling activation in Npc1−/− and Npc2−/− cells and mice.
a, Immunoblot analysis of proteins (left) in whole-brain lysates of BALB/c mice of the indicated genotypes (n = 3). b, qRT–PCR analysis of cholesterol-synthesis gene (Hmgcr) expression in the cerebellums of mice of the indicated genotypes (BALB/c) (n = 3). c, Heat map showing the expression of ISGs in BALB/c Npc1+/+, Npc1−/− and Npc2−/− mouse cerebellum (n = 3). The mRNA expression of each ISG was measured by qRT–PCR. d, qRT–PCR analysis of the expression of some ISGs in the cerebellum of mice of the indicated genotypes (BALB/c) (n = 3). e, Immunoblot analysis of the STING signalling cascade. BALB/c Npc1+/+, Npc1−/− and Npc2−/− mouse BMDMs were stimulated with DMXAA (50 μg ml−1) for 0, 2 or 4 h. Phosphorylated and total proteins blotted are shown on the left. f, qRT–PCR analysis of the baseline expression of ISGs in Npc2WT, Npc2KD and Npc2KDSting1KD MEFs (n = 3). g, qRT–PCR analysis of the baseline expression of ISGs in BALB/c Npc2+/+ and Npc2−/− mouse BMDMs that were mock-treated or treated with STING inhibitor C-176 (0.5 μM) overnight (n = 3). b, d, f, g, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. Data are representative of at least two independent experiments.
Supplementary Material
Acknowledgements
We thank G. Barber for Sting1−/− mice, K. Fitzgerald and T. Taniguchi for Irf3−/− mice, Z. Chen for Cgas−/− and Mavs−/− mice and J. Dietschy for Npc2+/− mice; D. Ramirez, W. Li and A. Nawaby at the University of Texas Southwestern (UTSW) Whole Brain Microscopy Core Facility (RRID:SCR_017949) for assistance with tissue sectioning, histological staining, immunostaining and slide scanning; A. Lemoff at the UTSW Proteomics Core Facility for assistance with TMT-MS experiments; R. Hammer and M. Nguyen at the UTSW Transgenic Technology Center for creating Npc1−/−Sting1−/− mice; K. Phelps and A. Bugde at the UTSW Live Cell Imaging Facility, a Shared Resource of the Harold C. Simmons Cancer Center, supported in part by NCI Cancer Center Support Grant 1P30 CA142543–01 and National Institutes of Health (NIH) Shared Instrumentation Award 1S10 OD021684–01; I. Raman at the UTSW Medical Center Genomics and Microarray Core; and P. Tsai and members of the Yan laboratory for discussions. This work was supported by the NIH (AI151708 and NS122825 to N.Y.), the Cancer Prevention and Research Institute of Texas (CPRIT; RP180288 to N.Y.), the Burroughs Wellcome Fund (N.Y.) and the Ara Parseghian Medical Research Foundation (J.J.R.).
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03762-2.
Peer review information Nature thanks Veit Hornung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at http://www.nature.com/reprints.
Data availability
There are no restrictions on data availability in this manuscript. All of the information is included in the manuscript. Original western blots and TMT-MS raw data are included as Supplementary Fig. 1 and Supplementary Data, respectively.
References
- 1.Chin AC Neuroinflammation and the cGAS–STING pathway.J. Neurophysiol 121, 1087–1091 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Yan N Immune diseases associated with TREX1 and STING dysfunction.J.Interferon Cytokine Res. 37, 198–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sliter DA et al.Parkin and PINK1 mitigate STING-induced inflammation.Nature 561, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K,Huang R, Fujihira H,Suzuki T&Yan N N-glycanase NGLY1 regulates mitochondrial homeostasis and inflammation through NRF1. J. Exp. Med. 215, 2600–2616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu C-H et al.TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell 183, 636–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J et al.STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216, 867–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner JD et al.STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 214, 3279–3292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luksch H et al.STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144, 254–266.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motwani M et al.Hierarchy of clinical manifestations in SAVIN153S and V154M mouse models. Proc. Natl Acad. Sci. USA 116, 7941–7950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbs N et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barber GN STING-dependent signaling.Nat. Immunol 12,929–930(2011). [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Dobbs N, Yang K & Yan N Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonugunta VK et al. Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 21, 3234–3242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokatayev V et al. Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nat. Immunol. 21, 158–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung V et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui X et al.Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srikanth S et al.The Ca2+ sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 20, 152–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MS, Radhakrishnan A & Goldstein JL Retrospective on cholesterol homeostasis: the central role of Scap. Annu. Rev. Biochem. 87, 783–807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W et al. ER adaptor SCAP translocates and recruits IRF3 to perinuclear microsome induced by cytosolic microbial DNAs. PLoS Pathog. 12, e1005462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor AM, Liu B, Mari Y, Liu B & Repa JJ Cyclodextrin mediates rapid changes in lipid balance in Npc1−/− mice without carrying cholesterol through the bloodstream. J. Lipid Res. 53, 2331–2342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan EA, Brown MS, Goldstein JL & Sakai J Cleavage site for sterol-regulated protease localized to a Leu–Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272, 12778–12785 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Hua X et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl Acad. Sci. USA 90, 11603–11607 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X et al. Structure of human Niemann–Pick C1 protein. Proc. Natl Acad. Sci. USA 113, 8212–8217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu B-B et al. Cholesterol transport through lysosome–peroxisome membrane contacts. Cell 161, 291–306 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Fog CK & Kirkegaard T Animal models for Niemann–Pick type C: implications for drug discovery & development. Expert Opin. Drug Discov. 14, 499–509 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Elrick MJ et al. Conditional Niemann–Pick C mice demonstrate cell autonomous Purkinje cell neurodegeneration. Hum. Mol. Genet. 19, 837–847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guyenet SJ et al. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp. 39, e1787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin SDK et al. Interferon downstream signaling is activated early in pre-symptomatic Niemann–Pick disease type C. Neurosci. Lett 706, 43–50 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Parra J et al. Npc1 deficiency in the C57BL/6J genetic background enhances Niemann–Pick disease type C spleen pathology. Biochem. Biophys. Res. Commun. 413, 400–406 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Cougnoux A et al. Microglia activation in Niemann–Pick disease, type C1 is amendable to therapeutic intervention. Hum. Mol. Genet. 27, 2076–2089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sleat DE et al. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc. Natl Acad. Sci. USA 101, 5886–5891 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit SS et al. Loss of Niemann–Pick C1 or C2 protein results in similar biochemical changes suggesting that these proteins function in a common lysosomal pathway. PLoS ONE 6, e23677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decout A, Katz JD, Venkatraman S & Ablasser A The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 10.1038/s41577-021-00524-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no restrictions on data availability in this manuscript. All of the information is included in the manuscript. Original western blots and TMT-MS raw data are included as Supplementary Fig. 1 and Supplementary Data, respectively.